Highlight
The 14-3-3 protein GRF9 is involved in allocating carbon from shoot to root and enhancing proton secretion in the root growing zone, and is essential for root growth under normal or water stress conditions.
Key words: 14-3-3 protein, carbon allocation, GRF9, proton secretion, root growth, water stress.
Abstract
Plant 14-3-3 proteins are phosphoserine-binding proteins that regulate a wide array of targets via direct protein–protein interactions. In this study, the role of a 14-3-3 protein, GRF9, in plant response to water stress was investigated. Arabidopsis wild-type, GRF9-deficient mutant (grf9), and GRF9-overexpressing (OE) plants were treated with polyethylene glycol (PEG) to induce mild water stress. OE plant showed better whole-plant growth and root growth than the wild type under normal or water stress conditions while the grf9 mutant showed worse growth. In OE plants, GRF9 favours the allocation of shoot carbon to roots. In addition, GRF9 enhanced proton extrusion, mainly in the root elongation zone and root hair zone, and maintained root growth under mild water stress. Grafting among the wild type, OE, and grf9 plants showed that when OE plants were used as the scion and GRF9 was overexpressed in the shoot, it enhanced sucrose transport into the root, and when OE plants were used as rootstock and GRF9 was overexpressed in the root, it caused more release of protons into the root surface under water stress. Taken together, the results suggest that under PEG-induced water stress, GRF9 is involved in allocating more carbon from the shoot to the root and enhancing proton secretion in the root growing zone, and this process is important for root response to mild water stress.
Introduction
Water stress is a major limitation to crop production in agriculture. Much evidence has shown that plants have developed some adaptive strategies to cope with water stress through a complex network of biological processes (Davies and Zhang, 1991; Neumann, 2008). Among them, regulated root growth plays an important role in response to water stress (Baluska et al., 2010; Gowda et al., 2011). For example, plants may invest more carbon to roots and maintain a certain level of root growth in response to soil water stress (Sharp et al., 2004; Schachtman and Goodger, 2008; Petricka et al., 2012; Xu et al., 2013). This will help plant survival by tapping water at depth.
The 14-3-3 proteins are a large family of proteins found in virtually all eukaryotic organisms (Comparot et al., 2003; Roberts, 2003). In higher plants, 14-3-3 proteins are phosphoserine-binding proteins that regulate the activities of a wide array of targets via direct protein–protein interactions and play indispensable roles in plant response to abiotic and biotic stresses (Ferl, 1996; Roberts et al., 2002; Mayfield et al., 2007). Some recent studies suggest that plant 14-3-3 proteins might play a role in response to water stress. For instance, apart from their well-established role in regulating plasma membrane (PM) H+-ATPase, which affects root growth and water acquisition (Palmgren, 2001), 14-3-3 proteins are thought to be involved in carbohydrate metabolism and transport, which also affects root adaptation to water stress (Comparot et al., 2003; Gowda et al., 2011). In addition, in Arabidopsis plants, 14-3-3 proteins can regulate root growth and chloroplast development as components of the photosensory system (Mayfield et al., 2012).
In Arabidopsis and tomato, thirteen and twelve 14-3-3 protein isoforms have been found, respectively. GRF9 is one of the 14-3-3 gene family members identified in Arabidopsis plants (Roberts, 2003; Xu and Shi, 2007; Mayfield et al., 2007). In this study, Arabidopsis GRF9 mutant (grf9) or GRF9-overexpressing (OE) (GRF9 overexpressed in the grf9 mutant under the control of the 35S promoter) plants were used, and their performance in response to water stress was compared. The results show that GRF9 is involvegd in plant root response to water stress by participating in shoot carbon allocation and root proton secretion, which lead to more root growth under water stress.
Materials and methods
Plant materials, growth conditions, and stress treatment
Arabidopsis plants were grown hydroponically as described in Xu et al. (2012) with constant light. For water stress treatment, different concentrations of polyethylene glycol 8000 (PEG 8000; 5% at –0.47MPa, 7.5% at –0.91MPa, 10% at –1.48MPa, 12.5% at –2.20MPa, and 15% at –3.02MPa) were kept in the nutrient solution (Xu et al., 2013). Since PEG 8000 can induce oxygen deficiency and inhibit root growth in a hydroponic system (Verslues et al., 1998), the multipoints and multidirectional aeration system (four pieces of aeration equipment were fixed along the four walls of the hydroponic pot) was adopted to increase oxygen movement or oxygen availability in this hydroponic system. After 12-day-old plants were treated uner normal growth conditions (control conditions) or water stress (PEG treatment) for 12 d, the roots and shoots of the plants were then separated, frozen, and stored at –80 °C until later analysis. Each independent experiment was arranged with three replicates and each replicate contained 10 Arabidopsis plants. Furthermore, each replicate was harvested separately and analysed separately. In addition, independent experiments were repeated twice at different times with the same growth conditions.
Real-time RT–PCR
Real-time reverse transcription–PCR (RT–PCR) was assayed according to the method described in Xu and Shi (2006). Total RNA was extracted from Arabidopsis under control conditions and water stress. Gene-specific primers for real-time RT–PCR were designed using Primer 5 software (Supplementary Table S1 available at JXB online). At-ACT2 is a strongly and constitutively expressed ‘housekeeping’ gene in Arabidopsis, so the mRNA of At-ACT2 was defined as 100 REU (relative expression units). The expression level of genes corresponds to the ratio of the copy number of cDNA of the studied gene to the copy number of the ‘housekeeping’ gene multiplied by 100 REU.
Subcellular localization of GRF9 and generation of genetically modified Arabidopsis plants
The full-length coding sequence of GRF9 was obtained by PCR using primers 5'-CACCATGGGTTCTGGAAAAGAGCGTG-3' and 5'-ATTTGATTTACCCCGAGTAAAGG-3', and cloned into the entry vector (pENTRTM TOPO vector) for the Gateway system (Invitrogen). After sequencing, the correct entry clone was recombined with the Gateway destination vector pGWB5 by using the LR reaction kit (Invitrogen). The formed correct p35S::GRF9-GFP (green fluorescent protein) plasmid was transformed into Agrobacterium GV3101 cells. The transformed Agrobacterium cells were then used to infiltrate the young leaves of 4-week-old tobacco plants. About 40h after infiltration, expression of GRF9–GFP was analysed by using confocal microscopy (Olympus FV-1000 spectral type SPD mar/G/R IX81 FLUOVIEW laser confocal system) according to the method of Xu et al. (2013). For imaging GFP, the 488nm line was used for excitation, and emission was detected at 520nm. In addition, plants of the GRF9 mutant line (grf9) of Arabidopsis (Salk_004455) were obtained from the Arabidopsis Biological Resource Center (ABRC, Ohio State University, Columbus, OH, USA). The homozygous GRF9 mutant was identified by PCR using primers specific for the T-DNA of Salk_004455 (Supplementary Table S1 at JXB online). After the p35S::GRF9-GFP plasmid was transformed into Agrobacterium GV3101 cells, transformation of grf9 plants was carried out according to the floral dip method (Xu et al. 2012). T3 homozygous Arabidopsis lines were used for further studies.
