Abstract
Subtropical evergreen broad-leaved forest (EBLF) is one of the most important vegetation types in China. Inferences from palaeo-biome reconstruction (PBR) and phylogeography regarding range shift history of EBLF during the late Quaternary are controversial and should be reconciled. We compared phylogeographic patterns of three EBLF constituents in China, Castanopsis tibetana, Machilus thunbergii and Schima superba. Contrary to a chorus of previous phylogeographic studies and the results of species distribution modelling (SDM) of this study (in situ survival during the LGM), the three species displayed three different phylogeographic patterns that conform to either an in situ survival model or an expansion-contraction model. These results are partially congruent with the inference of PBR that EBLF was absent to the north of 24° N at the LGM. This study suggests that the constituents of EBLF could have responded idiosyncratically to climate changes during the Late Quaternary. The community assemblages of EBLF could have been changing over time, resulting in no palaeo-analogs to modern-day EBLF, which may be the main reason responsible for the failure of PBR to detect the occurrence of EBLF north of 24° N at the LGM.
Subtropical evergreen broad-leaved forest (EBLF) occurs from 24 to 32° N latitude and 99 to 123° E longitude and covers approximately 25% of China (Fig. 1a), making it one of the most important vegetation types in China1,2. This biome is heavily influenced by the East Asian monsoon and human expansion and is predicted to be vulnerable to future climate change3. However, the potential impacts of future climate change on the ecosystem are poorly quantified because of sparse long-term in situ measurements3 and the inherent difficulties in predictions of climate change scenarios on biodiversity4. Retrospective analyses based on fossil and genetic data have greatly advanced our understanding of species diversity and distribution in response to past climatic changes5, thus providing a framework for explaining biotic responses to anthropogenic climate change in the future6. However, the range shift of EBLF in response to the late Quaternary climate changes remains a subject of dispute, which may compromise the ability to make informed decisions regarding biodiversity conservation and natural resource management in the EBLF region in response to future climate change.
Figure 1.
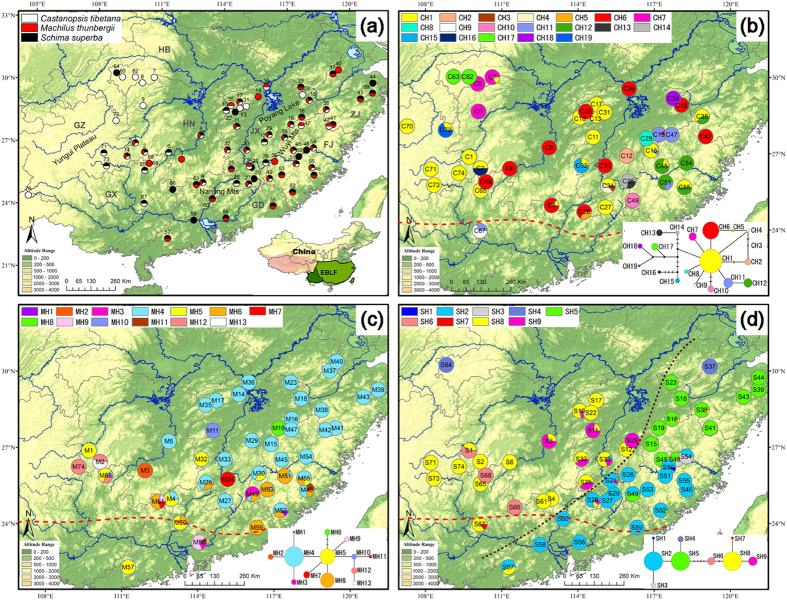
(a) Locations of sampled populations of Castanopsis tibetana, Machilus thunbergii, and Schima superba. The circle in the inset illustrates the distribution range of subtropical evergreen broad-leaved forest (EBLF) in China and the dashed line shows the boundary between two subregions of EBLF. (b) Geographic distribution and median-joining network of C. tibetana haplotypes (CH1–19). (c) Geographic distribution and median-joining network of M. thunbergii haplotypes (MH1–13). (d) Geographic distribution and median-joining network of S. superba haplotypes (SH1–9). Brown dashed lines denote lineage divergences identified by network and SAMOVA analysis. For each network in (b–d), the size of circles corresponds to the frequency of each haplotype. Vertical bars indicate unsampled or extinct haplotypes. Each solid line represents one mutational step that interconnects two haplotypes. Each population code in Supplementary Table S2 is preffixed by a letter C in (b), M in (c) and S in (d) that particularly denotes the sampled populations of Castanopsis tibetana, Machilus thunbergii and Schima superba, respectively. One population (M69) remote from others in Machilus thunbergii is not shown in (c). The red dashed lines show the approximate northern borders of EBLF during the LGM based on PBR (modified after Harrison et al., 2001). Maps were generated using ArcGIS version 9.3 (http://www.esri.com/software/arcgis/arcgis-for-desktop) and Adobe Illustrator CS3 13.0 and modified using Adobe Photoshop CS 8.0.
Over the past 3 million years, the climate on Earth has undergone periodic fluctuations, resulting in complex changes of vegetation distributions7. During the last glacial maximum (LGM), for example, the mean annual temperature of the EBLF region was cooler by c. 4–6 °C and drier by c. 400–600 mm/yr according to pollen analyses8. Palaeo-biome reconstructions (PBR) based on fossil pollen suggested that at the time of the LGM, the EBLF in subtropical China extended as far south as c. 24°N, a shift of ca. 1000 km relative to today9,10,11. These studies also indicated that EBLF subsequently expanded northward during the Holocene, covering a more extensive range than today, ca. 200 km north of the modern limit (glacial contraction and post-glacial expansion, EI model). In contrast, most previous phylogeographic studies, sometimes combined with species distribution modelling (SDM), have consistently concluded that the component species of EBLF survived the LGM in situ in multiple northern refugia and did not experience extensive northward spread from more southern refugia during the Holocene (in situ survival model, ISS model) (Supplementary Table S1).
Why are the results from PBR and phylogeography (sometimes coupled with SDM) so different ? On the one hand, the fossil pollen record in central east China (including the EBLF region) is so sparse that unambiguous reconstruction of biome boundaries in this region is difficult11. Furthermore, fossil pollen records are always limited in taxonomic resolution5, such that the reconstruction of entire biomes based on these data cannot produce a detailed picture of palaeodistributions of EBLF constituents12. On the other hand, previous phylogeographic studies of elements of the EBLF are heavily biased towards Fagaceae (e.g., Castanopsis eyrei13, Castanopsis fargesii14 and Quercus glauca15, Supplementary Table S1), and the conclusions may be phylogenetically biased and prone to phylogenetic niche conservatism (the tendency of species to retain ancestral ecological attributes)16,17. The remaining phylogeographic studies mostly involved species that are ecologically minor components of EBLF (e.g., Eurycorymbus cavaleriei18, Rhododendron simsii19 and Tetrastigma hemsleyanum20, Supplementary Table S1), and the significance of these results may therefore not be representative of the biome as a whole. Importantly, the investigated plants represent merely a small proportion of the total plant biodiversity in the EBLF region (up to 18,000 seed plant species21), and the conclusions of previous phylogeographic studies regarding the biome as a whole may therefore be premature. Additional phylogeographic surveys, especially rigorous comparisons among co-distributed dominant species within EBLF (i.e., comparative phylogeography), may provide insights into the divergent results offered by PBR, phylogeographic analyses, and SDM.
