Abstract
We performed a meta analysis to assess the relationship of FCGRs polymorphisms with the risk of SLE. Thirty-five articles (including up to 5741 cases and 6530 controls) were recruited for meta-analysis. The strongest association was observed between FCGR2B rs1050501 and SLE under the recessive genotypic model of C allele in the overall population (CC vs CT/TT, OR = 1.754, 95%CI: 1.422–2.165, P = 1.61 × 10−7) and in Asian population (CC vs CT/TT, OR = 1.784, 95%CI; 1.408–2.261, P = 1.67 × 10−6). We also found that FCGR3A rs396991 were significant association with the susceptibility to SLE in overall population in recessive model of T allele (TT vs TG/GG, OR = 1.263, 95%CI: 1.123–1.421, P = 9.62 × 10−5). The results also showed that significant association between FCGR2A rs1801274 and SLE under the allelic model in the overall population (OR = 0.879 per A allele, 95%CI: 0.819–0.943, P = 3.31 × 10−4). The meta-analysis indicated that FCGR3B copy number polymorphism NA1·NA2 was modestly associated with SLE in overall population (OR = 0.851 per NA1, 95%CI: 0.772–0.938, P = 1.2 × 10−3). We concluded that FCGR2B rs1050501 C allele and FCGR3A rs396991 T allele might contribute to susceptibility and development of SLE, and were under recessive association model. While, FCGR2A rs1801274 A allele and FCGR3B NA1 were associated with SLE and reduced the risk of SLE.
Systemic lupus erythematosus (SLE) is a kind of autoimmune disease with a strong genetic predisposition caused by complicated factors, it is also considered as an inflammatory disease caused by the mediation and deposition of immune complexes (ICs), leading to damage of multiple organs1. In different races or regions, the morbidity rate of SLE is quite different2,3, it is about 31-70/100,000 across China4, while it is 7-71/100,000 in Europeans5 and it increases to 200/100,000 in African population5. The etiology and pathogenesis of SLE is unclear yet, it is generally accepted that both genetic and environmental factors are involved in the development of this complex disease6. Since the end of last century, scientists were trying to use genetic linkage analysis to investigate the mechanism of SLE, a number of susceptibility area in SLE had been found such as 1q237, 1q418, 4p169, 11q1410, 12q2411. Linkage analysis for SLE had made some achievements, but it is not easy to find real susceptibility genes because of large positioning areas. Then, candidate gene association studies (CGASs), in which single-nucleotide polymorphisms (SNPs) were assayed in cases and controls, were widely used and found some valuable susceptibility genes such as IL-612, TLR213, VDR14, CTLA-415, FCGR2A16, FCGR2B17, PELI118, IKZF319. More recently, genome-wide association studies (GWAS) have been the powerful approach and found a lot of susceptibility genes and SNPs for SLE20,21,22,23,24,25,26,27.
Among these genes/proteins, FC gamma Receptor (FCγR) is a member of immunoglobulin superfamily, and it is very important to bind FCγR with the Fc protein of Immunoglobulin G (IgG), because FCγR binding may activate biological reaction, such as phagocytosis28. The human 1q21-23 locus contains 5 FCGR genes (FCGR2A, 2B, 2C, 3A and 3B) encoding the FCγRIIand FCγRIII receptor·families29. FCγRs mediate clearance of immune complexes and have been strongly implicated in the pathogenesis of SLE and lupus nephritis30. Thus the genes that encode these receptors have been the focus of many genetic studies in SLE31.
FCGRs were not genome-wide significantly identified by any GWAS above, and the results were not always consistent by candidate gene association study. The inconsistency of findings is related to many factors, such as the selecting of the sample, the size of sample and the dealing of the statistics, etc. Therefore, in order to reduce the limitations of single study and to overcome the possible random errors, we performed a large-scale meta-analysis involving different ethnics. Among all the studies, there were 5082 cases and 4951 controls to evaluate the relationship between FCGR2A rs1801274 and SLE and there were 2970 cases and 4197 controls for FCGR2B rs1050501. For FCGR3A rs396991 and FCGR3B NA1·NA2, there were 5694 cases and 6450 controls, 1692 cases and 1899 controls, respectively. The purpose of this study is to analyze whether the polymorphisms of FCGRs are susceptibility to SLE. We also made efforts to find the best-fit association model among the additive, recessive and dominant models for the polymorphisms.
Results
Studies included in the meta-analysis
In this meta-analysis, totally 436 relevant articles were found from PubMed, of which 337 were excluded because they were unrelated articles. Studies investigating other FCGR gene polymorphisms were also excluded17,32,33,34,35,36. One more article was also excluded because there was no detail genotyping data37. After filtering, 35 eligible articles were finally included16,33,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69. The flow chart of selecting articles process is presented in Fig. 1. Therefore, there were 34 studies with 5082 cases and 4951 controls to evaluate the relationship between FCGR2A rs1801274 polymorphism and SLE. For FCGR2B rs1050501 polymorphism, there were 13 studies involving a total of 2970 cases and 4197 controls. For FCGR3A rs396991 polymorphism and FCGR3B NA1·NA2 polymorphism, 26 studies (5694 cases and 6450 controls) and 11 studies (1692 cases and 1899 controls) were available, respectively. The basic information of these included studies genotype distributions and the allele frequencies are showed in Table 1.
Figure 1. The process of the articles selected in this meta-analysis.
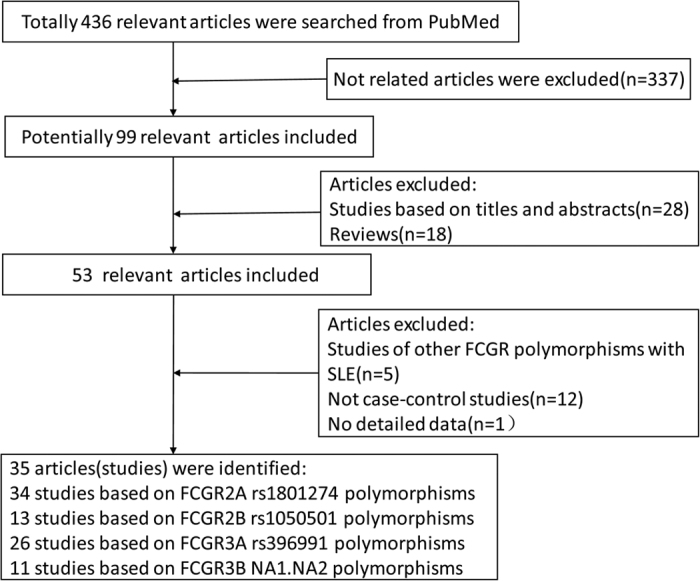
Table 1. The basic information of every studies included in this meta-analysis.
