Abstract
Triple-negative breast cancer (TNBC) and other molecularly heterogeneous malignancies present a significant clinical challenge due to a lack of high-frequency “driver” alterations amenable to therapeutic intervention. These cancers often exhibit genomic instability, resulting in chromosomal rearrangements that impact the structure and expression of protein-coding genes. However, identification of these rearrangements remains technically challenging. Using a newly developed approach that quantitatively predicts gene rearrangements in tumor-derived genetic material, we identified and characterized a novel oncogenic fusion involving the MER proto-oncogene tyrosine kinase (MERTK) and discovered a clinical occurrence and cell line model of the targetable FGFR3-TACC3 fusion in TNBC. Expanding our analysis to other malignancies, we identified a diverse array of novel and known hybrid transcripts, including rearrangements between non-coding regions and clinically relevant genes such as ALK, CSF1R, and CD274/PD-L1. The over 1000 genetic alterations we identified highlight the importance of considering non-coding gene rearrangement partners, and the targetable gene fusions identified in TNBC demonstrate the need to advance gene fusion detection for molecularly heterogeneous cancers.
INTRODUCTION
Despite advances in precision medicine, the treatment of many molecularly heterogeneous cancers remains challenging due to a lack of recurrent alterations amenable to therapeutic intervention. To make significant clinical advances against these recalcitrant cancers, integrative genomic and molecular analyses are required to understand their complexity and to identify targetable features arising from lower-frequency genetic events. Our laboratory has identified six molecular subtypes of one such heterogeneous disease, triple-negative breast cancer (TNBC), with each subtype displaying unique ontologies and differential response to standard-of-care chemotherapy(1,2). Ongoing genomic analysis of TNBC has identified a low frequency and widely varying clonality of therapeutically actionable alterations across subtypes, including mutations in PIK3CA and BRAF, amplification of EGFR, loss of PTEN, and expression of androgen receptor (AR)(3–5). However, a significant proportion of TNBC cases lack somatic alterations of any established “driver” gene, highlighting the importance of continued genomic analysis and discovery integrated with molecular profiling of the tumor microenvironment(3).
A defining feature of TNBC and other clinically challenging, molecularly heterogeneous cancers such as ovarian carcinoma and lung squamous cell carcinoma is copy number alteration (CNA), which is frequently accompanied by mutation or inactivation of the p53 tumor suppressor(4,6–8). The genomic instability that gives rise to CNA can also result in chromosomal rearrangements and gene fusions, which have long been recognized as oncogenic drivers and effective drug targets in a variety of hematological and solid malignancies(9). In recent years, the advent of next-generation sequencing (NGS) has enabled the identification of a number of gene fusion events in epithelial cancers, with varying frequency and therapeutic relevance(10–14).
In order to broaden our understanding of the somatic alterations underlying TNBC, we sought to identify known and novel gene rearrangements impacting the structure and/or expression level of protein-coding transcripts. While a number of computational approaches have been developed to identify hybrid RNA and DNA sequences using NGS data, numerous technical hurdles complicate accurate and efficient gene rearrangement detection(15). For instance, widespread regions of sequence homology and current limitations in read length result in sometimes-ambiguous alignment and a large number of false positives. To combat this, many detection methodologies employ filters designed to enrich for biologically relevant gene rearrangements, such as restricting both potential fusion partners to protein-coding loci(16). While these filters enrich for currently known gene fusions, they are not effective for the discovery of less canonical events, such as rearrangements between coding and non-coding regions of the genome. We developed a new algorithm, Segmental Transcript Analysis (STA), which uses exon-level expression estimates to rank a population of samples based on their likelihood of harboring a rearrangement in a given gene. We identified a number of known and novel rearrangements involving functionally diverse gene partners in TNBC, and expanded our analysis and discovery to a wider, multi-cancer cohort from The Cancer Genome Atlas (TCGA). The DNA-validated rearrangements that we identified include non-coding portions of the genome acting as both 5’ and 3’ partners in clinically relevant hybrid transcripts.
MATERIALS AND METHODS
Cell Culture
SUM185PE (Asterand, March 2010) and MCF10A cells (ATCC, June 2012) were cultured as previously described(1). Ba/F3 cells (provided by Dr. Christine Lovly, Vanderbilt University, November 2014) were cultured in RPMI + GlutaMAX (Gibco 61870) with 1 ng/mL IL-3 (Life Technologies) and 5% (v/v) FBS (Gemini). All cell lines were maintained in 100 U/mL penicillin and 100 μg/mL streptomycin (Gemini) and tested negative for mycoplasma (Lonza). Cell Line Genetics performed positive short tandem repeat DNA fingerprinting analysis on SUM185PE (March 2011); cells were cultured for fewer than 2 months post-identification. We also verified SUM185PE by manual identification of unique variants in NGS data. DNA fingerprinting analysis was not performed on MCF10A or Ba/F3 cells, but the Ba/F3 cell line displayed the previously published IL-3 dependence phenotype(17).
FGFR3/TACC3 siRNA transfection
SUM185PE cells (8000 cells/well) were reverse transfected in a 96-well format with RNAiMax (0.1 μL, Life Technologies) and siRNAs (1.25 pmole) targeting the FGFR3 exon 4/5 junction (Dharmacon D-003133-06), FGFR3 exon 11 (D-003133-05), TACC3 exon 13 (D-004155-03), TACC3 3’ UTR (D-004155-04), TACC3 exon 4 (D-004155-01), or TACC3 exon 5 (D-004155-02) (siRNA #1-6, respectively). AllStars Negative Control siRNA and AllStars Hs Cell Death Control siRNA (Qiagen) were included as experimental controls. Viability was assessed at 72 h by incubating cells with alamarBlue (Invitrogen). For each experiment, the mean fluorescence value (Ex/Em: 560/590 nm) of four wells per condition was normalized to the negative control.
IC50 determination
SUM185PE cells were seeded in quadruplicate (8000 cells/well) in 96-well plates. After overnight attachment, growth medium was replaced with medium (control) or medium containing half-log serial dilutions of the FGFR inhibitor PD173074 (Selleckchem). Viability was assessed at 72 h by incubating cells with alamarBlue (Invitrogen). Half-maximal inhibitory concentration (IC50) values were determined after normalization to untreated wells and double-log transformation of dose response curves as previously described(18).
Ba/F3 IL-3 withdrawal
Stably transfected Ba/F3 cells were seeded in 6-well plates (40,000 cells/well) in growth medium ± 1 ng/mL IL-3. Live cell counts were manually determined at three sequential 24 h timepoints by visual inspection using a hemocytometer and Trypan blue (Bio-Rad).
Cloning and generation of stable cell lines
The TMEM87B-MERTK expression construct, corresponding to amino acids 1–55 of TMEM87B (NM_032824) and amino acids 433-1000 of MERTK (NM_006343), was synthesized in a pMK-RQ-Bb vector using GeneOptimizer and GeneArt (Life Technologies) and cloned into pBABE-puro(19) (Addgene) by EcoRI/SalI restriction digest and ligation (New England BioLabs). The complete pBABE-puro-TMEM87B-MERTK expression construct sequence is deposited in XXXXXX under accession number XXXXXX. To generate stably transfected cell lines, pBABE-puro-TMEM87B-MERTK or pBABE-puro empty vector retroviruses were packaged in Phoenix cells (Orbigen) and Ba/F3 and MCF10A cells were transduced with virus for two 24 h intervals with 8 μg/mL polybrene (Sigma), then selected and maintained with 1.5 μg/mL (Ba/F3) or 0.5 μg/mL (MCF10A) puromycin (Sigma).