Comparative phylogeography (CP) seeks to understand how the geographic ranges of co-distributed species have changed over time22. By comparing phylogeographic patterns across species, it may be possible to discern whether changes in distribution and population size represent the influence of extrinsic factors that affected whole communities, or whether they can be ascribed to stochastic variation and other chance factors particular to each organism23. Some comparative studies have succeeded in identifying patterns common across many groups of organisms24,25,26. However, idiosyncratic phylogeographic patterns among co-distributed species are commonly reported27,28,29, suggesting that community composition is highly dynamic29. If the latter model applies to EBLF in subtropical China, it is conceivable that the absence of EBLF in subtropical China at the LGM might have been caused by alterations in community composition and the changes in relative abundance of EBLF components, rather than by the complete retreat to the south, thus settling the dispute between PBR and phylogeography. However, such a hypothesis has never been proposed and explicitly tested by phylogeographic researchers in China, possibly due to the difficulty of gathering large data sets from multiple co-distributed species.
To reconcile the two scenarios suggested by PBR and phylogeography, this study used the CP approach to test for congruent versus idiosyncratic phylogeographic patterns of multiple species from EBLF in subtropical China. The EBLF zone within China is divided into two subregions (Fig. 1a): the moist EBLF in eastern China influenced by the Pacific monsoon and the semi-moist EBLF in the west determined by the Indian monsoon2. Because the climate in the two subregions is quite different (e.g., the annual rainfall is 1000–2000 mm in the moist east and 900–1200 mm in the semi-moist west1), very few dominant tree species are shared across the two subregions. Therefore, we only focused on three tree species in the eastern subregion of EBLF, i.e., Castanopsis tibetana Hance (Fagaceae), Machilus thunbergii Sieb. et Zucc. (Lauraceae), and Schima superba Gardn. et Champ. (Theaceae) (Supplementary Table S2; Fig. 1). These species satisfy three criteria: 1) they are all dominant broad-leaved tree species in EBLF and thus play a fundamental role in the ecological functions of EBLF; 2) they belong to different families and genera that avoid the influence of conserved niche within lineages (and thus similar responses to climate change); and 3) they are components of the same forest communities. We surveyed chloroplast DNA (cpDNA) sequence variation in these three tree species and determined their phylogeographic and historical demographic patterns. We also used SDM to hindcast the geographical distribution of the three tree species at the time of the LGM. By providing quantitative and high-resolution predictions of individual palaeodistributions, this method can complement fossil and genetic evidence concerning the past distribution of species and forest communities30. By using molecular and SDM approaches, we address the following issues: i) whether the focal species conform to the ISS or the EI model during the late Quaternary; ii) whether they display congruent phylogeographic patterns and demographic histories across a shared landscape; and iii) whether CP is able to reconcile the dispute between PBR and previous phylogeographic studies.
Results
Sequence characteristics and genetic diversity
The combined alignments of the two chloroplast regions were 1826 bp (rpl32–trnL(UAG) 1313 bp + psbA–trnH 513 bp), 1450 bp (1048 bp + 402 bp), and 1399 bp (976 bp + 423 bp), comprising 29 (rpl32–trnL(UAG) 17 + psbA–trnH 12), 13 (7 + 6), and 13 (8 + 5) polymorphic sites and defining 19, 13, and 9 haplotypes in C. tibetana, M. thunbergii, and S. superba, respectively (Supplementary Table S3). All haplotype sequences are deposited in GenBank (accession numbers: KP892691–KP892749).
Estimates of haplotype diversity (h) and nucleotide diversity (π) for each population of each of the three species are summarized in Supplementary Table S2. The overall haplotype diversity was equivalent in C. tibetana and S. superba (hT = 0.797 and 0.765, respectively), but higher than that in M. thunbergii (hT = 0.644). Machilus thunbergii displays the lowest nucleotide diversity, while C. tibetana has the highest. The average population genetic diversity values are all considerably low in C. tibetana, M. thunbergii, and S. superba (hS = 0.151, 0.165, and 0.136, respectively).
Phylogeographic structure
In C. tibetana, samova failed to uncover any reliable population genetic grouping, and there was no apparent structure in the haplotype network (Fig. 1b). Except for two relatively widespread haplotypes, CH1 and CH6, which were found in 22 (49%) and 11 (24%) populations, respectively, most haplotypes were confined to a specific region. Fifteen polymorphic populations and 13 haplotypes (CH2–5, CH8–10, CH13–16 and CH18–19) private to a single population were scattered across the species’ range.
There was also no apparent association between haplotypes and geography according to the samova analysis in M. thunbergii. However, the haplotype distribution in M. thunbergii was distinct from that in C. tibetana (Fig. 1c). The most common haplotype, MH4, occurred in 32 (70%) populations and was fixed in 22 of them, especially those at higher latitudes ( ≥ 26°N). The second and third most common haplotypes, MH5 and MH6, were shared among 9 (20%) and 10 (22%) populations, respectively. Of the 10 rare haplotypes remaining, five (MH1, MH2, MH8, MH10, and MH13) were found in a single population (M52, M3, M19, M11, and M2, respectively), and five (MH3, MH7, MH9, MH11, and MH12) were shared by two or three populations. Most haplotypes (except for MH4, MH8, and MH10) and 14 polymorphic populations occurred at lower latitudes ( ≤ 26°N).
In the samova analysis of S. superba, the value of FCT reached a plateau at 0.894 when the number of groups was two (K = 2). As shown in Fig. 1d, the two groups largely correspond to western and eastern parts of the distribution range. The western group was dominated by SH8. The eastern group had two subgroups, the southern one nearly fixed for SH2 and the northern one for SH5. The haplotype network recovered all western haplotypes (SH6–9) as a clade, which was separated by five mutational steps from an eastern clade (containing SH1–5). Several populations in the contact area of the two groups showed a mixture of two clades (S21, S28, S33, and S57).
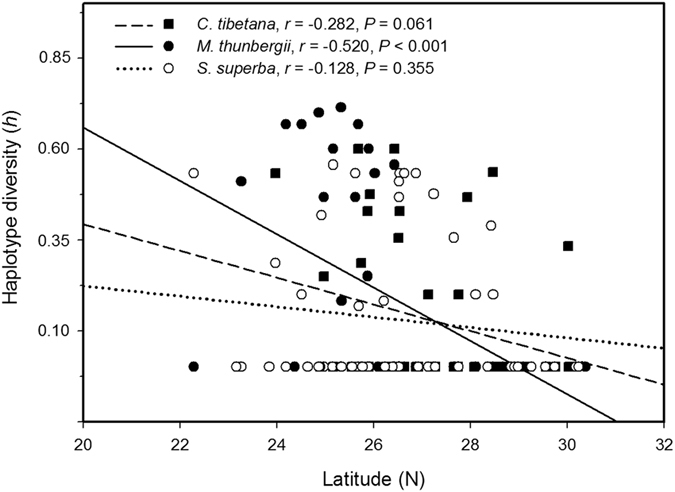
In the multiple regression analyses of genetic variation, haplotype diversity was correlated negatively with latitude in M. thunbergii (P < 0.001), but not in C. tibetana or S. superba (Fig. 2). Nonetheless, a U-test showed that NST was significantly larger than GST in S. superba (NST = 0.906; GST = 0.825; U = 2.81, P < 0.01), indicating phylogeographic structure of haplotype distribution, but this was not significant in either C. tibetana (NST = 0.851; GST = 0.815; U = 0.90, P > 0.05) or M. thunbergii (NST = 0.767; GST = 0.752; U = 0.18, P > 0.05).
