Abstract
The transcription factor T-box 16 (Tbx16/Spadetail) is an essential regulator of paraxial mesoderm development in zebrafish (Danio rerio). Mesodermal progenitor cells (MPCs) fail to differentiate into trunk somites in tbx16 mutants and instead accumulate within the tailbud in an immature state. The mechanisms by which Tbx16 controls mesoderm patterning have remained enigmatic, and we describe here the application of photoactivatable morpholino oligonucleotides to determine the Tbx16 transcriptome in MPCs. We identify 124 Tbx16-regulated genes that are expressed in zebrafish gastrulae, including several developmental signaling proteins and regulators of gastrulation, myogenesis, and somitogenesis. Unexpectedly, we observe that loss of Tbx16 function precociously activates posterior hox genes in MPCs, and overexpression of a single posterior hox gene is sufficient to disrupt MPC migration. Our studies support a model in which Tbx16 regulates the timing of collinear hox gene activation to coordinate the anterior-posterior fates and positions of paraxial MPCs.
INTRODUCTION
The spine is the defining characteristic of vertebrate organisms. Composed of repeating segments with distinct anterior-posterior identities, it protects the spinal cord and internal organs, provides structural support, and allows flexible motion. Fate mapping studies in vertebrate models have revealed that the spine originates from mesodermal progenitor cells (MPCs) that are induced during gastrulation.1 As the embryonic body axis forms and elongates, MPCs ingress from the epiblast into the paraxial mesoderm, where they differentiate and form segmented, paired blocks of tissue called somites. The somites then give rise to the vertebral column, skeletal muscle, and connective tissues.
Dysregulation of these processes can lead to scoliosis, spondylocostal dysostosis, and other congenital spine deformities.2 Studies of mouse, chick, and zebrafish embryos have uncovered critical roles for Notch, Wnt, fibroblast growth factor (FGF), and retinoic acid signaling in the segmentation process,3 and human spine abnormalities have been linked to congenital mutations in Notch pathway regulators.4–7 The molecular mechanisms that control anterior-posterior patterning within the paraxial mesoderm are less well understood, but a few key regulatory genes have been identified through mutant screens.8,9 In particular, zebrafish with loss-of-function mutations in T-box 16 (tbx16/spadetail) lack trunk somites and have abnormally large tailbuds.10,11 Consistent with these phenotypes, tbx16 expression is largely restricted to MPCs within the ventrolateral margin of zebrafish gastrulae and subsequently in the tailbud.12 As the MPCs migrate from these sites into the presomitic mesoderm, their tbx16 levels are downregulated.
Transplantation studies have shown that tbx16 mutant cells have impaired dorsal convergence and ingression during gastrulation, resulting in their accumulation within the tailbud.11,13 Based on these defects and the plasticity exhibited by similarly transplanted wild type cells, it is generally believed that the mislocalization of Tbx16-deficient MPCs precedes and directs changes in cell fate.13 Consistent with this idea, Tbx16 regulates the expression of protocadherin 8 (pcdh8),12,14 a cell adhesion molecule that promotes dorsal convergence, and mesogenin 1 (msgn1),15 a basic helix-loop-helix (bHLH) transcription factor that directly activates epithelial-mesenchymal transition (EMT) programs.16 These genes are transcribed in a Tbx16-dependent manner during gastrulation and early somitogenesis and then independently of Tbx16 function at late-somite stages.12,14,15
Mutant analyses have also revealed functional interactions between Tbx16 and morphogen signaling pathways during paraxial mesoderm development. Zebrafish double mutants lacking tbx16 and the Nodal receptor co-factor one-eyed pinhead (oep/teratocarcinoma-derived growth factor 1) are completely devoid of somites and exhibit larger tailbuds than those of tbx16 mutants.15 Similar phenotypes are observed in embryos with mutations in both tbx16 and the bone morphogenetic protein (BMP) antagonist chordin (chd).17 By transplanting cells overexpressing Nodal or BMP pathway regulators into wild type embryos, it has been shown that these signaling pathways can act prior to gastrulation to control MPC position along the anterior-posterior axis.18 Thus, Tbx16 may function in concert with Nodal and BMP morphogens to establish paraxial MPC fates.
Gene expression microarrays have provided additional insights into Tbx16 function. For example, transcriptomic analyses of wild type gastrulae and synchronous tbx16 morphants (embryos injected with antisense morpholino oligonucleotides) identified 15 genes that require Tbx16 for their expression.19 A complementary examination of tissues derived from 21-somite-stage wild type and tbx16 mutant embryos corroborated 8 of these targets and revealed another 6 Tbx16-dependent transcripts.20 While each survey identified Tbx16-regulated genes that contribute to myogenesis and somitogenesis, these targets likely represent a small fraction of the Tbx16 transcriptome in paraxial MPCs. The studies utilized embryos with constitutive, global loss of Tbx16 activity and analyzed the transcriptomes of either whole gastrulae or dissected middle tail regions from later-stage embryos. Thus, neither investigation was specifically designed to identify early Tbx16-dependent genes in presomitic MPCs.
We report here the application of photoactivatable caged morpholinos (cMOs) to elucidate the Tbx16 transcriptome during paraxial mesoderm development. cMOs enable greater spatiotemporal control of embryonic gene function than conventional mutant and morphant analyses,21,22 while avoiding the genetic compensatory mechanisms that can be induced by loss-of-function mutations.23 In our approach, zebrafish embryos are injected with a cMO and a photoactivatable lineage tracer, and the reagents are subsequently uncaged in specific cells of interest.24 Upon knockdown of the targeted gene, the embryos can then be dissociated into single cells, allowing the irradiated population to be isolated by fluorescence-activated cell sorting (FACS) and analyzed by mRNA profiling. Our studies specifically targeted ventral margin-derived MPCs that reside within a “no convergence, no extension” zone25 and contribute to trunk somites, allowing the roles of Tbx16 in MPC fate choice to be discerned from Tbx16-dependent movement. Through this optochemical strategy, we have identified 124 Tbx16-dependent genes, including developmental signaling proteins and regulators of gastrulation, myogenesis, and somitogenesis. Our results uncover an unexpected role for Tbx16 in posterior hox gene repression that is independent of Tbx16-dependent cell movement. We further observe that overexpression of a single posterior hox gene is sufficient to mislocalize MPCs. Taken together, our results support a model in which Tbx16 regulates the timing of collinear hox gene activation in MPCs, thereby establishing anterior-posterior pattern within the paraxial mesoderm.
RESULTS
Tbx16 acts during gastrulation to promote myogenesis
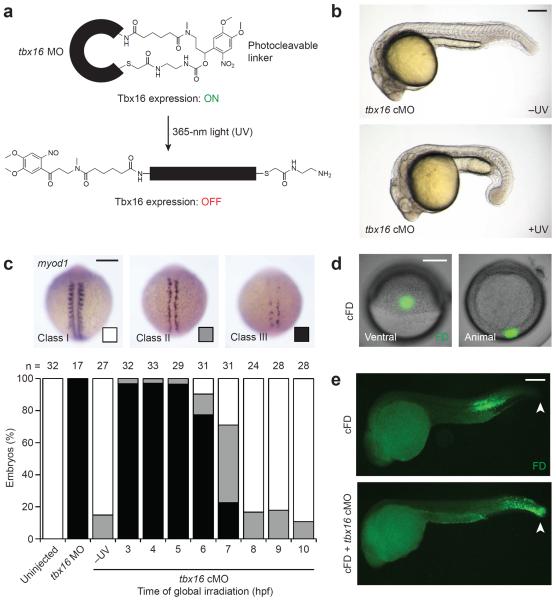
We recently developed a cyclic cMO that permits light-inducible knockdown of Tbx16 expression (Fig. 1a).26,27 Zebrafish embryos injected with this chemical probe and globally irradiated with ultraviolet (UV) light at the early blastula stage (3 hours post fertilization; hpf) failed to form trunk somites and acquired an enlarged tailbud, phenocopying the mesodermal defects observed in tbx16 mutants (Fig. 1b).11 In contrast, tbx16 cMO-injected embryos developed normally when cultured in the dark.
Figure 1. Optochemical regulation of Tbx16-dependent paraxial mesoderm development.
(a) Schematic representation of a cyclic tbx16 cMO and its photoactivation. (b) Phenotypes observed in embryos injected with the tbx16 cMO and cultured in the dark (−UV) or globally irradiated at 3 hpf (+UV). (c) Phenotypic classes of myod1 expression observed at 13 hpf and the distributions associated with global irradiation of tbx16 cMO-injected embryos at the designated times. (d) Live embryos injected with cFD and irradiated within the ventral margin at 6 hpf to target trunk somite progenitors. (e) Immunostaining of uncaged fluorescein in fixed 24-hpf embryos that were previously injected with cFD alone or a cFD/tbx16 cMO mixture and then irradiated as described for panel d. White arrowheads indicate the tailbud. Embryo orientations: a–b and e, lateral view, anterior left; c, dorsal view, anterior up; d, ventral and animal pole views. Scale bars: 200 μm.
Taking advantage of the conditionality afforded by cMOs, we characterized the timing by which Tbx16 promotes paraxial MPC differentiation into trunk somitic muscle. Wild type zebrafish transcribe myogenic differentiation 1 (myod1) in paraxial and adaxial cells during somitogenesis, and expression of this myogenic bHLH protein is largely extinguished in Tbx16-deficient embryos.28 We injected 1- to 4-cell-stage embryos with the tbx16 cMO and globally irradiated them at varying time points between 3 hpf and the end of gastrulation (10 hpf). myod1 expression phenotypes at 13 hpf were then assessed by whole-mount in situ hybridization and classified according to their severity (Fig. 1c): class I = wild type-like expression, class II = loss of paraxial but not adaxial expression; and class III = loss of expression in both mesodermal tissues. Uncaging of the tbx16 cMO by 6 hpf, the beginning of gastrulation, was required to recapitulate the near-complete loss of myod1-expressing cells in tbx16 mutants.28 We also determined by whole-mount immunostaining that Tbx16 protein is maximally depleted within 2 to 3 hours after cMO activation (Supplementary Results, Supplementary Fig. 1), indicating that Tbx16 acts during gastrulation to drive myogenesis.