Immunoblotting
SUM185PE cells were lysed 72 h after siRNA transfection. Ba/F3 cells were grown in suspension and incubated for 90 m in the indicated media before lysis. MCF10A cells seeded in 10 cm plates were incubated for 180 m in the indicated growth media before in-well lysis. All cells were lysed in RIPA buffer supplemented with protease and phosphatase inhibitors. Cell lysates (40 μg [SUM185PE] or 30 μg [Ba/F3 and MCF10A]) were separated on polyacrylamide gels and transferred to polyvinyl difluoride membranes (Millipore). Immunoblotting was performed using FGFR3 B-9 (1:200, Santa Cruz), GAPDH MAB374 (1:1000, Millipore), MERTK D21F11 (1:1000, Cell Signaling), phospho-Akt Ser473 D9E (1:2000, Cell Signaling), total Akt (1:1000, Cell Signaling 9272), phospho-Erk1/2 Thr202/Tyr204 D.13.14.4E (1:2000, Cell Signaling), and total Erk1/2 3A7 (1:2000, Cell Signaling).
Segmental Transcript Analysis algorithm
The median-normalized, exon-level RPKM vector for sample i in gene k is denoted as . The Fscore for sample i in gene k is defined as
where is the geometric mean of , ∀i ≠ i′ and d(x, y) is a distance measurement (i.e. Euclidean distance) between x and y. Di is the biggest difference of RPKM with lag 1 in sample i and is the standard deviation of for gene k. For ranking Fscore across genes, the normalized Fscore is defined as
For ranking Gscore between samples, the Segmental Transcript Analysis score is defined as
where is the mean of Gscore for all genes in sample i and is the standard deviation of Gscore for all genes in sample i.
Corresponding R code is included as a supplementary file. Detailed descriptions of file sources, processing, and filtering are available in the supplemental methods.
Transcript and protein annotation
Gene locus and exon data were determined by GENCODE and RefGene annotations as described in the supplemental methods. For exon-level expression plots, the exon order is based on sequential order of exons in the RefGene annotation. The presence of isoforms may cause deviation from the normal exon numbering scheme. For hybrid transcript frame calls, the RefGene annotation was processed to generate starting and ending frame values of 0, 1, 2 (CDS) or -1 (UTR) for each exon. Due to the potential for multiple reading frames at a given coordinate, a hybrid transcript between two exon boundaries featuring any concordance of starting and ending CDS frame was annotated as in-frame. A hybrid transcript between two exon boundaries with non-overlapping CDS frames was annotated as out-of-frame. An exon boundary with exclusively UTR status was annotated accordingly.
The domains and protein features depicted in schematics were obtained from UniProtKB(20). Features were exclusively selected from annotations with “Reviewed” status, and all coding features are to scale.
SUM185PE RNA sequencing
Total RNA was isolated from SUM185PE cells using RNeasy (Qiagen). RNA quality was assessed by NanoDrop (Thermo) and Bioanalyzer (Agilent). Two μg of total RNA were used for the TruSeq Stranded Total RNA Library Prep Kit (Illumina). Libraries were quantified by Qubit (Life Technologies) and qPCR, and library size and quality were assessed by Bioanalyzer (Agilent). The constructed RNA-seq library was sequenced at the Vanderbilt Technologies for Advanced Genomics (VANTAGE) core on an Illumina HiSeq 2500 using a paired-end 100-bp protocol. Reads were de-multiplexed and trimmed using SeqPrep. FASTQ files are deposited in the NCBI Read Sequence Archive under accession number SRPXXXXXX.
SUM185PE RNA-seq reads were aligned using the TCGA RNA-seq v2 pipeline (https://cghub.ucsc.edu/docs/tcga/UNC_mRNAseq_summary.pdf, 7/31/2013 revision) to ensure compatibility with TCGA data. Reference data and custom scripts for exon-level expression quantification were downloaded from the UNC database as referenced in the protocol. Alignment was conducted using the default TCGA workflow and MapSplice v12_07, RSEM v1.1.13, and UBU v1.2. Cell line RNA-seq data were grouped with the TCGA BRCA dataset for STA input and processed using the STA discovery pipeline described in the supplemental methods.
Shah et al. data acquisition
RNA-seq data for 80 TNBC clinical specimens(3) were downloaded from the European Genome-phenome Archive as EGAS00001000132 on 5/18/2014. Aligned BAM files were converted to fastq using bedtools v2.17.0 and were aligned and processed as described in the supplemental methods.
Statistical analysis
All analyses and graphical representations were performed using R v3.2.0. Post-siRNA viability ratios were plotted as the mean ± SEM of four independent experiments, and p-values were calculated using the Wilcoxon rank-sum test with Bonferroni multiple comparison adjustment. PD173074 IC50 values were plotted as the mean ± SEM of three independent experiments. Ba/F3 IL-3 withdrawal data were plotted as the mean ± SD of two conditions with three independent experiments across three days, and p-values were calculated using the restricted maximum likelihood (REML)-based mixed effects model.
Supplementary Information
Additional data (Supplementary Figs. S1–S9), their corresponding legends, and expanded details of sequence file processing are found as supplementary files.
RESULTS
Gene rearrangement prediction by STA
In order to prioritize downstream computation and minimize filters restricting the genomic location of putative rearrangement partners, we developed an algorithm known as Segmental Transcript Analysis (STA) that uses a distance matrix approach to generate aberrant transcript scores for a population of samples (Methods). Based on exon-level expression values across a gene, STA is used to assign a normalized score for each sample by quantifying deviation from the population in both magnitude and directionality of expression. This approach allows the detection of different structural classes of rearrangements – general up- or down-regulation of a transcript might accompany promoter or untranslated region (UTR) swapping, for instance, while abrupt gain or loss of expression from one exon to the next could result from a breakpoint within the intervening DNA. While past discovery approaches using microarray datasets have leveraged exon-level expression comparison in individual transcripts(21), the novel population-based comparison method employed in STA effectively controls for confounding issues such as alternative splicing and normalization artifacts that cause uneven exon-level expression values across genes and facilitates detection of modest but biologically relevant changes in transcript levels.
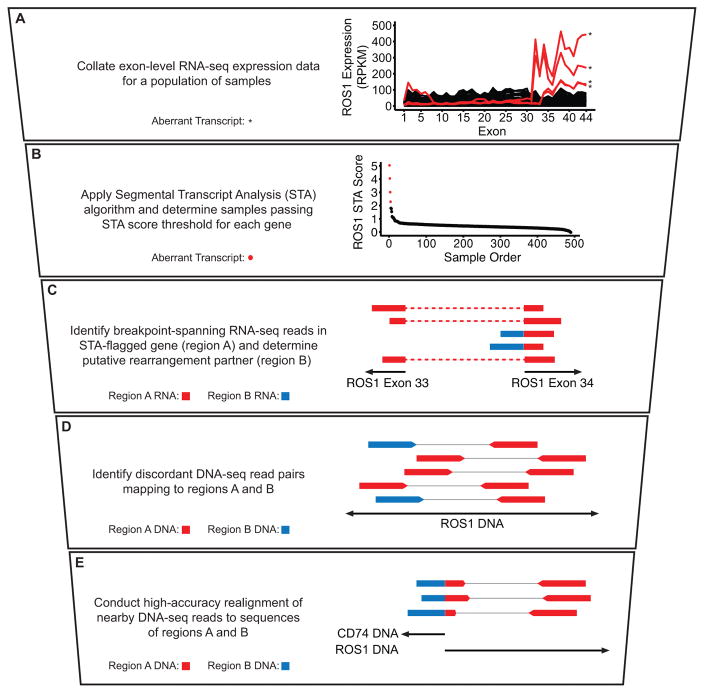
To evaluate the utility of STA as a prediction tool for both known and novel gene rearrangements, we developed a vertically integrated NGS analysis pipeline (Fig. 1). Briefly, exon-level expression data were collated for a given tumor type and STA scores were calculated for each sample on a per-gene basis (Fig. 1A–B). RNA-seq data for each aberrant transcript passing a defined STA score threshold were assessed for evidence of rearrangement between the candidate gene and another genomic region (Fig. 1C), followed by analysis and realignment of nearby whole-genome sequencing reads (WGS) to identify a DNA breakpoint with unique sequence spanning both rearrangement partners (Fig. 1D–E)(22). Samples displaying a discrete breakpoint upon realignment were classified as DNA-validated rearrangements. While reliant upon the availability and adequate sequencing depth of WGS data, this approach provides independent structural evidence for the validity of any detected rearrangements. As a consequence, rearrangements that might be omitted in an RNA sequencing-only approach due to concerns about homology and false positivity, such as hybrid transcripts between coding and non-coding regions of the genome, can be identified with greater confidence.