| Polymorphismsand study | Journal | Year | Ethnicity | Sample size |
Genotypes |
Allele frequencies (%) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases | Controls | Cases | Controls | Cases | Controls | ||||||||||
| rs1801274(FCGR2A) | AA | AG | GG | AA | AG | GG | A | G | A | G | |||||
| Vigato-Ferreira IC | Autoimmunity | 2014 | Caucasian | 157 | 160 | 23 | 59 | 75 | 35 | 43 | 82 | 0.334 | 0.666 | 0.353 | 0.647 |
| Dijstelbloem HM | Arthritis Rheum | 2000 | Caucasian | 230 | 154 | 54 | 108 | 68 | 42 | 80 | 32 | 0.470 | 0.530 | 0.532 | 0.468 |
| Zuñiga R | Arthritis Rheum | 2001 | Caucasian | 67 | 53 | 5 | 39 | 23 | 11 | 28 | 14 | 0.366 | 0.634 | 0.472 | 0.528 |
| Seligman VA | Arthritis Rheum | 2001 | Caucasian | 76 | 186 | 10 | 49 | 17 | 28 | 114 | 44 | 0.454 | 0.546 | 0.457 | 0.543 |
| Seligman VA | Arthritis Rheum | 2001 | Caucasian | 48 | 55 | 7 | 29 | 12 | 10 | 24 | 21 | 0.448 | 0.552 | 0.400 | 0.600 |
| Manger K | Ann Rheum Dis | 2002 | Caucasian | 140 | 187 | 46 | 55 | 39 | 53 | 84 | 50 | 0.525 | 0.475 | 0.508 | 0.492 |
| Botto M | Clin Exp Immunol | 1996 | Caucasian | 215 | 259 | 46 | 97 | 72 | 57 | 120 | 82 | 0.440 | 0.560 | 0.452 | 0.548 |
| Duits A | Arthritis Rheum | 1995 | Caucasian | 95 | 69 | 18 | 50 | 27 | 22 | 36 | 11 | 0.453 | 0.547 | 0.580 | 0.420 |
| Norsworthy P | Arthritis Rheum | 1999 | Caucasian | 195 | 283 | 32 | 96 | 67 | 62 | 131 | 90 | 0.410 | 0.590 | 0.451 | 0.549 |
| Smyth LJ | Ann Rheum Dis | 1997 | Caucasian | 81 | 66 | 10 | 49 | 22 | 12 | 38 | 16 | 0.426 | 0.574 | 0.470 | 0.530 |
| Smyth LJ | Ann Rheum Dis | 1997 | Caucasian | 42 | 52 | 14 | 16 | 12 | 20 | 24 | 8 | 0.524 | 0.476 | 0.615 | 0.385 |
| González-Escribano MF | Eur J Immunogenet | 2002 | Caucasian | 276 | 194 | 64 | 137 | 75 | 59 | 86 | 49 | 0.480 | 0.520 | 0.526 | 0.474 |
| Zhou XJ | Lupus | 2011 | Asian | 589 | 477 | 238 | 269 | 82 | 209 | 220 | 48 | 0.632 | 0.368 | 0.669 | 0.331 |
| Kobavashi T | J Periodontol | 2007 | Asian | 71 | 44 | 34 | 31 | 6 | 28 | 16 | 0 | 0.697 | 0.303 | 0.818 | 0.182 |
| Chu ZT | Tissue Antigens | 2004 | Asian | 163 | 129 | 72 | 70 | 21 | 53 | 58 | 18 | 0.656 | 0.344 | 0.636 | 0.364 |
| Kyogoku C | Arthritis Rheum | 2002 | Asian | 193 | 303 | 113 | 72 | 8 | 197 | 95 | 11 | 0.772 | 0.228 | 0.807 | 0.193 |
| Siriboonrit U | Tissue Antigens | 2003 | Asian | 87 | 187 | 37 | 40 | 10 | 93 | 76 | 18 | 0.655 | 0.345 | 0.701 | 0.299 |
| Seligman VA | Arthritis Rheum | 2001 | Asian | 57 | 40 | 11 | 37 | 9 | 6 | 27 | 7 | 0.518 | 0.482 | 0.488 | 0.513 |
| Salmon JE | Arthritis Rheum | 1999 | Asian | 148 | 97 | 70 | 66 | 12 | 41 | 47 | 9 | 0.696 | 0.304 | 0.665 | 0.335 |
| Hatta Y | Genes Immun | 1999 | Asian | 81 | 217 | 49 | 30 | 2 | 139 | 71 | 7 | 0.790 | 0.210 | 0.804 | 0.196 |
| Hatta Y | Genes Immun | 1999 | Asian | 69 | 93 | 42 | 26 | 1 | 62 | 28 | 3 | 0.797 | 0.203 | 0.817 | 0.183 |
| Lee HS | Rheumatology | 2003 | Asian | 299 | 144 | 131 | 114 | 54 | 67 | 66 | 11 | 0.629 | 0.371 | 0.694 | 0.306 |
| Botto M | Clin Exp Immunol | 1996 | Asian | 46 | 49 | 18 | 23 | 5 | 24 | 20 | 5 | 0.641 | 0.359 | 0.694 | 0.306 |
| Yun HR | Lupus | 2001 | Asian | 300 | 197 | 132 | 114 | 54 | 82 | 99 | 16 | 0.630 | 0.370 | 0.668 | 0.332 |
| Yap S | Lupus | 1999 | Asian | 175 | 108 | 59 | 91 | 25 | 28 | 63 | 17 | 0.597 | 0.403 | 0.551 | 0.449 |
| Yap S | Lupus | 1999 | Asian | 50 | 50 | 20 | 26 | 4 | 21 | 21 | 8 | 0.660 | 0.340 | 0.630 | 0.370 |
| Chen JY | Ann Rheum Dis | 2004 | Asian | 329 | 311 | 125 | 155 | 49 | 130 | 144 | 37 | 0.616 | 0.384 | 0.650 | 0.350 |
| Zidan HE | Mol Biol Rep | 2014 | African | 90 | 90 | 20 | 45 | 25 | 22 | 50 | 18 | 0.472 | 0.528 | 0.522 | 0.478 |
| Seligman VA | Arthritis Rheum | 2001 | African | 30 | 31 | 9 | 12 | 9 | 6 | 15 | 10 | 0.500 | 0.500 | 0.435 | 0.565 |
| Botto M | Clin Exp Immunol | 1996 | African | 70 | 77 | 8 | 37 | 25 | 17 | 35 | 25 | 0.379 | 0.621 | 0.448 | 0.552 |
| Seligman VA | Arthritis Rheum | 2001 | mixed population | 216 | 318 | 38 | 131 | 47 | 50 | 185 | 83 | 0.479 | 0.521 | 0.448 | 0.552 |
| Seligman VA | Arthritis Rheum | 2001 | Non-Caucasian | 140 | 132 | 28 | 82 | 30 | 22 | 71 | 39 | 0.493 | 0.507 | 0.436 | 0.564 |
| Salmon J | J Clin Invest | 1996 | African Americans | 43 | 39 | 4 | 23 | 16 | 14 | 15 | 10 | 0.360 | 0.640 | 0.551 | 0.449 |
| Salmon J | J Clin Invest | 1996 | African Americans | 214 | 100 | 37 | 97 | 80 | 27 | 50 | 23 | 0.400 | 0.600 | 0.520 | 0.480 |
| rs1050501(FCGR2B) | CC | CT | TT | CC | CT | TT | C | T | C | T | |||||
| Pradhan V | Indian J Med Res | 2011 | Asian | 80 | 80 | 16 | 49 | 15 | 10 | 52 | 18 | 0.506 | 0.494 | 0.450 | 0.550 |
| Koga M | J Hum Genet | 2011 | Asian | 282 | 222 | 29 | 103 | 150 | 9 | 85 | 128 | 0.285 | 0.715 | 0.232 | 0.768 |
| Willcocks LC | PNAS | 2010 | Asian | 819 | 1026 | 60 | 284 | 475 | 57 | 404 | 565 | 0.247 | 0.753 | 0.252 | 0.748 |
| Kobavashi T | J Periodontol | 2007 | Asian | 71 | 44 | 4 | 26 | 41 | 0 | 6 | 38 | 0.239 | 0.761 | 0.068 | 0.932 |
| Ji-Yih Chen | Arthritis Rheum | 2006 | Asian | 351 | 372 | 39 | 123 | 189 | 22 | 144 | 206 | 0.286 | 0.714 | 0.253 | 0.747 |
| Chu ZT | Tissue Antigens | 2004 | Asian | 108 | 85 | 11 | 48 | 49 | 4 | 30 | 51 | 0.324 | 0.676 | 0.224 | 0.776 |
| Kyogoku C | Arthritis Rheum | 2002 | Asian | 193 | 303 | 21 | 66 | 106 | 16 | 104 | 183 | 0.280 | 0.720 | 0.224 | 0.776 |
| Siriboonrit U | Tissue Antigens | 2003 | Asian | 79 | 165 | 12 | 29 | 38 | 12 | 56 | 97 | 0.335 | 0.665 | 0.242 | 0.758 |
| Magnusson V | Arthritis Rheum | 2004 | Caucasian | 263 | 228 | 7 | 67 | 189 | 4 | 53 | 171 | 0.154 | 0.846 | 0.134 | 0.866 |
| Willcocks LC | PNAS | 2010 | Caucasian | 326 | 1296 | 9 | 48 | 269 | 13 | 232 | 1051 | 0.101 | 0.899 | 0.100 | 0.900 |
| Li X | Arthritis Rheum | 2003 | Caucasian | 148 | 137 | 6 | 30 | 112 | 4 | 27 | 106 | 0.142 | 0.858 | 0.128 | 0.872 |
| Zidan HE | Mol Biol Rep | 2014 | African | 90 | 90 | 32 | 39 | 19 | 17 | 44 | 29 | 0.572 | 0.428 | 0.433 | 0.567 |
| Li X | Arthritis Rheum | 2003 | African-American | 160 | 149 | 14 | 49 | 97 | 17 | 53 | 79 | 0.241 | 0.759 | 0.292 | 0.708 |
