Abstract
Genetic differences that specify unique aspects of human evolution have typically been identified by comparative analyses between the genomes of humans and closely related primates1, including more recently the genomes of archaic hominins2,3. Not all regions of the genome, however, are equally amenable to such study. Recurrent copy number variation (CNV) at chromosome 16p11.2 accounts for ~1% of autism cases4,5 and is mediated by a complex set of segmental duplications, many of which arose recently during human evolution. We reconstructed the evolutionary history of the locus and identified BOLA2 (bolA family member 2) as a gene duplicated exclusively in Homo sapiens. We estimate that a 95 kbp segment containing BOLA2 duplicated across the critical region ~282 thousand years ago (kya), one of the latest among a series of genomic changes that dramatically restructured the locus during hominid evolution. All humans examined carry one or more copies of the duplication, which nearly fixed early in the human lineage—a pattern unlikely to have arisen so rapidly in the absence of selection (p < 0.0097). We show that the duplication of BOLA2 led to a novel, human-specific in-frame fusion transcript and that BOLA2 copy number correlates with both RNA expression (r = 0.36) and protein level (r = 0.65), with the greatest expression difference between human and chimpanzee in experimentally derived stem cells. Analyses of 152 patients carrying a chromosome 16p11.2 rearrangement showed that >96% of breakpoints occur within the Homo sapiens-specific duplication. In summary, the duplicative transposition of BOLA2 at the root of the Homo sapiens lineage ~282 kya simultaneously increased copy number of a gene associated with iron homeostasis and predisposed our species to recurrent rearrangements associated with disease.
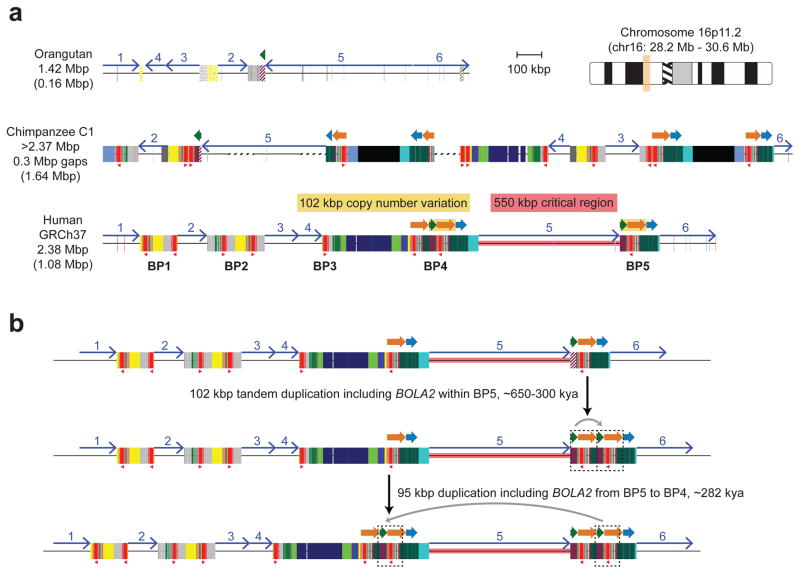
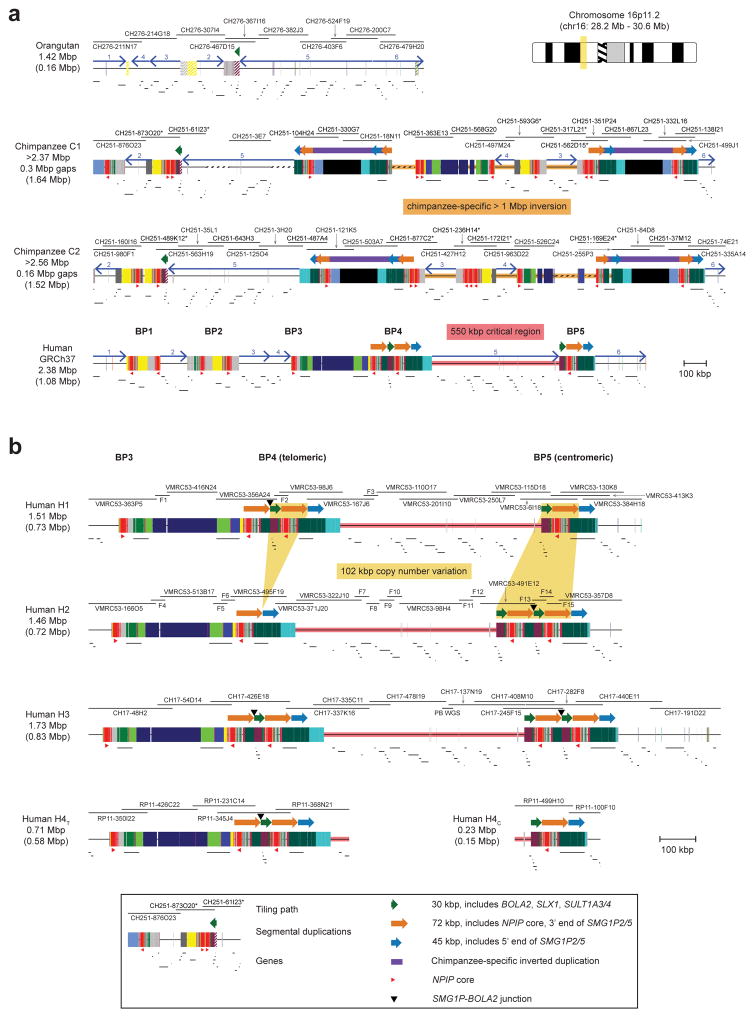
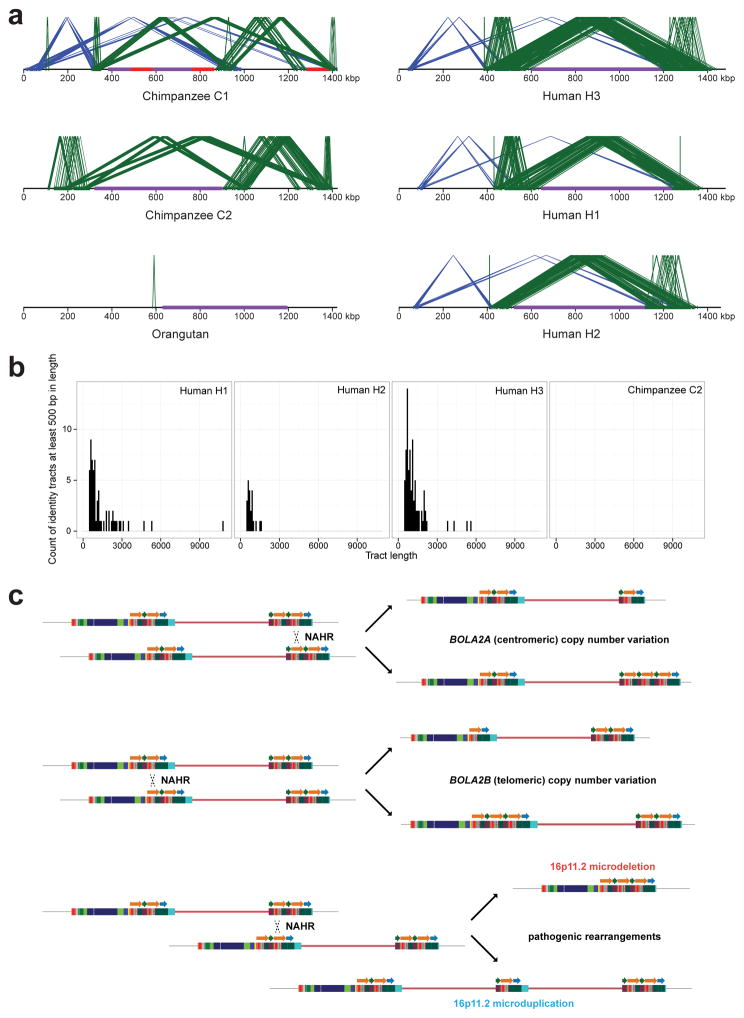
To reconstruct the evolutionary history of the chromosome 16p11.2 region, we generated complete, reference-quality genome sequence6 (Table S1) for one orangutan, two chimpanzee and three human haplotypes (Fig. 1a and Extended Data Fig. 1). Comparison with mouse establishes the orangutan configuration as ancestral. In both humans and chimpanzees, the region has been independently restructured, nearly doubling in length primarily by the differential accumulation of segmental duplications (Fig. 1a and Extended Data Fig. 1a). We find six inversions have occurred in the African great apes within chromosome 16p11.2 (Extended Data Figs. 2–4 and Tables S2, S3), a nonrandom clustering (p < 1 × 10−6), with breakpoints mapping near an ~20 kbp LCR16a (low copy repeat 16a) core duplicon. The core encodes a positively selected gene family (NPIP) that emerged on the human-African great ape lineage7. Only within the human lineage do large (>100 kbp) segmental duplications exist in a direct orientation flanking the autism critical region at breakpoint regions BP4 and BP5 (Extended Data Fig. 5a and Table S4)8, implying that susceptibility to large-scale CNV associated with disease4,5,9 arose specifically within the human species.
Figure 1. Comparative sequence analysis of chromosome 16p11.2 among apes and the evolution of BOLA2 duplications in humans.
