Highlight
Two SNPs in the promoter of GS5 are responsible for expression variation controlling grain size. Enhanced expression of GS5 competitively inhibits the interaction between OsBAK1 and OsMSBP1, promoting grain size.
Key words: cis-element, competitive interaction, differential regulation, expression variation, grain size, GS5.
Abstract
Grain weight is a major determinant of grain yield. GS5 is a positive regulator of grain size such that grain width, filling, and weight are correlated with its expression level. Previous work suggested that polymorphisms of GS5 in the promoter region might be responsible for the variation in grain size. In this study, two single nucleotide polymorphisms (SNPs) between the wide-grain allele GS5-1 and the narrow-grain allele GS5-2 in the upstream region of the gene that were responsible for the differential expression in developing young panicles were identified. These two polymorphs altered the responses of the GS5 alleles to abscisic acid (ABA) treatments, resulting in higher expression of GS5-1 than of GS5-2 in developing young panicles. It was also shown that SNPs in light-responsive elements of the promoter altered the response to light induction, leading to higher expression of GS5-2 than GS5-1 in leaves. Enhanced expression of GS5 competitively inhibits the interaction between OsBAK1-7 and OsMSBP1 by occupying the extracellular leucine-rich repeat (LRR) domain of OsBAK1-7, thus preventing OsBAK1-7 from endocytosis caused by interacting with OsMSBP1, providing an explanation for the positive association between grain size and GS5 expression. These results advanced our understanding of the molecular mechanism by which GS5 controls grain size.
Introduction
Grain weight is one of the most important yield traits in rice, and is determined by grain size and the degree of grain filling. Grain size is measured by grain length, width, and thickness. With the rapid advance of rice genome research, a number of quantitative trait loci (QTLs) for grain length (GS3) (Fan et al., 2006), grain width (GW2, qSW5/GW5, GS5, and GW8/OsSPL16) (Song et al., 2007; Shomura et al., 2008; Weng et al., 2008; Li et al., 2011; Wang et al., 2012), grain filling (GIF1) (Wang et al., 2008), and grain weight (TGW6) (Ishimaru et al., 2013) have been isolated in the past decade. Functional analyses of these genes have brought to light the molecular mechanisms by which the genes regulate grain size.
For example, GS3, a major negative regulator of grain length, encodes four putative domains functioning differentially in grain size regulation. It contains a plant-specific organ size regulation (OSR) domain in the N-terminus, which is both necessary and sufficient for functioning as a negative regulator. However, the tumour necrosis factor receptor/nerve growth factor receptor (TNFR/NGFR) family cysteine-rich domain and the von Willebrand factor type C (VWFC) domain in the C-terminus show an inhibitory effect on OSR function (Mao et al., 2010). Two major genes negatively controlling grain width, GW2 and qSW5/GW5, are likely to function in the ubiquitin–proteasome pathway, as GW2 encodes a RING-type E3 ubiquitin ligase (Song et al., 2007) and qSW5/GW5 may physically interact with polyubiquitin (Weng et al., 2008). GIF1 encodes a cell wall invertase required for carbon partitioning during early grain filling, and TGW6 encodes an indole-3-acetic acid (IAA)-glucose hydrolase affecting the transition from the syncytial to the cellular phase of the endosperm, both of which regulate the source–sink relationship during grain filling, eventually affecting the final grain weight (Wang et al., 2008; Ishimaru et al., 2013). The allelic variation at the GW8/OsSPL16 locus is a 10bp deletion in the promoter, which significantly reduces the expression level of the gene and thus the reduction in grain width (Wang et al., 2012), whereas allelic variations of all other genes are caused by mutations of the structural genes that change protein sequences. Despite this progress, the details are still lacking for almost all the genes regarding the mechanistic understanding of how they regulate grain size.
Using populations derived from a cross between Zhenshan 97 and H94, Li et al. (2011) cloned a minor QTL, GS5, on the short arm of chromosome 5 for grain width, filling, and weight. The grains of the near-isogenic line NIL(ZS97) are 8.7% wider and 7.0% heavier than those of NIL(H94), and the grain-filling rate is significantly higher in NIL(ZS97), leading to a 7.4% increase in grain yield per plant (Li et al., 2011).
GS5 encodes a putative serine carboxypeptidase-like (SCPL) protein, a member of a large family characterized by a conserved serine–aspartate–histidine catalytic triad (Fraser et al., 2005; Feng and Xue, 2006; Tripathi and Sowdhamini, 2006). The alleles from both Zhenshan 97 (wide grain) and H94 (narrow grain) are predicted to be full length, and overexpression of either allele could increase grain width (Li et al., 2011). Based on this result, it was concluded that GS5 is a positive regulator of grain size, and a higher expression level is correlated with increased grain width.
The study reported herein was attempted in order to investigate the functional relationship between grain size and the transcription level of GS5. Two single nucleotide polymorphisms (SNPs) located in the flanking sequence of a putative gibberellin- (GA) responsive element in the promoter were identified that altered the response of the GS5 alleles to abscisic acid (ABA) suppression, causing differential expression of the GS5 alleles in young panicles. Subcellular localization and protein–protein interaction assay indicated that enhanced expression of GS5 competitively inhibits the interaction between OsBAK1-7 and OsMSBP1 by occupying the extracellular leucine-rich repeat (LRR) domain of OsBAK1-7, thus preventing OsBAK1-7 from endocytosis caused by interacting with OsMSBP1. These results provided an explanation for the positive association between grain size and the level of GS5 expression.
Materials and methods
Field planting and trait measurement
Rice plants were grown and examined under natural field conditions in the experimental station of Huazhong Agriculture University, Wuhan, China. The planting density was 16.5cm between plants in a row and the rows were 26cm apart. Harvested grains were air-dried and stored at room temperature before testing. Thirty randomly chosen, fully filled grains from each plant were used for grain size measurement. Every 10 grains were lined up length-wise along a vernier caliper to measure grain length and then arranged by breadth to measure grain width.
Constructs and transformation
The coding sequences (CDS) of GS5 from H94 and Zhenshan 97, and that of OsBAK1-7 and OsMSBP1 from Zhenshan 97 were amplified by reverse transcription–PCR (RT–PCR). Both GS5 sequences were inserted into a modified plant binary vector pU1301 (Sun and Zhou, 2008) that contains a maize ubiquitin gene promoter and a 3× FLAG-tag located downstream in-frame to construct the P Ubi ::GS5-FLAG vectors. The GS5 and OsBAK1-7 CDSs were fused in-frame at their C-terminus with green fluorescent protein (GFP) under the control of the Cauliflower mosaic virus (CaMV) 35S promoter in another modified plant binary vector pDX2181 (Ye et al., 2012) to construct the P 35S ::GS5-GFP and P 35S ::OsBAK1-7-GFP vectors. For co-expression of OsBAK1-7 and OsMSBP1, the OsMSBP1 CDS was fused with red fluorescent protein (RFP) at its C-terminus and inserted downstream of a CaMV35S promoter; the P 35S ::OsMSBP1-RFP fragment was then cloned into the P 35S ::OsBAK1-7-GFP vector. For promoter strength analysis, the truncated GS5 promoter fragments were amplified from H94 and Zhenshan 97 and fused to β-glucuronidase (GUS) in pDX2181. The fraction-replaced promoter fragments were generated by restriction enzyme digestion and ligation at HindIII (–2002bp), BanII (–1139bp), EcoT22I (–931bp), XbaI (–604bp), and BamHI (–1bp) sites based on the constructs HH and ZZ, which carried the 2kb promoter fragment of H94 and Zhenshan 97, respectively. The mutated promoter fragments were generated by PCR site-directed mutagenesis. All these constructs were confirmed by sequencing, introduced into the Agrobacterium tumefaciens strain EHA105 by electroporation, and then introduced into Zhonghua 11 by Agrobacterium-mediated transformation as described in Lin and Zhang (2005) with minor modifications, or infiltrated into Nicotiana tabacum epidermal cells as previously described (Sparkes et al., 2006).