Measurement of dry weight, root length, and proton extrusion
Plants (roots and shoots) were separated, and then placed in an oven at 105 °C for 0.5h to inactivate the enzymes. To obtain the dry weight, harvested plant parts were dried to a constant weight at 70 °C. The root length of plants was measured using a root analysis instrument (WinRHIZO; Regent Instruments Inc., Quebec, ON, Canada) according to the method described (Xu and Shi, 2007). The proton extrusion rate of plant roots was analysed following the method described in Xu et al. (2012).
Carbohydrate determination
Plant samples were extracted in 0.5ml of 80% ethanol (v/v) for 20min at 70 °C, and the sucrose content was determined as described (Stitt et al., 1989). Starch was extracted from the residue by grinding in 0.5ml of 50mM sodium acetate (pH 4.8) containing α-amylase (10U) and amyloglucosidase (6U).
Enzyme measurements
Sucrose phosphate synthase (SPS) activity and starch synthase (SS) activity were determined as described in Sicher and Kremer (1984) and Zuk et al. (2005), respectively. PM H+-ATPase activity and protein concentration were determined as described in Shen et al. (2006) and Xu and Shi (2007), respectively.
Measurement of net H+ flux
Net fluxes of H+ in the root tip of plants were measured non-invasively using SIET (scanning ion-selective electrode technique, SIET system BIO-003A; Younger USA Science and Technology Corporation; Applicable Electronics Inc.; Science Wares Inc., Falmouth, MA, USA). The principle of this method and instrument are detailed in Xu et al. (2013).
Collection and analysis of phloem exudates
Phloem exudates were measured according to Fan et al. (2009). After Arabidopsis leaves were cut, the tip of the petiole was recut in EDTA buffer (5mM Na2EDTA, pH 7.5, osmotically adjusted to 270 mosmol with sorbitol) with fresh razor blades. During phloem sap exudation, the leaves were illuminated and incubated in a chamber. The buffer solution containing phloem exudates was analysed for sucrose content.
Grafting of Arabidopsis plants
Grafting of 6-day-old Arabidopsis plants grown on sterile vertical agar plates was performed as described previously (Xu et al., 2012). Seeds of Arabidopsis plants were surface-sterilized, and stratified at 4 °C for 3 d. The seeds were then placed onto sterile half-strength Murashige and Skoog (MS) agar (0.8%) plates. The plates were kept vertically in constant light (~120 μE) at 21 °C for 3 d in an Arabidopsis growth chamber, and then kept at 25 °C under an 8h photoperiod (60 μE) for another 3 d. The seedlings were grafted using silicon tubing collars, and grafted plants were kept on identical agar plates and under the same growth conditions for another 6 d until the graft junction had healed. Successfully grafted plants (12 d old) were transferred to the hydroponic system and were treated with control conditions and water stress (10% PEG 8000) for 12 d. Then, Arabidopsis plants were used for experimental analysis.
Statistical analyses
Data were subjected to analysis of variance (ANOVA), and post-hoc comparisons were done with Duncan’s multiple range test at the P<0.05 level. The statistical software program used was SPSS version 13.0. The values are the means and SD of six replicates from two independent experiments.
Results
Identification and selection of genetically modified Arabidopsis
Under PEG-induced water stress, GRF9 is 1.8-fold up-regulated (Fig. 1A) and GRF6 is 2-fold up-regulated (Supplementary Table S2 at JXB online). Regarding GRF9, the study by Mayfield et al. (2012) shows that GRF9 (14-3-3 mu) regulates root growth which is important for water use in plants, and thus it might also play important roles in plant response to water stress. With regards to GRF6, Yan et al. (2004) showed that overexpression of GRF6 (GF14λ) in cotton improves drought tolerance. Since the role of GRF6 in plants has thus been clarified (Yan et al., 2004), the focus therefore was on GRF9 in this study. Figure 1B shows that GRF9 can be located in the nucleus, cytosol, and plasma membrane, and was induced by water stress. GRF9 transcript was not found in the homozygous GRF9 mutant line (grf9) of Arabidopsis (Supplementary Fig. S1). GRF9 was thus overexpressed in the grf9 mutant under the control of the 35S promoter. According to the results (Supplementary Fig. S1), wild-type (Col-0), and six Arabidopsis transgenic lines (from OE-g1 to OE-g6) were confirmed to contain GRF9 transcript, and the GRF9 mRNA level of the overexpressing (OE) lines was significantly higher than that of the wild type (~4-fold). Further, no significant change was found in terms of the expression levels for other endogenous 14-3-3 genes between the wild-type and OE plants under control or water stress conditions (Supplementary Table S2). Thus, the wild-type Arabidopsis plants (Col-0), the homozygous GRF9 mutant (grf9, Salk_004455), and two GRF9 OE lines (OE-g1 and OE-g6) were selected for further study.
Fig. 1.
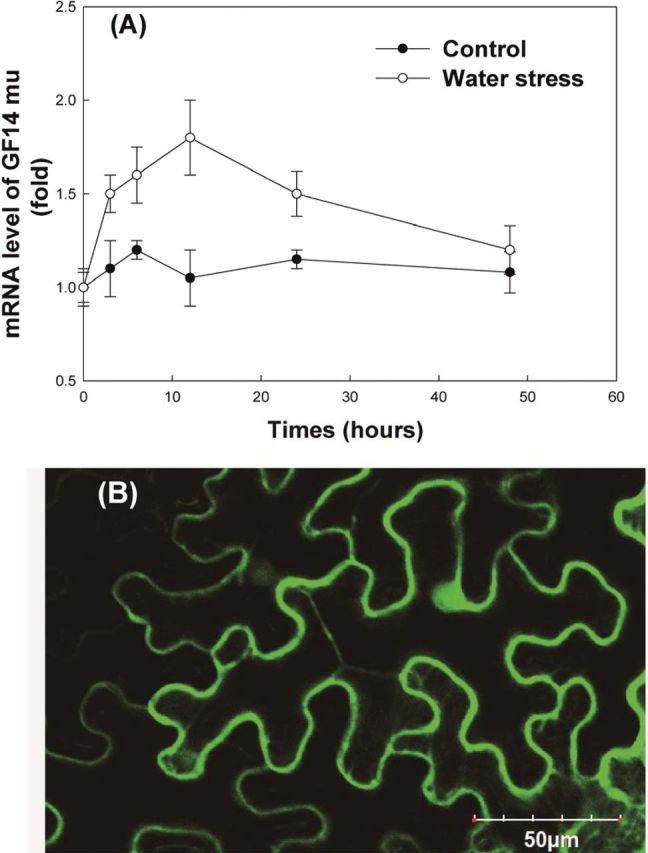
The mRNA level of GRF9 in Arabidopsis plants under control conditions or water stress (A) and subcellular localization of GRF9 in the tobacco system (B). Twelve-day-old Arabidopsis plants (Col-0) were subjected to control conditions or water stress (10% PEG 8000) for 0, 3, 6, 12, 24 and 48h under a hydroponic system. The mRNA level of GRF9 in the whole plant of Col-0 at 0h was taken as 1-fold.
