Significance
Odor detection in the mouse nose is mediated by ∼1,000 different odorant receptors (ORs) and 14 trace amine-associated receptors (TAARs). Different OR combinations generate different odor perceptions. However, a few TAARs are associated with innate odor attraction or aversion, suggesting they signal through hard-wired neural circuits resistant to combinatorial receptor inputs. Contrary to this prediction, we find that different ligands for a given TAAR can be attractive or aversive, or instead neutral. In addition, some attractive and aversive odorants block one another’s behavioral effects. Odor blocking can occur without receptor antagonism in the nose and can require sensory input from one receptor. Thus, innate odor-induced behaviors can be context-dependent and modulated by interactions in the brain among signals derived from different receptors.
Keywords: olfaction, behavior, TAAR, odorants
Abstract
The mechanisms by which odors induce instinctive behaviors are largely unknown. Odor detection in the mouse nose is mediated by >1, 000 different odorant receptors (ORs) and trace amine-associated receptors (TAARs). Odor perceptions are encoded combinatorially by ORs and can be altered by slight changes in the combination of activated receptors. However, the stereotyped nature of instinctive odor responses suggests the involvement of specific receptors and genetically programmed neural circuits relatively immune to extraneous odor stimuli and receptor inputs. Here, we report that, contrary to expectation, innate odor-induced behaviors can be context-dependent. First, different ligands for a given TAAR can vary in behavioral effect. Second, when combined, some attractive and aversive odorants neutralize one another’s behavioral effects. Both a TAAR ligand and a common odorant block aversion to a predator odor, indicating that this ability is not unique to TAARs and can extend to an aversive response of potential importance to survival. In vitro testing of single receptors with binary odorant mixtures indicates that behavioral blocking can occur without receptor antagonism in the nose. Moreover, genetic ablation of a single receptor prevents its cognate ligand from blocking predator odor aversion, indicating that the blocking requires sensory input from the receptor. Together, these findings indicate that innate odor-induced behaviors can depend on context, that signals from a single receptor can block innate odor aversion, and that instinctive behavioral responses to odors can be modulated by interactions in the brain among signals derived from different receptors.
The mammalian olfactory system detects a multitude of volatile chemicals perceived as odors as well as social cues that elicit instinctive behaviors (1–3). The olfactory epithelium of the mouse contains ∼1,000 different ORs and 14 trace amine-associated receptors (TAARs), each expressed by a different subset of olfactory sensory neurons (4–13). An accessory olfactory structure, the vomeronasal organ, also detects social cues but using different receptors (1–3). ORs are used in a combinatorial fashion to detect structurally diverse odorants, allowing the discrimination of myriad odorants as having different scents (7, 14).
Like ORs, TAARs are evolutionarily conserved in vertebrates (15–17), suggesting that TAARs may have a distinct function. Moreover, sensory neurons expressing TAARs appear to comprise a separate lineage (18–20), and genes encoding ORs and TAARs have different nuclear locations (20), which may contribute to TAARs’ segregated expression in different neurons. At least some TAARs specifically recognize volatile amines, and two are required for behavioral attraction or avoidance to animal-derived amines (10, 21–23). In addition, one zebrafish TAAR ligand is aversive to fish (24), and one human TAAR detects an odorant aversive to humans as well as rotten fish (17, 25, 26). These observations have raised the possibility that the TAAR family may have an evolutionarily conserved ability to induce innate responses, particularly aversion. Alternatively, or in addition, TAAR conservation could derive from an ability for high-sensitivity detection of amines (23).
In mice, attractive and aversive responses can be either learned by association with other attractive or aversive stimuli or innate. Innate odor responses, such as aversive responses to predator odors (3, 21, 23, 27–32), are similar among individuals. The stereotyped nature of these responses suggests the existence of genetically determined neural circuits in which olfactory sensory input from a given receptor or combination of receptors has a high probability of eliciting a specific response. Their similarity among individuals also suggests that these responses may be immune to other olfactory inputs. For example, whereas changes in the combination of activated ORs can change a perceived odor, predator odor activation of a particular receptor may induce instinctive aversive behavior regardless of what other receptors are simultaneously activated by other odorants in the environment. One receptor in the vomeronasal organ has been linked to mating behavior (33). However, the only nasal receptors demonstrated to play a role in innate odor responses thus far are the two mouse TAARs mentioned above (22, 23), suggesting that further study of the TAAR family could provide added insight into the mechanisms underlying innate odor responses.
Here, we initially set out to investigate whether other mouse TAARs might also induce attraction or aversion. We conducted a high-throughput screen to identify additional mouse TAAR ligands and then tested individual ligands for the ability to cause attractive or aversive behavior in mice. Many, but not all, TAAR ligands elicited attraction or aversion. However, different ligands for the same TAAR often had different effects. Even for a TAAR known to be involved in innate attraction, one ligand could be attractive and another one neutral, despite similar affinity interactions with the receptor. One potential explanation was that innate behavioral responses to odors can be context-dependent and influenced by the combination of receptors activated by an odorant. Consistent with this model, some attractive and aversive odorants blocked each other’s behavioral effects. In vitro studies of ligand–receptor interactions and behavioral studies of receptor knockout (KO) mice indicated that the observed blocking effects can occur without receptor antagonism in the nose and can require sensory input from a single receptor.
Results
Identification of Additional TAAR Ligands.
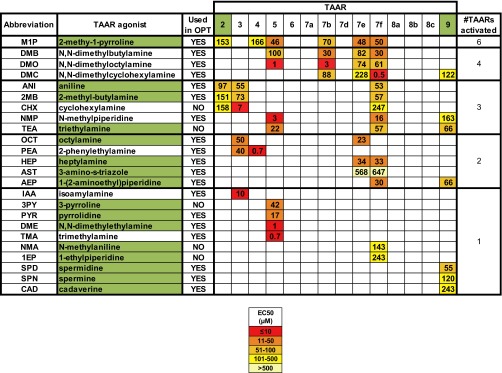
We first sought to identify additional mouse TAAR ligands beyond the small number already known (10, 34). To this end, we conducted a high-throughput chemical screen for TAAR activators using TAARs expressed individually in HEK293 tissue culture cells (Fig. 1A and Fig. S1). We identified 16 additional TAAR agonists, all volatile amines. Single odorants activated one to six TAARs, and, consistent with previous findings (10, 34), some TAARs responded to more than one odorant.
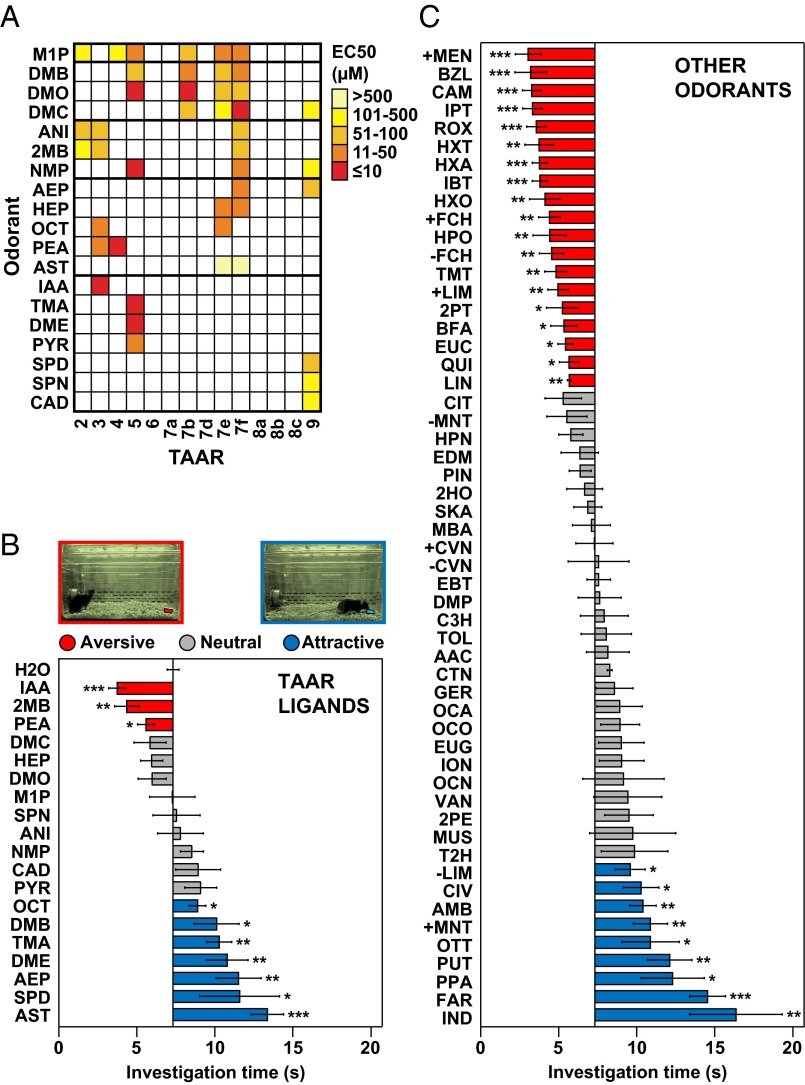
Fig. 1.
