Abstract
Corals rely on photosynthesis by their endosymbiotic dinoflagellates (Symbiodinium spp.) to form the basis of tropical coral reefs. High sea surface temperatures driven by climate change can trigger the loss of Symbiodinium from corals (coral bleaching), leading to declines in coral health. Different putative species (genetically distinct types) as well as conspecific populations of Symbiodinium can confer differing levels of thermal tolerance to their coral host, but the genes that govern dinoflagellate thermal tolerance are unknown. Here we show physiological and transcriptional responses to heat stress by a thermo-sensitive (physiologically susceptible at 32 °C) type C1 Symbiodinium population and a thermo-tolerant (physiologically healthy at 32 °C) type C1 Symbiodinium population. After nine days at 32 °C, neither population exhibited physiological stress, but both displayed up-regulation of meiosis genes by ≥ 4-fold and enrichment of meiosis functional gene groups, which promote adaptation. After 13 days at 32 °C, the thermo-sensitive population suffered a significant decrease in photosynthetic efficiency and increase in reactive oxygen species (ROS) leakage from its cells, whereas the thermo-tolerant population showed no signs of physiological stress. Correspondingly, only the thermo-tolerant population demonstrated up-regulation of a range of ROS scavenging and molecular chaperone genes by ≥ 4-fold and enrichment of ROS scavenging and protein-folding functional gene groups. The physiological and transcriptional responses of the Symbiodinium populations to heat stress directly correlate with the bleaching susceptibilities of corals that harbored these same Symbiodinium populations. Thus, our study provides novel, foundational insights into the molecular basis of dinoflagellate thermal tolerance and coral bleaching.
Keywords: symbiodinium, dinoflagellate, thermal tolerance, acclimation, coral, bleaching, transcriptomics.
Introduction
Corals and their dinoflagellate endophotosymbionts of the genus Symbiodinium create the foundation of tropical coral reefs, which support hundreds of thousands of plant and animal species (Reaka-Kudla et al. 1996). Tropical reef-building corals require metabolites provided by Symbiodinium for their nutrition and high rates of calcification (Muscatine and Porter 1977; Barnes and Chalker 1990; Gordon and Leggat 2010). Efficient recycling of nutrients between Symbiodinium and corals allows entire ecosystems to flourish in low nutrient waters (Roth 2014). Rising sea surface temperatures due to climate change cause the breakdown of the Symbiodinium-coral symbiosis resulting in the loss of Symbiodinium from the coral host (i.e., coral bleaching) and, consequently, drastic declines in coral health and cover worldwide (Hoegh-Guldberg 1999; Hoegh-Guldberg et al. 2007). Climate change impact models predict that many coral reefs will be irreversibly damaged in a matter of decades (Carpenter et al. 2008; Pandolfi et al. 2011). While the exact mechanistic role that Symbiodinium plays in coral bleaching has yet to be uncovered, increased production of ROS, such as superoxide and hydrogen peroxide, by Symbiodinium cells in response to heat stress is considered to be a key factor (Suggett et al. 2008; McGinty et al. 2012). Leakage of excess ROS from Symbiodinium cells when inside the coral tissues (in hospite) may exacerbate stress-induced oxidative damage of coral tissues and lead to Symbiodinium expulsion (Downs et al. 2002; Krueger et al. 2015).
The genus Symbiodinium is highly diverse, and substantial physiological differences exist among and even within “types”, i.e., genetic variants typically designated by the nuclear ribosomal DNA internal transcribed spacer 2 (ITS2) to notionally represent species (Arif et al. 2014). Different Symbiodinium can strongly influence coral gene expression and bleaching susceptibility (DeSalvo et al. 2010; Oliver and Palumbi 2011; Howells et al. 2012; Yuyama et al. 2012), and it is generally thought that Symbiodinium are more vulnerable to heat stress than their coral host (Fitt et al. 2001). Unraveling the molecular basis of variation in Symbiodinium thermal tolerance is thus an essential step required to understand variation in coral bleaching susceptibility.
Although Symbiodinium physiological responses to heat stress are well studied (Warner et al. 1999; Tchernov et al. 2004; Suggett et al. 2008; Howells et al. 2012; McGinty et al. 2012), the underlying gene regulation is still unresolved. Much of the evidence to date suggests that Symbiodinium lack a transcriptional response to heat stress (Leggat et al. 2011; Putnam et al. 2013; Barshis et al. 2014; Krueger et al. 2015), which contradicts the strong evidence in other organisms that physiological changes are largely driven by regulation of mRNA synthesis and degradation (Arbeitman et al. 2002; Wilusz and Wilusz 2004; Rossouw et al. 2009; Harb et al. 2010). In Symbiodinium, translational regulation and post-translational modifications have been hypothesized to primarily drive changes in the proteome under heat stress (Barshis et al. 2014), as only a small collection of transcription factors have been identified in the transcriptome and genome of Symbiodinium (Bayer et al. 2012; Shoguchi et al. 2013). Symbiodinium transcriptomes have also been found to contain microRNAs (Baumgarten et al. 2013), molecules that repress translation of mRNA into proteins as well as direct and accelerate mRNA degradation (Valencia-Sanchez et al. 2006; Wu et al. 2006). Regulation of mRNA abundance may, therefore, be an important contributor to physiological responses by Symbiodinium.
Several previous gene expression studies in Symbiodinium have applied acute heat stress on the scale of hours to a few days (Baumgarten et al. 2013; Barshis et al. 2014; Rosic et al. 2014; Krueger et al. 2015), but a study on mRNA stability in the dinoflagellate Karenia brevis found dinoflagellate mRNA half-lives to be considerably longer than in other organisms (Morey and Van Dolah 2013). The majority of transcripts involved in the stress response, metabolism, and transcriptional regulation had half-lives over 24 h, and in some cases over four days (e.g., catalase/peroxidase, thioredoxin, and chaperone protein DnaJ) (Morey and Van Dolah 2013). Thus, some dinoflagellate genes may simply require longer periods of time to develop significant, detectable mRNA expression changes. However, Morey and Van Dolah (2013) did not measure mRNA half-lives under temperature stress, which can significantly alter mRNA stability (Castells-Roca et al. 2011; Chiba et al. 2013).
In this study, we used two heterogeneous populations of type C1 Symbiodinium, an ecologically important and globally distributed type associated with a diverse range of coral species (LaJeunesse et al. 2004; LaJeunesse 2005; Tonk et al. 2013). Despite having identical ITS1 and ITS2 sequences, the populations exhibit different thermal tolerances. Physiological and transcriptional analyses were conducted for each population at ambient (27 °C) and elevated (32 °C) temperatures in culture in order to investigate the molecular basis of Symbiodinium thermal tolerance. The populations were originally isolated from the coral Acropora tenuis at two separate sites on the Great Barrier Reef: South Molle Island (SM; 20°16′33″S, 148°49′36″E) and Magnetic Island (MI; 19°9′6″S, 146°51′56″E) that have average summer daily maximums of 28.2 °C and 30.1 °C, respectively. Corals harboring the thermo-sensitive SM population were previously shown to bleach after 11 days at 32 °C, whereas corals harboring the thermo-tolerant MI population remained unaffected (Howells et al. 2012). A significant reduction in photosynthetic capacity due to heat stress, a diagnostic trait of Symbiodinium thermal sensitivity and coral bleaching (Warner et al. 1999), accompanied loss of the SM population from its coral host at 32 °C (Howells et al. 2012). The susceptibility of each population to elevated temperature in hospite correlated with thermal tolerance in culture (Howells et al. 2012).
Here we report on thousands of differentially expressed genes (DEGs) in both populations exposed to elevated temperature (32 °C) that align with physiological responses. Our findings demonstrate how distinct transcriptomic plasticity and regulation of hallmark thermal tolerance genes and functional gene groups (i.e., gene ontology categories) can allow allopatric, conspecific Symbiodinium populations to exhibit contrasting thermal tolerances.
Results and Discussion
Physiological Responses of Symbiodinium to Heat Stress
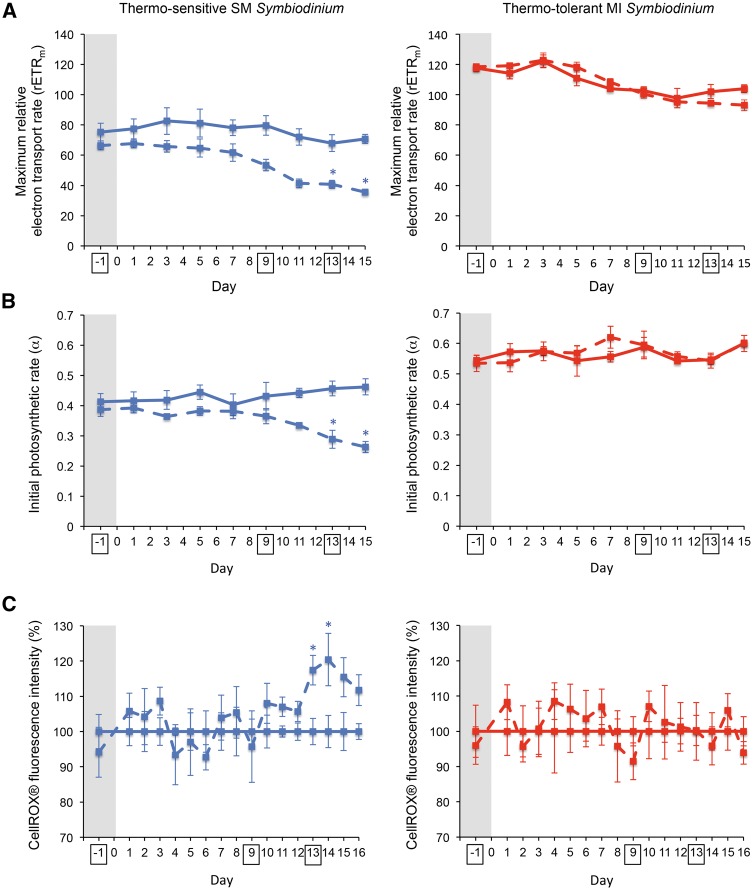
Each population was cultured at 27 °C and 32 °C in two replicate incubators (supplementary table S1, Supplementary Material online) to avoid potential artifacts from individual incubators in our results. Physiological measurements for detection of cellular heat stress were used to determine sampling time points for transcriptomics that were anticipated to identify DEGs between temperature treatments (fig. 1A–C and supplementary fig. S1A–D, Supplementary Material online). On day 13, both the maximum relative electron transport rate for photosynthesis (rETRm) and initial photosynthetic rate (α) significantly decreased (P < 0.05) at 32 °C compared with 27 °C in the SM population only (fig. 1A and B). Decreased photosynthetic ability of Symbiodinium has been strongly connected to Symbiodinium thermal sensitivity and coral bleaching susceptibility (Warner et al. 1999; Takahashi et al. 2009; Ragni et al. 2010; Howells et al. 2012). Additionally, a significant increase (P < 0.05) in general ROS leakage out of Symbiodinium cells was detected in the SM population at 32 °C beginning on day 13 (fig. 1C), an observation that is consistent with evidence that coral bleaching is largely driven by increased ROS inside coral tissues (Downs et al. 2002; Suggett et al. 2008). Therefore, day 13 was chosen as a sampling time point for transcriptomics, along with day −1 to account for any pre-experimental DEGs between groups. Day 9, the potential start of the declining trend in rETRm in the SM population, was also selected as a sampling time point for transcriptomics to determine if the transcriptional response to heat stress precedes significant physiological damage. The overall lower photosynthetic efficiency of the SM population may be due to the lower amounts of photosynthetic pigments (chlorophyll a and β−carotene) in cells from the SM population compared to those from the MI population (Howells et al. 2012).