Targeting Tbx16 function in trunk somite progenitors
Having established that Tbx16 is required during gastrulation for muscle cell formation, we sought a method for specifically targeting Tbx16 function within trunk somite progenitors at this stage. Zebrafish fate-mapping studies have shown that mesodermal and endodermal progenitors reside near the blastoderm margin,29 with those located more than four cell diameters above the margin differentiating exclusively into mesoderm.30 To identify a MPC population destined to become trunk muscle, we injected 1- to 4-cell-stage embryos with caged fluorescein dextran (cFD), and UV-irradiated individual 100-μm-diameter regions at 6 hpf. Ventral cells located approximately five cell layers above the margin consistently gave rise to trunk somites by 24 hpf (Fig. 1d–e). We next injected zebrafish embryos with a cFD/tbx16 cMO mixture and irradiated the ventral cells described above at 6 hpf. Rather than contributing to trunk somites, the targeted cells became mislocalized posteriorly by 24 hpf and did not acquire the elongated morphology of skeletal muscle fibers (Fig. 1e). Tbx16 activity was also significantly reduced in these cells and their derivatives by 8 hpf, as determined by whole-mount immunostaining (Supplementary Fig. 2).
To examine the dynamic behaviors of the targeted ventral cells, we then conducted time-lapse imaging of embryos injected with mRNA encoding Kaede-NLS, a nuclear photoconvertible protein.31 UV irradiation of these cells at 6 hpf labeled two cell populations: epiblast cells that moved toward the vegetal pole during gastrulation and localized to the margin by 9 hpf, and hypoblast cells that moved immediately toward the animal pole (Supplementary Fig. 3 and Supplementary Movie 1). Equivalently targeted cells in embryos co-injected with Kaede-NLS mRNA and the tbx16 cMO exhibited comparable distributions and movements. Tbx16 is therefore dispensable for the migration of either ventral cell population during gastrulation, consistent with their residence within the “no convergence, no extension” zone.25
At later developmental stages, the ventral epiblast-derived cells exhibited Tbx16-dependent movement. In zebrafish embryos injected with Kaede-NLS mRNA alone, these irradiated cells contributed to the posterior domain of the tailbud at 10.5 hpf, ingressed into the presomitic mesoderm, and then differentiated into somites by 17 hpf (Supplementary Fig. 4 and Supplementary Movie 2). In contrast, the equivalent irradiated population in embryos injected with a Kaede-NLS mRNA/tbx16 cMO mixture failed to enter the presomitic mesoderm and accumulated within the tailbud. Notably, none of these regiospecific perturbations grossly affected overall somite patterning by 24 hpf, suggesting that paraxial MPCs from non-irradiated regions were able to compensate for the Tbx16-deficient progenitors retained in the tailbud (Supplementary Fig. 5). Moreover, the targeted ventral cells did not display Tbx16-dependent movement at a time when Tbx16 is required for myogenesis, suggesting that paraxial MPC fate can be independent of cell position.
Determination of the Tbx16 transcriptome in paraxial MPCs
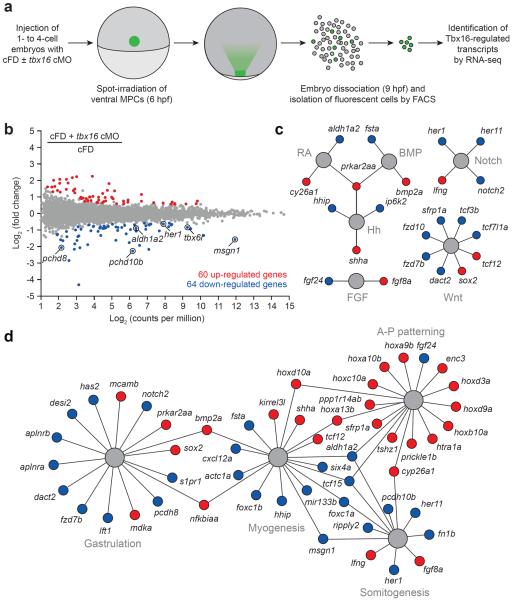
We next applied our optochemical probes to identify Tbx16-dependent transcripts in ventral margin-derived MPCs (Fig. 2a). We injected 1- to 4-cell-stage embryos with a cFD/tbx16 cMO mixture and UV-irradiated a 100-μm-diameter region above the ventral margin at 6 hpf as before. The embryos were cultured until 9 hpf, by which time Tbx16 levels in the targeted cells would be significantly reduced, and then dissociated them into single cells. Fluorescein-positive cells were isolated by FACS (Supplementary Fig. 6), and their transcripts were analyzed by RNA-seq. Embryos injected with cFD alone and subjected to identical conditions were used to obtain comparison controls.
Figure 2. Optochemical analysis of the Tbx16-dependent transcriptome in trunk somite progenitors.
(a) cMO-based profiling of the Tbx16 transcriptome in ventral margin-derived MPCs. (b) Whole-transcriptome comparison of genes expressed in the targeted MPCs after cFD or cFD/tbx16 cMO photoactivation. Differentially expressed genes were identified by EdgeR statistical analysis (fold-change ≥ 1.5 and false discovery rate < 0.01). Tbx16 targets identified in a previous microarray studies19,20 are circled. (c) Tbx16-regulated genes that participate in developmental signaling pathways. (d) Tbx16-regulated genes with established roles in gastrulation, myogenesis, somitogneesis, and anterior-posterior patterning. Functional networks depicted in c and d are based on gene descriptions available through Ingenuity Pathway Analysis (IPA) tools and previous studies, and genes that are up- or downregulated upon loss of Tbx16 function are depicted as red or blue circles, respectively. IPA gene set terms and statistical enrichment values: gastrulation (“epithelial-mesenchymal transition”; P-value = 3.97E-06), myogenesis (“formation of muscle”; P-value = 1.01E-09), somitogenesis (“development of somites”; P-value = 1.85E-06), and anterior-posterior patterning (“patterning of rostrocaudal axis”; P-value = 8.18E-13).
Through this approach, we identified 64 genes that are downregulated upon loss of Tbx16 function (fold change ≥ 1.5 and false discovery rate < 0.01; Fig. 2b and Supplementary Table 1), including 6 of the 21 Tbx16 targets identified in the two previous microarray-based screens.19,20 We also observed 60 upregulated genes (Fig. 2b and Supplementary Table 2), consistent with the ability of Tbx16 to act as a transcriptional repressor.32 These candidate Tbx16 targets include regulators of BMP, FGF, Hedgehog (Hh), Notch, retinoic acid, and Wnt signaling (Fig. 2c). Functional network analyses also revealed enrichment for genes involved in gastrulation, myogenesis, and somitogenesis (Fig. 2d).
To confirm that these genes are transcribed in a Tbx16-dependent manner, we compared the expression patterns of representative targets in 10-hpf wild type and tbx16 morphant embryos by whole-mount in situ hybridization. We analyzed 33 of the 124 genes and confirmed Tbx16-regulated transcript levels in every case (Supplementary Figs. 7 and 8). Consistent with the movement of ventral MPCs to the tailbud by 10 hpf, several of the Tbx16-dependent genes are expressed in this region. We also observed Tbx16-regulated mRNAs within the adaxial and paraxial mesoderm, perhaps due to the concomitant targeting of underlying hypoblast cells by our optochemical approach.
Tbx16 suppresses posterior hox gene activation
Our studies also uncovered a Tbx16-regulated ensemble of anterior-posterior patterning genes, including eight hox transcription factors that were upregulated upon Tbx16 knockdown (Fig. 2d). Vertebrates have 13 groups of Hox paralogs, which in zebrafish are distributed among seven distinct genomic clusters (hoxaa, hoxab, hoxba, hoxbb, hoxca, hoxcb, and hoxda).33 Hox genes are collinearly activated within each cluster in a 3'-to-5' order, generating nested expression domains that are increasingly restricted to the posterior end.34 Posterior 5' Hox genes (e.g., Hox13 paralogs) are functionally dominant over their anterior 3' counterparts (e.g., Hox1 paralogs), and the Hox code at a given location dictates specific rostrocaudal fates.
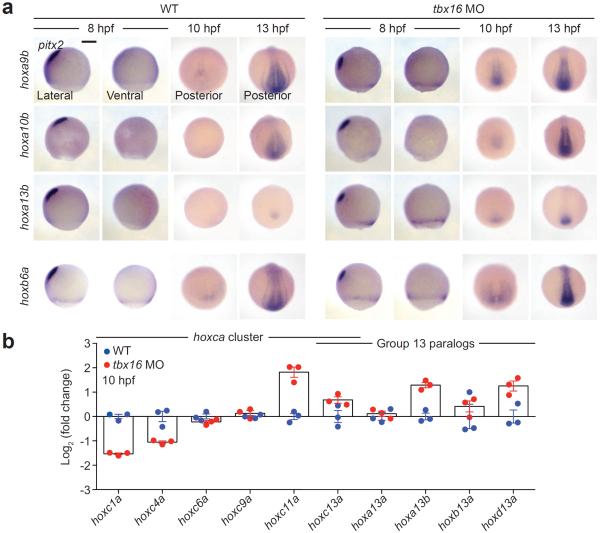
Of the eight Tbx16-regulated hox genes identified by RNA-seq, seven are considered posterior in class (groups 9–13), suggesting that Tbx16 might suppress collinear hox gene activation. To characterize the expression dynamics of these genes, we evaluated their mRNA levels in wild type and tbx16 morphant embryos. Posterior hox gene transcripts were undetectable in 8-hpf wild type embryos by in situ hybridization and could only be detected at low levels within the tailbud at 10 hpf (Fig. 3a and Supplementary Fig. 9). However, hoxa9b, hoxa13b, hoxb10a, and hoxc10a were visibly expressed in the ventrolateral margin of 8-hpf tbx16 morphants, and all seven posterior hox genes were transcribed within the tailbud by 10 hpf. In comparison, transcripts encoding the more anterior hox gene hoxb6a could be detected in both wild type and tbx16 morphant embryos at 8 hpf, with slightly higher levels observed in the latter. Genetic loss of tbx16 function similarly induced early expression of posterior hox genes (Supplementary Fig. 10). The upregulated hox genes were still expressed in a nested manner (e.g., 10-hpf tbx16 morphants in Fig. 3a and Supplementary Fig. 9), suggesting that Tbx16 regulates the timing of collinear activation rather than the independent expression of individual hox genes.
Figure 3. Tbx16 regulates hox gene activation during gastrulation.
