Abstract
Assaying effects on pre-rRNA processing and ribosome assembly upon depleting individual ribosomal proteins (r-proteins) provided an initial paradigm for assembly of eukaryotic ribosomes in vivo—that each structural domain of ribosomal subunits assembles in a hierarchical fashion. However, two features suggest that a more complex pathway may exist: (i) Some r-proteins contain extensions that reach long distances across ribosomes to interact with multiple rRNA domains as well as with other r-proteins. (ii) Individual r-proteins may assemble in a stepwise fashion. For example, the globular domain of an r-protein might assemble separately from its extensions. Thus, these extensions might play roles in assembly that could not be revealed by depleting the entire protein. Here, we show that deleting or mutating extensions of r-proteins L7 (uL30) and L35 (uL29) from yeast reveal important roles in early and middle steps during 60S ribosomal subunit biogenesis. Detailed analysis of the N-terminal terminal extension of L8 (eL8) showed that it is necessary for late nuclear stages of 60S subunit assembly involving two major remodeling events: removal of the ITS2 spacer; and reorganization of the central protuberance (CP) containing 5S rRNA and r-proteins L5 (uL18) and L11 (uL5). Mutations in the L8 extension block processing of 7S pre-rRNA, prevent release of assembly factors Rpf2 and Rrs1 from pre-ribosomes, which is required for rotation of the CP, and block association of Sda1, the Rix1 complex, and the Rea1 ATPase involved in late steps of remodeling.
Keywords: ribosome assembly, ribosomal protein extensions, rRNA, pre-ribosome remodeling
INTRODUCTION
The assembly of ribosomes requires the concurrent folding, modification, and processing of pre-rRNA, and the assembly of r-proteins with rRNA, along with the binding, function, and release of assembly factors. During construction of these complex ribonucleoprotein (RNP) particles, numerous protein–protein, protein–rRNA and rRNA–rRNA interactions are established and remodeled (Woolford and Baserga 2013; de la Cruz et al. 2015; Nerurkar et al. 2015). Determination of the crystal structures of ribosomes from prokaryotes, archaea, and eukaryotes revealed characteristics of r-proteins that could play critical roles to enable formation or rearrangement of these molecular interactions. One striking feature is that many r-proteins contain N- and/or C-terminal extensions or internal loops that extend from their conserved globular core and reach across the surface or penetrate the core of ribosomes, contacting other r-proteins and rRNA domains (Ben-Shem et al. 2011; Klinge et al. 2011; Jenner et al. 2012; Melnikov et al. 2012; Wilson and Doudna Cate 2012; Yusupova and Yusupov 2014). Most of these extensions are predicted to contain disordered domains. Thus, these sequences have properties ideally suited for RNP biogenesis, including: (i) flexibility, and thus the capability to access a large surface area of the assembling particles, and (ii) folding upon binding properties that enable them to stabilize their ligands. Such extended domains are enriched in RNA interacting proteins, suggesting roles in the assembly, structure, and function of many different RNPs (Cumberworth et al. 2013; Van Der Lee et al. 2014). For example, 38 of the 78 r-proteins in yeast contain such extensions (Peng et al. 2014). In the ribosome, disordered regions of r-proteins have been suggested to function as stabilizers of the tertiary structure of rRNA, as RNA chaperones to avoid kinetic traps, and as binding sites of dedicated assembly chaperones (Tompa 2003; Tompa and Csermely 2004; Van Der Lee et al. 2014).
The roles of yeast r-proteins have been systematically examined by depleting each of 62 of the 78 r-proteins and assaying effects on pre-rRNA processing and pre-ribosome composition. It was found that r-proteins that bind to the same rRNA domain as each other, to create an rRNP neighborhood, have similar depletion phenotypes. Each of these neighborhoods of r-protein and rRNA assembles hierarchically, roughly 5′–3′ with respect to the rRNA (Ferreira-Cerca et al. 2005; Gamalinda et al. 2014). In the 40S ribosomal subunit, the body domain, containing sequences more at the 5′ end of 18S rRNA, is formed initially, followed by assembly of the head domain, containing sequences more at the 3′ end of 18S rRNA (Neueder et al. 2010; Jakob et al. 2012). In the 60S ribosomal subunit, domains I, II, and VI of 25S rRNA are stabilized first, followed by further stabilization of domains I, III and the proximal stem. Finally, domains IV and V, as well as the central protuberance containing the 5S rRNA, are fully assembled (Gamalinda et al. 2014).
Because eukaryote-specific extensions of many r-proteins interact with multiple different domains of the ribosome, they may enable r-proteins to function in more than one step of assembly by completing their stable association with nascent ribosomes in a stepwise fashion. Yet, depletion of an r-protein can only reveal the earliest step in assembly for which it is required. Thus, targeting mutations to individual domains of r-proteins, including extensions, may reveal multiple roles for these r-proteins in assembly.
A limited number of studies are beginning to uncover specific functions for extensions of r-proteins. In prokaryotes, extensions of S4 and S12 were shown to have a role in restructuring 16S rRNA during 30S ribosomal subunit assembly in vivo and in vitro (Mayerle et al. 2011; Mayerle and Woodson 2013; Calidas et al. 2014; Kim et al. 2014). Extensions of L22 and L4 were shown to be dispensable, while the N-terminal extension of L20 was found to be required for bacterial 50S ribosomal subunit assembly (Timsit et al. 2009). The C-terminal sequence of yeast S14 was shown to function at late stages of 40S subunit assembly, whereas depletion of S14 blocked early steps of assembly (Jakovljevic et al. 2004). Recently, the internal loop of yeast L4 was shown to bind the assembly chaperone Acl4; mutations in this loop resulted in a late defect in ribosome assembly, while depletions of L4 resulted in an early defect (Gamalinda and Woolford 2014; Pillet et al. 2015; Stelter et al. 2015). Together, these studies indicate that the extensions of r-proteins may have distinct functions during ribosome assembly. Nevertheless, the precise mechanisms by which these extensions participate in assembly remain largely unexplored.
In order to better understand the hierarchy of 60S ribosomal subunit assembly at higher resolution than evident from depleting each r-protein, we specifically investigated the functions of the N- and/or C-terminal domains of r-proteins L7, L8, and L35 from Saccharomyces cerevisiae (Fig. 1A–C; Supplemental Fig. S1–S3). These r-proteins are named uL30, eL8, and uL29, respectively, in the new r-protein nomenclature (Ban et al. 2015). We tested the importance of the N-terminal extension of L7 (80 amino acids), the N- and C-terminal extensions of L8 (70 amino acids and 23 amino acids, respectively) and the C-terminal extension of L35 (50 amino acids) by introducing both serial truncations and missense mutations in them (Fig. 1C). Together, these proteins interact with four different rRNA domains that form two major neighborhoods of eukaryote-specific elements of the large subunit. Based on recently determined cryo-EM structures of pre-ribosomes, extensions of L7, L8, and L35 appear to have already adapted mature-like conformations at late nuclear stages of assembly (Supplemental Fig. S4; Wu et al. 2016). We assayed effects of removing these extensions on cell growth, pre-rRNA processing, and association of assembly factors with pre-ribosomes (Supplemental Table 1). Our data indicate that eukaryote-specific extensions of L7, L8, and L35 are essential for ribosome assembly. Mutations in the extensions of L7 and L35 affect ribosome assembly at the same stage as their depletions. Strikingly, in contrast to an early block in assembly when L8 is depleted, we found that removal of the N-terminal extension of L8 blocks late nuclear stages of assembly involving two major remodeling events: removal of internal transcribed spacer 2 (ITS2) and rearrangements of the central protuberance involving the release of assembly factors Rpf2, Rrs1, and Rsa4.
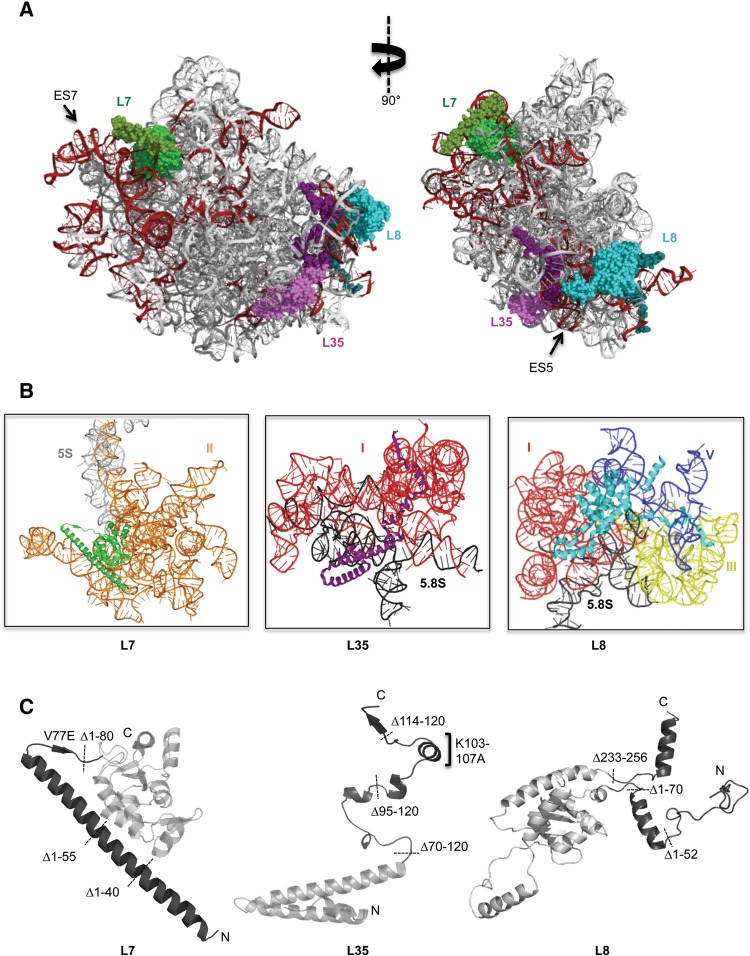
FIGURE 1.
