Summary
Bacteria of the SAR11 clade constitute up to one half of all microbial cells in the oxygen-rich surface ocean. DNA sequences from SAR11 are also abundant in oxygen minimum zones (OMZs) where oxygen falls below detection and anaerobic microbes play important roles in converting bioavailable nitrogen to N2 gas. Evidence for anaerobic metabolism in SAR11 has not yet been observed, and the question of how these bacteria contribute to OMZ biogeochemical cycling is unanswered. Here, we identify the metabolic basis for SAR11 activity in anoxic ocean waters. Genomic analysis of single cells from the world’s largest OMZ revealed diverse and previously uncharacterized SAR11 lineages that peak in abundance at anoxic depths, but are largely undetectable in oxygen-rich ocean regions. OMZ SAR11 contain adaptations to low oxygen, including genes for respiratory nitrate reductases (Nar). SAR11 nar genes were experimentally verified to encode proteins catalyzing the nitrite-producing first step of denitrification and constituted ~40% of all OMZ nar transcripts, with transcription peaking in the zone of maximum nitrate reduction rates. These results redefine the ecological niche of Earth’s most abundant organismal group and suggest an important contribution of SAR11 to nitrite production in OMZs, and thus to pathways of ocean nitrogen loss.
Introduction
Alphaproteobacteria of the SAR11 clade form one of the most ecologically dominant organism groups on the planet, representing up to half of the total microbial community in the oxygen-rich surface ocean1–5. All characterized SAR11 isolates, including the globally ubiquitous Pelagibacter genus, are aerobic heterotrophs adapted for scavenging dissolved organic carbon and nutrients under the oligotrophic conditions of the open ocean6–9. Gene-based surveys have also revealed diverse SAR11 lineages at high abundance in the deep waters of the meso- and bathypelagic realms10–13. However, the functional properties that distinguish SAR11 living in distinct ocean regions remain unclear. All known SAR11 genomes are small (typically less than 1.5 Mbp), with genomic streamlining as a potential adaptation to the nutrient limiting conditions of the open ocean.11 It has been hypothesized that adaptations in SAR11 do not involve large variations in gene content6,8, suggesting that SAR11’s contribution to ocean biogeochemistry is primarily through its role in aerobic oxidation of organic carbon.
Although genetic or biochemical evidence of anaerobic metabolism has not been reported for SAR11, high abundances of SAR11-related genes have been detected under anoxic conditions in marine oxygen minimum zones (OMZs). Permanent OMZs extend over ~8% of the oceanic surface area (O2 < 20 µM)14, with the largest and most intense OMZs in upwelling regions of the Eastern Pacific. In the cores of these regions microbial respiration of high surface primary production combines with low ventilation to deplete oxygen (O2) from mid-water depths, resulting in O2 concentrations below detection (~10 nM) over a major portion (~100-700 m) of the water column15. In the absence of O2, respiratory nitrate (NO3-) reduction to nitrite (NO2-) becomes the dominant process for organic matter oxidation 16, with respiratory NO3-reductases (Nar) being among the most abundant and highly expressed enzymes in OMZs 17–19. NO3- respiration results in a substantial accumulation of NO2- in OMZs, often to micromolar concentrations20. This NO2- pool is actively cycled through NO2--consuming microbial metabolisms, notably the anaerobic processes of denitrification and anaerobic ammonium oxidation (anammox)21,22, which together in OMZs account for 30-50% of the loss of bioavailable nitrogen from the ocean as either gaseous dinitrogen (N2) or nitrous oxide (N2O)21,22. Surprisingly, SAR11 bacteria are often the most abundant organisms in the NO2--enriched N-loss zone of OMZs where O2 is undetectable, representing ~20% (range: 10-40%) of all 16S rRNA genes and protein-coding metagenome sequences in the 0.2 to 1.6 µm biomass fraction 18,19,23,24. Such high abundances imply that SAR11 make up a substantial fraction of the OMZ community and raise the question of SAR11’s role in OMZ biogeochemistry.
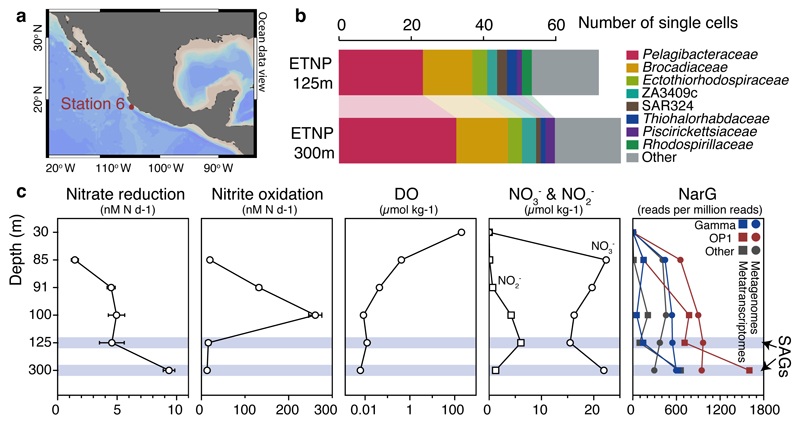
Here, we analyzed single amplified genomes (SAG) to identify the metabolic basis for SAR11's dominance in anoxic OMZs. We focused on SAR11 SAGs obtained from the Eastern Tropical North Pacific (ETNP) OMZ off Mexico, the world’s largest OMZ accounting for 41% of global OMZ surface area14 (Fig. 1a). Oxygen concentration ([O2]) at this site declined from ~200 μM at the surface to ~400 nM at the bottom of the oxycline (30-85 m) and was typically at or below the detection limit (~10 nM) from ~90 m to 700 m. At the time of sample collection, NO3- reduction rates increased with depth into the OMZ, peaking at ~9.5 nM N d-1 at 300 m19, paralleling an increase in the abundance of sequences encoding Nar-type NO3- reductases in coupled metagenomes and metatranscriptomes (Fig. 1c). In contrast, aerobic NO2- oxidation peaked at 100 m (260 nM N d-1) where trace O2 was available and NO2- was abundant, before declining 20-fold with depth into the OMZ (Fig. 1c). However, NO2- oxidation rates are likely overestimated due to slight O2 contamination in incubations20. These data highlight a transition to anoxia within the ETNP OMZ15,19, with in situ [O2] at least an order of magnitude lower than the inhibitory threshold for NO3- reduction, denitrification, and anammox25,26, consistent with micromolar accumulations of NO2- from NO3- reduction in this zone.
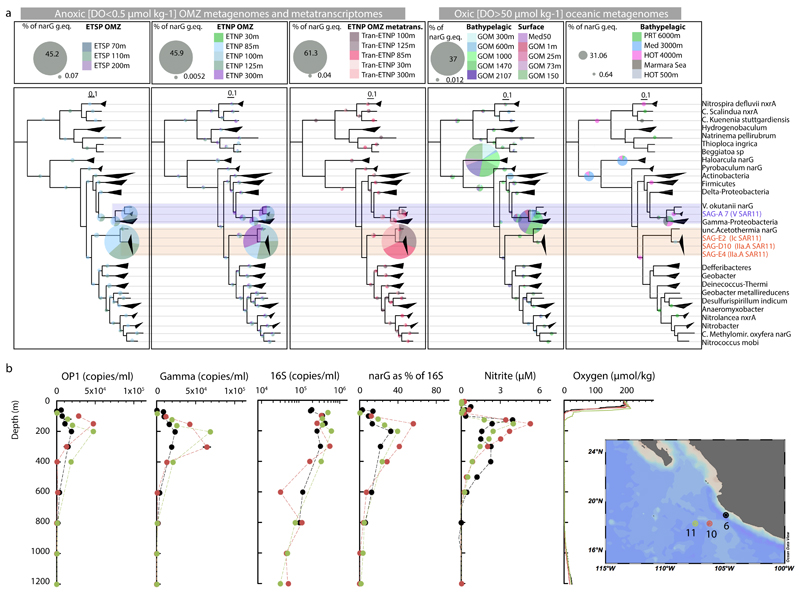
Figure 1. Site description and phylogenetic affiliation of single cells.
a, Location of station 6 (red) in the ETNP from which samples were obtained. Map was created with Ocean Data View (Schlitzer, R., odv.awi.de, 2015). b, Taxonomic classification of sorted single cells, based on their 16S rRNA genes. c, Nitrate reduction and nitrite oxidation rates relative to dissolved O2 (DO), nitrate, and nitrite concentrations and narG read abundance in metagenomes and metatranscriptomes. Error bars represent standard error from triplicate measurements. Note that a log10 scale is used for the DO plot and that 0.01 µmol kg-1 represents the detection limit of the STOX sensor oxygen data presented here. DO at 300 m was below the detection limit.
Diverse SAR11 single cell genomes from anoxic waters
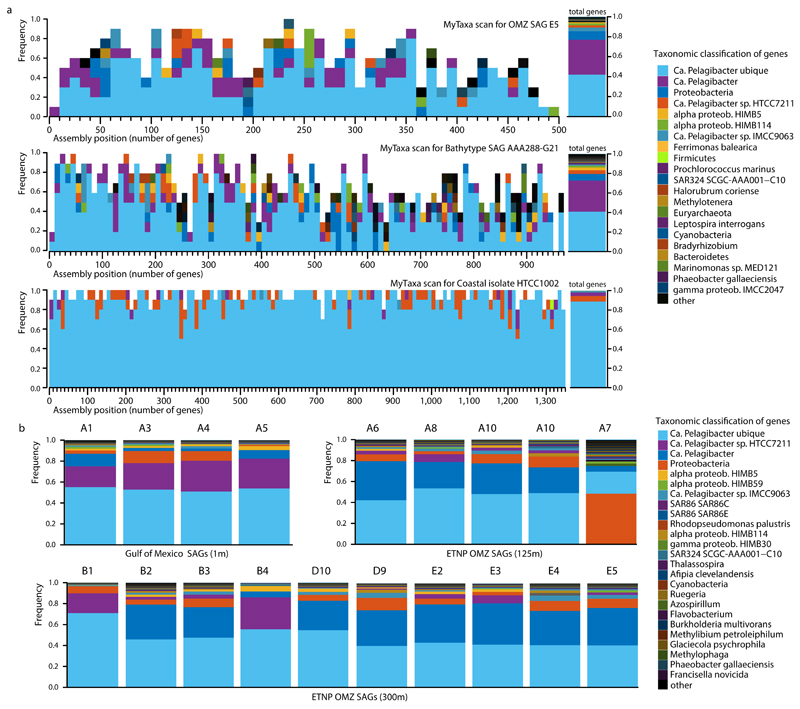
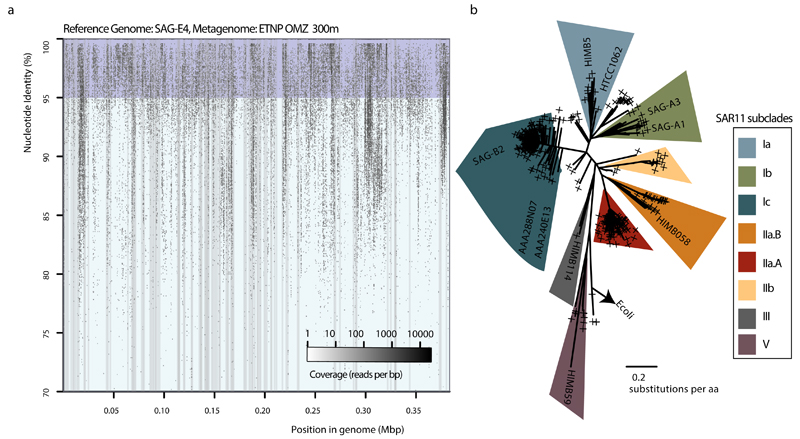
Samples for SAG analysis were obtained from two depths in the anoxic zone: at 125 m at the NO2- maximum (6 μM), and at 300 m in the core of the NO3- reduction zone. Single prokaryotic cells were isolated by fluorescence-activated sorting, subjected to genome amplification27, and screened by 16S rRNA gene fragment (470 bp) PCR and Sanger sequencing. From this screen, 23% and 32% of SAGs from 125 and 300 m, respectively, were confidently assigned to the SAR11 Family Pelagibacteraceae (Fig. 1b), thus confirming SAR11’s substantial numerical abundance in the OMZ. From this SAR11 subset, 10 SAGs from 125 m and 12 SAGs from 300 m were randomly selected for shotgun sequencing (Illumina), along with 5 technical control SAR11 SAGs from the oxic surface waters of the Gulf of Mexico (GoM). Following sequencing, quality filtering, and assembly, a total of 19 SAGs were used for analysis: 15 OMZ SAGs (5 from 125 m, 10 from 300 m) and 4 GoM control SAGs (Supplementary Table 1). These genomes exhibited varying levels of completeness (~2-90%; average 30%) and no detectable contamination (Extended Data Fig. 1), as assessed by the presence of single-copy housekeeping genes28,29, 16S rRNA gene identities, and the taxonomic assignment of SAG contigs (Supplementary Tables 1, 2, and Supplementary Discussion).