| rs396991(FCGR3A) | TT | TG | GG | TT | TG | GG | T | G | T | G | |||||
| Brambila-Tapia AJ | Rheumatol Int | 2011 | Caucasian | 94 | 98 | 61 | 5 | 28 | 52 | 8 | 38 | 0.676 | 0.324 | 0.571 | 0.429 |
| Dong C | Arthritis Rheumatol | 2014 | Caucasian | 834 | 1185 | 392 | 370 | 72 | 517 | 564 | 104 | 0.692 | 0.308 | 0.674 | 0.326 |
| Dijstelbloem HM | Arthritis Rheum | 2000 | Caucasian | 230 | 154 | 92 | 108 | 30 | 66 | 73 | 15 | 0.635 | 0.365 | 0.666 | 0.334 |
| Zuñiga R | Arthritis Rheum | 2001 | Caucasian | 67 | 53 | 25 | 38 | 4 | 15 | 26 | 12 | 0.657 | 0.343 | 0.528 | 0.472 |
| Seligman VA | Arthritis Rheum | 2001 | Caucasian | 78 | 207 | 37 | 30 | 11 | 55 | 102 | 50 | 0.667 | 0.333 | 0.512 | 0.488 |
| Seligman VA | Arthritis Rheum | 2001 | Caucasian | 55 | 57 | 25 | 15 | 15 | 30 | 21 | 6 | 0.591 | 0.409 | 0.711 | 0.289 |
| Manger K | Ann Rheum Dis | 2002 | Caucasian | 140 | 187 | 55 | 64 | 21 | 62 | 75 | 50 | 0.621 | 0.379 | 0.532 | 0.468 |
| Wu J | J Clin Invest | 1997 | Caucasian | 200 | 113 | 87 | 92 | 21 | 29 | 69 | 15 | 0.665 | 0.335 | 0.562 | 0.438 |
| González-Escribano MF | Eur J Immunogenet | 2002 | Caucasian | 276 | 194 | 101 | 131 | 44 | 66 | 104 | 24 | 0.603 | 0.397 | 0.608 | 0.392 |
| Dai M | Int J Rheum Dis | 2013 | Asian | 732 | 886 | 376 | 308 | 48 | 381 | 427 | 78 | 0.724 | 0.276 | 0.671 | 0.329 |
| Kobavashi T | J Periodontol | 2007 | Asian | 71 | 44 | 43 | 22 | 6 | 24 | 15 | 5 | 0.761 | 0.239 | 0.716 | 0.284 |
| Chu ZT | Tissue Antigens | 2004 | Asian | 163 | 129 | 76 | 74 | 13 | 48 | 63 | 18 | 0.693 | 0.307 | 0.616 | 0.384 |
| Kyogoku C | Arthritis Rheum | 2002 | Asian | 193 | 303 | 110 | 76 | 7 | 145 | 132 | 26 | 0.767 | 0.233 | 0.696 | 0.304 |
| Siriboonrit U | Tissue Antigens | 2003 | Asian | 87 | 187 | 42 | 35 | 10 | 64 | 96 | 27 | 0.684 | 0.316 | 0.599 | 0.401 |
| Seligman VA | Arthritis Rheum | 2001 | Asian | 59 | 41 | 22 | 29 | 8 | 12 | 22 | 7 | 0.619 | 0.381 | 0.561 | 0.439 |
| Salmon JE | Arthritis Rheum | 1999 | Asian | 148 | 97 | 44 | 81 | 23 | 19 | 64 | 14 | 0.571 | 0.429 | 0.526 | 0.474 |
| Hatta Y | Genes Immun | 1999 | Asian | 81 | 217 | 43 | 34 | 4 | 100 | 99 | 18 | 0.741 | 0.259 | 0.689 | 0.311 |
| Hatta Y | Genes Immun | 1999 | Asian | 69 | 93 | 37 | 29 | 3 | 46 | 38 | 9 | 0.746 | 0.254 | 0.699 | 0.301 |
| Lee EB | Rheum Int | 2002 | Asian | 145 | 75 | 89 | 51 | 5 | 40 | 29 | 6 | 0.790 | 0.210 | 0.727 | 0.273 |
| Lee HS | Rheumatology | 2003 | Asian | 299 | 144 | 90 | 163 | 46 | 52 | 77 | 15 | 0.574 | 0.426 | 0.628 | 0.372 |
| Yun HR | Lupus | 2001 | Asian | 300 | 197 | 90 | 164 | 46 | 71 | 104 | 22 | 0.573 | 0.427 | 0.624 | 0.376 |
| Chen JY | Ann Rheum Dis | 2004 | Asian | 302 | 311 | 119 | 138 | 45 | 133 | 146 | 32 | 0.623 | 0.377 | 0.662 | 0.338 |
| Dong C | Arthritis Rheumatol | 2014 | African-American | 648 | 953 | 289 | 283 | 76 | 413 | 431 | 109 | 0.664 | 0.336 | 0.659 | 0.341 |
| Seligman VA | Arthritis Rheum | 2001 | mixed population | 233 | 348 | 97 | 96 | 40 | 108 | 172 | 68 | 0.622 | 0.378 | 0.557 | 0.443 |
| Seligman VA | Arthritis Rheum | 2001 | Non-Caucasian | 155 | 141 | 60 | 66 | 29 | 53 | 70 | 18 | 0.600 | 0.400 | 0.624 | 0.376 |
| Seligman VA | Arthritis Rheum | 2001 | African | 35 | 36 | 11 | 19 | 5 | 7 | 25 | 4 | 0.586 | 0.414 | 0.542 | 0.458 |
| NA1/NA2 | NA1·NA1 | NA1·NA2 | NA2·NA2 | NA1·NA1 | NA1·NA2 | NA2·NA2 | NA1 | NA2 | NA1 | NA2 | |||||
| Kobavashi T | J Periodontol | 2007 | Asian | 71 | 44 | 20 | 46 | 5 | 20 | 19 | 5 | 0.606 | 0.394 | 0.670 | 0.330 |
| Chu ZT | Tissue Antigens | 2004 | Asian | 163 | 129 | 46 | 90 | 29 | 41 | 74 | 14 | 0.552 | 0.448 | 0.605 | 0.395 |
| Kyogoku C | Arthritis Rheum | 2002 | Asian | 193 | 303 | 62 | 98 | 33 | 116 | 145 | 42 | 0.575 | 0.425 | 0.622 | 0.378 |
| Siriboonrit U | Tissue Antigens | 2003 | Asian | 87 | 187 | 30 | 39 | 18 | 85 | 82 | 20 | 0.569 | 0.431 | 0.674 | 0.326 |
| Pradhan V | Int J Rheum Dis | 2010 | Asian | 80 | 80 | 20 | 32 | 28 | 18 | 32 | 30 | 0.450 | 0.550 | 0.425 | 0.575 |
| Hatta Y | Genes Immun | 1999 | Asian | 81 | 217 | 23 | 38 | 20 | 92 | 100 | 25 | 0.519 | 0.481 | 0.654 | 0.346 |
| Hatta Y | Genes Immun | 1999 | Asian | 69 | 93 | 18 | 33 | 18 | 44 | 39 | 10 | 0.500 | 0.500 | 0.683 | 0.317 |
| Chen JY | Ann Rheum Dis | 2004 | Asian | 302 | 311 | 117 | 132 | 53 | 119 | 145 | 47 | 0.606 | 0.394 | 0.616 | 0.384 |
| Dijstelbloem HM | Arthritis Rheum | 2000 | Caucasian | 230 | 154 | 42 | 101 | 87 | 27 | 66 | 61 | 0.402 | 0.598 | 0.390 | 0.610 |
| Manger K | Ann Rheum Dis | 2002 | Caucasian | 140 | 187 | 13 | 87 | 40 | 20 | 87 | 80 | 0.404 | 0.596 | 0.340 | 0.660 |
| González-Escribano MF | Eur J Immunogenet | 2002 | Caucasian | 276 | 194 | 30 | 77 | 169 | 20 | 75 | 99 | 0.248 | 0.752 | 0.296 | 0.704 |
Meta-analysis results
FCGR2A rs1801274 polymorphism and SLE risk
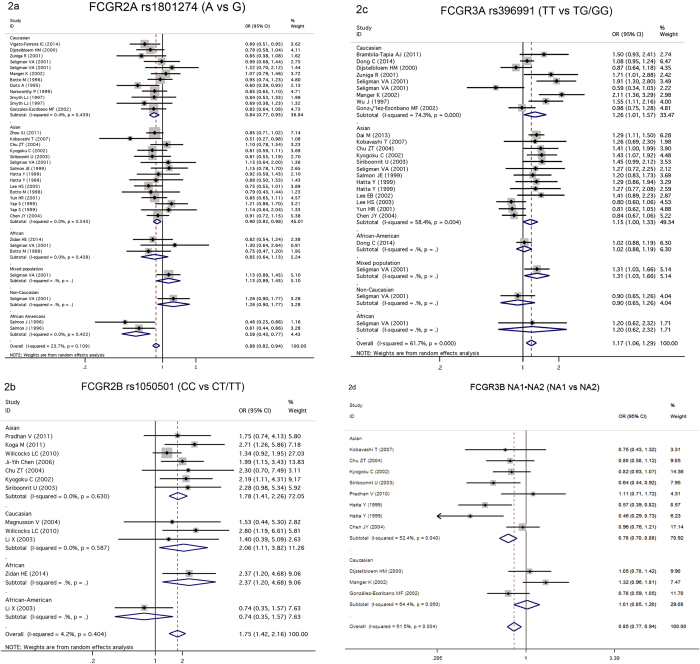
Test of heterogeneity in the overall population is not significant (P = 0.109, I2 = 23.70%), suggesting fixed effect model could be used. A strong association was found between rs1801274 and SLE under the allelic model in the overall population (OR = 0.879 per A allele, 95%CI: 0.819–0.943, P = 3.31 × 10−4, Table 2, Fig. 2a). Stratification analysis by ethnicity showed significant association between rs1801274 and SLE in Caucasian (OR = 0.845 per A allele, 95%CI: 0.766–0.932, P = 8.08 × 10−4, Table 2, Fig. 2a). And we also observed association between this polymorphism and SLE in African Americans (OR = 0.575 per A allele, 95%CI; 0.429–0.774, P = 2.73 × 10−4, Table 2, Fig. 2a) and in Asian population (OR = 0.896 per A allele, 95%CI: 0.822–0.977, P = 0.013, Table 2, Fig. 2a). No significant association was found in this meta-analysis between the polymorphism and the risk of SLE in African population (OR = 0.853 per A allele, 95%CI: 0.642–1.132, P = 0.271, Table 2, Fig. 2a). We also tested the dominant and recessive models of A allele in the overall, European, Asian and African populations, these results showed that the association was more significant in the recessive model than the dominant model in the overall population (Table 2, Supplementary Fig. S1a, Fig. S2a).
