Abstract
Accumulated studies have provided controversial evidences of the association between signal transducer and activator of transcription proteins 3 (STAT3) expression and survival of human solid tumors. To address this inconsistency, we performed a meta-analysis with 63 studies identified from PubMed, Medline and EBSCO. We found STAT3 overexpression was significantly associated with worse 3-year overall survival (OS) (OR = 2.06, 95% CI = 1.57 to 2.71, P < 0.00001) and 5-year OS (OR = 2.00, 95% CI = 1.53 to 2.63, P < 0.00001) of human solid tumors. Similar results were observed when disease free survival (DFS) were analyzed. Subgroup analysis showed that elevated STAT3 expression was associated with poor prognosis of gastric cancer, lung cancer, gliomas, hepatic cancer, osteosarcoma, prostate cancer, pancreatic cancer but better prognosis of breast cancer. The correlation between STAT3 and survival of solid tumors was related to its phosphorylated state. High expression level of STAT3 was also associated with advanced tumor stage. In conclusion, elevated STAT3 expression is associated with poor survival in most solid tumors. STAT3 is a valuable biomarker for prognosis prediction and a promising therapeutic target in human solid tumors.
Keywords: STAT3, solid tumors, prognosis, overall survival, disease free survival
INTRODUCTION
Signal transducer and activator of transcription proteins 3 (STAT3) is well demonstrated to play a crucial role in tumor development and cancer-related inflammation [1]. STAT3 is also linked to inflammation-related oncogenesis initiated by genetic alterations and environmental factors [2–4], and is constitutively activated in various cancers [5, 6]. Persistent activation of STAT3 is involved in promoting tumor cell proliferation, survival, tumor invasion, angiogenesis and immunosuppression, inducing and maintaining a pro-carcinogenic inflammatory microenvironment [7]. Growing studies identified novel tumor-promoting functions of STAT3 in mitochondria metabolism [8], drug resistance [9, 10], epigenetic regulation [11], cancer stem cells [12, 13] and pre-metastatic niches [14, 15]. Given the pivotal role in tumor development, STAT3 represents an attractive therapeutic target for solid tumors. Recently, accumulating studies have demonstrated STAT3-targeted therapy could effectively restrain tumor development in various solid tumors [16–21]. However, the prognostic value of STAT3 overexpression in human solid tumors is still controversial.
A plenty of studies showed that elevated STAT3 expression in tumor tissue was correlated with poor survival of patients with various solid tumors such as gastric cancer [22–29], lung cancer [30–37], gliomas [38–42], colorectal cancer [43], ovarian cancer [44], cervical cancer [45], hepatocellular carcinoma [46, 47], melanoma [48], esophageal cancer [49], osteosarcoma [50, 51], pancreatic cancer [52, 53], thymic epithelial tumor [54], astrocytomas [55], lingual squamous cell carcinoma [56], nasopharyngeal carcinoma [57], prostate cancer [58], renal cell carcinoma [59] and Wilms' tumor [60]. However, other studies reported that overexpression of STAT3 was correlated with favorable outcome of patients with breast cancer [61–65], gastric cancer [66], lung cancer [67–69], colorectal cancer [70, 71] and melanoma [72].
Therefore, we carried out a meta-analysis combining available evidences to evaluate the prognostic value of STAT3 expression in solid tumors. We also evaluated whether the clinical outcome of patients with solid tumors differed between STAT3 phosphorylation state and between different tumor types. This meta-analysis intended to assess the role of STAT3 in relation to survival in solid tumors, thereby supporting more rational development of therapeutic strategies against STAT3.
RESULTS
Search results and study characteristics
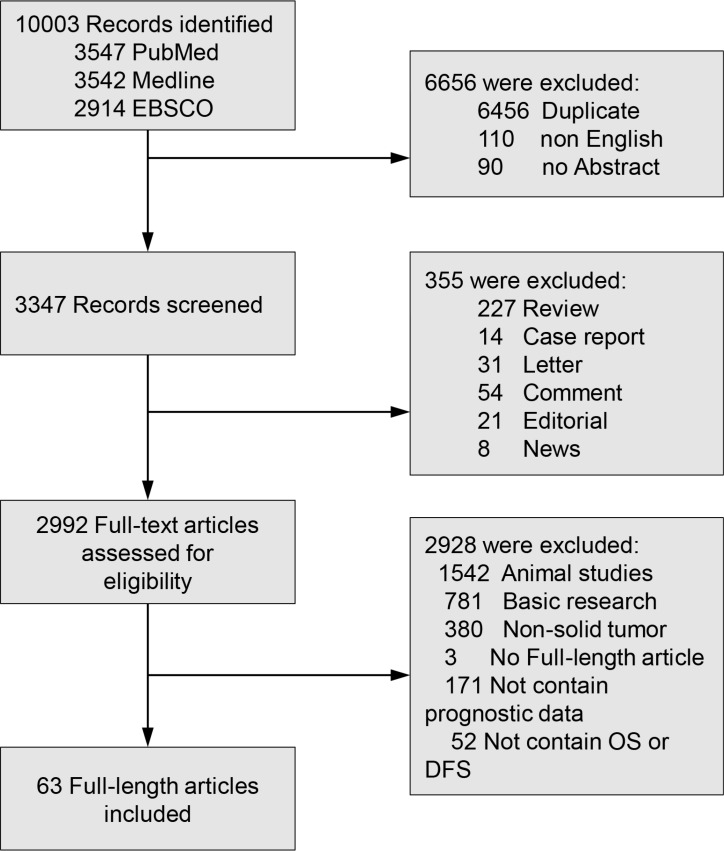
Sixty-three studies with a total of 9449 patients were included (Figure 1). Characteristics of included studies are shown in Table 1. Eleven studies evaluated lung cancer [30–37, 67–69], nine evaluated gastric cancer [22–29, 66], five evaluated breast cancer [61–65], five evaluated gliomas [38–42], four evaluated colorectal cancer [43, 70, 71, 73], three evaluated ovarian cancer [44, 74, 75], three evaluated cervical cancer [45, 76, 77], two evaluated hepatocellular carcinoma [46, 47], two evaluated melanoma [48, 72], two evaluated esophageal cancer [49, 78], two evaluated osteosarcoma [50, 51], two evaluated pancreatic cancer [52, 53], two evaluated thymic epithelial tumours [54, 79], two oral cancer [80, 81], and one each evaluated astrocytomas [55], chordoma [82], head and neck squamous cell carcinoma [83], lingual squamous cell carcinoma [56], nasopharyngeal carcinoma [84], pharyngeal cancer [57], prostate cancer [58], renal cell carcinoma [59], and Wilms' tumor [60]. Of these 63 studies, 20 studies evaluated STAT3, 37 studies evaluated p-STAT3, and 6 studies evaluated both STAT3 and p-STAT3. As for the region, 39 studies were conducted in Asia, 13 studies in America, 10 studies in Europe, and 1 study in Austria.
Figure 1. Flow diagram of study selection.
STAT3: Signal transducer and activator of transcription protein 3; OS: overall survival; DFS: disease-free survival.
Table 1. Characteristics of studies included in the meta-analysis.
| References | Country | Type of cancer | Patient No. | Age, median (range) | Male/Female | Stage | Follow-Up,months (Range) | STAT3 (+/–) NO. | 3-yearOS (+/–)% | 5-yearOS (+/–)% | NOS Score |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Studies including OS | |||||||||||
| Abou-Ghazal, M., et al. (2008) | USA | Gliomas | 128 | 44 (4–91) | NR | II-IV | NR | 65/63 | 36.7/48.9 | 34.7/41.7 | 7 |
| Ai, T., et al. (2012) | China | NSCLC | 65 | NR | 50/15 | I-IV | 29.9 ± 15.7 | 47/18 | 49.8/77.9 | NR | 8 |
| Birner, P., etal. (2010) | Bulgaria | Gliomas | 111 | 58.0 ± 11.6 | 57/54 | NR | 11.1 ± 0.8 | 65/46 | 0/17.9 | NR | 7 |
| Chang, K. C., et al. (2006) | China | TET | 118 | 52.7 (25–77) | 65/53 | I-IV | NR | 38/80 | 58.7/56.7 | 32.9/30 | 7 |
| Chatterjee, Devasis., et al. (2008) | USA | GC | 143 | 71.1 (31–96) | 75/68 | IA-IV | 34 (12–180) | 40/103 | 45.1/70.1 | 0/60.7 | 8 |
| Chen, C. C., et al. (2010) | China | NC | 95 | NR | NR | I-IV | 112.8 (31.2–240) | 34/61 | 39/61.2 | 31.3/53 | 6 |
| Chen, H. H., et al. (2012) | China | CC | 165 | 69 (29–89) | 0/165 | I-IV | NR | 36/129 | 53.8/60 | 47.3/48.5 | 8 |
| Cortas, T., et al. (2007) | USA | NSCLC | 145 | 70 (40–88) | 64/81 | I-III | 35 (4–85) | 50/84 | 70/60.7 | 55.5/52 | 7 |
| Deng, J. Y., et al. (2010) | China | GC | 53 | 55 (31–78) | 37/16 | I-IV | 38 (2–108) | 26/27 | 11.5/85.2 | 3.8/85.2 | 8 |
| Deng, J., et al. (2013) | China | GC | 114 | NR | 76/38 | NR | NR | 89/25 | 24.6/80 | 9.9/50.6 | 7 |
| Denley, S. M., et al. (2013)- Tyr705 | UK | PDA | 86 | NR | 43/43 | NR | 22 | 29/57 | 9/21.9 | 0/11 | 7 |
| Denley, S. M., et al. (2013)- Ser727 | UK | PDA | 86 | NR | 43/43 | NR | 22 | 30/56 | 83/22.2 | 0/11.2 | 7 |
| Dolled-F. M., et al. (2003)-M1-C | USA | BC | 286 | NR | 0/286 | NR | NR | 198/88 | 94.6/87.3 | 85.5/80 | 6 |
| Dolled-F. M., et al. (2003)-M1-N | USA | BC | 286 | NR | 0/286 | NR | NR | 66/220 | 94.6/92.1 | 93.1/81.2 | 6 |
| Dolled-F. M., et al. (2003)-M2-C | USA | BC | 286 | NR | 0/286 | NR | NR | 56/229 | 92.5/92.5 | 86.7/83.7 | 6 |
| Dolled-F. M., et al. (2003)-M2-N | USA | BC | 286 | NR | 0/286 | NR | NR | 124/161 | 95.7/89.9 | 91.3/78.7 | 6 |
| Galleges Ruiz, M. I., et al. (2009) | USA | NSCLC | 178 | NR | 127/51 | I-III | NR | 51/111 | 54.5/45 | 48.8/36.5 | 7 |
| Gordziel, C., et al. (2013)-C | Germany | CRC | 414 | NR | NR | I-III | 37 (0–146) | 132/282 | 82.8/70.6 | 74/61 | 6 |
| Gordziel, C., et al. (2013)-N | Germany | CRC | 414 | NR | NR | I-III | 37 (0–146) | 124/290 | 78.6/72 | 71.2/62.1 | 6 |
| Haura, Eric B., et al. (2005) | USA | NSCLC | 176 | 69 (45–84) | 97/79 | I | 72 (36–108) | 94/82 | 77.4/74.3 | 57.8/52.2 | 8 |
| Hbibi, A. Tadlaoui., et al. (2008)-M1 | France | CRC | 126 | 68.1 | NR | I-IV | NR | 62/38 | NR | 61.3/49.2 | 7 |
| Hbibi, A. Tadlaoui., et al. (2008)-M2 | France | CRC | 126 | 68.1 | NR | I-IV | NR | 27/73 | NR | 59.1/55.6 | 7 |
| Horiguchi, Akio., et al. (2002) | Japan | RCC | 48 | 63 (24–85) | 39/9 | I-IV | 15.9 (1–101 | 24/24 | 53.5/89.7 | 53.5/89.7 | 7 |
