Abstract
We herein demonstrate an approach to gas phase ion manipulation that provides MS3-level CID spectra of phospholipid radical cations that are almost independent of the original charging adduct ions. In the MS2 He-MAD spectra of the protonated, sodiated and potassiated adducts of POPC, the different adducts induce different primary fragmentation pathways and provide significantly different spectra, as is commonly observed by other activation methods. In separate experiments, the even-electron adduct ions ([M+H]+, [M+Na]+, [M+K]+) of 1-palmitoyl-2-oleoyl-phosphatidylcholine (POPC) were first converted to radical cations [POPC]+• by using helium metastable atom-activated dissociation (He-MAD) to eject the charging adduct ions, then exposed to low-energy collision induced dissociation (CID) to induce extensive fragmentation along the acyl chains. Such charge-remote fragmentation is generally inaccessible through low-energy CID of the even-electron precursor ions. The combination of He-MAD and CID provides radical-induced spectra that show very major similarities and only minor differences, and therefore overcomes major differences in chemistry that are otherwise observed by the original adducting species. Collisional activation of even-electron [POPC+H]+ required higher CID amplitudes than odd-electron [POPC]+• to effect fragmentation—as expected—and the latter provided fragments within the acyl chains that were influenced by the double bond position.
Introduction
Lipids are the major building blocks of cellular membranes and possess crucial relevance in signal transduction and the storage of energy in biological systems [1]. Changes in lipid profiles and their distribution are found to be closely related to many pathological conditions such as Alzheimer’s disease, Down syndrome, and diabetes [2]. Because of the biological relevance of lipids in organisms, substantial efforts have been devoted to the study of lipids, or lipidomics [3, 4].
Mass spectrometry is a powerful method that has become an indispensable tool in the study of biomolecules. Mass spectrometric applications in lipid analysis started in the early 1970s with electron ionization mass spectrometry (EI-MS) [5, 6] and fast atom bombardment mass spectrometry (FAB-MS) [7]. Matrix-assisted laser desorption ionization mass spectrometry (MALDI-MS) and electrospray ionization mass spectrometry (ESI-MS) became available in the 1990s [2, 8]. Being soft ionization techniques, MALDI and ESI exhibit a sensitivity that is 2 to 3 orders of magnitude greater than that achieved by FAB-MS [8].
The aforementioned soft ionization techniques excel in preserving intact molecular ions, but at the expense of useful structural information, that is, they provide molecular ions but no fragment ions. To enhance the structural information, tandem mass spectrometry (MSn) experiments are often performed, and the most common of which is collision-induced dissociation (CID) [8–12]. CID-based mass spectra of phospholipids are typically dependent on the adduct form of precursor ion. For example, the CID spectrum of protonated adducts of phosphatidylcholines are dominated by a phosphocholine ion at m/z 184, whereas CID of alkali metal-adducted ions produces several fragment ions that allow elucidation of the identities and positions of fatty acid substituents [10]. CID of proton-bound dimers has recently been shown to distinguish cis- and trans-isomers of double bonds in addition to their position in the acyl chains [13].
In addition to CID, other methods of tandem mass spectrometry—such as post source decay (PSD) [14], ozone induced dissociation (OzID) [15–20]—have also been applied in lipid analysis. In OzID, mass-selected lipid cations are exposed to ozone vapor to initiate the gas-phase ion-molecule reaction. Subsequent ozonolysis results in diagnostic fragment ions that can unambiguously identify C=C double bond location(s). Alternative fragmentation methods, such as infrared multiphoton dissociation (IRMPD) [21], ultraviolet photodissociation (UVPD) [22], electron transfer dissociation (ETD) [23], and electron impact excitation of ions from organics (EIEIO) [24] have also been recently employed in the characterization of glycerolipids. These high energy and radical-based methods typically activate more pathways than even electron low energy rearrangements, so they provide complementary fragments to conventional CID [25].
Metastable-atom activated dissociation (MAD) is another developing tandem MS fragmentation method [26–32]. To date, MAD has been used in a variety of studies concerning peptide structures, including multiply charged cations and anions, 1+ cations, phosphorylated cations, disulfide bonds, to cleave the amide ring structure of proline and to differentiate isoleucine from leucine [30–32]. In addition to fragmenting peptides, MAD has been shown to provide high energy and radical-induced fragmentation of lipid cations [33]. In contrast to CID, which exclusively proceeds through even-electron mechanism, MAD produces both even-electron and odd-electron fragments.
To explore the unique features of MAD, we herein demonstrate the ability to acquire CID spectra of radical cations that are independent of the charging adduct ions. Isolated, even-electron adduct ions ([M+H]+, [M+Na]+, [M+K]+) are first converted to odd-electron molecular ions [M]+• through He-MAD. Low-energy collisional activation of the isolated radical cations ions then induces extensive fragmentation along the acyl chains of the lipids through mechanisms that are not achievable from CID of even-electron precursor ions. Distinctive radical fragments are also observed and described, which illustrate the potential utility of this type of gas-phase ion manipulation.
Experimental
Instrumentation
All experiments were performed on a modified Bruker amaZon ETD mass spectrometer (Bruker Daltonics, Bremen, Germany). The modification method, the connection between electronic components and the working principles are described elsewhere [32, 33].
Materials
The lipid used in this study was 1-hexadecanoyl-2-(9Z-octadecenoyl)-sn-glycero-3-phosphocholine PC(16:0/18:1(9Z)), which is abbreviated as POPC. POPC was diluted to a final concentration of 60 µM using a 9:1 (v/v) mixture of methanol to water containing 1% (v/v) acetic acid to obtain the protonated ions and 0.05 M NaCl or KCl to obtain metal-adducted ions. Ultra high purity helium (Airgas, Parkersburg, WV) was used in the FAB gun and was further purified using a noble gas purifier (HP2, VICI, Houston, TX) to remove impurity gases that can rapidly quench metastable atoms.
Methods
Singly charged lipid precursor ions were generated through electrospray ionization (ESI) using an electronic syringe pump (#1725, Hamilton Company Reno, Nevada, NV) with a flow rate of 160 µL/h. The ion of interest was usually isolated using an isolation window of 4 Da before exposure to the helium metastable beam. Occasionally, isolation windows of 1 Da were used to obtain the mono-isotopic form of the precursor ion and help differentiate 13C isotope peaks from H-atom transfer peaks. The low mass cut off (LMCO) during MAD activation was typically set to m/z 155 to prevent the accumulation of Penning-ionized pump oil. The ESI injection conditions and ion injection times (0.2–5 ms) were adjusted to ensure that the trap was filled to capacity with isolated precursor ions before exposure to He-MAD. The metastable beam was created by a FAB gun that was pulsed on for 30 ms at an anode voltage of 7 kV, and ions and electrons were removed from the beam using deflection electrodes. Ions were stored for at least 50 ms during MAD and CID, even though the FAB gun was only on for 30 ms of the 50 ms storage time. The FAB gun is attached to the main vacuum chamber lid, so the FAB gas pressure is indirectly measured by reading the pressure of the ion trap gauge in the main vacuum region. In all experiments the FAB gas supply was adjusted to provide a reading of ~1.20×10−5 mbar.
