Summary
Antimicrobial peptides (AMPs) provide a primordial source of immunity, conferring upon eukaryotic cells resistance against bacteria, protozoa, and viruses. Despite a few examples of anionic peptides, AMPs are usually relatively short positively charged polypeptides, consisting of a dozen to about a hundred amino acids, and exhibiting amphipathic character. Despite significant differences in their primary and secondary structures, all AMPs discovered to date share the ability to interact with cellular membranes, thereby affecting bilayer stability, disrupting membrane organization, and/or forming well-defined pores. AMPs selectively target infectious agents without being susceptible to any of the common pathways by which these acquire resistance, thereby making AMPs prime candidates to provide therapeutic alternatives to conventional drugs. However, the mechanisms of AMP actions are still a matter of intense debate. The structure-function paradigm suggests that a better understanding of how AMPs elicit their biological functions could result from atomic resolution studies of peptide-lipid interactions. In contrast, more strict thermodynamic views preclude any roles for three-dimensional structures. Indeed, the design of selective AMPs based soley on structural parameters has been challenging. In this chapter, we will focus on selected AMPs for which studies on the corresponding AMP-lipid interactions have helped reach an understanding of how AMP effects are mediated. We will emphasize the roles of both liquid- and solid-state NMR spectroscopy for elucidating the mechanisms of action of AMPs.
Keywords: Antimicrobial peptides, solution NMR, solid-state NMR, lipid membranes
1. Introduction
The increasing resistance of bacteria to conventional antibiotics represents a serious global emergency for human health that can only be addressed by the discovery and clinical implementation of new antimicrobial drugs (1–8). Over the past 40 years, however, only three new classes of antibiotics have been found that could be used as drugs: lipopeptides, oxazolidinones, and streptogramins (9). Since antimicrobial peptides (AMPs) are not susceptible to any of the common mechanisms whereby organisms acquire drug resistance, representatives of the class of AMPs offer an alternative to standard antimicrobial therapy.
AMPs constitute one of the first evolved chemical defense mechanisms of eukaryotic cells against bacteria, protozoa, fungi, parasites, and viruses (10–12). AMPs have been isolated from a variety of organisms, including humans, and are present in virtually all living species from bacteria to invertebrates to vertebrates (13). The first antimicrobial peptide, nisin, was isolated in 1947 (14, 15), but systematic research on AMPs started only in early 1980s, upon publication of Hans Boman’s paper on the isolation, purification, sequencing, and specificity of two antibacterial peptides, cecropins A and B, involved in insect immunity (10). Since this pioneering research, more than 1800 AMPs have been identified and isolated from several organisms, and structural analogues have been synthesized and tested for clinical purposes (16–22). A comprehensive database on isolated or synthesized AMPs is available at http://aps.unmc.edu/AP/main.php (22).
Properties of AMPs in their role as antibiotics include a broad spectrum of antibacterial activity, high selectivity, and the ability to disrupt bacterial cell membranes (2, 11, 23, 24). In addition, several AMPs can modulate immune and inflammatory responses, inducing phagocytosis, promotion of immune cell recruitment, regulation of angiogenesis, and stimulation of prostaglandin release (13, 19, 20, 25–29)
AMPs can be grouped into diverse classes based on the pathways of their biosynthesis, their structural properties, and their biological activities. AMPs are either ribosomally synthesized oligopeptides, when produced by mammals, birds, amphibians, insects, plants, or certain microorganisms (30, 31) or non-ribosomally synthesized peptides when produced by bacteria and fungi, i.e., bacteriocins. Additionally, AMPs can be subdivided as antiviral, antifungal, anticancer, antiparasital, insecticidal, spermicidal, anti-HIV, and/or as having chemotactic activity (22). Finally, AMPs can be classified on the basis of molecular structure as (a) linear amphipathic α-helical, e.g., cathelicidins, magainins, and cecropins (32–38), (b) amphiphilic β-sheet structures containing disulphide bonds, e.g., defensins (39–41), and (c) turns and extended structures, e.g. protegrins (42–45).
AMPs are usually cationic, relatively short (less than a hundred amino acid residues), and amphipathic, with approximately half of the residues hydrophobic so as to enable their associations with cell membranes (13, 46–54) In fact, to elicit their biological functions, it is necessary for AMPs to interact with the bacterial cytoplasmic membrane, under which conditions AMPs often adopt a secondary structure (48, 54–58). From a physical chemical standpoint, antibacterial activity of AMPs seems to be related to amino acid composition, which may also dictate their selectivity towards bacterial cell membranes with specific lipid composition and structure (59, 60). However, a unified mechanism of action for AMPs has not been identified. It is possible that each class of AMPs has specific mechanism of action.
2. Proposed mechanisms of action
The biological actions of AMPs are not well understood. Researchers agree that the primary target for many AMPs is the outer cell membrane (61–64). Two main membrane interaction mechanisms have been proposed: (a) pore formation across the membrane and (b) a carpet-like mechanism (65, 66). These two models share some similarities and involve four major steps: attraction, attachment, insertion, and membrane permeation (13, 50). The initial attraction of the peptide toward the membrane is driven by electrostatic interactions between the positively charged residues (generally Arg and Lys) and the negative lipid headgroups of the bacterial membrane (67–69). The attachment step depends on the distribution of hydrophobic and hydrophilic residues of AMPs. After reaching the membrane, AMPs pass a concentration threshold after which cell membrane permeability is altered (48, 62, 70–73). At low peptide/lipid ratio (P/L) ratio, AMPs are positioned with their backbones parallel to the surface of the lipid bilayer, so that the AMPs interact with the lipid headgroups. With this topology, AMPs often insert into the membrane, modifying its thickness and curvature (74–80). As the P/L ratio increases, AMPs orient perpendicularly to the membrane and insert into the membrane bilayer while forming pores (81). Amino acid composition, charge, and amphipathicity drive AMP insertion. In addition, several studies show that membrane partitioning causes AMPs to adopt a regular secondary structure; this is referred to as the folding-upon-binding mechanism (82, 83). The latter effect causes a substantial reduction of the free energy of insertion, due to the formation of intramolecular hydrogen bonds (82, 84, 85). Within the lipid bilayer, AMPs can assume different architectures, including toroidal, carpet-like or barrel stave (65, 86–88). In the toroidal model, the polar faces of AMPs associate with lipid polar head groups and insert into the membrane in a manner that involves induced bending of the lipid monolayers and concomitant pore formation (50, 89). In the carpet-like model, AMPs coat the bilayer outer surface, just like a carpet; after reaching a certain concentration threshold, the AMPs disrupt the membrane in a detergent-like manner, causing membrane lysis (38, 50, 87, 90). In the barrel-stave model exemplified by alamethicin, AMPs aggregate to form a bundle of monomers (staves in a barrel), forming a pore in the membrane by thinning and bending of the inner and outer membrane leaflets (66, 91).
Disruption of the membrane dissipates the electrochemical gradients necessary for cells to thrive, while also causing the loss of essential cytoplasmic constituents. It has been reported the relative preference of barrel-stave versus toroidal mechanism depends on peptide length (57, 75, 92–94). Modifications of the peptide sequence involving changes in charge distribution, hydrophobicities, and surface areas of different amino acid side-chains have been shown to modulate the specificities and mechanisms of action for AMPs (49, 53, 70, 95, 96).
Over the past few years, a more thermodynamic approach has been applied to explain membrane permeabilization by AMPs. This viewpoint is based on the efflux of fluorescent dye observed with lipid vesicles, and precludes any structural factors (48, 62, 89, 97–101). Such studies suggest two possible mechanisms for cell permeation: (a) graded or (b) all-or-none (89, 101–104). In the graded mode, all vesicles release part of their content, while in the all-or-none mode only a fraction of the vesicles release their contents while the remainder do not lose any dye. In essence, the graded mechanism can be understood in terms of a destabilizing detergent-like effect on all vesicles, whereas the all-or-none mode of depletion suggests the formation of multimeric pores or the disruption of the vesicle structure (89, 101–104).
To what extent do structures affect the activities and selectivities of AMPs? Is is possible that millions of years of evolution did not encode specificity and function into these peptides as for larger proteins? At this writing, the questions just posed remain unanswered. While there are clear examples of structure-function relationship for several classes of AMPs, the proponents of a more thermodynamic viewpoint invoke the many examples where AMPs with all D-amino acid residues or scrambled sequences still manifest antimicrobial activity. In favor of the structure-activity relationship is the possibility that the landscape of the AMPs structural propensities as well as their interactions with lipid membranes is still limited to selected cases.
3. Bacterial membrane composition and membrane mimicking systems for structural studies
The plasma membrane contains both neutral and zwitterionic lipids such as phosphatidylcholine (PC), or acidic lipid such as phosphatidylserine (PS), cardiolipin, and phosphatidylglycerol (PG) (65, 105, 106). While it is relatively easy to reproduce the relevant lipid composition with synthetic lipids, it is very challenging to mimic the complexity of cell membrane organization. The latter constitutes a significant concern for both functional and structural studies. For the latter, several different model systems have been employed. As a general guideline, the membrane mimetic systems should not only represent the natural environment as closely as possible, but it should also be compatible with structural techniques such as NMR, CD, and fluorescence measurements. Originally, NMR researchers used aqueous solutions of organic solvents such as trifluoroethanol (TFE) or hexafluoroisopropanol (HFIP). Under such conditions, most known AMPs are forced to adopt α-helical conformations, which often do not correlate with biological activities (107–110). To overcome this problem, detergent micelles, such as those formed from DPC (dodecylphosphocholine) plus SDS (sodium dodecylsulfate), have been used in order to better simulate the membrane interface. Micelles are definitively a superior medium to study AMPs, since they provide a water/lipid interface similar to the lipid membranes.
Usually, micelles are spherical monolayers with a diameter of ~ 3 nm that can assume elliptical or rod-like shapes, depending on detergent concentration and chain length (111, 112). Their small size and fast tumbling enable solution NMR spectroscopy of micelle-bound AMPs and small membrane proteins (34, 113–117). Detergent micelles, however, are only a rough approximation to a membrane bilayer. The monolayer structure and small curvature radius of micelles can cause peptides and proteins to adopt incorrectly folded conformations or to aggregate (118). Recently, discoidal micelles or bicelles have been used in NMR spectroscopy of both membrane peptides and proteins (112, 118–122). Generally, bicelles are formed by long-chain phospholipids (DMPC, DMPG) and amphiphilic molecules such as CHAPSO, or a short-chain lipid, such as DHPC. The structures and shapes of bicelles depend on the lipid to detergent ratio (called q). At high detergent concentrations (q ~ 0.1–0.8), bicelles assume a discoidal shape and tumble rapidly in solution. At low detergent concentrations (q ~ 2.8 – 6), bicelles may resemble perforated bilayer sheets (123–125). While AMPs and membrane proteins can be analyzed by solution NMR techniques in isotropic bicelles, anisotropic bicelles align spontaneously in magnetic fields, enabling peptides and proteins to be analyzed using oriented solid-state NMR approaches (126–130). In addition to magnetically oriented bicelles, AMPs can be reconstituted in mechanically aligned membrane bilayers supported on glass plates (131–133). This method of sample preparation, originally developed by Seelig and co-workers (134–136), has been used widely to analyze membrane peptides and proteins (133, 137–142). This approach, however, is quite laborious and prone to artifacts since AMPs have a tendency to aggregate and disrupt the organization of lipid bilayers, making it difficult to discern the mechanism of action. Therefore, many studies are being carried out in lipid vesicles, which are amenable to magic angle spinning (MAS) NMR techniques (143, 144).
4. NMR spectroscopic approaches to study AMPs
Classical solution NMR techniques have been used successfully to determine three-dimensional structures of bioactive peptides in mixtures of water with fluorinated organic solvents (107, 110, 145, 146), in detergent micelles (34, 38, 115, 147, 148), and in isotropic bicelles (119, 149, 150). NOESY-based techniques have been used to measure distance restraints for structure calculations. In the case of recombinant-expressed peptides, weak alignment of peptides using acrylamide gels has been used to obtain residual dipolar couplings for orientational-dependent restraints (151–153). The positioning of peptides with respect to micelle surfaces has been inferred by paramagnetic relaxation enhancement mapping (34, 38, 147, 154, 155), water exposure measurements based on exchange peaks with the water signal (156–158), detection of NOEs between protonated micelles and peptides (159–162), or saturation transfer techniques (163–165).