Table 2. Meta-analysis of the association between FCGR2A rs1801274 polymorphism and SLE risk.
| Population | N | A vs. G(allele model) |
AA vs. AG+GG(recessive model) |
AA+AG vs. GG(dominant model) |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| OR(95%CI) | POR | Ph | OR(95%CI) | POR | Ph | OR(95%CI) | POR | Ph | ||
| Overall | 34 | 0.879(0.819–0.943) | 3.31 × 10−4 | 0.109 | 0.867(0.784–0.960) | 6.14 × 10−3 | 0.214 | 0.843(0.739–0.961) | 0.011 | 0.074 |
| Caucasian | 12 | 0.845(0.766–0.932) | 8.08 × 10−4 | 0.439 | 0.775(0.655–0.917) | 3.08 × 10−3 | 0.522 | 0.883(0.756–1.032) | 0.117 | 0.427 |
| Asian | 15 | 0.896(0.822–0.977) | 0.013 | 0.543 | 0.932(0.830–1.046) | 0.232 | 0.658 | 0.767(0.604–0.975) | 0.030 | 0.179 |
| African | 3 | 0.853(0.642–1.132) | 0.271 | 0.438 | 0.836(0.428–1.633) | 0.601 | 0.192 | 0.802(0.515–1.250) | 0.331 | 0.688 |
| Mixed population | 1 | 1.133(0.887–1.448) | 0.318 | — | 1.144(0.721–1.817) | 0.568 | — | 1.27(0.844–1.911) | 0.252 | — |
| Non-Caucasian | 1 | 1.259(0.898–1.765) | 0.181 | — | 1.250(0.674–2.317) | 0.479 | — | 1.538(0.887–2.666) | 0.125 | — |
| African Americans | 2 | 0.575(0.427–0.774) | 2.73 × 10−4 | 0.422 | 0.368(0.126–1.078) | 0.068 | 0.100 | 0.519(0.324–0.831) | 6.33 × 10–3 | 0.786 |
OR odd ratio, 95%CI confidence interval, POR P value for the test of association, Ph P value for heterogeneity analysis.
Figure 2. Forest plot for the meta-analysis of the association between FCGRs polymorphisms and SLE.
(a) FCGR2A rs1801274 and SLE (A vs G); (b) FCGR2B rs1050501 and SLE (CC vs CT/TT); (c) FCGR3A rs396991 and SLE (TT vs TG /GG); (d) FCGR3B NA1·NA2 and SLE (NA1 vs NA2).
FCGR2B rs1050501 polymorphism and SLE risk
To assess the association of FCGR2B rs1050501 polymorphism with SLE, 13 studies were included in this meta-analysis with 2970 cases and 4197 controls, however, we identified publication bias while the study by Kobavashi T et al.59 was included (Begg’s Test: Z = 2.14, P = 0.033), therefore, this study was removed in the final analysis with 2899 cases and 4153 controls. After exclusion, the Begg’s test showed no deviation (Z = 1.58, P = 0.115) (Supplementary Table S1).
A very significant association was identified between rs1050501 and SLE under the recessive genotypic model of C allele in the overall population (CC vs CT/TT, OR = 1.754, 95%CI: 1.422–2.165, P = 1.61 × 10−7, Fig. 2b, Table 3) and in Asian population (CC vs CT/TT, OR = 1.784, 95%CI; 1.408–2.261, P = 1.67 × 10−6, Table 3, Fig. 2b), these associations were not significant under dominant model, suggesting the recessive association model was fit for rs1050501_C (Table 3). In allelic test model, Significant association between rs1050501 and SLE was identified in the overall population (OR = 1.236 per C allele, 95%CI: 1.069–1.429, P = 6.93 × 10−3, Table 3, Supplementary Fig. S2b), and in the Asian population (OR = 1.326 per C allele, 95%CI: 1.095–1.604, P = 6.14 × 10−3, Table 3, Supplementary Fig. S2b) and in African population (OR = 1.749 per C allele, 95%CI: 1.153–2.655, P = 8.54 × 10−3, Table 3, Supplementary Fig. S2b).
Table 3. Meta-analysis of the association between FCGR2B rs1050501 polymorphism and SLE risk.
| Population | N | C vs. T(allele model) |
CC vs. CT+TT(recessive model) |
CC+CT vs. TT(dominant model) |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| OR(95%CI) | POR | Ph | OR(95%CI) | POR | Ph | OR(95%CI) | POR | Ph | ||
| Overall | 12 | 1.236(1.069–1.429) | 0.007 | 0.030 | 1.754(1.422–2.165) | 1.61 × 10−7 | 0.404 | 1.093(0.952–1.255) | 0.205 | 0.140 |
| Asian | 7 | 1.326(1.095–1.604) | 0.006 | 0.065 | 1.784(1.408–2.261) | 1.67 × 10−6 | 0.630 | 1.149(0.957–1.380) | 0.137 | 0.121 |
| Caucasian | 3 | 1.087(0.888–1.331) | 0.420 | 0.812 | 2.055(1.106–3.817) | 0.023 | 0.587 | 1.019(0.812–1.279) | 0.872 | 0.592 |
| African | 1 | 1.749(1.153–2.655) | 0.009 | — | 2.369(1.198–4.685) | 0.013 | — | 1.777(0.907–3.479) | 0.094 | — |
| African-American | 1 | 0.769(0.537–1.099) | 0.149 | — | 0.745(0.353–1.569) | 0.438 | — | 0.733(0.467–1.152) | 0.178 | — |
OR odd ratio, 95%CI confidence interval, POR P value for the test of association, Ph P value for heterogeneity analysis.
FCGR3A rs396991 polymorphism and SLE risk
There were 26 studies with 5694 cases and 6450 controls in our meta-analysis to evaluate the relationship between FCGR3A rs396991 polymorphism and SLE. Firstly, we tested the dominant and recessive models to estimate the relation between rs396991 and SLE risk (Table 4). We found that rs396991 were significant association with the susceptibility to SLE in overall population in recessive model of T allele (TT vs TG/GG, OR = 1.263, 95%CI: 1.123–1.421, P = 9.62 × 10−5, Table 4, Fig. 2c), and in Caucasian population (TT vs TG/GG, OR = 1.394, 95%CI: 1.087–1.789, P = 9.05 × 10−3) and in mixed population (TT vs TG/GG, OR = 1.585, 95%CI: 1.122–2.239, P = 9.05 × 10−3). Similarly, recessive model is the best fit for the association of rs396991_T, because we didn’t observe any association under dominant model in any populations (Table 4). We also tested the allelic model to observe the relationship between rs396991 and SLE. The significant association was seen between rs396991 and SLE in the overall population (OR = 1.17 per T allele, 95%CI: 1.059–1.291, P = 1.94 × 10−3, Table 4, Supplementary Fig. S2c). And we also found trend of association between this polymorphism and SLE in the stratified analysis of ethnicity: (Caucasian, OR = 1.259 per T allele, P = 0.039; Asian population, OR = 1.152 per T allele, P = 0.05, Table 4, Fig. 2c).
Table 4. Meta-analysis of the association between FCGR3A rs396991 polymorphism and SLE risk.
| Population | N | T vs. G(allele model) |
TT vs. TG+GG(recessive model) |
TT+TG vs. GG(dominant model) |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| OR(95%CI) | POR | Ph | OR(95%CI) | POR | Ph | OR(95%CI) | POR | Ph | ||
| Overall | 26 | 1.17(1.059–1.291) | 0.002 | 0.000 | 1.263(1.123–1.421) | 9.62 × 10−5 | 0.003 | 1.114(0.933–1.331) | 0.232 | 0.004 |
| Caucasian | 9 | 1.259(1.012–1.566) | 0.039 | 0.000 | 1.394(1.087–1.789) | 9.05 × 10−3 | 0.008 | 1.187(0.830–1.699) | 0.347 | 0.004 |
| Asian | 13 | 1.152(0.999–1.328) | 0.051 | 0.004 | 1.211(1.022–1.434) | 0.027 | 0.036 | 1.164(0.884–1.533) | 0.280 | 0.049 |
| African-American | 1 | 1.022(0.880–1.186) | 0.776 | — | 1.053(0.861–1.287) | 0.617 | — | 0.972(0.712–1.327) | 0.858 | — |
| Mixed population | 1 | 1.308(1.029–1.662) | 0.028 | — | 1.585(1.122–2.239) | 9.05 × 10−3 | — | 1.172(0.761–1.804) | 0.471 | — |
| Non-Caucasian | 1 | 0.903(0.649–1.258) | 0.548 | — | 1.049(0.656–1.677) | 0.843 | — | 0.636(0.336–1.204) | 0.164 | — |
| African | 1 | 1.196(0.616–2.324) | 0.597 | — | 1.899(0.638–5.654) | 0.249 | — | 0.750(0.184–3.060) | 0.688 | — |
OR odd ratio, 95%CI confidence interval, POR P value for the test of association, Ph P value for heterogeneity analysis.
