Abstract
The common age-related neurodegeneration of Parkinson's disease can result from dominant causes like increased dosage of vesicle-associated alpha-synuclein (SNCA) or recessive causes like deficiency of mitophagy factor PINK1. Interactions between these triggers and their convergence onto shared pathways are crucial, but currently conflicting evidence exists. Here, we crossed previously characterized mice with A53T-SNCA overexpression and with Pink1 deletion to generate double mutants (DMs). We studied their lifespan and behavior, histological and molecular anomalies at late and early ages. DM animals showed potentiated phenotypes in comparison with both single mutants (SMs), with reduced survival and strongly reduced spontaneous movements from the age of 3 months onwards. In contrast to SMs, a quarter of DM animals manifested progressive paralysis at ages >1 year and exhibited protein aggregates immunopositive for pSer129-SNCA, p62 and ubiquitin in spinal cord and basal brain. Brain proteome quantifications of ubiquitination sites documented altered degradation of SNCA and the DNA-damage marker H2AX at the age of 18 months. Global brain transcriptome profiles and qPCR validation experiments identified many consistent transcriptional dysregulations already at the age of 6 weeks, which were absent from SMs. The observed downregulations for Dapk1, Dcaf17, Rab42 and the novel SNCA-marker Lect1 as well as the upregulations for Dctn5, Mrpl9, Tmem181a, Xaf1 and H2afx reflect changes in ubiquitination, mitochondrial/synaptic/microtubular/cell adhesion dynamics and DNA damage. Thus, our study confirmed that SNCA-triggered neurotoxicity is exacerbated by the absence of PINK1 and identified a novel molecular signature that is detectable early in the course of this double pathology.
INTRODUCTION
The most common neurodegenerative disorders of old age are Alzheimer's disease (AD) with preferential affection of the cerebral cortex, and Parkinson's disease (PD) with preferential affection of the midbrain/brainstem. Multiple underlying genetic factors were identified for both entities over the past decades (1–3). For AD, mutations in amyloid precursor protein, in presenilin-1 and in presenilin-2 were all shown to modulate the beta-amyloid 42/40 ratio as the shared trigger of pathology (4). Whether such a common pathogenesis pathway exists also for PD is a matter of scientific controversy and intense investigation.
Among the autosomal dominant causes of PD, the increased dosage and missense mutations of alpha-synuclein (SNCA) leading to its gain-of-function and to its accumulation are prominent, because the subsequent aggregation of alpha-synuclein in ‘Lewy bodies’ and ‘Lewy neurites’ is observed not only in the brains of monogenic PD cases but also of most sporadic PD cases. SNCA mutations may lead to particularly early and severe manifestations with complete penetrance (5,6). SNCA is modulated by various post-translational events such as phosphorylation and ubiquitination (7), and its accumulation is well established to act as a neurotoxic stressor (8). Among the autosomal recessive causes of PD, a loss-of-function of the serine–threonine kinase PINK1 or of the ubiquitin-E3-ligase Parkin can lead to juvenile PD (9,10), through hampering cellular stress responses by impaired elimination of dysfunctional mitochondria via fission, trafficking and the autophago-lysosomal pathway (11,12).
Since most PD patients do not suffer from a monogenic disorder, but rather from polygenic interactions with environmental stressors, it is essential to explore the potential interaction of distinct genetic mechanisms in order to identify converging downstream pathways and putative molecular targets of neuroprotective therapy. In Drosophila melanogaster fly mutants as invertebrate models of PD, it was observed that PINK1 suppressed SNCA-triggered phenotypes of impaired mobility, cell degeneration and reduced lifespan (13,14). Similarly, it could be demonstrated in flies that Parkin counteracted SNCA-induced reductions of climbing activity and degenerative cell loss (15,16). In a vertebrate PD model, the medaka fish (Oryzias latipes), DMs with loss of PINK1 and Parkin showed phenotypes with late-onset locomotor dysfunction, a decrease in dopamine levels and a selective degeneration of dopaminergic neurons, whereas the single mutants (SMs) did not exhibit these features (17).
In mice, some early work seemed to be consistent with these findings in flies and fish, given that DM animals combining SNCA overexpression and Parkin deficiency in a >94% C57BL/6/<6% 129SvJ genetic background displayed potentiated pathology including enhanced mitochondrial damage and exacerbated respiration deficits in midbrain tissue (18). However, a knockout targeting another Parkin exon in a C57BL/6J genetic background paradoxically delayed the motor decline caused by SNCA accumulation, in a dose-dependent manner (19,20). Curiously, in DM mice, the knockout (KO) of DJ-1, which models another autosomal recessive variant of PD, failed to modify the course of SNCA-mediated neurotoxicity (21). Furthermore, in triple mouse mutants combining the loss of PINK1, Parkin and DJ-1, no neuron degeneration was detectable in the midbrain even at old ages (22). These data led to concerns among researchers regarding the cost and the utility of these mouse lines as suitable PD models.
Hoping to advance towards a mouse model of polygenic PD, and in order to clarify the role of PINK1 in SNCA-triggered neurotoxicity, we generated DM mice and studied them throughout their entire lifespan. For this purpose, we employed a previously well-characterized mouse line (PrPmtA with inbred FVB/N background), where the 1.5-fold overexpression of human A53T-mutant SNCA in the midbrain, striatum and cortex under control of the heterologous prion promoter leads to a progressive motor deficit and synaptic dysfunction by the age of 18 months, in the absence of protein aggregation or neuron loss (23–26). These mice were crossed to double homozygosity with a widely available Pink1−/− line of inbred 129Sv/Ev background (B6;129-Pink1tm1Aub/J at Jackson depository), which also shows a progressive motor deficit by age 16 months, again in the absence of protein aggregates or neuronal loss, with a mitochondrial dysfunction in neurons after exposure to stressors (27). As corresponding controls, we (I) aged wild-type (WT) mice derived from SM littermates with the appropriate inbred background in parallel to SM animals and (II) generated F1-hybrids from such WT FVB/N and 129Sv/Ev mice, which were aged in parallel with DM animals. The data generated indicate potentiated neurotoxicity in the DM mice and identify molecular targets, where both disease pathways converge and which are dysregulated before toxic protein aggregates become visible.
RESULTS
Crossbreeding of the two SM lines (homozygous A53T-SNCA-overexpressing PrPmtA animals with homozygous Pink1−/− animals) generated mice with homozygosity for both genotypes. A colony of such DM mice was established and made available through the Jackson depository [FVB;129-Pink1tm1Aub Tg(Prnp-SNCA*A53T)AAub/J]. In these DM, we investigated the maximal phenotype and pathology at ages >1 year and documented the preceding molecular changes in global unbiased surveys of the proteome at age 1 year and the transcriptome profile at the ages of 6 months and 6 weeks.
Potentiated phenotype in survival curve of DMs versus SMs
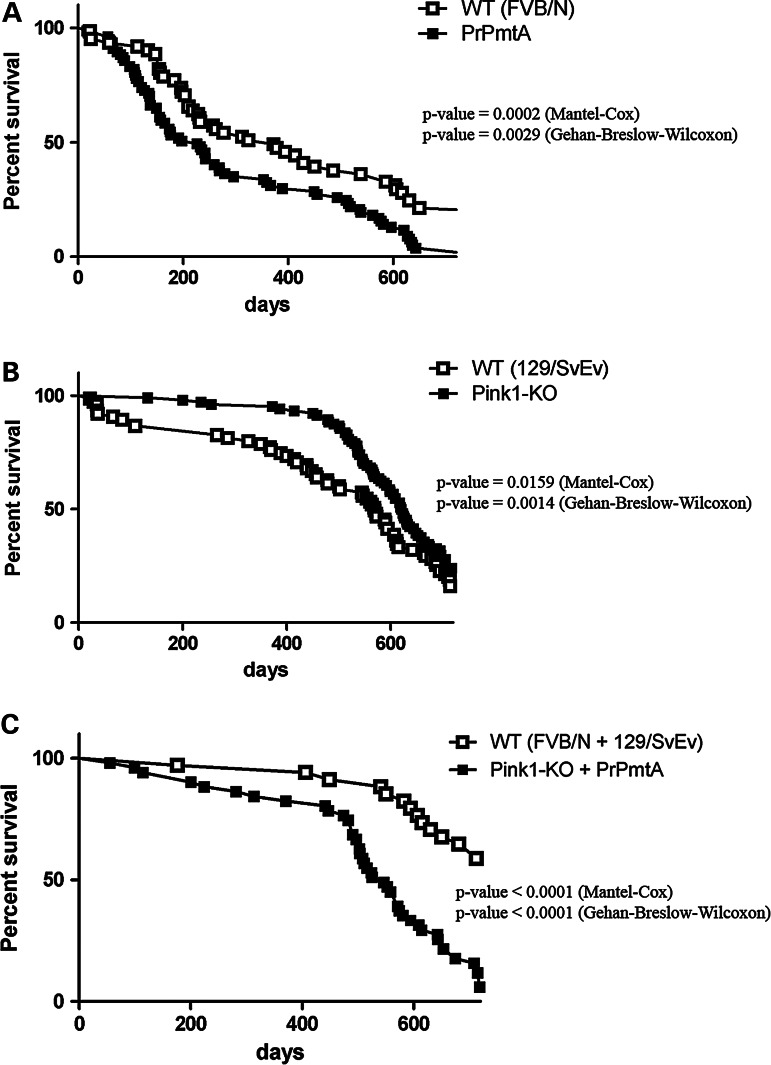
To document the unbiased organism phenotype, all animals were followed with regular weight analyses and neurological examinations (SHIRPA scores) from weaning age across their entire lifespan up to the maximum of 2 years, documenting their age at death by either natural causes or by the ethical decision of animal caretakers, who were blinded to genotypes. Kaplan–Meier survival curves were plotted and analyzed separately for the three mutant cohorts with their corresponding WT animals (Fig. 1). In the inbred FVB/N background with high aggressiveness and with old age multimorbidity, the PrPmtA genotype conferred an increased mortality rate in the first 180 days without further changes throughout the entire adult lifespan (Fig. 1A). In contrast, the Pink1−/− mutation not only resulted in a significantly reduced mortality up to an age of ∼ 450 days (note the different genetic background of inbred 129/SvEv; Fig. 1B) but also displayed an age-related sharp increase of the mortality rate not present in the background controls. Across all ages, the Pink1−/− triggered deficiency in stress responses provided a survival advantage (Fig. 1B). This effect might be explained by the stress-free environment in our animal facility—with food ad libitum, automated light–dark cycles, individually ventilated cages and regular health monitoring—where the absence of Pink1-dependent stress responses may constitute an advantage rather than a disadvantage. Under conditions of stress even at young age, the disadvantage of Pink1−/− genotype becomes apparent, e.g. when Pink1−/− heart tissue exhibits increased vulnerability after ischemia-reperfusion injury (28). The DM genotype showed a steeply progressive mortality rate with late onset at around 450 days, and this phase was preceded by a smaller but constant increase of mortality rate not present in the SMs. The comparison was made with the appropriate WT-F1-hybrid with the longer lifespan typical of outbreeding (Fig. 1C). In summary, the DM line showed a novel biphasic pattern of mortality rates.
Figure 1.
Survival curves of both SM versus DM. (A) While male FVB/N mice (starting n = 61) showed aggressive behavior at young age and inbreeding multimorbidity at advanced age resulting in constant high mortality across all ages independent from WT or mutant genotype, the homozygous PrPmtA line with overexpression of human A53T-SNCA (n = 77) resulted in some early mortality (significant P-value = 0.0002 in Mantel–Cox test, median survival reduced from 372 to 226 days). (B) WT 129/SvEv mice (n = 75) showed a moderate number of animal deaths at young age and slowly progressive mortality, as expected from an inbred line. The SM animals with Pink1−/− (n = 104) exhibited an extended lifespan (significant P-value = 0.0159, median survival lengthened from 545 to 592 days). (C) WT-F1-hybrids with a 50 : 50 ratio of FVB/N and 129/SvEv background (n = 34) showed few deaths during young age owing to aggressive behavior, and a much increased lifespan as often seen after outbreeding. In comparison, DM animals (n = 51) showed some mortality early on, which increased in a steeply progressive manner after age 450 days (significant P-value < 0.0001, median survival reduced from 723 to 546 days).