Fig. 1.
Physiological detection of Symbiodinium heat stress. Intact lines represent the 27 °C temperature treatment, and dashed lines represent the 32 °C temperature treatment. Before heating, all samples were kept at 27 °C (values in the grey regions). (A) rETRm (mean ± sem, n = 4). (B) α (mean ± sem, n = 4). (C) Leakage of ROS out of cells (mean ± sem, n = 4); unitless fluorescence intensities of CellROX® Orange reagent for oxidative stress detection were normalized across days by setting the fluorescence intensities of the 27 °C samples to 100%. Asterisks indicate statistically significant (PERMANOVA) differences between temperature treatments at P < 0.05. Sampling time points for transcriptomics are boxed. Additional physiological measurements are shown in supplementary fig. S1, Supplementary Material online.
Plasticity of Symbiodinium Transcriptomes under Heat Stress
The de novo assembled transcriptomes from the SM and MI populations were composed of 106,097 and 93,377 putative genes, respectively. However, the number of genes in each transcriptome likely overestimates the number of genes expressed by a single genotype because our study used heterogeneous populations rather than clonal cultures. Each population consisted of an unknown diversity of individuals within type C1 and, therefore, an unknown diversity of transcript variants and alleles. The SM and MI populations, rather than clonal cultures, were chosen in our study as their bleaching responses at 32 °C have been characterized in hospite (Howells et al. 2012) and as heterogeneous populations are more representative of symbiont communities inhabiting Great Barrier Reef corals. Average transcript lengths (SM: 858.1 bp and MI: 911.4 bp; supplementary table S2, Supplementary Material online) for the SM and MI transcriptomes were in range with those for previously published Symbiodinium transcriptomes (Bayer et al. 2012; Parkinson et al. 2016). Quantitative assessment of conserved eukaryotic orthologs (Simão et al. 2015) revealed that the SM and MI transcriptomes are the most complete Symbiodinium transcriptomes of the publicly accessible, published Symbiodinium transcriptomes to date (Bayer et al. 2012; Ladner et al. 2012; Baumgarten et al. 2013; Rosic et al. 2015; Xiang et al. 2015; Parkinson et al. 2016) (supplementary table S3, Supplementary Material online). The biological coefficient of variance (BCV) for gene expression across replicates in each population was found to be < 0.2 on all time points, well below the commonly accepted variance threshold of 0.4 (McCarthy et al. 2012; Chen et al. 2014).
For differential gene expression analysis, we defined significant biological relevance as ≥ 4-fold differential expression between temperature treatments combined with a conservative false discovery rate (FDR) ≤ 0.001. On day −1 prior to heat treatment, only one DEG (TR83958|c0_g1, a putative 10 kDa chaperonin) in the SM population and no DEGs in the MI population were found between the experimental groups of each population that had been pre-assigned to the different temperature treatments. TR83958|c0_g1 from the SM population was not differentially expressed on either of the later time points. The lack of DEGs between experimental groups in both populations before heating corroborates that DEGs detected on days 9 and 13 were in response to the temperature treatment and that differential expression cutoffs (fold ≥ 4 and FDR ≤ 0.001 between temperature treatments) and replication (n = 4) were adequate to achieve a high signal to noise ratio.
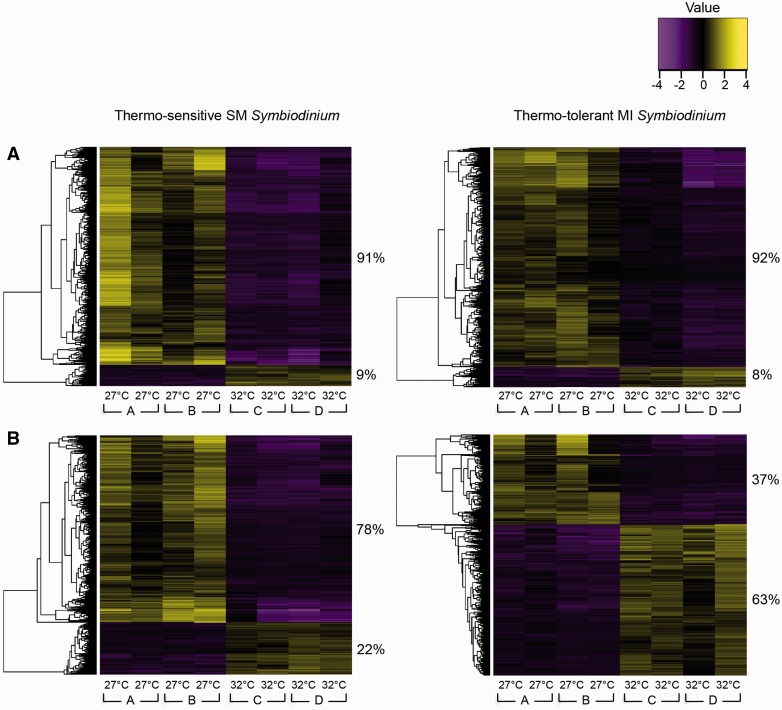
On day 9, a total of 4,608 and 2,379 DEGs were identified between the temperature treatments in the SM and MI populations, respectively. The vast majority of DEGs in the SM population (4,199 or 91%) and MI population (2,179 or 92%) were down-regulated at 32 °C relative to expression levels at 27 °C (fig. 2A). Down-regulation of the majority of DEGs in response to elevated temperature has been previously observed in marine organisms including Symbiodinium and corals (Baumgarten et al. 2013; Yampolsky et al. 2014; Bay and Palumbi 2015) and may be a strategy to conserve energy when confronted with environmental stress (Yampolsky et al. 2014).
Fig. 2.
Hierarchical clustering of DEGs. Heat maps show genes (rows) with differential expression (Trinity/edgeR: fold ≥ 4, FDR ≤ 0.001) between 27 °C and 32 °C samples (columns) for each population on (A) day 9 and (B) day 13. Expression values (fpkm) are log2-transformed and then median-centered by gene. Heat map values were calculated by subtracting each gene’s median log2(fpkm) value from its log2(fpkm) value in each sample. The proportions (%) of DEGs that were up- or down-regulated due to heat stress are noted to the right of the two main gene clusters of each heat map. Genes are independently clustered for each population at each time point. Samples from replicate cultures at each temperature treatment are presented in the same order for each time point. The experimental incubator (A, B, C, or D) that housed each sample is noted below the temperature treatment.
On day 13, a total of 4,272 and 3,513 DEGs were identified between the temperature treatments in the SM and MI populations, respectively. The SM population responded similarly to 32 °C on day 13 as on day 9 by down-regulating the majority of DEGs (3,341 or 78%). Conversely, the MI population up-regulated the majority of DEGs (2,201 or 63%) at 32 °C, suggesting acclimation to 32 °C (fig. 2B). Our results demonstrate that some Symbiodinium do exhibit transcriptomic plasticity and are capable of up-regulating a large number of genes in response to elevated temperature.
In the SM population at 32 °C, 239 and 1,925 genes remained up- and down-regulated, respectively, on both days 9 and 13. In the MI population at 32 °C, 113 and 585 genes remained up- and down-regulated, respectively, on both days 9 and 13. Interestingly, 353 genes in the MI population at 32 °C that were down-regulated on day 9 became up-regulated on day 13, whereas no genes switched from down- to up-regulation in the SM population at 32 °C. No up-regulated genes on day 9 became down-regulated on day 13 in either population at 32 °C.
Gene Ontology (GO) Analysis of DEGs to Identify Functional Gene Groups Involved in Thermal Tolerance
GO analysis (FDR < 0.05) of genes at 32 °C further supported that only the MI population acclimated to elevated temperature (fig. 3 and supplementary dataset S1A–M, Supplementary Material online), consistent with only the SM population suffering physiological damage after 13 days of heat stress. Acclimation to stressful conditions through transcriptional changes has been observed in other marine organisms including corals (Nymark et al. 2009; Yampolsky et al. 2014; Bay and Palumbi 2015; Hennon et al. 2015), but never before in Symbiodinium, or to our knowledge, in any dinoflagellate species.
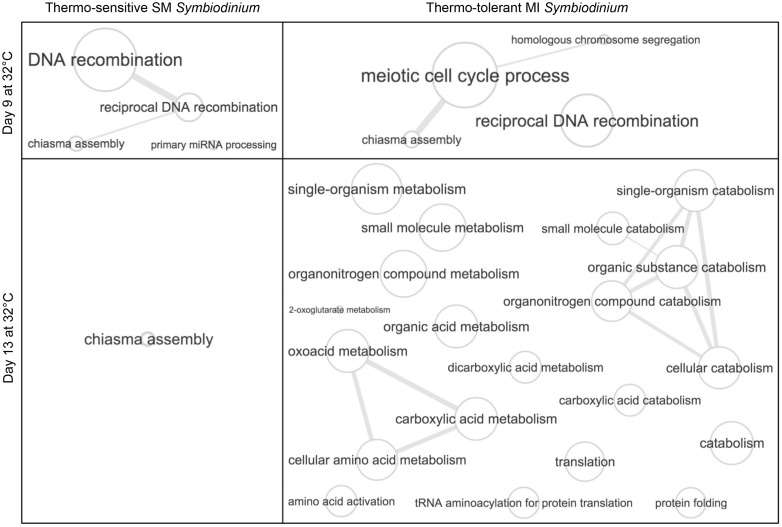
Fig. 3.
