ABSTRACT
Determining the function and regulation of paralogues is important in understanding microbial functional genomics and environmental adaptation. Heme homeostasis is crucial for the survival of environmental microorganisms. Most Shewanella species encode two paralogues of ferrochelatase, the terminal enzyme in the heme biosynthesis pathway. The function and transcriptional regulation of two ferrochelatase genes, hemH1 and hemH2, were investigated in Shewanella loihica PV-4. The disruption of hemH1 but not hemH2 resulted in a significant accumulation of extracellular protoporphyrin IX (PPIX), the precursor to heme, and decreased intracellular heme levels. hemH1 was constitutively expressed, and the expression of hemH2 increased when hemH1 was disrupted. The transcription of hemH1 was regulated by the housekeeping sigma factor RpoD and potentially regulated by OxyR, while hemH2 appeared to be regulated by the oxidative stress-associated sigma factor RpoE2. When an oxidative stress condition was mimicked by adding H2O2 to the medium or exposing the culture to light, PPIX accumulation was suppressed in the ΔhemH1 mutant. Consistently, transcriptome analysis indicated enhanced iron uptake and suppressed heme synthesis in the ΔhemH1 mutant. These data indicate that the two paralogues are functional in the heme synthesis pathway but regulated by environmental conditions, providing insights into the understanding of bacterial response to environmental stresses and a great potential to commercially produce porphyrin compounds.
IMPORTANCE Shewanella is capable of utilizing a variety of electron acceptors for anaerobic respiration because of the existence of multiple c-type cytochromes in which heme is an essential component. The cytochrome-mediated electron transfer across cellular membranes could potentially be used for biotechnological purposes, such as electricity generation in microbial fuel cells and dye decolorization. However, the mechanism underlying the regulation of biosynthesis of heme and cytochromes is poorly understood. Our study has demonstrated that two ferrochelatase genes involved in heme biosynthesis are differentially regulated in response to environmental stresses, including light and reactive oxygen species. This is an excellent example showing how bacteria have evolved to maintain cellular heme homeostasis. More interestingly, the high yields of extracellular protoporphyrin IX by the Shewanella loihica PV-4 mutants could be utilized for commercial production of this valuable chemical via bacterial fermentation.
INTRODUCTION
Heme is a critical cofactor involved in a wide range of important biological processes, such as respiration, detoxification, gas sensing and transport, and signal transduction. The biosynthesis of heme has been well characterized. The intermediates in the heme biosynthesis pathway are conserved across prokaryotes and eukaryotes, possibly due to the fundamental nature of many of the biochemical processes that require the involvement of heme (1). An incomplete heme synthesis pathway usually results in heme auxotrophy, such as in Haemophilus influenzae or Buchnera species (2). In humans, abnormal heme synthesis can lead to anemia or porphyria (3, 4).
Despite the high level of conservation, variations in the enzymes that actually carry out heme biosynthesis have been observed among different organisms. Eukaryotic cells use oxygen-dependent coproporphyrinogen oxidase encoded by hemF, while some bacteria use an oxygen-independent counterpart encoded by hemN (5). More recently, two new subpathways for heme biosynthesis were validated: one involves the production of heme from siroheme, and the other does not use protoporphyrin as an intermediate (6, 7). However, while most heme biosynthesis or heme homeostasis studies in prokaryotes were conducted in pathogenic or symbiotic microorganisms, relevant knowledge in environmentally important microorganisms is still lacking. Therefore, study of genes involved in the heme biosynthesis pathway in environmental microorganisms is very important for our understanding of the cellular response to fluctuation in environmental conditions and for manipulating microorganisms for industrial, medical, and environmental applications.
Shewanella species, which are frequently isolated from redox-stratified environments, are renowned for their respiratory versatility. Members of the Shewanella genus are capable of carrying out dissimilatory reduction of various organic compounds, metals, and nitrate, which are critical steps in the global cycling of carbon, metals, and nitrogen (8, 9). The ability to use such a diverse group of electron acceptors is largely attributed to the great variety of c-type cytochromes, many of which contain more than one heme (8, 9). Therefore, there is a strong demand for heme for the biosynthesis of c-type cytochromes and other proteins that require heme as a prosthetic group. However, excessive heme and its closely related porphyrin compounds are toxic (10–12), resulting in tight control of the intracellular heme/porphyrin pool in Shewanella species. Protoporphyrin IX (PPIX) is a photosensitizer, and previously, it was shown that PPIX can induce cell death through reactive oxygen species (ROS) production; also, bacteria employ various strategies, such as regulation of heme biosynthesis or sequestration, uptake, export, and degradation, to control intracellular levels of heme (13). Shewanella species can serve as an ideal system to study heme biosynthesis and homeostasis for environmental microorganisms.
Shewanella loihica PV-4 was isolated from iron-rich microbial mats at the active deep-sea hydrothermal Naha Vent (1,325 m below sea level), located on the South Rift of Lō'ihi Seamount, HI, in the Pacific Ocean. Two hemH genes, hemH1 (Shew_2229) and hemH2 (Shew_1140), are annotated in the PV-4 genome. Both genes encode the enzyme ferrochelatase, which catalyzes the last step of heme synthesis by inserting a ferrous ion into the porphyrin ring of protoporphyrin IX to form heme b. These two ferrochelatase paralogues share 47% amino acid sequence identity, although the DNA sequences vary significantly. We hypothesized that both hemH paralogues functioned in heme biosynthesis but might be differentially regulated for sustaining heme homeostasis in Shewanella strains, which encode a number of c-type cytochromes for flexible respiration. We conducted molecular genetics analyses, including transposon mutant library screening, genetic complementation, in-frame gene deletion, and functional genomics analyses, on ΔhemH1 and ΔhemH2 mutants by transcriptional profiling based on quantitative reverse transcription-PCR and microarray technology, heme staining assays, chemical analyses, and comparative genomics analyses to test our hypothesis. The disruption of hemH1 resulted in extremely high concentrations of extracellular PPIX, which were not observed when hemH2 was disrupted. The biosynthesis of heme and cytochromes were still observed and obviously driven by HemH2 in the ΔhemH1 mutant, and the double mutant could not yet be generated. More importantly, the transcription of two hemH paralogues was differentially regulated in response to environmental stresses. These data provide important insights into the mechanisms underlying bacterial adaptation to changing environmental conditions via differential regulation of paralogues for concerted gene expression, as well as the understanding of genetic redundancy, a phenomenon widely found in all life domains and in almost every gene category. In addition, the PPIX-overproducing mutants could potentially be exploited for commercial production of this porphyrin compound.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Bacterial strains were cultured in Luria-Bertani medium (5 g/liter yeast extract, 10 g/liter tryptone, 10 g/liter NaCl [pH 7.0]) or modified M1 minimal medium (supplemented with 15 μg/ml gentamicin or 50 μg/ml kanamycin when necessary) (14). The genome of S. loihica strain PV-4 was analyzed by the Joint Genome Institute (http://genome.jgi-psf.org/she_p/she_p.home.html). The primers used in this study are listed in Table 2.
TABLE 1.