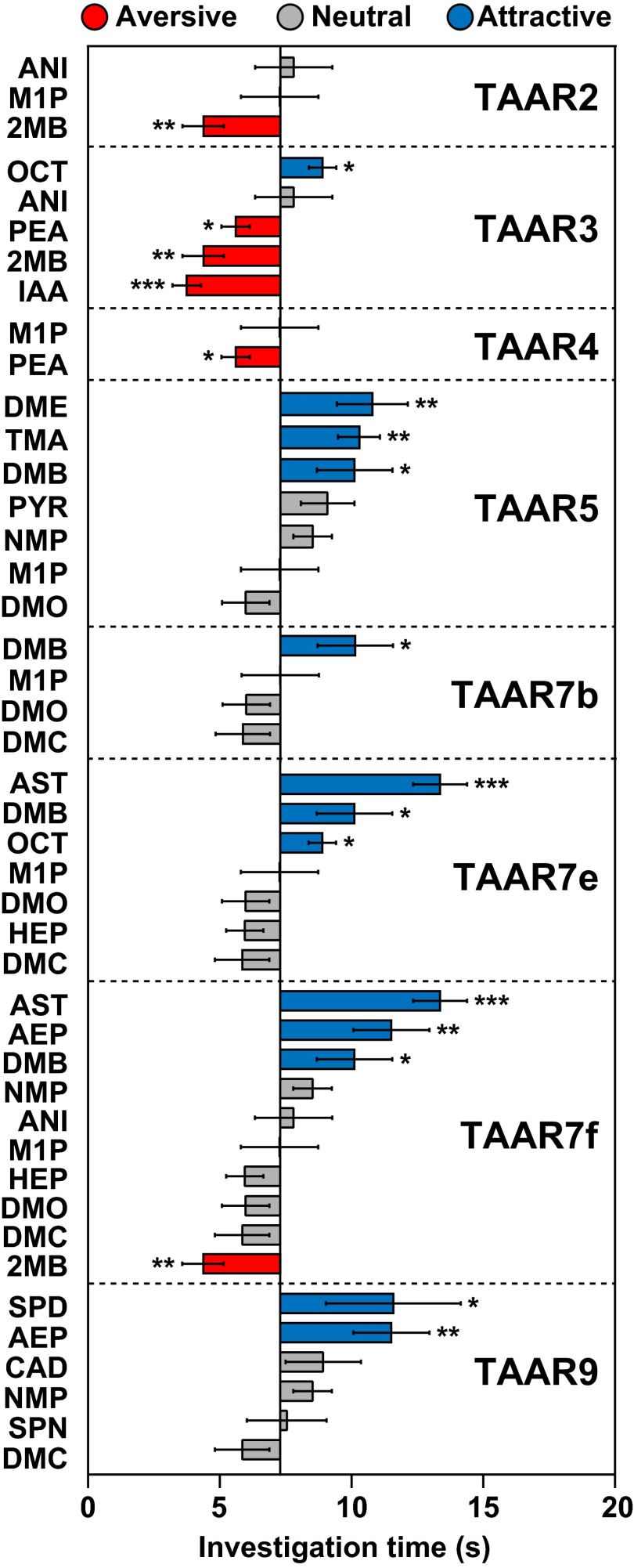
A number of TAAR ligands and other odorants elicit innate attraction or aversion. (A) Individual mouse TAARs were expressed in HEK293 cells and tested for responses to different odorants. Shown here are 19 of 24 odorants, all amines, which activated one or more TAARs (see Materials and Methods for full names). Colored boxes indicate EC50 values. Individual odorants activated one to six TAARs. (B and C) The olfactory preference test was used to assess the ability of 19 TAAR ligands (B) and 54 other odorants (C) to elicit attractive or aversive behavior. The test measures the time an animal spends investigating filter paper containing odorant or water during a 3-min period. Odorant abbreviations are shown at left (see Materials and Methods for full names). Bars indicate mean investigation time and error bars show SEM (n = 5–10 animals/odorant or water). Asterisks indicate responses significantly different from water (unpaired t test; two-tailed): *P < 0.05; **P < 0.01; ***P < 0.001. Bars are colored to indicate aversion (red), attraction (blue), or a neutral response (gray). Images of mice displaying aversive and attractive responses to filter paper (pseudocolored in red or blue) are seen above in B.
Fig. S1.
EC50 values of TAAR agonists. Individual mouse TAARs were expressed in HEK293 cells and tested for responses to different odorants. Shown here are EC50 values for 24 odorants that activated one or more TAARs, as determined in dose–response experiments. The abbreviation for each TAAR agonist is shown at left, followed by its chemical name. Those used in the olfactory preference test (OPT) are indicated. Some odorants activated only one TAAR, whereas others activated two to six TAARs. TAAR ligands newly identified in these studies are highlighted by green boxes.
Numerous TAAR Ligands, as Well as Other Odorants, Induce Innate Attraction or Aversion.
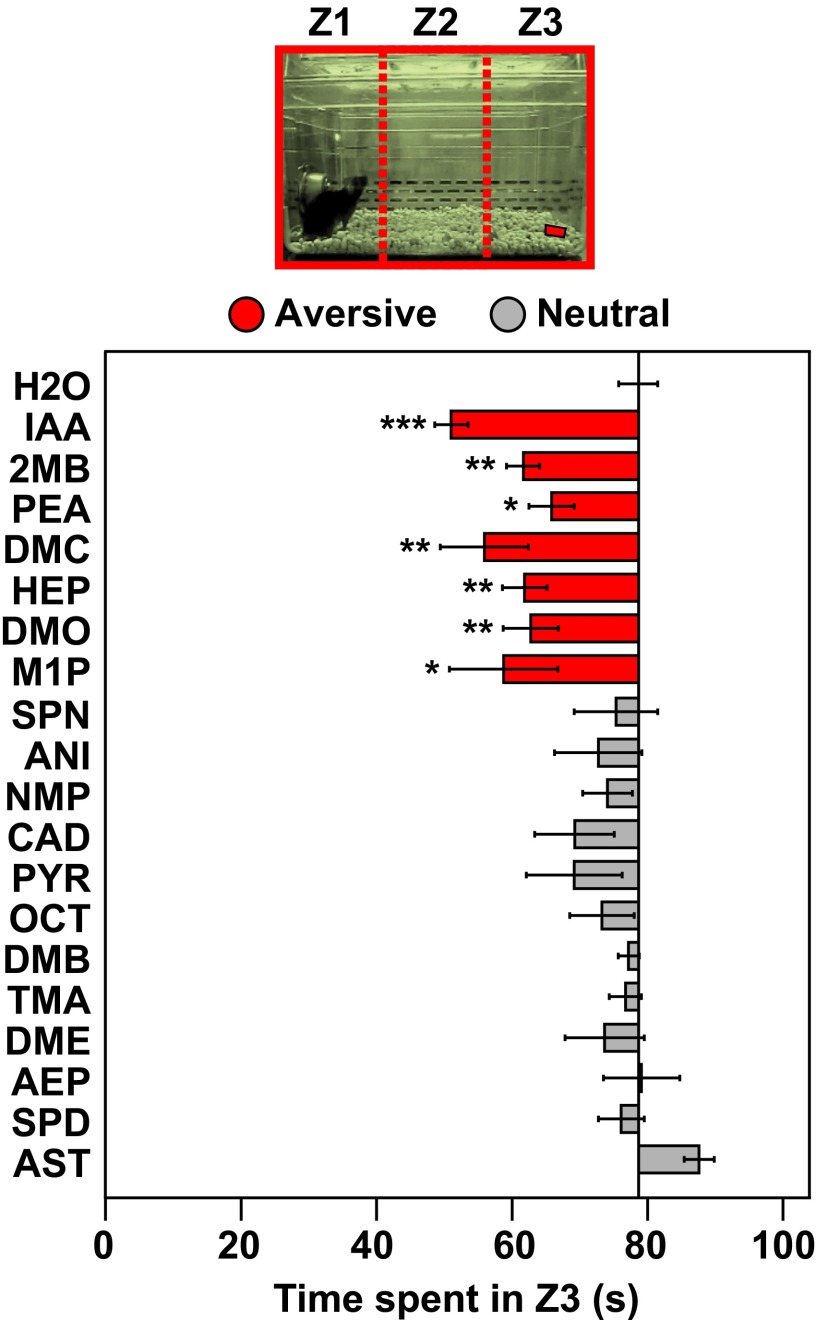
We next examined individual TAAR ligands for the ability to stimulate attractive or aversive behavior in naïve male mice. We used the olfactory preference test, which measures whether animals spend significantly more time investigating filter paper containing an odorant versus water (attraction) or significantly less time (aversion) (30). Strikingly, over half of TAAR ligands tested (10 of 19) induced a behavioral response, including three previously reported to do so (Fig. 1B and Fig. S2) (21–23). Seven ligands stimulated attraction, and three caused aversion. Aversion to the latter three ligands was also seen using an innate avoidance test, in which animals spent a significantly lower proportion of time in the third of the cage containing an aversive odorant (Fig. S3). Interestingly, four neutral TAAR agonists [N,N-dimethylcyclohexylamine (DMC), heptylamine (HEP), N,N-dimethyloctylamine (DMO), 2-methyl-1-pyrroline (M1P)] also elicited avoidance by this measure, indicating differences in the two assays. These differences presumably result from the analysis of close proximity to the odorant in the olfactory preference test versus analysis of time spent in a relatively large area of the cage in the innate avoidance test.
Fig. S2.
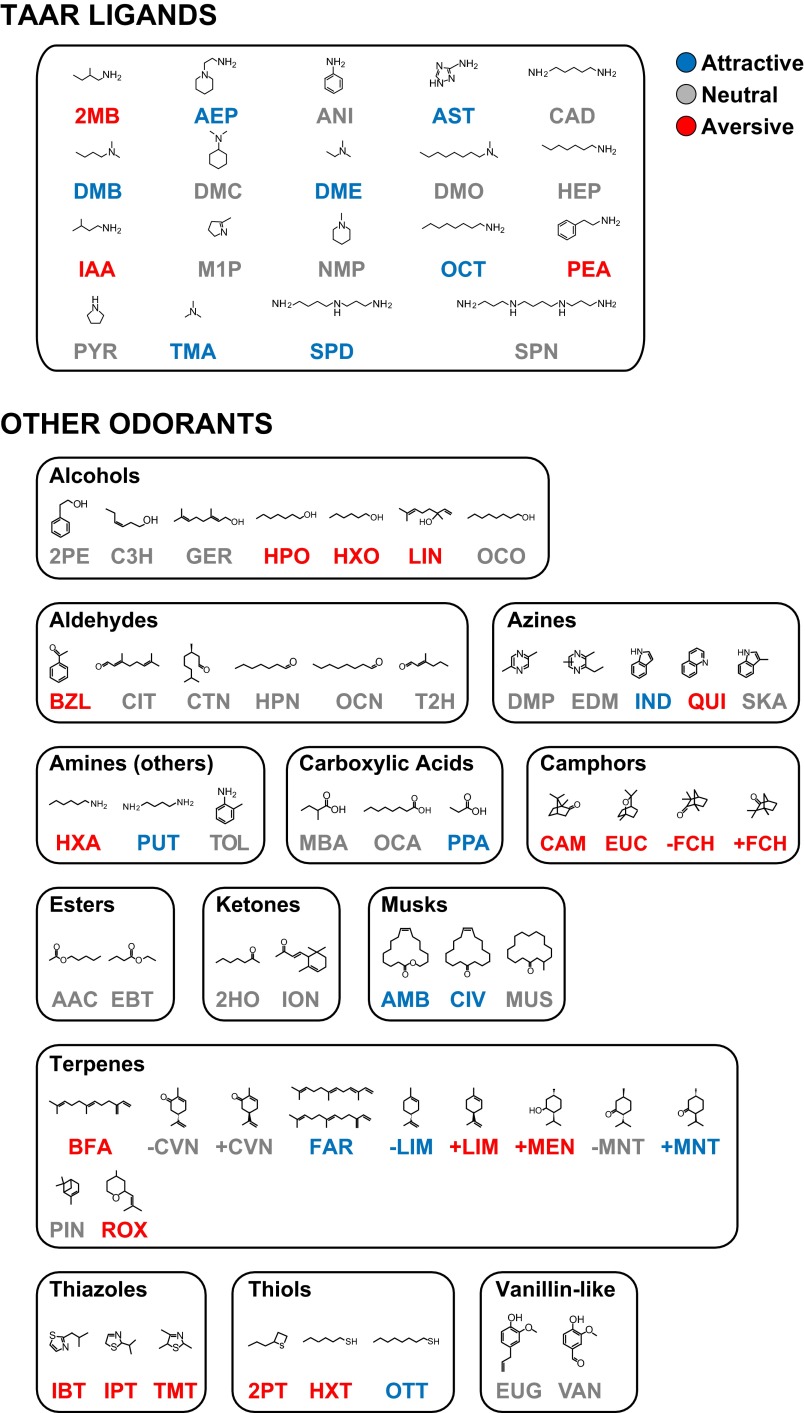
Structures and behavioral effects of TAAR ligands and other odorants. Shown here are the structures of 19 TAAR ligands and 54 other odorants. The other odorants are assorted into 13 structural classes. The abbreviation of each odorant is colored according to whether it elicited an attractive, neutral, or aversive behavioral response, as indicated (Fig. 1; see Materials and Methods for full names of odorants.)