Figure 2. The relationship between haplotype diversity (h) and latitude (°N) in three broad-leaved evergreen tree species in subtropical China.
Historical demography
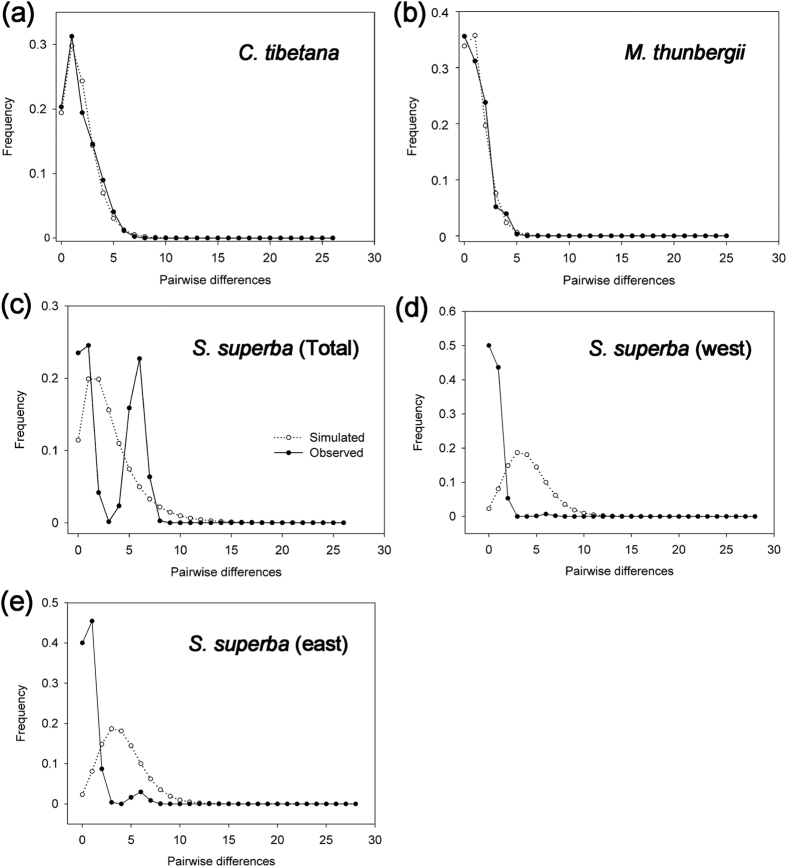
For C. tibetana, the neutrality tests showed that both Tajima’s D (D = −1.011, P = 0.157) and FS (FS = −2.888, P = 0.026) were negative. The mismatch distribution for haplotypes was unimodal (Fig. 3a) that was statistically consistent with the expansion model (P values > 0.05 for SSD and HRag; Table 1). Despite a slight recent decrease in population size detected by our BSP analysis (Supplementary Fig. S1a), the data suggest [we prefer] that C. tibetana likely experienced past expansion. Based on the corresponding τ value, the demographic expansion event was estimated to have occurred at 9,600 (95% highest posterior density, HPD: 0–53,000) years bp, although this estimate should be treated with caution given the wide confidence interval (Table 1).
Figure 3.
Distribution of the number of pairwise nucleotide differences for cpDNA sequence data in (a) Castanopsis tibetana as a whole, (b) Machilus thunbergii as a whole, (c) Schima superba as a whole, (d) eastern populations of Schima superba and (e) western populations of Schima superba. The solid line shows observed distributions of differences among haplotypes whereas the dashed line represents simulated distributions under a model of sudden (stepwise) population expansion (Rogers & Harpending, 1992).
Table 1. Mismatch distribution analysis (MDA) of cpDNA sequence data for three broadleaved evergreen tree species.
| Regions | τ | t (y) | Observed modality of mismatch distribution | SSD | P | HRag | P |
|---|---|---|---|---|---|---|---|
| C. tibetana | 0.625 (0–3.438) | 9,600 (0–53,000) | unimodal | 0.023 | 0.251 | 0.040 | 0.640 |
| M. thunbergii | 0.438 (0–3.828) | 3,700 (0–33,000) | unimodal | 0.009 | 0.544 | 0.042 | 0.822 |
| S. superba | 0.082 (0–1.458) | NC | bimodal | 0.188 | 0.040 | 0.491 | 0.020 |
| west | 0.082 (0–1.547) | 6,100 (0–115,100) | unimodal | 0.051 | 0.231 | 0.178 | 0.517 |
| east | 0.844 (0.717–1.045) | NC | bimodal | 0.017 | 0.003 | 0.146 | 0 |
Goodness-of-fit of observed to theoretical mismatch distributions under a demographic expansion model60 are tested with the sum of squared deviations (SSD) and raggedness index (HRag). Upper and lower 95% confidence limits around estimates of τ, and associated ranges of t (in years bp) are in parentheses.
NC, not calculated.
For M. thunbergii, the negative Tajima’s D (D = −0.746, P = 0.252) and FS (FS = −2.467, P = 0.041), unimodal mismatch distribution and positive values of SSD and HRag (P > 0.05) consistently suggested a past expansion event (Table 1, Fig. 3b). The expansion event was estimated to have occurred approximately 3,700 (95% HPD: 0–33,000) years bp based on the τ value. Our BSP analysis also showed that M. thunbergii populations experienced a recent sharp increase in population size at c. 4 ka (Supplementary Fig. S1b).
For S. superba, the neutrality tests gave contrasting results, with a non-significant positive Tajima’s D (D = 1.93, P = 0.979) and a significant positive FS (FS = 4.656, P = 0.013). Despite a good statistical fit of the expansion model (P values > 0.05 for SSD and HRag; Table 1), the mismatch distribution for S. superba was bimodal, which may reflect the regional population structure of the species due to the west-east divergence (Fig. 3c). In support of this explanation, when analyzed separately, the western populations showed a unimodal distribution (Fig. 3b) that was statistically consistent with the expansion model (P values > 0.05 for SSD and HRag; Table 1). This expansion event was dated at 6,100 (95% HPD: 0–115,100). When examined separately, the eastern populations showed a bimodal distribution (Fig. 3c), because this group was further structured into two subgroups (Fig. 3c). A BSP analysis detected a slight population increase in both the western and eastern groups (Supplementary Fig. S1c, S1d). Taken together, the data suggest that S. superba may have experienced regional post-glacial expansion from localized refugia.
Species distribution modeling
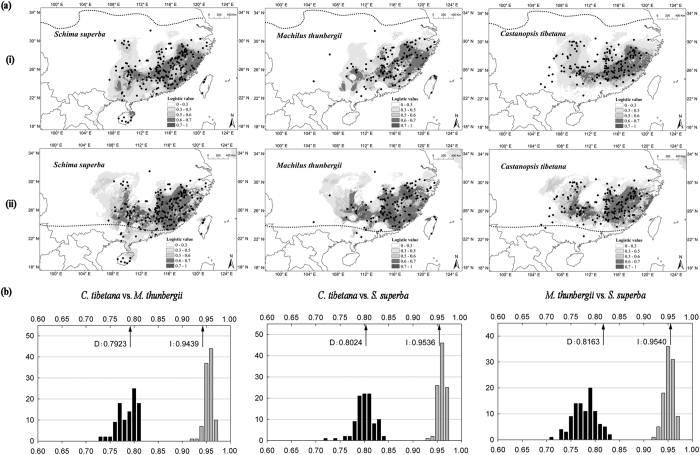
For each species at present and LGM, the areas under the ‘Receiver Operating Characteristic (ROC) Curve’ (AUC) all have values > 0.95, indicating good predictive model performance. In general, the predicted distributions of the three species under present conditions are similar to their actual distributions in mainland China, each with a potentially patchy range in mountainous areas of subtropical China and a slightly more continuous range for C. tibetana. At the LGM, the potential ranges of the three species contracted a little to the south in the eastern part of their ranges. However, most of the current occurrences are assigned high suitability at the LGM, suggesting overall range stability (Fig. 4a). Therefore, the three species did not retreat entirely to the tropical south during the LGM. SDM analyses found that the niches for the three species were very similar (Fig. 4b). ENMTOOLS showed that empirically observed values for I and D were comparable to those expected from pseudoreplicated data sets in all paired analyses (C. tibetana vs. M. thunbergii, C. tibetana vs. S. superba and M. thunbergii vs. S. superba) (Fig. 4b).