(a) hox gene expression in wild type and tbx16 morphant embryos during mid-gastrulation (8 hpf), tailbud formation (10 hpf), and early somitogenesis (13 hpf) as determined by in situ hybridization. Representative posterior hox genes identified in our transcriptome-wide survey for Tbx16 targets are shown, and the trunk hox gene hoxb6a is included for comparison. pitx2 expression in the prechordal mesoderm was used as a dorsal marker. Embryo orientations: lateral view, dorsal left; ventral view, anterior up; and posterior view, dorsal up. Scale bar: 200 μm. (b) Transcript levels of representative hoxca cluster genes and group 13 paralogs in whole wild type and tbx16 morphant embryos (10 hpf) as determined by quantitative RT-PCR. Average relative expression levels ± s.e.m. for three biological replicates are shown.
To assess the generality of these Tbx16/hox interactions, we examined the expression of representative anterior, trunk, and posterior genes within the hoxca cluster (hoxc1a, hoxc4a, hoxc6a, hoxc9a, hoxc11a, and hoxc13a) and group 13 paralogs across several hox clusters (hoxa13a, hoxa13b, hoxb13a, hoxc13a, and hoxd13a). Using quantitative RT-PCR, we compared transcript levels for each hox gene in wild type or tbx16 morphant embryos at 10 hpf. Tbx16 knockdown suppressed 3' genes and activated 5' genes within the hoxca cluster, while upregulating hox13 paralogs across multiple clusters (Fig. 3b). We also investigated whether Tbx16 primarily regulates hox gene expression in mesodermal cells, focusing on hox10 paralogs (hoxa10b, hoxb10a, hoxc10a, and hoxd10a) that are transcribed in both the neural tube and paraxial mesoderm. In wild type embryos, the anterior limit of expression for a given hox10 paralog is comparable between both tissues at 24 hpf (Supplementary Fig. 11), and loss of Tbx16 function shifted the anterior boundary in somitic but not neural tissues. Taken together, these results reveal an early role for Tbx16 in establishing hox codes in paraxial MPCs.
Non-cell-autonomous signals contribute to Tbx16 function
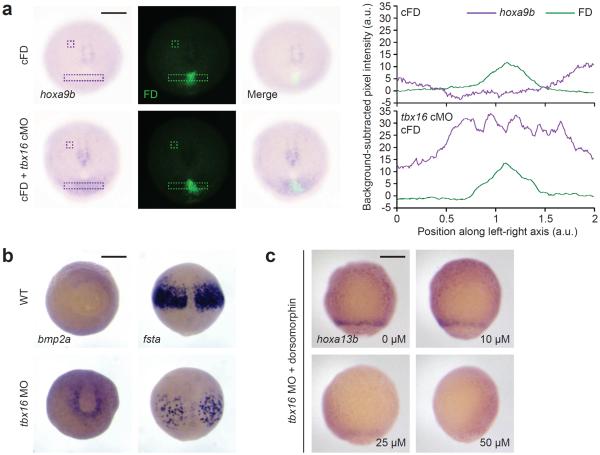
In principle, Tbx16 could regulate hox gene activation through cell-autonomous and/or non-cell-autonomous mechanisms. To determine whether posterior hox gene upregulation is strictly restricted to MPCs lacking Tbx16 function, we injected 1- to 4-cell-stage embryos with either cFD alone or a cFD/tbx16 cMO mixture and irradiated a 100-μm-diameter region within the ventral margin at 6 hpf. In comparison to embryos injected only with cFD, those injected with the cFD/tbx16 cMO mixture upregulated hoxa9b in the developing tailbud at 10 hpf (Fig. 4a). hoxa9b expression in these embryos extended beyond the fluorescein-positive irradiated cells, suggesting that non-cell-autonomous signals can couple Tbx16 activity to hox gene expression in neighboring tissues.
Figure 4. Non-cell-autonomous signals propagate posterior hox gene activation in Tbx16-deficient embryos.
(a) hoxa9b expression in 10-hpf zebrafish embryos that were previously injected with cFD alone or a cFD/tbx16 cMO mixture and irradiated within the ventral margin at 6 hpf. Contrast-enhanced micrographs are shown (left), and unprocessed images were used to determine integrated pixel-intensity profiles (right) across the indicated regions (dashed rectangles) and background signals (dashed squares). Note that the hoxa9b expression domain in cFD/tbx16 cMO-injected embryos extends beyond the irradiated cells, which are labeled with anti-fluorescein antibodies. (b) Staining of bmp2a transcripts in the ventral lateral margin and fsta transcripts in the anterior paraxial mesoderm of wild type and tbx16 morphant embryos at 9 hpf. (c) hoxa13b expression in 8-hpf tbx16 morphants that were treated with the indicated doses of dorsomorphin from 4 hpf onward. In comparison, wild type embryos do not express hoxa13b at this stage (see Fig. 3a). Embryo orientations: a, vegetal pole view, dorsal up; b, bmp2a, vegetal pole view and dorsal up; fsta, dorsal view and animal pole up; c, ventral view, animal pole up. Scale bars: 200 μm.
BMP signaling is one likely mediator of such intercellular interactions, given the synergistic effects of tbx16 and chd mutations on paraxial mesoderm development17 and the effects of BMP pathway perturbations on MPC distributions along the anterior-posterior axis.18 Our transcriptomic analyses also demonstrate that loss of Tbx16 function upregulates bmp2a and downregulates the BMP antagonist follistatin-a (fsta) (Fig. 2c and Supplementary Tables 1 and 2). We validated this functional interaction by in situ hybridization, observing increased bmp2a expression in the ventral lateral margin of tbx16 morphants at 8 hpf and diminished fsta expression within the anterior paraxial mesoderm (Fig. 4b). Genetic loss of tbx16 function elicited similar transcript level changes (Supplementary Fig. 12). In addition, the BMP receptor antagonist dorsomorphin35 suppressed hoxa13b upregulation in tbx16 morphants (Fig. 4c), suggesting that Tbx16 controls hox gene activation at least in part by suppressing BMP signaling.
hoxa13b expression alters paraxial MPC movement
The timing and location of posterior hox gene activation in Tbx16-deficient embryos argues against the prevailing hypothesis that paraxial MPC mislocalization precedes and directs changes in cell fate. Ectopic hox gene expression can be observed by mid-gastrulation, and ventral-most cells in the margin exhibit the highest transcript levels, even though their movements at this developmental stage are unaffected by Tbx16 knockdown. Our observations support an alternative paradigm: Tbx16 controls the timing of collinear hox gene activation in MPCs, and the resulting hox codes determine the anterior-posterior fates and subsequent positions of these cells. According to this model, ectopic posterior hox gene expression should impede MPC ingression into the presomitic mesoderm, leading to posterior accumulation of these cells.
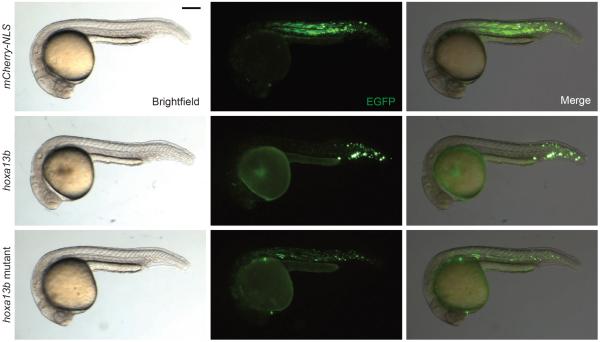
To test this hypothesis, we prepared plasmid vectors that utilize a 7.5-kb region of the tbx16 promoter36 to mosaically co-express enhanced green fluorescent protein (EGFP) and a gene of interest in MPCs, linked through a ribosomal “skipping” P2A sequence37 (Supplementary Fig. 13). Embryos injected with a tbx16:EGFP-P2A-mCherry-NLS construct at the 1- to 4-cell stage had somitic EGFP-positive cells along the entire anterior-posterior axis by 24 hpf. In contrast, EGFP-positive cells in embryos injected with a tbx16:EGFP-P2A-hoxa13b construct were predominantly localized to the posterior third of the body axis, failed to differentiate into muscle, and appeared to aggregate (Fig. 5). These hoxa13b-dependent changes in MPC behavior could be reversed by mutating residues within the DNA-binding domain. Overexpressing hoxa10b or hoxd9a but not hoxa9b could also posteriorize the MPC distribution along the body axis (Supplementary Fig. 14).
Figure 5. hoxa13b expression is sufficient to posteriorize paraxial MPCs.
Representative anterior-posterior distributions of EGFP-positive MPCs that co-express either mCherry-NLS, hoxa13b, or hoxa13b mutant transgenes. 22- to 24-hpf embryos are shown in lateral view, anterior left. Scale bar: 200 μm.
Precocious posterior hox gene activation did not appear to alter the mesodermal specification of MPCs. For example, hoxa13b-overexpressing cells continued to intermix with wild type MPCs and express Tbx16 at 12 hpf, at least seven hours after the initiation of transgene expression (Supplementary Fig. 15). Time-lapse imaging of the mosaic embryos further revealed that overexpression of hoxa13b but not mCherry-NLS impedes MPC movement into the presomitic mesoderm within this time frame (Supplementary Fig. 16 and Supplementary Movie 3). These results demonstrate the ability of individual posterior hox genes to alter the anterior-posterior identities and distributions of paraxial MPCs. The relative effects of hoxa9b, hoxa10b, and hoxa13b overexpression further suggest that these hox gene-induced changes may increase in a collinear manner.
DISCUSSION
The tbx16 mutant was the first reported zebrafish line with developmental defects, characterized by a striking loss of trunk mesoderm and an enlarged spade-like tailbud containing undifferentiated MPCs. Since its discovery in 1989 and molecular characterization nine years later,10,12 the tbx16 gene has been recognized as a critical regulator of paraxial mesoderm development. However, how this T-box transcription factor regulates paraxial mesoderm patterning has remained elusive. Earlier investigations of the Tbx16 transcriptome utilized whole gastrulae or dissected tissues of later-stage embryos, revealing a total of 21 Tbx16-dependent transcripts.19,20 Genes identified in both studies included cell migration regulators such as pcdh8, pchd10, and msgn1, consistent with the prevailing hypothesis that inappropriate MPC migration underlies the mesodermal defects in Tbx16-deficient embryos. According to this model, the posterior mislocalization of these progenitor cells precedes corresponding changes in fate.
