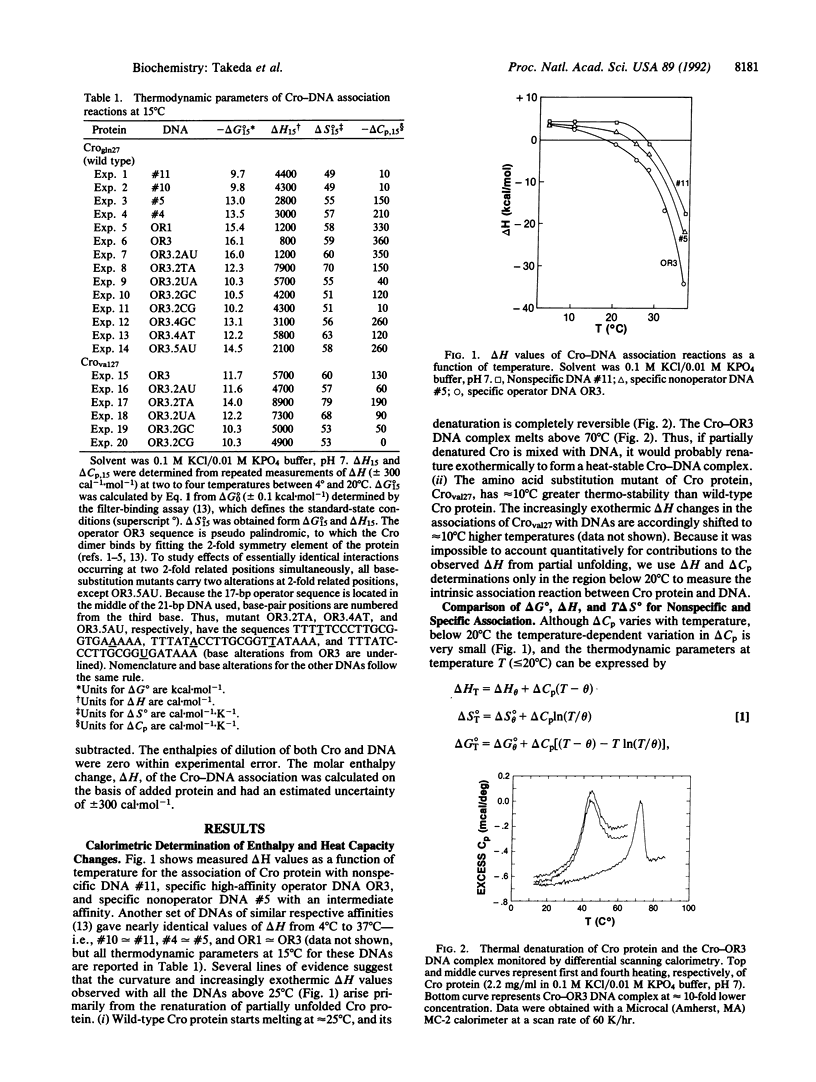
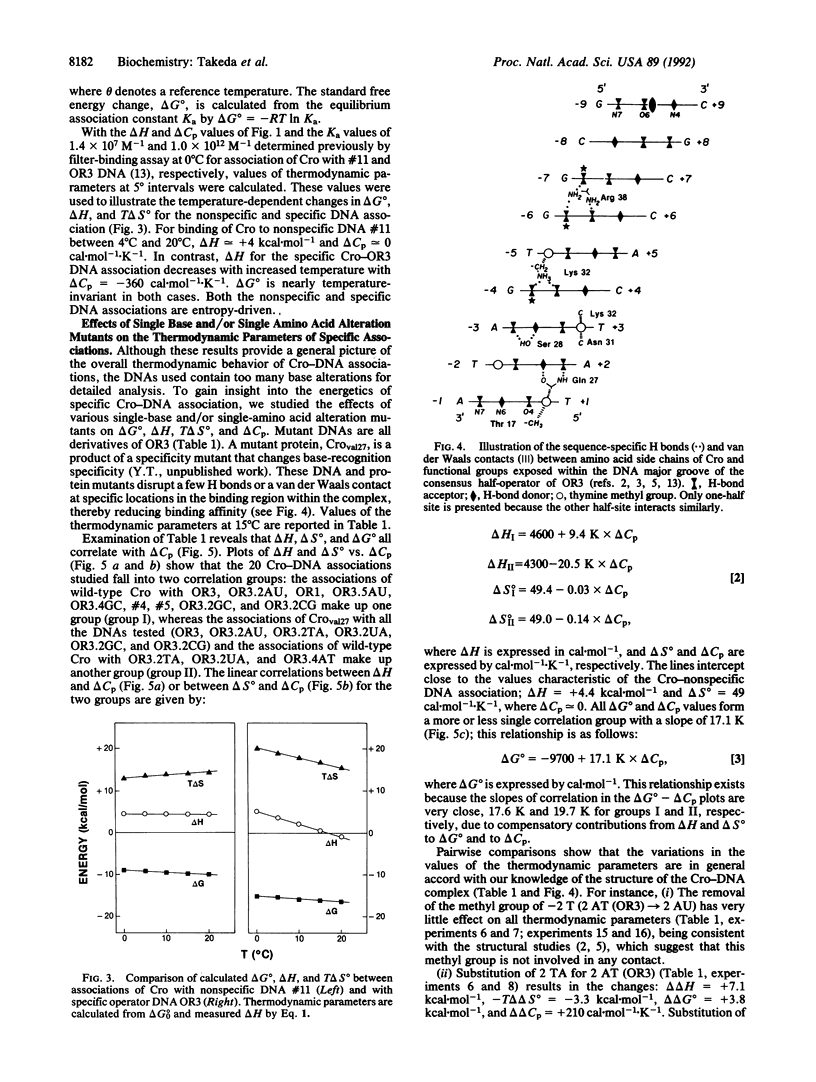
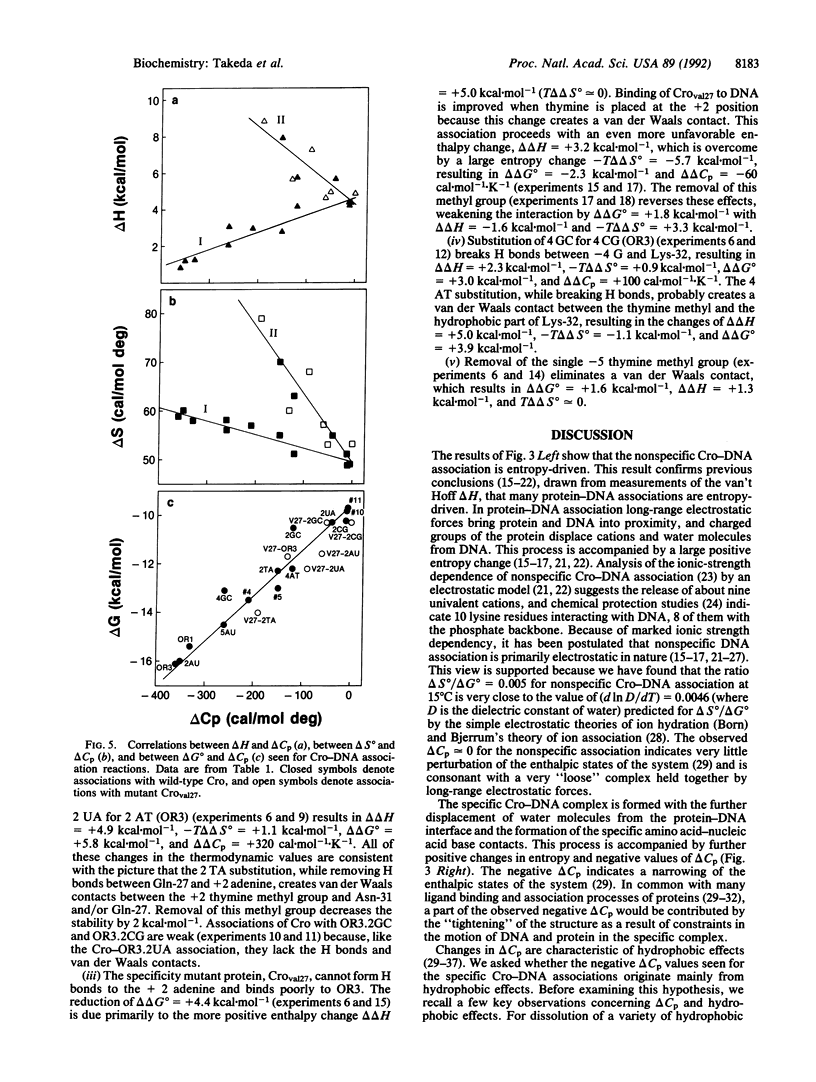
Abstract
Using a highly sensitive pulsed-flow microcalorimeter, we have measured the changes in enthalpy and determined the thermodynamic parameters delta H, delta S degree, delta G degree, and delta C(p) for Cro protein-DNA association reactions. The reactions studied include sequence-nonspecific DNA association and sequence-specific DNA associations involving single- and multiple-base alterations and/or single-amino acid alteration mutants. (i) The association of Cro protein with nonspecific DNA at 15 degrees C is characterized by delta H = +4.4 kcal.mol-1 (1 cal = 4.18J), delta S degrees = 49 cal.mol-1.K-1, delta G degrees = -9.7 kcal.mol-1, and delta Cp congruent to 0; the association with specific high-affinity operator OR3 DNA is characterized by delta H = +0.8 kcal.mol-1, delta S degree = 59 cal.mol-1.K-1, delta G degree = -16.1 kcal.mol-1, and delta Cp = -360 cal.mol-1.K-1, respectively. Both nonspecific and specific Cro-DNA associations are entropy-driven. (ii) Plots of delta H vs. delta Cp and delta S degree vs. delta Cp for the 20 association reactions studied fall into two correlation groups with linear slopes of +9.4 K and -20.5 K and of -0.03 and -0.14, respectively. These regression lines have common intercepts, at the delta H and delta S degree values of nonspecific association (where delta Cp congruent to 0). The results suggest that there are, at least, two distinct conformational subclasses in specific Cro-DNA complexes, stabilized by different combinations of enthalpic and entropic contributions. The delta G degree and delta Cp values form an approximately single linear correlation group as a consequence of compensatory contributions from delta H and delta S degree to delta G degree and to delta Cp. Cro protein-DNA associations share some similar thermodynamic properties with protein folding, but their overall energetics