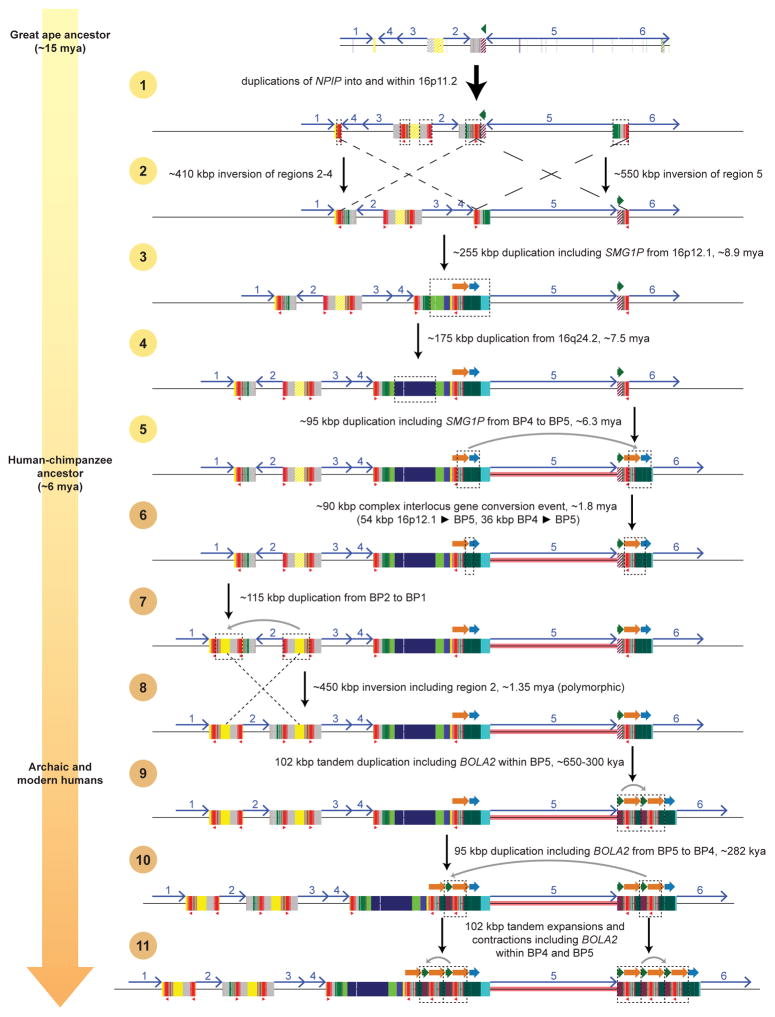
a) Schematic depicts the genomic organization of chromosome 16p11.2 for one orangutan and one chimpanzee haplotype along with the human reference haplotype (GRCh37 chr16:28195661–30573128). Blocks of segmental duplications within this locus mediate recurrent rearrangements in humans and have thus been defined as breakpoint regions BP1–BP5 (ref. 8). Colored boxes and thick arrows indicate the extent and orientation of segmental duplications (different colors denote duplicons from different ancestral genomic loci, and hashed boxes indicate sequence duplicated in humans but not in the species represented). Thin numbered arrows show orientations of gene-rich regions of unique sequence. Red triangles indicate locations and orientations of NPIP cores. Numbers (left) indicate the size of each haplotype, with the number of segmentally duplicated base pairs shown in parentheses. For chimpanzee, the size is a lower bound due to gaps (dotted line sections) and the contig not reaching unique region 1. Regions of human copy number variation (yellow highlight) occur on both sides of the critical region and involve the same 102 kbp unit: a 30 kbp block (green arrow) containing BOLA2, SLX1, and SULT1A3 and a 72 kbp block (orange arrow) harboring SMG1P. Expansion and contraction of this cassette underlie hundreds of kbp of structural diversity between human haplotypes. b) A model for the emergence of BOLA2 duplications during Homo sapiens evolution. Schematic depicts structural changes over time leading to the present-day human architecture. A full evolutionary model detailing the dynamic evolution of chromosome 16p11.2 in great apes is provided in the Supplementary Information and Extended Data Figs. 3, 4.
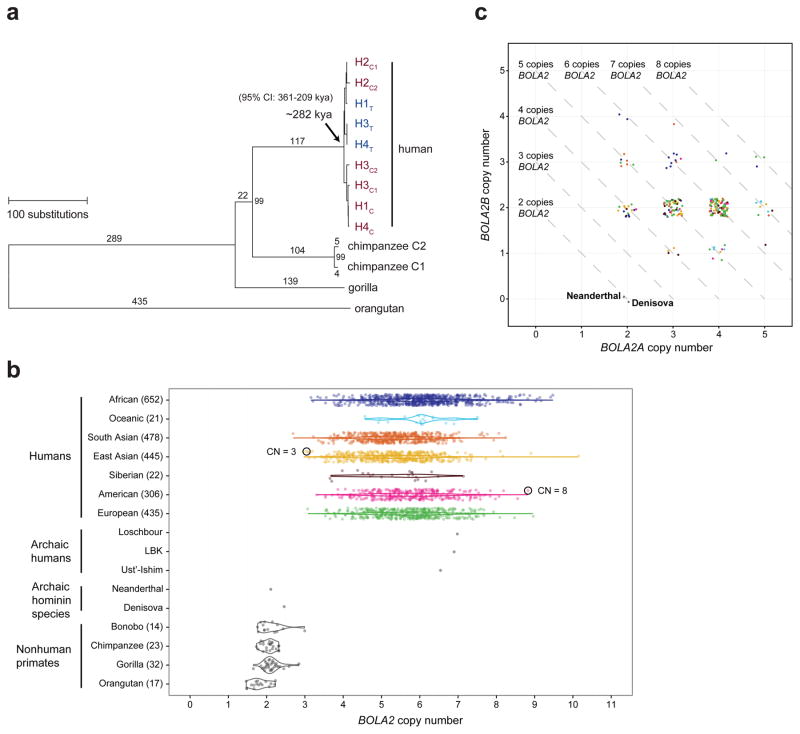
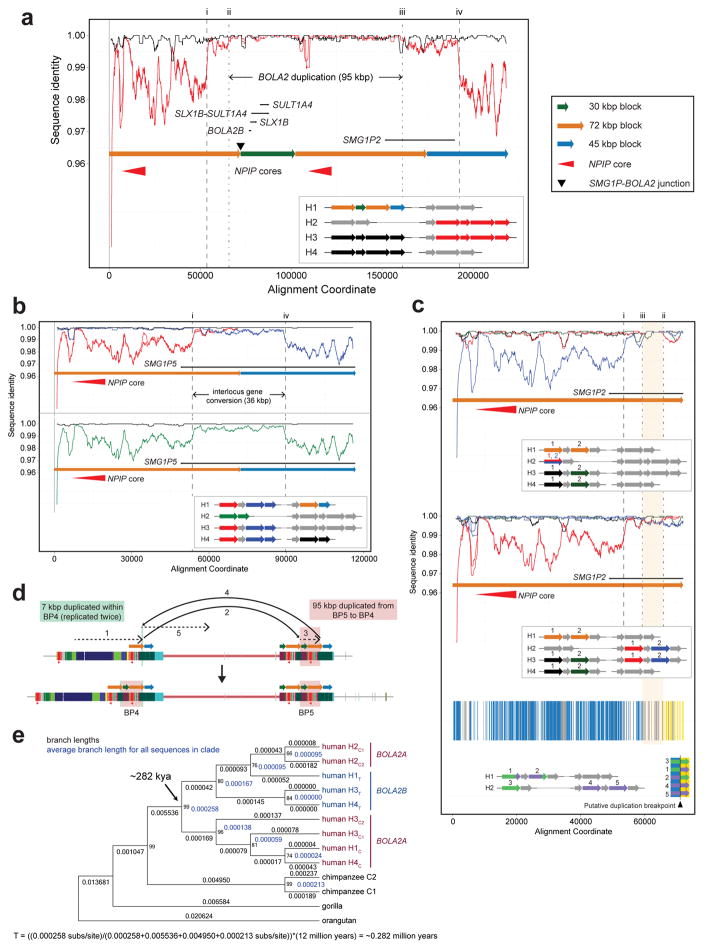
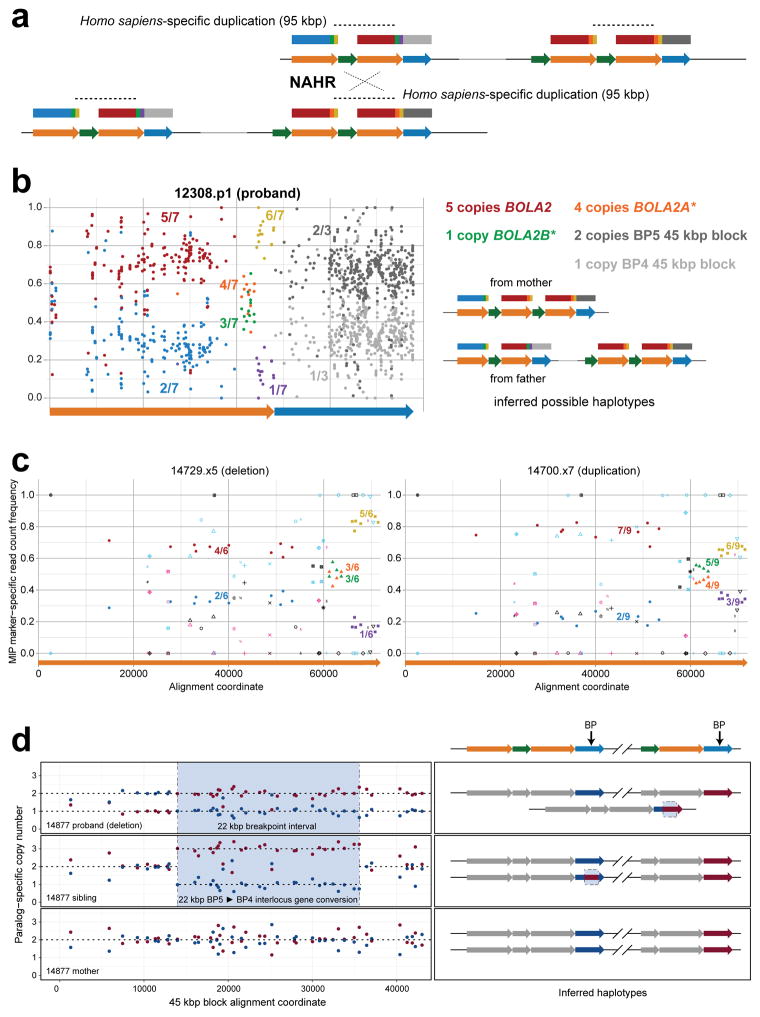
Structural differences between human haplotypes are largely restricted to integral changes in the copy number of a 102 kbp block within both the proximal and distal breakpoint regions (Extended Data Fig. 1b). This block is composed of two different segmental duplications originating from chromosome 16: a 72 kbp segment duplicated from chromosome 16p12.1 carrying NPIP and a portion of the SMG1 serine-threonine kinase gene (SMG1P) and a 30 kbp segment carrying three intact genes: BOLA2, SLX1 and SULT1A3 (Fig. 1a and Extended Data Fig. 1b). More than one dozen large-scale structural changes, including six duplicative transpositions (>830 kbp) from elsewhere on chromosome 16, are required to reconcile the organization of human and chimpanzee chromosome 16p11.2 (Extended Data Figs. 3, 4 and Table S3). Assuming a human–chimpanzee divergence time of 6 million years ago (mya)10 and a constant substitution rate, we estimate that a 95 kbp segment including BOLA2 duplicated across the critical region ~282 kya (95% confidence interval: 361–209 kya), around the time when Homo sapiens emerged as a species11 (Fig. 1b, Fig. 2a, Extended Data Fig. 6, and Tables S5–S7).
