Abstract
The male-specific regions of primate Y-chromosomes (MSY) are enriched for multi-copy genes highly expressed in the testis. These genes are located in large repetitive sequences arranged as palindromes, inverted-, and tandem repeats termed amplicons. In humans, these genes have critical roles in male fertility and are essential for the production of sperm. The structure of human and chimpanzee amplicon sequences show remarkable difference relative to the remainder of the genome, a difference that may be the result of intense selective pressure on male fertility. Four subspecies of common chimpanzees have undergone extended periods of isolation and appear to be in the early process of subspeciation. A recent study found amplicons enriched for testis-expressed genes on the primate X-chromosome the target of hard selective sweeps, and male-fertility genes on the Y-chromosome may also be the targets of selection. However, little is understood about Y-chromosome amplicon diversity within and across chimpanzee populations. Here, we analyze nine common chimpanzee (representing three subspecies: Pan troglodytes schweinfurthii, Pan troglodytes ellioti, and Pan troglodytes verus) and two bonobo (Pan paniscus) male whole-genome sequences to assess Y ampliconic copy-number diversity across the Pan genus. We observe that the copy number of Y chromosome amplicons is variable among chimpanzees and bonobos, and identify several lineage-specific patterns, including variable copy number of azoospermia candidates RBMY and DAZ. We detect recurrent switchpoints of copy-number change along the ampliconic tracts across chimpanzee populations, which may be the result of localized genome instability or selective forces.
Keywords: chimpanzee, bonobo, Y chromosome, amplicon, DAZ
Introduction
In mammals, males paternally inherit the Y chromosome, which contains the genetic content for canalizing embryos toward masculine development. As would be expected given the male-specific inheritance, many Y-linked genes are spermatogenesis factors with testis-specific expression (Lahn et al. 2001). These spermatogenesis genes are multi-copy and found in long stretches of repetitive DNA termed amplicons (Skaletsky et al. 2003; Hughes et al. 2010). Interchromosomal duplications of amplicons on the Y resulted in the formation of amplicon families, which share a high degree of identity among members. Amplicons are found on the Y as tandem repeats, inverted repeats, and palindromes. These formations can have mutational consequences for the genes they harbor. Palindromes and inverted repeats introduce large hairpin formations that physically align homologous sequence. In this arrangement, gene conversion between amplicon arms can occur, which may reverse deleterious mutations found within the multi-copy genes (Rozen et al. 2003). The majority of the Y does not undergo recombination with the X. Therefore, in the absence of a mechanism for rescue, the transmission and persistence of Y-linked deleterious mutations in a population would be inevitable (Bachtrog 2013). The amplicons are thought have to evolved in response to strong selection of spermatogenesis traits in mammals as protection against deleterious mutation. On the other hand, tandem repeats introduce a susceptibility of amplification or deletion of the interstitial sequence through non-allelic homologous recombination (NAHR), which is a frequent cause of gain or loss of genes in the human genome (Gu et al. 2008; Kidd et al. 2010).
Amplicons undergo constant change in the human lineage with structural polymorphisms accruing at ∼10,000 times the rate of nucleotide polymorphism (Repping et al. 2006). Many of these structural polymorphisms are located in the amplicon regions termed AZF b and c (azoospermia factor regions b and c). Large deletions of the AZFc (1.6–3.5 Mb) are acquired through NAHR and strongly associated with male infertility (Rozen et al. 2012). The genes within the AZFc include specific copies of DAZ (Deleted in Azoospermia), CDY (Chromodomain Y), BPY2 (Basic Protein Y, 2), and PRY (PTPN13-Like, Y-Linked). Each of these genes is found in a different amplicon family named after the colors used to denote them on initial Y-chromosome assemblies: DAZ is found in the red amplicon, CDY in yellow, BPY2 in green, and PRY in blue (Hughes and Page 2015). The most penetrant AZFc deletion reported is rare (1/2,320) and is caused by a NAHR event between two blue arms (b2/b4) and increases the risk of azoospermia by 145-fold (Rozen et al. 2012). The critical genes in the AZFb region are copies of RBMY (RNA-Binding Motif Protein Y) located in the teal amplicon and additional copies of CDY. While deletions within the AZFb appear to be more rare in human populations and contribute less to the overall genetic contribution of azoospermia, AZFb deletions have been reported in patients with spermategenic failure (Longepied et al. 2010). These epidemiological studies suggest that the amplicons harbor genes essential for the progression of germ cells to haploid stages and that the copy number of these genes has functional consequences.
The sequencing of a Western Chimpanzee (Pan troglodyte verus) Y revealed gross structural differences with the human sequence, specifically in the amplicon regions (Kuroki et al. 2006). The chimpanzee amplicon region is richer in content, in terms of the number unique families and copy number of members. The combined length of the chimpanzee amplicon regions are 14.7 Mb compared with the human amplicon regions, which is ∼10.2 Mb in sequence (Hughes et al. 2010). Interestingly, some of the AZFc amplicons and genes were not equivalent in copy number in the two primate species, including: BPY2 (human reference: 3, chimp reference: 2), CDY (human reference: 4, chimpanzee reference: 5), and PRY (human reference: 2, chimpanzee reference: 0). In contrast, DAZ and RBMY were found at the same copy number in the chimpanzee and human reference assembly, 4 and 6, respectively.
The Western chimpanzee is one of four recognized subspecies, which based on a recent Y-chromosome inferred phylogeny diverged from a common ancestor 1.15 Ma (Hallast et al. 2016). The Central chimpanzee of Central Africa (Pan troglodytes troglodytes) shows genetic evidence of a long-standing large population size (Wegmann and Excoffier 2010) and is thought to be the oldest subspecies. The Eastern chimpanzee (Pan troglodytes schweinfurthii) emerged from the Central chimpanzees ancestors by a founder event and is the most recent sub-speciation event among chimpanzees (Hey 2010). Moving westward in Africa from the Central-Eastern chimpanzee distribution, two related subspecies are recognized: Nigerian-Cameroon chimpanzee (Pan troglodytes ellioti) and Western chimpanzee (Gonder et al. 2011). The four chimpanzee subspecies show enough genetic distance for speculation that they may be in the early stages of speciation (Hey 2010). Additionally, bonobos (Pan paniscus) are a sister species to the common chimpanzee in the Pan genus, and reside in the Congo Basin between the current Eastern and Central chimpanzee distributions.
