Abstract
Francisella tularensis is composed of a number of subspecies with varied geographic distribution, host ranges, and virulence. In view of these marked differences, comparative functional genomics may elucidate some of the molecular mechanism(s) behind these differences. In this study a shared probe microarray was designed that could be used to compare the transcriptomes of Francisella tularensis subsp. tularensis Schu S4 (Ftt), Francisella tularensis subsp. holarctica OR960246 (Fth), Francisella tularensis subsp. holarctica LVS (LVS), and Francisella novicida U112 (Fn). To gain insight into expression differences that may be related to the differences in virulence of these subspecies, transcriptomes were measured from each strain grown in vitro under identical conditions, utilizing a shared probe microarray. The human avirulent Fn strain exhibited high levels of transcription of genes involved in general metabolism, which are pseudogenes in the human virulent Ftt and Fth strains, consistent with the process of genome decay in the virulent strains. Genes encoding an efflux system (emrA2 cluster of genes), siderophore (fsl operon), acid phosphatase, LPS synthesis, polyamine synthesis, and citrulline ureidase were all highly expressed in Ftt when compared to Fn, suggesting that some of these may contribute to the relative high virulence of Ftt. Genes expressed at a higher level in Ftt when compared to the relatively less virulent Fth included genes encoding isochorismatases, cholylglycine hydrolase, polyamine synthesis, citrulline ureidase, Type IV pilus subunit, and the Francisella Pathogenicity Island protein PdpD. Fth and LVS had very few expression differences, consistent with the derivation of LVS from Fth. This study demonstrated that a shared probe microarray designed to detect transcripts in multiple species/subspecies of Francisella enabled comparative transcriptional analyses that may highlight critical differences that underlie the relative pathogenesis of these strains for humans. This strategy could be extended to other closely-related bacterial species for inter-strain and inter-species analyses.
Introduction
Francisella tularensis is a highly virulent Gram negative bacterium. A pulmonary exposure to as few as 20 bacteria is believed to cause a fatal human disease [1]. Tularemia, the disease caused by F. tularensis, can present with different clinical symptoms depending on the route of entry into the human host. Ulcero-glandular tularemia is responsible for most of the natural cases of F. tularensis infection, but other clinical forms include occulo-glandular, gastrointestinal, and pulmonary tularemia. All forms can progress to a systemic infection that includes severe prostration, multi-system organ failure, and in some cases death. The threat of bioterrorism has sparked resurgence in research on this pathogen, in order to develop novel therapies and effective vaccine strategies [2, 3].
F. tularensis is classified further into several sub-species, including F. tularensis subsp. tularensis (Ftt), F. tularensis subsp. holarctica (Fth), and F. tularensis subsp. mediasiatica (Ftm). The bacterium F. novicida is alternately classified as a separate species, F. novicida (Fn) or as a Ft subspecies F.tularensis subsp. novicida (Ftn) [4]. While genomic relatedness might indicate that novicida should be classified as a Ft subspecies, arguments have been made to maintain these bacteria as a separate species, and we will utilize this separate species nomenclature here (Fn) [5]. An older classification system divided F. tularensis into highly virulent Type A strains, which now broadly corresponds to Ftt, and less virulent Type B strains, which correspond to Fth [6–8]. A live vaccine strain (LVS) was derived from Fth in the Soviet Union by repeated passage, and has been used to vaccinate humans. This strain is attenuated for virulence in humans but still can cause a lethal disease in mice. Ftm causes a mild disease and is geographically restricted to parts of Asia. Fn is considered non-pathogenic for healthy humans, but can cause a fatal disease in mice.
F. tularensis is known to infect a variety of cells such as macrophages, hepatic cells, endothelial cells, HeLa cells, mouse fibroblasts, and even amoebae [1, 9]. Its ability to infect and replicate in macrophages is critical for disease. The Francisella pathogenicity island (FPI), a cluster of genes that are essential for phagosome escape, intramacrophage replication, and virulence, encodes a Type VI secretion system [10–12]. There are two copies of the FPI in the Ftt and Fth strains, but only one copy in Fn. The FPI is virtually identical in the various F. tularensis strains, with the exception of the pdpD gene, which contains a deletion in Fth and a truncation in Ftt when compared to Fn pdpD [10]. The DNA binding protein PigR/FevR interacts with RNA polymerase complexed with the regulatory proteins MglA and SspA to activate transcription of the FPI genes[13–16]. Comprehensive transposon mutagenesis has identified additional genes that contribute to intra-macrophage survival, including genes involved in LPS O-antigen, capsule, siderophore, biotin, and DNA synthesis [17–23].
The first full genome annotation of F. tularensis to be published was that of the Ftt strain Schu S4 [24]. The genome is interspersed with multiple insertion sequences. The majority of insertion sequences, isftu1, belong to the IS630 Tc-1 Mariner class of transposases, while a smaller subset, isftu2, belong to the IS5 class. 30 percent of the annotated genes are hypothetical proteins, and a large number of these are unique to F. tularensis. 10 percent of coding sequences are predicted to be pseudogenes. The genome sequence of Ftt strain FSC198 differs from strain Schu S4 by only a few SNPs [25]. The genome sequence of Ftt strain WY96-3418 is also almost identical to Schu S4, but in contrast to FSC198, there is a marked difference in genomic organization [26]. These differences led to the division of Ftt strains into two clades: Type AI, which includes strains similar to strain Schu S4, and Type AII, which include strains similar to WY96-3418.
The genomes of Fth strains OSU18 [27], and FSC200 [28], differ by only a few SNPs. Fth shares extensive DNA sequence identity with Ftt, but shows a striking amount of genomic rearrangement in comparison with Ftt. Comparison of the genome of the Fth LVS strain with other Fth strains revealed 35 genes that have mutations predicted to alter protein sequence in LVS; some of these, including deletions in the genes encoding a Type IV pilus subunit (pilA) and an outermembrane protein (FTT0918), are likely the cause of virulence attenuation in this strain [28, 29]
The genome sequence of Fn strain U112 has given further insight into the evolution of F. tularensis [30]. It is clear that Fn, despite having ~98% identity at the nucleotide level with the more human virulent Ftt and Fth strains, contains only a fraction of pseudogenes compared to the other strains and has therefore more functional genes. It is hypothesized that the human pathogenic Ftt and Fth lineage emerged from a common ancestor with human non-pathogenic Fn, and during evolution, gene decay caused by IS transpositions and nucleotide substitutions led to increased pathoadaptation specific to human virulence in the Ftt and Fth strains [30].
In spite of the availability of large amounts of comparative genomics data, a comparative transcriptomics analysis for F. tularensis has not been published. The current study was designed to identify transcription differences that might contribute to differences between the virulence of the F. tularensis subspecies. The results discussed below give a summary of these findings.
Methods
Bacteria
Ftt strain Schu S4, Fth strains OR960246 and LVS, and Fn strain U112 were grown in Chamberlain’s medium [31] to an optical density of 0.6. The Fth strain OR960246 was obtained from the Centers for Disease Control; its genome was sequenced by Baylor Center for Bioinformatics and Computational Biology (https://wiki.umiacs.umd.edu/cbcb/index.php/Francisella_tularensis_holarctica_OR960246). OR960246 is the Fth strain we maintain in our laboratory and thus was chosen for analysis in the present study.
Design of the Shared Probeset Microarray
Fig 1 gives a simplified illustration of the design of the shared probe Nimble Express Affymetrix microarray. The four genome sequences used initially for the design of the microarray were Ftt strain Schu S4, Fth strains LVS and OSU18, and Fn strain U112 (AJ749949, AM233362, CP000439, and CP000437 respectively). Genome sequences of the four strains were formatted into strain specific raw sequence databases using “formatdb” of the NCBI tool box, and then aligned using megablast with the expectation value set to 1 x 10−4. Custom scripts were written to parse the output files to give alignment of genome sequences of the four strains. In places where more than one region of a target sequence could be aligned to an input sequence, the alignment that allowed the least amount of breaks in the raw sequences was selected.