Unsuccessful versus successful acclimation to elevated temperature. GO relationship graphs for enriched biological process GO categories (Goseq: FDR < 0.05) at 32 °C were generated using REVIGO (Supek et al. 2011). Bubble size indicates the frequency of the GO category in the UniProt database relative to the other GO categories that are in the same section. Lines link similar GO categories, and the line width indicates the degree of similarity between the GO categories relative to others in the same section. Redundant GO categories (similarity > 0.9) were collapsed into the category that is most frequent in the UniProt database. Graphs for enriched molecular function GO categories (e.g., unfolded protein binding and glutamate dehydrogenase activity) and enriched cellular component GO categories (e.g., oxidoreductase complex and motile cilium) at 32 °C are not included in this figure. Full GO analysis results are listed in supplementary dataset S1A–M, Supplementary Material online.
On day 9, the down-regulated genes in the SM population at 32 °C were enriched for 133 GO categories consisting of 26 metabolic and biosynthetic categories, whereas the down-regulated genes in the MI population at 32 °C were enriched for 311 GO categories that included 45 metabolic and biosynthetic categories (supplementary dataset S1B and D, Supplementary Material online). Reduced metabolic and biosynthetic activity has been shown to correlate with increased survival time of organisms under stress, as it allows for substantial energetic savings (Hand and Hardewig 1996). Specifically in the case of heat stress, such metabolic compensation is considered an acclimatory mechanism to elevated temperature in the zooplankton Daphnia pulex (Yampolsky et al. 2014).
The small number of significantly up-regulated genes in the SM and MI populations at 32 °C on day 9 was enriched for six and seven GO categories, respectively (fig. 3 and supplementary dataset S1A and C, Supplementary Material online). The majority of enriched GO categories in both populations were specific to meiosis, suggesting that Symbiodinium cells were participating in sexual rather than strictly asexual reproduction under heat stress. Potential sexual reproduction by Symbiodinium is particularly noteworthy since meiosis creates genetic diversity through chromosomal modifications and recombination, therefore promoting adaptation (Tamburini and Tyler 2005; D'Souza and Michiels 2010; Becks and Agrawal 2012). Meiosis-specific genes have been previously identified in Symbiodinium (Chi et al. 2014; Rosic et al. 2015), but so far, sexual reproduction has not been directly observed. However, recent studies have hypothesized that recombination during meiosis may be a mechanism of adaptation in Symbiodinium (Chi et al. 2014; Wilkinson et al. 2015). Other dinoflagellate species can rapidly increase genetic diversity by switching from mitosis to meiosis and can enter a sexual cyst life cycle stage when exposed to stressful conditions in order to survive and adapt (Figueroa et al. 2010; Bravo and Figueroa 2014), though no visually apparent Symbiodinium cysts were observed in our study.
On day 13, the dramatic increase in up-regulated genes in the MI population at 32 °C was characterized by enrichment of 60 GO categories (fig. 3 and supplementary dataset S1G, Supplementary Material online), which included nine metabolic categories along with several of the corresponding catabolic categories for maintaining cellular homeostasis. Importantly, GO categories for unfolded protein binding, protein folding, glutamate dehydrogenase (NAD+) activity and the oxidoreducatase complex—all of which are involved in stress tolerance (Srivastava and Singh 1987; Singh and Grover 2008; Bita and Gerats 2013; Tercé-Laforgue et al. 2015)—also became enriched in the up-regulated genes in the MI population at 32 °C. The subset of 353 up-regulated genes at 32 °C, which had been down-regulated at 32 °C on day 9, was enriched for 29 GO categories including seven for metabolism and biosynthesis, one for oxidoreductase activity, and one for motile cilium (supplementary dataset S1M, Supplementary Material online). The down-regulated genes in the MI population at 32 °C on day 13 were enriched for 160 GO categories covering a broad array of processes, though only 13 metabolic and biosynthetic GO categories were present (supplementary dataset S1H, Supplementary Material online). The largely reduced metabolic compensation at 32 °C on day 13 (down-regulated genes enriched for 13 metabolic and biosynthetic GO categories) relative to day 9 (down-regulated genes enriched for 45 metabolic and biosynthetic GO categories), along with up-regulation of genes enriched for metabolic and stress tolerance GO categories on day 13, suggests that the MI population had acclimated to 32 °C.
In contrast, GO enrichment analysis indicated that the SM population was unable to acclimate to 32 °C by day 13. The down-regulated genes in the SM population at 32 °C on day 13 were enriched for 135 GO categories, including 19 for metabolism and biosynthesis and one for the oxidoreductase complex (supplementary dataset S1F, Supplementary Material online). Only three GO categories were enriched in the up-regulated genes in the SM population at 32 °C, one of which was the meiosis GO category, chiasma assembly, potentially signifying a continued attempt to adapt by producing genetic diversity through sexual recombination (fig. 3 and supplementary dataset S1E, Supplementary Material online). No metabolic, biosynthetic, or stress tolerance GO categories became enriched in the up-regulated genes at 32 °C. The extended duration of metabolic compensation experienced by the SM population compared with the MI population under heat stress could potentially cause starvation of its coral host, which may contribute to the higher bleaching susceptibility of corals harboring the SM population (Howells et al. 2012).
Regulation of Hallmark Genes Involved in Adaptation and Thermal Tolerance
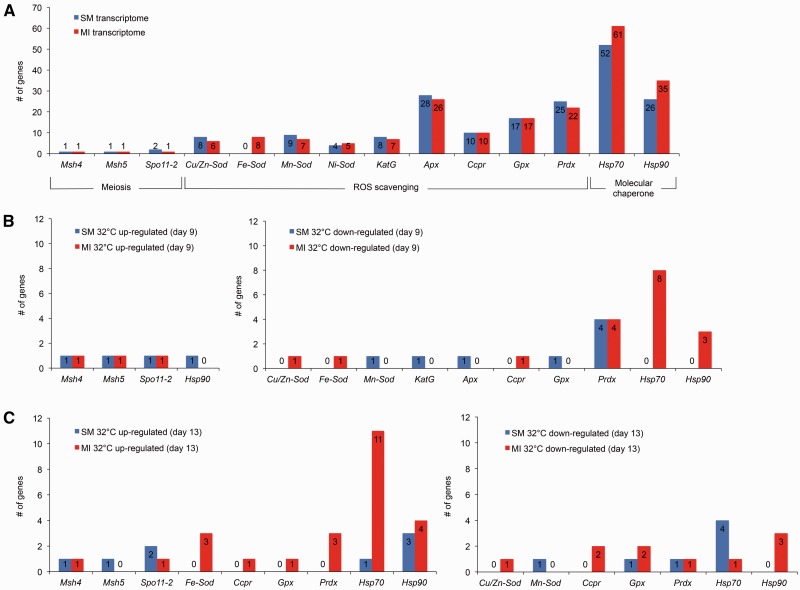
Although the responses of the two Symbiodinium populations to heat stress differed, both transcriptomes contained comparable suites of meiosis-specific and thermal tolerance genes (fig. 4A and supplementary fig. S2A, Supplementary Material online) that are consistent with gene content found in other Symbiodinium (Bayer et al. 2012; Chi et al. 2014; Krueger et al. 2015; Rosic et al. 2015). However, a striking difference in gene content between the transcriptomes of the SM and MI populations was the expression of eight iron superoxide dismutase (Fe-Sod) genes in the MI transcriptome, whereas no Fe-Sod genes were expressed in the SM transcriptome, implying that these genes are either absent from the SM population or their expression is suppressed through epigenetic regulation. Detectable Fe-Sod gene expression is inconsistent among other Symbiodinium (Krueger et al. 2015), and phylogenetic evidence that the acquisition of several ROS scavenging genes by Symbiodinium has resulted from horizontal gene transfer (Krueger et al. 2015) indicates that some Symbiodinium genomes may lack Fe-Sod genes entirely. Successful PCR amplification of the most highly expressed Fe-Sod gene (TR20255|c0_g1, open reading frame: 674-78[-]) from the genomic DNA of the MI population but not the SM population highlights the robustness of our transcriptome assemblies and supports that some gene content varies between the populations (supplementary fig. S3A, Supplementary Material online). However, our PCR results cannot confirm that no Fe-Sod genes are present in the SM population, as the primers were specific to the open reading frame of TR20255|c0_g1. Differences in nucleotide sequence between the open reading frame of TR20255|c0_g1 and the other seven Fe-Sod genes in the MI population, as well as the Fe-Sod genes identified by Krueger et al. (2015) in types B1, E, and F1 Symbiodinium (supplementary fig. S3B, Supplementary Material online), suggest that alternative Fe-Sod genes could be in the SM population but be silenced or expressed below the detectable level. Though, it should also be noted that Krueger et al. (2015) failed to find any Fe-Sod genes expressed in types C1, C3, C15, and D Symbiodinium.
Fig. 4.
Regulation of meiosis, ROS scavenging, and molecular chaperone genes. (A) The number of genes for gene types involved in sexual reproduction or thermal tolerance in the SM and MI transcriptomes. The number of DEGs at 32 °C from each gene type (Trinity/edgeR: fold ≥ 4 and FDR ≤ 0.001 relative to 27 °C) are shown for each population on (B) day 9 and (C) day 13. Gene types that had no DEGs in either population are excluded from (B) and (C). Gene abbreviations are as follows: mutS protein homolog 4 (Msh4), mutS protein homolog 5 (Msh5), meiotic recombination protein Spo11-2 (Spo11-2), copper/zinc superoxide dismutase (Cu/Zn-Sod), iron superoxide dismutase (Fe-Sod), manganese superoxide dismutase (Mn-Sod), nickel superoxide dismutase (Ni-Sod), catalase-peroxidase (KatG), ascorbate peroxidase (Apx), cytochrome c peroxidase (Ccpr), glutathione peroxidase (Gpx), peroxiredoxin (Prdx), heat shock protein 70 (Hsp70), heat shock protein 90 (Hsp90). Additional genes are shown in supplementary fig. S2, Supplementary Material online. DEG annotation and differential expression details are provided in supplementary tables S4–S7, Supplementary Material online.