Bacterial strains and plasmids
| Strain/plasmid | Descriptiona | Source or reference |
|---|---|---|
| Strains | ||
| Escherichia coli WM3064 | thrB1004 pro thi rpsL hsdS lacZΔM15 RP4-1360 (araBAD)567 dapA1341::[erm pir(wt)] | W. Metcalf |
| E. coli TOP10 | F− mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80dlacZΔM15 ΔlacX74 deoR recA1 araD139 Δ(ara-leu)7697 galU galK rpsL (Strr) endA1 nupG | Invitrogen |
| E. coli EC100D+ | F− mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80dlacZΔM15 ΔlacX74 recA1 endA1 araD139 Δ(ara, leu)7697 galU galK λ− rpsL (Strr) nupG pir+(DHFR) | Epicentre Technologies |
| E. coli DH5α | F− endA1 glnV44 thi-1 recA1 relA1 gyrA96 deoR nupG ϕ80dlacZΔM15 Δ(lacZYA-argF)U169 hsdR17(rK− mK+) phoA λ− | TaKaRa |
| Shewanella loihica PV-4 | Isolated from iron-rich microbial mats at a hydrothermal vent of Lō'ihi Seamount, Pacific Ocean | 9 |
| Modified S. loihica PV-4 | pstI (Shew_0993) and pstM (Shew_0992) double-deletion mutant derived from PV-4 | This study |
| PV-4 ΔhemH1 | hemH1 (Shew_2229) deletion mutant derived from PV-4 | This study |
| PV-4 ΔhemH2 | hemH2 (Shew_1140) deletion mutant derived from PV-4 | This study |
| Plasmids | ||
| pDS3.0 | Suicide vector derived from pCDV224; Ampr Gmr sacB | 16 |
| pDS3.0-PV-hemH1ko | Suicide plasmid for deletion of hemH1 derived from PV-4 | This study |
| pDS3.0-PV-hemH2ko | Suicide plasmid for deletion of hemH2 derived from PV-4 | This study |
| pHERD30T | Shuttle vector with pBAD promoter, Gmr | 17 |
| pHERD30T-PV-hemH1 | pHERD30T containing the hemH1 derived from PV-4, Gmr | This study |
| pHERD30T-PV-hemH2 | pHERD30T containing the hemH2 derived from PV-4, Gmr | This study |
| pHERD30T-PV-rpoE2 | pHERD30T containing the rpoE2 derived from PV-4, Gmr | This study |
| pHERD30T-MR-hemH1 | pHERD30T containing the hemH1 derived from MR-1, Gmr | This study |
| pHERD30T-MR-hemH2 | pHERD30T containing the hemH2 derived from MR-1, Gmr | This study |
| pHERD30T-MR-rpoE2 | pHERD30T containing the rpoE2 derived from MR-1, Gmr | This study |
| pHERD30T-PV-gltX | pHERD30T containing the gltX derived from PV-4, Gmr | This study |
| pHERD30T-PV-hemA | pHERD30T containing the hemA derived from PV-4, Gmr | This study |
wt, wild type; Strr, streptomycin resistance; DHFR, dihydrofolate reductase; Ampr, ampicillin resistance; Gmr, gentamicin resistance.
TABLE 2.
Primers used in this study
Transposon mutagenesis, in-frame deletion mutant generation, and complementation of the in-frame deletion mutant.
Genetic manipulation of the wild-type PV-4 strain is difficult possibly due to the presence of a PstI restriction-modification system. To facilitate genetic manipulation, a ΔpstI ΔpstM in-frame double-deletion mutant was created and used as a parental strain (here, PV-4 refers to the parental strain, unless otherwise noted) for transposon mutagenesis and in-frame deletions (see Fig. S1 in the supplemental material). The mariner transposon mutant library (pminiHmar RB1, courtesy of Daâd Saffarini, University of Wisconsin, Milwaukee, WI) preparation, mutant screening, and insertion site mapping were conducted as previously described (15). In-frame deletion mutants were generated using the suicide vector pDS3.0 (R6K replicon, sacB, gentamicin resistant [Gmr])-based constructs, as previously described (16). For complementation, the target genes were PCR amplified and cloned into the inducible pHERD30T vector with the appropriate amount of arabinose added for induction (17). The resultant constructs were transformed into PV-4 or mutant strains via conjugation using Escherichia coli WM3064 as a donor strain. Transformation of the empty vector was used as a control.
RNA isolation and qRT-PCR analysis.
Total RNA was extracted using RNAiso Plus (TaKaRa) and the RNAprep pure kit for cells/bacteria (Tiangen Biotech Co. Ltd., Beijing, China), followed by DNase I treatment. To prepare cDNA, 2 μg of total RNA was reverse transcribed using the PrimeScript RT reagent kit with genomic DNA (gDNA) eraser (TaKaRa) and the TIANScript RT kit (Tiangen Biotech Co. Ltd.). Reverse transcription-PCR (RT-PCR) was carried out as described previously (18). Quantitative real-time PCR (qRT-PCR) was performed in 20-μl total volume with 1 μl of template cDNA from 10-fold-diluted reverse transcription products. Relative gene expression levels were quantified using SYBR Premix dimer erase (TaKaRa) on a Roche LightCycler 480 II real-time PCR system (Roche Diagnostics, Penzberg, Germany) with triplicates. Gene expression was normalized against the 16S rRNA gene using the 2−ΔCT calculation: ΔCT = CTgene of interest − CT16S rRNA (CT, threshold cycle). The primers are listed in Table S2 in the supplemental material.
Microarray hybridization and data analysis.
The whole-genome microarray of S. loihica PV-4 covers 3,857 of the 3,869 annotated coding sequences. Sixty-five-mer nucleotide probes were designed with CommonOligo 2.0 (19). Three probes were designed for each open reading frame (ORF), if possible, and each probe was randomly localized on the array with 3 replicates. The microarray was synthesized by Roche NimbleGen (Madison, WI). Cell cultures grown in LB were collected at late-log phase when substantial PPIX accumulation occurred. Genomic DNA of the PV-4 strain was extracted using a bacterial genomic DNA extraction kit (Sigma, St. Louis, MO). Cy5-labeled cDNA (2 μg of total RNA per sample/labeling reaction mixture) and Cy3-labeled gDNA (150 ng of gDNA/labeling reaction mixture, with 1/12 of the labeling product used for each hybridization) were cohybridized in each array. Cy3-labeled gDNA was used as a control for microarray hybridization and data analysis. Labeling, hybridization, array scanning, and data processing were performed as previously described (20, 21).
Chemical and spectral analyses.
The samples and a PPIX standard (Sigma-Aldrich, St. Louis, MO) were dissolved in a solution containing 90% acetone and 10% 0.1 M NH4OH. The absorbance was measured every 5 nm with a spectrometer (Biowave II WPA) and quartz cuvettes. The UV-visible spectrograms were plotted. The fluorescence spectra of the PPIX standard and samples were measured by a Nicolet Magna-760 (Fourier transform infrared spectroscopy [FTIR]) system at the Oak Ridge National Laboratory, TN. High-performance liquid chromatography (HPLC) and mass spectrometry (MS) analyses were conducted at the Health Sciences Center of the University of Oklahoma, OK, using a Michrom Bioresources Paradigm MSRB capillary HPLC and HCT Ultra ion trap (Bruker Daltonics): column, Magic MS C18, 5 μm, 100 Å, 0.5 by 150 mm; solvent A, 0.09% formic acid, 0.01% trifluoroacetic acid (TFA), 2% CH3CN, 97.9% water; solvent B, 0.09% formic acid, 0.0085% TFA, 95% CH3CN, 4.9% water; and gradient, 30% to 100% solvent B over 15 min, hold 3 min, 100% to 30% over 2 min. For standard and sample preparation, 1.3 mg of standard was dissolved in 1.3 ml of 100% methanol and diluted 10 times with the solution containing 0.1% formic acid, 50% acetonitrile, and 50% water, and 10 μl was loaded on an HPLC-UV-MS system. Two hundred microliters of sample was dried with Speed-Vac and then reconstituted with 200 μl of solution containing 0.1% formic acid, 50% acetonitrile, and 50% water. Ten microliters was loaded on a 40-μl loop into an HPLC-UV-MS system (flow rate, 20 μl/min; UV wavelength, 216 nm). The mass spectrometry system used was a Bruker Daltonics HCT Ultra ion trap; mode, positive (target mass, m/z 500).
Determination of transcription start sites.
Terminal deoxynucleotidyl transferase (TdT; TaKaRa) was used to catalyze the incorporation of single deoxynucleotides (dATPs) into the 3′-OH terminus of cDNA to make the deoxyribosyladenine (dA)-tailed cDNA, according to the manufacturer's protocol. Touchdown and nested-PCR were used to amplify the dA-tailed cDNA by using an oligo(dT) (5′-GCCAGTCTTTTTTTTTTTTTTTTT-3′) primer and a gene-specific primer (22). The PCR product was cloned into the pMD18-T vector (TaKaRa, Dalian, China) for sequencing.