FCGR3B NA1·NA2 copy number polymorphism and SLE risk
Totally, 11 studies included 1692 cases and 1899 controls were in our meta-analysis to assess the relation between FCGR3B NA1·NA2 copy number polymorphism and SLE. The meta-analysis indicated that NA1·NA2 was modestly associated with SLE in overall population (allele genetic model: OR = 0.851 per NA1, 95%CI: 0.772–0.938, P = 1.2 × 10−3, Table 5, Fig. 2d; recessive model of NA1: OR = 0.799, 95%CI: 0.685–0.933, P = 0.005, Table 5, Supplementary Fig. S2d). Analysis by population showed that NA1·NA2 was modestly associated with SLE in Asian by three models (allele genetic model: OR = 0.785, 95%CI: 0.697–0.883, P = 6.07 × 10−5, Table 5, Fig. 2d; dominant model: OR = 0.684, 95%CI: 0.549–0.853, P = 7.2 × 10−4, Table 5, Supplementary Fig. S1d; recessive model: OR = 0.756, 95%CI: 0.635–0.898, P = 0.002, Table 5, Supplementary Fig. S2d).
Table 5. Meta-analysis of the association between FCGR3B copy number polymorphism NA1·NA2 and SLE risk.
| Population | N | NA1 vs. NA2(allele model) |
NA1·NA1 vs. NA1·NA2+NA2·NA2 (recessive model) |
NA1·NA1+NA2·NA2 vs. NA2·NA2 (dominant model) |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| OR(95%CI) | POR | Ph | OR(95%CI) | POR | Ph | OR(95%CI) | POR | Ph | ||
| Overall | 11 | 0.851(0.772–0.938) | 1.2 × 10−3 | 0.004 | 0.799(0.685–0.933) | 0.005 | 0.182 | 0.825(0.702–0.969) | 0.019 | 0.001 |
| Asian | 3 | 0.785(0.697–0.883) | 6.07 × 10−5 | 0.040 | 0.756(0.635–0.898) | 0.002 | 0.116 | 0.684(0.549–0.853) | 7.2 × 10−4 | 0.103 |
| Caucasian | 8 | 1.013(0.851–1.205) | 0.888 | 0.060 | 1.006(0.709–1.426) | 0.974 | 0.885 | 1.021(0.806–1.292) | 0.866 | 0.003 |
OR odd ratio, 95%CI confidence interval, POR P value for the test of association, Ph P value for heterogeneity analysis.
Allele frequency of the 3 SNPs and comparing to the 1000 genome population
In Table 6, we showed the distinct difference of allele frequencies in Asian, Caucasian, African and African American population in the meta-analysis of the 3 SNPs. The allele frequencies of the 3 SNPs in Asian, Caucasian, African and African American population in the meta–analysis were consistent with the allele frequencies in 1000 Genome Project EUR (European ancestry), ASN (Asian ancestry), AFR (African ancestry), ASW (Americans of African Ancestry), respectively.
Table 6. The allele frequency comparison between the meta-analysis and 1000 Genomes Project.
| Polymorphism | Populations | Meta-analysis(alleles frequencies) |
|
||||
|---|---|---|---|---|---|---|---|
| Cases |
Controls |
1000 Genomes(Alleles frequencies) |
|||||
| A | G | A | G | A | G | ||
| SNP rs1801274 | Caucasian | 0.445 | 0.555 | 0.474 | 0.526 | 0.500(EUR) | 0.5(EUR) |
| Asian | 0.652 | 0.348 | 0.697 | 0.303 | 0.722(ASN) | 0.278(ASN) | |
| African | 0.568 | 0.432 | 0.602 | 0.398 | 0.512(AFR) | 0.488(AFR) | |
| African Americans | 0.393 | 0.607 | 0.529 | 0.471 | 0.525(ASW) | 0.475(ASW) | |
| Mixed population | 0.479 | 0.521 | 0.448 | 0.552 | |||
| Non-Caucasian | 0.493 | 0.507 | 0.436 | 0.564 | |||
| All | 0.563 | 0.437 | 0.595 | 0.405 | 0.57(ALL) | 0.43(ALL) | |
| SNP rs1050501 | C | T | C | T | C | T | |
| Asian | 0.280 | 0.720 | 0.248 | 0.752 | 0.255(ASN) | 0.745(ASN) | |
| Caucasian | 0.128 | 0.872 | 0.107 | 0.893 | 0.123(EUR) | 0.877(EUR) | |
| African | 0.572 | 0.428 | 0.433 | 0.567 | 0.248(AFR) | 0.752(AFR) | |
| African-American | 0.241 | 0.759 | 0.292 | 0.708 | 0.213(ASW) | 0.787(ASW) | |
| All | 0.249 | 0.751 | 0.198 | 0.802 | 0.188(ALL) | 0.812(ALL) | |
| SNP rs396991 | T | G | T | G | T | G | |
| Caucasian | 0.659 | 0.341 | 0.629 | 0.371 | 0.731(EUR) | 0.269(EUR) | |
| Asian | 0.673 | 0.327 | 0.657 | 0.343 | 0.731(ASN) | 0.269(ASN) | |
| African-American | 0.664 | 0.336 | 0.659 | 0.341 | 0.713(ASW) | 0.287(ASW) | |
| Mixed population | 0.622 | 0.378 | 0.557 | 0.443 | |||
| Non-Caucasian | 0.600 | 0.400 | 0.624 | 0.376 | |||
| African | 0.586 | 0.414 | 0.542 | 0.458 | 0.785(AFR) | 0.215(AFR) | |
| All | 0.663 | 0.337 | 0.641 | 0.359 | 0.755(ALL) | 0.245(ALL) | |
EUR European ancestry, ASN Asian ancestry, AFR African ancestry, ASW Americans of African Ancestry, ALL All individuals from phase 1 of the 1000 Genomes Project.
Publication bias and Sensitivity analysis
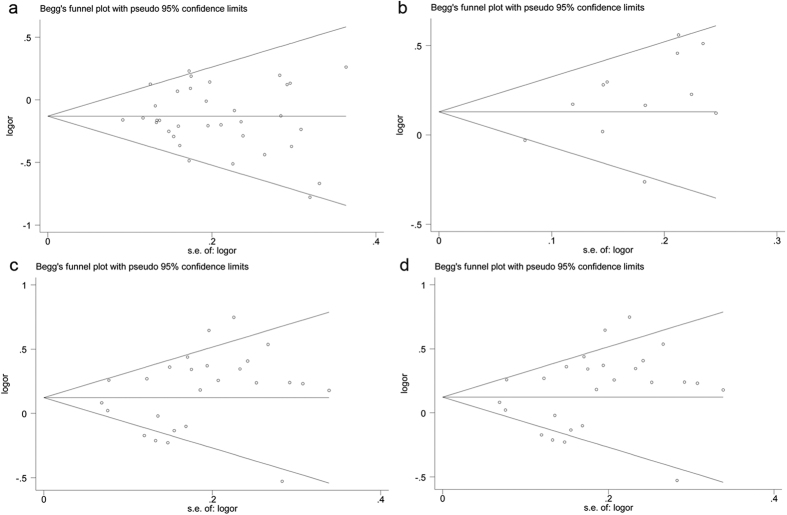
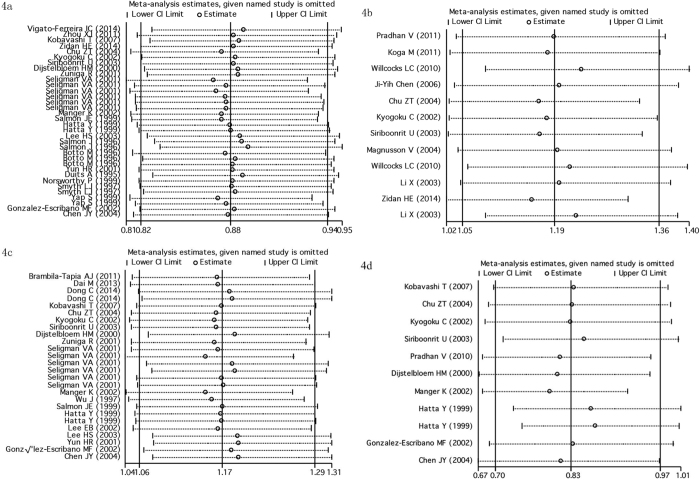
Begg’s funnel plot and Egger’s test were performed to estimate publication bias. There was no obvious evidence of symmetry from the shapes of the funnel plots (Fig. 3), and showed no evidence of publication bias in rs1801274 polymorphism (P = 0.594), rs396991 polymorphism (P = 0.252), NA1·NA2 polymorphism (P = 0.213), and rs1050501 polymorphism (P = 0.115, after excluded the study by Kobavashi T et al.59) under allele genetic model in our meta-analysis (Fig. 3a–d). We also conducted sensitivity analysis to assess the influence of individual studies on the pooled ORs. We found the pooled OR was not substantially altered, when any one study was deleted (Fig. 4a–d).
Figure 3. Begg’s funnel plot of publication bias in the meta-analysis of the association of FCGRs polymorphisms with SLE risk under allele genetic model.
(a) FCGR2A rs1801274 and SLE (A vs G); (b) FCGR2B rs1050501 and SLE (C vs T); (c) FCGR3A rs396991 and SLE (T vs G); (d) FCGR3B NA1·NA2 and SLE (NA1 vs NA2).
Figure 4. Sensitivity analysis to assess the stability of the meta-analysis.