| Huang, C., et al. (2012) | China | PDA | 71 | 67 (40–80) | 50/21 | I-IV | 33.7 (3–60) | 39/32 | 0/24.3 | 0/14.2 | 8 |
| Jia, Yanfei., et al. (2013) | China | GC | 48 | 66 (45–83) | 34/14 | I-IV | NR | 19/29 | 54.4/86.7 | 12.7/47.6 | 7 |
| Kim, D. Y., et al. (2009) | Korea | GC | 71 | NR | 48/23 | I-IV | 30 (11–83) | 27/44 | 59/86.4 | 40/81.8 | 6 |
| Kim, Yeon-Joo., et al. (2011) | Korea | NC | 38 | 48 (25–74) | 30/8 | I-IV | 43.7 (0.71–60) | 10/28 | NR | 41/77 | 8 |
| Kusaba, T., et al. (2006) | Japan | CRC | 108 | 65.6 (44–86) | 66/42 | I-IV | NR | 62/46 | 61.6/90.1 | 48.8/90.1 | 8 |
| Lee, I., et al. (2012) | USA | Melanoma | 299 | 56 (13–85) | 212/87 | IV | NR | 236/63 | 44.1/41.8 | 22.3/25.4 | 7 |
| Lee, J., et al. (2009) | China | GC | 303 | NR | 206/97 | II-III | 61.5 (12–134) | 79/224 | 68.4/78.4 | 59.5/70.5 | 7 |
| Li, Chao., et al. (2013) | China | TET | 80 | 46.5 (19–70) | 47/33 | I-IV | NR | 36/44 | 70.1/100 | 46.8/97.6 | 7 |
| Lin, G. S., et al. (2014) | China | Gliomas | 90 | 55 (18–79). | 54/36 | NR | 46.4 (1.2–109.6) | 73/17 | 14/31.4 | NR | 8 |
| Mano, Y., et al. (2013) | Japan | HC | 101 | NR | 81/20 | NR | NR | 36/65 | 71.3/84.9 | 60.7/84.7 | 8 |
| Min, Hao., et al. (2009)-M1 | China | OC | 50 | 50.6 (22–73) | 0/50 | I-IV | NR | 44/6 | 53.5/75.1 | 29/0 | 6 |
| Min, Hao., et al. (2009)-M2 | China | OC | 50 | 50.6 (22–73) | 0/50 | I-IV | NR | 29/21 | 35/88.1 | 0/58.7 | 6 |
| Monnien, F., et al. (2010) | France | CRC | 104 | 66 (37–80) | 76/28 | NR | 13 (8–48) | 39/65 | 82.2/78 | 71.8/65.5 | 7 |
| Pectasides, Eirini., et al. (2010)-1 | Greece | HNSC | 107 | NR | 87/20 | I-IV | 64 (1–120) | 23/47 | 90.4/45.9 | 72.4/38.3 | 7 |
| Pectasides, Eirini., et al. (2010)-2 | Greece | HNSC | 107 | NR | 87/20 | I-IV | 64 (1–120) | 12/25 | NR | 68.8/51.4 | 7 |
| Piperi, Christina., et al. (2011) | Greece | Gliomas | 97 | 59 (19–82) | 60/37 | II-IV | 63 (3–180) | 89/8 | 0/14 | NR | 6 |
| Rosen, D. G., et al. (2006) | USA | OC | 303 | 58.2 (20–86) | 0/303 | I-IV | 52 | 215/88 | 49.9/60 | 32.1/49.1 | 8 |
| Ryu, Keinosuke., et al. (2010) | USA | Osteosarcoma | 51 | 20.5 (5–61) | 38/13 | NR | NR | 31/20 | 48.5/75 | 35.2/75 | 8 |
| Schoppmann, S. F., et al. (2012) | Austria | EC | 324 | 63 | 252/72 | NR | NR | 144/180 | 32.6/57.6 | 24.9/53 | 7 |
| Sheen-Chen, et al. (2008) | China | BC | 102 | 48.2 (26–76) | 0/102 | I-III | NR | 27/75 | NR | 59/77.2 | 6 |
| Slinger, E., et al. (2010) | Sweden | Gliomas | 21 | NR | NR | NR | NR | 7/14 | 0/13.6 | NR | 8 |
| Sonnenblick, A., et al. (2012) | Israel | BC | 125 | NR | 0/125 | NR | 50 | 35/90 | 100/91 | 94.4/76.6 | 6 |
| Sonnenblick, A., et al. (2013)-1 | Israel | BC | 375 | 50 | 0/375 | NR | NR | 47/82 | 97.9/96.3 | 94/94 | 7 |
| Sonnenblick, A., et al. (2013)-2 | Israel | BC | 375 | 50 | 0/375 | NR | NR | 184/150 | 99/91.9 | 94.1/80.2 | 7 |
| Takemoto, S., et al. (2009) | Japan | CC | 125 | 47 (19–77) | 0/125 | I-II | NR | 71/54 | 80.9/96.8 | 78.5/94.5 | 7 |
| Tam, L., et al. (2007)-N | UK | PC | 50 | 70 (64–73) | 50/0 | NR | 29.5 (15–54) | 22/28 | 72.8/92.5 | 57.9/84.2 | 8 |
| Tam, L., et al. (2007)-C | UK | PC | 50 | 70 (64–73) | 50/0 | NR | 29.5 (15–54) | 19/31 | 58.1/100 | 42.5/92.7 | 8 |
| van Cruijsen, H., et al. (2009) | USA | NSCLC | 164 | 64.5 | NR | I-III | NR | 116/48 | 49/58.7 | 33.1/50.3 | 7 |
| Wang, M., et al. (2011) | China | NSCLC | 208 | 59.8 (35–76) | NR | I-III | 67 (1–78.2) | 128/80 | 53.9/73.2 | 24.7/39.8 | |
| Wang, Y. C., et al. (2011) | China | Osteosarcoma | 76 | NR | 25/51 | NR | 37 | 36/40 | 25.8/60.4 | 25.8/60.4 | 6 |
| Wang, Y., et al. (2011) | China | Gliomas | 68 | 45 (15–68) | 41/27 | NR | 51 (1–72) | 47/21 | 0/15.4 | NR | 7 |
| Woo, S., et al. (2011) | Korea | GC | 285 | 54.4 | 193/92 | I-IV | 39.7 (4–84) | 101/179 | 79/61.6 | 74.9/54.5 | 7 |
| Wu, Z.S., et al. (2011) | China | Melanoma | 90 | NR | 52/38 | I-IV | NR | 51/39 | 80.5/97.6 | 50.8/76.7 | 8 |
| Xiong, Hua., et al. (2012)-M1 | China | GC | 262 | 59.3 (23–79) | 176/86 | I-IV | 90 (2–273) | 248/14 | 44/64.3 | 28.4/42.9 | 8 |
| Xiong, Hua., et al. (2012)-M2 | China | GC | 262 | 59.3 (23–79) | 176/86 | I-IV | 90 (2–273) | 136/126 | 25.3/65.5 | 11.5/47.3 | 8 |
| Yakata, Yuichi., et al. (2007) | Japan | GC | 111 | 68.9 (38–89) | 63/48 | NR | 120 | 55/56 | 37.7/83.4 | 37.7/78.6 | 8 |
| Yamashita, H., et al. (2006) | Japan | BC | 506 | NR (22–91) | 0/506 | NR | NR | 206/300 | 92/87.9 | 86/81.6 | 7 |
| Yang, C., et al. (2013) | USA | OC | 49 | 61 (41–87) | 0/49 | I-IV | NR | 25/24 | 70.6/73.4 | 33.5/57.6 | 7 |
| Yang, Cao., et al. (2009) | USA | Chordoma | 70 | 59.5 (29–88) | 51/19 | NR | 16.8 (0.8–69.2) | 35/35 | 82.5/90.2 | 73.1/90.2 | 8 |
| Yin, Z., et al. (2012) | China | NSCLC | 76 | NR | 48/28 | I-IV | NR | 42/34 | 49.6/59.3 | 41.8/57.9 | 7 |
| Yu, Y., et al. (2015) | China | NSCLC | 82 | NR | 48/34 | I-IV | NR | 76/24 | 28.7/76.2 | 20.3/48.3 | 8 |
| Zhang, C. H., et al. (2012)-M1 | China | HC | 100 | 55.1 (28–77) | 80/20 | I-IV | 15.4 | 72/28 | 53.5/57.8 | 14/32.2 | 8 |
| Zhang, C. H., et al. (2012)-M2 | China | HC | 100 | 55.1 (28–77) | 80/20 | I-IV | 15.4 | 58/42 | 35.5/81.2 | 19/25.7 | 8 |
| Zhang, L. J., et al. (2013) | China | Wilms’ tumor | 58 | 31 (3–132) | 38/20 | I-IV | ≥ 78 | 17/41 | 45.6/72.1 | 45.6/72.1 | 7 |
| Zhao, X., et al. (2012)-M1 | China | SCLC | 128 | NR | 66/62 | I-IV | 67 (1–78.2) | 71/57 | 29.7/81.6 | 0/9.9 | 7 |
| Zhao, X., et al. (2012)-M2 | China | SCLC | 128 | NR | 66/62 | I-IV | 67 (1–78.2) | 62/66 | 43.4/62.3 | 0/3.9 | 7 |
| Zhao, Yan., et al. (2012) | China | LSCC | 163 | NR | NR | I-IV | NR | 100/63 | 75/86.7 | 41.8/78.6 | 8 |
| Studies including DFS | |||||||||||
| Choi, Chel Hun., et al. (2010) | Korea | CC | 29 | NR | 0/29 | I-II | NR | 20/9 | 49.9/84.6 | 49.9/84.6 | 8 |
| Lee, J., et al. (2009) | China | GC | 303 | NR | 206/97 | II-III | 61.5 (12–134) | 79/224 | 61.7/73.7 | 58.3/67.7 | 7 |
| Li, X., et al. (2015) | China | NSCLC | 164 | NR | 115/40 | I-III | NR | 107/57 | 57.4/87.6 | NR | 8 |
| Macha, Muzafar A., et al. (2011) | Canada | Oral cancer | 94 | NR | 70/24 | I-IV | NR | 63/31 | 18.6/53.9 | 7.1/53.9 | 7 |
| Mano, Y., et al. (2013) | Japan | HC | 101 | NR | 81/20 | NR | NR | 36/65 | 12.5/55.6 | 12.5/31.5 | 8 |
| Schoppmann, S. F., et al. (2012) | Austria | EC | 324 | 63 | 252/72 | NR | NR | 144/180 | 25.8/48.3 | 20.2/47.2 | 7 |
| Takemoto, S., et al. (2009) | Japan | CC | 125 | 47 (19–77) | 0/125 | I-II | NR | 71/54 | 82/97.8 | 78/95.3 | 7 |
| Wang, Y. C., et al. (2011) | China | Osteosarcoma | 76 | NR | 25/51 | NR | 37 | 36/40 | 33.8/67.1 | 24.9/56.3 | 6 |
| Yamashita, H., et al. (2006) | Japan | BC | 506 | NR (22–91) | 0/506 | NR | NR | 206/300 | 83.2/74.7 | 72.7/65.3 | 7 |
| Zhang, L. J., et al. (2013) | China | Wilms’ tumor | 58 | 31 (3–132) | 38/20 | I-IV | ≥ 78 | 17/41 | 31.4/74.1 | 31.4/74.1 | 7 |
M1: Marker1, STAT3; M2: Marker 2, p-STAT3; N: nuclear expression; C: cytoplasmic expression; 1: Cohort 1; 2: Cohort 2; NSCLC: Non-Small Cell Lung Cancer; GC: Gastric cancer; CRC: Colorectal Cancer; BC: Breast cancer; CC: Cervical Carcinoma; OC: Ovarian Carcinoma; PDA: Pancreatic Ductal Adenocarcinoma; TET: Thymic Epithelial Tumours; RCC: Renal Cell Carcinoma; HC: Hepatocellular Carcinoma; HNSCC: Head and Neck Squamous Cell Carcinoma; EC: Esophageal Cancer; OSCC: Oral Squamous Cell Carcinoma; SCLC: Small Cell Lung Cancer; NC: Nasopharyngeal carcinoma; LSCC: Lingual Squamous Cell Carcinoma; PC: Prostate Cancer; NR: Not Reported; DFS: disease-free survival, STAT3: Signal transducer and activator of transcription protein 3; NOS: newcastle–Ottawa Scale; OS: overall survival.
Evaluation and expression of STAT3
Antibodies, detection and definition method, and cut-off values of STAT3 expression used in the included studies is summarized in Table 2. Diverse antibodies were used for the assessment of STAT3 expression by IHC. For anti-STAT3 antibody, three studies used clone sc-8019, one study each used clone RB-9237, F-2, sc-7179, 79D7, 124H6, and sixteen studies did not report the antibody clone. For anti-p-STAT3 antibody, eight studies used clone D3A7, four studies used clone sc-7993, two studies used clone 9131, one study each used sc-483, sc-8001, sc-8059, ZP-0647, and twenty studies did not report the antibody clone. The median expression of STAT3 in solid tumors was 47.79%, range from 19.65% to 94.66%.