Despite the results from solution NMR, accurate descriptions of interactions between any given peptide and the membrane within which it exerts bioactivity require use of lipid membranes (35, 70, 128, 129, 131, 141, 142, 166–177). Meeting this need, solid-state NMR techniques are well suited for studying membrane-embedded peptides and proteins. The two major approaches are: (a) static or oriented solid-state NMR (170, 178–182), wherein nuclear anisotropic interactions are obtained from aligning samples with respect to the direction of the static magnetic field, and (b) magic angle spinning (MAS) NMR, whereby samples are spun at the magic angle (θ ~ 54.7°) to remove the effects of chemical shift anisotropy and dipolar couplings (70, 173, 183–185). With fast spinning at the magic angle, resonances can reach line widths similar to those observed in solution NMR spectra.
In the following, we will focus on selected antimicrobial peptides (magainins, pardaxins, distinctin, and cathelicidins) that our group has studied, and in so doing, highlight the role of NMR in understanding structure-function relationships.
4.1 Magainins
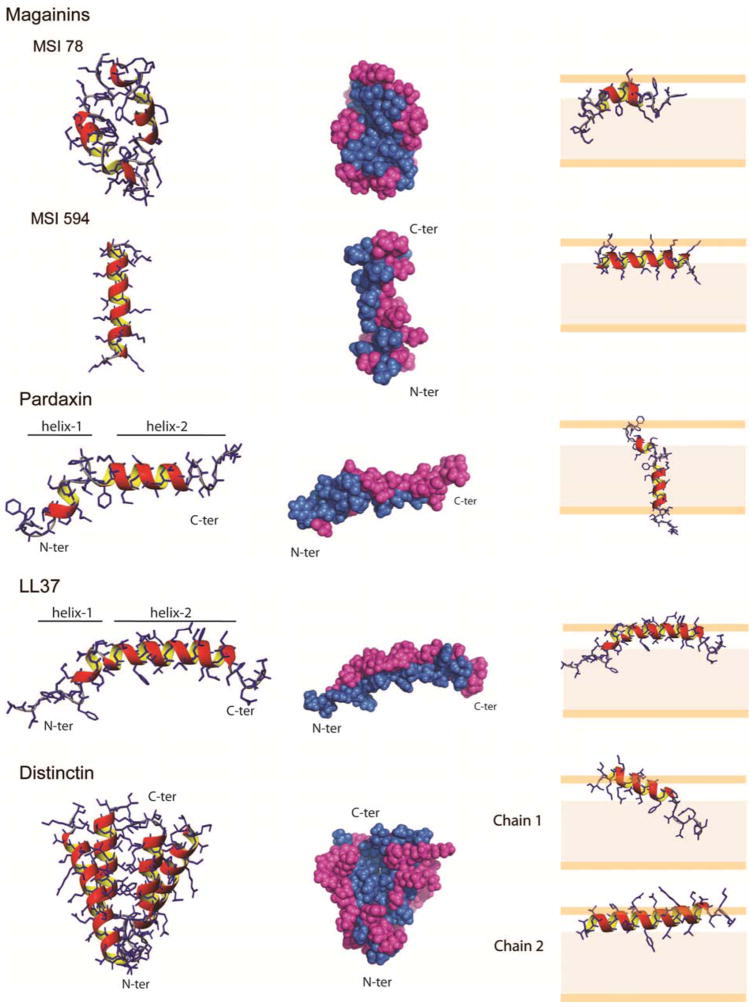
This class of AMPs is expressed by amphibians as a defense against microbes. Magainins were first identified by Zasloff in the early 1980’s (36, 186). In general, magainins are helical and amphipathic, and exhibit a broad spectrum of antimicrobial activities (33, 34, 86, 107, 108, 172). Their structures and orientations with respect to membrane bilayers have been studied extensively, as has been reviewed by the Zasloff, Opella, and Bechinger groups (35, 108, 172, 187–192). In our laboratory, we determined the high-resolution structures and lipid interactions of two synthetic variants of magainins (MSI-78 and MSI-594) that were originally designed by Genaera Corporation for clinical use (193). MSI-78, an analog of magainin 2, is a 22-residue polypeptide known as pexiganan that is being tested in phase II and III clinical trials for treatment of diabetic foot ulcers. MSI-78 interacts strongly with model bacterial cell membranes, selectively inducing bacterial membrane disruption while leaving mammalian cells essentially unperturbed (194). MSI-594 is a 24-residue peptide with high efficacy against herpes simplex virus I (195). The primary sequences of MSI-78 and MSI-594 differ significantly only at the C-terminus (Figure 1). In dodecylphosphocholine (DPC) micelles, each peptide gives a high-resolution spectral fingerprint that enables the sequential assignment of nearly all of the backbone and side chain resonances. In DPC micelles, MSI-78 assumes a distorted α-helical conformation throughout the entire length of the polypeptide chain. A significant feature of MSI-78 is its ability to form stable antiparallel dimers, as defined by head to tail NOEs contacts. The latter may explain its resistance to proteolysis and its higher efficacy with respect to MSI-594. As it turns out, MSI-594 adopts a well-defined helical conformation, but does not dimerize, a property that may account for the significantly lower antibacterial activity displayed by this molecule. Dimerization is a common feature for the majority of magainins, as exemplified by magainin 2 which forms stable dimers in PC vesicles (196).
Figure 1.
Primary sequences of the helical AMPs studied by our groups.
MSI-78 dimerization seems to be encoded in its primary sequence (Figure 1). In fact, the central hydrophobic core of the MSI-78 dimer is formed by a “phenylalanine zipper” that hold the two protomers together, with charged residues (e.g., lysines) pointing toward the bulk solvent. In contrast, MSI-594 lacks phenylalanine residues in position 13 and 16, preventing formation of a stable dimer. Moreover, the GIG motif located in the middle of the primary sequence enables MSI-78 to assume a curved structure, thereby inducing conformational dynamics (reported by broader NMR lines) that further hampers the dimerization process. The Ramamoorthy laboratory performed MAS solid-state NMR experiments on the magainin analogs reconstituted in POPC and 3:1 POPC/POPG MLVs. Peptides that had been 13C-labeled at the α-carbonyl displayed isotropic chemical shift values of ~176–179 ppm that are characteristic of α-helical conformations (197), and confirmatory of the helical structure determined by solution NMR. These authors also found that the line widths of MSI-594 are broader than the corresponding resonances of MSI-78, signifying that conformational exchange dynamics are more accentuated for MSI-78. Again, solution and solid-state NMR data were consistent with each other, pointing towards the same conclusions.
Based on NMR studies both in micelles and in the solid-state, we proposed that differences observed for MSI-78 and MSI-594, in terms of both function and selectivity, may be due to their differing tendencies to self-associate. These results support the in vivo antimicrobial activity of MSI-78, which has been proposed to oligomerize into toroidal-type structures that permeate bacterial cells. In contrast, the behavior of MSI-594 is completely different, suggesting an alternative mechanism of action that must await further studies for elucidation (34).
4.2 Pardaxins
This class of small shark-repellent AMPs are isolated from the sole fish of genus Pardachirus (198, 199). Pardaxins interact with cell membranes, causing disruption of ionic transport and presynaptic activity by forming voltage-dependent ion-selective channels (200–205). Pardaxins are proposed to follow the barrel-stave model, with an aggregation number of 6 (198). To test this hypothesis, we studied pardaxin 4 (Pa4), both in micelles and lipid bilayers, using a combination of solution and solid-state NMR spectroscopy (147). Fast tumbling in the DPC micelle/Pa4 complex enabled us to obtain high-quality solution NMR data and to assign the majority of resonances for both the backbone and the side chains. We found that Pa4 adopts a bend-helix-bend-helix secondary structure motif, with an angle of ~122° between the two helical domains. The topological orientation of Pa4 in micelles was assessed by paramagnetic quenching experiments, establishing that Pa4 lies on the surface of the micelle.
This model is supported by solid-state NMR data in lipid membranes. Specifically, 13C-15N rotational echo double resonance experiments (REDOR) carried out in vesicles of different composition (DMPC, POPC and POPE:POPG 3:1 mixture) containing 3% Pa4 are consistent with peptide helical structure when embedded in membranes (206, 207). 2H and 31P NMR experiments performed on d31-POPC multilamellar vesicles (MLVs) in the presence and absence of Pa4 revealed that upon peptide binding, disorder was increased both in headgroups and in acyl chains located in the hydrophobic core of the lipid membrane (197). Interestingly, in DMPC, the C-terminal helix of Pa4 adopts a transmembrane orientation, while in POPC this domain is oriented with the helical axis approximately parallel to the bilayer normal. The latter suggest that membrane composition plays a pivotal role in the mechanism of action of Pa4. Taken together, the data from both solution and solid state NMR corroborate the hypothesized “barrel stave” mechanism of action of pardaxin, with the N-terminal domain involved in insertion into the bilayer, and the C-terminal helical portion involved in putative ion-channel formation.
4.3 Distinctin
This 47-residue peptide extracted from Phyllomedusa distincta, a tree frog from the Brazilian forests, interacts with negatively charged membranes and is active against both Gram-positive and Gram-negative bacteria. The primary sequence of distinctin comprises two linear chains of 22 (chain 1) and 25 (chain 2) residues, linked by a disulfide bridge between Cys19 of chain 1 and Cys23 of chain 2 (208–210). Unlike other antimicrobial peptides, distinctin adopts a well-folded conformation in aqueous environment, with a non-covalent parallel four-helical bundle that confers upon this peptide stability against proteolysis (211). Scaloni and co-workers also showed that distinctin forms voltage-dependent ion channels in POPC/POPE planar bilayers (211), which were modeled as pentameric pores using molecular dynamic calculations (212). The mechanisms of pore formation and membrane permeabilization have also been investigated using electrochemical measurements with two different mercury-supported biomimetic membranes, i.e., a self assembled monolayer (SAM) and a tethered bilayer lipid membrane (tBLM). In SAM, distinctin forms selective ion channels for Tl+ and Cd2+ ions; while the formation of K+ permeable ion channels in tBLM occurred only at non-physiological potentials (209).
Using a combination of site-specific 15N and 2H NMR spectroscopy, Bechinger and co-workers showed that distinctin’s helical domains orient approximately parallel to the surface of mechanically oriented POPC (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine) lipid bilayers. In a parallel study, we confirmed Bechinger’s findings and showed that distinctin interacts more strongly with membranes containing charged lipid headgroups (PA, PS, and PG). We also discovered that both chains of distinctin adopt an approximately parallel orientation with respect to the membrane plane, with a slight angle between chains 1 and 2. However, in 1:1 POPC:DOPA lipid bilayers and 50:1 lipid to protein molar ratio, distinctin is unable to disrupt lipid bilayers. The chemical shift anisotropy and dipolar couplings from separated local field experiments were implemented in a hybrid simulated annealing protocol (213) to determine the orientation of the distinctin heterodimer more quantitatively. After energy minimization, the tilt angles with respect to the bilayer plane were calculated to be ~24° and ~5° for chains 1 and 2, respectively (129).
Our findings in mechanically oriented bilayers are supported by experiments carried out on distinctin reconstituted in DMPC/DHPC bicelles. Under these experimental conditions, distinctin adopts a dual topology for chain 2, indicating that the peptide can be either absorbed on the surface of the bilayer or exist in a trans-membrane topology (Verardi et al. in preparation). These new findings support the electrophysiological data, and explain how distinctin forms ion-conducting pores. In all, the data suggest that cell disruption may occur via formation in solution of stable tetramers that dissociate into monomers upon membrane interaction. Following an increase of concentration of the peptide on the membrane surface, distinctin organizes into ion-conducting pores. The propensity of distinctin to assume a transmembrane orientation even at low concentrations may suggest that stochastic formation of transient pores is also possible.