Transient expression in tobacco BY-2 protoplasts
The GS5 CDSs of H94 and Zhenshan 97 were cloned into the pM999-EGFP vector under the control of the CaMV35S promoter, fused in-frame at their C-terminus with GFP. The closed circular plasmid DNA was purified by equilibrium centrifugation in CsCl–ethidium bromide gradients as described in Sambrook et al. (1989). Tobacco BY-2 protoplast generation and purification, transformation of plasmid DNA into protoplasts via electroporation, and incubation of protoplasts for protein analysis were carried out as previously described (Miao and Jiang, 2007).
Gene expression analysis
For expression analysis, fresh tissues of NIL(H94) and NIL(ZS97) were harvested at 17:00h to 19:00h and stored at –70 °C before testing. For light conditions and plant hormone treatments, seeds of NIL(H94) and NIL(ZS97) were soaked in water at 30 °C for 2 d, and grown hydroponically to the trefoil stage at 26 °C. For diurnal expression analysis, half of the plants were incubated under long-day conditions (14h light/10h darkness) in an illumination incubator and the other half were incubated under short-day conditions (10h light/14h darkness) in another illumination incubator. For the hormone treatments, seedlings were incubated under long-day conditions and 10 μM GA3, ABA, or brassinosteroid (BR) was added to the nutrient solution with ddH2O as negative control. The third leaf blades were harvested from three different plants for each treatment at the specified time points and stored in liquid nitrogen. RNA isolation, reverse transcription, and quantitative real-time PCR were carried out as previously described (Mao et al., 2010). GS5qF and GS5qR were used to amplify the transcript of GS5, and Act1F and Act1R for Actin1 as the internal control.
Protein expression analysis
Expression of GS5-FLAG or GS5–GFP in young panicles of transgenic plants or tobacco leaves was analysed using monoclonal anti-FLAG M2 primary antibody (Sigma-Aldrich) and goat anti-mouse IgG secondary antibody (SouthernBiotech) or anti-GFP primary antibody (Abcam) and goat anti-rabbit IgG secondary antibody (Southern Biotech) following Huang et al. (2007). Fluorescence signals in rice lemma or tobacco leaf epidermal cells and BY-2 protoplasts were observed using a Leica TCS SP2 AOBS confocal microscope (Leica Microsystems) according to the user manual. Plasmolysis was induced by the addition of 1M mannitol solution to the tobacco leaf lower epidermis slice. For quantitative analysis of GUS activity, young panicles of 20cm in length from transgenic plants were harvested; total protein was extracted and quantified as described in Ye et al. (2012) and used for fluorometric assay according to the method described by Jefferson et al. (1987) with an Infinite 200 photometer (Tecan).
Yeast two-hybrid assay
Total RNA from young panicles of 2cm in length from Zhenshan 97 was isolated for generation of a cDNA library with BD Matchmaker™ Library Construction & Screening Kits (Clontech). The putative A-chain CDS of GS5 was cloned into the pGBKT7 vector and tested for transcriptional activation and toxicity as described in the user manual. Screening for two-hybrid interactions was carried out by yeast mating, and positive interactions were verified and analysed by series strategies according to the user manual of the kit. The putative B-chain CDS of GS5, the LRR domain CDS, and the kinase domain CDS of BAK1 homologues were cloned into pGADT7 and co-transformed with pGBKT7 A-chain to retest the interactions in yeast.
Results
Differential expression pattern of GS5
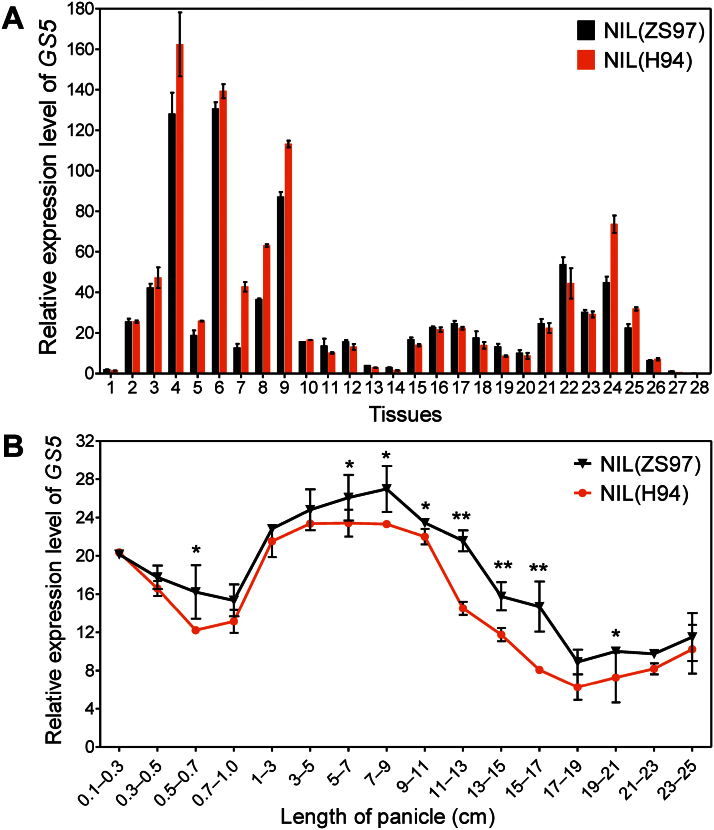
Li et al. (2011) showed that GS5-controlled grain size variation is positively correlated with its expression level. To understand how the difference in expression levels between the GS5 alleles is related to grain size, the temporal and spatial expression patterns of GS5 were assayed and it was found that in general it had a much higher expression level in green tissues than in non-green tissues, such as culm, root, young panicle, and endosperm (Fig. 1A). In non-green tissues, the highest expression of GS5 was detected in young panicles of 1–11cm in length (Fig. 1A), during which increase the volume of the lemmas/paleae increased rapidly. After that the GS5 transcript decreased gradually and reached the lowest level in 20cm long panicles. The lemmas/paleae reached almost their final size in the 20cm long panicle, and gradually turned green afterwards. The expression of GS5 in the green lemmas/paleae was remarkably up-regulated at the heading stage (Fig. 1A).
Fig. 1.
Expression pattern of the two GS5 alleles. Fresh tissues of NIL(ZS97) and NIL(H94) grown under natural field conditions were used. (A) Expression pattern of GS5 during the entire life cycle of the rice plant. 1, Embryo at 72h after imbibition; 2, plumule at 48h after emergence; 3, leaf from seedlings at the single-leaf stage; 4, leaf blade at the trefoil stage; 5, young leaf blade at the tillering stage; 6, mature leaf blade at the tillering stage; 7, flag leaf blade at the heading stage; 8, leaf sheath at the trefoil stage; 9, leaf sheath at the tillering stage; 10, tiller bud at the tillering stage; 11, radicle at 48h after emergence; 12, root at the trefoil stage; 13, young culm at the booting stage; 14, young culm at the heading stage; 15, developing panicle of 0.1–1cm in length; 16, developing panicle of 1–5cm in length; 17, developing panicle of 5–10cm in length; 18, developing panicle of 10–15cm in length; 19, developing panicle of 15–20cm in length; 20, developing panicle of 20–25cm in length; 21, panicle at the heading stage; 22, panicle on the day of flowering; 23, hull at 2 days after pollination (DAP); 24, hull at 5 DAP; 25, hull at 8 DAP; 26, endosperm at 2 DAP; 27, endosperm at 5 DAP; 28, endosperm at 8 DAP. (B) Comparison of GS5 transcripts in NIL(ZS97) and NIL(H94) during young panicle development. All data are presented as the mean ±SE (n ≥3). *P<0.05; **P<0.01, t-test. (This figure is available in colour at JXB online.)
