Abstract
Given their association with cardiovascular disease protection, there has been intense interest in understanding the biology of high density lipoproteins (HDL). HDL is actually a family of diverse particle types, each made up of discrete - but as yet undetermined – combinations of proteins drawn from up to 95 lipophilic plasma proteins. The abundant apolipoproteins (apo) of the A class (apoA-I, apoA-II and apoA-IV) have been proposed to act as organizing platforms for auxiliary proteins, but this concept has not been systematically evaluated. We assessed the impact of genetic knock down of each platform protein on the particle size distribution of auxiliary HDL proteins. Loss of apoA-I or apoA-II massively reduced HDL lipids and changed the plasma size pattern and/or abundance of several plasma proteins. Surprisingly though, many HDL proteins were not affected, suggesting they assemble on lipid particles in the absence of apoA-I or apoA-II. In contrast, apoA-IV ablation had minor effects on plasma lipids and proteins, suggesting that it forms particles that largely exclude other apolipoproteins. Overall, the data indicate that distinct HDL subpopulations exist that do not contain, nor depend on, apoA-I, apoA-II or apoA-IV and these contribute substantially to the proteomic diversity of HDL.
Keywords: High density lipoprotein, proteomics, lipoprotein, apolipoprotein, mass spectrometry, mouse model
Introduction
The name high density lipoprotein (HDL) was given to those plasma lipoproteins that float within the density range of 1.063 to 1.210 g/ml during density gradient ultracentrifugation (UC) thus distinguishing them from low (1.019–1.063 g/ml, LDL) and very low density lipoproteins (0.95–1.006 g/ml, VLDL). In the laboratory, the HDL family is well known for its heterogeneity in terms of size, charge and composition - so much so that efforts have been made to standardize the nomenclature of HDL particles [1]. In the clinic, on the other hand, HDL has historically been treated as a single entity and tracked by its cholesterol content (HDL-C). Indeed, HDL-C levels in plasma are associated with protection from cardiovascular disease (CVD) and this clinical test has been used for many years as a tool to assess individual risk [2]. However, recent work has shown that across-the-board elevation of plasma HDL-C levels, either as a consequence of natural genetic mutation [3] or by pharmacologic manipulation [4, 5], does not necessarily provide the cardio protective benefits predicted by the epidemiology. Nevertheless, the importance of an intact HDL pathway has recently been shown to be important in humans as non-functional SR-BI, the cell surface protein responsible for selective cholesteryl ester uptake into the liver and a critical component of reverse cholesterol transport, results in high HDL-C levels, but increased CVD risk [6].
Recent applications of modern mass spectrometry, tracked by the HDL Proteome Watch website [7], have shown that UC isolated HDL preparations contain about 95 consensus proteins [8–10]. These include the ‘major’ proteins like apolipoprotein (apo)A-I and apoA-II which together account for around 70% of total protein mass in UC-derived HDL. These are sometimes referred to as scaffold or platform proteins because of their key roles in maintaining HDL particle structure [11] and the possibility that they coordinate the binding of other proteins to the particles [12, 13]. Numerous ‘minor’ HDL-associated proteins with known roles in lipid transport, innate immunity, coagulation, regulation of the complement system, metal ion transport and even glucose metabolism have also been identified (reviewed in [10, 14]). As first argued in the classic paper of Kostner and Alupovic [15], all of these auxiliary proteins cannot reside on the same particle due to the limited size of HDL. Work by our laboratory and others has shown that these proteins distribute throughout the HDL family in distinct patterns across particle density [16–18], size [19, 20] and ionic character [21, 22]. This strongly argues that HDL is actually a diverse collection of subspecies that vary widely in protein composition, and presumably function.
Unfortunately, little is known about the molecular basis underlying why proteins segregate among HDL subspecies. In the current work, we tested the hypothesis that the most abundant apolipoproteins in HDL act as organizing scaffolds that mediate the recruitment of specific subsets of other HDL proteins. We asked a simple question: if a given scaffold protein, say apoA-I, is responsible for mediating the assembly of other proteins into HDL, then the HDL particle size distribution of those proteins should be perturbed when apoA-I is genetically ablated. Conversely, proteins that do not depend on apoA-I should not be affected in its absence. Having recently demonstrated that the lipoproteome of both LDL and HDL-sized lipoproteins in the mouse exhibit a similar overall complexity to humans [23], we took advantage of the mouse model to test this hypothesis. We tracked the size distribution and relative protein levels of the plasma lipoprotein proteome in mice lacking each of the three most abundant HDL proteins, apoA-I, apoA-II and apoA-IV, and compared them to WT controls.
Experimental Section
Animal models and plasma collection
All mouse strains were on the C57BL/6J background and were fed a standard chow diet at all times post weaning. Mice were maintained in American Association for Accreditation of Laboratory Animal Care-approved pathogen-free animal facilities, and the institutional laboratory animal medical services at University of Cincinnati approved all experimental protocols. ApoA-I knock out (KO) mice and age matched wild type (WT) controls were purchased from Jackson Laboratories. ApoA-II KO mice with age matched WT controls were provided by Dr. Catherine Reardon at the University of Chicago. The apoA-I KO and apoA-II KO cohorts were between 6 and 10 weeks of age at the time of blood sampling. The apoA-IV KO mice, derived from the original mouse line generated by the Breslow lab [24], were back-crossed for >10 generations on the C57BL6 background in the Tso lab [25]. They were 32 weeks old with an independent age-matched WT group. Blood was collected from ketamine anesthetized mice (n=3 for each KO and control group) by cardiac puncture using citrate as the anticoagulant. Cellular components were pelleted by centrifugation at ~1,590 × g for 15 minutes in a Horizon mini-E (Quest Diagnostics) at room temperature. Plasma was stored at 4°C until gel filtration separation, usually within 16 h. The samples were never frozen.
Plasma separation by gel filtration chromatography
370 µL of plasma from each mouse was applied to three Superdex 200 gel filtration columns (10/300 GL; GE Healthcare) arranged in series on an ÄKTA™ FPLC system (GE Healthcare) [20]. The sample was separated at a flow rate of 0.3 mL/min in standard Tris buffer (STB) (10mM Tris, 0.15M NaCl, 1mM EDTA, 0.2% NaN3). Eluate was collected as 1.5 mL fractions on a Frac 900 fraction collector (GE Healthcare) maintained at 4°C. Each fraction was assessed for protein by modified Lowry assay [26], and choline-containing phospholipid and total cholesterol by colorimetric kits from Wako (Richmond, VA).
Isolation of phospholipid-containing particles using calcium silica hydrate (CSH)
The fractions were then passed through a calcium silica hydrate (CSH) resin to bind components that contain phospholipid (PL), as previously described [20]. All PL-containing particles tightly associate with the CSH while non-lipid associated proteins are washed through. Briefly, in a 96 well filter plate (Millipore), 45 µg of CSH (from 100 mg/mL stock solution in 50 mM ammonium bicarbonate (AB)) per 1 µg of PL in 400 µL of fraction were mixed gently for 30 minutes at room temperature. The resin was eluted using a vacuum manifold and then washed with 50 mM AB buffer. Lipid-containing proteins that remained associated with the CSH were trypsinized with 1.5 µg of sequencing grade trypsin (Promega) overnight at 37°C and the peptides were washed off the resin, reduced with dithiothreitol (200 mM; 30 min. at 37°C) and carbamidomethylated with iodoacetamide (800mM; 30 min. at room temperature). The peptides were then vacuum pelleted and stored at −20 °C until MS analysis.
Mass spectrometry analysis of fractions
Dried peptides were reconstituted in 15 µL of 0.1% formic acid in water. An Agilent 1100 series autosampler/HPLC was used to draw 0.5 µL of sample and inject it onto a C18 reverse phase column (GRACE; 150 × 0.500mm) where an acetonitrile concentration gradient (5–30% in water with 0.1% formic acid) was used to elute peptides for on-line ESI-MS/MS by a QStar XL mass spectrometer (Applied Biosystems). Column cleaning was performed automatically with 2 cycles of a 5–85% acetonitrile gradient lasting 15 min each between runs.
MS data analysis
To identify the protein composition of lipid-containing particles in the gel filtration fractions, peak lists generated from an analysis of each fraction were scanned against the UniProtKB/Swiss-Prot Protein Knowledgebase (release 57.0, 03/2009) using the Mascot (version 2.1) search engine. Search criteria included: mouse taxonomy, variable modifications of Met oxidation and carbamidomethylation, both peptide tolerance and MS/MS tolerance were set to ± 35 PPM, and up to 3 missed tryptic cleavage sites were allowed. Scaffold software (version Scaffold_2_04_00, Proteome Software) was used to validate MS/MS based peptide and protein identifications. Peptide identification required a value of 90% probability (using data from both Mascot and X!Tandem) using the Peptide Prophet algorithm [27]. Positive protein identification also required a value of 90% probability by the Protein Prophet algorithm [28]. Also, a minimum of 2 peptides were required unless the protein in question was found with single peptide hits in multiple consecutive fractions that were consistent across animal subjects. Since equal volumes of sample were applied to the MS analysis, the relative amount of a given protein present in a given fraction is proportional to the number of spectral counts (i.e. the number of MS/MS spectra assigned to a particular protein) in each fraction. In no case were conclusions made about the relative abundance of two different proteins on the basis of peptide counting. We have previously demonstrated that this approach provides a semi-quantitative abundance pattern across each fraction that matches well with patterns derived from immunological analyses [18].