(A) Positions of r-proteins L7 (green), L35 (purple), and L8 (cyan) are shown in the mature yeast 60S ribosomal subunit. Eukaryote-specific extensions of r-proteins are indicated in darker shades of the corresponding colors. Expansion segments of 25S rRNA are colored dark red. The structure on the left corresponds to the solvent-exposed side of the 60S ribosomal subunit of S. cerevisiae (PDB 3U5H and 3U5I). (B) Cartoon representations of r-proteins L7, L35, and L8, showing the 25S rRNA domains that are contacted by globular regions and extensions. Domains I (red), II (orange), III (yellow), and V (blue) are indicated. (C) Globular domains of the proteins are indicated in light gray and extensions are shown in dark gray. Positions of the truncation mutations, and important point mutations investigated in this study are shown.
RESULTS
To examine the effects of mutations in r-protein extensions on large ribosomal subunit assembly in yeast, we constructed strains in which we could turn off expression of the wild-type protein to reveal the phenotypes of plasmid-borne mutant alleles. In each case, we confirmed that expression of the wild-type protein from the plasmid vector rescued the lethal phenotypes of depletion, while strains containing only the empty vector were not viable when expression of the wild-type gene was turned off (Fig. 2A,B).
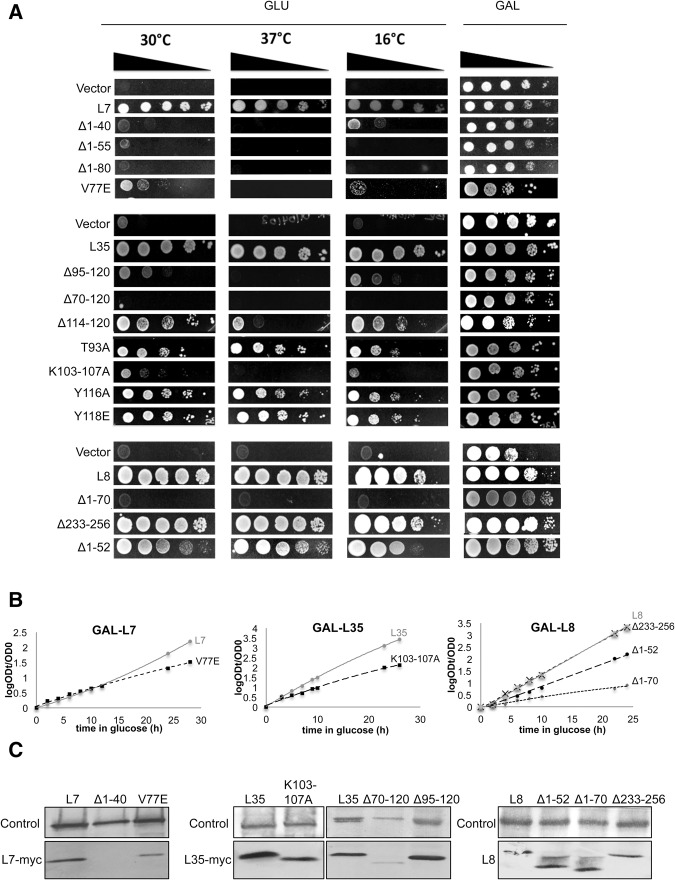
FIGURE 2.
(A) Growth on solid medium of yeast expressing wild-type or mutant forms of the r-proteins expressed from plasmids, at 30°C, 37°C, and 16°C. Empty vectors are included as negative controls for growth. (B) Growth of L7V77E, L35K103-107A and L8Δ1-52, L8Δ1-70 and L8Δ233-256 strains in liquid medium at 30°C, compared to strains expressing the corresponding wild-type proteins from the pRS315 plasmid. Cultures were diluted to ensure exponential growth. OD610 values are plotted as log (ODt/OD0), where ODt is the OD after t hours and OD0 is the starting OD. (C) Incorporation of mutant versions of the ribosomal proteins into pre-ribosomes was tested by TAP-purification of pre-ribosomes followed by Western blot assays. A MYC-tag was used for detection of r-proteins L7 and L35.
The extension of L7 is required for optimal processing of pre-rRNA during large ribosomal subunit assembly
In mature large ribosomal subunits, the conserved globular core of L7 binds domain II of 25S rRNA, as well as 5S rRNA. Similarly, the N-terminal extension of L7 interacts with ES7, the largest rRNA expansion segment in the 60S subunit, in domain II of 25S rRNA (Fig. 1A,B). This L7 extension contains a long α-helix (T22-G73) that contacts the globular domain of L7 through hydrophobic interactions, and a linker sequence (G73-Q80) that forms a shared β-sheet with the extension of r-protein L21 (Fig. 1C). Interestingly, the conserved valine77 within this linker sequence interacts with two other valine residues in r-proteins L20 and L21. In order to understand the role of the N-terminal extension of L7, we introduced three sequential, in-frame truncation mutations in the α-helix (L7Δ1-40, L7Δ1-55, L7Δ1-80), and one substitution mutation (V77E) of the conserved valine, which is predicted to disrupt its two-way hydrophobic interactions with L20 and L21 (Fig. 1C).
Each of the three truncations of the N-terminal extension impaired growth when wild-type L7 was depleted and only mutant L7 was expressed (Fig. 2A). However, these truncations resulted in destabilization of L7 (data not shown). Thus, as observed for depletion of L7, each truncation caused accumulation of 27SA3 pre-rRNA (Jakovljevic et al. 2012) (data not shown). This result was not surprising considering that this unusual extension is likely to affect folding of the globular region of L7. Moreover, the N-terminus of L7 was previously shown to contain monopartite and bipartite nuclear localization sequences, deletion of which could prevent nuclear import and assembly of L7, leading to its turnover (Chou et al. 2010; Tai et al. 2013). Since destabilization of L7 is the same as its depletion, these truncation mutations do not provide any novel information about potential roles of the L7 extension and were not studied further.
The L7V77E mutant impaired growth; however, unlike the truncated L7 protein, this mutant protein is stable (Fig. 2B, data not shown). Therefore, we focused on this mutant for further analysis. Western blotting of affinity-purified pre-ribosomes demonstrated that the L7V77E mutant protein assembles into pre-60S ribosomal particles (Fig. 2C), prompting us to investigate the role of its interactions with L20 and L21 in 60S subunit biogenesis. Interestingly, the L7V77E mutation had a different effect on pre-rRNA processing compared to depletion of L7. Depletion of L7 led to accumulation of 27SA2 and 27SA3 pre-rRNAs and decreased levels of 27SBS pre-rRNA (Jakovljevic et al. 2012). In contrast, the L7V77E mutation resulted in accumulation of the early 27SA2 and 27SA3 pre-rRNAs, but the ratio of 27SBL to 27SBS remained closer to wild type than for L7 depletion (Fig. 3A; Supplemental Fig. S5). We also observed a slight decrease in levels of the 7S pre-rRNA and 5.8S rRNA (Fig. 3B). Such changes in amounts of pre-rRNA processing intermediates suggest decreased rates of turnover of misassembled subunits relative to L7-depleted cells. We previously showed that as assembly of the 60S ribosomal subunit proceeds, the intermediates become progressively more stable. Blocking assembly at different successive steps leads to ever slower rates of pre-rRNA turnover (Gamalinda et al. 2014). Considering that the L7V77E mutation does not affect association of the globular part of L7 with pre-ribosomes, these particles are likely to be more stable compared to when L7 is completely absent. Moreover, it was previously shown that misfolding of rRNA is sufficient to perturb early steps of biogenesis, while it is the lack of A3 assembly factors that results in turnover of early pre-ribosomes (Talkish et al. 2014). Thus, we checked the levels in pre-ribosomes of Nop7, one of the six interdependent A3 factors, and Has1, a DEAD box protein whose association depends on the A3 factors, and did not observe a significant decrease (Fig. 4A). This result is consistent with decreased pre-ribosome turnover rates in the L7V77E mutant compared to depletion of L7. The slight decrease in the levels of assembly factors that are involved in later stages (Nsa2 and Bud20) is likely to reflect the mild defect in earlier steps.
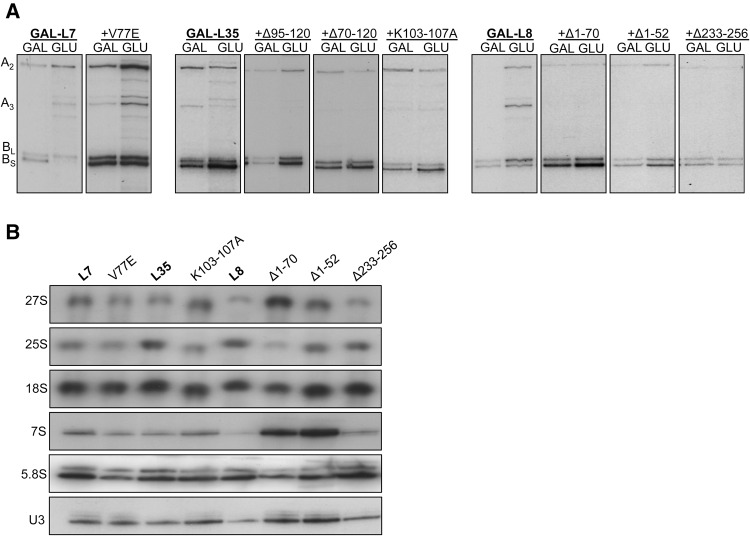
FIGURE 3.