Similar to humans, copy number variation (CNV) of amplicon genes has been observed in chimpanzees and bonobos using florescence in situ hybridization (FISH) experiments (Repping et al. 2006; Schaller et al. 2010). However, these experiments focused on copy number of CDY and DAZ. Furthermore, a recent analysis of primate X-chromosome revealed that amplicons enriched for testis-specific genes are the target of hard-selective sweeps, suggesting that male-fertility traits may be the targets of strong selective forces in chimpanzees (Nam et al. 2015). Despite these recent advancements, there are limited data of Y-linked amplicon content in the Pan lineage and given the dramatic structural changes identified in humans, we suspected the subspecies might harbor lineage-specific amplicon content not present in the panTro4 reference.
A challenge in the completion of a mammalian genome sequence is an accurate assembly of the amplicon regions of the Y-chromosome. Among the great apes, all four genera (Gorilla, Pongo, Pan, and Homo) have high-resolution reference genome assemblies, however, only the chimpanzee and human have fully sequenced Y-chromosomes (Skaletsky et al. 2003; Hughes et al. 2010; Locke et al. 2011; Scally et al. 2012). The amplicon regions of Y-chromosomes have been assembled with single-haplotype iterative mapping and sequencing (SHIMS), a clone-by-clone approach that identifies overlapping inserts through sequence-tagged sites (STS) content assayed with PCR, sanger sequencing of BAC end sequences, and restriction enzyme patterns (Hughes and Page 2015). At this point, the extensive work required to assemble the amplicon sequence of Y-chromosomes limits our perspective of Y-chromosome diversity across great ape species and populations despite recent sequencing efforts (Prado-Martinez et al. 2013). Alternatively, a draft gorilla Y chromosome has been mostly assembled through flow sorting Y-chromosomes and applying short- and long-read genome and transcriptome sequencing (Tomaszkiewicz et al. 2016). However, with this approach the ampliconic structure of the Y-chromosome remains poorly resolved.
To address this gap in our understanding of chimpanzee Y-chromosomes, we developed a method to estimate copy number of amplicons and their nested genes from next-generation sequencing data. Our method utilizes short read data by counting a pre-defined set of amplicon k-mers (oligmers of length k) identified in the reference that match strict stringency thresholds for conservation within amplicon families and a lack of non-specificity. We first applied our method to an exploratory set of high coverage males from the 1000 Genomes (1KG) data set (Genomes Project et al. 2012). We validated computationally derived copy number estimates of amplicons with digital droplet PCR (ddPCR) and a found strong concordance in results (table 1). We next applied our method to a comprehensive analysis of a diverse set of 11 chimpanzee and bonobo male samples sequenced by the Great Ape Genome Project GAGP (Prado-Martinez et al. 2013). We show that three chimpanzee subspecies evaluated here (Western Chimpanzee, Eastern Chimpanzee, and Nigerian-Cameroon Chimpanzee) and bonobos have distinctive Y-chromosome amplicon content, including variable copy number of AZFb and AZFc orthologs, RBMY and DAZ.
Table 1.
ddPCR Validation of Quick-mer AZFc Copy-Number Estimates in 1KG Samples
| Sample | Haplogroup | Blue Amplicon (Quick-mer Median/ddPCR) | Green Amplicon (Quick-mer Median/ddPCR) | Red Amplicon (Quick-mer Median/ddPCR) | Teal Amplicon (Quick-mer Median/ddPCR) |
|---|---|---|---|---|---|
| NA11994 | R1b1a2a1a2b1a2a1 | 3.99/3.78 | 3.05/2.81 | 3.84/3.83 | 1.99/1.82 |
| NA12155 | R1a1a1a | 3.88/3.81 | 3.08/2.88 | 3.77/3.87 | 1.94/1.97 |
| NA12342 | R1b1a2a1a2b3b | 3.97/3.95 | 2.95/3.14 | 3.84/3.85 | 1.93/1.97 |
| NA18622 | O3a2c1a | 3.9/3.94 | 3.34/2.84 | 3.79/3.93 | 2/1.95 |
| NA18623 | O3a2c1a | 3.85/3.99 | 3.22/3.01 | 3.81/3.88 | 1.92/1.99 |
| NA18636 | O2a1 | 3.82/3.9 | 3.18/2.82 | 3.74/3.88 | 1.86/1.89 |
| NA19119 | E1b1a1a1g1a | 3.87/3.87 | 2.95/2.84 | 3.66/3.76 | 1.83/1.96 |
| NA19213 | E1b1a1a1f1a1d | 3.88/3.78 | 3.02/3 | 3.82/3.87 | 1.91/1.89 |
| NA18519 | E1b1a1a1f1a1 | 3.87/3.68 | 2.98/2.83 | 3.73/3.78 | 1.95/1.95 |
Materials and Methods
Sample Selection and Data Processing
We analyzed 11 individuals previously sequenced by the Great Ape Genome Project with evidence of minimal sequence contamination (Prado-Martinez et al. 2013). Selected samples included two bonobos (Pan paniscus: A919_Desmond, A925_Bono), and nine chimpanzees from three subspecies including four Nigerian-Cameroon chimpanzees (Pan troglodytes ellioti: Akwaya_Jean, Basho, Damian, Koto), two Eastern chimpanzees (Pan troglodytes schweinfurthii: 100037_Vincent, A910_Bwambale), two Western chimpanzees (Pan troglodytes verus: 9668_Bosco, Clint), and one Western/Central-hybrid (9730_Donald) (supplementary table S2, Supplementary Material online). Analysis utilized reads mapped to the panTro-2.1.4 (UCSC panTro4) assembly processed as previously described (Prado-Martinez et al. 2013). We included human sample HGDP00222 (Pathan) (Martin et al. 2014) as an out-group. Human reads were mapped to panTro4 using bwa (Li and Durbin 2009) (version 0.5.9) with options aln –q 15 –n 0.01 and sampe -o 1000 and processed as previously described using Picard (version 1.62) and the Genome Analysis Toolkit (version 1.2-65) (McKenna et al. 2010). Our human data additionally includes publically available read alignments (merge of mapped and unmapped bam files) from nine human individuals from the 1000 genomes project (NA12342, NA11994, NA12155, NA18623, NA18622, NA18636, NA19213, NA18519, and NA19119). Reads from were aligned to human reference assembly hg19 and processed following 1KG consortium guidelines (Genomes Project et al. 2012).
