ABSTRACT
The clustered regularly interspaced short palindromic repeat (CRISPR)-associated (Cas) systems are adaptive immune systems of bacteria. A type II CRISPR-Cas9 system from Streptococcus pyogenes has recently been developed into a genome engineering tool for prokaryotes and eukaryotes. Here, we present a single-plasmid system which allows efficient genome editing of Bacillus subtilis. The plasmid pJOE8999 is a shuttle vector that has a pUC minimal origin of replication for Escherichia coli, the temperature-sensitive replication origin of plasmid pE194ts for B. subtilis, and a kanamycin resistance gene working in both organisms. For genome editing, it carries the cas9 gene under the control of the B. subtilis mannose-inducible promoter PmanP and a single guide RNA (sgRNA)-encoding sequence transcribed via a strong promoter. This sgRNA guides the Cas9 nuclease to its target. The 20-nucleotide spacer sequence at the 5′ end of the sgRNA sequence, responsible for target specificity, is located between BsaI sites. Thus, the target specificity is altered by changing the spacer sequences via oligonucleotides fitted between the BsaI sites. Cas9 in complex with the sgRNA induces double-strand breaks (DSBs) at its target site. Repair of the DSBs and the required modification of the genome are achieved by adding homology templates, usually two PCR fragments obtained from both sides of the target sequence. Two adjacent SfiI sites enable the ordered integration of these homology templates into the vector. The function of the CRISPR-Cas9 vector was demonstrated by introducing two large deletions in the B. subtilis chromosome and by repair of the trpC2 mutation of B. subtilis 168.
IMPORTANCE In prokaryotes, most methods used for scarless genome engineering are based on selection-counterselection systems. The disadvantages are often the lack of a suitable counterselection marker, the toxicity of the compounds needed for counterselection, and the requirement of certain mutations in the target strain. CRISPR-Cas systems were recently developed as important tools for genome editing. The single-plasmid system constructed for the genome editing of B. subtilis overcomes the problems of counterselection methods. It allows deletions and introduction of point mutations. It is easy to handle and very efficient, and it may be adapted for use in other firmicutes.
INTRODUCTION
A clustered regularly interspaced short palindromic repeat (CRISPR)-associated (Cas) system is a prokaryotic adaptive immune system against invading mobile elements, like phages and plasmids (1, 2). The system consists of CRISPRs and a variable set of Cas genes. The intervening sequences (spacers) are collected from unsuccessful intruders and serve as a memory for defense against future attacks by the same elements. The clusters are transcribed into pre-CRISPR RNAs (pre-crRNAs), which are processed into single spacer and repeat units. The crRNAs recruit effector complexes and guide them to the target DNA, the protospacer DNA. Finally, after base pairing, the invading element is targeted by a Cas endonuclease generating a double-strand break (DSB). To distinguish between its own and foreign DNA by the CRISPR-Cas system, an additional sequence next to the protospacer sequence is necessary in the invading DNA, the so-called protospacer adjacent motif (PAM) sequence (3, 4). In the case of the type II CRISPR-Cas9 system of Streptococcus pyogenes (5), the PAM consists of the triplet NGG. Although the PAM sequence is essential for cleavage, the high frequency of such short motifs in DNA make sure that nearly any DNA can be targeted by the crRNA; therefore, CRISPR-Cas systems have been successfully engineered for genome editing. The crRNA assisted by a trans-activating crRNA (tracrRNA) guides an endonuclease encoded by the gene cas9 to bind to the protospacer with an adjacent PAM sequence NGG and cleaves the double-stranded DNA (dsDNA). This system was further engineered by fusion of crRNA and tracrRNA to the single guide RNA (sgRNA) (6). Only an exchange of about 20 bp at the 5′ end of the sgRNA-encoding sequence is needed to reprogram the target site. In eukaryotic cells, the DSB introduced by Cas9 can be repaired by the erroneous nonhomologous DNA end joining (NHEJ) recombination system, resulting in a mutation at the target site (7, 8). NHEJ does not work efficiently or is not present in most bacteria. This may be seen as a disadvantage to using CRISPR-Cas9 technology for the inactivation of genes by small deletions or insertions (indels). On the other hand, for precise gene editing, DSBs may be repaired by homologous recombination using an engineered homologous template. The latter process is more efficient in prokaryotes than in eukaryotes. Here, a strong NHEJ might disturb the outcome and lead to off-target effects.
So far, the CRISPR-Cas9 system has been applied in very few bacterial species to edit their genomes (9–15). An example is the two-plasmid-based system constructed for engineering Escherichia coli (10). In principle, cas9 and sgRNA under the control of constitutive promoters are separated on two different plasmids. The two plasmids are finally combined in the host for genome engineering. The editing template homologous to the chromosomal target locus is integrated into the sgRNA-encoding plasmid or can be provided separately as a PCR fragment. In the latter case, the CRISPR-Cas9 system has to be combined with the λ Red system. E. coli degrades linear double-strand DNA very rapidly via the RecBCD and SbcCD exonucleases. Gam, one of the three proteins of λ, prevents the degradation of the linear DNA by inhibiting the nucleases, Exo generates single-stranded DNA, and Beta binds to this single-stranded DNA and facilitates recombination to homologous chromosomal regions (16). In Bacillus subtilis, such a system is not necessary. The DNA becomes single stranded during uptake by the competent cells and is efficiently recombined with homologous chromosomal regions.