Protein abundance determination
The abundance of a given protein in mouse plasma was determined by summing readouts of protein abundance from each fraction. To allow better comparisons between the mice strains, we used the spectral counts from contaminating albumin (which was not changed by any of the genetic manipulations) to allow for normalization of samples from each mouse. Our approach was to use these normalized spectral counts (from Scaffold) as a first pass to identify protein abundance changes between a given KO model and its corresponding WT control. A simple two-tailed student’s t-test was used to identify differences with a p value of <0.05 indicating significance. Then, these hypothesized differences were tested by MS1 full scan filtering using the Skyline Targeted Proteomics Environment [29]. Mass spectrometer files (*.wiff) and corresponding Mascot DAT files were imported into Skyline. Using a spectral library cut-off score of 95%, fixed modification of Cys with carbamidomethylation and variable modification of Met by oxidation, we created a spectral library with the 113 proteins previously identified in WT C57BL6 mice [30] (max 2 missed trypsin cleavages). Skyline is designed for analyzing multiple replicates of similarly prepared samples as a hypothesis testing tool. Thus, our use of chromatographically separated samples presented some challenges for data analysis. For example, the software was quite effective at locating peptides for each protein in the ‘sweet spot’ of its size gradient, i.e. where a particular protein was abundant with many MS/MS identifications, but it tended to erroneously pick peptides in regions where that protein was absent. Given the thousands of peptides, 18 size fractions, 6 mice per group and 3 groups of mice, manual annotation of the entire dataset was not practical. Therefore, we selected 3 peptides for each protein hypothesized to be different by spectral counting for manual annotation. Selected peptides had a balance of high signal intensity and lack of interfering peaks. Total areas for each of the 3 peptides were summed and compared between KO and WT using a student’s t-test with significance set at p<0.05. Protein differences identified by both spectral counting AND confirmed differences in ion intensity of at least 2 of 3 peptides were considered high confidence abundance changes. Note: due to an unexplained variation in mouse to mouse replicates in the apoA-II KO experiment, the Skyline analysis was not performed. In this case, we relied only on the peptide counting approach to identify overall protein abundance differences.
Protein elution profile data analysis
To quantitatively evaluate the impact of a genetic deletion on the size distribution of other known HDL proteins, we developed a novel elution pattern shift analysis that derives a lag-score (L-score) reflecting a quantitative shift in MS1 intensity pattern for a given protein between the WT and knockout mice. The L-score was calculated based on time-lag between the summed normalized protein abundance (see above) for each protein from WT and KO mice. Time-lag is a concept in electronic signal processing to measure the shift between two signals. We applied this concept here to identify whether a protein shifted to a different size after gene knockout. Assuming m represents WT mice and n represents the gene KO mice, an L-score for any given protein can be derived using the formula below:
Where Γ is a time-lag function, and, , i, j ∈ [1, 3] are abundance profiles of any protein from ith WT mouse and jth gene KO mouse. Larger L-score indicate a more significant shift in a protein in the KO compared to WT. This L-score was used to combat the shift bias generated from outliers and data noise from MS. A shift threshold (|Lx|> 0) was then applied to indicate the proteins with a significant shift. In a limited number of cases, i.e. when a protein was present in two peaks of unequal intensity and the gene KO affected the intensity but not the elution time of one of these peaks, the lag analysis was less reliable and was supplemented by manual analysis performed by two experienced data analysts. Furthermore, if a protein became undetectable in one model, but readily detectible in the others, this was also counted as a “shift” in size.
Results
Lipoprotein size patterns
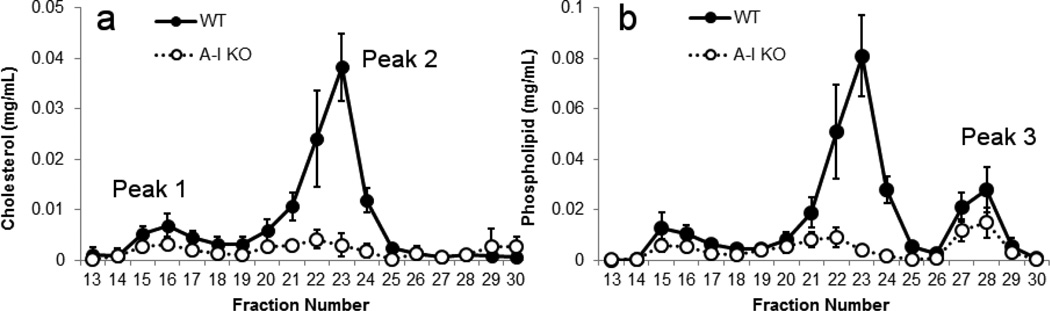
Mouse plasma was separated by size exclusion chromatography and the collected fractions were assayed for phospholipid (PL) and total cholesterol (CH) content. Figure 1 compares the elution profiles for apoA-I KO mice and age matched WT controls. In previous work with this system [20], we established that the apoB-containing lipoproteins VLDL and LDL elute as a single peak between fractions 14 and 18 (Peak 1). Since this system was specifically optimized to spread HDL-sized particles, these elute in a broader peak between fractions 19 and 26 (Peak 2). Smaller, lipid-poor proteins elute near the total volume of the system at fractions 27 to 30 (Peak 3). Fig. 1a shows that WT mice carried most of their cholesterol in particles in the HDL size range with minor amounts in the VLDL/LDL range as shown in hundreds of prior FPLC studies (see [31] for one of the earliest examples). PL was also most abundant in the HDL size range (Fig. 1b). However, a third peak of PL consistently appeared at fractions 27–30 (Peak 3). This is thought to be lysophospholipids associated with soluble proteins such as albumin [32]. In the apoA-I KO animals, modest reductions in both cholesterol and PL levels were noted in Peaks 1 and 3. However in the HDL size range, both lipids were reduced by ~70% (area under the curve) in the apoA-I KO compared to WT. In addition, peak 2 in the apoA-I KO animals appeared to be centered on fraction 22 whereas the WT peak centered on fraction 23, suggesting that the lipoproteins were of slightly larger size in the KO mice. These results are consistent with previous FPLC analyses of these animals [33].
Figure 1.
Size distribution profiles for mouse plasma separated by gel filtration chromatography – WT vs. apoA-I KO. 370 microliters of mouse plasma was analyzed by a triple Superdex setup as described in Methods. Total cholesterol (panel a) and choline-containing phospholipids (panel b) were measured in each fraction by colorimetric assay and plotted vs. the fraction number (1.5 ml fractions). WT animals are indicated with filled symbols and apoA-I KO with open symbols. Error bars represent 1 sample standard deviation from n=3 measurements (i.e. 3 animals) per group.
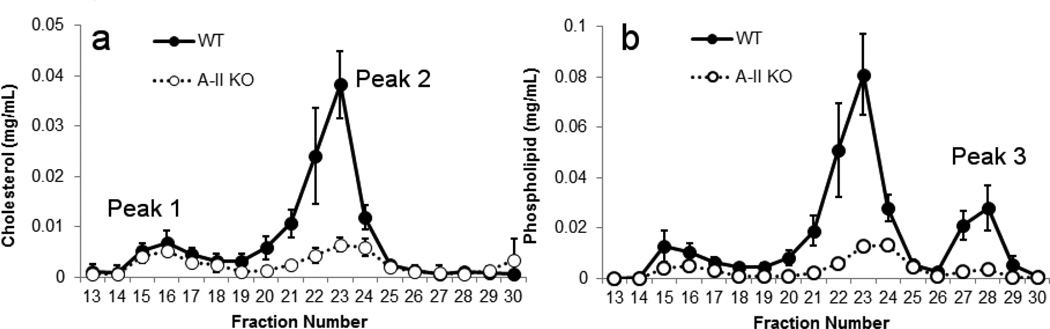
The apoA-II KO animals exhibited similar cholesterol levels in the VLDL/LDL (peak 1) as the WT, though the PL levels in this peak trended lower in the knockout (Fig. 2a and b). However, as was the case with apoA-I KO, apoA-II KO mice exhibited a massive decrease in both CH and PL in the HDL size range (~80% decrease in PL and 62% decrease in cholesterol), again consistent with the original characterization of these animals [34]. The lipid that remained in the apoA-II KO mice was associated with smaller sized particles than those in apoA-I KO mice and was centered at about fraction 23–24. Interestingly, these animals showed an almost complete absence of PL in Peak 3, unlike the apoA-I KO.
Figure 2.
Size distribution profiles for mouse plasma separated by gel filtration chromatography – WT vs. apoA-II KO. This figure is set up exactly like Fig. 2, except that the KO strain is deficient in apoA-II.
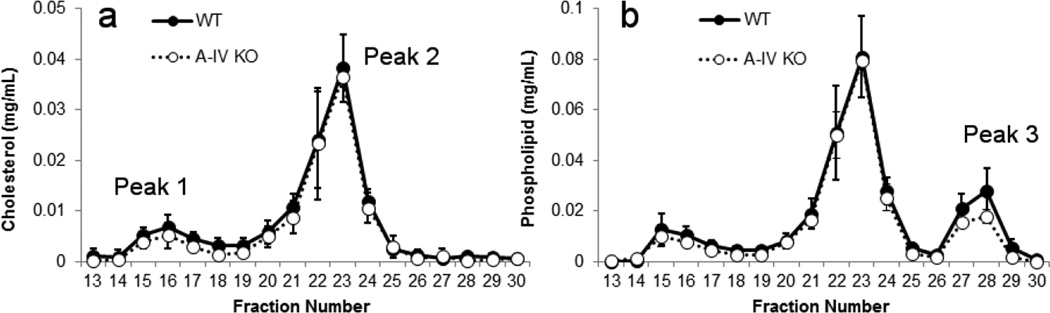
The deletion of apoA-IV in mice exhibited minimal effects on the plasma lipoprotein profile compared to apoA-I or apoA-II ablation. Fig. 3a and b shows that both the cholesterol and PL profiles were largely unchanged in both the VLDL/LDL and HDL range. Although a trend toward a reduction of PL levels in peak 3 was evident in the apoA-IV KO, this did not reach statistical significance.
Figure 3.
Size distribution profiles for mouse plasma separated by gel filtration chromatography – WT vs. apoA-IV KO. This figure is set up exactly like Fig. 2, except that the KO strain is deficient in apoA-IV.