(a) FCGR2A rs1801274 in SLE; (b) FCGR2B rs1050501 in SLE; (c) FCGR3A rs396991 in SLE; (d) FCGR3B NA1·NA2 in SLE).
Discussion
In this study, we conducted a meta-analysis of the association between FCGR2A, 2B, 3A and 3B polymorphisms and SLE susceptibility. We found that C allele of rs1050501 (FCGR2B) and T allele of rs396991 (FCGR3A) strongly increase the risk of SLE. We also found significant association between FCGR2A rs1801274, FCGR3B copy number polymorphism NA1·NA2, and SLE in the overall population.
SNP rs1801274 is a missense mutation in FCGR2A gene on chromosome 1q23.3 (161479745), which encodes substitution of histidine (H) by arginine (R) in the IgG-binding domain of FcgRIIa and it was reported that FcgRIIa-R has a lower binding affinity for IgG than FcgRIIa-H68. In our study, we found FCGR2A rs1801274 contributes to SLE susceptibility in overall population. And in the subgroup analysis, the polymorphism was associated with SLE in Asian, Caucasian, and African Americans but not in African population, however, there were only 3 studies for African population in this meta-analysis, consisting only 190 cases and 198 controls, and the effect direction of A allele in African population is the same as that in the overall population. Previous study such as by Karassa FB et al.70 presented the association between FCGR2A rs1801274 and SLE of Caucasian descent, but it was less clear in subjects of Asian or African descent. Another study71 found a significant association of rs1801274 G allele and increased SLE risk in all groups, and a clear effect of G allele on SLE was shown in European and Asian, these results were consistent with our study. We also confirmed the findings from Zhou XJ65 that investigated the association between rs1801274 and SLE in Chinese population. In many ways, we suggest that rs1801274 was associated with SLE, especially in Caucasian and Asian population. As for other populations, more studies were needed to evaluate association between the polymorphism and SLE. It’s likely that such differences may, at less in part, be attributable to the ethnic difference.
GWAS have found that there were significant associations between FCGR2A rs1801274 and Kawasaki disease72 and Inflammatory bowel disease (P = 2.12 × 10−38, OR = 1.12)73 and there were genome-wide significant associations between the SNP and Ulcerative colitis in European74, and Japanese population75. There was only one genome-wide association study between FCGR2A and SLE, however, SNP rs1801274 was not genome-wide significant27.
FcgRIIb is an inhibitory receptor mediating B-cell function via an immune receptor tyrosine-based inhibitory motif 59. FcgRIIb is the only FcgR that transmits an inhibitory signal and is expressed in B cells and myelomonocytic cells57. FCGR2B rs1050501 (c.695T > C) codes a non-synonymous substitution, Ile232Thr (I232T) on chromosome 1q23.3 (161644048), our meta-analysis showed that C allele significantly increased the risk of SLE under recessive association model and allelic test model in overall population (Table 3, Fig. S2a; Supplementary Fig. S2b). By subgroup analysis, the association was also found under allelic genetic model and recessive model in Asian populations, but not in Caucasians under allelic genetic model. In 2004, Chu ZT et al.57 had found rs1050501 was significant associated with SLE in Chinese population. These results were in agreement with Lee YH et al.76 that indicated the C allele significantly increased the risk of SLE in Asian population. Therefore, it was suggested that the association between FCGR2B rs1050501 and SLE was on the basis of ethnicity, and the C allele is a risk for SLE in Asian.
FcγRIIIa is expressed on the surfaces of natural killer (NK) cells, monocytes and macrophages and binds to IgG1 and IgG3 subclasses66. FCGR3A rs396991 is a missense mutation on chromosome 1q23.3 (161514792), leading to a valine (V) substitution for phenylalanine (F) at amino acid residue 176 (including the leader sequence)66. In our meta-analysis, it suggested that a significant association between FCGR3A rs396991_T and SLE in overall population under recessive association models and allele genetic model (Table 4, Fig. 2c; Supplementary Fig. S2c). Previous study77 had suggested a modest trend of SLE predisposition for FCGR3A rs396991 in 1,261 SLE patients and 1,455 disease-free controls but with significant between-study heterogeneity. In addition, we observed trend of association between this polymorphism and SLE in the stratified analysis of ethnicity in Caucasian and Asian population, which was consistent with the study of Li et al.78. However, the association was not confirmed in the population of African and African American.
The copy number variation (NA1·NA2) in FCGR3B has shown to influence the interaction between FcγRIIIb and human IgG61. Individuals who are homozygous for NA1 allele has greater phagocytosis of IgG opsonized targets than that of NA2 homozygous individuals. Our meta-analysis illustrated a modest association between this copy number polymorphism and SLE in overall population by allele genetic model and recessive model. Analysis by population showed that NA1·NA2 was associated with SLE in Asians by three models. This association was not observed in a small sample size of 165 Chinese patients with SLE and 129 healthy controls by Chu ZT et al.57. To further explain the differences, we compared frequency between our meta analysis and those from Chu ZT et al.57 in Table 1, From this table, we could tell the frequencies were consistent between the two, the sample size might have been responsible for the different results. Besides, we didn’t find an association between FCGR3B NA1·NA2 polymorphism and SLE in Caucasian.
Though we tried to control the potential bias of publications and populations. There were still have several limitations to be taken into consideration in this meta-analysis. Firstly, although the overall sample size is large, the size of each study is relatively small, with the smallest sample of 30 cases and 31 controls. Secondly, the meta-analysis for ethnicity included data more from population with Caucasian and Asian origin, and the findings are applicable to only these populations, more studies are required in other populations. Furthermore, the mechanism of SLE is considered to be sophisticated, including gene-gene and gene-environment interactions. More studies with enough statistical power are needed for deeply evaluation. Lastly, publication bias might affect the results, because the studies that found any negative results may not have been published.
Despite the limitations, this meta-analysis illustrated that C allele of FCGR2B rs1050501 and T allele of FCGR3A rs396991 might contribute to susceptibility and development of SLE, and were under recessive association model. While, A allele of FCGR2A rs1801274 and FCGR3B NA1 were associated with SLE and reduced the risk of SLE. Considering the limited samples in Africans and African Americans in this meta-analysis, studies with larger sample size including diverse ethnic populations are still required to investigate the association between FCGRs genes polymorphisms and SLE in the future.
Methods
Identification of eligible studies
We aimed to analyze the association between FCGR2A (SNP rs1801274), FCGR2B (SNP rs1050501), FCGR3A (SNP rs396991), FCGR3B copy number polymorphism (NA1/NA2) polymorphisms and SLE. Therefore, all published literatures before December 2015 that investigated the association between these polymorphisms and SLE risk were searched using the PubMed engine (National Center for Biotechnology, National Library of Medicine). We looked for the articles with keywords “FCGR2A”, “FCGR2B”, “FCGR3A”, “FCGR3B”, “FCγRs”, “polymorphism” in combination with “Systemic Lupus erythematosus” or “SLE”. Finally, we extracted data from the published articles, not from conference abstracts or any meetings.
Data extraction
All studies should meet the following conditions: 1) case-control study; 2) with original data to calculate genotype counts and odds ratio (OR); 3) the diagnosis of SLE patients according to the American College of Rheumatology criteria79,80. The following information is shown in our study: first author, year of publication, ethnicity, sample size of cases and controls, allele frequency and genotype frequency.
Statistical analysis
The allele frequencies of polymorphisms from each study were calculated by the allele counting method. Pooled ORs and 95% confidence intervals (CIs) were used to evaluate the strength of association between polymorphisms and SLE risk for every eligible study. Heterogeneity was evaluated using the I2 metric, which ranges between 0 and 100% (25%, low heterogeneity; 50%, moderate; 75%, high heterogeneity)81. If the P value for heterogeneity test was higher than 0.01, the fixed effect model was used to weight of each study. Moreover, the random effect model was also used. In this meta-analysis, P value of less than 0.05 was considered a statistically significant.
In order to get better search results, we evaluated possible publication bias by Egger’s linear regression text82. P value < 0.05 was considered representative of statistical publication bias82. We also used a funnel plot to evaluate the publication bias by Begg’s test83. For sensitivity analysis, removed one study from the total and tested residual studies. Statistical analysis was carried out using the software program STATA10.1 (Stata Corporation, College Station, Texas).
Additional Information
How to cite this article: Zhu, X.-W. et al. Comprehensive Assessment of the Association between FCGRs polymorphisms and the risk of systemic lupus erythematosus: Evidence from a Meta-Analysis. Sci. Rep. 6, 31617; doi: 10.1038/srep31617 (2016).
Supplementary Material
Acknowledgments
This study was supported by the National Natural Science Foundation of China (81501145), the State Key Development Program for Basic Research of China (973 Program, 2014CB541701), the HZNUARI-Pilot Research Grant, the Medical Science and Technology Program of Shandong Province (2014WS0191) and the Science and Technology Program of Binzhou City (2014ZC0125). The funding agencies had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. We thank the peer reviewers for their thorough and helpful review of this manuscript.
Footnotes
Author Contributions Conceived and designed the study: H.-F.Z. Performed the literature searching: X.-W.Z., Y.W. and Y.-H.W. Contributed crosschecking the literatures: Y.W., Y.-H.W., J.-J.R., P.-P.Z., X.-B.W., W.-Y.Z., X.-W.Z., L.W., X.-W.Z. and H.-F.Z. Analyzed the data: H.-F.Z. Wrote the manuscript: X.-W.Z. and H.-F.Z.