(A) Pre-RNA processing defects in r-protein extension mutants. The 27SA2, 27SA3, 27SBS, 27SBL pre-rRNAs were detected by primer extension and used as representatives of different steps of pre-rRNA processing. (B) The steady-state levels of pre-rRNAs (27S, 7S) and mature rRNAs (25S, 18S, and 5.8S) were tested by Northern blots. U3 snoRNA was used as the loading control.
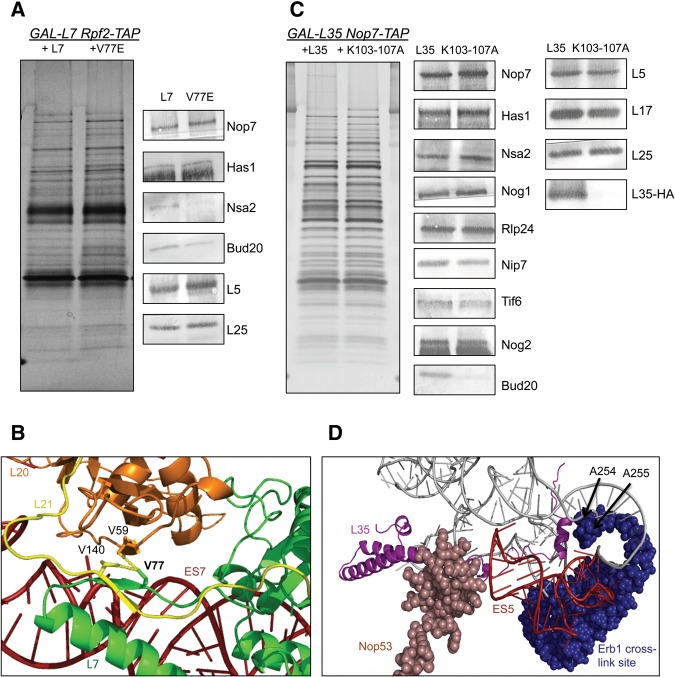
FIGURE 4.
(A) TAP-tagged assembly factor Rpf2 was used as the bait to assay the protein composition of pre-ribosomes in the L7V77E mutant strain. Protein levels were initially tested by SDS-PAGE, followed by silver staining (left panel). Antibodies against the indicated proteins were used to test their abundance by Western blotting. (B) Zoomed-in representation of the hydrophobic interactions mediated by the V77 residue of L7 (green), the V140 residue of L21 (yellow), and the V59 residue of L20 (orange). PyMol generated from the cryo-EM structure of Nog2 containing pre-ribosomes (PDB accession number 3JCT). (C) TAP-tagged assembly factor Nop7 was used as the bait to assay the protein composition of pre-ribosomes in the L35K103-107A mutant strain. Protein levels were initially tested by SDS-PAGE, followed by silver staining (left panel). Antibodies against the indicated proteins were used to test their abundance by Western blotting. (D) Zoomed-in representation of the C-terminal extension of L35 (purple). Assembly factor Nop53 was colored light pink while the cross-linking site of assembly factor Erb1 is shown in dark blue. ES5 is indicated in red. PyMol generated from the cryo-EM structure of Nog2 containing pre-ribosomes (PDB accession number 3JCT).
Since V77 interacts both with r-proteins L21 and L20 (Fig. 4B), and L21 is known to stabilize later stages of ribosome assembly (Ohmayer et al. 2013), we tested whether the defect in L7V77E reflects a combination of early and late defects in assembly. We disturbed the β-sheet of L21 by two substitution mutations and confirmed that the interaction of L7 and L21 is dispensable for viability in yeast (Supplemental Fig. S6). Alternatively, these substitution mutations may not have completely abolished the interaction between L7 and L21. However, a cluster of substitution mutations in L20 result in the same pre-rRNA processing defect and modest rates of pre-rRNA turnover as observed for L7V77E (S Biedka, pers. comm.). Our findings suggest that the V77 residue of L7 contributes to the efficiency of 60S ribosomal assembly, and that weakening its hydrophobic interactions with L20 alters the local structure of domain II, slowing down the assembly pathway (Fig. 4B).
An α-helix in the C-terminal extension of L35 that interacts with 25S rRNA is required for processing of 27SB pre-rRNA
The globular domain of L35 predominantly binds 5.8S rRNA while its C-terminal extension interacts with several conserved regions in domain I surrounding the polypeptide exit tunnel (PET), as well as expansion segment ES5 (Fig. 1A,B). The globular domain of L35 also contacts the assembly factor Nop53, which is required for cleavage of ITS2, while the L35 extension contacts rRNA where it cross-links to assembly factor Erb1 (Granneman et al. 2011; Thoms et al. 2015; Wu et al. 2016). Thus, L35 is positioned in a functionally important site, perhaps to promote long-range communication among 5.8S rRNA, the PET, and ITS2. Consistent with this location, depletion of L35 prevents cleavage of ITS2, resulting in accumulation of 27SB pre-rRNA (Babiano and de la Cruz 2010; Gamalinda et al. 2013). The C-terminal extension of L35 is rich in arginine and lysine residues and exhibits a high level of sequence conservation among eukaryotes, suggesting that its function may require sequence specificity (Supplemental Fig. S3).
In order to investigate whether the L35 extension has a role in ribosome assembly, we constructed three serial truncation mutations (L35Δ114-120, L35Δ95-120, L35Δ70-120), along with four substitution mutations in residues predicted to bind rRNA (T93A, K103-107A) or r-protein L13 (Y116A, I118E). The L35Δ114-120 mutation had a mild growth defect at 37°C, L35Δ95-120 was slow growing, and L35Δ70-120, in which the entire extension was removed, was lethal (Fig. 2A). The K103-107A mutation severely affected viability at all three temperatures while the T93A, Y116A, and I118E mutations had a very small or no effect on cell growth (Fig. 2A,B). None of these seven different rpl35− mutations affected the stability of L35 protein (data not shown). All of the mutant L35 proteins, except for L35Δ70-120, assembled into pre-ribosomes at wild-type levels (Fig. 2C).
Analysis of pre-rRNA processing confirmed that all three rpl35− mutants that had significant growth impairment (L35Δ70-120, L35Δ95-120, and L35K103-107A) are defective in subunit biogenesis. As is the case for other proteins located in this neighborhood, depletion of L35 results in accumulation of 27SBS pre-rRNA, accompanied by turnover of pre-60S particles before the next step in assembly can occur (Babiano and de la Cruz 2010; Gamalinda et al. 2013, 2014). The three slow growing rpl35− mutants had the same initial pre-rRNA processing defect as the depletion of L35, namely accumulation of 27SB pre-rRNA (Fig. 3A,B). Since levels of assembly factors Nsa2 and Nog2 decrease significantly when L35 is depleted, we checked the levels of these factors in the L35K103-107A mutant. We observed a slight decrease in the levels of Nog2 and a significant decrease in Bud20, while the amounts of Nsa2 did not change, suggesting an increase in the stability of preribosomes compared to the L35 depletion (Fig. 4C). Because the short α-helix formed by residues K103-K107 interacts with both ES5 and nucleotides A254 and A255 of 25S rRNA, we suggest that it has an important role in stabilizing tertiary interactions between ES5 and nucleotides A254 and A255 (Fig. 4D). These nucleotides are part of the rRNA binding site of assembly factor Erb1 (Granneman et al. 2011; Thoms et al. 2015). Thus, it is tempting to speculate that this stabilization mediated by the L35 extension may be involved in Erb1 removal, which is coupled with cleavage of ITS2 (Thomson and Tollervey 2005; Bassler et al. 2010).
The N-terminal extension of L8 is involved in late steps of 60S subunit biogenesis
The globular domain of L8 contacts mostly domain I of 25S rRNA, which is stabilized early in the assembly pathway, while the N-terminal extension interacts predominantly with domain V, which is stabilized at late stages (Fig. 1B; Gamalinda et al. 2014). The α-helix in this extension (V56-I66) bridges domain V and the proximal stem formed by base-pairing of the 3′ end of 5.8S rRNA with the 5′ end of 25S rRNA (Fig. 1B). Thus, L8 provides a model to understand multiple roles of r-proteins during sequential stabilization of different rRNA domains in vivo. We constructed two truncation mutations of the N terminus of L8 (L8Δ1-52, L8Δ1-70), and one of the C terminus (L8Δ233-256) (Fig. 1C). We observed a slight growth defect with the L8Δ1-52 mutant, while the L8Δ233-256 mutant grew at wild-type rates. In contrast, the L8Δ1-70 truncation mutant was lethal, and exhibited a dominant phenotype at 37°C when the wild-type protein was also present (Fig. 2A,B, data not shown). None of the L8 truncation mutations affected the stability or assembly of L8, enabling us to further investigate the roles of this essential N-terminal extension in subunit assembly (Fig. 2C).