Throughout development of the young panicle, the GS5 transcript in the developing panicle was more abundant in NIL(ZS97) than in NIL(H94) (Fig. 1B), consistent with the fact that NIL(ZS97) produced wider grains. Interestingly, in leaves, the GS5 transcript had a lower level in NIL(ZS97) (Fig. 1A), implying that the expression of GS5 was differentially regulated in green leaves and developing panicles by different regulatory elements.
Induction of GS5 expression by light
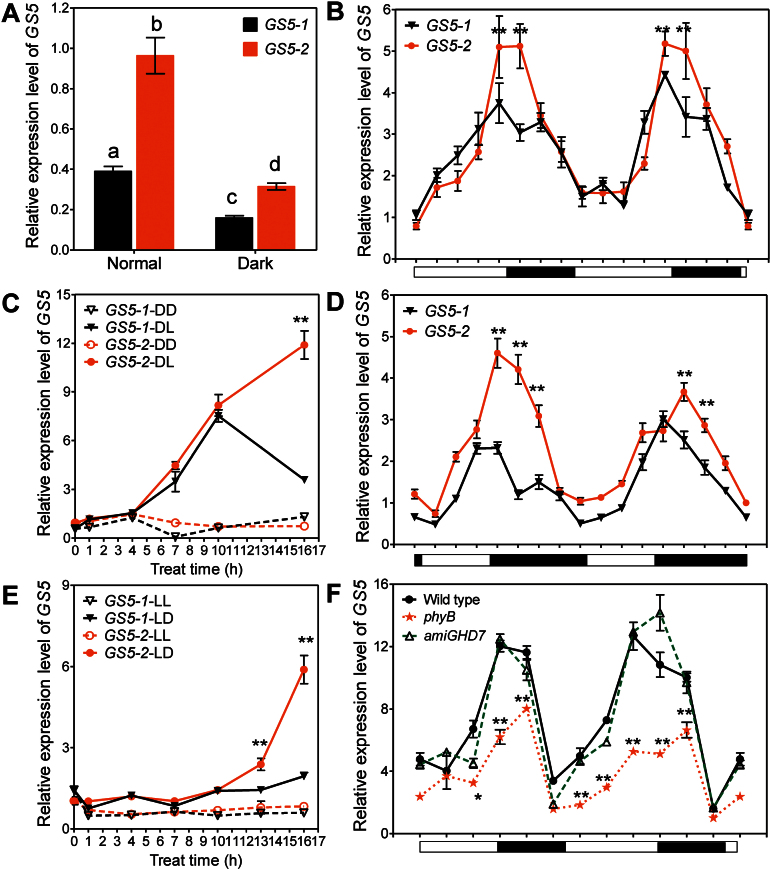
In order to determine the regulatory elements of GS5 expression, the promoter region (~2kb fragment upstream of the translation start site) was analysed using PlantCARE (Lescot et al., 2002). Many light-responsive elements were identified, and the polymorphisms between the two GS5 alleles caused different numbers of light-responsive elements (Supplementary Table S1 available at JXB online). Seedlings cultivated in the dark had significantly less GS5 transcript than those cultivated under normal light conditions (Fig. 2A), suggesting that light may contribute to the higher expression level in green tissues. For ease of description, the GS5 allele from Zhenshan 97 was designated GS5-1 and that from H94 was designated GS5-2.
Fig. 2.
Light induction of GS5 expression in the leaf. Fifteen-day-old seedlings of NIL(H94) and NIL(ZS97) grown under different light conditions were used for the various light treatments presented. (A) Expression of GS5 in seedlings cultivated under long days (14h light/10h darkness) (Normal) and continuous darkness (Dark). (B, D) Diurnal expression patterns of GS5 in leaves under long-day (B) and short-day (10h light/14h darkness) (D) conditions. (C, E) Induced expression of GS5 in leaves by light. (C) DD, seedlings were grown in continuous darkness; DL, after 15 d of continuous darkness, seedlings were shifted to continuous illumination at time point 0. (E) LL, seedlings were grown under continuous illumination; LD, after 15 d of continuous illumination, seedlings were shifted to continuous darkness at time point 0. (F) Suppression of GS5 transcript in a phyB mutant. All data are presented as the mean ±SE (n ≥3). *P<0.05; **P<0.01; different letters above the bars, P<0.01, t-test. (This figure is available in colour at JXB online.)
The diurnal expression pattern of GS5 was analyzed during a 24h period in leaves. The GS5 transcript increased gradually in the daytime, reached its peak value at dusk, and then declined gradually until it fell to the minimum at dawn (Fig. 2B). Both GS5 alleles had circadian rhythms, though GS5-2 had a higher peak value. The length of the photoperiod had no impact on the circadian rhythms of GS5 (Fig. 2B, D). The circadian rhythm disappeared when the seedlings were cultivated under continuous darkness or illumination, under which conditions GS5 expression only stayed at its basal level (Fig. 2C, E). Changing the light conditions could induce expression of GS5 in several hours, either from continuous darkness to illumination or from continuous illumination to darkness; the up-regulation of GS5-2 expression was more dramatic than that of GS5-1 (Fig. 2C, E). These results were consistent with the fact that GS5-2 had more light-responsive elements, and thus may be more sensitive to light induction. It was also found that the GS5 transcript in leaves was significantly suppressed in a phyB mutant, but not influenced in a GHD7-silenced transgenic plant (Weng et al., 2014) (Fig. 2F), suggesting that GS5 might function downstream in the PHYB pathway.
Although GS5 positively regulated mitosis in the panicle (Li et al., 2011), the differential expression in leaves between the NILs did not affect leaf size (Table 1), implying that GS5 may have a different cellular function in leaves. Interestingly, the phyB mutant exhibited increased grain size (both grain width and length), and high grain chalkiness with reduced plumpness (Supplementary Fig. S1 at JXB online), suggesting that the PHYB pathway somehow influenced both grain size and filling.
Table 1.
Grain and leaf size of transgenic positive and negative plants in the T1 generation
| Genotype | No. of plants | 10-Grain width (mm) | 10-Grain length (mm) | Flag leaf length (cm) | Flag leaf width (mm) |
|---|---|---|---|---|---|
| P Ubi ::GS5-1-FLAG (+) | 45 | 35.43±0.07 | 74.95±0.36 | 26.44±0.40 | 10.57±0.08 |
| P Ubi ::GS5-1-FLAG (–) | 16 | 33.60±0.03 | 73.52±0.65 | 26.06±0.37 | 11.18±0.10 |
| P-value | 7.75E-33 | 0.06 | 0.82 | 0.20 | |
| P Ubi ::GS5-2-FLAG (+) | 43 | 35.30±0.06 | 75.28±0.43 | 26.06±0.41 | 10.90±0.14 |
| P Ubi ::GS5-2-FLAG (–) | 22 | 33.71±0.05 | 74.71±0.31 | 26.57±0.43 | 11.16±0.12 |
| P-value | 4.44E-30 | 0.46 | 0.38 | 0.89 | |
| P 35S ::GS5-1-GFP (+) | 44 | 35.29±0.22 | 76.82±0.65 | – | – |
| P 35S ::GS5-1-GFP (–) | 18 | 33.64±0.16 | 75.31±0.39 | – | – |
| P-value | 3.42E-10 | 0.06 | |||
| P 35S ::GS5-2-GFP (+) | 48 | 35.37±0.19 | 75.28±0.41 | – | – |
| P 35S ::GS5-2-GFP (–) | 22 | 33.53±0.12 | 75.62±0.63 | – | – |
| P-value | 3.00E-11 | 0.73 |
(+) and (–) represent transgenic positive and negative plants, respectively.