References
- Harley J. B., Kelly J. A. & Kaufman K. M. Unraveling the genetics of systemic lupus erythematosus. Springer Semin Immunopathol 28, 119–130 (2006). [DOI] [PubMed] [Google Scholar]
- Hopkinson N. D., Doherty M. & Powell R. J. Clinical features and race-specific incidence/prevalence rates of systemic lupus erythematosus in a geographically complete cohort of patients. Ann Rheum Dis 53, 675–680 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau C. S., Yin G. & Mok M. Y. Ethnic and geographical differences in systemic lupus erythematosus: an overview. Lupus 15, 715–719 (2006). [DOI] [PubMed] [Google Scholar]
- Zeng Q. Y. et al. Rheumatic diseases in China. Arthritis research & therapy 10, R17 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danchenko N., Satia J. A. & Anthony M. S. Epidemiology of systemic lupus erythematosus:a comparison of worldwide disease burden. Lupus 15, 308–318. (2006). [DOI] [PubMed] [Google Scholar]
- Ardoin S. P. & Pisetsky D. S. Developments in the scientific understanding of lupus. Arthritis Rheum 10, 218 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsao B. P. et al. Linkage and interaction of loci on 1q23 and 16q12 may contribute to susceptibility to systemic lupus erythematosus. Arthritis and rheumatism 46, 2928–2936 (2002). [DOI] [PubMed] [Google Scholar]
- Tsao B. P. et al. Evidence for linkage of a candidate chromosome 1 region to human systemic lupus erythematosus. J Clin Invest 99, 725–731 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray-McGuire C. et al. Genome scan of human systemic lupus erythematosus by regression modeling: evidence of linkage and epistasis at 4p16-15.2. Am J Hum Genet 67, 1460–1469 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawalha. et al. Genetic linkage of systemic lupus erythematosus with chromosome 11q14 (SLEH1) in African-American families stratified by a nucleolar antinuclear antibody pattern. Genes Immun 3 Suppl 1, S31–34 (2002). [DOI] [PubMed] [Google Scholar]
- Nath S. K. et al. Linkage at 12q24 with Systemic Lupus Erythematosus (SLE) Is Established and Confirmed in Hispanic and European American Families. Am J Hum Genet 74, 73–82 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linker-Israeli M. et al. Association of IL-6 gene alleles with systemic lupus erythematosus (SLE) and with elevated IL-6 expression. Genes Immun 1, 45–52 (1999). [DOI] [PubMed] [Google Scholar]
- Devaraju P. G. R., Antony P. T., Mithun C. B. & Negi V. S. Susceptibility to SLE in South Indian Tamils may be influenced by genetic selection pressure on TLR2 and TLR9 genes. Mol Immunol 64, 123–126 (2015). [DOI] [PubMed] [Google Scholar]
- Sakulpipatsin W. et al. Vitamin D receptor gene BsmI polymorphisms in Thai patients with systemic lupus erythematosus. Arthritis Res Ther 8, R48 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi F. N. et al. The CTLA-4-1661A/G and-1772T/C dimorphisms in Japanese patients with systemic sclerosis. Rheumatol Int 27, 785–787 (2007). [DOI] [PubMed] [Google Scholar]
- Smyth L. J. et al. Fc gamma RIIa polymorphism in systemic lupus erythematosus. Ann Rheum Dis 56, 744–746 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan F. et al. Association of Fcgamma receptor IIB gene polymorphism with genetic susceptibility to systemic lupus erythematosus in Chinese populations–a family-based association study. J Dermatolo Sci 43, 35–41 (2006). [DOI] [PubMed] [Google Scholar]
- Chen F. R. et al. Association of PELI1 polymorphisms in systemic lupus erythematosus susceptibility in a Chinese population. Lupus (2015). [DOI] [PubMed] [Google Scholar]
- Cai X. et al. Association between polymorphisms of the IKZF3 gene and systemic lupus erythematosus in a Chinese Han population. PloS one 9, e108661 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hom G. et al. Association of systemic lupus erythematosus with C8orf13-BLK and ITGAM-ITGAX. N Engl J Med 358, 900–909 (2008). [DOI] [PubMed] [Google Scholar]
- International Consortium for Systemic Lupus Erythematosus. et al. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet 40, 204-210 (2008). [DOI] [PMC free article] [PubMed]
- Yang W. et al. Asian Lupus Genetics, Consortium Genome-wide association study in Asian populations identifies variants in ETS1 and WDFY4 associated with systemic lupus erythematosus. PLoS Genet 6, e1000841 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S. A. et al. Differential genetic associations for systemic lupus erythematosus based on anti-dsDNA autoantibody production. PLoS Genet 7, e1001323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y. et al. A Genome-Wide Association Study Identified AFF1 as a Susceptibility Locus for Systemic Lupus Eyrthematosus in Japanese. PLoS Genet 8 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. H., Bae. S. C., Choi S. J., Ji J. D. & Song G. G. Genome-wide pathway analysis of genome-wide association studies on systemic lupus erythematosus and rheumatoid arthritis. Mol Biol Rep 39, 10627–10635 (2012). [DOI] [PubMed] [Google Scholar]
- Han J. W. et al. Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus. Nat Genet 41, 1234–1237 (2009). [DOI] [PubMed] [Google Scholar]
- Armstrong D. L. et al. GWAS identifies novel SLE susceptibility genes and explains the association of the HLA region. Genes Immun 15, 347–354 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravetch J. V. B. & IgG Fc S. receptors. Annual review of immunology 19, 275–290 (2001). [DOI] [PubMed] [Google Scholar]
- Smith K. G. C. & FcgammaRIIB M. R. in autoimmunity and infection: evolutionary and therapeutic implications. Nat Rev Immunol 10, 328–343 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y. T. B. P. Genetic susceptibility to systemic lupus erythematosus in the genomic era. Nat Rev Immunol 6, 683–692 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Criswell L. A. The genetic contribution to systemic lupus erythematosus. Bull NYU Hosp Jt Dis 66, 176–183 (2008). [PubMed] [Google Scholar]
- Su K. et al. Promoter Haplotype of the Immunoreceptor Tyrosine-Based Inhibitory Motif-Bearing Fc RIIb Alters Receptor Expression and Associates with Autoimmunity. I. Regulatory FCGR2B Polymorphisms and Their Association with Systemic Lupus Erythematosus. J Immunol 172, 7186–7191 (2004). [DOI] [PubMed] [Google Scholar]
- Magnusson V. et al. Polymorphisms of the Fc gamma receptor type IIB gene are not associated with systemic lupus erythematosus in the Swedish population. Arthritis Rheum 50, 1348–1350 (2004). [DOI] [PubMed] [Google Scholar]
- Blank M. C. et al. Decreased transcription of the human FCGR2B gene mediated by the -343 G/C promoter polymorphism and association with systemic lupus erythematosus. Hum Genet 117, 220–227 (2005). [DOI] [PubMed] [Google Scholar]
- Pan F. et al. Genetic susceptibility and haplotype analysis between Fcgamma receptor IIB and IIIA gene with systemic lupus erythematosus in Chinese population. Lupus 17, 733–738 (2008). [DOI] [PubMed] [Google Scholar]
- Zhong H. et al. eplicated associations of TNFAIP3, TNIP1 and ETS1 with systemic lupus erythematosus in a southwestern Chinese population. Arthritis Res Ther 13, R186 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jönsen A., Bengtsson A. A., Sturfelt G. & Truedsson L. Analysis of HLA DR, HLA DQ, C4A, FcgammaRIIa, FcgammaRIIIa, MBL, and IL-1Ra allelic variants in Caucasian systemic lupus erythematosus patients suggests an effect of the combined FcgammaRIIa R/R and IL-1Ra 2/2 genotypes on disease susceptibility. Arthritis Res Ther 6, R557–562 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duits A. J. et al. Skewed distribution of IgG Fc receptor IIa(CD32) polymorphism is associated with renal disease in systemic lupus erythematosus patients. Arthritis Res Ther 38, 1832–1836 (1995). [DOI] [PubMed] [Google Scholar]
- Botto M. et al. Fc gamma RIIa polymorphism in systemic lupus erythematosus (SLE)_ no association with disease. Clin Exp Immunol 104, 264–268 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon J. E. et al. Fc gamma RIIA alleles are heritable risk factors for lupus nephritis in African Americans. J Clin Invest 97, 1348–1354 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J. E. et al. A novel polymorphism of FcgammaRIIIa (CD16) alters receptor function and predisposes to autoimmune disease. J Clin Invest 100, 1059–1070 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatta Y. et al. Association of Fc gamma receptor IIIB, but not of Fc gamma receptor IIA and IIIA polymorphisms with systemic lupus erythematosus in Japanese. Genes Immun 1, 53–60 (1999). [DOI] [PubMed] [Google Scholar]