Amplicon Analysis
The Y chromosome sequence assembly in panTro4 differs slightly from that originally reported (Hughes et al. 2010). We therefore determined the boundaries of amplicon and palindrome units defined by Hughes et al. using the dotplot software Gepard (Krumsiek et al. 2007). We follow the original color-based naming of amplicons, but additionally provide a unique number for each amplicon family (see supplementary table S2, Supplementary Material online). We assessed copy number of each amplicon family using Quick-mer, a novel pipeline for paralog-specific copy-number analysis. Briefly, QuicK-mer utilizes the Jellyfish-2 (Marcais and Kingsford 2011) program to efficiently tabulate the depth of coverage of a predefined set of k-mer sequences (here, k = 30) in a set of sequencing reads. The resulting k-mer counts are normalized to account for effects of local GC percentage on read depth (Alkan et al. 2009). For this analysis, we utilized two sets of k-mers: 30 mers determined to be unique throughout the genome, and k-mers determined to be specific to each individual amplicon family. For example, as there are four copies of the “red” amplicon in panTro4, we identified 93,200 k-mers present in each copy of the red amplicon and otherwise absent from the reference (Shen and Kidd 2015).
To identify candidate k-mers specific to each amplicon family, the sequences of the amplicon family were extracted from the reference sequence and then blatted to confirm that the sequence was found at the expected copy number in the reference. Next, 30-bp k-mers found across the amplicon set were tabulated using Jellyfish-2 and those k-mers present at the expected copy number were extracted. Genome wide unique k-mer candidates were identified based on Jellyfish analysis of the genome reference assembly. In all cases, Jellyfish-2 was ran in such a way that a k-mer and its reverse complement were considered to be identical. We applied a series of three quality control filters to remove k-mers that share identity with other sequence in the reference genome. First, we predefined a mask consisting of 15mers that are overrepresented in the genome. Any candidate 30mer intersecting with this mask was eliminated. Second, all matches against the reference genome within two substitutions were identified using mrsFAST (Hach et al. 2010) and any k-mer with >100 matches was eliminated. Finally, all mapping locations within an edit distance of 2, including indels, were identified using mrFAST (Alkan et al. 2009; Xin et al. 2013) and any k-mer with >100 matches was filtered from the analysis. For RBMY, we were unable to identify k-mers at the expected copy number in the reference due to prevalent pseudogenization. We increased the expected copy number in the panTro4 reference to 10 from 6 to maximize k-mer coverage.
Depth for genome-wide unique and amplicon-specific k-mers was then determined using Quick-mer (Shen and Kidd 2015). GC normalization curves were calculated based on autosomal regions not previously identified as copy-number variable. K-mer depths were converted to copy-number estimates by dividing by the average depth of Y chromosome k-mers that passed the callable genome mask filter in the X-degenerate region on the chimpanzee Y. Prior to analysis all Y chromosome k-mers with a depth >0 in the female chimpanzee sample Julie_A959 (SAMN01920535) were removed. K-mer by position plots and heirachical clustering was created with R.
ddPCR Validation of Quick-Mer
CNV calls for select amplicons were validated in genomic DNA from human samples NA12342, NA11994, NA12155, NA18623, NA18622, NA18636, NA19213, NA18519, and NA19119 (Coriell, Camden, NJ) using Droplet Digital PCR technology (ddPCR) (BioRad, Hercules, CA). We digested 1 μg of DNA at 37 °C overnight in a 50-μl reaction of 1xNEB4 with 2.5 U of BsmI and HindIII. The following morning, an additional 2 U of each enzyme was added and the digest continued for another hour. After incubation, the reaction was diluted with 50 μl of H2O and 8 μl was used for ddPCR. Primer/probe assays were designed with Primer3Plus software http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi, last accessed February 23, 2016 using settings recommended for ddPCR by BioRad and synthesized at manufacturer’s recommended primer/probe ratio with a 6-FAM (Fluorescein) label on the target probes, a HEX label on the reference probes, and an Iowa Black Quencher on both probes (Integrated DNA Technologies, Coralville, IA). Multiple primer/probe sets were designed for each locus and reference then the optimal set selected based on droplet cluster formation with minimal rain and agreement with locus CNV in a reference genome (supplementary table S1, Supplementary Material online). For ddPCR workflow, a 20-μl mixture containing 0.9 μM primer, 0.25 μM probe, 8 μl (80 ng) of digested DNA, and 1× Bio-Rad ddPCR Supermix for Probes (no dUTP) (BioRad, Hercules, CA) was emulsified, droplets were transferred to 96-well reaction plate, and plate was heat-sealed with foil using manufacturer’s recommended reagents and conditions. We amplified the samples with the following PCR cycling protocol: 10 min at 95 °C, 40 cycles of 30 s at 94 °C and a 1-min extension at 60 °C, followed by 10 min at 98 °C and a hold at 8 °C. After PCR, droplets were read on droplet reader and data was analyzed using manufacturer’s recommended reagents. “Positive” droplets were identified by fluorescence intensity and thresholds were determined manually for each experiment. We set thresholds based on the ddPCR workflow previously described by (Handsaker et al. 2015). Thresholds were set above the cluster of droplets that were negative for target and reference fluorophores using QuantaCell software. Droplets above this minimum amplitude threshold were counted as positive.