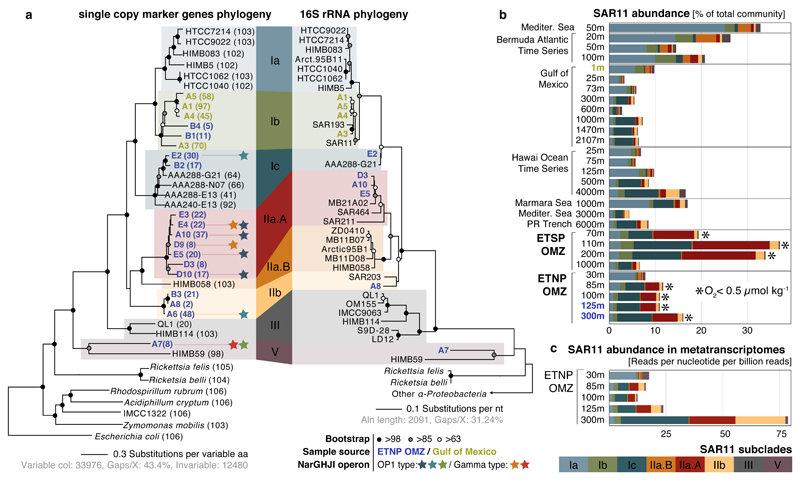
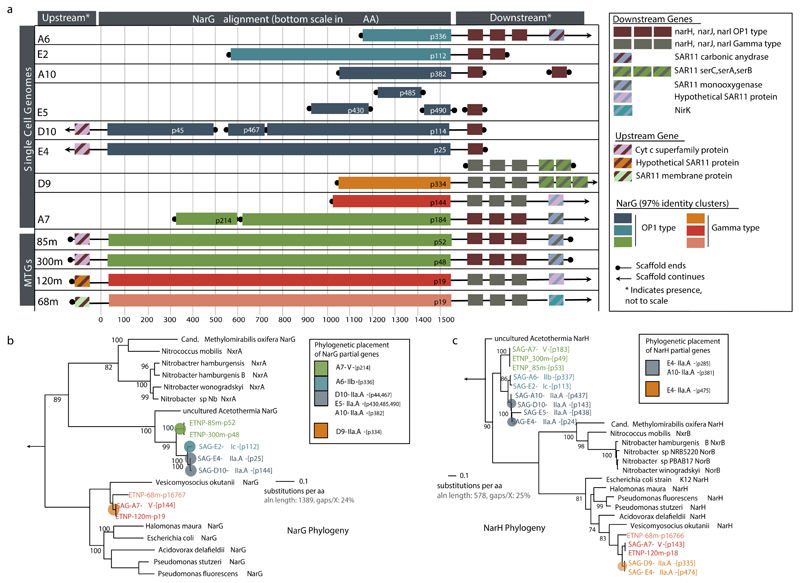
The identified SAGs represented a diverse and novel SAR11 community in the OMZ. Phylogenetic reconstructions based on either 16S rRNA genes or single-copy housekeeping proteins placed the 19 SAGs in 5 subclades of SAR11 (Fig. 2a). Average amino acid identity (AAI) comparisons among all available SAR11 genomes (Supplementary Table 3) further corroborated this classification, placing (i) 7 OMZ SAGs within the previously uncharacterized deep-branching monophyletic group of subclade IIa (hereafter designated subclade IIa.A), distinct (>5% 16S divergence) from SAG HIMB058 from the tropical North Pacific (hereafter designated subclade IIa.B), (ii) 3 OMZ SAGs within the deep-branching subclade IIb, (iii) 2 OMZ SAGs within subclade Ic, which includes recently described single-cell genomes from the bathypelagic ocean6, (iv) 2 OMZ and all 4 GoM surface SAGs within subclade Ib, which thus far lacks genome representatives, and (v) OMZ SAG A7 as most closely related to HIMB59, a member of the divergent SAR11 subclade V8,30,31. Note that the exact placement of subclade V in the SAR11 phylogeny is unstable depending on the marker gene and outgroup used32,33. The average estimated genome size of OMZ SAGs was 1.33 Mbp (Supplementary Table 1), consistent with prior reports of genome streamlining in SAR11.
Figure 2. Diversity, abundance, and transcription of nitrate-reducing SAR11.
a, Maximum likelihood phylogeny based on the concatenated alignment of single copy housekeeping (left) and 16S rRNA (right) genes in SAGs from this study, SAR11 and representative alphaproteobacterial genomes. Values in parentheses denote the number of housekeeping genes used per genome. For the 16S-based tree, only full-length sequences from the genomes in the left tree were included. Star symbols of the same color represent closely related narG genes (>97% aa identity), encoding the catalytic subunit of the respiratory nitrate reductase of the DMSO family. b, Abundance of SAR11 subclades (left) in selected oceanic metagenomes. Note that the major nar-encoding clade IIa.A peaks in abundance at oxygen-depleted OMZ depths. Dataset descriptions are available in Supplementary Table 1. c, Normalized average coverage of SAR11 subclades in ETNP metatranscriptomes. Transcription by nar-encoding lineages increases from the base of the oxycline (85 m) to spike at the OMZ core (300 m), but is negligible in the overlying oxic zone (30 m).
OMZ SAR11 abundance peaks under oxygen depletion
To estimate the in situ abundance and activity of OMZ SAR11, metagenome and metatranscriptome reads from OMZ sites and from diverse oxic ocean regions (Supplementary Table 4) were recruited to 39 available SAR11 genomes (Supplementary Table 1). Metagenomic read recruitment, performed essentially as described previously34, showed that each OMZ SAR11 subclade represents a sequence-discrete (and hence tractable) population (Supplementary Discussion), but with each population encompassing substantial intra-population variation (~92-100% average nucleotide identity between members of the population vs. <90% between populations), as well as gene content variability (Extended Data Fig. 2). We therefore estimated SAR11 abundance at the subclade level, based on the average coverage of 507 genes shared between genomes from all SAR11 subclades. Based on this analysis, SAR11 subclades Ic, IIa.A, and IIb together comprised about 10 to 30% of the bacterial community in ETNP and ETSP metagenomes and metatranscriptomes from depths with undetectable O2 (Fig. 2b, 2c), consistent with the high abundance of SAR11 in the pool of cells sorted for SAG analysis (Fig. 1b). Subclade IIa.A, composed exclusively of 7 SAGs from this study, was particularly abundant, making up to 15% of the community in anoxic samples. All OMZ subclades were absent from or much less abundant (<5%) in metagenomes from oxic sites, including those from above the ETNP OMZ (Fig. 2b). Together, these results identify newly described SAR11 subclades whose distribution is linked to an oxygen-depleted niche.
Metabolic adaptations to low oxygen in SAR11 genomes
OMZ and GoM SAGs were then analyzed for evidence of microaerobic or anaerobic metabolism. Surprisingly, in 8 of the 15 OMZ SAGs, belonging to SAR11 subclades Ic, IIa.A, IIb and V, protein family-based classification detected genes encoding the respiratory NO3-reductase (Nar) of the DMSO reductase superfamily (Fig. 2a). Evidence of a complete canonical nar operon (narGHJI) –encoding the α subunit that catalyzes NO3- reduction to NO2- (NarG), the iron-sulfur-containing β subunit (NarH) that transfers electrons to the molybdenum cofactor of NarG, the transmembrane cytochrome b-like γ subunit (NarI) involved in electron transfer from membrane quinols to NarH, and the NarJ chaperone involved in enzyme formation– was found within a single assembled contig in 4 SAGs (A6, E4, D9, A7), while partial narG and narH fragments were identified in another 4 SAGs (Extended Data Fig. 3). In all SAR11 SAGs containing nar on a contig, we identified other genes upstream or downstream on the same contig taxonomically assigned to SAR11 reference genomes (Supplementary Table 5, Supplementary Discussion), further confirming the association of nar with SAR11. Genes encoding the NO3-/ NO2- transporter NarK and proteins for biosynthesis of the essential molybdenum cofactor (moeA, mobA) were also identified in 8 and 5 of the SAGs, respectively (Supplementary Table 1). In only 4 of the 15 OMZ SAGs were Nar or cofactor synthesis genes not detected, presumably due to sequencing gaps (completeness of these SAGs: 4-20%; Supplementary Table 1). In contrast, these genes were not detected in any of the 4 control SAGs from the oxic GoM, despite high completeness of those genomes (average 61%). Genes encoding for downstream steps of denitrification or other dissimilatory anaerobic metabolisms were not found in any of the SAGs. However, in contrast to all previously analyzed SAR11 genomes, 3 of the OMZ SAGs, all from subclade IIa.A, also contained genes encoding high-affinity O2-utilizing bd-type terminal oxidases (Supplementary Table 1). Compared to the coxI-type oxidases present in all known SAR11 genomes, including the OMZ SAGs analyzed here, bd-type oxidases have a much higher affinity for O2 (3-8 nM; Supplementary Discussion), suggesting a potential for microaerobic respiration by OMZ SAR11. These results provide the first indication of adaptation to low oxygen in SAR11 and the ability to respire NO3- to NO2- in the absence of oxygen, consistent with the distribution of these bacteria in the OMZ water column.