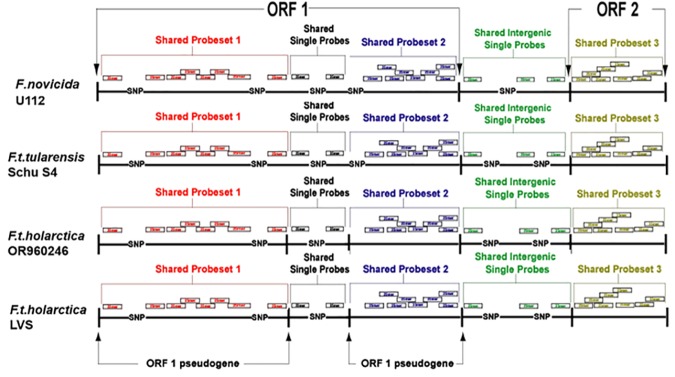
Fig 1. Design Summary of Multi Strain Shared Probe Sets.
An example of how multiple shared probe sets were created in regions of ambiguity where a full length open reading frame (ORF) is present in one strain and the homologous ORF in the other strains is broken into fragments. In the hypothetical example shown, ORF1 is an ORF that is intact in Fn U112 and Ftt Schu S4, but the orthologs in both Fth strains are pseudogenes separated into two putative coding regions. Two probe sets were designed to cover each of the two fragments, such that the larger intact ORFs in Fn and Ftt were monitored by two probe sets, while each pseudogene in Fth was monitored by one probe set. The intergenic region between ORF1 and ORF2 also had shared probes. ORF2 is intact in all four strains so a single consensus probe set was designed. Probes were also designed for the anti-sense regions of the ORFs (not shown).
Regions with a length of 25 nucleotides on both strands of the whole genome alignment were chosen for probe selection using custom scripts. If a dissimilar nucleotide was encountered in the alignment, a probe was not designed across the region of dissimilarity. By this methodology, consensus probes were designed throughout the length of all the four genomes. Every effort was made to design single shared probes for all the four strains; however if a single shared probe could not be designed that matched all four strains, then probes were designed that were shared for the maximum number of strains in the alignment, and strain-specific non-consensus probes were designed for the differing strains. Unique probes of 25 nucleotides were designed for the unique regions of each strain. This strategy resulted in complete coverage of the entire genome of all four strains, with a maximum gap in probe coverage of 10 nucleotides in the initial stage of design. The probe selection regions were extended up to a maximum of five nucleotides in either direction in some cases to allow for a selection of probes of better quality.
Next, due to the maximum probe set limit of the Nimble Express Affymetrix platform, steps were taken to reduce the number of probe sets that would need to be tiled on the microarray. In order to use similar annotation parameters for all four strains, Glimmer 2.0 was used to predict open reading frames (ORF) using default settings [32]. All ORFs with > 99% identity in all four strains and which had no paralogs in any of the strains were selected for “traditional” probe sets (11 probe pairs per gene: 11 match probes and 11 mis-match probes); 994 ORFs met these criteria. The “tiled” probe sets corresponding to insertion elements with a high copy number (isftu1 and isftu2) were removed and replaced by a single “traditional” probe set for isftu1; isftu2 was not included due to inability of the Affymetrix design pipeline to design a reliable probe set. The rest of the probes representing the remaining portions of the genomes were designed as single probe pair “tiled probe sets”. Seven additional sequences from plasmid pFNLTP, pFNLTP6_orf3, pFNLTP6_repA, pFNLTP6_orf2, pFNLTP6_kanR, pFNLTP6_ampR, and GFP, were selected for “traditional” probe sets. The sequences of the probe selection regions were subjected to hard pruning and soft pruning by the Affymetrix design team, to minimize cross-hybridization with host (human, mouse and rat) contaminant RNA.
The virulent Fth strain OR960246 was used for expression analysis. To maintain the shared probe set nature of the microarray, the draft genome sequence of OR960246 (Dr. Joseph Petrosino, Baylor College of Medicine; https://wiki.umiacs.umd.edu/cbcb/index.php/Francisella_tularensis_holarctica_OR960246) was used to identify the probes affected by subtle differences between OSU18 and OR960246, which were only a few single nucleotide polymorphisms (SNPs) and indels. The probes that were affected by these differences were corrected. Accession numbers of the orthologs from OSU18 are used to describe the corresponding ORFs of OR960246.
Microarrays were manufactured by Affymetrix using a custom manufacturing process. Two custom library files, one for monitoring ORF expression and another for monitoring intergenic expression, were designed in a format that is compatible with Gene chip operating system (GCOS). The library files were loaded into GCOS using the Library files update software provided by Affymetrix. The probe sequences, library files, and the complete design of the microarray were made public by submission to GEO database (GPL20119).
Processing of Samples for Microarray Analysis
Ft strains LVS, Schu S4, and OR960246 and Fn strain U112 were grown to 0.6 optical density at 600 nm in Chamberlains defined medium (CDM) at 37°C [31]. RNA isolation, cDNA synthesis and microarray processing was performed as described previously [33, 34]. Total RNA was isolated using TRIZOL (Invitrogen) and treated with Turbo DNA-free DNase (Ambion). cDNA was synthesized and labeled according to the Affymetrix standard prokaryotic protocol. Labeled cDNA was hybridized to the custom Nimble Express Affymetrix microarray. All samples were performed in triplicate. GCOS (Affymetrix) was used to normalize the microarray data, setting the mean of the core probe sets (994) to 1000; the data was further analyzed using GeneSpring 7.2 and GeneSpring GX (Agilent Technologies). Genes with >/ = 5-fold difference in expression level between strains (p </ = 0.05) were selected for further analysis. The p-values were obtained using GeneSpring (One-Way ANOVA, parametric test assuming equal variance, Benjamin–Hochberg multiple testing corrections, no post-hoc testing). All of the raw data has been submitted to GEO database (GSE68478).
Results and Discussion
Shared Probeset Microarray
We designed a shared probe Francisella whole genome microarray, utilizing four different genome sequences (Ftt strain Schu S4, Fth strains LVS and OSU18, and Fn strain U112). As described above, this microarray has a core set of shared probes to sequences that are common among the four strains (species/subspecies), as well as specific probes to those genes that are unique to a subset of these species/subspecies. Our goal was to create a single microarray for these species/subspecies that would enable comparative expression studies between Ftt, Fth, and/or Fn. All the strains in the study were grown to identical optical densities in Chamberlain’s medium (CDM), and their transcriptomes were analyzed by the shared probe microarray, as described above. Chamberlain’s medium was chosen due to the fact that it is the defined medium that can support growth of Francisella strains, and that we and others have already identified the proteome of various strains grown in this medium and can thus compare transcriptome to proteome [34]. An example of whole genome transcription in Fn strain U112 from this approach is shown in Fig 2, indicating that the expression of ORFs can be monitored using this microarray.
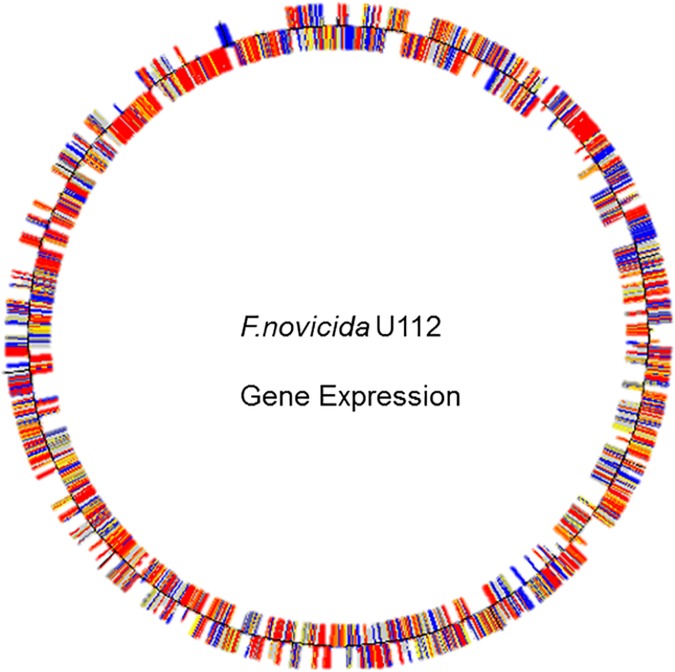
Fig 2. Expression of Open Reading Frames in Fn U112.