On days 9 and 13, both populations maintained up-regulation of meiosis-specific genes at 32 °C (fig. 4B and C), although statistically significant up-regulation (fold ≥ 4, FDR ≤ 0.001) of mutS homolog 5 (Msh5) was limited to day 9 in the MI population. The heterodimer partners mutS homolog 4 (Msh4) and Msh5 are members of the Msh gene family. Unlike the other Msh genes that are involved in mismatch repair, DNA damage repair, and mitotic recombination (Modrich and Lahue 1996; Wang and Qin 2003; Stojic et al. 2004), studies in a wide range of organisms (including humans, mice, yeast, Caenorhabditis elegans, Arabidopsis thaliana, and Tetrahymena thermophile) show that Msh4 and Msh5 genes are essential and specific to meiosis (Hollingsworth et al. 1995; Bocker et al. 1999; Kelly et al. 2000; Kneitz et al. 2000; Novak et al. 2001; Argueso et al. 2004; Higgins et al. 2004; Shodhan et al. 2014). MSH4 and MSH5 proteins form a meiosis-specific sliding clamp that holds and pairs homologous chromosomes during meiosis (Kneitz et al. 2000; Snowden et al. 2004). Mutations to Msh4 and Msh5 genes have both been shown to affect crossing over of homologous chromosomes but not to affect mismatch repair (Ross-Macdonald and Roeder 1994; Hollingsworth et al. 1995). However, some studies indicate that MSH4 and/or MSH5 proteins may have additional functions outside of meiosis processes such as DNA damage response (Her et al. 2003; Sekine et al. 2007; Tompkins et al. 2009). To consider whether Symbiodinium Msh4 and Msh5 genes may atypically function in non-meiotic pathways with other Msh genes, we investigated the gene expression of Msh1, 2, 3, and 6 in the Symbiodinium transcriptomes. Interestingly, none of the Msh genes besides Msh4 and Msh5 were up-regulated at 32 °C, supporting that meiosis-specific processes were induced rather than mismatch repair, DNA damage repair, or mitotic recombination.
The meiotic recombination protein Spo11-2 (Spo11-2) gene was also up-regulated in both Symbiodinium transcriptomes on days 9 and 13 at 32 °C. Spo11-2 and its paralog, the meiotic recombination protein Spo11-1 (Spo11-1) gene, are the meiosis-specific members of the Spo11 gene family. SPO11-1 and SPO11-2 proteins create meiosis-specific double-strand breaks in DNA and form the synaptonemal complex to initiate meiosis (Cao et al. 1990; Keeney et al. 1997; Tsubouchi and Roeder 2005; Keeney 2007). Mutations to Spo11-2 in Arabidopsis thaliana cause sterility and aneuploidy (Stacey et al. 2006; Hartung et al. 2007). Although we are not aware of any examples in which the Spo11-2 gene acts outside of meiosis, future detailed studies will be important to confirm that the meiosis-specific functions of Spo11-2, as well as Msh4 and Msh5, are conserved in Symbiodinium.
Superoxide dismutases are key scavengers of superoxide, peroxidases are engaged in the removal of hydrogen peroxide, and molecular chaperones are essential for refolding damaged proteins (Vierling 1991; Gill and Tuteja 2010)—making them key contributors to thermal tolerance. Despite the many examples that up-regulation of these genes confers thermal tolerance in numerous photosynthetic species (Van Breusegem et al. 1999; Tang et al. 2006; Singh and Grover 2008; Bita and Gerats 2013), many studies report no notable differential expression of these genes in Symbiodinium at elevated temperature (Leggat et al. 2011; Putnam et al. 2013; Barshis et al. 2014; Krueger et al. 2015). Yet, limited evidence suggests that the transcriptional heat stress response of Symbiodinium may involve up-regulation of some genes classically associated with thermal tolerance. The first study demonstrated through qPCR that cytochrome P450 (Cyp450) gene expression by type C3 Symbiodinium increased at 26 °C and 29 °C compared with 23–24 °C, whereas exposure to 32 °C resulted in decreased Cyp450 expression (Rosic et al. 2010). The next study also used qPCR and showed that heat shock protein 70 (Hsp70) expression in type C1 Symbiodinium was slightly increased at approximately 30 °C, but down-regulation of Hsp70 occurred at 32 °C (Rosic et al. 2011). An RNA-seq study of type A1 Symbiodinium found up-regulation of one peroxiredoxin (Prdx) gene, one Hsp gene, and one chaperone protein DnaJ (DnaJ) gene from exposure to 34 °C for 12 h (Baumgarten et al. 2013). However, the importance of the differential gene expression at this extreme temperature was not substantiated by sample replication, correspondence to a physiological heat stress response, or relation to a coral bleaching response (Baumgarten et al. 2013). Finally, a recent RNA-seq study detected minor up-regulation of Hsp90 by in hospite Symbiodinium after 24 h of exposure to 30 °C relative to 23–24 °C, but not after 72 h of exposure to 30 °C (Rosic et al. 2014).
In our study, general down-regulation of thermal tolerance genes was observed on day 9 in both populations at 32 °C (fig. 4B and supplementary fig. S2B and tables S4 and S5, Supplementary Material online). One Hsp90 gene, one Cyp450 gene, and two DnaJ genes were up-regulated by the SM population at 32 °C compared with just one DnaJ gene up-regulated in the MI population at 32 °C. Hsp genes were uniquely found to be down-regulated in the MI population at 32 °C, although the MI population also showed no signs of physiological heat stress throughout the study. Elevated temperature has previously been shown to reduce the expression of Hsp genes and Cyp450 genes in Symbiodinium (Rosic et al. 2010; Rosic et al. 2011) as well as the expression of Hsp genes and ROS scavenging genes in corals (Rosic et al. 2014; Bay and Palumbi 2015). Down-regulation of some thermal tolerance genes may be attributed to the general down-regulation of > 90% of all DEGs in both populations at 32 °C on day 9. The down-regulated genes in each population were not enriched for GO categories related to thermal tolerance (e.g., unfolded protein binding, the oxidoreductase complex), supporting the notion that down-regulation of thermal tolerance genes may simply reflect a non-targeted, global reduction in transcription to conserve energy at 32 °C.
On day 13, only one glutaredoxin (Glrx) gene was up-regulated in the SM population at 32 °C. In contrast, three Fe-Sod genes, one cytochrome c peroxidase (Ccpr) gene, one glutathione peroxidase (Gpx) gene, three Prdx genes, two thioredoxin (Txn) genes, and one Cyp450 gene were significantly up-regulated in the MI population at 32 °C, highlighting the importance of ROS scavenging genes in type C1 Symbiodinium thermal tolerance. Additionally, 11 Hsp70 genes, four Hsp90 genes, and eight DnaJ genes were up-regulated by the MI population at 32 °C compared with only one Hsp70 gene, three Hsp90 genes, and four DnaJ genes up-regulated in the SM population at 32 °C (fig. 4C and supplementary fig. S2C and tables S6 and S7, Supplementary Material online). Up-regulation of Hsp90 genes by both populations under heat stress is consistent with findings that Symbiodinium HSP90 protein abundance increases under heat stress (Ross 2014).
Linking Symbiodinium Transcriptional Heat Stress Responses to Thermal History, Physiological Heat Stress Responses, and Coral Bleaching Susceptibility
The SM and MI Symbiodinium populations have been kept in culture for more than 4 years at approximately 27 °C, and their relative thermal tolerances, reported in 2012 (Howells et al. 2012), were confirmed in our current study conducted in 2015. The marked difference in their transcriptional responses to elevated temperature may, therefore, be driven by stable, heritable stress memory due to the different thermal regimes of the SM and MI reefs. Stress memory (or “priming”) is the process in which previous exposure to a particular stress causes epigenetic and/or chromosomal modifications. The modifications allow for a faster and stronger acclimation response to subsequent exposures and can be stably passed on to future generations (Bruce et al. 2007). The warmer MI reef reaches ≥ 32 °C on approximately 12% of summer days, unlike the cooler SM reef where no summer days reach ≥ 32 °C (Howells et al. 2012), suggesting that only the MI population has been primed and/or genetically adapted for efficient acclimation to 32 °C. Successful PCR amplification of a Fe-Sod gene from only the genomic DNA of the MI population (supplementary fig. S3A, Supplementary Material online) indicates that genetic adaptation is involved in acclimation to 32 °C, but epigenomic and genomic analysis will be necessary to determine whether stress memory also contributes to the transcriptional acclimation response.
Acclimation to elevated temperature by the MI population highlights the importance of up-regulating hallmark thermal tolerance genes. Particularly, significant up-regulation of genes for unfolded protein binding, protein folding, and the oxidoreductase complex likely minimizes damage to photosynthetic apparatuses and ROS leakage from cells—both of which were observed in the heat stressed SM population. We hypothesize that the observed transcriptional response by the MI population in culture may also allow the MI population to maintain symbiosis with its coral host at elevated temperature (Howells et al. 2012). Conversely, the observed leakage of ROS out of cells in the SM population due to unsuccessful acclimation to elevated temperature may cause oxidative damage to the coral host, resulting in bleaching as previously seen with corals harboring the SM population when exposed to heat stress (Howells et al. 2012) (fig. 5). Although Hsp gene expression has been found to be indistinguishable between Symbiodinium in culture and in hospite (Rosic et al. 2011), more extensive temporal studies of in hospite Symbiodinium gene expression will be necessary to determine the effect of symbiosis on the comprehensive collection of DEGs identified here. Metabolomics should also be utilized to determine if metabolic compensation of in hospite Symbiodinium over extended periods of heat stress factors into the breakdown of Symbiodinium-coral symbiosis.
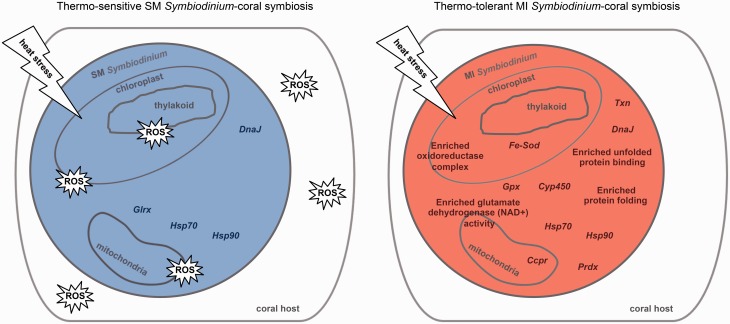
Fig. 5.
Model of the molecular basis of Symbiodinium thermal tolerance and its impacts on Symbiodinium-coral symbiosis. Schematics of Symbiodinium cells from the SM population and MI population after 13 days at 32 °C hypothesize the impacts of their respective up-regulated thermal tolerance genes (Trinity/edgeR: fold ≥ 4 and FDR ≤ 0.001 relative to 27 °C) and enriched thermal tolerance GO categories (Goseq: FDR < 0.05) on their coral hosts. The main organelles contributing to ROS production are depicted. The shape containing “ROS” represents oxidative damage to the Symbiodinium cell or the coral host. Gene abbreviations are as follows: iron superoxide dismutase (Fe-Sod), cytochrome c peroxidase (Ccpr), glutathione peroxidase (Gpx), peroxiredoxin (Prdx), heat shock protein 70 (Hsp70), heat shock protein 90 (Hsp90), glutaredoxin (Glrx), thioredoxin (Txn), cytochrome P450 (Cyp450), chaperone protein DnaJ (DnaJ).