A single-plasmid system (pCRISPomyces-2) was constructed for the genome editing of Streptomyces species (9). The components of this shuttle vector were (i) a temperature-sensitive pSG5 replicon for Streptomyces, which was used for clearing the plasmid after genome editing; (ii) a ColE1 replicon for constructing and modifying the vector in E. coli; (iii) an apramycin resistance gene for selection in both species; (iv) the cas9 gene codon optimized according to the high GC content of Streptomyces; and (v) a programmable sgRNA expression cassette. Remarkably, the custom-designed spacer sequences are inserted between BbsI restriction sites to replace the lacZ α fragment and allow blue/white screening for successful insertions. Using this vector, a series of deletions were introduced in Streptomyces lividans, Streptomyces viridochromogenes, and Streptomyces albus.
In this study, a single-plasmid system was constructed for the genome editing of Bacillus subtilis that was similar to the pCRISPomyces-2. Soil-dwelling B. subtilis is a model organism for studying the various facets of bacterial life (17, 18) and an important producer strain for industrial enzymes and valuable low-molecular-weight substances (19). Various tools, like markerless gene deletion and insertion systems, are already available for genome editing. The CRISPR-Cas9 system described here may be a worthwhile amendment of the genetic toolbox of B. subtilis, with advantages and limitations.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
For DNA cloning and plasmid preparation, the E. coli strain JM109 (20) was used. For transformation of B. subtilis, plasmid DNA was isolated from the recA-proficient strain E. coli NM538 (21). E. coli strains were transformed using the transformation and storage solution (TSS) heat shock method (22). B. subtilis 168 (DSM 23778 trpC2, obtained from Leibniz-Institut DSMZ-Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH) was transformed according to the conventional two-step procedure with Spizizen's minimal medium as described previously (23). B. subtilis strain KM0 is derived from B. subtilis 168 by repair of the mutated trpC2 gene (24). Strains were propagated in LB liquid medium and on LB agar plates at 37°C, if not otherwise indicated. To select plasmids in E. coli, ampicillin was used at a final concentration of 100 μg ml−1 and kanamycin was used at 30 μg ml−1. For B. subtilis, kanamycin was used at a final concentration of 5 μg ml−1. To detect amylase production, colonies were streaked on starch containing LB agar plates (1% starch, soluble; Sigma-Aldrich), and the plates were overlaid with Lugol's solution (0.2% iodine in 2% potassium iodide) after 2 days of incubation at 37°C.
DNA manipulations.
Standard molecular techniques were carried out according to the methods of Sambrook and Russell (25). Oligonucleotides were obtained from Eurofins MWG Operon (Ebersberg, Germany) (Table 1). When DNA fragments were amplified by PCR from genomic or plasmid DNA, the Q5 high-fidelity DNA polymerase (New England BioLabs, Frankfurt am Main, Germany) was used. Colony PCR was done with DreamTaq DNA polymerase as described before (26). Chromosomal DNA was extracted from B. subtilis with a DNeasy blood and tissue kit (catalog no. 69506; Qiagen, Hilden, Germany). Plasmid DNA was isolated from E. coli using the innuPrep plasmid minikit (Analytic Jena AG, Jena, Germany). Digested DNA fragments from agarose gel and amplified DNAs in PCRs were isolated with the NucleoSpin gel and PCR cleanup kit (Macherey-Nagel, Düren, Germany). DNA samples were sequenced by GATC Biotech (Constance, Germany).
TABLE 1.
Oligonucleotides used in this study
| Name | Sequence | Purpose |
|---|---|---|
| s10631 | 5′-AATTCTTGTCGACGGCCAACGAGGCCCGGGCCAATAAGGCCTTTCTAGATTTGTACAGAA | SfiI sites in CRISPR vector |
| s10632 | 5′-TGTACAAATCTAGAAAGGCCTTATTGGCCCGGGCCTCGTTGGCCGTCGACAA | |
| s10625 | 5′-AAAAAGTCGACGAGCGTTCACCGACAAAC | sgRNA in CRISPR vector |
| s10626 | 5′-AAAAGAATTCTTGACCGTGATTAGAGAATTGAGTAAAA | |
| s10741 | 5′-CGAATTCGAGCTCGAGGT | Kanamycin resistance gene in CRISPR vector |
| s10742 | 5′-GCATGCCTGCAGTTGAAATCCCCTCAAAAACCC | |
| s10629 | 5′-AAAAATGTACACCAATGTCATGACATTTTTATC | Insertion of the 5′ end of cas9 in CRISPR vector |
| s10630 | 5′-AAAAATCTAGACTTCTTGGCTAGCTC | |
| s10781 | 5′-AAAAACTGAGACCGATTCATTAATGCAGCTG | Insertion of lacZ α in CRISPR vector |
| s10794 | 5′-TTTACGCGAGACCTCATTCGCCATTCAGGCTG | |
| s10627 | 5′-AAAAACTTAAGTTAGGAGGCAAAAATGGATAA | Fusion of cas9 to PmanPA |
| s10628 | 5′-AAAAATGTACATCTAGACTTCTTGGCTAGCTCCCC | |
| s10621 | 5′-AATTGAGTAAAATGTACCTACGCGAGACCG | Semisynthetic sgRNA promoter |
| s10622 | 5′-GATCCGGTCTCGCGTAGGTACATTTTACTC | |
| s10623 | 5′-AAAAGGATCCGGTCTCAGTTTTAGAGCTAGAAATAGCAAG | PCR sgRNA of pCRISPomyces-2 |
| s10624 | 5′-AAAAAGCTTTAGATAAAAAACGCCCGGCG | |
| s10784 | 5′-TACGTGAAGATCAGGCTATCACTG | Target sequence for amyE deletion |
| s10785 | 5′-AAACCAGTGATAGCCTGATCTTCA | |
| s10782 | 5′-AAGGCCAACGAGGCCATCGGCTTTTTTCTCAG | PCR of homology template for amyE deletion |
| s10715 | 5′-AAGGCCATGTTGGCCTTGCCGAATTGTTTCTATAA | |
| s10783 | 5′-AAGGCCTTATTGGCCTGAGGAAGCGTCTAAATCA | |
| s10716 | 5′-AAGGCCAACATGGCCTCCGTGATGACGAAAAACA | |
| s10893 | 5′-TACGTCCGGAGCTCCGATAAAAAA | Target sequence for pulcherrimin gene deletion |
| s10894 | 5′-AAACTTTTTTATCGGAGCTCCGGA | |
| s10889 | 5′-AAGGCCAACGAGGCCCAACAGGAAAAGCTTTCG | PCR of homology template for pulcherrimin deletion |
| s10890 | 5′-AAGGCCATGTTGGCCCATGCGGGTGAGACTC | |
| s10891 | 5′-AAGGCCAACATGGCCGCGGGCAGCTTTTTTTA | |
| s10892 | 5′-AAGGCCTTATTGGCCTGGTACGCTGTCCCATG | |
| s10962 | 5′-TACGTATTGATTCTCTTCAAGTAG | Target sequence for trpC2 gene repair |
| s10963 | 5′-AAACCTACTTGAAGAGAATCAATA | |
| s10964 | 5′-AAGGCCAACGAGGCCGGTTTTTCATCACCGTG-3′ | PCR trpC wild-type sequence |
| s10965 | 5′-AAGGCCTTATTGGCCGTGACGGTTTAGATGA-3′ | |
| s11056 | 5′-CTACAGGTTGAAGAATCAAGAAGAATCGGAGCGGAT-3′ | Site-specific mutagenesis of trpC |
| s11057 | 5′-AGAATCAATAATAAAATCTTTTCTAAGTACAGG-3′ |
Construction of the plasmid pJOE8999.