Fig. S3.
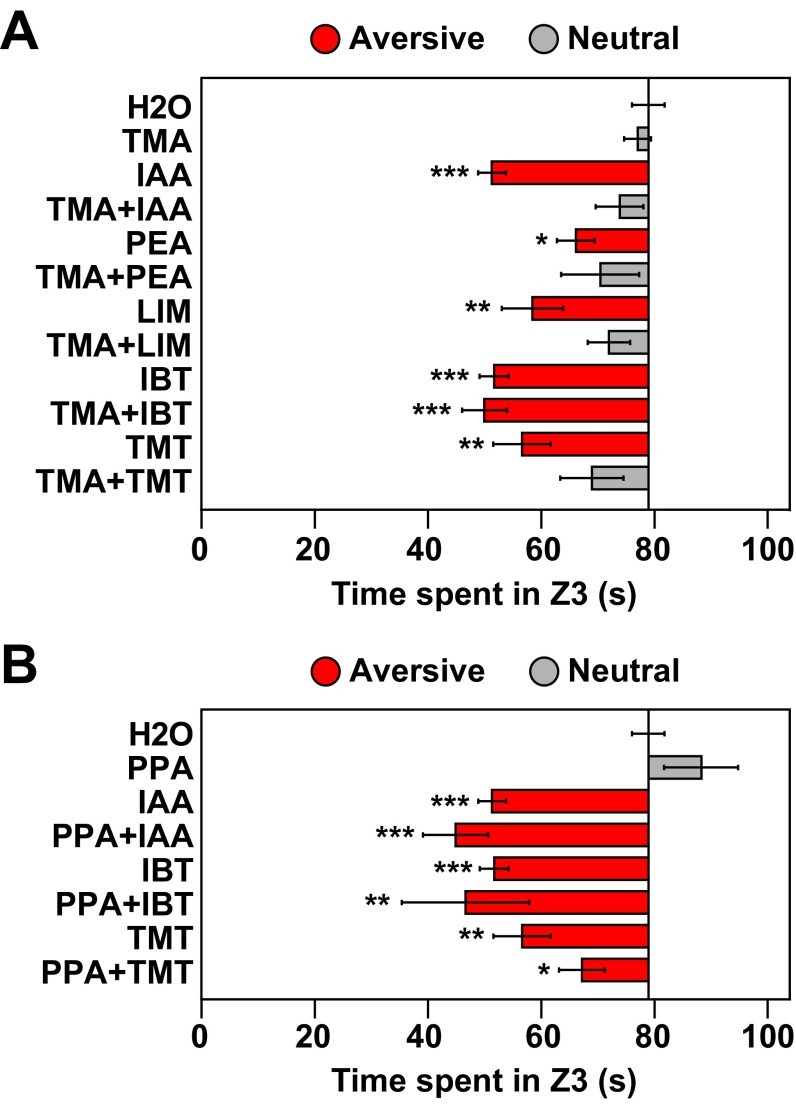
Innate avoidance test using TAAR ligands. Video recordings of mice exposed to TAAR ligands in the olfactory preference test (Fig. 1B) were additionally analyzed in a three-compartment innate avoidance test. Using Ethovision XT11 software, the test cage was divided into three zones of equal size [Zone 1 (Z1), Zone 2 (Z2), and Zone 3 (Z3)], and the time an animal spent in Z3 during a 3-min period was measured. Filter paper containing odorant or water was located in Z3. If an animal spent significantly less time in Z3 when exposed to an odorant versus water, it was considered an avoidance response. Bars indicate mean investigation time and error bars show SEM (n = 5–9 animals/odorant or water). Asterisks indicate responses significantly different from water (unpaired t test, two-tailed): *P < 0.05; **P < 0.01; ***P < 0.001. Bars are colored to indicate avoidance (red) or a neutral response (gray). Attractive responses were not seen with this assay. Seven TAAR agonists elicited aversion, including the three that caused aversion in the innate olfactory preference test (Fig. 1B).
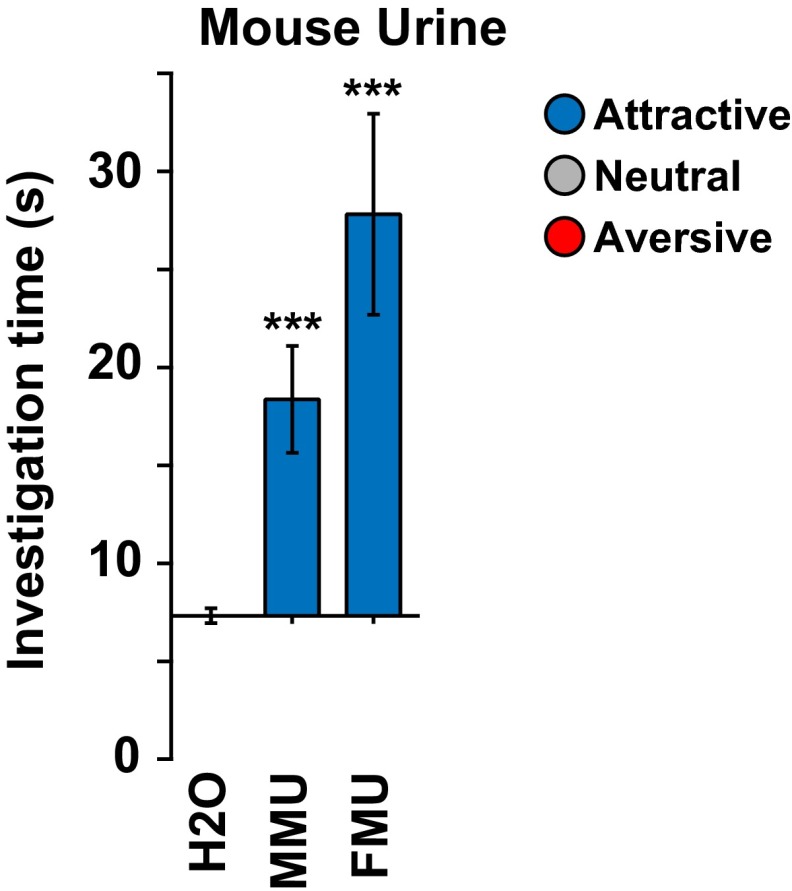
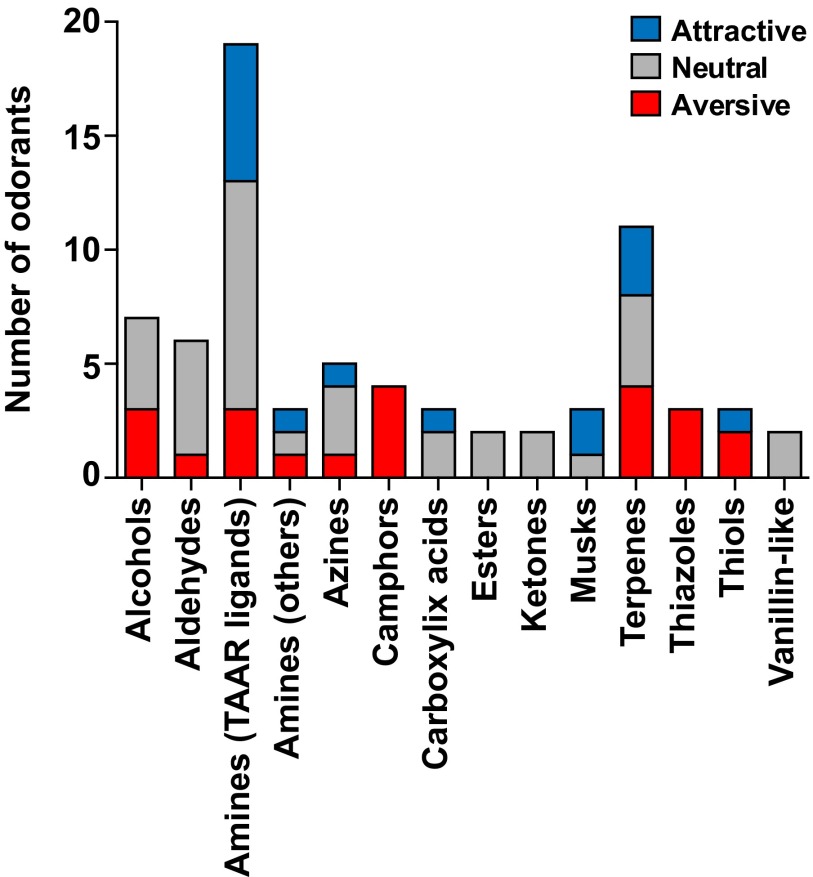
For comparison, we also tested 54 other odorants (Fig. 1C and Fig. S2). Based on their structures, we assigned these odorants to 13 odorant classes: alcohols, aldehydes, other amines, azines, camphors, carboxylic acids, esters, ketones, musks, terpenes, thiazoles, thiols, and vanillin-like compounds (14). Surprisingly, many of these odorants also elicited attraction (9 of 54) or aversion (19 of 54). Consistent with previous studies, the fear-inducing fox predator odor, 2,5-dihydro-2,4,5-trimethylthiazoline (TMT) caused aversion, and male and female mouse urine (MMU and FMU) both stimulated attraction (Fig. 1C and Fig. S4) (29–32, 35). Some odorant classes (azines, terpenes, thiols) included odorants with different behavioral effects whereas others (camphors and thiazoles) included only aversive odorants and still others (esters, ketones, and vanillin-like compounds) were uniformly neutral (Figs. S2 and S5).
