Abstract
EPB41L4A-AS2 is a novel long non-coding RNA of unknown function. In this study, we investigated the expression of EPB41L4A-AS2 in breast cancer tissues and evaluated its relationship with the clinicopathological features and prognosis of patients with breast cancer. This entailed conducting a meta-analysis and prognosis validation study using two cohorts from the Gene Expression Omnibus (GEO). In addition, we assessed EPB41L4A-AS2 expression and its relationship with the clinicopathological features of renal and lung cancers using the Cancer Genome Atlas cohort and a GEO dataset. We also clarified the role of EPB41L4A-AS2 expression in mediating cancer cell proliferation in breast, renal, and lung cancer cell lines transfected with an EPB41L4A-AS2 expression vector. We found that high EPB41L4A-AS2 expression is associated with favorable disease outcomes. Gene ontology enrichment analysis revealed that EPB41L4A-AS2 may be involved in processes associated with tumor biology. Finally, overexpression of EPB41L4A-AS2 inhibited tumor cell proliferation in breast, renal, and lung cancer cell lines. Our clinical and in vitro results suggest that EPB41L4A-AS2 inhibits solid tumor formation and that evaluation of this long non-coding RNA may have prognostic value in the clinical management of such malignancies.
Keywords: antisense lncRNA, EPB41L4A-AS2, prognostic value, proliferation, breast cancer
INTRODUCTION
Up to 30% of node-negative and 70% of node-positive breast cancer patients develop distant metastases during the course of their illness [1, 2]. Once distant metastases occur, breast cancer is no longer considered curable [3]. Despite a large amount of information on breast cancer risk factors, it is not understood how these risks influence metastases [4, 5]. Known risk factors account for only a small proportion of the overall incidence in high-risk populations; therefore, investigations into other risk factors may reduce breast cancer morbidity and mortality while providing high-quality screening, diagnosis, and treatment for patients who are at risk for developing breast cancer.
Long non-coding RNAs (lncRNA) are transcripts longer than 200 nucleotides with little or no protein-coding capacity [6]. LncRNAs regulate gene expression at various levels, including chromatin modifications, transcription, and post-transcriptional processing [7–11]. Recent studies have demonstrated that lncRNAs are involved in tumorigenesis and can be used as cancer biomarkers [12–21]. The lncRNAs PCA3 and UCA1 were identified as sensitive, specific, and unique diagnostic markers for human prostate cancer and bladder cancer, respectively [22–24]. In addition, SChLAP1 is an lncRNA biomarker associated with metastatic progression in prostate cancer [20]. The lncRNA, BCAR4, induces antiestrogen resistance in breast cancer, whereas PVT1 and MRUL induce multidrug-resistance in gastric cancer [25–28]. Moreover, several lncRNAs influence prognosis in solid tumors, such as GAPLINC in gastric cancer, UBC1 and GAS5 in bladder cancer, HOTTIP in hepatocellular carcinoma, and HOTAIR in breast cancer and colorectal cancer [29–36]. For single-gene disorders, a therapy for Angelman syndrome has been successfully performed by targeting an lncRNA UBE3A-ATS [37].
In searching approximately 100 breast cancer microarray datasets on the NCBI Gene Expression Omnibus (GEO) website, we found that a novel lncRNA, EPB41L4A-AS2, was closely associated with several vital tumor pathophysiological processes including tumorigenesis, chemoresistance, and estrogen regulation (http://www.ncbi.nlm.nih.gov/geoprofiles). EPB41L4A-AS2 is located in the 5q22.2 region of the genome and is an antisense lncRNA [38]. There is nothing in the literature about the biological function or association of EPB41L4A-AS2 with cancer or other diseases. To investigate the clinical relevance of EPB41L4A-AS2 and its biological role in breast cancer, we measured the expression of EPB41L4A-AS2 and its association with clinical and pathological features in 250 breast tumor samples, and further validated our findings using The Cancer Genome Atlas (TCGA) dataset. Next, we examined the association of EPB41L4A-AS2 expression with survival of different solid tumor patients in our breast cancer patient cohort along with two other GEO cohorts (renal cancer and lung cancer). To validate the prognostic value of EPB41L4A-AS2 expression and confirm our clinical findings, we conducted a meta-analysis in a fourth GEO cohort with 3699 breast cancer patients. We also performed gene ontology term enrichment analysis, and found that EPB41L4A-AS2 is involved with many tumor-associated biological processes. Finally, we showed that overexpression of EPB41L4A-AS2 inhibits tumor cell growth in breast cancer, renal cancer, and lung cancer cell lines. A flowchart of this study is shown in Supplementary Figure S1. Overall, our findings indicate that EPB41L4A-AS2 may be a prognostic biomarker and can act as a tumor suppressor in mediating the proliferation of solid tumors.