Figure 4.
(a) Predicted distributions of Castanopsis tibetana, Machilus thunbergii and Schima superba based on species distribution modeling (i) at present (1950–2000), (ii) at the Last Glacial Maximum (LGM; c. 21 kya). Species distribution models were established with bioclimatic variables on the basis of extant occurrence points (black dots) of the three species using maxent version 3.3.3 k (http://www.cs.princeton.edu/~schapire/maxent/). The dashed lines indicate the northern borders of EBLF today and during the LGM inferred from PBR (modified after Harrison et al., 2001). Maps were generated using ArcGIS version 9.3 (http://www.esri.com/software/arcgis/arcgis-for-desktop). (b) The results of identity tests using enmtools version 1.3. The black Bar indicates the null distributions of D, and gray bar indicates the null distributions of I. Both are generated from 100 randomizations. X-axis indicates value of I and D, Y-axis indicates number of randomizations. The arrow indicates the value in actual maxent runs. The Histograms were drawn using SigmaPlot 10.0.
Discussion
Idiosyncratic responses of three dominant trees in EBLF to the late Quaternary climate changes
Recently, phylogeographic patterns of subtropical EBLF constituents in China have received increasing attention (Supplementary Table S1). These studies consistently revealed limited or regional post/inter-glacial expansion from northern localized refugia (ISS model), rather than extensive northward spread from more southerly located refugia (EI model) as inferred from palaeo-biome reconstruction (PBR) based on the fossil pollen record9,10,11. However, we found that three dominant tree species in EBLF, C. tibetana, M. thunbergii and S. superba, have different range shift trajectories during the late Quaternary, which cannot be predicted purely from either the ISS or EI model.
Castanopsis tibetana (Fagaceae), displays a pattern of multiple northern refugia (private haplotypes and polymorphic populations occurring in the north, Fig. 1b) and regional postglacial expansion, which is consistent with the ISS model. It exhibits the highest genetic diversity among the three focal species, suggesting that more glacial refugia could have maintained a larger historical population size. All members of Fagaceae in EBLF examined so far show similar phylogeographic patterns (i.e., Castanopsis eyrei13, Castanopsis fargesii14 and Quercus glauca15). The phylogeographic congruence among members of Fagaceae may be explained by shared population histories because all these species are common in EBLF and always constitute the canopy of the forest31. However, an alternative explanation for the congruence may be niche conservatism among closely related species17. Based on a meta-analysis of phylogenies from many studies in the Southern Hemisphere, Crisp et al.32 concluded that closely related species are more ecologically similar than would be expected by chance. Therefore, similar ecological requirements due to phylogenetic relatedness among members of Fagaceae in EBLF may account for their phylogeographic congruence, at least in part.
In contrast to C. tibetana, Machilus thunbergii has the lowest genetic diversity of the three focal species and shows a significant negative correlation between genetic diversity and latitude (Fig. 2). The results suggest that it conforms to a typical pattern of pioneer or leading-edge expansion, which is expected by the classical EI model33. This is the first study that reports the EI model for constituents of EBLF in subtropical mainland China, although evidence of extensive range expansion has been implicated in two case studies of Taiwanese plants. Wu et al.34 found that Machilus kusanoi in Taiwan (most of which is covered by EBLF, Fig. 1a) experienced strong post-glacial range expansion. Machilus thunbergii was also investigated in Wu et al.34: range expansion is also evident in this species because of low genetic diversity among populations and the presence of a widespread haplotype, similar to the pattern observed in mainland China in our study. For another member of Lauraceae, Cinnamomum kanehirae, Liao et al.35 also detected significant spatial range expansion in Taiwan. Together, these studies suggest that some constituents of EBLF in subtropical China (e.g., several species of Lauraceae), may have retreated to the south during the LGM and recolonized the present EBLF region during the Holocene. This conclusion is congruent with recent findings of warm-temperate deciduous forest species in subtropical China. While most species of this type of vegetation exhibit a pattern of multiple refugia and limited range expansion (reviewed by Liu et al.36), some species show evidence of extensive range expansion37,38.
All populations of Schima superba were divided into two groups by the samova analysis (Fig. 1d). Furthermore, the eastern group can be subdivided into two subgroups. This pattern is an expected consequence of long-term isolation among multiple refugia and subsequent localized range expansion. However, compared to C. tibetana, S. superba shows significant phylogeographic structure because the major groups correspond to two highly differentiated clades (Fig. 1d). The east-west divergence in eastern subtropical China has also been observed in Rhododendron simsii19 and Tsuga39 and may reflect the strong landscape effects of the Wuyi Mountains and the Poyang Lake valley as natural barriers to dispersal. However, such barriers may be semi-permeable and not applicable to all species, because similar divergence has not been observed in C. tibetana and M. thunbergii, which occupy the same region.
Taken together, in contrast to phylogeographic congruence of EBLF constituents in subtropical China in previous reports13,14,15, this study revealed diverse phylogeographic patterns in three dominant trees, i.e., multiple refugia (across the range) without significant phylogeographic structure in C. tibetana, glacial retreat and post-glacial recolonization in M. thunbergii, and multiple refugia with significant phylogeographic structure in S. superba. This observation agrees with the suggestion of many CP studies that congruent phylogeographic patterns are not necessarily the rule for co-distributed species because of idiosyncrasies in their biological and ecological characteristics, short duration of sharing geographical distribution, and/or different evolutionary histories of genes in the species studied (reviewed by Gutiérrez-García & Vázquez-Domínguez40). The results of this study also support the view that communities are not cohesive units that may be broken up and reformed in different configurations repeatedly and regularly on time scales of a few thousand years41.
Reconciling palaeo-biome reconstruction and phylogeography
Although there are contradictory results in a few cases42,43, the fossil record and phylogeography are largely synergistic in the inferences of past distribution of organisms5. Therefore, the distinct scenarios of the past distribution of EBLF in subtropical China inferred from PBR and phylogeography are quite unusual and need to be reconciled. In this study, we found that the phylogeographic patterns of C. tibetana and S. superba are congruent with the results of previous phylogeographic studies of EBLF constituents. However, M. thunbergii displays a typical pioneer or leading-edge expansion pattern, which conforms to the inference of PBR9,10,11, although there is a small difference in the northern limit at the LGM between M. thunbergii phylogeography (26°N) and PBR (24°N). These results suggest that the inference of previous phylogeographic studies could be biased by stochastic variance among species and phylogenetic niche conservatism within Fagaceae. More phylogeographic surveys, particularly CP studies with the same sampling strategy and molecular markers, may provide an opportunity to reconcile the discrepancy between PBR and phylogeography.