We have used optochemical probes, flow cytometry, and whole-transcriptome sequencing to specifically interrogate the Tbx16 transcriptome in paraxial MPCs. Our cMO technology enabled us to define gastrulation as the time of Tbx16 action in myogenesis and to identify a ventral margin-derived population of MPCs that gives rise to trunk somites without undergoing dorsal convergence and extension. Through this targeted approach, we have identified 124 Tbx16-regulated transcripts that are independent of Tbx16-controlled cell movement, with comparable numbers of upregulated and downregulated genes.
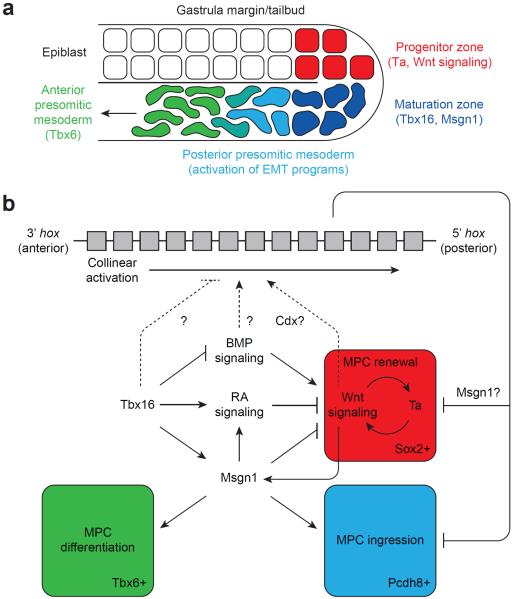
Previous studies have revealed an autoregulatory feedback loop involving the Brachyury/T homolog Ta (also known as No tail-a) and Wnt signaling that maintains an undifferentiated, self-renewing pool of MPCs in the zebrafish tailbud epiblast (Fig. 6).38 These mesodermal progenitors express the neural stem cell marker Sox2, indicative of their bipotential character.39 Msgn1 is required to suppress this renewal program and permit MPCs to undergo EMT, ingress into the hypoblast, and differentiate into Tbx6-expressing presomitic cells.16,40 BMP and retinoic acid signaling have also been found to promote and inhibit Ta/Wnt-dependent MPC renewal, respectively.41–44 Our transcriptomic analysis indicates that Tbx16 inhibits this Ta/Wnt feedback loop through each of these regulatory nodes. Loss of Tbx16 function abrogates the initial phase of msgn1 expression, increases BMP signaling (modulation of bmp2a and fsta), and decreases retinoic acid signaling (modulation of aldh1a2 and cyp26a1). Tbx16 knockdown may also enhance Wnt signaling, as it downregulates the pathway antagonists sfrp1a and dact2. Accordingly, Tbx16-deficient MPCs upregulate sox2.
Figure 6. A model for Tbx16-dependent paraxial mesoderm patterning.
(a) Schematic representation of paraxial MPCs as they transition from proliferative to myogenic programs and migrate into the anterior presomitic mesoderm. Key factors associated with each developmental phase are indicated. (b) A proposed signaling network for the regulation of paraxial MPC renewal, movement, and differentiation. Our studies support a model in which Tbx16 suppresses the Ta/Wnt-dependent proliferation of epiblast MPCs and promotes their ingression into the presomitic mesoderm and differentiation into muscle. Tbx16 also regulates the timing of collinear hox gene activation, and the resulting hox codes coordinate the rates of MPC renewal, movement, and differentiation in space and time.
Our studies further reveal an unanticipated role for Tbx16 in regulating the collinear activation of hox genes. Several posterior hox genes are prematurely expressed in Tbx16-deficient gastrulae, appearing in nested domains along the dorsoventral (future anteroposterior) axis. These changes occur in ventral margin-derived MPCs during a period of Tbx16-independent movement, and the resulting MPC hox codes and anterior-posterior fates are therefore specified prior to any change in cell localization. In addition, overexpressing a single posterior hox gene in MPCs is sufficient to caudalize their distribution along the body axis.
These findings support an alternative model of Tbx16 function that integrates hox codes and previously characterized mechanisms of MPC self-renewal, migration, and differentiation (Fig. 6). In this paradigm, hox genes are collinearly activated in paraxial MPCs, establishing a spatiotemporal gradient of hox codes along the dorsoventral axis of zebrafish gastrulae and anteroposterior axis of somite-stage embryos. As hox gene activation proceeds in a 3'-to-5' manner, MPCs adopt increasingly posterior fates and enter the presomitic mesoderm at increasingly slower rates, thereby ensuring that cell fate and position are intrinsically linked. Tbx16 suppresses the onset and/or rate of collinear hox activation, and loss of this T-box factor leads to precocious expression of posterior hox genes and reduced MPC ingression. At the same time, MPC renewal is unconstrained in Tbx16-deficient embryos, at least until msgn1 is expressed through Tbx16-independent mechanisms during late somitogenesis. Assuming that body axis elongation and somitogenesis proceed at relatively normal rates, these changes in MPC proliferation, ingression, and differentiation rates can account for the selective loss of trunk somites and tailbud accumulation of immature MPCs in tbx16 mutants. A similar role for Hox genes has been observed during avian mesoderm development. Hox genes are collinearly activated in epiblast cells lateral to the primitive streak in gastrula-stage chick embryos.45 Furthermore, when Hox-overexpressing cells are introduced to this region by tissue grafting or cDNA electroporation, they enter the underlying presomitic mesoderm with rates that inversely correlate with hox group number.45,46 Our studies therefore provide a mechanistic framework for paraxial mesoderm development that may be evolutionarily conserved across vertebrates.
How Tbx16 regulates hox gene activation remains to be determined. Cell transplantation studies demonstrate that this transcription factor acts cell-autonomously in MPCs to control directed migration and non-cell-autonomously to effect migration speed and differentiation.13,47 Tbx16 may promote hox gene activation through either or both mechanisms, as localized tbx16 cMO photoactivation upregulates posterior hox genes in the targeted cells and flanking regions. Consistent with a non-cell-autonomous mode of action, Tbx16 knockdown increases BMP signaling, and BMP pathway inhibition can suppress posterior hox gene expression in tbx16 morphants. These signaling interactions could explain the dramatically enhanced tailbud phenotype observed in tbx16;chd double mutants.17 It has also been shown that a BMP-Wnt-Cdx-Hox pathway specifies hematopoietic fates in zebrafish,48 and tbx16 mutants have been reported to express higher levels of the caudal-type homeobox factors cdx1a and cdx4.49 While these findings suggest that a BMP-Wnt-Cdx-Hox pathway could be operative in paraxial MPCs, we do not observe increased expression of cdx1a, cdx4, or the Wnt target gene axin2 in Tbx16-deficient MPCs. A BMP-Wnt-Cdx-Hox signaling axis could still contribute to paraxial mesoderm patterning in wild type embryos, and Wnt pathway components regulate msgn1 transcription in zebrafish.16 One intriguing possibility is that proliferating MPCs use Wnt pathway activity to gate both collinear hox gene activation and msgn1 expression.
Understanding how Hox transcription factors regulate paraxial mesoderm development also awaits further study. Interestingly, overexpressing posterior Hox genes in chick epiblast cells was found to downregulate Brachyury/T expression, repress Wnt signaling, and promote retinoic acid signaling in a collinear fashion.46 These changes indicate that posterior Hox genes can block MPC renewal and promote migratory and differentiation pathways. On the other hand, we observe that Tbx16 knockdown promotes both posterior hox gene activation and Ta/Wnt-dependent MPC renewal. These seemingly contradictory findings can be reconciled if Hox proteins suppress the Ta/Wnt autoregulatory feedback loop through Msgn1-dependent mechanisms (Fig. 6). The initial loss of msgn1 expression in Tbx16-deficient embryos would consequently preclude the ability of posterior hox genes to suppress MPC renewal while leaving their effects on MPC migration intact. Indeed, integrating Hox and Msgn1 activities could embody how vertebrates modulate paraxial MPC behaviors in space and time to form distinct regions of the spine.
ONLINE METHODS
Zebrafish husbandry
Adult zebrafish (Danio rerio; AB strain) were obtained from the Zebrafish International Resource Center (ZIRC), and adult tbx16b104/+ zebrafish10 were generously provided by Sharon Amacher. Adult fish (3 to 18 months old) were raised according to standard protocols,50 and embryos were obtained through natural matings and then staged as described previously.51 All zebrafish experiments were approved by the Stanford University Administrative Panel on Laboratory Animal Care.
Morpholino reagents and lineage tracers
tbx16 MO (5'-CTCTGATAGCCTGCATTATTTAGCC-3') was purchased from Gene-Tools, LLC and used at a 1.5 ng/embryo dose. A photoactivatable tbx16 cMO with the same oligonucleotide sequence was synthesized as described22,27 and used at a 1 ng/embryo dose. Caged fluorescein-conjugated dextran (cFD) was synthesized as reported24 and 3 nL of a 0.2% (w/v) cFD aqueous solution was injected per embryo.
To generate Kaede-NLS mRNA, the Kaede coding region with an upstream Kozak sequence was PCR-amplified from the HuC:Kaede plasmid52 (a gift from Hitoshi Okamoto) using the following primers: (5'-TACAAGCTACTTGTTCTTTTTGCAGGATCCTCCACCATGAGTCTGATTAAAC-3') and (5'-TCTTTTTTGGATCGGACTTGACGTTGTCCGGCA-3'). A nuclear localization signal (NLS) sequence was also PCR-amplified from the 8xGliBS-IVS2-mCherry-NLS-polyA-Tol2 plasmid53 using the following primers: (5'-GGACAACGTCAAGTCCGATCCAAAAAAGAAGAG-3') and (5'-AGTTCTAGAGGCTCGAGAGGCCTTGAATTCCATAAAATGAATGCAATTGTTGTTG-3'). pCS2+ (German Resource Center for Genome Research) was digested with BamHI and EcoRI, and pCS2+ Kaede-NLS was assembled from the digested pCS2+ plasmid and Kaede and NLS PCR products using Gibson Assembly Master Mix (NEB). The construct was then linearized with NotI and purified by phenol-chloroform extraction. Kaede mRNA was in vitro transcribed from pCS2+ Kaede-NLS using the mMessage mMachine SP6 Transcription Kit (Ambion) and used at a 75 pg/embryo dose.