Growth of Arabidopsis plants under water stress
The growth of Col-0, grf9, OE-g1, and OE-g6 plants was studied for water stress tolerance by growing them hydroponically under control conditions or different concentrations of PEG 8000 (5%, 7.5%, 10%, 12.5%, and 15%). Figure 2 shows that when PEG concentrations were >10%, the dry weight of Col-0 was significantly higher than that of grf9 but lower than that of OE-g1 or OE-g6. Figure 2 also shows that when PEG concentrations were >10%, the total root length of OE plants (OE-g1 or OE-g6) was significantly higher than that of Col-0. The root length of grf9 was always the lowest. The relative root length inhibitory rates among the three genotypes (Col-0, grf9, OE-g1, and OE-g6) were also analysed. If the root length under 0% PEG is set as 100%, the relative inhibition under 10% PEG was 28.8, 35, and 37.4%, respectively, for OE, Col-0, and grf9 plants. The statistics show that the relative inhibition by 10% PEG on OE plants was significantly less than that for Col-0, 28.8% versus 35%, although grf9 was inhibited to almost the same extent as the wild type, 37.4% versus 35%. These results suggest that GRF9 is essential to maintain root growth under normal conditions, and overexpression of GRF9 is useful for root growth under water stress conditions.
Fig. 2.
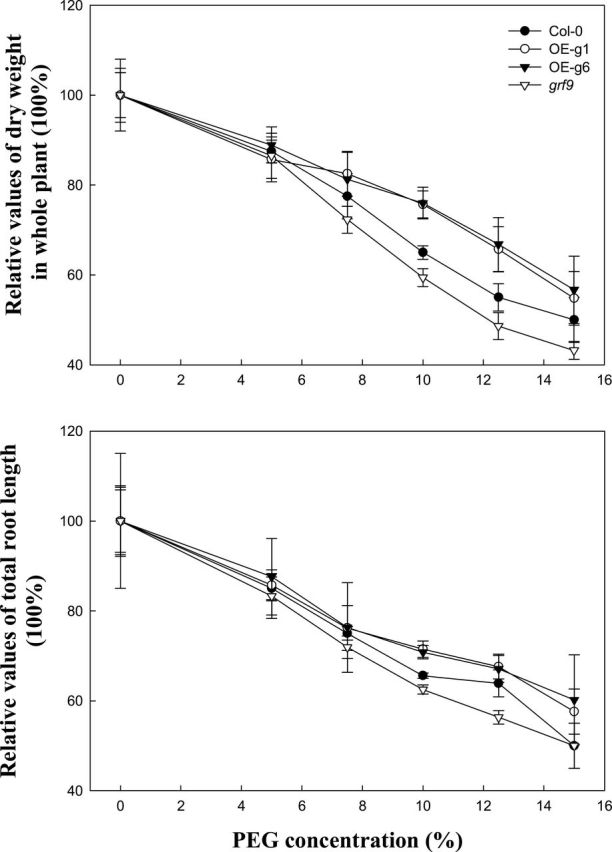
Dry weight of the whole plant and the total root length in Arabidopsis wild-type (Col-0), 14-3-3 protein GRF9 mutant plants (grf9), and the two GRF9-overexpressing lines (OE-g1 and OE-g6) under different PEG concentration treatments (water stress). Twelve-day-old Arabidopsis plants (Col-0, grf9, OE-g1, and OE-g6) were subjected to control conditions or treatment with different PEG 8000 concentrations for 12 d under a hydroponic system. Absolute values of dry weight of Col-0 (2mg), OE-g1 (2.1mg), OE-g6 (2.08mg), and grf9 (1.85mg) at 0% PEG, and root length of Col-0 (20cm), OE-g1 (21cm), OE-g6 (21.5cm), and grf9 (15.98cm) at 0% PEG are considered as 100%, respectively, for the plant groups concerned.
Activities of starch synthase (SS) and sucrose phosphate synthase (SPS) under water stress
It has been suggested that SS is one of the targets of 14-3-3 proteins in carbohydrate metabolism (Sehnke et al., 2001; Cao et al., 2007). Under short-term water stress, plants have increased sucrose and decreased starch levels in the shoot, and the sucrose loading into phloem is also higher in the shoot. Thus, these responses can help plant to adapt to water stress. However, these adaptations are inhibited under long-term water stress, and then plant growth is impaired (Neumann, 2008). Thus, the activity of SS (Fig. 3) was investigated, and SPS was used as a control. Under control conditions or water stress, no significant difference in the SPS activity was observed between wild-type (Col-0), transgenic (OE-g1 and OE-g6), and grf9 mutant Arabidopsis plants, while the activity of SS in grf9 was significantly higher than that in Col-0 or OE plants under water stress(Fig. 3). Moreover, SS activity in Col-0 was significantly higher than that in OE plants. The sucrose/starch ratio in the leaves of OE plants was significantly higher than that in Col-0 or mutant plants, and the sucrose/starch ratio in Col-0 was significantly higher than that in grf9 (Fig. 3; Supplementary Fig. S2 at JXB online).
Fig. 3.