Table 2. Evaluation of human STAT3/p-STAT3 by IHC in the selected studies.
| References | Type of cancer | Marker | Cutoff | Antibody (Clone) |
|---|---|---|---|---|
| Abou-Ghazal, M., et al. (2008) | Gliomas | p-STAT3 | NR | anti-p-STAT3 (Tyr705), Cell Signaling Technology |
| Ai, T., et al. (2012) | NSCLC | STAT3 | IHC > 51% | anti-STAT3, Cell Signaling Technology |
| Birner, P., etal. (2010) | Gliomas | p-STAT3 | IHC ≥ 5% | anti-p-STAT3 (Tyr705), clone D3A7, Cell Signaling |
| Chang, K. C., et al. (2006) | TET | STAT3 | IHC ≥ 10% | anti-Stat3 F-2: sc-8019, Santa Cruz Biotechnology, Inc. |
| Chatterjee, Devasis., et al. (2008) | GC | STAT3-nuclear | IHC scores ≥ 4 | anti-STAT3, Santa Cruz Biotechnology, Inc. |
| Chen, H. H., et al. (2012) | CC | STAT3 | IHC ≥ 20% | anti-STAT3, Santa Cruz Biotechnology, Inc. |
| Chen, C. C., et al. (2010) | NC | p-STAT3 | IHC > 10% | NR |
| Choi, Chel Hun., et al. (2010) | CC | p-STAT3 | IHC > 51% | anti-p-STAT3 (ser727), Santa Cruz Biotechnology |
| Cortas, T., et al. (2007) | NSCLC | p-STAT3 | IHC ≥ 5% | anti-p-STAT3 (sc-8059), Santa Cruz Biotechnology |
| Deng, J. Y., et al. (2010) | GC | p-STAT3 | ≥ 10% | anti-p-STAT3 (sc-483) |
| Deng, J., et al. (2013) | GC | p-STAT3 | IHC > 25% | anti-p-STAT3, Santa, sc-8001-R |
| Denley, S. M., et al. (2013) | PDA | p-STAT3 | IHC ≥ 2% | anti-pStat3 Tyr 705, 9131, Cell Signaling Technology |
| anti-pStat3 (Ser 727), 9134, Cell Signaling Technology | ||||
| Dobi, E., et al. (2013) | CRC | p-STAT3 | IHC > 15% | anti-p-STAT3, sc-7993, Santa Cruz Biotechnology |
| Dolled-Filhart, M., et al. (2003) | BC | STAT3-cytoplasmic | IHC score ≥ 1 | anti-STAT3, Cell Signaling Technology |
| STAT3-nuclear | anti-STAT3, Cell Signaling Technology | |||
| p-STAT3-cytoplasmic | anti-p-STAT3 (Tyr 705), Cell Signaling Technology | |||
| p-STAT3-nuclear | anti-p-STAT3 (Tyr 705), Cell Signaling Technology | |||
| Galleges Ruiz, M. I., et al. (2009) | NSCLC | p-STAT3-nuclear | IHC score > 210 | anti–p-STAT3 |
| Gordziel, C., et al. (2013) | CRC | STAT3-cytoplasmic | IHC score ≥ 2 | anti-STAT3: Stat3 (79D7), Cell Signaling Technology |
| STAT3-nuclear | ||||
| Haura, Eric B., et al. (2005) | NSCLC | p-STAT3-nuclear | IHC score ≥ 1 | anti-p-Stat3 (Tyr 705), Cell Signaling Technology |
| Hbibi, A. Tadlaoui., et al. (2008) | CRC | p-STAT3 | IHC score ≥ 6 | anti-P-STAT3 (Tyr 705), Cell Signaling Technology |
| STAT3 | anti-STAT3, Cell Signaling | |||
| Horiguchi, Akio., et al. (2002) | RCC | p-STAT3 | IHC ≥ 10% | anti-p-STAT3, (Tyr 705), Cell Signaling Technology |
| Huang, C., et al. (2012) | PDA | p-STAT3 | IHC ≥ 25% | anti-p-STAT3, Cell Signaling Technology |
| Jia, Yanfei., et al. (2013) | GC | STAT3 | NR | anti-STAT3, Santa Cruz Biotechnology |
| Kim, D. Y., et al. (2009) | GC | STAT3 | NR | anti-STAT3, Chemicon International |
| Kim, Yeon-Joo., et al. (2011) | NC | STAT3 | IHC ≥ 10% | anti-STAT3, Epitomics |
| Kusaba, T., et al. (2006) | CRC | p-STAT3 | IHC > 15% | anti-p-STAT3 (Tyr705), Santa Cruz Biotechnology |
| Lee, I., et al. (2012) | Melanoma | p-STAT3 | IHC ≥ 1% | anti-p-STAT3 (Tyr705), Santa Cruz Biotechnology |
| Lee, J., et al. (2009) | GC | p-STAT3 | IHC ≥ 1% | anti-p-STAT3 (Tyr705), Cell Signaling Technology |
| Li, Chao., et al. (2013) | TET | STAT3 | IHC > 10% | anti-STAT3, Santa Cruz Biotechnology |
| Li, X., et al. (2015) | NSCLC | STAT3 | IHC score ≥ 4 | anti-STAT3, Santa Cruz Biotechnology |
| Lin, G. S., et al. (2014) | Gliomas | p-STAT3 | IHC > 5% | anti-p-STAT3 (Tyr705), D3A7, Cell Signaling |
| Macha, Muzafar A., et al. (2011) | Oral cancer | p-STAT3 | NR | anti-p-STAT3 (Tyr 705), Cell Signaling |
| Mano, Y., et al. (2013) | HC | p-STAT3 | NR | anti-p-STAT3 (Tyr 705), D3A7, Cell Signaling |
| Min, Hao., et al. (2009) | OC | STAT3 | IHC ≥ 10% | anti-Stat3, (SC-8019), Santa Cruz Biotechnology |
| p-STAT3 | IHC ≥ 10% | anti-p-Stat3 (Tyr 705), ZP-0647, Abzoom Biotechnology | ||
| Monnien, F., et al. (2010) | CRC | p-STAT3 | IHC > 15% | anti-p-Stat3 (Tyr 705), sc-7993, Santa Cruz |
| Pectasides, Eirini., et al. (2010) | HNSCC | STAT3-nuclear | NR | anti-Stat3, clone 124H6; Cell Signaling Technology |
| Piperi, Christina., et al. (2011) | Gliomas | p-STAT3 | IHC ≥ 6% | anti-p-STAT3 (Tyr 705), D3A7 XP, Cell Signaling |
| Rosen, D. G., et al. (2006) | OC | p-STAT3 | IHC > 10% | anti-p-Stat3, (SC-7993-R), Santa Cruz Biotechnology |
| Ryu, Keinosuke., et al. (2010) | Osteosarcoma | p-STAT3 | IHC > 51% | anti-p-STAT, Cell Signaling Technology |
| Schoppmann, Sebastian F., et al. (2012) | EC | p-STAT3 | IHC > 10% | anti-p-STAT3 (Tyr 705), D3A7, Cell Signaling |
| Shah, N. G., et al. (2006) | OSCC | STAT3-nuclear | IHC > 10% | anti-STAT3, Santa Cruz Biotechnology |
| Slinger, E., et al. (2010) | Gliomas | p-STAT3 | IHC > 30% | anti-p-STAT3, (Tyr 705), Cell Signaling |
| Sheen-Chen, Shyr-Ming., et al. (2008) | BC | STAT3 | IHC score ≥ 3 | anti-STAT3 (RB-9237), NeoMarkers |
| Sonnenblick, A., et al. (2012) | BC | p-STAT3 | IHC ≥ 25% | anti-p-STAT3, (Tyr 705), Cell Signaling |
| Sonnenblick, A., et al. (2013) | BC | p-STAT3 | IHC ≥ 10% | anti-p-STAT3, (Tyr 705), Cell Signaling |
| Takemoto, S., et al. (2009) | CC | p-STAT3 | IHC ≥ 5% | anti-p-Stat3 (Tyr 705), sc-7993, Santa Cruz Biotechnology |
| Tam, L., et al. (2007) | PC | p-STAT3-cytoplasmic | ICCC > 0.7 | anti-p-STAT3 (Tyr 705), 9131, Cell Signaling |
| p-STAT3-nuclear | ||||
| van Cruijsen, H., et al. (2009) | NSCLC | p-STAT3 | NR | anti-p-STAT3 (Tyr 705), clone D3A7, Cell Signaling |
| Wang, M., et al. (2011) | NSCLC | p-STAT3 | IHC > 25% | anti-p-STAT3, Cell Signaling Technology |
| Wang, Y., et al. (2011) | Gliomas | p-STAT3 | IHC score > 4 | anti-p-STAT3 (Tyr 705), clone D3A7, Cell Signaling |
| Wang, Y. C., et al. (2011) | Osteosarcoma | STAT3 | IHC > 5% | anti-STAT3, Santa Cruz Biotechnology |
| Wu, Zheng-Sheng., et al. (2011) | Melanoma | p-STAT3 | NR | anti-p-STAT3, Santa Cruz Biotechnology |
| Woo, S., et al. (2011) | GC | p-STAT3 | IHC ≥ 1% | anti-p-STAT3, (Tyr 705), Cell Signaling |
| Xiong, Hua., et al. (2012) | GC | STAT3 | IHC > 15% | anti-STAT3 |
| p-STAT3 | anti-p-STAT3 (Tyr 705) | |||
| Yakata, Yuichi., et al. (2007) | GC | p-STAT3 | IHC > 10% | anti-p-STAT3, Santa Cruz Biotechnology |
| Yamashita, H., et al. (2006) | BC | STAT3 | IHC score ≥ 2 | anti-STAT3, (F-2), Santa Cruz Biotechnology |
| Yang, C., et al. (2013) | OC | p-STAT3 | IHC > 50% | anti-p-STAT3, (Tyr 705), Cell Signaling Technology |
| Yang, Cao., et al. (2009) | Chordoma | p-STAT3 | IHC score ≥ 4 | anti-p-STAT3, Cell Signaling Technology |
| Yin, Z., et al. (2012) | NSCLC | STAT3 | IHC ≥ 50% | anti-STAT3, (sc-8019); Santa Cruz |
| You, Z., et al. (2012) | EC | p-STAT3 | IHC score ≥ 2 | anti-p-STAT3, (Tyr 705), Cell Signaling Technology |
| Yu, Y., et al. (2015) | NSCLC | pSTAT3 | IHC score ≥ 3 | NR |
| Zhang, C. H., et al. (2012) | HC | STAT3 | IHC > 10% | anti-STAT3, Santa Cruz Biotechnology |
| p-STAT3 | anti-p-STAT3, (Tyr 705), Cell Signaling Technology | |||
| Zhang, L. J., et al. (2013) | Wilms' tumor | STAT3 | IHC > 51% | anti-STAT3, (sc-7179), Santa Cruz Biotechnology |
| Zhao, X., et al. (2012) | SCLC | STAT3 | IHC ≥ 25% | anti-STAT3, Wuhan Boster Company |
| p-STAT3 | anti-p-STAT3, clone B-7, Wuhan Boster Company | |||
| Zhao, Yan., et al. (2012) | LSCC | STAT3 | IHC ≥ 10% | anti-STAT3, Santa Cruz Biotechnology |
NSCLC: Non-Small Cell Lung Cancer; GC: Gastric cancer; CRC: Colorectal Cancer; BC: Breast cancer; CC: Cervical Carcinoma; OC: Ovarian Carcinoma; PDA: Pancreatic Ductal Adenocarcinoma; TET: Thymic Epithelial Tumours; RCC: Renal Cell Carcinoma; HC: Hepatocellular Carcinoma; HNSCC: Head and Neck Squamous Cell Carcinoma; EC: Esophageal Cancer; OSCC: Oral Squamous Cell Carcinoma; SCLC: Small Cell Lung Cancer; NC: Nasopharyngeal carcinoma; LSCC: Lingual Squamous Cell Carcinoma; PC: Prostate Cancer; ICCH: Interclass Correlation Coefficient; NR: Not Reported.