Figure 1. Quantitative prediction by STA facilitates an integrated fusion detection pipeline.
A–E, Stepwise description of STA discovery pipeline with accompanying schematics and example data. A, Exon-level expression values for a population of samples plotted as continuous lines. Samples passing STA score threshold (B) are plotted in red and denoted by asterisks; samples below the threshold are plotted in black. B, ROS1 STA scores for each sample plotted in descending order. Samples with an STA score of 2 or above are plotted in red; samples with an STA score below 2 are plotted in black. C, Schematic of RNA-seq reads. Dotted lines denote continuous read segments. D, Schematic of discordant whole-genome sequencing (WGS) read pairs. Thin lines represent denote read pairs. E, Schematic of breakpoint-spanning WGS reads identified after realignment. In C–E, colors indicate alignment location of individual read segments, as depicted in the description at left. Schematics are not to scale.
Using test RNA-seq and WGS data from TCGA, we confirmed the ability of STA to identify known gene fusions across multiple cancer types, including CD74-ROS1 in lung adenocarcinoma, NFASC-NTRK1 in glioblastoma, and TMPRSS2-ETV4 in prostate adenocarcinoma (Fig. 1 and Supplementary Fig. S1A–B)(23–25). The algorithm also reliably identified therapeutically actionable anaplastic lymphoma kinase (ALK) fusions in lung adenocarcinoma, although DNA validation was not available in all cases due to incomplete availability of WGS data (Supplementary Fig. S1C)(11). Numerous aberrant transcripts were detected for the RET receptor tyrosine kinase, which undergoes rearrangement in 10–20% of sporadic papillary thyroid cancers(26)(Supplementary Fig. S1D). When possible, we obtained DNA validation for RET fusions in these samples, including known rearrangements with CCDC6, ERC1, and NCOA4 (Supplementary Table S1). To evaluate the ability of STA to predict fusions previously identified in TCGA RNA-seq, we compared the STA score distribution of fusion transcripts across all genes(27) (Supplementary Fig. S2A) and recurrently fused kinases(28) (Supplementary Fig. S2B) to the distribution for all genes and samples evaluated. In both cases, STA scores were significantly higher in the rearrangement-harboring transcripts (p < 2.2×10−6).
Due to the ability of STA to detect aberrant loss of expression in addition to gain, we hypothesized that inactivation of tumor suppressors would constitute a substantial portion of STA-predicted rearrangements. Indeed, the algorithm displayed a robust ability to identify intragenic loss of expression of known tumor suppressors. In a lung squamous cell carcinoma sample, we identified a rearrangement resulting in early truncation of the SWI/SNF subunit ARID1A (Supplementary Fig. S3A)(29). We additionally validated rearrangements with non-coding DNA in bladder and endometrial carcinoma resulting in the loss of functional domains of the PI3K negative regulator PTEN and the p300 histone acetyltransferase, respectively (Supplementary Fig. S3B–C)(30,31). In a kidney chromophobe tumor, we identified a rearrangement between a non-coding portion of chromosome 5 and the gene encoding the miRNA-processing enzyme DROSHA that results in its early truncation (Supplementary Fig. S3D)(32). Interestingly, inactivating DROSHA mutations have recently been reported in over 10% of cases of Wilms tumor, a pediatric kidney cancer(33).
A novel oncogenic kinase fusion in TNBC
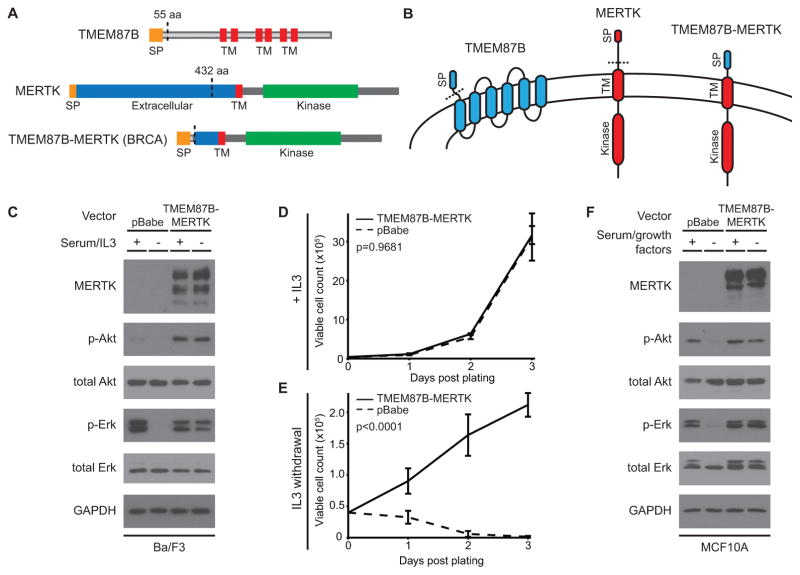
To discover known and novel gene rearrangements in clinical TNBC cases, we used NGS data from TCGA and performed STA prediction on 173 TNBC tumors. We identified and validated at the DNA level a previously uncharacterized fusion involving the MER proto-oncogene tyrosine kinase (MERTK)(34). In this rearrangement, a nearby gene encoding the transmembrane protein TMEM87B acts as 5’ partner, breaking shortly after its signal peptide and fusing with the late extracellular domain-coding portion of MERTK (Fig. 2A). The resulting fusion transcript displays increased expression and retains the full transmembrane and intracellular kinase domains of MERTK, which is overexpressed or ectopically expressed in numerous cancers (Fig. 2B)(35). In order to assess if this truncated form of MERTK retains its ability to activate the oncogenic MAPK/Erk and Akt signaling pathways(36), we engineered a retroviral expression construct encoding the tumor-derived TMEM87B-MERTK fusion. In the IL3-dependent Ba/F3 mouse lymphocyte cell line, stable expression of TMEM87B-MERTK led to constitutively elevated levels of phospho-Akt and retention of robust Erk and Akt signaling even after serum starvation and withdrawal of IL3 (Fig. 2C)(17). Accordingly, while the TMEM87B-MERTK and empty vector control cells grew similarly in the presence of IL3, the fusion protein conferred a clear survival advantage after IL3 withdrawal (Fig. 2D–E). Whereas the control cells died by day 3 after IL3 withdrawal, the TMEM87B-MERTK-expressing cells proliferated under the same conditions (Fig. 2E) and could be cultured in the absence of IL3 for at least one month (data not shown). These results are consistent with the survival-promoting role of full-length MERTK in melanoma, glioblastoma, and other cancers(36–38). To verify that the fusion protein-modulated signaling could be replicated in breast-derived, basal epithelial cells, we expressed TMEM87B-MERTK in immortalized MCF10A cells and observed similar activation of Erk and Akt after serum starvation and growth factor withdrawal (Fig. 2F). Of note, we identified an identical TMEM87B-MERTK fusion in the lung squamous cell carcinoma RNA-seq data set from TCGA, along with a BCL2L11-MERTK RNA transcript with the same breakpoint in bladder carcinoma, but WGS data were not available for DNA validation (data not shown). An independent RNA-seq fusion analysis of TCGA samples corroborated the TMEM87B-MERTK fusion in TNBC and identified identical rearrangements in cervical carcinoma and lung adenocarcinoma(27), demonstrating selection for a recurrently truncated form of MERTK in multiple cancer types.