Phylogenetic Analysis
To identify a robust set of variants, we imposed a series of regional and site-level filters similar to the procedure utilized for the human MSY (Poznik et al. 2013). First, we generated a regional callability mask, which defines the regions across the chromosomes that yield reliable genotype calls with short read sequence data. We calculated average filtered read depth and the MQ0 ratio (the number of reads with a mapping quality of zero divided by total read depth) in contiguous 1-kb windows based on the output of the GATK Unified Genotyper ran in emit all sites mode. We computed an exponentially weighted moving average (EWMA) across the windows and removed regions that deviated from a narrow envelope that excluded the tails of the read depth and MQ0 distributions.
We next applied a series of site-level filters to the individual positions that passed the regional masking (supplementary fig. S2, Supplementary Material online). We excluded positions with MQ0 fraction < 0.10, that contained missing genotypes in at least one sample, or contained a site where the maximum likelihood genotype was heterozygous in at least one sample. Of the remaining sequence, we plotted the distribution of site-level depths and removed positions with depths at the tail ends of the distribution. We excluded all tri-allelelic sites and further filtered variants within 5 bp of indels. Separate regional callability masks and site-level filters were generated for the human and the combined chimp-bonobo samples and positions that failed either mask were dropped from the analysis. The combined call set consisted of 4,233,540 callable basepairs on the Y-chromosome. A neighbor-joining tree (500 bootstraps) of the MSY callable sites was generated in MEGA 6.06 using the Tamura–Nei substitution model (Tamura et al. 2013).
Results
Validation in Human Reference Populations
We first applied our method to five AZFb and AZFc amplicons, noted for recurrent deletions in azoospermia cases. We mapped the positions of the yellow, red, blue, teal, and green amplicons from a Y chromosome (hg19) self-alignment represented as a dot-plot. Stretches of self identify (∼150–500 kb) within the AZFc region were annotated as amplicons (supplementary table S2A, Supplementary Material online). In total, we identified two copies of the yellow amplicon, four copies of the red amplicon, four copies of the blue amplicon, two copies of the teal amplicon, and three copies of the green amplicon. Our amplicon definitions are nearly identical to those previously reported by Hughes et al., though one minor difference is that we mapped amplicon boundaries where homology is shared across all members of the family (i.e., all members within a family are close to equal in amplicon length and the copy number of an amplicon should be consistent by position based on the hg19 assembly). We extracted sequence within our amplicon boundaries and generated a set of k-mers conserved within and specific to each amplicon family, gene, and spacer (see Materials and Methods). This predefined reference set of k-mers was used as input for the paralog sensitive k-mer counting pipeline, Quick-mer (Shen and Kidd 2015). Quick-mer counts the depth of a set of k-mers from short-read data and estimates the copy number relative to a specified reference sequence of a known copy number. As our copy-number reference sequence, we used the X-degenerate region found on the Y-chromosome, where massive CNV is infrequent and we expected to be at a copy number of one on most Y-haplotypes.
We applied Quick-mer and our pre-defined set of amplicon k-mers to a diverse set of high coverage samples (>6×) from the 1KG data sets. Our 1KG set includes nine samples representing three clades of the Y-chromosome tree: E, R, and O. The E, R, and O Y chromosomes are commonly found among European, African, and Asian peoples, respectively. While DNA or short read data from the biological reference sample (RP-11) is not publically available, data reported in the literature confirms that most individuals carry the same copy number as the reference AZFc haplotype (Repping et al. 2006). Applying Quick-mer to the study population revealed estimates matching the reference AZFc structure in nine of nine samples (supplementary fig. S1, Supplementary Material online). To validate our Quick-mer estimates, we used digital droplet PCR (ddPCR) to replicate our results. We designed our ddPCR primers within blocks of contiguous amplicon k-mers and used X-degenerate sequence as a copy number one control. An abundance of mid-level amplitude droplets, termed “rain”, is the result of poor target accessibility. After excluding yellow amplicon ddPCR results due to excessive “rain”, we were able to validate Quick-mer results in the remaining four amplicon families (table 1).
Pan Amplicon Diversity
We applied the same procedure outlined above for mapping amplicons on the panTro4 version of the Y-chromosome. In total, we identified the 51 units across 10 amplicon families previously annotated by Hughes et al. 2010 (supplementary table S2B, Supplementary Material online). A numbering scheme for the amplicon families has been added as an alternative to the color naming system (fig. 1). Additionally, we mapped the unique “spacer” sequence between palindrome arms and ampliconic-gene positions (based on refSeq and ensembl) from the UCSC genome browser (Karolchik et al. 2003) (supplementary table S2C, Supplementary Material online). As a control, we compared Quick-mer amplicon copy number estimates estimated from Clint to the panTro4 reference (tables 2 and 3). After rounding median k-mer values to the nearest integer, we find strong agreement between the values, suggesting that Quick-mer provides accurate estimates of amplicon copy number. However, a few disagreements between our estimate of Clint’s copy number and the reference sequence are evident. Quick-mer estimates Clint’s copy number of “violet arms” based on the median depth to be 11.54 (SD = 2.53) compared with the 13 represented in the reference assembly. Our copy number estimates are also different from the reference in the CDY (Clint: 3.98 vs. panTro4: 5), VCY (Clint: 0.77 vs. panTro4: 2), and RBMY (Clint 6 vs. panTro4: 10.91) genes. These differences likely arise from truncated copies of the genes and noise in our analysis due to the relatively short length of these genes. However, assembly errors in the panTro4 reference may also be a contributing factor. We also measured copy number a human sample (HGDP00222) based on the same 30-mers used to estimate chimpanzee copy number. Many of our copy number estimates of HGDP00222 match the structures present in the human reference sequence as reported (Hughes et al. 2010). Observed differences between the copy number present in the human assembly (hg19) and HGDP00222 reflect a loss of chimpanzee-human homology for some 30-mers or amplicon polymorphism differences between HGDP00222 and the hg19 reference.