In this study, we have detailed gene regulation by a thermo-sensitive type C1 Symbiodinium population and a thermo-tolerant type C1 Symbiodinium population in response to heat stress that parallels their respective physiological responses to heat stress and previously described bleaching responses in hospite (Howells et al. 2012). Furthermore, our study is the first to identify individual genes as well as overarching functional gene groups that influence dinoflagellate thermal tolerance. Our results provide critical insights into the impacts of Symbiodinium gene regulation on coral bleaching and present genes (e.g., Msh4, Msh5, and Spo11-2) that could be used to detect heat stress in Symbiodinium before potential physiological damage occurs.
Materials and Methods
Culture Maintenance and Genotyping
The SM and MI heterogeneous Symbiodinium populations (aims-aten-C1-WSY and aims-aten-C1-MI, respectively) were provided by the Symbiont Culture Facility at the Australian Institute of Marine Science and are the same as reported in Howells et al. (2012). Following isolation from Acropora tenuis, the Symbiodinium populations were initially cultured in filtered seawater supplemented with Daigo IMK (Wako Pure Chemical Industries, Ltd.) and bacterial antibiotics for one month, which minimized the bacterial community to prevent bacterial overgrowth. Cultures were then routinely subcultured in media without antibiotics and monitored regularly by microscopy to ensure no increase in the remaining bacterial community was observed. Complete removal of all bacteria originating from the coral holobiont was not desirable, as optimal growth of dinoflagellate cultures has been shown to require associated bacteria (Alavi et al. 2001; Croft et al. 2005). Dinoflagellate cultures may depend on bacteria to provide necessary components, including but not limited to vitamin B12, in order to thrive (Croft et al. 2005; Ritchie 2012). Unlike the free-living life cycle stage in which Symbiodinium naturally live without a coral host (Yamashita and Koike 2013; Granados-Cifuentes et al. 2015), we are not aware of any stage in which Symbiodinium naturally live without associated bacteria. Therefore, complete removal of Symbiodinium-associated bacteria may have unnatural effects on Symbiodinium transcriptomes.
For genotyping of the Symbiodinium populations, DNA from cultured cells in exponential growth phase was extracted using the DNeasy Plant Mini Kit (Qiagen). The ITS1 region was amplified with the PCR primers and conditions from van Oppen et al. (2001). The partial 5.8S rDNA, ITS2, and partial 28S rDNA region was amplified with the PCR primers and conditions from Stat et al. (2009). Purified PCR products were sequenced by the Australian Genome Research Facility. The ITS1 and ITS2 sequences of each population were reconfirmed to be type C1 and to be identical between the SM and MI populations, as previously reported by Howells et al. (2012). In future studies, alternative molecular markers such as the non-coding region from the psbA minicircle could be assessed through next generation sequencing to investigate finer-scale evolutionarily divergence that may exist between the two C1 populations as well as within each C1 population (LaJeunesse and Thornhill 2011). Increased genetic resolution may provide valuable insight into the different transcriptional responses to heat stress observed in our study.
Experimental Setup
Each population (∼1 × 106 cells/ml, 50 ml total volume) was added to eight replicate culture flasks (n = 4 for each temperature treatment, supplementary table S1, Supplementary Material online). Two flasks per population were randomly assigned to each of four experimental incubators and acclimated at 27 °C. Light was provided to cells at an intensity of 30 µmol quanta m−2 s−1 (Crompton 36W cool white fluorescent tubes, 4000 K) with a 12:12 h light:dark cycle. After 10 days of acclimation, fresh media was supplied to the cultures. Following an additional four days of acclimation (two weeks of acclimation total), two incubators were ramped on day 0 at 0.5 °C/h to 32 °C for the heat stress temperature treatment, whereas two incubators remained at 27 °C for the control temperature treatment. Temperature and light intensity in the incubators were monitored with HOBO data loggers (Onset Computer Corporation). Cultures remained in exponential growth phase, determined by the average of three replicate haemocytometer counts for each sample recorded throughout the experiment (supplementary fig. S1D, Supplementary Material online).
Photosynthesis Measurements
A Mini-PAM fluorometer (Walz, Germany) was used to measure effective quantum yield (supplementary fig. S1A, Supplementary Material online) and rapid light curves (RLCs) (fig. 1A and B and supplementary fig. S1B, Supplementary Material online). RLCs are ideal for providing quick snapshots of Symbiodinium responses to a range of irradiances with results that are reasonably comparable to steady-state light curves (Suggett et al. 2015). With RLCs, < 90 s of exposure to high irradiances is applied per sample, which was short enough to avoid significant long-term damage that could greatly affect the physiology and gene expression of the Symbiodinium and allowed for the concurrent analysis of all 16 samples within an equivalent period of the light cycle on each measurement day.
A RLC protocol adapted from Ralph et al. (2002) was used in our study. After 7 h of light exposure, the fiber–optic cable of the Mini-PAM fluorometer was held against the bottom of each culture flask where the Symbiodinium cells had settled. Symbiodinium were exposed to nine steps of increasing actinic light (0–1,775 μmol m−2 s−1 PAR) for 10 s each, separated by a saturating pulse (0.8 s, > 4,000 μmol m−2 s−1 PAR). The light responses of each population at each temperature were determined by fitting the RLCs to the model by Platt et al. (1980). The variables rETRm, α, and Ek (fig. 1A and B and supplementary fig. S1B, Supplementary Material online) were calculated using SigmaPlot as per Hill et al. (2004).
ROS Measurements
Cultures were gently agitated to evenly distribute cells in the media, and aliquots (300 µl per sample) were centrifuged at 3,000g × 5 min. Media (for measuring ROS leakage) were collected without disturbing the cell pellet and incubated with CellROX® Orange reagent (5 µM, Thermo Fisher Scientific) for oxidative stress detection in a 96-well black clear bottom plate (Costar) for 2 h at 27 °C in the dark. CellROX® reagent is irreversibly converted to a fluorescent state in the presence of ROS without requiring the activity of intracellular esterases, making it an appropriate dye for measuring general ROS content in media. Fluorescence intensity of the CellROX® reagent was measured at excitation 540 nm and emission 565 nm with an EnSpire® Multimode Plate Reader (PerkinElmer). The use of CellROX® reagent with Symbiodinium culture media was validated through CellROX® reagent signal quenching from addition of antioxidant chemicals (supplementary fig. S4, Supplementary Material online).
Culture Viability Measurements
Culture viability was measured with SYTOX® Green nucleic acid stain (Life Technologies), which is unable to penetrate live Symbiodinium cells. Cultures were gently agitated to evenly distribute cells in the media, and aliquots (50 µl per sample) were incubated with SYTOX® Green nucleic acid stain (1 μM) in the dark for 15 min. An Olympus fv1000 confocal microscope with a 488 nm argon-ion laser was used to quantify the proportion of live cells in each sample based on counts of stained and unstained cells averaged across three separate fields of view (supplementary fig. S1C, Supplementary Material online).
Statistical Analysis of Physiological Measurements
The PRIMER software with the PERMANOVA+ package was used to determine significant differences (P < 0.05) between temperature treatments for each physiological measurement using PERMANOVA with two replicate incubators as a nested factor within each level of the factor temperature (27 °C and 32 °C) and two flasks of each population in each incubator for each temperature. Where the effect of incubators was not significant (P > 0.2), the incubator factor was pooled, and each temperature treatment within each population (n = 4) was compared using a one-way PERMANOVA.
Preparation and Sequencing of RNA Samples
Precisely after 6 h of light exposure, cultures were gently agitated to evenly distribute cells in the media. Aliquots containing 2–4 × 106 cells per sample were immediately snap frozen in liquid nitrogen within 10 s of removal from the experimental incubators. Instant snap freezing of Symbiodinium cells that were still in media (rather than the standard method of pelleting by centrifugation for 5–10 min, removing media, and then snap freezing, Rosic and Hoegh-Guldberg 2010; Baumgarten et al. 2013; Krueger et al. 2015) caused no sign of cell lysis or loss of RNA integrity (supplementary fig. S5, Supplementary Material online). We developed this method to ensure that the effects of experimental temperature treatments on gene expression remained unaltered during sample preservation because gene expression can be affected by centrifugation and extended handling (Baldi and Hatfield 2002). Our method is the only one of which we are aware to immediately preserve Symbiodinium RNA since the compatibility of RNAlater (Thermo Fisher Scientific) with Symbiodinium has not yet been validated. Samples were stored at −80 °C until completion of the heat stress experiment and were processed together on the same day to prevent batch effect.
Snap frozen cells were thawed at room temperature and pelleted at 4 °C (3,000g × 5 min). Media were removed, and pellets were lysed in buffer RLT (RNeasy Plant Mini Kit, Qiagen) containing β-mercaptoethanol by bead beating with 0.3 g of 710–1,180 µm acid-washed glass beads (Sigma) using a TissueLyser II (Qiagen) for 90 s at 30 Hz. RNA was then extracted and purified using the RNeasy Plant Mini Kit (Qiagen) with an added on-column DNase I treatment (Qiagen). Total RNA (150–500 ng) of each sample was sent to the Australian Genome Research Facility for confirmation of high quality RNA using an Agilent 2100 bioanalyzer, polyA-purification, Illumina TruSeq stranded library preparation, and sequencing with an Illumina HiSeq2500 (single end 100 bp, ∼107 reads per sample, supplementary table S2, Supplementary Material online).