A typical sequence of data acquisition consisted of 0.5 minutes of mass acquisition of isolated precursor ion signal, 2 minutes of He-MAD of the precursor ion, and 0.5 minutes of He-MAD background signal (ESI source off with He-MAD source on). The isolated precursor signal was stored to verify the width and intensity of the isotope envelope of the precursor ion(s). Before spectral interpretation, an averaged He-MAD background spectrum was subtracted from an averaged He-MAD tandem mass spectrum to obtain a background subtracted He-MAD spectrum. A typical MS3 MAD/CID experiment included an additional 0.5 minutes of mass acquisition of the isolation of precursor ion at MS3 stage, and an additional 4 minutes of MS3-stage CID reaction with an amplitude of 0.35 V. A conventional MS2 CID experiment included 0.5 minutes of storage of isolation spectra and 4 minutes of CID product ion spectra.
Results and discussion
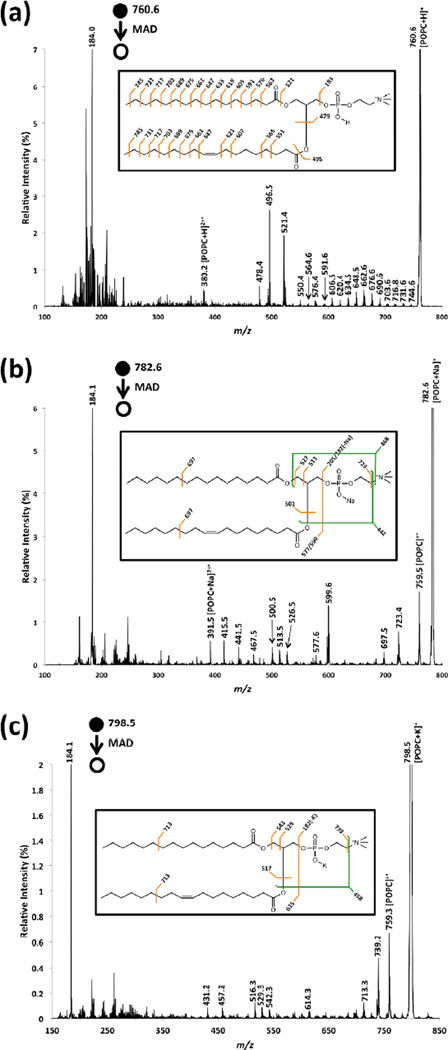
Figure 1 shows the He-MAD spectra of the protonated, sodiated and potassiated forms of POPC. Upon the interaction with helium metastable atoms, the protonated form of POPC produces a variety of cleavages along the acyl chains. The 14 Da increments resemble high-energy CID (HE-CID) spectra, which suggests that a charge-remote-like mechanism is taking place in this process [33–35]. Conversely, MAD of the metal-adducted forms rarely cleaves the acyl chains, but instead forms abundant fragment ions arising from the glycerol backbone and head group. These results show how the adducting species of the precursor ion has a significant influence on the MAD fragmentation patterns, as has been observed qualitatively for CID of phosphocholines [11, 12]. Relative to the protonated precursor, the presence of metal adducts (i.e. Na+ or K+) appears to restrict the number of fragments within the acyl chains (i.e. in the region m/z 550–750).
Figure 1.
He-MAD spectra of (a) protonated form of POPC, (b) sodiated form of POPC, (c) potassiated form of POPC. Insets show possible cleavages and theoretical masses for fragmentations without hydrogen rearrangements.
He-MAD spectra of all the three forms show a predominant peak at m/z 184.0 (or 184.1), corresponding to the formation of the low-energy phosphocholine ion [12]. Distinctive peaks at m/z 380.2 and 391.5 are observed from both protonated and sodiated forms of POPC, respectively, which are the characteristic 2+ Penning ionized product ions [32, 33]. The 2+ Penning ionized product is not observed in the He-MAD spectrum of potassiated POPC.
In the He-MAD spectrum of protonated POPC, ions at m/z 478.4, 496.5 and 521.4 are also observed, which correspond to the loss of the sn-2 fatty acid (18:1, −282 Da) as well as the loss of acyl chains as ketenes from sn-2 (18:1, −264 Da) and sn-1 (16:0, −239 Da) positions, respectively. These fragments are commonly observed in low energy CID experiments [12] and also observed in lipidomic analyses using an LC-MSE approach [36]. He-MAD of metal-adducted POPC produces similar sn-1 and sn-2 cleavages as well. MAD product ions are also observed which correspond to two covalent cleavages, such as the simultaneous loss of the sn-2 acyl chain and either the trimethylamine (N(CH3)3) moiety or the phosphocholine head group for both sodiated and potassiated POPC.