Figure 2. The TMEM87B-MERTK gene fusion in TNBC promotes constitutive oncogenic signaling and cell survival.
A, Diagram of the TMEM87B and MERTK proteins and the DNA-validated gene fusion protein product. Protein features are labeled. B, Protein schematics indicating membrane topology (not to scale). Colors indicate protein sequences as indicated. In A and B, dotted lines represent protein regions encoded by the gene fusion transcript. C, Immunoblot analysis of the indicated proteins from Ba/F3 cells transfected with an empty vector or one expressing the TMEM87B-MERTK fusion gene. Cell lines were grown in the continuous presence of 5% FBS and 1 ng/mL IL3 (+) or switched to 0.5% FBS and no IL3 (−) for 90 min. D–E, Graphs depicting growth curves of Ba/F3 cells transfected with the TMEM87B-MERTK fusion gene (solid line) or empty vector (dotted line). Cells were grown in the continuous presence of 1 ng/mL IL3 (D) or switched to no-IL3 media at day 0 (E) and viable cell counts were obtained by hemocytometer with trypan blue exclusion at the indicated timepoints. Error bars represent standard deviation of three replicates and p-values comparing the two conditions are specified at top left. F, Immunoblot analysis of the indicated proteins from MCF10A cells transfected with constructs identical to C. Cells were grown in complete growth media with 2.5% horse serum (+) or switched to base media with 0.5% horse serum and no growth factor additives for 180 min (−). aa: amino acid; BRCA: breast invasive carcinoma; SP: signal peptide; TM: transmembrane.
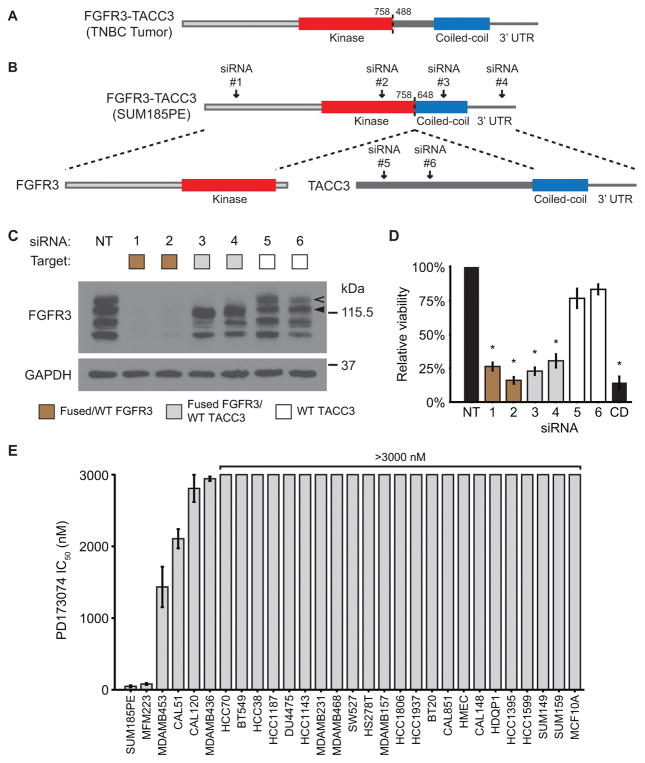
FGFR3-TACC3 gene fusion is a targetable driver alteration in TNBC
In order to identify and evaluate an endogenous model of oncogenic gene fusions in TNBC, we expanded our analysis to RNA-seq data from 80 additional TNBC tumors(3) and 28 TNBC cell line models. In both a tumor specimen and the SUM185PE cell line, we discovered FGFR3-TACC3 fusions similar to the oncogenic rearrangements recently observed in glioblastoma and bladder carcinoma, which result in the fusion of the FGFR3 kinase domain to the coiled-coil domain of TACC3 (Fig. 3A–B)(12,39). To determine if FGFR3-TACC3 is a targetable ‘driver alteration’ in TNBC, we conducted knockdown and pharmaceutical inhibition of the fusion protein in SUM185PE cells. Immunoblotting for FGFR3 in cell lysates produced distinct bands consistent with the predicted molecular weights of the wild-type and fused proteins (Fig. 3C). Two siRNAs targeting FGFR3 (Fig. 3B, siRNA #1–2) reduced expression of both forms of the protein and decreased cell viability after 72 hr to levels comparable to a cell-death control (Fig. 3C–D). To verify that the viability decrease was due to the loss of fused FGFR3 rather than wild-type, we assessed two siRNAs targeting the fused portion of TACC3 (Fig. 3B, siRNA #3–4) and two siRNAs targeting sequences outside the recombined region (Fig. 3B, siRNA #5–6). The two TACC3 siRNAs targeting a portion of the transcript contained within the fusion decreased expression of the resulting hybrid protein and reduced viability to a level similar to FGFR3 knockdown, whereas addition of the siRNAs targeting a portion of TACC3 outside the fused region did not significantly decrease viability or fusion protein expression (Fig. 3C–D). Additionally, SUM185PE cells displayed pronounced sensitivity to the FGFR inhibitor PD173074 (IC50 = 48 ± 13 nM), whereas the majority of other cell lines tested displayed micromolar or greater half-maximal inhibitory concentrations (Fig. 3E)(40). The only other line with similar sensitivity, MFM223, harbors a previously reported amplification of FGFR2(41).
Figure 3. The FGFR3-TACC3 gene fusion is a targetable driver alteration in TNBC.
A–B, diagram of the protein products of the FGFR3-TACC3 gene fusions found in a tumor sample from TNBC patient TTR0001024(3) (A) and the SUM185PE TNBC cell line (B). Protein features are labeled and dotted lines outline protein regions encoded by the gene fusion transcript. Numbers indicate amino acid position in the wild-type proteins and arrows indicate targeting locations of the siRNAs used in the experiments. C–D, Immunoblot analysis of the indicated proteins from SUM185PE lysate (C) and relative viability of the cells (D) after 72-hr treatment with the indicated siRNAs (depicted in B), a non-targeting control (NT), or a cell death-inducing positive control (CD). In C, the legend indicates proteins expected to undergo knockdown based on siRNA target location. Wild-type (WT) and fused forms of FGFR3 are denoted by a filled and hollow arrow, respectively. In D, viability as assessed by alamarBlue is normalized to the non-targeting control. Error bars represent standard error of the mean of four independent experiments. Asterisks indicate p < 0.001 when compared to NT control. E, Half-maximal inhibitory concentrations (IC50) of the FGFR inhibitor PD173074 for the indicated cell lines as assessed by alamarBlue assay. Error bars represent standard error of the mean of three independent experiments.