Despite the fact that some species in EBLF such as M. thunbergii retreated to the south during the LGM, the complete absence of EBLF to the north of 24°N at that time suggested by PBR is still contrary to the phylogeographic patterns of most plant species characteristic of this vegetation. Although additional paleopalynological investigations are needed to verify the conclusion of previous PBR, the absence of EBLF to the north of 24°N during the LGM cannot be simply ascribed to the sparse pollen record. In fact, by comparing fossil records of individual forest plants and palaeo-biome maps in North America, Williams et al.41 also found that the results of PBR sometimes did not match with the original fossil records of individual plants. This observation was attributed to information loss in PBR about the distribution and abundance of individual taxa during the categorization of pollen taxa to one or more plant functional types (PFTs) and biomes44 and the violation of the assumption of community stability. According to our CP results, EBLF may have survived in the north during the LGM, but with reduced distribution and abundance (as suggested by the strong evidence for regional postglacial expansion in both C. tibetana and S. superba) as well as with a different species composition (as suggested by the evidence that M. thunbergii retreated to the south). This kind of community dynamics makes it likely that PBR would omit some EBLF components in the fossil record or misidentify the “EBLF” of the LGM as other types of vegetation because the LGM “EBLF” has no counterpart in the present vegetation41. This conclusion suggests that CP at the community level is capable of providing a full understanding of historical community dynamics22, avoiding the stochastic variance inherent in single-species phylogeographic studies and identifying the artifacts specific to PBR. Therefore, CP offers an insightful perspective for testing competing biogeographic hypotheses evoked by different disciplines.
The difference between comparative phylogeography and species distribution modeling
Our predicted distributions of three dominant tree species showed that three species survived in situ during the LGM (Fig. 4a), consistent with the SDM results of a few EBLF constituents13,14,15. However, these predictions are only partially congruent with the patterns observed in the CP study conducted here, because M. thunbergii exhibits an expansion-contraction pattern. As with any model, SDMs rely on many underlying assumptions and thus incorporate uncertainties45. One of the most important assumptions in SDMs is niche stability over the study period30. However, niche stability may be affected by either changes in biotic interactions in the community or evolutionary adaptation to the biotic and abiotic environment (i.e. changes in realized niches and fundamental niches)4. While fundamental niches are less likely to evolve over relatively short periods, such as the LGM to the present, shifts in the realized niche may occur due to changes in biotic interactions30. For example, Stewart et al.46 suggested that extinction of competitors owing to environmental change and small patch size in glacial refugia could change community structure and thus alter a species’ realized niche.
The three species studied here are canopy trees that occupy similar niches (Fig. 4b), implying that competition may exist among them. Such competition could have become much more severe when the EBLF retreated to small glacial refugia because of the low carrying capacity within refugia47. It is well known that Fagaceae-dominated vegetation types are climax communities in the EBLF region48, indicating that these taxa may have the competitive advantage over other EBLF associates49. Therefore, it is reasonable to postulate that M. thunbergii might have been a weak competitor that could have been eliminated from glacial refugia by stronger competitors such as C. tibetana. Thus, the different results from phylogeography (Fig. 1c) and SDMs (Fig. 4a) for M. thunbergii may reflect the fact that it had fewer opportunities to occupy the full extent of its fundamental niche during the LGM because of its poor competitive ability. Future studies using SDMs that account for biotic interactions50 might provide projected distributions with higher biological realism that might be better reconciled with phylogeographic inferences.
The limitations of this study
Comparative phylogeography is a powerful tool for addressing community dynamics in distribution and abundance at the regional scale. However, the conclusions of this study might be subject to two limitations. First, because of financial constraints, this study was limited to two cpDNA markers to allow for a large number of individuals and populations to be surveyed. However, a given spatial genetic structure inferred by cpDNA reflects the history of maternal lineages, as recorded by seed movement, whereas pollen flow, as measured by the nuclear genome, may be more extensive than seed movement, and stochastic processes may lead to a wide range of genetic patterns27. Therefore, the patterns observed here should ideally be confirmed using nuclear molecular markers. Second, the three species studied here have a variety of congeners in the EBLF of subtropical China31, and hybridization among them is a point of concern for phylogeographic and population genetic studies51. However, this situation is difficult to avoid because almost all dominant tree genera in EBLF are species-rich taxa31. To limit the potential influence of hybridization on our work, we only included samples from individuals exhibiting typical morphological traits identified by experienced botanists, and we abandoned individuals or populations where morphologically similar species co-occurred. Further investigations on the influence of hybridization by comparing phylogeographic structures of congeneric species (e.g., Saeki et al.)51 in EBLF may complement the conclusions of this study. Despite these caveats, we believe that the basic conclusions of this study are robust because the phylogeographic pattern of multiple refugia in subtropical China observed in C. tibetana and S. superba has been repeatedly revealed in the past (see review in Liu et al.)36. Furthermore, the pattern of glacial retreat and post-glacial recolonization, although rarely reported in the past, has been stressed in a few recent studies37,38. The main finding of idiosyncratic responses of EBLF constituents in China to the late Quaternary climate changes suggests that the community assemblages of EBLF have changed over time and there is no palaeo-analog to the modern-day EBLF, which explains the discrepancy between PBR and phylogeography.
Materials and Methods
Population sampling
Our sampling strategy for phylogeographic analyses was to collect a few individuals (1–13 spaced at least 50 m for each population) from many localities to sufficiently cover the entire geographic range of each species: 332 individuals/45 populations of C. tibetana, 392/46 of M. thunbergii, and 515/54 of S. superba, were sampled (Supplementary Table S2; Fig. 1). All voucher specimens were deposited in the Herbarium of Jiangxi Agricultural University (JXAU).
DNA extraction, PCR amplification, and sequencing
Total genomic DNA was extracted from ca. 20 mg of silica-dried leaf material using a modified cetyltrimethyl ammonium bromide (CTAB) protocol52. After preliminary screening of several chloroplast fragments, we chose the psbA–trnH and rpl32–trnL(UAG) intergenic spacer (IGS) regions for the full phylogeographic survey because they could be successfully amplified in all three species and contained relatively rich polymorphic sites.
PCRs were performed in 20-μl volume containing 50–100 ng genomic DNA, 0.5 μmol/L of each primer, and 10 μl 2 × Taq PCR MasterMix [0.1 U Taq polymerase/μl, 0.5 mmol/L dNTP each, 20 mmol/L Tris-HCL (pH 8.3), 100 mmol/L KCl, and 3 mmol/L MgCl2; Tiangen]. PCR amplifications were conducted under the following conditions: 5 min at 80 °C, 31–35 cycles of 1 min at 95 °C, 1 min of annealing at 50–58 °C, followed by a ramp rate of 0.3 °C/s to 65 °C, and extension at 65 °C for 4 min, with a final 5-min extension at 65 °C. PCR products were purified using TIANgel Midi Purification Kit (Beijing Tiangen Biotech Co., LTD) prior to sequencing. Sequencing was carried out from both directions using the amplification primers. Sequences containing microsatellites and singletons in some individuals were amplified and sequenced twice to exclude the possibility of PCR errors.
Phylogeographic and population demography analyses
All sequences for each region were edited with sequencher (GeneCodes Corporation, Ann Arbor, MI, USA) and were aligned using clustal_x 1.8153. Indels and inversions were treated as single mutations. Haplotype relationships were assessed using the median-joining network method54 in network 4.1.0.8 (http://www.fluxus-engineering.com).