RESULTS
Low EPB41L4A-AS2 expression is correlated with adverse clinicopathological features in breast cancer
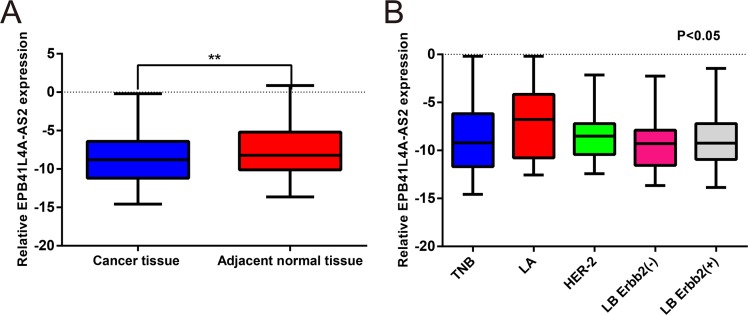
To assess the expression of EPB41L4A-AS2 in breast cancer, 250 breast cancer tissues and 50 adjacent normal tissues (Cohort I) were examined by quantitative real-time PCR (qRT-PCR). Expression of EPB41L4A-AS2 in breast cancer tissues was categorized as high or low according to the median value. EPB41L4A-AS2 levels were downregulated in breast cancer tissues compared with corresponding normal tissues (p = 0.005; Figure 1A). Among five breast cancer molecular subtypes, EPB41L4A-AS2 was more highly expressed in the luminal A subtype than the other four subtypes (p < 0.001; Figure 1B). We then investigated the relationship between EPB41L4A-AS2 expression and clinicopathological characteristics in our cohort. Larger tumor size (p = 0.026), higher stage and grade (p = 0.016 and 0.014, respectively), and ER and PR negative expression (p = 0.013 and 0.021, respectively) were correlated with low EPB41L4A-AS2 expression. However, EPB41L4A-AS2 expression was not associated with age, HER-2 status, or Ki67 status (p > 0.05; Table 1).
Figure 1. EPB41L4A-AS2 expression in human breast cancer tissues and adjacent non-cancerous tissues by qRT-PCR.
(A) Relative expression of EPB41L4A-AS2 in 250 breast cancer tissues and 50 adjacent non-cancerous tissues. (B) Relative expression of EPB41L4A-AS2 among five breast cancer molecular subtypes. qRT-PCR amplification was performed in triplicate and the levels of EPB41L4A-AS2 were normalized against control GAPDH expression. LA: Luminal A subtype; LB: Luminal B subtype; TNB: Triple negative subtype; ****p < 0.0001; ***p < 0.001; **p < 0. 01; *p < 0.05.
Table 1. Associations of EPB41L4A-AS2 expression with clinicpathological factors.