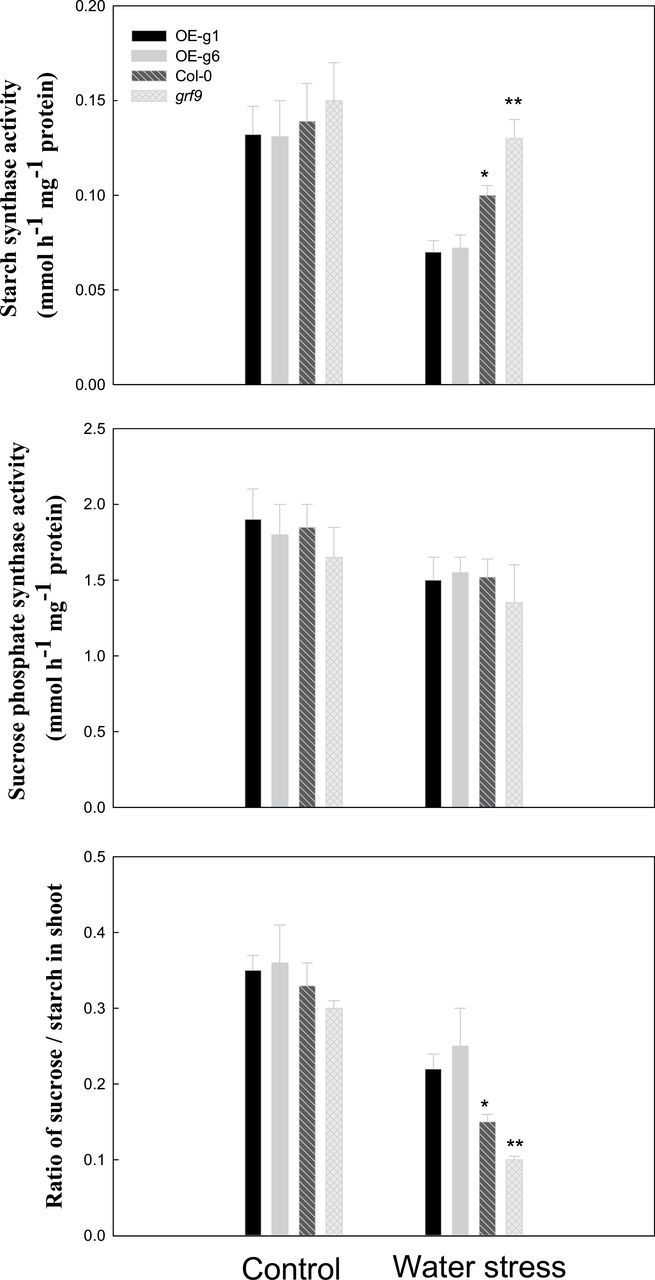
The activities of starch synthase and sucrose phosphate synthase, and the sucrose/starch ratio in Arabidopsis wild-type (Col-0), GRF9-overexpressing (OE-g1 and OE-g6), and grf9 mutant plants under water stress. Twelve-day-old Arabidopsis plants (Col-0, mutant, OE-g1, and OE-g6) were subjected to control conditions or water stress (10% PEG 8000) for 12 d under a hydroponic system.
Sucrose transport and carbohydrate content in Arabidopsis under water stress
The phloem sucrose transport and sucrose content were determined in Col-0, OE-g1,OE-g6, and grf9 mutant Arabidopsis plants (Fig. 4). Under water stress, the phloem sucrose transport, root sucrose content, and root sucrose/shoot sucrose ratio in OE-g1 or OE-g6 were significantly higher than those in Col-0 or grf9. At the same time, these parameters in Col-0 were also significantly higher than in grf9 under water stress (Fig. 4).
Fig. 4.
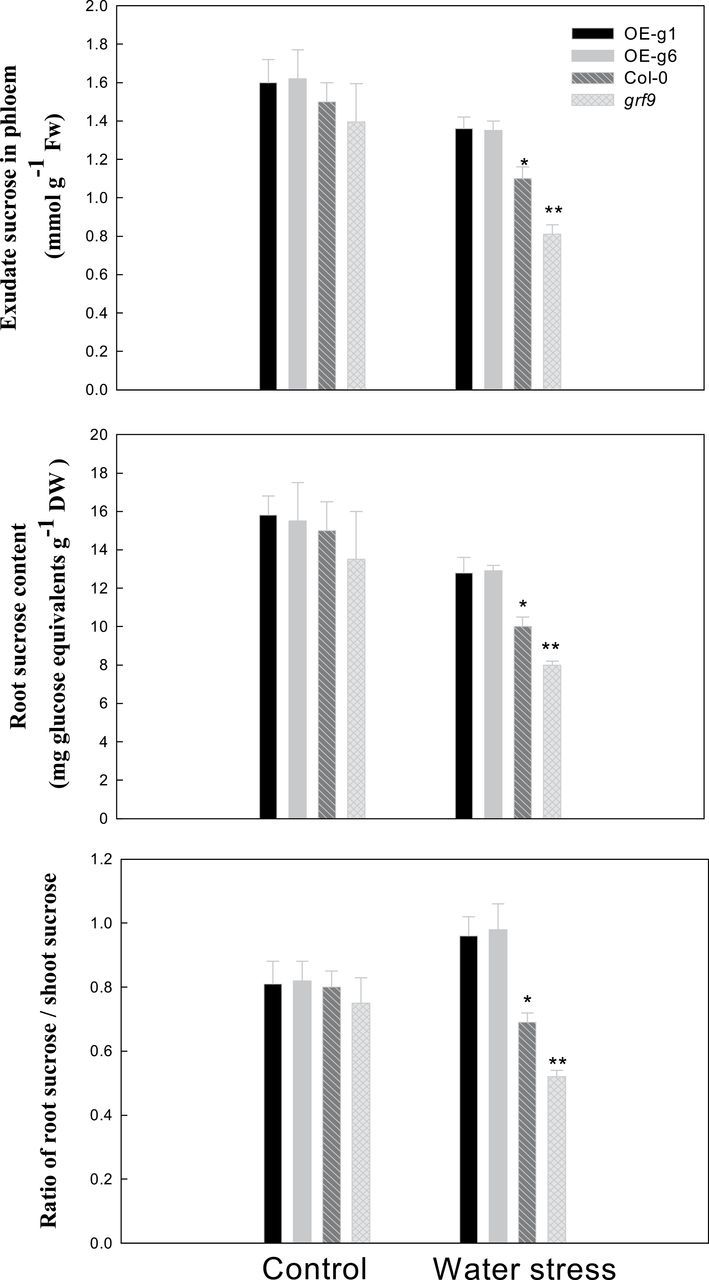
Phloem exudate sucrose or root sucrose content in wild-type (Col-0), GRF9-overexpressing (OE-g1 and OE-g6), and grf9 mutant Arabidopsis plants under water stress. Twelve-day-old Arabidopsis plants (Col-0, mutant, OE-g1, and OE-g6) were treated as described in Fig. 3.