Expression of predicted genes (open reading frames) in Fn U112 genome. Genes in the outer circle are on the plus strand while those in the inner circle are on the minus strand. Red indicates genes with high expression, yellow indicates genes with intermediate expression and blue indicates genes with low expression. The 12 O’clock position corresponds to the start of the genome.
Expression of Pseudogenes and Transposases in Francisella
The Ftt and Fth genomes contain a relatively large number of ORFs annotated as pseudogenes, when compared to the Fn genome, and this genomic decay has been ascribed to pathoadaptation by the more virulent strains [30]. Many of the ORFs annotated as pseudogenes in Ftt, Fth and Fn have high levels of transcription (S1, S2, S3, S4, S5 and S6 Tables). We previously observed that peptide fragments from pseudogenes are present in the Ftt proteome [34]. The biological significance of these transcribed and/or translated pseudogenes is not known.
The most prevalent insertion sequence (IS) element in the Ftt and Fth genomes is isftu1; Ftt strain Schu S4 has 50 copies, Fth strain OSU18 has 59 copies, whereas Fn strain U112 has only a single copy. Carlson et al showed that the transposase encoded in isftu1, along with that in another IS element isftu2, are transcribed in Ftt Schu S4 and Fth LVS, and their transcription increases in response to spermine [35]. Comparative transcriptional analysis performed with the shared probe microarray demonstrated that isftu1 was highly transcribed in both virulent sub-species (Ftt and Fth) during growth in CDM, in contrast to Fn, which showed no significant expression (S7 Table). Because all copies of isftu1 are identical, we could not determine if the high level of transcription is due to the cumulative effect resulting from the high copy number in Ftt/Fth, or if only a subset of isftu1 elements is transcribed to high levels. Transcription of the other prevalent IS element isftu2 could not be monitored, due to the lack of reliable probe sets on the microarray. The less prevalent IS elements, (isftu3, isftu4, isftu5, or isftu6) did not have comparable high expression levels in any of the strains. The high level transcription of the isftu1 transposase in Ftt and Fth suggests that these genomes have a potential for ongoing rearrangements.
Differential RNA Expression in Strains of Francisella
Because the Francisella genomes largely share the same genes, and the microarray was designed to detect the shared gene set, transcription levels can be directly compared between the three Francisella species/subspecies grown under identical conditions. Interestingly, we found that transcription levels of many genes are notably different among the three species/subspecies of Francisella (Fig 3, Table 1, Table 2 and Table 3, S1, S2, S3, S4, S5 and S6 Tables). In general, transcription of relatively more genes was higher in Fn compared to Ftt or Fth. In addition to unique genes in each species/subspecies, pseudogene formation in Ftt and Fth due to possible genome decay may have contributed to significant transcriptional differences. The large number of pseudogenes in the Ftt and Fth genomes may be one cause of the difference in expression between Ftt/Fth and Fn, because the pseudogene may not be expressed to the same level as the corresponding gene in Fn, or the presence of a pseudogene may influence transcription of downstream genes. Likewise, transposons in Ftt and Fth may negatively impact transcription of genes by inserting into that gene or its promoter, or downstream genes within an operon. Finally genomic reorganization in Ftt and Fth may cause breaks within operons that place genes in a different genomic context. However, differential expression for a number of genes could not be easily attributed to one of these possibilities, and thus transcriptional differences between the strains may be due to differences in regulatory genes and/or networks elsewhere in the genome (i.e. trans effects).
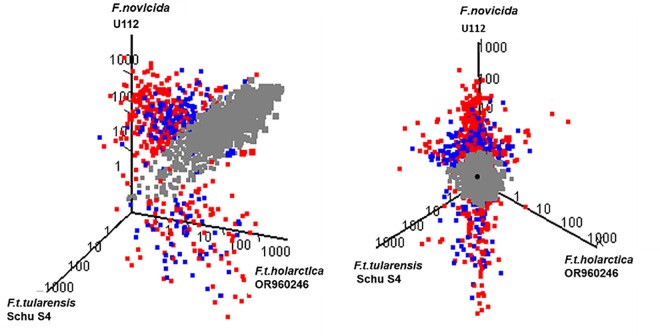
Fig 3. Comparison of Transcriptomes among Species/Subspecies of F.tularensis.
The left and right panels show the same 3 dimensional graph visualized from two different angles. Each point represents a probe set monitoring the transcription of an ORF. The values on x, y and z axes represent the geometric mean of normalized intensity of the three biological replicates of Ftt Schu S4, Fth OR960246 and Fn U112 respectively. Probe sets that have differential expression of less than five fold and/or a p-value greater than 0.05 in both of the one way comparisons of Ftt Schu S4 and Fth OR960246 with Fn U112 are colored grey. Of the remaining probe sets, those representing genes which are intact in all of the three strains are colored red, while the probe sets representing genes with at least one of the orthologs annotated as a pseudogene are colored blue.
Table 1. Genes with High Expression in Ftt Schu S4 Compared to Fn U112**.
| Locus in Ftt | Locus in Fn | Fold Difference* | Product |
|---|---|---|---|
| FTT0027 | FTN_1684 | 16 | pyridoxal-dependent decarboxylase (lys1) |
| FTT0028 | FTN_1683 | 6 | drug:H+ antiporter-1 (DHA1) family protein |
| FTT0029 | FTN_1682 | 17 | siderophore biosynthesis protein (fslA) |
| FTT0103 | FTN_1612 | 5 | transposase |
| FTT0221 | FTN_0090 | 5 | acid phosphatase (precursor)(acpA) |
| FTT1140 | FTN_1122 | 9 | hypothetical protein |
| FTT1242 | FTN_1260 | 10 | hypothetical protein |
| FTT1653 | FTN_0030 | 99 | hypothetical protein |
| FTT1654 | FTN_0029 | 190 | HlyD family secretion protein(emrA2) |
| FTT1655 | FTN_0028 | 72 | hypothetical protein |
Table 2. Genes with High Expression in Ftt Schu S4 Compared to Fth OR960246**.
| Locus in Ftt | Locus in Fth | Fold Difference* | Product |
|---|---|---|---|
| FTT0127 | FTH_1591 | 5 | major facilitator superfamily (MFS) transport protein |
| FTT0442 | FTH_1570 | 8 | major facilitator superfamily (MFS) transport protein |
| FTT0707 | FTH_1479 | 6 | nicotinamide mononucleotide transport (NMT) family protein |
| FTT0784 | FTH_1400 | 6 | hypothetical protein |
| FTT0815 | FTH_1370 | 5 | chitin binding protein |
| FTT1004 | FTH_1172 | 6 | DMT superfamily drug/metabolite transporter |
| FTT1089 | FTH_1087 | 5 | isochorismatase hydrolase family protein |
| FTT1090 | FTH_1086 | 8 | possible NMC family Nicotinamide mononucleotide uptake permease PnuC |
| FTT1175 | FTH_0772 | 5 | hypothetical protein |
| FTT1234 | FTH_0713 | 18 | choloylglycine hydrolase family protein |
| FTT1414 | FTH_0648 | 7 | hypothetical protein |
| FTT1784 | FTH_1866 | 11 | hypothetical protein |
*≥ 5 fold
p ≤ 0.05
** The table only lists the genes that are intact in both Ftt and Fth. Please see S3 Table for a complete list that includes pseudogenes and missing genes.