Figure 2. Homo sapiens-specific BOLA2 duplication and copy number diversity.
a) A phylogenetic tree representing the last interspersed segmental duplication from BP5 to BP4 in humans. The unrooted neighbor-joining tree was constructed from a 21,102 bp multiple sequence alignment including allelic, paralogous, and orthologous copies of the BOLA2-containing segmental duplications. Human taxon labels denote haplotypes and locations of different copies (telomeric, T, blue; centromeric, C, red, with C1 closer to the critical region than C2). The number of substitutions (above each branch) and bootstrap support (at nodes) are indicated. Timing estimates assume human-chimpanzee divergence 6 mya10. b) Diploid copy number estimates (points) for BOLA2 based on sequence read depth12 are shown for 2,359 humans, three archaic humans13,14, a Neanderthal2, a Denisovan3, and 86 nonhuman primates, with violin plots overlaid. c) Paralog-specific BOLA2 copy number genotypes (points, jittered around their integer values) were inferred from WGS read depth over informative markers for 222 individuals sequenced to high coverage. Colors correspond to different populations as in panel b.
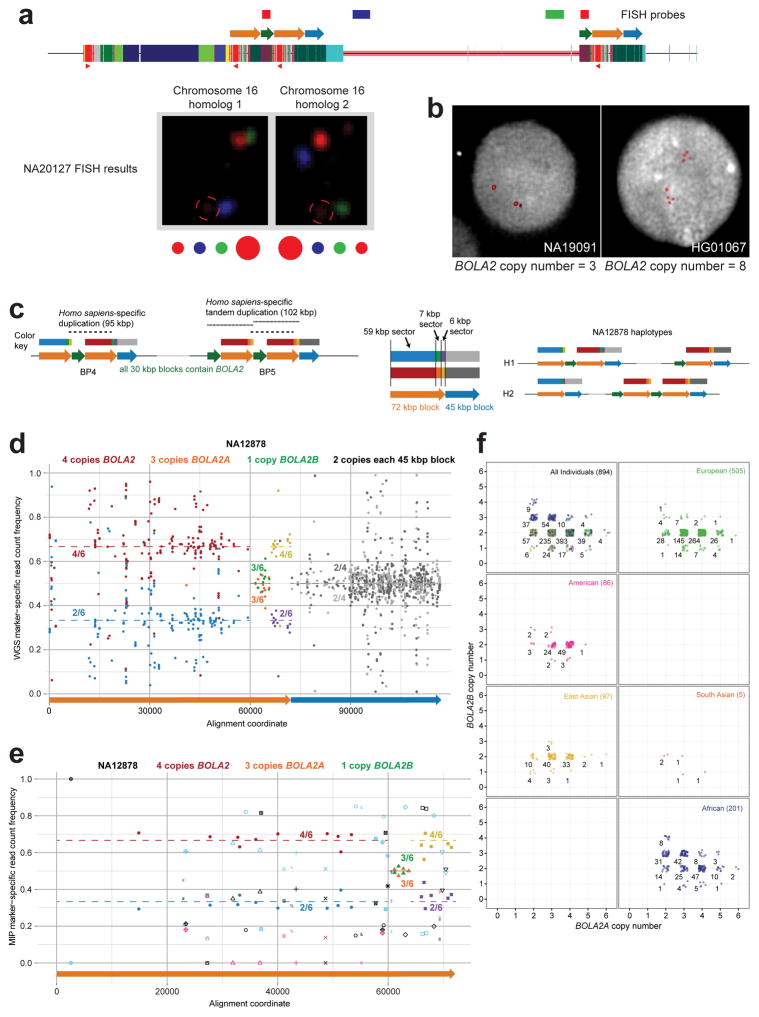
We examined copy number diversity12 of the duplicated genes mapping to the 102 kbp cassette—BOLA2, SLX1, and SULT1A3—in humans, archaic humans, and apes (Fig. 2b–c, Extended Data Fig. 7, and Tables S8–S10). We found that BOLA2 is duplicated in all Homo sapiens individuals examined, including archaic representatives of Neolithic and Mesolithic populations13, as well as the oldest sequenced archaic human, Ust’-Ishim, estimated to have lived 45 kya14. In sharp contrast, BOLA2 is single copy (i.e., diploid copy number = 2) in nonhuman primates and the archaic hominins Neanderthal2 and Denisova3 (Fig. 2b–c and Table S8), consistent with our phylogenetic point estimate of the duplication age. Human genomes contain from 3 to 8 diploid BOLA2 copies, with at least one copy of the distal duplicate BOLA2B (range = 1–4; mean and median = 2 copies) and at least two copies of the proximal ancestral BOLA2A (range = 2–5 copies; mean and median = 4 copies, Fig. 2c and Table S8).
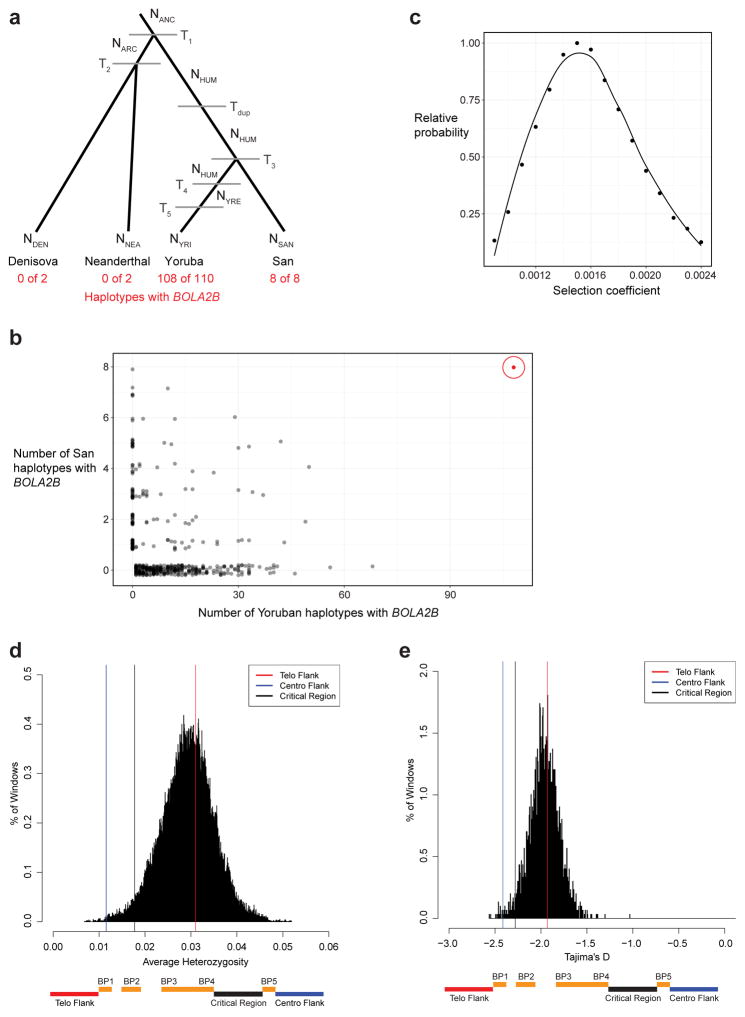
In light of its recent origin and its potential to promote disease-causing rearrangement, we considered it remarkable that 99.8% of humans carry four or more copies of this segment. Ancient humans such as Ust’-Ishim as well as some of the oldest branches of modern humans (e.g., San and Biaka pygmy15) typically carry five or six copies, indicating that it spread rapidly early in human history. We modeled various evolutionary scenarios by simulation based on the observed genotypes and a realistic model of human demographic history (Extended Data Fig. 8a), assuming neutral evolution16–18. The observed genotypes or genotypes with higher BOLA2B frequencies only in humans were improbable (p < 0.0097, Extended Data Fig. 8b), even when the duplication age parameter was varied by an order of magnitude. Scenarios incorporating recurrent duplication were also deemed unlikely (p < 0.0062). We next implemented a model incorporating the 282 kya age estimate but varying the selection coefficient (s) as an input parameter, yielding a maximum likelihood estimate of s = 0.0015 (Extended Data Fig. 8c). Interestingly, the unique ~550 kbp critical region flanked by BOLA2 duplications showed signatures consistent with a region under positive selection: the absence of archaic introgression19, low diversity (bottom 2.7%) and an excess of rare variants (Extended Data Fig. 8d–e).
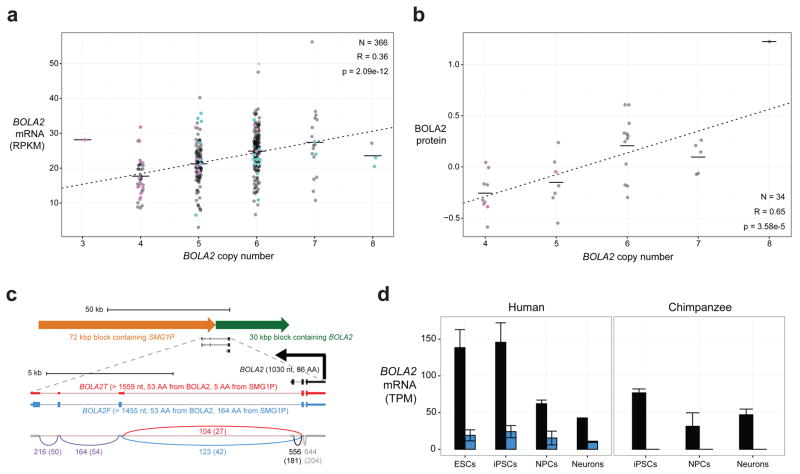
Because humans show extensive copy number variation, we assessed whether copy number correlated with mRNA and protein levels. We found a significant correlation between BOLA2 copy number and expression at the RNA level based on analysis of 366 lymphoblastoid cell lines (LCLs)20 (r = 0.36, p = 2.09 × 10−12, Fig. 3a and Tables S11, S12) and at the protein level based on analysis of whole-protein lysates from 34 LCLs (r = 0.64, p = 4.34 × 10−5, Fig. 3b and Tables S13, S14).