Interestingly, the L8Δ1-70 mutant was blocked at later steps of assembly than when L8 was depleted. In this mutant, there was a modest increase in amounts of 27SB pre-rRNA but a significant increase in 7S pre-rRNA relative to wild-type levels (Fig. 3A,B). In contrast, depletion of L8 resulted in accumulation of 27SA3 pre-rRNA and decreased amounts of 27SB and 7S pre-rRNAs (Jakovljevic et al. 2012). Processing of 27S pre-rRNAs occurs in the nucleolus, whereas 7S pre-rRNA processing takes place later, in the nucleoplasm, just prior to nuclear export of pre-60S particles. Thus, the globular domain and N-terminal extension of L8 function in different stages of 60S subunit assembly that occur in different nuclear subcompartments.
Absence of the N-terminal extension of L8 blocks assembly following association of Nog2 and before the Rix1/Rea1-dependent remodeling step
To investigate, in more detail, the role of the N-terminal extension of L8 in late stages of 60S subunit assembly, we first focused on a broad range of assembly intermediates by affinity-purifying pre-rRNPs using TAP-tagged Rpf2. This assembly factor joins pre-ribosomes very early in the pathway and exits during late nuclear stages, and thus is present in several consecutive assembly intermediates (Zhang et al. 2007). Very few changes in the SDS-PAGE profile of proteins in Rpf2-containing pre-ribosomes were evident in the L8Δ1-70 mutant. Western blotting revealed, for example, that levels of assembly factors Nog2 and Has1 as well as r-protein L17 were unchanged (Supplemental Fig. S7). In contrast, when L8 is depleted, amounts of nine early-acting assembly factors, as well as the later-acting assembly factors Nsa2, Nog2, Nug1, Nop53, and Arx1 decrease. Additionally, r-proteins L13, L15, and L36, which lie adjacent to L8 in rRNA domain I, are greatly diminished, as are r-proteins that become more stably associated at later stages in assembly, e.g., L17 (Jakovljevic et al. 2012).
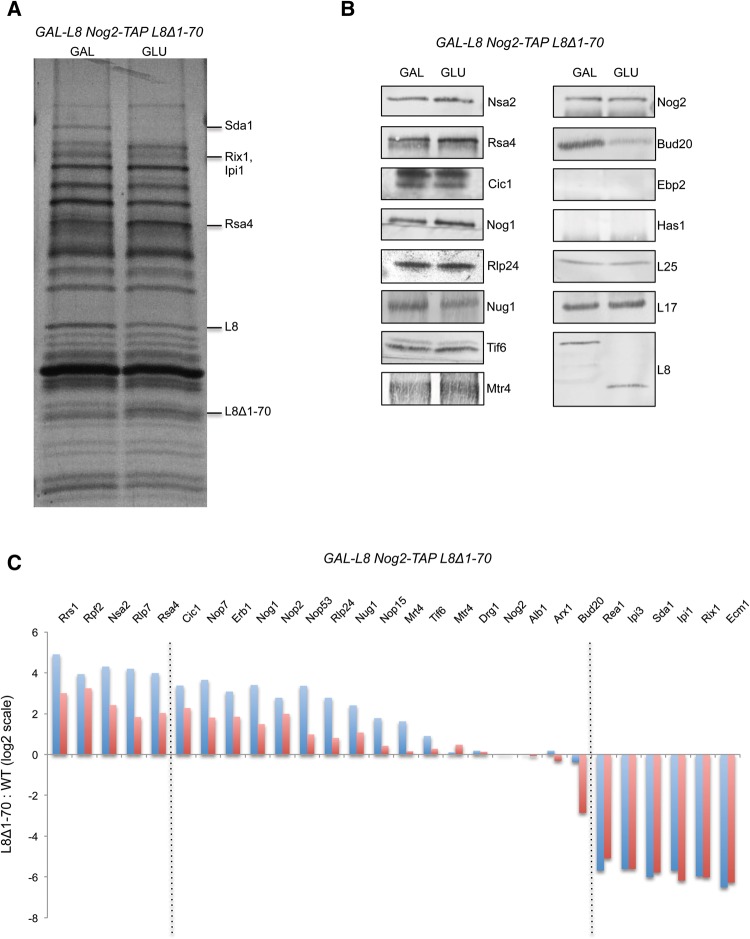
Because the L8Δ1-70 mutation blocks late nuclear steps of pre-rRNA processing and does not affect association of early-acting assembly factors and r-proteins, we specifically examined whether the composition of late nuclear assembly intermediates was altered in the L8Δ1-70 mutant. We used TAP-tagged Nog2 to affinity-purify these pre-rRNPs, since its association with pre-ribosomes was not affected in the mutant. Nog2 enters pre-ribosomes in the nucleolus, after 27SB pre-rRNA is formed, and exits from pre-60S particles just before their export from the nucleoplasm to the cytoplasm (Saveanu et al. 2001). Using Nog2 as the bait protein has two more advantages compared to Rpf2: (i) Nog2 particles are less complex and more homogeneous than Rpf2 particles. (ii) The cryo-EM structure of Nog2 containing pre-ribosomes was recently determined at near atomic resolution (Wu et al. 2016). SDS-PAGE, Western blotting, and iTRAQ mass spectrometry demonstrated that Nog2-containing pre-ribosomes in the L8Δ1-70 mutant have significantly decreased amounts of the assembly factor Sda1, the remodeling complex Rix1-Ipi1-Ipi3 and Rea1, and nuclear export factors Bud20 and Ecm1 (Fig. 5A–C). In contrast, amounts of assembly factors Rpf2, Rrs1, and Rsa4 were increased in this mutant. Levels of the other late-acting factors Nug1, Nop53, and Mtr4 were not significantly altered. Taken together, these results indicate that the N-terminal extension of L8 is involved in the recruitment, function, and release of assembly factors previously implicated in late nuclear steps of 60S ribosomal subunit maturation, including pre-rRNP remodeling, 7S pre-rRNA processing, and nuclear export.
FIGURE 5.
(A) TAP-tagged assembly factor Nog2 was used as the bait to purify late nuclear pre-60S particles. Protein composition was assayed by SDS-PAGE, followed by silver staining, when wild-type L8 or the L8Δ1-70 mutant protein was expressed as the sole version of L8. (B) The levels of indicated assembly factors and r-proteins were tested by Western blots. As expected, Ebp2 and Has1 were not detected in Nog2-associated particles; these assembly factors mostly dissociate before Nog2 assembles into pre-ribosomes. (C) Changes in levels of large ribosomal subunit assembly factors in the L8Δ1-70 mutant compared to wild-type L8 were determined by semiquantitative iTRAQ analysis. Relative levels of each protein were normalized to the ratio of the Nog2 bait protein, and results were reported on a log2 scale. The blue and red bars correspond to the ratio of each assembly factor in mutant versus wild-type (mutant: WT) for duplicate experiments. The dashed lines indicate arbitrarily chosen cutoffs where the average ratio of duplicated experiments increases to greater than three, or decreases to less than five.
Modifications at structurally important sites of ITS2 interfere with pre-rRNA processing in the L8 mutant
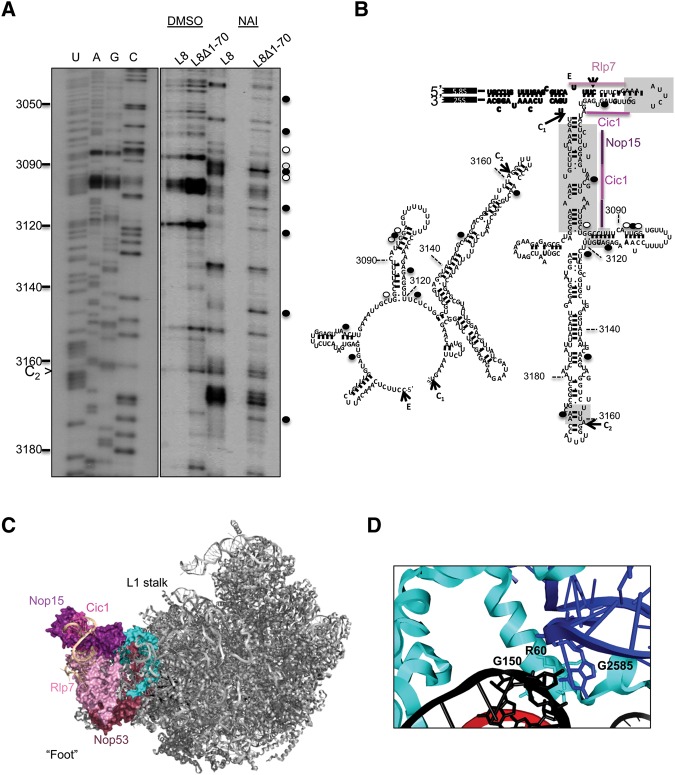
Previous studies suggested that certain evolutionarily conserved structural elements of ITS2 are important for cleavage at the C2 site and for removal of this spacer sequence (van Nues et al. 1995; Coleman 2003, 2015). ITS2 is thought to undergo a conformational change from a “ring structure” to a “hairpin structure” as assembly progresses, possibly during or after dissociation of assembly factors Cic1, Rlp7, and Nop15 (Granneman et al. 2011). Even though it is still unclear how this conformational change enables processing of ITS2 from 7S pre-rRNA, some studies indicate that both conformations of this spacer sequence are important for progression of assembly (Côté et al. 2002; Sahasranaman et al. 2011). The absence of the N-terminal extension of L8 slows down cleavage at the C2 site and blocks subsequent removal of ITS2, which involves formation of the proximal stem. Thus, we tested whether the absence of the L8 extension results in a conformational change at the proximal stem and structurally important sites of ITS2, using in vivo RNA structure probing with NAI followed by primer extension (Fig. 6A,B). The DMSO lanes were used as controls to determine the natural stops that occur due to the flexibility of the rRNA backbone. Some nucleotides at structurally important sites were modified, indicating that the N-terminal extension of L8 is involved in forming the ITS2 structure (Fig. 6A,B). The cryo-EM structure of Nog2-TAP particles contains ∼59 nucleotides (nt) of ITS2, which includes parts of the binding sites of the assembly factors Rlp7, Cic1, and Nop15 (Fig. 6C). Interestingly, we observe increased modification of 2 nt in the Cic1 interaction site, which may indicate a change in the binding of Cic1. Moreover, we observe changes in reactivity at a common subset of nucleotides that would be consistent with a shift from ring structure to hairpin structure of ITS2 (Granneman et al. 2011; Babiano et al. 2013; Dembowski et al. 2013). Thus, in the absence of the N-terminal extension of L8, the binding of some assembly factors that interact with ITS2 are distorted, which may cause ITS2 to form a structure that inhibits its processing after C2 cleavage.