Fig. S4.
Male and female mouse urine elicit attraction. The olfactory preference test was used to assess the response of naive adult male mice to urine from adult male or female mice (MMU or FMU) (see legend to Fig. 1). Filter paper containing mouse urine was placed at one end of the animal's cage at the beginning of the test and the observed responses compared with those obtained when animals were exposed to water. Bars indicate mean investigation time and error bars show SEM (n = 5–9 animals/urine or water). Asterisks indicate responses significantly different from the response to water (unpaired t test, two-tailed): ***P < 0.001. The mice showed attraction to both male and female urine.
Fig. S5.
Behavioral effects of odorants assigned to different structural classes. The 19 TAAR ligands used in these studies are all amines whereas the 54 other odorants can be assorted into 13 structural classes. This diagram shows the number of odorants in each class that elicited attraction (blue), aversion (red), or a neutral response (gray) using the olfactory preference test (see Fig. 1 for responses and Materials and Methods for names and abbreviations of odorants in each structural class). Odorants in some classes varied in their behavioral effects whereas the camphors and thiazoles tested uniformly caused aversion and the esters, ketones, and vanillin-like compounds all elicited a neutral response.
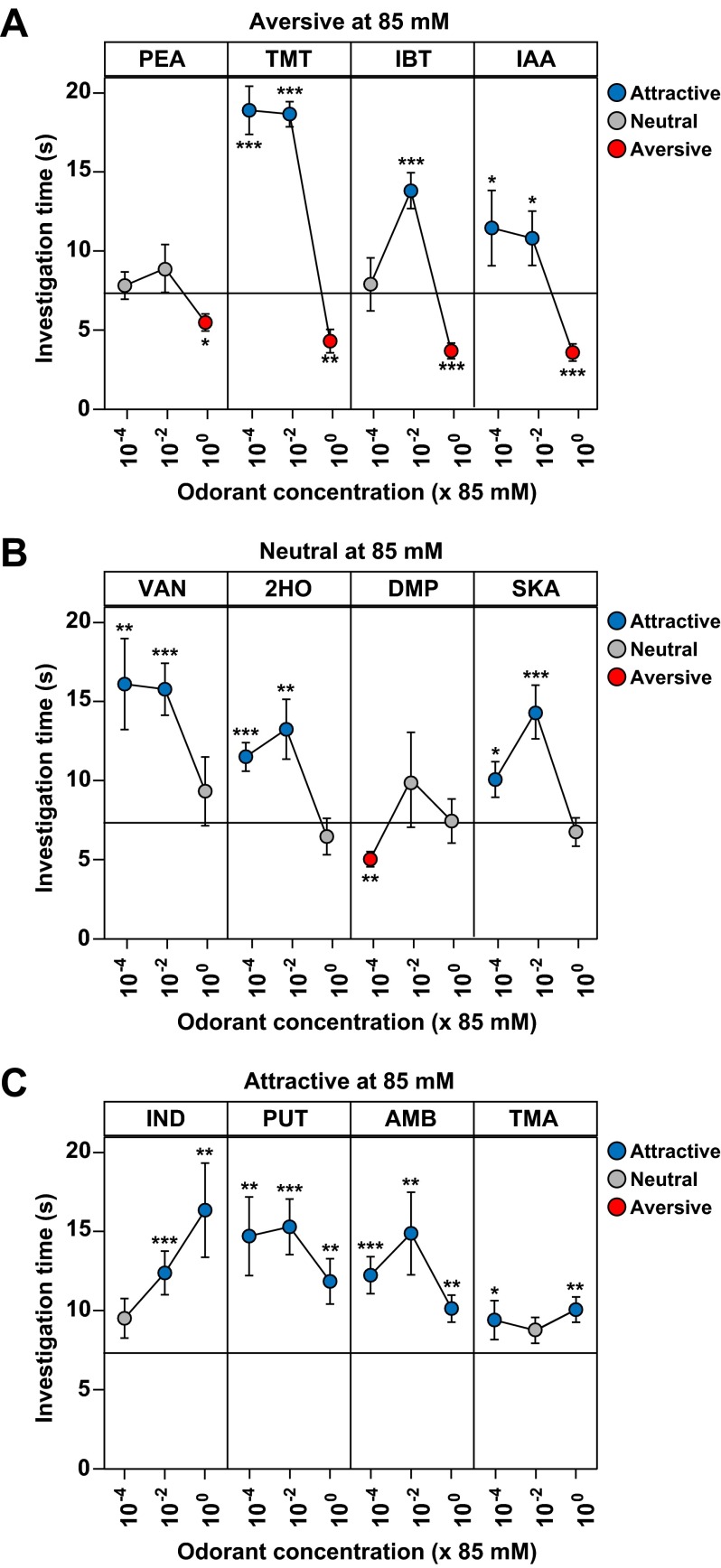
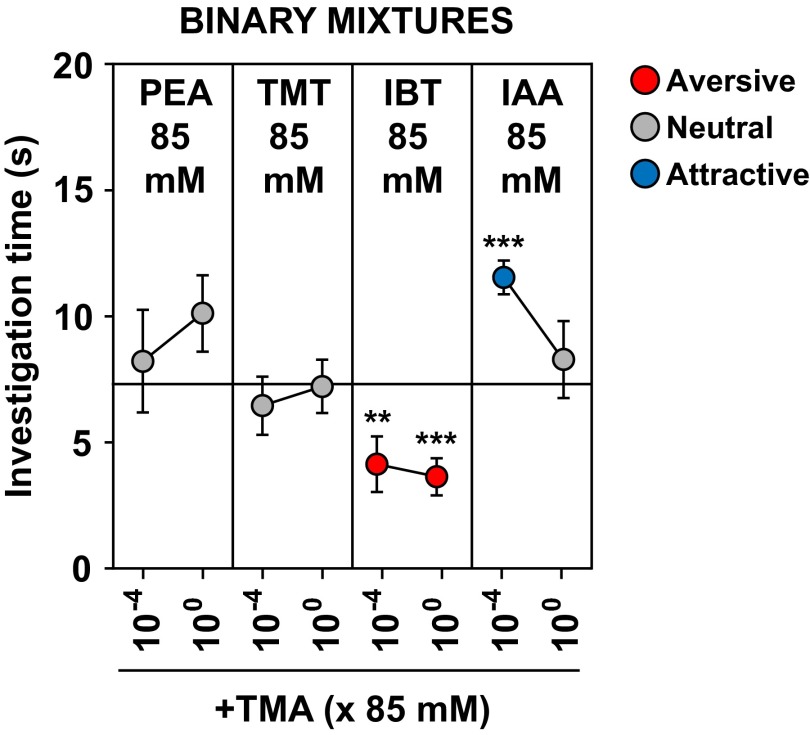
Further testing showed that the behavioral responses, whether they are attractive, aversive, or neutral, can be concentration-dependent (Fig. S6). Whereas a few odorants maintained their behavioral effects at lower concentrations, others lost their behavioral effects or induced a different behavior. For example, TMT and the aversive TAAR ligand, isoamylamine (IAA), both became attractive when their concentration was lowered by 102- or 104-fold (Fig. S6A).
Fig. S6.
Concentration effects on behavioral responses to odorants. The olfactory preference test (see legend to Fig. 1) was used to assess the effects of concentration on responses to specific odorants that were aversive (A), neutral (B), or attractive (C) at 85 mM, as seen in Fig. 1. Shown here are results obtained using 85 mM (10−0) and 102- and 104-fold dilutions. Balls and error bars show mean investigation time ± SEM (n = 5–9 animals/odorant or water) and are colored, as indicated, according to whether they reflected attraction, aversion, or a neutral response. Asterisks indicate responses significantly different from water (unpaired t test, two-tailed): *P < 0.05; **P < 0.01; ***P < 0.001.
These results are consistent with the idea that TAAR ligands, and likely TAARs, can elicit attractive or aversive behaviors. However, the results also reveal that TAAR ligands are not unique in this regard. A number of other odorants likely to be detected by ORs can also stimulate these behaviors. It is likely that the observed behavioral responses to many or all of these odorants are innate, not learned, because animals were first exposed to them during the behavioral assay. However, it cannot be excluded that animals had previous experience with some odorants, because of their association with, for example, littermates, bedding, or food.
Ligands for the Same TAAR Have Varied Behavioral Effects.
As already noted, innate behavioral responses to odorants are stereotyped and occur without prior learning. These characteristics suggest the involvement of specific olfactory receptors that feed into hard-wired neural circuits that may operate independently of signals generated by other receptors.
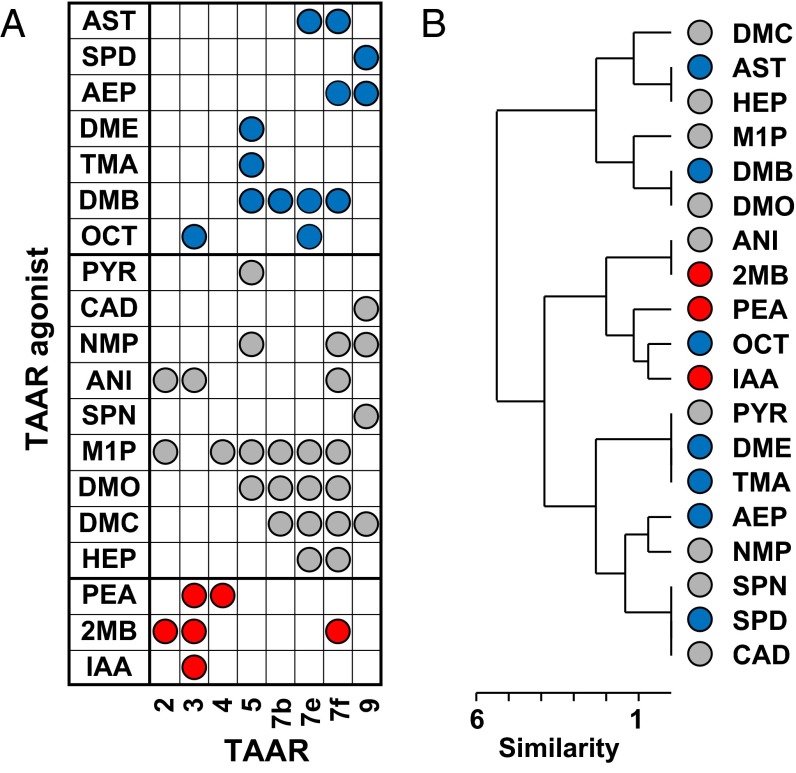
Our studies showed that ligands for the same TAAR, identified using a heterologous system, can have varied effects on behavior (Fig. 2A and Fig. S7). Different ligands for the same TAAR were attractive or neutral for four TAARs; aversive or neutral for two TAARs; and attractive, neutral, or aversive for another two TAARs.