Effects of platform protein knockout on lipid associated protein abundance and size distribution
In our previous characterization of the mouse lipoproteome [30], we identified 113 lipid-associated proteins in the WT C57Bl6 mice. We detected 114, 104 and 103 proteins in the apoA-I KO, apoA-IV KO, and apoA-II KO groups (summing WT controls and knockouts (KO)), respectively (n = 6 per group). No additional proteins were detected in any of the KO models compared to the corresponding WT group. For most of the detected proteins, the total peptide counts found in the apoA-II KO and WT cohorts were 30–50% lower than those in the apoA-I KO, and apoA-IV KO cohorts, despite the equal volumes of plasma sampled. We also noted increased mouse to mouse variability in the apoA-II KO comparison, most notably in the apoA-II KO group for reasons that were unclear.
Knockout of apoA-I
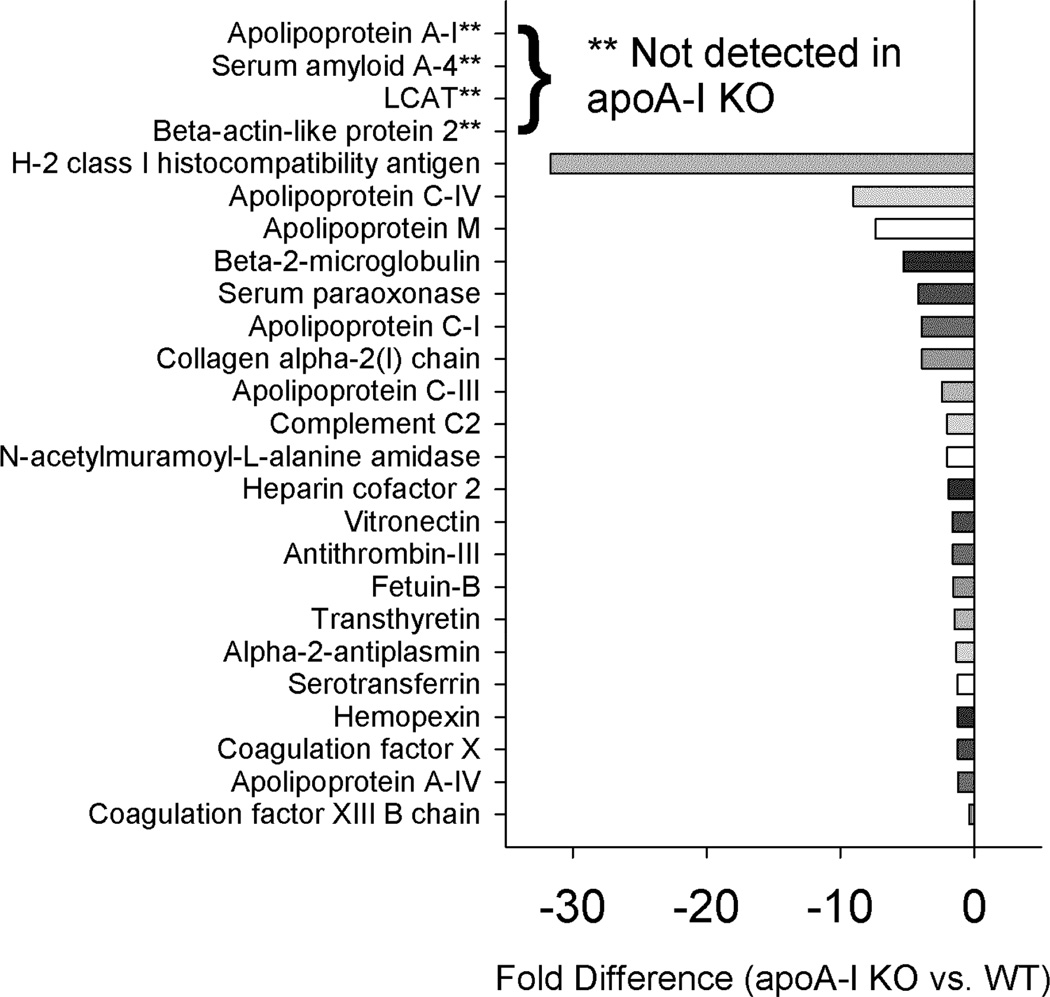
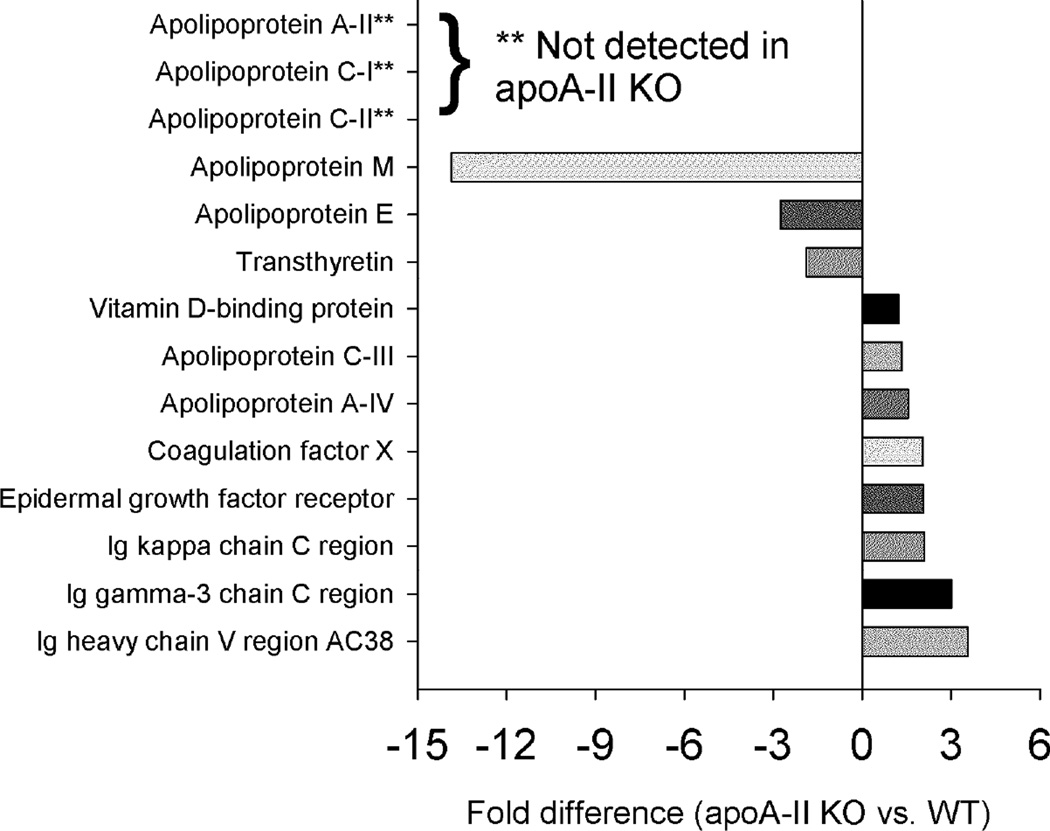
Statistically significant differences in the protein abundance summed across fractions for the WT vs. apoA-I KO mice are shown in Fig. 4. As expected, apoA-I was abundant in the WT but absent in the KO. Similarly, we were unable to detect β-actin-like protein 2, serum amyloid A-4 (SAA-4) and lecithin: cholesterol acyl transferase (LCAT) in the apoA-I KO mice. Many of the classic lipid associated “apos” were significantly decreased in the apoA-I KO animals including apos C-I, C-III, C-IV, M and A-IV. Additionally we saw clear decreases in levels of other well-recognized HDL proteins such as paraoxonase, heparin cofactor 2, vitronectin, transthyretin and hemopexin. While not known as a lipoprotein constituent, we noticed a striking reduction of H-2 class 1 histocompatibility antigen in the apoA-I KO mice.
Figure 4.
Changes in overall protein abundance – WT vs. apoA-I KO. Normalized peptide counts (see Methods) for each fraction were summed and compared with a t-test p<0.05. These were the tested by MS1 integration for three representative peptides, also compared with a t-test p<0.05. Results are shown only if they were significant by both peptide counting and MS1 integration strategies. Bars to the left reflect proteins that were lower in abundance in the KO.