Spontaneous motor activity reduced from the age of 3 months
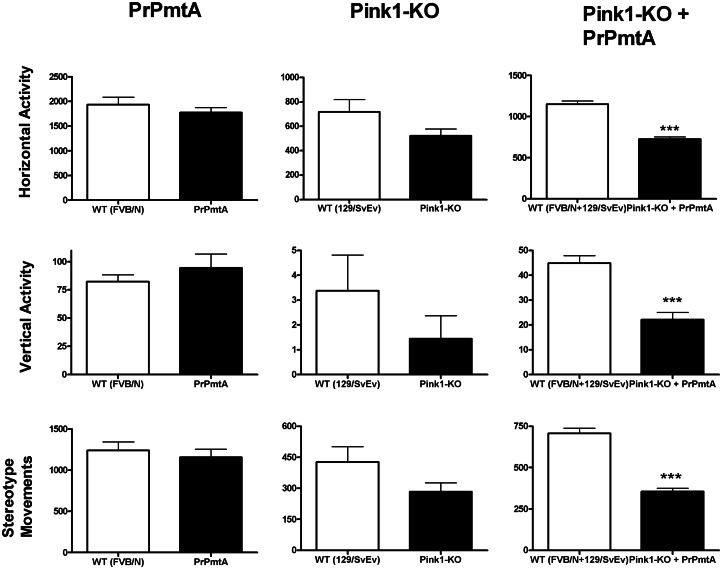
To document putative movement deficits, the open-field paradigm was used to observe and quantify the spontaneous motor activity at different adult ages. As previously published, inbred WT mice with FVB/N background are much more active than WT mice with 129/SvEv background, but for both SMs (PrPmtA and Pink1−/−), a significantly reduced horizontal and vertical locomotion in comparison with appropriate WT controls is evident by age 18 months (23,24,27). In comparison, the DM cohort exhibited highly significant reductions in the parameters ‘horizontal activity’, ‘vertical activity’ as well as ‘stereotype movement counts’ already at the age of 3 months in comparison with their appropriate WT (Fig. 2 shows the comparison with SMs; Supplementary Material, Fig. S1 shows the dynamics of this motor deficit across the DM lifespan). Gender analysis by two-way ANOVA showed females to exhibit slightly higher horizontal exploration, independent of genotype. The early onset motor dysfunction in DM mice is consistent with the increased mortality rate at young age in this line. The relative deficit of spontaneous movement in comparison with the appropriate WT animals and to both SMs persisted at ages 6, 9, 12 and 18 months, without evidence for accelerated progression in DM mice (Supplementary Material, Fig. S1). At ages >1 year, a lethal motor deficit appeared in some DM animals, guiding us to conduct further tissue analyses in midbrain and spinal cord. While it is known that the design of transgenic mice with the heterologous prion or Thy-1 promoters usually drives a strong overexpression only in the spinal cord and hindbrain (19,29), our PrPmtA line shows exceptionally strong transgene levels in dopaminergic (DAergic) midbrain neurons (23), leading to deficient dopamine release and impaired synaptic plasticity in the striatum (24,26), as well as to altered expression of synaptic vesicle cycling factors like Cplx1, Rabgef1 and Rabl2a in midbrain tissue (30).
Figure 2.
Spontaneous motor activity of DM and SM mice at the age of 3 months. Documentation of spontaneous open-field behavior as the most sensitive and reliable measure of abnormal movements in PD mouse models revealed highly significant marked reductions of horizontal activity, vertical activity and stereotype movements for the DM mice (n = 45) relative to their appropriate WT controls (n = 43), already at the age of 3 months (columns at the right side). These deficits were not observed in both SM lines versus their appropriate WT controls (n = 10 PrPmtA, 22 FVB/N, 16 Pink1−/− and 16 129Sv/Ev).
Progressive paralysis of aged DM mice
At ages beyond 1 year, within a total cohort of sixty-four DM animals under weekly observation, 16 manifested an initially unilateral, later bilateral flaccid paresis of hindlimbs, progressing within days to full paraplegia (Supplementary Material, Movie 1). Eighteen animals from the same cohort are presently alive without paralysis in this age range. Nineteen animals were sacrificed for experiments, and eight were found dead, possibly after developing a rapidly progressive pathology. At present, three DM animals at such advanced age exhibited kyphosis, falls and rigidity. Similar phenotypes were never observed in the SM lines PrPmtA (ageing studies performed since year 2000) or Pink1−/− (ageing studies performed since 2007). These aged DM animals were dissected, and their brain tissue underwent immunohistochemical studies in comparison with age- and sex-matched appropriate WT and SM controls.
Immunohistochemistry in spinal cord at the ages of 15–17 months
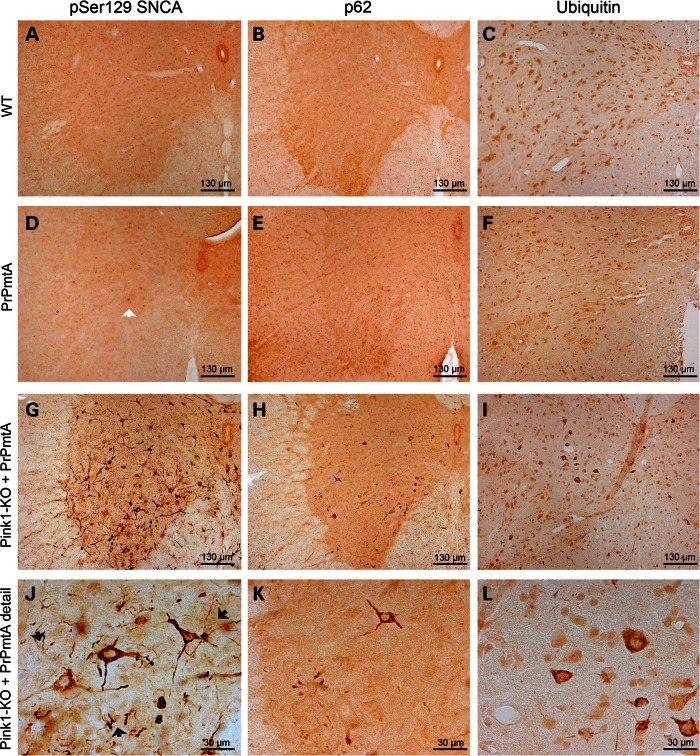
In the lumbar spinal cord of paralyzed DM mice (Fig. 3), we detected prominent pSer129-SNCA immunoreactivity, mostly throughout the gray matter, distributed in granules or threads within the cytoplasm of neurons and in neurites with neurodegenerative changes such as corkscrew morphology (Fig. 3G and J). Immunohistochemistry of the consecutive tissue sections with the universal aggregophagy marker p62 (31) (Fig. 3H and K) and that of the protein degradation tag ubiquitin (32) (Fig. 3I and L) produced similar staining patterns with less sensitivity. As in the spinal cord of late-stage PD patients (33), neurons in the dorsal horn, the autonomous preganglionic layer and particularly in the alpha-motoneuron layer were affected by aggregate formation. Thoracic and cervical spinal cord of paralytic DM mice showed milder pathology than lumbar levels (Supplementary Material, Fig. S2A). Non-paralytic DM mice of corresponding age exhibited some pSer129-SNCA immunoreactivity in the cytoplasm of lumbar spinal cord neurons, but very little pathology in neurites (Supplementary Material, Fig. S2D). Immunohistochemical analyses of WT-F1-hybrids and those of PrPmtA mice at the corresponding age did not detect aggregates with any of the three stainings, only scant pSer129-SNCA immunoreactivity in neuronal nuclei reflecting the overexpression of human A53T-SNCA in PrPmtA mice (Fig. 3D, open arrow). Similarly, the immunohistochemistry of age-matched Pink1−/− tissue was devoid of aggregate staining (Supplementary Material, Fig. S3 upper panels). Thus, protein aggregates with molecular composition and with morphology, which were reminiscent of the neuronal Lewy pathology in human PD, were observed exclusively in DM mice.
Figure 3.
Immunohistochemistry in spinal cord at ages 15–17 months. Bright-field immunohistochemical stainings were performed in age-matched WT-F1-hybrids, PrPmtA and DM mouse (for Pink1−/−, see Supplementary Material, Fig. S3 upper panels) spinal cord (focused on one anterior horn with ventral surface below and midline central canal to the right) in comparison. Protein aggregation was detected by antibodies against phospho-Serine129-alpha-synuclein (pSer129-SNCA), p62 or ubiquitin. In WT spinal cord at old age, no protein aggregates were detected. In PrPmtA, the pSer129-SNCA antibody detected few neuronal nuclei owing to the overexpression of SNCA (3D, open arrow). In DM, strong immunoreactivity throughout the gray substance in neuronal cytoplasm with extension along neurites can be observed. In high-magnification panels (panel row below), the granular and thread-like composition of the signals and the corkscrew-appearance of some immunopositive neurites become apparent (3J, black arrows), as well as the higher sensitivity of the pSer129-SNCA antibody for this pattern.
Immunohistochemistry in midbrain at the ages of 15–17 months
The immunohistochemical investigation of brain tissue from DM mice with paralysis again demonstrated protein aggregates in granula or threads throughout the cytoplasm of neurons and neurites, detectable with high sensitivity by pSer129-SNCA and with less sensitivity by p62 or ubiquitin staining (Fig. 4G–L). The staining of appropriate WT, PrPmtA or Pink1−/− mice with corresponding age (Fig. 4A–F, Supplementary Material, Fig. S3 lower panels) did not reveal specific immunoreactivity except the neuronal nuclear pSer129-SNCA signals in PrPmtA mice owing to the A53T-SNCA overexpression (Fig. 4D, open arrow). The regional distribution of the aggregates resembled the pattern previously reported for other mouse lines with very strong SNCA overexpression driven by the heterologous prion promoter (29), extending in basal brain from the ventral tegmental area (VTA), along the substantia nigra and the subthalamic nucleus to the zona incerta, with some paramedian cortical neurons also affected. Some aggregates appeared in white matter as coiled bodies, which are usually formed in oligodendrocytes in human PD (34). In paralyzed DM mice, the motor cortex showed very discrete lesions, and the striatum remained apparently normal (Supplementary Material, Fig. S2B and C). Only scant pathology was detectable in the VTA of non-paralytic DM mice within this age range (Supplementary Material, Fig. S2E).
Figure 4.
Immunohistochemistry in midbrain at ages 15–17 months. Bright-field immunohistochemical stainings were performed in age-matched WT-F1-hybrids, PrPmtA and DM mouse (for Pink1−/−, see Supplementary Material, Fig. S3 lower panels) midbrain (focused on a region between VTA and SNc, with ventral surface below and midline to the left) in comparison. Protein aggregation was detected by antibodies against phospho-Serine129-alpha-synuclein (pSer129-SNCA), p62 and ubiquitin. In WT spinal cord at old age, no protein aggregates were detected. In PrPmtA, the pSer129-SNCA antibody again detected few neuronal nuclei owing to the overexpression of SNCA in the basal brain (4D, open arrow). In DM, strong immunoreactivity throughout these areas in neuronal cytoplasm with extension along neurites can be observed. In high-magnification panels (panel row below), the granular and thread-like composition of the signals and the corkscrew-appearance of some immunopositive neurites become apparent (4J, black arrows), as well as the higher sensitivity of the pSer129-SNCA antibody for this pattern.