Construction of the plasmid pJOE8999 started with the plasmid pJOE8829.1 (unpublished), consisting of the pUC18 minimal origin (20), the temperature-sensitive minimal replicon from plasmid pE194ts (27), the spectinomycin resistance gene aad9 from pDG1730 (28), and a chloramphenicol resistance gene from pMTLBS72 (29). In the first step, the aad9 gene was replaced by a sequence consisting of two SfiI sites, separated by a SmaI site and a T7 promoter sequence. The plasmid was cut with BglI and EcoRI, and the vector backbone was ligated with the two complementary oligonucleotides s10631 and s10632 to plasmid pJOE8917.1. In the next step, an sgRNA-encoding sequence with a constitutive promoter was added by cutting pJOE8917.1 with EcoRI and SalI and insertion of a DNA fragment which was obtained by PCR with the oligonucleotides s10625 and s10626 and pJOE8928.1 as the template (see below for construction of pJOE8928.1). In the new plasmid (pJOE8946.16), the chloramphenicol resistance gene was replaced with a kanamycin resistance gene due to the difficulties of selection in E. coli. The plasmid pJOE8946.16 was cleaved with PstI and SacI, and the vector backbone was ligated with a fragment cleaved by the same enzymes and obtained by PCR using the oligonucleotides s10741/s10742, with pMG9 (30) as the template. The new plasmid, pJOE8958.1, carrying the kanamycin resistance gene, was then combined with the cas9 gene under the control of the B. subtilis mannose manP promoter (PmanP). For technical reasons, i.e., to avoid mutations from PCR amplification of large fragments, this was done in two steps. First, PmanP and the 5′ end of cas9 were added by cleavage of vector pJOE8958.1 with XbaI and BsrGI and ligation with a PCR fragment amplified from the template pJOE8930.8 (see below) using oligonucleotides s10629 and s10630 to construct plasmid pJOE8964.1. In the next step, the cas9 gene was completed by cutting pJOE8964.1 and pCas (10) with NheI and XbaI and ligation of the cas9 3′ end into pJOE8964.1 to obtain plasmid pJOE8973.1. Finally, the lacZ α fragment with the promoter and operator sequences from vector pJOE4786.1 (31) was amplified with oligonucleotides s10781 and s10794, and the PCR fragment was inserted between the BsaI sites of pJOE8964.1 into the final vector, pJOE8999.
The plasmid pJOE8930.8 with the 5′ end of cas9 under the mannose promoter was obtained by amplification of the corresponding cas9 gene from pCas without its own promoter and ribosomal binding site with the oligonucleotides s10627/s10628 and ligation between the AflII and BsrGI site of pMW168 (32).
The plasmid pJOE8928.1 with the sgRNA-encoding sequence and a semisynthetic promoter was constructed in several steps starting from the E. coli vanillate-inducible expression vector pJOE7879.1, which contains the Corynebacterium glutamicum vanR regulatory gene and a vanABK optimized promoter sequence fused to the eGFP gene (33). A MluI/EcoRI fragment with the vanR gene and the −35 region of the vanABK promoter was inserted into the rhamnose expression vector pJOE5751.1 (34), which was cut with the same enzymes to give pJOE8924.1. In a second step, the plasmid was cut with EcoRI and BamHI, and the two complementary oligonucleotides, s10621/s10622, were added to complete the new promoter sequence and to introduce a BsaI site (pJOE8925.1). From pCRISPomyces-2, the sgRNA-encoding sequence together with the oop transcription terminator sequence was amplified using oligonucleotides s10623/s10624, and the PCR fragment as well as the vector pJOE8925.1 was cut with BamHI and HindIII and ligated to give pJOE8928.1.
Construction of the vectors pJOE9012.1, pJOE9150.2, pJOE9151.1, pJOE9202.1, and pJOE9194.1.