are quite different. Although the nonspecific complex is stabilized predominantly by electrostatic forces, it appears that H bonds, van der Waals contacts, hydrophobic effects, and charge interactions all contribute to the stability (delta G degree and delta Cp) of the specific complex. (iii) The variations in the values of the thermodynamic parameters are in general accord with our knowledge of the structure of the Cro-DNA complex.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aggarwal A. K., Rodgers D. W., Drottar M., Ptashne M., Harrison S. C. Recognition of a DNA operator by the repressor of phage 434: a view at high resolution. Science. 1988 Nov 11;242(4880):899–907. doi: 10.1126/science.3187531. [DOI] [PubMed] [Google Scholar]
- Anderson W. F., Ohlendorf D. H., Takeda Y., Matthews B. W. Structure of the cro repressor from bacteriophage lambda and its interaction with DNA. Nature. 1981 Apr 30;290(5809):754–758. doi: 10.1038/290754a0. [DOI] [PubMed] [Google Scholar]
- Boschelli F. Lambda phage cro repressor. Non-specific DNA binding. J Mol Biol. 1982 Dec 5;162(2):267–282. doi: 10.1016/0022-2836(82)90526-5. [DOI] [PubMed] [Google Scholar]
- Brennan R. G., Matthews B. W. The helix-turn-helix DNA binding motif. J Biol Chem. 1989 Feb 5;264(4):1903–1906. [PubMed] [Google Scholar]
- Brennan R. G., Roderick S. L., Takeda Y., Matthews B. W. Protein-DNA conformational changes in the crystal structure of a lambda Cro-operator complex. Proc Natl Acad Sci U S A. 1990 Oct;87(20):8165–8169. doi: 10.1073/pnas.87.20.8165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha J. H., Spolar R. S., Record M. T., Jr Role of the hydrophobic effect in stability of site-specific protein-DNA complexes. J Mol Biol. 1989 Oct 20;209(4):801–816. doi: 10.1016/0022-2836(89)90608-6. [DOI] [PubMed] [Google Scholar]
- Harrison S. C., Aggarwal A. K. DNA recognition by proteins with the helix-turn-helix motif. Annu Rev Biochem. 1990;59:933–969. doi: 10.1146/annurev.bi.59.070190.004441. [DOI] [PubMed] [Google Scholar]