Activity of PM H+-ATPase and H+ flux in the root of Arabidopsis plants under water stress
In plants, 14-3-3 proteins can activate PM H+-ATPase by protein–protein interactions (Palmgren, 2001). Figure 5 shows that under water stress, the PM H+-ATPase activity and root proton extrusion in GRF9-overexpressing Arabidopsis lines (OE-g1 and OE-g6) were significantly higher than those in wild-type Arabidopsis (Col-0) or GRF9 mutant Arabidopsis plants (grf9), and the two parameters in Col-0 were also significantly higher than those in grf9. Further, the proton (H+) flux in the meristem zone (MZ), transition zone (TZ), elongation zone (EZ), and root hair zone (RHZ) was analysed in the root tip of Arabidopsis plants (Fig. 6). Under control conditions, no significant difference in proton flux was found in the root tip of these Arabidopsis plants. Under water stress, in the EZ and RHZ of the root tip, proton efflux in OE-g1 or OE-g6 was significantly higher than that in Col-0 or grf9, and proton efflux in Col-0 was also significantly higher than that in grf9.
Fig. 5.
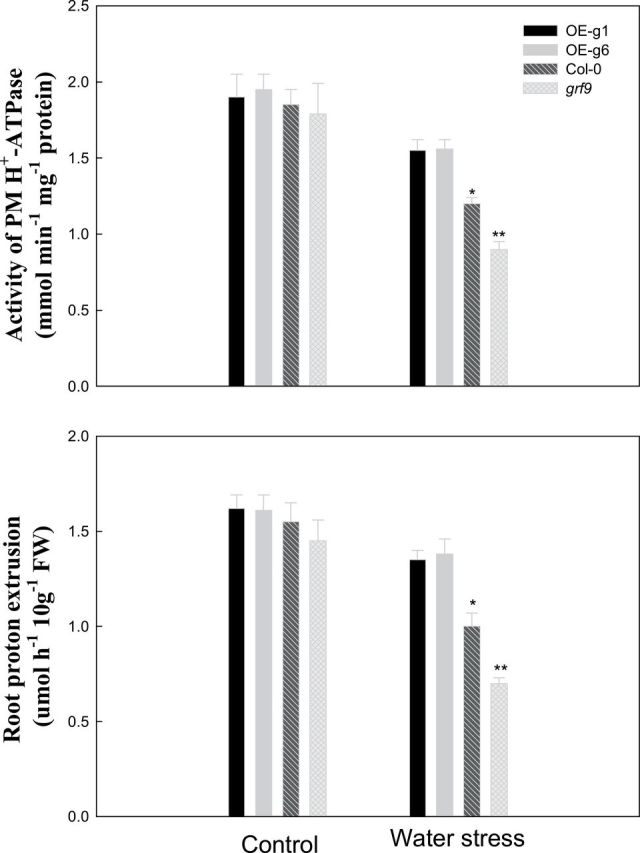
PM H+-ATPase activity or proton extrusion in roots of wild-type (Col-0), GRF9-overexpressing (OE-g1 and OE-g6), and grf9 mutant Arabidopsis plants under water stress. Twelve-day-old Arabidopsis plants (Col-0, mutant, OE-g1, and OE-g6) were treated as described in Fig. 3.
Fig. 6.
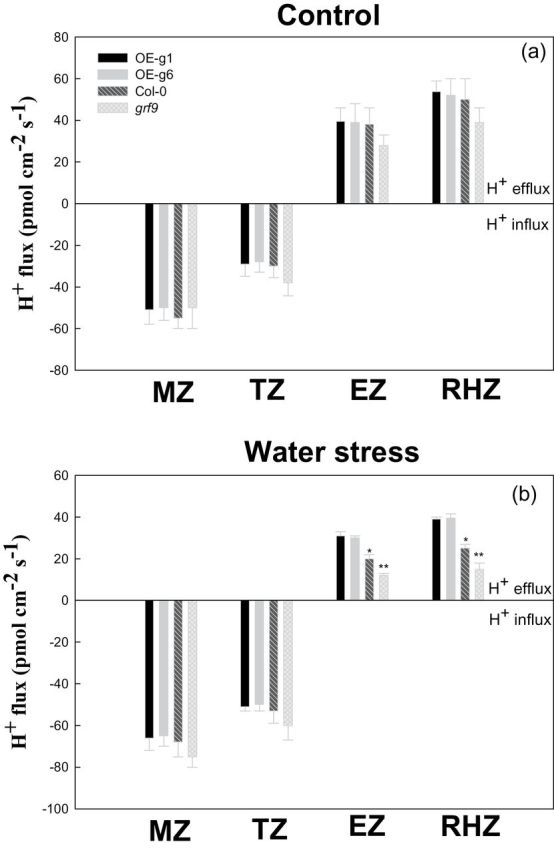
Proton (H+) flux in the meristem zone (MZ), transition zone (TZ), elongation zone (EZ), and root hair zone (RHZ) in the root tip of wild-type (Col-0), GRF9-overexpressing (OE-g1 and OE-g6), and grf9 mutant Arabidopsis plants under water stress. Twelve-day-old Arabidopsis plants (Col-0, mutant, OE-g1, and OE-g6) were treated as described in Fig. 3.
Physiological parameters in grafted Arabidopsis plants
Reciprocal grafting was performed using wild-type Arabidopsis plants (WT), GRF9-overexpressing plants (OE), and GRF9 mutant plants (MN). Seven combinations of grafted plants were generated: overexpressing plant shoot to overexpressing plant root (designated as OES/OER), wild-type shoot to wild-type root (WTS/WTR), mutant shoot to mutant root (MNS/MNR), overexpressing plant shoot to wild-type root (OES/WTR), wild-type shoot to overexpressing plant root (WTS/OER), mutant shoot to wild-type root (MNS/WTR), and wild-type shoot to mutant root (WTS/MNR). Under control conditions or water stress, total root length, phloem exudate sucrose, and root proton extrusion were measured in these grafted plants. There was no significant difference in the total root length, phloem exudate sucrose, or root proton extrusion among these grafted plants under control conditions (Supplementary Table S3 at JXB online). The following effects were observed under water stress (Fig. 7): (i) the total root length, phloem exudate sucrose, and root proton extrusion in OES/OER were significantly higher than those in WTS/WTR or MNS/MNR; (ii) the three parameters in WTS/WTR were also significantly higher than those in MNS/MNR; (iii) the total root length in OES/WTR or WTS/OER was higher than that in WTS/WTR, MNS/WTR, WTS/MNR, or MNS/MNR, but lower than that in OES/OER; and (iv) the total root length in MNS/WTR or WTS/MNR was only higher than that in MNS/MNR. Additionally, no significant difference in phloem exudate sucrose was observed between OES/OER and OES/WTR under water stress. In addition, phloem exudate sucrose in WTS/WTR, WTS/OER, or WTS/MNR was lower than that in OES/OER or OES/WTR but higher than that in MNS/MNR or MNS/WTR. Further, no significant difference in root proton extrusion was observed between OES/OER and WTS/OER under water stress. Moreover, root proton extrusion in WTS/WTR, OES/WTR, or MNS/WTR was lower than that in OES/OER or WTS/OER but higher than that in MNS/MNR or WTS/MNR. Furthermore, under water stress, the total root length or phloem exudate sucrose in OES/WTR was higher than that in MNS/WTR, but no significant difference in root proton extrusion was observed between OES/WTR and MNS/WTR. On the other hand, under water stress, the total root length or root proton extrusion in WTS/OER was higher than that in WTS/MNR, but no significant difference in phloem exudate sucrose was observed between WTS/OER and WTS/MNR.