We used the program haplonst55 to calculate the total and within-population haplotype diversity (hT and hS), as well as population differentiation based on ordered (NST) and unordered (GST) haplotypes. The values of NST and GST were then compared using U-statistics to test for the presence of phylogeographic structure. To test the EI model for each species, we calculated the correlation of genetic variation with latitude by performing a multiple regression analysis in spss version 13.0 (SPSS Inc, Chicago, IL). Spatial genetic structure of each species was analyzed by the program samova (Spatial Analysis of Molecular Variation)56, which seeks for the best grouping (K) of populations that are geographically homogeneous and maximally differentiated from each other.
Tajima’s D57 and Fu’s FS58 were tested for each species to infer potential population growth and expansion. Mismatch distribution analyses59 were implemented in arlequin (ver. 3.1)60 under the model of demographic expansion. The goodness-of-fit was tested with the sum of squared deviations (SDD) between observed and expected mismatch distributions, and raggedness index (HRag), using 1000 parametric bootstrap replicates. The expansion time (in generations) for expanding species or groups was calculated using the formula T = τ/2 μkg59, where μ is the substitution rate in substitution/site/year, k is the average sequence length used for analysis (C. tibetana: 1826 bp; M. thunbergii: 1450 bp; S. superba: 1399 bp), and g is the generation time in years. The μ of C. tibetana was set as 0.71 × 10−9 substitutions per site year−1 (s/s/y), which is the mean substitution rate of two chloroplast intergenic spacers (ndhJ-trnF and atpI-atpH) in a closely related genus Fagus we previously studied61. This rate is similar to μ of three chloroplast intergenic spacers (trnH-psbA, trnT-trnL, and atpI-atpH) in another Fagaceae member, Quercus glauca (0.96 × 10−9 or 0.69 × 10−9 s/s/y)15. To approximate the substitution rate of chloroplast genomic non-coding regions in M. thunbergii,a phylogenetic tree of Lauraceae was reconstructed based on the chloroplast sequences (rpl16, trnL-F, and psbA-trnH) of species in Fig. 3 of Nie et al.62, and the substitution rates across the trees were computed by beast v.1.7.1 using the same settings as that study used. We chose the local substitution rate (4 × 10−9 s/s/y) of clade D of Fig. 3 in Nie et al.62 to represent the substitution rate of psbA–trnH and rpl32–trnL(UAG) in M. thunbergii, because substitution rates varied among clades and clade D is a minimally monophyletic group that includes M. thunbergii when only chloroplast sequences were used for phylogenetic reconstruction. Similarly, we reconstructed the phylogenetic tree of Theaceae in Fig. 2 of Li et al.63 using beast v.1.7.1 to approximate the substitution rate of chloroplast genome in S. superba. The local substitution rate (0.6 × 10−9 s/s/y) of the clade C1 [see Fig. 2 in Li et al.63] in which the genus Schima was embedded, was adopted in this study. There is no accurate record for the first reproduction age of the three species. For C. tibetana, 25-year was used based on the generation time reported in another Castanopsis species64. In Lauraceae, Ocotea tenera takes at least 5 years to reach its reproductive maturity65, but the age at maturity of Sassafras was reported as 10 years66. Natural populations are likely to take a longer time to reach reproductive maturity. Therefore, we used 10 years as the generation time for M. thunbergii. We used 8 years to approximate the generation time of S. superba based on observations on age of first reproduction of cultivated trees at the arboretum of Jiangxi Agricultural University (Z.Y. Zhang, personal observation).
We used the Bayesian skyline plots (BSPs) method67 as implemented in beast v. 1.7.1 for estimating fluctuations in the effective population size of each species using the above substitution rates accordingly. MCMC chains were run for 20,000,000 generations for C. tibetana, M. thunbergii and S. superba under the GTR, GTR + G and HKY model chosen by jModelTest 2.1.5 respectively68.
Species distribution modeling
We employed the maximum entropy approach (maxent)69 to predict the distribution of the three species at the present time and at the time of the LGM (0.021–0.018 Ma). Nineteen environmental variables for present and the time of the LGM (MIROC 3.2 scenario) were compiled from the WORLDCLIM database with a resolution of 2.5 arc-min (http://www.worldclim.org)70 for each environmental layer. To avoid over-fitting of niche models, seven variables with pairwise Pearson correlation coefficients r ≤ 0.70 (annual mean temperature, BIO1, mean diurnal range, BIO2, isothermality, BIO3, temperature seasonality, BIO4, annual precipitation, BIO12, precipitation seasonality, BIO15 and precipitation of coldest quarter, BIO19) were retained for subsequent analyses.
SDMs were constructed using 205/127/169 presence records of Castanopsis tibetana, Machilus thunbergii, and Schima superba, respectively, including all sites recorded in the field work of this study and for which specimen records with GPS (Chinese Virtual Herbarium) data are available. Each model was run 10 times using the default parameters (convergence threshold of 10−5, maximum iterations of 500 and regularization multiplier of 1) and the following user-selected features: application of a random seed, duplicate presence records removal and logistic probabilities used for the output. 80% of species records were used to train the model and 20% to test the model. Model accuracy was measured by the area under the ‘Receiver Operating Characteristic (ROC) Curve’ (AUC)71,72. A score between 0.7 and 1.0 indicates that the model performs better than random and was considered acceptable discrimination71.
We then measured niche differences between each species’ ENM following the suggestions of Warren et al.73. enmtools, version 1.3, was used to calculate Schoener’s D74 and a standardized version of Hellinger distance (calculated as I)73. Both D and I ranged from 0 (no niche overlap) to 1 (identical niches). Next, we conducted an identity test to build niche models based on a set of pseudoreplicates generated from a random sampling of data points pooled for each pair of species. A total of 100 replicates were used to generate the pseudoreplicated data sets. The observed measures of niche similarity between species were compared with the null distribution.
Additional Information
How to cite this article: Fan, D. et al. Idiosyncratic responses of evergreen broad-leaved forest constituents in China to the late Quaternary climate changes. Sci. Rep. 6, 31044; doi: 10.1038/srep31044 (2016).
Supplementary Material
Acknowledgments
We are grateful to the many people who provided help with the fieldwork. We also appreciate the valuable comments by D. B. Marchant (University of Florida). This study was supported by grants from the National Natural Science Foundation of China (grant nos 31160082 and 31160043) and China Scholarship Council (201208360115).
Footnotes
Author Contributions D.F. produced data, analyzed data and wrote manuscript; W.H. and M.Z. conducted experiments and analyzed data; B.L. collected samples. A.B.M., D.E.S. and P.S.S. wrote manuscript. Z.Z. designed research, collected samples and wrote manuscript.