For deletion of the amyE chromosomal region, the plasmid pJOE8999 was first cut with BsaI and the lacZ α fragment was replaced by the two complementary oligonucleotides s10784/s10785 to obtain plasmid pJOE9007.2. Two PCR fragments, obtained with B. subtilis 168 chromosomal DNA as the template and the oligonucleotides s10782/s10715 and s10783/s10716, were cut with SfiI and inserted between the two SfiI sites of pJOE9007.2 to give pJOE9012.1. The pulcherrimin biosynthesis genes were deleted with the plasmid pJOE9150.2. First, the lacZ α fragment of pJOE8999.1 was replaced by the complementary oligonucleotides s10893/s10894 (pJOE9137.1). Again, two PCR fragments, obtained with B. subtilis 168 chromosomal DNA as the template and the oligonucleotides s10889/s10890 and s10891/s10892, were cut with SfiI and inserted between the two SfiI sites of pJOE9007.2 to give pJOE9150.2. To repair the trpC2 mutation of B. subtilis 168, the oligonucleotides s10962/s10963 targeting the mutated region were inserted between the BsaI sites of pJOE8999 (pJOE9146.1). The wild-type trpC sequence was obtained by PCR using KM0 DNA and the oligonucleotides s10964/s10965, and the 1.5-kb PCR fragment was cleaved by SfiI and ligated with pJOE9146.1 DNA cut by the same enzyme (pJOE9151.1). For site-specific mutagenesis of the trpC wild-type sequence, a SalI/XbaI fragment from pJOE9151.1 containing the 1.5-kb PCR fragment was inserted into pUC18 (pJOE9193.1). This whole new plasmid was amplified with the primers s11056/s11057, and the PCR fragment was purified and treated with polynucleotide kinase to phosphorylate the 5′ ends, which were again purified and treated with DpnI to remove the original template DNA. Finally, the PCR fragment was circularized by T4 DNA ligase and used to transform JM109. The plasmid with the requested sequence was identified by nucleotide sequencing (pJOE9197.3). Finally, the SfiI fragment with the modified trpC sequence was introduced again in pJOE9146.1 to get pJOE9198.1. Plasmid pJOE9202.1, used as a control for homologous recombination, was obtained by cloning the modified trpC sequence from pJOE9198.1 between the SfiI sites of pJOE8999.
RESULTS
Construction of the vector pJOE8999 to enable CRISPR-Cas9-based genome editing.
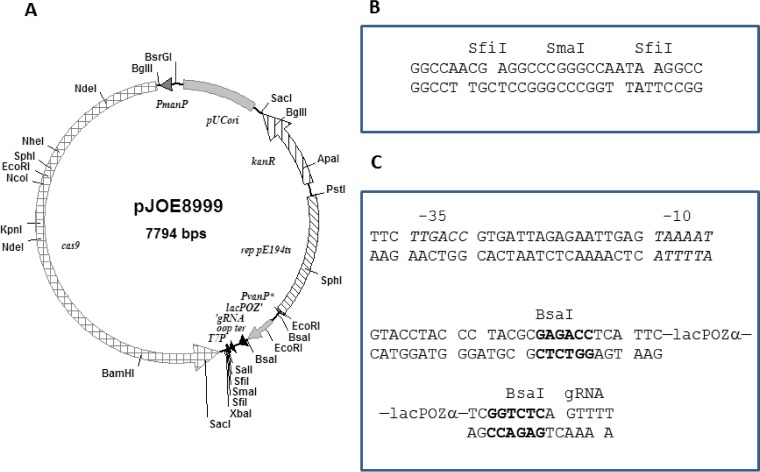
The vector pJOE8999 (Fig. 1A) is a shuttle vector containing the minimal pUC origin of replication in E. coli and the temperature-sensitive replicon from pE194ts for replication in B. subtilis. A kanamycin resistance gene allows the selection of the plasmid in both species. For cas9 expression, the S. pyogenes native promoter of the cas9 gene was replaced by the B. subtilis mannose-inducible promoter PmanP. This promoter is not active in E. coli, and in B. subtilis, it is activated by ManR in the presence of mannose and in the absence of glucose (35). For constitutive sgRNA expression, a semisynthetic promoter with high identity to the improved PvanABK promoter of Corynebacterium glutamicum was used (Fig. 1C). The sgRNA-encoding sequence was amplified by PCR from pCRISPomyces-2 together with the λ oop terminator sequence. For rapid and easy exchange of the spacer sequence, a lacZ α fragment flanked by BsaI sites instead of the BbsI sites, as found in pCRISPomyces-2, was inserted into the empty CRISPR array. For the cloning of homology templates to repair the double-strand breaks caused by the endonuclease Cas9, two SfiI sites separated by a SmaI site were inserted (Fig. 1B). This allows a single-step assembly of multiple fragments with a designed order, a method referred to as ordered gene assembly in B. subtilis (OGAB) (36). Besides the SfiI sites, a T7 promoter sequence was inserted to allow DNA sequencing of the cloned homology templates.
FIG 1.
CRISPR-Cas9 vector pJOE8999. (A) Physical map of vector pJOE8999 containing the pUC18 minimal origin, the temperature-sensitive replication origin from pE194ts, a kanamycin resistance gene (kanR), cas9 under the control of the PmanP promoter, the sgRNA transcribed from the semisynthetic promoter PvanP* interrupted by the lacZ α fragment (lacPOZ'), the λ oop terminator, and the T7 promoter (T7P). (B) Cloning sites in the vector pJOE8999. (C) Promoter sequence (PvanP*) upstream of the sgRNA with the −35 and −10 sequences and the BsaI restriction sites (bold) for insertion of the spacer sequence.
Deletion of two B. subtilis chromosomal regions employing the vector pJOE8999.