Double-immunofluorescence histochemistry in the midbrain with pSer129-SNCA and with the DAergic neuron marker tyrosine hydroxylase (TH) confirmed the overexpression of human A53T-SNCA (selectively detectable with the 4B12 antibody) throughout the DAergic neurons of the substantia nigra (Supplementary Material, Fig. S4 above) but showed no aggregate formation in DAergic neurons. However, we detected aggregates immunopositive for pSer129-SNCA, p62 and ubiquitin in non-DAergic cells located in a band just dorsal to the substantia nigra pars compacta (SNc) (Supplementary Material, Fig. S4). Attempts to define the affected cell type in its differentiation pattern by double-immunofluorescence indicated that the aggregates occurred not in GFAP-positive astroglia, but in NeuN-positive neurons, with features of parvalbumin-positive interneurons and rarely of VGLUT2-positive glutamatergic neurons, but not of GAD65-positive GABAergic neurons (Supplementary Material, Fig. S5). The selective early pathology in this neuron population may reflect the abnormal SNCA-overexpression pattern owing to the heterologous prion promoter rather than the established progression stages of idiopathic PD (35) or may mirror a selective early vulnerability of these neurons.
Immunoblots in protein extracts from brain hemisphere at the age of 1 year
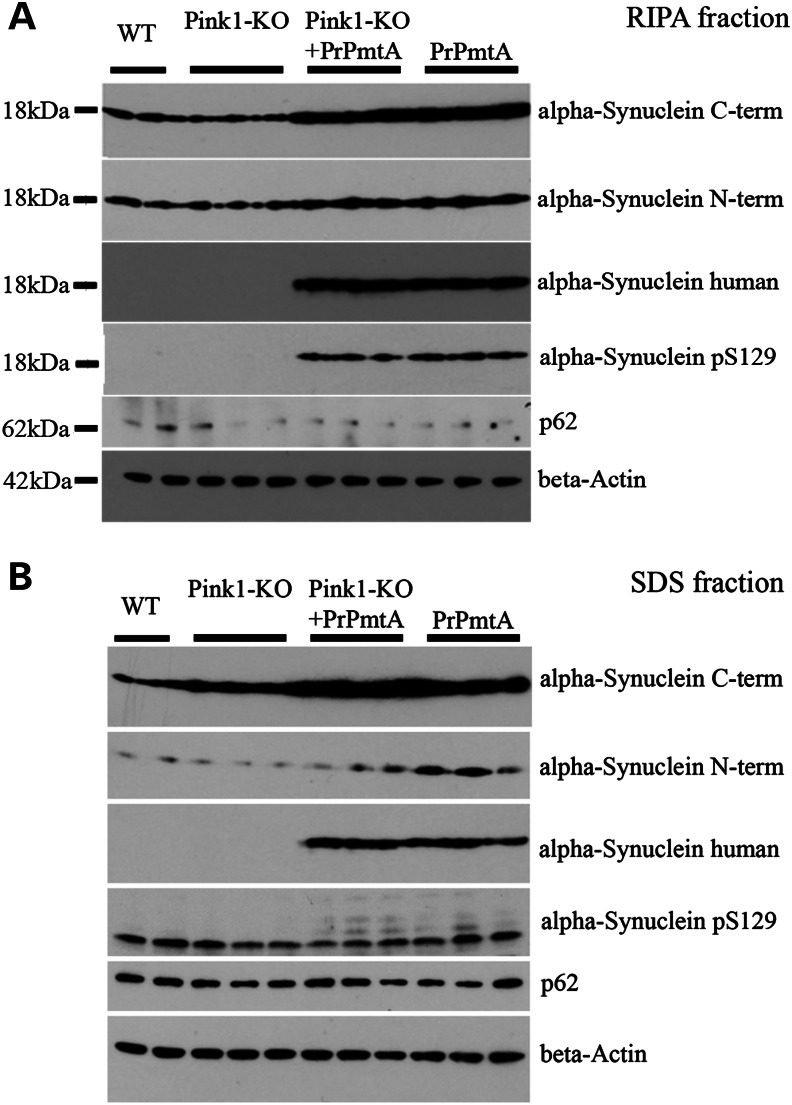
In view of the affection of diverse neuron populations throughout midbrain, basal forebrain and cortex, we sought to define pathological events in a global analysis of brain hemispheres at the age 1 year. To characterize the abundance and processing of SNCA in comparison between the DM animals without paralysis and their appropriate WT controls, as well as both SM lines, quantitative immunoblots were performed with beta-actin as loading control. Because gain-of-function mutations of SNCA lead to its progressive insolubility, the RIPA-soluble protein fraction was compared with the fraction requiring high concentrations of the strong detergent SDS for solubilization. For both fractions, an induction of the protein aggregate and Lewy body marker p62/SQSTM1 (36) could not be substantiated at this age (Fig. 5). The increased abundance of SNCA owing to the presence of transgenic human A53T-SNCA was evident in the DM and PrPmtA samples in both fractions. Interestingly, immunoreactivity against the phosphorylation at Serine129 in alpha-synuclein (pSer129-SNCA) as a marker of inclusion body formation (37) was detectable in the RIPA fraction of DM and PrPmtA samples (Fig. 5A) and exhibited additional bands of higher molecular weight in the SDS fraction of DM and PrPmtA samples (Fig. 5B). A C-terminal epitope of SNCA appeared slightly accumulated in DM lanes in the SDS fraction (Fig. 5B). Thus, homogenized brain provided only subtle hints at altered solubility and aggregation of SNCA during this age in animals without paralysis.
Figure 5.
Immunoblots in protein extracts from brain hemisphere at the age of 1 year. Quantitative comparison of proteins with soluble (RIPA fraction) or more insoluble (SDS fraction) distribution, in brain hemispheres from WT (F1-hybrids between FVB/N and 129/SvEv), both SM lines and DM mice. Altered abundance of p62 could not be detected in any condition; beta-actin was used as a loading control. (A) RIPA fraction shows the increased dosage of alpha-synuclein owing to the overexpression of human A53T-SNCA in the DM and the PrPmtA mice, with the appearance of phospho-Serine129-alpha-synuclein immunoreactivity (pSer129) in these two lines. (B) SDS fraction confirms the increased abundance of alpha-synuclein owing to the presence of human A53T-SNCA in these two lines and documents the appearance of additional pSer129 immunoreactive bands of higher molecular for both lines. The antibody against the alpha-synuclein C-term epitope detected a putative light increase of immunoreactivity in DM tissue.
Global ubiquitination changes of DM brain hemispheres at the age of 18 months
To obtain molecular insights into the late-stage digenic pathology that leads to protein aggregates, brain hemispheres were dissected from DM mice without paralysis and appropriate WT controls (three versus three, sex-matched) at the advanced age of 18 months. Protein extraction, proteolysis and quantitative assessment of post-translational ubiquitination motifs by mass spectrometry (MS) (UbiScan®) were performed. This global proteomics approach identified increased ubiquitination at four motifs within SNCA (on average 6.5-fold at lysines K12, K23, K43 and K45, a consistent increase at all sites in all three animal pairs) as the prominent effect (Supplementary Material, Table S1A). Among many other effects with nominal significance, but weaker fold-changes and less consistency, the reduced ubiquitination of H2AX [gene symbol H2afx, also called HIST2H2AB, a variant H2A histone involved in repair that is an established marker of DNA-damage foci, see (38)] at K96/K119 stands out biomathematically as the only change with consistency among all three animal pairs and with a fold change exceeding 2.8-fold in every pair. The complete data set including quality controls is provided in Supplementary Material, Table S1A; additional bioinformatic information is shown in Supplementary Material, Table S1B. These results document enhanced cellular efforts to eliminate SNCA but suggest that these compensatory mechanisms are insufficient in view of putative incipient genotoxicity, which appears to result in H2AX stabilization.
However, in view of the multimorbidity at such advanced age and the small number of animals surveyed, biological variability rather than Parkinson pathology might underlie these observations. Thus, further follow-up studies at very advanced age in independent cohorts of animals will become necessary. In the meantime, we made a second global molecular screen with a more established and sensitive survey technique at initial disease stages, employing two age groups, more animals and distinct areas of the nervous system.
Global transcriptome changes of DM brain hemispheres at the ages of 6 weeks and 6 months
For this purpose, the transcriptome profile was documented by oligonucleotide microarrays (Affymetrix®) at the age of 6 weeks and 6 months, in three brain regions, for every condition in four DM versus four appropriate WT mice of matched sex. Significant expression dysregulations were determined for the two ages in three areas (all areas are affected by PINK1 absence; but abundance of transgenic human A53T-SNCA is prominent in midbrain/brainstem and striatum) and filtered for consistency. Many ubiquitous effects with similar dysregulations appeared in cerebellum, midbrain/brainstem and striatum at both early ages (Supplementary Material, Table S2), whereas previous transcriptome profiling of Pink1−/− SM brains at ages 6 and 18 months in the absence of a neurotoxic stressor had failed to detect these changes. The dysregulated levels of several transcripts with a role for the PTEN/PI3K-pathway (Pten-Pigo-Ndfip2-Rragd-Ube2m-Dapk1-Mapk8), for ubiquitin degradation (Dcaf17-Ube2m-Otub1-Vps37c-Ndfip2), for mitochondrial biology (the Parkin neighbor gene Qk and Pnpt1-Lars2-Bcat2-Xaf1-Ndufv1-Acaa1a/b-Mrpl9-Slc25a12), microtubular transport (Kif5c-Dynlt1a-Dynlt1d-Dynlt1f-Dctn5), vesicle dynamics (Ergic-Cisd2-Htt-Nisch), synaptic signaling (Gabrb3-Creb3) and DNA-damage repair (Atm-Mlh1-Ssbp2-Tmem181a) showed significance in these regions and ages, with consistency (Supplementary Material, Table S2). Among the significant effects with preferential appearance in midbrain/brainstem and striatum, the dysregulated levels of neurotransmitter receptors/receptor scaffolds (Homer1-Adora2b) had been observed previously in the PrPmtA SM mice (39). Similarly, the dysregulated levels of members of the Rab family of vesicle regulators (Rab2b-Rab42) are possibly a consequence of the overexpression of A53T-SNCA and its association with presynaptic vesicles, because similar dysregulation of the Rab cycle factors Rabgef1 and Rabl2a was already observed in the SM PrPmtA midbrain tissue (30). The dysregulation of March5 levels in the DM midbrain and striatum appears immediately credible, because MARCH5 ubiquitinates mitofusin during mitophagy (40), in parallel to PINK1/Parkin (41–44).