Association of STAT3 with OS
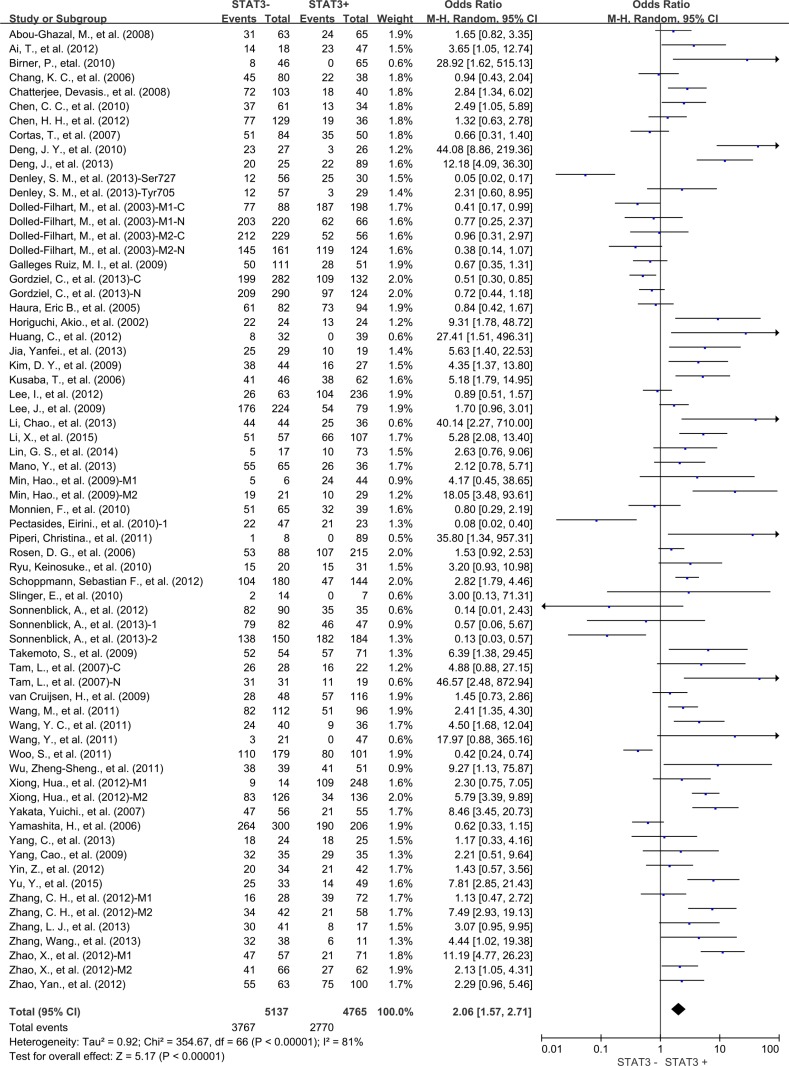
The combined analysis of 54 studies showed that STAT3 overexpression in tumor tissue was associated with worse 3-year OS of solid tumors (OR = 2.06, 95% CI = 1.57 to 2.71, P < 0.00001) (Figure 2). There was significant heterogeneity among studies (Cochran's Q P < 0.00001, I2 = 81%), so we conducted meta-regression analysis and subgroup meta-analysis to investigate the possible source of the heterogeneity among studies.
Figure 2. Three-year overall survival (OS) by STAT3 expression.
M1: Marker 1, STAT3; M2: Marker 2, p-STAT3; 1: Cohort 1; 2: Cohort 2; N: nuclear expression; C: cytoplasmic expression.
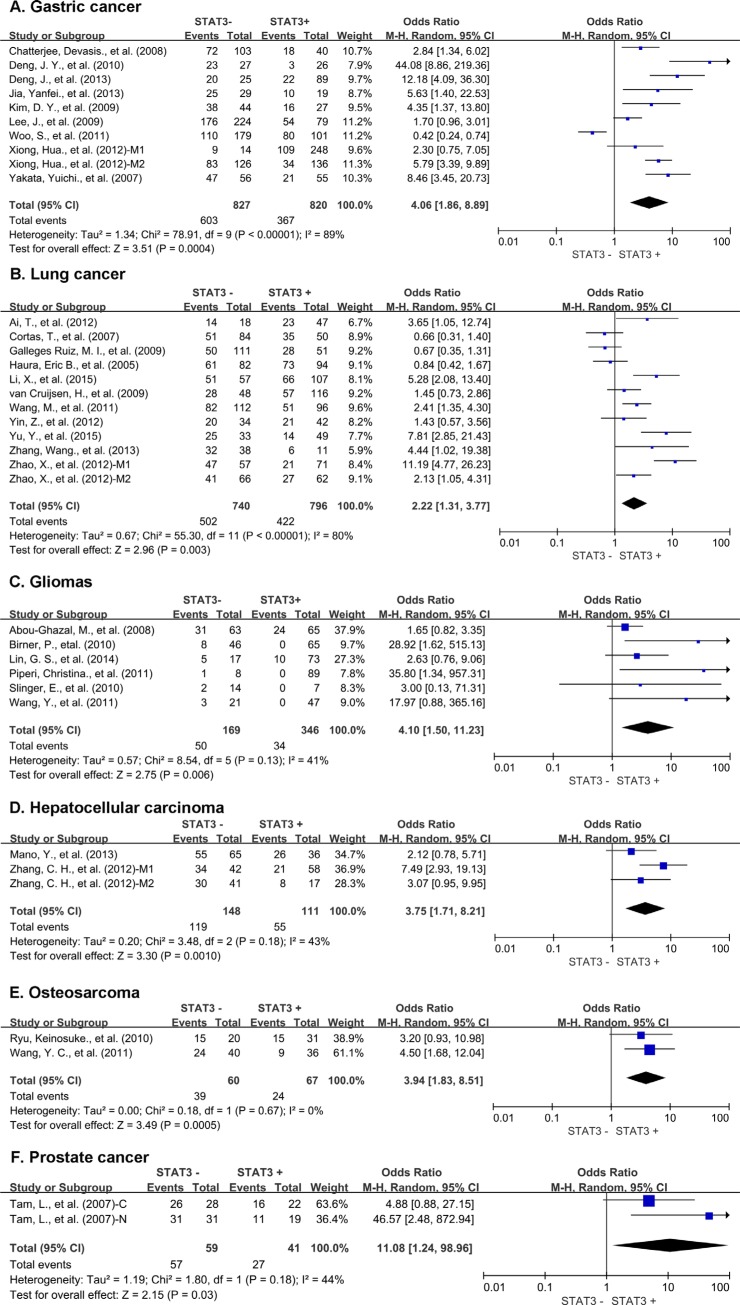
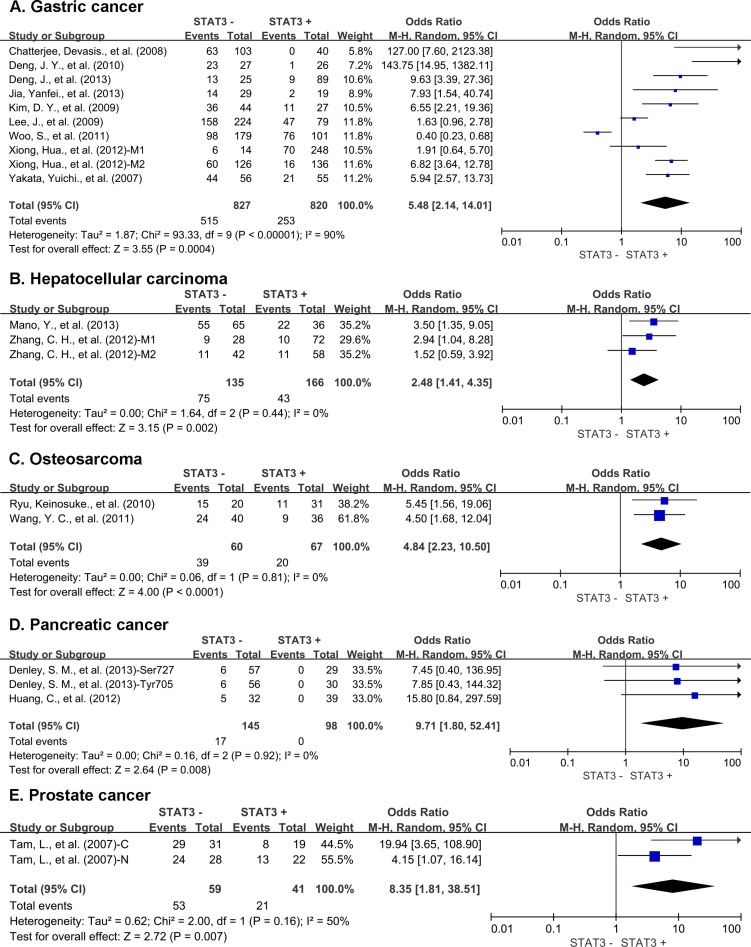
In the stratified analysis by tumor types, STAT3 expression was associated with worse 3-year OS of gastric cancer (OR = 4.06, 95% CI = 1.86 to 8.89, P = 0.0004), lung cancer (OR = 2.22, 95% CI = 1.31 to 3.77, P = 0.003), gliomas (OR = 4.10, 95% CI = 1.50 to 11.23, P = 0.006), hepatic cancer (OR = 3.75, 95% CI = 1.71 to 8.21, P = 0.001), osteosarcoma (OR = 3.94, 95% CI = 1.83 to 8.51, P = 0.0005) and prostate cancer (OR = 11.08, 95% CI = 1.24 to 98.96, P = 0.03) (Figure 3). There was no significant association between STAT3 expression and 3-year OS of colorectal cancer, ovarian cancer, pancreatic cancer, cervical cancer, melanoma and thymic epithelial tumor (Supplementary Figure S1). Interestingly, STAT3 overexpression was associated with favorable 3-year OS of breast cancer (OR = 0.51, 95% CI = 0.35 to 0.74, P = 0.0004) (Supplementary Figure S2).
Figure 3. Subgroup analysis of 3-year OS by STAT3 expression in different tumor types.
(A) gastric cancer; (B) lung cancer; (C) gliomas; (D) hepatic cancer; (E) osteosarcoma; (F) prostate cancer. M1: Marker 1, STAT3; M2: Marker 2, p-STAT3; 1: Cohort 1; 2: Cohort 2 N: nuclear expression; C: cytoplasmic expression.
Meta-regression analysis showed that publication year, country, gender and NOS score did not contribute to the heterogeneity (data not shown).
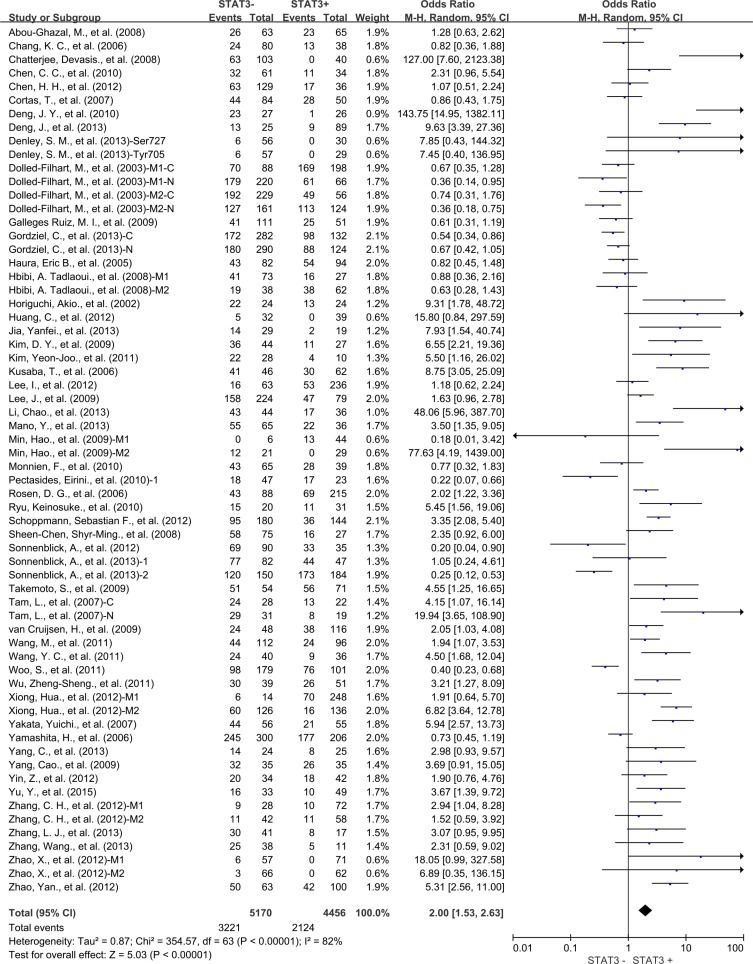
Analysis of 49 studies showed STAT3 expression was also associated with worse 5-year OS (OR = 2.00, 95% CI = 1.53 to 2.63, P < 0.00001) (Figure 4) of solid tumors. There was also high heterogeneity among studies for 5-year OS (Cochran's Q P < 0.00001, I2 = 82%), so we conducted subgroup meta-analysis.