Photoactivation of morpholinos and lineage tracers
Embryos injected with photoactivatable MOs and/or lineage tracers were globally or regiospecifically irradiated as described.27 For global irraditions, embryos were placed in a single-well of a 6-well microplate containing E3 medium, agitated continuously by vortexing, and irradiated for 15 minutes using a 365–370 nm light-emitting diode (LED) (Stanford Photonics) at an intensity of 10.6 mW/cm2. For regiospecific spot irradiations, chorionated 6-hpf embryos were arrayed in an agarose template (560-μm × 960-μm wells) filled with E3 medium, oriented with the ventral margin facing upward. The embryos were then irradiated using a Leica DM4500B upright compound microscope equipped with a mercury lamp, an A4 filter (Ex: 360/40 nm), and a 20x/0.5 NA water-immersion objective. An iris diaphragm was used to limit the targeted region to a 100-μm-diameter circle positioned approximately five cell layers above the margin, and the light intensity at the focal point was measured to be 40.9 mW/cm2. Embryos injected with cFD alone or in combination with the tbx16 cMO were irradiated for 15 seconds in this manner, and embryos injected with Kaede-NLS mRNA alone or in combination with the tbx16 cMO were irradiated for 60 seconds. Phenotype statistics for experiments using these photoactivatable probes are provided in Supplementary Table 3.
Whole-mount immunostaining and in situ hybridization
Whole-mount immunostaining was performed on paraformaldehyde-fixed embryos as previously described.27 The following antibodies were used: mouse monoclonal anti-Tbx16 (1:100 dilution, ZIRC, catalog number ZDB-ATB-081002-3), mouse monoclonal anti-fluorescein (1:200 dilution, Roche, catalog number 1426320), and rabbit polyclonal anti-fluorescein (1:50 dilution, Molecular Probes, catalog number A-889). Micrographs of the stained embryos were imaged with either a Leica DM4500B microscope equipped with 5x/0.12 N Plan and 10x/0.30 HC PL Fluotar objectives, GFP and TXR filters, and a Retiga-SRV Fast 1394 camera, a Leica M205FA microscope equipped with a 1.0x Planapochromatic objective, GFP2 510 LP and TXR 610 LP filters, and a Leica DFC500 camera, or a Zeiss Axio Imager Z1 upright microscope equipped with a 20x/0.5 NA water-immersion objective, fluorescence and transmission photomultiplier tube detectors, and an LSM700 laser-scanning confocal head.
Whole-mount in situ hybridization of RNA transcripts was performed according to standard procedures (digoxigenin-labeled riboprobes, anti-digoxigenin antibodies conjugated with alkaline phosphatase, and alkaline phosphatase-NBT/BCIP staining).54 The construct used to prepare myod1 probes has been described,28 and additional probes were prepared in the following manner. Zebrafish cDNA was prepared from RNA extracted from bud-stage (10 hpf), 8-somite (13 hpf), and 26-somite (22 hpf) embryos with an RNAqueous-Micro Kit (Ambion) and reverse-transcribed with the SuperScript III First-Strand Synthesis System (Life Technologies). PCR products containing a T7 promoter and gene-specific sequences were amplified with the designated primers (T7 sequence underlined):
actc1a (5'-CGTGTGCGACAATGGTTCTG-3' and 5'-CGTAATACGACTCACTATAGGGACGCATGATGGCATGAGGAA-3'), adam8a (5'-CTCAGTCAGCGTGGGACTTT-3' and 5'-CGTAATACGACTCACTATAGGGTCCTCAACAGTTCCGCAGTC-3'), aplnra (5'-GAAGGACTCAAAGCCAACGC-3' and 5'-CGTAATACGACTCACTATAGGGCAGTCCTGCAATCCAGAGGG-3'), aplnrb (5'-CTCGCTGACCTGACCTTTGT-3' and 5'-CGTAATACGACTCACTATAGGGTCACCACATGGAAGGGCATC-3', atp2a2a (5'-CACTGACACATAGTTCTTTTGGGG-3' and 5'-CGTAATACGACTCACTATAGGGGGTGGACTTGATGGCACTGA-3'), bmp2a (5'-CTCCAGTGGACTCGTTCCTC-3' and 5'-CGTAATACGACTCACTATAGGGTGTGAAGGGACCGACTTACG-3'), cxcl12a (5'-TGCCAAATATGCGTCCCAGT-3' and 5'-CGTAATACGACTCACTATAGGGGAGCGTGAAGCAACAGTGTG-3'), cyp26a1 (5'-GGTTTGAGGGCACGCAATTT-3' and 5'-CGTAATACGACTCACTATAGGGTAGTGCCCTCGTTTTCGCTT-3'), foxc1b (5'-TAAGCAAGGCTGGCAGAACA-3' and 5'-CGTAATACGACTCACTATAGGGACTGGACGGGAAAGCCATTT-3'), fsta (5'-TAACGTCACCTGGAAAGGGC-3' and 5'-CGTAATACGACTCACTATAGGGTGGGCAGCATTGGATTGTCT-3'), fzd10 (5'-TCATTGATCCCCAGCGCTTT-3' and 5'-CGTAATACGACTCACTATAGGGATGCAGCCTTCCTCCTTGTC-3'), fzd7b (5'-CGTCCCAGAGCATGGTTTCT-3' and 5'-CGTAATACGACTCACTATAGGGCGCAGTTTTTCGCTCCCAAA-3'), her11 (5'-GGCTTCCTCTTATCCCCACC-3' and 5'-CGTAATACGACTCACTATAGGGCCACTGCTGCACATACCAGT-3'), hivep2a (5'-TTCCATCCCCTCACTCGCTA-3' and 5'-CGTAATACGACTCACTATAGGGTGGAGCTACTGCTTATGGAGC-3'), hoxa9b (5'-ACTCACACCTTCCACACGAG-3' and 5'-CGTAATACGACTCACTATAGGGCATGCAGCCAGTTGGACAAA-3'), hoxa10b (5'-AATAACGGCTGTGTTCCGGT-3' and 5'-CGTAATACGACTCACTATAGGGACTCGCGGATTCGGTTTTCT-3'), hoxa13b (5'-TGTTCGGCCGTGCAAAATAC-3' and 5'-CGTAATACGACTCACTATAGGGACCAGATGGTGACTTGTCGC-3'), hoxb6a (5'-CCTCCGGATACACAGACCCT-3' and 5'-CGTAATACGACTCACTATAGGGTTCATTCGCCGGTTTTGGAAC-3'), hoxb10a (5'-CTGAACAGTGCAGACGACCA-3' and 5'-CGTAATACGACTCACTATAGGGGATTTGTGCGCTGAGTTCGG-3'), hoxc10a (5'-TGTTGAGGCACTAGCGTCAG-3' and 5'-CGTAATACGACTCACTATAGGGTTTGCAGGGTGTACCAGTCC-3'), hoxd3a (5'-CTACATCAGTCCGGCAAGCA-3' and 5'-CGTAATACGACTCACTATAGGGCGTACACGGGGCTACTTTGT-3'), hoxd9a (5'-GTCCTCGGTTCATCAGCCAT-3' and 5'-CGTAATACGACTCACTATAGGGGCATTAGCTTCTTGCACGCT-3'), hoxd10a (5'-TAGAGCAACCGCTCAACCAG-3' and 5'-CGTAATACGACTCACTATAGGGTGCGTGCTCACGAAAAAGTG-3'), meis1 (5'-TTTGCTCGATCCTTGTCGCT-3' and 5'-CGTAATACGACTCACTATAGGGTAGCGGCTTTTCTGCTCGAA-3'), mespab (5'-GCCAGATGCAAACTCGAACC-3' and 5'-CGTAATACGACTCACTATAGGGGCATGAGTCCCTGTTGTCCA-3'), msgn1 (5'-GGACATGGCGCAAATCGAC-3' and 5'-CGTAATACGACTCACTATAGGGGAGCGTCTGGATCTT GGTGA-3'), notch2 (5'-GCGAGTGTCCTCAGGGTTTC-3' and 5'-CGTAATACGACTCACTATAGGGGTGGTTTGCAGTGGCATACG-3'), pcdh10b (5'-CTTGAGCTTTATCACACGCCA-3' and 5'-CGTAATACGACTCACTATAGGGGTCGTTGATGTCCACCACCT-3'), rem1 (5'-CTCTGTGGGCAAAGTGGGAG-3') and 5'-CGTAATACGACTCACTATAGGGAAAGAGCTCGTGGACGTTGT-3'), ripply2 (5'-ACGCGGGGATTCTGATCG-3' and 5'-CGTAATACGACTCACTATAGGGACATTTACATTCAGCGTCCAGT-3'), s1pr1 (5'-GCTTCATAGCATGTTGGGCG-3' and 5'-CGTAATACGACTCACTATAGGGAGGGAGCAATCCCCGAAAAG-3'), tagln3b (5'-GACTGAGTCGTGAGGTGCAG-3' and 5'-CGTAATACGACTCACTATAGGGGAGATGCCCCACGGTTACTG-3'), tbx6l (5'-AAGGGCACTTTGCAGGTGAT-3' and 5'-CGTAATACGACTCACTATAGGGACTTCGTTTTGGTTTCTTGTTGAC-3'), tcf12 (5'-TGTATGAGTAGGGAGCGGGTT-3' and 5'-CGTAATACGACTCACTATAGGGGGTCCTCCATCCGAGACAGA-3'), and pitx2 (5'-CCTCCAGTCCAGAGTCCGTA-3' and 5'-CGTAATACGACTCACTATAGGGTCCATCACAGGATTGGACGC-3').
For each RNA transcript, in situ hybridization conditions (e.g., embryo clutch, concentration of the digoxigenin-labeled riboprobe, duration of alkaline phosphatase-NBT/BCIP staining, etc.) were identical across treatment protocols (e.g., wild type embryos versus tbx16 morphants) to enable qualitative comparisons. Phenotype statistics for in situ hybridization analyses are provided in Supplementary Table 3.
Images were digitally processed using Photoshop software (Adobe) to enhance contrast and/or brightness if necessary, and all adjustments were applied uniformly to the entire image without altering gamma settings. To quantify hoxa9b in situ hybridization and fluorescein dextran signals in Fig. 4a, Photoshop was used to convert the color brightfield and fluorescence micrographs to grayscale, and pixel intensities for the brightfield images were then inverted. For each designated region of interest, integrated pixel intensities for each position along the left-right axis were quantified using ImageJ, and corresponding regions without hoxa9b expression or fluorescein fluorescence were used to determine average background signals.