Multiple divergent nitrate reductases in OMZ SAR11
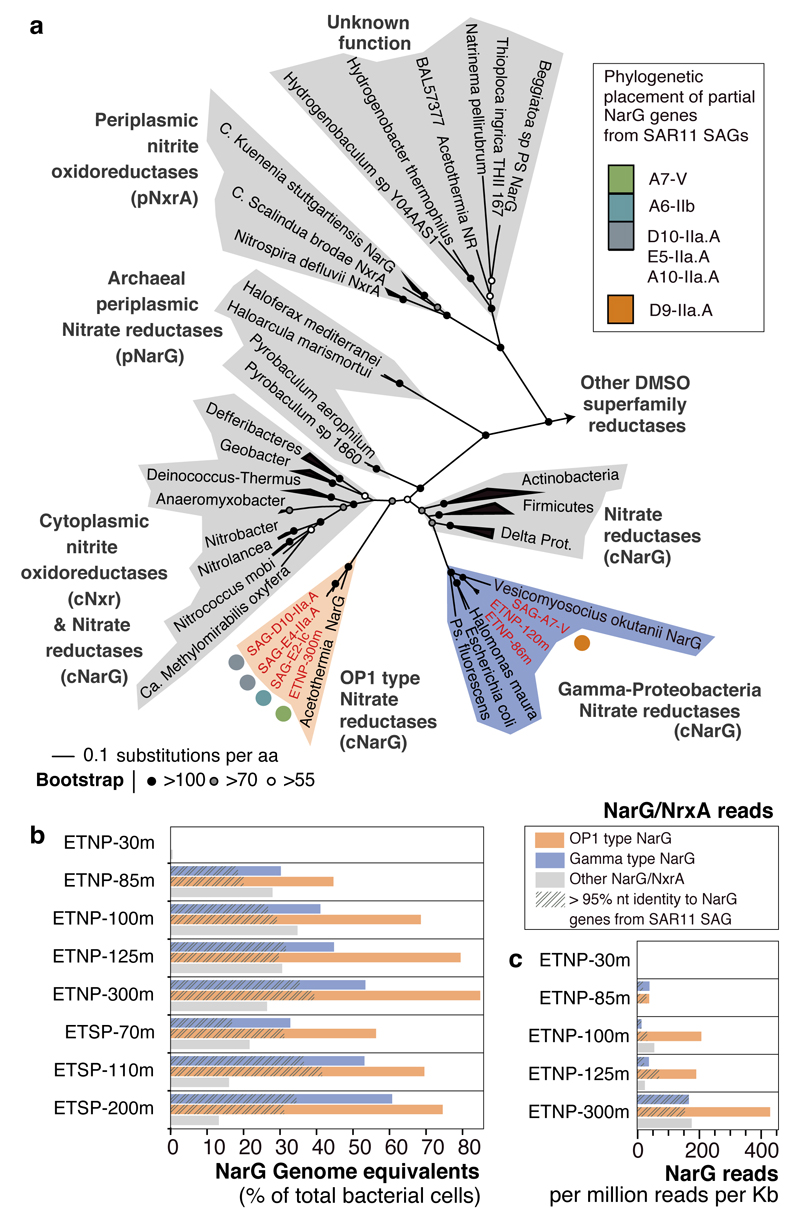
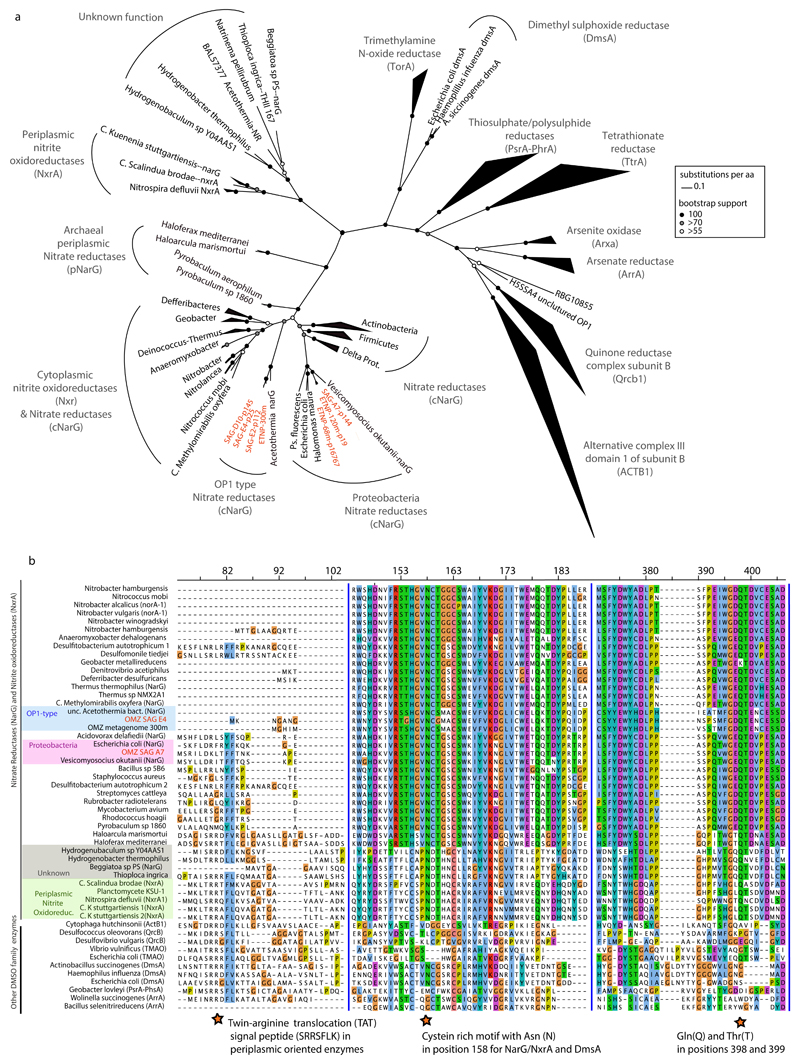
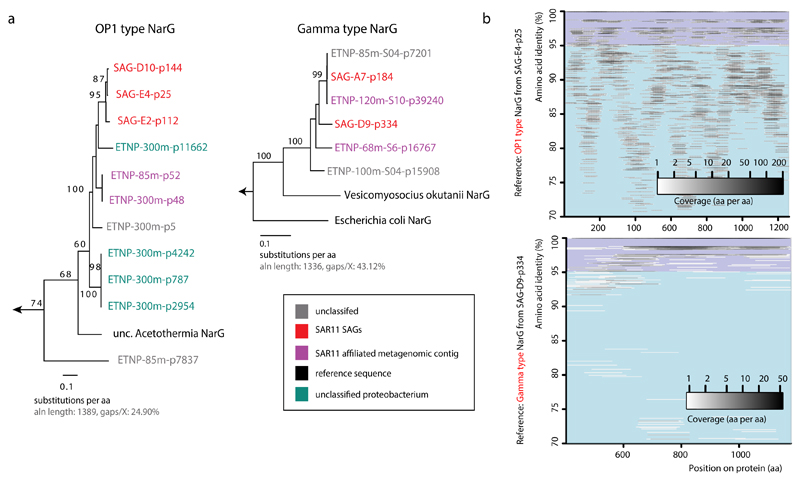
Phylogenetic placement of all identified narG and narH genes and partial fragments revealed two divergent nar variants in OMZ SAGs (Fig. 3a, Extended Data Fig. 3): (i) an “OP1-type” in which all 4 nar genes and an upstream cytochrome c protein were most similar (56-78% amino acid identity) to homologs from ‘Candidatus Acetothermus autotrophicum’ (Supplementary Table 5), a putative anaerobic acetogen of the candidate bacterial phylum OP135, and (ii) a “Gamma-type” variant most similar (51-78% identity) to Nar from a denitrifying Gammaproteobacteria endosymbiont (Ca. Vesicomyosocius okutanii strain HA)36. At least two of the OMZ SAR11 SAGs from subclade IIa.A, as well as SAG A7 from subclade V, encoded both OP1- and Gamma-type nar variants, suggesting that divergent nar copies (~42% AAI) co-occur in the same genome (Supplementary Discussion). Multiple nar operons per genome have been reported for diverse bacteria and are hypothesized to be related to adaptation to different oxygen conditions, with one variant constitutively expressed at low baseline levels37–39. For both OP1- and Gamma-type variants, the sequence divergence among recovered sequences was consistent with the phylogenetic placement of the SAGs. For example, OP1-type narG fragments represented 3 distinct 97% amino acid identity clusters (Fig. 2a). Sequences from clade IIa.A SAGs fell within the same cluster, sharing ~96.5% identity with sequences of the closely related Ic and IIb subclades, and ~90% with sequences from the more distant A7 SAG (Extended Data Figure 3). This pattern suggests diversification of nar operons in parallel with its genomic background, and also confirms that these sequences are not a systemic contaminant (Supplementary Discussion).
Figure 3. Diversity, abundance, and transcription of nitrate reductase enzymes in the OMZ.
a, Phylogenetic reconstruction of NarG sequences identified in the SAR11 SAGs and metagenomic SAR11 contigs (ETNP prefix), along with reference NO3- reductase and NO2- oxidoreductase enzymes. Partial gene sequences (represented with colored pies) were subsequently added to the pre-constructed tree with phylogenetic placement. b, Relative abundance of NarG/NxrA enzymes in OMZ metagenomic datasets. Abundance was normalized to the rpob gene abundance and thus represents genome equivalents, or the portion of OMZ bacterial cells that encode the enzyme. c, Relative expression of NarG/NxrA proteins in the ETNP transcriptomes.
Sequence-based and experimental characterization of SAR11 nitrate reductases
We sought to further characterize the biochemical function of SAR11 nar genes. Phylogenetic reconstruction based on 392 proteins of the diverse DMSO superfamily revealed that both OP1- and Gamma-type NarG fall within the clade of membrane-bound cytoplasm-oriented NO3-reductases (Nar) and NO2-oxidoreductases (Nxr), and were most closely related to Nar from known NO3-reducing bacteria (Fig. 3a)40. The lack of a TAT peptide motif at the N-terminus corroborated the probable cytoplasmic orientation of the NarG active site41, similar to experimentally verified Nar in Escherichia coli42. Additionally, the identified NarG sequences contain diagnostic functional domains found in NarG but not in other oxidoreductases of the DMSO reductase superfamily (Extended Data Fig. 4)40.
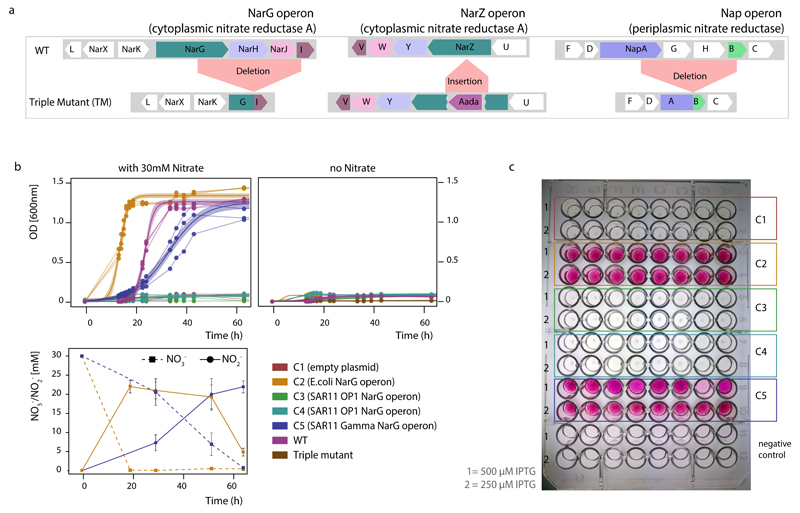
In order to verify NO3-reduction potential in SAR11 we introduced full-length SAR11 nar operons into a NO3-reductase-deficient Escherichia coli mutant and tested for enzyme activity. The Gamma-type nar operon was successfully expressed in E. coli, yielding Nar proteins of the predicted size range and enabling growth of the mutant under anoxic conditions in the presence of NO3-, coupled with simultaneous NO3-reduction to NO2-(Extended Data Figure 5), thereby providing direct evidence for the function of this enzyme in vivo. The OP1-type operon did not reverse the E. coli mutant phenotype, presumably due to the much greater divergence of this variant from the E. coli nar operon. Given the high similarity of Nar and Nxr protein sequences43–45, and the reversibility of the NO3-reduction reaction, it is possible that either or both OP1- and Gamma-type proteins could also function in situ to aerobically oxidize NO2-. While enticing, this possibility is remote given the experimental and phylogenetic evidence, a positive relationship between NO3-reduction rates and the abundance of OP1 and Gamma-type genes and transcripts in the anoxic OMZ depths (Fig. 1c), and prior results showing O2 sensitivity of OP1-type nar transcription25 (Supplementary Discussion). Rather, the results strongly suggest that the identified SAR11 narG genes encode functional NO3-reductases.