Nitrate reduction and H2O2 tolerance test.
Nitrate reduction was examined in PV-4 and mutants cultured in modified M1 medium, with sodium nitrate as the electron acceptor. The cultures were incubated under anaerobic or microoxic conditions (without shaking). Nitrate and nitrite concentrations were measured with a standard colorimetric assay (23). H2O2 sensitivity was tested by monitoring the optical density at 600 nm (OD600) of relevant strains cultured in LB broth with a gradient of H2O2 concentrations (0, 0.1, 0.3, 0.5, 0.7, and 1 mM) (24).
SDS-PAGE and heme staining.
Mid-log-phase cell cultures (OD600, 0.6 to ∼0.8) grown in LB were collected and homogenized by applying high pressure (a low-temperature ultra-high-pressure continuous-flow cell disrupter [JN-02C; Juneng Biology and Technology Co., Guangzhou, China] or an ultrasonic cell disruption system [Scientz-IID; Ningbo Xingzhi Biotechnology Co., China]). After centrifugation at 13,000 rpm at 4°C, the supernatants containing the cellular protein fraction were resuspended in SDS loading buffer and separated by SDS-PAGE using 12% polyacrylamide gels. Heme stains were performed using 3,3′,5,5′-tetramethylbenzidine dihydrochloride as the substrate, as previously described (25). Images of the stained gels were taken with the Gel Doc XR+ system (Bio-Rad Laboratories, Inc., United Kingdom).
Bioinformatics analysis.
Amino acid and nucleotide sequences were retrieved from the NCBI database using the BLAST search tool. The ClustalW2 package (http://www.ebi.ac.uk/Tools/msa/clustalw2/) was used for amino acid and nucleotide sequence alignments, and WebLogo (http://weblogo.berkeley.edu) was applied to nucleotide sequences for promoter motif identification.
Accession number(s).
The microarray data have been deposited in the NCBI GEO database under the accession no. GSE81743.
RESULTS
Disruption of hemH1 resulted in a red-colored phenotype.
Genetic manipulations of PV-4 strain were very difficult possibly due to the PstI-like restriction-modification system (26). To facilitate genetic manipulation, we deleted the putative PstI-like endonuclease (designated PstI, Shew_0993) and DNA methylase (designated PstM, Shew_0992) genes from the PV-4 strain, facilitating the generation of both transposon insertional and in-frame deletion mutants of the parental PV-4 strain (see Fig. S1 in the supplemental material). The wild-type PV-4 and the parental PV-4 (ΔpstI ΔpstM double mutant) are orange in color, as are most Shewanella species. Interestingly, red-colored colonies were obtained from a screening of a transposon mutant library of PV-4. The transposon insertions in the red-colored mutants were mapped to the hemH1 gene, encoding a ferrochelatase (see Fig. S2 in the supplemental material), which catalyzes the insertion of ferrous iron into protoporphyrin IX (PPIX [7,12-diethenyl 3,8,13,17-tetramethyl-21II,23II-porphine 2,18 dipropanoic acid]) to form heme b. Interestingly, none of the red-colored transposon mutants contained an insertion of transposon in hemH2, suggesting that hemH1 might play a more prominent role in heme biosynthesis in PV-4. Despite the highly conserved biosynthesis pathways for protoporphyrin IX, heme, and c-type cytochromes in Escherichia and Shewanella, the red-colored colony phenotype was not obtained in Escherichia coli, although an accumulation of PPIX was observed in E. coli hemH (visA) mutants (10, 11).
To further confirm the direct link between hemH1 and the red-colored phenotype, in-frame deletion mutants of hemH1 or hemH2 were generated. Red pigmentation was observed in the ΔhemH1 in-frame deletion mutant but not in the ΔhemH2 mutant. Reintroduction of a wild-type copy of the hemH1 gene into the ΔhemH1 mutant fully reversed the red-colored phenotype (Fig. 1). Besides the red-colored phenotype in the ΔhemH1 mutant, no other growth deficiencies were observed in the ΔhemH1 or ΔhemH2 mutant when cultured in the LB medium under aerobic conditions. Double-deletion mutants of hemH1 and hemH2 could not be generated, suggesting that the disruption of heme biosynthesis in PV-4 is lethal and that both hemH paralogues function in the heme biosynthesis pathway, with hemH1 playing a major role.
FIG 1.
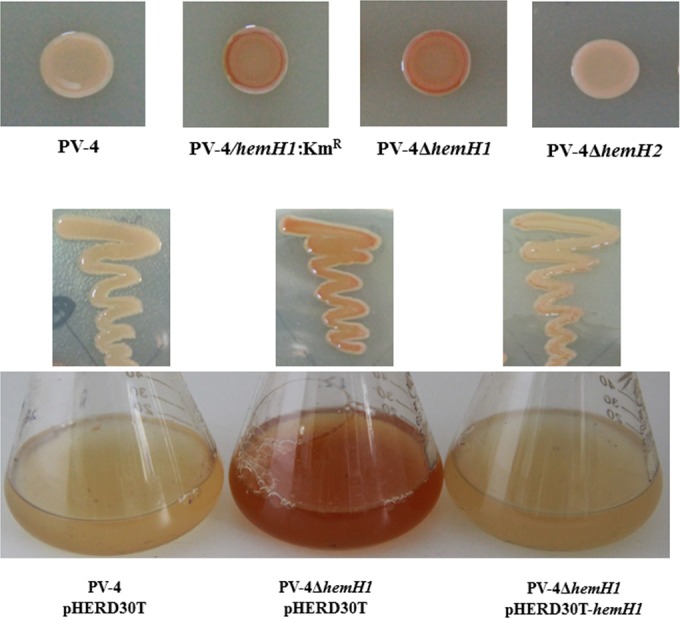
Effects of hemH1 disruption and deletion on phenotypes in PV-4. Inactivation of a ferrochelatase gene resulted in the overproduction of protoporphyrin IX in Shewanella loihica PV-4. The transposon insertional mutant (hemH1::Kmr) and in-frame deletion mutant (PV-4 ΔhemH1) exhibited similar phenotypes, while the deletion of another paralog, hemH2, did not cause the same phenotypes. In genetic complementation analyses on the hemH1 deletion mutant PV-4 ΔhemH1, the plasmid-borne wild-type hemH1 gene restored the phenotype of the mutant to that of the wild-type strain carrying the empty vector.
Accumulation of PPIX in ΔhemH1 mutant led to red-colored phenotype.