Transcriptome Assembly and Differential Gene Expression Analysis
Illumina Truseq (TruSeq3-SE) adapters were removed from RNA sequence reads using Trimmomatic (Bolger et al. 2014). Prinseq (Schmieder and Edwards 2011) was then used to remove poly-A tails (min tail: 6-A) and to filter out short (min length: 60 bp), low quality (min mean quality score: 20, base window: 1, base step: 1), and low complexity sequences (dust method threshold: 7). The sequence reads for the 24 samples per population (four replicates, two temperature treatments, three time points) that remained after quality filtering were combined for de novo assembly of the SM population transcriptome and MI population transcriptome using Trinity (Grabherr et al. 2011; Haas et al. 2013) (version: 2.0.6). Minimum transcript length for de novo assembly was set to 150 bp. To focus on transcripts with higher coverage, only transcripts ≥ 250 bp were retained for analysis, as in Baumgarten et al. (2013). Redundant transcripts (99% sequence similarity over 99% of the shorter transcript) in each de novo assembly were collapsed into the longest representative transcript using cd-hit-est (Huang et al. 2010) (supplementary table S2, Supplementary Material online). Completeness of the SM, MI, and other publicly accessible, published Symbiodnium transcriptomes (Bayer et al. 2012; Ladner et al. 2012; Baumgarten et al. 2013; Rosic et al. 2015; Xiang et al. 2015; Parkinson et al. 2016) was assessed using BUSCO with the set of 429 conserved eukaryotic orthologs that have been found to be present in > 90% of surveyed eukaryotic species (though the surveyed species currently lack protist representatives leading BUSCO to be biased towards lower metrics for protists than would otherwise be expected) (Simão et al. 2015) (supplementary table S3, Supplementary Material online). Non-redundant (nr) genes (transcript clusters determined by Trinity based on shared sequence content) were then analyzed for differential expression (fold ≥ 4 and FDR ≤ 0.001 between temperature treatments) according to the standard Trinity pipeline (Haas et al. 2013) (https://github.com/trinityrnaseq/trinityrnaseq/wiki, last accessed June, 2016) using RSEM (Li and Dewey 2011) and edgeR (Robinson et al. 2010). Additionally, the BCV of expression counts for all genes across replicates at each time point was separately calculated in edgeR according to Chen et al. (2014).
Annotation and GO Analysis
Transcriptomes were functionally annotated with Trinotate (http://trinotate.github.io/, last accessed June, 2016), using the SwissProt and UniRef90/TrEMBL databases (NCBI BLAST+, e-value ≤ 10−5) and the Pfam-A database (HMMR, domain noise cutoff). Top hits from SwissProt were used to annotate transcripts. If a hit was not generated against SwissProt, then the top hit from UniRef90/TrEMBL determined annotation. In the absence of a UniRef90/TrEMBL hit, Pfam-A annotation was used. GOseq (Young et al. 2010), which corrects for transcript length bias, was used as detailed with Trinity (https://github.com/trinityrnaseq/trinityrnaseq/wiki/Running_GOSeq, last accessed June, 2016) for GO analysis (FDR < 0.05, ancestral terms included) of DEGs (fold ≥ 4 and FDR ≤ 0.001 between temperature treatments). SwissProt was used to assign GO categories. In the absence of a SwissProt assignment, GO categories provided by Pfam-A were used.
In the SM and MI populations, 33% and 34% of genes received a hit from SwissProt, 46% and 49% of genes received a hit from UniRef90/TrEMBL, and 34% and 36% of genes received a hit from Pfam-A; respectively. In total, 50% of genes in the SM population and 52% of genes in the MI population received annotation from at least one database, and 35% of genes in the SM population and 36% of genes in the MI population were annotated with GO categories—similar to what has been previously reported for annotation of other Symbiodinium transcriptomes (Baumgarten et al. 2013; Rosic et al. 2015; Parkinson et al. 2016). Raw sequence reads, assembled transcriptomes, gene read count matrices, and transcript annotation results are available on NCBI GEO (GEO: GSE72763).
Isolation of a Fe-Sod Gene
The SM and MI populations were cultured for one week in sterile media with 300 μg/ml of ampicillin, followed by one week in sterile media with 300 μg/ml kanamycin, and finally for one week in sterile media with 100 μg/ml spectinomycin. Afterwards, Symbiodinium cells were pelleted (3,000g × 5 min) and washed three times in sterile media. Genomic DNA was extracted with a PureLink® Genomic DNA Mini Kit (Thermo Fisher Scientific). To confirm that the genomic DNA was of high quality and amplifiable, ITS2 PCR primers (Stat et al. 2011) were successfully used to amplify the ITS2 region from 25 ng of SM or MI genomic DNA. Primers for amplification of a full-length Symbiodinium Fe-Sod gene were based on TR20255|c0_g1 (open reading frame: 674-78[-]) from the MI population (forward: 5′ ATG GCC TTC TCC ATC CCA CCG 3′; reverse: 5′ TCA CAG GTT GGA CTC GGC GAA C 3′) and used for PCR reactions containing 125 ng of SM or MI genomic DNA (supplementary fig. S3A, Supplementary Material online). The purified Fe-Sod PCR product that was amplified from the MI genomic DNA was sequenced by the Australian Genome Research Facility and confirmed to match TR20255|c0_g1 (open reading frame: 674-78[-]). The sequence of TR20255|c0_g1 (open reading frame: 674-78[-]) was aligned to the sequences of Symbiodinium Fe-Sod genes identified by Krueger et al. (2015) using ClustalW (Thompson et al. 2002). Alignments were visualized with UCSF Chimera (Pettersen et al. 2004) (supplementary fig. S3B, Supplementary Material online).
Supplementary Material
Supplementary material figures S1–S5, tables S1–S7 and dataset S1 is available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The Biomedical Imaging Facility at The University of New South Wales covered expenses for the use of confocal microscopes and plate readers. Iveta Slapetova provided technical support for imaging. Alexandra Campbell and Ezequiel Marzinelli advised on statistical analysis with PRIMER/PERMANOVA+. Duncan Smith provided technical support and training for the Katana computational cluster at The University of New South Wales. Brian Haas, Cristina Diez-Vives, Maxine Lim, Peter Davey, Cheong Xin Chan, and Zhiliang Chen provided technical support for RNA-seq analysis. David Suggett provided valuable feedback on the writing of this manuscript. The Centre for Marine Bio-Innovation at The University of New South Wales, the Australian Institute of Marine Science, and the Linnean Society of New South Wales contributed financial support for this study. Raw and processed transcriptomics data are available through NCBI GEO (GSE72763).
References
- Alavi M, Miller T, Erlandson K, Schneider R, Belas R. 2001. Bacterial community associated with Pfiesteria‐like dinoflagellate cultures. Environ Microbiol 3:380–396. [DOI] [PubMed] [Google Scholar]
- Arbeitman MN, Furlong EEM, Imam F, Johnson E, Null BH, Baker BS, Krasnow MA, Scott MP, Davis RW, White KP. 2002. Gene expression during the life cycle of Drosophila melanogaster. Science 297:2270–2275. [DOI] [PubMed] [Google Scholar]
- Argueso JL, Wanat J, Gemici Z, Alani E. 2004. Competing crossover pathways act during meiosis in Saccharomyces cerevisiae. Genetics 168:1805–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arif C, Daniels C, Bayer T, Banguera–Hinestroza E, Barbrook A, Howe CJ, LaJeunesse TC, Voolstra CR. 2014. Assessing Symbiodinium diversity in scleractinian corals via next‐generation sequencing‐based genotyping of the ITS2 rDNA region. Mol Ecol 23:4418–4433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldi P, Hatfield GW. 2002. DNA microarrays and gene expression: from experiments to data analysis and modeling. Cambridge: Cambridge University Press. [Google Scholar]
- Barnes DDJ, Chalker BBE. 1990. Calcification and photosynthesis in reef-building corals and algae In: Dubinsky Z, editor. Ecosystems of the world. Vol. 25 Coral Reefs. Amsterdam: Elsevier; p. 109–131. [Google Scholar]
- Barshis DJ, Ladner JT, Oliver TA, Palumbi SR. 2014. Lineage-specific transcriptional profiles of Symbiodinium spp. unaltered by heat stress in a coral host. Mol Biol Evol 31:1343–1352. [DOI] [PubMed] [Google Scholar]
- Baumgarten S, Bayer T, Aranda M, Liew YJ, Carr A, Micklem G, Voolstra CR. 2013. Integrating microRNA and mRNA expression profiling in Symbiodinium microadriaticum, a dinoflagellate symbiont of reef-building corals. BMC Genomics 14:704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bay RA, Palumbi SR. 2015. Rapid acclimation ability mediated by transcriptome changes in reef-building corals. Genome Biol Evol 7:1602–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer T, Aranda M, Sunagawa S, Yum LK, DeSalvo MK, Lindquist E, Coffroth MA, Voolstra CR, Medina M. 2012. Symbiodinium transcriptomes: genome insights into the dinoflagellate symbionts of reef-building corals. PLoS One 7:e35269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becks L, Agrawal AF. 2012. The evolution of sex is favoured during adaptation to new environments. PLoS Biol 10:e1001317.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bita CE, Gerats T. 2013. Plant tolerance to high temperature in a changing environment: scientific fundamentals and production of heat stress-tolerant crops. Front Plant Sci 4:273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocker T, Barusevicius A, Snowden T, Rasio D, Guerrette S, Robbins D, Schmidt C, Burczak J, Croce CM, Copeland T. 1999. hMSH5 a human MutS homologue that forms a novel heterodimer with hMSH4 and is expressed during spermatogenesis. Cancer Res 59:816–822. [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo I, Figueroa RI. 2014. Towards an ecological understanding of dinoflagellate cyst functions. Microorganisms 2:11–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce TJA, Matthes MC, Napier JA, Pickett JA. 2007. Stressful “memories” of plants: evidence and possible mechanisms. Plant Sci 173:603–608. [Google Scholar]
- Cao L, Alani E, Kleckner N. 1990. A pathway for generation and processing of double-strand breaks during meiotic recombination in S. cerevisiae. Cell 61:1089–1101. [DOI] [PubMed] [Google Scholar]
- Carpenter KE, Abrar M, Aeby G, Aronson RB, Banks S, Bruckner A, Chiriboga A, Cortés J, Delbeek JC, De Vantier L. 2008. One-third of reef-building corals face elevated extinction risk from climate change and local impacts. Science 321:560–563. [DOI] [PubMed] [Google Scholar]
