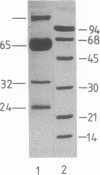
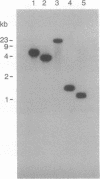
Abstract
Oxygenic photosynthesis of chloroplasts and cyanobacteria involves two photosystems, which originate from different prokaryotic ancestors. The reaction center of photo-system 2 (PS2) is related to the well-characterized reaction center of purple bacteria, while the reaction center of photosystem 1 (PS1) is related to the green sulfur bacteria, as is convincingly documented here. An operon encoding the P840 reaction center of Chlorobium limicola f.sp. thiosulfatophilum has been cloned and sequenced. It contains two structural genes, coding for proteins of 730 and 232 amino acids. The first protein resembles the large subunits of the PS1 reaction center. Putative binding elements for the primary donor, P840 in Chlorobium and P700 in PS1, and for the acceptors A0, A1, and FeS center X are conserved. The second protein is related to the PS1 subunit carrying the FeS centers A and B. An adjacent third gene, not belonging to the reaction center, encodes a protein related to dolichyl-phosphate-D-mannose synthase from yeast. The different origins of PS1 and PS2 are discussed.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beanland T. J. Evolutionary relationships between "Q-type" photosynthetic reaction centres: hypothesis-testing using parsimony. J Theor Biol. 1990 Aug 23;145(4):535–545. doi: 10.1016/s0022-5193(05)80487-4. [DOI] [PubMed] [Google Scholar]
- Bengis C., Nelson N. Purification and properties of the photosystem I reaction center from chloroplasts. J Biol Chem. 1975 Apr 25;250(8):2783–2788. [PubMed] [Google Scholar]
- Biggins J. Evaluation of selected benzoquinones, naphthoquinones, and anthraquinones as replacements for phylloquinone in the A1 acceptor site of the photosystem I reaction center. Biochemistry. 1990 Aug 7;29(31):7259–7264. doi: 10.1021/bi00483a014. [DOI] [PubMed] [Google Scholar]
- Cozens A. L., Walker J. E. The organization and sequence of the genes for ATP synthase subunits in the cyanobacterium Synechococcus 6301. Support for an endosymbiotic origin of chloroplasts. J Mol Biol. 1987 Apr 5;194(3):359–383. doi: 10.1016/0022-2836(87)90667-x. [DOI] [PubMed] [Google Scholar]
- Deisenhofer J., Michel H. Nobel lecture. The photosynthetic reaction centre from the purple bacterium Rhodopseudomonas viridis. EMBO J. 1989 Aug;8(8):2149–2170. doi: 10.1002/j.1460-2075.1989.tb08338.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fish L. E., Kück U., Bogorad L. Two partially homologous adjacent light-inducible maize chloroplast genes encoding polypeptides of the P700 chlorophyll a-protein complex of photosystem I. J Biol Chem. 1985 Feb 10;260(3):1413–1421. [PubMed] [Google Scholar]
- Hauska G. Phylloquinone in photosystem I: are quinones the secondary electron acceptors in all types of photosynthetic reaction centers? Trends Biochem Sci. 1988 Nov;13(11):415–416. doi: 10.1016/0968-0004(88)90206-X. [DOI] [PubMed] [Google Scholar]