Fig. 7.
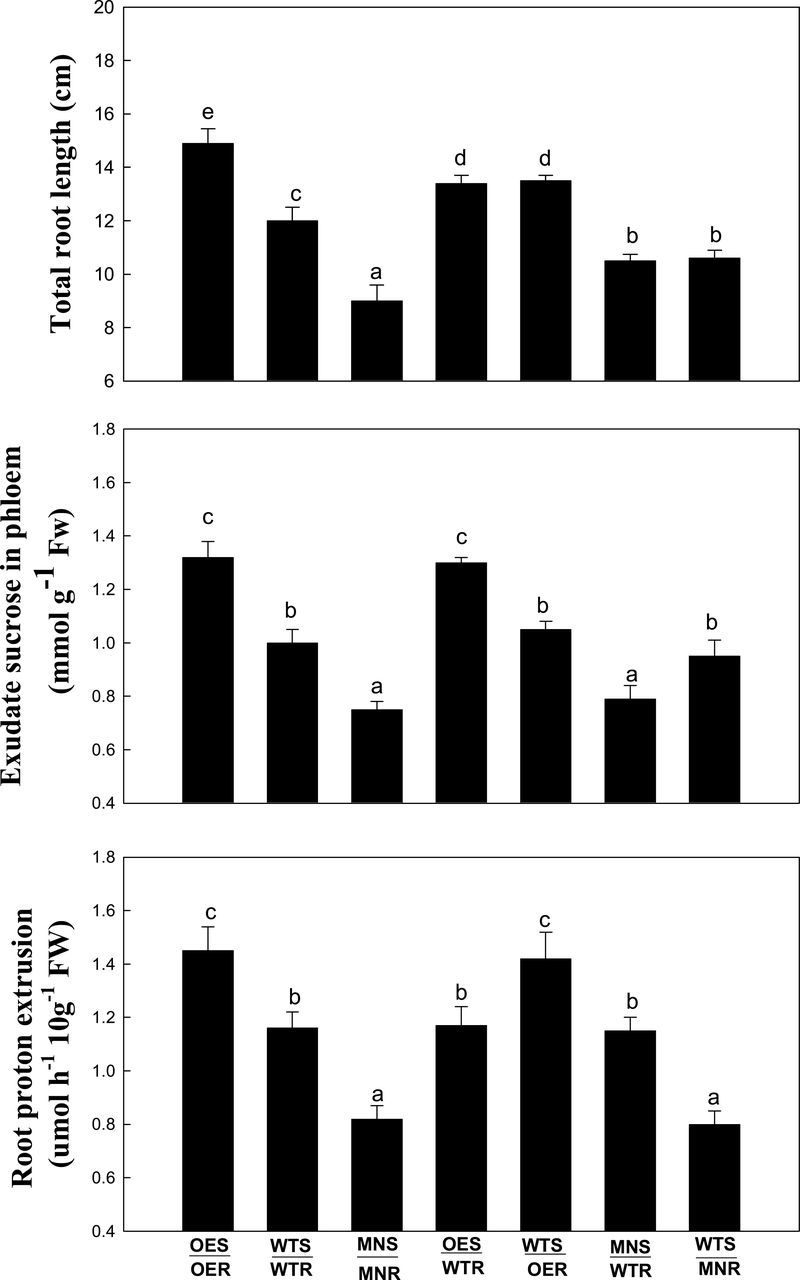
Total root length, phloem exudate sucrose, and root proton extrusion of grafted plants between Arabidopsis wild-type, GRF9-overexpressing, and grf9 mutant plants under water stress. Twelve-day-old grafted Arabidopsis plants were subjected to control conditions or water stress (10% PEG 8000) for 12 d under a hydroponic system. OES, shoot of GRF9-overexpressing plants; OER, root of GRF9-overexpressing plants; WTS, wild-type shoot; WTR, wild-type root; MNS, shoot of grf9 mutant plants; MNR, root of grf9 mutant plants.
Discussion
Plants, sessile in their environments, have evolved to adapt to soil drying by morphological and physiological modulations. These include an enhanced ability to draw water from deep soil via an extended root system and a control of water loss from the shoot through reduced stomatal opening. It has been known for a long time that a primary adaptation of plants to drought stress is the inhibition of shoot growth and improvement of root growth (Sharp et al., 2004; Gowda et al., 2011). Such physiological regulation should require a change in carbon partitioning; that is, more photosynthate to be transported from growth-inhibited shoot to stress-adaptive root.The present results show that the 14-3-3 protein GRF9 may play a regulatorye role for such a response (Fig. 3). Enhanced carbon flow into the roots may also stimulate more proton secretion in the root tip to maintain root growth under water stress conditions.
Efficient sucrose transport in phloem (from shoot to root) is very important for root growth (Salerno and Curatti, 2003). The present results suggest that Arabidopsis 14-3-3 protein GRF9 is involved in water stress response by regulating phloem sucrose transport to allocate more carbon from shoot to root (Fig. 4). In addition, root elongation and growth occur as a result of cell wall extension at the cellular level, during which protons are the major wall-loosening factor (Rayle and Cleland, 1992). In this study, it was found that under water stress, compared with wild-type Arabidopsis, proton secretion in GRF9-overexpressing Arabidopsis plants was less inhibited, while proton secretion in GRF9-deficient mutant Arabidopsis plants was more inhibited (Fig. 5). It was also found that the effect of GRF9 on proton secretion was not on proton influx but on proton efflux, especially in the elongation zone or root hair zone of the root tip (Fig. 6). Thus, it is proposed that the Arabidopsis 14-3-3 protein GRF9 is also involved in plant water stress response by enhancing proton extrusion to maintain root growth.