Table 3. Genes with Low Expression in Ftt Schu S4 Compared to Fth OR960246**.
| Locus in Ftt | Locus in Fth | Fold Difference* | Product |
|---|---|---|---|
| FTT0845 | FTH_0338 | 9 | hypothetical protein |
| FTT1143 | FTH_0806 | 14 | hypothetical protein |
| FTT0642 | FTH_0895 | 6 | acetolactate synthase small subunit |
| FTT0158 | FTH_1669 | 9 | hypothetical membrane protein |
*≥ 5 fold
p ≤ 0.05
** The table only lists the genes that are intact in both Ftt and Fth. Please see S4 Table for a complete list that includes pseudogenes and missing genes.
Comparison of transcription in Ftt and Fn
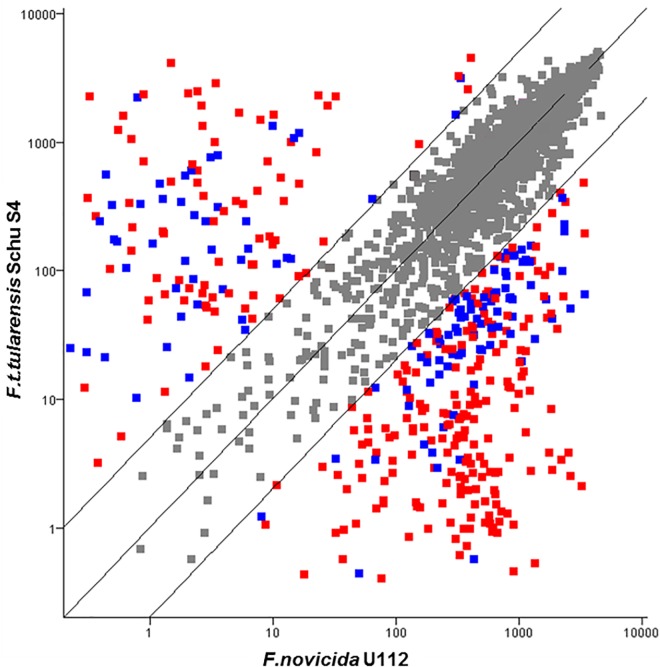
When transcription in the human virulent Ftt Schu S4 strain is compared to transcription in the human avirulent Fn U112 strain (Fig 4; S1 and S2 Tables), the largest group of shared genes falls into similar levels of expression (<5-fold). However, the majority of the genes that falls outside this range are transcribed at higher levels in Fn compared to Ftt. A number of these genes have low expression in Ftt either due to pseudogene formation or polar effects of pseudogenes and transposases. In contrast, most of the genes with high expression in Ftt are actually unique to Ftt and missing from Fn.
Fig 4. Comparison of Transcriptomes of Ftt Schu S4 and Fn U112.
Despite high genomic identity, Ftt and Fn have large scale transcriptional differences when grown under identical conditions. Each point represents a probe set monitoring the transcription of an ORF. The values on x and y axes represent the geometric mean of three biological replicates. Probe sets that have differential expression of less than five fold and/or a p-value greater than 0.05 in one way comparisons of Ftt Schu S4 with Fn U112 are colored grey. Of the remaining probe sets, those representing genes which are intact in both strains are colored red, while the probe sets representing genes with one of the orthologs annotated as a pseudogene are colored blue.].
Of particular interest are shared genes that are more highly transcribed in Ftt when compared to Fn, as these may give insight into the higher virulence of Ftt for humans. Some of these include an acid phosphatase, an Emr secretion protein, an iron acquisition gene cluster (fsl operon), transposases (described above) and genes encoding hypothetical proteins (Table 1). EmrA proteins are typically the membrane fusion protein components (MFP) of tripartite efflux assemblies, which encompass multidrug efflux systems (RND) and Type I secretion systems (T1SS) [36, 37]. MFPs span the periplasm and connect the efflux transporter in the cytoplasmic membrane with a specific outermembrane porin to facilitate secretion across both membranes in one step [38]. MFPs are generally involved in transport of various substances (e.g. hemolysins, multidrug resistance). The OM porin TolC known to be utilized for MDR and T1S in other bacteria is involved in efflux of detergents, dyes and antibiotics in Francisella, and the transporter/MFP pair AcrAB also function in resistance to antibiotics; both contribute to the virulence of Francisella [39, 40]. Also, the EmrA1 MFP in Fth (LVS) has been shown to be required for resistance to oxidative stress and virulence [41]. The emrA2 (FTT1654) gene in Francisella is flanked by two ORFs in an apparent operon: a predicted lipoprotein with a conserved domain of unknown function (FTT1653; DUF3568), and a putative inner membrane protein involved in fusaric acid resistance (FTT1655). All of these genes are highly transcribed in Ftt Schu S4 and in both of the Fth strains (OR960246 and LVS), but expressed at low levels in Fn U112.
The fsl gene cluster is likewise highly expressed in Ftt and both Fth strains, but transcribed at low levels in Fn. The fsl genes are required for siderophore synthesis in Francisella [42–44]. Siderophores facilitate iron uptake in bacteria by scavenging ferric iron in the surrounding environment and transporting it to the cytoplasm. The fslA gene product is a siderophore synthetase, and the other genes within the operon (fslBCDEF) are also required for siderophore synthesis/utilization [42, 44]. Mutations in these genes leads to poor growth in iron depleted medium [44], however there is another uptake system for ferrous iron (feo) that has been shown to compensate for the lack of siderophore synthesis in vivo, at least in Fth [45]. The high level of transcription of the ferric-siderophore operon in Ftt and Fth strains when compared to Fn during growth in CDM suggests that Ftt and Fth are experiencing iron limitation, but that Fn may have additional mean(s) to acquire iron under these conditions.
Acid phosphatases have been hypothesized to contribute to the virulence of Francisella by inhibition of the oxidative burst inside the phagosome [46]. There are 5–6 genes encoding acid phosphatases in the different Francisella species. It has been shown that four functional acid phosphatases AcpA, AcpB, AcpC and HapA contribute 90–99% of the acid phosphatase activity in Fn and Ftt and together are required for intramacrophage survival and virulence [47–49]. However it has also been reported that acid phosphatases do not contribute to Ftt virulence [50]. AcpA, which provides most of the acid phosphatase activity, is secreted in vitro [51], and translocated into the host macrophage cytosol by Fn and Ftt [52]. Higher level transcription of acpA in Ftt compared to Fn may influence the relative virulence these strains have for humans. Further investigation is necessary to definitively identify the role of AcpA in the virulence of different species/subspecies of Francisella.
FTT1140 (198 bp) and FTT1242 (1263 bp) are both annotated as encoding hypothetical proteins. FTT1140 is predicted to encode a short peptide of 65 aa, whereas the Fn ortholog (FTN1122) is predicted to be 129 aa. Interestingly, the Ftt protein appears to have a truncation at the N-terminus with respect to the Fn protein, and this region is predicted to contain a transmembrane segment (HMMTOP), suggesting that these proteins would have different subcellular locations within the two strains. FTT1242 contains a conserved domain (COG0172) found in seryl-tRNA synthetases. Interestingly, Fn contains two FTT1242 orthologs (FTN1260 and FTN1261) that share 65% identity; FTT1242 shares the closest identity to FTN1260 (95%) than to FTN1261 (61%). This suggests that the FTN1261 gene was deleted from Ftt/Fth during their evolutionary divergence from Fn. The relevance of these two hypothetical genes being transcribed at higher levels in Ftt is not clear.
Comparison of transcription of Ftt and Fth
Ftt strain Schu S4 and Fth (wildtype) strain OR960246 are more closely related to each other than to Fn, however a number of differences in gene expression were observed when these strains were grown under identical conditions. Since Ftt is known to be more virulent in humans than Fth, some of these transcription differences may underlie their relative virulence (S3 and S4 Tables). Genes that are similar between these two strains but with significant transcription differences are noted in Tables 2 and 3.