4.4 Cathelicidins
This well-known family of structurally different antimicrobial peptides (37, 44, 214, 215) includes peptides that comprise an N-terminal signal peptide that shares a highly conserved cathelin-like domain and a variable cationic C-terminal domain, which is responsible for the antimicrobial activity. LL-37 is the only cathelicidin-derived peptide found in humans (LL-37-hCAP18) (44, 216–218). LL-37-hCAP18 is expressed in epythelia, monocytes, and lymphocytes. During infection, inflammatory processes, or wound healing a 100-residues pro-peptide is expressed, containing the signal peptide, the cathelin domains, and the 37-residue mature antimicrobial peptide located at the C-terminal region (37, 214, 217, 219). LL-37 has a broad-spectrum of bactericidal activity, may play a role in cystic fibrosis remediation, and has been found to inhibit HIV-1 infection in vitro (37, 220, 221).
Using DPC micelles, we were able to obtain high-resolution spectra of the mature LL-37 peptide (38). The overall structure resembles that of Pa4, with a helix-break-helix motif and an angle between the two helical domains of ~120°. The N-terminal helix ends with a break at K12, and is more dynamic than the C-terminal helix (residues 13-33) and ends with a break at K12 (Figure 2). The kink starting at residue 12 may be due to a hydrophobic cluster of residues located in the concave face of the peptide (I13, F17 and I20) and appears to be facilitated by a groove created by G14. Dynamic light scattering measurements show that addition of LL-37 to DPC micelles does not change the overall organization of micelles, which display an average hydrodynamic radius of 24 ± 2 and 26 ± 2 Å in the absence and presence of LL-37, respectively. The latter result suggests that LL-37 associates on the micelle surface without changing the overall micellar shape. These data were supported by paramagnetic quenching experiments carried out with Gd3+ and by the detection of several peptide-to-micelle NOEs. The overall topology for LL-37 is common to other natural occurring helical AMPs, with the hydrophilic residues pointing toward the bulk solvent and the hydrophobic toward the inner hydrocarbon core of the micelles. The structural topology of LL-37 in micelle is reported in Figure 2. The peptide concave face containing the hydrophobic cluster points toward the micelle interior with the hydrophilic residues pointing outward. The helix-kink-helix motif recurs in other AMPs (i.e., pardaxin), as already discussed. It has been hypothesized that the hinge region is required in order to confer structural flexibility for membrane insertion and pore formation (200, 202, 222, 223). The large curvature present in LL-37 has also been observed in the magainin-derived peptides (MSI-78 and MSI-594) as well as in other lysine-rich peptides (57, 95, 224). On the other hand, this curvature may be artificial and due to the interaction between the peptides and curved surfaces of the micelles. As described earlier in this article, micellar systems represent only a coarse approximation of membrane bilayers. Synthetic lipid bilayers such as vesicles or planar bilayers are preferable for testing lipid/peptide interactions. Despite structural similarities between LL-37 and pardaxins, their mechanisms of action appear to be different. Specifically, pardaxins are thought to disrupt cell membranes via a “barrel stave” mechanism, while LL-37 operates according to a “carpet-like” mechanism. The latter conclusion is supported by our helical model, as well as solid-state NMR studies from Ramamoorthy’s laboratory (197, 225, 226). Therefore, structure is important to modulate function, but amino acid composition is an important component that helps dictate the mechanism of action.
Figure 2.
Structures and membrane orientations (topology) of the helical AMPs studied by our groups. Note that the structures of MSI-78, MSI-594, Pa4, and LL-37 were obtained in detergent micelles, while the oligomeric structure of distinctin was obtained by in aqueous buffer. Left: backbone and side chain average structures of the AMPs from the NMR structural ensembles. Center: space filled model of the structures with the hydrophilic residues colored purple and the hydrophobic in blue. Right: topology of the AMPs deduced from ssNMR experiments carried out in lipid membranes.
5. Conclusions and perspectives
Antimicrobial peptides (AMPs) are emerging therapeutics with considerable potential. However, progress in the rational design of AMPs as powerful and selective drugs has been slow. A significant problem continues to be how to define the structural determinants for activities and specificities of action. Until this is understood, identification of the sequence features important for antimicrobial activity cannot be done on anything other than a trial-and-error basis. NMR is clearly playing a fundamental role in understanding the role of structure for AMPs, as reported in the recent literature both from our laboratory (129, 209) and others (52, 227–230). The essential interactions of AMPs with membranes preclude the use of x-ray crystallography to determine high-resolution structural information. However, structure does not seem to be the only determining factor to account for how AMPs elicit their biological functions. Primary sequence (amino acid content and nature), interactions with lipids, and flexibility are important factors to define specificity. Perhaps a most neglected aspect of research on AMPs is the characterization of their structural flexibilities. This is an area for which NMR is expected to have a special niche, pending the development of robust recombinant methods that will offer affordable combinations of isotope-labeled materials for solution and solid-state NMR.
Our working hypothesis is that AMPs are metamorphic polypeptides, capable of adopting different shapes. Thus, special structure may not be important per se. Instead, we suggest that the biological activities of AMPs may be encoded in their structural flexibility an ability to adopt several conformations and topologies upon homotropic (peptide-peptide) or heterotropic (peptide-lipid) interactions.
Acknowledgments
This work is partially supported by the National Institute of Health (GM 64742 to G.V.).
References
- 1.Bommarius B, Kalman D. Antimicrobial and host defense peptides for therapeutic use against multidrug-resistant pathogens: new hope on the horizon. I Drugs. 2009;12:376–380. [PubMed] [Google Scholar]
- 2.Bragonzi A. Fighting back: peptidomimetics as a new weapon in the battle against antibiotic resistance. Sci Transl Med. 2010;2:21ps29. doi: 10.1126/scitranslmed.3000889. [DOI] [PubMed] [Google Scholar]
- 3.Giamarellou H, Poulakou G. Multidrug-resistant Gram-negative infections: what are the treatment options? Drugs. 2009;69:1879–1901. doi: 10.2165/11315690-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 4.Gryllos I, Tran-Winkler HJ, Cheng MF, Chung H, Bolcome R, 3rd, Lu W, Lehrer RI, Wessels MR. Induction of group A Streptococcus virulence by a human antimicrobial peptide. Proc Natl Acad Sci U S A. 2008;105:16755–16760. doi: 10.1073/pnas.0803815105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ho J, Tambyah PA, Paterson DL. Multiresistant Gram-negative infections: a global perspective. Curr Opin Infect Dis. 2010;23:546–553. doi: 10.1097/QCO.0b013e32833f0d3e. [DOI] [PubMed] [Google Scholar]
- 6.Paterson DL. Clinical experience with recently approved antibiotics. Curr Opin Pharmacol. 2006;6:486–490. doi: 10.1016/j.coph.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 7.Cornaglia G, Rossolini GM. Forthcoming therapeutic perspectives for infections due to multidrug-resistant Gram-positive pathogens. Clin Microbiol Infect. 2009;15:218–223. doi: 10.1111/j.1469-0691.2009.02740.x. [DOI] [PubMed] [Google Scholar]
- 8.Wu G, Li X, Fan X, Wu H, Wang S, Shen Z, Xi T. The activity of antimicrobial peptide S-thanatin is independent on multidrug-resistant spectrum of bacteria. Peptides. 2011;32:1139–1145. doi: 10.1016/j.peptides.2011.03.019. [DOI] [PubMed] [Google Scholar]
- 9.Marr AK, Gooderham WJ, Hancock RE. Antibacterial peptides for therapeutic use: obstacles and realistic outlook. Curr Opin Pharmacol. 2006;6:468–472. doi: 10.1016/j.coph.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 10.Steiner H, Hultmark D, Engstrom A, Bennich H, Boman HG. Sequence and specificity of two antibacterial proteins involved in insect immunity. Nature. 1981;292:246–248. doi: 10.1038/292246a0. [DOI] [PubMed] [Google Scholar]
- 11.Wiesner J, Vilcinskas A. Antimicrobial peptides: the ancient arm of the human immune system. Virulence. 2010;1:440–464. doi: 10.4161/viru.1.5.12983. [DOI] [PubMed] [Google Scholar]
- 12.Zasloff M. Antimicrobial peptides, innate immunity, and the normally sterile urinary tract. J Am Soc Nephrol. 2007;18:2810–2816. doi: 10.1681/ASN.2007050611. [DOI] [PubMed] [Google Scholar]
- 13.Jenssen H, Hamill P, Hancock RE. Peptide antimicrobial agents. Clin Microbiol Rev. 2006;19:491–511. doi: 10.1128/CMR.00056-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berridge NJ. Preparation of the antibiotic nisin. Biochem J. 1949;45:486–493. doi: 10.1042/bj0450486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mattick AT, Hirsch A. Further observations on an inhibitory substance (nisin) from lactic streptococci. Lancet. 1947;2:5–8. doi: 10.1016/s0140-6736(47)90004-4. [DOI] [PubMed] [Google Scholar]
- 16.Ambatipudi K, Joss J, Raftery M, Deane E. A proteomic approach to analysis of antimicrobial activity in marsupial pouch secretions. Dev Comp Immunol. 2008;32:108–120. doi: 10.1016/j.dci.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 17.Schittek B, Paulmann M, Senyurek I, Steffen H. The role of antimicrobial peptides in human skin and in skin infectious diseases. Infect Disord Drug Targets. 2008;8:135–143. doi: 10.2174/1871526510808030135. [DOI] [PubMed] [Google Scholar]