Transcriptome validation by qPCR in DM midbrain at the age of 6 months
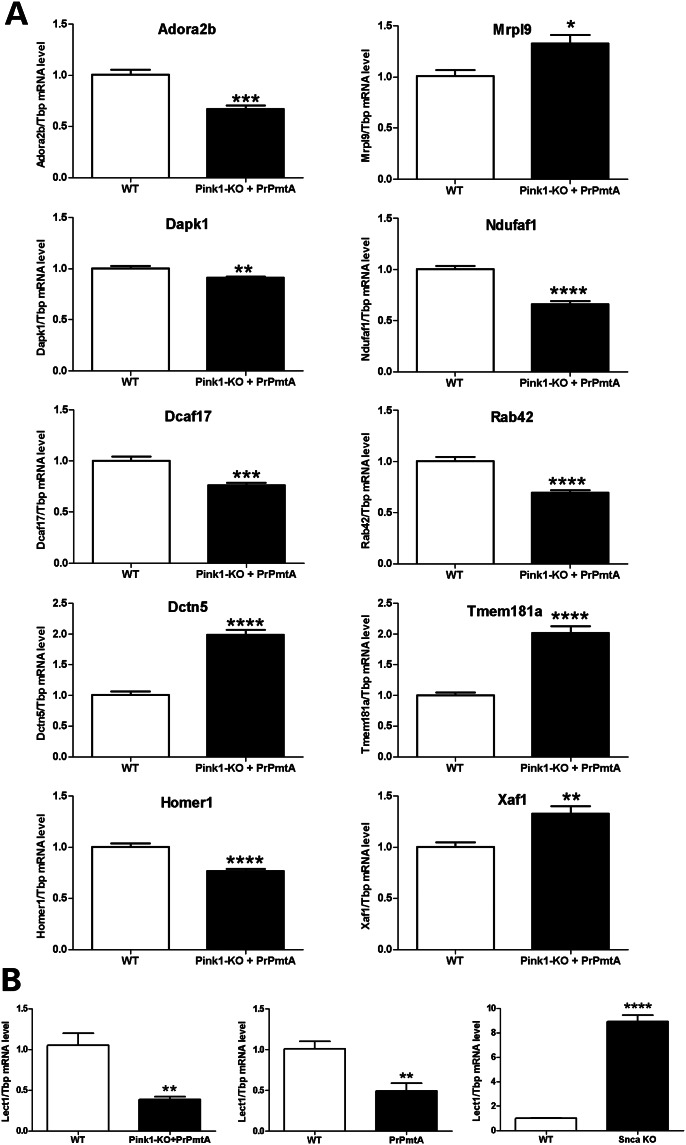
For independent validation of these microarray dysregulations (representing mostly mRNA 3′UTR sequences), we studied midbrain from additional six DM and six appropriate WT mice at the age of 6 months, employing qPCR assays (usually designed to target exon junctions). In spite of the change in target sequences and the changed brain region specificity, this approach reproduced (Fig. 6A) significant downregulations for the synaptic receptor mRNA Adora2b (0.67-fold), the autophagosome dynamics kinase mRNA Dapk1 (0.91-fold), the ubiquitination cofactor mRNA Dcaf17 (0.76-fold), the glutamate receptor scaffold mRNA Homer1 (0.77-fold), the putative vesicle cycling factor mRNA Rab42 (0.70-fold) and significant upregulations for the microtubular transport activator mRNA Dctn5 (1.98-fold), the mitochondrial ribosomal subunit mRNA Mrpl9 (1.33-fold), the Mig-14 Wnt-binding domain-containing factor (apparently involved in DNA toxicity of cytolethal distending toxins) mRNA Tmem181a (2.02-fold) and the mitochondrial apoptosis activator mRNA Xaf1 (1.33-fold).
Figure 6.
Transcriptome validation by qPCR in DM midbrain at the age of 6 months. (A) The global transcriptome survey was assessed by the independent technique qPCR in midbrain of additional animals (six DM versus six WT-F1-hybrids). Tissues from an earlier age were employed to define expression dysregulations that play a role at disease stages before the formation of aggregates. This demonstrated significant downregulations for Adora2b, Dapk1, Dcaf17, Homer1, Rab42 and significant upregulations for Dctn5, Mrpl9, Tmem181a, Xaf1 mRNA levels. The bar graphs show average 2−ΔΔCt values ± SEM, relative to Tbp loading control, normalized to WT. These data identify factors with a role in early pathogenesis, which reflect anomalies in mitochondrial translation/apoptosis, ubiquitination, microtubular membrane transport, synaptic vesicles/receptors and DNA-damage repair. (B) On the basis of transcriptome bioinformatics identifying significant Lect1 transcript downregulation in the DM midbrain, qPCR was employed to demonstrate Lect1 levels to correlate inversely with the gain-of-function (left panel, five DM versus five F1-hybrid WT mice) (middle panel, four PrPmtA versus four FVB/N WT mice) and the loss-of-function (right panel, five Snca-KO versus five 129S6/SvEvTac WT mice) of alpha-synuclein in brain hemispheres of 6-month-old animals.
Thus, the validation confirmed several molecular pathway changes which precede the appearance of SNCA aggregates in brain hemispheres of DM animals and which may represent shared targets of autosomal recessive and dominant PD mechanisms.
Transcriptome bioinformatic analysis
While the genes identified above show striking expression changes in view of their consistency across brain regions and ages, we also endeavored to identify region-specific and progression-correlated anomalies, employing advanced bioinformatics of pathway dysregulation through several approaches. In an initial search for over-representation of strongly dysregulated transcripts per gene ontology molecular function, the dysregulations of 1-phosphatidylinositol 4-kinase activity exceeded expectations strongly and consistently (midbrain at 6 months with P-value 0.003 and odds ratio 61.7; midbrain at 6 weeks with P-value 0.004 and odds ratio 53.7; striatum at 6 months with P-value 0.003 and odds ratio 58.1; striatum at 6 weeks with P-value 0.004 and odds ratio 50.6; cerebellum at 6 months with P-value 0.004 and odds ratio 50.2; cerebellum at 6 weeks with P-value 0.004 and odds ratio 49.7). In keeping with the strong dysregulation of Pigo expression (Supplementary Material, Table S2), these findings may suggest that the phosphoinositol composition of inner membrane sides is altered, with consequences for endocytosis and vesicle dynamics, but such hypotheses require future experimental validations through metabolomics. In further over-representation analyses of strongly dysregulated transcripts per gene ontology biological process, the relevant pathways (each of them significant in five analyses among the three tissues at two ages) included the (I) regulation of microtubule polymerization or depolymerization, (II) long-chain fatty acid biosynthetic process and (III) regulation of translation.
Additional evaluation of the global transcriptome per brain region and age by the Gene Set Enrichment Analysis (GSEA) software (summarized in Supplementary Material, Table S3) revealed downregulations in the NOUSHMEHR_GBM_SILENCED_BY_METHYLATION signature (for cerebellum at 6 weeks with significance, for striatum at 6 weeks and for midbrain at both ages with statistical trends). The downregulated transcripts in these samples comprised Lect1, Fmod, Gjb2, Fzd6 and Cthrc1 (Supplementary Material, Fig. S6). This was particularly credible in light of previous converse results in a global transcriptome profiling effort of our team for mice with genetic Snca ablation (39,45), where the loss-of-function of alpha-synuclein had resulted in unpublished significant upregulations of Lect1 (7.5-fold, Affymetrix oligonucleotide spot 1460258_at) and Fmod (1.6-fold, Affymetrix oligonucleotide spot 1437718_x_at) transcript levels. To validate by qPCR that Lect1 levels show indeed an inverse correlation to alpha-synuclein levels, we studied the brain hemispheres of 6-month-old DM and SM PrPmtA versus Snca knockout animals (46). These experiments confirmed that Lect1 levels are a novel molecular marker of the gain-of-function or loss-of-function of alpha-synuclein in brain (Fig. 6B), in view of a reduction to 37% in DM brain (P-value 0.0025), a reduction to 49% in PrPmtA brain (P = 0.0078) and an induction to 892% in Snca-KO brain (P < 0.0001). This effect on Lect1 expression is possibly a correlate of the known alpha-synuclein effects on cell adhesion (47,48). Lect1 is known to be coregulated with Dusp5 and Wnt7b by NGF and p53 (49). A known phenotype of decreased Lect1 levels is elevated levels of H2AX phosphorylation (50). Further GSEA findings included significant upregulations in the NAGASHIMA_EGF_SIGNALING_UP signature for cerebellum at both ages (Supplementary Material, Figs S7 and S8), which were similarly observed for Dusp1/Dusp6 and c-Fos already in the SM PrPmtA line in striatum of 18-month-old mice (51).
Other prominent GSEA results concerned significant downregulations in the MITOCHONDRIAL_MATRIX signature for cerebellum at 6 months, significant upregulations in the QI_HYPOXIA_TARGETS_OF_HIF1A_AND_FOXA2 signature for midbrain at 6 months and significant upregulations in the PID_ATM_PATHWAY signature for midbrain at 6 months (Supplementary Material, Table S3 and Figs S9–S11). Importantly, for the interpretation of the reduced ubiquitination of H2AX observed at old age, this latter gene set contained a significant upregulation of the corresponding transcript H2afx, underscoring diverse cellular efforts in DM brain tissue through several age steps to upregulate the abundance of this DNA-damage repair factor. Apart from this upregulation, a significant increase of Cdc25a transcript levels contained in the DNA-damage-response pathways was noted in midbrain at 6 months and already in midbrain at 6 weeks. We chose to ignore several significant changes in the LEIN_CHOROID_PLEXUS_MARKERS signature representing several transcripts with specific expression in the choroid plexus of brain ventricles (Supplementary Material, Table S3), because our tissue dissection procedures had not controlled the presence or absence of these extraneural tissues, before the molecular surveys were performed.
DISCUSSION
To improve our understanding of the interactions between diverse PD causes and their potential convergence onto shared substrates and pathways, we generated a DM mouse line, where the dominant action of SNCA neurotoxicity is combined with the recessive loss of PINK1 neuroprotection. These animals exhibit potentiated pathology in a shortened survival curve and earlier deficits of spontaneous motor activity. More than 25% of the DM animals aged >1 year exhibited penetrance of a progressive hindlimb paralysis, possibly in dependence on genetic background variance, and this lethal phenotype was accompanied by accumulation of protein aggregates immunopositive for pSer129-SNCA, p62 and ubiquitin. This exacerbated toxicity in our digenic PD model was surprising, because two previous PD modeling attempts in DM mice involving DJ1-SNCA and Parkin-SNCA had not shown any potentiation (19–21). The aggregation pathology did not occur preferentially in dopaminergic neurons, but in diverse neuron populations of the spinal cord, midbrain, basal brain and cerebral cortex. On global surveys of post-translational protein modifications and of the transcriptome, we also detected activation of cellular protein degradation efforts for SNCA, as well as very early perturbations of a spectrum of pathways that range from mitochondrial dynamics, synaptic adaptation, microtubular trafficking, DNA-damage repair, to cell adhesion. As a novel early tissue-specific marker of alpha-synuclein function, an inverse correlation to Lect1 expression was identified. The role of LECT1 protein for neurons is poorly studied, but it shares the beta-sheet-aggregation-inhibitory BRICHOS domain (52) with BRI2, a protein which is mutated in familial British and Danish dementias, influences mitochondrial membrane potential, modulates oxidative stress, interacts with amyloid precursor protein, is deposited in AD plaques and reduces amyloid beta deposition (53,54). BRI2-deficient mice display synaptic and memory loss in the absence of neurofibrillary tangles or amyloid plaques (55).
On the one hand, our evidence points to PINK1 as a relevant modulator of SNCA aggregation and Lewy pathology. Indeed, Lewy pathology has been observed in PD patients with PINK1 mutations (56,57), but the mechanism is unclear. It is well established that PINK1 and its downstream effector Parkin are crucial requisites for the autophagic pathway, which selectively eliminates damaged mitochondria (mitophagy) (2,58). In addition, PINK1 could impact mitochondrial degradation through its recently reported phosphorylation of ubiquitin and activation of the Parkin E3 ligase activity (59). It is therefore conceivable that the absence of PINK1 could lead to the accumulation of dysfunctional mitochondria with low membrane potential, to enhanced generation of oxygen radicals and to potentiated formation of SNCA fibrils. Moreover, it is also known that PINK1 is regulated during nonselective autophagy and plays a role for the initiation and for the composition of the autophagy machinery (60–62). PINK1 was shown to modulate the vesicular elimination of cargo proteins through the lysosomal pathway (63). Thus, the absence of PINK1 might impair the autophagic degradation of protein aggregates in any cell. Differential vulnerability of specific neuron populations might arise owing to a high ratio of transgenic versus endogenous SNCA levels and low cell-specific efficacy of aggregophagy and compensation efforts.