SAR11 nar genes and transcripts are abundant in anoxic OMZ waters
We next examined the abundance of SAR11-affiliated nar genes within the OMZ to evaluate the contribution of SAR11 cells to NO3-reduction. We first identified nar sequence reads in OMZ metagenomes using a similarity search-trained model that discriminates NO3- reductase (or NO2-oxidoreductase) reads from those of other genes of the DMSO superfamily (Supplementary Discussion). These narG reads were then classified within a reference phylogeny containing 320 NarG proteins, including OP1- and Gamma-type sequences. Remarkably, the majority of narG reads from OMZ metagenomes were classified as OP1- or Gamma-type NO3- reductases (Fig. 3b, Extended Data Fig. 6a), with the two variants accounting for 70% of total narG sequences at anoxic depths (Supplementary Table 4). Such high representation is consistent with qPCR-based counts of OP1- and Gamma-type narG copies at the collection site, where the two variants (summed) spiked at the OMZ NO2- maximum at >200,000 copies ml-1 (Extended Data Fig. 6b). The average number of nar genes per cell (i.e., genome equivalents) was estimated by comparing the abundance of nar sequences with those of rpoB, a universal single-copy gene. Based on those estimations, Gamma and OP1 nar variants occur in up to 61 and 85% of OMZ bacteria, respectively (Fig. 3b, Extended Data Fig. 6b), assuming each nar type occurs once per genome. Such high values are striking but consistent with prior results based on BLAST-based taxonomic assignments18. These values also exceed the estimated SAR11 abundances in the metagenomes, or those calculated directly from SAG 16S screening (up to 32% of the community), indicating that these gene variants occur in multiple copies per genome or in diverse bacteria (Supplementary Discussion). Metagenomic evidence suggests that the majority of these nar operons are found in SAR11 genomes within the OMZ. First, while our SAG collection captured only a fraction of total nar diversity, additional nar operons were identified in metagenomic contigs classified as SAR11 (Extended Data Figure 3, 7, Supplementary Table S6). Second, the majority of the metagenomic narG reads showed >95% nucleotide identity with the narG genes encoded by the SAGs, suggesting that SAR11 cells are among the major contributors of NO3- reductases in the OMZ (Fig. 2b).
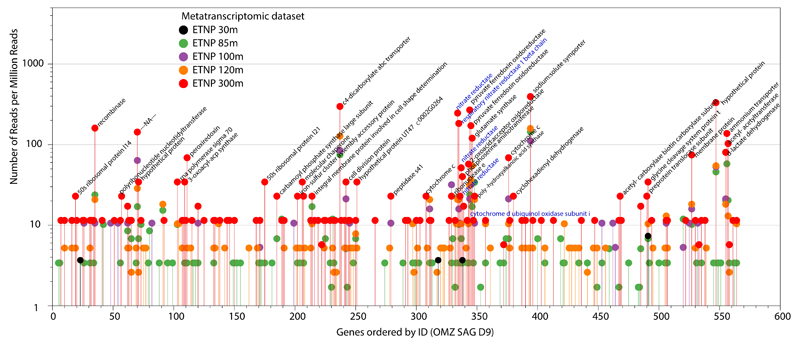
Metatranscriptome sequencing confirmed that SAR11-affiliated nar genes are transcribed in the OMZ. The abundance of both OP1- and Gamma-type variants in ETNP metatranscriptomes increased steadily from the lower oxycline (85 m) to the OMZ core (300 m), directly paralleling the abundance of the respective genes and the depth trend in NO3- reduction rates (Fig. 1c). Notably, within the ETNP OMZ, an average of 39% of all narG transcripts shared >95% nucleotide identity with the OP1- or Gamma-type sequences detected in SAR11 SAGs (Fig 3c), a conservative lower-bound estimate of the contribution of SAR11 bacteria to the total nar transcripts within the OMZ. Accordingly, within the anoxic OMZ depths, nar genes are among the most transcriptionally active genes in the SAG genomes (Extended Data Figure 8). The high transcriptional activity of SAR11 nar operons, interpreted alongside their distribution relative to NO3- reduction rates, suggests that SAR11 bacteria contribute substantially to community NO3- respiration.
Conclusions
Collectively, our findings identify diverse and abundant SAR11 lineages whose genome content and environmental distribution reflect adaptation to an anoxic niche, unlike all other SAR11 bacteria characterized to date. The experimentally verified NO3- reductase activity in the Gamma-type SAR11 nar variant, along with the high expression levels of divergent SAR11 nar genes in the functionally anoxic core of the OMZ, suggest that persistence in this niche is linked to NO3- respiration, consistent with the fundamental importance of this process in OMZs. Nitrate respiration in OMZs constitutes the primary mode for organic carbon mineralization and the main production route of NO2-, a critical substrate for the major nitrogen loss processes of anammox and denitrification. The presence and activity of nar operons in SAR11, as well as the high abundance of nar-associated SAR11 clades in the OMZ, implicate these versatile organisms as major contributors to the initiation of OMZ nitrogen loss. Together, these findings redefine the ecological niche of one of the planet's most dominant group of organisms, providing a set of genomic references to establish SAR11 as a model for studies of nitrogen and carbon cycling in OMZs.
Methods
Collection of ETNP and GoM samples for SAG analysis
Samples were collected from the ETNP OMZ during the Oxygen Minimum Zone Microbial Biogeochemistry Expedition (OMZoMBiE) cruise (R/V Horizon, 13-28 June, 2013). Seawater for single cell sorting and single amplified genome (SAG) analysis was collected from two depths within the OMZ (125 and 300 m) at Station 6 (18° 54.0N, 104° 54.0W) on June 19th (Fig. 1). Additional ("control") samples were collected from a depth profile (1-2107 m) of the Gulf of Mexico (GoM) on May 29, 2012 aboard the R/V Endeavor (cruise EN509) at station 5, with samples for SAG analysis preserved from the oxic surface (1 m). Collections were made using Niskin bottles on a rosette containing a Conductivity-Temperature-Depth profiler (Sea-Bird SBE 911plus). Water samples were prepared by cryopreservation according to the protocol recommended by the Bigelow Single Cell Genomics Center. Briefly, triplicate 1 ml samples of bulk seawater (no prefiltration) were gently mixed with 100 µl of a glycerol TE stock solution (20 ml 100X TE pH 8.0, 60 ml sterile water, 100 ml glycerol) and frozen at -80ºC.
ETNP OMZ rate measurements, and oxygen and nutrient analysis
Samples for oxygen and nutrient measurements were collected on the same date and casts as those for single cell sorting described above. Samples for rate measurements and metagenomics/transcriptomics (below) were collected a few hours later on the same day. Detailed collection and analysis procedures for those samples have been previously described19. Briefly, oxygen concentrations were determined using rosette-mounted sensors, including a SBE43 dissolved oxygen sensor for micromolar sensitivity and a high-resolution Switchable Trace amount OXygen (STOX) sensor for nanomolar level measurements46. CTD-based oxygen measurements (SBE43) from 3 casts spanning this sampling period revealed no significant movement in the oxycline, indicating stability in water column conditions.
Metagenome and metatranscriptome samples
Metadata, sequencing statistics, and accession numbers of all analyzed metagenome and metatranscriptome datasets are in Supplementary Table 4. Here, we summarize the OMZ and GoM datasets at the core of our analysis. ETNP OMZ metatranscriptomic and metagenomic datasets were generated via MiSeq Illumina sequencing in 19 and 47 respectively, for 5 depths at station 6: the upper oxycline (30 m), lower oxycline (85 m), secondary chlorophyll maximum (100 m), secondary nitrite maximum OMZ (125 m), and OMZ core (300 m) (Supplementary Table 4). Metagenome datasets from the ETSP were generated by Roche 454 pyrosequencing as previously described18 for 4 depths at an OMZ site (20o 05S, 70o 48W) off the coast of Iquique, Chile: the suboxic (<10 μM) upper OMZ just below the oxycline (70 m), the anoxic OMZ core (110 m, 200 m), and the oxic zone below the OMZ (1000 m). The ETNP and ETSP datasets analyzed here reflect the 0.2-1.6 µm biomass size fraction; this fraction was shown to contain the vast majority of bacterioplankton and SAR11 cells19. We also included two additional metagenomes, sampled on May 5, 2014 from the same site (station 6) in the ETNP, in order to obtain full-length nar operons for cloning purposes (see below). These metagenomes were obtained from depths of 68 m within the oxycline and 120 m within the OMZ. For the 9 GoM metagenomes released with this study, samples were collected from Niskin bottles (60 L per depth), and filtered onboard using the same filtration systems as for the ETNP and ETSP metagenomes (0.2-1.6 µm fraction). DNA was extracted with the same protocol as for the OMZ samples18 and libraries were prepared and sequenced in two lanes on an Illumina HiSeq (150 bp paired reads).
All metagenomic and metatranscriptomic datasets were quality trimmed as described below for the SAG datasets. The metatranscriptomic datasets were further filtered to remove rRNA transcripts using the SortMeRNA algorithm48. 454 metagenomic datasets were filtered to remove duplicate sequences. The quality trimmed reads from the OMZ metagenomes (ETNP and ETSP), were assembled with IDBA49 and genes were predicted on contigs longer than 500 bp with MetaGeneMark.hmm50. Taxonomic classification of metagenomic contigs was performed with MyTaxa51. Nar operons were identified on metagenomic contigs as described below for the SAG assemblies.
Single cell sorting, multiple displacement amplification, and SAG 16S rRNA gene screening
Single amplified genomes (SAGs) were generated from individual bacterial cells52, according to standard procedures in the Department of Energy Joint Genome Institute workflow53. Briefly, individual cells sorted on a BD Influx (BD Biosciences) were treated with Ready-Lyse lysozyme (Epicentre; 5U/µL final conc.) for 15 min at room temperature prior to the addition of lysis solution. Whole genome amplification was performed with the REPLI-g Single Cell Kit (Qiagen) in 2 μL reactions set up with an Echo acoustic liquid handler (Labcyte). Only the lysis and stop reagents from the REPLI-g kit received UV treatment since the amplification cocktail was pre-treated by the manufacturer. Amplification reactions were terminated after 6 hr. PCR amplification and Sanger sequencing of a ~470 bp region of the 16S rRNA gene (amplified using primers 926wF (5′-AAACTYAAAKGAATTGRCGG3′) and 1392R (5′-ACGGGCGGTGTGTRC3′) for archaea and bacteria was used to assign a preliminary taxonomic identification to each of the SAGs, via comparisons to the Greengenes rRNA database.
SAG sequencing
A total of 27 SAR11 classified SAGs identified were selected for sequencing, including 10 and 12 SAGs from 125 m and 300 m in the ETNP, respectively, and 5 "control" SAR11 SAGs from surface water (1 m) in the GoM. SAG DNA was prepared using the NexteraXT DNA Sample Prep kit (Illumina, San Diego, CA, USA) following the manufacturer’s instructions. Libraries were pooled and sequenced at Georgia Tech on two runs of an Illumina MiSeq using a 500 cycle (paired end 250 x 250 bp) kit. Of the initial 27 SAGs, 8 were recovered in very low abundance in the read data or were removed due to potential contamination (>5%) as estimated with CheckM (see below) or the presence of 18S rRNA gene fragments, yielding the final set of 19 SAGs analyzed here (Supplementary Table 1).
SAG sequence quality control assembly and functional gene annotation
Coupled reads were merged, when overlapping, using PEAR54. Both merged and un-merged reads were trimmed using SolexaQA++55 with a PHRED score cutoff of 20 and a minimum fragment length of 50 bp. Illumina adaptors were clipped using Scythe (https://github.com/vsbuffalo/scythe) and reads were re-filtered for length (50 bp). Quality-trimmed reads were assembled with SPAdes56. Percentage of contamination and genome completeness were assessed based on recovery of lineage-specific marker gene sets using CheckM29. From the total of 27 SAG assemblies, 7 were excluded from the analysis due to low coverage (i.e., less than 70 kb) or the presence of 18S rRNA sequences and BLASTP top matches to eukaryotic sequences reflecting contamination. For the remaining SAGs that passed the original quality control thresholds (Supplementary Table 1), when multiple fragments of a bacterial single-copy marker gene were identified, manual inspection of alignments revealed that multiplicity was due to assembly breaking points rather than contamination from divergent sequences, and such cases were retained for analysis (Supplementary Table 2). Evidence for contamination was detected in only one SAG, SAG A2 from the GoM, as multiplicity of divergent and nearly full-length marker genes. This SAG was excluded from further analysis.
For the final dataset of 19 SAGs, coding sequences were predicted on scaffolds longer than 500 bp with GeneMark.hmm50 and 16S rRNA gene sequences were identified using RNAmmer57. 16S rRNA sequences identified in the assemblies (4/4 GoM SAGs, and 8/15 OMZ SAGs) were compared to the 470 bp 16S fragment obtained during the initial SAG screening and confirmed to be identical. As an additional quality control step, all predicted genes from the 19 SAGs were taxonomically annotated using MyTaxa51 and the taxonomic distributions of adjacent genes in the concatenated assembly (10 gene windows) were inspected for possible contamination. As discussed in Supplementary Discussion, a contaminant genome in the assembled contigs can be visualized in the MyTaxa scan plots (Extended Data Fig. 1).