For demonstration of the functionality of the CRISPR-Cas9 system in B. subtilis, two deletions were introduced into the genome, a very large deletion with 25.1 kb containing the amyE gene and a shorter deletion with 4.1 kb containing the pulcherrimin biosynthesis genes. In the meantime, a range of sequence features in and around the target sequences that predict sgRNA efficiency have been identified (37). With the help of the sgRNA Designer tool provided by the Broad Institute (38), candidate sgRNA sequences were identified in the amyE sequence, and a high-scoring sequence was chosen for oligonucleotide synthesis. In the first step, the two complementary oligonucleotides s10784 and s10785 with protruding ends were inserted into the BsaI cleaved vector pJOE8999 (plasmid pJOE9007.2). In a second step, two fragments flanking the target sequence with lengths of about 700 bp were amplified by PCR from B. subtilis 168 chromosomal DNA as the template. On both sides of the fragments, SfiI sites were introduced by the oligonucleotides used for PCR and were ligated with SfiI-cut pJOE9007.2 DNA to construct pJOE9012.1, ready for transformation of B. subtilis.
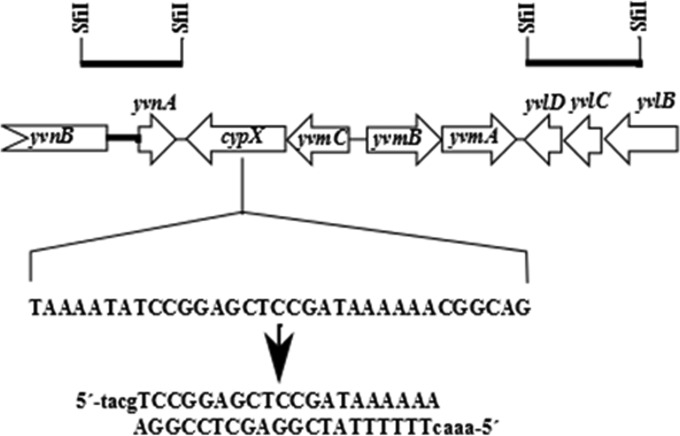
In the same way, the plasmid pJOE9150.2 for the deletion of pulcherrimin biosynthesis and regulatory genes was constructed. The gene yvmC, encoding the cyclodipeptide synthase, was checked with the sgRNA Designer software, and a high-scoring 20-nucleotide (nt) sequence was identified. The sequence was synthesized into two complementary oligonucleotides and inserted between the BsaI sites of pJOE8999 to construct pJOE9037.1. Two 700-bp fragments flanking the target region were amplified by PCR and inserted between the SfiI sites of pJOE9037.1 to get pJOE9150.2 (Fig. 2).
FIG 2.
B. subtilis chromosomal region carrying the pulcherrimin biosynthesis genes. The bars on the top indicate the fragments amplified by PCR and integrated between the SfiI sites of pJOE8999 to delete the genes cypX and yvmA. The sequence in cypX was selected as a target for the Cas9/sgRNA complex, and the oligonucleotides inserted into pJOE8999 are shown below. Lowercase letters indicate the deoxynucleotides fitting to the BsaI sites.
B. subtilis 168 was transformed with pJOE9012.1, pJOE9150.1, and, as a control, the vector pWAL275 (24), a pUB110/pUC shuttle plasmid without CRISPR-Cas9 components. The LB agar plates containing 5 μg ml−1 kanamycin and 0.2% mannose for induction of cas9 under the control of PmanP were first incubated at 30°C. Colonies appearing within 2 days were placed with a toothpick on LB plates without antibiotics and incubated at 50°C; on the next day, they were streaked on LB plates to obtain single colonies at 42°C. Finally, the colonies were tested again for plasmid loss by transferring single colonies to LB agar plates with kanamycin. Usually, about 90% of all colonies tested had lost the plasmids (Table 2). The plasmid-cured cells were checked by colony PCR using the outer primers from the 700-bp homology templates. In the case of a successful deletion, the fragment size should be reduced from 26.5 kb to 1.5 kb with pJOE9012.1 and from 5.6 kb to 1.5 kb with pJOE9150.1. In addition, cells transformed with plasmid pJOE9012.1 should have lost the amyE gene. Therefore, these colonies were also streaked on starch-containing LB agar plates, and amylase production was checked by iodine staining. The results are summarized in Table 2. The transformation rate of the CRISPR-Cas9-containing plasmids was slightly lower than that of pWAL275. Notably, the colonies appeared to be delayed compared to the control and were heterogeneous in size. About 90% of colonies treated with pJOE9012.1 had lost the amyE gene according to the amylase test on the agar plates, and this result was confirmed by colony PCR. Most colonies showed the expected amplified fragment of about 1.5 kb; only 3 amylase-negative colonies showed no amplified DNA, and one colony gave a smaller fragment. DNA sequence analysis of three PCR fragments showed that the deletion occurred as expected. In the case of pJOE9050.1, 97% of the colonies showed the expected deletion of the pulcherrimin biosynthesis genes as tested by colony PCR. Only 1 of 46 colonies produced no amplified fragment. Again, 3 fragments were sequenced and showed the expected sequence fusion caused by the deletion.
TABLE 2.
Numbers of transformants and mutations obtained with the various CRISPR-Cas9 plasmids
| Plasmid | No. of transformants/μg | % kanamycin-resistant colonies (total no. of colonies tested)a | % ΔamyE or trpC+ mutants (total no. of colonies tested) | % of PCR fragments of expected size (total no. of colonies tested) |
|---|---|---|---|---|
| pJOE9012.1 | 1,033 ± 246 | 6.9 (72) | 93.9 (72) | 89 (47) |
| pJOE9150.2 | 1,222 ± 379 | 15.2 (46) | 97 (46) | |
| pJOE9198.1 | 1,206 ± 368 | 2.7 (72) | 100 (72) | 100 (44) |
| pJOE9202.1 | 2,167 ± 529 | 1.5 (66) | 22.9 (66) | 100 (24) |
| pWAL275 | 1,594 ± 575 |
Colonies were tested after growth at 50°C and 42°C.