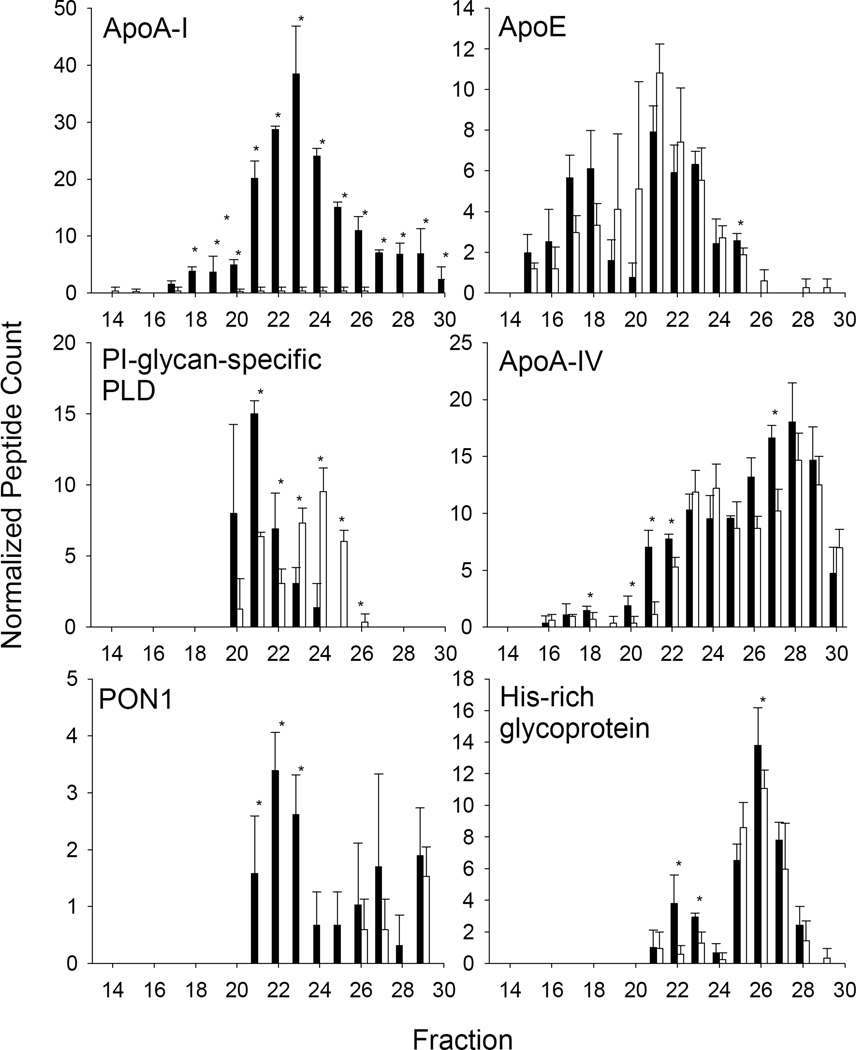
We also noted several proteins that underwent a clear shift in size distribution between the WT profile and apoA-I KO profile irrespective of whether they showed a change in plasma abundance. These included apoE, PI-glycan-specific phospholipase D, apoA-IV, paraoxonase 1 (PON1), and histidine-rich glycoprotein (Fig. 5). Other proteins that underwent a size shift included β-2-microglobulin and complement C2 (not shown). In the case of PI-glycan-specific PLD, most co-eluted with larger sized HDL particles in WT mice. However in the apoA-I KO, most of this protein shifted to a smaller sized pattern without a net loss of protein. PON1 on the other hand, shifted from predominantly HDL sized fractions to fractions containing lipid-poor proteins implying that is no longer lipoprotein-associated in the absence of an apoA-I scaffold. Histidine-rich glycoprotein eluted in two size ranges in the WT mice, a large species centered on fraction 22 and a smaller one centered on fraction 26. In the absence of apoA-I, the larger species was reduced without major changes in the smaller one. ApoA-IV and apoE did not show dramatic differences in overall abundance between the WT and apoA-I KO animals, but we did note subtle differences in the larger HDL fractions (19–21) with apoA-IV being reduced in those fractions but apoE actually becoming more abundant in those fractions in the apoA-I KO.
Figure 5.
Examples of proteins undergoing a size shift in response to apoA-I ablation. The normalized peptide counts (determined as in Fig. 5) for a given protein across each fraction are shown. For all figures, WT mouse data is represented by filled bars, apoA-I KO data is represented by open bars. Data represents a mean peptide count from 3 animals in each group. Stars indicate a significant difference between the groups by t-test p<0.05.
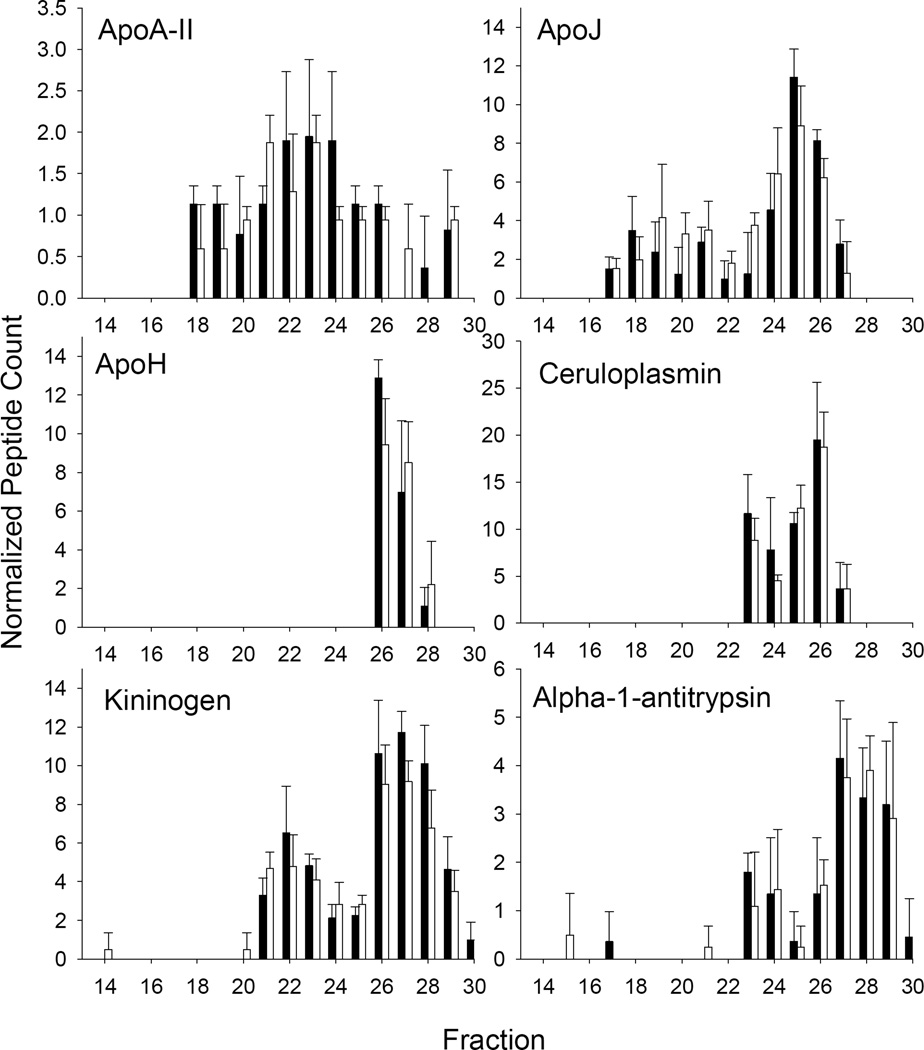
Despite these changes, we noted that other classically HDL-associated proteins did not differ remarkably in either abundance or elution pattern between WT and apoA-I KO (Fig. 6). Particularly striking was apoA-II which has long been thought to associate with apoA-I in HDL. Importantly, these similarities were noted not only using the peptide counting approach (as shown), but also with MS1 integration (not shown).
Figure 6.
Examples of proteins NOT undergoing a size shift in response to apoA-I ablation. The figure is set up as for Fig. 6.
Knockout of apoA-II
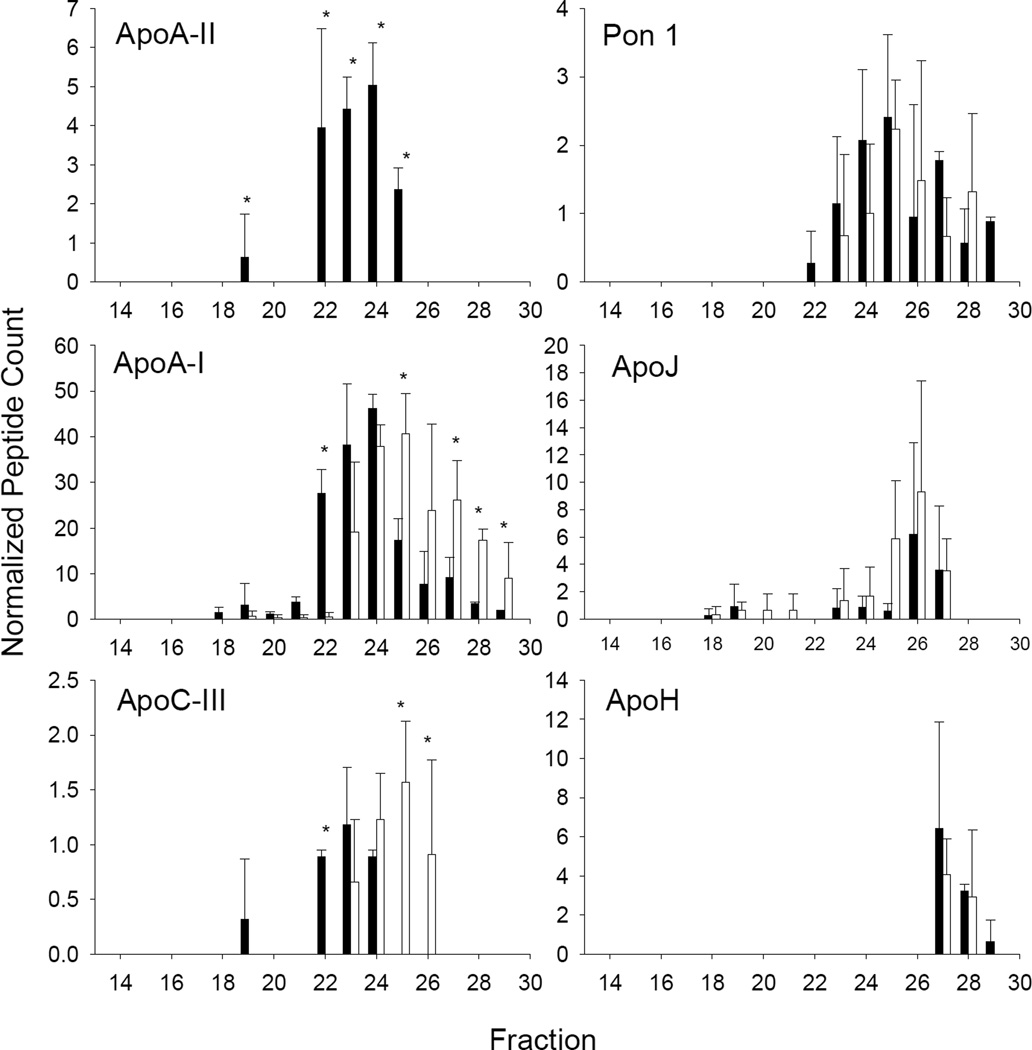
Genetic ablation of apoA-II also had profound effects on the abundance of several proteins. Along with apoA-II, we were unable to detect apoC-I and apoC-II in apoA-II KO animals (Fig. 7). Like in the apoA-I KO, circulating levels of apoM were substantially reduced in the absence of apoA-II. Interestingly, other proteins showed increased abundance when apoA-II was missing. These included apoC-III, apoA-IV and several IgG molecules. Most of the impact of apoA-II ablation involved changing abundance levels of proteins rather than changing their elution patterns. Some exceptions included apoA-I (Fig. 8, left), which while surprisingly abundant despite the lack of CH and PL seen in Fig. 1, shifted to smaller sized fractions in the absence of apoA-II. ApoC-III showed a similar shift to smaller particles. As was the case for the apoA-I KO, there were numerous proteins that were indifferent to the absence of apoA-II (examples shown in Fig. 8, right).