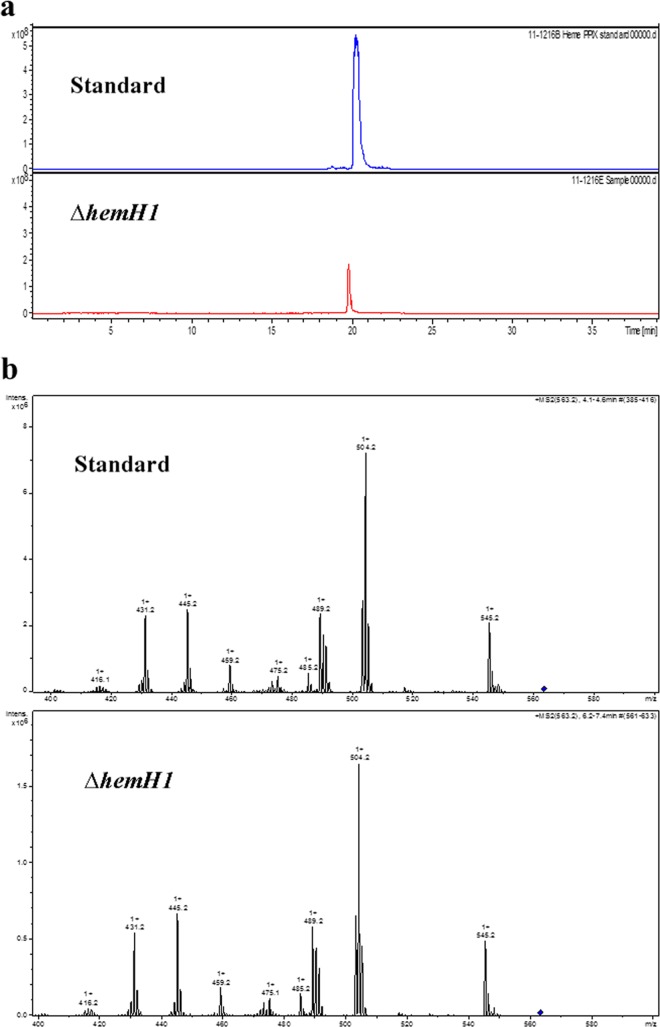
Red-pigmented aggregates accumulated in the culture broth of the ΔhemH1 mutant, suggesting that the red pigment was secreted into the medium. To determine whether the red pigment was PPIX, acetone-NH4OH extracts of the cell pellets were analyzed with a commercial PPIX standard as the control. High-performance liquid chromatography (HPLC) showed that the ΔhemH1 mutant extracts and the PPIX standard eluted at the same time (Fig. 2a). Further analysis with electrospray ionization-tandem mass spectrometry (ESI-MS/MS) clearly indicated that the ΔhemH1 mutant sample had the same fragment profile as the commercial PPIX standard (Fig. 2b). Additionally, UV-visible absorbance spectra of the ΔhemH1 mutant extract were very similar to those of the PPIX standard, with the maximum absorbance at 405 nm. However, the wild-type strain extract did not show an absorbance peak at 405 nm, indicating that there was no PPIX accumulated in the wild-type cells (see Fig. S3 in the supplemental material). Furthermore, Fourier transform infrared spectroscopy (FTIR) analyses also showed the same fluorescence spectrum pattern (see Fig. S4 in the supplemental material). Taken together, these data indicate that the red pigment in the ΔhemH1 mutant was PPIX. With HPLC separation and fluorescence detection (excitation at 405 nm and fluorescence at 630 nm), the yield of PPIX in the ΔhemH1 mutant was estimated to be ∼3 mg/g (dry weight).
FIG 2.
Chemical analyses of the extracellular compound. (a) Comparison of high-performance liquid chromatography-mass spectrometry extract ion chromatograms (EIC; m/z 563.2 ± 0.3). (b) Electrospray ionization-tandem mass spectrometry (ESI-MS/MS; precursor ion, m/z 563.2, PPIX [blue diamond]) analyses showing almost identical structures of the PPIX standard and the bacterial sample.
Impaired heme synthesis and defective nitrate reduction in ΔhemH1 mutant.
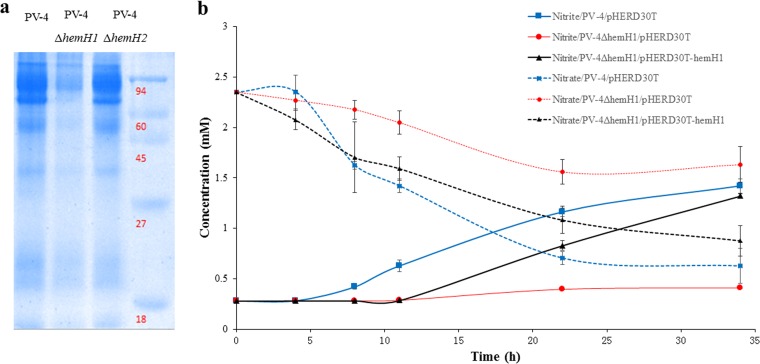
In order to determine if the accumulation of PPIX affected the intracellular heme levels, heme staining of the cellular proteins was carried out. Compared to levels in wild-type PV-4, levels of c-type cytochromes decreased in the ΔhemH1 mutant but remained similar in the ΔhemH2 mutant (Fig. 3a), suggesting that the deletion of hemH1 substantially affected cytochrome c synthesis.
FIG 3.
Effects of hemH1 disruption and deletion on c-type cytochrome synthesis and nitrate reduction in PV-4. (a) Heme staining analyses of c-type cytochromes in the wild-type strain and hemH1 and hemH2 single mutants of S. loihica PV-4. After cell disruption, the supernatants containing the cellular protein fraction were resuspended in the SDS loading buffer without β-mercaptoethanol and then incubated at 37°C for 1 h. Molecular mass markers (in kilodaltons) are shown on the right. (b) Nitrate reduction of wild-type S. loihica PV-4 strain and the hemH1 deletion mutant. The parental strain carrying empty vector (PV-4/pHERD30T), the hemH1 deletion mutant carrying empty vector (PV4 ΔhemH1/pHERD30T), and the complementation strain carrying plasmid-borne hemH1 (PV4 ΔhemH1/pHERD30T-hemH1) were cultivated in the modified M1 minimal medium supplemented with 2 mM sodium nitrate under microoxic conditions (in tightly capped tubes without shaking). The blank represents the used culture medium without bacterial inoculation. The error bars represent the standard deviation (SD).
Since heme and cytochrome c are critical components involved in anaerobic respiration, nitrate reduction was examined using nitrate as the sole electron acceptor in the anaerobic M1 medium. Compared with PV-4, the ΔhemH1 mutant was deficient in nitrate reduction, as reflected by the much lower rate of nitrate reduction. Complementation of PV-4 ΔhemH1 with plasmid-borne hemH1 partially restored nitrate reduction (Fig. 3b).
Taken together, the deletion of hemH1 in PV-4 resulted in dramatic extracellular accumulation of PPIX, decreased intracellular cytochrome c levels, and impaired nitrate reduction. The deletion of hemH2, on the contrary, did not lead to any of these significant changes under the same conditions tested. However, it appeared that hemH2 also encodes a functional ferrochelatase, because (i) the ΔhemH1 mutant showed normal growth (see Fig. S5 in the supplemental material) and still produced cytochromes despite being present at a lower level, indicating that some PPIX was still converted to heme; (ii) plasmid-borne hemH2 fully restored heme synthesis despite the loss of hemH1 (Fig. 4b; see also Fig. S6 in the supplemental material); and (iii) we were unable to generate a ΔhemH1 ΔhemH2 double mutant even with supplementation of hemin, indicating that heme synthesis is essential for the growth of this bacterium. The transcription levels of the two paralogues in the wild-type and mutant strains were compared to further elucidate their cellular functions.
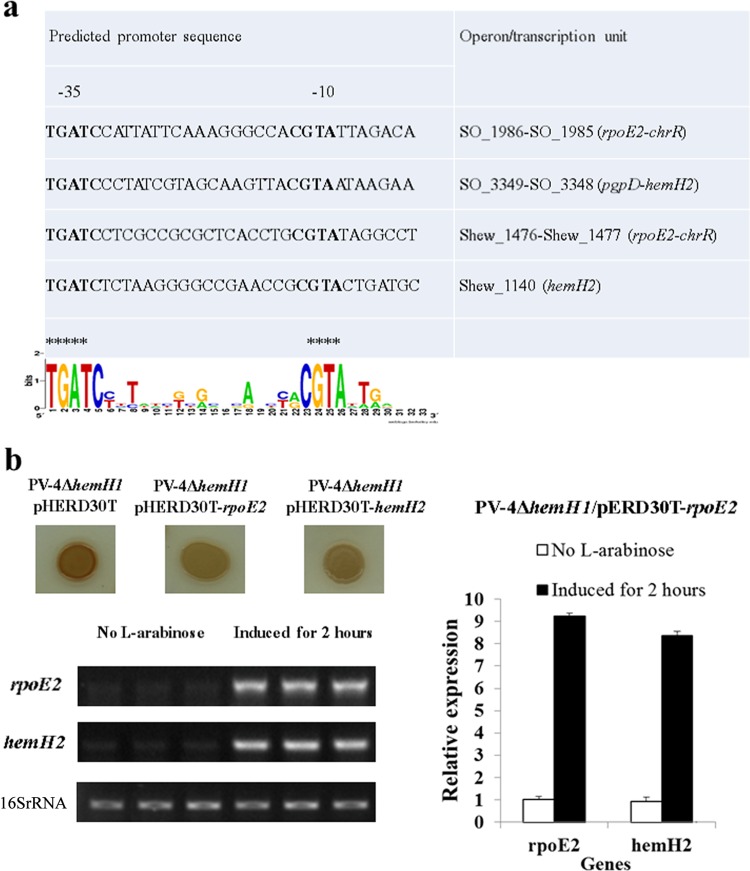
FIG 4.
