Abstract
Design of high-quality primers for multiple target sequences is essential for qPCR experiments, but is challenging due to the need to consider both homology tests on off-target sequences and the same stringent filtering constraints on the primers. Existing web servers for primer design have major drawbacks, including requiring the use of BLAST-like tools for homology tests, lack of support for ranking of primers, TaqMan probes and simultaneous design of primers against multiple targets. Due to the large-scale computational overhead, the few web servers supporting homology tests use heuristic approaches or perform homology tests within a limited scope. Here, we describe the MRPrimerW, which performs complete homology testing, supports batch design of primers for multi-target qPCR experiments, supports design of TaqMan probes and ranks the resulting primers to return the top-1 best primers to the user. To ensure high accuracy, we adopted the core algorithm of a previously reported MapReduce-based method, MRPrimer, but completely redesigned it to allow users to receive query results quickly in a web interface, without requiring a MapReduce cluster or a long computation. MRPrimerW provides primer design services and a complete set of 341 963 135 in silico validated primers covering 99% of human and mouse genes. Free access: http://MRPrimerW.com.
INTRODUCTION
Polymerase chain reaction (PCR) is a widely adopted technique for fast mass duplication of specific DNA sequences. As a standard laboratory technique, PCR is used in a wide variety of applications including phylogenetic analysis (1–3), genetic testing (4) and DNA cloning (5). In particular, quantitative PCR (qPCR), also known as real-time PCR, is commonly used to confirm the results of high-throughput experiments by validating changes in the expression of multiple selected genes (6).
Optimal primer design is essential for best results in all PCR applications. Manual design of primers is time-consuming and may easily yield incorrect results due to the need to simultaneously consider a large number of filtering constraints on each primer and primer pair (7). Another important consideration in primer design is homology testing, i.e. verifying that the designed primers will only amplify the target sequence(s) rather than off-target sequences; such tests usually require an additional BLAST-like tool. Fast automatic design of high-quality primers that satisfy both filtering constraints and homology tests remains a challenge that has not yet been completely solved, especially when simultaneously designing a large number of primers for qPCR that satisfy the same set of stringent and uniform constraints. For qPCR experiments, in addition to the above SYBR Green primers, TaqMan probes are also commonly used to detect products and they can significantly increase the specificity of detection; however, this requires extreme care in the design of both probes and primers to ensure they satisfy both the filtering constraints and the homology tests (7).
To aid in designing primers for PCR experiments, many websites have been developed, including Primer3Plus (8,9), BatchPrimer3 (10), Primique (11), QuantPrime (12), primer-BLAST (13) and PrimerBank (6,7). Primer3Plus, a web interface of Primer3, is one of the most widely used tools; it allows users to specify a set of filtering constraints for a single target gene. BatchPrimer3, which adopts the Primer3 core algorithm, can design primers in batches for multiple target genes. However, neither server performs homology tests on off-target sequences, requiring users to perform time-consuming homology tests on each candidate primer pair using extrinsic alignment tools. Primique performs homology tests using BLAST in a limited scope, i.e. only on a small secondary set of off-target sequences uploaded by the user. Due to a high-computation overhead of homology testing, the maximum size of this secondary database is limited to 10 MB, much smaller than a whole genome sequence database and therefore too small for the design of high-quality primers. QuantPrime performs homology testing for primer pairs designed by Primer3 against the whole transcriptome (mRNA) and genome database using BLAST. Both Primique and QuantPrime rely on a local alignment algorithm for homology testing. However, a heuristic approach based on local alignment cannot accurately count the number of mismatches between a primer and an off-target sequence (13); as a result, these methods could yield suboptimally specific primer pairs. On the contrary, Primer-BLAST performs homology tests with a global alignment algorithm to ensure full primer-target alignment; accordingly, Primer-BLAST tends to return more target-specific primer pairs. Although Primer-BLAST exhibits better performance in terms of homology testing, it does not rank the designed primer pairs by their penalty scores, but ranks them by their specificity; moreover, it cannot support batch design for multi-target qPCR due to the large computational overhead required for more accurate homology tests. Some websites, including PrimerBank (6,14), RTPrimerDB (15–17) and qPrimerDepot (18), simply search a database of pre-designed primers, rather than designing primers in real time in response to user queries. In particular, PrimerBank is one of the largest databases of primers built and updated over the past several years. Because the specificities of the primers of PrimerBank have been experimentally validated under uniform conditions, these primers are fairly effective in real PCR experiments. However, because PrimerBank relies on the pre-designed primers, it does not allow users to adjust the filtering constraints, which might be important in the context of qPCR experiments requiring a full set of primer pairs that satisfy the same constraints.