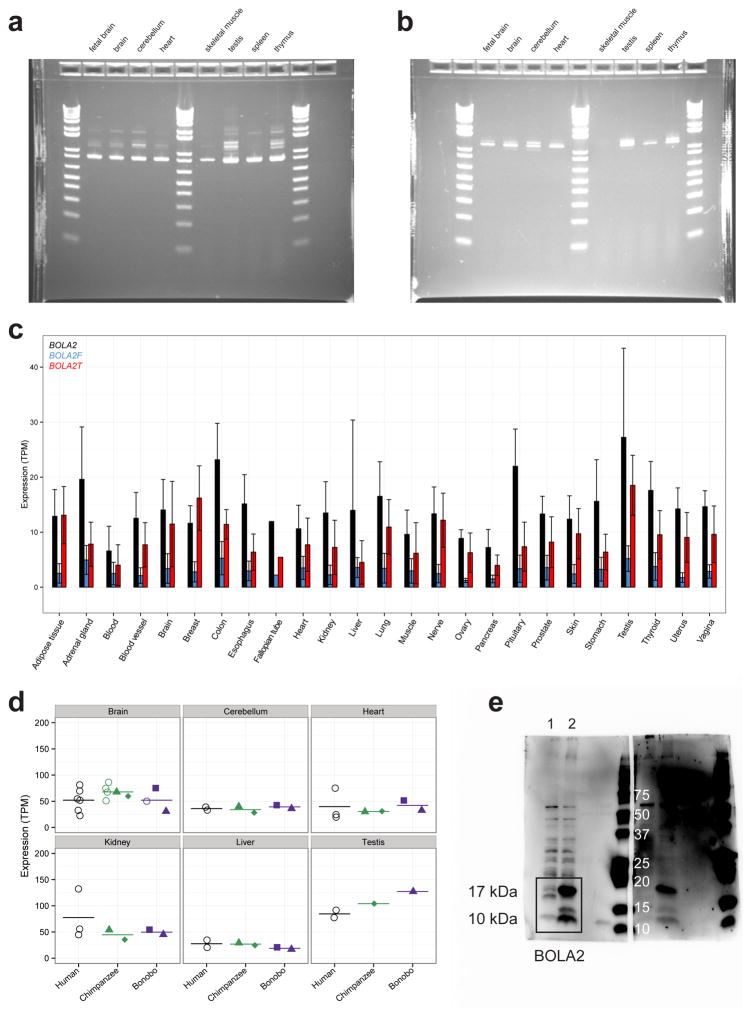
Figure 3. BOLA2 expression analyses.
a) Normalized BOLA2 mRNA expression quantifications20 in 366 LCLs from individuals genotyped for BOLA2 paralog-specific copy number. Points indicate expression levels and copy number (jittered) for each cell line, and horizontal lines show the mean expression level for each copy number. Line shows least squares regression. Point colors indicate BOLA2B copy number (pink = 1 copy, black = 2 copies, cyan = 3 copies). Groups with the same aggregate BOLA2 copy number but different combinations of paralog-specific copy number do not exhibit differential expression, consistent with both BOLA2A and BOLA2B producing mRNA. b) Plot layout is the same as in panel a, but data show BOLA2 protein expression quantified by Western blot densitometry on protein lysates from 34 LCLs. Though the sample size is small, no evidence indicates differential protein expression of distinct BOLA2 paralogs. c) BOLA2 gene models, predicted protein products, and support from RNA-seq data from human iPSCs. RT-PCR, cloning, and capillary sequencing experiments identified three BOLA2 isoforms: the canonical isoform (BOLA2, black) encoding an 86 residue protein and two fusion isoforms consisting of the first two exons from canonical BOLA2 fused with three exons from SMG1P. One of the fusion isoforms (BOLA2F, blue) maintains the BOLA2 ORF well beyond the fusion junction and is predicted to encode a 217 residue protein deriving primarily from SMG1P, whereas a third isoform (BOLA2T, red) contains a premature stop codon within the first SMG1P-derived exon. Numbers next to curved lines indicate mean counts of RNA-seq reads from two human iPSCs (two independent clones each) supporting each exon-exon junction, with standard errors in parentheses. d) RNA-seq quantification of BOLA2 mRNA expression through in vitro differentiation of primate iPSCs into neurons. Data from two human and two chimpanzee cell lines (two independent clones each, except for neurons) reveal significantly higher levels of BOLA2 transcripts in human iPSCs than in chimpanzee iPSCs and that BOLA2 RNA levels decrease through neuronal differentiation. Bar heights indicate mean expression levels for each species and differentiation stage in transcripts per million (TPM), with error bars showing standard errors. Bar colors correspond to different BOLA2 isoforms as in panel c. BOLA2 expression in human ESCs (two cell lines) is consistent with data from human iPSCs, suggesting the iPSC data accurately reflect BOLA2 expression at early stages in development.
We also performed RT-PCR and identified an alternate gene structure composed of the first two exons from BOLA2 joined with three novel 3′ exons from an older segmental duplication containing SMG1P (Fig. 3c). This fusion isoform contains an open reading frame (ORF) predicted to encode a 217 residue protein including 53 residues from BOLA2 and 164 residues from SMG1P. Both canonical and fusion transcripts are co-expressed in a wide variety of tissues and developmental stages (Extended Data Fig. 9). Although the predicted fusion protein cannot be detected by existing antibodies, it is interesting that ribosome profiling data provide evidence that the mRNA is translated (Table S15). Importantly, since the ancestral BOLA2 at BP5 lacked the SMG1P duplication downstream, the origin of the fusion product must have coincided with the juxtaposition of BOLA2 and SMG1P by the tandem 102 kbp segmental duplication ~650–300 kya at BP5. We conclude that this fusion isoform is Homo sapiens-specific.
BOLA2 was previously identified as one of the top 50 genes differentially expressed between humans and nonhuman apes in induced pluripotent stem cells (iPSCs)21, implying that this gene might be particularly relevant early in development. Based on our characterization of the different BOLA2 isoforms, we revisited this observation by quantifying BOLA2 mRNA levels by RNA-seq in human and chimpanzee iPSCs, iPSC-derived neural progenitor cells (NPCs), and eight-week-old neurons. Remarkably, we found the greatest differences in canonical BOLA2 expression at the iPSC state (2-fold) and to a lesser extent in NPCs (1.5-fold) (Fig. 3d and Table S16). Quantification of BOLA2 expression in two primary human embryonic stem cell (ESC) lines revealed transcript levels comparable to human iPSCs (Fig. 3d and Table S16). In contrast, examination of a panel of adult tissues22 revealed no substantial differences in BOLA2 mRNA levels between human and chimpanzee (Extended Data Fig. 9d). As expected, expression of the fusion BOLA2-SMG1P transcript was detected exclusively in human.
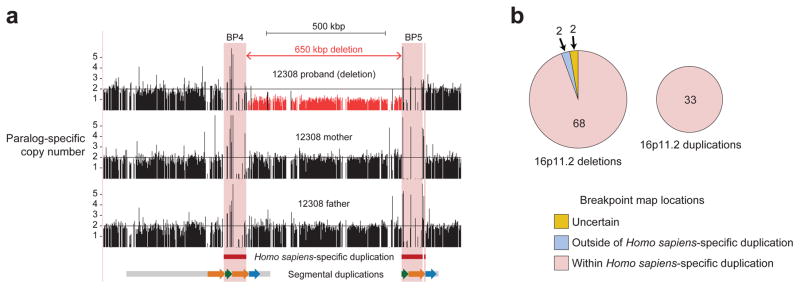
The duplication of BOLA2 across the critical region expanded by threefold the size of flanking high-identity, directly oriented sequence blocks (Extended Data Fig. 5a–b and Tables S4, S17, S18), theoretically predisposing the locus to recurrent CNV via unequal crossover (Extended Data Fig. 5c) specifically in the human lineage. To test this, we refined breakpoint locations in autism and developmental delay patients carrying either the chromosome 16p11.2 microduplication or microdeletion event23. Using whole-genome sequence (WGS) data and a molecular inversion probe (MIP) assay24, we localized breakpoints in 152 patients corresponding to 105 independent rearrangement events (Fig. 4a, Extended Data Fig. 10, and Table S19). We found 96% (101 of 105) of the disease-causing rearrangement breakpoints map within the Homo sapiens-specific duplication containing BOLA2 (Fig. 4b). Thus, the expansion of this segment rendered the chromosome 16p11.2 locus susceptible to recurrent rearrangement.
Figure 4. Refinement of chromosome 16p11.2 rearrangement breakpoints.
a) Results of whole-genome sequencing of a family with a de novo chromosome 16p11.2 microdeletion in a child with autism. Normalized read depth at unique 30-mer positions in the human reference genome GRCh37 is depicted for the proband, her mother, and her father. Read-depth signatures reveal a deletion in the proband extending between but not beyond the Homo sapiens-specific duplicated sequences (highlighted in pink). b) Summary of results across 105 independent microdeletion and microduplication events from 152 individuals. ~96% of breakpoints map to the Homo sapiens-specific segmental duplication.