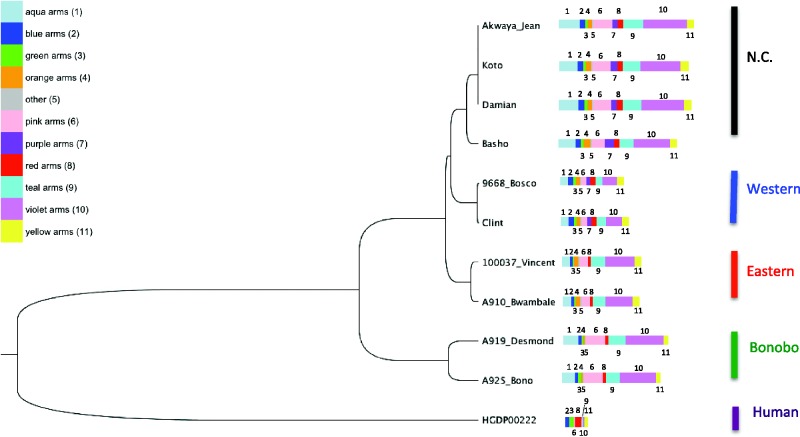
Fig. 1.—
Y chromosome tree and amplicon copy number. The phylogenetic relationship among sequenced Pan and human Y chromosomes is illustrated with neighbor joining tree using a Tamura–Nei substitution model. The size of the color bars to the right of the tree indicates relative median copy number of each amplicons estimated per sample. A numbering scheme of amplicons is included in parenthesis in the legend. Note: The order of the color bars does not reflect the spatial distribution of amplicons. N.C., Nigerian-Cameroon.
Table 2.
Copy Number of Amplicons by Pan Population
| Hughes et al. (2010) (Human) | Hughes et al. (2010) (Chimpanzee) | Clint | Human | Eastern Averagea | Western Averagea | Nigerian- Cameroon Averagea | Bonobo Averagea | |
|---|---|---|---|---|---|---|---|---|
| Aqua arms | 0 | 6 | 5.49 | 0 | 5.9 | 5.53 | 13.43 | 10.16 |
| Aqua spacer | 0 | 3 | 3.22 | 0 | 2.91 | 3.11 | 6.93 | 6.3 |
| Blue arms | 4 | 4 | 3.84 | 2.66 | 1.98 | 3.85 | 3.86 | 2.2 |
| Green arms | 3 | 2 | 1.87 | 3.43 | 1.04 | 1.91 | 1.93 | 1.72 |
| Orange arms | 0 | 3 | 2.7 | 0 | 3.16 | 2.77 | 3.73 | 1.05 |
| Other | 0.93 | 0 | 0.93 | 0.92 | 0.92 | 0.94 | ||
| Pink arms | 0 | 4 | 3.86 | 0.95 | 5.99 | 3.81 | 11.87 | 13.47 |
| Purple armsb | 0 | 3 | 2.82 | 0 | 0 | 2.78 | 4.74 | 0 |
| Red arms | 4 | 4 | 3.92 | 4.39 | 2.01 | 3.89 | 3.92 | 2.26 |
| Red spacer | 2 | 2 | 1.87 | 1.52 | 1.06 | 1.92 | 1.96 | 1.09 |
| Teal arms | 2 | 7 | 6.68 | 0.95 | 9.66 | 5.46 | 12.4 | 11.13 |
| Teal spacer | 0 | 3 | 2.84 | 0 | 4.89 | 2.21 | 5.71 | 4.76 |
| Violet arms | 0 | 13 | 11.54 | 1.14 | 20.08 | 10.63 | 28.57 | 26.44 |
| Violet spacer | 0 | 7 | 6.84 | 0 | 10.29 | 6.22 | 14.72 | 13.52 |
| Yellow arms | 2 | 5 | 4.87 | 2.65 | 5.02 | 4.9 | 5.08 | 3.34 |
| Yellow spacer | 0 | 2 | 1.83 | 2.3 | 2.09 | 1.95 | 1.95 | 1.04 |
| Yellow spacer | 0 | 3 | 2.69 | 2.72 | 3.1 | 2.72 | 3.15 | 2.1 |
Note.—aAverage median value, bAdditional purple amplicon mapped in the panTro4 assembly.
Table 3.
Copy Number of Ampliconic Genes by Pan Population
| Hughes et al. (2010) (Human) | Hughes et al. (2010) (Chimpanzee) | Clint | Human | Eastern (Average) | Western (Average) | Nigerian-Cameroon (Average) | Bonobo (Average) | |
|---|---|---|---|---|---|---|---|---|
| BPY2 | 3 | 2 | 1.81 | 3.61 | 1.1 | 1.86 | 2.07 | 1.79 |
| CDY | 4 | 5 | 3.98 | 4 | 5.86 | 4.51 | 5.41 | 3.59 |
| DAZ | 4 | 4 | 4.37 | 0.19 | 1.76 | 4.11 | 3.85 | 2.26 |
| RBMY | 6 | 6 | 10.91 | 10.32 | 14.34 | 9.16 | 24.12 | 32.28 |
| TSPY | 35 | 6 | 5.46 | 38.85 | 16.6 | 10.27 | 26.35 | 32.1 |
| VCY | 2 | 2 | 0.77 | 0 | 0.44 | 1.01 | 1.41 | 0.05 |
UCSC Genome Browser Gene Identifier: CDY (NM_001145039), DAZ (ENSPTRG00000022523), BPY2 (NM_004678.2), RBMY (NM_001006121.2), TSPY (NM_001077697.2), and VCY (NM_004679.2).