The average values with standard error are shown (n ≥3).
The P-values are derived from Student’s t-tests between transgenic positive and negative plants.
Key nucleotide polymorphisms responsible for differential expression of GS5 in young panicles
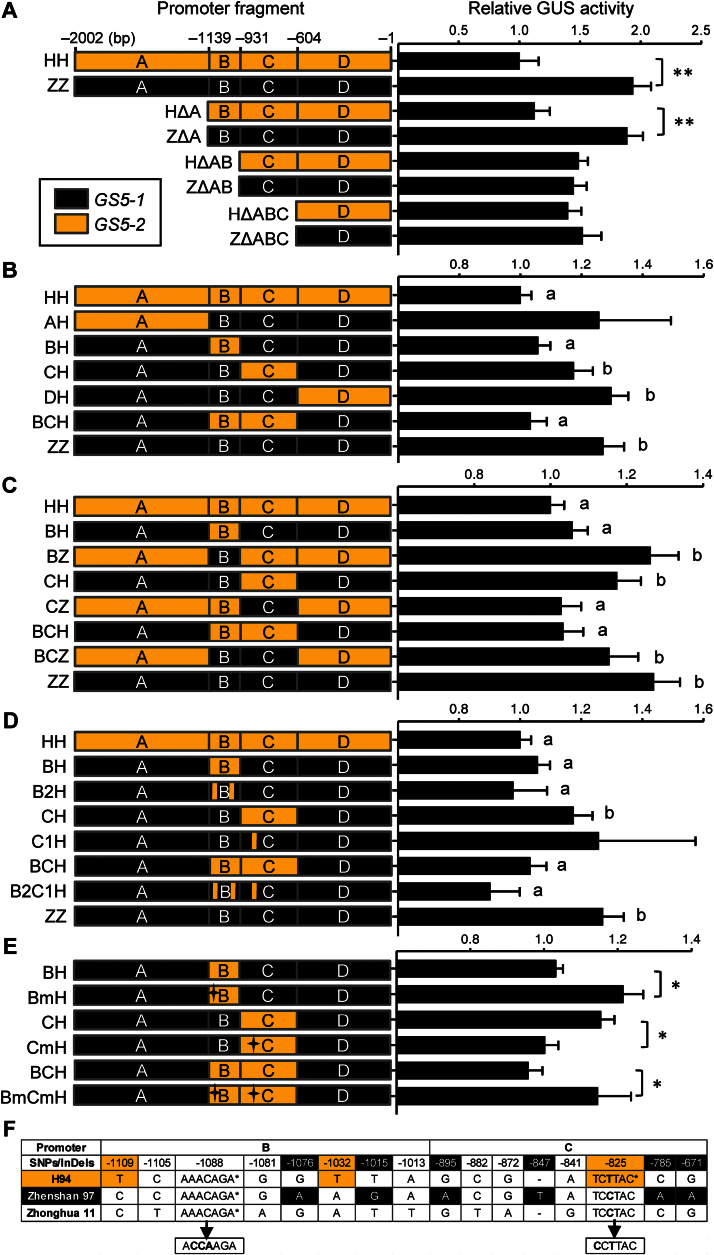
To investigate the nucleotides responsible for up-regulation of GS5-1 in the young panicle, the 2kb promoter was divided into four fractions (A, B, C, and D) (Fig. 3A), to construct serial 5′ deletions fused with the GUS reporter gene, which were introduced into Zhonghua 11, an Oryza sativa L. ssp. japonica variety suitable for transformation. Young panicles of 20cm in length from transgenic plants were harvested and used for GUS activity assay. The results showed that the expression difference between the two alleles was retained in deletions up to –1139bp, but was lost at –931bp (Fig. 3A), indicating that polymorphisms in fraction B must be responsible for the higher expression of GS5-1.
Fig. 3.
Activity of the truncated and chimeric promoter fragments of GS5. (A–E) Relative GUS activity of the truncated and chimeric promoter fragments of GS5. Constructs HH and ZZ carrying the 2kb promoter fragments from H94 and Zhenshan 97, respectively, were used as references. (A) The 2kb promoter was divided into four fractions, A (–2002bp to –1139bp), B (–1139bp to –931bp), C (–931bp to –604bp,) and D (–604bp to –1bp), and constructs with 5′ deletions were prepared accordingly. (B) The fractions A, B, C, and D of ZZ were replaced with the corresponding sequences from HH to construct AH, BH, CH, and DH, respectively. In construct BCH, fractions B and C of ZZ were replaced with the corresponding sequences from HH. (C) Either or both fractions B and C of HH were replaced with the corresponding sequences from ZZ to construct BZ, CZ, and BCZ. (D) SNP_–1109 and SNP_–1032 in fraction B of constructs B2H and B2C1H and SNP_–825 in fraction C of constructs C1H and B2C1H were changed from ZZ to the GS5-2 genotype. (E) The putative GA-responsive element in fraction B of BH and BCH and the light-responsive element in fraction C of CH and BCH were mutated to constructs BmH, CmH, and BmCmH. The mutated cis-acting elements are indicated by an asterisk. All data are presented as the mean ±SE (n ≥15). *P<0.05; **P<0.01; different letters on the bars, P <0.05, t-test. (F) Polymorphisms in fractions B and C among H94, Zhenshan 97, and Zhonghua 11. The specific polymorphisms of H94 and Zhenshan 97 are highlighted in grey and black, respectively. The putative GA-responsive element (–1088, AAACAGA) and light-responsive element (–825, TCTTAC) are indicated by asterisks and the mutated forms are shown at the bottom. (This figure is available in colour at JXB online.)
To validate these results, the four fractions of GS5-1 were replaced with the corresponding sequences from GS5-2, and it was found that the replacements of fraction B reduced the GS5-1 promoter strength significantly (Fig. 3B). Conversely, replacement of fraction B of GS5-2 with the corresponding sequences from GS5-1 raised the GS5-2 promoter strength (Fig. 3C). As most of the light-responsive elements were located in fraction C, which must be responsible for the light-related expression in leaves, the impact of fraction C on the promoter strength in panicles was also tested. The results showed no significant change when fraction C was replaced with the counterpart from the other genotype (Fig. 3B, C); thus the promoter strength in the panicle was only related to the sequence of fraction B.
There were four SNPs in fraction B between GS5-1 and GS5-2 (Fig. 3F). To narrow down the range of candidate sites, the 2kb promoter of GS5-1 and GS5-2 was compared with the GS5 allele from Zhonghua 11, GS5-3, another wide grain variety (Li et al., 2011). Seven polymorphisms occurred between GS5-2 and the other two varieties; two of them (SNP_–1109 and SNP_–1032) were in fraction B and one (SNP_–825) in fraction C (Fig. 3F). These three sites of GS5-1 were mutated into the GS5-2 genotype (Fig. 3D). The results showed that the mutated GS5-1 promoter strength was reduced in the same way as replacements of the entire fraction (Fig. 3D), indicating that SNPs at –1109 and –1032 in fraction B were the key nucleotides responsible for differential expression of GS5-1 and GS5-2 in developing young panicles.