In summary, the level of genetic difference between humans and chimpanzees for chromosome 16p11.2 stands in sharp contrast to the oft-quoted 99% genetic identity between the species. The region has undergone extensive inversion and duplication, including a 95 kbp segment containing BOLA2 that duplicated after our divergence with ancient hominins. This event contributes more derived sequence specific to Homo sapiens than 35,500 previously reported human-specific single-nucleotide variants and indels combined2. The rapid rise and dispersal of this duplicated segment at the root of Homo sapiens (~282 kya) are unlikely to have occurred under neutral evolution but rather are consistent with modest positive selection (s = 0.0015). The estimated strength of selection on the BOLA2 duplication is an order of magnitude weaker than what is typically observed for recent positive selection (such as the emergence of lactase persistence ~10 kya25) but an order of magnitude stronger than nearly neutral mutations. Remarkably, the BOLA2 duplication rapidly rose to high frequency in humans despite predisposing our species to recurrent CNV associated with disease. The expansion of this segment resulted in the formation of a novel fusion transcript and dramatic BOLA2 expression differences between chimpanzee and human iPSCs. Although the phenotypic consequences of increased BOLA2 expression and the novel fusion transcript await future in vivo characterization, it is known that BOLA2 physically interacts in a heterotrimeric complex with GLRX3 (glutaredoxin 3)26. This complex is conserved from prokaryotes to humans27 and was shown to have a role in iron sensing in yeast28. In vertebrates, BOLA2 has been hypothesized to play important roles in iron regulation29 and iron-sulfur protein biogenesis30. We speculate that the expansion of this conserved gene may enhance iron utilization and homeostasis, especially during human embryonic development.
METHODS1–30
Single-molecule, real-time (SMRT) sequencing was used to generate high-quality sequence6 from bacterial artificial chromosome (BAC) clones obtained from genomic libraries. Clone sequences were assembled using HGAP and error-corrected using Quiver31. Contig assembly was performed using Sequencher (Gene Codes Corporation, Ann Arbor, MI) and validated by FISH. Copy number genotyping of genes and segmental duplications was performed using a read-depth method12 and WGS data from humans32,33, nonhuman primates34, and archaic genomes2,3,13,14, as well as single molecule MIPs (smMIPs)35 targeted to paralogous sequence variants24. We estimated evolutionary timing of segmental duplication events based on comparative sequencing and phylogenetic analyses (neighbor-joining method), adjusting branch lengths for trees that failed the Tajima’s relative rate test and assuming divergence times of 6 mya (human–chimpanzee)10 and 15 mya (human–orangutan). Evolutionary conservation analysis of BOLA2 was performed by maximum likelihood (PAML). Likelihoods of BOLA2B fixation under different scenarios were assessed using the coalescent simulators ms17 and msms18, adapting a previously published demographic model16. BOLA2 copy number estimates were correlated (Pearson’s r) using RNA-seq quantifications20 (PEER-normalized RPKM) and Western blot BOLA2 densities in human LCLs grown in complete RPMI medium and lysed in RIPA buffer. After SDS-PAGE and transfer to PVDF membrane, blots were incubated with an anti-BOLA2 antibody (Santa Cruz Biotechnology, Dallas, TX) and an anti-actin antibody (Sigma) for normalization purposes. Band densities were quantified using the Bio1D software. BOLA2 coding DNA sequence (CDS) was cloned using the Gateway system (Invitrogen, Carlsbad, CA). HeLa cells were transfected with cytomegalovirus-BOLA2 CDS (both 10 and 17 kDa forms) and analyzed by Western blotting. BOLA2 gene models were established via RT-PCR, cloning, and capillary sequencing. RNA-seq data were generated from previously described ESC and iPSC lines21, as well as iPSC lines differentiated into NPCs and neurons. BOLA2 mRNA expression was quantified in transcripts per million (TPM) with Kallisto36 (version 0.42.1) using a custom catalog of transcripts including all human RefSeq transcripts with the three BOLA2 isoforms. Breakpoints of chromosome 16p11.2 rearrangements were refined using Illumina whole-genome shotgun sequencing37,38 and smMIP analysis24,35,37 of patient DNA obtained from the Simons Variation in Individuals Project (Simons VIP)23 and Simons Simplex Collection (SSC)39. All procedures for clinical assessment and blood extraction were approved by the institutional review boards (IRBs) of participating institutions, and informed consent was obtained for participation in this research.
Extended Data
Extended Data Figure 1. Comparative sequence analysis of chromosome 16p11.2 among apes.
a) Schematic depicts the genomic organization of chromosome 16p11.2 for one orangutan and two chimpanzee haplotypes along with the human reference haplotype (GRCh37 chr16:28195661–30573128; see ideogram for approximate chromosomal location). Blocks of segmental duplications within this locus mediate recurrent rearrangements in humans; thus, these blocks have been defined as breakpoint regions BP1–BP5 (ref. 8). The ~550 kbp critical region (pink) and a >1 Mbp chimpanzee-specific inversion polymorphism (orange) are highlighted. Tiling paths of sequenced clones are indicated above each haplotype, with chimpanzee clones that could not be fully resolved marked with asterisks. Colored boxes and thick arrows indicate the extent and orientation of segmental duplications (with different colors denoting duplicons from different ancestral genomic loci, and hashed boxes indicating sequence duplicated in humans but not in the species represented). Thin numbered arrows show orientations of gene-rich regions of unique sequence. Numbers (left) indicate the size of each orthologous haplotype, with the number of segmentally duplicated base pairs shown in parentheses. Note that, for chimpanzee, these sizes are lower bounds due to gaps in the contigs (dotted line sections) and the contigs not reaching unique sequence beyond BP1 (i.e., unique region 1). b) Schematic depicts distinct human structural haplotypes over the chromosome 16p11.2 critical region and flanking sequences (three complete haplotypes extending from unique sequence distal to BP3 to unique sequence proximal to BP5 and one partial haplotype including BP3–BP4 and BP5 sequence contigs). High-quality sequence for each haplotype was generated by sequencing a total of 40 BACs and 15 fosmids from three different human genomic libraries. Regions of copy number variation (highlighted in yellow along the first two haplotypes) occur on both sides of the critical region and involve the same 102 kbp unit in direct orientation, including a 30 kbp block containing BOLA2 and two other genes and a 72 kbp block harboring a partial segmental duplication of SMG1 (SMG1P). Expansion and contraction of this cassette underlie hundreds of kbp of structural diversity between human haplotypes. BOLA2 paralog-specific copy number genotype data suggest that H1 and H3 likely represent the most common haplotype structures in humans.
Extended Data Figure 2. Comparison of chromosome 16p11.2 structure between apes.
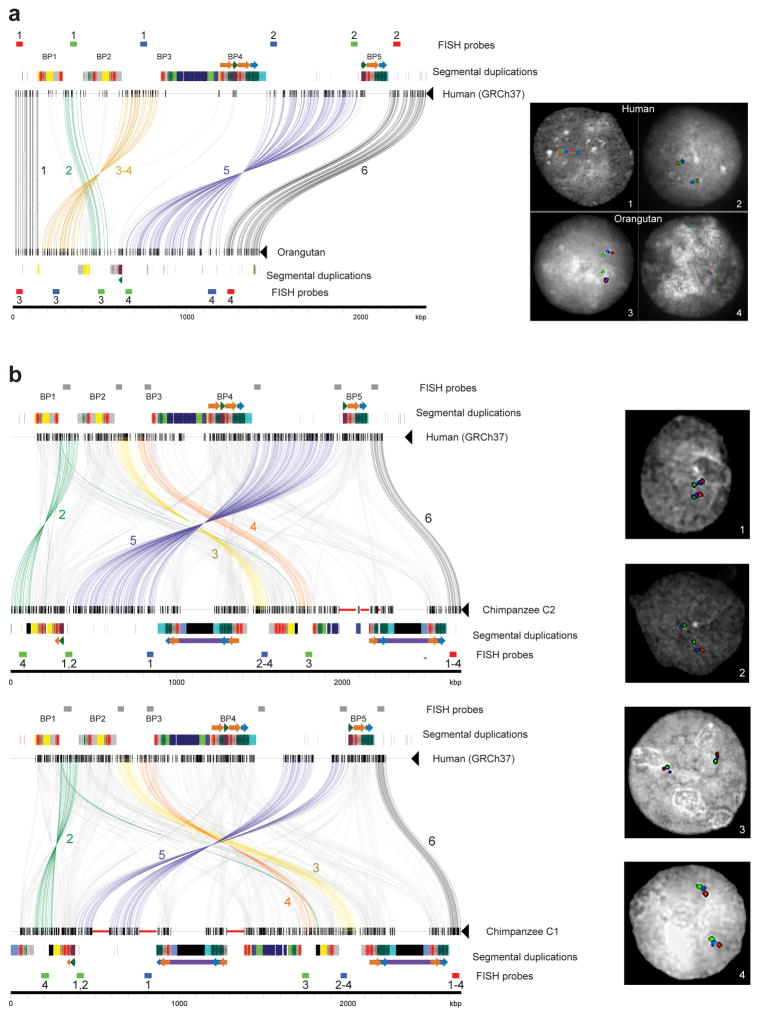
a) Sequences (thin horizontal lines) from human (GRCh37 chr16:28195661–30573128) and orangutan (contig sequence) at chromosome 16p11.2 are compared using Miropeats (s = 1,000) and annotated with locations of human segmental duplications and FISH probes used to validate the organization of the region. Lines connecting the sequences show regions of homology, and line colors highlight differences in the order and orientation of distinct gene-rich regions of unique sequence across the locus (numbered 1–6). Numbers below FISH probes correspond to numbers within the images on the right, specifying which probes were used in each experiment. Experiment 1 used the same probes as experiment 3, and experiment 2 used the same probes as experiment 4. Three-color interphase FISH on human and orangutan chromosomes confirms the accuracy of our assembled orangutan contig. b) Sequences (thin horizontal lines) from human (GRCh37 chr16:28195661–30573128) and two chimpanzee structural haplotypes at chromosome 16p11.2 are compared using Miropeats (s = 1,500) and annotated with locations of human segmental duplications and FISH probes used to validate the organization of the region. Thick red horizontal lines indicate gaps in the chimpanzee contigs, and black boxes correspond to chimpanzee-specific segmental duplications (i.e., sequences not duplicated in humans). Lines connecting the sequences show regions of homology, and line colors highlight differences in the order and orientation of distinct gene-rich regions of unique sequence across the locus (numbered 2–6). Numbers below FISH probes correspond to numbers within the images on the right, specifying which probes were used in each experiment. Gray rectangles show mapping locations of FISH probes in human. Three-color interphase FISH on chimpanzee chromosomes confirms the accuracy of our assembled contigs.
Extended Data Figure 3. Dynamic evolution of human chromosome 16p11.2.