FIGURE 6.
(A) In vivo structure probing of rRNA, using NAI followed by primer extensions, was conducted for L8 wild-type and L8Δ1-70 mutant yeast. Nucleotides with increased modification in the mutant are marked with solid black circles, while hollow circles indicate the nucleotides with decreased modification. (B) Changes in the nucleotide reactivity are shown in the ring conformation and the hairpin conformation of ITS2. The gray boxes correspond to structures necessary for progression of 60S subunit assembly (Côté et al. 2002). The assembly factors that interact with the first 59 nt of ITS2 in the cryo-EM structure of Nog2-associated particles were indicated on the hairpin conformation (PDB accession number 3JCT). (C) The position of L8 (cyan) in Nog2-associated pre-60S ribosomal subunits is shown from the subunit interface. The proteins that bind the “foot” structure are colored different shades of pink. ITS2 is colored beige. (D) The R60 residue in the N-terminal extension of L8 interacts with both 5.8S rRNA in the proximal stem (nucleotide G150) and domain V (nucleotide G2585) of 25S rRNA.
DISCUSSION
Examining the effects of depleting r-proteins set the ground rules to understand the hierarchy of the intertwined processes that occur during ribosome assembly (Ferreira-Cerca et al. 2005; O'Donohue et al. 2010; Gamalinda et al. 2014). However, the design of these experiments limits our understanding about potential multiple roles of r-proteins in ribosome biogenesis. When an r-protein is depleted, only the earliest defect in assembly can be detected. Yet, it has been suggested that r-proteins establish several different stepwise interactions with rRNA during the assembly pathway, and thus may be involved in multiple stages of pre-RNP remodeling and pre-rRNA processing (Adilakshmi et al. 2008; Kim et al. 2014; Napper and Culver 2015). According to this model, the initial binding of an r-protein to rRNA forms an “encounter complex,” which is then converted to more stable complexes by establishing greater numbers of contacts with rRNA. This multistage binding mechanism seems especially likely for assembly of r-proteins containing extended portions that contact different rRNA domains than those bound by the globular body of the protein. Thus, eukaryote-specific extensions of r-proteins that contain disordered domains are good starting points to dissect multiple roles of r-proteins, as their structural flexibility may be important to coordinate dynamic remodeling of pre-ribosomes during assembly. The elongated structure of these extensions facilitates their interaction with multiple partners, possibly forming a coordination network (Gunasekaran et al. 2003; Dyson and Wright 2005; Cumberworth et al. 2013).
By specifically examining the effects of mutating r-protein extensions, this article reveals roles of extensions of three strategically positioned r-proteins during ribosome assembly. We found that the N-terminal extensions of L7 (80 amino acids long) and of L8 (70 amino acids long), and the C-terminal extension of L35 (50 amino acids long), are essential for optimal growth in yeast. On the other hand, the C-terminal extension of L8 (23 amino acids long) is not essential.
Substitution mutations in the N-terminal extension of L7 and the C-terminal extension of L35 inhibited the same pre-rRNA processing steps as depletions of each protein. Both of these extensions interact with the same rRNA domains as their globular regions. Thus, our results further confirm that pre-rRNA processing and hierarchical stabilization of rRNA domains in vivo during ribosome assembly is directly dependent on the timely establishment of rRNA–protein interactions. Even though the protein or rRNA contacts made by the extensions of L7 and L35 are required for optimal growth and efficient pre-rRNA processing, the remaining portions of each of these proteins can stabilize the pre-ribosomes enough to prevent their rapid turnover. Truncations of L7 and L35 extensions either destabilized the entire protein or decreased their incorporation levels to pre-ribosomes. These observations strongly suggest that dedicated assembly chaperones may exist to facilitate the incorporation of L7 and L35 to pre-ribosomes.
Depletion of L8 results in an early block of 60S subunit maturation through destabilization of nucleolar assembly intermediates containing 27SA3 pre-rRNA (Jakovljevic et al. 2012). This early defect most likely reflects the loss of binding of the globular core of L8, which contacts domain I of 25S rRNA that undergoes structural stabilization early in the 60S subunit assembly. However, more than half of the rRNA contacts of L8 occur through its N-terminal extension binding to rRNA domain V, which is stabilized at late stages. This could explain why removal of this 70 amino acid long extension of L8 causes defects in assembly of late nuclear intermediates. We note, however, that deletion of the 52 N-terminal amino acids of L8 caused only a modest growth defect. Taken together, these results suggest a critical role in late stages of 60S ribosomal subunit assembly for the conserved α-helix between W52 and K70 in the N-terminal extension of L8. Interestingly, residues V56-R60 of this α-helix interact with the 3′end of 5.8S rRNA (G150), as well as rRNA domain V of 25S rRNA (G2585) (Fig. 6C,D).
The question arises as to how deleting the extension of L8 might prevent late stages of assembly. This stage includes two major remodeling events: removal of the 235 nt-long ITS2, and reorganization of the central protuberance containing 5S rRNA, and r-proteins L5 and L11 (Bradatsch et al. 2012; Greber et al. 2012; Barrio-Garcia et al. 2016). Cleavage of the C2 site in ITS2 occurs after assembly factors Nog2, Nop53, and Rsa4 bind to pre-ribosomes, and is mediated by the Las1 endonuclease (Tang et al. 2008; Kressler et al. 2012; Schillewaert et al. 2012; Talkish et al. 2012; Gasse et al. 2015). This study shows that Nog2, Nop53, and Rsa4 are present in pre-ribosomes when the N-terminal extension of L8 is deleted; however, we could not determine whether Las1 binding is affected. The assembly events that trigger recruitment of Las1 to the C2 site are currently unclear. Yet, previous studies showed that cleavage of the C2 site in ITS2 requires the proper formation of the proximal stem by base-pairing of the 3′end of 5.8S rRNA with the 5′end of 25S rRNA, which are separated in pre-rRNA by ITS2 (van Nues et al. 1995; Côté and Peculis 2001; Côté et al. 2002). Formation of this structure may require the presence of assembly factors Cic1, Nop15, and Rlp7 that are bound to it (Granneman et al. 2011). In the L8 mutant, all three of these assembly factors are present. However, our RNA structure-probing experiments revealed that the structure of ITS2 is altered, especially at sites that are known to be important for the processing of ITS2 (van Nues et al. 1995).
Because much more 7S pre-rRNA accumulates than 27SB pre-rRNA in the L8 extension mutant, it seems likely that a proximal role of the L8 extension is in the nucleolytic removal of ITS2 sequences from the 7S pre-rRNA, rather than C2 cleavage. Presumably, Nop15, Cic1, and Rlp7 dissociate from the 7S pre-rRNA before or during its processing. Following cleavage at the C2 site in ITS2, a complex of exonucleases and the rRNA helicase Mtr4 are recruited to pre-ribosomes to mediate processing of 7S pre-rRNA (Thoms et al. 2015). Recruitment of Mtr4 to ITS2 and 5.8S rRNA (H8, H9, and H10) is mediated by assembly factor Nop53. However, both iTRAQ and Western blotting suggest that the absence of the N-terminal extension of L8 does not block recruitment of either Nop53 or Mtr4 to pre-ribosomes. Yet, structure-probing shows some alterations in the structure in the CRAC-based binding sites of Mtr4, suggesting that in the L8 mutant, the processing machinery may be recruited to pre-ribosomes but not properly positioned to carry out its function. Thus, interaction of the N-terminal extension of L8 with the 3′ end of 5.8S rRNA and the RNA binding sites of Mtr4 may help position the proximal stem and coordinate efficient processing of ITS2 after C2 cleavage takes place.