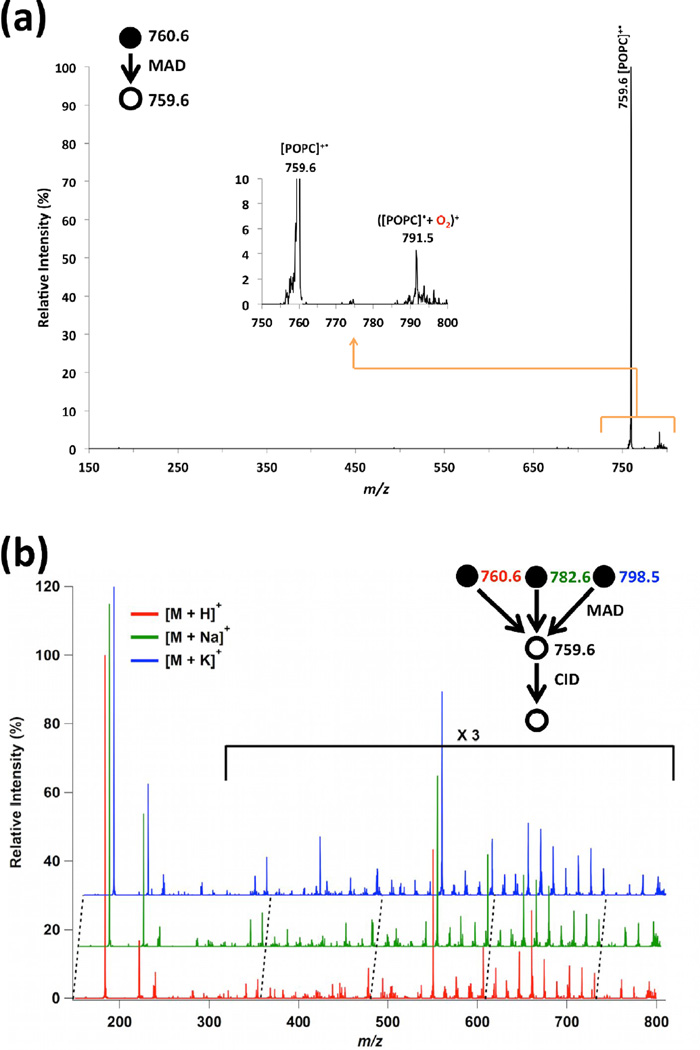
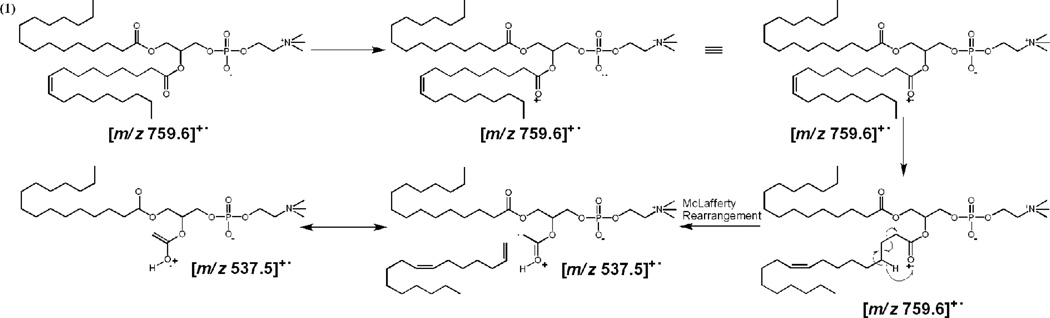
He-MAD of the sodiated and potassiated precursors of POPC both give strong molecular ions at m/z 759.6. The [POPC]+• molecular ion is not readily observable in the product ion spectrum of the protonated precursor, which could be due to several reasons: 1) the mass resolution is compromised because of the space charge effects of the abundant precursor ion at m/z 760.6 [37], and 2) the loss of a hydrogen is far less favorable than the loss of sodium or potassium. Although the product at m/z 759.6 ([POPC]+•) is obscured by the more dominant [POPC+H]+ ion at m/z 760.6 in the He-MAD spectrum of [POPC+H]+, the product ion at m/z 759.6 was effectively isolated using the standard isolation procedure of the Bruker ion trap. Mass isolation uses slower scan speeds than mass acquisition, so achieves superior mass resolution. Evidence for the formation of [POPC]+• from [POPC+H]+, via the loss of H•, is presented in Figure 2a. The reaction presumably proceeds via the following generic mechanism:
| (1) |
where X = H, Na or K; HeM is the helium metastable atom and He0 is the helium ground state atom. In reaction 1, the electron released from Penning ionization of the lipid is captured by the charging cation, which subsequently dissociates because of the significantly reduced binding affinity.
Figure 2.
Panel (a): Example of an isolation spectrum for the [POPC]+• cation at m/z 759.6 from He-MAD of protonated form of POPC. Inset in panel (a) shows the magnified spectrum of the m/z range of interest. Panel (b): Waterfall plot to show the similarities of the MS3 CID spectra of the same intermediate in panel a from the protonated, sodiated and potassiated forms of POPC.
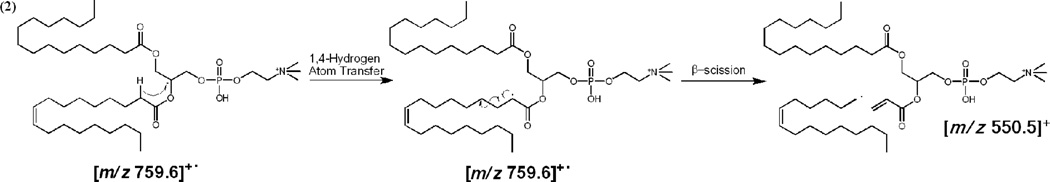
Reaction (1) is also in competition with the straightforward Penning ionization product shown in reaction (2), in which the Penning ionized electron is not captured by the charging cation.
| (2) |
The 2+ product ion of reaction (2) is a prominent feature in the He-MAD spectra when X = H+ and Na+, but is not observed when X = K+. The preference for reaction (1) vs reaction (2) is probably dependent on the combined effects of electron affinity, electron capture cross section and binding affinity. The potassiated adduct clearly favors internal electron capture through reaction (1) despite the fact that it has the lowest electron affinity of the three cations. At present, we can only speculate that the binding energy of the cations is a dominant factor in these reactions [38] and that conformational differences are a possible secondary factor [39]. Potassium has the lowest binding affinity for POPC of the three cations and is therefore most readily lost [38].
Isolation and collisional activation of [POPC+• + Na+]2+• at m/z 391.5 resulted in charge separation and a dominant radical molecular ion of [POPC]+• at m/z 759.6, as shown in reaction (3) (See Supplemental Figure S1).
| (3) |
Collisional activation of the protonated radical dication at m/z 380.2 yielded many abundant fragments (See Supplemental Figure S2), but did not provide the radical molecular ion through reaction (3). These observations indicate that a variety of competing reactions occur during He-MAD, and the adducting cations show evidence of both capturing and not capturing the free electron from Penning ionization of the lipid. Potassium seems to have the largest electron capture cross section despite its smallest electron affinity.
Figure 2a shows the isolated [POPC]+• product peak at m/z 759.6 following He-MAD of [POPC+H]+. The observation of a ‘companion’ product ion at m/z 791.5 in the isolation spectrum of [POPC]+• is evidence for the oxidation of the radical cation with background oxygen in the trap. McLuckey and coworkers have documented the enhanced reactivity of radical peptide cations towards oxidation with residual oxygen [40]. The observation of ([POPC]•+O2)+ ion is indicative of a distonic radical ion structure for at least some portion of the [POPC]+• population, which indicates that the radical is distributed among a variety of locations that are remote from the charge site.