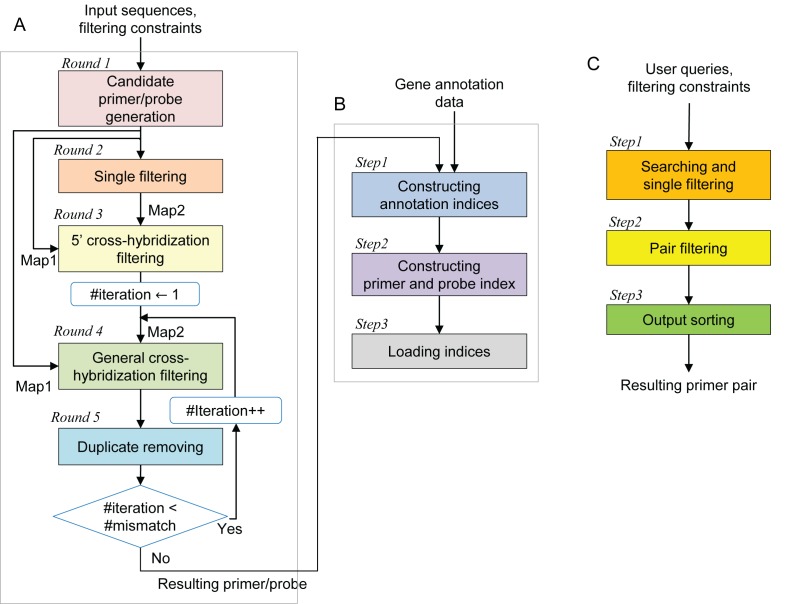
Here, we describe a new website, MRPrimerW, for batch design of primers for qPCR experiments. This tool checks filtering constraints, performs rigorous homology testing against a whole genome database, and ranks the resultant primer pairs according to their penalty scores to pick the best one for each target sequence. MRPrimerW supports the design of not only SYBR Green primers, but also TaqMap probes. A comparison of MRPrimerW with other existing tools is summarized in Table 1. MRPrimerW is an online processing method based on our previously proposed offline processing method MRPrimer (18), which returns all feasible and valid primer pairs for a DNA database at once. MRPrimer performs a fairly complex, large-scale computation based on the MapReduce framework, resulting in design of very high-quality primers. Through qPCR analysis using 343 primer pairs and corresponding sequencing and comparative analyses, we showed that the primer pairs designed by MRPrimer are very stable and effective in qPCR experiments. However, although MRPrimer can design very high-quality primers, routine use is inconvenient because it runs on a cluster of computers and requires several hours of runtime when the filtering constraints are adjusted. MRPrimerW solved this problem completely. On the MRPrimerW website, users can rapidly design primers of the same high quality without using their own computer cluster, typically within a minute, while instantly and freely adjusting filtering constraints. To achieve this level of performance, we adopted an approach based on Google's search system. In particular, we reorganized the complex MRPrimer algorithm, which consists of seven MapReduce rounds, into two parts: offline processing and online processing (Figure 1). We built index structures using the results of offline processing and loaded them into the MRPrimerW web server. Using these indices, the online processing stage can quickly design high-quality primers against a user-specified target, as in a Google keyword search.
Table 1. Comparison among websites for primer design.
| Method | Batch designing | Filtering constraints | Homology test | Scoring (Ranking) | TaqMan probes |
|---|---|---|---|---|---|
| Primer3Plus (9) | X | O | X | O | O |
| BatchPrimer3 (10) | O | O | X | O | O |
| Primique (11) | O | O | Δ | O | X |
| QuantPrime (12) | O | O | Δ | O | X |
| Primer-BLAST (13) | X | O | O | Δa | O |
| PrimerBank (6,7) | X | X | O | X | X |
| MRPrimerW | O | O | O | O | O |
O: fully supported.
Δ: partially supported.