Fig. 2.
Ligands for the same TAAR elicit different behaviors. (A) A graphical display emphasizes that different agonists for the same TAAR vary in their behavioral effects. Attractive, aversive, and neutral responses are indicated in blue, red, and gray, respectively. (B) Hierarchical clustering analysis of TAARs that detected different odorants failed to reveal a consistent correlation between TAARs activated by different odorants and the odorants' behavioral effects. Attractive, aversive, and neutral responses are indicated in blue, red, and gray, respectively.
Fig. S7.
Behavioral effects of different ligands for individual TAARs. Odorants detected by individual TAARs are shown together with the behavioral responses to those ligands seen in Fig. 1. Asterisks indicate responses significantly different from water (unpaired t test, two-tailed): *P < 0.05; **P < 0.01; ***P < 0.001. Eight TAARs recognized more than one odorant. In every case, ligands for the same TAAR elicited different behaviors. Aversive, neutral, and attractive behavioral responses are shown in different colors, as indicated.
Hierarchical clustering failed to reveal consistent correlations between the behavioral effects of TAAR ligands and the receptors they activated (Fig. 2B). Although a degree of clustering was seen for some ligands that caused the same behavioral response, ligands that elicited different responses were largely interspersed.
One possible explanation for the varied behavioral effects of ligands for a given TAAR is that the TAAR has no effect on behavior. For example, some ligands for that TAAR might induce innate aversion by activating ORs whose identities are unknown. However, this possibility is unlikely to be the case for TAAR5, because ablation of this receptor causes a loss of innate attraction to its ligand, trimethylamine (TMA) (20). Nonetheless, some TAAR5 ligands were attractive, whereas others were neutral. Moreover, attractive and neutral TAAR5 ligands had EC50 values of 0.7–100 µM and 1–46 µM, respectively, in in vitro experiments, suggesting that different affinity interactions between TAAR5 and its ligands were not responsible for the behavioral differences.
Another possible explanation for these results is that, like odor perceptions generated by ORs, innate behavioral responses to TAAR ligands can be context-dependent in that they can be influenced by the specific combination of TAARs, and possibly ORs, activated. In this model, signals from different receptors activated by the same odorant would have the ability to modulate one another’s behavioral effects.
Odorants Can Block Each Other’s Behavioral Effects.
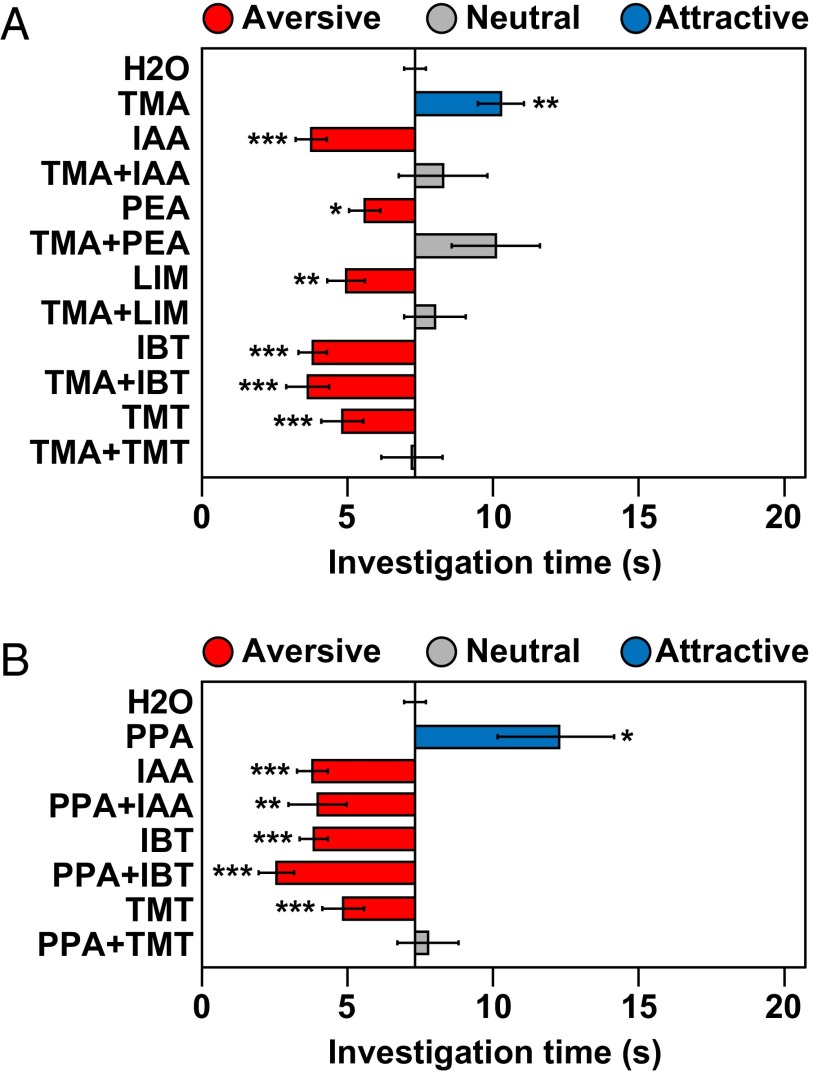
To test this model, we examined responses of mice to binary combinations of attractive and aversive odorants (Fig. 3A). We first combined the attractive TAAR5 ligand, TMA, with several different aversive odorants, including the fear-inducing predator odor, TMT.
Fig. 3.
Odorants can block one another’s effects on behavior. The effects of binary odorant mixtures on behavior were assessed using the olfactory preference test and compared with responses to single odorants in Fig. 1 (see legend to Fig. 1). Bars and error bars show mean investigation time ± SEM (n = 5–11 animals/odorant or water). Bars are colored, as indicated, according to whether odorants elicited attraction, aversion, or a neutral response. Asterisks indicate responses significantly different from water (unpaired t test, two-tailed): *P < 0.05; **P < 0.01; ***P < 0.001. The TAAR5 ligand TMA (A) and the attractive common odorant PPA (B) differed in their ability to block avoidance to specific aversive odorants.
TMA blocked aversion to two other TAAR ligands, 2-phenylethylamine (PEA) and IAA, as well as the aversive common odorant, (+)-limonene (+LIM or LIM). Surprisingly TMA also blocked aversion to TMT. In each case, the behavioral response to the paired odorants was neutral, indicating that the aversive odorants also blocked attraction to TMA. The neutral behavioral response was also seen using the innate avoidance test (Fig. S8A). These results are consistent with the idea that innate behavioral responses to odorants can be influenced by the combination of receptors activated in the nose and thus context-dependent.
Fig. S8.
Innate avoidance test of responses to odorant mixtures. Video recordings of mice exposed to single odorants versus binary odorant mixtures in the olfactory preference test (Figs. 1 and 3) were also analyzed in the three-compartment innate avoidance test using Ethovision XT11 software (see legend to Fig. S3). Bars indicate mean investigation time and error bars show SEM (n = 5–11 animals/odorant or water). Asterisks indicate responses significantly different from water (unpaired t test, two-tailed): *P < 0.05; **P < 0.01; ***P < 0.001. Bars are colored to indicate avoidance (red) or a neutral response (gray). No attractive responses were observed with this assay. As in the olfactory preference test (Fig. 3A), TMA blocked aversion to IAA, PEA, LIM, and TMT, but not IBT (A). However, PPA blocked aversion to TMT in the olfactory preference test (Fig. 3B) but not the innate avoidance test (B).
To examine whether odor blocking is concentration-dependent, we tested whether TMA could block aversion when present at 0.01% the concentration used in the above experiments (Figs. S6C and S9). Even at this much lower concentration, TMA induced an attractive response and also blocked aversion to IAA, PEA, and TMT.
Fig. S9.
Effects of odorant concentration on responses to odorant mixtures. The olfactory preference test (see legend to Fig. 1) was used to test whether lowering the concentration of TMA would affect its ability to block aversion to other odorants. Responses obtained using 85 mM each odorant (Fig. 3A) are compared here with responses obtained using 85 mM each aversive odorant, but a 104-fold lower concentration of TMA. Circles indicate mean investigation time and error bars show SEM (n = 5–11 animals/odorant or water). Circles are colored to indicate aversion (red), attraction (blue), or a neutral response (gray) compared with water exposure. Asterisks indicate responses significantly different from water (unpaired t test, two-tailed): **P < 0.01; ***P < 0.001. Lowering the concentration of TMA 10,00-fold did not alter its effect on aversion to PEA, TMT, IBT, or IAA.
We next tested binary odorant pairs containing an attractive common odorant, propionic acid (PPA) (Figs. 1B and 3B). Like TMA, PPA blocked aversion to TMT, and TMT blocked attraction to PPA. However, unlike TMA, PPA had no effect on aversion to IAA. Neither TMA nor PPA affected aversion to 2-isobutylthiazole (IBT). Similar results were obtained using the innate avoidance test, except that the mixture of PPA and TMT remained mildly aversive by that measure (Fig. S8B).
These results indicate that attractive and aversive TAAR ligands, as well as other odorants, can neutralize each other’s innate effects on behavior. They further indicate that a given attractive odorant, be it a TAAR ligand or common odorant, can block aversion to some odorants, but not others. In addition, different attractive odorants can block behavioral responses to different sets of aversive odorants, suggesting that odor blocking involves mechanisms more complex than a simple mutual cancellation of positive and negative signals. Unlike in Drosophila, where attractive and aversive odorants may have predictable mixture effects (36), the ability of specific pairs of odorants to block each other’s behavioral effects in mice is presently unpredictable.
Behavioral Blocking Can Occur Without Receptor Antagonism.