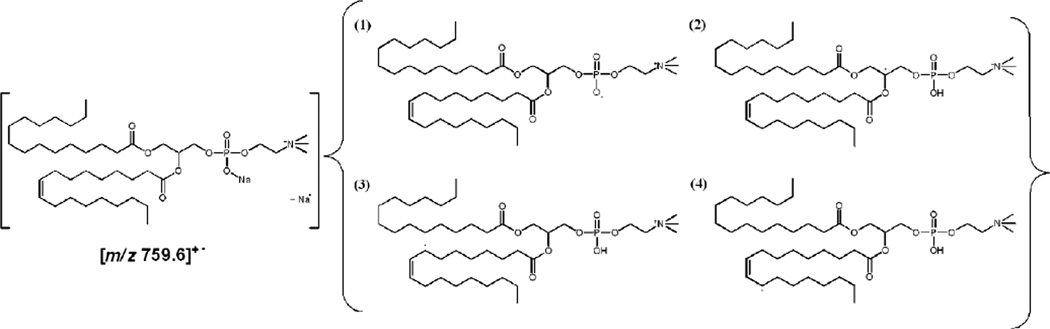
Blanksby and coworkers [41, 42] have reported the study of lipids and fatty acid derivatives using radical-directed dissociation (RDD) in which a variety of radical sites were proposed to rationalize various fragments arising from radical-directed processes. Scheme 1 shows four such examples of the possible isomeric structures of [POPC]+• ion using He-MAD. Isomeric structure (1) rationalizes the formation of the fragment at m/z 537.5. Isomeric structure (2) rationalizes the fragment at m/z 550.5. Isomers (3) and (4) are consistent with the formation of fragment ion pair at m/z 632.5 through an α-allylic H-abstraction pathway and m/z 634.4 through a ω-allylic H-abstraction pathway, vide infra [41].
Scheme 1.
Examples of possible isomeric structures of radical ion at m/z 759.6.
Different from their adducted precursor ions, radical intermediates at m/z 759.6 produce almost indistinguishable fragmentation patterns at the MS3 level of fragmentation, regardless of whether the intermediate originates from the protonated, sodiated or potassiated forms of POPC (Figure 2b). Some minor differences are observed, however, such as the peak at m/z 414 in the MS3 spectrum from the potassiated precursor. The similarities in the CID spectra of the m/z 759.6 intermediate indicate that the intermediates are constitutionally similar radical cations, but that minor differences in the distribution of isomeric forms may exist before/during collisional activation. Whereas the adduct ions clearly affect the distribution of first-generation fragment ions in MAD (Figure 1), the isolated [POPC]+• ions produced via MAD apparently have considerably less influence from the original adducting species.
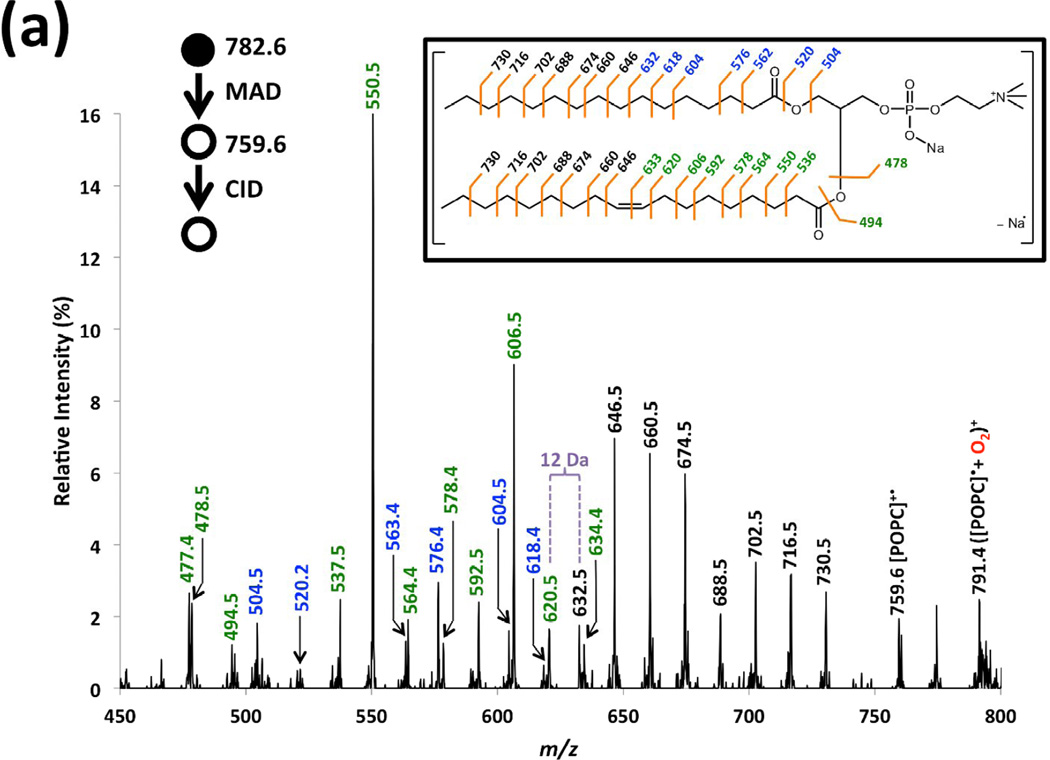
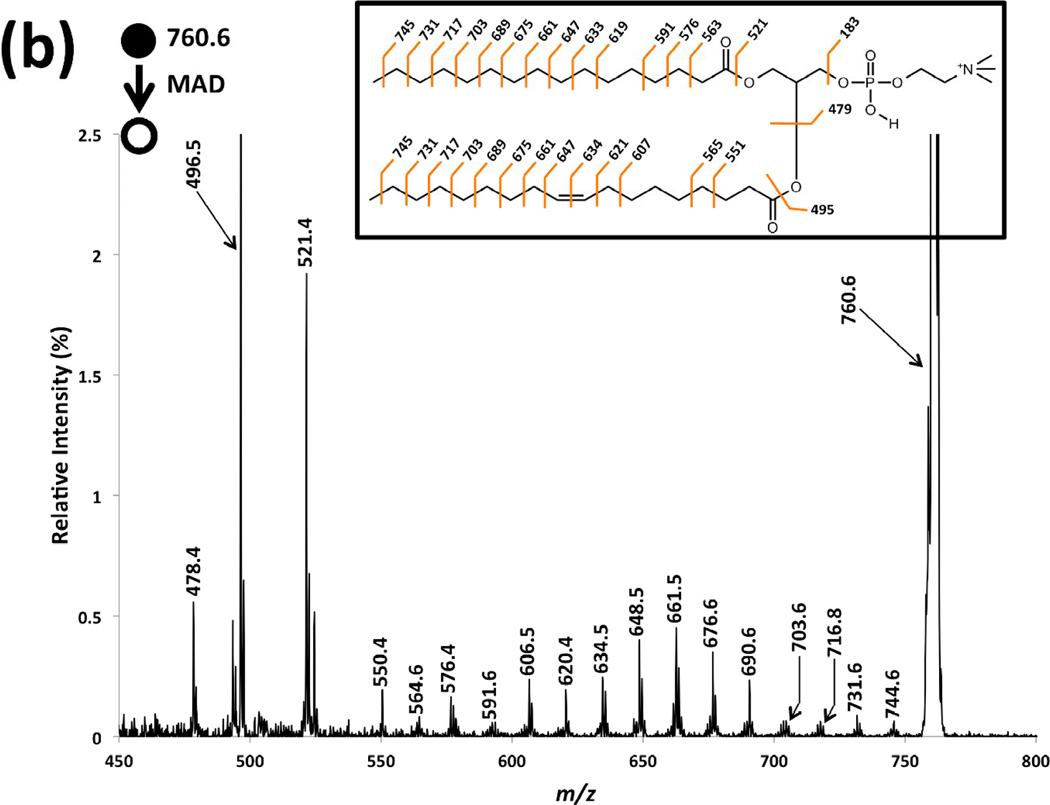
Figure 3a is a magnification of the MAD spectrum of sodiated POPC in Figure 2b and serves as an exemplar CID spectrum of [POPC]+• (m/z 759.6). Figure 3b shows the He-MAD spectrum of [POPC+H]+ (m/z 760.6), without CID. The two spectra are similar in that they both display fragmentation within the acyl chains, but distinct in that the relative distribution of fragments is vastly different. The series of peaks labeled in black font from m/z 646.5–730.5 show clear 14 Da increments, and both acyl chains can contribute to these fragment ions. The double bond at the Δ9 position of the sn-2 acyl chain makes the acyl fragments differ in mass below m/z 646.5, so the fragments can be linked to each acyl chain. For example, the fragments labeled in blue font correspond to the losses from the saturated sn-1 chain, and the fragments labeled in green are for the unsaturated sn-2 chain.
Figure 3.
Magnified spectra of (a) MS3 CID of radical cation at m/z 759.6 from He-MAD of sodiated form of POPC, (b) He-MAD of protonated form of POPC.
The fragmentation pattern described above has also been observed in EIEIO spectra of phosphatidylcholines reported by Baba and coworkers [24]. For the saturated sn-1 chain, the near complete fragmentation pattern was highly analogous to that of collisional activation of [FAMEs]+• derived from EI [43, 44]. The minor differences are that He-MAD spectra show a lack of cleavage of the C4-C5 bond and include a McLafferty rearrangement fragment at m/z 563.4.