- Heijne G. The distribution of positively charged residues in bacterial inner membrane proteins correlates with the trans-membrane topology. EMBO J. 1986 Nov;5(11):3021–3027. doi: 10.1002/j.1460-2075.1986.tb04601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Komiya H., Yeates T. O., Rees D. C., Allen J. P., Feher G. Structure of the reaction center from Rhodobacter sphaeroides R-26 and 2.4.1: symmetry relations and sequence comparisons between different species. Proc Natl Acad Sci U S A. 1988 Dec;85(23):9012–9016. doi: 10.1073/pnas.85.23.9012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel H., Weyer K. A., Gruenberg H., Dunger I., Oesterhelt D., Lottspeich F. The 'light' and 'medium' subunits of the photosynthetic reaction centre from Rhodopseudomonas viridis: isolation of the genes, nucleotide and amino acid sequence. EMBO J. 1986 Jun;5(6):1149–1158. doi: 10.1002/j.1460-2075.1986.tb04340.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitschke W., Feiler U., Rutherford A. W. Photosynthetic reaction center of green sulfur bacteria studied by EPR. Biochemistry. 1990 Apr 24;29(16):3834–3842. doi: 10.1021/bi00468a005. [DOI] [PubMed] [Google Scholar]
- Nitschke W., Rutherford A. W. Photosynthetic reaction centres: variations on a common structural theme? Trends Biochem Sci. 1991 Jul;16(7):241–245. doi: 10.1016/0968-0004(91)90095-d. [DOI] [PubMed] [Google Scholar]
- Nitschke W., Sétif P., Liebl U., Feiler U., Rutherford A. W. Reaction center photochemistry of Heliobacterium chlorum. Biochemistry. 1990 Dec 18;29(50):11079–11088. doi: 10.1021/bi00502a010. [DOI] [PubMed] [Google Scholar]
- Oh-oka H., Takahashi Y., Kuriyama K., Saeki K., Matsubara H. The protein responsible for center A/B in spinach photosystem I: isolation with iron-sulfur cluster(s) and complete sequence analysis. J Biochem. 1988 Jun;103(6):962–968. doi: 10.1093/oxfordjournals.jbchem.a122394. [DOI] [PubMed] [Google Scholar]
- Orlean P., Albright C., Robbins P. W. Cloning and sequencing of the yeast gene for dolichol phosphate mannose synthase, an essential protein. J Biol Chem. 1988 Nov 25;263(33):17499–17507. [PubMed] [Google Scholar]
- Reznikoff W. S., Siegele D. A., Cowing D. W., Gross C. A. The regulation of transcription initiation in bacteria. Annu Rev Genet. 1985;19:355–387. doi: 10.1146/annurev.ge.19.120185.002035. [DOI] [PubMed] [Google Scholar]
- Schägger H., von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987 Nov 1;166(2):368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- Shinozaki K., Ohme M., Tanaka M., Wakasugi T., Hayashida N., Matsubayashi T., Zaita N., Chunwongse J., Obokata J., Yamaguchi-Shinozaki K. The complete nucleotide sequence of the tobacco chloroplast genome: its gene organization and expression. EMBO J. 1986 Sep;5(9):2043–2049. doi: 10.1002/j.1460-2075.1986.tb04464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J. M., Dowson C. G., Spratt B. G. Localized sex in bacteria. Nature. 1991 Jan 3;349(6304):29–31. doi: 10.1038/349029a0. [DOI] [PubMed] [Google Scholar]
- Tanaka M., Haniu M., Yasunobu K. T., Evans M. C., Rao K. K. Amino acid sequence of ferredoxin from a photosynthetic green bacterium, Chlorobium limicola. Biochemistry. 1974 Jul 2;13(14):2953–2959. doi: 10.1021/bi00711a026. [DOI] [PubMed] [Google Scholar]
- Trost J. T., Blankenship R. E. Isolation of a photoactive photosynthetic reaction center-core antenna complex from Heliobacillus mobilis. Biochemistry. 1989 Dec 26;28(26):9898–9904. doi: 10.1021/bi00452a003. [DOI] [PubMed] [Google Scholar]
- Webber A. N., Malkin R. Photosystem I reaction-centre proteins contain leucine zipper motifs. A proposed role in dimer formation. FEBS Lett. 1990 May 7;264(1):1–4. doi: 10.1016/0014-5793(90)80749-9. [DOI] [PubMed] [Google Scholar]
- Woese C. R. Bacterial evolution. Microbiol Rev. 1987 Jun;51(2):221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]