Novel and non-canonical rearrangements in TNBC
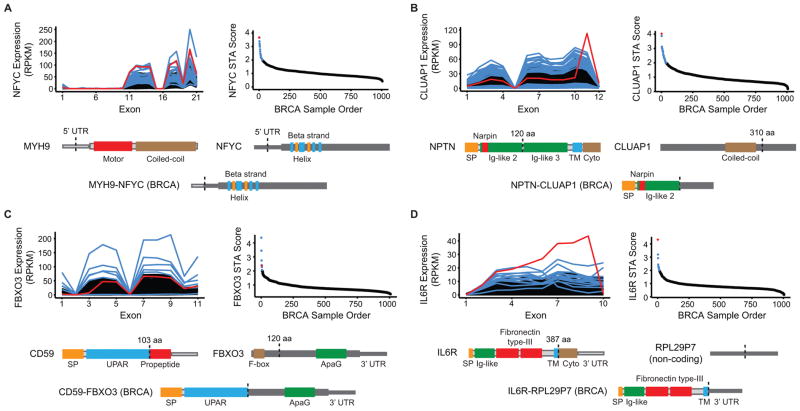
Additional STA predictions from the TCGA TNBC clinical data set led to the identification and DNA validation of a structurally and functionally diverse array of gene rearrangements. In one sample, dual 5’ UTR breakpoints result in promoter swapping between the myosin heavy chain gene MYH9 and the histone modifier gene NFYC, which was recently identified as an oncogene in choroid plexus carcinoma(42). The resulting fusion transcript is highly expressed and retains the entire NFYC open reading frame (Fig. 4A). In another rearrangement, the 55 kDa isoform of the transmembrane glycoprotein neuroplastin is fused to the C-terminus of the cilia-associated transcript CLUAP1, leading to a transcript encoding the signal peptide and a single extracellular Ig-like domain from neuroplastin (Fig. 4B). Importantly, a small portion of the retained Ig-like domain was previously demonstrated to be sufficient for the FGFR1 activation exhibited by the full-length protein(43), implying that the NPTN-CLUAP1 gene fusion may lead to the secretion of a paracrine FGFR1 activator.
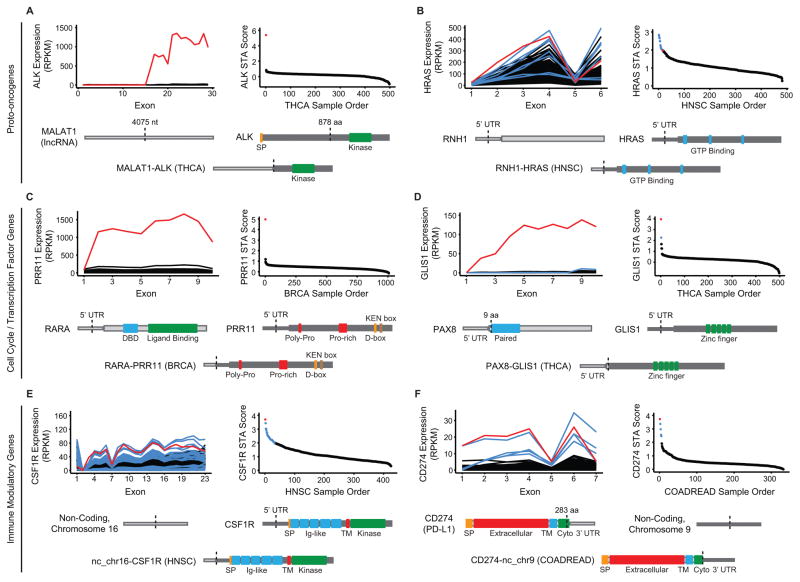
Figure 4. Triple-negative breast cancers harbor a functionally diverse array of gene rearrangements.
A–D, Four examples of STA-predicted rearrangements in triple-negative breast cancers from TCGA. Each panel features an exon-level expression diagram and STA score plot for the gene and cancer type analyzed. Red indicates the representative DNA-validated rearrangement that is depicted at the bottom of each panel as a schematic of the resulting aberrant protein. Blue indicates additional aberrant transcripts meeting STA score threshold. Black indicates background population below threshold. Protein features and untranslated regions (UTRs) are labeled and dotted lines indicate hybrid transcript junctions. aa: amino acid; BRCA: breast invasive carcinoma; Cyto: cytoplasmic domain; nt: nucleotide; SP: signal peptide; TM: transmembrane.
We also identified rearrangements in TNBC involving immune-related proteins. In one case, FBXO3 undergoes rearrangement with the gene encoding the membrane attack complex inhibitor CD59, with resultant expression of a transcript encoding the functional domains of CD59. Interestingly, the RNA breakpoint for CD59 occurs at a non-annotated splice site within the coding sequence that precisely mimics the cleavage site of the mature protein from a glycosylphosphatidylinositol (GPI) anchor addition signal (Fig. 4C)(44). The consequences of GPI anchor loss and the retention of a portion of FBXO at the C-terminus were not assessed; however, soluble forms of CD59 have been previously noted to retain their complement-mediated cytotoxicity-suppressive function(45). In a final example, the gene encoding the interleukin 6 receptor (IL6R) breaks at the junction between its transmembrane and cytoplasmic domains and undergoes rearrangement with the non-coding pseudogene RPL29P7 (Fig. 4D). Intriguingly, previous studies have demonstrated that the cytoplasmic domain of IL6R is dispensable for its interaction with the gp130 transactivator(46), and the resulting fusion transcript is highly expressed. We speculate that the IL6R-RPL29P7 fusion protein retains the IL6-binding and transactivation capacity of wild-type IL6R, and is overexpressed due to loss of the negative-regulatory IL6R 3’ UTR. This rearrangement illustrates a mechanism by which 3’ hybrid transcript formation with non-coding regions of the genome can lead to increased expression of tumor-promoting genes, similar to the increased expression resulting from 3’ UTR loss in the FGFR3-TACC3 gene fusion(47) and confirming findings from previous gene fusion discovery efforts in breast cancer tumors and cell lines(48,49).
Functionally and structurally diverse rearrangements across cancer
Given the ability of STA to identify novel classes of rearrangements in TNBC, we broadened our discovery efforts using 14 additional NGS datasets from TCGA. In total, we analyzed 5461 tumor samples with RNA-seq across 14 cancer types, of which 1264 (23%) had accompanying WGS available (Supplementary Fig. S4). We validated 1178 gene rearrangements at the RNA and DNA levels (Supplementary Table S1). Of note, our attempts to validate newly discovered rearrangements at the DNA level were confounded in part by variable WGS availability and sequencing depth across TCGA studies. We found a clear correlation between both the validation rate and frequency of rearrangements detected per sample and WGS file size (Supplementary Figs. S5 and S6).
Fusions between two protein-coding genes constituted 40% of the validated rearrangements. Among the remaining rearrangements of coding genes with non-coding DNA, a majority featured the protein-coding gene as the 5’ partner, as inferred by RNA breakpoints. For 3% of total rearrangements detected, however, non-coding regions of the genome acted as 5’ hybrid transcript partners and caused deregulated expression of coding genes (Supplementary Fig. S7A). To characterize the function of genes undergoing each rearrangement type, we classified rearrangement partners according to a previously published oncogene and tumor suppressor prediction method(50). The enrichment pattern of genes in these categories was consistent with tumor-promoting gain or loss of function (Supplementary Fig. S7B–C). Coding genes acting as the 3’ transcript partner in out-of-frame fusions included a higher proportion of tumor suppressors, for example, whereas in-frame fusions and rearrangement at UTRs included more oncogenes (Supplementary Fig. S7C).