Repair of the trpC2 mutation.
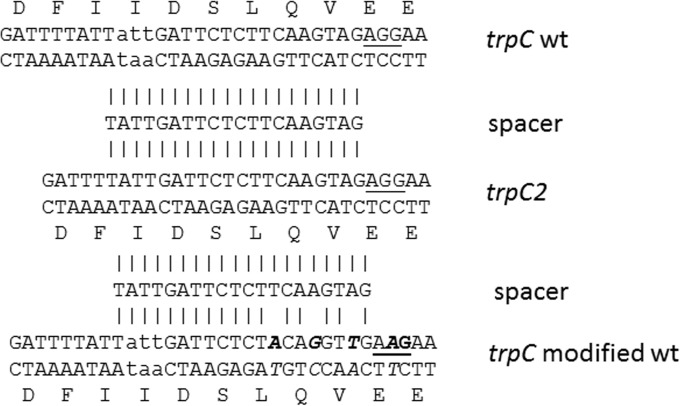
Another important aim of genome editing is the exchange of single or a very low number of nucleotides to repair mutations or to improve or change enzyme properties. Regarding the repair of mutations, the trpC2 mutation of B. subtilis 168 became a good candidate for which to screen. The trpC2 mutation is due to a deletion of the triplet ATT, which leads to a lack of an isoleucine residue in TrpC. Inspection of the DNA sequence around the mutation revealed only one possible PAM sequence (AGG) that was accessible for the Cas9 enzyme, located 16 bp downstream from the missing triplet sequence. Unfortunately, the deduced spacer sequence directed against the trpC2 mutant sequence also completely matched the trpC wild-type sequence (Fig. 3). This means that the target sites for Cas9 were in the trpC2 gene on the chromosome as well as in the editing template of the plasmid. Nevertheless, the spacer sequence was integrated between the BsaI sites of pJOE8999 (plasmid pJOE9146.1). The trpC wild-type sequence was obtained from B. subtilis KM0 DNA by PCR, and the amplified fragment was integrated between the SfiI sites of pJOE9146.1 to give pJOE9151.1. When B. subtilis 168 was transformed with this plasmid, no colonies were obtained. In a second attempt, the transformation was repeated and the cells were plated on LB agar plates containing 0.5% glucose in addition to kanamycin, which should reduce cas9 expression due to catabolite repression. In this way, colonies were obtained; however, they could not be further propagated on kanamycin-containing LB agar plates. Obviously, even with a reduced expression of cas9, there was enough Cas9 produced to slowly cleave the target sites. This should lead to plasmid loss, unrepaired DSBs in the chromosome, antibiotic sensitivity, and finally inhibition of cell growth. It further demonstrated that in E. coli the mannose promoter is completely inactive.
FIG 3.
Nucleotide and amino acid sequences of the trpC2 gene of B. subtilis 168 around the missing ATT codon (middle), corrected sequence (top) with the added codon (lowercase letters), and new modified sequence (bottom, lowercase and bold letters). The spacer sequence used in the CRISPR-Cas9 vectors to correct the mutation is aligned (vertical lines). wt, wild type.
To succeed in the repair of trpC2, a strong difference between wild-type trpC and trpC2 was needed. By site-specific mutagenesis, 3 bp were exchanged between the trpC2 mutation site and the PAM motif within the trpC wild-type sequence. The changes were conducted in the codon wobble positions to leave the amino acid sequence unaltered. Care was taken to do the base changes proximal to the PAM sequence, the so-called seed sequence, which has the most influence on target sequence recognition (6). In addition, the first G residue in the PAM sequence was changed to an A residue (AAG). The fragment was inserted into pJOE9146.1 to get pJOE9198.1. Transformation rates now reached the same level as those obtained with the control plasmid pWAL275. After heat treatment for plasmid curing, most colonies were kanamycin sensitive and all colonies were tryptophan positive (Table 2). Amplification of the trpC region with the primers used before for cloning of the 1.5-kb trpC wild-type sequence gave the expected size, and DNA sequence analysis showed the modified trpC wild-type sequence. To see that this high frequency of repair is caused mainly by Cas9's action and not just by homologous recombination, the modified trpC-containing fragment was further inserted into the vector pJOE8999 without the corresponding spacer sequence (pJOE9202.2). Using pJOE9202.1, B. subtilis 168 was transformed as before, the plasmid was cured, and the colonies were tested. Only a few colonies were still kanamycin resistant, and only about 20% were tryptophan prototrophs, demonstrating the strong selection of repair by the CRISPR-Cas9 system.