- 18.Steffen H, Rieg S, Wiedemann I, Kalbacher H, Deeg M, Sahl HG, Peschel A, Gotz F, Garbe C, Schittek B. Naturally processed dermcidin-derived peptides do not permeabilize bacterial membranes and kill microorganisms irrespective of their charge. Antimicrob Agents Chemother. 2006;50:2608–2620. doi: 10.1128/AAC.00181-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hata TR, Gallo RL. Antimicrobial peptides, skin infections, and atopic dermatitis. Semin Cutan Med Surg. 2008;27:144–150. doi: 10.1016/j.sder.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lai Y, Gallo RL. AMPed up immunity: how antimicrobial peptides have multiple roles in immune defense. Trends Immunol. 2009;30:131–141. doi: 10.1016/j.it.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Menousek J, Mishra B, Hanke ML, Heim CE, Kielian T, Wang G. Database screening and in vivo efficacy of antimicrobial peptides against methicillin-resistant Staphylococcus aureus USA300. Int J Antimicrob Agents. 2012;39:402–406. doi: 10.1016/j.ijantimicag.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang G, Li X, Wang Z. APD2: the updated antimicrobial peptide database and its application in peptide design. Nucleic Acids Res. 2009;37:D933–937. doi: 10.1093/nar/gkn823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abbanat D, Morrow B, Bush K. New agents in development for the treatment of bacterial infections. Curr Opin Pharmacol. 2008;8:582–592. doi: 10.1016/j.coph.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 24.Hancock RE. Peptide antibiotics. Lancet. 1997;349:418–422. doi: 10.1016/S0140-6736(97)80051-7. [DOI] [PubMed] [Google Scholar]
- 25.Diamond G, Beckloff N, Weinberg A, Kisich KO. The roles of antimicrobial peptides in innate host defense. Curr Pharm Des. 2009;15:2377–2392. doi: 10.2174/138161209788682325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hancock RE, Diamond G. The role of cationic antimicrobial peptides in innate host defences. Trends Microbiol. 2000;8:402–410. doi: 10.1016/s0966-842x(00)01823-0. [DOI] [PubMed] [Google Scholar]
- 27.Barak O, Treat JR, James WD. Antimicrobial peptides: effectors of innate immunity in the skin. Adv Dermatol. 2005;21:357–374. doi: 10.1016/j.yadr.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 28.Izadpanah A, Gallo RL. Antimicrobial peptides. J Am Acad Dermatol. 2005;52:381–390. doi: 10.1016/j.jaad.2004.08.026. quiz 391–382. [DOI] [PubMed] [Google Scholar]
- 29.Yin M, Gentili C, Koyama E, Zasloff M, Pacifici M. Antiangiogenic treatment delays chondrocyte maturation and bone formation during limb skeletogenesis. J Bone Miner Res. 2002;17:56–65. doi: 10.1359/jbmr.2002.17.1.56. [DOI] [PubMed] [Google Scholar]
- 30.Nissen-Meyer J, Nes IF. Ribosomally synthesized antimicrobial peptides: their function, structure, biogenesis, and mechanism of action. Arch Microbiol. 1997;167:67–77. [PubMed] [Google Scholar]
- 31.Papagianni M. Ribosomally synthesized peptides with antimicrobial properties: biosynthesis, structure, function, and applications. Biotechnol Adv. 2003;21:465–499. doi: 10.1016/s0734-9750(03)00077-6. [DOI] [PubMed] [Google Scholar]
- 32.Berkowitz BA, Bevins CL, Zasloff MA. Magainins: a new family of membrane-active host defense peptides. Biochem Pharmacol. 1990;39:625–629. doi: 10.1016/0006-2952(90)90138-b. [DOI] [PubMed] [Google Scholar]
- 33.Boggs JM, Jo E, Polozov IV, Epand RF, Anantharamaiah GM, Blazyk J, Epand RM. Effect of magainin, class L, and class A amphipathic peptides on fatty acid spin labels in lipid bilayers. Biochim Biophys Acta. 2001;1511:28–41. doi: 10.1016/s0005-2736(00)00379-5. [DOI] [PubMed] [Google Scholar]
- 34.Porcelli F, Buck-Koehntop BA, Thennarasu S, Ramamoorthy A, Veglia G. Structures of the dimeric and monomeric variants of magainin antimicrobial peptides (MSI-78 and MSI-594) in micelles and bilayers, determined by NMR spectroscopy. Biochemistry. 2006;45:5793–5799. doi: 10.1021/bi0601813. [DOI] [PubMed] [Google Scholar]
- 35.Ramamoorthy A, Thennarasu S, Lee DK, Tan A, Maloy L. Solid-state NMR investigation of the membrane-disrupting mechanism of antimicrobial peptides MSI-78 and MSI-594 derived from magainin 2 and melittin. Biophys J. 2006;91:206–216. doi: 10.1529/biophysj.105.073890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zasloff M. Magainins, a class of antimicrobial peptides from Xenopus skin: isolation, characterization of two active forms, and partial cDNA sequence of a precursor. Proc Natl Acad Sci U S A. 1987;84:5449–5453. doi: 10.1073/pnas.84.15.5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nijnik A, Hancock RE. The roles of cathelicidin LL-37 in immune defences and novel clinical applications. Curr Opin Hematol. 2009;16:41–47. doi: 10.1097/moh.0b013e32831ac517. [DOI] [PubMed] [Google Scholar]
- 38.Porcelli F, Verardi R, Shi L, Henzler-Wildman KA, Ramamoorthy A, Veglia G. NMR structure of the cathelicidin-derived human antimicrobial peptide LL-37 in dodecylphosphocholine micelles. Biochemistry. 2008;47:5565–5572. doi: 10.1021/bi702036s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bals R, Wang X, Wu Z, Freeman T, Bafna V, Zasloff M, Wilson JM. Human beta-defensin 2 is a salt-sensitive peptide antibiotic expressed in human lung. J Clin Invest. 1998;102:874–880. doi: 10.1172/JCI2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Howell MD. The role of human beta defensins and cathelicidins in atopic dermatitis. Curr Opin Allergy Clin Immunol. 2007;7:413–417. doi: 10.1097/ACI.0b013e3282a64343. [DOI] [PubMed] [Google Scholar]
- 41.Tang M, Waring AJ, Lehrer RI, Hong M. Orientation of a beta-hairpin antimicrobial peptide in lipid bilayers from two-dimensional dipolar chemical-shift correlation NMR. Biophys J. 2006;90:3616–3624. doi: 10.1529/biophysj.105.062075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Steinstraesser L, Kraneburg UM, Hirsch T, Kesting M, Steinau HU, Jacobsen F, Al-Benna S. Host defense peptides as effector molecules of the innate immune response: a sledgehammer for drug resistance? Int J Mol Sci. 2009;10:3951–3970. doi: 10.3390/ijms10093951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boman HG. Peptide antibiotics and their role in innate immunity. Ann Rev Immunol. 1995;13:61–92. doi: 10.1146/annurev.iy.13.040195.000425. [DOI] [PubMed] [Google Scholar]
- 44.Guani-Guerra E, Santos-Mendoza T, Lugo-Reyes SO, Teran LM. Antimicrobial peptides: general overview and clinical implications in human health and disease. Clin Immunol. 2010;135:1–11. doi: 10.1016/j.clim.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 45.Buffy JJ, Hong T, Yamaguchi S, Waring AJ, Lehrer RI, Hong M. Solid-state NMR investigation of the depth of insertion of protegrin-1 in lipid bilayers using paramagnetic Mn2+ Biophys J. 2003;85:2363–2373. doi: 10.1016/s0006-3495(03)74660-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chekmenev EY, Jones SM, Nikolayeva YN, Vollmar BS, Wagner TJ, Gor’kov PL, Brey WW, Manion MN, Daugherty KC, Cotten M. High-field NMR studies of molecular recognition and structure-function relationships in antimicrobial piscidins at the water-lipid bilayer interface. J Am Chem Soc. 2006;128:5308–5309. doi: 10.1021/ja058385e. [DOI] [PubMed] [Google Scholar]
- 47.Bonev BB, Chan WC, Bycroft BW, Roberts GC, Watts A. Interaction of the lantibiotic nisin with mixed lipid bilayers: a 31P and 2H NMR study. Biochemistry. 2000;39:11425–11433. doi: 10.1021/bi0001170. [DOI] [PubMed] [Google Scholar]
- 48.Almeida PF, Pokorny A. Mechanisms of antimicrobial, cytolytic, and cell-penetrating peptides: from kinetics to thermodynamics. Biochemistry. 2009;48:8083–8093. doi: 10.1021/bi900914g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bechinger B. Insights into the mechanisms of action of host defence peptides from biophysical and structural investigations. J Pept Sci. 2011;17:306–314. doi: 10.1002/psc.1343. [DOI] [PubMed] [Google Scholar]
- 50.Brogden KA. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat Rev Microbiol. 2005;3:238–250. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- 51.Dathe M, Wieprecht T. Structural features of helical antimicrobial peptides: their potential to modulate activity on model membranes and biological cells. Biochim Biophys Acta. 1999;1462:71–87. doi: 10.1016/s0005-2736(99)00201-1. [DOI] [PubMed] [Google Scholar]
- 52.Bechinger B, Salnikov ES. The membrane interactions of antimicrobial peptides revealed by solid-state NMR spectroscopy. Chem Phys Lipids. 2012 doi: 10.1016/j.chemphyslip.2012.01.009. [DOI] [PubMed] [Google Scholar]
- 53.Blazyk J, Wiegand R, Klein J, Hammer J, Epand RM, Epand RF, Maloy WL, Kari UP. A novel linear amphipathic beta-sheet cationic antimicrobial peptide with enhanced selectivity for bacterial lipids. J Biol Chem. 2001;276:27899–27906. doi: 10.1074/jbc.M102865200. [DOI] [PubMed] [Google Scholar]
- 54.Epand RF, Epand RM, Monaco V, Stoia S, Formaggio F, Crisma M, Toniolo C. The antimicrobial peptide trichogin and its interaction with phospholipid membranes. Eur J Biochem. 1999;266:1021–1028. doi: 10.1046/j.1432-1327.1999.00945.x. [DOI] [PubMed] [Google Scholar]
- 55.Boland MP, Separovic F. Membrane interactions of antimicrobial peptides from Australian tree frogs. Biochim Biophys Acta. 2006;1758:1178–1183. doi: 10.1016/j.bbamem.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 56.Epand RF, Umezawa N, Porter EA, Gellman SH, Epand RM. Interactions of the antimicrobial beta-peptide beta-17 with phospholipid vesicles differ from membrane interactions of magainins. Eur J Biochem. 2003;270:1240–1248. doi: 10.1046/j.1432-1033.2003.03484.x. [DOI] [PubMed] [Google Scholar]
- 57.Fernandez DI, Sani MA, Gehman JD, Hahm KS, Separovic F. Interactions of a synthetic Leu-Lys-rich antimicrobial peptide with phospholipid bilayers. Eur Biophys J. 2011;40:471–480. doi: 10.1007/s00249-010-0660-5. [DOI] [PubMed] [Google Scholar]
- 58.Gazit E, Boman A, Boman HG, Shai Y. Interaction of the mammalian antibacterial peptide cecropin P1 with phospholipid vesicles. Biochemistry. 1995;34:11479–11488. doi: 10.1021/bi00036a021. [DOI] [PubMed] [Google Scholar]
- 59.Jiang Z, Kullberg BJ, van der Lee H, Vasil AI, Hale JD, Mant CT, Hancock RE, Vasil ML, Netea MG, Hodges RS. Effects of hydrophobicity on the antifungal activity of alpha-helical antimicrobial peptides. Chem Biol Drug Des. 2008;72:483–495. doi: 10.1111/j.1747-0285.2008.00728.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dathe M, Wieprecht T, Nikolenko H, Handel L, Maloy WL, MacDonald DL, Beyermann M, Bienert M. Hydrophobicity, hydrophobic moment and angle subtended by charged residues modulate antibacterial and haemolytic activity of amphipathic helical peptides. FEBS Lett. 1997;403:208–212. doi: 10.1016/s0014-5793(97)00055-0. [DOI] [PubMed] [Google Scholar]
- 61.Hancock RE, Rozek A. Role of membranes in the activities of antimicrobial cationic peptides. FEMS Microbiol Lett. 2002;206:143–149. doi: 10.1111/j.1574-6968.2002.tb11000.x. [DOI] [PubMed] [Google Scholar]
- 62.Almeida PF, Pokorny A. Binding and permeabilization of model membranes by amphipathic peptides. Methods Mol Biol. 2010;618:155–169. doi: 10.1007/978-1-60761-594-1_11. [DOI] [PubMed] [Google Scholar]
- 63.Epand RF, Pollard JE, Wright JO, Savage PB, Epand RM. Depolarization, bacterial membrane composition, and the antimicrobial action of ceragenins. Antimicrob Agents Chemother. 2010;54:3708–3713. doi: 10.1128/AAC.00380-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Epand RF, Schmitt MA, Gellman SH, Epand RM. Role of membrane lipids in the mechanism of bacterial species selective toxicity by two alpha/beta-antimicrobial peptides. Biochim Biophys Acta. 2006;1758:1343–1350. doi: 10.1016/j.bbamem.2006.01.018. [DOI] [PubMed] [Google Scholar]