Suppression of GS5 expression by plant hormones
SNP_–1109 and SNP_–1032 in fraction B were located in the flanking sequence of a putative GA-responsive element (Fig. 3F). To examine how these two sites in fraction B affect the expression of GS5, the putative GA-responsive element in construct BH was mutated (Fig. 3E, F). Compared with construct BH, construct BmH with a mutated GA-responsive element resulted in higher GUS activity, implying that the putative GA-responsive element might function as a transcription repressor (Fig. 3E). Interestingly, SNP_–825 in fraction C resulted in a change of the light-responsive element, thus this cis-acting element was missing in GS5-1 (Fig. 3F). This light-responsive element was also mutated in construct CH, which carried fraction C from GS5-2 in the GS5-1 promoter backbone, and it was found that construct CmH with the element mutated had lower GUS activity than construct CH (Fig. 3E, F), indicating that the light-responsive element promoted activation of transcription, which was in agreement with the previous results. When both cis-acting elements were mutated in construct BmCmH, increased GUS activity was observed, suggesting that the putative GA-responsive element was the limiting factor for GS5 transcription in young panicles (Fig. 3E).
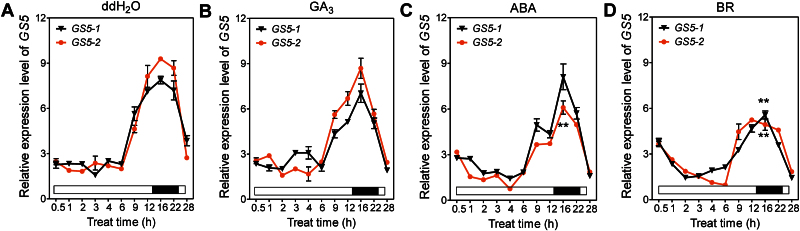
To determine the role of the putative GA-responsive element, the expression of GS5 was examined after plant hormone treatments. The results showed that both GS5-1 and GS5-2 failed to respond to GA (Fig. 4A, B). However, the expression of GS5-2, but not GS5-1, was obviously suppressed by ABA, resulting in a lower transcript level than GS5-1 (Fig. 4A, C). Published results had proved that transcription activated by GA could be inhibited by ABA via a GA-responsive element (Jacobsen and Beach, 1985; Skriver et al., 1991; Gubler and Jacobsen, 1992; Washio and Ishikawa, 1994). Therefore, it was concluded that SNP_–1109 and SNP_–1032 in fraction B altered the response of GS5 to ABA.
Fig. 4.
Expression level of GS5 in response to plant hormone treatments. Fifteen-day-old seedlings of NIL(ZS97) and NIL(H94) grown under long-day conditions were treated with ddH2O (A), 10 μM GA3 (B), 10 μM ABA (C), or 10 μM BR (D). All data are presented as the mean ±SE (n ≥3). *P<0.05; **P<0.01, t-test. (This figure is available in colour at JXB online.)
It was also detected that BR suppressed the expression of both GS5-1 and GS5-2 (Fig. 4A, D). It was previously demonstrated that BZR1 and BES1/BZR2 were the key transcription factors for BR-regulated gene expression (He et al., 2005; Yin et al., 2005). It was found that BZR1 binds to the BR response element (BRRE; CGTGT/CG) (He et al., 2005) and BES1/BZR2 binds to the E-box (CANNTG) (Yin et al., 2005). The BRREs were usually present in BR-repressed genes and the E-boxes were present in both BR-repressed and BR-induced genes (Yu et al., 2011). Eight E-boxes were found in fractions A and B in the promoter region of GS5 (Supplementary Table S1 at JXB online), suggesting that GS5 was probably one of the target genes of the BR signalling pathway.
Secretion of GS5–GFP to the cell surface
Epitope tags were used for visualization of the GS5 proteins. To ensure that addition of epitope tags would not affect the function of the GS5 protein, constructs for both GS5-1 and GS5-2 fused to FLAG-tag driven by the ubiquitin promoter were generated, and they were expressed in Zhonghua 11. The grain width of transgenic positive plants of both constructs significantly increased, and also co-segregated with the higher expression level of GS5 in the T1 progeny (Table 1), just like the original GS5 CDSs (Li et al., 2011). An increase in grain width in transgenic plants harbouring P 35S ::GS5-1-GFP or P 35S ::GS5-2-GFP was also observed (Table 1). Thus, higher expression of both epitope tag constructs produced wider grains just like the GS5 proteins, indicating that the GS5–epitope tag fusion proteins were functionally equivalent to GS5 in planta.
Phylogenetic analysis suggested that GS5 belonged to SCPL group II proteins, the same group as Arabidopsis BRS1 (Supplementary Fig. S2 at JXB online), a secreted and active serine carboxypeptidase which plays a role in the BR signalling pathway (Li et al., 2001; Zhou and Li, 2005). Using an anti-GFP antibody to analyse the total protein extracts from transgenic plants, two specific bands were detected, indicating that GS5 could be cleaved into two chains (A and B) after a predicted processing step as in many SCPL-II proteins such as BRS1 (Zhou and Li, 2005). The 80kDa band represented the intact GS5–GFP protein and the 47kDa band represented the B-chain, the C-terminal fragment of GS5 attached to the GFP-tag after processing (Fig. 5A), while the A-chain could not be detected. A similar result was also obtained in a western blot assay of GS5-FLAG transgenic plants using anti-FLAG antibody (Supplementary Fig. S3A). These results indicated that the GS5–GFP fusion protein had functional integrity and the subcellular localization of GS5 could be viewed using GS5–GFP.
Fig. 5.
Subcellular localization of the GS5 protein. (A) Western blot assay of GS5–GFP transgenic plants using anti-GFP antibody showing the cleavage GS5–GFP into two chains (A and B) as in many SCPL-II proteins. 1, P 35S ::GS5-1-GFP; N, negative control; 2, P 35S ::GS5-2-GFP; the white arrow indicates the full-length GS5–GFP protein; the red arrow indicates the C-terminus of GS5 with the B-chain–GFP peptide. (B) Localization of GS5-1–GFP and GS5-2–GFP on the spikelet epidermal cell surface of transgenic plants. Scales bars=20 μm. (C) Localization of transiently expressed GS5–GFP in tobacco leaf epidermal cells showing the distribution both on the cell surface and in the internal space, whereas OsBAK1-7–GFP is found only on the cell surface (left panel). After induction of plasmolysis, the GS5–GFP signal is mostly found in the plasma membrane and a portion of the signal is detected outside the plasma membrane, whereas OsBAK1-7–GFP is found only on the plasma membrane (right panel). Scales bars=20 μm.
To investigate the subcellular localization of GS5, lemmas from young panicles of 15cm in length from GS5–GFP transgenic plants were used for confocal microscopy assay. Green fluorescence signals of GS5-1–GFP and GS5-2–GFP were detected on the cell surface (Fig. 5B). Since it was difficult to induce plasmolysis in rice lemma epidermal cells, P 35S ::GS5-GFP was transiently expressed in tobacco leaf epidermal cells, using P 35S ::OsBAK1-7-GFP, a homologue of plasma membrane protein bri1-associated receptor kinase 1 (BAK1) as a reference (Li et al., 2002; Nam and Li, 2002). The majority of green fluorescence signal of GS5–GFP was localized on the cell surface, and there was also an obvious net-like signal inside the cell (Fig. 5C, left panel). After induction of plasmolysis, the OsBAK1-7–GFP signal moved with the plasma membrane (Fig. 5C, right panel), indicating that OsBAK1-7–GFP was localized in the plasma membrane, whereas the GS5–GFP signal was mostly found aggregating in the plasma membrane and a portion of the signal was detected outside the plasma membrane (Fig. 5C, right panel).