Predicted genes were functionally annotated using the blast2go pipeline58 for assignment to metabolic pathways, and screened manually for evidence of anaerobic energy metabolism. Detected genes of anaerobic metabolism, including nitrate reductase (nar) genes, as well as terminal oxidase genes and the single-copy marker gene rpoB, were further verified using HMMER3 (http://hmmer.janelia.org/) with default settings and recommended cutoffs for a match against available Pfam models59. Statistics of SAG quality control, assemblies, and contamination testing are in Supplementary Table 1 and 2.
Phylogenetic placement of SAGs
The evolutionary relatedness of SAR11 SAGs was assessed using the identified full or almost full-length 16S rRNA gene sequences from the assembled SAGs. For the SAGs from which no full-length 16S rRNA fragments were assembled, the shorter fragments obtained during screening were used in pairwise comparisons with full-length sequence references (Supplementary Table 3, 16S matrix). The 16S rRNA sequences from publicly available SAR11 genomes, as well as previously published 16S sequences6,13 from subclades with no genome representatives, were included in the alignment to aid in the classification of the SAR11 subclades. Additionally genome representatives of divergent alphaproteobacteria classes, as well as a beta- and gammaproteobacterium were included to facilitate the rooting of the tree. Maximum likelihood phylogenetic reconstruction was performed with RAxML with 1000 bootstraps and the GTR model for nucleotides60. Additionally, Hidden Markov Models (HMMs) of 106 housekeeping genes found in single copy in bacterial genomes were used to identify marker genes in available SAGs and reference genomes using HMMER3 (http://hmmer.janelia.org/) with default settings and the recommended cutoff28. The identified marker genes (Supplementary Table 1) were aligned using Clustal Omega61 and the protein alignments concatenated using Aln.cat.rb from the enve-omics collection (http://enve-omics.ce.gatech.edu/) to remove invariable sites and maintain protein coordinates. The concatenated alignment was used to build a maximum likelihood phylogeny with RAxML, using 1000 bootstraps, and the PROTGAMMAAUTO function, which identifies the best amino acid substitution model for each protein. SAGs where assigned to SAR11 subclades based on the consensus categorization of both 16S rRNA and marker gene phylogenies, in accordance with previously published subclade identification sequences6,13. OMZ-derived SAR11 SAGs from the SAR11 IIa lineage were further categorized as subclade IIa.A, to differentiate them from the currently available reference SAR11 IIa representative (HIMB058), classified here as subclade IIa.B. Average amino acid identities (AAI) were estimated as described previously62.
Nar functional gene validation and phylogeny
Reference nitrate reductase and nitrite oxidoreductase protein sequences (n=697) representing divergent bacterial and archaeal phyla were downloaded from UniProt/Swiss-Prot63, together with representatives of other DMSO family oxidoreductases (n=71), using as a guide the reference tree from 64. From this 697-sequence set, 321 full-length NarG/NxrA sequences were selected to represent all the clades, along with the 71 additional non-NarG/NxrA proteins. The NarG/NxrA subset included the closest relatives to the SAG OP1 and Gamma-type Nar variants, as determined by BLAST. All protein sequences (n=392), including the full-length NarG identified in the SAGs, were aligned with Clustal Omega, and a maximum likelihood phylogeny was reconstructed with RAxML with 1000 bootstraps and the PROTGAMMAAUTO model. Partial fragments of the NarG protein were then added to the alignment using MAFFT’s “addfragments”65, and the Evolutionary Placement Algorithm (EPA) implemented in RAxML was used to place them within the reference tree66. The same procedure was followed for the phylogenetic reconstruction and placement of identified NarH protein sequences.
Quantification of narG-encoding reads from the metagenomes and metatranscriptomes was done using BLAST searches against a manually curated NarG database and the software ROCker67. Using Receiver-Operator Curve (ROC) analysis, ROCker identifies the most discriminant BLAST bit-score per position in a reference alignment (NarG database) given a certain read length by simulating in silico metagenomic datasets that include the reference genes. This strategy permits the accurate estimation of abundance of target genes in short-read datasets, minimizing false negatives and positives derived from closely related proteins or conserved domains, a critical challenge in the detection of narG due to the ubiquity of other closely related DMSO oxidoreductases. The NarG database was manually curated and confirmed by the phylogenetic reconstruction of all available nitrate reductase and nitrite oxidoreductase sequences and visual inspection of the multi-sequence alignment for conservation of known functional domains and motifs. The final NarG database consisted of 697 nitrate reductases/nitrite oxidoreductases (positive set) and 71 representative non-NarG/NxrA DMSO family proteins (negative set for identification of false positive BLAST matches). All datasets, as well as the ROCker models built for narG quantifications in metagenomes with different read lengths, are available at http://enve-omics.ce.gatech.edu/rocker/. Additionally, the model for the identification of rpoB fragments in metagenomes was used to estimate coverage of rpoB in metagenomes.
The abundance of narG sequences in meta-omic datasets was estimated as genomic equivalents for each sample, by normalizing the coverage of narG for the gene length (reads per nucleotide of narG), and dividing the normalized value by the rpoB-normalized coverage (reads per nucleotide of rpoB) as shown in Supplementary Table 4. In order to quantify the abundance of the narG variants (OP1, Gamma-type), protein fragments were predicted in all identified (from ROCker) narG reads using FragGeneScan68 and placed in the reference DMSO tree using RAxML-EPA. The abundances of the OP1-type or Gamma-type variants were estimated based on the number of reads that were placed in the terminal or internal nodes of the aforementioned clades on the reference tree, using JPlace.to_iToL.rb from the enve-omics collection. The NarG metagenomic reads (predicted orfs) placed within those nodes, were used to construct the recruitment plots shown in Extended Data Fig. 8b. BLASTP was used to map the reads against the reference NarG sequences, and the recruitment plots were constructed with the BlastTab.catsbj.pl and BlastTab.recplot.R scripts from the enve-omics collection.
Thus, the reported abundances of OP1 and Gamma-type narG in metagenomes/metatranscriptomes are based on phylogenetic assignment of nar reads, rather than a strict sequence similarity cutoff. In order to estimate a lower limit for the abundance of NarG sequences presumably encoded by SAR11 genomes, the number of reads with more than 95% nucleotide identity to the reference NarG sequences found in the SAGs was estimated, and shown in Extended Data Fig. 6b and c. The figure shows abundance estimates for reads that are phylogenetically assigned to OP1 and Gamma nodes, with partitioning of the data into reads that share less than and greater than 95% nucleotide identity with the SAG OP1 and Gamma-type references.
NarG divergence in reference closed genomes
Identification of NarG in all closed genomes available from GOLD (27,461 bacterial and 685 archaeal genomes)70 was performed using HMMER3 with default settings. The results were further refined by a competitive BLAST search70 against the custom-made NarG reference database (used for ROCker), which included DMSO family oxidoreductase enzyme reference sequences. Matches with best hit against NarG sequences and a bit score higher than 900 were annotated as nitrate reductases or nitrite oxidoreductases. When found in multiple copies (up to 6), a reciprocal BLASTP search was performed to estimate sequence divergence, measured as amino acid identity.
Quantification of SAR11 clades in metagenomes and metatranscriptomes
For each metagenome/metatranscriptome, reads potentially derived from SAR11 genomes were identified by a competitive BLAST best-match approach. A custom database was built using all available closed genomes from NCBI-ftp (2638 bacterial, 165 archaeal) and 39 genome representatives of the SAR11 lineage, including 20 published isolate or SAG sequences and the 19 SAG sequences produced in this study (Supplementary Table 1). Metagenomic and metatranscriptomic reads (predicted orfs with FragGeneScan) were then compared against the database using BLASTP, and the subset of reads with a best match against any of the SAR11 genomes and an e-value < 0.001 was classified as “SAR11 reads” (Supplementary Table 4). To quantify the relative abundance of distinct SAR11 subclades, the “SAR11 reads” were further classified as follows. We used the coverage of universal marker genes that could be found in all the subclades to more accurately estimate the abundance of distinct subclades and overcome both the biased representation of SAR11 subclades in the available genomes, and the partial nature of SAG genomes. For all 39 available SAR11 genomes, 5707 orthologous genes (OGs) were identified by reciprocal best match and Markov Clustering with inflation 1.5 using ogs.mcl.rb from the enve-omics collection. From the identified OGs, 507 were represented at least once in each of the 8 subclades. All metagenomic and metatranscriptomic reads (SAR11 subsets) were mapped against the database containing all protein sequences from the 507 OGs (which were tagged according to subclade of origin) using the BLASTX option from Diamond71 and only the best matches for each read were kept. The coverage of each OG for each subclade was estimated based on that competitive best match result, normalized for the gene length (reads per bp of each OG), and the average coverage of all 507 OGs was used to estimate the abundance of subclades. Additionally, the number of rpoB reads for each metagenome was identified (for either the total dataset or the subset of the SAR11 reads), and the coverage of rpoB was used as a normalization factor to estimate the abundance of SAR11 subclades over the total bacterial community.
Functional characterization of SAR11 nar operons
A previously constructed NO3- reductase deficient Escherichia coli strain72 was used as the genetic system for heterologous expression of SAR11 nar genes. We used whole genome sequencing (Illumina MiSeq) to confirm that this strain lacked all three NO3- reductases (ΔnarGI ΔnapAB narZ::Ω; Extended Data Figure 5). The phenotype of this strain, hereafter referred to as the triple mutant, was verified by a lack of NO2- production and an absence of growth with NO3- under anaerobic conditions, compared to the wild type MC4100 E. coli strain (Extended Data Figure 5).
Complete sequences from one OP1-type, and one Gamma type nar operon, containing upstream and downstream sequences, were identified from the ETNP-300 m and ETNP-120 m metagenomes (see above). These sequences were confirmed to be identical to the operons in SAG A7 (which was lacking part of the N-terminus of the NarG gene; Extended Data Figure 3). Purified DNA from the ETNP-300 m and ETNP-120 m metagenomic samples was used as template for PCR amplification. In addition, we used genomic DNA from E. coli strain K12 MG1655 as a positive control. Because metagenomic samples are usually fragmented and the entire nar operon is 6.9 Kb, primers were designed to amplify the OP1-type, Gamma type and E. coli wild-type operon in two blocks. The first block spanned from the native NarG ribosome binding site to the end of the narG gene, and the second block included the end of the narG gene to the narI stop codon. The resulting PCR products were gel purified, assembled and cloned into pBbA1K, a low copy vector including the IPTG-inducible pTrc promoter 73 by In-Fusion cloning (Clontech, Mountain View, CA). The cloning reactions were transformed into TOP10 cells, and inserts were sequence-verified by Pacbio sequencing (Pacific Biosciences, Menlo Park, CA). The final nar sequences were identical (OP1 operon, and NarG,I proteins of Gamma operon) or nearly identical with silent substitutions (99% and 98% aa identity for the Gamma-type NarG and H proteins) compared to the sequences from SAG A7 (GenBank: KX275213, KX275214). Correct clones were isolated for each operon type, and purified plasmid was used to electroporate the triple mutant E. coli strains described above to generate recombinant strains expressing the heterologous nar operons for functional characterization.
For anaerobic cultures performing NO3- respiration, strains were first induced in LB medium with 0.5mM IPTG for 5h, and 20 μl of inoculum was subsequently introduced in gas tight tubes under N2 atmosphere. The medium was prepared as previously described74, composed from potassium phosphate buffer (100 mM, pH 7.4), 15 mM (NH4)2SO4, 9 mM NaCl, 2 mM MgSO4, 5 µM Na2MoO4, 10 µM Mohr's salt, 100 µM CaCl2, 0.5% casaminoacids and 0.01% thiamine. Glycerol (40 mM) was used as the sole carbon, and NO3- was added at 30 mM. IPTG (0.5 mM), kanamycin (30µg/ml) and streptomycin (30 µg/ml ) were used with the recombinant strains. Samples for NO3- and NO2- concentrations were obtained at regular time intervals during incubations, filtered through 0.2 µm porosity filters and injected into a Dionex DX ion chromatography unit with the Dionex IonPac AS14A analytical column75. Growth in incubations was assessed as optical density (OD; 600 nm). Growth curve data from replicated cultures (triplicate) were fitted to a logistic model with variables r (specific growth rate), P_0 (initial population), and K (carrying capacity), using nonlinear least-squares estimates and prediction of OD per time point with confidence intervals as implemented in enve.growthcurve from the enve-omics collection (http://enve-omics.ce.gatech.edu/).