References
- Song Y. C. The essential characteristics and main types of the broad-leaved evergreen forest in China. Phytocoenologia 16, 105–123 (1988). [Google Scholar]
- Wang X. H., Kent M. & Fang X. F. Evergreen broad-leaved forest in Eastern China: its ecology and conservation and the importance of resprouting in forest restoration. Forest. Ecol. Manag. 245, 76–87 (2007). [Google Scholar]
- Zhao D., Wu S. & Yin Y. Responses of terrestrial ecosystems’ net primary productivity to future regional climate change in China. PloS One 8, e60849 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearman P. B. et al. Prediction of plant species distributions across six millennia. Ecol. Lett. 11, 357–369 (2008). [DOI] [PubMed] [Google Scholar]
- Gavin D. G. et al. Climate refugia: joint inference from fossil records, species distribution models and phylogeography. New Phylot. 204, 37–54 (2014). [DOI] [PubMed] [Google Scholar]
- Riddle B. R. The molecular phylogeographic bridge between deep and shallow history in continental biotas. Trends Ecol. Evol. 11, 207–211 (1996). [DOI] [PubMed] [Google Scholar]
- Hewitt G. M. The genetic legacy of the Quaternary ice ages. Nature 405, 907–913 (2000). [DOI] [PubMed] [Google Scholar]
- Zheng Z., Yuan B. Y. & Petit-Maire N. Palaeoenvironments in China during the Last Glacial Maximum and the Holocene Optimum. Episodes 21, 152–158 (1998). [Google Scholar]
- Yu G. et al. Palaeovegetation of China: a pollen data-based synthesis for the mid-Holocene and last glacial maximum. J. Biogeogr. 27, 635–666 (2000). [Google Scholar]
- Harrison S. P., Yu G., Takahara H. & Prentice I. C. Palaeovegetation diversity of temperate plants in East Asia. Nature 413, 129–130 (2001). [DOI] [PubMed] [Google Scholar]
- Ni J., Cao X., Jeltsch F. & Herzschuh U. Biome distribution over the last 22,000 yr in China. Palaeogeogr. Palaeoclim. Palaeoecol. 409, 33–47 (2014). [Google Scholar]
- Qian H. & Ricklefs R. E. Palaeovegetation: diversity of temperate plants in East Asia–reply. Nature 413, 130 (2001). [DOI] [PubMed] [Google Scholar]
- Shi M. M., Michalski S. G., Welk E., Chen X. Y. & Durka W. Phylogeography of a widespread Asian subtropical tree: genetic east-west differentiation and climate envelope modelling suggest multiple glacial refugia. J. Biogeogr. 41, 1710–1720 (2014). [Google Scholar]
- Sun Y., Hu H. Q., Huang H. W. & Vargas-Mendoza C. F. Chloroplast diversity and population differentiation of Castanopsis fargesii (Fagaceae): a dominant tree species in evergreen broad-leaved forest of subtropical China. Tree Genet. Genomes 10, 1531–1539 (2014). [Google Scholar]
- Xu J. et al. Phylogeography of Quercus glauca (Fagaceae), a dominant tree of East Asian subtropical evergreen forests, based on three chloroplast DNA interspace sequences. Tree Genet. Genomes 11, 805 (2015). [Google Scholar]
- Webb C. O., Ackerly D. D., McPeek M. A. & Donoghue M. J. Phylogenies and community ecology. Annu. Rev. Ecol. Syst. 33, 475–505 (2002). [Google Scholar]
- Wiens J. J. & Graham C. H. Niche conservatism: integrating evolution, ecology, and conservation biology. Annu. Rev. Ecol. Evol. Syst. 36, 519–539 (2005). [Google Scholar]
- Wang J., Gao P. X., Kang M., Lowe A. J. & Huang H. W. Refugia within refugia: the case study of a canopy tree Eurycorymbus cavaleriei in subtropical China. J. Biogeogr. 36, 2156–2164 (2009). [Google Scholar]
- Li Y., Yan H. F. & Ge X. J. Phylogeographic analysis and environmental niche modelling of widespread shrub Rhododendron simsii in China reveals multiple glacial refugia during the last glacial maximum. J. Syst. Evol. 50, 362–373 (2012). [Google Scholar]
- Wang Y. H. et al. Molecular phylogeography and ecological niche modelling of a widespread herbaceous climber, Tetrastigma hemsleyanum (Vitaceae): insights into Plio-Pleistocene range dynamics of evergreen forest in subtropical China. New Phytol. 206, 852–867 (2015). [DOI] [PubMed] [Google Scholar]
- López-Pujol J., Zhang F. M., Sun H. Q., Ying T. S. & Ge S. Centres of plant endemism in China: places for survival or for speciation? J. Biogeogr. 38, 1267–1280 (2011). [Google Scholar]
- Arbogast B. S. & Kenagy G. J. Comparative phylogeography as an integrative approach to historical biogeography. J. Biogeogr. 28, 819–825 (2001). [Google Scholar]
- Avise J. C. Phylogeography: retrospect and prospect. J. Biogeogr. 36, 3–15 (2009). [Google Scholar]
- Hewitt G. M. Postglacial recolonization of European biota. Biol. J. Linn. Soc. 68, 87–112 (1999). [Google Scholar]
- Lapointe F. J. & Rissler L. J. Congruence, consensus, and the comparative phylogeography of codistributed species in California. Amer. Natur. 166, 290–299 (2005). [DOI] [PubMed] [Google Scholar]
- Soltis D. E., Morris A. B., Mclachlan J. S., Manos P. S. & Soltis P. S. Comparative phylogeography of unglaciated eastern North America. Mol. Ecol. 15, 4261–4293 (2006). [DOI] [PubMed] [Google Scholar]
- Carstens B., Brunsfeld S. J., Demboski J. R., Good J. & Sullivan J. Investigating the evolutionary history of the Pacific Northwest mesic forest ecosystem: hypothesis testing within a comparative phylogeographic framework. Evolution 59, 1639–1652 (2005). [PubMed] [Google Scholar]
- Sunnucks P. et al. A tale of two flatties: different responses of two terrestrial flatworms to past environmental climatic fluctuations at Tallaganda in montane southeastern Australia. Mol. Ecol. 15, 4513–4531 (2006). [DOI] [PubMed] [Google Scholar]
- Moussalli A., Moritz C., Williams S. E. & Carnaval A. C. Variable responses of skinks to a common history of rainforest fluctuation: concordance between phylogeography and palaeo-distribution models. Mol. Ecol. 18, 483–499 (2009). [DOI] [PubMed] [Google Scholar]
- Svenning J. C., Flójgaard C., Marske K. A. & Nógues-Bravo D. Applications of species distribution modelling to palaeobiology. Quat. Sci. Rev. 30, 2930–2947 (2011). [Google Scholar]
- Wu C. Y. Vegetation of China. (Science Press, Beijing, 1980). [Google Scholar]
- Crisp M. D. et al. Phylogenetic biome conservatism on a global scale. Nature 458, 754–756 (2009). [DOI] [PubMed] [Google Scholar]
- Hewitt G. M. Some genetic consequences of ice ages, and their role in divergence and speciation. Biol. J. Linn. Soc. 58, 247–276 (1996). [Google Scholar]
- Wu S. H. et al. Contrasting phylogeographical patterns of two closely related species, Machilus thunbergii and Machilus kusanoi (Lauraceae), in Taiwan. J. Biogeogr. 33, 936–947 (2006). [Google Scholar]
- Liao P. C. et al. Historical spatial range expansion and a very recent bottleneck of Cinnamomum kanehirae Hay. (Lauraceae) in Taiwan inferred from nuclear genes. BMC Evol. Biol. 10, 124 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. Q., Sun Y. S., Ge X. J., Gao L. M. & Qiu Y. X. Phylogeographic studies of plants in China: Advances in the past and directions in the future. J. Syst. Evol. 50, 267–275 (2012). [Google Scholar]
- Li X. H., Shao J. W., Lu C., Zhang X. P. & Qiu Y. X. Chloroplast phylogeography of a temperate tree Pteroceltis tatarinowii (Ulmaceae) in China. J. Syst. Evol. 50, 325–333 (2012). [Google Scholar]