To demonstrate whether the net-like signal was endoplasmic reticulum (ER), the GS5–GFPs were co-expressed with the ER marker RFP–HDEL (Supplementary Fig. S4A at JXB online) or the cis-Golgi marker Man1–RFP (Supplementary Fig. S4B) in tobacco BY-2 protoplasts. The results confirmed that GS5–GFPs were co-located with RFP–HDEL (Supplementary Fig. S4A). These observations indicated that GS5 was secreted from the ER to the cell surface, and the plasma membrane signal might result from its interaction with some plasma membrane protein(s).
Interaction between GS5 and the extracellular domain of BAK1 homologues
To identify interacting proteins, the A-chain of GS5, which included a putative substrate-binding region (Feng and Xue, 2006) (Supplementary Fig. S5 at JXB online), was used to screen a yeast two-hybrid library generated using the total RNA from young panicles of 2cm in length from Zhenshan 97. A B-chain of a SCPL protein was identified, indicating that the A-chain folded correctly in yeast and could be used as a bait (Fig. 6A). A homologue of the membrane protein BAK1 was detected. Interactions were also detected between the A-chain of GS5 and the extracellular LRR domains of three BAK1 homologues (Supplementary Fig. S6), but not their intracellular kinase domains or GAL4 activation domain (Fig. 6A).
Fig. 6.
Protein–protein interaction between GS5 and OsBAK1. (A) Yeast two-hybrid assay showing interactions of the A-chain of GS5 (BD-A chain) with the B-chain (AD-B chain) and the extracellular LRR domains of three BAK1 homologues, OsBAK1-4, OsBAK1-7, and OsBAK1-9 (AD-LRR-4, AD-LRR-7, and AD-LRR-9), but not with the GAL4 activation domain (AD) or the kinase domains of OsBAK1 (AD-KD-4, AD-KD-7, and AD-KD-9). BD, GAL4 DNA-binding domain; AD, GAL4 activation domain. (B) Co-expression of OsBAK1-7–GFP and OsMSBP1–RFP in tobacco leaf epidermal cells showing co-localization of the two proteins in the vesicle-like compartments inside the cell (top panel). Co-expression of OsBAK1-7–GFP and OsMSBP1–RFP with GS5-FLAG in tobacco leaf epidermal cells showing the distinct localization of OsBAK1-7–GFP and OsMSBP1–RFP (bottom panel). The experiments were repeated three times, yielding similar results, and representative images are shown. Scales bars=20 μm. The data for the statistical analysis of the fluorescent signals in the cytoplast beneath the images are presented as the mean ±SE (n ≥10). **P<0.01, t-test. (C) Western blot assay of GS5-FLAG expression in the experiment described in (B) using anti-FLAG antibody. Top panel, western blot assay; bottom panel, Ponceau-stained protein sample; black arrow, full-length GS5-FLAG protein.
In Arabidopsis thaliana, membrane steroid-binding protein 1 (MSBP1) specifically interacts with the extracellular domain of BAK1 in vivo which accelerates the endocytosis of BAK1, resulting in suppressed BR signalling (Song et al., 2009). It was thus suspected that enhanced GS5 expression would competitively inhibit the interaction between OsBAK1 and OsMSBP1 by occupying the extracellular LRR domain of OsBAK1, thus preventing endocytosis of OsBAK1.
To test this hypothesis, the MSBP1 homologue in rice (LOC_Os10g35870) was first obtained by BLAST, and then P 35S ::OsMSBP1-RFP was constructed and co-expressed with P 35S ::OsBAK1-7-GFP in tobacco leaf epidermal cells. OsBAK1-7–GFP was localized at the plasma membrane when expressed alone (Fig. 5C). However, when it was co-expressed with OsMSBP1–RFP, a large part of the OsBAK1-7–GFP signal appeared in the vesicle-like compartments, co-localized with OsMSBP1–RFP just as in A. thaliana (Fig. 6B, top panel). Next P 35S ::OsBAK1-7-GFP, P 35S ::OsMSBP1-RFP, and P Ubi ::GS5-FLAG were co-expressed in tobacco leaf epidermal cells, and the expression of GS5-FLAG was detected by western blot using anti-FLAG antibody (Fig. 6C). The results showed that when GS5-FLAG was present, there was no overlap between the subcellular localizations of OsBAK1-7–GFP and OsMSBP1–RFP. The signal of OsBAK1-7–GFP appeared only at the plasma membrane, while OsMSBP1–RFP was found in numerous vesicle–like small compartments (Fig. 6B, bottom panel), which were obviously different from those when co-expressed with OsBAK1-7–GFP only (Fig. 6B, top panel). This suggested that the secreted GS5 protein could competitively interact with the extracellular LRR domains of OsBAK1 on the cell surface, which could explain the correlation between grain size and the expression level of GS5.
Discussion
The expression of GS5 is differentially regulated in different tissues
Most of the differences in phenotypes result from variations in protein function or gene expression. In the case of GS5, phenotypic variation is due to the expression difference caused by the polymorphisms in the promoter, while both the wide-grain allele GS5-1 and the narrow-grain allele GS5-2 encode a functional SCPL protein and higher expression of both proteins produces wider grains. This is similar to the situation of another positive grain-width regulator GW8/OsSPL16, whose expression is reduced because of a 10bp deletion in the promoter (Wang et al., 2012). However, GS5 showed a more complex spatial and temporal expression pattern in the life cycle of rice; it is regulated by multiple elements in the promoter whose polymorphisms caused differential expression of the gene in different tissues. In addition to the two key nucleotides in fractions B (SNP_–1109 and SNP_–1032) of the GS5 promoter that alter the response of GS5 to ABA, a number of light-responsive elements are also involved, which regulate activation of transcription and light-induced expression of GS5 in leaves. Together these variations lead to the result that the GS5-2 transcript is more abundant in green tissues, while GS5-1 has a higher expression level in developing panicles.
As a positive regulator of mitosis, differential expression of GS5 during young panicle development regulates grain size variation. However, the GS5 transcript in leaves does not affect leaf size, suggesting that GS5 may have a different cellular function in the leaf. GS5 also regulates grain filling, which involves the accumulation, distribution, and transportation of a number of substances (Wang et al., 2008; Wu et al., 2008; Ishimaru et al., 2013).
A proposed model for GS5 regulation of grain size
In this study, it was found that the GS5 protein on the cell surface and OsMSBP1 competitively interact with the extracellular LRR domain of OsBAK1-7, and they have opposite effects on the localization of OsBAK1-7; OsMSBP1 accelerated the endocytosis of OsBAK1-7, whereas GS5 kept OsBAK1-7 on the cell surface. In A. thaliana, enhanced expression of MSBP1 suppressed BR signalling by shifting the equilibrium of BAK1 toward endosomes and reducing the BRI1–BAK1 association at the plasma membrane, which can be recovered by overexpressing BAK1 (Song et al., 2009). Furthermore, BR-related genes are shown to have effects on grain weight. Loss-of-function mutants of the genes for BR biosynthesis and the signalling pathway usually have shorter grains (Yamamuro et al., 2000; Hong et al., 2003, 2005; Tanabe et al., 2005; Morinaka et al., 2006; Nakamura et al., 2006). Modification of the endogenous expression level of OsBAK1, a member of the same protein family as OsBAK1-7, can also alter grain size (Li et al., 2009). BR also regulates grain filling by stimulating the flow of assimilates from the source to the sink (Wu et al., 2008). Transgenic plants expressing a sterol C-22 hydroxylase, which controls BR levels using a promoter active only in the stems, leaves, and roots, produce heavier grains, but the enzyme has no apparent effect on grain weight when expressed in the embryos or endosperms (Wu et al., 2008).