The FTT1089-FTT1091 operon is highly expressed in Ftt in comparison to Fth. This operon encodes two isochorismatases (FTT1089 and FTT1091) and a transporter (FTT1090). Isochorismatases catalyze one of the steps in the conversion of isochorismate to 2-3-Dihydroxy Benzoic acid (2-3-DHBA), which itself can act as an iron chelator or can be diverted to form further complex siderophores [53]. It is unknown whether 2-3-DHBA is involved in iron uptake in Francisella. This operon appears intact in Ftt, whereas in Fth (both OR960246 and LVS), one of the isochorismatases (the ortholog of FTT1091) is a pseudogene due to insertion of a transposase, and in Fn, the transporter (the ortholog of FTT1090) is a pseudogene. In Fth the operon is not expressed, in contrast to Ftt Schu S4. Due to the differences in transcription and translation between Ftt and Fth, as well as the differences in siderophore (fsl) transcription mentioned above, this operon warrants further investigation.
Four different genes predicted to encode transporters were transcribed at higher levels in Ftt Schu S4 than in Fth OR960246: FTT0127, FTT0442, FTT0707, and FTT1004. Transposon insertions upstream of the FTT1004 and FTT1234 orthologs in Fth may explain their lower transcription, but it is unclear why the other genes are also expressed at lower levels in Fth. FTT1234 encodes a cholylglycine hydrolase that has been implicated in virulence in Brucella and Listeria [54, 55]. FTT0784, FTT1175, FTT1414, and FTT1784 encode hypothetical proteins whose orthologs are transcribed at lower levels in Fth; the presence of pseudogenes upstream in Fth possibly explains this differential expression. Orthologs to hypothetical genes FTT0185, FTT0845, and FTT1143, as well as a gene involved in valine biosynthesis, FTT0642, were transcribed at higher levels in Fth OR960246 compared to Ftt Schu S4 (Table 3).
There are interesting differences in the presence and expression of genes predicted to be involved in polyamine synthesis between the various Ft strains (S1 and S3 Tables). The response of bacterial pathogens to polyamines has been associated with various aspects of virulence. It has been shown that the Fth and Ftt response to exogenous spermine and spermidine alters global gene expression patterns, including an upregulation of transcription from ISFtu1 and ISFtu2 insertion elements, which also increases expression of genes adjacent to these elements [35]. Growth of Ft in spermine or spermidine downregulates expression of TNFα and IL-12 by infected macrophages. CDM contains spermine, and was utilized in the studies presented here.
Putrescine and spermidine are the major endogenous polyamines in bacteria, and they can synthesize putrescine either directly from ornithine or indirectly via arginine, and then convert putrescine to spermidine; bacteria do not synthesize spermine. Ftt Schu S4 contains a cluster of genes, speD (S-adenosylmethionine carboxylase), speE (spermidine synthase), speA (arginine decarboxylase), FTT0434 (agmatine deiminase; aguA) and FTT0435 (N-carbamoylputrescine amidohydrolase; aguB) in an apparent operon. In Fth strains, speD and speE are intact while speA, aguA and aguB are pseudogenes, and the entire region is absent in Fn U112. Thus Fth strains might be predicted to not be able to convert arginine to putrescine because they lack the three genes necessary, but to maintain the ability to convert putrescine to spermidine due to the presence of speD and speE, whereas Fn lacks this entire pathway. Alternatively, bacteria can convert ornithine directly to putrescine by ornithine decarboxylase (speC), but no speC ortholog has been annotated in the Ft genome. The gene annotated as diaminopimelate decarboxylase (lysA; FTT0027) may fulfill this function as SpeC. As mentioned above, this gene lies within the siderophore synthesis operon and is also annotated as fslC for its role in iron uptake. The Ft siderophore resembles rhizoferrin and has a putrescine backbone [42], and FTT0027 is a member of the pyridoxal-dependent decarboxylases (pfam02784) which include ornithine decarboxylases. Thus this gene may provide synthesis of putrescine directly from ornithine.
Interestingly, even though speD and speE are intact in the Fth strains, their expression is significantly lower than their orthologs in Ftt Schu S4 when grown under identical conditions. The lower expression of speD and speE, and absence of speA, aguA and aguB in Fth compared to Ftt may contribute to the relative lower virulence of this subspecies in humans. Considering that Fn lacks all five genes and exhibits the lowest virulence of all three subspecies in humans, this suggests that endogenous polyamine synthesis may contribute to Ft virulence. Endogenous polyamines influence many aspects of bacteria, including transcription, translation, cell growth, and stress resistance, and polyamines have been shown to modulate the virulence of a variety of pathogens, including Shigella, Yersinia, and Salmonella [56].
Additional genes more highly expressed in Ftt compared to Fth may contribute to enhanced virulence (S3 Table). For example, NADPH-quinone reductase (mdaB) catalyzes the reduction of quinones and may help protect against damage by free radicals and reactive oxygen species within the macrophage more effectively. The tlyC gene, which encodes a protein with cystathionine beta-synthase (CBS) domains found in transporters, is more highly expressed in Ftt, but it is not clear whether this protein has any hemolytic activity despite being annotated as a hemolysin [57]. The pdpD gene within the Francisella pathogenicity island is expressed higher in Ftt than in Fth. Differences in this gene distinguish the different Francisella subspecies, with Fn expressing a PdpD protein that is 50 aa larger than that in Ftt. In contrast, the pdpD gene is essentially missing from Fth, with only a small portion of the C-terminal coding sequence remaining [10]. The shared probes within the microarray were designed to this portion, and transcription was reduced 12-fold in Fth vs Ftt, while the transcription of the downstream genes (iglABCD) was not altered. PdpD contributes to the virulence of Fn in chicken embryos and mice, but it is not required for intracellular growth in vitro [58]. Francisella spp have Type IV pilus genes that are involved in secretion and pilus fiber expression [59]. The high transcription of pilT and pilA in Ftt Schu S4 compared to Fth may contribute to its higher virulence.
Comparison of transcription of Fth OR960246 and Fth LVS
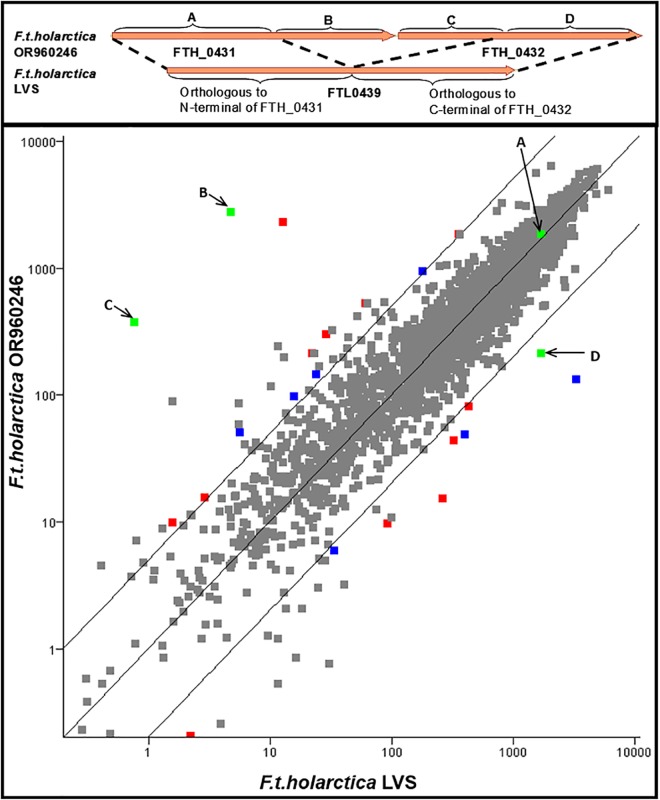
The Fth LVS strain is highly attenuated for virulence and was derived by repeated passage in the laboratory. The LVS genome differs from that of the wildtype Fth OR960246 strain by a number of SNPs and indels. Not surprisingly, comparative transcriptional analysis revealed that the two Fth strains have very similar expression profiles, mirroring their highly similar genome architecture (Fig 5, Tables 4 and 5). There are two paralogs, FTH_0431 (fopC) and FTH_0432, in Fth that share 50.4% homology and that have undergone recombination to form a fusion gene in LVS, FTL_0439. The difference in architecture of the gene between the two Fth strains is shown in Fig 5. In Fth OR960246 and in Ftt Schu4, the two genes in the orthologous genomic region have different levels of expression; FTH_0431 (fopC) is highly expressed and FTH_0432 has a much lower expression. Due to the fusion of the C-terminal region of FTH_0432 (probe set region D) to the N-terminal region of FTH_0431 (probe set region A) in LVS, the two regions (probe set regions A and D) have the same level of expression.