For the unsaturated sn-2 chain, alkylene moiety fragments at m/z 620.5 and 632.5 are characteristically spaced by 12 Da, whereas the rest of the acyl chain fragments exhibit a spacing of 14 Da. This diagnostic feature—the discrepancy in spacing between fragment derived from saturated and unsaturated moieties—serves as an unambiguous identification of the double bond location [41, 42, 45, 46]. An interesting fragment ion pair at m/z 632.5 and 634.4 was also observed, which correspond to a losses of 127 or 125, respectively, which are unsaturated •C9H19 and •C9H17 losses, respectively. The similar “doubled peak” pairs have been reported in the study using RDD for differentiation of lipid isomers, the formation of which has been rationalized via 1,4-hydrogen transfer, 1,6-elimination and β-scission [41].
The MS3 CID spectrum of [POPC]+• is dominated by fragments of the sn-2 unsaturated chain, including peaks at m/z 477.4, 478.5, 550.5 and 606.6. The peak at m/z 550.5 is consistent with a 1,4-hydrogen shift and gamma cleavage relative to the sn-2 carbonyl group (Scheme 2). The peak at m/z 537.5 probably originates from McLafferty rearrangement of the proposed radical structure (2) in Scheme 1. The corresponding fragmentation pathway is shown in Scheme 3. The two similar products from the saturated acyl chain in the sn-1 position were observed at m/z 576.4 and m/z 563.4.
Scheme 2.
Proposed fragmentation pathways for the formation of product ion at m/z 550.5.
Scheme 3.
Proposed fragmentation pathway for the formation of product ion at m/z 537.5.
In contrast to CID of the radical ion [POPC]+•, the He-MAD spectrum of [POPC+H]+ shows dominant sn-1 and sn-2 glycerol backbone cleavages at m/z 496.5 and 521.4, which has been observed elsewhere [33]. It is noteworthy that the MS3 He-MAD/CID spectrum in Figure 3a has superior signal-to-noise ratio than the MS2 He-MAD spectrum in Figure 3b. The MS3 He-MAD/CID spectrum selectively promotes radical-induced fragmentations over competing mechanisms/pathways and background chemical noise.
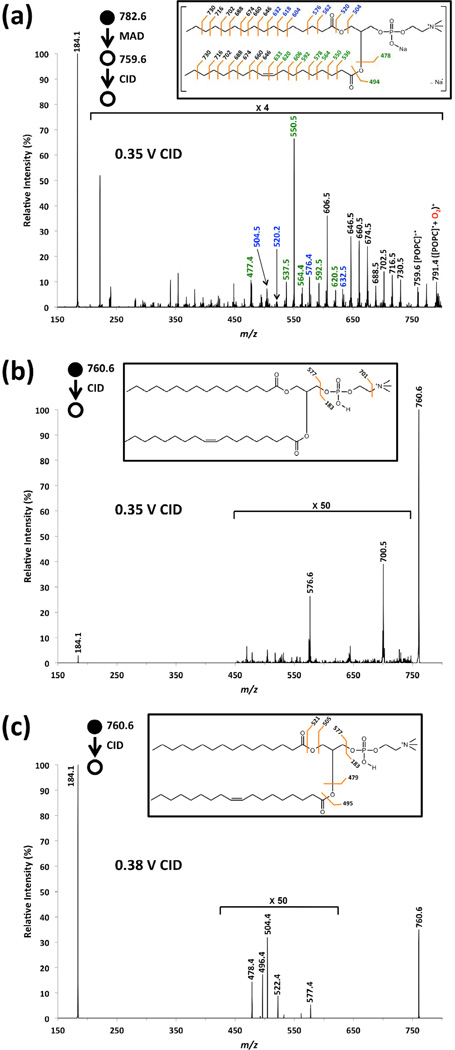
Figure 4 compares a CID spectrum of the radical cation of POPC with a CID spectrum of even-electron [POPC+H]+ cations at two different CID amplitudes. Using a CID amplitude of 0.35 V, the odd-electron [POPC]+• precursor produced near-complete fragmentation of the lipid (as discussed in Figs 2 and 3). However, using the same CID amplitude of 0.35 V, the even electron [POPC+H]+ precursor only produced two weak product ions: the loss of trimethylamine (N(CH3)3) and the loss of entire head group. Using a CID amplitude of 0.38 V (Fig 4c), the even-electron [POPC+H]+ ion dissociated into more fragment ions and with better efficiency, but with a significantly different distribution of product ions from the radical intermediate. When subjected to collisional activation, the odd-electron ion clearly has a significantly lower activation barrier than the even-electron precursor. Similar differences in collisional activation energies have been observed for the radical [M+2H]+• precursors versus even-electron [M+H]+ precursors of peptide ions [47].