X: not supported.
aPrimer-BLAST ranks the designed primer pairs not by penalty scores, but by specificity.
Figure 1.
Overall flow of the MRPrimerW method. MRPrimerW mainly consists of offline processing part, (A and B), and online processing part (C). (A) Offline processing part, which is irrelevant to queries, designs validated candidate primers and probes (five MapReduce rounds). (B) Then the resultant primers, probes and gene annotation data are converted into indices and the indices are loaded into database. (C) Online processing part, which the website performs, searches primers applied user-defined filtering constraints and then outputs the best primer pairs for the targets.
MATERIALS AND METHODS
Offline processing by MRPrimerW, which is independent of user queries, generates all validated candidate SYBR Green primers and TaqMan probes satisfying homology tests. Homology testing on an entire sequence database can be achieved by a large-scale self-join computation without specifying a target sequence. Because this stage of processing performs homology tests for every candidate primer and probe against the entire sequence database via a non-heuristic approach, the resultant primers and probes are all target gene–specific, and at the same time no valid (i.e. target gene specific) primers and probes are missed. Offline processing takes at least several hours on a cluster of computers (e.g. 10 PCs). On the other hand, the online processing stage is responsive to user queries, i.e. a specified set of target genes. This stage quickly searches for the best primer pairs for the target genes and shows them to the user, and in particular returns the best pair among all valid primers that satisfy user-specified filtering constraints for the corresponding target gene. Along with SYBR Green primer pairs, online processing returns TaqMan probes for the target gene, if applicable. As with MRPrimer, the criteria used for ranking the primers in MRPrimerW are the same as those used in Primer3Plus (18).
Offline processing
Offline processing by MRPrimerW takes as input a DNA sequence database and several filtering constraints, and yields as output all possible primers that satisfy both homology testing and given filtering constraints (Figure 1A). As an input source DNA sequence database for MRPrimerW, we used the consensus coding sequence (CCDS) database for human and mouse genes (https://www.ncbi.nlm.nih.gov/CCDS/) (Table 2). We selected these templates because the gene annotations have been defined by extensive manual curation and are represented consistently in the NCBI, Ensembl and UCSC Genome Browsers (19–21). The most up-to-date CCDS datasets contain 31 394 human gene sequences (Release 18, the last update was 12 May 2015) and 24 833 mouse gene sequences (Release 19, the last update was 30 July 2015). About 1% of human and mouse genes in CCDS do not have any target gene-specific primers; as a result, the offline processing stage produced valid primers for 31 376 human genes (99%) and 24 797 mouse genes (99%), comprising 165 923 450 and 176 039 685 distinct primers, respectively.
Table 2. Statistics of MRPrimerW primers.
| Human | Mouse | Both species | |
|---|---|---|---|
| Total number of genes | 31 394 | 24 833 | 56 227 |
| Number of covered genes (%) | 31 376 (99%) | 24 797 (99%) | 56 173 (99%) |
| Number of valid primers | 165 923,450 | 176 039 685 | 341 963 135 |
For filtering constraints, MRPrimerW considers eight parameters for each primer and five parameters for each pair, as in MRPrimer (Table 3). Most of these constraints are checked during online processing. However, four parameters (primer length, melting temperature, GC content and contiguous residue) are checked during offline processing, because primers with values out of the appropriate range (e.g. primer length 10 bp) are non-functional in general; consequently, they do not need to be considered during online processing. Table 3 shows the list of filtering constraints used in offline and online processing. The parameter ranges in the ‘Online’ column indicate the default settings, which can be adjusted in each web search.
Table 3. List of filtering constraints used in offline and online processing of MRPrimerW.
| Parameter | Value range | ||
|---|---|---|---|
| Offline | Online (default)** | ||
| Each primer | primer length | 17–27 bp | 19–23 bp |
| melting temperature (TM)* | 56–64°C | 58–62°C | |
| GC content | 30–70% | 40–60% | |
| self-complementarity | - | <5-mer | |
| 3′ self-complementarity | - | <4-mer | |
| Contiguous residue | <5-mer | <6-mer | |
| Gibbs free energy (ΔG) | - | ≥-9 kcal/mol | |
| Hairpin | <3-mer | ||
| Primer pair | length difference | - | ≤5-mer |
| TM difference | - | ≤3°C | |
| product size | - | 100–250 bp | |
| pair-complementarity | - | <5-mer | |
| 3′ pair-complementarity | - | <4-mer | |
-indicates not applicable.