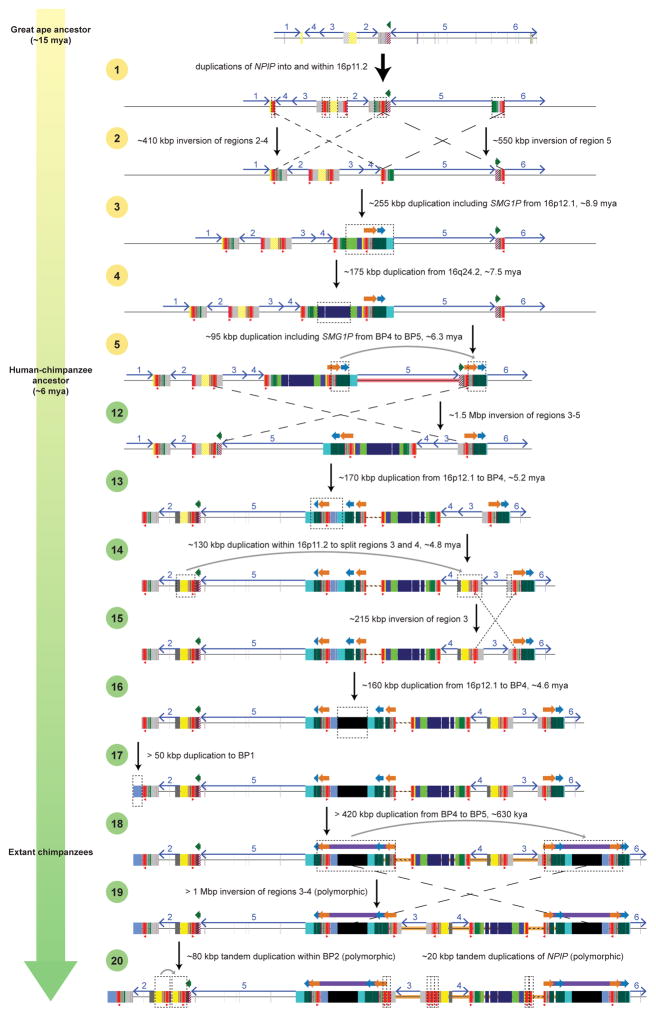
a) A model for the evolution of the chromosome 16p11.2 BP1–BP5 region (ref. 8) during great ape evolution. The schematic depicts structural changes over time leading to the present-day human architecture (see Supplementary Information for details). The orangutan structure (top) is largely devoid of segmental duplications and deemed to represent the ape ancestral organization because it is conserved with mouse. Subsequent steps were inferred based on phylogenetic reconstruction, origins of the duplicated sequences, and the most parsimonious path with respect to changes in gene order (inversions). (See Supplementary Information for a detailed discussion of all supporting evidence and confidence levels for each step.) Note that, without access to genomes containing intermediate chromosome 16p11.2 structures, it is impossible to know with certainty the entire step-by-step evolutionary history. Some details presented here may not be accurate.
Extended Data Figure 4. Dynamic evolution of chimpanzee chromosome 16p11.2.
A model for the evolution of the chromosome 16p11.2 BP1–BP5 region (ref. 8) during great ape evolution. The schematic depicts structural changes over time leading to the present-day chimpanzee architecture (see Supplementary Information for details and discussion of all supporting evidence and confidence levels for each step.).
Extended Data Figure 5. Comparison of duplications around the chromosome 16p11.2 autism critical region among apes and NAHR model underlying CNV at human chromosome 16p11.2.
a) Local directly oriented (green) and inversely oriented (blue) intrachromosomal segmental duplications flanking the chromosome 16p11.2 autism critical region (purple) are visualized using Miropeats (s = 1,000). Gaps in the chimpanzee C1 contig are shown in red. The smaller size (<50 kbp) and lower average sequence identity (at most 98.6%) of directly oriented duplications flanking the critical region in chimpanzee compared to human haplotypes including BOLA2 on both sides of the critical region (at least 147 kbp of directly oriented duplications having at least 99.3% average sequence identity) suggest that susceptibility to NAHR resulting in microdeletions and microduplications at this locus evolved specifically in humans. b) Perfect sequence identity tract lengths (>500 bp) within directly oriented duplications flanking the critical region for human vs. chimpanzee. Histograms show counts of tracts of perfect sequence identity (lacking single-nucleotide variants and indels) between directly oriented segmental duplications of interest within each indicated haplotype and the distribution of these tracts over different size ranges. Human haplotypes having BOLA2 on both sides of the critical region (bottom panels) contain the highest number of such tracts and the longest such tracts, including one tract spanning 10,774 bp. In contrast, the longest tract of perfect sequence identity between duplications of interest in chimpanzee (considering both the C1 and C2 haplotypes) spans 450 bp. c) NAHR model underlying normal and disease-associated CNV at human chromosome 16p11.2.
Extended Data Figure 6. Sequence refinement of interspersed BOLA2 duplication breakpoints, inference of BOLA2 duplication mechanism, and phylogenetic BOLA2 duplication timing.
a) H1 human BP4 sequence (orange, green, orange, and blue arrows in inset) was aligned to its allelic (black arrows in inset) and paralogous (red arrows in inset) counterparts. The sequence identity for each alignment was computed and plotted over 2 kbp windows, sliding by 100 bp. Black lines indicate sequence identity for allelic comparisons, whereas red lines correspond to paralogous comparisons. While the allelic comparisons exhibit uniform, near-perfect sequence identity across the entirety of the alignment, paralogous comparisons reveal three distinct levels of sequence identity, with the highest level in the middle. This pattern suggests that the BOLA2 duplication (highest-identity region, 95 kbp) landed within an evolutionarily older segmental duplication having paralogs at BP4 and BP5. Dashed vertical lines (numbered i–iv) indicate putative breakpoints for events that occurred after this older segmental duplication. Junction sequence from the BP5 102 kbp tandem duplication (i.e., the SMG1P-BOLA2 junction) was clearly included in the 95 kbp duplication from BP5 to BP4. b) Alignment of BP4 sequences containing the putative left (red arrows in inset) and right (dark blue arrows in inset) BOLA2 duplication breakpoints to the BP5 paralog associated with the evolutionarily older segmental duplication (orange and light blue arrows in inset) and sliding window sequence identity analysis supports the hypothesis outlined above. Sequence identity lines for comparisons involving left and right BP4 sequences intersect in the vicinity of the hypothesized BOLA2 duplication breakpoints. Comparing this result with the same analysis of the human H2 BP4 sequence lacking BOLA2 (green arrows in inset and green identity line) suggests this BP4 sequence represents the ancestral state of BP4 before the BOLA2 duplication arrived. Thus, two levels of sequence identity existed between BP4 and BP5 before the BOLA2 duplication, consistent with an interlocus gene conversion event. c) Alignment of BP4 sequences (orange arrows in insets) containing the putative BOLA2 duplication breakpoints to their ancestral BP4 (top plot) and their ancestral BP5 (middle plot) counterparts and sliding window sequence identity analysis reveals an ~7 kbp window (highlighted in orange) defining the BOLA2 duplication breakpoints. Analysis of the underlying multiple sequence alignment (Table S5) identified positions with signatures informative for breakpoint localization (blue vertical lines, left BP4 72 kbp block outside of the BOLA2 duplication and right BP4 72 kbp block within the BOLA2 duplication; yellow vertical lines, left BP4 72 kbp block within the BOLA2 duplication and right BP4 72 kbp block outside of the BOLA2 duplication). Gray vertical lines indicate positions showing signatures of interlocus gene conversion. As both left and right 72 kbp block BP4 sequences within the ~7 kbp window are more highly identical to ancestral BP4 sequence (20/24 informative positions match the ancestral BP4 sequence) than to ancestral BP5 sequence, it is likely that this interval was involved in the BOLA2 duplication but duplicated only within BP4. Its boundaries define the most likely BOLA2 duplication breakpoints, and this pattern of sequence identity suggests a template switching replicative mechanism as most likely underlying the BOLA2 duplication event. d) Template-switching model for the formation of BOLA2B. This mechanism was inferred from the sequence identity analyses in panels a–c and from analysis of a multiple sequence alignment (Table S5). e) Phylogenetic characterization of the 95 kbp duplication containing BOLA2 from BP5 to BP4. Cladogram representation of an unrooted neighbor-joining phylogenetic tree based on a 21,102 bp multiple sequence alignment spanning BOLA2 and most of the 30 kbp block including human sequences from BP4 and BP5 and single-copy orthologous sequences from chimpanzee, gorilla, and orangutan. Branch lengths (substitutions per site) are shown on each branch (black decimal numbers), and bootstrap support is indicated (black integers at nodes). Blue numbers correspond to nodes and indicate average branch lengths for all sequences in corresponding clades. Branch lengths were used to estimate the time corresponding to the 95 kbp duplication containing BOLA2 from BP5 to BP4 as shown.