Figure 4. Five-year OS by STAT3 expression.
M1: Marker 1, STAT3; M2: Marker 2, p-STAT3; 1: Cohort 1; 2: Cohort 2 N: nuclear expression; C: cytoplasmic expression.
Subgroup analysis showed that STAT3 expression was associated with worse 5-year OS of gastric cancer (OR = 5.48, 95% CI = 2.14 to 14.01, P = 0.0004), hepatic cancer (OR = 2.48, 95% CI = 1.41 to 4.35, P = 0.002), osteosarcoma (OR = 4.84, 95% CI = 2.23 to 10.50, P < 0.0001), pancreatic cancer (OR = 9.71, 95% CI = 1.80 to 52.41, P = 0.008) and prostate cancer (OR = 8.35, 95% CI = 1.81 to38.51, P = 0.007) (Figure 5). There was no significant association between STAT3 expression and the 5-year OS of colorectal cancer, lung cancer, ovarian cancer, cervical cancer, melanoma and thymic epithelial tumor (Supplementary Figure S3). STAT3 overexpression was associated with favorable 5-year OS of breast cancer (OR = 0.57, 95% CI = 0.37 to 0.89, P = 0.01) (Supplementary Figure S4).
Figure 5. Subgroup analysis of 5-year OS by STAT3 expression in different tumor types.
(A) gastric cancer; (B) hepatic cancer; (C) osteosarcoma; (D) pancreatic cancer; (E) prostate cancer. M1: Marker 1, STAT3; M2: Marker 2, p-STAT3; 1: Cohort 1; 2: Cohort 2 N: nuclear expression; C: cytoplasmic expression.
Twenty studies evaluated STAT3, 38 studies evaluated p-STAT3 and 5 studies evaluated both STAT3 and p-STAT3. Our result showed that both STAT3 and p-STAT3 overexpression were associated with worse OS of solid tumors. However, elevated p-STAT3 (OR = 2.45, 95% CI = 1.73 to 3.46, P < 0.00001) expression in tumor tissue seemed to be more significantly associated with worse 3-year OS than STAT3 expression (OR = 1.72, 95% CI = 1.10 to 2.70, P = 0.02) (Supplementary Figure S5). Similar result was observed for 5-year OS analysis (Supplementary Figure S6). A subgroup meta-analysis of studies evaluated both STAT3 and p-STAT3 shown that p-STAT3 expression was associated with worse 3-year and 5-year OS of solid tumor, but not STAT3 (Supplementary Figure S7).
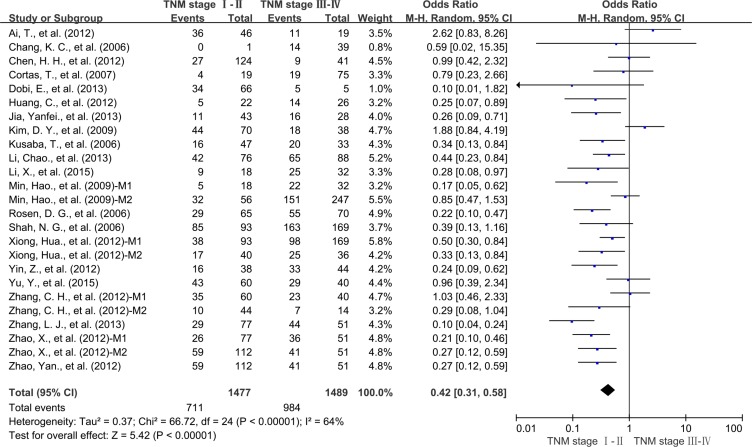
We also evaluated the correlation between STAT3 overexpression and the TNM stage of tumor. High expression level of STAT3 was significantly associated with advanced TNM stage (OR = 0.42, 95% CI = 0.31 to 0.58, P < 0.00001) (Figure 6).
Figure 6. Subgroup analysis the correlation of STAT3 expression and tumor stage.
M1: Marker 1, STAT3; M2: Marker 2, p-STAT3.
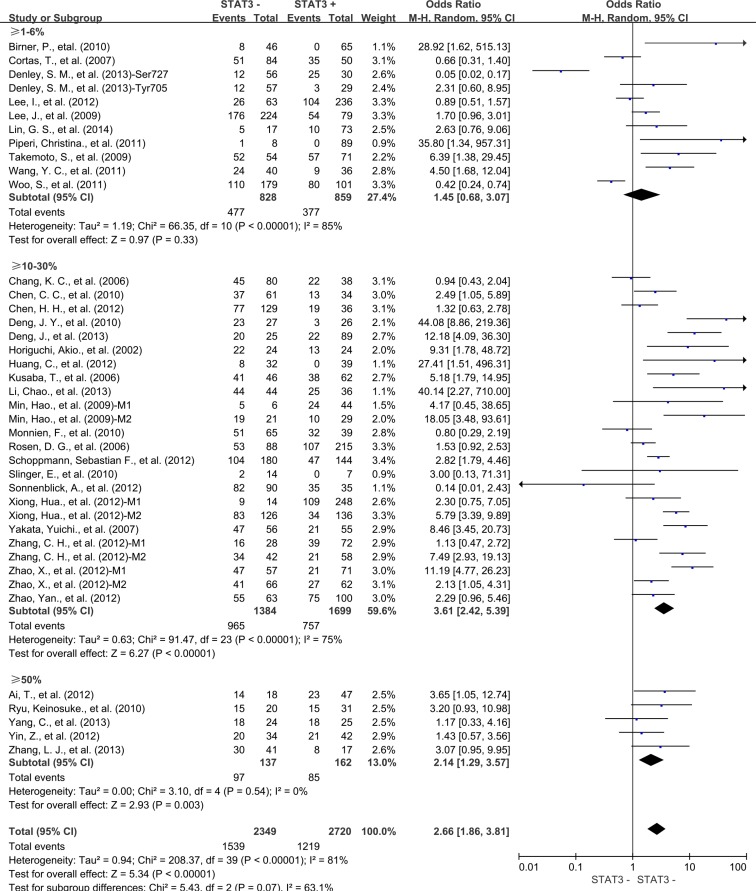
Next, we conducted subgroup analysis according to STAT3 expression level. Results showed STAT3 expression was associated with poor 3-year OS in the studies using cutoff values of 10%–30% (OR = 3.61, 95% CI = 2.42 to 5.39, P < 0.00001) and 50% (OR = 2.14, 95% CI = 1.29 to 3.57, P = 0.003) (Figure 7) to determine STAT3 positivity. Similar result was observed in 5-year OS (Supplementary Figure S6). However, the studies used cutoff value of STAT3 overexpression as more than 1%-6% tumor cells positive was not associated with 3-year and 5-year OS of solid tumors.
Figure 7. Subgroup analysis the correlation between STAT3 overexpression and 3-year OS of solid tumors according to cut-off values determining STAT3 positivity.
M1: Marker 1, STAT3; M2: Marker 2, p-STAT3.
Association of STAT3 with DFS
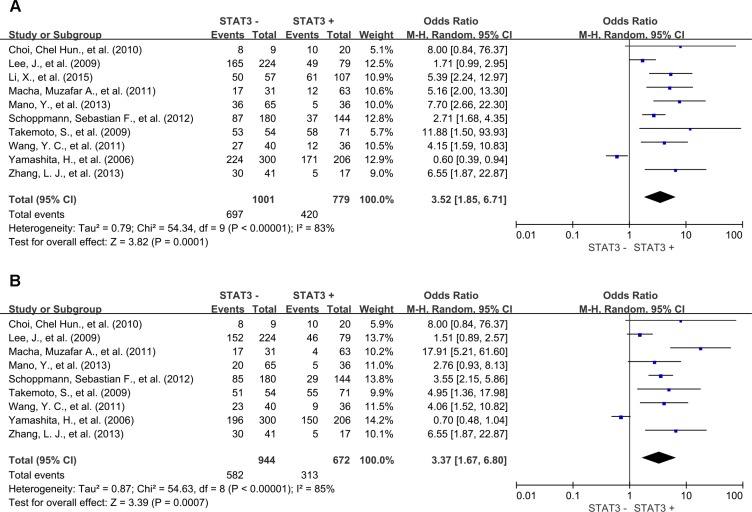
Meta-analysis of 10 studies showed that STAT3 expression was associated with statistically significant poor 3-year DFS (OR = 3.52, 95% CI = 1.85 to 6.71, P = 0.0001) (Figure 8A) and poor 5-year DFS (OR = 3.37, 95% CI = 1.67 to 6.80, P = 0.0007) (Figure 8B).
Figure 8. Three and five-year DFS by STAT3 expression.
(A) 3-year DFS; (B) 5-year DFS. M1: Marker 1, STAT3; M2: Marker 2, p-STAT3.
Sensitivity analyses
Removal of the studies that was an outlier (score, IRS, > 50% vs 1%–6% for other studies) or no report (NR) with regard to the cutoff of STAT3 overexpression by IHC did not influence results for 3- or 5-year OS (OR = 2.45, 95% CI = 1.73 to 3.48, p < 0.00001; OR = 2.08, 95% CI = 1.46 to 2.96, p < 0.0001; respectively). Exclusion of these studies did not reduce heterogeneity for 3- or 5-year OS (Cochran's Q P < 0.00001, I2 = 82%; Cochran's Q P < 0.00001, I2 = 84%, respectively).
Removal of studies with NOS score 6 did not influence results for 3- or 5-year OS (OR = 2.49, 95% CI = 1.86 to 3.34, p < 0.00001; OR = 2.33, 95% CI = 1.74 to 3.12, p < 0.00001, respectively). Exclusion of these studies did not reduce heterogeneity for 3- or 5-year OS (Cochran's Q P < 0.00001, I2 = 80%; Cochran's Q P < 0.00001, I2 = 82%, respectively).
Publication bias
Funnel plot analysis showed that there was no statistical evidence of publication bias in our meta-analysis (data not shown).
DISCUSSION
This meta-analysis is the most comprehensive assessment of the literatures regarding STAT3 expression and tumor prognosis to date. We systematically evaluated survival data for 9449 solid tumor patients included in 63 different studies. Our study demonstrated that the expression of STAT3 is a marker of poor prognosis in solid tumors, with consistent results of OS at 3 and 5 years. Regarding to the tumor types, elevated STAT3 expression in tumor tissues were associated with worse OS of gastric cancer, lung cancer, gliomas, hepatic cancer, osteosarcoma, prostate cancer and pancreatic cancer. However, elevated STAT3 expression was associated with better prognosis of breast cancer. In addition, expression level of phosphorylated STAT3 was more significantly associated with worse outcome of solid tumors than unphosphorylated STAT3.
Our study found there is no significant correlation between STAT3 overexpression and OS of colorectal cancer and ovarian cancer. And STAT3 overexpression in breast cancer tissue is associated with favorable OS. However, recent studies demonstrated that STAT3-targeted inhibitor could restrain tumor development in various solid tumor models including breast cancer [16, 19, 85, 86], melanoma [87] and ovarian cancer [16, 88]. These divergences suggest that further study is needed to shed more light on the underling mechanism of STAT3 signal pathway in pro-tumor microenvironment in different tumor types.
There are several important implications in this meta-analysis. First, it shows that STAT3 expression is related to adverse outcome of most solid tumors. Second, it identifies a subgroup of tumors with unfavorable outcome in gastric cancer, lung cancer, hepatic cancer, prostate cancer and glioblastoma, but with favorable outcome in breast cancer. Finally, it emphasizes the potential of STAT3 to developing a valuable therapeutic target and prognostic biomarker for solid tumor.
This study also has some limitations. First, from the literature we could only extract summarized population-level data rather than individual patient-level data. Second, the method for assessing STAT3 expression and definition of STAT3 positivity are inconsistent. Finally, substantial heterogeneity observed across included studies cannot be fully accounted for by our use of appropriate meta-analytic techniques with random-effects modeling.