- Hinz H. J. Thermodynamics of protein-ligand interactions: calorimetric approaches. Annu Rev Biophys Bioeng. 1983;12:285–317. doi: 10.1146/annurev.bb.12.060183.001441. [DOI] [PubMed] [Google Scholar]
- Jordan S. R., Pabo C. O. Structure of the lambda complex at 2.5 A resolution: details of the repressor-operator interactions. Science. 1988 Nov 11;242(4880):893–899. doi: 10.1126/science.3187530. [DOI] [PubMed] [Google Scholar]
- KAUZMANN W. Some factors in the interpretation of protein denaturation. Adv Protein Chem. 1959;14:1–63. doi: 10.1016/s0065-3233(08)60608-7. [DOI] [PubMed] [Google Scholar]
- Lin S., Riggs A. D. The general affinity of lac repressor for E. coli DNA: implications for gene regulation in procaryotes and eucaryotes. Cell. 1975 Feb;4(2):107–111. doi: 10.1016/0092-8674(75)90116-6. [DOI] [PubMed] [Google Scholar]
- Manning G. S. The molecular theory of polyelectrolyte solutions with applications to the electrostatic properties of polynucleotides. Q Rev Biophys. 1978 May;11(2):179–246. doi: 10.1017/s0033583500002031. [DOI] [PubMed] [Google Scholar]
- Matthew J. B., Ohlendorf D. H. Electrostatic deformation of DNA by a DNA-binding protein. J Biol Chem. 1985 May 25;260(10):5860–5862. [PubMed] [Google Scholar]
- Mondragón A., Harrison S. C. The phage 434 Cro/OR1 complex at 2.5 A resolution. J Mol Biol. 1991 May 20;219(2):321–334. doi: 10.1016/0022-2836(91)90568-q. [DOI] [PubMed] [Google Scholar]
- Mudd C. P., Berger R. L. A computer-controlled all-tantalum stopped-flow microcalorimeter with microjoule resolution. J Biochem Biophys Methods. 1988 Nov;17(3):171–191. doi: 10.1016/0165-022x(88)90002-4. [DOI] [PubMed] [Google Scholar]
- Murphy K. P., Privalov P. L., Gill S. J. Common features of protein unfolding and dissolution of hydrophobic compounds. Science. 1990 Feb 2;247(4942):559–561. doi: 10.1126/science.2300815. [DOI] [PubMed] [Google Scholar]