Our evidence from this long-term DM assessment of PINK1 as a modulator of SNCA aggregation is complemented by a very recently published short-term study of SM Pink1−/− mice, which were subjected to stereotactic injections of adeno-associated virus into the substantia nigra (64), mediating an 8- to 30-fold SNCA overexpression level during an observation period of 9 weeks (65). An increased presence of SNCA aggregates and higher loss of nigral dopaminergic and SNCA-positive neurons in Pink1−/− mice were observed (64). Spinal cord and behavior effects were not studied. Jointly, both approaches corroborate the role PINK1 as a modulator of SNCA-triggered neurotoxicity.
On the other hand, our data also indicate that SNCA overexpression in neurons acts as a stressor, which is useful to elucidate the mechanism of PINK1-dependent stress responses and neuroprotective actions. In the past, it was difficult to identify the downstream targets of PINK1, because the absence of PINK1 resulted in mild phenotypes under normal conditions (22,27,66,67). In vitro investigations advanced considerably, once CCCP (carbonyl cyanide m-chlorophenylhydrazone) as uncoupling agent and mitochondrial stressor was introduced to reveal PINK1 actions in peripheral cells such as HeLa (41,68–70), but CCCP proved to be of limited value in neuronal cells or in brain tissue (71,72). Our observation of ubiquitous expression changes in DM brain at very early ages, which had not been observed in SM Pink1−/− brain or in PrPmtA brain, suggests that SNCA accumulation might act as the appropriate chronic mitochondrial stress in neurons, in agreement with previous claims (73,74). It will be interesting to explore the value of PINK1-dependent neuroprotective responses for future preventive therapies of PD.
METHODS
Breeding and ageing of DM mice with homozygosity for Pink1−/− and for PrPmtA
Our generation and characterization of the Pink1−/− mouse line in a 129/SvEv background as well as of the PrPmtA line with A53T-SNCA overexpression in a FVB/N background were described previously (23,24,27). Double-mutant mice were generated by crossing these lines to obtain animals with homozygosity for both genotypes and establishing a colony by interbreeding such animals. Given that the genetic background of these DM mice would contain 129/SvEv and FVB/N in a 50 : 50 distribution on average, as WT control mice we aged F1-hybrids that had resulted from crossbreeding of 129/SvEv and FVB/N mice that had been derived from littermates of SM animals. To compensate for background variation effects, the cohort of DM animals followed over time was larger than the inbred cohorts of both SMs. All animals were maintained in individually ventilated cages under 12 h light cycle with food and water ad libitum. Sentinel mice and regular health monitoring including blood tests for viral and parasite infections uncovered no pathology. Housing of animals was in accordance with the German Animal Welfare Act, the Council Directive of 24 November 1986 (86/609/EWG) with Annex II and the ETS123 (European Convention for the Protection of Vertebrate Animals). The mice under investigation were bred and aged at the FELASA-certified Central Animal Facility (ZFE) of the Frankfurt University Medical School. After decapitation, the organs were removed and immediately frozen in liquid nitrogen. Other animal were perfused for fixation and histological analyses.
Survival curves
Age at death (by natural causes or by veterinarians' decision to curb animal suffering, at the latest around 2 years of life) was documented for each animal. For statistical analysis of each mouse line, the GraphPad Prism software package was used and Kaplan–Meier survival curves were generated. Significance was determined with log rank (Mantel–Cox) tests.
Spontaneous motor activity
All tests were performed in a cohort of 43 DM and 45 appropriate WT (FVB/N + 129/SvEv) mice at 3, 6 and 12 months of age, in comparison with a cohort of 16 SM Pink1−/− and 16 appropriate WT (129/SvEv) mice and to a cohort of 10 PrPmtA and 21 appropriate WT (FVB/N) controls. General health and neurological performance were evaluated with the SHIRPA test (75), obvious pathology was documented in video by a Panasonic HCV707M camera and the digital file edited with Microsoft Windows 7 Movie Maker. The weight as a measure of metabolism and the production of urine/feces during 5-min open-field tests as a measure of anxiety were recorded. The spontaneous movement activity of mice in a 20 × 20 cm arena was registered by infrared beams in a Digiscan monitor (AccuScan, Columbus, OH, USA) as previously described for both SMs (23,24,27).
Immunoblots
Brains from mice at age 1 year were dissected to use one hemisphere for protein extraction and immunoblotting, performed as previously described (76). Fifty milligrams of tissue were homogenized with a 7-ml Dounce tissue grinder (Wheaton) in 10 vol. RIPA buffer [50 mm Tris–HCl (pH 8.0), 150 mm NaCl, 1 mm EDTA, 1 mm EGTA, 1% Igepal CA-630 (Sigma), 0.5% sodium deoxycholate, 0.1% SDS and one tablet Complete Protease Inhibitor Cocktail (Roche)] followed by 15 min of incubation on ice. After centrifugation at 16 000g at 4°C for 20 min, the supernatant (RIPA fraction) was preserved and 1/2 vol. 2× SDS-lysis buffer (137 mm Tris–HCl, pH 6.8; 4% SDS; 20% glycerol) was added to the pellet, which was then sonicated and centrifuged at full-speed for 10 min. The second supernatant represented the SDS fraction. Protein concentration was determined using the Bradford Method kit (Roth). 10–20 µg of proteins were loaded and separated on NOVEX 4–12% NuPage gels and transferred to Protran BA83 nitrocellulose membranes (Whatman). Incubation was done with the following primary antibody and titers: anti-α-synuclein N-term (1 : 500, Millipore #04-1053), anti-α-synuclein C-term (1 : 2000, Cell Signaling #2642), anti-human-α-synuclein (1 : 2000, Covance clone 4B12), anti-phospho-α-synuclein (1 : 2000, Abcam EP1536Y), anti-p62 (1 : 2000, Novus Biologicals NBP1-49956) and anti-β-actin as loading control (1:10 000, Sigma). The adequate secondary antibodies (anti-mouse IgG NA931V, anti-rabbit IgG NA934V, GE Healthcare UK Limited) were incubated for 1 h at the recommended titers. Detection was performed with SuperSignal West Pico (Thermo Scientific). For densitometry, the Image Master Total Lab software (Amersham Pharmacia) was used.
Immunohistochemistry
Tissue preparation for histology
For immunohistochemical analysis, animals were transcardially perfused. For this, mice were deeply anesthetized using sodium pentobarbital i.p. (Nacoren, Merial) and perfusion-fixed with buffered ice-cold, 4% paraformaldehyde (PFA) (pH 7.4) (Sigma–Aldrich). In order to ensure a good perfusion quality, heparin (Ratiopharm) was injected transcardially just before PFA perfusion. 4% paraformaldehyde solution was applied with a peristaltic pump (Minipuls 3, Abimed Gilson) at a speed of 20 ml/min during 7 min. Thereafter, brain and spinal cord were rapidly extracted and post-fixed in the same fixative solution for 12 h at 4°C followed by cryoprotection in ice-cold phosphate-buffered sucrose (25% sucrose in 0.1 m PB) for 12 h at 4°C.
pSer129-SNCA and p62 bright-field immunohistochemistry in free-floating sections
For immunohistochemistry of phospho-Ser129-alpha-synuclein and p62, brains were cut into 30-µm and lumbar (with some cervical and thoracic) spinal cord into 40-µm coronal sections on a vibrating blade microtome (Leica VT1000 S). Free-floating sections were rinsed in phosphate-buffered saline (PBS) at 4°C, followed by 1 h of incubation at 4°C in blocking solution [10% normal goat serum (Vector Laboratories), 0.5% Triton X-100 (AppliChem), PBS] to prevent unspecific staining. Thereafter, sections were incubated with the respective first antibody, either phospho-Ser129-alpha-synuclein (Abcam, dilution 1 : 500) or p62 (Novus Biologicals, dilution 1 : 200) in carrier solution [1% normal goat serum (Vector Laboratories), 0.5% Triton X-100 (AppliChem), PBS] for 48 h at 4°C. Then, sections were washed in PBS and incubated with the appropriate biotinylated secondary antibody for 2 h at room temperature (Vectastain anti-rabbit dilution 1 : 500; Vector Laboratories). Tissue-bound antibodies were visualized by incubating for 90 min with avidin–biotin–peroxidase complex (Vectastain ABC-kit, Vector Laboratories) and Peroxidase substrate Vector NovaRED Substrate Kit (SK-4800 kit; Vector Laboratories). Sections were dehydrated in ascending ethanol concentrations followed by xylene; they were cover-slipped using VectaMount permanent mounting medium (H-5000; Vector Laboratories).
Ubiquitin bright-field immunohistochemistry in paraffin-embedded sections
For immunohistochemistry with an anti-ubiquitin antibody, perfusion-fixed brains and lumbar spinal cord were dehydrated in ascending ethanol concentrations followed by xylene and molted paraffin (Paraplast X-TRA tissue-embedding medium; McCormick scientific) (brain 24 h incubation in each concentration, spinal cord 2 h). Thereafter, tissue was embedded in paraffin (Paraplast X-TRA tissue-embedding medium; McCormick scientific) and stored at room temperature until sectioning. Coronal sections of 5 µm thickness were cut using a microtome (Jung GmbH) and floated in a 50°C water bath containing distilled water. After mounting the sections, immunohistochemistry for ubiquitin was performed. For this, sections were deparaffinated in xylene and rehydrated in descending series of ethanol concentrations. Antigens retrieval was performed by boiling the sections in 0.01 m citrate buffer, pH 6.0, five times for 3 min in a microwave. To reduce endogenous peroxidase staining, sections were incubated in PBS containing 3% H2O2 and 20% ethanol for 5 min. Thereafter, nonspecific epitopes were blocked by incubation with TRIS containing 5% BSA, 20% normal goat serum (Vector Laboratories) and 0.5% Triton X-100 (AppliChem) for 1 h at room temperature. Sections were incubated in carrier solution [1% normal goat serum (Vector Laboratories), 0.5% Triton X-100 (AppliChem), PBS] containing the primary ubiquitin antibody (DAKO; dilution 1 : 1000) at 4°C overnight, followed by 90 min of incubation with the biotinylated secondary antibody (Vectastain anti-rabbit dilution 1 : 500; Vector Laboratories) at room temperature. Tissue-bound antibodies were visualized as described for free-floating sections by incubating for 90 min with avidin–biotin–peroxidase complex (Vectastain ABC-kit, Vector Laboratories) and a Peroxidase substrate Vector NovaRED Substrate Kit (SK-4800 kit; Vector Laboratories). Sections were dehydrated in ascending ethanol concentrations followed by xylene and cover-slipped using VectaMount permanent mounting medium (H-5000; Vector Laboratories).