Nitrate reductase activity was further verified in cell lysates from cells grown anaerobically for 12 days. Cells resuspended in 100 mM sodium phosphate buffer (pH 7.2) containing 0.02% Tween 80 were lysed by sonication in a Bioruptor UCD-200 (Diagenode). Protein concentration of the cell lysate was determined using a Qubit 2.0 fluorometer (Thermo Fisher Scientific) and 100 µg of protein was added to a reaction containing 100 mM NaNO3 and benzyl viologen as electron donor. The reaction was bubbled with N2 for 2 min before initiation with the addition of 50 µl of 30 mM sodium dithionite in 10 mM NaOH (final volume: 500 µl). Aliquots (50 µl) were removed at 20 min intervals and NO2- concentration determined colorimetrically after the addition of 50 μl Griess reagent (prepared with equal volumes of 0.1% N-1-napthylethylenediamine dihydrochloride in water and 1% sulfanilamide in 5% phosphoric acid). All assays were performed in triplicate. Finally, NO2- production from NO3 was further confirmed using whole cell assays with of 8 replicate clones (per recombinant strain) grown aerobically on 96-well plates in 70 μl Luria-Bertani (LB) broth supplemented with 30 mM NO3- and various IPTG concentrations. Nitrite production was identified via the Griess reaction as described above.
Quantitative PCR of 16S rRNA genes and SAR11 nar variants
Quantitative PCR (qPCR) was used to count OP1- and Gamma-type narG and total bacterial 16S rRNA gene copies. Seawater samples for qPCR were collected in 2014 from three sites in the ETNP, including station 6 from which the SAG samples were obtained.
Primers for narG PCR were designed based on alignments of narG sequences recovered from OMZ SAGs, targeting sites inclusive of all OMZ SAR11-affiliated nar variants and exclusive of narG from the closest database reference sequences. Primer selection resulted in the following: GammaF – 5’- GCG TAA AAT AAT TTC TTC TCC TAC ATG GA -3’ and GammaR – 5’- AGT TCA ATC CAG TCA TTA TCT TCT ACA TC -3’ amplifying a 401 nt fragment of the Gamma-type nar, and OP1F – 5’- ACC ATC AAG GAA TAA GAG AAT TAG G -3’ and OP1R – 5’- TGG ATT CCG TTT TCA CAA TAC ATT TC -3’ amplifying a 288 nt fragment of the OP1-type nar. PCR reactions were performed with DNA template from the OMZ 300m sample (station 6) and the oxic Gulf of Mexico as a negative control with the following conditions: incubation at 50°C for 2 min, 95 °C for 10 min, followed by 40 cycles of denaturation at 95°C (15 sec) and annealing at 53°C (for OP1) and 54°C (for Gamma) (1 min each). Amplicons with the expected length were observed only in the OMZ sample and were purified and concentrated using the QIAquick PCR purification kit (Qiagen). Clone libraries were prepared with the TOPO TA cloning kit (Life Technologies) following the manufacturer’s protocol, and plasmids from overnight grown selected colonies were isolated with the PureLink Quick Plasmid Miniprep Kit (Life Technologies). Inserts were purified using the QIAquick PCR Purification kit and sequenced on an Applied Biosystems 3730xl DNA Analyzer using BigDye Terminator v3.1 cycle Sanger sequencing (Life Technologies). Sequencing recovered 14 sequences generated using OP1 primers and 12 generated using Gamma primers. All OP1-like sequences were most closely related (via BLASTX against the NCBI-nr database) to narG of an uncultured Acetothermia bacterium OP1 (dbj|BAL57372.1|), whereas all Gamma-like sequences were most closely related to the gammaproteobacterial endosymbiont of Calyptogena okutanii (C. Vesicomyosocius okutanii; ref|WP_011930032.1|), consistent with the phylogenetic classification of the recovered SAG nar sequences as described in the main text and confirming the specificity of the primer sets. However, sequences within each clone set shared on average 96% (OP1 set) and 93% (Gamma set) nucleotide identity, raising the possibility that our primer sets may not amplify all OP1 and Gamma-type nar variants in the community. We therefore consider our abundance estimates to be lower bounds.
The OP1 and Gamma primer sets, along with universal bacterial 16S rRNA gene primers 1055f and 1392r, were used for SYBR® Green-based qPCR. Ten-fold serial dilutions of DNA from a plasmid carrying narG amplicons (described above) and a single copy of the 16S rRNA gene (from Dehalococcoides mccartyi) were included on each qPCR plate and used to generate standard curves, with a detection limit of ~30 and 10-15 gene copies ml-1 for 16S rRNA and narG variants, respectively. Assays were run on a 7500 Fast PCR System and a StepOnePlus ™ Real-Time PCR System (Applied Biosystems). All samples were run in triplicate with conditions as follows: 2 min incubation at 50°C, followed by 10 min at 95 °followed by 40 cycles of denaturation at 95°C (15 sec) and annealing at 60°C (1 min).
Data availability
SAR11 SAG sequences from the ETNP and GOM can be found under the NCBI BioProjects PRJNA290513 and PRJNA291283 respectively. The two OMZ metagenomes sequenced for this study can be found under the JGI Project IDs 1059848 and 1059863. Metagenomes from the Gulf of Mexico are available under the NCBI BioProject PRJNA291283. Sample accession numbers and further information on all metagenomics and SAG datasets used in this study are provided in Supplementary Tables 4 and 1 respectively. The mutant E.coli genome sequenced here can be found under the BioProject PRJNA322349. Sequences of the clones SAR11 Nar operons have been deposited in NCBI with Genebank accession numbers KX275213-KX275214.
Extended Data
Extended Data Figure 1. Evaluation of contamination based on MyTaxa taxonomic affiliations.
a, Representative MyTaxa plots to test for contamination based on taxonomic affiliations of predicted genes. The MyTaxa algorithm 51 predicts the taxonomic affiliation based on a weighted classification scheme that takes into account the phylogenetic signal of each protein family. Each gene is assigned to the deepest taxonomic resolution (out of phylum, genus, and species) for which a high confidence value can be obtained (score 0.5). Each MyTaxa scan represents taxonomic distributions of all the predicted genes for one genome, given in windows of 10 genes, and sorted based on their position in the concatenated assembly of the genome (when a partial genome is used). White space in the histograms represents genes that could not be assigned to a given taxon due to (a) lack of BLASTP hits against the reference database (a collection of closed and draft genomes) or (b) lack of high confidence scores. Notice that for the representative OMZ SAG E5, more than 80% of the genes can be classified as Candidatus Pelagibacter (SAR11), with an additional 10% assigned to Proteobacteria. Note there are no genome representatives for this taxon (i.e., SAR11 subclade IIa.A) in the database upon which MyTaxa is based. Similar results are obtained for the bathytype SAR11 SAG 6, as this genome also lacks representatives. The closed genome from a coastal isolate HTCC1002 is shown for comparison to demonstrate a typical pattern for cases when close relatives of the query genome are available in the reference database, as is the case for this isolate. b, Taxonomic classifications of genes from the 19 SAGs analyzed here. Each distribution was obtained from the MyTaxa scans performed for each SAG. The percentage of the total genes that could be taxonomically classified with MyTaxa was on average ~60%, and varied depending on the completeness of the genome (i.e., partial genes are less likely to be assigned taxonomy with high confidence). These values are also reported in Supplementary Table 1. Of the genes that could be classified, the majority (>90%) were classified to SAR11 taxa.
Extended Date Figure 2. Microdiversity within the SAR11 populations.
a, Recruitment plot of metagenomic reads from the ETNP OMZ 300 m sample, against scaffolds from SAG E4. Notice that the recruited reads vary in identities from 100 down to 85%, indicating the presence of closely affiliated clades, as well as extensive microdiversity within the same clade (i.e., reads sharing >95% identity) b, Phylogenetic reconstruction of reference RpoB protein sequences from SAR11 genomes, and placement of identified RpoB metagenomic sequences (denoted with the cross symbols). The alignment length was 1406 columns with 5.9% gaps or undetermined sites. The presence of multiple divergent rpoB reads within the same subclade (predominantly for subclades IIa.A and Ic) suggests high abundance but also extensive microdiversity within those populations (rather than clonal populations).
Extended Date Figure 3. nar genes encoded by SAR11 populations of OMZs.
a, nar operon and adjacent genes identified in SAR11 single amplified genomes (SAGs) from the ETNP OMZ, and in assemblies from the 85 m and 300 m ETNP OMZ metagenomes. narG sequences with at least 97% amino acid similarity are represented with the same color. b,c, Representative maximum likelihood phylogeny to show sequence variation among full or near full-length narG (b) and narH (c) amino acid sequences identified in the SAGs. A subset of cytoplasm-oriented nitrate reductases and nitrate oxidoreductases from publicly available genomes is also included. A comprehensive phylogeny showing the placement of SAR11 nar sequences relative to enzymes (n=392) of the DMSO family is in Fig. 2a. Colored pies represent the placement of shorter narG/narH gene fragments identified in the SAGs. Bootstrap values over 50 are shown. Outgroups (arrows) are Escherichia coli dmsA (b) and dmsB (c). Note: the Gamma-type nar-containing contig recovered in E4 (Fig. 2a) contains narHJI, but not narG; E4 Gamma-type is therefore not represented in Fig. 3b. All genes co-localized in the nar-containing contigs are listed in Supplementary Table 5. The p-numbers are gene IDs given by the gene prediction software, consistent with those in Supplementary Table 5.
Extended Data Figure 4. Identified NarG in SAR11 SAGs are members of the DMSO superfamily of oxidoreductases.
a, Phylogenetic reconstruction of NarG and DMSO enzymes. The tree shown in Figure 2 is presented here but has been expanded to include diverse DMSO oxidoreductases for direct comparison with the NarG/NxrA enzymes. Notice that both OP1 (green, blue, grey) and Gamma-type (red, orange) variants cluster within the cytoplasmically oriented nitrate reductases and nitrite oxidoreductases. 697 NarG/NxrA proteins were identified from UniRef 63, and from those 321 full length sequences were selected to represent all the diverse clades. An additional 71 non-NarG/NxrA proteins, representative of the diverse enzymes of the DMSO superfamily were also included in the collection. The full-length amino acid sequences were aligned with Clustal Omega 61 and the phylogenetic tree was constructed by maximum likelihood and 1000 bootstraps using RAxML 60. The alignment length was 1803 columns, out of which 31.2% were gaps or undetermined. Partial NarG sequences identified in the SAGs were placed on the tree using the epa algorithm from RAxML 66. The same collection of proteins was used to train the Rocker models and quantify the narG metagenomic fragments, and can be found in the enve-omics website (http://enve-omics.ce.gatech.edu/rocker/models). b, Alignment of NarG sequences from OMZ SAR11 with representative sequences from the DMSO superfamily of oxidoreductases. The protein motifs in the second and third panels are present in all functional nitrate reductases (NarG) and nitrite oxidoreductases (NxrA) but not in closely related enzymes of the DMSO superfamily. The first panel shows the presence/absence of the TAT signal peptide (SRRSFLK), whose presence typically denotes a protein excreted to the outer membrane 40,41. SAR11 NarG is instead oriented toward the cytoplasm (lack of TAT). The second panel shows the cysteine-rich motif typically found in the N-terminus of the type-II DMSO superfamily oxidoreductases 76 and believed to enable the formation of a [4Fe–4S] cluster in these proteins 77. The Asn (N) in position 158 of the alignment is typically found in catalytic subunits of nitrite reductases and DMSO oxidoreductases (DmsA) but not in other DMSO family enzymes. The third panel shows the Gln(Q) and Thr(T) in positions 398 and 399 within the putative substrate entry channel of the protein, which differentiate the Nar proteins from all other oxidoreductases of the DMSO family 40.