- Ohlendorf D. H., Anderson W. F., Fisher R. G., Takeda Y., Matthews B. W. The molecular basis of DNA-protein recognition inferred from the structure of cro repressor. Nature. 1982 Aug 19;298(5876):718–723. doi: 10.1038/298718a0. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z., Schevitz R. W., Zhang R. G., Lawson C. L., Joachimiak A., Marmorstein R. Q., Luisi B. F., Sigler P. B. Crystal structure of trp repressor/operator complex at atomic resolution. Nature. 1988 Sep 22;335(6188):321–329. doi: 10.1038/335321a0. [DOI] [PubMed] [Google Scholar]
- Pabo C. O., Sauer R. T. Protein-DNA recognition. Annu Rev Biochem. 1984;53:293–321. doi: 10.1146/annurev.bi.53.070184.001453. [DOI] [PubMed] [Google Scholar]
- Privalov P. L., Gill S. J. Stability of protein structure and hydrophobic interaction. Adv Protein Chem. 1988;39:191–234. doi: 10.1016/s0065-3233(08)60377-0. [DOI] [PubMed] [Google Scholar]
- Record M. T., Jr, Anderson C. F., Lohman T. M. Thermodynamic analysis of ion effects on the binding and conformational equilibria of proteins and nucleic acids: the roles of ion association or release, screening, and ion effects on water activity. Q Rev Biophys. 1978 May;11(2):103–178. doi: 10.1017/s003358350000202x. [DOI] [PubMed] [Google Scholar]
- Revzin A., von Hippel P. H. Direct measurement of association constants for the binding of Escherichia coli lac repressor to non-operator DNA. Biochemistry. 1977 Nov 1;16(22):4769–4776. doi: 10.1021/bi00641a002. [DOI] [PubMed] [Google Scholar]
- Riggs A. D., Bourgeois S., Cohn M. The lac repressor-operator interaction. 3. Kinetic studies. J Mol Biol. 1970 Nov 14;53(3):401–417. doi: 10.1016/0022-2836(70)90074-4. [DOI] [PubMed] [Google Scholar]
- Ross P. D., Subramanian S. Thermodynamics of protein association reactions: forces contributing to stability. Biochemistry. 1981 May 26;20(11):3096–3102. doi: 10.1021/bi00514a017. [DOI] [PubMed] [Google Scholar]