Extended Data Figure 7. Analyses of BOLA2 aggregate and paralog-specific copy number variation in humans.
a) Interphase FISH confirms both BOLA2A and BOLA2B show copy number variation. Previous interphase FISH analysis (data not shown) suggests the individual NA20127 has six total copies of BOLA2. Diagram outlines a three-color FISH assay including two probes (blue, green) targeting sequences within the autism critical region and one probe (red) targeting ~18 kbp of sequence (including BOLA2) over the 30 kbp duplication block. Signals from the red probe are detected on the telomeric (BP4) and centromeric (BP5) sides of the critical region (adjacent to the blue and green probes, respectively) on both chromosome 16 homologs. However, the red probe signal intensity is strongest adjacent to the green probe for one homolog but, in contrast, is strongest adjacent to the blue probe for the other chromosome 16 homolog, consistent with higher BOLA2A copy number in the first case and higher BOLA2B copy number in the second case. These data indicate that individual NA20127 has three copies each of BOLA2A and BOLA2B. This differential signal intensity pattern does not result from an inversion of the chromosome 16p11.2 critical region in this individual, as data from another FISH experiment (data not shown) refute this possibility. Information on probes used in these FISH experiments is provided in Table S2. b) Interphase FISH experiments using a probe targeting BOLA2 and surrounding sequence for individuals having the lowest (3) and highest (8) confirmed aggregate BOLA2 copy numbers. c) Left and middle schematics detail three distinct sectors of the 72 kbp blocks (orange arrows). Each block has paralogous sequence variants that are informative for particular region(s) when compared to others in chromosome 16p11.2. These markers are color-coded into three sectors within the 72 kbp block of paralogy (a 59 kbp sector, blue and red boxes; a 7 kbp sector, green and orange boxes; and a 6 kbp sector, purple and yellow boxes), indicating which particular regions they distinguish. Right schematic shows known haplotype structures for individual NA12878. d) Analyzing WGS data from NA12878 yields copy number estimates for BOLA2A and BOLA2B that match the known BOLA2 PSCN for this individual. Each point shows a relative marker-specific read count frequency (y-axis) and its position within the duplication blocks (x-axis). Point colors correspond to different marker sets for each sector, as diagramed in panel c. Fractions indicate the relative copy number of each marker set. Estimates of 4/6 (red marker set) vs. 2/6 (blue marker set) for the 59 kbp sector confirms the sequenced architecture (panel c) with an aggregate of 4 BOLA2 copies, and the estimate of 3/6 (orange marker set) confirms three copies of BOLA2A. WGS analysis also yields accurate PSCN estimates for the 45 kbp block. e) Using MIPs, we employed the same relative read-depth strategy. Genotyping results for the same sample as in panel d are shown, with additional markers (points not color-coded as in panels c–d) added based on polymorphic variants (symbols indicate different patterns of presence/absence among 72 kbp blocks, considering all such blocks from our four contiguous human haplotypes). MIP genotypes confirm WGS estimates (in panel d). f) BOLA2 PSCN genotypes (points, jittered around their integer values for clarity) were inferred from MIP sequence data for 894 humans. Numbers indicate total counts of individuals in each population having a particular BOLA2 PSCN genotype. Low-confidence estimates were excluded.
Extended Data Figure 8. Population genetic modeling of the BOLA2B duplication and critical region analyses.
a) Demographic model (adapted from ref. 16) used to simulate BOLA2B evolution under different scenarios. NANC, effective population size of Homo ancestor, 21,600. NARC, effective population size of Neanderthal-Denisova ancestor, 500. NHUM, effective population size of human ancestor, 24,000. NYRE, effective size of Yoruban population after expansion, 45,000. NDEN, effective population size of Denisova, 500. NNEA, effective population size of Neanderthal, 500. NYRI, effective size of extant Yoruban population, 10,000. NSAN, effective size of extant San population, 10,000. T1, time of archaic hominin divergence from modern humans, 650,000 years. T2, time of Neanderthal-Denisova divergence, 525,000 years. Tdup, time of formation of BOLA2B, 282,000 years. T3, time of Yoruban-San divergence, 200,000 years. T4, time of Yoruban population expansion, 157,500 years. T5, time of Yoruban population decline, 37,500 years. b) Simulation results (n = 1,000,000) assuming that the duplication that formed BOLA2B occurred once, 282 kya, along the modern human ancestral lineage and evolved under neutrality compared to the observed genotype frequencies of BOLA2B in 8 San and 110 Yoruban haplotypes. Nearly all (999,531) simulations resulted in BOLA2B being lost from both populations; results from the remaining 469 simulations (black) are shown alongside the observed data (red, circled). Under this simple neutral model incorporating BOLA2B age, the observed BOLA2B frequency is never approached. c) Simulation was repeated exploring a range of selection coefficients from 0.0009 to 0.0024 (increments of 0.0001), and the relative probability of the observed data under each scenario was calculated as the proportion of simulations yielding the observed BOLA2B genotypes among simulations where BOLA2B was not lost relative to the maximum such proportion for any single selection coefficient considered. The maximum likelihood estimate for the selection coefficient was s = 0.0015. Smoothed line is LOESS regression curve. d) Low average heterozygosity of the chromosome 16p11.2 BP4–BP5 critical region. Distribution of average heterozygosity values for 100,000 ~550 kbp regions of unique sequence randomly sampled with replacement from the autosomal genome compared to average heterozygosity values for the critical region (black line) and flanking unique sequences (colored lines). The critical region lies in the bottom 2.6% of the distribution, showing low diversity consistent with potential positive selection. Bottom schematic indicates locations of the critical region and flanking unique regions in relation to segmental duplications across the locus—note that BOLA2A is located at BP5 and BOLA2B at BP4. e) Low Tajima’s D score for the chromosome 16p11.2 BP4–BP5 critical region. Distribution of Tajima’s D scores for 2,987 non-overlapping ~550 kbp regions across the genome compared to Tajima’s D scores for the critical region (black line) and flanking unique sequences (colored lines). The critical region lies in the bottom 2.7% of the distribution, consistent with possible positive selection. The distribution is centered near −2 rather than 0 because most SNVs in the 1000 Genomes dataset are rare variants having arisen during the large expansions of human populations over the past 100,000 years. Bottom schematic indicates locations of the critical region and flanking unique regions in relation to segmental duplications across the locus.
Extended Data Figure 9. BOLA2 expression and antibody validation.
a) RT-PCR expression profile for canonical BOLA2. The expected product size for canonical BOLA2 (838 bp) was observed in all eight human tissues. 1 kb + DNA ladder (Thermo Fisher). b) RT-PCR expression profile for BOLA2-SMG1 fusion product. The expected product size for the BOLA2 fusion transcript (1,239 bp) was observed as a doublet in all tissues except skeletal muscle. Intensity of upper band differs between tissues. 1 kb + DNA ladder (Thermo Fisher). c) BOLA2 RNA-seq expression analysis. Canonical (BOLA2) and fusion transcripts (BOLA2F, BOLA2T) were assessed across 25 humans from GTEx RNA-seq data. Bar heights indicate mean expression levels for each tissue in TPM with standard errors shown (error bars). Colors correspond to different BOLA2 isoforms as indicated. d) BOLA2 expression among primates in six adult tissues. Each point indicates a BOLA2 expression estimate from a single tissue sample, with samples obtained from a total of 18 humans, 6 chimpanzees, and 3 bonobos. Open circles correspond to individuals analyzed in a single experiment, while closed shapes denote data from multiple experiments involving the same individual, with each distinct color + shape pattern showing all experiments for a particular individual. Horizontal lines show mean expression values for each species and tissue. Combined with our expression analyses of iPSCs, these data show BOLA2 expression differs substantially between human, chimpanzee, and bonobo only in stem cells. e) Western blotting of HeLa cells transfected with the human BOLA2 annotated CDS and probed with an anti-BOLA2 antibody (Sc-163747). Whole-cell lysate of HeLa cells non-transfected with the overexpression construct (lane 1) and transfected with the human BOLA2 annotated CDS (lane 2) were probed with anti-BOLA2 antibody. Two bands with molecular weights of 10 and 17 kDa are identified and more abundant in transfected cells and correspond to two BOLA2 protein isoforms arising from different translation start sites.
Extended Data Figure 10. Chromosome 16p11.2 rearrangement breakpoint refinement.
a) Schematic depicts NAHR between directly oriented segmental duplications at BP4 and BP5. This unequal crossover results in chromosome 16p11.2 microdeletions and microduplications (Extended Data Fig. 5c). Colored arrows and boxes correspond to duplication blocks and sectors within them color-coded as in Extended Data Fig. 7c. Unequal crossover could occur in eight distinct regions with regard to duplication block and sector boundaries. Three such regions are located within the ~95 kbp Homo sapiens-specific duplication (dashed lines). Only unequal crossover events outside the Homo sapiens-specific duplication produce recombinants having a sector with non-uniform marker-specific copy number across its extent. b) Plot shows relative marker-specific read count frequencies (points) determined from WGS analysis for a microdeletion proband. Fractions indicate relative marker-specific copy numbers, as in Extended Data Fig. 7d, and diagrams adjacent to the plot show inferred haplotype structures for each chromosome 16 homolog for this individual. Though the data in the plot provide only diploid genotypes (and not resolved haplotypes), the haplotypes suggested here reflect this genotype information together with data from the parents (not shown) and the assumption (supported by our PSCN data) that haplotypes having two BOLA2A copies and a single BOLA2B copy are the most common. Because marker-specific copy number is uniform across each sector, unequal crossover breakpoints must have occurred within the Homo sapiens-specific duplication. c) Breakpoint refinement based on MIP PSCN marker data. Plots show relative marker-specific read count frequencies (points) determined using MIPs for a typical microdeletion patient (left) and a typical microduplication patient (right). Shapes and color code designate different markers, and fractions indicate relative marker-specific copy numbers (as in Extended Data Fig. 7). Because marker-specific copy number is uniform across each sector for both individuals, in both cases, unequal crossover breakpoints must have occurred within the Homo sapiens-specific duplication. d) Data from an atypical patient where the breakpoints are inferred to map outside of the Homo sapiens-specific segmental duplication. The plots show paralog-specific copy number for a chromosome 16p11.2 microdeletion proband, his sibling, and his mother over a 45 kbp duplication block shared between BP4 and BP5. Paralog-specific copy number was estimated using a MIP assay targeting 54 informative markers over this region, with data from 43 markers fixed among haplotypes H1–H4 shown (points). Dashed lines indicate calls inferred using an automated caller, which were also confirmed by visual inspection. Adjacent schematics indicate the inferred haplotypes for each individual based on these data, with approximate breakpoint locations shown (arrows). The results demarcate the location of the unequal crossover interval based on the reciprocal copy number transition between the BP5 (red) and BP4 (blue) 45 kbp block segmental duplications. In this case, the breakpoints clearly map to a 22 kbp region outside of the typical hotspot. Analysis of the sibling suggests that this region was the site of an interlocus gene conversion event from BP5 to BP4, and data from the mother imply that chromosomes having this event were present in the paternal germline. DNA from the father was not available for testing.