Extended Data Figure 5. Functional characterization of the SAR11 nar operons in the Escherichia coli heterologous expression system.
a, Genotype of the E. coli triple mutant confirmed by whole genome sequencing. The triple mutant lacks complete functional operons of all three NO3- reductases, and thus is incapable of NO3- reduction. b, Anaerobic growth of triple mutant clones, complemented with the SAR11 nar operons. For each strain three independent clones were monitored, and data from the replicate growth curves were fitted into a logistic model. Shaded areas represent the 95% confidence intervals of optical density readings (OD600nm) in the fitted logistic growth models. NO3- and NO2- were measured in parallel with ion chromatography. Note that the Gamma-type SAR11 operon complements the triple mutant phenotype, growing anaerobically by reducing NO3- to NO2-. E. coli encodes functional nitrite reductases, thus the accumulated NO2- can be further reduced to ammonia, accounting for the non-stoichiometric NO2- production. c, Whole cell NO2- production assays under aerobic conditions. Eight independent clones (columns A-H) of each type (C1-C5) were inoculated in LB supplemented with 30mM NO3- and different IPTG concentrations, and the well plate was incubated for 2 days at room temperature. Griess reagent was added, and development of pink color indicated NO2- production.
Extended Data Figure 6. Relative abundance of narG variants in ETNP OMZ metagenomes and metatranscriptomes and various other ocean metagenomes.
a, Relative abundance and diversity of NarG/NxrA enzymes as revealed by phylogenetic placement of identified narG metagenomic reads (colored pies). All identified short metagenomic narG reads from various oceanic metagenomes were placed within a reconstructed reference NarG tree in order to estimate the abundance of the different narG variants. The results of the placement are presented in 5 separate trees, based on the origin of the analyzed metagenomic reads (ETSP metagenomes, ETNP metagenomes and metatranscriptomes, oxic bathypelagic and oxic surface metagenomes) for clarity. In each of the 5 trees, the colored pies represent the abundance (normalized for dataset size) of the short metagenomic reads clustering in the respective node. Specifically, the pie radius reflects read abundance as a percentage of the total narG genome equivalents identified (i.e., number of narG reads compared to number of rpoB reads, normalized for gene length and total number of reads in each metagenome), with the size of grey pies in the legends representing the highest and lowest relative abundance, respectively. The reference tree is the same as in Figure 3a. Scale bars represent substitutions per amino acid. Notice that the two narG variants affiliated with the SAR11 SAGs (highlighted in orange for the OP1 type and blue for the Gamma type) are only abundant in the metagenomes and metatranscriptomes from the OMZ, where they comprise more than 70% of the total narG read pool, as can also be observed in Figure 3b and c. The number of narG reads of the OP1 or Gamma type are also given in Supplementary Table 1. b, qPCR-based abundance of SAR11 affiliated narG genes in the ETNP OMZ relative to nitrite, nitrate, and oxygen concentrations OMZ and qPCR-based counts of 16S rRNA. Counts of total bacterial 16S rRNA, OP1-type narG, and Gamma-type narG genes at three stations (map on legend) west of Manzanillo, Mexico in May 2014. Map was created with Ocean Data View (Schlitzer, R., odv.awi.de, 2015). All assays were performed in triplicates, and the bars represent standard errors. Note: counts of OP1 and Gamma-type narG variants are likely underestimates given the observed microdiversity in the community (Extended Data Fig. 2 and 7), and therefore the possibility that our primers did not match all OP1 and Gamma-type variants.
Extended Data Figure 7. Diversity of OP1 and Gamma-type narG amino acid sequences in the ETNP OMZ metagenome.
a, Phylogenies showing all full-length narG sequences recovered in the ETNP OMZ metagenomes (85, 100, 125, 300 m), as well as those from the SAR11 SAGs and corresponding narG reference sequences, with the left tree showing OP1-type variants and the right tree showing Gamma-type variants. NarG sequences are color-coded based on the taxonomic classification of adjacent genes in the same metagenomic scaffolds, as show in Supplementary Table 6. b, Recruitment of metagenomic reads (predicted open reading frames) from the OMZ 300 m sample, against OP1 (left) or Gamma (right) type narG sequences from the SAR11 SAGs. The metagenomic reads used for recruitment were identified as “narG” using the ROCker pipeline, and their identity further confirmed by phylogenetic placement within the narG clade on a reference DMSO superfamily protein tree, in order to minimize non-specific recruitments in conserved protein regions. Notice that based on this analysis, the OP1 type narG variants are highly diverse in the OMZ metagenome.
Extended Data Figure 8. Transcriptional profile of predicted genes from the SAR11 OMZ SAG-D9.
Transcriptomic reads with >99% identity matches were counted for each gene, and the counts were normalized for the dataset size. Note that the nar operon genes are among the most actively transcribed in the ETNP 300m OMZ sample.
Supplementary Material
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Acknowledgments
This work was supported by the National Science Foundation (1151698 to FJS and 1416673 to KTK), the NASA Exobiology Program (NNX14AJ87G to JBG and FJS), the Sloan Foundation (RC944 to FJS), a Community Science Program grant from the Department of Energy (to FJS and KTK). The work conducted by the U.S. Department of Energy Joint Genome Institute, a DOE Office of Science User Facility, is supported under Contract No. DE-AC02-05CH11231. LAB, ML and BT were supported by a European Research Council Advanced Grant (OXYGEN, 267233) and by the Danish National Research Foundation (DNRF53). D.T acknowledges the support of Onassis Foundation Fellowship. We are grateful for the generosity of Dr. Jim Cole, Dr. Alex Magalon, Dr. Chuck Sohaskey, and Dr. Frank Sargent for providing nitrate reductase deficient E. coli strains, Dr. Spyros Pavlostathis for the ion chromatography methods and Dr. Jim Spain for his suggestions on the heterologous expression experiment. The authors declare no conflicts of interest in relation to this work.
Footnotes
Author Contributions: D.T. conducted bioinformatics analyses. J.W., P.R., and N.S. conducted next-generation sequencing. C.C.P. and B.K.S. conducted qPCR analyses. L.M.R. developed additional bioinformatic methods for SAG contamination evaluation. R.M. and T.W. conducted cell sorting and SAG generation. L.A.B. and B.T. conducted process rate measurements. M.L. conducted STOX oxygen measurements. D.T., S.D., S.N., and A.S.B. conducted the heterologous expression experiments. F.J.S. and K.T.K designed the study. F.J.S. and D.T. analyzed the data and wrote the paper. All authors discussed the results and helped edit the manuscript.
Author Information: All data generated and analyzed in this paper are publically available. Accession numbers of all datasets are listed in Supplementary Tables 1 and 5. Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests in association with this study.
References
- 1.Brown MV, Schwalbach MS, Hewson I, Fuhrman JA. Coupling 16S-ITS rDNA clone libraries and automated ribosomal intergenic spacer analysis to show marine microbial diversity: development and application to a time series. Environ Microbiol. 2005;7:1466–1479. doi: 10.1111/j.1462-2920.2005.00835.x. [DOI] [PubMed] [Google Scholar]
- 2.Carlson CA, et al. Seasonal dynamics of SAR11 populations in the euphotic and mesopelagic zones of the northwestern Sargasso Sea. ISME J. 2008;3:283–295. doi: 10.1038/ismej.2008.117. [DOI] [PubMed] [Google Scholar]
- 3.Eiler A, Hayakawa DH, Church MJ, Karl DM, Rappé MS. Dynamics of the SAR11 bacterioplankton lineage in relation to environmental conditions in the oligotrophic North Pacific subtropical gyre. Environ Microbiol. 2009;11:2291–2300. doi: 10.1111/j.1462-2920.2009.01954.x. [DOI] [PubMed] [Google Scholar]
- 4.Morris RM, et al. SAR11 clade dominates ocean surface bacterioplankton communities. Nature. 2002;420:806–810. doi: 10.1038/nature01240. [DOI] [PubMed] [Google Scholar]
- 5.Salter I, et al. Seasonal dynamics of active SAR11 ecotypes in the oligotrophic Northwest Mediterranean Sea. ISME J. 2015;9:347–360. doi: 10.1038/ismej.2014.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thrash JC, et al. Single-cell enabled comparative genomics of a deep ocean SAR11 bathytype. ISME J. 2014;8:1440–1451. doi: 10.1038/ismej.2013.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giovannoni SJ, et al. Genome streamlining in a cosmopolitan oceanic bacterium. Science. 2005;309:1242–1245. doi: 10.1126/science.1114057. [DOI] [PubMed] [Google Scholar]
- 8.Grote J, et al. Streamlining and Core Genome Conservation among Highly Divergent Members of the SAR11 Clade. mBio. 2012;3:e00252–12. doi: 10.1128/mBio.00252-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tripp HJ. The unique metabolism of SAR11 aquatic bacteria. J Microbiol Seoul Korea. 2013;51:147–153. doi: 10.1007/s12275-013-2671-2. [DOI] [PubMed] [Google Scholar]
- 10.Konstantinidis KT, Braff J, Karl DM, DeLong EF. Comparative Metagenomic Analysis of a Microbial Community Residing at a Depth of 4,000 Meters at Station ALOHA in the North Pacific Subtropical Gyre. Appl Environ Microbiol. 2009;75:5345–5355. doi: 10.1128/AEM.00473-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Swan BK, et al. Potential for chemolithoautotrophy among ubiquitous bacteria lineages in the dark ocean. Science. 2011;333:1296–1300. doi: 10.1126/science.1203690. [DOI] [PubMed] [Google Scholar]
- 12.King GM, Smith CB, Tolar B, Hollibaugh JT. Analysis of composition and structure of coastal to mesopelagic bacterioplankton communities in the northern gulf of Mexico. Front Microbiol. 2012;3:438. doi: 10.3389/fmicb.2012.00438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vergin KL, et al. High-resolution SAR11 ecotype dynamics at the Bermuda Atlantic Time-series Study site by phylogenetic placement of pyrosequences. ISME J. 2013;7:1322–1332. doi: 10.1038/ismej.2013.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paulmier A, Ruiz-Pino D. Oxygen minimum zones (OMZs) in the modern ocean. Prog Oceanogr. 2009;80:113–218. [Google Scholar]
- 15.Tiano L, Garcia-Robledo E, Revsbech NP. A New Highly Sensitive Method to Assess Respiration Rates and Kinetics of Natural Planktonic Communities by Use of the Switchable Trace Oxygen Sensor and Reduced Oxygen Concentrations. PLoS ONE. 2014;9:e105399. doi: 10.1371/journal.pone.0105399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kalvelage T, et al. Nitrogen cycling driven by organic matter export in the South Pacific oxygen minimum zone. Nat Geosci. 2013;6:228–234. [Google Scholar]
- 17.Stewart FJ, Ulloa O, DeLong EF. Microbial metatranscriptomics in a permanent marine oxygen minimum zone. Environ Microbiol. 2012;14:23–40. doi: 10.1111/j.1462-2920.2010.02400.x. [DOI] [PubMed] [Google Scholar]
- 18.Ganesh S, Parris DJ, DeLong EF, Stewart FJ. Metagenomic analysis of size-fractionated picoplankton in a marine oxygen minimum zone. ISME J. 2014;8:187–211. doi: 10.1038/ismej.2013.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ganesh S, et al. Size-fraction partitioning of community gene transcription and nitrogen metabolism in a marine oxygen minimum zone. ISME J. 2015;9:2682–2696. doi: 10.1038/ismej.2015.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ulloa O, Canfield DE, DeLong EF, Letelier RM, Stewart FJ. Microbial oceanography of anoxic oxygen minimum zones. Proc Natl Acad Sci. 2012;109:15996–16003. doi: 10.1073/pnas.1205009109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Codispoti LA, et al. The oceanic fixed nitrogen and nitrous oxide budgets: Moving targets as we enter the anthropocene? Sci Mar. 2001;65:85–105. [Google Scholar]