Quantitative RT-PCR
Total RNA was isolated from 15 wild-type or tbx16 morphant embryos at 10 hpf using the RNAqueous-Micro Kit, and 1.5 μg was converted into first-strand cDNA using the SuperScript III First-Strand Synthesis System. The resulting 20-μL solution of cDNA was diluted 1:10 with water, and 4.5 μL was used as the template for analyses with hoxc1a (Dr03125125_m1), hoxc4a (Dr03086656_m1), hoxc6a (Dr03112038_m1), hoxc9a (Dr03074138_m1), hoxc11a (Dr03074043_m1), hoxc13a (Dr03112201_m1), hoxa13a (Dr03124342_g1), hoxa13b (Dr03125011_m1), hoxb13a (Dr03078278_m1), hoxd13a (Dr03432701_m1), and eef1a1l1 (Dr03432748_m1) TaqMan quantitative PCR probes (Life Technologies) and a Light Cycler 480II (Roche). hox gene expression levels were normalized to that of eef1a1l1 (eukaryotic translation elongation factor 1, alpha 1, like 1), and biological triplicates and technical duplicates were used for each experimental condition.
Time-lapse microscopy
Chorionated embryos that had been injected with the tbx16 cMO and/or photoactivatable tracers and spot-irradiated at 6 hpf were placed in an agarose template (560-μm × 960-μm wells) filled with E3 medium, oriented with the dorsal shield facing upward. Imaging was performed with a Leica DMI6000B inverted compound microscope equipped with a UVI 6.3x/0.13 objective, GFP and TX2 filters, and a CoolSnap HQ2 monochrome camera controlled by MetaMorph microscopy automation and image analysis software (Molecular Devices). Fluorescent micrographs were acquired at rate of 1 frame per 5 minutes, with exposure times automatically adjusted every five frames to account for progressive photobleaching. ImageJ software (NIH) was used to normalize signal brightness and generate annotated movie montages.
RNA-sequencing library preparation and analysis
1-to 4-cell-stage zebrafish embryos were injected with cFD or a cFD/tbx16 cMO mixture and cultured until 6 hpf. For each experimental condition, 25 to 30 embryos were then individually spot-irradiated to photoactivate the probes within a 100-μm-diameter region approximately five cell layers above the ventral margin. Care was taken to alternate between cFD- and cFD/tbx16 cMO-injected embryos every five irradiations to distribute temporal variability between the two conditions. The embryos were then dechorionated with Pronase treatment at 9 hpf and dissociated into single cells as previously described.24 A BD FACSAria sorter was then used to isolate the population of viable, single fluorescein-positive cells, which were identified through forward, side-scatter, and fluorescence (Ex: 488 nm; Em: 530 nm) gating. An average of 6,300 fluorescein-positive cells were purified per sample and sorted directly into a 1.5-mL microfuge tube containing 0.5 mL of chilled lysis buffer from the RNAqueous-Micro Kit. Cell lysates were then generated by vortexing and passage through an 18–22 gauge needle, flash frozen in liquid nitrogen, and stored at −80 °C. Five paired biological replicates (cFD- and cFD/tbx16 cMO-injected embryos) were prepared in this manner.
After all five paired replicates were collected, the lysates were thawed and total RNA was isolated from each sample using the RNAqueous-Micro Kit. Agilent Bioanalyzer analysis was used to assess RNA quantity and quality, which averaged 24.8 ng and an RNA integrity number of 9.0, respectively. Library preparation proceeded according to the CEL-Seq protocol.55 Reverse-transcription reactions were performed using 20 ng of each total RNA preparation and the following primers, which contain an anchored polyT, unique barcode (underlined), a 5' Illumina adapter, and a T7 promoter. Fluorescein-positive cells isolated from irradiated, cFD-injected embryos: replicate 1 (5'-CGATTGAGGCCGGTAATACGACTCACTATAGGGGTTCAGAGTTCTACAGTCCGACGATCCATCACGCTTTTTTTTTTTTTTTTTTTTTTTTVN-3'), replicate 2 (5'-CGATTGAGGCCGGTAATACGACTCACTATAGGGGTTCAGAGTTCTACAGTCCGACGATCGTCGTTCCTTTTTTTTTTTTTTTTTTTTTTTTVN-3'), replicate 3 (5'-CGATTGAGGCCGGTAATACGACTCACTATAGGGGTTCAGAGTTCTACAGTCCGACGATCGTCGTGAATTTTTTTTTTTTTTTTTTTTTTTTVN-3'), replicate 4 (5'-CGATTGAGGCCGGTAATACGACTCACTATAGGGGTTCAGAGTTCTACAGTCCGACGATCTCACACGCTTTTTTTTTTTTTTTTTTTTTTTTVN-3'), replicate 5 (5'-CGATTGAGGCCGGTAATACGACTCACTATAGGGGTTCAGAGTTCTACAGTCCGACGATCTCACAGAGTTTTTTTTTTTTTTTTTTTTTTTTVN-3'); Fluorescein-positive cells isolated from irradiated, cFD/tbx16 cMO-injected embryos: replicate 1 (5'-CGATTGAGGCCGGTAATACGACTCACTATAGGGGTTCAGAGTTCTACAGTCCGACGATCTGATGCGCTTTTTTTTTTTTTTTTTTTTTTTTVN-3'), replicate 2 (5'-CGATTGAGGCCGGTAATACGACTCACTATAGGGGTTCAGAGTTCTACAGTCCGACGATCACGACTCCTTTTTTTTTTTTTTTTTTTTTTTTVN-3'), replicate 3 (5'-CGATTGAGGCCGGTAATACGACTCACTATAGGGGTTCAGAGTTCTACAGTCCGACGATCACGACGAATTTTTTTTTTTTTTTTTTTTTTTTVN-3'), replicate 4 (5'-CGATTGAGGCCGGTAATACGACTCACTATAGGGGTTCAGAGTTCTACAGTCCGACGATCATGTGCGCTTTTTTTTTTTTTTTTTTTTTTTTVN-3'), replicate 5 (5'-CGATTGAGGCCGGTAATACGACTCACTATAGGGGTTCAGAGTTCTACAGTCCGACGATCCGTGTGAGTTTTTTTTTTTTTTTTTTTTTTTTVN-3'). The resulting products were pooled and linearly amplified by T7-mediated in vitro transcription. Amplified RNA was then fragmented (peak size of approximately 500 bp), and 780 ng of the sheared RNA was treated with calf intestinal phosphatase and T4 polynucleotide kinase, ligated to a 3' Illumina adapter, reverse transcribed, and PCR amplified (11 cycles) using a TruSeq Small RNA Sample Preparation Kit (Illumina). The PCR product was purified using AMPure XP beads (Beckman Coulter), and Bioanalyzer analysis of the resulting library indicated an oligonucleotide concentration of 39.77 ng/μL and average size of 392 bp.
High-throughput sequencing was conducted on two lanes of an Illumina HiSeq 2000. Basecalls were made using the HiSeq control software (version 1.5.15). Paired-end reads passing the initial quality control from both lanes were concatenated head-to-tail. Data analysis was performed according to the CEL-seq protocol using a cloud-based instance of the Galaxy bioinformatics platform (galaxyproject.org) available through Amazon Web Services (aws.amazon.com) and custom scripts. The paired reads were de-multiplexed by a unique 8-bp barcode on read 1 followed by filtering and truncation of read 2 (quality score > 20; 35 bp). Read 2 was then aligned to the zebrafish Zv8 genome assembly, and feature counting was performed. Count files were analyzed using EdgeR (Bioconductor), and differentially expressed genes were statistically identified assuming batch effects. False discovery rate (< 0.01) and expression fold-change (absolute value ≥ 1.5) were used as thresholds. Differentially expressed genes were analyzed by Ingenuity Pathway Analysis software (Ingenuity Systems) to identify functional enrichments.
Mesoderm-targeted posterior hox gene expression
Using the zebrafish cDNA described above, the hoxa13b PCR product was amplified using the following primers: (5'-AAAAGAGCTGCTGGGCTCC-3' and 5'-GGTGCCTGAAAGCAACTTCG-3'). The resulting product was used as template for a second round of PCR amplification with the following nested primers, which contain BamHI and EcoRI sites (underlined): [5'-ACGTACGGATCCCACCATGACAGCGTCTTTACTCCTCC-3' (BamHI) and 5'-CATGCAGAATTCTTAACTGATGCCCTTGTACT-3' (EcoRI)]. This PCR product was then digested with BamHI and EcoRI and ligated into pCS2+ to generate a pCS2+ hoxa13b expression construct. A PCR fragment of the hoxa13b mutant containing N258A, R259A, and V261A point mutations within the conserved DNA-binding homeodomain was then cloned from the pCS2+ hoxa13b construct with following primers (mutated codons underlined): (5'-ATGACAGCGTCTTTACTCCTC-3' and 5'-ACTGATGCCCTTGTACTTGTTGACCACTTTCTTTTCTTTTACCGCCCTGGCCGCAAACCAGAT-3'). PCR fragments containing the hoxa9b, hoxa10b, or hoxd9a coding sequences were generated from the zebrafish cDNA using the following primers: hoxa9b (5'-TCAACGGATTCTGCTTTTGA-3' and 5'-CGCAGAACCTATTTCCCTGA-3'); hoxa10b (5'-CAATCTTCTTCCACCGTCAAA-3' and 5'-TGACGACTGGGTTGTCAAAT-3'); hoxd9a (5'-GAAGGTGAAGGCAGCAAAAA-3' and 5'-GAAACGCGCATTAGCTTCTT-3').
The pDest-Tol2-pA2 backbone was cloned from the pDest-Tol2-mpeg1-mCherry-pA2 plasmid56 (a gift from Graham Lieschke) using the following primers containing BamHI and EcoRI sites (underlined): [5'-CAAAGGGATCCAGACATGATAAGATAC-3' (BamHI) and 5'-CAAAGGAATTCCTCGAGGGCCCATCTGGCCTGTGTTTC-3' (EcoRI)]. The resulting PCR product was then digested with BamHI and EcoRI. pEGFP-N1-tbx16pr36 (a gift from Michael Lardelli) was digested with BamHI and EcoRI to obtain the 7.5-kb tbx16 promoter sequence, which was resolved by agarose gel electrophoresis, purified using a QIAquick Gel Extraction Kit (QIAGEN) and then ligated into BamHI- and EcoRI-digested pDest-Tol2-pA2 to create pDest-Tol2-tbx16-pA2.