How do attractive and aversive odorants neutralize each other’s behavioral effects? One possibility is that these odorants do so by receptor antagonism in the nose. This mechanism would be consistent with previous reports of odorants that can act as antagonists to block the activation of certain ORs or nasal neurons by other odorants (37–39).
We investigated this possibility by exposing HEK293 cells expressing TAAR3, TAAR4, or TAAR5 to different concentrations of their cognate ligands together with varied concentrations of other odorants that neutralized behavioral responses to those ligands.
We found no evidence for receptor antagonism (Fig. 4 A–C). Although TMA blocks aversion to IAA and PEA, it does not block activation of TAAR3 by IAA or of TAAR4 by PEA (Fig. 4 A and B). Furthermore, whereas IAA, PEA, IBT, and TMT all block attraction to TMA, they do not affect TMA activation of TAAR5, which is required for attraction to TMA (22) (Fig. 4C). These results indicate that the behavioral blocking effects we observed are likely to be attributable to events in the brain, not the nose.
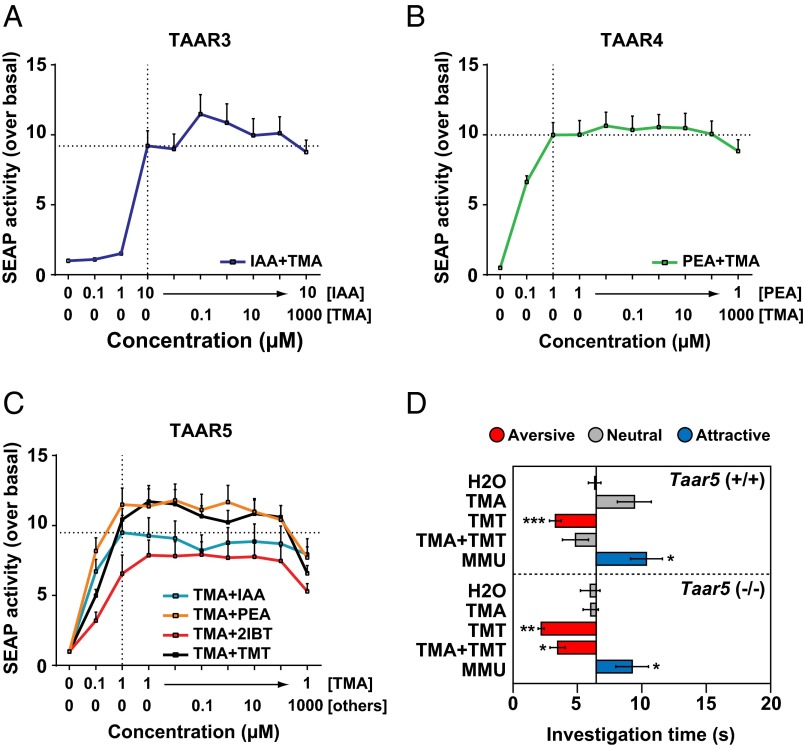
Fig. 4.
Odor blocking can occur within the brain. (A–C) HEK293 cells expressing TAAR3, TAAR4, or TAAR5 were tested with different concentrations of their respective ligands (IAA, PEA, or TMA) paired with varied concentrations of odorants that blocked the ligand's behavioral effects. Receptor activation was scored using a cAMP reporter assay that measures SEAP activity. SEAP activity over basal (no odorant) is shown in relative fluorescence units. Error bars indicate SEM (n = 8–24). Responses of TAARs to their cognate ligands were not significantly altered by pairing those ligands with another odorant (two-way ANOVA followed by post hoc Fisher LSD tests). (D) KO mice lacking TAAR5 [Taar5(−/−)] or their WT littermates [Taar5(+/+)] were tested for behavioral attraction or aversion to single or paired odorants using the olfactory preference test (see legend to Fig. 1) [n = 6–10 per condition for Taar5(−/−) mice and n = 5–11 per condition for Taar5(+/+) mice]. Asterisks indicate responses significantly different from responses to water: *P < 0.05; **P < 0.01; ***P < 0.001 (unpaired t test, two tailed). WhereasTaar5(+/+) mice showed a neutral response to TMT+TMA, Taar5(−/−) mice lacking Taar5 showed aversion to TMT+TMA, similar to that seen with TMT alone. Thus, TAAR5 is required for TMA to block aversion to TMT.
Behavioral Blocking by One Odorant Requires Signals from a Specific TAAR.
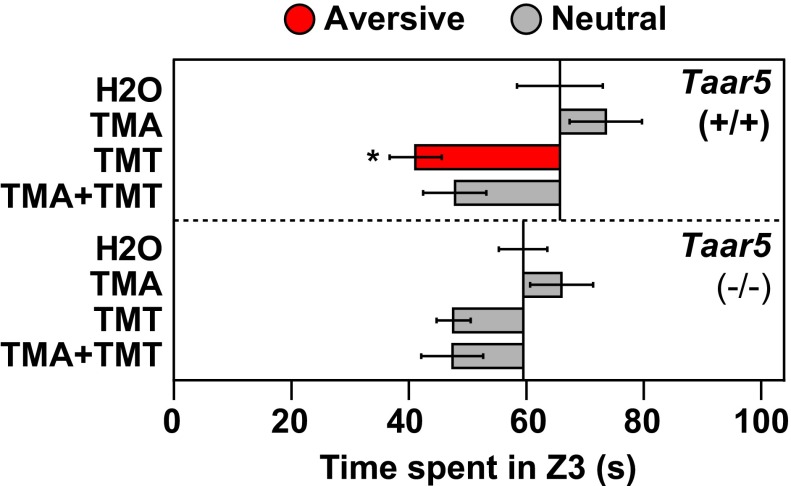
To further explore the mechanisms underlying odor blocking, we examined KO mice that lack TAAR5, which is required for TMA attraction (Fig. 4D). Like the isogenic WT mice used in the above experiments, Taar5 WT littermates [Taar5(+/+)] showed significant aversion to TMT but not to TMT plus TMA (TMT+TMA), even though the mice’s attraction to TMA was not statistically significant (P = 0.059). In contrast, Taar5 KO animals [Taar5(−/−)] showed aversion to both TMT and TMT+TMA. A similar trend was seen using the innate avoidance test, although aversion to TMT and TMT+TMA by the KO mice was not statistically significant using this test (Fig. S10).
Fig. S10.
Innate avoidance test in TAAR5 KO animals. The innate avoidance test (Fig. S3) was used to assess the behavioral effects of TMA, TMT, and the binary mixture TMA+TMT in KO mice lacking TAAR5. Bars indicate mean investigation time and error bars show SEM (n = 5–11 animals/odorant or water). Asterisks indicate responses significantly different from water (unpaired t test, two-tailed): *P < 0.05. Bars are colored to indicate avoidance (red), attraction (blue), or a neutral response (gray). Similar to the results obtained in the innate olfactory preference test (Fig. 4D), Taar5 WT(+/+) animals displayed avoidance to TMT, but not to the TMT+TMA mixture. In contrast, TAAR5 KO(−/−) animals did not display aversion to TMT or to TMT+TMA, although they showed aversion to both in the olfactory preference test.
These results indicate that TAAR5 is required for TMA to block TMT aversion. This finding clearly implies that TAAR5-derived sensory signals act within the brain to block TMT-induced signals from stimulating aversive behavior. The results also suggest that odor blocking can occur via inputs from a single receptor in the nose.
Discussion
Instinctive behavioral responses to odors can be important to individual survival and species perpetuation. In mice, these responses are involved in intraspecies social interactions as well as aversion to predator odors. The similarity of these responses among individuals suggests that they involve selected olfactory receptors and genetically programmed neural circuits, but the underlying mechanisms are largely unknown. Previous studies indicate that at least two TAARs in the mouse nose are involved in innate attraction or aversion to identified animal-derived amines. In addition, studies of fish and human TAARs suggest that TAARs may play an evolutionarily conserved role in innate behavioral responses to volatile amines, particularly avoidance.
To investigate this possibility, we conducted chemical screens to identify additional TAAR ligands and then tested individual ligands for behavioral effects. Numerous TAAR ligands, but not all, induced behavioral attraction or aversion and, unexpectedly, a number of common odorants also did so. However, different ligands for the same TAAR often had different behavioral effects, even in the case of one TAAR clearly associated with attractive behavior. One potential explanation was that the different ligands activated not only a given TAAR but also other receptors and that the constellation of activated receptors influenced behavioral output. Consistent with this idea, some attractive and aversive odorants neutralized each other’s behavioral effects. In vitro analyses of individual TAARs with these odorants indicated that the observed odor blocking can occur without receptor antagonism in the nose. Moreover, an attractive TAAR ligand no longer blocked aversion to a predator odor when the gene encoding its cognate receptor was ablated, indicating that sensory input from the receptor was required for the blocking effect.
Numerous TAAR Ligands and Other Odorants Induce Innate Responses.
Our previous studies identified TAARs as a second family of chemosensory receptors in the mouse nose and showed that TAAR expression patterns in the nose resemble those of ORs (10). By screening individual TAARs with >200 varied odorants, we identified ligands for several TAARs, all of which were volatile amines. Ligands for a few TAARs were reportedly present in mouse urine, a rich source of social cues, suggesting that TAARs might be involved in social cue recognition. Consistent with this idea, TAAR5 responded preferentially to mouse urine from adult males (10). One TAAR ligand we identified (TMA for TAAR5) was later found to be enriched in male mouse urine and another (PEA for TAAR4) elevated in some predator urines (21, 22). Moreover, TMA-induced attraction required TAAR5 and PEA-induced aversion required TAAR4 (22, 23). Together with the finding that individual fish and human TAARs recognize volatile amines aversive to those species (17, 24–26), these findings have suggested that the evolutionary conservation of the TAAR family may be linked to an ability to induce innate behaviors to volatile amines.
In the present studies, we conducted high throughput chemical screens on TAARs expressed in tissue culture cells and identified additional volatile amines that function as TAAR ligands. We then tested individual ligands for the ability to induce attraction or aversion in naïve mice. Of 19 TAAR ligands tested, seven caused attraction and three caused aversion. Altogether, odorants detected by eight different TAARs induced attractive or aversive behavior. Because some odorants activated more than one TAAR and each TAAR ligand might also activate members of the OR family, assignment of attractive or aversive functions to specific TAAR family members will require analyses of mice lacking specific Taar genes.
Little is known about the ability of various odorants to elicit attraction or aversion in mice. To compare the behavioral effects of TAAR ligands versus other odorants, we tested 54 other odorants with diverse structures and perceived scents in humans. These odorants belonged to 13 different structural classes. Unexpectedly, like TAAR ligands, many of these odorants elicited behavioral attraction or aversion. Odorants belonging to the same structural class varied in their behavioral effects, except that all those classified as camphors and thiazoles were aversive and those classified as esters, ketones, or vanillin-like were neutral.