*To calculate the melting temperature, we adopted the nearest-neighbor thermodynamic model (22).
**The value ranges in this column indicate the default setting, which can be freely adjusted by users.
Offline processing consists of five MapReduce rounds (Figure 1A). The first and second rounds generate all possible subsequences satisfying the four filtering constraints described in the ‘Offline’ column of Table 3 for forward and reverse primers. The next three rounds perform homology tests on the resultant candidate primers. The 5′ cross-hybridization filtering round (Round 3) eliminates candidate primers that are the same as any subsequence of an off-target sequence at the 3′ end and has only a few mismatches (up to four mismatches) at the 5′ end, and thus might cross-hybridize with an off-target sequence due to their high complementarity, especially at the 3′ end. The general cross-hybridization filtering round (Round 4) eliminates candidate primers that are similar to any subsequence of an off-target sequence (up to two mismatches anywhere). The duplicate removal round (Round 5) eliminates false-positive primers that still violate the general cross-hybridization filtering constraint. Rounds 4 and 5 are iterated until the checking of the general cross-hybridization filtering constraint is completed. The details of offline processing algorithm flow are shown in Supplementary Figure S1. For TaqMap probes, we performed the same offline processing algorithm flow with a different set of filtering constraints (Supplementary Table S1). The large-scale computation of each round of homology testing relies on distributed data processing in MapReduce. Details and examples of Rounds 3–5 are described in a previous publication (22).
Building indices
After offline processing, we create a set of indices based on the results, which are then loaded into the main memory of the web server for online processing (Figure 1B). The detailed structures of the indices are illustrated in Supplementary Figure S1. Nine indices are built: six partial annotation indices, one full annotation index, one primer index and one probe index. All indices follow the structure of a key-value database, in which each row is a pair of key and value. Annotation data were downloaded from GenBank ftp (ftp://ftp.ncbi.nlm.nih.gov/genomes/).
Online processing supports six kinds of queries (i.e. ‘Search by’ options) including NCBI gene symbol, NCBI CCDS ID, NCBI gene ID, GenBank accession number, GenBank alias and keyword. Accordingly, six partial annotation indices are used for the various query types (Supplementary Figure S2A and B). The key portions of the indices are used for matching with user queries. For instance, if a user sets the query type to ‘NCBI Gene Symbol’ and inputs ‘Adcy6 Anxa2 Cacna1c’ in the text field of the website, those three symbols are used to match with key portions of the corresponding index. The value portions of the indices are single sequence IDs (sids) or lists of sids in which the key occurs in the full annotation index and the primer index.
The full annotation index, which simply combines all six kinds of annotation information, is used to generate the resulting web page (Supplementary Figure S2C). The primer index contains primer sequences and positions (Supplementary Figure S2D). The key portion is a pair of sid and primer length, and the value portion is a pair consisting of the primer sequence and the <sid, pos> where the primer sequence occurs. For example, when a user inputs gene symbol ‘Olfr156,’ MRPrimerW first accesses the partial annotation index for NCBI Gene Symbol and finds a sid corresponding to ‘Olfr156’. Then, using the sid as the key, MRPrimerW retrieves a set of candidate primers, especially their sequences and positions, from the primer index, which are subjected to online processing.
Online processing
Online processing consists of three steps that check the filtering constraints provided by the users and rank the primers to return the top-1 best primers (Figure 1C). The first step takes the user query and uses the indices to retrieve all candidate primers containing the query. While extracting the candidates, MRPrimerW applies eight filtering constraints for each primer, described in Table 3. Here, the constraints for length, melting temperature, GC content and contiguous residue must be within the range pre-defined in offline processing. The second step applies five filtering constraints for primer pairs, described in Table 3. For this purpose, MRPrimerW performs a self-join computation on each group of candidate primers from the same target sequence, i.e. it joins forward primers and backward primers into the same group. This pair-filtering step may take a long time if the length of the input query (i.e. the number of gene symbols) is long or the number of candidate primers retrieved is very large.