- 22.Gruber N. In: The Ocean Carbon Cycle and Climate. Follows M, Oguz T, editors. Springer; Netherlands: 2004. pp. 97–148. [Google Scholar]
- 23.Stewart FJ, Sharma AK, Bryant JA, Eppley JM, DeLong EF. Community transcriptomics reveals universal patterns of protein sequence conservation in natural microbial communities. Genome Biol. 2011;12:R26. doi: 10.1186/gb-2011-12-3-r26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lüke C, Speth DR, Kox MAR, Villanueva L, Jetten MSM. Metagenomic analysis of nitrogen and methane cycling in the Arabian Sea oxygen minimum zone. PeerJ. 2016;4:e1924. doi: 10.7717/peerj.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dalsgaard T, et al. Oxygen at Nanomolar Levels Reversibly Suppresses Process Rates and Gene Expression in Anammox and Denitrification in the Oxygen Minimum Zone off Northern Chile. mBio. 2014;5:e01966–14. doi: 10.1128/mBio.01966-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kalvelage T, et al. Oxygen Sensitivity of Anammox and Coupled N-Cycle Processes in Oxygen Minimum Zones. PLoS ONE. 2011;6:e29299. doi: 10.1371/journal.pone.0029299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dean FB, et al. Comprehensive human genome amplification using multiple displacement amplification. Proc Natl Acad Sci USA. 2002;99:5261–5266. doi: 10.1073/pnas.082089499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dupont CL, et al. Genomic insights to SAR86, an abundant and uncultivated marine bacterial lineage. ISME J. 2012;6:1186–1199. doi: 10.1038/ismej.2011.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thrash JC, et al. Phylogenomic evidence for a common ancestor of mitochondria and the SAR11 clade. Sci Rep. 2011;1 doi: 10.1038/srep00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luo H. Evolutionary origin of a streamlined marine bacterioplankton lineage. ISME J. 2015;9:1423–1433. doi: 10.1038/ismej.2014.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodríguez-Ezpeleta N, Embley TM. The SAR11 Group of Alpha-Proteobacteria Is Not Related to the Origin of Mitochondria. PLoS ONE. 2012;7:e30520. doi: 10.1371/journal.pone.0030520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Viklund J, Martijn J, Ettema TJG, Andersson SGE. Comparative and Phylogenomic Evidence That the Alphaproteobacterium HIMB59 Is Not a Member of the Oceanic SAR11 Clade. PLoS ONE. 2013;8:e78858. doi: 10.1371/journal.pone.0078858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Konstantinidis KT, DeLong EF. Genomic patterns of recombination, clonal divergence and environment in marine microbial populations. ISME J. 2008;2:1052–1065. doi: 10.1038/ismej.2008.62. [DOI] [PubMed] [Google Scholar]
- 35.Takami H, et al. A deeply branching thermophilic bacterium with an ancient acetyl-CoA pathway dominates a subsurface ecosystem. PloS One. 2012;7:e30559. doi: 10.1371/journal.pone.0030559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuwahara H, et al. Reduced genome of the thioautotrophic intracellular symbiont in a deep-sea clam, Calyptogena okutanii. Curr Biol CB. 2007;17:881–886. doi: 10.1016/j.cub.2007.04.039. [DOI] [PubMed] [Google Scholar]
- 37.Iobbi C, Santini C-L, Bonnefoy V, Giordano G. Biochemical and immunological evidence for a second nitrate reductase in Escherichia coli K12. Eur J Biochem. 1987;168:451–459. doi: 10.1111/j.1432-1033.1987.tb13438.x. [DOI] [PubMed] [Google Scholar]
- 38.Iobbi-Nivol C, Santini CL, Blasco F, Giordano G. Purification and further characterization of the second nitrate reductase of Escherichia coli K12. Eur J Biochem FEBS. 1990;188:679–687. doi: 10.1111/j.1432-1033.1990.tb15450.x. [DOI] [PubMed] [Google Scholar]
- 39.Philippot L. Denitrifying genes in bacterial and Archaeal genomes. Biochim Biophys Acta. 2002;1577:355–376. doi: 10.1016/s0167-4781(02)00420-7. [DOI] [PubMed] [Google Scholar]
- 40.Martinez-Espinosa RM, et al. Look on the positive side! The orientation, identification and bioenergetics of ‘Archaeal’ membrane-bound nitrate reductases. FEMS Microbiol Lett. 2007;276:129–139. doi: 10.1111/j.1574-6968.2007.00887.x. [DOI] [PubMed] [Google Scholar]
- 41.Rothery RA, Workun GJ, Weiner JH. The prokaryotic complex iron–sulfur molybdoenzyme family. Biochim Biophys Acta BBA - Biomembr. 2008;1778:1897–1929. doi: 10.1016/j.bbamem.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 42.Yoshimatsu K, Iwasaki T, Fujiwara T. Sequence and electron paramagnetic resonance analyses of nitrate reductase NarGH from a denitrifying halophilic euryarchaeote Haloarcula marismortui. FEBS Lett. 2002;516:145–150. doi: 10.1016/s0014-5793(02)02524-3. [DOI] [PubMed] [Google Scholar]
- 43.Lücker S, et al. A Nitrospira metagenome illuminates the physiology and evolution of globally important nitrite-oxidizing bacteria. Proc Natl Acad Sci U S A. 2010;107:13479–13484. doi: 10.1073/pnas.1003860107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Starkenburg SR, et al. Genome sequence of the chemolithoautotrophic nitrite-oxidizing bacterium Nitrobacter winogradskyi Nb-255. Appl Environ Microbiol. 2006;72:2050–2063. doi: 10.1128/AEM.72.3.2050-2063.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sorokin DY, et al. Nitrification expanded: discovery, physiology and genomics of a nitrite-oxidizing bacterium from the phylum Chloroflexi. ISME J. 2012;6:2245–2256. doi: 10.1038/ismej.2012.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Revsbech NP, et al. Determination of ultra-low oxygen concentrations in oxygen minimum zones by the STOX sensor: STOX oxygen sensor. Limnol Oceanogr Methods. 2009;7:371–381. [Google Scholar]
- 47.Glass JB, et al. Meta-omic signatures of microbial metal and nitrogen cycling in marine oxygen minimum zones. Front Microbiol. 2015;6:998. doi: 10.3389/fmicb.2015.00998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kopylova E, Noé L, Touzet H. SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinforma Oxf Engl. 2012;28:3211–3217. doi: 10.1093/bioinformatics/bts611. [DOI] [PubMed] [Google Scholar]
- 49.Peng Y, Leung HCM, Yiu SM, Chin FYL. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinforma Oxf Engl. 2012;28:1420–1428. doi: 10.1093/bioinformatics/bts174. [DOI] [PubMed] [Google Scholar]
- 50.Zhu W, Lomsadze A, Borodovsky M. Ab initio gene identification in metagenomic sequences. Nucleic Acids Res. 2010;38:e132. doi: 10.1093/nar/gkq275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Luo C, Rodriguez-R LM, Konstantinidis KT. MyTaxa: an advanced taxonomic classifier for genomic and metagenomic sequences. Nucleic Acids Res. 2014;42:e73. doi: 10.1093/nar/gku169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Raghunathan A, et al. Genomic DNA amplification from a single bacterium. Appl Environ Microbiol. 2005;71:3342–3347. doi: 10.1128/AEM.71.6.3342-3347.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rinke C, et al. Insights into the phylogeny and coding potential of microbial dark matter. Nature. 2013;499:431–437. doi: 10.1038/nature12352. [DOI] [PubMed] [Google Scholar]
- 54.Zhang J, Kobert K, Flouri T, Stamatakis A. PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinforma Oxf Engl. 2014;30:614–620. doi: 10.1093/bioinformatics/btt593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cox MP, Peterson DA, Biggs PJ. SolexaQA: At-a-glance quality assessment of Illumina second-generation sequencing data. BMC Bioinformatics. 2010;11:485. doi: 10.1186/1471-2105-11-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bankevich A, et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lagesen K, et al. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35:3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Conesa A, et al. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 59.Finn RD, et al. Pfam: the protein families database. Nucleic Acids Res. 2014;42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinforma Oxf Engl. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 61.Sievers F, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7 doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Konstantinidis KT, Tiedje JM. Prokaryotic taxonomy and phylogeny in the genomic era: advancements and challenges ahead. Curr Opin Microbiol. 2007;10:504–509. doi: 10.1016/j.mib.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 63.Suzek BE, Wang Y, Huang H, McGarvey PB, Wu CH. UniRef clusters: a comprehensive and scalable alternative for improving sequence similarity searches. Bioinformatics. 2015;31:926–932. doi: 10.1093/bioinformatics/btu739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Castelle CJ, et al. Extraordinary phylogenetic diversity and metabolic versatility in aquifer sediment. Nat Commun. 2013;4 doi: 10.1038/ncomms3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Katoh K, Standley DM. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Berger SA, Krompass D, Stamatakis A. Performance, Accuracy, and Web Server for Evolutionary Placement of Short Sequence Reads under Maximum Likelihood. Syst Biol. 2011;60:291–302. doi: 10.1093/sysbio/syr010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Orellana LH, Rodriguez-R LM, Konstantinidis KT. ROCker: a pipeline for accurate detection and quantification of target genes in short-read metagenomic datasets. Nucleic Acids Res. doi: 10.1093/nar/gkw900. under review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rho M, Tang H, Ye Y. FragGeneScan: predicting genes in short and error-prone reads. Nucleic Acids Res. 2010;38:e191. doi: 10.1093/nar/gkq747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Reddy TBK, et al. The Genomes OnLine Database (GOLD) v.5: a metadata management system based on a four level (meta)genome project classification. Nucleic Acids Res. 2015;43:D1099–1106. doi: 10.1093/nar/gku950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 71.Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2015;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 72.Potter LC, Millington P, Griffiths L, Thomas GH, Cole JA. Competition between Escherichia coli strains expressing either a periplasmic or a membrane-bound nitrate reductase: does Nap confer a selective advantage during nitrate-limited growth? Biochem J. 1999;344:77–84. [PMC free article] [PubMed] [Google Scholar]
- 73.Khlebnikov A, Keasling JD. Effect of lacYExpression on Homogeneity of Induction from the Ptac and Ptrc Promoters by Natural and Synthetic Inducers. Biotechnol Prog. 2002;18:672–674. doi: 10.1021/bp010141k. [DOI] [PubMed] [Google Scholar]
- 74.Alberge F, et al. Dynamic subcellular localization of a respiratory complex controls bacterial respiration. eLife. 2015;4:e05357. doi: 10.7554/eLife.05357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hajaya MG, Pavlostathis SG. Fate and effect of benzalkonium chlorides in a continuous-flow biological nitrogen removal system treating poultry processing wastewater. Bioresour Technol. 2012;118:73–81. doi: 10.1016/j.biortech.2012.05.050. [DOI] [PubMed] [Google Scholar]
- 76.Bender KS, et al. Identification, characterization, and classification of genes encoding perchlorate reductase. J Bacteriol. 2005;187:5090–5096. doi: 10.1128/JB.187.15.5090-5096.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jormakka M, Richardson D, Byrne B, Iwata S. Architecture of NarGH Reveals a Structural Classification of Mo-bisMGD Enzymes. Structure. 2004;12:95–104. doi: 10.1016/j.str.2003.11.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
SAR11 SAG sequences from the ETNP and GOM can be found under the NCBI BioProjects PRJNA290513 and PRJNA291283 respectively. The two OMZ metagenomes sequenced for this study can be found under the JGI Project IDs 1059848 and 1059863. Metagenomes from the Gulf of Mexico are available under the NCBI BioProject PRJNA291283. Sample accession numbers and further information on all metagenomics and SAG datasets used in this study are provided in Supplementary Tables 4 and 1 respectively. The mutant E.coli genome sequenced here can be found under the BioProject PRJNA322349. Sequences of the clones SAR11 Nar operons have been deposited in NCBI with Genebank accession numbers KX275213-KX275214.