An EGFP coding sequence flanked by Gibson adapters was cloned from pEGFP-N1-tbx16pr using the following primers: (5'-AGGCAATATCCGGATCACTCTGGAGGATCCACCGGTCGCCACCATGGT-3' and 5'-TAGTAGCTCCGCTTCCCTTGTACAGCTCGTCCATGCC-3'). mCherry-NLS, hoxa13b, hoxa13b mutant, hoxa9b, hoxa10b, and hoxd9a coding sequences flanked by Gibson adapters and a 5' P2A linker were cloned using the following templates and primers: mCherry-NLS (8xGliBS-IVS2-mCherry-NLS-polyA-Tol2, 5'-CGAGCTGTACAAGGGAAGCGGAGCTACTAACTTCAGCCTGCTGAAGCAGGCTGGAGACGTGGAGGAGAACCCTGGACCTATGGTGAGCAAGGGCGAG-3' and 5'-AACTCATCAATGTATCTTATCATGTCTGTTTACTTCTTGTACCCTACCTTTCTCTTC-3'); hoxa13b (pCS2+ hoxa13b, 5'-CGAGCTGTACAAGGGAAGCGGAGCTACTAACTTCAGCCTGCTGAAGCAGGCTGGAGACGTGGAGGAGAACCCTGGACCTATGACAGCGTCTTTACTC-3' and 5'- AACTCATCAATGTATCTTATCATGTCTGTTTAACTGATGCCCTTGTACTTG-3'); hoxa13b mutant (hoxa13b PCR product, hoxa13b primers described above); hoxa9b (hoxa9b PCR product, 5'-CGAGCTGTACAAGGGAAGCGGAGCTACTAACTTCAGCCTGCTGAAGCAGGCTGGAGACGTGGAGGAGAACCCTGGACCTATGTCGACATTGGGAACAC-3' and 5'-AACTCATCAATGTATCTTATCATGTCTGTTTAAATGTCTTTTGGGCGATC-3'); hoxa10b (hoxa10b PCR product, 5'-CGAGCTGTACAAGGGAAGCGGAGCTACTAACTTCAGCCTGCTGAAGCAGGCTGGAGACGTGGAGGAGAACCCTGGACCTATGTCATGCTCCGATAGTCC-3' and 5'-AACTCATCAATGTATCTTATCATGTCTGTTTATGAAAAACTGAAGTTGGCTGAC-3'); hoxd9a (hoxd9a PCR product, 5'-CGAGCTGTACAAGGGAAGCGGAGCTACTAACTTCAGCCTGCTGAAGCAGGCTGGAGACGTGGAGGAGAACCCTGGACCTATGTCGACTAGTTCAGCTC-3' and 5'-AACTCATCAATGTATCTTATCATGTCTGTTTACGGATCTTTGCTGCTCCTC-3'). pDestTol2-tbx16-pA2 was linearized with BamHI and assembled with the EGFP, mCherryNLS, hoxa13b, and hoxa13b mutant PCR products using the Gibson Assembly Master Mix to generate the following constructs: pDest-Tol2-tbx16-EGFP-P2A-mCherryNLS-pA2, pDest-Tol2-tbx16-EGFP-P2A-hoxa13b-pA2, and pDest-Tol2-tbx16-EGFP-P2A-hoxa13bmutant-pA2. The final constructs were injected into 1-to 2-cell-stage wild-type zebrafish (6 pg/embryo), and the distribution of EGFP-positive cells along the anterior-posterior axis at 20 hpf was evaluated.
To quantify MPC distributions along the zebrafish body axis (Supplementary Fig. 13), embryos were fixed at 22 to 24 hpf with 4% PFA in PBS for 3 hours at room temperature. Each embryo was then mounted in agar and imaged on the Leica DM4500B microscope. Images were acquired using the auto-exposure function in the MetaMorph control software to minimize pixel saturation, and ImageJ was then used to quantify pixel intensity. To determine the signal background for each image, a 50-pixel × 50-pixel box was positioned within the zebrafish head, a region with minimal EGFP accumulation. The mean pixel intensity from this region plus 15 arbitrary units were then subtracted from all pixels in the image, and the resulting pixel values were defined as the EGFP signal. Anterior, trunk, and posterior regions were delineated using the yolk extension as a morphological marker. Each region was then circumscribed using the ImageJ polygon tool, and the total EGFP fluorescence intensity within each domain was calculated using the integrated density measurement “RawIntDen”.
Dorsomorphin treatment of tbx16 morphants
Wild type embryos were injected with tbx16 MO at the 1- to 4-cell stage, cultured until 4 hpf in E3 medium, and then transferred into 12-well microplate wells containing 2 mL of E3 medium and either 10 uM, 25, uM, or 50 uM dorsomorphin (Calbiochem) or an equivalent amount of DMSO vehicle (1% (v/v) final DMSO concentration). The embryos were cultured until 8 hpf and then fixed in 4% (w/v) paraformaldehyde in PBS, and hoxa13b transcripts were detected by in situ hybridization as described above.
Statistical analyses
For all experiments performed, no statistical methods were used to determine sample size per condition. At least two breeding tanks, each containing 3 to 4 males and 4 to 5 females from separate stocks, were set up to generate embryos per each experiment. To evenly distribute sample variability, embryos obtained within the first 15 minutes of natural mating were collected from all breeding tanks, pooled, and then randomly distributed across the tested conditions. After embryo collection and initial treatment, pre-established exclusion criteria were applied to remove unfertilized and developmentally abnormal embryos from further study. Blinding was not applied when assigning conditions or during phenotypic analysis.
To quantify MPC distributions along the zebrafish body axis (Supplementary Fig. 13), the normality of each distribution was assessed using the Shapiro-Wilk test, and variance was determined using GraphPad Prism 6 software. As these tests determined that the distributions were non-normal and had wide degrees of variances between samples, P-values were calculated using the two-tailed Mann-Whitney test for non-parametric, pair-wise comparisons, with each condition compared against the mCherry-NLS control. P-values were not adjusted for multiple comparison testing, and five planned comparisons were conducted for each defined region (anterior, trunk, and posterior).
Database depositions
Raw RNA sequencing data, count files, and EdgeR outputs have been uploaded into the Gene Expression Omnibus (GEO) database (NCBI) under accession number GSE70623.
Supplementary Material
ACKNOWLEDGEMENTS
We thank C. Crumpton, B. Gomez, and O. Herman of the Stanford Shared FACS Facility for technical assistance with flow cytometry, Z. Weng of the Stanford Sequencing Service Center for assistance with RNA library sequencing, I. Yanai and F. Wagner for assistance with sequence-read processing, S. Amacher for tbx16b104/+ zebrafish, M. Lardelli, G. Lieschke, and H. Okamoto for plasmids, and K. Mruk for helpful discussions. This work was supported by the NIH (DP1 HD075622, R01 GM087292, and R01 GM108952 to J.K.C.; P50 GM107615 to the Stanford Center for Systems Biology), an A. P. Giannini Foundation Fellowship for Medical Research (L.E.M.), and a Japan Society for the Promotion of Science Fellowship (S.Y.).
Footnotes
ACCESSION CODES Raw RNA sequencing data, count files, and EdgeR outputs have been uploaded into the Gene Expression Omnibus (GEO) database (NCBI) under accession number GSE70623.
AUTHOR CONTRIBUTIONS A.Y.P, L.E.M., and J.K.C. designed the experiments; A.Y.P., L.E.M., and W.J.W. performed the experiments; A.Y.P, L.E.M., and J.K.C. analyzed data; S.Y. synthesized reagents for cMO preparation; A.Y.P. and J.K.C. wrote the manuscript.
COMPETING FINANCIAL INTERESTS The authors declare no competing financial interests.