The final step calculates the penalty scores of the primers obtained in the previous step and sorts the primers according to their scores. Then, it returns the top-1 best primer, i.e. the primer with the lowest penalty score, for each target sequence. The penalty score is calculated according to the method used in Primer3Plus (8). If the user inputs 12 target genes, MRPrimerW shows the 12 top hits, which can be used for qPCR experiment in most cases. However, if some of the target genes have no top-1 best primers, the user can relax the filtering constraints (i.e. using Advanced Settings) and click the search button to design a set of top primers that satisfy the same stringent filtering constraints and are target gene-specific. If a user selects the TaqMan probe design option, MRPrimerW returns a TaqMan probe for each target gene, where the probe is located between forward and reverse primers. Since in many cases users do not change default settings on filtering constraints, the response time can be improved by using the cached top-1 primer pairs index, which stores the top-1 primer pairs under the default setting for sequences of the database in key-value format in the main memory of the web server (Supplementary Figure S2E).
WEB SERVER
The MRPrimerW web server is implemented using Redis (http://redis.io/), an in-memory key-value store, for data management. Redis supports various kinds of data structures for various types of values, including string, hash, list, set and sorted set. Among these, MRPrimerW uses hash and set for annotation and primer indices (Supplementary Figure S1). In detail, for the server side, we adopted phpredis (https://github.com/phpredis/phpredis) for communication between Redis and PHP, and AJAX (asynchronous JavaScript and XML) for client-server communication. For the client side, MRPrimerW generates web pages using HTML with CSS and bootstrap (http://getbootstrap.com/) for styling interactive user-interface components. For dynamic HTML behavior, we used JavaScript and jQuery. MRPrimerW supports most major web browsers including Microsoft Internet Explorer, Google Chrome, Apple Safari and Opera.
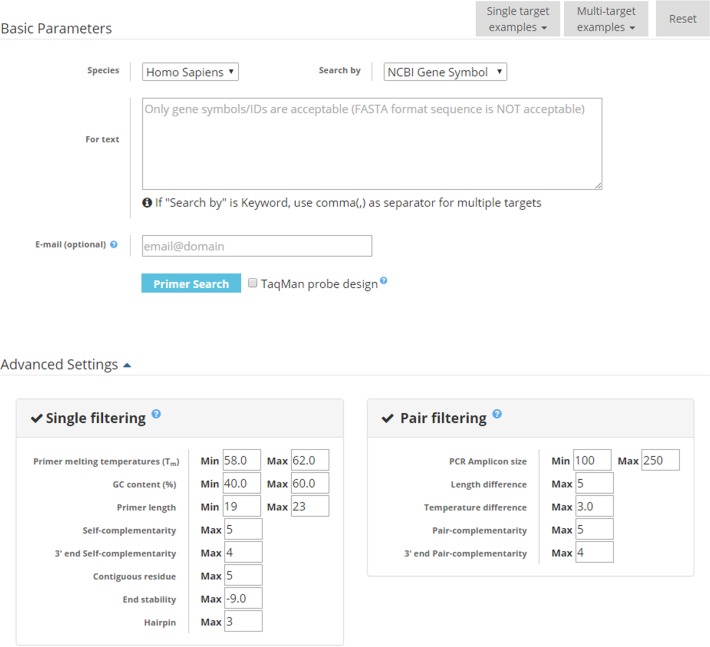
Figure 2 illustrates an example query of MRPrimerW. MRPrimerW allows the user to choose species (human or mouse) and query type (NCBI gene symbol, NCBI CCDS ID, NCBI gene ID, GenBank accession number, GenBank aliases or keyword), and then enter the input query. The user can select the TaqMan probe design option to design TaqMan probes as well as SYBR Green primers. MRPrimerW also provides a feature that sends the query result to a user via email. If a user enters his/her email address in the query web page, the web server sends an email containing a link to the result page to the user after query processing is completed. The result page accessible via the link in the email is available for 2 weeks (i.e. 14 days). In Advanced Settings, the user can adjust single- and pair-filtering constraints. MRPrimerW provides six example queries for single target genes and another six example queries for multiple target genes, in particular, 24 genes related to signaling molecules.
Figure 2.
Input interface of MRPrimerW. MRPrimerW takes as input species, query type (NCBI gene symbol, GenBank accession, NCBI CCDS ID, NCBI Gene ID, aliases or keyword) and query (a set of target genes). The interface provides the TaqMan probe design option and a text box to enter the email address to which a link to the query result should be sent. The user can adjust filtering constraints in the Advanced Settings.