- 65.Shai Y. Mode of action of membrane active antimicrobial peptides. Biopolymers. 2002;66:236–248. doi: 10.1002/bip.10260. [DOI] [PubMed] [Google Scholar]
- 66.Shai Y. Mechanism of the binding, insertion and destabilization of phospholipid bilayer membranes by alpha-helical antimicrobial and cell non-selective membrane-lytic peptides. Biochim Biophys Acta. 1999;1462:55–70. doi: 10.1016/s0005-2736(99)00200-x. [DOI] [PubMed] [Google Scholar]
- 67.Papo N, Shai Y. Can we predict biological activity of antimicrobial peptides from their interactions with model phospholipid membranes? Peptides. 2003;24:1693–1703. doi: 10.1016/j.peptides.2003.09.013. [DOI] [PubMed] [Google Scholar]
- 68.Teixeira V, Feio MJ, Bastos M. Role of lipids in the interaction of antimicrobial peptides with membranes. Prog Lipid Res. 2012;51:149–177. doi: 10.1016/j.plipres.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 69.Epand RM, Epand RF. Bacterial membrane lipids in the action of antimicrobial agents. J Pept Sci. 2011;17:298–305. doi: 10.1002/psc.1319. [DOI] [PubMed] [Google Scholar]
- 70.Aisenbrey C, Bertani P, Bechinger B. Solid-state NMR investigations of membrane-associated antimicrobial peptides. Methods Mol Biol. 2010;618:209–233. doi: 10.1007/978-1-60761-594-1_14. [DOI] [PubMed] [Google Scholar]
- 71.Bechinger B. Membrane association and pore formation by alpha-helical peptides. Adv Exp Med Biol. 2010;677:24–30. doi: 10.1007/978-1-4419-6327-7_3. [DOI] [PubMed] [Google Scholar]
- 72.Huang HW. Molecular mechanism of antimicrobial peptides: the origin of cooperativity. Biochim Biophys Acta. 2006;1758:1292–1302. doi: 10.1016/j.bbamem.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 73.Huang HW. Action of antimicrobial peptides: two-state model. Biochemistry. 2000;39:8347–8352. doi: 10.1021/bi000946l. [DOI] [PubMed] [Google Scholar]
- 74.Bechinger B. Biophysical investigations of membrane perturbations by polypeptides using solid-state NMR spectroscopy (review) Mol Membr Biol. 2000;17:135–142. doi: 10.1080/09687680050197365. [DOI] [PubMed] [Google Scholar]
- 75.Chen FY, Lee MT, Huang HW. Evidence for membrane thinning effect as the mechanism for peptide-induced pore formation. Biophys J. 2003;84:3751–3758. doi: 10.1016/S0006-3495(03)75103-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pabst G, Grage SL, Danner-Pongratz S, Jing W, Ulrich AS, Watts A, Lohner K, Hickel A. Membrane thickening by the antimicrobial peptide PGLa. Biophys J. 2008;95:5779–5788. doi: 10.1529/biophysj.108.141630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nomura K, Corzo G. The effect of binding of spider-derived antimicrobial peptides, oxyopinins, on lipid membranes. Biochim Biophys Acta. 2006;1758:1475–1482. doi: 10.1016/j.bbamem.2006.04.022. [DOI] [PubMed] [Google Scholar]
- 78.Nomura K, Corzo G, Nakajima T, Iwashita T. Orientation and pore-forming mechanism of a scorpion pore-forming peptide bound to magnetically oriented lipid bilayers. Biophys J. 2004;87:2497–2507. doi: 10.1529/biophysj.104.043513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Smith PE, Brender JR, Ramamoorthy A. Induction of negative curvature as a mechanism of cell toxicity by amyloidogenic peptides: the case of islet amyloid polypeptide. J Am Chem Soc. 2009;131:4470–4478. doi: 10.1021/ja809002a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wieprecht T, Beyermann M, Seelig J. Thermodynamics of the coil-alpha-helix transition of amphipathic peptides in a membrane environment: the role of vesicle curvature. Biophys Chem. 2002;96:191–201. doi: 10.1016/s0301-4622(02)00025-x. [DOI] [PubMed] [Google Scholar]
- 81.Wi S, Kim C. Pore structure, thinning effect, and lateral diffusive dynamics of oriented lipid membranes interacting with antimicrobial peptide protegrin-1: 31P and 2H solid-state NMR study. J Phys Chem B. 2008;112:11402–11414. doi: 10.1021/jp801825k. [DOI] [PubMed] [Google Scholar]
- 82.White SH, Wimley WC. Membrane protein folding and stability: physical principles. Annu Rev Biophys Biomol Struct. 1999;28:319–365. doi: 10.1146/annurev.biophys.28.1.319. [DOI] [PubMed] [Google Scholar]
- 83.Seelig J. Thermodynamics of lipid-peptide interactions. Biochim Biophys Acta. 2004;1666:40–50. doi: 10.1016/j.bbamem.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 84.Wimley WC. Describing the mechanism of antimicrobial peptide action with the interfacial activity model. ACS Chem Biol. 2010;5:905–917. doi: 10.1021/cb1001558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hristova K, Wimley WC. A look at arginine in membranes. J Membr Biol. 2011;239:49–56. doi: 10.1007/s00232-010-9323-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Oren Z, Shai Y. Mode of action of linear amphipathic alpha-helical antimicrobial peptides. Biopolymers. 1998;47:451–463. doi: 10.1002/(SICI)1097-0282(1998)47:6<451::AID-BIP4>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 87.Shai Y, Oren Z. From “carpet” mechanism to de-novo designed diastereomeric cell-selective antimicrobial peptides. Peptides. 2001;22:1629–1641. doi: 10.1016/s0196-9781(01)00498-3. [DOI] [PubMed] [Google Scholar]
- 88.Avrahami D, Oren Z, Shai Y. Effect of multiple aliphatic amino acids substitutions on the structure, function, and mode of action of diastereomeric membrane active peptides. Biochemistry. 2001;40:12591–12603. doi: 10.1021/bi0105330. [DOI] [PubMed] [Google Scholar]
- 89.Pokorny A, Almeida PF. Kinetics of dye efflux and lipid flip-flop induced by delta-lysin in phosphatidylcholine vesicles and the mechanism of graded release by amphipathic, alpha-helical peptides. Biochemistry. 2004;43:8846–8857. doi: 10.1021/bi0497087. [DOI] [PubMed] [Google Scholar]
- 90.Oren Z, Lerman JC, Gudmundsson GH, Agerberth B, Shai Y. Structure and organization of the human antimicrobial peptide LL-37 in phospholipid membranes: relevance to the molecular basis for its non-cell-selective activity. Biochem J. 1999;341(Pt 3):501–513. [PMC free article] [PubMed] [Google Scholar]
- 91.He K, Ludtke SJ, Worcester DL, HWH Neutron scattering in the plane of membranes: structure of alamethicin pores. Biophys J. 1996;70:2659–2666. doi: 10.1016/S0006-3495(96)79835-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gehman JD, Luc F, Hall K, Lee TH, Boland MP, Pukala TL, Bowie JH, Aguilar MI, Separovic F. Effect of antimicrobial peptides from Australian tree frogs on anionic phospholipid membranes. Biochemistry. 2008;47:8557–8565. doi: 10.1021/bi800320v. [DOI] [PubMed] [Google Scholar]
- 93.Ludtke SJ, He K, Heller WT, Harroun TA, Yang L, Huang HW. Membrane pores induced by magainin. Biochemistry. 1996;35:13723–13728. doi: 10.1021/bi9620621. [DOI] [PubMed] [Google Scholar]
- 94.Yang L, Harroun TA, Weiss TM, Ding L, Huang HW. Barrel-stave model or toroidal model? A case study on melittin pores. Biophys J. 2001;81:1475–1485. doi: 10.1016/S0006-3495(01)75802-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Epand RF, Maloy L, Ramamoorthy A, Epand RM. Amphipathic helical cationic antimicrobial peptides promote rapid formation of crystalline states in the presence of phosphatidylglycerol: lipid clustering in anionic membranes. Biophys J. 2010;98:2564–2573. doi: 10.1016/j.bpj.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Epand RF, Maloy WL, Ramamoorthy A, Epand RM. Probing the “charge cluster mechanism” in amphipathic helical cationic antimicrobial peptides. Biochemistry. 2010;49:4076–4084. doi: 10.1021/bi100378m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pag U, Oedenkoven M, Sass V, Shai Y, Shamova O, Antcheva N, Tossi A, Sahl HG. Analysis of in vitro activities and modes of action of synthetic antimicrobial peptides derived from an alpha-helical ‘sequence template’. J Antimicrob Chemother. 2008;61:341–352. doi: 10.1093/jac/dkm479. [DOI] [PubMed] [Google Scholar]
- 98.Wieprecht T, Apostolov O, Beyermann M, Seelig J. Membrane binding and pore formation of the antibacterial peptide PGLa: thermodynamic and mechanistic aspects. Biochemistry. 2000;39:442–452. doi: 10.1021/bi992146k. [DOI] [PubMed] [Google Scholar]
- 99.Yandek LE, Pokorny A, Floren A, Knoelke K, Langel U, Almeida PF. Mechanism of the cell-penetrating peptide transportan 10 permeation of lipid bilayers. Biophys J. 2007;92:2434–2444. doi: 10.1529/biophysj.106.100198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ladokhin AS, Wimley WC, Hristova K, White SH. Mechanism of leakage of contents of membrane vesicles determined by fluorescence requenching. Methods Enzymol. 1997;278:474–486. doi: 10.1016/s0076-6879(97)78025-x. [DOI] [PubMed] [Google Scholar]
- 101.Ladokhin AS, Wimley WC, White SH. Leakage of membrane vesicle contents: determination of mechanism using fluorescence requenching. Biophys J. 1995;69:1964–1971. doi: 10.1016/S0006-3495(95)80066-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gregory SM, Cavenaugh A, Journigan V, Pokorny A, Almeida PF. A quantitative model for the all-or-none permeabilization of phospholipid vesicles by the antimicrobial peptide cecropin A. Biophys J. 2008;94:1667–1680. doi: 10.1529/biophysj.107.118760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gregory SM, Pokorny A, Almeida PF. Magainin 2 revisited: a test of the quantitative model for the all-or-none permeabilization of phospholipid vesicles. Biophys J. 2009;96:116–131. doi: 10.1016/j.bpj.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wimley WC, Selsted ME, White SH. Interactions between human defensins and lipid bilayers: evidence for formation of multimeric pores. Protein Sci. 1994;3:1362–1373. doi: 10.1002/pro.5560030902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Epand RM, Vogel HJ. Diversity of antimicrobial peptides and their mechanisms of action. Biochim Biophys Acta. 1999;1462:11–28. doi: 10.1016/s0005-2736(99)00198-4. [DOI] [PubMed] [Google Scholar]
- 106.Koprivnjak T, Peschel A. Bacterial resistance mechanisms against host defense peptides. Cell Mol Life Sci. 2011;68:2243–2254. doi: 10.1007/s00018-011-0716-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Marion D, Zasloff M, Bax A. A two-dimensional NMR study of the antimicrobial peptide magainin 2. FEBS Lett. 1988;227:21–26. doi: 10.1016/0014-5793(88)81405-4. [DOI] [PubMed] [Google Scholar]
- 108.Bechinger B. The structure, dynamics and orientation of antimicrobial peptides in membranes by multidimensional solid-state NMR spectroscopy. Biochim Biophys Acta. 1999;1462:157–183. doi: 10.1016/s0005-2736(99)00205-9. [DOI] [PubMed] [Google Scholar]