Recently, a study by Sun et al. (2014) showed that the grf9 mutant (GF14μ is GRF9) is more tolerant to drought stress under soil drying compared with wild-type plants. However, the results reported here show that the total root growth of the grf9 mutant was less than that of wild-type plants under PEG-induced water stress. Some studies have shown that PEG-induced water stress is very different from soil drying (Lilley and Ludlow, 1996; Uga et al., 2013). For example, Kinandang Patong rice is more tolerant to soil drying than IR64 rice. However, under PEG-induced water stress, IR64 rice was shown to be more tolerant to osmotic stress (dehydration) than Kinandang Patong rice. It is understandable that PEG-induced water stress represents a drastic or ‘immediate no-water stress’, while soil drying is gradual from ‘enough water’ to ‘no water’ over time. Under PEG-induced water stress, the grf9 mutant with shorter roots is not tolerant to osmotic stress (dehydration) compared with wild-type plants. So, just like Kinandang Patong, the grf9 mutant can also be more tolerant to soil drying compared with wild-type plants.
Many genes useful in plant drought responses have been identified by molecular and genetic studies using, in most cases, the model plant Arabidopsis thaliana. For example, one of them, Arabidopsis HD-START protein, can confer drought tolerance in higher plants by improving root growth (Yu et al., 2008). While paying more attention to the discovery of new genes, efforts should also be focused on the regulatory genes that can fine-tune the functions of other important genes for drought response in higher plants (Umezawa et al., 2004). The 14-3-3 proteins are the indispensable regulators in plant growth and development, and also play important roles in response to environmental stress (Roberts et al., 2002; Yan et al., 2004; Campo et al., 2012). In this study, it was found that the Arabidopsis 14-3-3 protein GRF9 is involved in plant adaptation to water stress by regulating shoot SS and root PM H+-ATPase (Figs 2–6). It is concluded that the Arabidopsis 14-3-3 protein GRF9 is useful for the improvement of plant drought tolerance.
Plants have developed diverse responses to cope with water stress. These responses are possibly achieved by the co-ordination of the adaptive network comprising local and systemic responses (Yamaguchi and Sharp, 2010). The localized responses mainly occur in roots and are partially involved in root proton exudation, while the systemic responses mainly occur in shoot and are partially associated with carbohydrate metabolic pathways. Grafting was used to generate chimeric Arabidopsis plants that overexpress or lack GRF9 in the shoots and in roots (Fig. 7). The results showed that compared with self-grafted wild-type plants, as long as GRF9 is overexpressed in shoots, plants show higher tolerance to water stress with higher sucrose transport in phloem, but no significant difference in root proton extrusion was found. Similarly, as long as GRF9 is overexpressed in roots, plants show higher root proton extrusion and better tolerance to water stress, but no significant difference in phloem exudate sucrose was found. Furthermore, when GRF9 was mutated in either the shoot or the root, plants showed the same lower tolerance to water stress with lower phloem sucrose transport or lower root proton extrusion, respectively. Therefore, it is concluded that GRF9 can possibly be synchronously regulated from the local and systemic response under water stress. In shoot, GRF9 is involved in the systemic response to water stress by regulating shoot carbon allocation and increasing phloem sucrose transport to promote root growth. In root, GRF9 is directly involved in the local response to water stress by activating root PM H+-ATPase to release more protons.
In conclusion, using genetically modified GRF9-overexpressing or mutant Arabidopsis plants, it was found that GRF9, an Arabidopsis 14-3-3 gene family member, plays a role in response to water stress by involvement in shoot carbon allocation and root proton secretion. This process helps plant drought tolerance and explains the long observed phenomenon that plants have relatively more root growth than shoot growth when they are exposed to soil drying.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Expression of the GRF9 gene in Arabidopsis plants (wild-type plant, Col-0; Arabidopsis 14-3-3 protein GRF9 mutant, grf9; GRF9-overexpressing lines, from OE-g1 to OE-g6).
Figure S2. Shoot sucrose or starch content in wild-type (Col-0), GRF9-overexpressing (OE-g1 2 and OE-g6), and grf9 mutant Arabidopsis plants under water stress.
Table S1. Gene-specific primers used in real-time RT–PCR for 14-3-3 genes in Arabidopsis.
Table S2. Expression of Arabidopsis endogenous 14-3-3 genes (except GRF9) in wild-type Arabidopsis plants and GRF9-overexpressing transgenic Arabidopsis plants under water stress for 3 d.
Table S3. Total root length, phloem exudate sucrose, and root proton extrusion of grafted plants in wild-type, GRF9-overexpressing, and grf9 mutant Arabidopsis plants under control conditions.
Acknowledgements
We are grateful for grant support from the strategic leading special science and technology project of the Chinese Academy of Sciences (XDB15030201), the National Basic Research Program of China (nos 2014CB954500, 2013CB127402, and 2012CB114300), the NSFC Major Research Plan project (91317307), the National Natural Science Foundation of China (nos 31422047 and 31272229), the Hong Kong Research Grant Council (AoE/M-05/12), and Shenzhen Overseas Talents Innovation & Entrepreneurship Funding Scheme (The Peacock Scheme).