Figure 4.
(a) MS3 CID spectrum of the radical cation at m/z 759.6 from He-MAD of sodiated POPC ([POPC+Na]+); (b) CID spectra of protonated form of POPC ([POPC+H]+) with the same activation voltage (0.35 V)* as that in (a); (c) same as (b), but with higher activation voltage (0.38 V).*See experimental for details.
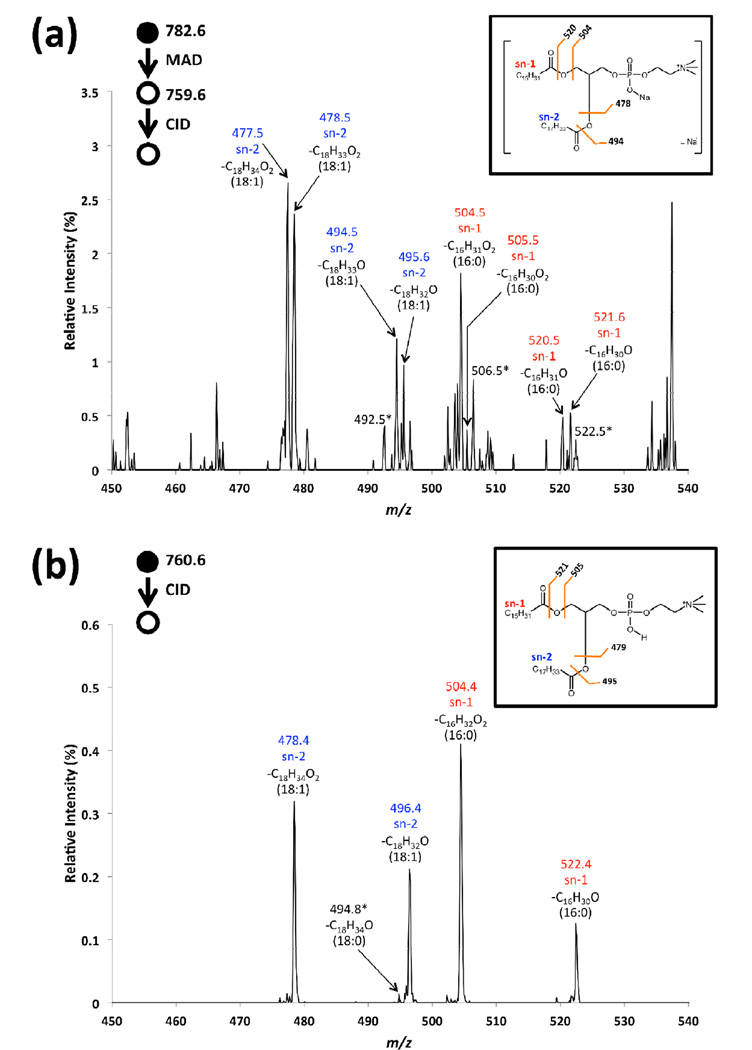
Figure 5 shows the magnified mass spectra of both MS3 CID of [POPC]+• and CID of [POPC+H]+. When starting with even-electron precursor [POPC+H]+ (Fig 5b), CID only produces even mass even electron fragments in this region. The commonly observed fragments at m/z 478.4 and 496.4 are from sn-2 cleavages and the fragments at m/z 504.4 and 522.4 are from sn-1 cleavages. CID of the radical precursor [POPC]+• provides similar cleavages around sn-1 and sn-2 positions, but with the presence of odd mass/radical fragments. In the case of potassiated and sodiated POPC, the more complicated spectra and poorer signal to noise ratios in the MS3 spectrum makes conventional CID a superior tool for assessing the acyl chain lengths and degrees of unsaturation. However, the ability to first convert the protonated, sodiated and potassiated precursor ions to a generic radical molecular ion intermediate, then perform CID to fragment within the acyl chains provides a rather unique gas-phase ion manipulation tool for the structural interrogation of phospholipids.
Figure 5.
Magnified mass spectra corresponding to (a) MS3 CID of [POPC]+• from MAD of sodiated POPC, (b) CID of [POPC+H]+.
Supplementary Material
Highlights.