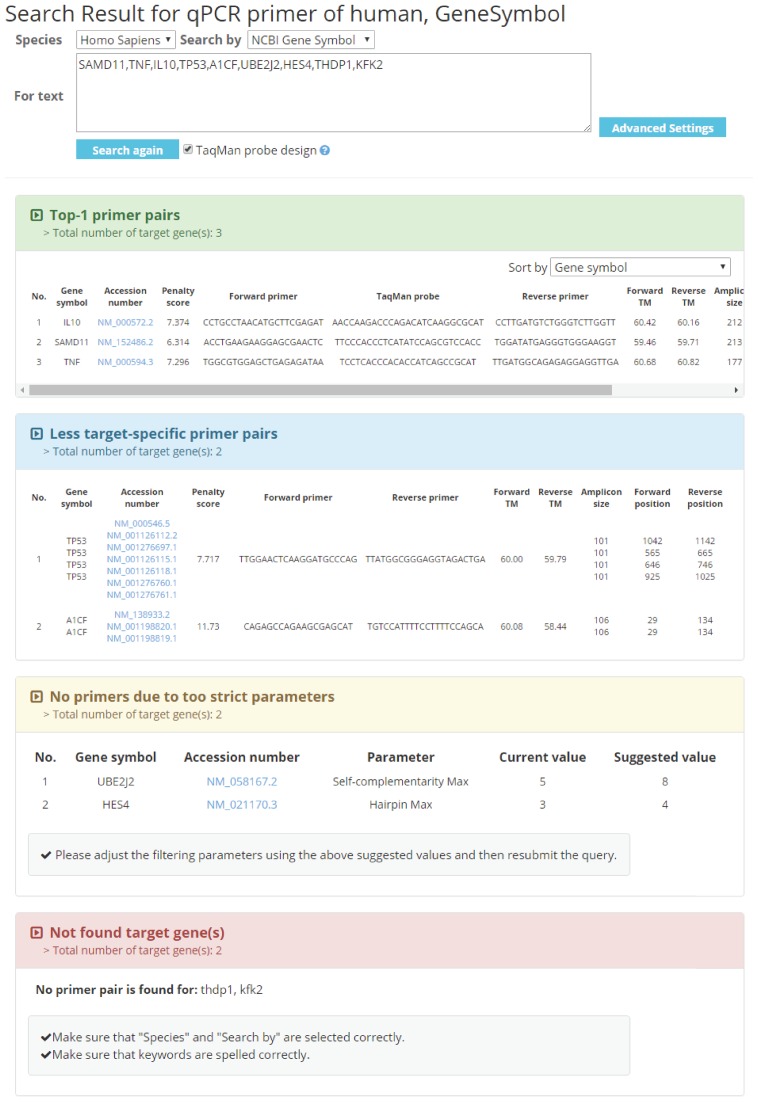
Figure 3 illustrates the results of the example query for nine target genes (SAMD11, TNF, IL10, TP53, A1CF, UBE2J2, HES4, THDP1, KFK2), where the species is human and the search type is NCBI Gene Symbol. Among nine target genes, we assume that three target genes will have primer pairs that amplify each of them solely (case 1: IL10, SAMD11, TNF), two target genes will have only less target-specific primer pairs that may amplify multiple targets (case 2: TP53, A1CF), two target genes will have target-specific primer pairs, but the given filtering constraints are too strict to return them (case 3: UBE2J2, HES4) and two target genes will have typos in their symbols (case 4: THDP1, KFK2). Then, resultant web page shows four tables, each of which contains the primers for each of the above cases. In detail, the first table (for case 1) shows three top-1 primer pairs satisfying the same stringent and uniform constraints. The table shows forward and backward primer sequences, TaqMan probe sequences, gene symbol, GenBank accession number (with a link to detailed gene information from GenBank and primer information), penalty score, melting temperatures (TM), amplicon size and primer positions. The second table (for case 2) shows a set of less target-specific primers that may amplify multiple targets for two target genes. The format of the second table is the same as that of the first table. The third table (for case 3) shows the set of genes that may be amplified by relaxing filtering constraints and how the constraints should be adjusted for each gene. The fourth table (for case 4) shows the set of genes that may be wrong or have typos. With this information, users can modify their queries or input parameters to obtain primers for query genes having no results.