- 109.Hauge HH, Mantzilas D, Eijsink VG, Nissen-Meyer J. Membrane-mimicking entities induce structuring of the two-peptide bacteriocins plantaricin E/F and plantaricin J/K. J Bacteriol. 1999;181:740–747. doi: 10.1128/jb.181.3.740-747.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Luo P, Baldwin RL. Mechanism of helix induction by trifluoroethanol: a framework for extrapolating the helix-forming properties of peptides from trifluoroethanol/water mixtures back to water. Biochemistry. 1997;36:8413–8421. doi: 10.1021/bi9707133. [DOI] [PubMed] [Google Scholar]
- 111.Prosser RS, Evanics F, Kitevski JL, Al-Abdul-Wahid MS. Current applications of bicelles in NMR studies of membrane-associated amphiphiles and proteins. Biochemistry. 2006;45:8453–8465. doi: 10.1021/bi060615u. [DOI] [PubMed] [Google Scholar]
- 112.Warschawski DE, Arnold AA, Beaugrand M, Gravel A, Chartrand E, Marcotte I. Choosing membrane mimetics for NMR structural studies of transmembrane proteins. Biochim Biophys Acta. 2011;1808:1957–1974. doi: 10.1016/j.bbamem.2011.03.016. [DOI] [PubMed] [Google Scholar]
- 113.Bourbigot S, Fardy L, Waring AJ, Yeaman MR, Booth V. Structure of chemokine-derived antimicrobial Peptide interleukin-8alpha and interaction with detergent micelles and oriented lipid bilayers. Biochemistry. 2009;48:10509–10521. doi: 10.1021/bi901311p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Khandelia H, Kaznessis YN. Molecular dynamics simulations of helical antimicrobial peptides in SDS micelles: what do point mutations achieve? Peptides. 2005;26:2037–2049. doi: 10.1016/j.peptides.2005.03.058. [DOI] [PubMed] [Google Scholar]
- 115.Mascioni A, Porcelli F, Ilangovan U, Ramamoorthy A, Veglia G. Conformational preferences of the amylin nucleation site in SDS micelles: an NMR study. Biopolymers. 2003;69:29–41. doi: 10.1002/bip.10305. [DOI] [PubMed] [Google Scholar]
- 116.Buffy JJ, Buck-Koehntop BA, Porcelli F, Traaseth NJ, Thomas DD, Veglia G. Defining the intramembrane binding mechanism of sarcolipin to calcium ATPase using solution NMR spectroscopy. J Mol Biol. 2006;358:420–429. doi: 10.1016/j.jmb.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 117.Sherman PJ, Jackway RJ, Gehman JD, Praporski S, McCubbin GA, Mechler A, Martin LL, Separovic F, Bowie JH. Solution structure and membrane interactions of the antimicrobial peptide fallaxidin 4.1a: an NMR and QCM study. Biochemistry. 2009;48:11892–11901. doi: 10.1021/bi901668y. [DOI] [PubMed] [Google Scholar]
- 118.Poget SF, Cahill SM, Girvin ME. Isotropic bicelles stabilize the functional form of a small multidrug-resistance pump for NMR structural studies. J Am Chem Soc. 2007;129:2432–2433. doi: 10.1021/ja0679836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sanders CR, 2nd, Landis GC. Reconstitution of membrane proteins into lipid-rich bilayered mixed micelles for NMR studies. Biochemistry. 1995;34:4030–4040. doi: 10.1021/bi00012a022. [DOI] [PubMed] [Google Scholar]
- 120.Sanders CR, Prosser RS. Bicelles: a model membrane system for all seasons? Structure. 1998;6:1227–1234. doi: 10.1016/s0969-2126(98)00123-3. [DOI] [PubMed] [Google Scholar]
- 121.Marcotte I, Wegener KL, Lam YH, Chia BC, de Planque MR, Bowie JH, Auger M, Separovic F. Interaction of antimicrobial peptides from Australian amphibians with lipid membranes. Chem Phys Lipids. 2003;122:107–120. doi: 10.1016/s0009-3084(02)00182-2. [DOI] [PubMed] [Google Scholar]
- 122.Dittmer J, Thogersen L, Underhaug J, Bertelsen K, Vosegaard T, Pedersen JM, Schiott B, Tajkhorshid E, Skrydstrup T, Nielsen NC. Incorporation of antimicrobial peptides into membranes: a combined liquid-state NMR and molecular dynamics study of alamethicin in DMPC/DHPC bicelles. J Phys Chem B. 2009;113:6928–6937. doi: 10.1021/jp811494p. [DOI] [PubMed] [Google Scholar]
- 123.Jiang Y, Wang H, Kindt JT. Atomistic simulations of bicelle mixtures. Biophys J. 2010;98:2895–2903. doi: 10.1016/j.bpj.2010.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.van Dam L, Karlsson G, Edwards K. Morphology of magnetically aligning DMPC/DHPC aggregates-perforated sheets, not disks. Langmuir. 2006;22:3280–3285. doi: 10.1021/la052988m. [DOI] [PubMed] [Google Scholar]
- 125.Bechinger B. Detergent-like properties of magainin antibiotic peptides: a 31P solid-state NMR spectroscopy study. Biochim Biophys Acta. 2005;1712:101–108. doi: 10.1016/j.bbamem.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 126.Cardon TB, Tiburu EK, Lorigan GA. Magnetically aligned phospholipid bilayers in weak magnetic fields: optimization, mechanism, and advantages for X-band EPR studies. J Magn Reson. 2003;161:77–90. doi: 10.1016/s1090-7807(02)00109-x. [DOI] [PubMed] [Google Scholar]
- 127.Diller A, Loudet C, Aussenac F, Raffard G, Fournier S, Laguerre M, Grelard A, Opella SJ, Marassi FM, Dufourc EJ. Bicelles: A natural ‘molecular goniometer’ for structural, dynamical and topological studies of molecules in membranes. Biochimie. 2009;91:744–751. doi: 10.1016/j.biochi.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.De Angelis AA, Grant CV, Baxter MK, McGavin JA, Opella SJ, Cotten ML. Amphipathic antimicrobial piscidin in magnetically aligned lipid bilayers. Biophys J. 2011;101:1086–1094. doi: 10.1016/j.bpj.2011.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Verardi R, Traaseth NJ, Shi L, Porcelli F, Monfregola L, De Luca S, Amodeo P, Veglia G, Scaloni A. Probing membrane topology of the antimicrobial peptide distinctin by solid-state NMR spectroscopy in zwitterionic and charged lipid bilayers. Biochim Biophys Acta. 2012;1808:34–40. doi: 10.1016/j.bbamem.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Mote KR, Gopinath T, Traaseth NJ, Kitchen J, Gor’kov PL, Brey WW, Veglia G. Multidimensional oriented solid-state NMR experiments enable the sequential assignment of uniformly 15N labeled integral membrane proteins in magnetically aligned lipid bilayers. J Biomol NMR. 2011;51:339–346. doi: 10.1007/s10858-011-9571-8. [DOI] [PubMed] [Google Scholar]
- 131.Bechinger B, Kim Y, Chirlian LE, Gesell J, Neumann JM, Montal M, Tomich J, Zasloff M, Opella SJ. Orientations of amphipathic helical peptides in membrane bilayers determined by solid-state NMR spectroscopy. J Biomol NMR. 1991;1:167–173. doi: 10.1007/BF01877228. [DOI] [PubMed] [Google Scholar]
- 132.Opella SJ, Ma C, Marassi FM. Nuclear magnetic resonance of membrane-associated peptides and proteins. Methods Enzymol. 2001;339:285–313. doi: 10.1016/s0076-6879(01)39319-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Valentine KG, Liu SF, Marassi FM, Veglia G, Opella SJ, Ding FX, Wang SH, Arshava B, Becker JM, Naider F. Structure and topology of a peptide segment of the 6th transmembrane domain of the Saccharomyces cerevisae alpha-factor receptor in phospholipid bilayers. Biopolymers. 2001;59:243–256. doi: 10.1002/1097-0282(20011005)59:4<243::AID-BIP1021>3.0.CO;2-H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Killian JA, Borle F, de Kruijff B, Seelig J. Comparative 2H- and 31P-NMR study on the properties of palmitoyllysophosphatidylcholine in bilayers with gramicidin, cholesterol and dipalmitoylphosphatidylcholine. Biochim Biophys Acta. 1986;854:133–142. doi: 10.1016/0005-2736(86)90073-8. [DOI] [PubMed] [Google Scholar]
- 135.Klocek G, Schulthess T, Shai Y, Seelig J. Thermodynamics of melittin binding to lipid bilayers. Aggregation and pore formation. Biochemistry. 2009;48:2586–2596. doi: 10.1021/bi802127h. [DOI] [PubMed] [Google Scholar]
- 136.Seelig J, MacDonald PM. Phospholipids and Proteins in Biological Membranes. 2H NMR as a Method To Study Structure, Dynamics, and Interactions. Acc Chem Res. 1987;20:221–228. [Google Scholar]
- 137.Sharma M, Yi M, Dong H, Qin H, Peterson E, Busath DD, Zhou HX, Cross TA. Insight into the mechanism of the influenza A proton channel from a structure in a lipid bilayer. Science. 2010;330:509–512. doi: 10.1126/science.1191750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Mascioni A, Karim C, Barany G, Thomas DD, Veglia G. Structure and orientation of sarcolipin in lipid environments. Biochemistry. 2002;41:475–482. doi: 10.1021/bi011243m. [DOI] [PubMed] [Google Scholar]
- 139.Mascioni A, Karim C, Zamoon J, Thomas DD, Veglia G. Solid-state NMR and rigid body molecular dynamics to determine domain orientations of monomeric phospholamban. J Am Chem Soc. 2002;124:9392–9393. doi: 10.1021/ja026507m. [DOI] [PubMed] [Google Scholar]
- 140.Traaseth NJ, Buffy JJ, Zamoon J, Veglia G. Structural dynamics and topology of phospholamban in oriented lipid bilayers using multidimensional solid-state NMR. Biochemistry. 2006;45:13827–13834. doi: 10.1021/bi0607610. [DOI] [PubMed] [Google Scholar]
- 141.Traaseth NJ, Shi L, Verardi R, Mullen DG, Barany G, Veglia G. Structure and topology of monomeric phospholamban in lipid membranes determined by a hybrid solution and solid-state NMR approach. Proc Natl Acad Sci U S A. 2009;106:10165–10170. doi: 10.1073/pnas.0904290106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Verardi R, Shi L, Traaseth NJ, Walsh N, Veglia G. Structural topology of phospholamban pentamer in lipid bilayers by a hybrid solution and solid-state NMR method. Proc Natl Acad Sci U S A. 2011;108:9101–9106. doi: 10.1073/pnas.1016535108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Epand RM, Epand RF. Liposomes as models for antimicrobial peptides. Methods Enzymol. 2003;372:124–133. doi: 10.1016/S0076-6879(03)72007-2. [DOI] [PubMed] [Google Scholar]
- 144.Marquette A, Lorber B, Bechinger B. Reversible liposome association induced by LAH4: a peptide with potent antimicrobial and nucleic acid transfection activities. Biophys J. 2010;98:2544–2553. doi: 10.1016/j.bpj.2010.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Georgescu J, Munhoz VH, Bechinger B. NMR structures of the histidine-rich peptide LAH4 in micellar environments: membrane insertion, pH-dependent mode of antimicrobial action, and DNA transfection. Biophys J. 2010;99:2507–2515. doi: 10.1016/j.bpj.2010.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Malliavin TE, Giudice E. Analysis of peptide rotational diffusion by homonuclear NMR. Biopolymers. 2002;63:335–342. doi: 10.1002/bip.10128. [DOI] [PubMed] [Google Scholar]
- 147.Porcelli F, Buck B, Lee DK, Hallock KJ, Ramamoorthy A, Veglia G. Structure and orientation of pardaxin determined by NMR experiments in model membranes. J Biol Chem. 2004;279:45815–45823. doi: 10.1074/jbc.M405454200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Park SH, Kim YK, Park JW, Lee B, Lee BJ. Solution structure of the antimicrobial peptide gaegurin 4 by H and 15N nuclear magnetic resonance spectroscopy. Eur J Biochem. 2000;267:2695–2704. doi: 10.1046/j.1432-1327.2000.01287.x. [DOI] [PubMed] [Google Scholar]