- Castells-Roca L, García-Martínez J, Moreno J, Herrero E, Bellí G, Pérez-Ortín JE. 2011. Heat shock response in yeast involves changes in both transcription rates and mRNA stabilities. PLoS One 6:e17272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Lun ATL, Smyth GK. 2014. Differential expression analysis of complex RNA-seq experiments using edgeR In: Datta S, Nettleton D, editors. Statistical analysis of next generation sequencing data. New York: Springer; p. 51–74. [Google Scholar]
- Chi J, Parrow MW, Dunthorn M. 2014. Cryptic sex in Symbiodinium (Alveolata, Dinoflagellata) is supported by an inventory of meiotic genes. J Euk Microbiol 61:322–327. [DOI] [PubMed] [Google Scholar]
- Chiba Y, Mineta K, Hirai MY, Suzuki Y, Kanaya S, Takahashi H, Onouchi H, Yamaguchi J, Naito S. 2013. Changes in mRNA stability associated with cold stress in Arabidopsis cells. Plant Cell Physiol 54:180–194. [DOI] [PubMed] [Google Scholar]
- Croft MT, Lawrence AD, Raux-Deery E, Warren MJ, Smith AG. 2005. Algae acquire vitamin B12 through a symbiotic relationship with bacteria. Nature 438:90–93. [DOI] [PubMed] [Google Scholar]
- D'Souza TG, Michiels NK. 2010. The costs and benefits of occasional sex: theoretical predictions and a case study. J Hered 101:S34–S41. [DOI] [PubMed] [Google Scholar]
- De Salvo MK, Sunagawa S, Fisher PL, Voolstra CR, Iglesias‐Prieto R, Medina M. 2010. Coral host transcriptomic states are correlated with Symbiodinium genotypes. Mol Ecol 19:1174–1186. [DOI] [PubMed] [Google Scholar]
- Downs CA, Fauth JE, Halas JC, Dustan P, Bemiss J, Woodley CM. 2002. Oxidative stress and seasonal coral bleaching. Free Radical Biol Med 33:533–543. [DOI] [PubMed] [Google Scholar]
- Figueroa RI, Rengefors K, Bravo I, Bensch S. 2010. From homothally to heterothally: mating preferences and genetic variation within clones of the dinoflagellate Gymnodinium catenatum. Deep Sea Res Part II: Top Stud Oceanogr 57:190–198. [Google Scholar]
- Fitt WK, Brown BE, Warner ME, Dunne RP. 2001. Coral bleaching: interpretation of thermal tolerance limits and thermal thresholds in tropical corals. Coral Reefs 20:51–65. [Google Scholar]
- Gill SS, Tuteja N. 2010. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol Biochem 48:909–930. [DOI] [PubMed] [Google Scholar]
- Gordon BR, Leggat W. 2010. Symbiodinium—invertebrate symbioses and the role of metabolomics. Mar Drugs 8:2546–2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29:644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granados-Cifuentes C, Neigel J, Leberg P, Rodriguez-Lanetty M. 2015. Genetic diversity of free-living Symbiodinium in the Caribbean: the importance of habitats and seasons. Coral Reefs 34:927–939. [Google Scholar]
- Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M. 2013. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc 8:1494–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hand SC, Hardewig I. 1996. Downregulation of cellular metabolism during environmental stress: mechanisms and implications. Ann Rev Physiol 58:539–563. [DOI] [PubMed] [Google Scholar]
- Harb A, Krishnan A, Ambavaram MMR, Pereira A. 2010. Molecular and physiological analysis of drought stress in Arabidopsis reveals early responses leading to acclimation in plant growth. Plant Physiol 154:1254–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung F, Wurz-Wildersinn R, Fuchs J, Schubert I, Suer S, Puchta H. 2007. The catalytically active tyrosine residues of both SPO11-1 and SPO11-2 are required for meiotic double-strand break induction in Arabidopsis. Plant Cell 19:3090–3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennon GMM, Ashworth J, Groussman RD, Berthiaume C, Morales RL, Baliga NS, Orellana MV, Armbrust EV. 2015. Diatom acclimation to elevated CO2 via cAMP signalling and coordinated gene expression. Nat Clim Change 5:761–765. [Google Scholar]
- Her C, Wu X, Griswold MD, Zhou F. 2003. Human MutS homologue MSH4 physically interacts with von Hippel-Lindau tumor suppressor-binding protein 1. Cancer Res 63:865–872. [PubMed] [Google Scholar]
- Higgins JD, Armstrong SJ, Franklin FCH, Jones GH. 2004. The Arabidopsis MutS homolog AtMSH4 functions at an early step in recombination: evidence for two classes of recombination in Arabidopsis. Genes Dev 18:2557–2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill R, Schreiber U, Gademann R, Larkum AWD, Kühl M, Ralph PJ. 2004. Spatial heterogeneity of photosynthesis and the effect of temperature-induced bleaching conditions in three species of corals. Mar Biol 144:633–640. [Google Scholar]
- Hoegh-Guldberg O. 1999. Climate change, coral bleaching and the future of the world's coral reefs. Mar Freshwater Res 50:839–866. [Google Scholar]
- Hoegh-Guldberg O, Mumby PJ, Hooten AJ, Steneck RS, Greenfield P, Gomez E, Harvell CD, Sale PF, Edwards AJ, Caldeira K. 2007. Coral reefs under rapid climate change and ocean acidification. Science 318:1737–1742. [DOI] [PubMed] [Google Scholar]
- Hollingsworth NM, Ponte L, Halsey C. 1995. MSH5, a novel MutS homolog, facilitates meiotic reciprocal recombination between homologs in Saccharomyces cerevisiae but not mismatch repair. Genes Dev 9:1728–1739. [DOI] [PubMed] [Google Scholar]
- Howells EJ, Beltran VH, Larsen NW, Bay LK, Willis BL, Van Oppen MJH. 2012. Coral thermal tolerance shaped by local adaptation of photosymbionts. Nat Clim Change 2:116–120. [Google Scholar]
- Huang Y, Niu B, Gao Y, Fu L, Li W. 2010. CD-HIT Suite: a web server for clustering and comparing biological sequences. Bioinformatics 26:680–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney S. 2007. Spo11 and the formation of DNA double-strand breaks in meiosis In: Egel R, Lankenau D, editors. Recombination and meiosis. New York: Springer; p. 81–123. [Google Scholar]
- Keeney S, Giroux CN, Kleckner N. 1997. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88:375–384. [DOI] [PubMed] [Google Scholar]
- Kelly KO, Dernburg AF, Stanfield GM, Villeneuve AM. 2000. Caenorhabditis elegans msh-5 is required for both normal and radiation-induced meiotic crossing over but not for completion of meiosis. Genetics 156:617–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneitz B, Cohen PE, Avdievich E, Zhu L, Kane MF, Hou H, Kolodner RD, Kucherlapati R, Pollard JW, Edelmann W. 2000. MutS homolog 4 localization to meiotic chromosomes is required for chromosome pairing during meiosis in male and female mice. Genes Dev 14:1085–1097. [PMC free article] [PubMed] [Google Scholar]
- Krueger T, Fisher PL, Becker S, Pontasch S, Dove S, Hoegh-Guldberg O, Leggat W, Davy SK. 2015. Transcriptomic characterization of the enzymatic antioxidants FeSOD, MnSOD, APX and KatG in the dinoflagellate genus Symbiodinium. BMC Evol Biol 15:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladner JT, Barshis DJ, Palumbi SR. 2012. Protein evolution in two co-occurring types of Symbiodinium: an exploration into the genetic basis of thermal tolerance in Symbiodinium clade D. BMC Evol Biol 12:217.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaJeunesse TC. 2005. “Species” radiations of symbiotic dinoflagellates in the Atlantic and Indo-Pacific since the Miocene–Pliocene transition. Mol Biol Evol 22:570–581. [DOI] [PubMed] [Google Scholar]
- LaJeunesse TC, Thornhill DJ. 2011. Improved resolution of reef-coral endosymbiont (Symbiodinium) species diversity, ecology, and evolution through psbA non-coding region genotyping. PLoS One 6:e29013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaJeunesse TC, Thornhill DJ, Cox EF, Stanton FG, Fitt WK, Schmidt GW. 2004. High diversity and host specificity observed among symbiotic dinoflagellates in reef coral communities from Hawaii. Coral Reefs 23:596–603. [Google Scholar]
- Leggat W, Seneca F, Wasmund K, Ukani L, Yellowlees D, Ainsworth TD. 2011. Differential responses of the coral host and their algal symbiont to thermal stress. PLoS One 6:e26687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy DJ, Chen Y, Smyth GK. 2012. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res 40:4288–4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinty ES, Pieczonka J, Mydlarz LD. 2012. Variations in reactive oxygen release and antioxidant activity in multiple Symbiodinium types in response to elevated temperature. Microb Ecol 64:1000–1007. [DOI] [PubMed] [Google Scholar]
- Modrich P, Lahue R. 1996. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu Rev Biochem 65:101–133. [DOI] [PubMed] [Google Scholar]
- Morey JS, Van Dolah FM. 2013. Global analysis of mRNA half-lives and de novo transcription in a dinoflagellate, Karenia brevis. PLoS One 8:e66347.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muscatine L, Porter JW. 1977. Reef corals: mutualistic symbioses adapted to nutrient-poor environments. Bioscience 27:454–460. [Google Scholar]
- Novak JE, Ross-Macdonald PB, Roeder GS. 2001. The budding yeast Msh4 protein functions in chromosome synapsis and the regulation of crossover distribution. Genetics 158:1013–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nymark M, Valle KC, Brembu T, Hancke K, Winge P, Andresen K, Johnsen G, Bones AM. 2009. An integrated analysis of molecular acclimation to high light in the marine diatom Phaeodactylum tricornutum. PLoS One 4:e7743.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver TA, Palumbi SR. 2011. Many corals host thermally resistant symbionts in high-temperature habitat. Coral Reefs 30:241–250. [Google Scholar]
- Pandolfi JM, Connolly SR, Marshall DJ, Cohen AL. 2011. Projecting coral reef futures under global warming and ocean acidification. Science 333:418–422. [DOI] [PubMed] [Google Scholar]
- Parkinson JE, Baumgarten S, Michell CT, Baums IB, LaJeunesse TC, Voolstra CR. 2016. Gene expression variation resolves species and individual strains among coral-associated dinoflagellates within the genus Symbiodinium. Genome Biol Evol 8:665–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. [DOI] [PubMed] [Google Scholar]
- Platt T, Gallegos CL, Harrison WG. 1980. Photoinhibition of photosynthesis in natural assemblages of marine phytoplankton. J Mar Res 38:687–701. [Google Scholar]
- Putnam HM, Mayfield AB, Fan TY, Chen CS, Gates RD. 2013. The physiological and molecular responses of larvae from the reef-building coral Pocillopora damicornis exposed to near-future increases in temperature and pCO2. Mar Biol 160:2157–2173. [Google Scholar]
- Ragni M, Airs RL, Hennige SJ, Suggett DJ, Warner ME, Geider RJ. 2010. PSII photoinhibition and photorepair in Symbiodinium (Pyrrhophyta) differs between thermally tolerant and sensitive phylotypes. Mar Ecol Prog Ser 406:57–70. [Google Scholar]