REFERENCES
- 1.Scaal M. Early development of the vertebral column. Semin. Cell Dev. Biol. 2016;49:83–91. doi: 10.1016/j.semcdb.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 2.Eckalbar WL, Fisher RE, Rawls A, Kusumi K. Scoliosis and segmentation defects of the vertebrae. Wiley Interdiscip. Rev. Dev. Biol. 2012;1:401–423. doi: 10.1002/wdev.34. [DOI] [PubMed] [Google Scholar]
- 3.Hubaud A, Pourquie O. Signalling dynamics in vertebrate segmentation. Nat. Rev. Mol. Cell Biol. 2014;15:709–721. doi: 10.1038/nrm3891. [DOI] [PubMed] [Google Scholar]
- 4.Turnpenny PD, et al. Novel mutations in DLL3, a somitogenesis gene encoding a ligand for the Notch signalling pathway, cause a consistent pattern of abnormal vertebral segmentation in spondylocostal dysostosis. J. Med. Genet. 2003;40:333–339. doi: 10.1136/jmg.40.5.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Whittock NV, et al. Mutated MESP2 causes spondylocostal dysostosis in humans. Am. J. Hum. Genet. 2004;74:1249–1254. doi: 10.1086/421053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sparrow DB, et al. Mutation of the LUNATIC FRINGE gene in humans causes spondylocostal dysostosis with a severe vertebral phenotype. Am. J. Hum. Genet. 2006;78:28–37. doi: 10.1086/498879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sparrow DB, Guillen-Navarro E, Fatkin D, Dunwoodie SL. Mutation of Hairy-and-Enhancer-of-Split-7 in humans causes spondylocostal dysostosis. Hum. Mol. Genet. 2008;17:3761–3766. doi: 10.1093/hmg/ddn272. [DOI] [PubMed] [Google Scholar]
- 8.Holley SA. Anterior-posterior differences in vertebrate segments: specification of trunk and tail somites in the zebrafish blastula. Genes Dev. 2006;20:1831–1837. doi: 10.1101/gad.1453706. [DOI] [PubMed] [Google Scholar]
- 9.Mallo M, Vinagre T, Carapuco M. The road to the vertebral formula. Int. J. Dev. Biol. 2009;53:1469–1481. doi: 10.1387/ijdb.072276mm. [DOI] [PubMed] [Google Scholar]
- 10.Kimmel CB, Kane DA, Walker C, Warga RM, Rothman MB. A mutation that changes cell movement and cell fate in the zebrafish embryo. Nature. 1989;337:358–362. doi: 10.1038/337358a0. [DOI] [PubMed] [Google Scholar]
- 11.Ho RK, Kane DA. Cell-autonomous action of zebrafish spt-1 mutation in specific mesodermal precursors. Nature. 1990;348:728–730. doi: 10.1038/348728a0. [DOI] [PubMed] [Google Scholar]
- 12.Griffin KJ, Amacher SL, Kimmel CB, Kimelman D. Molecular identification of spadetail: regulation of zebrafish trunk and tail mesoderm formation by T-box genes. Development. 1998;125:3379–3388. doi: 10.1242/dev.125.17.3379. [DOI] [PubMed] [Google Scholar]
- 13.Ho RK. Cell movements and cell fate during zebrafish gastrulation. Dev. Suppl. 1992:65–73. [PubMed] [Google Scholar]
- 14.Yamamoto A, et al. Zebrafish paraxial protocadherin is a downstream target of spadetail involved in morphogenesis of gastrula mesoderm. Development. 1998;125:3389–3397. doi: 10.1242/dev.125.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Griffin KJ, Kimelman D. One-Eyed Pinhead and Spadetail are essential for heart and somite formation. Nat. Cell Biol. 2002;4:821–825. doi: 10.1038/ncb862. [DOI] [PubMed] [Google Scholar]
- 16.Fior R, et al. The differentiation and movement of presomitic mesoderm progenitor cells are controlled by Mesogenin 1. Development. 2012;139:4656–4665. doi: 10.1242/dev.078923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O'Neill K, Thorpe C. BMP signaling and spadetail regulate exit of muscle precursors from the zebrafish tailbud. Dev. Biol. 2013;375:117–127. doi: 10.1016/j.ydbio.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Szeto DP, Kimelman D. The regulation of mesodermal progenitor cell commitment to somitogenesis subdivides the zebrafish body musculature into distinct domains. Genes Dev. 2006;20:1923–1932. doi: 10.1101/gad.1435306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garnett AT, et al. Identification of direct T-box target genes in the developing zebrafish mesoderm. Development. 2009;136:749–760. doi: 10.1242/dev.024703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mueller RL, Huang C, Ho RK. Spatio-temporal regulation of Wnt and retinoic acid signaling by tbx16/spadetail during zebrafish mesoderm differentiation. BMC Genomics. 2010;11:492. doi: 10.1186/1471-2164-11-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shestopalov IA, Sinha S, Chen JK. Light-controlled gene silencing in zebrafish embryos. Nat. Chem. Biol. 2007;3:650–651. doi: 10.1038/nchembio.2007.30. [DOI] [PubMed] [Google Scholar]
- 22.Yamazoe S, Shestopalov IA, Provost E, Leach SD, Chen JK. Cyclic caged morpholinos: conformationally gated probes of embryonic gene function. Angew. Chem. Int. Ed. Engl. 2012;51:6908–6911. doi: 10.1002/anie.201201690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rossi A, et al. Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature. 2015;524:230–233. doi: 10.1038/nature14580. [DOI] [PubMed] [Google Scholar]
- 24.Shestopalov IA, Pitt CL, Chen JK. Spatiotemporal resolution of the Ntla transcriptome in axial mesoderm development. Nat. Chem. Biol. 2012;8:270–276. doi: 10.1038/nchembio.772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Myers DC, Sepich DS, Solnica-Krezel L. Bmp activity gradient regulates convergent extension during zebrafish gastrulation. Dev. Biol. 2002;243:81–98. doi: 10.1006/dbio.2001.0523. [DOI] [PubMed] [Google Scholar]
- 26.Yamazoe S, Liu Q, McQuade LE, Deiters A, Chen JK. Sequential gene silencing using wavelength-selective caged morpholino oligonucleotides. Angew. Chem. Int. Ed. Engl. 2014;53:10114–10118. doi: 10.1002/anie.201405355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Payumo AY, Walker WJ, McQuade LE, Yamazoe S, Chen JK. Optochemical Dissection of T-box Gene-Dependent Medial Floor Plate Development. ACS Chem. Biol. 2015;10:1466–1475. doi: 10.1021/cb5010178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weinberg ES, et al. Developmental regulation of zebrafish MyoD in wild-type, no tail and spadetail embryos. Development. 1996;122:271–280. doi: 10.1242/dev.122.1.271. [DOI] [PubMed] [Google Scholar]
- 29.Kimmel CB, Warga RM, Schilling TF. Origin and organization of the zebrafish fate map. Development. 1990;108:581–594. doi: 10.1242/dev.108.4.581. [DOI] [PubMed] [Google Scholar]
- 30.Warga RM, Nusslein-Volhard C. Origin and development of the zebrafish endoderm. Development. 1999;126:827–838. doi: 10.1242/dev.126.4.827. [DOI] [PubMed] [Google Scholar]
- 31.Ando R, Hama H, Yamamoto-Hino M, Mizuno H, Miyawaki A. An optical marker based on the UV-induced green-to-red photoconversion of a fluorescent protein. Proc. Natl. Acad. Sci. U. S. A. 2002;99:12651–12656. doi: 10.1073/pnas.202320599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bouldin CM, et al. Wnt signaling and tbx16 form a bistable switch to commit bipotential progenitors to mesoderm. Development. 2015;142:2499–2507. doi: 10.1242/dev.124024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Amores A, et al. Zebrafish hox clusters and vertebrate genome evolution. Science. 1998;282:1711–1714. doi: 10.1126/science.282.5394.1711. [DOI] [PubMed] [Google Scholar]
- 34.Montavon T, Soshnikova N. Hox gene regulation and timing in embryogenesis. Semin. Cell Dev. Biol. 2014;34:76–84. doi: 10.1016/j.semcdb.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 35.Yu PB, et al. Dorsomorphin inhibits BMP signals required for embryogenesis and iron metabolism. Nat. Chem. Biol. 2008;4:33–41. doi: 10.1038/nchembio.2007.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wells S, Nornes S, Lardelli M. Transgenic zebrafish recapitulating tbx16 gene early developmental expression. PLoS One. 2011;6:e21559. doi: 10.1371/journal.pone.0021559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim JH, et al. High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PLoS One. 2011;6:e18556. doi: 10.1371/journal.pone.0018556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martin BL, Kimelman D. Regulation of canonical Wnt signaling by Brachyury is essential for posterior mesoderm formation. Dev. Cell. 2008;15:121–133. doi: 10.1016/j.devcel.2008.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martin BL, Kimelman D. Canonical Wnt signaling dynamically controls multiple stem cell fate decisions during vertebrate body formation. Dev. Cell. 2012;22:223–232. doi: 10.1016/j.devcel.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yabe T, Takada S. Mesogenin causes embryonic mesoderm progenitors to differentiate during development of zebrafish tail somites. Dev. Biol. 2012;370:213–222. doi: 10.1016/j.ydbio.2012.07.029. [DOI] [PubMed] [Google Scholar]
- 41.Neave B, Holder N, Patient R. A graded response to BMP-4 spatially coordinates patterning of the mesoderm and ectoderm in the zebrafish. Mech. Dev. 1997;62:183–195. doi: 10.1016/s0925-4773(97)00659-x. [DOI] [PubMed] [Google Scholar]
- 42.Row RH, Kimelman D. Bmp inhibition is necessary for post-gastrulation patterning and morphogenesis of the zebrafish tailbud. Dev. Biol. 2009;329:55–63. doi: 10.1016/j.ydbio.2009.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harvey SA, Tumpel S, Dubrulle J, Schier AF, Smith JC. no tail integrates two modes of mesoderm induction. Development. 2010;137:1127–1135. doi: 10.1242/dev.046318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martin BL, Kimelman D. Brachyury establishes the embryonic mesodermal progenitor niche. Genes Dev. 2010;24:2778–2783. doi: 10.1101/gad.1962910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Iimura T, Pourquie O. Collinear activation of Hoxb genes during gastrulation is linked to mesoderm cell ingression. Nature. 2006;442:568–571. doi: 10.1038/nature04838. [DOI] [PubMed] [Google Scholar]
- 46.Denans N, Iimura T, Pourquie O. Hox genes control vertebrate body elongation by collinear Wnt repression. Elife. 2015;4:e04379. doi: 10.7554/eLife.04379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Manning AJ, Kimelman D. Tbx16 and Msgn1 are required to establish directional cell migration of zebrafish mesodermal progenitors. Dev. Biol. 2015;406:172–185. doi: 10.1016/j.ydbio.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lengerke C, et al. BMP and Wnt specify hematopoietic fate by activation of the Cdx-Hox pathway. Cell Stem Cell. 2008;2:72–82. doi: 10.1016/j.stem.2007.10.022. [DOI] [PubMed] [Google Scholar]
- 49.Warga RM, Mueller RL, Ho RK, Kane DA. Zebrafish Tbx16 regulates intermediate mesoderm cell fate by attenuating Fgf activity. Dev. Biol. 2013;383:75–89. doi: 10.1016/j.ydbio.2013.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
METHODS-ONLY REFERENCES
- 50.Westerfield M. The Zebrafish Book. University of Oregon Press; Oregon: 1995. [Google Scholar]
- 51.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 52.Sato T, Takahoko M, Okamoto H. HuC:Kaede, a useful tool to label neural morphologies in networks in vivo. Genesis. 2006;44:136–142. doi: 10.1002/gene.20196. [DOI] [PubMed] [Google Scholar]
- 53.Mich JK, Payumo AY, Rack PG, Chen JK. In vivo imaging of Hedgehog pathway activation with a nuclear fluorescent reporter. PLoS One. 2014;9:e103661. doi: 10.1371/journal.pone.0103661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thisse B, Thisse C. High Throughput Expression Analysis of ZF-Models Consortium Clones. ZFIN Direct Data Submission. 2005 http://zfin.org.
- 55.Hashimshony T, Wagner F, Sher N, Yanai I. CEL-Seq: single-cell RNA-Seq by multiplexed linear amplification. Cell Rep. 2012;2:666–673. doi: 10.1016/j.celrep.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 56.Ellett F, Pase L, Hayman JW, Andrianopoulos A, Lieschke GJ. mpeg1 promoter transgenes direct macrophage-lineage expression in zebrafish. Blood. 2011;117:e49–56. doi: 10.1182/blood-2010-10-314120. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.