Ability to form radical dications from protonated and sodiated phosphatidycholine
Ability to remove charging adduct ions and form radical molecular ions
Ability to acquire MS3-level CID spectra from MS2–level He-MAD intermediates
Ability to identify double bond position in acyl chains
Acknowledgments
The authors acknowledge financial support form the National Institutes of Health (NIH) (1R01GM114494-01). The opinions, findings, and conclusions or recommendations expressed in this publication are those of the author(s) and do not necessarily reflect the views of NIH. This manuscript was significantly enhanced by the thoughtful critiques of several reviewers for which the authors are very grateful.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pulfer M, Murphy RC. Electrospray mass spectrometry of phospholipids. Mass Spectrom. Rev. 2003;22:332–364. doi: 10.1002/mas.10061. [DOI] [PubMed] [Google Scholar]
- 2.Jackson SN, Woods AS. Direct profiling of tissue lipids by MALDI-TOFMS. J. Chrom. B. 2009;877:2822–2829. doi: 10.1016/j.jchromb.2008.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blanksby SJ, Mitchell TW. Advances in mass spectrometry for lipidomics. Ann. Rev. Anal. Chem. 2010;3:433–465. doi: 10.1146/annurev.anchem.111808.073705. [DOI] [PubMed] [Google Scholar]
- 4.Li M, Zhou Z, Nie H, Bai Y, Liu H. Recent advances of chromatography and mass spectrometry in lipidomics. Anal. Bioanal. Chem. 2011;399:243–249. doi: 10.1007/s00216-010-4327-y. [DOI] [PubMed] [Google Scholar]
- 5.Klein RA. Mass spectrometry of phosphatidylcholines - dipalmitoyl, dioleoyl, and stearoyl-oleoyl glycerylphosphorylcholines. J. Lipid Res. 1971;12:123-&. [PubMed] [Google Scholar]
- 6.Klein RA. Mass spectrometry of phosphatidylcholines - fragmentation processes for dioleoyl and stearoyl-oleoyl glycerylphosphorylcholine. J. Lipid Res. 1971;12:628-&. [PubMed] [Google Scholar]
- 7.Murphy RC, Harrison KA. Fast-atom-bombardment mass-spectrometry of phospholipids. Mass Spectrom. Rev. 1994;13:57–75. [Google Scholar]
- 8.Hsu FF, Turk J. Structural determination of sphingomyelin by tandem mass spectrometry with electrospray ionization. J. Am. Soc. Mass Spectrom. 2000;11:437–449. doi: 10.1016/S1044-0305(99)00150-6. [DOI] [PubMed] [Google Scholar]
- 9.Hsu FF, Turk J. Studies on phosphatidylglycerol with triple quadrupole tandem mass spectrometry with electrospray ionization: Fragmentation processes and structural characterization. J. Am. Soc. Mass Spectrom. 2001;12:1036–1043. [Google Scholar]
- 10.Hsu FF, Turk J. Electrospray ionization/tandem quadrupole mass spectrometric studies on phosphatidylcholines: The fragmentation processes. J. Am. Soc. Mass Spectrom. 2003;14:352–363. doi: 10.1016/S1044-0305(03)00064-3. [DOI] [PubMed] [Google Scholar]
- 11.Hsu FF, Turk J. Structural characterization of unsaturated glycerophospholipids by multiple-stage linear ion-trap mass spectrometry with electrospray ionization. J. Am. Soc. Mass Spectrom. 2008;19:1681–1691. doi: 10.1016/j.jasms.2008.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ho YP, Huang PC. A novel structural analysis of glycerophosphocholines as TFA/K+ adducts by electrospray ionization ion trap tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2002;16:1582–1589. doi: 10.1002/rcm.751. [DOI] [PubMed] [Google Scholar]
- 13.Pham HT, Prendergast MB, Dunstan CW, Trevitt AJ, Mitchell TW, Julian RR, Blanksby SJ. Dissociation of proton-bound complexes reveals geometry and arrangement of double bonds in unsaturated lipids. Int. J. Mass Spectrom. 2015 [Google Scholar]
- 14.Al-Saad KA, Siems WF, Hill HH, Zabrouskov V, Knowles NR. Structural analysis of phosphatidylcholines by post-source decay matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. J. Am. Soc. Mass Spectrom. 2003;14:373–382. doi: 10.1016/S1044-0305(03)00068-0. [DOI] [PubMed] [Google Scholar]
- 15.Pham HT, Maccarone AT, Campbell JL, Mitchell TW, Blanksby SJ. Ozone-induced dissociation of conjugated lipids reveals significant reaction rate enhancements and characteristic odd-electron product ions. J. Am. Soc. Mass Spectrom. 2013;24:286–296. doi: 10.1007/s13361-012-0521-9. [DOI] [PubMed] [Google Scholar]
- 16.Pham HT, Maccarone AT, Thomas MC, Campbell JL, Mitchell TW, Blanksby SJ. Structural characterization of glycerophospholipids by combinations of ozone- and collision-induced dissociation mass spectrometry: the next step towards "top-down" lipidomics. Analyst. 2014;139:204–214. doi: 10.1039/c3an01712e. [DOI] [PubMed] [Google Scholar]
- 17.Poad BL, Pham HT, Thomas MC, Nealon JR, Campbell JL, Mitchell TW, Blanksby SJ. Ozone-induced dissociation on a modified tandem linear ion-trap: observations of different reactivity for isomeric lipids. J. Am. Soc. Mass Spectrom. 2010;21:1989–1999. doi: 10.1016/j.jasms.2010.08.011. [DOI] [PubMed] [Google Scholar]
- 18.Thomas MC, Mitchell TW, Blanksby SJ. Ozonolysis of phospholipid double bonds during electrospray ionization: a new tool for structure determination. J. Am. Chem. Soc. 2006;128:58–59. doi: 10.1021/ja056797h. [DOI] [PubMed] [Google Scholar]
- 19.Thomas MC, Mitchell TW, Harman DG, Deeley JM, Murphy RC, Blanksby SJ. Elucidation of double bond position in unsaturated lipids by ozone electrospray ionization mass spectrometry. Anal. Chem. 2007;79:5013–5022. doi: 10.1021/ac0702185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thomas MC, Mitchell TW, Harman DG, Deeley JM, Nealon JR, Blanksby SJ. Ozone-induced dissociation: elucidation of double bond position within mass-selected lipid ions. Anal. Chem. 2008;80:303–311. doi: 10.1021/ac7017684. [DOI] [PubMed] [Google Scholar]
- 21.Zehethofer N, Scior T, Lindner B. Elucidation of the fragmentation pathways of different phosphatidylinositol phosphate species (PIPx) using IRMPD implemented on a FT-ICR MS. Anal. Bioanal. Chem. 2010;398:2843–2851. doi: 10.1007/s00216-010-4184-8. [DOI] [PubMed] [Google Scholar]