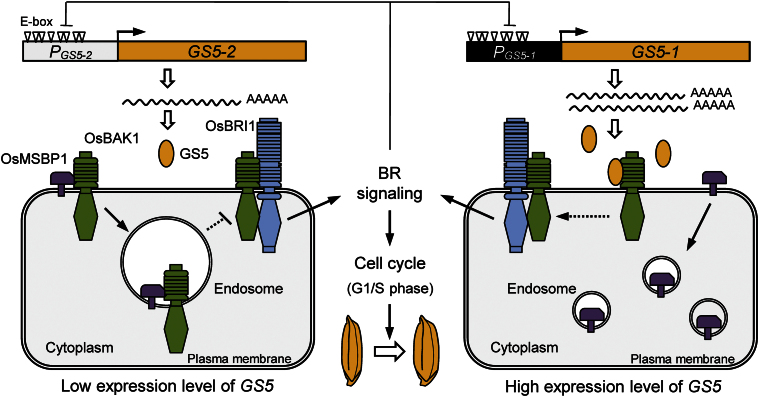
Based on the above results, it was reasonable to assume that the increased expression of the secreted GS5 protein on the cell surface kept OsBAK1 at the plasma membrane, where it could interact with OsBRI1 and enhance BR signalling. Then the enhanced BR signalling would have a dual effect on the GS5-regulated grain size: (i) it would promote the cell cycle thus increasing grain size; but at the same time (ii) it would suppress GS5 transcription via the E-boxes in the promoter by feedback regulation. The final grain size is thus the outcome of a balance between them (Fig. 7).
Fig. 7.
The hypothetical molecular mechanism of GS5 regulation of grain size. The two key SNPs in promoter fraction B result in different expression levels of the GS5 alleles (GS5-1 from Zhenshan 97 and GS5-2 from H94) in developing young panicles. The secreted GS5 protein and the membrane steroid-binding protein 1 homologue (OsMSBP1) can competitively interact with the extracellular leucine-rich repeat (LRR) domain of OsBAK1. When the GS5 expression level is low, the membrane protein OsBAK1 interacts with OsMSBP1 and the interaction accelerates the endocytosis of OsBAK1, reducing the OsBRI1–OsBAK1 complex at the plasma membrane. With increased expression of GS5, the large amount of GS5 protein on the cell surface occupies the extracellular domain of OsBAK1, preventing it from interacting with OsMSBP1 and keeping it at the plasma membrane, thus facilitating the OsBRI1–OsBAK1 interaction. The OsBRI1–OsBAK1 interaction enhances BR signalling, which promotes mitotic division in the lemma/palea, resulting in wider grains, and also feedback suppression of the GS5 transcription via the E-boxes (indicated by the triangle) in the promoter of GS5. (This figure is available in colour at JXB online.)
Interestingly, there is a secreted SCPL protein, BRS1, which shared high sequence similarity with GS5, involved in BR signalling in A. thaliana. The effect of BRS1 is selective: enhanced expression of BRS1 can suppress the bri1 extracellular domain mutants, but overexpression in either the wild type or the kinase-dead bri1 mutant results in no phenotypic alterations (Li et al., 2001), and the degree of suppression of the bri1 mutant is also positively correlated with the BRS1 expression level (Zhou and Li, 2005). These studies suggest that BRS1 probably acts at an early step in BR signalling by processing some rate-limiting protein(s), but the actual molecular mechanism is still unclear (Li et al., 2001; Zhou and Li, 2005). In addition to the similar characteristics between GS5 and BRS1, it was also found that overexpression of BRS1 in rice can increase grain width, and the grain width is positively correlated with the BRS1 expression level just as in the case of GS5 (Supplementary Fig. S7 at JXB online), strongly suggesting that BRS1 may function similarly to GS5. These results strongly suggest a potential link among GS5, the BR-related pathway, and grain size regulation, although more research is still needed to pinpoint the exact underlying mechanism.
What makes GS5 a minor gene for grain size?
GS5 is a minor QTL controlling grain size; grains of NIL(ZS97) are only 8.7% wider than those of NIL(H94) (Li et al., 2011). The slight variation of grain size stemmed from the limited expression difference caused by the polymorphisms in the promoter region. Since an elevated expression level of GS5 increases grain size, the question then arises of how much of a grain size increase can be obtained by manipulating GS5. Can overexpressing the gene with a stronger promoter further increase the grain size? It is clear from the present results that the answer to this question is no. Overexpression of GS5 increases the grain width by only ~6% (Table 1), clearly indicating that the GS5 protein has only a limited effect on grain size determination. This is probably because the effect of competitive interaction of the proteins is dependent on the quantity of the other two proteins, and is also subject to feedback regulation of BR signalling. Also, excess GS5 proteins may get stuck in the ER. Therefore, it is not possible to increase grain size beyond a certain range by excess expression of GS5.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Grain size and chalkiness of the phyB mutant.
Figure S2. Phylogenetic tree of members of the serine carboxypeptidase-like protein family.
Figure S3. Assay of GS5-FLAG protein in P Ubi ::GS5-FLAG transgenic positive plants.
Figure S4. Co-localization assay of GS5 with the endoplasmic reticulum marker inside the cell.
Figure S5. Amino acid alignment of group II serine carboxypeptidase-like proteins.
Figure S6. Phylogenetic tree of the rice BAK1 homologues.
Figure S7. Grain size and leaf size of P Ubi :: AtBRS1-FLAG transgenic plants.
Table S1. Polymorphisms and cis-acting elements in the 2kb promoter of GS5.
Table S2. Primers used in this work.
Acknowledgements
We thank Professor Jian Xu from the National University of Singapore for kindly providing the pM999-EGFP vector, Professor Liwen Jiang from The Chinese University of Hong Kong for providing tobacco BY-2 suspension-cultured cells and marker constructs RFP–HDEL and Man1–RFP, and Professor Hongwei Xue from Shanghai Institutes for Biological Sciences for providing the phyB mutant. This work was supported by grants from the 863 Project (grant no. 2012AA10A303) and the 973 Project (grant no. 2014CB943400) of China, the National Natural Science Foundation of China (grant no. 31300992), and the Bill and Melinda Gates Foundation.