Figure 7.
Changes in overall protein abundance – WT vs. apoA-II KO. Normalized peptide counts (see Methods) for each fraction were summed and compared with a t-test p<0.05. These were the tested by MS1 integration for three representative peptides, also compared with a t-test p<0.05. Results are shown only if they were significant by both peptide counting and MS1 integration strategies. Bars to the left reflect proteins that were lower in the KO, bars to the right indicate higher abundance in the KO.
Figure 8.
Proteins patterns in response to apoA-II ablation. The figure is set up as for Fig. 6.
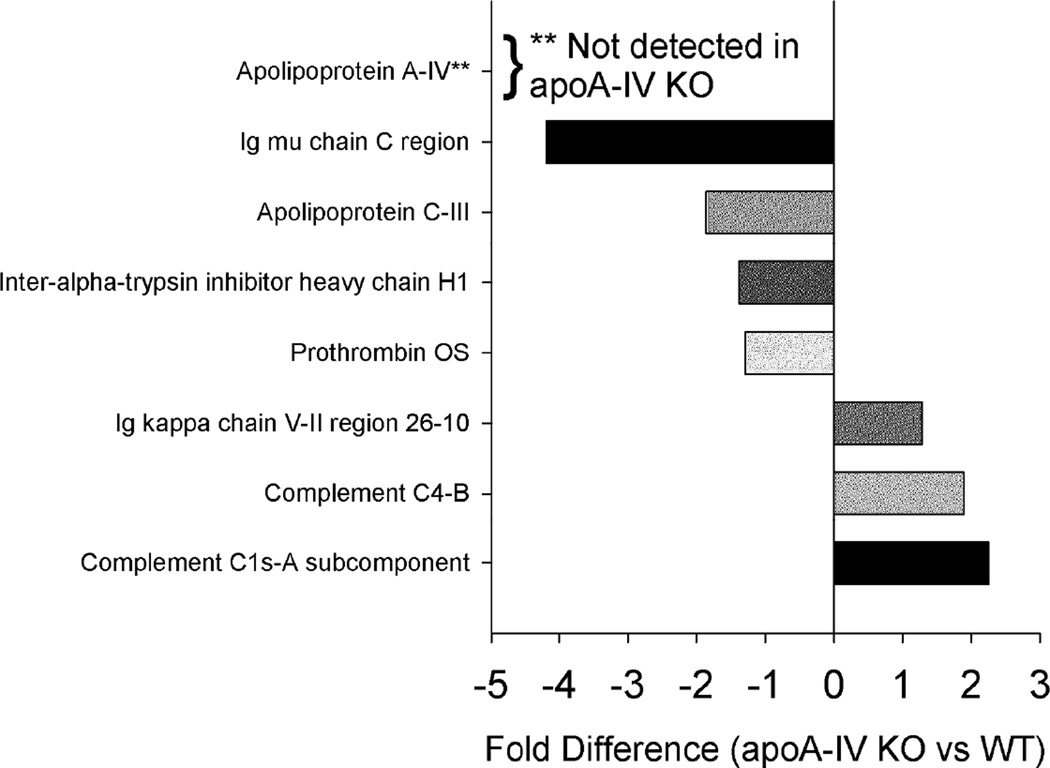
Knockout of apoA-IV
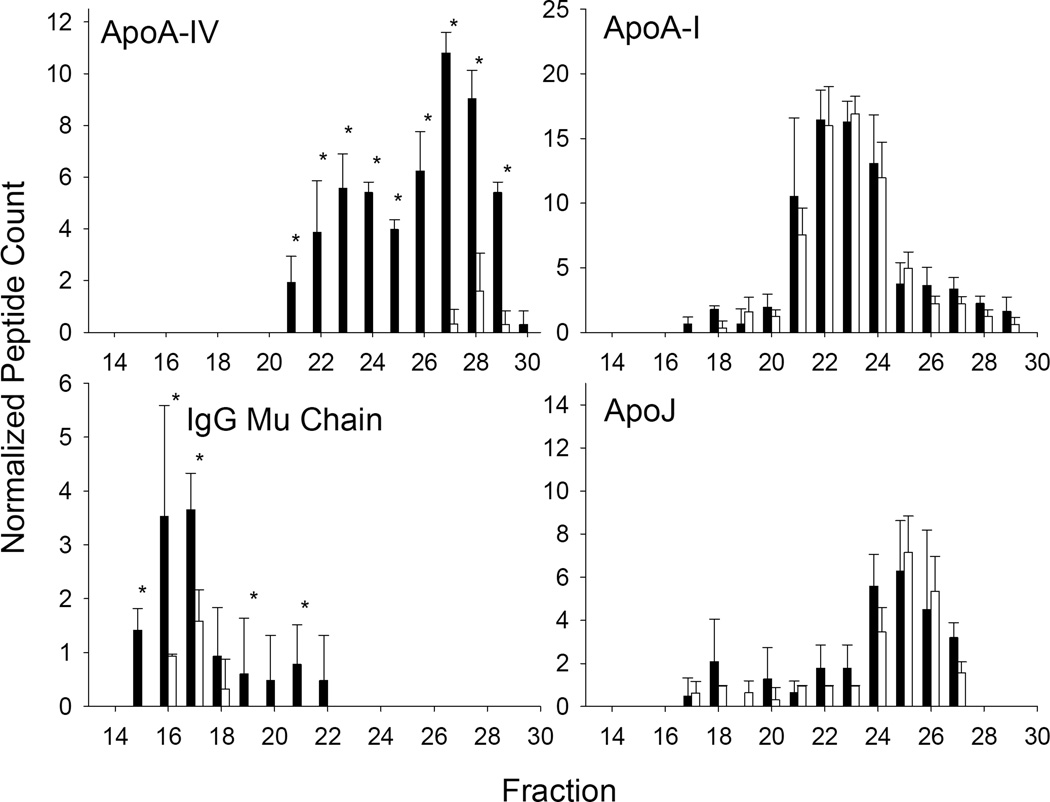
Of the three KO lines studied, the absence of apoA-IV had the most subtle effects on the remaining plasma lipoproteome. We noted significant differences in 7 proteins (Fig. 9) with some decreasing and others increasing slightly in the absence of apoA-IV. One striking observation was the reduction of a particular IgG molecule, the Mu chain. Despite little overlap between IgG Mu chain and apoA-IV in the WT animals (Fig. 10, left), the absence of apoA-I had a profound effect on the abundance of this much larger species. However, for the most part, the loss of apoA-IV had minimal effects on the remainder of the proteins detected. Some examples are shown in Fig. 10, right.
Figure 9.
Changes in overall protein abundance – apoA-IV KO vs. WT. Normalized peptide counts (see Methods) for each fraction were summed and compared with a t-test p<0.05. These were the tested by MS1 integration for three representative peptides, also compared with a t-test p<0.05. Results are shown only if they were significant by both peptide counting and MS1 integration strategies. Bars to the left reflect proteins that were lower in the KO, bars to the right indicate higher abundance in the KO.
Figure 10.
Proteins patterns in response to apoA-IV ablation. The figure is set up as for Fig. 6.
Discussion
The classic studies of Cheung and Albers [19] introduced the concept that HDL particles can be classified in terms of apoA-I and apoA-II content, specifically as LpA-I/A-II (with both proteins) and LpA-I (with apoA-I but not apoA-II). This suggested that apoA-I and apoA-II act as molecular scaffolds that sequester extracellular lipid into structures that allow the docking of additional proteins required for lipid metabolism, regulation of inflammation, innate immune functions, etc. These auxiliary proteins may either dock directly with the scaffold itself or with surface lipids once appropriately emulsified. This view is supported by the known interactions of apoA-I with plasma lipid modifying factors such as LCAT, for example [35]. However, our studies using gel filtration chromatography [20] and ultracentrifugation [18] have shown that most of these lower abundance proteins are found in the smaller, denser range of HDL particles. Recent understanding of the structure of apoA-I on small HDL3c particles [36] indicates that even two molecules of apoA-I can monopolize nearly 90% of the particle surface area, leaving little room for anything else. Thus, it is difficult to understand how apoA-I or even apoA-II can fulfill a scaffold role on these particles, unless they are predominantly making protein:protein contacts with the auxiliary proteins. Additionally, the summed molecular weights of many of these proteins quickly surpass the measured mass of an HDL3c particle when apoA-I is invoked as a scaffold. These observations led us to hypothesize a third class of lipoproteins that do not contain apoA-I or perhaps even apoA-II. Since most lipophilic proteins adopt a hydrated density that is overall similar to HDL when associated with even small amounts of lipid [37], these particles would co-isolate with HDL by ultracentrifugation. In the absence of alternative separation methods, such species would be ‘lumped in’ with LpA-I or LpA-I/A-II particles. We took advantage of the genetic tractability of the mouse to ascertain the role of the major structural apolipoproteins of the A class on the distribution of the minor proteins across plasma lipoproteins.