Analysis of the RpoE2-recognizing regulated promoter in Shewanella strains. (a) Sequence logos of the predicted RpoE2-dependent promoter motifs and RpoE2-dependent hemH2 gene in the S. oneidensis MR-1 and S. loihica PV-4 strains. Asterisks highlight TGATC and CGTA. (b) The induced expression of plasmid-borne rpoE2 suppressed the PPIX-overproducing phenotype of the PV4 ΔhemH1 strain and enhanced the transcription of the chromosomal hemH2 gene in the S. loihica PV-4 strain. l-Arabinose (0.01% [wt/vol]) was added to induce the expression of rpoE2, and the bacterial culture without l-arabinose was used as a control. The experiments were performed at least three times. The error bars represent the standard deviations (SD) of the results from triplicate independent samples.
Different gene expression patterns of hemH1 and hemH2.
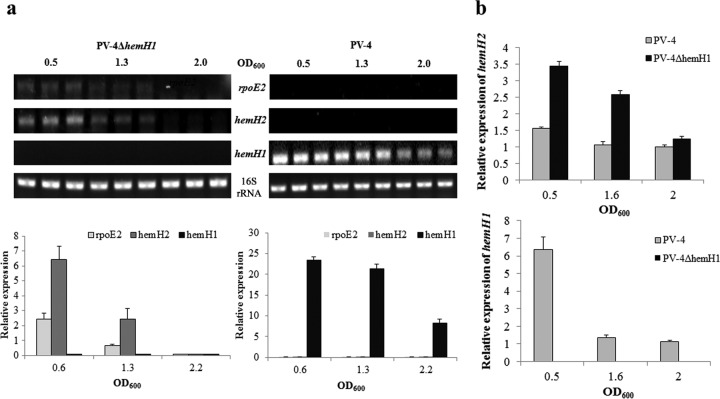
Temporal expression patterns of hemH1 and hemH2 were monitored with RT-PCR and qRT-PCR. In PV-4, hemH1 showed constitutive expression, while the expression of hemH2 was undetectable (Fig. 5a). Previously, we demonstrated that the pgpD-hemH2 operon was regulated by the RpoE2 sigma factor in the Shewanella oneidensis MR-1 strain (18). Interestingly, the expression of rpoE2 was not detected in the wild-type PV-4 strain (Fig. 5a). However, in the ΔhemH1 mutant, the expression of hemH2 and rpoE2 was detected in both exponential (OD600, 0.6) and early stationary (OD600, 1.3) phases. The qRT-PCR analysis validated the results of the RT-PCR, showing a mild increase (∼2-fold) in hemH2 expression in the ΔhemH1 mutant compared to the basal-level expression in PV-4 (Fig. 5b). These results indicated that hemH1 is the dominant gene under standard growth conditions and that hemH2 may play a supplementary role when hemH1 is disrupted.
FIG 5.
Transcriptional analyses of rpoE2 and hemH paralogues in the wild-type strain and the hemH1-null mutants of S. loihica PV-4. (a) Semiquantitative RT-PCR analyses of rpoE2 and hemH2 transcripts in S. loihica PV-4 and PV-4 ΔhemH1 strains. (b) Real-time PCR analyses of hemH2 transcripts in S. loihica PV-4 and PV-4 ΔhemH1 strains. Transcription of the 16S rRNA genes was analyzed and used as the loading control. The assays were performed in triplicate, and the error bars represent the standard deviations (SD) of the results from triplicate independent samples.
Differential regulation of hemH1 and hemH2.
To further determine how the expression of hemH1 and hemH2 is regulated, the upstream regions of each gene were examined to identify potential transcriptional regulator binding motifs. A transcriptional start site was identified using a primer extension assay. The +1 site was determined to be nucleotide A downstream of the −10 (TAGCCT) and −35 (GGCTTT) promoter motifs (see Fig. S7a in the supplemental material). Sequence analysis identified an RpoD binding motif (−10/−15 sites) and a putative OxyR (shew_1035) binding motif (TTCTCATATTTGAAAAAGC) upstream of the −10/−15 sites (Fig. S7b). Therefore, the expression of hemH was predicted to be dependent on the housekeeping sigma factor RpoD and possibly regulated by the oxidative stress response regulator OxyR (Fig. S7b). Our previous study in S. oneidensis MR-1 identified the gene pgpD, encoding glutathione peroxidase, as belonging to a regulon of RpoE2, an extracytoplasmic function (ECF) sigma factor involved in the oxidative stress response (18). Coincidentally, hemH2 in MR-1 is in the same operon as pgpD, and a similar RpoE2-dependent promoter motif was identified upstream of hemH2 in PV-4 (Fig. 4a). A primer extension assay also showed that hemH2 was transcribed from the nucleotide A (+1) downstream of the −35 (TGATCT) and −10 (CGTACT) promoter motifs, which are recognized by RpoE2 (see Fig. S9 in the supplemental material).
To verify the regulatory role of OxyR on hemH1, we induced the expression of oxyR in trans in the parent strain. The result showed that hemH1 transcription is significantly induced due to a higher level of oxyR expression (see Fig. S8a in the supplemental material). To validate the regulation of hemH2 by RpoE2, the expression of hemH2 was monitored in PV-4 harboring a vector pHERD30T with a native rpoE2 gene driven by an arabinose-inducible promoter. Under control conditions, both rpoE2 and hemH2 exhibited basal-level expression. After a 2-h induction with arabinose, the expression of rpoE2 increased 9-fold, and the expression of hemH2 increased about 8-fold (Fig. 4b). Induction of rpoE2 in the ΔhemH1 mutant completely reversed the red-colored phenotype of the ΔhemH1 mutant (see Fig. S6 in the supplemental material). These results suggested that expression of hemH2 was regulated by RpoE2.
Functions of hemH paralogues under oxidative stress.
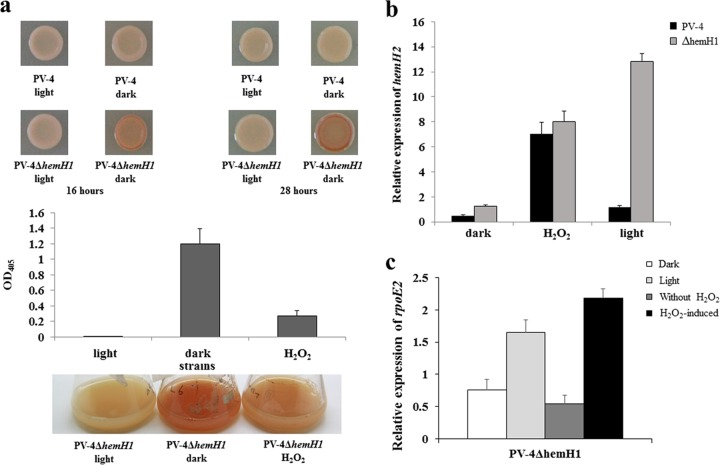
Protoporphyrin IX is a long-known photosensitizer and produces reactive oxygen species (ROS) upon light exposure, leading to cell damage or even death (12, 27). To uncover the roles of hemH paralogues under oxidative stress, gene expression and phenotypes in PV-4 or the ΔhemH1 mutant were examined under visible light (about 700 to ∼1,000 lx) or by adding H2O2 to the medium. The introduction of H2O2 to the medium partially suppressed the red-colored phenotype of the ΔhemH1 mutant. Interestingly, light exposure completely reversed the red-colored phenotype (Fig. 6a). The qRT-PCR results demonstrated an increased expression of rpoE2 and hemH2 in the ΔhemH1 mutant but not in the parental strain under light exposure. Also, increased expression of rpoE2 and hemH2 was observed in both PV-4 and the ΔhemH1 mutant after the addition of H2O2 (Fig. 6b and c; see also Fig. S10 in the supplemental material). Additionally, the ΔhemH1 mutant appeared to be more sensitive to H2O2 than the ΔhemH2 mutant or PV-4 (see Fig. S11 in the supplemental material). Under the oxidative stress conditions, hemH1 exhibited significant upregulation (see Fig. S8b in the supplemental material), which was likely dependent on the transcription factor OxyR. Overexpression of oxyR led to the upregulation of hemH1 in the PV-4 strain (Fig. S8a). These results suggested that hemH1 plays a more dominant role in the oxidative stress response, while the expression of hemH2, mediated by rpoE2, is induced under oxidative stress due to either the addition of H2O2 or the ROS produced from PPIX under light exposure. HemH2 might act as a backup to sustain heme homeostasis under oxidative stresses.