Supplementary Material
Acknowledgments
We thank families at the participating Simons VIP and SSC sites, as well as the Simons VIP Consortium. Approved researchers can obtain the Simons VIP dataset, the SSC dataset, and/or biospecimens by applying at https://base.sfari.org. We thank M. Chaisson for SMRT WGS data, B. Vernot for archaic introgression data, B.J. Nelson and K. Munson for technical assistance, M.L. Gage for editorial comments, and T. Brown for assistance with manuscript preparation. This work was supported by the Paul G. Allen Foundation (grant #11631 to E.E.E.), the Simons Foundation Autism Research Initiative (SFARI #303241 to E.E.E. and #274424 to A.R.), the U.S. National Institutes of Health (NIH grant 2R01HG002385 to E.E.E.), the Swiss National Science Foundation (31003A_160203 and CRSII33-133044 to A.R.), and funds from NIH TR01 MH095741, the Helmsley Charitable Fund, the Mathers Foundation, and the JPB Foundation (to F.H.G.). X.N. was supported by a U.S. National Science Foundation Graduate Research Fellowship under grant #DGE-1256082. G.G. was awarded a Pro-Women Scholarship from the Faculty of Biology and Medicine, University of Lausanne. M.H.D. is supported by U.S. National Institute of Mental Health grant 1F30MH105055-01. L.B. is supported by the EC grant N653706, project iNEXT. S.C.B. and F.C. were supported by Ente Cassa di Risparmio grant (ID no. 2013/7201). E.E.E. is an investigator of the Howard Hughes Medical Institute. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions
X.N., G.G., M.H.D., A. Reymond, and E.E.E. designed the study. X.N., G.G., M.H.D., M.M., J.H., L.D., L.H., C. Baker, A. Raja, and K.P. contributed to sequencing and assembly of haplotypes. X.N. developed the evolutionary model, with input from G.G. P.H.S. genotyped aggregate copy number from WGS data. X.N. and M.H.D. performed MIP experiments and analyzed WGS data to genotype paralog-specific copy number and refine rearrangement breakpoints. N.J. performed massively parallel sequencing. J.G.S., M.H.D., and X.N. performed population genetic simulations, with input from J.M.A. G.G. analyzed RNA-seq data from LCLs, performed Western blots, and assessed the correlation of expression with copy number. I.N., C. Benner, and M.C.N.M. performed and analyzed RNA-seq experiments over in vitro differentiation of experimentally derived primate stem cells, with supervision from F.H.G. O.P., G.G., and X.N. analyzed RNA-seq data from different human and nonhuman primate tissues. J.H. performed inversion density simulations using data provided by F.A. and M.V. G.C. and F.A. performed FISH experiments. F.C., S.C.B., H.A.F.S., and L.B. performed functional experiments and provided insights into potential effects of increased BOLA2 dosage. W.J.T. and C.T.A. constructed a BAC library. X.N. and E.E.E. wrote the paper, with input and approval from all coauthors.
Author Information
Clone sequences, haplotype contig sequences, MIP data, and RNA-seq data for NPCs and neurons are available at NCBI BioProject (accession number PRJNA325679 in the NCBI BioProject database, https://www.ncbi.nlm.nih.gov/bioproject/). Patient WGS and MIP data are available at SFARI base (accession numbers SFARI_SVIP_WGS_1 and SFARI_SVIP_MIPS_1). Reprints and permission information is available at www.nature.com/reprints. The authors declare competing financial interests: details accompany the full-text HTML version of the paper at www.nature.com/nature.
COMPETING FINANCIAL INTERESTS
E.E.E. is on the scientific advisory board (SAB) of DNAnexus, Inc., and is a consultant for the Kunming University of Science and Technology (KUST) as part of the 1000 China Talent Program.
References
- 1.King MC, Wilson AC. Evolution at two levels in humans and chimpanzees. Science (New York, NY) 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- 2.Prufer K, et al. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature. 2014;505:43–49. doi: 10.1038/nature12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meyer M, et al. A high-coverage genome sequence from an archaic Denisovan individual. Science (New York, NY) 2012;338:222–226. doi: 10.1126/science.1224344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weiss LA, et al. Association between microdeletion and microduplication at 16p11.2 and autism. The New England journal of medicine. 2008;358:667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 5.Kumar RA, et al. Recurrent 16p11.2 microdeletions in autism. Human molecular genetics. 2008;17:628–638. doi: 10.1093/hmg/ddm376. [DOI] [PubMed] [Google Scholar]
- 6.Huddleston J, et al. Reconstructing complex regions of genomes using long-read sequencing technology. Genome research. 2014;24:688–696. doi: 10.1101/gr.168450.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson ME, et al. Positive selection of a gene family during the emergence of humans and African apes. Nature. 2001;413:514–519. doi: 10.1038/35097067. [DOI] [PubMed] [Google Scholar]
- 8.Zufferey F, et al. A 600 kb deletion syndrome at 16p11.2 leads to energy imbalance and neuropsychiatric disorders. Journal of medical genetics. 2012;49:660–668. doi: 10.1136/jmedgenet-2012-101203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jacquemont S, et al. Mirror extreme BMI phenotypes associated with gene dosage at the chromosome 16p11.2 locus. Nature. 2011;478:97–102. doi: 10.1038/nature10406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patterson N, Richter DJ, Gnerre S, Lander ES, Reich D. Genetic evidence for complex speciation of humans and chimpanzees. Nature. 2006;441:1103–1108. doi: 10.1038/nature04789. [DOI] [PubMed] [Google Scholar]
- 11.Auton A, et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sudmant PH, et al. Diversity of human copy number variation and multicopy genes. Science (New York, NY) 2010;330:641–646. doi: 10.1126/science.1197005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lazaridis I, et al. Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature. 2014;513:409–413. doi: 10.1038/nature13673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fu Q, et al. Genome sequence of a 45,000-year-old modern human from western Siberia. Nature. 2014;514:445–449. doi: 10.1038/nature13810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tishkoff SA, et al. The genetic structure and history of Africans and African Americans. Science (New York, NY) 2009;324:1035–1044. doi: 10.1126/science.1172257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang MA, Harris K, Slatkin M. The projection of a test genome onto a reference population and applications to humans and archaic hominins. Genetics. 2014;198:1655–1670. doi: 10.1534/genetics.112.145359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hudson RR. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics (Oxford, England) 2002;18:337–338. doi: 10.1093/bioinformatics/18.2.337. [DOI] [PubMed] [Google Scholar]
- 18.Ewing G, Hermisson J. MSMS: a coalescent simulation program including recombination, demographic structure and selection at a single locus. Bioinformatics (Oxford, England) 2010;26:2064–2065. doi: 10.1093/bioinformatics/btq322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vernot B, et al. Excavating Neandertal and Denisovan DNA from the genomes of Melanesian individuals. Science (New York, NY) 2016;352:235–239. doi: 10.1126/science.aad9416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lappalainen T, et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 2013;501:506–511. doi: 10.1038/nature12531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marchetto MC, et al. Differential L1 regulation in pluripotent stem cells of humans and apes. Nature. 2013;503:525–529. doi: 10.1038/nature12686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brawand D, et al. The evolution of gene expression levels in mammalian organs. Nature. 2011;478:343–348. doi: 10.1038/nature10532. [DOI] [PubMed] [Google Scholar]
- 23.Simons Variation in Individuals Project (Simons VIP): a genetics-first approach to studying autism spectrum and related neurodevelopmental disorders. Neuron. 2012;73:1063–1067. doi: 10.1016/j.neuron.2012.02.014. [DOI] [PubMed] [Google Scholar]
- 24.Nuttle X, et al. Rapid and accurate large-scale genotyping of duplicated genes and discovery of interlocus gene conversions. Nature methods. 2013;10:903–909. doi: 10.1038/nmeth.2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bersaglieri T, et al. Genetic signatures of strong recent positive selection at the lactase gene. American journal of human genetics. 2004;74:1111–1120. doi: 10.1086/421051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H, Mapolelo DT, Randeniya S, Johnson MK, Outten CE. Human glutaredoxin 3 forms [2Fe-2S]-bridged complexes with human BolA2. Biochemistry. 2012;51:1687–1696. doi: 10.1021/bi2019089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H, Outten CE. Monothiol CGFS glutaredoxins and BolA-like proteins: [2Fe-2S] binding partners in iron homeostasis. Biochemistry. 2012;51:4377–4389. doi: 10.1021/bi300393z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumanovics A, et al. Identification of FRA1 and FRA2 as genes involved in regulating the yeast iron regulon in response to decreased mitochondrial iron-sulfur cluster synthesis. The Journal of biological chemistry. 2008;283:10276–10286. doi: 10.1074/jbc.M801160200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haunhorst P, et al. Crucial function of vertebrate glutaredoxin 3 (PICOT) in iron homeostasis and hemoglobin maturation. Molecular biology of the cell. 2013;24:1895–1903. doi: 10.1091/mbc.E12-09-0648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Banci L, Camponeschi F, Ciofi-Baffoni S, Muzzioli R. Elucidating the Molecular Function of Human BOLA2 in GRX3-Dependent Anamorsin Maturation Pathway. Journal of the American Chemical Society. 2015;137:16133–16143. doi: 10.1021/jacs.5b10592. [DOI] [PubMed] [Google Scholar]
- 31.Chin CS, et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nature methods. 2013;10:563–569. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 32.Abecasis GR, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sudmant PH, et al. Global diversity, population stratification, and selection of human copy-number variation. Science (New York, NY) 2015;349:aab3761. doi: 10.1126/science.aab3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prado-Martinez J, et al. Great ape genetic diversity and population history. Nature. 2013;499:471–475. doi: 10.1038/nature12228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hiatt JB, Pritchard CC, Salipante SJ, O'Roak BJ, Shendure J. Single molecule molecular inversion probes for targeted, high-accuracy detection of low-frequency variation. Genome research. 2013;23:843–854. doi: 10.1101/gr.147686.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bray N, Pimentel H, Melsted P, Pachter L. Near-optimal RNA-Seq quantification. 2015 doi: 10.1038/nbt.3519. arXiv, 1505.02710. [DOI] [PubMed] [Google Scholar]
- 37.Nuttle X, Itsara A, Shendure J, Eichler EE. Resolving genomic disorder-associated breakpoints within segmental DNA duplications using massively parallel sequencing. Nature protocols. 2014;9:1496–1513. doi: 10.1038/nprot.2014.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Antonacci F, et al. Palindromic GOLGA8 core duplicons promote chromosome 15q13.3 microdeletion and evolutionary instability. Nature genetics. 2014;46:1293–1302. doi: 10.1038/ng.3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fischbach GD, Lord C. The Simons Simplex Collection: a resource for identification of autism genetic risk factors. Neuron. 2010;68:192–195. doi: 10.1016/j.neuron.2010.10.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.