To confirm the Y-chromosomes of our chimpanzee samples are consistent with the autosomal inferred phylogeny, we constructed a SNP callset from the panTro4 Y chromosome. Following the procedure reported by Poznik et al. (2013), we used alignment quality scores to identify regions of the Y chromosome amenable to variant calling. We identified 4.2 Mb of sequence in the X-degenerate regions for variant calling with chimpanzee and human samples. In total, we identified 23,946 and 78,022 SNVs when excluding and including the human sequence, respectively. A neighbor-joining tree confirms that the three chimpanzee subspecies form monophyletic clades (fig. 1). The Western Chimpanzee Donald from the GAGP was previously reported to have Nigerian-Cameroon Chimpanzee admixture. However, from our phylogenetic tree we are able to verify his Y chromosome is of Western Chimpanzee origin and therefore we included him in our analysis as a Western Chimpanzee. We also observe a deep divergence between the bonobo samples, nearly equal to the common ancestor of the chimpanzee in age. As point of comparison, our tree closely resembles a recent MSY inferred chimpanzee phylogeny (Hallast et al. 2016).
We see substantial variance in the copy number of amplicons, spacers, and ampliconic-genes across the chimpanzee subspecies and bonobos (tables 2 and 3 and supplementary table S3A and B, Supplementary Material online). However, within individual Pan lineages we observe an overall conservation of copy number in the ampliconic regions (fig. 1). The teal and violet palindromes found within the 2.2-Mb palindrome array near the centromere are highly variable in copy number (Hughes et al. 2010). This correlated with an increase of the RBMY gene across samples (median k-mer depth; Western: 9.16, Eastern: 14.34, Nigerian-Cameroon: 24.12, bonobos: 32.28) (supplementary fig. S3J and K, Supplementary Material online). We note that our RBMY estimates are likely inflated due to psuedogenization of the gene on the chimpanzee Y-chromosome, as evident in the panTro4 assembly (see Materials and Methods). The Western chimpanzees are also comparably lacking in pink arms and the adjacent TSPY gene: the median copy number of the pink amplicon (3.81) and the TSPY gene (10.27) is one-fourth and one-half of the estimates in Nigerian-Cameroon chimpanzees (pink amplicon: 11.87, TSPY: 26.35) and bonobos (pink amplicon: 13.47, TSPY: 32.10), respectively (supplementary fig. S3T, Supplementary Material online). However, copy number TSPY increases by position in all samples, therefore it is likely our estimates of TSPY copy number (based on k-mer counts) may also include truncated of copies of the gene.
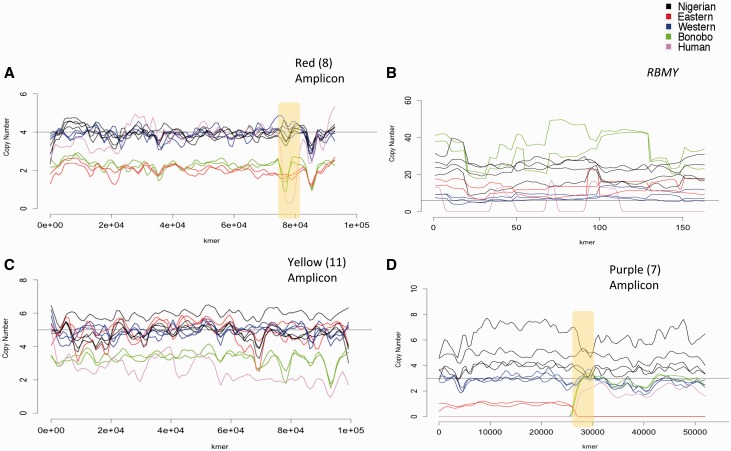
We find that DAZ and the red amplicon ranges in copy number between two (Eastern and bonobos) and four (Nigerian-Cameroon and Westerns) copies. Interestingly, within the red amplicon we observe two exon-spanning deletions in the human and bonobo samples that share a common break point located in intron 15 of DAZ (fig. 2A and supplementary fig. S3Q, Supplementary Material online). A similar event is observed in the purple arms (fig. 2D), where there is amplification of copy number in the Eastern chimpanzees and reduction in the bonobos on the 5′ of a breakpoint, and the reverse of the pattern on the 3′ end.
Fig. 2.—
Chimpanzee and bonobo amplicon copy number by k-mer. Amplicon copy numbers are plotted against k-mer position. Each line represents a single sample colored by species/subspecies and smoothed with lowess function. Copy number of the PanTro4 reference is drawn with a solid black line. Breakpoints of interest mentioned in the text are boxed in yellow. (A) Red amplicon, (B) RBMY, (C) yellow amplicon, and (D) purple amplicon.
Discussion
Our data reveal that amplicons continue to evolve in chimpanzees, leading to distinct gene content found in the Eastern, Western, and Nigerian-Cameroon chimpanzee Y-chromosomes. This finding represents an important step in our understanding of the evolution of sex chromosomes, specifically in the early stages of primate speciation. Although identifying the specific causes of functional diversity is challenging, we suggest that meiotic drive during spermatogenesis may be increasing DAZ, RBMY, and TSPY diversity among chimpanzees and may be a contributor to their ongoing subspeciation (Bachtrog 2014). As a point of comparison, there are no subspecies classifications for bonobos and here we find limited amplicon copy number diversity between individuals, consistent with a previous report (Schaller et al. 2010). This is a surprising observation in light of the high level of nucleotide diversity we detect on the bonobo Y-chromosome (fig. 1).