Across tumor types, we noticed a trend of tumor-promoting gene overexpression by a structurally diverse set of gene rearrangements. In a thyroid carcinoma sample, we identified a rearrangement between a 5’ portion of the long non-coding RNA (lncRNA) MALAT1 and the recurrently fused ALK(11), leading to extremely high expression of a transcript retaining the ALK kinase domain (Fig. 5A). Of note, the MALAT1-ALK rearrangement occurs upstream of ALK exon 16 rather than the most common exon 19 and 20 breakpoints, but numerous in-frame methionines in the exon 16–19 region could allow translation initiation after the non-coding, 5’ MALAT1 portion of the hybrid transcript (data not shown). We also identified abnormally high expression of the MAPK activator HRAS in a head and neck squamous cell carcinoma sample. WGS analysis revealed a rearrangement between RNH1 and a DNA breakpoint upstream of the HRAS transcriptional start site. The 5’ UTR of RNH1 is subsequently spliced to exon 2 of HRAS, resulting in elevated transcription of a complete HRAS open reading frame (Fig. 5B). We noted a similar overexpression of embryonic stem cell-expressed Ras (ERAS) by the PQBP1 promoter in lung squamous cell carcinoma, but the DNA breakpoint for that sample occurred within the first intron of ERAS (Supplementary Table S1). Although KRAS fusions have been described in metastatic prostate cancer(10), we believe these are the first DNA-validated fusions involving HRAS and ERAS to be reported. A similar rearrangement leading to overexpression of the entire HRAS coding sequence was identified in a murine Moloney leukemia virus-induced cell line and showed the ability to transform NIH 3T3 fibroblasts(51).
Figure 5. Overexpression of oncogenic transcripts across cancer types results from gene rearrangement with coding and non-coding DNA.
A–F, Six examples of STA-predicted rearrangements from additional tumor types in TCGA, representing the categories described at left. Each panel features an exon-level expression diagram and STA score plot for the gene and cancer type analyzed. Red indicates the representative DNA-validated rearrangement that is depicted at the bottom of each panel as a schematic of the resulting aberrant protein. Blue indicates additional aberrant transcripts meeting STA score threshold. Black indicates background population below threshold. Protein features and untranslated regions (UTRs) are labeled and dotted lines indicate hybrid transcript junctions. aa: amino acid; BRCA: breast invasive carcinoma; COADREAD: colorectal carcinoma; Cyto: cytoplasmic domain; HNSC: head and neck squamous cell carcinoma; nt: nucleotide; SP: signal peptide; THCA: thyroid carcinoma; TM: transmembrane.
DNA breakpoints upstream of the transcriptional start site, as seen in RNH1-HRAS, can lead to extreme overexpression that is easily detectable by STA. In an estrogen receptor (ER)-positive breast cancer sample, the ER-responsive gene RARA displayed rearrangement with a region upstream of PRR11, leading to more than 10-fold increase of PRR11 transcript compared to the population average (Fig. 5C). While PRR11 is a relatively understudied cell cycle progression gene with normally periodic expression, it is overexpressed in breast and lung cancer and PRR11 knockdown inhibits cancer cell proliferation(52,53). A similar rearrangement and consequent increase in transcript levels occurred in a thyroid carcinoma, where PAX8, the 5’ partner in the recurrent PAX8-PPARG thyroid fusion, is spliced to the 5’ UTR of GLIS1, a transcription factor whose expression was previously correlated with Wnt pathway activation and epithelial to mesenchymal transition in a mouse model of breast cancer (Fig. 5D)(54,55).
The non-canonical rearrangements identified using STA also include clinically relevant immune-modulatory genes. In a head and neck squamous cell carcinoma sample, we identified a hybrid transcript in which a non-coding portion of chromosome 16 is fused to the 5’ UTR of the gene encoding the colony stimulating factor 1 receptor (CSF1R), leading to overexpression of a complete CSF1R-coding transcript (Fig. 5E)(56). Additionally, a colorectal cancer sample with abnormally high expression of CD274, which encodes the T-cell suppressor PD-L1, harbors a rearrangement between the second-to-last exon of CD274 and a non-coding region of chromosome 9 (Fig. 5F)(57). The resulting hybrid transcript encodes a near-complete copy of PD-L1; as in the previous IL6R-RPL29P7 rearrangement, we speculate that loss of the endogenous negative-regulatory 3’ UTR of CD274 leads to the increase in transcript levels.
DISCUSSION
The wide spectrum of rearrangements identified using STA demonstrates the ability of oncogenic selection to exploit the modular architecture of the human genome to ensure tumor proliferation and survival. While some novel rearrangements emerging from our analysis consisted of classic receptor tyrosine kinase fusions, such as TMEM87B-MERTK in TNBC (Fig. 2), we also observed overexpression of entire coding transcripts resulting from promoter and UTR swapping, including oncogenic Ras family members (Fig. 5B). Importantly, our ability to assess rearrangements with non-coding regions led to the detection of not only inactivating truncations in tumor suppressors, but also gain-of-function events. For instance, the gene encoding the tumor suppressor PD-L1, which has emerged as a prime target in the rapidly advancing field of cancer immunotherapy, underwent rearrangement and overexpression of a transcript harboring a non-coding sequence in place of its endogenous, negative-regulatory 3’ UTR (Fig. 5F)(58). We observed similar rearrangements multiple times in our analyses, including the IL6R-RPL29P7 fusion in TNBC (Fig. 4D), and we hypothesize that overexpression by this mechanism may explain many of the rearrangements occurring between oncogenes and non-coding regions of the genome (Supplementary Fig. S7B). The MALAT1-ALK lncRNA-gene fusion and the increased expression of tumor-associated macrophage drug target CSF1R by a non-coding 5’ hybrid transcript partner additionally demonstrate the clinical relevance of gene rearrangements involving non-coding regions (Fig. 5A,E)(56). Further consideration of these non-canonical events in future gene rearrangement discovery will be critical for our understanding and treatment of TNBC and other molecularly heterogeneous cancers.
The targetable gene fusions we identified across many cancer types demonstrate the need to rapidly advance gene fusion detection for molecularly heterogeneous cancers. Although the frequency of individual gene rearrangements in these cancers may be low, the two rearrangements we validated with biological relevance for TNBC involve specific molecular targets for therapies already in clinical investigation(14) or development(36). If readily detectable, several of the gene fusions we identified would provide an immediate opportunity for patient alignment to targeted therapy and would serve as biomarkers for patient selection in basket trials such as the NCI-MATCH (Molecular Analysis of Therapy Choice) Program.
Our results provide evidence for the effectiveness of STA as a quantitative prediction tool for both known and novel gene rearrangements. Although our analysis made use of RNA and DNA sequencing files already processed by the TCGA Research Network, a pipeline based on focused assembly of STA-predicted transcripts could increase the sensitivity, efficiency, and breadth of rearrangement detection, accelerating discovery of diagnostic and prognostic markers. As tumor sequencing efforts continue, we look forward to optimizing and expanding the use of STA to comprehensively catalogue gene rearrangements across cancer.
The novel approach employed in STA enabled the discovery and analysis of known and novel gene rearrangements on a genome-wide scale, which has clear relevance to the development and repurposing of targeted therapies. Moreover, analysis of the rearrangements we identified provides insight into the multi-modular architecture of proteins and the diverse functional and regulatory domains selected for and against during tumorigenesis. Continued advances in detection methods such as STA will be critical for the treatment of diseases such as TNBC and the ability of the field to apply mechanistic insight from “exceptional responders” to individual tumor genomes containing unique mutations and rearrangements.