Fig 5. Comparison of Transcriptomes of Strains of Fth OR960246 and Fth LVS.
The genomic region of Fth LVS FTL0439 compared to the orthologous region FTH0431 and FTH0432 in Fth OR960246 is shown in the top panel. The bottom panel shows the comparison of gene expression in Fth OR960246 and Fth LVS strains. Each point represents a probe set monitoring the transcription of a unique computationally predicted gene (average of three biological replicates). Probe sets that have differential expression of less than five fold and/or a p-value greater than 0.05 in one way comparisons of Fth OR960246 with Fth LVS are colored grey. Of the remaining probe sets, those representing genes which are intact in both strains are colored red, while the probe sets representing genes with one of the orthologs annotated as a pseudogene are colored blue. The probe sets covering the FTL0439/FTH0431-0432 region are colored green and indicated by letter corresponding to top panel.
Table 4. Genes with High Expression in Fth OR960246 Compared to Fth LVS.
| Locus in Fth OR960246 | Locus in Fth LVS | Fold Difference* | Product |
|---|---|---|---|
| FTH_0160 | FTL_0167 | 10 | DNA helicase |
| FTH_0384 | None | 182 | pilA |
| FTH_0431 | None | 494 | fopC |
| FTH_0432 | None | 583 | hypothetical protein |
| FTH_0752 | FTL_0750 | 5 | hypothetical protein |
* ≥ 5 fold
p ≤ 0.05
Table 5. Genes with Low Expression in Fth OR960246 Compared to Fth LVS.
| Locus in Fth OR960246 | Locus in Fth LVS | Fold Difference* | Product |
|---|---|---|---|
| FTH_0061 | None | 6 | predicted pseudogene |
| FTH_0383 | None | 25 | predicted pseudogene |
| FTH_1250 | FTL_1277 | 5 | nagC |
| FTH_1328 | FTL_1363 | 7 | Esterase |
*≥ 5 fold
p ≤ 0.05
FopC (FupC) is a paralog of figE (fslE) of the fig operon described in the sections above. It has been implicated in the high affinity uptake of ferrous iron [60]. The ortholog of FTH_0431 in Ftt Schu S4 (FTT_0918) was shown to be required for virulence and a strain with a mutation in this gene was effective as a live vaccine [61]. Reintroduction of the FTH_0431 (fopC) along with pilA (FTH_0384) into LVS restored virulence to a level indistinguishable from the wild type Fth strains [29]. FopC has also been shown to help in avoiding IFN gamma-mediated immune defenses [62]. No studies have been performed to understand the biological function and immunogenicity of the downstream FTH_0432. It is probable that one of the major factors responsible for the attenuation of LVS is the lack of the C-terminal region of FTH0431.
In addition to FTL_0431, the other gene that is highly expressed in OR960246 when compared to LVS is a pilA gene. Pilus genes have been shown previously to be required for virulence [63]. The genes that are highly expressed in LVS and expressed at lower levels in OR960246 are an esterase and nagC. NagC is involved in the synthesis and catabolism of glucosaminoglycans and chitin [64]. The biological role of these genes and their effect on virulence is not yet known.
Limitations
This study represents a comparison of the transcriptome of multiple strains of Francisella grown under identical conditions in the chemically-defined Chamberlain’s medium. We have noted the genes with 5-fold or greater differences in transcription (p < 0.05) between the strains, and suggested that some of these differences may contribute to the relative virulence of these strains. However this technique was utilized as a discovery tool, and further analyses like 1) RT-PCR confirmation 2) comparative proteomic analyses 3) growth analyses in different media and cells, and 4) animal infection studies will be required to validate these expression differences as the bases of differences in virulence. Additionally, different growth conditions (e.g. medium, temperature, etc) are likely to highlight additional differences in gene expression between species/subspecies. There are also multiple genes whose expression differed from 2- to 5-fold between strains (p <0.05) that were not discussed but that could also be of significance. Of course numerous other factors not addressed here may also contribute to relative expression of the corresponding proteins, such as message half-life, translation efficiency, and post translational modifications.
Summary and Future Directions
The Francisella shared probe set microarray designed for this study allowed us to perform comparative transcription studies of Ftt, Fth, and Fn strains grown under identical conditions. In the past, microarray studies have been performed with probes designed and optimized for Ftt, but utilized for additional Francisella strains [43, 65]. These approaches were useful, but had significant limitations due to the variability of genome sequences across the Francisella subspecies. However, due to high similarity in the genomes of these subspecies, we were able to design a consensus microarray for Ftt, Fth, and Fn that represented the core set of shared genes, as well as the species-specific genes and the inter-genic regions. Our studies allowed us to perform direct comparisons of relative transcript levels across species/subspecies, and highlighted potentially interesting differences in gene expression that may be the basis for further studies to identify the underlying cause of the relative virulence of these strains in humans.
Supporting Information
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
We would like to acknowledge Dr. George M. Hilliard for providing guidance and lab space for this work at the University of Tennessee Health Science Center.
Data Availability
All files are already publicly available from the GEO database (accession numbers,GPL20119, GSE68478).