The DAZ locus is a candidate fertility factor and thought to be at least partially responsible for the azoosperima phenotype in AZFc deletions. Molecular clock analyses of the DAZ locus have revealed considerable evolution as its transposition to the primate Y chromosome from chromosome 3 38.5 Ma (Saxena et al. 1996). Subsequently, the Y chromosome paralog duplicated 33 Ma, prior to the Old World Monkey—Ape split, and today it is found in two copies in the rhesus macaque (Hughes et al. 2012). Comparisons between the chimpanzee and human reference sequence revealed four copies in both assemblies. However, comparative analysis of the bordering palindrome sequences reveals that these are the result of independent amplifications in chimpanzee and human (Skaletsky et al. 2003; Hughes et al. 2010). As noted in a previous study, copy number of DAZ is polymorphic among the common chimpanzees (Schaller et al. 2010). Here we find that Eastern chimpanzees and bonobos each have two copies of DAZ and the red amplicon, while Western and Nigerian-Cameroon have four copies. With respect to copy number, we hypothesize that the bonobos and Eastern represent the ancestral state of the DAZ locus in the Pan lineage. Our results support a scenario where the second DAZ duplication event evident in the panTro4 assembly is the result of a duplication that occurred early in the formation of the Western/Nigerian-Cameroon clade. Furthermore, the DAZ RNA-recognition motif shows substantial positive selection in the chimpanzee reference assembly relative to the human and rhesus macaque assemblies (Hughes et al. 2012). However, if the amplification of DAZ is unique to the Western/Nigerian-Cameroon lineages, we suspect that Y-chromosome selection may be higher in these clades. Hierarchical clustering of the chimpanzee and bonobo samples (based on fold change of amplicon copy number) reveals a Western/Nigerian-Cameroon chimpanzee and bonobo/eastern Chimpanzee divergence that supports this hypothesis (supplementary fig. S4, Supplementary Material online). Interestingly, this tree structure persists even after the removal of the red amplicon from the analysis as this result is also driven by the blue and purple amplicons (table 2). However, we are limited in power to infer phylogeny from our copy-number data due to low counts of unique amplicon structures (n = 10) and samples (n = 11). Although we focused on samples from Prado-Martinez et al. without reported sequence contamination, low levels of cross-sample contamination may exist in this data set. Such contamination would manifest as increased noise in our estimates, particularly if it involved samples with different underlying amplicon structures or copy numbers. Additionally, such analyses are complicated by the potential for recurrent copy number changes along these lineages. If male fertility has influenced the diversification of the common chimpanzee, the DAZ duplication we observe in the Western and Nigerian-Cameroon clade may have played a critical role. Further studies are required to determine if the DAZ is under positive selection in the Central/Eastern chimpanzees and bonobos.
Additionally, copy number of RBMY displayed conspicuous variability across our samples (table 3). While copy number of the RBMY k-mers in the common chimpanzees is relatively constant by position, we observe abrupt changes in bonobo copy number at switch points within the gene (fig. 2B). In chimpanzees, multiple copies of RBMY are nested within the “palindrome array” of violet and teal amplicons, which display amplification in chimpanzee populations and bonobos relative to Western chimpanzees (supplementary table S3A, Supplementary Material online). These results suggest that the palindrome array is subject to structural rearrangement on the Pan Y-chromosome. Furthermore, the conservation of RBMY on the Y-chromosome as marsupials (divergence from placental mammals was ∼160 Ma (Luo et al. 2011) suggests that this locus may have an important physiological role (Delbridge et al. 1997). This stands in contrast to DAZ, which moved from the autosomes to the Y-chromosome exclusively in the primate lineage (Gromoll et al. 1999). Additionally, the testis exhibit extensive alternative splicing of RBMY and it is one of the few genes identified to activate testis-specific splicing events (Yeo et al. 2004; Liu et al. 2009). The RBMY CNVs may result in variable expression of testis-specific isoforms across Pan and could contribute to population-specific male-fertility traits.
We also characterized unique Western chimpanzee sequence within the non-repetitive sequence in the ampliconic region termed “other” (supplementary fig. S7E, Supplementary Material online). We speculate that there may be subspecies-specific amplicons not assayed here. Future studies of the chimpanzee Y-chromosomes would benefit from high-quality MSY assemblies specific for subspecies to identify patterns of gain and loss of unique sequence. Given the potential contribution of ampliconic genes to reproductive phenotypes, incorporating measures of Y-chromosome amplicon structure could complement existing markers used to guide conservation efforts focused on maintaining Pan diversity.
We found that our k-mer counting approach is optimal for detecting CNV of amplicons and more limited with small genes and highly repetitive arrays such as the TSPY. Our method of assaying CNVs of ampliconic genes is also limited in that it does not distinguish between intact and pseudogenized copies. Furthermore, divergence between the reference and the analyzed samples may reduce the accuracy of our approach. However, the bonobos represent a divergence from the panTro4 reference of ∼1–2 Ma and we find that the k-mer by position plots show a minimal increase in noise (Prado-Martinez et al. 2013). In contrast, the analyzed human sample, which represents a ∼6-Ma divergence, displays uneven copy number by position across amplicons and genes (fig. 2C). While the relative quantity of annotated amplicons generated from short read data is reported here, assemblies would provide the spatial diversity of amplicons. With assemblies in hand, one could recreate the non-homologous recombination events that gave rise to the structural diversity within Pan and further clarify the contribution of ampliconic sequence to subspecies evolution.
Supplementary Material
Supplementary figures S1–S4 and S7 and tables S1–S3 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
This work was supported by a grant from the National Institutes of Health, Office of the Director (grant number 1DP5OD009154). We thank Brenna Henn and Jacob Mueller for thoughtful comments on manuscript drafts.