Being far and away the most abundant protein found in classically isolated HDL, knocking out apoA-I resulted in a dramatic drop in cholesterol and phospholipid levels in the HDL size range. In response, several of the classic “apo” proteins showed either substantial decreases in concentration or elution pattern changes. ApoM, SAA-4 and LCAT were below our threshold of detection in the apoA-I KO mice. This may indicate that an apoA-I-containing particle is required to recruit or stabilize these proteins on lipoproteins. Since we only measured lipid bound proteins (via the PL binding resin), we do not know if lipid-free versions were circulating in the KO animal or if they were cleared. The interaction of LCAT and apoA-I is well known, but while SAA-4 and apoM are also established HDL-associated proteins [38, 39], we are unaware of reports linking them specifically to apoA-I. Other proteins such as PON1, PI-glycan-specific PLD (GPI-PLD) and His-rich glycoprotein shifted their elution pattern from one of a lipid bound particle in the HDL size range to smaller species that may be lipid-poor. This strongly suggests that these proteins, or at least a fraction of their circulating forms, normally associate with apoA-I. This agrees with previous studies showing that PON1 binds to reconstituted LpA-I HDL particles with higher affinity than those containing apoA-II or apoA-IV [40]. Additionally, the majority of GPI-PLD was shown to associate with LpA-I particles in human plasma [41]. Still other proteins such as apoE and apoA-IV showed minor alterations in the apoA-I KO, specifically in larger sized HDL particles in the fraction 20–21 range. This may indicate that the lion’s share of these proteins exist on particles that are independent of apoA-I, but that there may be low abundance subpopulations containing both proteins. Again, this is largely consistent with 2-D gel electrophoresis experiments performed by Asztalos and colleagues showing that areas that react with anti-apoA-I antibodies are largely distinct from those which react with apoE and apoA-IV antibodies [22].
Perhaps more interesting from an HDL particle subclass viewpoint were the proteins that failed to exhibit a shift when apoA-I was ablated. This was surprising given the massive loss of PL in the apoA-I KO. Strikingly, apoA-II, well known to coexist with apoA-I in HDL, retained its overall plasma abundance and general elution pattern when apoA-I was ablated. This observation held up both when the protein levels were evaluated by peptide counting and by MS1 integration (though apoA-II levels in the apoA-I KO mice trended about 30% lower than WT by MS1). How apoA-II (8 kDa) maintains its size distribution in the absence of most of the HDL phospholipid and cholesterol is unclear. Given the well-known hydrophobicity of apoA-II, it may be that the protein self-associates into protein-rich particles of similar size to HDL in the absence of apoA-I and most of the plasma phospholipid. Other notable proteins that failed to undergo a change in pattern were apoJ and apoH. In humans, apoJ exists in two major populations of about 70–90 kDa (presumably lipid-poor) and in an HDL sized particle of 200 kDa or larger [42], similar to what we saw in mice. Immunoprecipitation studies showed that isolated apoJ-HDL was largely devoid of apoA-I [43]. ApoH is a 50 kDa glycoprotein that has been shown to be associated with a variety of lipoproteins. It eluted significantly larger than its molecular weight peaking around fraction 26 (67 kDa albumin eluted at fraction 28) suggesting its association with lipid containing particles that likely do not contain apoA-I. We did not see elution shifts in many of the more abundant plasma proteins like complement C3, serotransferin, gelsolin, vitamin D-binding protein, and many others. While these have been consistently found in HDL preparations, the exact distribution of these between lipid-free and lipid-bound is not fully known. Our data suggests that most of these proteins probably exhibit subpopulations that are associated with PL to an extent that allows them to float in the HDL density range (and bind our lipid binding resin), but these likely do not require cohabitation with apoA-I. Given the large number of known HDL-associating proteins that were not affected by the loss of apoA-I, we argue that a significant degree of the proteomic diversity attributed to HDL may be contributed by low abundance ‘particles’ that lack apoA-I.
Like apoA-I, knockout of apoA-II also had a profound effect on circulating plasma lipids, reducing them to a degree comparable to the apoA-I KO. This was noted in the original studies of these mice [44]. The authors speculated that apoA-II is an inhibitor of hepatic lipase (HL), and in the absence of apoA-II, HL runs amok reducing the HDL PL pool. This was supported by studies in apoA-II and HL double KO mice which showed a substantial recovery of HDL cholesterol [34]. Excess HL activity could be envisioned to affect a majority of PL-associated proteins, even ones that do not normally require apoA-II residence on the particle. Thus, the case of the apoA-II KO is more complicated than whether a given protein requires apoA-II to form their normal sized HDL particle “home”. For these reasons, we are reluctant to suggest that a shift in the size distribution or abundance of a given protein in the context of apoA-II ablation can necessarily be interpreted as evidence for their physical co-localization on lipoproteins. However, we did note a few interesting points. First, despite the lack of HDL PL, apoM levels were reduced, but not completely eliminated as they were in the apoA-I KO animals. This may be due to the relatively abundant apoA-I that remains in the plasma of apoA-II KO mice (Fig. 10). It is possible that even poorly lipidated apoA-I in these animals can stabilize apoM levels to some extent, further suggesting an interaction between these proteins. Also, the overall size patterns of apoJ and apoH were not greatly disturbed in the apoA-II KO suggesting that apoA-II may not cohabitate with these proteins. This also may mean that apoJ and apoH containing particles are relatively resistant to HL action. As with apoA-I, many other proteins were not affected by apoA-II KO either in abundance or pattern, again suggesting that these particles form independently of both proteins.
The absence of apoA-IV, on the other hand, did not affect the size distribution of many proteins nor lipids. Furthermore, in the absence of apoA-I and apoA-II, apoA-IV elution patterns were not profoundly affected, other than slight changes in abundance. These findings suggest that apoA-IV-containing HDL particles do not host apoA-I, apoA-II or many of the minor HDL proteins. Thus, apoA-IV may be present largely in isolation on its HDL subspecies. This is consistent with the studies of Asztalos et al [22] who have shown that apoA-IV migrates independently of apoA-I and apoA-II in a 2-D electrophoresis analysis of human plasma. The plasma distribution between lipid-poor apoA-IV and these HDL subspecies raises intriguing questions as to the physiological roles of each.
One striking result was the decrease in the mu chain of immunoglobulins (Ig). This chain was not significantly affected in either the apoA-I or apoA-II KO studies. Proteomic studies of HDL routinely find Ig molecules and it is not clear whether they are truly associated with the particle or are contaminants due to their high plasma abundance. The difference in the mu chain is particularly intriguing given the large differences in size (elution volume) between the Ig and apoA-IV-containing particles. While this observation may be a statistical anomaly needing further validation, the fact that IgM antibodies play an important role in primary defense mechanisms is in line with a host of other HDL proteome members involved in immunity.
In summary, we provided the first comprehensive analysis of the impact of genetic ablation of the class A apolipoproteins on the size and abundance of HDL auxiliary proteins. Our results indicate that apoA-I and apoA-II are important HDL structural proteins that serve as an assembly scaffold for a subset of auxiliary HDL proteins, particularly the C’s and apoM. Whether this requires specific protein:protein interactions or more subtle effects such as modulating lipid content or surface characteristics of the particles is an area of active investigation in our laboratory. The data also indicate that distinct HDL subpopulations exist that do not contain, nor depend on, apoA-I, apoA-II or apoA-IV and these contribute substantially to the proteomic diversity of HDL. Indeed, many lipid-containing complexes appear to assemble just fine in their absence. Unlike LDL, which can be defined by the presence of a single copy of apoB with additional proteins associated, HDL should not be defined solely by the presence of apoA-I (or apoA-II). Since the density of a given macromolecular complex is a weighted average of the density of its components, a simple calculation shows that any protein (d~1.3 g/ml) [45] only needs to bind 70–90% of its mass in phospholipid (d~1.04 g/ml) [46] to fall within the HDL density range and thereby co-isolate with apoA-I-containing HDL particles by density ultracentrifugation. Are these complexes generated by similar mechanisms, i.e. ATP binding cassette transporter A1 [47–49], that are known to generate apoA-I containing particles? Do these complexes play lipid transport roles like apoA-I containing particles, or do they represent a lower abundant class that are tasked to alternative functions like anti-inflammation, proteolysis, immunity support or cell proliferation/apoptosis? Further study may result in the identification of particles with superior cardio protective properties or altogether new HDL functions that may relate to other chronic inflammatory diseases. It may be that manipulation of these specialized species could prove more fruitful than non-specific HDL-raising strategies which have proven disappointing to date.
Supplementary Material
Significance.
Plasma levels of high density lipoproteins (HDL) are inversely correlated with cardiovascular disease. These particles are becoming known as highly heterogeneous entities that have diverse compositions and functions that may impact disease. Unfortunately, we know little about the forces that maintain the composition of each particle in plasma. It has been suggested that certain ‘scaffold’ proteins, such as apolipoprotein (apo) A-I, apoA-II and apoA-IV, may act as organizing centers for the docking of myriad accessory proteins. To test this hypothesis, we took advantage of the genetic tractability of the mouse model and ablated these three proteins individually. We then tracked the abundance and size profile of the remaining HDL proteins by gel filtration chromatography combined with mass spectrometry. The results clearly show that certain cohorts of proteins depend on each scaffold molecule to assemble normal sized HDL particles under wild-type conditions. This work forms the basis for more detailed studies that will define the specific compositions of HDL subspecies with the possibility of connecting them to specific functions or roles in disease.
Highlights.
Plasma levels of high density lipoproteins (HDL) are inversely correlated with cardiovascular disease, but little is understood about how they assemble.
Certain platform or scaffold apolipoproteins have been proposed to mediate HDL particle assembly.
Genetic ablation of apoA-I and apoA-II had dramatic impacts on circulating plasma lipids as well as the size pattern of several other apolipoproteins.