- 22.Madsen JA, Cullen TW, Trent MS, Brodbelt JS. IR and UV photodissociation as analytical tools for characterizing lipid structures. Anal. Chem. 2011;83:5107–5113. doi: 10.1021/ac103271w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liang X, Liu J, LeBlanc Y, Covey T, Ptak AC, Brenna JT, McLuckey SA. Electron transfer dissociation of doubly sodiated glycerophosphocholine lipids. J. Am. Soc. Mass Spectrom. 2007;18:1783–1788. doi: 10.1016/j.jasms.2007.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Campbell JL, Baba T. Near-complete structural characterization of phosphatidylcholines using electron impact excitation of ions from organics. Anal. Chem. 2015;87:5837–5845. doi: 10.1021/acs.analchem.5b01460. [DOI] [PubMed] [Google Scholar]
- 25.Pham HT, Julian RR. Radical delivery and fragmentation for structural analysis of glycerophospholipids. Int. J. Mass Spectrom. 2014;370:58–65. [Google Scholar]
- 26.Misharin AS, Silivra OA, Kjeldsen F, Zubarev RA. Dissociation of peptide ions by fast atom bombardment in a quadrupole ion trap. Rapid Commun. Mass Spectrom. 2005;19:2163–2171. doi: 10.1002/rcm.2038. [DOI] [PubMed] [Google Scholar]
- 27.Berkout VD. Fragmentation of protonated peptide ions via interaction with metastable atoms. Anal. Chem. 2006;78:3055–3061. doi: 10.1021/ac060069a. [DOI] [PubMed] [Google Scholar]
- 28.Berkout VD. Fragmentation of singly protonated peptides via interaction with metastable rare gas atoms. Anal. Chem. 2009;81:725–731. doi: 10.1021/ac802214e. [DOI] [PubMed] [Google Scholar]
- 29.Berkout VD, Doroshenko VM. Fragmentation of phosphorylated and singly charged peptide ions via interaction with metastable atoms. Int. J. Mass Spectrom. 2008;278:150–157. doi: 10.1016/j.ijms.2008.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cook SL, Zimmermann CM, Singer D, Fedorova M, Hoffmann R, Jackson GP. Comparison of CID, ETD and metastable atom-activated dissociation (MAD) of doubly and triply charged phosphorylated tau peptides. J. Mass Spectrom. 2012;47:786–794. doi: 10.1002/jms.3023. [DOI] [PubMed] [Google Scholar]
- 31.Cook SL, Jackson GP. Metastable atom-activated dissociation mass spectrometry of phosphorylated and sulfonated peptides in negative ion mode. J. Am. Soc. Mass Spectrom. 2011;22:1088–1099. doi: 10.1007/s13361-011-0123-y. [DOI] [PubMed] [Google Scholar]
- 32.Cook SL, Collin OL, Jackson GP. Metastable atom-activated dissociation mass spectrometry: leucine/isoleucine differentiation and ring cleavage of proline residues. J. Mass Spectrom. 2009;44:1211–1223. doi: 10.1002/jms.1598. [DOI] [PubMed] [Google Scholar]
- 33.Deimler RE, Sander M, Jackson GP. Radical-induced fragmentation of phospholipid cations using metastable atom-activated dissociation mass spectrometry (MAD-MS) Int. J. Mass Spectrom. 2015 doi: 10.1016/j.ijms.2015.08.009. http://dx.doi.org/10.1016/j.ijms.2015.1008.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adams J, Gross ML. Charge-remote fragmentations of closed-shell ions - a thermolytic analogy. J. Am. Chem. Soc. 1989;111:435–440. [Google Scholar]
- 35.Deterding LJ, Gross ML. Tandem mass spectrometry for identifying fatty-acid derivatives that undergo charge-remote fragmentations. Org. Mass Spectrom. 1988;23:169–177. [Google Scholar]
- 36.Castro-Perez JM, Kamphorst J, DeGroot J, Lafeber F, Goshawk J, Yu K, Shockcor JP, Vreeken RJ, Hankemeier T. Comprehensive LC–MSE lipidomic analysis using a shotgun approach and Its application to biomarker detection and identification in osteoarthritis patients. J. Proteome Res. 2010;9:2377–2389. doi: 10.1021/pr901094j. [DOI] [PubMed] [Google Scholar]
- 37.March RE. An introduction to quadrupole ion trap mass spectrometry. J. Mass Spectrom. 1997;32:351–369. [Google Scholar]
- 38.Ruan C, Huang H, Rodgers MT. A simple model for metal cation-phosphate interactions in nucleic acids in the gas phase: Alkali metal cations and trimethyl phosphate. J. Am. Soc. Mass Spectrom. 2008;19:305–314. doi: 10.1016/j.jasms.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 39.Fales B, Fujamade N, Oomens J, Rodgers MT. Infrared multiple photon dissociation action spectroscopy and theoretical studies of triethyl phosphate complexes: Effects of protonation and sodium cationization on structure. J. Amer. Soc. Mass Spectrom. 2011;22:1862–1871. doi: 10.1007/s13361-011-0208-7. [DOI] [PubMed] [Google Scholar]
- 40.Xia Y, Chrisman PA, Pitteri SJ, Erickson DE, McLuckey SA. Ion/molecule reactions of cation radicals formed from protonated polypeptides via gas-phase ion/ion electron transfer. J. Am. Chem. Soc. 2006;128:11792–11798. doi: 10.1021/ja063248i. [DOI] [PubMed] [Google Scholar]
- 41.Pham HT, Ly T, Trevitt AJ, Mitchell TW, Blanksby SJ. Differentiation of complex lipid isomers by radical-directed dissociation mass spectrometry. Anal. Chem. 2012;84:7525–7532. doi: 10.1021/ac301652a. [DOI] [PubMed] [Google Scholar]
- 42.Pham HT, Trevitt AJ, Mitchell TW, Blanksby SJ. Rapid differentiation of isomeric lipids by photodissociation mass spectrometry of fatty acid derivatives. Rapid Commun. Mass Spectrom. 2013;27:805–815. doi: 10.1002/rcm.6503. [DOI] [PubMed] [Google Scholar]
- 43.Zirrolli JA, Murphy RC. Low-energy tandem mass spectrometry of the molecular ion derived from fatty acid methyl esters: A novel method for analysis of branched-chain fatty acids. J. Am. Soc. Mass Spectrom. 1993;4:223–229. doi: 10.1016/1044-0305(93)85085-C. [DOI] [PubMed] [Google Scholar]
- 44.Ran-Ressler RR, Lawrence P, Brenna JT. Structural characterization of saturated branched chain fatty acid methyl esters by collisional dissociation of molecular ions generated by electron ionization. J. Lipid Res. 2012;53:195–203. doi: 10.1194/jlr.D020651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dobson G, Christie WW. Spectroscopy and spectrometry of lipids - Part 2 - Mass spectrometry of fatty acid derivatives. Eur. J. Lipid Sci. Tech. 2002;104:36–43. [Google Scholar]
- 46.Griffiths WJ, Yang Y, Lindgren JA, Sjovall J. Charge remote fragmentation of fatty acid anions in 400 eV collisions with xenon atoms. Rapid Commun. Mass Spectrom. 1996;10:21–28. [Google Scholar]
- 47.Swaney DL, McAlister GC, Wirtala M, Schwartz JC, Syka JE, Coon JJ. Supplemental activation method for high-efficiency electron-transfer dissociation of doubly protonated peptide precursors. Anal. Chem. 2007;79:477–485. doi: 10.1021/ac061457f. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.