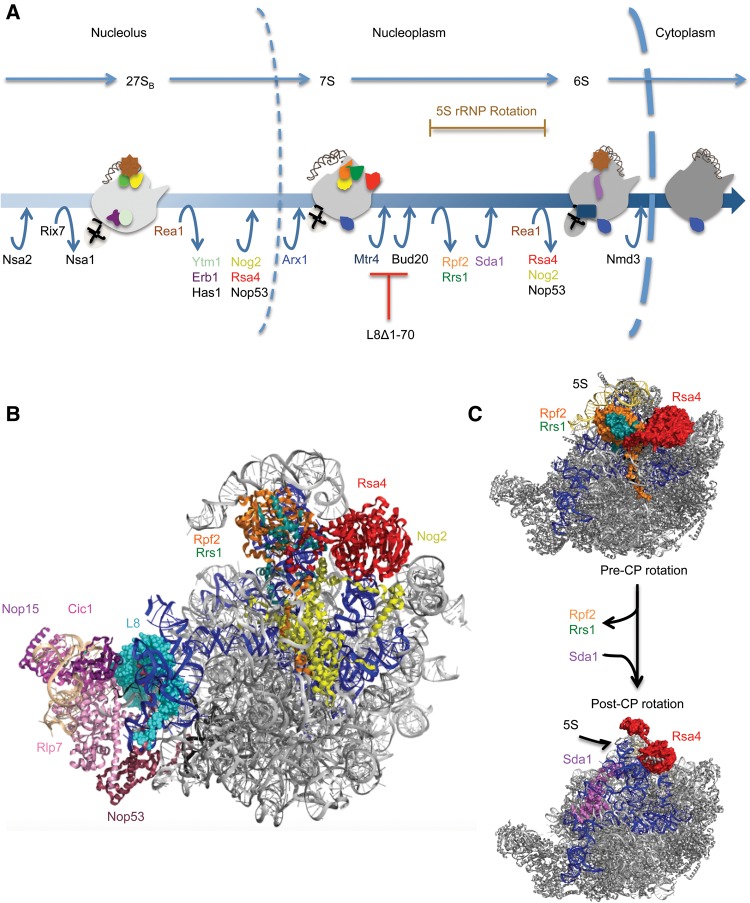
A second major remodeling event in late stages of assembly is the reorganization of the central protuberance and the peptidyl transferase center. This includes a 180° rotation of 5S rRNA bound to L5 and L11, and large scale movement of the A-site finger (H38) and H89 of 25S rRNA (Bradatsch et al. 2012; Leidig et al. 2014). The cryo-EM structures of pre–ribosomes purified using Nog2, Arx1, or/and Rix1 suggest that this rotation is triggered by the removal of Rpf2 and Rrs1 and by recruitment of Sda1, Rea1, and the Rix1-subcomplex (Rix1, Ipi1, Ipi3) (Bradatsch et al. 2012; Greber et al. 2012; Barrio-Garcia et al. 2016; Wu et al. 2016). Subsequently, the Rea1 ATPase catalyzes removal of Rsa4 and Nog2 (Fig. 7A; Bassler et al. 2010; Kressler et al. 2012; Baßler et al. 2014). When the N-terminal extension of L8 was truncated, we observed decreased amounts of Sda1, the Rix1-subcomplex, and Rea1 in Nog2-containing pre-ribosomes. Furthermore, there were increased amounts of Rpf2, Rrs1, and Rsa4 in these mutant pre-ribosomes. These results indicate that the block in ribosome assembly in this rpl8− mutant occurs after Nog2, Rsa4, and Nop53 have associated. Thus, a second proximal effect of deleting the N-terminal extension of L8 may be on the release of Rpf2 and Rrs1, and/or recruitment of Sda1, the Rix1-subcomplex and Rea1 (Fig. 7A–C). Consistent with the 7S pre-rRNA accumulation in the rpl8− mutant, it was previously shown that the depletions of Sda1 or the Rix1-subcomplex also result in a block of 7S pre-rRNA processing (Supplemental Fig. S8; Dez et al. 2006; Barrio-Garcia et al. 2016). We also observed decreased levels of export factors Bud20 and Ecm1 (Yao et al. 2010; Bassler et al. 2012). Both of these nonessential proteins interact with FG-rich nucleoporins, and their absence seems unlikely to cause a block of 60S assembly prior to nuclear export.
FIGURE 7.
(A) A working model for late nuclear steps of yeast 60S ribosomal subunit assembly. Entry and exit points for assembly factors in wild-type cells are indicated. In the L8Δ1-70 strain, remodeling events downstream from the association of Mtr4 are blocked, Rpf2 and Rrs1 cannot dissociate from pre-ribosomes, and Sda1, Rea1, and Ipi1-Rix1-Ipi3 cannot associate with assembling ribosomes. (B) The high-resolution cryo-EM structure of Nog2-associated particles shown from the subunit-interface (PDB accession number 3JCT). Domain V is colored dark blue while 5.8S rRNA and 5S rRNA are colored black and dark gray, respectively. (C) The pre- and post-rotated structures of the central protuberance (5S rRNA, L11, and L5) and assembly factor Rsa4 are modeled on the cryo-EM structure of Nog2-associated pre-ribosomes. In the post-rotated state of the central protuberance, assembly factors Rpf2 and Rrs1, which are present in the pre-rotated state, are no longer present and assembly factor Sda1 associates with pre-ribosomes. Note that the binding site of Sda1 overlaps with that of Rpf2.
Interestingly, Rpf2, Rrs1, and Rsa4 form a network that drives formation of the final structure of H89 in rRNA domain V (Kharde et al. 2015; Madru et al. 2015, Wu et al. 2016). Since all of these assembly factors interact with rRNA domain V in the assembling 60S subunit, our results further confirm the role of the N-terminal extension of L8 in structuring domain V (Fig. 7B,C).
Truncation of the L8 N-terminal extension is one of the first examples of a mutation that blocks ITS2 processing even though the association of late nuclear factors Nsa2 and Nog2 is not perturbed. Thus, this mutant provides a novel gateway to investigate networks that enable remodeling after C2 cleavage during late nuclear steps of 60S subunit assembly. Our findings lead to a “communication” paradigm in which 25S rRNA domain I is stabilized early in the assembly of 60S ribosomal subunits, by binding to the globular part of L8, while subsequent processing of ITS2 requires the presence of the N-terminal extension of L8. This extension may help ITS2 adapt its proper conformation for recruitment of the exosome, and connect the release of Rpf2 and Rrs1 with processing of 7S pre-rRNA and remodeling of domain V via incorporation of late assembly factors. The causative relationship between C2 cleavage, 7S pre-rRNA processing and release of Rpf2 and Rrs1 remains unclear. However, we speculate that Sda1 may be one of the key players due to its remarkable positioning between ITS2 and the Rpf2-Rrs1 binding domains (Fig. 7C).
All of the phenotypes that we observed upon mutating r-protein extensions are in accordance with our initial hypothesis that these eukaryotic extensions are involved in the stabilization of rRNA domains with which they interact (see Introduction). This article provides an example of how rearrangements of local structures can be propagated through different rRNA domains. Interdomain r-proteins that form long-range rRNA interactions can stabilize multiple different rRNA domains with which they interact. Also, this article further demonstrates the significance of studying such regions within ribosomal proteins and contributes to a more detailed understanding of the hierarchical assembly of the large ribosomal subunit, especially at late nuclear stages of assembly.
MATERIALS AND METHODS
Construction of mutant r-protein genes
Plasmids containing r-protein genes obtained from the Yeast Genomic Tiling Collection (Open Biosystems) were used as templates for a two-step PCR protocol, to incorporate unique restriction sites upstream and downstream from the corresponding open reading frames. The following restriction enzyme sites were inserted ∼800 nt upstream and ∼50 nt downstream from the respective genes; BamHI and HindIII for RLP8 and RPL25; BamHI and SalI for RPL35; and SacII and StuI for RPL7. PCR products digested with restriction enzymes were ligated into plasmid pRS315 containing the LEU2 gene. Deletion and substitution mutations, and addition of a MYC-tag at the N or C terminus of each protein, were constructed using the QuickChange Site-directed Mutagenesis Kit (Stratagene). Proper insertion of the genes and construction of mutations were confirmed by sequencing (Genewiz).
Generation of yeast strains
Yeast strains conditional for expression of RPL7, RPL8, or RPL35 were previously constructed by replacing their endogenous promoters with the GAL1 promoter (Jakovljevic et al. 2012; Ohmayer et al. 2013; Gamalinda et al. 2014). The pRS315 vector or plasmids containing the respective r-protein gene mutations were transformed into these yeast strains conditional for expression of the corresponding r-proteins.
Growth assays
Yeast strains were grown in galactose-containing medium to OD600 of 0.5. Cells were serially diluted (from 10 to 10−5), and ten microliters of each dilution were spotted on galactose- and glucose-containing medium.
Steady-state analysis of mature rRNAs and pre-rRNA intermediates
Analysis of mature rRNAs and pre-rRNA processing intermediates was carried out by agarose gel electrophoresis and Northern blotting or primer extensions, as previously described (Horsey et al. 2004). Probes for Northern blots are available upon request. An oligonucleotide complementary to ITS2 was used for primer extension reactions.
In vivo SHAPE analysis of rRNA structure
For in vivo structure probing using 2-methylnicotinic acid imidazolide (NAI), 200 mL of cells were grown to OD600 of 0.6. After washing with PBS, NAI was added to the cells at a final concentration of 100 mM and incubated for 20 min at 30°C. RNA was extracted and primer extension was conducted with transcriptor reverse transcriptase (Roche) as previously described (Talkish et al. 2014). An oligonucleotide complementary to ITS2 (5′ TATTCTTCTCTCGCAGATCCG 3′) was used for primer extensions to assay modifications by NAI.
Affinity purification of pre-ribosomes followed by SDS-PAGE, silver staining, Western blotting, and iTRAQ analysis
Pre-ribosomes were purified by tandem affinity purification (TAP) with magnetic Dynabeads (Invitrogen) as explained previously (Sahasranaman et al. 2011). TAP-tagged Nop7, Rpf2, or Nog2 were used as the bait to isolate pre-ribosomes from wild-type cells or rpl− mutants.
The protein composition of the TAP-purified pre-ribosomes was determined by SDS-PAGE (4%–10% Tris–glycine and 4%–12% Bis–Tris, Invitrogen) followed by silver staining (Sahasranaman et al. 2011). Incorporation of mutated proteins into pre-ribosomes was tested by Western blotting.