Literature Cited
- Alkan C, et al. 2009. Personalized copy number and segmental duplication maps using next-generation sequencing. Nat Genet. 41:1061–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtrog D. 2013. Y-chromosome evolution: emerging insights into processes of Y-chromosome degeneration. Nat Rev Genet. 14:113–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtrog D. 2014. Signs of genomic battles in mouse sex chromosomes. Cell 159:716–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delbridge ML, et al. 1997. A human candidate spermatogenesis gene, RBM1, is conserved and amplified on the marsupial Y chromosome. Nat Genet. 15:131–136. [DOI] [PubMed] [Google Scholar]
- Genomes Project C, et al. 2012. An integrated map of genetic variation from 1,092 human genomes. Nature 491:56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonder MK, et al. 2011. Evidence from Cameroon reveals differences in the genetic structure and histories of chimpanzee populations. Proc Natl Acad Sci U S A. 108:4766–4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromoll J, et al. 1999. The Old World monkey DAZ (Deleted in AZoospermia) gene yields insights into the evolution of the DAZ gene cluster on the human Y chromosome. Hum Mol Genet. 8:2017–2024. [DOI] [PubMed] [Google Scholar]
- Gu W, Zhang F, Lupski JR. 2008. Mechanisms for human genomic rearrangements. Pathogenetics 1:4.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hach F, et al. 2010. mrsFAST: a cache-oblivious algorithm for short-read mapping. Nat Methods 7:576–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallast P, et al. 2016. Great ape Y Chromosome and mitochondrial DNA phylogenies reflect subspecies structure and patterns of mating and dispersal. Genome Res. 26:427–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handsaker RE, et al. 2015. Large multiallelic copy number variations in humans. Nat Genet. 47:296–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey J. 2010. The divergence of chimpanzee species and subspecies as revealed in multipopulation isolation-with-migration analyses. Mol Biol Evol. 27:921–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes JF, et al. 2010. Chimpanzee and human Y chromosomes are remarkably divergent in structure and gene content. Nature 463:536–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes JF, Page DC. 2015. The Biology and Evolution of Mammalian Y Chromosomes. Annu Rev Genet. 49:507–527. [DOI] [PubMed] [Google Scholar]
- Hughes JF, Skaletsky H, Page DC. 2012. Sequencing of rhesus macaque Y chromosome clarifies origins and evolution of the DAZ (Deleted in AZoospermia) genes. Bioessays 34:1035–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D, et al. 2003. The UCSC Genome Browser Database. Nucleic Acids Res. 31:51–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd JM, et al. 2010. A human genome structural variation sequencing resource reveals insights into mutational mechanisms. Cell 143:837–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumsiek J, Arnold R, Rattei T. 2007. Gepard: a rapid and sensitive tool for creating dotplots on genome scale. Bioinformatics 23:1026–1028. [DOI] [PubMed] [Google Scholar]
- Kuroki Y, et al. 2006. Comparative analysis of chimpanzee and human Y chromosomes unveils complex evolutionary pathway. Nat Genet. 38:158–167. [DOI] [PubMed] [Google Scholar]
- Lahn BT, Pearson NM, Jegalian K. 2001. The human Y chromosome, in the light of evolution. Nat Rev Genet. 2:207–216. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, et al. 2009. The germ cell nuclear proteins hnRNP G-T and RBMY activate a testis-specific exon. PLoS Genet. 5:e1000707.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke DP, et al. 2011. Comparative and demographic analysis of orang-utan genomes. Nature 469:529–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longepied G, et al. 2010. Complete deletion of the AZFb interval from the Y chromosome in an oligozoospermic man. Hum Reprod. 25:2655–2663. [DOI] [PubMed] [Google Scholar]
- Luo ZX, Yuan CX, Meng QJ, Ji Q. 2011. A Jurassic eutherian mammal and divergence of marsupials and placentals. Nature 476:442–445. [DOI] [PubMed] [Google Scholar]
- Marcais G, Kingsford C. 2011. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 27:764–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin AR, et al. 2014. Transcriptome sequencing from diverse human populations reveals differentiated regulatory architecture. PLoS Genet. 10:e1004549.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, et al. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20:1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam K, et al. 2015. Extreme selective sweeps independently targeted the X chromosomes of the great apes. Proc Natl Acad Sci U S A. 112:6413–6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poznik GD, et al. 2013. Sequencing Y chromosomes resolves discrepancy in time to common ancestor of males versus females. Science 341:562–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado-Martinez J, et al. 2013. Great ape genetic diversity and population history. Nature 499:471–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repping S, et al. 2006. High mutation rates have driven extensive structural polymorphism among human Y chromosomes. Nat Genet. 38:463–467. [DOI] [PubMed] [Google Scholar]
- Rozen S, et al. 2003. Abundant gene conversion between arms of palindromes in human and ape Y chromosomes. Nature 423:873–876. [DOI] [PubMed] [Google Scholar]
- Rozen SG, et al. 2012. AZFc deletions and spermatogenic failure: a population-based survey of 20,000 Y chromosomes. Am J Hum Genet. 91:890–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena R, et al. 1996. The DAZ gene cluster on the human Y chromosome arose from an autosomal gene that was transposed, repeatedly amplified and pruned. Nat Genet. 14:292–299. [DOI] [PubMed] [Google Scholar]
- Scally A, et al. 2012. Insights into hominid evolution from the gorilla genome sequence. Nature 483:169–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller F, et al. 2010. Y chromosomal variation tracks the evolution of mating systems in chimpanzee and bonobo. PLoS One 5:1–6. doi: 10.1371/journal.pone.0012482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen F, Kidd J. 2015. QuicK-mer: a rapid paralog sensitive CNV detection pipeline. bioRxiv. doi: 10.1101/028225. [Google Scholar]
- Skaletsky H, et al. 2003. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature 423:825–837. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomaszkiewicz M, et al. 2016. A time- and cost-effective strategy to sequence mammalian Y Chromosomes: an application to the de novo assembly of gorilla Y. Genome Res. 26:530–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegmann D, Excoffier L. 2010. Bayesian inference of the demographic history of chimpanzees. Mol Biol Evol. 27:1425–1435. [DOI] [PubMed] [Google Scholar]
- Xin H, et al. 2013. Accelerating read mapping with FastHASH. BMC Genomics 14(Suppl 1):S13.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo G, Holste D, Kreiman G, Burge CB. 2004. Variation in alternative splicing across human tissues. Genome Biol. 5:R74.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.