FIG 6.
Effects of hydrogen peroxide (H2O2) and visible light on the PV-4 ΔhemH1 mutant. (a) Suppression of protoporphyrin IX (PPIX) overproduction by addition of hydrogen peroxide (H2O2) and visible light irradiance in the Shewanella loihica PV-4 ΔhemH1 mutant. PPIX production in the LB broth and measurement of relative PPIX yield in terms of optical density at 405 nm (OD405). (b) Effects of hydrogen peroxide (H2O2) and visible light on the transcription of the hemH2 gene in the Shewanella loihica PV-4 wild-type strain and the PV-4 ΔhemH1 mutant. (c) Effects of hydrogen peroxide (H2O2) and visible light on the transcription of the rpoE2 gene in the PV-4 ΔhemH1 mutant. The error bars represent the standard deviations (SD).
Increased expression of iron uptake genes and decreased expression of PPIX synthesis genes in the ΔhemH1 mutant.
Genome-wide gene expression changes in the ΔhemH1 mutant at the late-log phase were examined with microarray analysis. Compared with expression in PV-4 (see Table S1 in the supplemental material), genes involved in iron uptake exhibited significantly increased expression (log2R > 3) in the ΔhemH1 mutant. These included two siderophore receptor genes (Shew_3097 and Shew_3841), one ferric iron binding gene (Shew_0861), two ferrous iron transporter genes (Shew_2517 [feoA] and Shew_2518 [feoB]), and a bacterioferritin-associated ferredoxin gene (Shew_0552). Considering the substantial accumulation of PPIX at the late-log phase and the need for ferrous iron in the biosynthesis of heme from PPIX, it was reasonable that iron uptake was enhanced in order to counteract PPIX accumulation in the ΔhemH1 mutant.
In contrast to increased expression of iron uptake genes, the expression of genes involved in the heme biosynthesis pathway prior to the PPIX production step significantly decreased. These included glutamyl-tRNA reductase (Shew_2913) and delta-aminolevulinic acid dehydratase (Shew_3382) genes, which are involved in the synthesis of early precursors of heme (5-aminolevulinate and porphobilinogen, respectively), and hemF (Shew_0073) and hemG (Shew_0025), which are involved in the synthesis of protoporphyrinogen and PPIX, respectively. In addition, the levels of expression of several cytochrome c genes, including two decaheme cytochrome c genes, omcA and mtrC (Shew_0918 and Shew_2525, respectively), were significantly decreased. These results suggest that a potential negative feedback effect of accumulation of PPIX on heme biosynthesis occurs. Significant repression of flagellar synthesis genes was also detected, possibly as a cellular response to stresses imposed by PPIX accumulation.
DISCUSSION
The great respiratory flexibility found in Shewanella species is achieved through the expression of multiple c-type cytochromes (around 40), which requires sufficient heme supply (28). Therefore, heme homeostasis is critical in Shewanella species, which predominantly conduct a respiratory lifestyle (29). There are two ferrochelatase genes in 19 out of 24 sequenced Shewanella genomes (see Table S2 in the supplemental material). Heme homeostasis might be controlled by the differential expression of the paralogues, which in turn has a profound impact on critical cellular processes, like respiration and tolerance to stresses. In this study, we demonstrated that two hemH paralogues, hemH1 and hemH2, both encode a functional ferrochelatase but are under different transcriptional regulation for sustaining heme homeostasis in S. loihica PV-4.
Although it has long been known that PPIX might accumulate due to inactivation of ferrochelatase in Escherichia coli (30, 31), the level of PPIX accumulation in the PV-4 ΔhemH1 mutant was two orders of magnitude higher, conferring a red color to bacterial colonies and broth, than that found in previous studies (10, 11, 32). The high yields of PPIX by the PV-4 mutants could potentially be exploited for commercial production. The photosensitivity of PPIX is utilized in photodynamic therapy (PDT) against different forms of cancers. Protoporphyrin IX disodium salt is also used as a health care supplement and hepatic protectant (e.g., the Prolmon tablets sold in Japan) for patients with infective hepatitis and chronic liver diseases. Currently, the commercial production of PPIX and related products is mainly based on extraction from livestock blood samples. Bacterial fermentation-based bioprocess may reduce costs and is expected to be more environmentally friendly, because the extraction and purification of the secreted PPIX will be simple, use fewer chemical reagents, and/or may be achieved by simple physical processes.
Interestingly, the PPIX-accumulating phenotype was observed in the ΔhemH1 mutant but not in the ΔhemH2 mutant, indicating that the expression of hemH1 alone might be sufficient to meet the demand for heme under the tested conditions. However, multiple lines of evidence showed that hemH2 is also functional, as the hemH2 expression in trans might complement the loss of hemH1. The fact that a deletion mutant of both hemH1 and hemH2 could not be achieved in PV-4 suggested that heme might be essential for the bacterial growth of PV-4. Alignment of the amino acid sequences of the ferrochelatase paralogues in PV-4 with those in other organisms (see Fig. S12 in the supplemental material) revealed conserved motifs and amino acid residues, such as the invariant residues His235 and Glu314 of the Saccharomyces cerevisiae ferrochelatase (33) and the conserved residues His183 and Glu264 from Bacillus subtilis (34). However, a third well-conserved residue (Ser118) in Bacillus subtilis was not found in PV-4 HemH2 (Shew_1140), where an Ala167 was present (Fig. S12). In addition, an essential [2Fe-2S] cluster coordinated by cysteine residues in the C terminus of the mammalian ferrochelatases was not present in the bacterial homologues (35). These results indicate that HemH1 and HemH2 are functional ferrochelatases and have conserved motifs and amino acid residues, providing a forward direction for study on the crystal structures of HemH1 and HemH2, especially protein structure-activity analysis.
The different roles of hemH paralogues in PV-4 are further supported by dissimilar regulatory control, leading to their distinctive expression patterns. The hemH1 gene is under more classical regulation by RpoD and OxyR, consistent with its constitutive expression. The expression of hemH2 is controlled by ECF sigma factor RpoE2, which operates with an anti-sigma factor ChrR (Shew_1477) and is self-regulated (36). Sequence analysis and transcriptional start site mapping revealed the existence of a conserved RpoE2 regulatory motif upstream of rpoE2 and hemH2 in multiple Shewanella strains other than PV-4, indicating that this is a common feature in this genus (18). Under nonstress conditions, the anti-sigma factor ChrR binds to RpoE2, preventing RpoE2 from binding to recognition sites on the genome. In the case of oxidative stress, ChrR undergoes a conformational change, and RpoE2 is released, resulting in induction in its regulon members, including rpoE2 itself (37). In accordance with previous studies demonstrating that RpoE2 is an oxidative stress response regulator, hemH2 expression is conditionally induced under oxidative stress due to H2O2 or ROS generated from light exposure upon PPIX producing the ΔhemH1 mutant. Without a functioning hemH paralogue, light radiance in PPIX export-deficient Streptococcus agalactiae mutants or the PPIX-accumulating E. coli hemH mutant is harmful or even lethal (10, 11, 30, 38). Because hemH2 “backed up” the loss of hemH1, instead of killing the cells, light exposure led the ΔhemH1 mutant to revert to wild-type-like growth.