Funding Statement
Portions of the study were funded by NIH AI057986 and NIH-NIAID-DMID-05-22 (P.I. Lyons) to Karl E. Klose. Portions of the study were also funded by NIAID grant AI059642 to Dr. George M. Hilliard. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Oyston PC, Sjostedt A, Titball RW. Tularaemia: bioterrorism defence renews interest in Francisella tularensis. Nat Rev Microbiol. 2004;2:967–78. [DOI] [PubMed] [Google Scholar]
- 2.Tarnvik A, Berglund L. Tularemia. Eur Respir J. 2003;21:361–73. [DOI] [PubMed] [Google Scholar]
- 3.Petersen JM, Schriefer ME. Tularemia: emergence/re-emergence. Vet Res. 2005;36(3):455–67. 10.1051/vetres:2005006 . [DOI] [PubMed] [Google Scholar]
- 4.Johansson A, Celli J, Conlan W, Elkins KL, Forsman M, Keim PS, et al. Objections to the transfer of Francisella novicida to the subspecies rank of Francisella tularensis. Int J Syst Evol Microbiol. 2010;60(Pt 8):1717–8; author reply 8–20. 10.1099/ijs.0.022830-0 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kingry LC, Petersen JM. Comparative review of Francisella tularensis and Francisella novicida. Front Cell Infect Microbiol. 2014;4:35 10.3389/fcimb.2014.00035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rodionova IV. [Differentiation of geographic races of Francisella tularensis on the basis of citrulline ureidase activity]. Lab Delo. 1970;1:42–3. . [PubMed] [Google Scholar]
- 7.Rodionova IV. [Citrulline ureidase activity in geographical races of Francisella tularensis]. Dokl Akad Nauk SSSR. 1968;179(2):457–60. . [PubMed] [Google Scholar]
- 8.Sandstrom G, Sjostedt A, Forsman M, Pavlovich NV, Mishankin BN. Characterization and classification of strains of Francisella tularensis isolated in the central Asian focus of the Soviet Union and in Japan. J Clin Microbiol. 1992;30(1):172–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellis J, Oyston PCF, Green M, Titball R. Tularemia. Clin Microbiol Rev. 2002;15:631–46. PubMed Central PMCID: PMC12364373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nano FE, Zhang N, Cowley SC, Klose KE, Cheung KK, Roberts MJ, et al. A Francisella tularensis Pathogenicity Island Required for Intramacrophage Growth. J Bacteriol. 2004;186:6430–6. PubMed Central PMCID: PMC15375123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Golovliov I, Ericsson M, Sandstrom G, Tarnvik A, Sjostedt A. Identification of proteins of Francisella tularensis induced during growth in macrophages and cloning of the gene encoding a prominently induced 23-kilodalton protein. Infect Immun. 1997;65:2183–9. PubMed Central PMCID: PMC 9169749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barker JR, Chong A, Wehrly TD, Yu J-J, Rodriguez SA, Liu J, et al. The Francisella tularensis pathogenicity island encodes a secretion system that is required for phagosome escape and virulence. Molecular microbiology. 2009;74:1459–70. PubMed Central PMCID: PMC 20054881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lauriano CM, Barker JR, Yoon SS, Nano FE, Arulanandam BP, Hassett DJ, et al. MglA regulates transcription of virulence factors necessary for Francisella tularensis intra-amoebae and intra-macrophage survival. Proc Natl Acad Sci USA. 2004;101:4246–9. PubMed Central PMCID: PMC15010524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mohapatra NP, Soni S, Bell BL, Warren R, Ernst RK, Muszynski A, et al. Identification of an orphan response regulator required for the virulence of Francisella spp. and transcription of pathogenicity island genes. Infection and immunity. 2007;75(7):3305–14. 10.1128/IAI.00351-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Charity JC, Costante-Hamm MM, Balon EL, Boyd DH, Rubin EJ, Dove SL. Twin RNA Polymerase-associated proteins control virulence gene expression in Francisella tularensis. PLoS Pathog. 2007;3:e84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rohlfing AE, Dove SL. Coordinate control of virulence gene expression in Francisella tularensis involves direct interaction between key regulators. J Bacteriol. 2014;196(19):3516–26. 10.1128/JB.01700-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weiss DS, Brotcke A, Henry T, Margolis JJ, Chan K, Monack DM. In vivo negative selection screen identifies genes required for Francisella virulence. Proc Natl Acad Sci U S A. 2007;104(14):6037–42. 0609675104 [pii] 10.1073/pnas.0609675104 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Su J, Yang J, Zhao D, Kawula TH, Banas JA, Zhang JR. Genome-wide identification of Francisella tularensis virulence determinants. Infection and immunity. 2007;75(6):3089–101. 10.1128/IAI.01865-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kraemer PS, Mitchell A, Pelletier MR, Gallagher LA, Wasnick M, Rohmer L, et al. Genome-wide Screen in Francisella novicida for Genes required for pulmonary and systemic infection in mice. Infection and immunity. 2009;77:232–44. 10.1128/IAI.00978-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schulert GS, McCaffrey RL, Buchan BW, Lindemann SR, Hollenback C, Jones BD, et al. Francisella tularensis genes required for inhibition of the neutrophil respiratory burst and intramacrophage growth identified by random transposon mutagenesis of strain LVS. Infection and immunity. 2009;77(4):1324–36. 10.1128/IAI.01318-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buchan BW, McLendon MK, Jones BD. Identification of Differentially regulated Francisella tularensis genes by use of a newly developed Tn5-based transposon delivery system. Appl Environ Microbiol. 2008;74:2637–45. 10.1128/AEM.02882-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maier TM, Casey MS, Becker RH, Dorsey CW, Glass EM, Maltsev N, et al. Identification of Francisella tularensis Himar1-based transposon mutants defective for replication in macrophages. Infection and immunity. 2007;75(11):5376–89. 10.1128/IAI.00238-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qin A, Mann BJ. Identification of transposon insertion mutants of Francisella tularensis tularensis strain Schu S4 deficient in intracellular replication in the hepatic cell line HepG2. BMC Microbiol. 2006;6:69 10.1186/1471-2180-6-69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Larsson P, Oyston PC, Chain P, Chu MC, Duffield M, Fuxelius HH, et al. The complete genome sequence of Francisella tularensis, the causative agent of tularemia. Nat Genet. 2005;37:153–9. [DOI] [PubMed] [Google Scholar]
- 25.Chaudhuri RR, Ren CP, Desmond L, Vincent GA, Silman NJ, Brehm JK, et al. Genome sequencing shows that European isolates of Francisella tularensis subspecies tularensis are almost identical to US laboratory strain Schu S4. PloS one. 2007;2(4):e352 10.1371/journal.pone.0000352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beckstrom-Sternberg SM, Auerbach RK, Godbole S, Pearson JV, Beckstrom-Sternberg JS, Deng Z, et al. Complete genomic characterization of a pathogenic A.II strain of Francisella tularensis subspecies tularensis. PloS one. 2007;2(9):e947 10.1371/journal.pone.0000947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Petrosino JF, Xiang Q, Karpathy SE, Jiang H, Yerrapragada S, Liu Y, et al. Chromosome rearrangement and diversification of Francisella tularensis revealed by the type B (OSU18) genome sequence. J Bacteriol. 2006;188(19):6977–85. 10.1128/JB.00506-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rohmer L, Brittnacher M, Svensson K, Buckley D, Haugen E, Zhou Y, et al. Potential source of Francisella tularensis live vaccine strain attenuation determined by genome comparison. Infection and immunity. 2006;74(12):6895–906. 10.1128/IAI.01006-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Salomonsson E, Kuoppa K, Forslund AL, Zingmark C, Golovliov I, Sjostedt A, et al. Reintroduction of two deleted virulence loci restores full virulence to the live vaccine strain of Francisella tularensis. Infection and immunity. 2009;77:3424–31. PubMed Central PMCID: PMC19506014. 10.1128/IAI.00196-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rohmer L, Fong C, Abmayr S, Wasnick M, Larson Freeman TJ, Radey M, et al. Comparison of Francisella tularensis genomes reveals evolutionary events associated with the emergence of human pathogenic strains. Genome Biol. 2007;8(6):R102 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chamberlain RE. Evaluation of Live Tularemia Vaccine Prepared in a Chemically Defined Medium. Appl Microbiol. 1965;13:232–5. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Delcher AL, Harmon D, Kasif S, White O, Salzberg SL. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 1999;27(23):4636–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abdelrahman YM, Rose LA, Belland RJ. Developmental expression of non-coding RNAs in Chlamydia trachomatis during normal and persistent growth. Nucleic Acids Res. 2011;39(5):1843–54. 10.1093/nar/gkq1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Waldo RH, Cummings ED, Sarva ST, Brown JM, Lauriano CM, Rose LA, et al. Proteome cataloging and relative quantification of Francisella tularensis tularensis strain Schu4 in 2D PAGE using preparative isoelectric focusing. J Proteome Res. 2007;6(9):3484–90. 10.1021/pr070107m . [DOI] [PubMed] [Google Scholar]