- Norsworthy P. et al. Overrepresentation of the Fcgamma receptor type IIA R131_R131 genotype in caucasoid systemic lupus erythematosus patients with autoantibodies to C1q and glomerulonephritis. Arthritis Rheum 42, 1828–1832 (1999). [DOI] [PubMed] [Google Scholar]
- Salmon J. E. et al. Altered distribution of Fcgamma receptor IIIA alleles in a cohort of Korean patients with lupus nephritis. Arthritis and rheumatism 42, 818–819 (1999). [DOI] [PubMed] [Google Scholar]
- Yap S. N., Phipps M. E., Manivasagar M., Tan S. Y. & Bosco J. J. Human Fc receptor IIA (Fc RIIA) genotyping and association with systemic lupus erythematosus (SLE) in Chinese and Malays in Malaysia. Lupus 8, 305–310 (1999). [DOI] [PubMed] [Google Scholar]
- Dijstelbloem H. M. et al. Fcgamma receptor polymorphisms in systemic lupus erythematosus: association with disease and in vivo clearance of immune complexes. Arthritis Rheum 43, 2793–2800 (2000). [DOI] [PubMed] [Google Scholar]
- Seligman V. A. et al. The Fcgamma receptor IIIA-158F allele is a major risk factor for the development of lupus nephritis among Caucasians but not non-Caucasians. Arthritis Rheum 44, 618–625 (2001). [DOI] [PubMed] [Google Scholar]
- Yun H. R. et al. FcgammaRIIa/IIIa polymorphism and its association with clinical manifestations in Korean lupus patients. Lupus 10, 466–472 (2001). [DOI] [PubMed] [Google Scholar]
- Zuniga R. et al. Low-binding alleles of Fcgamma receptor types IIA and IIIA are inherited independently and are associated with systemic lupus erythematosus in Hispanic patients. Arthritis Rheum 44, 361–367 (2001). [DOI] [PubMed] [Google Scholar]
- González-Escribano M. F., Aguilar F., Sánchez-Román J. & Núñez-Roldán A. FcgammaRIIA, FcgammaRIIIA and FcgammaRIIIB polymorphisms in Spanish patients with systemic lupus erythematosus. Eur J Immunogenet 29, 301–306 (2002). [DOI] [PubMed] [Google Scholar]
- Kyogoku C. et al. Fcgamma receptor gene polymorphisms in Japanese patients with systemic lupus erythematosus: contribution of FCGR2B to genetic susceptibility. Arthritis Rheum 46, 1242–1254 (2002). [DOI] [PubMed] [Google Scholar]
- Lee E. B. et al. Fcγ receptor IIIA polymorphism in Korean patients with systemic lupus erythematosus. Rheumatol Int 21, 222–226 (2002). [DOI] [PubMed] [Google Scholar]
- Manger K. et al. Fcgamma receptor IIa, IIIa, and IIIb polymorphisms in German patients with systemic lupus erythematosus: association with clinical symptoms. Ann Rheum Dis 61, 786–792 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. S. et al. Independent association of HLA-DR and FCgamma receptor polymorphisms in Korean patients with systemic lupus erythematosus. Rheumatology 42, 1501–1507 (2003). [DOI] [PubMed] [Google Scholar]
- Siriboonrit U. et al. Association of Fcgamma receptor IIb and IIIb polymorphisms with susceptibility to systemic lupus erythematosus in Thais. Tissue antigens 61, 374–383 (2003). [DOI] [PubMed] [Google Scholar]
- Chen J. Y. et al. Fcgamma receptor IIa, IIIa, and IIIb polymorphisms of systemic lupus erythematosus in Taiwan. Ann Rheum Dis 63, 877–880 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu Z. T. et al. Association of Fcgamma receptor IIb polymorphism with susceptibility to systemic lupus erythematosus in Chinese: a common susceptibility gene in the Asian populations. Tissue antigens 63, 21–27 (2004). [DOI] [PubMed] [Google Scholar]
- Ye D. et al. A novel single-nucleotide polymorphism of the Fc gamma receptor IIIa gene is associated with genetic susceptibility to systemic lupus erythematosus in Chinese populations: a family-based association study. Clin Exp Dermatol 31, 553–557 (2006). [DOI] [PubMed] [Google Scholar]
- Kobayashi T. et al. The combined genotypes of stimulatory and inhibitory Fc gamma receptors associated with systemic lupus erythematosus and periodontitis in Japanese adults. J Periodontol 78, 467–474 (2007). [DOI] [PubMed] [Google Scholar]
- Morris D. et al. Evidence for both copy number and allelic (NA1/NA2) risk at the FCGR3B locus in systemic lupus erythematosus. Eur J Hum Genet 18, 1027–1031 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradhan V. et al. Fc gamma R IIIB polymorphisms: their association with clinical manifestations and autoantibodies in SLE patients from Western India. Int J Rheum Dis 13, 138–143 (2010). [DOI] [PubMed] [Google Scholar]
- Willcocks L. C. et al. A defunctioning polymorphism in FCGR2B is associated with protection against malaria but susceptibility to systemic lupus erythematosus. Pro Natl Acad USA 107, 7881–7885 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brambila-Tapia A. J. et al. FCGR3A V(176) polymorphism for systemic lupus erythematosus susceptibility in Mexican population. Rheumatol Int 31, 1065–1068 (2011). [DOI] [PubMed] [Google Scholar]
- Pradhan V., Patwardhan M., Nadkarni A. & Ghosh K. Fc γ R IIB gene polymorphisms in Indian systemic lupus erythematosus (SLE) patients. Indian J Med Res 134, 181–185 (2011). [PMC free article] [PubMed] [Google Scholar]
- Zhou X. J. et al. Is FCGR2A a susceptibility gene to systemic lupus erythematosus in Chinese? Lupus 20, 1198–1202 (2011). [DOI] [PubMed] [Google Scholar]
- Dai M., Zhou Z., Wang X., Qian X. & Huang X. Association of FcγRIIIa-158V_F with systemic lupus erythematosus in a Chinese population. Int J Rheum Dis 16, 685–691 (2013). [DOI] [PubMed] [Google Scholar]
- Dong C. et al. Fc gamma Receptor IIIa Single-Nucleotide Polymorphisms and Haplotypes Affect Human IgG Binding and Are Associated With Lupus Nephritis in African Americans. Arthritis Rheumatol 66, 1291–1299 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigato-Ferreira I. C. et al. FcgammaRIIa and FcgammaRIIIb polymorphisms and associations with clinical manifestations in systemic lupus erythematosus patients. Autoimmunity 47, 451–458 (2014). [DOI] [PubMed] [Google Scholar]
- Zidan H. E. et al. Association of FcgammaRIIB and FcgammaRIIA R131H gene polymorphisms with renal involvement in Egyptian systemic lupus erythematosus patients. Mol Biol Rep 41, 733–739 (2014). [DOI] [PubMed] [Google Scholar]
- Karassa F. B., Trikalinos T. A., Ioannidis J. P. & FcgammaRIIa-SLE Meta-Analysis Investigators. Role of the Fcgamma receptor IIa polymorphism in susceptibility to systemic lupus erythematosus and lupus nephritis: a meta-analysis. Arthritis Rheum 46, 1563–1571 (2002). [DOI] [PubMed] [Google Scholar]
- Yuan H. et al. Meta analysis on the association between FcgammaRIIa-R/H131 polymorphisms and systemic lupus erythematosus. Mol Biol Rep 36, 1053–1058 (2009). [DOI] [PubMed] [Google Scholar]
- Khor C. C. et al. Genome-wide association study identifies FCGR2A as a susceptibility locus for Kawasaki disease. Nat Genet 43, 1241–1246 (2011). [DOI] [PubMed] [Google Scholar]
- Jostins L. et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 491, 119–124 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson C. A. et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat Genet 43, 246–252 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano K. et al. A genome-wide association study identifies three new susceptibility loci for ulcerative colitis in the Japanese population. Nat Genet 41, 1325–1329 (2009). [DOI] [PubMed] [Google Scholar]
- Lee Y. H., Ji J. & Song G. G. Fcgamma receptor IIB and IIIB polymorphisms and susceptibility to systemic lupus erythematosus and lupus nephritis: a meta-analysis.pdf. Lupus 18, 727–734 (2009). [DOI] [PubMed] [Google Scholar]
- Karassa F. B., Trikalinos T. A., Ioannidis J. P. & Fc gamma RIIIA-SLE meta-analysis investigators Fc gamma RIIIA-SLE meta-analysis investigators. The Fc gamma RIIIA-F158 allele is a risk factor for the development of lupus nephritis: a meta-analysis. Kidney Int 63, 1475–1482 (2003). [DOI] [PubMed] [Google Scholar]
- Li L. H. et al. Role of the Fcgamma receptor IIIA-V/F158 polymorphism in susceptibility to systemic lupus erythematosus and lupus nephritis: a meta-analysis. Scand J Rheumatol 39, 148–154 (2010). [DOI] [PubMed] [Google Scholar]
- Tan E. M. et al. The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum 25, 1271–1277 (1982). [DOI] [PubMed] [Google Scholar]
- Hochberg M. C. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum 40, 1725 (1997). [DOI] [PubMed] [Google Scholar]
- Higgins J. P., Thompson S. G., Deeks J. J. & Altman D. G. Measuring inconsistency in meta-analysis. BMJ 327, 557–560 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger M., Davey S. G., Schneider M. & Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ 315, 629–634 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begg C. B. & Mazumdar M. Operating Characteristics of a Rank CorrelationTest for publcation bias. Biometrics 50, 1088–1101 (1994). [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.