Genetic ablation of apoA-IV had minimal impact on plasma lipids and affected the abundance of only a few proteins.
Many lipid associated proteins did not show abundance or size differences in the absence of these platform proteins.
Acknowledgments
This work was supported by NIH grants HL67093 (WSD), HL104136 (WSD) and HL111829 (LJL) as well as a MicroMouse grant from the National Mouse Metabolic Phenotyping Center (WSD) and a pre-doctoral fellowship from the Great Rivers Affiliate of the American Heart Association (SMG).
Abbreviations
- Apo
apolipoprotein
- CVD
cardiovascular disease
- CSH
calcium silica hydrate
- HDL
high density lipoprotein
- HDL-C
high density lipoprotein cholesterol
- LDL
low density lipoprotein
- MS
mass spectrometry
- PL
phospholipid
- STB
standard Tris buffer
- UC
ultracentrifugation
- VLDL
very low density lipoprotein
- WT
wild-type
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Scott M. Gordon, Email: Scott.Gordon@nih.gov.
Hailong Li, Email: hailong.li@cchmc.org.
Xiaoting Zhu, Email: xiaoting.zhu@cchmc.org.
Patrick Tso, Email: Patrick.Tso@uc.edu.
Catherine A. Reardon, Email: reardon@uchicago.edu.
Amy S. Shah, Email: Amy.Shah@cchmc.org.
L. Jason Lu, Email: long.lu@cchmc.org.
References
- 1.Rosenson RS, Brewer HB, Jr, Chapman MJ, Fazio S, Hussain MM, Kontush A, Krauss RM, Otvos JD, Remaley AT, Schaefer EJ. HDL measures, particle heterogeneity, proposed nomenclature, and relation to atherosclerotic cardiovascular events. Clin Chem. 2011;57(3):392–410. doi: 10.1373/clinchem.2010.155333. [DOI] [PubMed] [Google Scholar]
- 2.Gordon T, Castelli WP, Hjortland MC, Kannel WB, Dawber TR. High density lipoprotein as a protective factor against coronary heart disease. The Framingham Study. Am.J.Med. 1977;62(5):707–714. doi: 10.1016/0002-9343(77)90874-9. [DOI] [PubMed] [Google Scholar]
- 3.Voight BF, Peloso GM, Orho-Melander M, Frikke-Schmidt R, Barbalic M, Jensen MK, Hindy G, Holm H, Ding EL, Johnson T, Schunkert H, Samani NJ, Clarke R, Hopewell JC, Thompson JF, Li M, Thorleifsson G, Newton-Cheh C, Musunuru K, Pirruccello JP, Saleheen D, Chen L, Stewart A, Schillert A, Thorsteinsdottir U, Thorgeirsson G, Anand S, Engert JC, Morgan T, Spertus J, Stoll M, Berger K, Martinelli N, Girelli D, McKeown PP, Patterson CC, Epstein SE, Devaney J, Burnett MS, Mooser V, Ripatti S, Surakka I, Nieminen MS, Sinisalo J, Lokki ML, Perola M, Havulinna A, de FU, Gigante B, Ingelsson E, Zeller T, Wild P, de Bakker PI, Klungel OH, Maitland-van der Zee AH, Peters BJ, de BA, Grobbee DE, Kamphuisen PW, Deneer VH, Elbers CC, Onland-Moret NC, Hofker MH, Wijmenga C, Verschuren WM, Boer JM, van der Schouw YT, Rasheed A, Frossard P, Demissie S, Willer C, Do R, Ordovas JM, Abecasis GR, Boehnke M, Mohlke KL, Daly MJ, Guiducci C, Burtt NP, Surti A, Gonzalez E, Purcell S, Gabriel S, Marrugat J, Peden J, Erdmann J, Diemert P, Willenborg C, Konig IR, Fischer M, Hengstenberg C, Ziegler A, Buysschaert I, Lambrechts D, Van de Werf F, Fox KA, El Mokhtari NE, Rubin D, Schrezenmeir J, Schreiber S, Schafer A, Danesh J, Blankenberg S, Roberts R, McPherson R, Watkins H, Hall AS, Overvad K, Rimm E, Boerwinkle E, Tybjaerg-Hansen A, Cupples LA, Reilly MP, Melander O, Mannucci PM, Ardissino D, Siscovick D, Elosua R, Stefansson K, O'Donnell CJ, Salomaa V, Rader DJ, Peltonen L, Schwartz SM, Altshuler D, Kathiresan S. Plasma HDL cholesterol and risk of myocardial infarction: a mendelian randomisation study. Lancet. 2012;380(9841):572–580. doi: 10.1016/S0140-6736(12)60312-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Masana L, Cabre A, Plana N. HPS2-THRIVE results: bad for niacin/laropiprant, good for ezetimibe? Atherosclerosis. 2013;229(2):449–450. doi: 10.1016/j.atherosclerosis.2013.06.006. [DOI] [PubMed] [Google Scholar]
- 5.Schwartz GG, Olsson AG, Abt M, Ballantyne CM, Barter PJ, Brumm J, Chaitman BR, Holme IM, Kallend D, Leiter LA, Leitersdorf E, McMurray JJ, Mundl H, Nicholls SJ, Shah PK, Tardif JC, Wright RS. Effects of dalcetrapib in patients with a recent acute coronary syndrome. N.Engl.J Med. 2012;367(22):2089–2099. doi: 10.1056/NEJMoa1206797. [DOI] [PubMed] [Google Scholar]
- 6.Zanoni P, Khetarpal SA, Larach DB, Hancock-Cerutti WF, Millar JS, Cuchel M, DerOhannessian S, Kontush A, Surendran P, Saleheen D, Trompet S, Jukema JW, De Craen A, Deloukas P, Sattar N, Ford I, Packard C, Majumder A, Alam DS, Di Angelantonio E, Abecasis G, Chowdhury R, Erdmann J, Nordestgaard BG, Nielsen SF, Tybjaerg-Hansen A, Schmidt RF, Kuulasmaa K, Liu DJ, Perola M, Blankenberg S, Salomaa V, Mannisto S, Amouyel P, Arveiler D, Ferrieres J, Muller-Nurasyid M, Ferrario M, Kee F, Willer CJ, Samani N, Schunkert H, Butterworth AS, Howson JM, Peloso GM, Stitziel NO, Danesh J, Kathiresan S, Rader DJ, Consortium CHDE, Consortium CAE. C. Global Lipids Genetics, Rare variant in scavenger receptor BI raises HDL cholesterol and increases risk of coronary heart disease. Science. 2016;351(6278):1166–1171. doi: 10.1126/science.aad3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davidson WS. Davidson Lab Webpage: HDL Proteome Watch. 2012 [Google Scholar]
- 8.Vaisar T, Pennathur S, Green PS, Gharib SA, Hoofnagle AN, Cheung MC, Byun J, Vuletic S, Kassim S, Singh P, Chea H, Knopp RH, Brunzell J, Geary R, Chait A, Zhao XQ, Elkon K, Marcovina S, Ridker P, Oram JF, Heinecke JW. Shotgun proteomics implicates protease inhibition and complement activation in the antiinflammatory properties of HDL. J.Clin.Invest. 2007;117(3):746–756. doi: 10.1172/JCI26206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karlsson H, Leanderson P, Tagesson C, Lindahl M. Lipoproteomics II: mapping of proteins in high-density lipoprotein using two-dimensional gel electrophoresis and mass spectrometry. HDL composition. 2005;5(5):1431–1445. doi: 10.1002/pmic.200401010. [DOI] [PubMed] [Google Scholar]
- 10.Shah AS, Tan L, Long JL, Davidson WS. Proteomic diversity of high density lipoproteins: our emerging understanding of its importance in lipid transport and beyond. J Lipid Res. 2013;54(10):2575–2585. doi: 10.1194/jlr.R035725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Phillips MC. New insights into the determination of HDL structure by apolipoproteins: Thematic review series: high density lipoprotein structure, function, and metabolism. Journal of Lipid Research. 2013;54(8):2034–2048. doi: 10.1194/jlr.R034025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang Y, Wu Z, Riwanto M, Gao S, Levison BS, Gu X, Fu X, Wagner MA, Besler C, Gerstenecker G, Zhang R, Li XM, DiDonato AJ, Gogonea V, Tang WH, Smith JD, Plow EF, Fox PL, Shih DM, Lusis AJ, Fisher EA, Didonato JA, Landmesser U, Hazen SL. Myeloperoxidase, paraoxonase-1, and HDL form a functional ternary complex. J.Clin.Invest. 2013;123(9):3815–3828. doi: 10.1172/JCI67478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shiflett AM, Bishop JR, Pahwa A, Hajduk SL. Human high density lipoproteins are platforms for the assembly of multi-component innate immune complexes. Journal of Biological Chemistry. 2005;280(38):32578–32585. doi: 10.1074/jbc.M503510200. [DOI] [PubMed] [Google Scholar]
- 14.Heinecke JW. The HDL proteome: a marker--and perhaps mediator--of coronary artery disease. Journal of Lipid Research. 2009;50(Suppl):S167–S171. doi: 10.1194/jlr.R800097-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kostner G, Alaupovic P. Studies of the composition and structure of plasma lipoproteins. Separation and quantification of the lipoprotein families occurring in the high density lipoproteins of human plasma. Biochemistry. 1972;11(18):3419–3428. doi: 10.1021/bi00768a015. [DOI] [PubMed] [Google Scholar]
- 16.Cheung MC, Albers JJ. Characterization of lipoprotein particles isolated by immunoaffinity chromatography. Particles containing A-I and A-II and particles containing A-I but no A-II. Journal of Biological Chemistry. 1984;259(19):12201–12209. [PubMed] [Google Scholar]
- 17.Coetzee GA, Strachan AF, van der Westhuyzen DR, Hoppe HC, Jeenah MS, de Beer FC. Serum amyloid A-containing human high density lipoprotein 3. Density, size, and apolipoprotein composition. Journal of Biological Chemistry. 1986;261(21):9644–9651. [PubMed] [Google Scholar]