- 35.Carlson PE Jr., Horzempa J, O'Dee DM, Robinson CM, Neophytou P, Labrinidis A, et al. Global transcriptional response to spermine, a component of the intramacrophage environment, reveals regulation of Francisella gene expression through insertion sequence elements. J Bacteriol. 2009;191(22):6855–64. 10.1128/JB.00995-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Symmons MF, Marshall RL, Bavro VN. Architecture and roles of periplasmic adaptor proteins in tripartite e ffl ux assemblies. Front Microbiol. 2015;6:513 10.3389/fmicb.2015.00513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li XZ, Plesiat P, Nikaido H. The challenge of efflux-mediated antibiotic resistance in Gram-negative bacteria. Clin Microbiol Rev. 2015;28(2):337–418. 10.1128/CMR.00117-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zgurskaya HI, Weeks JW, Ntreh AT, Nickels LM, Wolloscheck D. Mechanism of coupling drug transport reactions located in two different membranes. Front Microbiol. 2015;6:100 10.3389/fmicb.2015.00100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gil H, Platz GJ, Forestal CA, Monfett M, Bakshi CS, Sellati TJ, et al. Deletion of TolC orthologs in Francisella tularensis identifies roles in multidrug resistance and virulence. Proc Natl Acad Sci U S A. 2006;103(34):12897–902. 10.1073/pnas.0602582103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bina XR, Lavine CL, Miller MA, Bina JE. The AcrAB RND efflux system from the live vaccine strain of Francisella tularensis is a multiple drug efflux system that is required for virulence in mice. FEMS Microbiol Lett. 2008;279(2):226–33. 10.1111/j.1574-6968.2007.01033.x . [DOI] [PubMed] [Google Scholar]
- 41.Ma Z, Banik S, Rane H, Mora VT, Rabadi SM, Doyle CR, et al. EmrA1 membrane fusion protein of Francisella tularensis LVS is required for resistance to oxidative stress, intramacrophage survival and virulence in mice. Molecular microbiology. 2014;91(5):976–95. 10.1111/mmi.12509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sullivan JT, Jeffery EF, Shannon JD, Ramakrishnan G. Characterization of the siderophore of Francisella tularensis and role of fslA in siderophore production. J Bacteriol. 2006;188(11):3785–95. 10.1128/JB.00027-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deng K, Blick RJ, Liu W, Hansen EJ. Identification of Francisella tularensis genes affected by iron limitation. Infection and immunity. 2006;74(7):4224–36. Epub 2006/06/23. 10.1128/IAI.01975-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kiss K, Liu W, Huntley JF, Norgard MV, Hansen EJ. Characterization of fig operon mutants of Francisella novicida U112. FEMS Microbiol Lett. 2008;285(2):270–7. 10.1111/j.1574-6968.2008.01237.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perez NM, Ramakrishnan G. The reduced genome of the Francisella tularensis live vaccine strain (LVS) encodes two iron acquisition systems essential for optimal growth and virulence. PloS one. 2014;9(4):e93558 10.1371/journal.pone.0093558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reilly TJ, Baron GS, Nano FE, Kuhlenschmidt MS. Characterization and sequencing of a respiratory burst-inhibiting acid phosphatase from Francisella tularensis. J Biol Chem. 1996;271(18):10973–83. . [DOI] [PubMed] [Google Scholar]
- 47.Mohapatra NP, Balagopal A, Soni S, Schlesinger LS, Gunn JS. AcpA is a Francisella acid phosphatase that affects intramacrophage survival and virulence. Infection and immunity. 2007;75(1):390–6. 10.1128/IAI.01226-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mohapatra NP, Soni S, Reilly TJ, Liu J, Klose KE, Gunn JS. Combined deletion of four Francisella novicida acid phosphatases attenuates virulence and macrophage vacuolar escape. Infection and immunity. 2008;76(8):3690–9. 10.1128/IAI.00262-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mohapatra NP, Soni S, Rajaram MV, Strandberg KL, Gunn JS. Type A Francisella tularensis acid phosphatases contribute to pathogenesis. PloS one. 2013;8(2):e56834 10.1371/journal.pone.0056834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Child R, Wehrly TD, Rockx-Brouwer D, Dorward DW, Celli J. Acid phosphatases do not contribute to the pathogenesis of type A Francisella tularensis. Infection and immunity. 2010;78(1):59–67. 10.1128/IAI.00965-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Konecna K, Hernychova L, Reichelova M, Lenco J, Klimentova J, Stulik J, et al. Comparative proteomic profiling of culture filtrate proteins of less and highly virulent Francisella tularensis strains. Proteomics. 2010;10(24):4501–11. 10.1002/pmic.201000248 . [DOI] [PubMed] [Google Scholar]
- 52.Dai S, Mohapatra NP, Schlesinger LS, Gunn JS. The acid phosphatase AcpA is secreted in vitro and in macrophages by Francisella spp. Infection and immunity. 2012;80(3):1088–97. 10.1128/IAI.06245-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ratledge C, Dover LG. Iron metabolism in pathogenic bacteria. Annu Rev Microbiol. 2000;54:881–941. 10.1146/annurev.micro.54.1.881 . [DOI] [PubMed] [Google Scholar]
- 54.Marchesini MI, Connolly J, Delpino MV, Baldi PC, Mujer CV, DelVecchio VG, et al. Brucella abortus choloylglycine hydrolase affects cell envelope composition and host cell internalization. PloS one. 2011;6(12):e28480 10.1371/journal.pone.0028480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dussurget O, Cabanes D, Dehoux P, Lecuit M, Buchrieser C, Glaser P, et al. Listeria monocytogenes bile salt hydrolase is a PrfA-regulated virulence factor involved in the intestinal and hepatic phases of listeriosis. Molecular microbiology. 2002;45(4):1095–106. . [DOI] [PubMed] [Google Scholar]
- 56.Di Martino ML, Campilongo R, Casalino M, Micheli G, Colonna B, Prosseda G. Polyamines: emerging players in bacteria-host interactions. Int J Med Microbiol. 2013;303(8):484–91. 10.1016/j.ijmm.2013.06.008 . [DOI] [PubMed] [Google Scholar]
- 57.Whitworth T, Popov VL, Yu XJ, Walker DH, Bouyer DH. Expression of the Rickettsia prowazekii pld or tlyC gene in Salmonella enterica serovar Typhimurium mediates phagosomal escape. Infection and immunity. 2005;73(10):6668–73. 10.1128/IAI.73.10.6668-6673.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ludu JS, de Bruin OM, Duplantis BN, Schmerk CL, Chou AY, Elkins KL, et al. The Francisella pathogenicity island protein PdpD is required for full virulence and associates with homologues of the type VI secretion system. J Bacteriol. 2008;190(13):4584–95. JB.00198-08 [pii] 10.1128/JB.00198-08 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zogaj X, Chakraborty S, Liu J, Thanassi DG, Klose KE. Characterization of the Francisella tularensis subsp. novicida type IV pilus. Microbiology. 2008;154(Pt 7):2139–50. 154/7/2139 [pii] 10.1099/mic.0.2008/018077-0 . [DOI] [PubMed] [Google Scholar]
- 60.Ramakrishnan G, Sen B, Johnson R. Paralogous outer membrane proteins mediate uptake of different forms of iron and synergistically govern virulence in Francisella tularensis tularensis. J Biol Chem. 2012;287(30):25191–202. 10.1074/jbc.M112.371856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Twine S, Bystrom M, Chen W, Forsman M, Golovliov I, Johansson A, et al. A mutant of Francisella tularensis strain Schu S4 lacking the ability to express a 58-kilodalton protein is attenuated for virulence and is an effective live vaccine. Infection and immunity. 2005;73:8345–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nallaparaju KC, Yu JJ, Rodriguez SA, Zogaj X, Manam S, Guentzel MN, et al. Evasion of IFN-gamma signaling by Francisella novicida is dependent upon Francisella outer membrane protein C. PloS one. 2011;6(3):e18201 10.1371/journal.pone.0018201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Forslund AL, Kuoppa K, Svensson K, Salomonsson E, Johansson A, Bystrom M, et al. Direct repeat-mediated deletion of a type IV pilin gene results in major virulence attenuation of Francisella tularensis. Molecular microbiology. 2006;59:1818–30. [DOI] [PubMed] [Google Scholar]
- 64.Miyashiro T, Klein W, Oehlert D, Cao X, Schwartzman J, Ruby EG. The N-acetyl-D-glucosamine repressor NagC of Vibrio fischeri facilitates colonization of Euprymna scolopes. Molecular microbiology. 2011;82(4):894–903. 10.1111/j.1365-2958.2011.07858.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brotcke A, Weiss DS, Kim CC, Chain P, Malfatti S, Garcia E, et al. Identification of MglA-regulated genes reveals novel virulence factors in Francisella tularensis. Infection and immunity. 2006;74(12):6642–55. IAI.01250-06 [pii] 10.1128/IAI.01250-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All files are already publicly available from the GEO database (accession numbers,GPL20119, GSE68478).