Figure 3.
Output interface of MRPrimerW. MRPrimerW displays the four tables. The first table shows top primer pairs for specific target genes. The second table shows a set of less target-specific primers for the query genes having no results in the first table. The third table shows the set of genes that may have target-specific primer pairs if the filtering constraints are relaxed and guidelines on how to adjust the constraints. The fourth table shows the set of genes that may be wrong or have typos.
CONCLUSIONS
We developed and launched the MRPrimerW web server, a straightforward but powerful tool for designing high-quality primer pairs that can be used simultaneously to detect multiple target genes in qPCR experiments. MRPrimerW overcomes the major drawbacks of existing web servers for primer design by enabling users to freely adjust filtering constraints, performing complete homology tests, supporting batch designing for qPCR, supporting TaqMan probe design and supporting ranking of primers (Table 1). These powerful features were achieved by performing large-scale computation for homology testing on all possible candidate primers in an exact manner, and then materializing the resultant valid primers in eight kinds of indices in the main memory of the web server. Based on these indices, the web server quickly performs online processing in three steps and returns a complete set of the top primer pairs corresponding to the user's query. Although the current web server provides services only for human and mouse, users can easily build and maintain their own web server for other species using the MRPrimerW source code. We believe that MRPrimerW will contribute to increasing the efficiency and specificity of experiments involving PCR.
Supplementary Material
Acknowledgments
We appreciate the allocation of computing nodes of the supercomputer iREMB Mark II of DGIST Supercomputing & Big-data Convergence Research Center, which is used for performing offline processing.
Footnotes
Present address: Min-Soo Kim, Department of Information and Communication Engineering, DGIST, 333, Techno Jungang Daero, Daegu 42988, South Korea.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Korean Health Technology R&D Project, Ministry for Health, Welfare & Family Affairs [HI13C0423]; DGIST R&D Program of the Ministry of Science, ICT & Technology of Korea [16-BD-06, 16-BD-0404]; IT R&D program of MSIP/KEIT [10041145, Self-Organized Software platform(SoSp) for Welfare Devices]. Funding for open access charge: Korean Health Technology R&D Project, Ministry for Health, Welfare & Family Affairs [HI13C0423]; DGIST R&D Program of the Ministry of Science, ICT & Technology of Korea [16-BD-06, 16-BD-0404]; IT R&D program of MSIP/KEIT [10041145, Self-Organized Software platform(SoSp) for Welfare Devices].
Conflict of interest statement. None declared.
REFERENCES
- 1.Dwivedi B., Schmieder R., Goldsmith D.B., Edwards R.A., Breitbart M. PhiSiGns: an online tool to identify signature genes in phages and design PCR primers for examining phage diversity. BMC Bioinformatics. 2012;13:37–47. doi: 10.1186/1471-2105-13-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gorron E., Rodriguez F., Bernal D., Rodriguez-Rojas L.M., Bernal A., Restrepo S., Tohme J. A new method for designing degenerate primers and its use in the identification of sequences in Brachiaria showing similarity to apomixis-associated genes. Bioinformatics. 2010;26:2053–2054. doi: 10.1093/bioinformatics/btq312. [DOI] [PubMed] [Google Scholar]
- 3.Rose T.M., Schultz E.R., Henikoff J.G., Pietrokovski S., McCallum C.M., Henikoff S. Consensus-degenerate hybrid oligonucleotide primers for amplification of distantly related sequences. Nucleic Acids Res. 1998;26:1628–1635. doi: 10.1093/nar/26.7.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chuang L.Y., Cheng Y.H., Yang C.H. Specific primer design for the polymerase chain reaction. Biotechnol. Lett. 2013;35:1541–1549. doi: 10.1007/s10529-013-1249-8. [DOI] [PubMed] [Google Scholar]
- 5.Huang J., Khan I., Liu R., Yang Y., Zhu N. Single primer-mediated circular PCR for hairpin DNA cloning and plasmid editing. Anal. Biochem. 2016 doi: 10.1016/j.ab.2015.12.022. doi:10.1016/j.ab.2015.12.022. [DOI] [PubMed] [Google Scholar]