Double-immunofluorescence in paraffin-embedded sections
In order to analyze the localization of human A53T-SNCA-, pSer129-SNCA-, p62- and ubiquitin-positive cells, we performed a double-immunofluorescence staining with TH. Sections of 5 µm thickness were deparaffinated in xylene and rehydrated in descending series of ethanol as described for bright-field immunohistochemistry. For the double-immunofluorescence of human A53T-SNCA (the antibody 4B12 recognizes the human isoform of SNCA selectively) and TH, antigen retrieval was performed by incubating the slices in TRIS pH 9 for 15 min at 90°C. For the double-immunofluorescence of pSer129-SNCA and TH, no antigen retrieval was performed, and to reduce endogenous peroxidase staining, sections were incubated in TRIS containing 20% methanol (Sigma–Aldrich) and 3% H2O2 for 30 min at room temperature. For the double-immunofluorescence of p62 and TH, antigen retrieval was performed by incubating the slices in TRIS pH 9 for 15 min at 90°C. For the double fluorescence of ubiquitin and TH, slices were boiled in 0.01 m citrate buffer, pH 6.0, five times for 3 min in a microwave. For all stainings except pSer129-SNCA with TH, endogenous peroxidase was reduced by incubating sections in PBS containing 3% H2O2 and 20% ethanol for 5 min at room temperature. In order to block nonspecific epitopes, all slices were incubated with TRIS containing 5% BSA, 20% normal goat serum (Vector Laboratories) and 0.5% Triton X-100 (AppliChem) for 1 h at room temperature. Thereafter, sections were incubated in carrier solution [1% normal goat serum (Vector Laboratories), 0.5% Triton X-100 (AppliChem), PBS] containing either the primary antibodies phospho-Ser129-alpha-synuclein (Abcam, dilution 1 : 500) with TH (Pel-freez Biologicals; dilution 1 : 1000), p62/SQSTM1 (Novus Biologicals, dilution 1 : 200) with TH (Pel-freez Biologicals; dilution 1 : 1000), ubiquitin (DAKO; dilution 1 : 1000) with TH (Pel-freez Biologicals; dilution 1 : 1000) or alpha-synuclein (4B12; Pierce Antibodies; dilution 1 : 500) with TH (Millipore; dilution 1 : 1000), at 4°C for 24 h, followed by 6 h of incubation with the appropriate secondary antibodies at room temperature. Secondary antibodies were anti-rabbit or anti-mouse Cy2- or Cy3-conjugated IgG (1 : 1000; Jackson ImmunoResearch). Slices were cover-slipped using fluorescence mounting medium (DAKO). The images for the double fluorescent immunostainings were collected on a confocal laser-scanning microscope (Nikon eclipse90i, Nikon) by using a 60× objective.
Double-immunofluorescence in free-floating sections
In order to characterize the pSer129-SNCA-, p62- and ubiquitin-positive but TH-negative cells of paralyzed DM mice in more detail, double-immunofluorescence with rabbit anti-pSer129-SNCA and respective mouse anti-GFAP, -NeuN, -GAD65, -VGLUT2 and -parvalbumin was performed. Free-floating sections of 30 µm were rinsed in PBS. Thereafter, sections were blocked for 1 h in blocking solution [10% normal goat serum (Vector Laboratories), 0.5% Triton X-100 (AppliChem), PBS] to prevent unspecific staining. The following primary antibody combinations were used: pSer129-SNCA (Abcam, dilution 1 : 500) and GFAP (Cell Signaling, dilution 1 : 500); pSer129-SNCA (Abcam, dilution 1 : 500) and NeuN (Chemicon, dilution 1 : 1000); pSer129-SNCA (Abcam, dilution 1 : 500) and GAD65 (Sigma, dilution 1 : 500); pSer129-SNCA (Abcam, dilution 1 : 500) and VGLUT2 (Millipore, dilution 1 : 500); pSer129-SNCA (Abcam, dilution 1 : 500) and Parvalbumin (Chemicon, dilution 1 : 500). Primary antibodies were incubated in carrier solution [1% normal goat serum (Vector Laboratories), 0.5% Triton X-100 (AppliChem), PBS] for 48 h at 4°C. Thereafter, sections were washed three times for 10 min in PBS before they were incubated for 6 h at room temperature with the appropriate secondary antibodies. Secondary antibodies were anti-rabbit Cy2-conjugated IgG (Jackson ImmunoResearch, dilution 1 : 1000) and anti-mouse Cy3-conjugated IgG (Jackson ImmunoResearch, dilution 1 : 1000). Slices were cover-slipped using fluorescence mounting medium (DAKO). Images were acquired using a confocal laser-scanning microscope (Nikon eclipse90i) and a ×60 objective and NIS-Elements Imaging Software 4.20.00.
Global ubiquitination survey
Brain hemispheres from mice at age 18 months (three DM versus three WT matched for sex, both groups with 50 : 50 FVB/N and 129/SvEv background) were dissected in parallel, snap frozen in liquid nitrogen, stored at −80°C and shipped on dry ice for the commercial UbiScan® procedure by Cell Signaling Technology, Inc. In short, tissue extracts were protease-digested and subjected to C18 solid-phase extraction. The lyophilized peptides were immunoprecipitated by protein-A-agarose-immobilized ubiquitin-branch-motif antibody #3925. Peptides were loaded directly onto a 10 cm × 75 μm PicoFrit capillary column packed with Magic C18 AQ reversed-phase resin. The column was developed with a 90-min linear gradient of acetonitrile in 0.125% formic acid delivered at 280 nl/min. The peptide identification with relative quantification by MS occurred by LC-MS/MS analysis using LTQ-Orbitrap-VELOS with ESI-CID Sorcerer search. The MS parameter settings were as follows: MS Run Time 96 min, MS1 Scan Range (300.0–1500.00), Top 20 MS/MS (Min Signal 500, Isolation Width 2.0, Normalized Coll. Energy 35.0, Activation-Q 0.250, Activation Time 20.0, Lock Mass 371.101237, Charge State Rejection Enabled, Charge State 1+ Rejected, Dynamic Exclusion Enabled, Repeat Count 1, Repeat Duration 35.0, Exclusion List Size 500, Exclusion Duration 40.0, Exclusion Mass Width Relative to Mass, Exclusion Mass Width 10 ppm). MS/MS spectra were evaluated using SEQUEST 3G and the SORCERER 2 platform from Sage-N Research (v4.0, Milpitas, CA, USA). Searches were performed against the most recent update of the NCBI Mus musculus database with mass accuracy of ±50 ppm for precursor ions and 1 Da for product ions.
Results were filtered with mass accuracy of ±5 ppm on precursor ions and presence of the intended motif (K-GG). With double injections of the 6 biological samples, 12 LC-MS/MS experiments were conducted and bioinformatically processed, using the maximum % coefficient of variation (% CV) to control replicate reproducibility. Using a 5% default false-positive rate used to filter the SORCERER results, this procedure yielded a total of 4224 redundant ubiquitinated peptide assignments (Details Tab in Supplementary Material, Table S1) to 2334 non-redundant ubiquitinated peptides (Summary Tab in Supplementary Material, Table S1).
Global transcriptomics
Cerebellum, midbrain plus brainstem and striatum from mice with ages 6 weeks or 6 months (for each analysis four DM versus four WT tissues) were sent to MFT Services (Tübingen, Germany) to be processed as published previously (77). In summary, after RNA extraction and verification of RNA integrity (RIN values above 9.0), the linear amplification and biotinylation of 100 ng of total RNA were performed with the GeneChip HT 3′IVT Express Kit (Affymetrix, Santa Clara, CA, USA). GeneChip HT Mouse Genome 430 2.0 Array Plates (Affymetrix) were used to hybridize 15 µg of labeled and fragmented cRNA, to wash, stain and scan automatically in a GeneTitan instrument (Affymetrix). Each of these oligonucleotide microarray chips is able to detect >39 000 transcripts with multiple probes for each mRNA. Visual inspection of scanned images was used to control for hybridization artifacts and proper grid alignment. AGCC 3.0 (Affymetrix) processed results were stored in CEL files. Further data analysis steps were carried out with the software platform R 2.14.0 and Bioconductor 2.14.0. First, the complete expression information from every chip was background corrected, quantile normalized and summarized with Robust Multichip Average. Empirical Bayes shrinkage of the standard errors was employed to derive the moderated F-statistic. The resulting P-values underwent multiple testing corrections according to ‘Benjamini-Hochberg’. A decision matrix was produced through the function ‘decide tests’ within the limma package, to attribute significant changes to individual contrasts. Thus, significant up- or down-regulations were encoded by values of 1 or −1, respectively, to compare the consistency of significant expression changes across tissues and ages. All original transcriptome data are being deposited at the public database Gene Expression Omnibus (DM data at GEO series accession # GSE60414, SM Pink1-KO data at # GSE60413, http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE60414).
qPCR transcriptome validation
Midbrain from mice aged 6 months were extracted with Trizol (Invitrogen) following manufacturer’s instructions. Before cDNA synthesis, 1 μg of RNA was digested with DNase I Amplification Grade (Invitrogen). Reverse transcription was performed with SuperScript III Reverse Transcriptase (Invitrogen). Subsequently, expression levels were measured with the StepOnePlus Real-Time PCR System (Applied Biosystems) using cDNA from 25 ng RNA, 10 μl of FastStart Universal Probe Master (Rox) Mix (04914058001, Roche) and 1 μl of one of the following TaqMan Assays (Applied Biosystems): Adora2b (Mm00839292_m1), Dapk1 (Mm01352536_m1), Dcaf17 (Mm01334341_m1), Dctn5 (Mm00727515_s1), Homer1 (Mm00516275_m1), Mrpl9 (Mm00659648_m1), Mrpl9 (Mm00659648_m1), Pink1 (Mm00550827_m1) as independent control of genotypes, Rab42 (Mm01187370_g1), Tmem181a (Mm02581460_g1), Xaf1 (Mm01248390_m1), Lect1 (Mm00495291_m1) and Tbp (Mm00446973_m1) as endogenous control. Expression changes were analyzed both with the 2−ΔΔCt method, as previously described (77).
Transcriptome bioinformatics
Gene ontology and KEGG pathway over-representation analysis
The Gene Ontology is a bioinformatic initiative to generate a vocabulary that describes and unifies genes and gene product attributes across all species (http://en.wikipedia.org/wiki/Gene_ontology). This vocabulary can be used to check whether there is a non-random accumulation of GO terms in lists of differentially regulated transcripts. The lists of differentially regulated transcripts were analyzed in the main branches ‘biological process’ and ‘molecular function’ with a conditional hypergeometric test using the Bioconductor package GOStats. GO categories with a P-value ≤0.01 were called significantly enriched.
In the same line, the lists were analyzed for over-representation of known signal transduction or biochemical pathways from the KEGG database (Kyoto Encyclopedia of Genes and Genomes; www.genome.jp/kegg/).
Gene set enrichment analysis
As an alternative approach, the microarray data were subjected to nonspecific filtering (see above) and the remaining genes were analyzed using GSEA using the Java-based version GSEA-P (78,79). For each comparison, the probe IDs were ranked according the t-test statistic. Probe IDs were collapsed to gene symbols. For duplicate entries, the maximum value was used. Permutations were performed on gene sets owing to the low number of biological replicates. We used the c2 (online pathway databases, Pubmed publications, expert of domain knowledge) and c5 (Gene Ontology categories) gene sets from the MSigDB database (v4.0, May 2013, http://www.broadinstitute.org/gsea/msigdb/index.jsp) to analyze the data sets.
Statistics
The statistical evaluation of data by unpaired two-tailed t-tests and their graphical documentation were carried out with the GraphPad Prism 5 for Windows software package (version 5.04). Significance was indicated by asterisks (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001).
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the German Federal Ministry of Education through the National Genome Research Network (NGFNplus, BMBF 01GS08138) and the GerontoMitoSys network (BMBF PTJ 0315584A); by the European Union through ERAnet-RePARK (DLR 01EW1012); and by the Dr. Senckenbergische Stiftung. N.B. was partially financed by a scholarship from the International Max Planck Research School (IMPRS) for Neural Circuits in Frankfurt. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript. Funding to pay the Open Access publication charges for this article was provided by the Goethe University Frankfurt.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to our technical assistants Birgitt Meseck-Selchow, Mekhman Azizov and Mohamed Bouzrou. Our thanks go also to the animal care team at the ZFE Frankfurt (in particular the veterinarians Dr. A. Theisen and Dr. C. Tandi, and the caretakers E. Daut and B. Janton).
Conflict of Interest statement. None declared.