- 149.Matsumori N, Murata M. 3D structures of membrane-associated small molecules as determined in isotropic bicelles. Nat Prod Rep. 2010;27:1480–1492. doi: 10.1039/c0np00002g. [DOI] [PubMed] [Google Scholar]
- 150.Yamamoto K, Vivekanandan S, Ramamoorthy A. Fast NMR data acquisition from bicelles containing a membrane-associated peptide at natural-abundance. J Phys Chem B. 2011;115:12448–12455. doi: 10.1021/jp2076098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Chou JJ, Delaglio F, Bax A. Measurement of one-bond 15N-13C’ dipolar couplings in medium sized proteins. J Biomol NMR. 2000;18:101–105. doi: 10.1023/a:1008358318863. [DOI] [PubMed] [Google Scholar]
- 152.Kubat JA, Chou JJ, Rovnyak D. Nonuniform sampling and maximum entropy reconstruction applied to the accurate measurement of residual dipolar couplings. J Magn Reson. 2007;186:201–211. doi: 10.1016/j.jmr.2007.01.022. [DOI] [PubMed] [Google Scholar]
- 153.Jaroniec CP, Boisbouvier J, Tworowska I, Nikonowicz EP, Bax A. Accurate measurement of 15N-13C residual dipolar couplings in nucleic acids. J Biomol NMR. 2005;31:231–241. doi: 10.1007/s10858-005-0646-2. [DOI] [PubMed] [Google Scholar]
- 154.Al-Abdul-Wahid MS, Verardi R, Veglia G, Prosser RS. Topology and immersion depth of an integral membrane protein by paramagnetic rates from dissolved oxygen. J Biomol NMR. 2011;51:173–183. doi: 10.1007/s10858-011-9551-z. [DOI] [PubMed] [Google Scholar]
- 155.Donghi D, Sigel RK. Metal ion-RNA interactions studied via multinuclear NMR. Methods Mol Biol. 2012;848:253–273. doi: 10.1007/978-1-61779-545-9_16. [DOI] [PubMed] [Google Scholar]
- 156.Glover KJ, Whiles JA, Vold RR, Melacini G. Position of residues in transmembrane peptides with respect to the lipid bilayer: a combined lipid Noes and water chemical exchange approach in phospholipid bicelles. J Biomol NMR. 2002;22:57–64. doi: 10.1023/a:1013817818794. [DOI] [PubMed] [Google Scholar]
- 157.Huang H, Melacini G. High-resolution protein hydration NMR experiments: probing how protein surfaces interact with water and other non-covalent ligands. Anal Chim Acta. 2006;564:1–9. doi: 10.1016/j.aca.2005.10.049. [DOI] [PubMed] [Google Scholar]
- 158.Melacini G, Boelens R, Kaptein R. Band-selective editing of exchange-relay in protein-water NOE experiments. J Biomol NMR. 1999;13:67–71. doi: 10.1023/A:1008350918005. [DOI] [PubMed] [Google Scholar]
- 159.Choutko A, Glattli A, Fernandez C, Hilty C, Wuthrich K, van Gunsteren WF. Membrane protein dynamics in different environments: simulation study of the outer membrane protein X in a lipid bilayer and in a micelle. Eur Biophys J. 2010;40:39–58. doi: 10.1007/s00249-010-0626-7. [DOI] [PubMed] [Google Scholar]
- 160.Fernandez C, Adeishvili K, Wuthrich K. Transverse relaxation-optimized NMR spectroscopy with the outer membrane protein OmpX in dihexanoyl phosphatidylcholine micelles. Proc Natl Acad Sci U S A. 2001;98:2358–2363. doi: 10.1073/pnas.051629298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Fernandez C, Hilty C, Wider G, Wuthrich K. Lipid-protein interactions in DHPC micelles containing the integral membrane protein OmpX investigated by NMR spectroscopy. Proc Natl Acad Sci U S A. 2002;99:13533–13537. doi: 10.1073/pnas.212515099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Fernandez C, Wuthrich K. NMR solution structure determination of membrane proteins reconstituted in detergent micelles. FEBS Lett. 2003;555:144–150. doi: 10.1016/s0014-5793(03)01155-4. [DOI] [PubMed] [Google Scholar]
- 163.Nishida N, Shimada I. An NMR method to study protein-protein interactions. Methods Mol Biol. 2011;757:129–137. doi: 10.1007/978-1-61779-166-6_10. [DOI] [PubMed] [Google Scholar]
- 164.Shimada I. NMR techniques for identifying the interface of a larger protein-protein complex: cross-saturation and transferred cross-saturation experiments. Methods Enzymol. 2005;394:483–506. doi: 10.1016/S0076-6879(05)94020-2. [DOI] [PubMed] [Google Scholar]
- 165.Takahashi H, Miyazawa M, Ina Y, Fukunishi Y, Mizukoshi Y, Nakamura H, Shimada I. Utilization of methyl proton resonances in cross-saturation measurement for determining the interfaces of large protein-protein complexes. J Biomol NMR. 2006;34:167–177. doi: 10.1007/s10858-006-0008-8. [DOI] [PubMed] [Google Scholar]
- 166.Cook GA, Zhang H, Park SH, Wang Y, Opella SJ. Comparative NMR studies demonstrate profound differences between two viroporins: p7 of HCV and Vpu of HIV-1. Biochim Biophys Acta. 2010;1808:554–560. doi: 10.1016/j.bbamem.2010.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Howard KP, Opella SJ. High-resolution solid-state NMR spectra of integral membrane proteins reconstituted into magnetically oriented phospholipid bilayers. J Magn Reson B. 1996;112:91–94. doi: 10.1006/jmrb.1996.0116. [DOI] [PubMed] [Google Scholar]
- 168.Gopinath T, Traaseth NJ, Mote K, Veglia G. Sensitivity enhanced heteronuclear correlation spectroscopy in multidimensional solid-state NMR of oriented systems via chemical shift coherences. J Am Chem Soc. 2010;132:5357–5363. doi: 10.1021/ja905991s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Gopinath T, Veglia G. Dual acquisition magic-angle spinning solid-state NMR-spectroscopy: simultaneous acquisition of multidimensional spectra of biomacromolecules. Angew Chem Int Ed Engl. 2012;51:2731–2735. doi: 10.1002/anie.201108132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Bechinger B, Gierasch LM, Montal M, Zasloff M, Opella SJ. Orientations of helical peptides in membrane bilayers by solid state NMR spectroscopy. Solid State Nucl Magn Reson. 1996;7:185–191. doi: 10.1016/0926-2040(95)01224-9. [DOI] [PubMed] [Google Scholar]
- 171.Bechinger B, Skladnev DA, Ogrel A, Li X, Rogozhkina EV, Ovchinnikova TV, O’Neil JD, Raap J. 15N and 31P solid-state NMR investigations on the orientation of zervamicin II and alamethicin in phosphatidylcholine membranes. Biochemistry. 2001;40:9428–9437. doi: 10.1021/bi010162n. [DOI] [PubMed] [Google Scholar]
- 172.Bechinger B, Zasloff M, Opella SJ. Structure and orientation of the antibiotic peptide magainin in membranes by solid-state nuclear magnetic resonance spectroscopy. Protein Sci. 1993;2:2077–2084. doi: 10.1002/pro.5560021208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Gustavsson M, Traaseth NJ, Veglia G. Probing ground and excited states of phospholamban in model and native lipid membranes by magic angle spinning NMR spectroscopy. Biochim Biophys Acta. 2012;1818:146–153. doi: 10.1016/j.bbamem.2011.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Tang M, Hong M. Structure and mechanism of beta-hairpin antimicrobial peptides in lipid bilayers from solid-state NMR spectroscopy. Mol Biosyst. 2009;5:317–322. doi: 10.1039/b820398a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Yamaguchi S, Huster D, Waring A, Lehrer RI, Kearney W, Tack BF, Hong M. Orientation and dynamics of an antimicrobial peptide in the lipid bilayer by solid-state NMR spectroscopy. Biophys J. 2001;81:2203–2214. doi: 10.1016/S0006-3495(01)75868-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Ramamoorthy A, Lee DK, Santos JS, Henzler-Wildman KA. Nitrogen-14 solid-state NMR spectroscopy of aligned phospholipid bilayers to probe peptide-lipid interaction and oligomerization of membrane associated peptides. J Am Chem Soc. 2008;130:11023–11029. doi: 10.1021/ja802210u. [DOI] [PubMed] [Google Scholar]
- 177.Thennarasu S, Lee DK, Poon A, Kawulka KE, Vederas JC, Ramamoorthy A. Membrane permeabilization, orientation, and antimicrobial mechanism of subtilosin A. Chem Phys Lipids. 2005;137:38–51. doi: 10.1016/j.chemphyslip.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 178.Ketchem RR, Hu W, Cross TA. High-resolution conformation of gramicidin A in a lipid bilayer by solid-state NMR. Science. 1993;261:1457–1460. doi: 10.1126/science.7690158. [DOI] [PubMed] [Google Scholar]
- 179.Quine JR, Brenneman MT, Cross TA. Protein structural analysis from solid-state NMR-derived orientational constraints. Biophys J. 1997;72:2342–2348. doi: 10.1016/S0006-3495(97)78878-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Bechinger B, Kinder R, Helmle M, Vogt TC, Harzer U, Schinzel S. Peptide structural analysis by solid-state NMR spectroscopy. Biopolymers. 1999;51:174–190. doi: 10.1002/(SICI)1097-0282(1999)51:3<174::AID-BIP2>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 181.Marassi FM, Ma C, Gesell JJ, Opella SJ. Three-dimensional solid-state NMR spectroscopy is essential for resolution of resonances from in-plane residues in uniformly (15)N-labeled helical membrane proteins in oriented lipid bilayers. J Magn Reson. 2000;144:156–161. doi: 10.1006/jmre.2000.2036. [DOI] [PubMed] [Google Scholar]
- 182.De Angelis AA, Nevzorov AA, Park SH, Howell SC, Mrse AA, Opella SJ. High-resolution NMR spectroscopy of membrane proteins in aligned bicelles. J Am Chem Soc. 2004;126:15340–15341. doi: 10.1021/ja045631y. [DOI] [PubMed] [Google Scholar]
- 183.Andronesi OC, Pfeifer JR, Al-Momani L, Ozdirekcan S, Rijkers DT, Angerstein B, Luca S, Koert U, Killian JA, Baldus M. Probing membrane protein orientation and structure using fast magic-angle-spinning solid-state NMR. J Biomol NMR. 2004;30:253–265. doi: 10.1007/s10858-004-3452-3. [DOI] [PubMed] [Google Scholar]
- 184.Elena B, Hediger S, Emsley L. Correlation of fast and slow chemical shift spinning sideband patterns under fast magic-angle spinning. J Magn Reson. 2003;160:40–46. doi: 10.1016/s1090-7807(02)00037-x. [DOI] [PubMed] [Google Scholar]
- 185.Griffin RG. Dipolar recoupling in MAS spectra of biological solids. Nat Struct Biol. 1998;5(Suppl):508–512. doi: 10.1038/749. [DOI] [PubMed] [Google Scholar]
- 186.Zasloff M, Martin B, Chen HC. Antimicrobial activity of synthetic magainin peptides and several analogues. Proc Natl Acad Sci U S A. 1988;85:910–913. doi: 10.1073/pnas.85.3.910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Bechinger B, Zasloff M, Opella SJ. Structure and dynamics of the antibiotic peptide PGLa in membranes by solution and solid-state nuclear magnetic resonance spectroscopy. Biophys J. 1998;74:981–987. doi: 10.1016/S0006-3495(98)74021-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Bechinger B, Zasloff M, Opella SJ. Structure and interactions of magainin antibiotic peptides in lipid bilayers: a solid-state nuclear magnetic resonance investigation. Biophys J. 1992;62:12–14. doi: 10.1016/S0006-3495(92)81763-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Gesell J, Zasloff M, Opella SJ. Two-dimensional 1H NMR experiments show that the 23-residue magainin antibiotic peptide is an alpha-helix in dodecylphosphocholine micelles, sodium dodecylsulfate micelles, and trifluoroethanol/water solution. J Biomol NMR. 1997;9:127–135. doi: 10.1023/a:1018698002314. [DOI] [PubMed] [Google Scholar]