- 6.Wang X., Spandidos A., Wang H., Seed B. PrimerBank: a PCR primer database for quantitative gene expression analysis, 2012 update. Nucleic Acids Res. 2012;40:D1144–D1149. doi: 10.1093/nar/gkr1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao H., Shockey J.M. Comparison of TaqMan and SYBR Green qPCR methods for quantitative gene expression in tung tree tissues. J. Agric. Food Chem. 2012;60:12296–12303. doi: 10.1021/jf304690e. [DOI] [PubMed] [Google Scholar]
- 8.Untergasser A., Cutcutache I., Koressaar T., Ye J., Faircloth B.C., Remm M., Rozen S.G. Primer3–new capabilities and interfaces. Nucleic Acids Res. 2012;40:e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Untergasser A., Nijveen H., Rao X., Bisseling T., Geurts R., Leunissen J.A. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007;35:W71–W74. doi: 10.1093/nar/gkm306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.You F.M., Huo N., Gu Y.Q., Luo M.C., Ma Y., Hane D., Lazo G.R., Dvorak J., Anderson O.D. BatchPrimer3: a high throughput web application for PCR and sequencing primer design. BMC Bioinformatics. 2008;9:253–265. doi: 10.1186/1471-2105-9-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fredslund J., Lange M. Primique: automatic design of specific PCR primers for each sequence in a family. BMC Bioinformatics. 2007;8:369–375. doi: 10.1186/1471-2105-8-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arvidsson S., Kwasniewski M., Riano-Pachon D.M., Mueller-Roeber B. QuantPrime–a flexible tool for reliable high-throughput primer design for quantitative PCR. BMC Bioinformatics. 2008;9:465–479. doi: 10.1186/1471-2105-9-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ye J., Coulouris G., Zaretskaya I., Cutcutache I., Rozen S., Madden T.L. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 2012;13:134–144. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spandidos A., Wang X., Wang H., Seed B. PrimerBank: a resource of human and mouse PCR primer pairs for gene expression detection and quantification. Nucleic Acids Res. 2010;38:D792–D799. doi: 10.1093/nar/gkp1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lefever S., Vandesompele J., Speleman F., Pattyn F. RTPrimerDB: the portal for real-time PCR primers and probes. Nucleic Acids Res. 2009;37:D942–D945. doi: 10.1093/nar/gkn777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pattyn F., Robbrecht P., De Paepe A., Speleman F., Vandesompele J. RTPrimerDB: the real-time PCR primer and probe database, major update 2006. Nucleic Acids Res. 2006;34:D684–D688. doi: 10.1093/nar/gkj155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pattyn F., Speleman F., De Paepe A., Vandesompele J. RTPrimerDB: the real-time PCR primer and probe database. Nucleic Acids Res. 2003;31:122–123. doi: 10.1093/nar/gkg011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cui W., Taub D.D., Gardner K. qPrimerDepot: a primer database for quantitative real time PCR. Nucleic Acids Res. 2007;35:D805–D809. doi: 10.1093/nar/gkl767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Farrell C.M., O'Leary N.A., Harte R.A., Loveland J.E., Wilming L.G., Wallin C., Diekhans M., Barrell D., Searle S.M., Aken B., et al. Current status and new features of the Consensus Coding Sequence database. Nucleic Acids Res. 2014;42:D865–D872. doi: 10.1093/nar/gkt1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harte R.A., Farrell C.M., Loveland J.E., Suner M.M., Wilming L., Aken B., Barrell D., Frankish A., Wallin C., Searle S., et al. Tracking and coordinating an international curation effort for the CCDS Project. Database. 2012;2012:bas008. doi: 10.1093/database/bas008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pruitt K.D., Harrow J., Harte R.A., Wallin C., Diekhans M., Maglott D.R., Searle S., Farrell C.M., Loveland J.E., Ruef B.J., et al. The consensus coding sequence (CCDS) project: Identifying a common protein-coding gene set for the human and mouse genomes. Genome Res. 2009;19:1316–1323. doi: 10.1101/gr.080531.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim H., Kang N., Chon K.W., Kim S., Lee N., Koo J., Kim M.S. MRPrimer: a MapReduce-based method for the thorough design of valid and ranked primers for PCR. Nucleic Acids Res. 2015;43:e130. doi: 10.1093/nar/gkv632. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.