For iTRAQ analysis, cell lysates from 1 L cultures of wild-type and rpl8Δ1-70 yeast containing TAP-tagged Nog2 were used to purify pre-ribosomes. Purified samples were sent to the Penn State Hershey Core Research Facilities for trypsin digestion and 4-plex labeling with iTRAQ reagents 114, 115, 116, 117 (Applied Biosystems). Peptides were separated by two-dimensional liquid chromatography and parent ions were identified on a Sciex/ABI 5800 matrix-assisted laser desorption ionization-tandem time of flight mass spectrometer. iTRAQ ratios as an average of all peptides for each protein were obtained using the Protein Pilot 4.0 program. Proteins identified with >99.9% confidence were used for further data analysis. For each pair-wise comparison, data were normalized to the change in ratio of the TAP-tagged protein (loading control), and fold change for each protein was reported in log2 scale. Each pair-wise comparison was done in two biological replicates. The order of changes is reported based on the average from duplicate experiments.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Woolford laboratory for critical discussions and reading of the manuscript. We also thank the following people for their generous gifts of antibodies: Jesus de la Cruz and Patrick Linder (Has1, Rsa4), Janine Maddock (Nog1), Michael McAlear (Ebp2), Cosmin Saveanu and Micheline Fromont-Racine (Rlp24, Nsa2, Tif6, and Nog2), Arlen Johnson (rpL8), Sabine Rospert (rpL17), K. Siegers (rpL25), Elizabeth Tosta (Cic1), Vikram Panse (Bud20, Nug1), and David Tollervey (Mtr4). This work was supported by National Institutes of Health grant R01 GM028301 to J.L.W.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.055798.115.
REFERENCES
- Adilakshmi T, Bellur DL, Woodson SA. 2008. Concurrent nucleation of 16S folding and induced fit in 30S subunit assembly. Nature 455: 1268–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiano R, de la Cruz J. 2010. Ribosomal protein L35 is required for 27SB pre-rRNA processing in Saccharomyces cerevisiae. Nucleic Acids Res 38: 5177–5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiano R, Badis G, Saveanu C, Namane A, Doyen A, Diaz-Quintana A, Jacquier A, Fromont-Racine M, de la Cruz J. 2013. Yeast ribosomal protein L7 and its homologue Rlp7 are simultaneously present at distinct sites on pre-60S ribosomal particles. Nucleic Acids Res 41: 9461–9470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban N, Beckmann R, Cate JH, Dinman JD, Dragon F, Ellis SR, Lafontaine DL, Lindahl L, Liljas A, Lipton JM, et al. 2015. A new system for naming ribosomal proteins. Curr Opin Struct Biol 24: 165–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrio-Garcia C, Thoms M, Flemming D, Kater L, Berninghausen O, Baßler J, Beckmann R, Hurt E. 2016. Architecture of the Rix1–Rea1 checkpoint machinery during pre-60S-ribosome remodeling. Nat Struct Mol Biol 23: 37–44. [DOI] [PubMed] [Google Scholar]
- Bassler J, Kallas M, Pertschy B, Ulbrich C, Thoms M, Hurt E, Baßler J. 2010. The AAA-ATPase Rea1 drives removal of biogenesis factors during multiple stages of 60S ribosome assembly. Mol Cell 38: 712–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassler J, Klein I, Schmidt C, Kallas M, Thomson E, Wagner MA, Bradatsch B, Rechberger G, Strohmaier H, Hurt E, et al. 2012. The conserved Bud20 zinc finger protein is a new component of the ribosomal 60S subunit export machinery. Mol Cell Biol 32: 4898–4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baßler J, Paternoga H, Holdermann I, Thoms M, Granneman S, Barrio-Garcia C, Nyarko A, Clark S, Schraivogel D, Kallas M, et al. 2014. A network of assembly factors is involved in remodeling rRNA elements during preribosome maturation. J Cell Biol 207: 481–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M. 2011. The structure of the eukaryotic ribosome at 3.0 Å resolution. Science 334: 1524–1529. [DOI] [PubMed] [Google Scholar]
- Bradatsch B, Leidig C, Granneman S, Gnädig M, Tollervey D, Böttcher B, Beckmann R, Hurt E. 2012. Structure of the pre-60S ribosomal subunit with nuclear export factor Arx1 bound at the exit tunnel. Nat Struct Mol Biol 19: 1234–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calidas D, Lyon H, Culver GM. 2014. The N-terminal extension of S12 influences small ribosomal subunit assembly in Escherichia coli. RNA 20: 321–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou GW, Tai LR, Kirby R, Lee IF, Lin A. 2010. Importin β3 mediates the nuclear import of human ribosomal protein L7 through its interaction with the multifaceted basic clusters of L7. FEBS 584: 4151–4156. [DOI] [PubMed] [Google Scholar]
- Coleman AW. 2003. ITS2 is a double-edged tool for eukaryote evolutionary comparisons. Trends Genet 19: 370–375. [DOI] [PubMed] [Google Scholar]
- Coleman AW. 2015. Nuclear rRNA transcript processing versus internal transcribed spacer secondary structure. Trends Genet 31: 157–163. [DOI] [PubMed] [Google Scholar]
- Côté CA, Peculis BA. 2001. Role of the ITS2-proximal stem and evidence for indirect recognition of processing sites in pre-rRNA processing in yeast. Nucleic Acids Res 29: 2106–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Côté CA, Greer CL, Peculis BA. 2002. Dynamic conformational model for the role of ITS2 in pre-rRNA processing in yeast. RNA 8: 786–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cumberworth A, Lamour G, Babu MM, Gsponer J. 2013. Promiscuity as a functional trait: intrinsically disordered regions as central players of interactomes. Biochem J 454: 361–369. [DOI] [PubMed] [Google Scholar]
- De la Cruz J, Karbstein K, Woolford JL. 2015. Functions of ribosomal proteins in assembly of eukaryotic ribosomes in vivo. Annu Rev Biochem 84: 93–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dembowski J, Ramesh M, McManus JC, Woolford JL. 2013. Identification of the binding site of Rlp7 on assembling 60S ribosomal subunits in Saccharomyces cerevisiae. RNA 19: 1639–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dez C, Houseley J, Tollervey D. 2006. Surveillance of nuclear-restricted pre-ribosomes within a subnucleolar region of Saccharomyces cerevisiae. EMBO J 25: 1534–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson HJ, Wright PE. 2005. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol 6: 197–208. [DOI] [PubMed] [Google Scholar]
- Ferreira-Cerca S, Pöll G, Gleizes P-E, Tschochner H, Milkereit P. 2005. Roles of eukaryotic ribosomal proteins in maturation and transport of pre-18S rRNA and ribosome function. Mol Cell 20: 263–275. [DOI] [PubMed] [Google Scholar]
- Gamalinda M, Woolford JL. 2014. Deletion of L4 domains reveals insights into the importance of ribosomal protein extensions in eukaryotic ribosome assembly. RNA 20: 1725–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamalinda M, Jakovljevic J, Babiano R, Talkish J, de la Cruz J, Woolford JL. 2013. Yeast polypeptide exit tunnel ribosomal proteins L17, L35 and L37 are necessary to recruit late-assembling factors required for 27SB pre-rRNA processing. Nucleic Acids Res 41: 1965–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamalinda M, Ohmayer U, Jakovljevic J, Kumcuoglu B, Woolford J, Mbom B, Lawrence L, Woolford JL. 2014. A hierarchical model for assembly of eukaryotic 60S ribosomal subunit domains. Genes Dev 28: 198–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasse L, Flemming D, Hurt E. 2015. Coordinated ribosomal ITS2 RNA processing by the Las1 complex integrating endonuclease, polynucleotide kinase, and exonuclease activities. Mol Cell 60: 808–815. [DOI] [PubMed] [Google Scholar]
- Granneman S, Petfalski E, Tollervey D. 2011. A cluster of ribosome synthesis factors regulate pre-rRNA folding and 5.8S rRNA maturation by the Rat1 exonuclease. EMBO J 30: 4006–4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greber BJ, Boehringer D, Montellese C, Ban N. 2012. Cryo-EM structures of Arx1 and maturation factors Rei1 and Jjj1 bound to the 60S ribosomal subunit. Nat Struct Mol Biol 19: 1228–1233. [DOI] [PubMed] [Google Scholar]
- Gunasekaran K, Tsai C-J, Kumar S, Zanuy D, Nussinov R. 2003. Extended disordered proteins: targeting function with less scaffold. Trends Biochem Sci 28: 81–85. [DOI] [PubMed] [Google Scholar]
- Horsey EW, Jakovljevic J, Miles TD, Harnpicharnchai P, Woolford JL. 2004. Role of the yeast Rrp1 protein in the dynamics of pre-ribosome maturation. RNA 10: 813–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakob S, Ohmayer U, Neueder A, Hierlmeier T, Perez-Fernandez J, Hochmuth E, Deutzmann R, Griesenbeck J, Tschochner H, Milkereit P. 2012. Interrelationships between yeast ribosomal protein assembly events and transient ribosome biogenesis factors interactions in early pre-ribosomes. PLoS One 7: e32552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakovljevic J, de Mayolo PA, Miles TD, Nguyen TM-L, Léger-Silvestre I, Gas N, Woolford JL. 2004. The carboxy-terminal extension of yeast ribosomal protein S14 is necessary for maturation of 43S preribosomes. Mol Cell 14: 331–342. [DOI] [PubMed] [Google Scholar]