References
- Fan C, Xing Y, Mao H, Lu T, Han B, Xu C, Li X, Zhang Q. 2006. GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theoretical and Applied Genetics 112, 1164–1171. [DOI] [PubMed] [Google Scholar]
- Feng Y, Xue Q. 2006. The serine carboxypeptidase like gene family of rice (Oryza sativa L. ssp. japonica). Functional and Integrative Genomics 6, 14–24. [DOI] [PubMed] [Google Scholar]
- Fraser CM, Rider LW, Chapple C. 2005. An expression and bioinformatics analysis of the Arabidopsis serine carboxypeptidase-like gene family. Plant Physiology 138, 1136–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler F, Jacobsen JV. 1992. Gibberellin-responsive elements in the promoter of a barley high-pI alpha-amylase gene. The Plant Cell 4, 1435–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J- X, Gendron JM, Sun Y, Gampala SSL, Gendron N, Sun CQ, Wang Z-Y. 2005. BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science 307, 1634–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Z, Ueguchi-Tanaka M, Fujioka S, Takatsuto S, Yoshida S, Hasegawa Y, Ashikari M, Kitano H, Matsuoka M 2005. The rice brassinosteroid-deficient dwarf2 mutant, defective in the rice homolog of Arabidopsis DIMINUTO/DWARF1, is rescued by the endogenously accumulated alternative bioactive brassinosteroid, dolichosterone. The Plant Cell 17, 2243–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Z, Ueguchi-Tanaka M, Umemura K, Uozu S, Fujioka S, Takatsuto S, Yoshida S, Ashikari M, Kitano H, Matsuoka M. 2003. A rice brassinosteroid-deficient mutant, ebisu dwarf (d2), is caused by a loss of function of a new member of cytochrome P450. The Plant Cell 15, 2900–2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Sun Q, Qin F, Li C, Zhao Y, Zhou D-X. 2007. Down-regulation of a SILENT INFORMATION REGULATOR2-related histone deacetylase gene, OsSRT1, induces DNA fragmentation and cell death in rice. Plant Physiology 144, 1508–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimaru K, Hirotsu N, Madoka Y, et al. 2013. Loss of function of the IAA-glucose hydrolase gene TGW6 enhances rice grain weight and increases yield. Nature Genetics 45, 707–711. [DOI] [PubMed] [Google Scholar]
- Jacobsen JV, Beach LR. 1985. Control of transcription of α-amylase and rRNA genes in barley aleurone protoplasts by gibberellin and abscisic acid. Nature 316, 275–277. [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. 1987. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO Journal 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S. 2002. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Research 30, 325–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Wang L, Wang M, Xu Y-Y, Luo W, Liu Y-J, Xu Z-H, Li J, Chong K. 2009. Engineering OsBAK1 gene as a molecular tool to improve rice architecture for high yield. Plant Biotechnology Journal 7, 791–806. [DOI] [PubMed] [Google Scholar]
- Li J, Lease KA, Tax FE, Walker JC. 2001. BRS1, a serine carboxypeptidase, regulates BRI1 signaling in Arabidopsis thaliana . Proceedings of the National Academy of Sciences, USA 98, 5916–5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wen J, Lease KA, Doke JT, Tax FE, Walker JC. 2002. BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110, 213–222. [DOI] [PubMed] [Google Scholar]
- Li Y, Fan C, Xing Y, et al. 2011. Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nature Genetics 43, 1266–1269. [DOI] [PubMed] [Google Scholar]
- Lin YJ, Zhang Q. 2005. Optimising the tissue culture conditions for high efficiency transformation of indica rice. Plant Cell Reports 23, 540–547. [DOI] [PubMed] [Google Scholar]
- Mao H, Sun S, Yao J, Wang C, Yu S, Xu C, Li X, Zhang Q. 2010. Linking differential domain functions of the GS3 protein to natural variation of grain size in rice. Proceedings of the National Academy of Sciences, USA 107, 19579–19584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y, Jiang L. 2007. Transient expression of fluorescent fusion proteins in protoplasts of suspension cultured cells. Nature Protocols 2, 2348–2353. [DOI] [PubMed] [Google Scholar]
- Morinaka Y, Sakamoto T, Inukai Y, Agetsuma M, Kitano H, Ashikari M, Matsuoka M. 2006. Morphological alteration caused by brassinosteroid insensitivity increases the biomass and grain production of rice. Plant Physiology 141, 924–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura A, Fujioka S, Sunohara H, et al. 2006. The role of OsBRI1 and its homologous genes, OsBRL1 and OsBRL3, in rice. Plant Physiology 140, 580–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam KH, Li J. 2002. BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110, 203–212. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual , 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Shomura A, Izawa T, Ebana K, Ebitani T, Kanegae H, Konishi S, Yano M. 2008. Deletion in a gene associated with grain size increased yields during rice domestication. Nature Genetics 40, 1023–1028. [DOI] [PubMed] [Google Scholar]
- Skriver K, Olsen FL, Rogers JC, Mundy J. 1991. cis-acting DNA elements responsive to gibberellin and its antagonist abscisic acid. Proceedings of the National Academy of Sciences, USA 88, 7266–7270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Shi Q-M, Yang X-H, Xu Z-H, Xue H-W. 2009. Membrane steroid-binding protein 1 (MSBP1) negatively regulates brassinosteroid signaling by enhancing the endocytosis of BAK1. Cell Research 19, 864–876. [DOI] [PubMed] [Google Scholar]
- Song X-J, Huang W, Shi M, Zhu M-Z, Lin H-X. 2007. A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nature Genetics 39, 623–630. [DOI] [PubMed] [Google Scholar]
- Sparkes IA, Runions J, Kearns A, Hawes C. 2006. Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nature Protocols 1, 2019–2025. [DOI] [PubMed] [Google Scholar]
- Sun Q, Zhou D-X. 2008. Rice jmjC domain-containing gene JMJ706 encodes H3K9 demethylase required for floral organ development. Proceedings of the National Academy of Sciences, USA 105, 13679–13684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe S, Ashikari M, Fujioka S, et al. 2005. A novel cytochrome P450 is implicated in brassinosteroid biosynthesis via the characterization of a rice dwarf mutant, dwarf11, with reduced seed length. The Plant Cell 17, 776–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi L, Sowdhamini R 2006. Cross genome comparisons of serine proteases in Arabidopsis and rice. BMC Genomics 7, 200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E, Wang J, Zhu X, et al. 2008. Control of rice grain-filling and yield by a gene with a potential signature of domestication. Nature Genetics 40, 1370–1374. [DOI] [PubMed] [Google Scholar]
- Wang S, Wu K, Yuan Q, et al. 2012. Control of grain size, shape and quality by OsSPL16 in rice. Nature Genetics 44, 950–954. [DOI] [PubMed] [Google Scholar]
- Washio K, Ishikawa K. 1994. Organ-specific and hormone-dependent expression of genes for serine carboxypeptidases during development and following germination of rice grains. Plant Physiology 105, 1275–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng J, Gu S, Wan X, et al. 2008. Isolation and initial characterization of GW5, a major QTL associated with rice grain width and weight. Cell Research 18, 1199–1209. [DOI] [PubMed] [Google Scholar]
- Weng X, Wang L, Wang J, Hu Y, Du H, Xu C, Xing Y, Li X, Xiao J, Zhang Q 2014. Grain number, plant height, and heading date 7 is a central regulator of growth, development, and stress response. Plant Physiology 164, 735–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C-y, Trieu A, Radhakrishnan P, et al. 2008. Brassinosteroids regulate grain filling in rice. The Plant Cell 20, 2130–2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamuro C, Ihara Y, Wu X, Noguchi T, Fujioka S, Takatsuto S, Ashikari M, Kitano H, Matsuoka M 2000. Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. The Plant Cell 12, 1591–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye R, Zhou F, Lin Y. 2012. Two novel positive cis-regulatory elements involved in green tissue-specific promoter activity in rice (Oryza sativa L ssp.). Plant Cell Reports 31, 1159–1172. [DOI] [PubMed] [Google Scholar]
- Yin Y, Vafeados D, Tao Y, Yoshida S, Asami T, Chory J. 2005. A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis . Cell 120, 249–259. [DOI] [PubMed] [Google Scholar]
- Yu X, Li L, Zola J, et al. 2011. A brassinosteroid transcriptional network revealed by genome-wide identification of BESI target genes in Arabidopsis thaliana . The Plant Journal 65, 634–646. [DOI] [PubMed] [Google Scholar]
- Zhou A, Li J. 2005. Arabidopsis BRS1 is a secreted and active serine carboxypeptidase. Journal of Biological Chemistry 280, 35554–35561. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.