REFERENCES
- 1.Karch C.M., Cruchaga C., Goate A.M. Alzheimer's disease genetics: from the bench to the clinic. Neuron. 2014;83:11–26. doi: 10.1016/j.neuron.2014.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Corti O., Lesage S., Brice A. What genetics tells us about the causes and mechanisms of Parkinson's disease. Physiol. Rev. 2011;91:1161–1218. doi: 10.1152/physrev.00022.2010. [DOI] [PubMed] [Google Scholar]
- 3.Trinh J., Farrer M. Advances in the genetics of Parkinson disease. Nat. Rev. Neurol. 2013;9:445–454. doi: 10.1038/nrneurol.2013.132. [DOI] [PubMed] [Google Scholar]
- 4.Hardy J., Bogdanovic N., Winblad B., Portelius E., Andreasen N., Cedazo-Minguez A., Zetterberg H. Pathways to Alzheimer's disease. J. Intern. Med. 2014;275:296–303. doi: 10.1111/joim.12192. [DOI] [PubMed] [Google Scholar]
- 5.Polymeropoulos M.H., Lavedan C., Leroy E., Ide S.E., Dehejia A., Dutra A., Pike B., Root H., Rubenstein J., Boyer R., et al. Mutation in the alpha-synuclein gene identified in families with Parkinson's disease. Science. 1997;276:2045–2047. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- 6.Singleton A.B., Farrer M., Johnson J., Singleton A., Hague S., Kachergus J., Hulihan M., Peuralinna T., Dutra A., Nussbaum R., et al. alpha-Synuclein locus triplication causes Parkinson's disease. Science. 2003;302:841. doi: 10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- 7.Oueslati A., Fournier M., Lashuel H.A. Role of post-translational modifications in modulating the structure, function and toxicity of alpha-synuclein: implications for Parkinson's disease pathogenesis and therapies. Prog. Brain Res. 2010;183:115–145. doi: 10.1016/S0079-6123(10)83007-9. [DOI] [PubMed] [Google Scholar]
- 8.el-Agnaf O.M., Irvine G.B. Aggregation and neurotoxicity of alpha-synuclein and related peptides. Biochem. Soc. Trans. 2002;30:559–565. doi: 10.1042/bst0300559. [DOI] [PubMed] [Google Scholar]
- 9.Valente E.M., Abou-Sleiman P.M., Caputo V., Muqit M.M., Harvey K., Gispert S., Ali Z., Del Turco D., Bentivoglio A.R., Healy D.G., et al. Hereditary early-onset Parkinson's disease caused by mutations in PINK1. Science. 2004;304:1158–1160. doi: 10.1126/science.1096284. [DOI] [PubMed] [Google Scholar]
- 10.Kitada T., Asakawa S., Hattori N., Matsumine H., Yamamura Y., Minoshima S., Yokochi M., Mizuno Y., Shimizu N. Mutations in the Parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- 11.Scarffe L.A., Stevens D.A., Dawson V.L., Dawson T.M. Parkin and PINK1: much more than mitophagy. Trends Neurosci. 2014;37:315–324. doi: 10.1016/j.tins.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Vries R.L., Przedborski S. Mitophagy and Parkinson's disease: be eaten to stay healthy. Mol. Cell Neurosci. 2013;55:37–43. doi: 10.1016/j.mcn.2012.07.008. [DOI] [PubMed] [Google Scholar]
- 13.Todd A.M., Staveley B.E. Pink1 suppresses alpha-synuclein-induced phenotypes in a Drosophila model of Parkinson's disease. Genome. 2008;51:1040–1046. doi: 10.1139/G08-085. [DOI] [PubMed] [Google Scholar]
- 14.Todd A.M., Staveley B.E. Expression of Pink1 with alpha-synuclein in the dopaminergic neurons of Drosophila leads to increases in both lifespan and healthspan. Genet. Mol. Res. 2012;11:1497–1502. doi: 10.4238/2012.May.21.6. [DOI] [PubMed] [Google Scholar]
- 15.Haywood A.F., Staveley B.E. Parkin counteracts symptoms in a Drosophila model of Parkinson's disease. BMC Neurosci. 2004;5:14. doi: 10.1186/1471-2202-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haywood A.F., Staveley B.E. Mutant alpha-synuclein-induced degeneration is reduced by Parkin in a fly model of Parkinson's disease. Genome. 2006;49:505–510. doi: 10.1139/g06-011. [DOI] [PubMed] [Google Scholar]
- 17.Matsui H., Gavinio R., Asano T., Uemura N., Ito H., Taniguchi Y., Kobayashi Y., Maki T., Shen J., Takeda S., et al. PINK1 and Parkin complementarily protect dopaminergic neurons in vertebrates. Hum. Mol. Genet. 2013;22:2423–2434. doi: 10.1093/hmg/ddt095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stichel C.C., Zhu X.R., Bader V., Linnartz B., Schmidt S., Lubbert H. Mono- and double-mutant mouse models of Parkinson's disease display severe mitochondrial damage. Hum. Mol. Genet. 2007;16:2377–2393. doi: 10.1093/hmg/ddm083. [DOI] [PubMed] [Google Scholar]
- 19.Fournier M., Vitte J., Garrigue J., Langui D., Dullin J.P., Saurini F., Hanoun N., Perez-Diaz F., Cornilleau F., Joubert C., et al. Parkin deficiency delays motor decline and disease manifestation in a mouse model of synucleinopathy. PLoS One. 2009;4:e6629. doi: 10.1371/journal.pone.0006629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fournier M., Roux A., Garrigue J., Muriel M.P., Blanche P., Lashuel H.A., Anderson J.P., Barbour R., Huang J., du Montcel S.T., et al. Parkin depletion delays motor decline dose-dependently without overtly affecting neuropathology in alpha-synuclein transgenic mice. BMC Neurosci. 2013;14:135. doi: 10.1186/1471-2202-14-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramsey C.P., Tsika E., Ischiropoulos H., Giasson B.I. DJ-1 deficient mice demonstrate similar vulnerability to pathogenic Ala53Thr human alpha-syn toxicity. Hum. Mol. Genet. 2010;19:1425–1437. doi: 10.1093/hmg/ddq017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kitada T., Tong Y., Gautier C.A., Shen J. Absence of nigral degeneration in aged parkin/DJ-1/PINK1 triple knockout mice. J. Neurochem. 2009;111:696–702. doi: 10.1111/j.1471-4159.2009.06350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gispert S., Del Turco D., Garrett L., Chen A., Bernard D.J., Hamm-Clement J., Korf H.W., Deller T., Braak H., Auburger G., et al. Transgenic mice expressing mutant A53 T human alpha-synuclein show neuronal dysfunction in the absence of aggregate formation. Mol. Cell Neurosci. 2003;24:419–429. doi: 10.1016/s1044-7431(03)00198-2. [DOI] [PubMed] [Google Scholar]
- 24.Kurz A., Double K.L., Lastres-Becker I., Tozzi A., Tantucci M., Bockhart V., Bonin M., Garcia-Arencibia M., Nuber S., Schlaudraff F., et al. A53T-alpha-synuclein overexpression impairs dopamine signaling and striatal synaptic plasticity in old mice. PLoS One. 2010;5:e11464. doi: 10.1371/journal.pone.0011464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kurz A., May C., Schmidt O., Muller T., Stephan C., Meyer H.E., Gispert S., Auburger G., Marcus K. A53T-alpha-synuclein-overexpression in the mouse nigrostriatal pathway leads to early increase of 14-3-3 epsilon and late increase of GFAP. J. Neural Transm. 2012;119:297–312. doi: 10.1007/s00702-011-0717-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Platt N.J., Gispert S., Auburger G., Cragg S.J. Striatal dopamine transmission is subtly modified in human A53Talpha-synuclein overexpressing mice. PLoS One. 2012;7:e36397. doi: 10.1371/journal.pone.0036397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gispert S., Ricciardi F., Kurz A., Azizov M., Hoepken H.H., Becker D., Voos W., Leuner K., Muller W.E., Kudin A.P., et al. Parkinson phenotype in aged PINK1-deficient mice is accompanied by progressive mitochondrial dysfunction in absence of neurodegeneration. PLoS One. 2009;4:e5777. doi: 10.1371/journal.pone.0005777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Siddall H.K., Yellon D.M., Ong S.B., Mukherjee U.A., Burke N., Hall A.R., Angelova P.R., Ludtmann M.H., Deas E., Davidson S.M., et al. Loss of PINK1 increases the heart's vulnerability to ischemia-reperfusion injury. PLoS One. 2013;8:e62400. doi: 10.1371/journal.pone.0062400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giasson B.I., Duda J.E., Quinn S.M., Zhang B., Trojanowski J.Q., Lee V.M. Neuronal alpha-synucleinopathy with severe movement disorder in mice expressing A53T human alpha-synuclein. Neuron. 2002;34:521–533. doi: 10.1016/s0896-6273(02)00682-7. [DOI] [PubMed] [Google Scholar]
- 30.Gispert S., Kurz A., Brehm N., Rau K., Walter M., Riess O., Auburger G. Complexin-1 and Foxp1 expression changes are novel brain effects of alpha-synuclein pathology. Mol. Neurobiol. 2014 doi: 10.1007/s12035-014-8844-0. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johansen T., Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 2011;7:279–296. doi: 10.4161/auto.7.3.14487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Atkin G., Paulson H. Ubiquitin pathways in neurodegenerative disease. Front Mol. Neurosci. 2014;7:63. doi: 10.3389/fnmol.2014.00063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Del Tredici K., Braak H. Spinal cord lesions in sporadic Parkinson's disease. Acta. Neuropathol. 2012;124:643–664. doi: 10.1007/s00401-012-1028-y. [DOI] [PubMed] [Google Scholar]
- 34.Seidel K., Mahlke J., Siswanto S., Kruger R., Heinsen H., Auburger G., Bouzrou M., Grinberg L., Wicht H., Korf H.W., den Dunnen W., Rüb U. The brainstem pathologies of Parkinson's disease and dementia with Lewy bodies. Brain Pathol. 2014 doi: 10.1111/bpa.12168. doi:10.1111/bpa.12168. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Braak H., Del Tredici K., Rub U., de Vos R.A., Jansen Steur E.N., Braak E. Staging of brain pathology related to sporadic Parkinson's disease. Neurobiol. Aging. 2003;24:197–211. doi: 10.1016/s0197-4580(02)00065-9. [DOI] [PubMed] [Google Scholar]
- 36.Kuusisto E., Parkkinen L., Alafuzoff I. Morphogenesis of Lewy bodies: dissimilar incorporation of alpha-synuclein, ubiquitin, and p62. J. Neuropathol. Exp. Neurol. 2003;62:1241–1253. doi: 10.1093/jnen/62.12.1241. [DOI] [PubMed] [Google Scholar]
- 37.Waxman E.A., Giasson B.I. Specificity and regulation of casein kinase-mediated phosphorylation of alpha-synuclein. J. Neuropathol. Exp. Neurol. 2008;67:402–416. doi: 10.1097/NEN.0b013e3186fc995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bekker-Jensen S., Mailand N. Assembly and function of DNA double-strand break repair foci in mammalian cells. DNA Repair (Amst.) 2010;9:1219–1228. doi: 10.1016/j.dnarep.2010.09.010. [DOI] [PubMed] [Google Scholar]
- 39.Kurz A., Rabbani N., Walter M., Bonin M., Thornalley P., Auburger G., Gispert S. Alpha-synuclein deficiency leads to increased glyoxalase I expression and glycation stress. Cell Mol. Life Sci. 2011;68:721–733. doi: 10.1007/s00018-010-0483-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park Y.Y., Lee S., Karbowski M., Neutzner A., Youle R.J., Cho H. Loss of MARCH5 mitochondrial E3 ubiquitin ligase induces cellular senescence through dynamin-related protein 1 and mitofusin 1. J. Cell Sci. 2010;123:619–626. doi: 10.1242/jcs.061481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gegg M.E., Cooper J.M., Chau K.Y., Rojo M., Schapira A.H., Taanman J.W. Mitofusin 1 and mitofusin 2 are ubiquitinated in a PINK1/parkin-dependent manner upon induction of mitophagy. Hum. Mol. Genet. 2010;19:4861–4870. doi: 10.1093/hmg/ddq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Glauser L., Sonnay S., Stafa K., Moore D.J. Parkin promotes the ubiquitination and degradation of the mitochondrial fusion factor mitofusin 1. J. Neurochem. 2011;118:636–645. doi: 10.1111/j.1471-4159.2011.07318.x. [DOI] [PubMed] [Google Scholar]