DISCUSSION
Various methods for genome engineering are available for B. subtilis (39–44, 26). The most useful ones are certainly the markerless systems, which allow the editing of the genome in such a way that no scars, such as antibiotic resistance genes or site-specific recombination sites, are left behind. Examples described so far for B. subtilis are the selection/counterselection systems. As with the CRISPR-Cas9 system described above, two fragments flanking the target sequence are inserted into a plasmid vector. The vector, constructed in E. coli, does not replicate in B. subtilis. After transformation of B. subtilis, the integration by a single-crossover event into the genome is selected by an antibiotic resistance gene. In a second step, excision of the vector is forced by a counterselection marker. On average, in about 50% of cells, the vector is excised again as it is integrated, and in the other 50%, the vector is excised together with the target sequence, leading to the desired modification. Less effort is needed for construction of the plasmids used in the selection/counterselection systems than in the CRISPR-Cas9 system since no spacer sequences are required. For changing single nucleotides, the requirement of a PAM sequence nearby limits the use of the CRISPR-Cas9 method or requires even more effort to prepare a proper homology template, as was demonstrated in this work for repairing the trpC2 mutation. Another slight disadvantage is the longer incubation time needed after transformation with the CRISPR-Cas9 plasmid. Presumably, due to the double-strand break caused by the sgRNA/Cas9 complex, a considerable proportion of the cells do not survive. On the other hand, there are important advantages of the CRISPR-Cas9 system. First of all, counterselection genes are rare, and a suitable gene has to be identified for every new strain. Furthermore, for most selection/counterselection systems, certain mutations in a host are a prerequisite. Examples are counterselection systems using the toxicity of mannose in a manA (mannose 6-phosphate isomerase) strain (26) or the toxicity of 5-fluorouracil in a upp mutant (43). Another advantage is the shorter time to achieve the mutations. The colonies appearing after transformation are already modified in the desired way and need only to be cured for the plasmid. Finally, although selection/counterselection systems should produce about 50% of cells with the desired modification, sometimes it can be very laborious to find the corresponding mutant, which may be caused by an unlucky distribution of χ sites, recognized by the homologous recombination system. In summary, this new B. subtilis genome editing tool provides some important advantages and should be useful for engineering B. subtilis. Except for the positively regulated B. subtilis mannose promoter, the replication origin of broad-host-range pE194, the kanamycin resistance gene, and the sgRNA promoter should be functional in other firmicutes as well, so the plasmid might be easily adapted for genome engineering of other firmicutes species.
ACKNOWLEDGMENTS
I appreciate the great technical support from Annette Schneck. I thank Kambiz Morrabi Heravi for critical reading of the manuscript and Ralf Mattes for support.
The work was funded by university budget funds.
Funding Statement
This project received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
REFERENCES
- 1.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 2.Selle K, Barrangou R. 2015. Harnessing CRISPR-Cas systems for bacterial genome editing. Trends Microbiol 23:225–232. doi: 10.1016/j.tim.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 3.Marraffini LA, Sontheimer EJ. 2010. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat Rev Genet 11:181–190. doi: 10.1038/nrg2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. 2013. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol 31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. 2011. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. 2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harrison MM, Jenkins BV, O'Connor-Giles KM, Wildonger J. 2014. A CRISPR view of development. Genes Dev 28:1859–1872. doi: 10.1101/gad.248252.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gratz SJ, Cummings AM, Nguyen JN, Hamm DC, Donohue LK, Harrison MM, Wildonger J, O'Connor-Giles KM. 2013. Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics 194:1029–1035. doi: 10.1534/genetics.113.152710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cobb RE, Wang Y, Zhao H. 2015. High-efficiency multiplex genome editing of Streptomyces species using an engineered CRISPR/Cas system. ACS Synth Biol 4:723–728. doi: 10.1021/sb500351f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang Y, Chen B, Duan C, Sun B, Yang J, Yang S. 2015. Multigene editing in the Escherichia coli genome via the CRISPR-Cas9 system. Appl Environ Microbiol 81:2506–2514. doi: 10.1128/AEM.04023-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pyne ME, Moo-Young M, Chung DA, Chou CP. 2015. Coupling the CRISPR/Cas9 system with lambda red recombineering enables simplified chromosomal gene replacement in Escherichia coli. Appl Environ Microbiol 81:5103–5114. doi: 10.1128/AEM.01248-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barrangou R, van Pijkeren J. 2016. Exploiting CRISPR-Cas immune systems for genome editing in bacteria. Curr Opin Biotechnol 37:61–68. doi: 10.1016/j.copbio.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 13.Oh J, van Pijkeren J. 2014. CRISPR-Cas9-assisted recombineering in Lactobacillus reuteri. Nucleic Acids Res 42:e131. doi: 10.1093/nar/gku623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Y, Zhang Z, Seo S, Choi K, Lu T, Jin Y, Blaschek HP. 2015. Markerless chromosomal gene deletion in Clostridium beijerinckii using CRISPR/Cas9 system. J Biotechnol 200:1–5. doi: 10.1016/j.jbiotec.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 15.Wang Y, Zhang Z, Seo S, Lynn P, Lu T, Jin Y, Blaschek HP. 26 April 2016. Bacterial genome editing with CRISPR-Cas9: deletion, integration, single nucleotide modification, and desirable “clean” mutant selection in Clostridium beijerinckii as an example. ACS Synth Biol doi: 10.1021/acssynbio.6b00060. [DOI] [PubMed] [Google Scholar]