Supplementary Material
Acknowledgments
We thank members of the Pietenpol lab for their critical review of the manuscript and members of the Lovly lab for guidance with the Ba/F3 cell line model. This work was conducted in part using the resources of the Advanced Computing Center for Research and Education at Vanderbilt University. The results published here are in part based upon data generated by TCGA Research Network: http://cancergenome.nih.gov. T.M.S. was supported by CA183531, GM008554, and HHMI MIG56006779. B.D.L. was supported by Komen CCR13262005. J.A.P. was supported by CA098131, CA105436, and Komen SAC110030. Shared resources were provided by CA068485.
Footnotes
Disclosure of Potential Conflicts of Interest: The authors have read and understood the AACR journal policy on reporting conflicts of interest and have no conflicts to disclose.
References
- 1.Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121(7):2750–67. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Masuda H, Baggerly KA, Wang Y, Zhang Y, Gonzalez-Angulo AM, Meric-Bernstam F, et al. Differential response to neoadjuvant chemotherapy among 7 triple-negative breast cancer molecular subtypes. Clin Cancer Res. 2013;19(19):5533–40. doi: 10.1158/1078-0432.CCR-13-0799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shah SP, Roth A, Goya R, Oloumi A, Ha G, Zhao Y, et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012;486(7403):395–9. doi: 10.1038/nature10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lehmann BD, Bauer JA, Schafer JM, Pendleton CS, Tang L, Johnson KC, et al. PIK3CA mutations in androgen receptor-positive triple negative breast cancer confer sensitivity to the combination of PI3K and androgen receptor inhibitors. Breast Cancer Res. 2014;16(4):406. doi: 10.1186/s13058-014-0406-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu X, Stern HM, Ge L, O'Brien C, Haydu L, Honchell CD, et al. Genetic alterations and oncogenic pathways associated with breast cancer subtypes. Mol Cancer Res. 2009;7(4):511–22. doi: 10.1158/1541-7786.MCR-08-0107. [DOI] [PubMed] [Google Scholar]
- 7.Cancer Genome Atlas Research N. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474(7353):609–15. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cancer Genome Atlas Research N. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489(7417):519–25. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rabbitts TH. Chromosomal translocations in human cancer. Nature. 1994;372(6502):143–9. doi: 10.1038/372143a0. [DOI] [PubMed] [Google Scholar]
- 10.Wang XS, Shankar S, Dhanasekaran SM, Ateeq B, Sasaki AT, Jing X, et al. Characterization of KRAS rearrangements in metastatic prostate cancer. Cancer Discov. 2011;1(1):35–43. doi: 10.1158/2159-8274.CD-10-0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448(7153):561–6. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 12.Williams SV, Hurst CD, Knowles MA. Oncogenic FGFR3 gene fusions in bladder cancer. Hum Mol Genet. 2013;22(4):795–803. doi: 10.1093/hmg/dds486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310(5748):644–8. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 14.Shaw AT, Hsu PP, Awad MM, Engelman JA. Tyrosine kinase gene rearrangements in epithelial malignancies. Nat Rev Cancer. 2013;13(11):772–87. doi: 10.1038/nrc3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davare MA, Tognon CE. Detecting and targetting oncogenic fusion proteins in the genomic era. Biol Cell. 2015;107(5):111–29. doi: 10.1111/boc.201400096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Annala MJ, Parker BC, Zhang W, Nykter M. Fusion genes and their discovery using high throughput sequencing. Cancer Lett. 2013;340(2):192–200. doi: 10.1016/j.canlet.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palacios R, Steinmetz M. Il-3-dependent mouse clones that express B-220 surface antigen, contain Ig genes in germ-line configuration, and generate B lymphocytes in vivo. Cell. 1985;41(3):727–34. doi: 10.1016/s0092-8674(85)80053-2. [DOI] [PubMed] [Google Scholar]
- 18.Bauer JA, Ye F, Marshall CB, Lehmann BD, Pendleton CS, Shyr Y, et al. RNA interference (RNAi) screening approach identifies agents that enhance paclitaxel activity in breast cancer cells. Breast Cancer Res. 2010;12(3):R41. doi: 10.1186/bcr2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morgenstern JP, Land H. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 1990;18(12):3587–96. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Magrane M Consortium U. UniProt Knowledgebase: a hub of integrated protein data. Database (Oxford) 2011;2011:bar009. doi: 10.1093/database/bar009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giacomini CP, Sun S, Varma S, Shain AH, Giacomini MM, Balagtas J, et al. Breakpoint analysis of transcriptional and genomic profiles uncovers novel gene fusions spanning multiple human cancer types. PLoS Genet. 2013;9(4):e1003464. doi: 10.1371/journal.pgen.1003464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Faust GG, Hall IM. YAHA: fast and flexible long-read alignment with optimal breakpoint detection. Bioinformatics. 2012;28(19):2417–24. doi: 10.1093/bioinformatics/bts456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rikova K, Guo A, Zeng Q, Possemato A, Yu J, Haack H, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131(6):1190–203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 24.Kim J, Lee Y, Cho HJ, Lee YE, An J, Cho GH, et al. NTRK1 fusion in glioblastoma multiforme. PLoS One. 2014;9(3):e91940. doi: 10.1371/journal.pone.0091940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tomlins SA, Mehra R, Rhodes DR, Smith LR, Roulston D, Helgeson BE, et al. TMPRSS2:ETV4 gene fusions define a third molecular subtype of prostate cancer. Cancer Res. 2006;66(7):3396–400. doi: 10.1158/0008-5472.CAN-06-0168. [DOI] [PubMed] [Google Scholar]
- 26.Grieco M, Santoro M, Berlingieri MT, Melillo RM, Donghi R, Bongarzone I, et al. PTC is a novel rearranged form of the ret proto-oncogene and is frequently detected in vivo in human thyroid papillary carcinomas. Cell. 1990;60(4):557–63. doi: 10.1016/0092-8674(90)90659-3. [DOI] [PubMed] [Google Scholar]
- 27.Yoshihara K, Wang Q, Torres-Garcia W, Zheng S, Vegesna R, Kim H, et al. The landscape and therapeutic relevance of cancer-associated transcript fusions. Oncogene. 2014 doi: 10.1038/onc.2014.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stransky N, Cerami E, Schalm S, Kim JL, Lengauer C. The landscape of kinase fusions in cancer. Nat Commun. 2014;5:4846. doi: 10.1038/ncomms5846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jones S, Wang TL, Shih Ie M, Mao TL, Nakayama K, Roden R, et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science. 2010;330(6001):228–31. doi: 10.1126/science.1196333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee JO, Yang H, Georgescu MM, Di Cristofano A, Maehama T, Shi Y, et al. Crystal structure of the PTEN tumor suppressor: implications for its phosphoinositide phosphatase activity and membrane association. Cell. 1999;99(3):323–34. doi: 10.1016/s0092-8674(00)81663-3. [DOI] [PubMed] [Google Scholar]