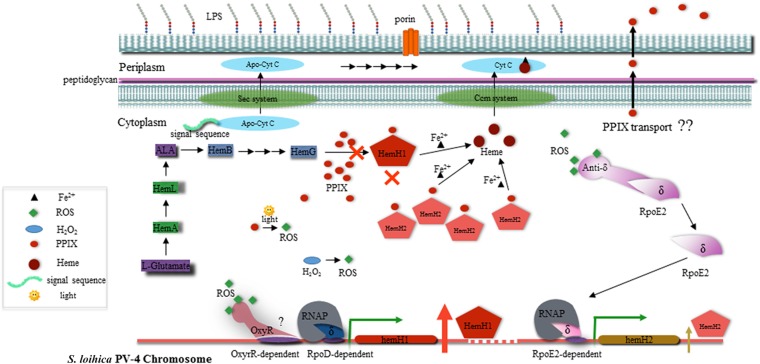
Genetic redundancy is a common phenomenon observed in all life domains and happens in almost every gene category, including housekeeping, functional, and regulatory genes (39). It remains an intriguing subject to understand the function, regulation, and fate of the paralogues during the evolutionary course. In terms of the hemH paralogues in PV-4, it seems that transcriptional reprogramming due to feedback effects (e.g., reduced heme and PPIX accumulation) or environmental stresses (e.g., oxidative stress) played a key role in maintaining heme homeostasis and thus the selection and necessity for the two paralogues. We propose a model to depict the roles of hemH1 and hemH2 in heme biosynthesis and their regulation in context with respiration and oxidative stress (Fig. 7). Under anaerobic/microoxic and dark conditions, the RpoD-dependent hemH1 gene is constitutively expressed and plays a primary role in heme and c-type cytochrome synthesis. Inactivation of hemH1 blocked the biosynthesis of heme and cytochromes and resulted in the accumulation of PPIX, which may mediate the production of ROS under aerobic and light-exposure conditions. The presence of ROS is sensed by ChrR, resulting in the release and activation of RpoE2 to drive the transcription of hemH2 for heme synthesis. Additionally, the synthesis of heme precursors was decreased, and the uptake of iron was increased by feedback regulation. Under oxidative stress, the OxyR transcriptional factor may also act to enhance the transcription of hemH1. Overproduction of PPIX in the ΔhemH1 mutant suggests the existence of a yet-to-be-identified PPIX/heme export system, similar to those reported in pathogens such as Streptococcus agalactiae, to prevent an excessive intracellular porphyrin pool (38), which is under investigation. The availability of abundant genome sequences and a mature mutagenesis platform make Shewanella an ideal model organism for studying genetic redundancy and its implication in the physiology, ecology, and stress response for environmental microbes using comparative genomics.
FIG 7.
Schematic diagram illustrating the biosynthesis pathway of heme and cellular function and transcriptional regulation of two ferrochelatase paralogues in S. loihica PV-4 strain. LPS, lipopolysaccharide; Sec, secretion; Ccm, cytochrome c maturation.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by DOE grant DE-FG02-07ER64383 to J.Z. and the Chinese Academy of Sciences grant Y15103-1-401 and One-Hundred Scholar Award to D.Q.
We thank Huaiwen Wang of the Laboratory for Molecular Biology and Cytometry Research, University of Oklahoma Health Sciences Center, for his help in the analyses of PPIX samples.
J.Z., D.Q., Z.H., and M.X. have a potential financial conflict of interest resulting from a published patent application (no. WO2014144329 A2) regarding the Shewanella-based production of protoporphyrin IX.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00203-16.
REFERENCES
- 1.Oborník M, Green BR. 2005. Mosaic origin of the heme biosynthesis pathway in photosynthetic eukaryotes. Mol Biol Evol 22:2343–2353. doi: 10.1093/molbev/msi230. [DOI] [PubMed] [Google Scholar]
- 2.Baumann P, Baumann L, Lai C-Y, Rouhbakhsh D, Moran NA, Clark MA. 1995. Genetics, physiology, and evolutionary relationships of the genus Buchnera: intracellular symbionts of aphids. Annu Rev Microbiol 49:55–94. doi: 10.1146/annurev.mi.49.100195.000415. [DOI] [PubMed] [Google Scholar]
- 3.Iolascon A, De Falco L, Beaumont C. 2009. Molecular basis of inherited microcytic anemia due to defects in iron acquisition or heme synthesis. Haematologica 94:395–408. doi: 10.3324/haematol.13619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moore MR. 1998. The biochemistry of heme synthesis in porphyria and in the porphyrinurias. Clin Dermatol 16:203–223. doi: 10.1016/S0738-081X(97)00201-0. [DOI] [PubMed] [Google Scholar]
- 5.Panek H, O'Brian MR. 2002. A whole genome view of prokaryotic haem biosynthesis. Microbiology 148:2273–2282. doi: 10.1099/00221287-148-8-2273. [DOI] [PubMed] [Google Scholar]
- 6.Bali S, Lawrence AD, Lobo SA, Saraiva LM, Golding BT, Palmer DJ, Howard MJ, Ferguson SJ, Warren M. 2011. Molecular hijacking of siroheme for the synthesis of heme and d1 heme. Proc Natl Acad Sci U S A 108:18260–18265. doi: 10.1073/pnas.1108228108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dailey HA, Gerdes S, Dailey TA, Burch JS, Phillips JD. 2015. Noncanonical coproporphyrin-dependent bacterial heme biosynthesis pathway that does not use protoporphyrin. Proc Natl Acad Sci U S A 112:2210–2215. doi: 10.1073/pnas.1416285112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hau HH, Gralnick JA. 2007. Ecology and biotechnology of the genus Shewanella. Annu Rev Microbiol 61:237–258. doi: 10.1146/annurev.micro.61.080706.093257. [DOI] [PubMed] [Google Scholar]
- 9.Fredrickson JK, Romine MF, Beliaev AS, Auchtung JM, Driscoll ME, Gardner TS, Nealson KH, Osterman AL, Pinchuk G, Reed GL, Rodionov DA, Rodrigues G, Saffarini DA, Serres MH, Spormann AM, Zhulin IB, Tiedje JM. 2008. Towards environmental systems biology of Shewanella. Nat Rev Microbiol 6:592–603. doi: 10.1038/nrmicro1947. [DOI] [PubMed] [Google Scholar]
- 10.Nakahigashi K, Nishimura K, Miyamoto K, Inokuchi H. 1991. Photosensitivity of a protoporphyrin-accumulating, light-sensitive mutant (visA) of Escherichia coli K-12. Proc Natl Acad Sci U S A 88:10520–10524. doi: 10.1073/pnas.88.23.10520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miyamoto K, Nishimura K, Masuda T, Tsuji H, Inokuchi H. 1992. Accumulation of protoporphyrin IX in light-sensitive mutants of Escherichia coli. FEBS Lett 310:246–248. doi: 10.1016/0014-5793(92)81341-I. [DOI] [PubMed] [Google Scholar]
- 12.Cox GS, Bobilier C, Whitten DG. 1982. Photooxidation and singlet oxygen sensitization by protoporphyrin IX and its photooxidation products. Photochem Photobiol 36:401–407. doi: 10.1111/j.1751-1097.1982.tb04393.x. [DOI] [Google Scholar]