- Jakovljevic J, Ohmayer U, Gamalinda M, Talkish J, Alexander L, Linnemann J, Milkereit P, Woolford JL. 2012. Ribosomal proteins L7 and L8 function in concert with six A3 assembly factors to propagate assembly of domains I and II of 25S rRNA in yeast 60S ribosomal subunits. RNA 18: 1805–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenner L, Melnikov S, de Loubresse NG, Ben-Shem A, Iskakova M, Urzhumtsev A, Meskauskas A, Dinman J, Yusupova G, Yusupov M. 2012. Crystal structure of the 80S yeast ribosome. Curr Opin Struct Biol 22: 759–767. [DOI] [PubMed] [Google Scholar]
- Kharde S, Calviño FR, Gumiero A, Wild K, Sinning I. 2015. The structure of Rpf2-Rrs1 explains its role in ribosome biogenesis. Nucleic Acids Res 43: 7083–7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Abeysirigunawarden SC, Chen K, Mayerle M, Ragunathan K, Luthey-Schulten Z, Ha T, Woodson S. 2014. Protein-guided RNA dynamics during early ribosome assembly. Nature 506: 334–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinge S, Voigts-Hoffmann F, Leibundgut M, Arpagaus S, Ban N. 2011. Crystal structure of the eukaryotic 60S ribosomal subunit in complex with initiation factor 6. Science 334: 941–948. [DOI] [PubMed] [Google Scholar]
- Kressler D, Hurt E, Bergler H, Bassler J. 2012. The power of AAA-ATPases on the road of pre-60S ribosome maturation--molecular machines that strip pre-ribosomal particles. Biochim Biophys Acta 1823: 92–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidig C, Thoms M, Holdermann I, Bradatsch B, Berninghausen O, Bange G, Sinning I, Hurt E, Beckmann R. 2014. 60S ribosome biogenesis requires rotation of the 5S ribonucleoprotein particle. Nat Commun 5: 3491. [DOI] [PubMed] [Google Scholar]
- Madru C, Lebaron S, Blaud M, Delbos L, Pipoli J, Pasmant E, Rety S, Leulliot N. 2015. Chaperoning 5S RNA assembly. Genes Dev 29: 1432–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayerle M, Woodson SA. 2013. Specific contacts between protein S4 and ribosomal RNA are required at multiple stages of ribosome assembly. RNA 19: 574–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayerle M, Bellur DL, Woodson S. 2011. Slow formation of stable complexes during coincubation of minimal rRNA and ribosomal protein S4. J Mol Biol 412: 453–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnikov S, Ben-Shem A, Garreau de Loubresse N, Jenner L, Yusupova G, Yusupov M. 2012. One core, two shells: bacterial and eukaryotic ribosomes. Nat Struct Mol Biol 19: 560–567. [DOI] [PubMed] [Google Scholar]
- Napper N, Culver GM. 2015. Analysis of r-protein and RNA conformation of 30S subunit intermediates in bacteria. RNA 21: 1323–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nerurkar P, Altvater M, Gerhardy S, Schütz S, Fischer U, Weirich C, Panse VG. 2015. Eukaryotic ribosome assembly and nuclear export. Int Rev Cell Mol Biol 319: 107–140. [DOI] [PubMed] [Google Scholar]
- Neueder A, Jakob S, Pöll G, Linnemann J, Deutzmann R, Tschochner H, Milkereit P. 2010. A local role for the small ribosomal subunit primary binder rpS5 in final 18S rRNA processing in yeast. PLoS One 5: e10194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donohue M-F, Choesmel V, Faubladier M, Fichant G, Gleizes PE. 2010. Functional dichotomy of ribosomal proteins during the synthesis of mammalian 40S ribosomal subunits. J Cell Biol 190: 853–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmayer U, Gamalinda M, Sauert M, Ossowski J, Poll G, Linnemann J, Hierlmeier T, Perez-Fernandez J, Kumcuoglu B, Leger-Silvestre I, et al. 2013. Studies on the assembly characteristics of large subunit ribosomal proteins in S. cerevisae. PLoS One 8: e68412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Z, Oldfield CJ, Xue B, Mizianty MJ, Dunker K, Kurgan L, Uversky VN. 2014. A creature with a hundred waggly tails: intrinsically disordered proteins in the ribosome. Cell Mol Life Sci 71: 1477–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillet B, García-Gómez JJ, Pausch P, Falquet L, Bange G, de la Cruz J, Kressler D. 2015. The dedicated chaperone Acl4 escorts ribosomal protein Rpl4 to its nuclear pre-60S assembly site. PLoS Genet 11: e1005565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahasranaman A, Dembowski J, Strahler J, Andrews P, Maddock J, Woolford JL. 2011. Assembly of Saccharomyces cerevisiae 60S ribosomal subunits: role of factors required for 27S pre-rRNA processing. EMBO J 30: 4020–4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saveanu C, Bienvenu D, Namane A, Gleizes PE, Gas N, Jacquier A, Fromont-Racine M. 2001. Nog2p, a putative GTPase associated with pre-60S subunits and required for late 60S maturation steps. EMBO J 20: 6475–6484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schillewaert S, Wacheul L, Lhomme F, Lafontaine DLJ. 2012. The evolutionarily conserved protein LAS1 is required for pre-rRNA processing at both ends of ITS2. Mol Cell Biol 32: 430–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelter P, Huber FM, Kunze R, Flemming D, Hoelz A, Hurt E. 2015. Coordinated ribosomal L4 protein assembly into the pre-ribosome is regulated by its eukaryote-specific extension. Mol Cell 58: 854–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai LR, Chou CW, Lee IF, Kirby R, Lin A. 2013. The quantitative assessment of the role played by basic amino acid clusters in the nuclear uptake of human ribosomal protein L7. Exp Cell Res 319: 367–375. [DOI] [PubMed] [Google Scholar]
- Talkish J, Zhang J, Jakovljevic J, Horsey EW, Woolford JL. 2012. Hierarchical recruitment into nascent ribosomes of assembly factors required for 27SB pre-rRNA processing in Saccharomyces cerevisiae. Nucleic Acids Res 40: 8646–8661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talkish J, Campbell IW, Sahasranaman A, Jakovljevic J, Woolford JL. 2014. Ribosome assembly factors Pwp1 and Nop12 are important for folding of 5.8S rRNA during ribosome biogenesis in Saccharomyces cerevisiae. Mol Cell Biol 34: 1863–1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang L, Sahasranaman A, Jakovljevic J, Schleifman E, Woolford JL. 2008. Interactions among Ytm1, Erb1, and Nop7 required for assembly of the Nop7-subcomplex in yeast preribosomes. Mol Biol Cell 19: 2844–2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoms M, Thomson E, Baßler J, Gnädig M, Griesel S, Hurt E. 2015. The exosome is recruited to RNA substrates through specific adaptor proteins. Cell 162: 1029–1038. [DOI] [PubMed] [Google Scholar]
- Thomson E, Tollervey D. 2005. Nop53p is required for late 60S ribosome subunit maturation and nuclear export in yeast. RNA 11: 1215–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timsit Y, Acosta Z, Allemand F, Chiaruttini C, Springer M. 2009. The role of disordered ribosomal protein extensions in the early steps of eubacterial 50 S ribosomal subunit assembly. Int J Mol Sci 10: 817–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompa P. 2003. The functional benefits of protein disorder. J Mol Struct 666–667: 361–371. [Google Scholar]
- Tompa P, Csermely P. 2004. The role of structural disorder in the function of RNA and protein chaperones. FASEB J 18: 1169–1175. [DOI] [PubMed] [Google Scholar]
- Van Der Lee R, Buljan M, Lang B, Weatheritt RJ, Daughdrill GW, Dunker AK, Fuxreiter M, Gough J, Gsponer J, Jones D, et al. 2014. Classification of intrinsically disordered regions and proteins. Chem Rev 114: 6589–6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nues RW, Rientjes JMJ, Morre SA, Mollee E, Planta RJ, Venema J, Raue HA. 1995. Evolutionarily conserved structural elements are critical for processing of internal transcribed spacer 2 from Saccharomyces cerevisiae precursor ribosomal RNA. J Mol Biol 250: 24–36. [DOI] [PubMed] [Google Scholar]
- Wilson DN, Doudna Cate JH. 2012. The structure and function of the eukaryotic ribosome. Cold Spring Harb Perspect Biol 4: a011536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolford JL, Baserga SJ. 2013. Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics 195: 643–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Tutuncuoglu B, Yan K, Brown H, Zhang Y, Tan D, Gamalinda M, Yuan Y, Li Z, Jakovljevic J, et al. 2016. Diverse roles of assembly factors revealed by structures of late nuclear pre-60S particles. Nature 534: 133–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y, Demoinet E, Saveanu C, Lenormand P, Jacquier A, Fromont-Racine M. 2010. Ecm1 is a new pre-ribosomal factor involved in pre-60S particle export. RNA 16: 1007–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusupova G, Yusupov M. 2014. High-resolution structure of the eukaryotic 80S ribosome. Annu Rev Biochem 83: 467–486. [DOI] [PubMed] [Google Scholar]
- Zhang J, Harnpicharnchai P, Jakovljevic J, Tang L, Guo Y, Oeffinger M, Rout MP, Hiley SL, Hughes T, Woolford JL Jr. 2007. Assembly factors Rpf2 and Rrs1 recruit 5S rRNA and ribosomal proteins rpL5 and rpL11 into nascent ribosomes. Genes Dev 21: 2580–2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.