- Ralph P, Gademann R, Larkum A, Kühl M. 2002. Spatial heterogeneity in active chlorophyll fluorescence and PSII activity of coral tissues. Mar Biol 141:639–646. [Google Scholar]
- Reaka-Kudla ML, Wilson DE, Wilson EO. 1996. Biodiversity II: understanding and protecting our biological resources. Washington (DC: ): Joseph Henry Press. [Google Scholar]
- Ritchie KB. 2012. Bacterial symbionts of corals and Symbiodinium In: Rosenberg E, Gophna U, editors. Beneficial microorganisms in multicellular life forms. New York: Springer; p. 139–150. [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosic N, Kaniewska P, Chan C-KK, Ling EYS, Edwards D, Dove S, Hoegh-Guldberg O. 2014. Early transcriptional changes in the reef-building coral Acropora aspera in response to thermal and nutrient stress. BMC Genomics 15:1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosic N, Ling EYS, Chan C-KK, Lee HC, Kaniewska P, Edwards D, Dove S, Hoegh-Guldberg O. 2015. Unfolding the secrets of coral–algal symbiosis. ISME J 9:844–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosic NN, Hoegh-Guldberg O. 2010. A method for extracting a high-quality RNA from Symbiodinium sp. J Appl Phycol 22:139–146. [Google Scholar]
- Rosic NN, Pernice M, Dove S, Dunn S, Hoegh-Guldberg O. 2011. Gene expression profiles of cytosolic heat shock proteins Hsp70 and Hsp90 from symbiotic dinoflagellates in response to thermal stress: possible implications for coral bleaching. Cell Stress Chap 16:69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosic NN, Pernice M, Dunn S, Dove S, Hoegh-Guldberg O. 2010. Differential regulation by heat stress of novel cytochrome P450 genes from the dinoflagellate symbionts of reef-building corals. Appl Environ Microbiol 76:2823–2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross C. 2014. Nitric oxide and heat shock protein 90 co-regulate temperature-induced bleaching in the soft coral Eunicea fusca. Coral Reefs 33:513–522. [Google Scholar]
- Ross-Macdonald P, Roeder GS. 1994. Mutation of a meiosis-specific MutS homolog decreases crossing over but not mismatch correction. Cell 79:1069–1080. [DOI] [PubMed] [Google Scholar]
- Rossouw D, Olivares-Hernandes R, Nielsen J, Bauer FF. 2009. Comparative transcriptomic approach to investigate differences in wine yeast physiology and metabolism during fermentation. Appl Environ Microbiol 75:6600–6612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth MS. 2014. The engine of the reef: photobiology of the coral–algal symbiosis. Front Microbiol 5:422.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmieder R, Edwards R. 2011. Quality control and preprocessing of metagenomic datasets. Bioinformatics 27:863–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine H, Ferreira RC, Pan-Hammarström Q, Graham RR, Ziemba B, De Vries SS, Liu J, Hippen K, Koeuth T, Ortmann W. 2007. Role for Msh5 in the regulation of Ig class switch recombination. Proc Natl Acad Sci USA 104:7193–7198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shodhan A, Lukaszewicz A, Novatchkova M, Loidl J. 2014. Msh4 and Msh5 function in SC-independent chiasma formation during the streamlined meiosis of Tetrahymena. Genetics 198:983–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoguchi E, Shinzato C, Kawashima T, Gyoja F, Mungpakdee S, Koyanagi R, Takeuchi T, Hisata K, Tanaka M, Fujiwara M. 2013. Draft assembly of the Symbiodinium minutum nuclear genome reveals dinoflagellate gene structure. Curr Biol 23:1399–1408. [DOI] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31:3210–3212. [DOI] [PubMed] [Google Scholar]
- Singh A, Grover A. 2008. Genetic engineering for heat tolerance in plants. Physiol Mol Biol Plants 14:155–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snowden T, Acharya S, Butz C, Berardini M, Fishel R. 2004. hMSH4-hMSH5 recognizes Holliday Junctions and forms a meiosis-specific sliding clamp that embraces homologous chromosomes. Mol Cell 15:437–451. [DOI] [PubMed] [Google Scholar]
- Srivastava HS, Singh RP. 1987. Role and regulation of L-glutamate dehydrogenase activity in higher plants. Phytochemistry 26:597–610. [Google Scholar]
- Stacey NJ, Kuromori T, Azumi Y, Roberts G, Breuer C, Wada T, Maxwell A, Roberts K, Sugimoto ‐Shirasu K. 2006. Arabidopsis SPO11-2 functions with SPO11-1 in meiotic recombination. Plant J 48:206–216. [DOI] [PubMed] [Google Scholar]
- Stat M, Bird CE, Pochon X, Chasqui L, Chauka LJ, Concepcion GT, Logan D, Takabayashi M, Toonen RJ, Gates RD. 2011. Variation in Symbiodinium ITS2 sequence assemblages among coral colonies. PLoS One 6:e15854.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stat M, Pochon X, Cowie ROM, Gates RD. 2009. Specificity in communities of Symbiodinium in corals from Johnston Atoll. Mar Ecol Prog Ser 386:83–96. [Google Scholar]
- Stojic L, Brun R, Jiricny J. 2004. Mismatch repair and DNA damage signalling. DNA Repair 3:1091–1101. [DOI] [PubMed] [Google Scholar]
- Suggett DJ, Goyen S, Evenhuis C, Szabó M, Pettay DT, Warner ME, Ralph PJ. 2015. Functional diversity of photobiological traits within the genus Symbiodinium appears to be governed by the interaction of cell size with cladal designation. New Phytol 208:370–381. [DOI] [PubMed] [Google Scholar]
- Suggett DJ, Warner ME, Smith DJ, Davey P, Hennige S, Baker NR. 2008. And production of hydrogen peroxide by symbiodinium (Pyrrophyta) phylotypes with different thermal tolerances. J Phycol 44:948–956. Photosynthesis [DOI] [PubMed] [Google Scholar]
- Supek F, Bošnjak M, Škunca N, Šmuc T. 2011. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One 6:e21800.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S, Whitney SM, Badger MR. 2009. Different thermal sensitivity of the repair of photodamaged photosynthetic machinery in cultured Symbiodinium species. Proc Natl Acad Sci USA 106:3237–3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamburini BA, Tyler JK. 2005. Localized histone acetylation and deacetylation triggered by the homologous recombination pathway of double-strand DNA repair. Mol Cell Biol 25:4903–4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang L, Kwon S-Y, Kim SH, Kim JS, Choi JS, Cho KY, Sung CK, Kwak SS, Lee HS. 2006. Enhanced tolerance of transgenic potato plants expressing both superoxide dismutase and ascorbate peroxidase in chloroplasts against oxidative stress and high temperature. Plant Cell Rep 25:1380–1386. [DOI] [PubMed] [Google Scholar]
- Tchernov D, Gorbunov MY, de Vargas C, Yadav SN, Milligan AJ, Häggblom M, Falkowski PG. 2004. Membrane lipids of symbiotic algae are diagnostic of sensitivity to thermal bleaching in corals. Proc Natl Acad Sci USA 101:13531–13535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tercé-Laforgue T, Clément G, Marchi L, Restivo FM, Lea PJ, Hirel B. 2015. Resolving the role of plant NAD-glutamate dehydrogenase: III. Overexpressing individually or simultaneously the two enzyme subunits under salt stress induces changes in the leaf metabolic profile and increases plant biomass production. Plant Cell Physiol 56:1918–1925. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson T, Higgins DG. 2002. Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics 2.3.1-2.3.22. [DOI] [PubMed]
- Tompkins JD, Wu X, Chu Y-L, Her C. 2009. Evidence for a direct involvement of hMSH5 in promoting ionizing radiation induced apoptosis. Exp Cell Res 315:2420–2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonk L, Bongaerts P, Sampayo EM, Hoegh-Guldberg O. 2013. SymbioGBR: a web-based database of Symbiodinium associated with cnidarian hosts on the Great Barrier Reef. BMC Ecol 13:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubouchi T, Roeder GS. 2005. A synaptonemal complex protein promotes homology-independent centromere coupling. Science 308:870–873. [DOI] [PubMed] [Google Scholar]
- Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. 2006. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev 20:515–524. [DOI] [PubMed] [Google Scholar]
- Van Breusegem F, Slooten L, Stassart J-M, Moens T, Botterman J, Van Montagu M, Inzé D. 1999. Overproduction of Arabidopsis thaliana FeSOD confers oxidative stress tolerance to transgenic maize. Plant Cell Physiol 40:515–523. [DOI] [PubMed] [Google Scholar]
- van Oppen MJH, Palstra FP, Piquet AMT, Miller DJ. 2001. Patterns of coral-dinoflagellate associations in Acropora: significance of local availability and physiology of Symbiodinium strains and host–symbiont selectivity. Proc R Soc Lond B Biol Sci 268:1759–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierling E. 1991. The roles of heat shock proteins in plants. Annu Rev Plant Biol 42:579–620. [Google Scholar]
- Wang Y, Qin J. 2003. MSH2 and ATR form a signaling module and regulate two branches of the damage response to DNA methylation. Proc Natl Acad Sci USA 100:15387–15392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner ME, Fitt WK, Schmidt GW. 1999. Damage to photosystem II in symbiotic dinoflagellates: a determinant of coral bleaching. Proc Natl Acad Sci USA 96:8007–8012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson SP, Fisher PL, van Oppen MJH, Davy SK. 2015. Intra-genomic variation in symbiotic dinoflagellates: recent divergence or recombination between lineages? BMC Evol Biol 15:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz CJ, Wilusz J. 2004. Bringing the role of mRNA decay in the control of gene expression into focus. Trends Genet 20:491–497. [DOI] [PubMed] [Google Scholar]
- Wu L, Fan J, Belasco JG. 2006. MicroRNAs direct rapid deadenylation of mRNA. Proc Natl Acad Sci USA 103:4034–4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang T, Nelson W, Rodriguez J, Tolleter D, Grossman AR. 2015. Symbiodinium transcriptome and global responses of cells to immediate changes in light intensity when grown under autotrophic or mixotrophic conditions. Plant J 82:67–80. [DOI] [PubMed] [Google Scholar]
- Yamashita H, Koike K. 2013. Genetic identity of free‐living Symbiodinium obtained over a broad latitudinal range in the Japanese coast. Phycol Res 61:68–80. [Google Scholar]
- Yampolsky LY, Zeng E, Lopez J, Williams PJ, Dick KB, Colbourne JK, Pfrender ME. 2014. Functional genomics of acclimation and adaptation in response to thermal stress in Daphnia. BMC Genomics 15:859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young MD, Wakefield MJ, Smyth GK, Oshlack A. 2010. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol 11:R14. Method [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuyama I, Harii S, Hidaka M. 2012. Algal symbiont type affects gene expression in juveniles of the coral Acropora tenuis exposed to thermal stress. Mar Environ Res 76:41–47. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.