- Tian S. et al. Repeated range expansions and inter-/postglacial recolonization routes of Sargentodoxa cuneata (Oliv.) Rehd. et Wils. (Lardizabalaceae) in subtropical China revealed by chloroplast phylogeography. Mol. Phylogenet. Evol. 85, 238–246 (2015). [DOI] [PubMed] [Google Scholar]
- Cun Y. Z. & Wang X. Q. Phylogeography and evolution of three closely related species of Tsuga (hemlock) from subtropical eastern Asia: further insights into speciation of conifers. J. Biogeogr. 42, 315–327 (2015). [Google Scholar]
- Gutiérrez-García T. A. & Vázquez-Domínguez E. Comparative phylogeography: designing studies while surviving the process. Bioscience 61, 857–868 (2011). [Google Scholar]
- Williams J. W. & Shuman B. N., Webb lll, T., Bartlein, P.J. & Leduc, P.L. Late-Quaternary vegetation dynamics in north America: scaling from taxa to biomes. Ecol. Monogr. 74, 309–334 (2004). [Google Scholar]
- Morris A. B., Graham C. H., Soltis D. E. & Soltis P. S. Reassessment of phylogeographical structure in an eastern North American tree using Monmonier’s algorithm and ecological nich modelling. J. Biogeogr. 37, 1657–1667 (2010). [Google Scholar]
- Rodríguez-Sánchez F., Hampe A., Jordano P. & Arroyo J. Past tree range dynamics in the Iberian Peninsula inferred through phylogeography and palaeodistribution modelling: a review. Rev. Palaeobot. Palyno. 162, 507–521 (2010). [Google Scholar]
- Prentice I. C., Guiot J., Huntley B., Jolly D. & Cheddadi R. Reconstructing biomes from palaeoecological data: a general method and its application to European pollen data at 0 and 6 ka. Clim. Dynam. 12, 185–194 (1996). [Google Scholar]
- Wiens J. A., Stralberg D., Jongsomjit D., Howell C. A. & Snyder M. A. Niches, models, and climate change: assessing the assumptions and uncertainties. Proc. Nat. Acad. Sci. USA 106, 19729–19736 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart J. R., Lister A. M., Barnes I. & Dalén L. Refugia revisited: individualistic responses of species in space and time. Proc. Roy. Soc. London. Ser. B, Biol. Sci. 277, 6615–6671 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampe A. & Jump A. S. Climate relicts: past, present, future. Annu. Rev. Ecol. Evol. Syst. 42, 313–333 (2011). [Google Scholar]
- Castro-Díez P., Villar-Salvador P., Pérez-Rontomé C., Maestro-Martínez M. & Montserrat-Martí G. Leaf morphology and leaf chemical composition in three Querus (Fagaceae) species along a rainfall gradient in NE Spain. Trees 11, 127–134 (1997). [Google Scholar]
- Zhang G. S. Study on the intraspecific and interspecific competition in Machilus thunbergii community. J. Fujian Coll. For. 30, 179–182 (2010). [Google Scholar]
- Dullinger S. et al. Extinction debt of high-mountain plants under twenty-first-century climate change. Nat. Clim. Change 2, 619–622 (2012). [Google Scholar]
- Saeki I., Dick C. W., Barnes B. V. & Murakami N. Comparative phylogeography of red maple (Acer rubrum L.) and silver maple (Acer saccharinum L.): impacts of habitat specialization, hybridization and glacial history. J. Biogeogr. 38, 992–1005 (2011). [Google Scholar]
- Doyle J. J. & Doyle J. L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemistry 19, 11–15 (1987). [Google Scholar]
- Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F. & Higgins D. G. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucl. Acids Res. 25, 4876–4882 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandelt H. J., Forster P. & Rohl A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16, 37–48 (1999). [DOI] [PubMed] [Google Scholar]
- Pons O. & Petit R. J. Measuring and testing genetic differentiation with ordered versus unordered alleles. Genetics 144, 1237–1245 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupanloup I., Schneider S. & Excoffier L. A simulated annealing approach to define the genetic structure of populations. Mol. Ecol. 11, 2571–2581 (2002). [DOI] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123, 585–595 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y. X. & Li W. H. Statistical tests of neutrality of mutations. Genetics 133, 693–709 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers A. R. & Harpending H. C. Population growth makes waves in the distribution of pairwise genetic differences. Mol. Biol. Evol. 9, 552–569 (1992). [DOI] [PubMed] [Google Scholar]
- Excoffier L., Laval G. & Schneider S. Arlequin ver. 3.1: an integrated software package for population genetics data analysis. Evol. Bioinformatics Online 1, 47–50 (2005). [PMC free article] [PubMed] [Google Scholar]
- Zhang Z. Y. et al. Comparative phylogeography of two sympatric beeches in subtropical China: Species-specific geographic mosaic of lineages. Ecol. Evol. 3, 4461–4472 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie N. L., Wen J. & Sun H. Phylogeny and biogeography of Sassafras (Lauraceae) disjunct between eastern Asia and eastern North America. Plant Syst. Evol. 267, 191–203 (2007). [Google Scholar]
- Li M. M., Li J. H., Del Tredici P., Corajod J. & Fu C. X. Phylogenetics and biogeography of Theaceae based on sequences of plastid genes. J. Syst. Evol. 51, 396–404 (2013). [Google Scholar]
- Liu R. L., Wang L. & Du T. Z. Features of population ecological quantity field of Castanopsis fabri community in Jinggang Mountain. Sci. Silv. Sin. 44, 1–7 (2008). [Google Scholar]
- Wheelwright N., Sinclair J. P. & Hochwender C. Leaf size in three generations of a dioecious tropical tree, Ocotea tenera (Lauraceae): sexual dimorphism and changes with age. Am. J. Bot. 99, 1350–1355 (2012). [DOI] [PubMed] [Google Scholar]
- Verdú M. Age at maturity and diversification in woody angiosperms. Evolution 56, 1352–1361 (2002). [DOI] [PubMed] [Google Scholar]
- Drummond A. J., Rambaut A., Shapiro B. & Pybus O. G. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol. Biol. Evol. 22, 1185–1192 (2005). [DOI] [PubMed] [Google Scholar]
- Darriba D., Taboada G. L., Doallo R. & Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat. Meth. 9, 772 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips S. J., Anderson R. P. & Schapire R. E. Maximum entropy modelling of species geographic distributions. Ecol. Model. 190, 231–259 (2006). [Google Scholar]
- Hijmans R. J., Cameron S. E., Parra J. L., Jones P. G. & Jarvis A. Very high resolution interpolated climate surfaces for global land areas. Int. J. Climatol. 25, 1965–1978 (2005). [Google Scholar]
- Fielding A. H. & Bell J. F. A review of methods for the assessment of prediction errors in conservation presence/absence models. Environ. Conserv. 24, 38–49 (1997). [Google Scholar]
- Elith J. et al. Novel methods improve prediction of species’ distributions from occurrence data. Ecography 29, 129–151 (2006). [Google Scholar]
- Warren D. L., Glor R. E. & Turelli M. Environmental niche equivalency versus conservatism: quantitative approaches to niche evolution. Evolution 62, 2868–2883 (2008). [DOI] [PubMed] [Google Scholar]
- Schoener T. W. The Anolis lizards of Bimini: resource partitioning in a complex fauna. Ecology 49, 704–726 (1968). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.