- 190.Matsuzaki K, Mitani Y, Akada KY, Murase O, Yoneyama S, Zasloff M, Miyajima K. Mechanism of synergism between antimicrobial peptides magainin 2 and PGLa. Biochemistry. 1998;37:15144–15153. doi: 10.1021/bi9811617. [DOI] [PubMed] [Google Scholar]
- 191.Ramamoorthy A, Marassi FM, Zasloff M, Opella SJ. Three-dimensional solid-state NMR spectroscopy of a peptide oriented in membrane bilayers. J Biomol NMR. 1995;6:329–334. doi: 10.1007/BF00197814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Wieprecht T, Apostolov O, Seelig J. Binding of the antibacterial peptide magainin 2 amide to small and large unilamellar vesicles. Biophys Chem. 2000;85:187–198. doi: 10.1016/s0301-4622(00)00120-4. [DOI] [PubMed] [Google Scholar]
- 193.Islam K, Hawser SP. MSI-78 Magainin Pharmaceuticals. IDrugs. 1998;1:605–609. [PubMed] [Google Scholar]
- 194.Yang P, Ramamoorthy A, Chen Z. Membrane orientation of MSI-78 measured by sum frequency generation vibrational spectroscopy. Langmuir. 2011;27:7760–7767. doi: 10.1021/la201048t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Domadia PN, Bhunia A, Ramamoorthy A, Bhattacharjya S. Structure, interactions, and antibacterial activities of MSI-594 derived mutant peptide MSI-594F5A in lipopolysaccharide micelles: role of the helical hairpin conformation in outer-membrane permeabilization. J Am Chem Soc. 2010;132:18417–18428. doi: 10.1021/ja1083255. [DOI] [PubMed] [Google Scholar]
- 196.Wakamatsu K, Takeda A, Tachi T, Matsuzaki K. Dimer structure of magainin 2 bound to phospholipid vesicles. Biopolymers. 2002;64:314–327. doi: 10.1002/bip.10198. [DOI] [PubMed] [Google Scholar]
- 197.Henzler-Wildman KA, Martinez GV, Brown MF, Ramamoorthy A. Perturbation of the hydrophobic core of lipid bilayers by the human antimicrobial peptide LL-37. Biochemistry. 2004;43:8459–8469. doi: 10.1021/bi036284s. [DOI] [PubMed] [Google Scholar]
- 198.Lazarovici P, Primor N, Loew LM. Purification and pore-forming activity of two hydrophobic polypeptides from the secretion of the Red Sea Moses sole (Pardachirus marmoratus) J Biol Chem. 1986;261:16704–16713. [PubMed] [Google Scholar]
- 199.Oren Z, Shai Y. A class of highly potent antibacterial peptides derived from pardaxin, a pore-forming peptide isolated from Moses sole fish Pardachirus marmoratus. Eur J Biochem. 1996;237:303–310. doi: 10.1111/j.1432-1033.1996.0303n.x. [DOI] [PubMed] [Google Scholar]
- 200.Bhunia A, Domadia PN, Torres J, Hallock KJ, Ramamoorthy A, Bhattacharjya S. NMR structure of pardaxin, a pore-forming antimicrobial peptide, in lipopolysaccharide micelles: mechanism of outer membrane permeabilization. J Biol Chem. 2010;285:3883–3895. doi: 10.1074/jbc.M109.065672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Epand RF, Ramamoorthy A, Epand RM. Membrane lipid composition and the interaction of pardaxin: the role of cholesterol. Protein Pept Lett. 2006;13:1–5. [PubMed] [Google Scholar]
- 202.Hallock KJ, Lee DK, Omnaas J, Mosberg HI, Ramamoorthy A. Membrane composition determines pardaxin’s mechanism of lipid bilayer disruption. Biophys J. 2002;83:1004–1013. doi: 10.1016/S0006-3495(02)75226-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Hsu JC, Lin LC, Tzen JT, Chen JY. Pardaxin-induced apoptosis enhances antitumor activity in HeLa cells. Peptides. 2011;32:1110–1116. doi: 10.1016/j.peptides.2011.04.024. [DOI] [PubMed] [Google Scholar]
- 204.Lelkes PI, Lazarovici P. Pardaxin induces aggregation but not fusion of phosphatidylserine vesicles. FEBS Lett. 1988;230:131–136. doi: 10.1016/0014-5793(88)80656-2. [DOI] [PubMed] [Google Scholar]
- 205.Vad BS, Bertelsen K, Johansen CH, Pedersen JM, Skrydstrup T, Nielsen NC, Otzen DE. Pardaxin permeabilizes vesicles more efficiently by pore formation than by disruption. Biophys J. 2010;98:576–585. doi: 10.1016/j.bpj.2009.08.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Yang J, Parkanzky PD, Bodner ML, Duskin CA, Weliky DP. Application of REDOR subtraction for filtered MAS observation of labeled backbone carbons of membrane-bound fusion peptides. J Magn Reson. 2002;159:101–110. doi: 10.1016/s1090-7807(02)00033-2. [DOI] [PubMed] [Google Scholar]
- 207.Lee DK, Ramamoorthy A. Determination of the Solid-State Conformations of Polyalanine Using Magic-Angle Spinning NMR Spectroscopy. J Phys Chem B. 1999;103:271–275. [Google Scholar]
- 208.Batista CV, Scaloni A, Rigden DJ, Silva LR, Rodrigues Romero A, Dukor R, Sebben A, Talamo F, Bloch C. A novel heterodimeric antimicrobial peptide from the tree-frog Phyllomedusa distincta. FEBS Lett. 2001;494:85–89. doi: 10.1016/s0014-5793(01)02324-9. [DOI] [PubMed] [Google Scholar]
- 209.Becucci L, Papini M, Mullen D, Scaloni A, Veglia G, Guidelli R. Probing membrane permeabilization by the antimicrobial peptide distinctin in mercury-supported biomimetic membranes. Biochim Biophys Acta. 2011;1808:2745–2752. doi: 10.1016/j.bbamem.2011.07.033. [DOI] [PubMed] [Google Scholar]
- 210.Cirioni O, Ghiselli R, Orlando F, Silvestri C, De Luca S, Salzano AM, Mocchegiani F, Saba V, Scalise G, Scaloni A, Giacometti A. Efficacy of the amphibian peptide distinctin in a neutropenic mouse model of staphylococcal sepsis. Crit Care Med. 2008;36:2629–2633. doi: 10.1097/CCM.0b013e318184430d. [DOI] [PubMed] [Google Scholar]
- 211.Raimondo D, Andreotti G, Saint N, Amodeo P, Renzone G, Sanseverino M, Zocchi I, Molle G, Motta A, Scaloni A. A folding-dependent mechanism of antimicrobial peptide resistance to degradation unveiled by solution structure of distinctin. Proc Natl Acad Sci U S A. 2005;102:6309–6314. doi: 10.1073/pnas.0409004102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Dalla Serra M, Cirioni O, Vitale RM, Renzone G, Coraiola M, Giacometti A, Potrich C, Baroni E, Guella G, Sanseverino M, De Luca S, Scalise G, Amodeo P, Scaloni A. Structural features of distinctin affecting peptide biological and biochemical properties. Biochemistry. 2008;47:7888–7899. doi: 10.1021/bi800616k. [DOI] [PubMed] [Google Scholar]
- 213.Shi L, Traaseth NJ, Verardi R, Cembran A, Gao J, Veglia G. A refinement protocol to determine structure, topology, and depth of insertion of membrane proteins using hybrid solution and solid-state NMR restraints. J Biomol NMR. 2009;44:195–205. doi: 10.1007/s10858-009-9328-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Bals R, Wang X, Zasloff M, Wilson JM. The peptide antibiotic LL-37/hCAP-18 is expressed in epithelia of the human lung where it has broad antimicrobial activity at the airway surface. Proc Natl Acad Sci U S A. 1998;95:9541–9546. doi: 10.1073/pnas.95.16.9541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Wong CC, Zhang L, Ren SX, Shen J, Chan RL, Cho CH. Antibacterial peptides and gastrointestinal diseases. Curr Pharm Des. 2011;17:1583–1586. doi: 10.2174/138161211796197025. [DOI] [PubMed] [Google Scholar]
- 216.Wu WK, Wong CC, Li ZJ, Zhang L, Ren SX, Cho CH. Cathelicidins in inflammation and tissue repair: Potential therapeutic applications for gastrointestinal disorders. Acta Pharmacol Sin. 2010;31:1118–1122. doi: 10.1038/aps.2010.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Cederlund A, Gudmundsson GH, Agerberth B. Antimicrobial peptides important in innate immunity. FEBS J. 2011;278:3942–3951. doi: 10.1111/j.1742-4658.2011.08302.x. [DOI] [PubMed] [Google Scholar]
- 218.Tecle T, Tripathi S, Hartshorn KL. Review: Defensins and cathelicidins in lung immunity. Innate Immun. 2010;16:151–159. doi: 10.1177/1753425910365734. [DOI] [PubMed] [Google Scholar]
- 219.Yamasaki K, Gallo RL. Rosacea as a disease of cathelicidins and skin innate immunity. J Investig Dermatol Symp Proc. 2011;15:12–15. doi: 10.1038/jidsymp.2011.4. [DOI] [PubMed] [Google Scholar]
- 220.Amatngalim GD, Nijnik A, Hiemstra PS, Hancock RE. Cathelicidin Peptide LL-37 Modulates TREM-1 Expression and Inflammatory Responses to Microbial Compounds. Inflammation. 2010 doi: 10.1007/s10753-010-9248-6. [DOI] [PubMed] [Google Scholar]
- 221.Brown KL, Poon GF, Birkenhead D, Pena OM, Falsafi R, Dahlgren C, Karlsson A, Bylund J, Hancock RE, Johnson P. Host Defense Peptide LL-37 Selectively Reduces Proinflammatory Macrophage Responses. J Immunol. 2011;186:5497–5505. doi: 10.4049/jimmunol.1002508. [DOI] [PubMed] [Google Scholar]
- 222.Ramamoorthy A, Lee DK, Narasimhaswamy T, Nanga RP. Cholesterol reduces pardaxin’s dynamics-a barrel-stave mechanism of membrane disruption investigated by solid-state NMR. Biochim Biophys Acta. 2010;1798:223–227. doi: 10.1016/j.bbamem.2009.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Ramamoorthy A. Beyond NMR spectra of antimicrobial peptides: dynamical images at atomic resolution and functional insights. Solid State Nucl Magn Reson. 2009;35:201–207. doi: 10.1016/j.ssnmr.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Hong J, Oren Z, Shai Y. Structure and organization of hemolytic and nonhemolytic diastereomers of antimicrobial peptides in membranes. Biochemistry. 1999;38:16963–16973. doi: 10.1021/bi991850y. [DOI] [PubMed] [Google Scholar]
- 225.Durr UH, Sudheendra US, Ramamoorthy A. LL-37, the only human member of the cathelicidin family of antimicrobial peptides. Biochim Biophys Acta. 2006;1758:1408–1425. doi: 10.1016/j.bbamem.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 226.Henzler Wildman KA, Lee DK, Ramamoorthy A. Mechanism of lipid bilayer disruption by the human antimicrobial peptide, LL-37. Biochemistry. 2003;42:6545–6558. doi: 10.1021/bi0273563. [DOI] [PubMed] [Google Scholar]
- 227.Dubosclard V, Blondot ML, Eleouet JF, Bontems F, Sizun C. 1H, 13C, and 15N resonance assignment of the central domain of hRSV transcription antitermination factor M2-1. Biomol NMR Assign. 2011;5:237–239. doi: 10.1007/s12104-011-9308-3. [DOI] [PubMed] [Google Scholar]
- 228.Aisenbrey C, Pendem N, Guichard G, Bechinger B. Solid state NMR studies of oligourea foldamers: interaction of 15N-labelled amphiphilic helices with oriented lipid membranes. Org Biomol Chem. 2012;10:1440–1447. doi: 10.1039/c1ob06278f. [DOI] [PubMed] [Google Scholar]
- 229.Pius J, Morrow MR, Booth V. (2)H solid-state nuclear magnetic resonance investigation of whole Escherichia coli interacting with antimicrobial peptide MSI-78. Biochemistry. 2012;51:118–125. doi: 10.1021/bi201569t. [DOI] [PubMed] [Google Scholar]
- 230.Bertelsen K, Vad B, Nielsen EH, Hansen SK, Skrydstrup T, Otzen DE, Vosegaard T, Nielsen NC. Long-term-stable ether-lipid vs conventional ester-lipid bicelles in oriented solid-state NMR: altered structural information in studies of antimicrobial peptides. J Phys Chem B. 2012;115:1767–1774. doi: 10.1021/jp110866g. [DOI] [PubMed] [Google Scholar]