These studies indicate that mice can respond to a number of TAAR ligands as well as some other odorants with behavioral attraction or aversion. It is highly likely that these behavioral results reflect innate rather than learned odor responses because animals were not knowingly exposed to the odorants before the behavioral assays. However, we cannot exclude the possibility that some were attributable to previously learned responses to environmental stimuli, such as those associated with littermates, parents, bedding, or food.
Evidence Against the Idea That One Receptor Leads Irrevocably to a Given Behavior.
As noted above, innate odor responses do not require learning and are similar among individuals. These features suggest the involvement of hard-wired neural circuits that require the activation of one genetically determined receptor or combination of receptors in the nose or vomeronasal organ. They further suggest that innate odor responses are likely to be resistant to other olfactory inputs. For example, predator odor activation of a specific receptor may cause instinctive aversive behavior regardless of what other receptors are simultaneously activated by other odorants in the immediate environment.
However, we found that ligands for the same TAAR can have different behavioral effects. Whereas one or more ligands for a given TAAR were attractive or aversive, one or more others were neutral. Our studies show that the behavioral response to an odorant can be concentration-dependent. For example, TMT and IAA were both aversive when tested at 85 mM but were attractive when tested at a 100- or 10,000-fold lower concentration. This finding suggests the possibility that ligands for the same receptor that were aversive (or attractive) versus neutral might simply have different affinity interactions with the receptor. Although this possibility cannot be excluded, the in vitro EC50 values for a neutral and attractive or aversive ligand for the same receptor were in some cases similar, suggesting that there could be other explanations.
Because a single TAAR ligand might activate not only multiple TAARs, but also ORs (18, 40), one other potential explanation was that, although input from a given TAAR might induce an innate behavior, the behavioral response is not actually resistant to all other receptor inputs. For example, an aversive and neutral ligand for the same TAAR might both activate additional receptors, but one or more of those activated by the neutral ligand blocks the aversive response, whereas those activated by the aversive ligand do not. In this model, the behavioral response resulting from activation of the specific TAAR would not be irreversible. Rather, like odor perceptions, it would depend upon the combination of receptors activated and whether any of the additional receptors can interfere with the behavioral response.
Innate Odor Responses Can Be Modulated by Combinatorial Odorant Inputs.
Consistent with the idea that inputs from different receptors can modulate each other’s behavioral effects, we found that binary mixtures of attractive and aversive odorants sometimes elicited a neutral response. The results of these experiments argue against the possibility that this effect is attributable to a simple mutual cancellation of attractive and aversive signals in the brain. The attractive TAAR5 ligand TMA and the common odorant PPA both blocked aversion to the predator odor TMT. However, even though PPA was investigated for a longer time than TMA when they were tested alone, TMA also blocked aversion to IAA, whereas PPA did not.
These results suggest a complex scenario in which the ability of individual attractive and aversive odorants to block one another’s innate behavioral effects is largely unpredictable. Future insight into the neural circuits that mediate odor attraction and avoidance may provide insight into the mechanisms and logic underlying these interactions. Interestingly, it has been reported that rose oil and the woody odorant, hinokitiol, can suppress the stress hormone response to TMT (41, 42), raising additional questions about the relationship between the ability of odorants to block the behavioral versus stress hormone components of an instinctive fear response.
Odor Blocking Can Occur in the Brain.
Previous studies indicate that odorants can act not only as agonists but also antagonists for ORs (37–39). Single odorants have been shown to block activation of a specific OR or nasal neurons by another odorant. However, our studies indicate that at least some of the odor-blocking effects we observed were not attributable to receptor antagonism in the nose. For example, TAAR5 is required for attraction to TMA and TAAR4 is required for aversion to PEA. However, whereas TMA and PEA neutralized each other’s behavioral effects, in in vitro studies, TMA did not block PEA activation of TAAR4 and PEA did not block TMA activation of TAAR5. These results suggest that odor blocking effects on behavior can result from events occurring in the brain.
Experiments using mice lacking TAAR5 are consistent with this idea. In Taar5 KO mice, TMA failed to block aversion to TMT, suggesting that sensory signals provided to the brain by TAAR5 are required for TMA to block aversion to TMT.
In summary, our findings indicate that the innate effect of an odorant on behavior can be context-dependent and subject to modification by other odorants. It is conceivable that signals derived from a single receptor can elicit innate behavioral attraction or aversion and involve hard-wired neural circuits. However, our studies indicate that these behavioral responses can be modulated by sensory inputs from other receptors via the interactions of signals derived from the different receptors within the brain. In short, innate behavioral output can be influenced by interactions within the brain among signals derived from different receptors in the nose.
Materials and Methods
Mice.
All procedures using animals were approved by the Fred Hutchinson Cancer Research Center Institutional Animal Care and Use Committee. Adult male C57BL/6J mice were obtained from The Jackson Laboratory. Taar5 KO mice [Taar5tm1(KOMP)Vlcg mice] were as described previously (20).
Odorants.
Chemicals of the highest purity available were purchased from Sigma-Aldrich, except for TMT, which was purchased from Contech. Mouse urine was collected while applying gentle abdominal pressure from a pool of three to five adult (8- to 10-wk-old) group-housed mice. Urine used in behavioral assays was always collected immediately before the assay.
The TAAR ligands used, with their abbreviations in parentheses, were as follows: amines: 2-methyl-1-pyrroline (M1P); N,N-dimethylbutylamine (DMB); N,N-dimethyloctylamine (DMO); N,N-dimethylcyclohexylamine (DMC); aniline (ANI); 2-methylbutylamine (2MB); N-methylpiperidine (NMP); 1-(2-aminoethyl)piperidine (AEP); heptylamine (HEP); octylamine (OCT); 2-phenylethylamine (β-phenylethylamine) (PEA); 3-amino-s-triazole (AST); isoamylamine (IAA); trimethylamine (TMA); N,N-dimethylethylamine (DME); pyrrolidine (PYR); spermidine (SPD); spermine (SPN); and cadaverine (CAD).
Other odorants used, which were assigned to different structural classes as indicated, were as follows:
Alcohols: 2-phenylethanol (2PE); geraniol (GER); heptanol (HPO); hexanol (HXO); linalool (LIN); octanol (OCO); cis-3-hexenol (C3H)
Aldehydes: benzaldehyde (BZL); citral (CIT); citronellal (CTN); heptanal (HPN); octanal (OCN); trans-2-hexenal (T2H)
Amines (other): hexylamine (HXA); o-toluidine (TOL); putrescine (PUT)
Azines: 2,5-dimethylpirazine (DMP); 2-ethyl-3,5 (6)-dimethylpirazine (EDM); indole (IND); quinoline (QUI); skatole (SKA)
Camphors: (±)-camphor (CAM); (−)-fenchone (−FCH); (+)-fenchone (+FCH); eucalyptol (EUC)
Carboxylic acids: 2-methylbutyric acid (MBA); octanoic acid (OCA); propionic acid (PPA)
Esters: amyl acetate (AAC); ethyl butyrate (EBT)
Ketones: 2-heptanone (2HO); α-ionone (ION)
Musks: ambrettolide (AMB); civetone (CIV); muscone (MUS)
Terpenes: β-farnesene (BFA); (−)-carvone (-CVN); (+)-carvone (+CVN); farnesene mixed isomers (alpha+beta) (FAR); (−)-limonene (−LIM); (+)-limonene (+LIM or LIM); (+)-menthol (+MEN); (−)-menthone (−MNT); (+)-menthone (+MNT); α-pinene (PIN); rose oxide (ROX)
Thiazoles: 2-isobutylthiazole (IBT); 2-isopropyl-4-5-dihydrothiazole (IPT); 2,5-dihydro-2,4,5-trimethylthiazoline (TMT)
Thiols: 2-propylthietane (2PT); hexanethiol (HXT); octanethiol (OTT);
Vanillin-like compounds: eugenol (EUG); vanillin (VAN)
TAAR Functional Assays.
Functional analysis of TAARs was conducted in HEK293 cells grown in 96-well plates using methods previously described (10) with the following modifications. Each well contained 50,000 HEK293 cells (American Type Culture Collection) cotransfected (using Lipofectamine2000; Invitrogen) with 20 ng each of the Taar plasmid and a cAMP response element, secreted alkaline phosphatase (CRE-SEAP), reporter plasmid (BD Biosciences). Cells were incubated for 24 h at 37 °C in serum-free media with or without test compounds and then for 2 h at 65–70 °C. An aliquot of supernatant from each well was incubated (5–20 min; room temperature) with an equal volume of 1.2 mM 4-methylumbelliferyl phosphate (Sigma-Aldrich) in 2 M diethanolamine bicarbonate, pH 10.0, and fluorescence was then measured with a CytoFluor4000 plate reader (Applied Biosystems). We used a three-parameters nonlinear regression analysis to calculate EC50 values from each dose–response curve by using GraphPad Prism (v6.04).
Olfactory Preference Test.
The olfactory preference test was conducted as described previously (30), with minor modifications (see below). Adult male C57BL/6J mice (8–14 wk of age; The Jackson Laboratory) or Taar5(−/−) or Taar5(+/+) mice (7–20 wk of age) were each assayed only once to avoid possible bias attributable to learning. Animals were habituated to the institutional animal facility for at least 5 d after arrival and maintained in group-housed conditions on a 12:12 h light:dark schedule (lights on at 0700 hours), with experiments performed between 1700 and 1900 hours. On the day of testing, mice were brought to the experimental room and habituated for 5–10 min. Mice were then separated into new cages (one mouse per cage) and habituated again for ≥10 min. In the single odorant experiments, exposure to olfactory stimuli was done by gently dropping a piece of filter paper (1.5 × 2 cm) impregnated with 50 µL of double-distilled water (H2O) or odorant (85 mM in H2O, equivalent to 4.25 µmol), into one end of the cage. For the binary odorant mixture exposures, 25 µL of each odorant (4.25 µmol) was placed on a different end of the filter paper for a final concentration of 85 mM of each odorant. For the dose–response experiments, dilutions were made in water. Behavioral tests were video recorded for 3 min with a Canon PowerShot ELPH300HS camera. The videos were either randomized and scored blind (to the odorant or genotype) by the experimenter or scored double blind by a nonexperimenter. The total olfactory investigation times were measured during the initial 3 min of odorant exposure.