- 43.Rakovic A., Grunewald A., Kottwitz J., Bruggemann N., Pramstaller P.P., Lohmann K., Klein C. Mutations in PINK1 and Parkin impair ubiquitination of Mitofusins in human fibroblasts. PLoS One. 2011;6:e16746. doi: 10.1371/journal.pone.0016746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tanaka A., Cleland M.M., Xu S., Narendra D.P., Suen D.F., Karbowski M., Youle R.J. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J. Cell Biol. 2010;191:1367–1380. doi: 10.1083/jcb.201007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kurz A., Wohr M., Walter M., Bonin M., Auburger G., Gispert S., Schwarting R.K. Alpha-synuclein deficiency affects brain Foxp1 expression and ultrasonic vocalization. Neuroscience. 2010;166:785–795. doi: 10.1016/j.neuroscience.2009.12.054. [DOI] [PubMed] [Google Scholar]
- 46.Cabin D.E., Shimazu K., Murphy D., Cole N.B., Gottschalk W., McIlwain K.L., Orrison B., Chen A., Ellis C.E., Paylor R., et al. Synaptic vesicle depletion correlates with attenuated synaptic responses to prolonged repetitive stimulation in mice lacking alpha-synuclein. J. Neurosci. 2002;22:8797–8807. doi: 10.1523/JNEUROSCI.22-20-08797.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takenouchi T., Hashimoto M., Hsu L.J., Mackowski B., Rockenstein E., Mallory M., Masliah E. Reduced neuritic outgrowth and cell adhesion in neuronal cells transfected with human alpha-synuclein. Mol. Cell Neurosci. 2001;17:141–150. doi: 10.1006/mcne.2000.0923. [DOI] [PubMed] [Google Scholar]
- 48.Tsuboi K., Grzesiak J.J., Bouvet M., Hashimoto M., Masliah E., Shults C.W. Alpha-synuclein overexpression in oligodendrocytic cells results in impaired adhesion to fibronectin and cell death. Mol. Cell Neurosci. 2005;29:259–268. doi: 10.1016/j.mcn.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 49.Brynczka C., Labhart P., Merrick B.A. NGF-mediated transcriptional targets of p53 in PC12 neuronal differentiation. BMC Genomics. 2007;8:139. doi: 10.1186/1471-2164-8-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Paulsen R.D., Soni D.V., Wollman R., Hahn A.T., Yee M.C., Guan A., Hesley J.A., Miller S.C., Cromwell E.F., Solow-Cordero D.E., et al. A genome-wide siRNA screen reveals diverse cellular processes and pathways that mediate genome stability. Mol. Cell. 2009;35:228–239. doi: 10.1016/j.molcel.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brehm N., Bez F., Carlsson T., Kern B., Gispert S., Auburger G., Cenci M. A genetic mouse model of Parkinson's disease shows involuntary movements and increased post-synaptic sensitivity to apomorphine. Mol. Neurobiol. 2014 doi: 10.1007/s12035-014-8911-6. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 52.Knight S.D., Presto J., Linse S., Johansson J. The BRICHOS domain, amyloid fibril formation, and their relationship. Biochemistry. 2013;52:7523–7531. doi: 10.1021/bi400908x. [DOI] [PubMed] [Google Scholar]
- 53.Del Campo M., Hoozemans J.J., Dekkers L.L., Rozemuller A.J., Korth C., Muller-Schiffmann A., Scheltens P., Blankenstein M.A., Jimenez C.R., Veerhuis R., et al. BRI2-BRICHOS is increased in human amyloid plaques in early stages of Alzheimer's disease. Neurobiol. Aging. 2014;35:1596–1604. doi: 10.1016/j.neurobiolaging.2014.01.007. [DOI] [PubMed] [Google Scholar]
- 54.Todd K., Fossati S., Ghiso J., Rostagno A. Mitochondrial dysfunction induced by a post-translationally modified amyloid linked to a familial mutation in an alternative model of neurodegeneration. Biochim. Biophys. Acta. 2014 doi: 10.1016/j.bbadis.2014.09.010. doi:10.1016/j.bbadis.2014.09.010. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tamayev R., Giliberto L., Li W., d'Abramo C., Arancio O., Vidal R., D'Adamio L. Memory deficits due to familial British dementia BRI2 mutation are caused by loss of BRI2 function rather than amyloidosis. J. Neurosci. 2010;30:14915–14924. doi: 10.1523/JNEUROSCI.3917-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Samaranch L., Lorenzo-Betancor O., Arbelo J.M., Ferrer I., Lorenzo E., Irigoyen J., Pastor M.A., Marrero C., Isla C., Herrera-Henriquez J., et al. PINK1-linked parkinsonism is associated with Lewy body pathology. Brain. 2010;133:1128–1142. doi: 10.1093/brain/awq051. [DOI] [PubMed] [Google Scholar]
- 57.Poulopoulos M., Levy O.A., Alcalay R.N. The neuropathology of genetic Parkinson's disease. Mov. Disord. 2012;27:831–842. doi: 10.1002/mds.24962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Youle R.J., Narendra D.P. Mechanisms of mitophagy. Nat. Rev. Mol. Cell Biol. 2011;12:9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Koyano F., Okatsu K., Kosako H., Tamura Y., Go E., Kimura M., Kimura Y., Tsuchiya H., Yoshihara H., Hirokawa T., et al. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature. 2014;510:162–166. doi: 10.1038/nature13392. [DOI] [PubMed] [Google Scholar]
- 60.Klinkenberg M., Gispert S., Dominguez-Bautista J.A., Braun I., Auburger G., Jendrach M. Restriction of trophic factors and nutrients induces PARKIN expression. Neurogenetics. 2012;13:9–21. doi: 10.1007/s10048-011-0303-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Parganlija D., Klinkenberg M., Dominguez-Bautista J., Hetzel M., Gispert S., Chimi M.A., Drose S., Mai S., Brandt U., Auburger G., et al. Loss of PINK1 impairs stress-induced autophagy and cell survival. PLoS One. 2014;9:e95288. doi: 10.1371/journal.pone.0095288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Michiorri S., Gelmetti V., Giarda E., Lombardi F., Romano F., Marongiu R., Nerini-Molteni S., Sale P., Vago R., Arena G., et al. The Parkinson-associated protein PINK1 interacts with Beclin1 and promotes autophagy. Cell Death Differ. 2010;17:962–974. doi: 10.1038/cdd.2009.200. [DOI] [PubMed] [Google Scholar]
- 63.McLelland G.L., Soubannier V., Chen C.X., McBride H.M., Fon E.A. Parkin and PINK1 function in a vesicular trafficking pathway regulating mitochondrial quality control. Embo. J. 2014;33:282–295. doi: 10.1002/embj.201385902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Oliveras-Salva M., Macchi F., Coessens V., Deleersnijder A., Gérard M., Van der Perren A., Van den Haute C., Baekelandt V. Alpha-synuclein-induced neurodegeneration is exacerbated in PINK1 knockout mice. Neurobiol. Aging. 2014;35:2625–2636. doi: 10.1016/j.neurobiolaging.2014.04.032. [DOI] [PubMed] [Google Scholar]
- 65.Oliveras-Salva M., Van der Perren A., Casadei N., Stroobants S., Nuber S., D'Hooge R., Van den Haute C., Baekelandt V. rAAV2/7 vector-mediated overexpression of alpha-synuclein in mouse substantia nigra induces protein aggregation and progressive dose-dependent neurodegeneration. Mol. Neurodegener. 2013;8:44. doi: 10.1186/1750-1326-8-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wood-Kaczmar A., Gandhi S., Yao Z., Abramov A.Y., Miljan E.A., Keen G., Stanyer L., Hargreaves I., Klupsch K., Deas E., et al. PINK1 is necessary for long term survival and mitochondrial function in human dopaminergic neurons. PLoS One. 2008;3:e2455. doi: 10.1371/journal.pone.0002455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kitada T., Pisani A., Porter D.R., Yamaguchi H., Tscherter A., Martella G., Bonsi P., Zhang C., Pothos E.N., Shen J. Impaired dopamine release and synaptic plasticity in the striatum of PINK1-deficient mice. Proc. Natl. Acad. Sci. USA. 2007;104:11441–11446. doi: 10.1073/pnas.0702717104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vives-Bauza C., Zhou C., Huang Y., Cui M., de Vries R.L., Kim J., May J., Tocilescu M.A., Liu W., Ko H.S., et al. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc. Natl. Acad. Sci. USA. 2010;107:378–383. doi: 10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Matsuda N., Sato S., Shiba K., Okatsu K., Saisho K., Gautier C.A., Sou Y.S., Saiki S., Kawajiri S., Sato F., et al. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J. Cell Biol. 2010;189:211–221. doi: 10.1083/jcb.200910140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Narendra D.P., Jin S.M., Tanaka A., Suen D.F., Gautier C.A., Shen J., Cookson M.R., Youle R.J. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Van Laar V.S., Arnold B., Cassady S.J., Chu C.T., Burton E.A., Berman S.B. Bioenergetics of neurons inhibit the translocation response of Parkin following rapid mitochondrial depolarization. Hum. Mol. Genet. 2011;20:927–940. doi: 10.1093/hmg/ddq531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Geisler S., Holmstrom K.M., Skujat D., Fiesel F.C., Rothfuss O.C., Kahle P.J., Springer W. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat. Cell Biol. 2010;12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 73.Kamp F., Exner N., Lutz A.K., Wender N., Hegermann J., Brunner B., Nuscher B., Bartels T., Giese A., Beyer K., et al. Inhibition of mitochondrial fusion by alpha-synuclein is rescued by PINK1, Parkin and DJ-1. Embo. J. 2010;29:3571–3589. doi: 10.1038/emboj.2010.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Guardia-Laguarta C., Area-Gomez E., Rub C., Liu Y., Magrane J., Becker D., Voos W., Schon E.A., Przedborski S. Alpha-synuclein is localized to mitochondria-associated ER membranes. J. Neurosci. 2014;34:249–259. doi: 10.1523/JNEUROSCI.2507-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rogers D.C., Fisher E.M., Brown S.D., Peters J., Hunter A.J., Martin J.E. Behavioral and functional analysis of mouse phenotype: SHIRPA, a proposed protocol for comprehensive phenotype assessment. Mamm. Genome. 1997;8:711–713. doi: 10.1007/s003359900551. [DOI] [PubMed] [Google Scholar]
- 76.Damrath E., Heck M.V., Gispert S., Azizov M., Nowock J., Seifried C., Rub U., Walter M., Auburger G. ATXN2-CAG42 sequesters PABPC1 into insolubility and induces FBXW8 in cerebellum of old ataxic knock-in mice. PLoS Genet. 2012;8:e1002920. doi: 10.1371/journal.pgen.1002920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Heck M.V., Azizov M., Stehning T., Walter M., Kedersha N., Auburger G. Dysregulated expression of lipid storage and membrane dynamics factors in Tia1 knockout mouse nervous tissue. Neurogenetics. 2014;15:135–144. doi: 10.1007/s10048-014-0397-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Subramanian A., Kuehn H., Gould J., Tamayo P., Mesirov J.P. GSEA-P: a desktop application for gene set enrichment analysis. Bioinformatics. 2007;23:3251–3253. doi: 10.1093/bioinformatics/btm369. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.