- 16.Mosberg JA, Gregg CJ, Lajoie MJ, Wang HH, Church GM. 2012. Improving lambda red genome engineering in Escherichia coli via rational removal of endogenous nucleases. PLoS One 7(9):e44638. doi: 10.1371/journal.pone.0044638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mirouze N, Dubnau D. 2013. Chance and necessity in Bacillus subtilis development. Microbiol Spectr 1:TBS-0004-2012. doi: 10.1128/microbiolspectrum.TBS-0004-2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lopez D, Vlamakis H, Kolter R. 2009. Generation of multiple cell types in Bacillus subtilis. FEMS Microbiol Rev 33:152–163. doi: 10.1111/j.1574-6976.2008.00148.x. [DOI] [PubMed] [Google Scholar]
- 19.Schallmey M, Singh A, Ward OP. 2004. Developments in the use of Bacillus species for industrial production. Can J Microbiol 50:1–17. doi: 10.1139/w03-076. [DOI] [PubMed] [Google Scholar]
- 20.Yanisch-Perron C, Vieira J, Messing J. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 21.Frischauf AM, Lehrach H, Poustka A, Murray N. 1983. Lambda replacement vectors carrying polylinker sequences. J Mol Biol 170:827–842. doi: 10.1016/S0022-2836(83)80190-9. [DOI] [PubMed] [Google Scholar]
- 22.Chung CT, Niemela SL, Miller RH. 1989. One-step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. Proc Natl Acad Sci U S A 86:2172–2175. doi: 10.1073/pnas.86.7.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anagnostopoulos C, Spizizen J. 1961. Requirements for transformation in Bacillus subtilis. J Bacteriol 81:741–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rahmer R, Morabbi Heravi K, Altenbuchner J. 2015. Construction of a super-competent Bacillus subtilis 168 using the PmtlA-comKS inducible cassette. Front Microbiol 6:1431. doi: 10.3389/fmicb.2015.01431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambrook J, Russell DW. 2001. Molecular cloning. A laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 26.Wenzel M, Altenbuchner J. 2015. Development of a markerless gene deletion system for Bacillus subtilis based on the mannose phosphoenolpyruvate-dependent phosphotransferase system. Microbiology 161:1942–1949. doi: 10.1099/mic.0.000150. [DOI] [PubMed] [Google Scholar]
- 27.Arnaud M, Chastanet A, Débarbouillé M. 2004. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl Environ Microbiol 70:6887–6891. doi: 10.1128/AEM.70.11.6887-6891.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guérout-Fleury AM, Frandsen N, Stragier P. 1996. Plasmids for ectopic integration in Bacillus subtilis. Gene 180:57–61. doi: 10.1016/S0378-1119(96)00404-0. [DOI] [PubMed] [Google Scholar]
- 29.Lagodich AV, Cherva EA, Shtaniuk IV, Prokulevich VA, Fomichev IK, Prozorov AA, Titok MA. 2005. Construction of vector system for molecular cloning in Bacillus subtilis and Escherichia coli. Mol Biol 39:306–309. doi: 10.1007/s11008-005-0043-7. [DOI] [PubMed] [Google Scholar]
- 30.Gimpel M, Brantl S. 2012. Construction of a modular plasmid family for chromosomal integration in Bacillus subtilis. J Microbiol Methods 91:312–317. doi: 10.1016/j.mimet.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 31.Jeske M, Altenbuchner J. 2010. The Escherichia coli rhamnose promoter rhaP(BAD) is in Pseudomonas putida KT2440 independent of Crp-cAMP activation. Appl Microbiol Biotechnol 85:1923–1933. doi: 10.1007/s00253-009-2245-8. [DOI] [PubMed] [Google Scholar]
- 32.Wenzel M, Müller A, Siemann-Herzberg M, Altenbuchner J. 2011. Self-inducible Bacillus subtilis expression system for reliable and inexpensive protein production by high-cell-density fermentation. Appl Environ Microbiol 77:6419–6425. doi: 10.1128/AEM.05219-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morabbi Heravi K, Lange J, Watzlawick H, Kalinowski J, Altenbuchner J. 2015. Transcriptional regulation of the vanillate utilization genes (vanABK operon) of Corynebacterium glutamicum by VanR, a PadR-like repressor. J Bacteriol 197:959–972. doi: 10.1128/JB.02431-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang L, Watzlawick H, Fridjonsson O, Hreggvidsson G, Altenbuchner J. 2013. Improved soluble expression of the gene encoding amylolytic enzyme Amo45 by fusion with the mobile-loop-region of co-chaperonin GroES in Escherichia coli. Biocatal Biotransformation 31:335–342. doi: 10.3109/10242422.2013.858712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun T, Altenbuchner J. 2010. Characterization of a mannose utilization system in Bacillus subtilis. J Bacteriol 192:2128–2139. doi: 10.1128/JB.01673-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsuge K, Matsui K, Itaya M. 2003. One step assembly of multiple DNA fragments with a designed order and orientation in Bacillus subtilis plasmid. Nucleic Acids Res 31:e133. doi: 10.1093/nar/gng133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peng R, Lin G, Li J. 2016. Potential pitfalls of CRISPR/Cas9-mediated genome editing. FEBS J 283:1218–1231. doi: 10.1111/febs.13586. [DOI] [PubMed] [Google Scholar]
- 38.Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, Smith I, Tothova Z, Wilen C, Orchard R, Virgin HW, Listgarten J, Root DE. 2016. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol 34:184–191. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jeong D, Park S, Pan J, Kim E, Choi S. 2015. Genome engineering using a synthetic gene circuit in Bacillus subtilis. Nucleic Acids Res 43:e42. doi: 10.1093/nar/gku1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leenhouts K, Buist G, Bolhuis A, ten Berge A, Kiel J, Mierau I, Dabrowska M, Venema G, Kok J. 1996. A general system for generating unlabelled gene replacements in bacterial chromosomes. Mol Gen Genet 253:217–224. doi: 10.1007/s004380050315. [DOI] [PubMed] [Google Scholar]
- 41.Bloor AE, Cranenburgh RM. 2006. An efficient method of selectable marker gene excision by Xer recombination for gene replacement in bacterial chromosomes. Appl Environ Microbiol 72:2520–2525. doi: 10.1128/AEM.72.4.2520-2525.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bramucci MG, Nagarajan V. 1996. Direct selection of cloned DNA in Bacillus subtilis based on sucrose-induced lethality. Appl Environ Microbiol 62:3948–3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fabret C, Ehrlich SD, Noirot P. 2002. A new mutation delivery system for genome-scale approaches in Bacillus subtilis. Mol Microbiol 46:25–36. doi: 10.1046/j.1365-2958.2002.03140.x. [DOI] [PubMed] [Google Scholar]
- 44.Zhang X, Yan X, Cui Z, Hong Q, Li S. 2006. mazF, a novel counter-selectable marker for unmarked chromosomal manipulation in Bacillus subtilis. Nucleic Acids Res 34:e71. doi: 10.1093/nar/gkl358. [DOI] [PMC free article] [PubMed] [Google Scholar]