In summary, STAT3 expression in solid tumor tissues is associated with poor survival in most solid tumors, which suggests that STAT3 is a valuable prognostic biomarker and a promising therapeutic target for solid tumors.
MATERIALS AND METHODS
This meta-analysis was conducted according to the statement for reporting systematic reviews and meta-analyses [89]. This study summarized and analyzed the results of previous studies, so the ethical approval was not necessary.
Search strategy and study selection
An electronic search of Pubmed, Web of Science and EBSCO were undertaken for studies evaluating STAT3 or p-STAT3 expression and clinical outcome in solid tumors from 1994 to August 2015. The search was performed with subject heading terms including “signal transducer and activator of transcription 3” or “STAT3 transcription factor” or “STAT3” or “phosphorylated signal transducer and activator of transcription 3” or “phosphorylated STAT3 transcription factor” or “phospho-STAT3” and “neoplasms” and the results were limited to human studies of solid tumors. In addition, the entry “signal transducer and activator of transcription 3” or “STAT3 transcription factor” or “STAT3” or “phosphorylated signal transducer and activator of transcription 3” or “phosphorylated STAT3 transcription factor” or “phospho-STAT3” and the name of each specific solid tumor were used for additional studies. A total of 3547, 3542 and 2914 entries were identified, respectively. Inclusion criteria were the measurement of STAT3 and (or) p-STAT3 by immunohistochemistry (IHC), availability of survival data for at least 3 years, and original article written in English. Exclusion criteria were studies evaluating gene expression of STAT3 measured by polymerase chain reaction (PCR) and STAT3 expression in lymph node and myeloid cells. Citation lists of retrieved articles were manually screened to ensure sensitivity of the search strategy. Study selection was based on the association of STAT3 and survival. Two reviewers (Pin Wu and Dang Wu) evaluated independently all of the full articles for study eligibility. Inter-reviewer agreement was assessed using Cohen's kappa coefficient. Disagreement was resolved by consensus.
Data extraction
Overall survival (OS) and disease free survival (DFS) were the primary endpoints of interest. Data were extracted using predefined abstraction forms. The following details were extracted by two authors (Pin Wu and Dang Wu): name of first author, year of publication, country of publication, tumor type, patient number, tumor stage, antibodies used for the evaluation, method and score for STAT3 assessment, and cut-off values to determine STAT3 positivity. Data for 3 and 5 year of OS and DFS were extracted from tables or Kaplan–Meier curves for both STAT3 negative and STAT3 positive group.
The studies included in our meta-analysis were all cohort studies. Two independent authors evaluated the quality of each included study using Newcastle-Ottawa Scale (NOS) [90]. The studies with 6 scores or more were considered as high quality studies. A consensus NOS score for each item was achieved finally.
Data synthesis
The relative frequency of OS and DFS at 3 and 5 years between STAT3 negative and STAT3 positive group was presented as an odds ratio (OR) and its 95% confidence interval (CI). Sensitivity analyses were carried out for different analytical methods and cut-offs for defining STAT3 expression and NOS scores for quality assessment of included studies. Publication bias was assessed by visual inspection of the funnel plot.
Statistical analysis
Data were extracted from the primary publications and combined into a meta-analysis using RevMan 5.3 analysis software (Cochrane Collaboration, Copenhagen, Denmark). Estimates of ORs were weighted and pooled using the Mantel–Haenszel random effect model. Statistical heterogeneity was assessed using the Cochran's Q and I2 statistics. Differences between subgroups were assessed using methods as previous described by Deeks et al. [91]. Meta-regression analysis was conducted using Stata 12.0 software (StataCorp LP, College Station, TX). All statistical tests were two-sided, and statistical significance was defined as P less than 0.05. No correction was made for multiple statistical testing.
SUPPLEMENTARY MATERIALS FIGURES
ACKNOWLEDGMENTS AND FUNDING
We thank all the group members for helpful discussions. This work was supported by grants from the National Natural Science Foundation of China (81572800, PW), the Science and Technology Department of Zhejiang Province (2013c03044–7, YC), and Natural Science Foundation of Zhejiang Province (LY13H160016, YC; Y15H160094, GS and LY15H160041, PW).
Footnotes
CONFLICTS OF INTEREST
The authors declare no competing financial interest.
REFERENCES
- 1.Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer. 2009;9:798–809. doi: 10.1038/nrc2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ernst M, Najdovska M, Grail D, Lundgren-May T, Buchert M, Tye H, Matthews VB, Armes J, Bhathal PS, Hughes NR, Marcusson EG, Karras JG, Na S, et al. STAT3 and STAT1 mediate IL-11-dependent and inflammation-associated gastric tumorigenesis in gp130 receptor mutant mice. J Clin Invest. 2008;118:1727–1738. doi: 10.1172/JCI34944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gao SP, Mark KG, Leslie K, Pao W, Motoi N, Gerald WL, Travis WD, Bornmann W, Veach D, Clarkson B, Bromberg JF. Mutations in the EGFR kinase domain mediate STAT3 activation via IL-6 production in human lung adenocarcinomas. J Clin Invest. 2007;117:3846–3856. doi: 10.1172/JCI31871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kortylewski M, Xin H, Kujawski M, Lee H, Liu Y, Harris T, Drake C, Pardoll D, Yu H. Regulation of the IL-23 and IL-12 balance by Stat3 signaling in the tumor microenvironment. Cancer Cell. 2009;15:114–123. doi: 10.1016/j.ccr.2008.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crescenzo R, Abate F, Lasorsa E, Tabbo F, Gaudiano M, Chiesa N, Di Giacomo F, Spaccarotella E, Barbarossa L, Ercole E, Todaro M, Boi M, Acquaviva A, et al. Convergent mutations and kinase fusions lead to oncogenic STAT3 activation in anaplastic large cell lymphoma. Cancer Cell. 2015;27:516–532. doi: 10.1016/j.ccell.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fan QW, Cheng CK, Gustafson WC, Charron E, Zipper P, Wong RA, Chen J, Lau J, Knobbe-Thomsen C, Weller M, Jura N, Reifenberger G, Shokat KM, et al. EGFR phosphorylates tumor-derived EGFRvIII driving STAT3/5 and progression in glioblastoma. Cancer Cell. 2013;24:438–449. doi: 10.1016/j.ccr.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu H, Lee H, Herrmann A, Buettner R, Jove R. Revisiting STAT3 signalling in cancer: new and unexpected biological functions. Nat Rev Cancer. 2014;14:736–746. doi: 10.1038/nrc3818. [DOI] [PubMed] [Google Scholar]
- 8.Gough DJ, Corlett A, Schlessinger K, Wegrzyn J, Larner AC, Levy DE. Mitochondrial STAT3 supports Ras-dependent oncogenic transformation. Science. 2009;324:1713–1716. doi: 10.1126/science.1171721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee HJ, Zhuang G, Cao Y, Du P, Kim HJ, Settleman J. Drug resistance via feedback activation of Stat3 in oncogene-addicted cancer cells. Cancer Cell. 2014;26:207–221. doi: 10.1016/j.ccr.2014.05.019. [DOI] [PubMed] [Google Scholar]
- 10.Dai B, Meng J, Peyton M, Girard L, Bornmann WG, Ji L, Minna JD, Fang B, Roth JA. STAT3 mediates resistance to MEK inhibitor through microRNA miR-17. Cancer Res. 2011;71:3658–3668. doi: 10.1158/0008-5472.CAN-10-3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ernst M, Thiem S, Nguyen PM, Eissmann M, Putoczki TL. Epithelial gp130/Stat3 functions: an intestinal signaling node in health and disease. Semin Immunol. 2014;26:29–37. doi: 10.1016/j.smim.2013.12.006. [DOI] [PubMed] [Google Scholar]
- 12.Guryanova OA, Wu Q, Cheng L, Lathia JD, Huang Z, Yang J, MacSwords J, Eyler CE, McLendon RE, Heddleston JM, Shou W, Hambardzumyan D, Lee J, et al. Nonreceptor tyrosine kinase BMX maintains self-renewal and tumorigenic potential of glioblastoma stem cells by activating STAT3. Cancer Cell. 2011;19:498–511. doi: 10.1016/j.ccr.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim E, Kim M, Woo DH, Shin Y, Shin J, Chang N, Oh YT, Kim H, Rheey J, Nakano I, Lee C, Joo KM, Rich JN, et al. Phosphorylation of EZH2 activates STAT3 signaling via STAT3 methylation and promotes tumorigenicity of glioblastoma stem-like cells. Cancer Cell. 2013;23:839–852. doi: 10.1016/j.ccr.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deng J, Liu Y, Lee H, Herrmann A, Zhang W, Zhang C, Shen S, Priceman SJ, Kujawski M, Pal SK, Raubitschek A, Hoon DS, Forman S, et al. S1PR1-STAT3 signaling is crucial for myeloid cell colonization at future metastatic sites. Cancer Cell. 2012;21:642–654. doi: 10.1016/j.ccr.2012.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fukuda A, Wang SC, Morris JPt, Folias AE, Liou A, Kim GE, Akira S, Boucher KM, Firpo MA, Mulvihill SJ, Hebrok M. Stat3 and MMP7 contribute to pancreatic ductal adenocarcinoma initiation and progression. Cancer Cell. 2011;19:441–455. doi: 10.1016/j.ccr.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hedvat M, Huszar D, Herrmann A, Gozgit JM, Schroeder A, Sheehy A, Buettner R, Proia D, Kowolik CM, Xin H, Armstrong B, Bebernitz G, Weng S, et al. The JAK2 inhibitor AZD1480 potently blocks Stat3 signaling and oncogenesis in solid tumors. Cancer Cell. 2009;16:487–497. doi: 10.1016/j.ccr.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sen M, Thomas SM, Kim S, Yeh JI, Ferris RL, Johnson JT, Duvvuri U, Lee J, Sahu N, Joyce S, Freilino ML, Shi H, Li C, et al. First-in-human trial of a STAT3 decoy oligonucleotide in head and neck tumors: implications for cancer therapy. Cancer Discov. 2012;2:694–705. doi: 10.1158/2159-8290.CD-12-0191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hussain SF, Kong LY, Jordan J, Conrad C, Madden T, Fokt I, Priebe W, Heimberger AB. A novel small molecule inhibitor of signal transducers and activators of transcription 3 reverses immune tolerance in malignant glioma patients. Cancer Res. 2007;67:9630–9636. doi: 10.1158/0008-5472.CAN-07-1243. [DOI] [PubMed] [Google Scholar]