Innate Avoidance Test.
The same videos used in the olfactory preference test were analyzed for the three-compartment assay. The test cage was divided into equally sized three compartments, and the duration that any part of mice stayed inside the third that contain a filter was scored by using Ethovision XT11 software (Noldus Information Technology). The videos were not analyzed if a mouse carried the filter paper out of the analyzed third.
Data Analysis.
Statistical analyses were done using GraphPad Prism (v6.04). Hierarchical clustering analysis was performed using the statistics software package PAST (v2.17c) (folk.uio.no/ohammer/past). For this analysis, we used a binarized version of the data in Fig. 1A, with activation given a value of 1, and no activation given a value of 0. Euclidean distances and Ward’s classification were used for the hierarchical clustering analysis.
Acknowledgments
We thank members of the L.B.B. laboratory for helpful comments and discussions. This work was supported by the Howard Hughes Medical Institute (L.B.B.) and National Institutes of Health (NIDCD) Grants F32 DC011699 (to K.-h.Y.) and R01 DC009324 (to L.B.B). L.B.B. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1605973113/-/DCSupplemental.
References
- 1.Buck LB, Bargmann C. Smell and taste: The chemical senses. In: Kandel E, Schwartz J, Jessell T, Siegelbaum S, Hudspeth AJ, editors. Principles of Neuroscience. McGraw-Hill; New York: 2012. pp. 712–742. [Google Scholar]
- 2.Munger SD, Leinders-Zufall T, Zufall F. Subsystem organization of the mammalian sense of smell. Annu Rev Physiol. 2009;71:115–140. doi: 10.1146/annurev.physiol.70.113006.100608. [DOI] [PubMed] [Google Scholar]
- 3.Stowers L, Kuo TH. Mammalian pheromones: Emerging properties and mechanisms of detection. Curr Opin Neurobiol. 2015;34:103–109. doi: 10.1016/j.conb.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buck L, Axel R. A novel multigene family may encode odorant receptors: A molecular basis for odor recognition. Cell. 1991;65(1):175–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- 5.Ressler KJ, Sullivan SL, Buck LB. A zonal organization of odorant receptor gene expression in the olfactory epithelium. Cell. 1993;73(3):597–609. doi: 10.1016/0092-8674(93)90145-g. [DOI] [PubMed] [Google Scholar]
- 6.Vassar R, Ngai J, Axel R. Spatial segregation of odorant receptor expression in the mammalian olfactory epithelium. Cell. 1993;74(2):309–318. doi: 10.1016/0092-8674(93)90422-m. [DOI] [PubMed] [Google Scholar]
- 7.Malnic B, Hirono J, Sato T, Buck LB. Combinatorial receptor codes for odors. Cell. 1999;96(5):713–723. doi: 10.1016/s0092-8674(00)80581-4. [DOI] [PubMed] [Google Scholar]
- 8.Godfrey PA, Malnic B, Buck LB. The mouse olfactory receptor gene family. Proc Natl Acad Sci USA. 2004;101(7):2156–2161. doi: 10.1073/pnas.0308051100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Niimura Y, Nei M. Extensive gains and losses of olfactory receptor genes in mammalian evolution. PLoS One. 2007;2(8):e708. doi: 10.1371/journal.pone.0000708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liberles SD, Buck LB. A second class of chemosensory receptors in the olfactory epithelium. Nature. 2006;442(7103):645–650. doi: 10.1038/nature05066. [DOI] [PubMed] [Google Scholar]
- 11.Hanchate NK, et al. Single-cell transcriptomics reveals receptor transformations during olfactory neurogenesis. Science. 2015;350(6265):1251–1255. doi: 10.1126/science.aad2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tan L, Li Q, Xie XS. Olfactory sensory neurons transiently express multiple olfactory receptors during development. Mol Syst Biol. 2015;11(12):844. doi: 10.15252/msb.20156639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saraiva LR, et al. Hierarchical deconstruction of mouse olfactory sensory neurons: From whole mucosa to single-cell RNA-seq. Sci Rep. 2015;5:18178. doi: 10.1038/srep18178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nara K, Saraiva LR, Ye X, Buck LB. A large-scale analysis of odor coding in the olfactory epithelium. J Neurosci. 2011;31(25):9179–9191. doi: 10.1523/JNEUROSCI.1282-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lindemann L, et al. Trace amine-associated receptors form structurally and functionally distinct subfamilies of novel G protein-coupled receptors. Genomics. 2005;85(3):372–385. doi: 10.1016/j.ygeno.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 16.Hussain A, Saraiva LR, Korsching SI. Positive Darwinian selection and the birth of an olfactory receptor clade in teleosts. Proc Natl Acad Sci USA. 2009;106(11):4313–4318. doi: 10.1073/pnas.0803229106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horowitz LF, Saraiva LR, Kuang D, Yoon KH, Buck LB. Olfactory receptor patterning in a higher primate. J Neurosci. 2014;34(37):12241–12252. doi: 10.1523/JNEUROSCI.1779-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pacifico R, Dewan A, Cawley D, Guo C, Bozza T. An olfactory subsystem that mediates high-sensitivity detection of volatile amines. Cell Reports. 2012;2(1):76–88. doi: 10.1016/j.celrep.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson MA, et al. Neurons expressing trace amine-associated receptors project to discrete glomeruli and constitute an olfactory subsystem. Proc Natl Acad Sci USA. 2012;109(33):13410–13415. doi: 10.1073/pnas.1206724109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoon KH, et al. Olfactory receptor genes expressed in distinct lineages are sequestered in different nuclear compartments. Proc Natl Acad Sci USA. 2015;112(18):E2403–E2409. doi: 10.1073/pnas.1506058112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferrero DM, et al. Detection and avoidance of a carnivore odor by prey. Proc Natl Acad Sci USA. 2011;108(27):11235–11240. doi: 10.1073/pnas.1103317108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Q, et al. Synchronous evolution of an odor biosynthesis pathway and behavioral response. Curr Biol. 2013;23(1):11–20. doi: 10.1016/j.cub.2012.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dewan A, Pacifico R, Zhan R, Rinberg D, Bozza T. Non-redundant coding of aversive odours in the main olfactory pathway. Nature. 2013;497(7450):486–489. doi: 10.1038/nature12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hussain A, et al. High-affinity olfactory receptor for the death-associated odor cadaverine. Proc Natl Acad Sci USA. 2013;110(48):19579–19584. doi: 10.1073/pnas.1318596110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang J, Pacifico R, Cawley D, Feinstein P, Bozza T. Ultrasensitive detection of amines by a trace amine-associated receptor. J Neurosci. 2013;33(7):3228–3239. doi: 10.1523/JNEUROSCI.4299-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wallrabenstein I, et al. Human trace amine-associated receptor TAAR5 can be activated by trimethylamine. PLoS One. 2013;8(2):e54950. doi: 10.1371/journal.pone.0054950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Apfelbach R, Blanchard CD, Blanchard RJ, Hayes RA, McGregor IS. The effects of predator odors in mammalian prey species: A review of field and laboratory studies. Neurosci Biobehav Rev. 2005;29(8):1123–1144. doi: 10.1016/j.neubiorev.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 28.Takahashi LK. Olfactory systems and neural circuits that modulate predator odor fear. Front Behav Neurosci. 2014;8:72. doi: 10.3389/fnbeh.2014.00072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pérez-Gómez A, et al. Innate predator odor aversion driven by parallel olfactory subsystems that converge in the ventromedial mypothalamus. Curr Biol. 2015;25(10):1340–1346. doi: 10.1016/j.cub.2015.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kobayakawa K, et al. Innate versus learned odour processing in the mouse olfactory bulb. Nature. 2007;450(7169):503–508. doi: 10.1038/nature06281. [DOI] [PubMed] [Google Scholar]
- 31.Root CM, Denny CA, Hen R, Axel R. The participation of cortical amygdala in innate, odour-driven behaviour. Nature. 2014;515(7526):269–273. doi: 10.1038/nature13897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brechbühl J, et al. Mouse alarm pheromone shares structural similarity with predator scents. Proc Natl Acad Sci USA. 2013;110(12):4762–4767. doi: 10.1073/pnas.1214249110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haga S, et al. The male mouse pheromone ESP1 enhances female sexual receptive behaviour through a specific vomeronasal receptor. Nature. 2010;466(7302):118–122. doi: 10.1038/nature09142. [DOI] [PubMed] [Google Scholar]
- 34.Ferrero DM, et al. Agonists for 13 trace amine-associated receptors provide insight into the molecular basis of odor selectivity. ACS Chem Biol. 2012;7(7):1184–1189. doi: 10.1021/cb300111e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weiss J, et al. Loss-of-function mutations in sodium channel Nav1.7 cause anosmia. Nature. 2011;472(7342):186–190. doi: 10.1038/nature09975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thoma M, Hansson BS, Knaden M. Compound valence is conserved in binary odor mixtures in Drosophila melanogaster. J Exp Biol. 2014;217(Pt 20):3645–3655. doi: 10.1242/jeb.106591. [DOI] [PubMed] [Google Scholar]
- 37.Spehr M, et al. Identification of a testicular odorant receptor mediating human sperm chemotaxis. Science. 2003;299(5615):2054–2058. doi: 10.1126/science.1080376. [DOI] [PubMed] [Google Scholar]
- 38.Oka Y, Omura M, Kataoka H, Touhara K. Olfactory receptor antagonism between odorants. EMBO J. 2004;23(1):120–126. doi: 10.1038/sj.emboj.7600032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peterlin Z, et al. The importance of odorant conformation to the binding and activation of a representative olfactory receptor. Chem Biol. 2008;15(12):1317–1327. doi: 10.1016/j.chembiol.2008.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tazir B, Khan M, Mombaerts P, Grosmaitre X. The extremely broad odorant response profile of mouse olfactory sensory neurons expressing the odorant receptor MOR256-17 includes trace amine-associated receptor ligands. Eur J Neurosci. 2016;43(5):608–617. doi: 10.1111/ejn.13153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matsukawa M, Imada M, Murakami T, Aizawa S, Sato T. Rose odor can innately counteract predator odor. Brain Res. 2011;1381:117–123. doi: 10.1016/j.brainres.2011.01.053. [DOI] [PubMed] [Google Scholar]
- 42.Murakami T, et al. Stress-related activities induced by predator odor may become indistinguishable by hinokitiol odor. Neuroreport. 2012;23(18):1071–1076. doi: 10.1097/WNR.0b013e32835b373b. [DOI] [PubMed] [Google Scholar]