- Spolar R. S., Ha J. H., Record M. T., Jr Hydrophobic effect in protein folding and other noncovalent processes involving proteins. Proc Natl Acad Sci U S A. 1989 Nov;86(21):8382–8385. doi: 10.1073/pnas.86.21.8382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz T. A. Structural studies of protein-nucleic acid interaction: the sources of sequence-specific binding. Q Rev Biophys. 1990 Aug;23(3):205–280. doi: 10.1017/s0033583500005552. [DOI] [PubMed] [Google Scholar]
- Sturtevant J. M. Heat capacity and entropy changes in processes involving proteins. Proc Natl Acad Sci U S A. 1977 Jun;74(6):2236–2240. doi: 10.1073/pnas.74.6.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda Y., Kim J. G., Caday C. G., Steers E., Jr, Ohlendorf D. H., Anderson W. F., Matthews B. W. Different interactions used by Cro repressor in specific and nonspecific DNA binding. J Biol Chem. 1986 Jul 5;261(19):8608–8616. [PubMed] [Google Scholar]
- Takeda Y., Ohlendorf D. H., Anderson W. F., Matthews B. W. DNA-binding proteins. Science. 1983 Sep 9;221(4615):1020–1026. doi: 10.1126/science.6308768. [DOI] [PubMed] [Google Scholar]
- Takeda Y., Sarai A., Rivera V. M. Analysis of the sequence-specific interactions between Cro repressor and operator DNA by systematic base substitution experiments. Proc Natl Acad Sci U S A. 1989 Jan;86(2):439–443. doi: 10.1073/pnas.86.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vershon A. K., Liao S. M., McClure W. R., Sauer R. T. Bacteriophage P22 Mnt repressor. DNA binding and effects on transcription in vitro. J Mol Biol. 1987 May 20;195(2):311–322. doi: 10.1016/0022-2836(87)90652-8. [DOI] [PubMed] [Google Scholar]
- Whitson P. A., Olson J. S., Matthews K. S. Thermodynamic analysis of the lactose repressor-operator DNA interaction. Biochemistry. 1986 Jul 1;25(13):3852–3858. doi: 10.1021/bi00361a017. [DOI] [PubMed] [Google Scholar]
- deHaseth P. L., Lohman T. M., Record M. T., Jr Nonspecific interaction of lac repressor with DNA: an association reaction driven by counterion release. Biochemistry. 1977 Nov 1;16(22):4783–4790. doi: 10.1021/bi00641a004. [DOI] [PubMed] [Google Scholar]