- 19.Lin L, Hutzen B, Zuo M, Ball S, Deangelis S, Foust E, Pandit B, Ihnat MA, Shenoy SS, Kulp S, Li PK, Li C, Fuchs J, Lin J. Novel STAT3 phosphorylation inhibitors exhibit potent growth-suppressive activity in pancreatic and breast cancer cells. Cancer Res. 2010;70:2445–2454. doi: 10.1158/0008-5472.CAN-09-2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yan S, Li Z, Thiele CJ. Inhibition of STAT3 with orally active JAK inhibitor, AZD1480, decreases tumor growth in Neuroblastoma and Pediatric Sarcomas In vitro and In vivo. Oncotarget. 2013;4:433–445. doi: 10.18632/oncotarget.930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Siddiquee K, Zhang S, Guida WC, Blaskovich MA, Greedy B, Lawrence HR, Yip ML, Jove R, McLaughlin MM, Lawrence NJ, Sebti SM, Turkson J. Selective chemical probe inhibitor of Stat3, identified through structure-based virtual screening, induces antitumor activity. Proc Natl Acad Sci U S A. 2007;104:7391–7396. doi: 10.1073/pnas.0609757104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chatterjee D, Sabo E, Tavares R, Resnick MB. Inverse association between Raf Kinase Inhibitory Protein and signal transducers and activators of transcription 3 expression in gastric adenocarcinoma patients: implications for clinical outcome. Clin Cancer Res. 2008;14:2994–3001. doi: 10.1158/1078-0432.CCR-07-4496. [DOI] [PubMed] [Google Scholar]
- 23.Deng J, Liang H, Zhang R, Sun D, Pan Y, Liu Y, Zhang L, Hao X. STAT3 is associated with lymph node metastasis in gastric cancer. Tumour Biol. 2013;34:2791–2800. doi: 10.1007/s13277-013-0837-5. [DOI] [PubMed] [Google Scholar]
- 24.Deng JY, Sun D, Liu XY, Pan Y, Liang H. STAT-3 correlates with lymph node metastasis and cell survival in gastric cancer. World J Gastroenterol. 2010;16:5380–5387. doi: 10.3748/wjg.v16.i42.5380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jia Y, Liu D, Xiao D, Ma X, Han S, Zheng Y, Sun S, Zhang M, Gao H, Cui X, Wang Y. Expression of AFP, STAT3 is involved in arsenic trioxide-induced apoptosis and inhibition of proliferation in AFP-producing gastric cancer cells. PLoS One. 2013;8:e54774. doi: 10.1371/journal.pone.0054774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim DY, Cha ST, Ahn DH, Kang HY, Kwon CI, Ko KH, Hwang SG, Park PW, Rim KS, Hong SP. STAT3 expression in gastric cancer indicates a poor prognosis. J Gastroenterol Hepatol. 2009;24:646–651. doi: 10.1111/j.1440-1746.2008.05671.x. [DOI] [PubMed] [Google Scholar]
- 27.Lee J, Kang WK, Park JO, Park SH, Park YS, Lim HY, Kim J, Kong J, Choi MG, Sohn TS, Noh JH, Bae JM, Kim S, et al. Expression of activated signal transducer and activator of transcription 3 predicts poor clinical outcome in gastric adenocarcinoma. Apmis. 2009;117:598–606. doi: 10.1111/j.1600-0463.2009.02512.x. [DOI] [PubMed] [Google Scholar]
- 28.Xiong H, Du W, Wang J-L, Wang Y-C, Tang J-T, Hong J, Fang J-Y. Constitutive activation of STAT3 is predictive of poor prognosis in human gastric cancer. J Mol Med (Berl) 2012;90:1037–1046. doi: 10.1007/s00109-012-0869-0. [DOI] [PubMed] [Google Scholar]
- 29.Yakata Y, Nakayama T, Yoshizaki A, Kusaba T, Inoue K, Sekine I. Expression of p-STAT3 in human gastric carcinoma: significant correlation in tumour invasion and prognosis. Int J Oncol. 2007;30:437–442. [PubMed] [Google Scholar]
- 30.Ai T, Wang Z, Zhang M, Zhang L, Wang N, Li W, Song L. Expression and prognostic relevance of STAT3 and cyclin D1 in non-small cell lung cancer. Int J Biol Markers. 2012;27:e132–138. doi: 10.5301/JBM.2012.9146. [DOI] [PubMed] [Google Scholar]
- 31.Li X, Yu Z, Li Y, Liu S, Gao C, Hou X, Yao R, Cui L. The tumor suppressor miR-124 inhibits cell proliferation by targeting STAT3 and functions as a prognostic marker for postoperative NSCLC patients. Int J Oncol. 2015;46:798–808. doi: 10.3892/ijo.2014.2786. [DOI] [PubMed] [Google Scholar]
- 32.van Cruijsen H, Ruiz MG, van der Valk P, de Gruijl TD, Giaccone G. Tissue micro array analysis of ganglioside N-glycolyl GM3 expression and signal transducer and activator of transcription (STAT)-3 activation in relation to dendritic cell infiltration and microvessel density in non-small cell lung cancer. BMC Cancer. 2009;9:180. doi: 10.1186/1471-2407-9-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang M, Chen GY, Song HT, Hong X, Yang ZY, Sui GJ. Significance of CXCR4, phosphorylated STAT3 and VEGF-A expression in resected non-small cell lung cancer. Exp Ther Med. 2011;2:517–522. doi: 10.3892/etm.2011.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yin Z, Zhang Y, Li Y, Lv T, Liu J, Wang X. Prognostic significance of STAT3 expression and its correlation with chemoresistance of non-small cell lung cancer cells. Acta Histochem. 2012;114:151–158. doi: 10.1016/j.acthis.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 35.Yu Y, Zhao Q, Wang Z, Liu XY. Activated STAT3 correlates with prognosis of non-small cell lung cancer and indicates new anticancer strategies. Cancer Chemother Pharmacol. 2015;75:917–922. doi: 10.1007/s00280-015-2710-2. [DOI] [PubMed] [Google Scholar]
- 36.Zhang W, Pal SK, Liu X, Yang C, Allahabadi S, Bhanji S, Figlin RA, Yu H, Reckamp KL. Myeloid clusters are associated with a pro-metastatic environment and poor prognosis in smoking-related early stage non-small cell lung cancer. PLoS One. 2013;8:e65121. doi: 10.1371/journal.pone.0065121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao X, Sun X, Li XL. Expression and clinical significance of STAT3, P-STAT3, and VEGF-C in small cell lung cancer. Asian Pacific journal of cancer prevention: Asian Pac J Cancer Prev. 2012;13:2873–2877. doi: 10.7314/apjcp.2012.13.6.2873. [DOI] [PubMed] [Google Scholar]
- 38.Abou-Ghazal M, Yang DS, Qiao W, Reina-Ortiz C, Wei J, Kong L-Y, Fuller GN, Hiraoka N, Priebe W, Sawaya R, Heimberger AB. The incidence, correlation with tumor-infiltrating inflammation, and prognosis of phosphorylated STAT3 expression in human gliomas. Clin Cancer Res. 2008;14:8228–8235. doi: 10.1158/1078-0432.CCR-08-1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Birner P, Toumangelova-Uzeir K, Natchev S, Guentchev M. STAT3 tyrosine phosphorylation influences survival in glioblastoma. J Neurooncol. 2010;100:339–343. doi: 10.1007/s11060-010-0195-8. [DOI] [PubMed] [Google Scholar]
- 40.Lin GS, Yang LJ, Wang XF, Chen YP, Tang WL, Chen L, Lin ZX. STAT3 Tyr705 phosphorylation affects clinical outcome in patients with newly diagnosed supratentorial glioblastoma. Med Oncol. 2014;31:924. doi: 10.1007/s12032-014-0924-5. [DOI] [PubMed] [Google Scholar]
- 41.Slinger E, Maussang D, Schreiber A, Siderius M, Rahbar A, Fraile-Ramos A, Lira SA, Soderberg-Naucler C, Smit MJ. HCMV-encoded chemokine receptor US28 mediates proliferative signaling through the IL-6-STAT3 axis. Sci Signal. 2010;3:ra58. doi: 10.1126/scisignal.2001180. [DOI] [PubMed] [Google Scholar]
- 42.Wang Y, Chen L, Bao Z, Li S, You G, Yan W, Shi Z, Liu Y, Yang P, Zhang W, Han L, Kang C, Jiang T. Inhibition of STAT3 reverses alkylator resistance through modulation of the AKT and beta-catenin signaling pathways. Oncol Rep. 2011;26:1173–1180. doi: 10.3892/or.2011.1396. [DOI] [PubMed] [Google Scholar]
- 43.Kusaba T, Nakayama T, Yamazumi K, Yakata Y, Yoshizaki A, Inoue K, Nagayasu T, Sekine I. Activation of STAT3 is a marker of poor prognosis in human colorectal cancer. Oncol Rep. 2006;15:1445–1451. [PubMed] [Google Scholar]
- 44.Min H, Wei-hong Z. Constitutive activation of signal transducer and activator of transcription 3 in epithelial ovarian carcinoma. J Obstet Gynaecol Res. 2009;35:918–925. doi: 10.1111/j.1447-0756.2009.01045.x. [DOI] [PubMed] [Google Scholar]
- 45.Takemoto S, Ushijima K, Kawano K, Yamaguchi T, Terada A, Fujiyoshi N, Nishio S, Tsuda N, Ijichi M, Kakuma T, Kage M, Hori D, Kamura T. Expression of activated signal transducer and activator of transcription-3 predicts poor prognosis in cervical squamous-cell carcinoma. Br J Cancer. 2009;101:967–972. doi: 10.1038/sj.bjc.6605212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mano Y, Aishima S, Fujita N, Tanaka Y, Kubo Y, Motomura T, Taketomi A, Shirabe K, Maehara Y, Oda Y. Tumor-associated macrophage promotes tumor progression via STAT3 signaling in hepatocellular carcinoma. Pathobiology. 2013;80:146–154. doi: 10.1159/000346196. [DOI] [PubMed] [Google Scholar]
- 47.Zhang CH, Xu GL, Jia WD, Li JS, Ma JL, Ren WH, Ge YS, Yu JH, Liu WB, Wang W. Activation of STAT3 signal pathway correlates with twist and E-cadherin expression in hepatocellular carcinoma and their clinical significance. J Surg Res. 2012;174:120–129. doi: 10.1016/j.jss.2010.10.030. [DOI] [PubMed] [Google Scholar]
- 48.Wu Z-S, Cheng X-W, Wang X-N, Song N-J. Prognostic significance of phosphorylated signal transducer and activator of transcription 3 and suppressor of cytokine signaling 3 expression in human cutaneous melanoma. Melanoma Res. 2011;21:483–490. doi: 10.1097/CMR.0b013e32834acc37. [DOI] [PubMed] [Google Scholar]
- 49.Schoppmann SF, Jesch B, Friedrich J, Jomrich G, Maroske F, Birner P. Phosphorylation of signal transducer and activator of transcription 3 (STAT3) correlates with Her-2 status, carbonic anhydrase 9 expression and prognosis in esophageal cancer. Clin Exp Metastasis. 2012;29:615–624. doi: 10.1007/s10585-012-9475-3. [DOI] [PubMed] [Google Scholar]
- 50.Ryu K, Choy E, Yang C, Susa M, Hornicek FJ, Mankin H, Duan Z. Activation of signal transducer and activator of transcription 3 (Stat3) pathway in osteosarcoma cells and overexpression of phosphorylated-Stat3 correlates with poor prognosis. J Orthop Res. 2010;28:971–978. doi: 10.1002/jor.21088. [DOI] [PubMed] [Google Scholar]
- 51.Wang YC, Zheng LH, Ma BA, Zhou Y, Zhang MH, Zhang DZ, Fan QY. Clinical value of signal transducers and activators of transcription 3 (STAT3) gene expression in human osteosarcoma. Acta Histochem. 2011;113:402–408. doi: 10.1016/j.acthis.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 52.Denley SM, Jamieson NB, McCall P, Oien KA, Morton JP, Carter CR, Edwards J, McKay CJ. Activation of the IL-6R/Jak/stat pathway is associated with a poor outcome in resected pancreatic ductal adenocarcinoma. J Gastrointest Surg. 2013;17:887–898. doi: 10.1007/s11605-013-2168-7. [DOI] [PubMed] [Google Scholar]
- 53.Huang C, Huang R, Chang W, Jiang T, Huang K, Cao J, Sun X, Qiu Z. The expression and clinical significance of pSTAT3, VEGF and VEGF-C in pancreatic adenocarcinoma. Neoplasma. 2012;59:52–61. doi: 10.4149/neo_2012_007. [DOI] [PubMed] [Google Scholar]
- 54.Li C, Wang Z, Liu Y, Wang P, Zhang R. STAT3 expression correlates with prognosis of thymic epithelial tumors. J Cardiothorac Surg. 2013;8:92. doi: 10.1186/1749-8090-8-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Piperi C, Samaras V, Levidou G, Kavantzas N, Boviatsis E, Petraki K, Grivas A, Barbatis C, Varsos V, Patsouris E, Korkolopoulou P. Prognostic significance of IL-8-STAT-3 pathway in astrocytomas: correlation with IL-6, VEGF and microvessel morphometry. Cytokine. 2011;55:387–395. doi: 10.1016/j.cyto.2011.05.012. [DOI] [PubMed] [Google Scholar]
- 56.Zhao Y, Zhang J, Xia H, Zhang B, Jiang T, Wang J, Chen X, Wang Y. Stat3 is involved in the motility, metastasis and prognosis in lingual squamous cell carcinoma. Cell Biochem Funct. 2012;30:340–346. doi: 10.1002/cbf.2810. [DOI] [PubMed] [Google Scholar]