- 31.Gayther SA, Batley SJ, Linger L, Bannister A, Thorpe K, Chin SF, et al. Mutations truncating the EP300 acetylase in human cancers. Nat Genet. 2000;24(3):300–3. doi: 10.1038/73536. [DOI] [PubMed] [Google Scholar]
- 32.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425(6956):415–9. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 33.Torrezan GT, Ferreira EN, Nakahata AM, Barros BD, Castro MT, Correa BR, et al. Recurrent somatic mutation in DROSHA induces microRNA profile changes in Wilms tumour. Nat Commun. 2014;5:4039. doi: 10.1038/ncomms5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Graham DK, Dawson TL, Mullaney DL, Snodgrass HR, Earp HS. Cloning and mRNA expression analysis of a novel human protooncogene, c-mer. Cell Growth Differ. 1994;5(6):647–57. [PubMed] [Google Scholar]
- 35.Cummings CT, Deryckere D, Earp HS, Graham DK. Molecular pathways: MERTK signaling in cancer. Clin Cancer Res. 2013;19(19):5275–80. doi: 10.1158/1078-0432.CCR-12-1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schlegel J, Sambade MJ, Sather S, Moschos SJ, Tan AC, Winges A, et al. MERTK receptor tyrosine kinase is a therapeutic target in melanoma. J Clin Invest. 2013;123(5):2257–67. doi: 10.1172/JCI67816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Y, Moncayo G, Morin P, Jr, Xue G, Grzmil M, Lino MM, et al. Mer receptor tyrosine kinase promotes invasion and survival in glioblastoma multiforme. Oncogene. 2013;32(7):872–82. doi: 10.1038/onc.2012.104. [DOI] [PubMed] [Google Scholar]
- 38.Brandao LN, Winges A, Christoph S, Sather S, Migdall-Wilson J, Schlegel J, et al. Inhibition of MerTK increases chemosensitivity and decreases oncogenic potential in T-cell acute lymphoblastic leukemia. Blood Cancer J. 2013;3:e101. doi: 10.1038/bcj.2012.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Singh D, Chan JM, Zoppoli P, Niola F, Sullivan R, Castano A, et al. Transforming fusions of FGFR and TACC genes in human glioblastoma. Science. 2012;337(6099):1231–5. doi: 10.1126/science.1220834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mohammadi M, Froum S, Hamby JM, Schroeder MC, Panek RL, Lu GH, et al. Crystal structure of an angiogenesis inhibitor bound to the FGF receptor tyrosine kinase domain. EMBO J. 1998;17(20):5896–904. doi: 10.1093/emboj/17.20.5896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Turner N, Lambros MB, Horlings HM, Pearson A, Sharpe R, Natrajan R, et al. Integrative molecular profiling of triple negative breast cancers identifies amplicon drivers and potential therapeutic targets. Oncogene. 2010;29(14):2013–23. doi: 10.1038/onc.2009.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tong Y, Merino D, Nimmervoll B, Gupta K, Wang YD, Finkelstein D, et al. Cross-Species Genomics Identifies TAF12, NFYC, and RAD54L as Choroid Plexus Carcinoma Oncogenes. Cancer Cell. 2015;27(5):712–27. doi: 10.1016/j.ccell.2015.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Owczarek S, Kiryushko D, Larsen MH, Kastrup JS, Gajhede M, Sandi C, et al. Neuroplastin-55 binds to and signals through the fibroblast growth factor receptor. FASEB J. 2010;24(4):1139–50. doi: 10.1096/fj.09-140509. [DOI] [PubMed] [Google Scholar]
- 44.Sugita Y, Nakano Y, Oda E, Noda K, Tobe T, Miura NH, et al. Determination of carboxyl-terminal residue and disulfide bonds of MACIF (CD59), a glycosyl-phosphatidylinositol-anchored membrane protein. J Biochem. 1993;114(4):473–7. doi: 10.1093/oxfordjournals.jbchem.a124202. [DOI] [PubMed] [Google Scholar]
- 45.Brasoveanu LI, Fonsatti E, Visintin A, Pavlovic M, Cattarossi I, Colizzi F, et al. Melanoma cells constitutively release an anchor-positive soluble form of protectin (sCD59) that retains functional activities in homologous complement-mediated cytotoxicity. J Clin Invest. 1997;100(5):1248–55. doi: 10.1172/JCI119638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jones SA, Horiuchi S, Topley N, Yamamoto N, Fuller GM. The soluble interleukin 6 receptor: mechanisms of production and implications in disease. FASEB J. 2001;15(1):43–58. doi: 10.1096/fj.99-1003rev. [DOI] [PubMed] [Google Scholar]
- 47.Parker BC, Annala MJ, Cogdell DE, Granberg KJ, Sun Y, Ji P, et al. The tumorigenic FGFR3-TACC3 gene fusion escapes miR-99a regulation in glioblastoma. J Clin Invest. 2013;123(2):855–65. doi: 10.1172/JCI67144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Asmann YW, Necela BM, Kalari KR, Hossain A, Baker TR, Carr JM, et al. Detection of redundant fusion transcripts as biomarkers or disease-specific therapeutic targets in breast cancer. Cancer Res. 2012;72(8):1921–8. doi: 10.1158/0008-5472.CAN-11-3142. [DOI] [PubMed] [Google Scholar]
- 49.Edgren H, Murumagi A, Kangaspeska S, Nicorici D, Hongisto V, Kleivi K, et al. Identification of fusion genes in breast cancer by paired-end RNA-sequencing. Genome Biol. 2011;12(1):R6. doi: 10.1186/gb-2011-12-1-r6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Davoli T, Xu AW, Mengwasser KE, Sack LM, Yoon JC, Park PJ, et al. Cumulative haploinsufficiency and triplosensitivity drive aneuploidy patterns and shape the cancer genome. Cell. 2013;155(4):948–62. doi: 10.1016/j.cell.2013.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ihle JN, Smith-White B, Sisson B, Parker D, Blair DG, Schultz A, et al. Activation of the c-H-ras proto-oncogene by retrovirus insertion and chromosomal rearrangement in a Moloney leukemia virus-induced T-cell leukemia. J Virol. 1989;63(7):2959–66. doi: 10.1128/jvi.63.7.2959-2966.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ji Y, Xie M, Lan H, Zhang Y, Long Y, Weng H, et al. PRR11 is a novel gene implicated in cell cycle progression and lung cancer. Int J Biochem Cell Biol. 2013;45(3):645–56. doi: 10.1016/j.biocel.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 53.Zhou F, Liu H, Zhang X, Shen Y, Zheng D, Zhang A, et al. Proline-rich protein 11 regulates epithelial-to-mesenchymal transition to promote breast cancer cell invasion. Int J Clin Exp Pathol. 2014;7(12):8692–9. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 54.Kroll TG, Sarraf P, Pecciarini L, Chen CJ, Mueller E, Spiegelman BM, et al. PAX8-PPARgamma1 fusion oncogene in human thyroid carcinoma [corrected] Science. 2000;289(5483):1357–60. doi: 10.1126/science.289.5483.1357. [DOI] [PubMed] [Google Scholar]
- 55.Vadnais C, Shooshtarizadeh P, Rajadurai CV, Lesurf R, Hulea L, Davoudi S, et al. Autocrine Activation of the Wnt/beta-Catenin Pathway by CUX1 and GLIS1 in Breast Cancers. Biol Open. 2014;3(10):937–46. doi: 10.1242/bio.20148193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhu Y, Knolhoff BL, Meyer MA, Nywening TM, West BL, Luo J, et al. CSF1/CSF1R blockade reprograms tumor-infiltrating macrophages and improves response to T-cell checkpoint immunotherapy in pancreatic cancer models. Cancer Res. 2014;74(18):5057–69. doi: 10.1158/0008-5472.CAN-13-3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Iwai Y, Ishida M, Tanaka Y, Okazaki T, Honjo T, Minato N. Involvement of PD-L1 on tumor cells in the escape from host immune system and tumor immunotherapy by PD-L1 blockade. Proc Natl Acad Sci U S A. 2002;99(19):12293–7. doi: 10.1073/pnas.192461099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12(4):252–64. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.