- 13.Anzaldi LL, Skaar EP. 2010. Overcoming the heme paradox: heme toxicity and tolerance in bacterial pathogens. Infect Immun 78:4977–4989. doi: 10.1128/IAI.00613-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beliaev AS, Klingeman DM, Klappenbach JA, Wu L, Romine MF, Tiedje JM, Nealson KH, Fredrickson JK, Zhou J. 2005. Global transcriptome analysis of Shewanella oneidensis MR-1 exposed to different terminal electron acceptors. J Bacteriol 187:7138–7145. doi: 10.1128/JB.187.20.7138-7145.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bouhenni R, Gehrke A, Saffarini D. 2005. Identification of genes involved in cytochrome c biogenesis in Shewanella oneidensis, using a modified mariner transposon. Appl Environ Microbiol 71:4935–4937. doi: 10.1128/AEM.71.8.4935-4937.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wan XF, Verberkmoes NC, McCue LA, Stanek D, Connelly H, Hauser LJ, Wu LY, Liu XD, Yan TF, Leaphart A, Hettich RL, Zhou JZ, Thompson DK. 2004. Transcriptomic and proteomic characterization of the fur modulon in the metal-reducing bacterium Shewanella oneidensis. J Bacteriol 186:8385–8400. doi: 10.1128/JB.186.24.8385-8400.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qiu D, Damron FH, Mima T, Schweizer HP, Yu HD. 2008. PBAD-based shuttle vectors for functional analysis of toxic and highly regulated genes in Pseudomonas and Burkholderia spp. and other bacteria. Appl Environ Microbiol 74:7422–7426. doi: 10.1128/AEM.01369-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dai J, Wei H, Tian C, Damron FH, Zhou J, Qiu D. 2015. An extracytoplasmic function sigma factor-dependent periplasmic glutathione peroxidase is involved in oxidative stress response of Shewanella oneidensis. BMC Microbiol 15:34. doi: 10.1186/s12866-015-0357-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He Z, Gentry TJ, Schadt CW, Wu L, Liebich J, Chong SC, Huang Z, Wu W, Gu B, Jardine P, Criddle C, Zhou J. 2007. GeoChip: a comprehensive microarray for investigating biogeochemical, ecological and environmental processes. ISME J 1:67–77. doi: 10.1038/ismej.2007.2. [DOI] [PubMed] [Google Scholar]
- 20.Zhou A, Baidoo E, He Z, Mukhopadhyay A, Baumohl JK, Benke P, Joachimiak MP, Xie M, Song R, Arkin AP, Hazen TC, Keasling JD, Wall JD, Stahl DA, Zhou J. 2013. Characterization of NaCl tolerance in Desulfovibrio vulgaris Hildenborough through experimental evolution. ISME J 7:1790–1802. doi: 10.1038/ismej.2013.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tu Q, Yu H, He Z, Deng Y, Wu L, Van Nostrand JD, Zhou A, Voordeckers J, Lee YJ, Qin Y, Hemme CL, Shi Z, Xue K, Yuan T, Wang A, Zhou J. 2014. GeoChip 4: a functional gene-array-based high-throughput environmental technology for microbial community analysis. Mol Ecol Resour 14:914–928. doi: 10.1111/1755-0998.12239. [DOI] [PubMed] [Google Scholar]
- 22.Mendoza-Vargas A, Olvera L, Olvera M, Grande R, Vega-Alvarado L, Taboada B, Jimenez-Jacinto V, Salgado H, Juárez K, Contreras-Moreira B, Huerta AM, Collado-Vides J, Morett E. 2009. Genome-wide identification of transcription start sites, promoters and transcription factor binding sites in E. coli. PLoS One 4:e7526. doi: 10.1371/journal.pone.0007526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.China EPA. 2002. Water and wastewater monitoring methods, 4th ed Chinese Environmental Science Publishing House, Beijing, China. [Google Scholar]
- 24.Won SH, Lee BH, Lee HS, Jo J. 2001. An Ochrobactrum anthropi gene conferring paraquat resistance to the heterologous host Escherichia coli. Biochem Biophys Res Commun 285:885–890. doi: 10.1006/bbrc.2001.5268. [DOI] [PubMed] [Google Scholar]
- 25.Thomas PE, Ryan D, Levin W. 1976. An improved staining procedure for the detection of the peroxidase activity of cytochrome-P-450 on sodium dodecyl-sulfate polyacrylamide gels. Anal Biochem 75:168–176. doi: 10.1016/0003-2697(76)90067-1. [DOI] [PubMed] [Google Scholar]
- 26.Qiu D, Wei H, Tu Q, Yang Y, Xie M, Chen J, Pinkerton MH Jr, Liang Y, He Z, Zhou J. 2013. Combined genomics and experimental analyses of respiratory characteristics of Shewanella putrefaciens W3-18-1. Appl Environ Microbiol 79:5250–5257. doi: 10.1128/AEM.00619-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bagdonas S, Ma LW, Iani V, Rotomskis R, Juzenas P, Moan J. 2000. Phototransformations of 5-aminolevulinic acid-induced protoporphyrin IX in vitro: a spectroscopic study. Photochem Photobiol 72:186–192. doi:. [DOI] [PubMed] [Google Scholar]
- 28.Konstantinidis KT, Serres MH, Romine MF, Rodrigues JL, Auchtung J, McCue LA, Lipton MS, Obraztsova A, Giometti CS, Nealson KH, Fredrickson JK, Tiedje JM. 2009. Comparative systems biology across an evolutionary gradient within the Shewanella genus. Proc Natl Acad Sci U S A 106:15909–15914. doi: 10.1073/pnas.0902000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pinchuk GE, Geydebrekht OV, Hill EA, Reed JL, Konopka AE, Beliaev AS, Fredrickson JK. 2011. Pyruvate and lactate metabolism by Shewanella oneidensis MR-1 under fermentation, oxygen limitation, and fumarate respiration conditions. Appl Environ Microbiol 77:8234–8240. doi: 10.1128/AEM.05382-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miyamoto K, Nakahigashi K, Nishimura K, Inokuchi H. 1991. Isolation and characterization of visible light-sensitive mutants of Escherichia coli K12. J Mol Biol 219:393–398. doi: 10.1016/0022-2836(91)90180-E. [DOI] [PubMed] [Google Scholar]
- 31.Baysse C, Matthijs S, Pattery T, Cornelis P. 2001. Impact of mutations in hemA and hemH genes on pyoverdine production by Pseudomonas fluorescens ATCC 17400. FEMS Microbiol Lett 205:57–63. doi: 10.1111/j.1574-6968.2001.tb10925.x. [DOI] [PubMed] [Google Scholar]
- 32.Yang H, Sasarman A, Inokuchi H, Adler J. 1996. Non-iron porphyrins cause tumbling to blue light by an Escherichia coli mutant defective in hemG. Proc Natl Acad Sci U S A 19:2459–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karlberg T, Lecerof D, Gora M, Silvegren G, Labbe-Bois R, Hansson M, Al-Karadaghi S. 2002. Metal binding to Saccharomyces cerevisiae ferrochelatase. Biochemistry 41:13499–13506. doi: 10.1021/bi0260785. [DOI] [PubMed] [Google Scholar]
- 34.Hansson MD, Karlberg T, Rahardja MA, Al-Karadaghi S, Hansson M. 2007. Amino acid residues His183 and Glu264 in Bacillus subtilis ferrochelatase direct and facilitate the insertion of metal ion into protoporphyrin IX. Biochemistry 46:87–94. doi: 10.1021/bi061760a. [DOI] [PubMed] [Google Scholar]
- 35.Dailey HA, Finnegan MG, Johnson MK. 1994. Human ferrochelatase is an iron-sulfur protein. Biochemistry 33:403–407. doi: 10.1021/bi00168a003. [DOI] [PubMed] [Google Scholar]
- 36.Bashyam MD, Hasnain SE. 2004. The extracytoplasmic function sigma factors: role in bacterial pathogenesis. Infect Genet Evol 4:301–308. doi: 10.1016/j.meegid.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 37.Kazmierczak MJ, Wiedmann M, Boor KJ. 2005. Alternative sigma factors and their roles in bacterial virulence. Microbiol Mol Biol Rev 69:527–543. doi: 10.1128/MMBR.69.4.527-543.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fernandez A, Lechardeur D, Derré-Bobillot A, Couvé E, Gaudu P, Gruss A. 2010. Two coregulated efflux transporters modulate intracellular heme and protoporphyrin IX availability in Streptococcus agalactiae. PLoS Pathog 6:e1000860. doi: 10.1371/journal.ppat.1000860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang J. 2012. Genetic redundancies and their evolutionary maintenance. Adv Exp Med Biol 751:279–300. doi: 10.1007/978-1-4614-3567-9_13. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.