- 57.Chen CC, Chen WC, Lu CH, Wang WH, Lin PY, Lee KD, Chen MF. Significance of interleukin-6 signaling in the resistance of pharyngeal cancer to irradiation and the epidermal growth factor receptor inhibitor. Int J Radiat Oncol Biol Phys. 2010;76:1214–1224. doi: 10.1016/j.ijrobp.2009.09.059. [DOI] [PubMed] [Google Scholar]
- 58.Tam L, McGlynn LM, Traynor P, Mukherjee R, Bartlett JM, Edwards J. Expression levels of the JAK/STAT pathway in the transition from hormone-sensitive to hormone-refractory prostate cancer. Br J Cancer. 2007;97:378–383. doi: 10.1038/sj.bjc.6603871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Horiguchi A, Oya M, Shimada T, Uchida A, Marumo K, Murai M. Activation of signal transducer and activator of transcription 3 in renal cell carcinoma: a study of incidence and its association with pathological features and clinical outcome. J Urol. 2002;168:762–765. [PubMed] [Google Scholar]
- 60.Zhang LJ, Liu W, Gao YM, Qin YJ, Wu RD. The expression of IL-6 and STAT3 might predict progression and unfavorable prognosis in Wilms' tumor. Biochem Biophys Res Commun. 2013;435:408–413. doi: 10.1016/j.bbrc.2013.04.102. [DOI] [PubMed] [Google Scholar]
- 61.Sonnenblick A, Uziely B, Nechushtan H, Kadouri L, Galun E, Axelrod JH, Katz D, Daum H, Hamburger T, Maly B, Allweis TM, Peretz T. Tumor STAT3 tyrosine phosphorylation status, as a predictor of benefit from adjuvant chemotherapy for breast cancer. Breast Cancer Res Treat. 2013;138:407–413. doi: 10.1007/s10549-013-2453-x. [DOI] [PubMed] [Google Scholar]
- 62.Sheen-Chen S-M, Huang C-C, Tang R-P, Chou F-F, Eng H-L. Prognostic value of signal transducers and activators of transcription 3 in breast cancer. Cancer Epidemiol Biomarkers Prev. 2008;17:2286–2290. doi: 10.1158/1055-9965.EPI-08-0089. [DOI] [PubMed] [Google Scholar]
- 63.Dolled-Filhart M, Camp RL, Kowalski DP, Smith BL, Rimm DL. Tissue microarray analysis of signal transducers and activators of transcription 3 (Stat3) and phospho-Stat3 (Tyr705) in node-negative breast cancer shows nuclear localization is associated with a better prognosis. Clin Cancer Res. 2003;9:594–600. [PubMed] [Google Scholar]
- 64.Sonnenblick A, Shriki A, Galun E, Axelrod JH, Daum H, Rottenberg Y, Hamburger T, Mali B, Peretz T. Tissue microarray-based study of patients with lymph node-positive breast cancer shows tyrosine phosphorylation of signal transducer and activator of transcription 3 (tyrosine705-STAT3) is a marker of good prognosis. Clin Transl Oncol. 2012;14:232–236. doi: 10.1007/s12094-012-0789-z. [DOI] [PubMed] [Google Scholar]
- 65.Yamashita H, Nishio M, Ando Y, Zhang Z, Hamaguchi M, Mita K, Kobayashi S, Fujii Y, Iwase H. Stat5 expression predicts response to endocrine therapy and improves survival in estrogen receptor-positive breast cancer. Endocr Relat Cancer. 2006;13:885–893. doi: 10.1677/erc.1.01095. [DOI] [PubMed] [Google Scholar]
- 66.Woo S, Lee BL, Yoon J, Cho SJ, Baik TK, Chang MS, Lee HE, Park JW, Kim YH, Kim WH. Constitutive activation of signal transducers and activators of transcription 3 correlates with better prognosis, cell proliferation and hypoxia-inducible factor-1alpha in human gastric cancer. Pathobiology. 2011;78:295–301. doi: 10.1159/000321696. [DOI] [PubMed] [Google Scholar]
- 67.Cortas T, Eisenberg R, Fu P, Kern J, Patrick L, Dowlati A. Activation state EGFR and STAT-3 as prognostic markers in resected non-small cell lung cancer. Lung Cancer. 2007;55:349–355. doi: 10.1016/j.lungcan.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 68.Galleges Ruiz MI, Floor K, Steinberg SM, Grunberg K, Thunnissen FBJM, Belien JAM, Meijer GA, Peters GJ, Smit EF, Rodriguez JA, Giaccone G. Combined assessment of EGFR pathway-related molecular markers and prognosis of NSCLC patients. Br J Cancer. 2009;100:145–152. doi: 10.1038/sj.bjc.6604781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Haura EB, Zheng Z, Song L, Cantor A, Bepler G. Activated epidermal growth factor receptor-Stat-3 signaling promotes tumor survival in vivo in non-small cell lung cancer. Clin Cancer Res. 2005;11:8288–8294. doi: 10.1158/1078-0432.CCR-05-0827. [DOI] [PubMed] [Google Scholar]
- 70.Gordziel C, Bratsch J, Moriggl R, Knosel T, Friedrich K. Both STAT1 and STAT3 are favourable prognostic determinants in colorectal carcinoma. Br J Cancer. 2013;109:138–146. doi: 10.1038/bjc.2013.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Monnien F, Zaki H, Borg C, Mougin C, Bosset JF, Mercier M, Arbez-Gindre F, Kantelip B. Prognostic value of phosphorylated STAT3 in advanced rectal cancer: a study from 104 French patients included in the EORTC 22921 trial. J Clin Pathol. 2010;63:873–878. doi: 10.1136/jcp.2010.076414. [DOI] [PubMed] [Google Scholar]
- 72.Lee I, Fox PS, Ferguson SD, Bassett R, Kong LY, Schacherer CW, Gershenwald JE, Grimm EA, Fuller GN, Heimberger AB. The expression of p-STAT3 in stage IV melanoma: risk of CNS metastasis and survival. Oncotarget. 2012;3:336–344. doi: 10.18632/oncotarget.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hbibi AT, Lagorce C, Wind P, Spano JP, Des Guetz G, Milano G, Benamouzig R, Rixe O, Morere JF, Breau JL, Martin A, Fagard R. Identification of a functional EGF-R/p60c-src/STAT3 pathway in colorectal carcinoma: analysis of its long-term prognostic value. Cancer Biomark. 2008;4:83–91. doi: 10.3233/cbm-2008-4204. [DOI] [PubMed] [Google Scholar]
- 74.Rosen DG, Mercado-Uribe I, Yang G, Bast RC, Jr, Amin HM, Lai R, Liu J. The role of constitutively active signal transducer and activator of transcription 3 in ovarian tumorigenesis and prognosis. Cancer. 2006;107:2730–2740. doi: 10.1002/cncr.22293. [DOI] [PubMed] [Google Scholar]
- 75.Yang C, Lee H, Jove V, Deng J, Zhang W, Liu X, Forman S, Dellinger TH, Wakabayashi M, Yu H, Pal S. Prognostic significance of B-cells and pSTAT3 in patients with ovarian cancer. PLoS One. 2013;8:e54029. doi: 10.1371/journal.pone.0054029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen HH, Chou CY, Wu YH, Hsueh WT, Hsu CH, Guo HR, Lee WY, Su WC. Constitutive STAT5 activation correlates with better survival in cervical cancer patients treated with radiation therapy. Int J Radiat Oncol Biol Phys. 2012;82:658–666. doi: 10.1016/j.ijrobp.2010.11.043. [DOI] [PubMed] [Google Scholar]
- 77.Choi CH, Song SY, Kang H, Lee Y-Y, Kim C-J, Lee J-W, Kim T-J, Kim B-G, Lee J-H, Bae D-S. Prognostic significance of p-STAT3 in patients with bulky cervical carcinoma undergoing neoadjuvant chemotherapy. The J Obstet Gynaecol Res. 2010;36:304–310. doi: 10.1111/j.1447-0756.2009.01131.x. [DOI] [PubMed] [Google Scholar]
- 78.You Z, Xu D, Ji J, Guo W, Zhu W, He J. JAK/STAT signal pathway activation promotes progression and survival of human oesophageal squamous cell carcinoma. Clin Transl Oncol. 2012;14:143–149. doi: 10.1007/s12094-012-0774-6. [DOI] [PubMed] [Google Scholar]
- 79.Chang KC, Wu MH, Jones D, Chen FF, Tseng YL. Activation of STAT3 in thymic epithelial tumours correlates with tumour type and clinical behaviour. J Pathol. 2006;210:224–233. doi: 10.1002/path.2041. [DOI] [PubMed] [Google Scholar]
- 80.Shah NG, Trivedi TI, Tankshali RA, Goswami JA, Jetly DH, Kobawala TP, Shukla SN, Shah PM, Verma RJ. Stat3 expression in oral squamous cell carcinoma: association with clinicopathological parameters and survival. Int J Biol Markers. 2006;21:175–183. doi: 10.1177/172460080602100307. [DOI] [PubMed] [Google Scholar]
- 81.Macha MA, Matta A, Kaur J, Chauhan SS, Thakar A, Shukla NK, Gupta SD, Ralhan R. Prognostic significance of nuclear pSTAT3 in oral cancer. Head Neck. 2011;33:482–489. doi: 10.1002/hed.21468. [DOI] [PubMed] [Google Scholar]
- 82.Yang C, Schwab JH, Schoenfeld AJ, Hornicek FJ, Wood KB, Nielsen GP, Choy E, Mankin H, Duan Z. A novel target for treatment of chordoma: signal transducers and activators of transcription 3. Mol Cancer Ther. 2009;8:2597–2605. doi: 10.1158/1535-7163.MCT-09-0504. [DOI] [PubMed] [Google Scholar]
- 83.Pectasides E, Egloff A-M, Sasaki C, Kountourakis P, Burtness B, Fountzilas G, Dafni U, Zaramboukas T, Rampias T, Rimm D, Grandis J, Psyrri A. Nuclear localization of signal transducer and activator of transcription 3 in head and neck squamous cell carcinoma is associated with a better prognosis. Clin Cancer Res. 2010;16:2427–2434. doi: 10.1158/1078-0432.CCR-09-2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kim Y-J, Go H, Wu H-G, Jeon YK, Park SW, Lee SH. Immunohistochemical study identifying prognostic biomolecular markers in nasopharyngeal carcinoma treated by radiotherapy. Head Neck. 2011;33:1458–1466. doi: 10.1002/hed.21611. [DOI] [PubMed] [Google Scholar]
- 85.Bharadwaj U, Eckols TK, Kolosov M, Kasembeli MM, Adam A, Torres D, Zhang X, Dobrolecki LE, Wei W, Lewis MT, Dave B, Chang JC, Landis MD, et al. Drug-repositioning screening identified piperlongumine as a direct STAT3 inhibitor with potent activity against breast cancer. Oncogene. 2015;34:1341–1353. doi: 10.1038/onc.2014.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang X, Yue P, Page BD, Li T, Zhao W, Namanja AT, Paladino D, Zhao J, Chen Y, Gunning PT, Turkson J. Orally bioavailable small-molecule inhibitor of transcription factor Stat3 regresses human breast and lung cancer xenografts. Proc Natl Acad Sci U S A. 2012;109:9623–9628. doi: 10.1073/pnas.1121606109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kong LY, Abou-Ghazal MK, Wei J, Chakraborty A, Sun W, Qiao W, Fuller GN, Fokt I, Grimm EA, Schmittling RJ, Archer GE, Jr., Sampson JH, Priebe W, et al. A novel inhibitor of signal transducers and activators of transcription 3 activation is efficacious against established central nervous system melanoma and inhibits regulatory T cells. Clin Cancer Res. 2008;14:5759–5768. doi: 10.1158/1078-0432.CCR-08-0377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rath KS, Naidu SK, Lata P, Bid HK, Rivera BK, McCann GA, Tierney BJ, Elnaggar AC, Bravo V, Leone G, Houghton P, Hideg K, Kuppusamy P, et al. HO-3867, a safe STAT3 inhibitor, is selectively cytotoxic to ovarian cancer. Cancer Res. 2014;74:2316–2327. doi: 10.1158/0008-5472.CAN-13-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Liberati A, Altman DG, Tetzlaff J, Mulrow C, Gotzsche PC, Ioannidis JP, Clarke M, Devereaux PJ, Kleijnen J, Moher D. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: explanation and elaboration. PLoS Med. 2009;6:e1000100. doi: 10.1371/journal.pmed.1000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. 2010;25:603–605. doi: 10.1007/s10654-010-9491-z. [DOI] [PubMed] [Google Scholar]
- 91.Deeks JJ HJ, Altman DG. Analysing and presenting results. In: Higgins JPT, Green S, editors. Cochrane Handbook for Systematic Reviews of Interventions 425. Chichester, UK: John Wiley & Sons; 2006. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.