References
- Baluska F, Mancuso S, Volkmann D, Barlow PW. 2010. Root apex transition zone: a signaling-response nexus in the root. Trends in Plant Science 15, 402–408. [DOI] [PubMed] [Google Scholar]
- Campo S, Peris-Peris C, Montesinos L, Penas G, Messeguer J, San SB. 2012. Expression of the maize ZmGF14-6 gene in rice confers tolerance to drought stress while enhancing susceptibility to pathogen infection. Journal of Experimental Botany 63, 983–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao A, Jain A, Baldwin JC, Raghothama KG. 2007. Phosphate differentially regulates 14-3-3 family members and GRF9 play a role in Pi-starvation induced responses. Planta 226, 1219–1230. [DOI] [PubMed] [Google Scholar]
- Comparot S, Lingiah G, Martin T. 2003. Function and specificity of 14-3-3 proteins in the regulation of carbohydrate and nitrogen metabolism. Journal of Experimental Botany 54, 595–604. [DOI] [PubMed] [Google Scholar]
- Davies WJ, Zhang JH. 1991. Root signals and the regulation of growth and development of plants in drying soil. Annual Review of Plant Physiology and Plant Molecular Biology 42, 55–76. [Google Scholar]
- Fan SC, Lin CS, Hsu PK, Lin SH, Tsay YF. 2009. The Arabidopsis nitrate transporter NRT1.7, expressed in phloem, is responsible for source-to-sink remobilization of nitrate. The Plant Cell 21, 2750–2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferl RJ. 1996. 14-3-3 proteins and signal transduction. Annual Review of Plant Physiology and Plant Molecular Biology 47, 49–73. [DOI] [PubMed] [Google Scholar]
- Gowda VRP, Henry A, Yamauchi A, Shashidhar HE, Serraj R. 2011. Root biology and genetic improvement for drought avoidance in rice. Field Crops Research 122, 1–13. [Google Scholar]
- Lilley JM, Ludlow MM. 1996. Expression of osmotic adjustment and dehydration tolerance in diverse rice lines. Field Crops Research 48, 185–197. [Google Scholar]
- Mayfield JD, Folta KM, Paul AL, Ferl RJ. 2007. The 14-3-3 proteins μ and ν influence transition to flowering and early phytochrome response. Plant Physiology 145, 1692–1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayfield JD, Paul AL, Ferl RJ. 2012. The 14-3-3 proteins of Arabidopsis regulate root growth and chloroplast development as components of the photosensory system. Journal of Experimental Botany 63, 3061–3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann PM. 2008. Coping mechanisms for crop plants in drought-prone environments. Annals of Botany 101, 901–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmgren MG. 2001. Plant plasma membrane H+-ATPases: powerhouses for nutrient uptake. Annual Review of Plant Physiology and Plant Molecular Biology 52, 817–845. [DOI] [PubMed] [Google Scholar]
- Petricka JJ, Winter CM, Benfey PN. 2012. Control of Arabidopsis root development. Annual Review of Plant Biology 63, 563–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayle DL, Cleland RE. 1992. The acid growth theory of auxin-induced cell elongation is alive and well. Plant Physiology 99, 1271–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts MR. 2003. 14-3-3 proteins find new partners in plant cell signaling. Trends in Plant Science 8, 218–223. [DOI] [PubMed] [Google Scholar]
- Roberts MR, Salinas J, Collinge DB. 2002. 14-3-3 proteins and response to abiotic and biotic stress. Plant Molecular Biology 50, 1031–1039. [DOI] [PubMed] [Google Scholar]
- Salerno GL, Curatti L. 2003. Origin of sucrose metabolism in higher plants: when, how and why? Trends in Plant Science 8, 63–69. [DOI] [PubMed] [Google Scholar]
- Schachtman DP, Goodger JQD. 2008. Chemical root to shoot signaling under drought. Trends in Plant Science 13, 281–287. [DOI] [PubMed] [Google Scholar]
- Sehnke PC, Chung HJ, Wu K, Ferl RJ. 2001. Regulation of starch accumulation by granule-associated plant 14-3-3 proteins. Proceedings of the National Academy of Sciences, USA 98, 765–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp RE, Poroyko V, Hejlek LG, Spollen WG, Springer GK, Bohnert HJ, Nguyen HT. 2004. Root growth maintenance during water deficits: physiology to functional genomics. Journal of Experimental Botany 55, 2343–2351. [DOI] [PubMed] [Google Scholar]
- Shen H, Chen J, Wang Z, Yang C, Sasaki T, Yamamoto Y, Matsumoto H, Yan X. 2006. Root plasma membrane H+-ATPase activity is involved in the adaptation of soybean to phosphorus starvation. Journal of Experimental Botany 57, 1353–1362. [DOI] [PubMed] [Google Scholar]
- Sicher RC, Kremer DF. 1984. Changes of sucrose-phosphate synthase activity in barley primary leaves during light/dark transitions. Plant Physiology 76, 910–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitt M, Lilley RMC, Gerhardt R, Heldt HW. 1989. Metabolites in specific cells and subcellular compartments of plant leaves. Methods in Enzymology 174, 518–552. [Google Scholar]
- Sun X, Luo X, Sun M, et al. 2014. A Glycine soja 14-3-3 protein GsGF14o participates in stomata and root hair development and drought tolerance in Arabidopsis thaliana. Plant and Cell Physiology 55, 99–118. [DOI] [PubMed] [Google Scholar]
- Uga Y, Sugimoto K, Ogawa S, et al. 2013. Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nature Genetics 45, 1097–1102. [DOI] [PubMed] [Google Scholar]
- Umezawa T, Yoshida R, Maruyama K, Yamaguchi-Shinozaki K, Shinozaki K. 2004. SRK2C, a SNF1-related protein kinase 2, improves drought tolerance by controlling stress-responsive gene expression in Arabidopsis thaliana . Proceedings of the National Academy of Sciences, USA 101, 17306–17311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verslues PE, Ober ES, Sharp RE. 1998. Root growth and oxygen relations at low water potentials: impact of oxygen availability in polyethylene glycol solutions. Plant Physiology 116, 1403–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu WF, Jia L, Shi W, Zhou F, Li Q, Zhang J. 2013. Abscisic acid accumulation modulates auxin transport in the root tip to enhance proton secretion for maintaining root growth under moderate water stress. New Phytologist 197, 139–150. [DOI] [PubMed] [Google Scholar]
- Xu WF, Shi WM. 2006. Expression profiling of the 14-3-3 gene family in response to salt stress and potassium and iron deficiencies in young tomato (Solanum lycopersicum) roots: analysis by real-time RT–PCR. Annals of Botany 98, 965–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu WF, Shi WM. 2007. Mechanisms of salt tolerance in transgenic Arabidopsis thaliana constitutively overexpressing the tomato 14-3-3 protein TFT7. Plant and Soil 301, 17–28. [Google Scholar]
- Xu WF, Shi WM, Jia LG, Liang JS, Zhang JH. 2012. TFT6 and TFT7, two different members of tomato 14-3-3 gene family, play distinct roles in plant adaption to low phosphorus stress. Plant, Cell and Environment 35, 1393–1406. [DOI] [PubMed] [Google Scholar]
- Yamaguchi M, Sharp RE. 2010. Complexity and coordination of root growth at low water potentials: recent advances from transcriptomic and proteomic analyses. Plant, Cell and Environment 33, 590–603. [DOI] [PubMed] [Google Scholar]
- Yan J, He C, Wang J, Mao Z, Holaday SA, Allen RD, Zhang H. 2004. Overexpression of the Arabidopsis 14-3-3 proteins GF14λ in cotton leads to a ‘stay-green’ phenotype and improves stress tolerance under moderate drought conditions. Plant and Cell Physiology 45, 1007–1014. [DOI] [PubMed] [Google Scholar]
- Yu H, Chen X, Hong YY, Wang Y, Xu P, Ke SD, Liu HY, Zhu JK, Oliver DJ, Xiang CB. 2008. Activated expression of an Arabidopsis HD-START protein confers drought tolerance with improved root system and reduced stomatal density. The Plant Cell 20, 1134–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuk M, Weber R, Szopa J. 2005. 14-3-3 protein down-regulates key enzyme activity of nitrate and carbohydrate metabolism in potato plants. Journal of Agricultural and Food Chemistry 53, 3454–3460. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