- 18.Davidson WS, Silva RAGD, Chantepie S, Lagor WR, Chapman MJ, Kontush A. Proteomic Analysis of Defined HDL Subpopulations Reveals Particle-Specific Protein Clusters: Relevance to Antioxidative Function. Arterioscler Thromb Vasc Biol. 2009;29(6):870–876. doi: 10.1161/ATVBAHA.109.186031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheung MC, Albers JJ. Distribution of high density lipoprotein particles with different apoprotein composition: particles with A-I and A-II and particles with A-I but no A-II. Journal of Lipid Research. 1982;23(5):747–753. [PubMed] [Google Scholar]
- 20.Gordon SM, Deng J, Lu LJ, Davidson WS. Proteomic characterization of human plasma high density lipoprotein fractionated by gel filtration chromatography. J.Proteome.Res. 2010;9:5239–5249. doi: 10.1021/pr100520x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gordon SM, Deng J, Tomann AB, Shah AS, Lu LJ, Davidson WS. Multi-dimensional co-separation analysis reveals protein:protein interactions defining plasma lipoprotein subspecies. Mol Cell Proteomics. 2013 doi: 10.1074/mcp.M113.028134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Asztalos BF, Schaefer EJ. High-density lipoprotein subpopulations in pathologic conditions. Am.J.Cardiol. 2003;91(7A):12E–17E. doi: 10.1016/s0002-9149(02)03383-0. [DOI] [PubMed] [Google Scholar]
- 23.Gordon SM, Li H, Zhu X, Shah AS, Lu LJ, Davidson WS. A comparison of the mouse and human lipoproteome: suitability of the mouse model for studies of human lipoproteins. J.Proteome.Res. 2015;14(6):2686–2695. doi: 10.1021/acs.jproteome.5b00213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weinstock PH, Bisgaier CL, Hayek T, Aalto-Setala K, Sehayek E, Wu L, Sheiffele P, Merkel M, Essenburg AD, Breslow JL. Decreased HDL cholesterol levels but normal lipid absorption, growth, and feeding behavior in apolipoprotein A-IV knockout mice. Journal of Lipid Research. 1997;38:1782–1794. [PubMed] [Google Scholar]
- 25.Wang F, Kohan AB, Kindel TL, Corbin KL, Nunemaker CS, Obici S, Woods SC, Davidson WS, Tso P. Apolipoprotein A-IV improves glucose homeostasis by enhancing insulin secretion. Proc.Natl.Acad.Sci.U.S.A. 2012;109(24):9641–9646. doi: 10.1073/pnas.1201433109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Markwell MA, Haas SM, Bieber LL, Tolbert NE. A modification of the Lowry procedure to simplify protein determination in membrane and lipoprotein samples. Analytical Biochemistry. 1978;87(1):206–210. doi: 10.1016/0003-2697(78)90586-9. [DOI] [PubMed] [Google Scholar]
- 27.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal.Chem. 2002;74(20):5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 28.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal.Chem. 2003;75(17):4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 29.MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, Kern R, Tabb DL, Liebler DC, MacCoss MJ. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26(7):966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gordon SM, Li H, Zhu X, Shah AS, Lu LJ, Davidson WS. A comparison of the mouse and human lipoproteome: suitability of the mouse model for studies of human lipoproteins. J Proteome Res. 2015;14(6):2686–2695. doi: 10.1021/acs.jproteome.5b00213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiao S, Cole TG, Kitchens RT, Pfleger B, Schonfeld G. Genetic heterogeneity of lipoproteins in inbred strains of mice: analysis by gel-permeation chromatography. Metabolism. 1990;39(2):155–160. doi: 10.1016/0026-0495(90)90069-o. [DOI] [PubMed] [Google Scholar]
- 32.Switzer S, Eder HA. Transport of lysolecithin by albumin in human and rat plasma. Journal of Lipid Research. 1965;6(4):506–511. [PubMed] [Google Scholar]
- 33.Williamson R, Lee D, Hagaman J, Maeda N. Marked reduction of high density lipoprotein cholesterol in mice genetically modified to lack apolipoprotein A-I. Proc.Natl.Acad.Sci.U.S.A. 1992;89(15):7134–7138. doi: 10.1073/pnas.89.15.7134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weng W, Brandenburg NA, Zhong S, Halkias J, Wu L, Jiang XC, Tall A, Breslow JL. ApoA-II maintains HDL levels in part by inhibition of hepatic lipase. Studies In apoA-II and hepatic lipase double knockout mice. Journal of Lipid Research. 1999;40(6):1064–1070. [PubMed] [Google Scholar]
- 35.Soutar AK, Garner CW, Baker HN, Sparrow JT, Jackson RL, Gotto AM, Smith LC. Effect of the human plasma apolipoproteins and phosphatidylcholine acyl donor on the activity of lecithin: cholesterol acyltransferase. Biochemistry. 1975;14(14):3057–3064. doi: 10.1021/bi00685a003. [DOI] [PubMed] [Google Scholar]
- 36.Huang R, Silva RA, Jerome WG, Kontush A, Chapman MJ, Curtiss LK, Hodges TJ, Davidson WS. Apolipoprotein A-I structural organization in high-density lipoproteins isolated from human plasma. Nat.Struct.Mol.Biol. 2011;18(4):416–422. doi: 10.1038/nsmb.2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davidson WS, Jonas A, Clayton DF, George JM. Stabilization of alpha-synuclein secondary structure upon binding to synthetic membranes. Journal of Biological Chemistry. 1998;273(16):9443–9449. doi: 10.1074/jbc.273.16.9443. [DOI] [PubMed] [Google Scholar]
- 38.de Beer MC, Yuan T, Kindy MS, Asztalos BF, Roheim PS, de Beer FC. Characterization of constitutive human serum amyloid A protein (SAA4) as an apolipoprotein. Journal of Lipid Research. 1995;36(3):526–534. [PubMed] [Google Scholar]
- 39.Christoffersen C, Ahnstrom J, Axler O, Christensen EI, Dahlback B, Nielsen LB. The signal peptide anchors apolipoprotein M in plasma lipoproteins and prevents rapid clearance of apolipoprotein M from plasma. J Biol Chem. 2008;283(27):18765–18772. doi: 10.1074/jbc.M800695200. [DOI] [PubMed] [Google Scholar]
- 40.Gaidukov L, Tawfik DS. High affinity, stability, and lactonase activity of serum paraoxonase PON1 anchored on HDL with ApoA-I. Biochemistry. 2005;44(35):11843–11854. doi: 10.1021/bi050862i. [DOI] [PubMed] [Google Scholar]
- 41.Deeg MA, Bierman EL, Cheung MC. GPI-specific phospholipase D associates with an apoA-I- and apoA-IV-containing complex. J Lipid Res. 2001;42(3):442–451. [PubMed] [Google Scholar]
- 42.Jenne DE, Lowin B, Peitsch MC, Bottcher A, Schmitz G, Tschopp J. Clusterin (complement lysis inhibitor) forms a high density lipoprotein complex with apolipoprotein A-I in human plasma. Journal of Biological Chemistry. 1991;266(17):11030–11036. [PubMed] [Google Scholar]
- 43.Stuart WD, Krol B, Jenkins SH, Harmony JA. Structure and stability of apolipoprotein J-containing high-density lipoproteins. Biochemistry. 1992;31(36):8552–8559. doi: 10.1021/bi00151a024. [DOI] [PubMed] [Google Scholar]
- 44.Weng W, Breslow JL. Dramatically decreased high density lipoprotein cholesterol, increased remnant clearance, and insulin hypersensitivity in apolipoprotein A-II knockout mice suggest a complex role for apolipoprotein A-II in atherosclerosis susceptibility. Proc.Natl.Acad.Sci.U.S.A. 1996;93:14788–14794. doi: 10.1073/pnas.93.25.14788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Quillin ML, Matthews BW. Accurate calculation of the density of proteins. Acta Crystallogr.D.Biol.Crystallogr. 2000;56(Pt 7):791–794. doi: 10.1107/s090744490000679x. [DOI] [PubMed] [Google Scholar]
- 46.Small DM. Phospholipids, The Physical Chemistry of Lipids: From Alkanes to Phospholipids. New York: Plenum Press; 1986. pp. 475–517. [Google Scholar]
- 47.Rust S, Rosier M, Funke H, Real J, Amoura Z, Piette JC, Deleuze JF, Brewer HB, Duverger N, Denefle P, Assmann G. Tangier disease is caused by mutations in the gene encoding ATP-binding cassette transporter 1. Nat.Genet. 1999;22(4):352–355. doi: 10.1038/11921. [DOI] [PubMed] [Google Scholar]
- 48.Brooks-Wilson A, Marcil M, Clee SM, Zhang LH, Roomp K, van Dam M, Yu L, Brewer C, Collins JA, Molhuizen HO, Loubser O, Ouelette BF, Fichter K, Ashbourne-Excoffon KJ, Sensen CW, Scherer S, Mott S, Denis M, Martindale D, Frohlich J, Morgan K, Koop B, Pimstone S, Kastelein JJ, Hayden MR. Mutations in ABC1 in Tangier disease and familial high-density lipoprotein deficiency. Nat.Genet. 1999;22(4):336–345. doi: 10.1038/11905. [DOI] [PubMed] [Google Scholar]
- 49.Lawn RM, Wade DP, Garvin MR, Wang X, Schwartz K, Porter JG, Seilhamer JJ, Vaughan AM, Oram JF. The Tangier disease gene product ABC1 controls the cellular apolipoprotein-mediated lipid removal pathway. J.Clin.Invest. 1999;104(8):R25–R31. doi: 10.1172/JCI8119. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.