| Patient Features | Total No. (%) | Low EPB41L4A-AS2 No. (%) | High EPB41L4A-AS2 No. (%) | P value |
|---|---|---|---|---|
| Age (years) | 250 | 0.357 | ||
| < 50 | 160 (64.00) | 84 (52.50) | 76 (47.50) | |
| ≥ 50 | 90 (36.00) | 41 (45.56) | 49 (54.44) | |
| Tumor size | 250 | 0.026 | ||
| T1 | 65 (26.00) | 24 (36.92) | 41 (63.08) | |
| T2 | 138 (55.20) | 72 (52.17) | 66 (47.83) | |
| T3 | 47 (18.80) | 29 (61.70) | 18 (38.30) | |
| Tumor Stage | 250 | 0.016 | ||
| Stage I | 42 (16.80) | 14 (33.33) | 28 (66.67) | |
| Stage II | 168 (67.20) | 85 (50.60) | 83 (49.40) | |
| Stage III | 40 (16.00) | 26 (65.00) | 14 (35.00) | |
| Tumor Grade | 250 | 0.034 | ||
| Grade 1 | 44 (17.60) | 16 (36.36) | 28 (63.64) | |
| Grade 2 | 102 (40.80) | 48 (47.06) | 54 (52.94) | |
| Grade 3 | 104 (41.60) | 61 (58.65) | 43 (41.35) | |
| ER status | 250 | 0.013 | ||
| Positive | 156 (62.40) | 68 (43.59) | 88 (56.41) | |
| Negative | 94 (37.60) | 57 (60.64) | 37 (39.36) | |
| PR status | 250 | 0.021 | ||
| Positive | 147 (58.80) | 64 (43.54) | 83 (56.46) | |
| Negative | 103 (42.20) | 61 (59.22) | 42 (41.78) | |
| Her-2 status | 250 | 0.871 | ||
| Positive | 46 (18.40) | 24 (52.17) | 22 (47.83) | |
| Negative | 204 (72.60) | 101 (49.51) | 103 (50.49) | |
| Ki67 status | 250 | 0.591 | ||
| < 14% | 83 (33.20) | 39 (46.99) | 44 (63.01) | |
| ≥ 14% | 167 (67.80) | 86 (51.50) | 81 (48.50) |
EPB41L4A-AS2 expression is reduced in several malignant tumors
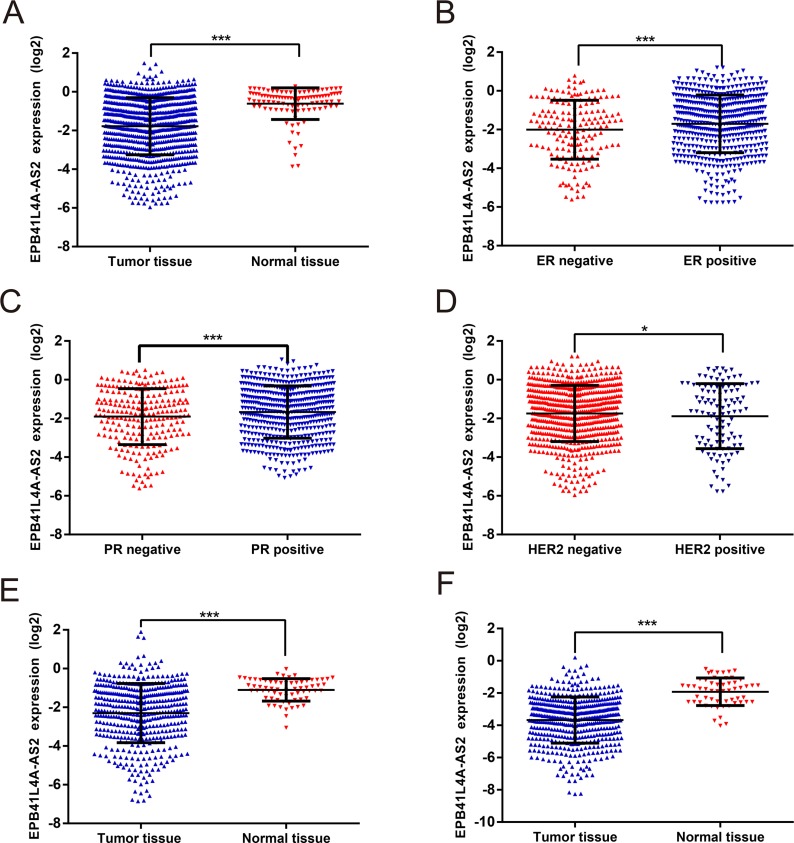
We next utilized data from the TCGA to determine whether the expression of EPB41L4A-AS2 was similar in a large cohort of patients with different solid tumors (Cohort II, https://tcga-data.nci.nih.gov/). In Cohort II, EPB41L4A-AS2 levels were again downregulated in breast cancer tissues compared with the corresponding normal tissues (p < 0.001; Figure 2A). Patients with positive hormone receptor expression had higher EPB41L4A-AS2 levels than those with negative receptor status (p < 0.001 for ER or PR; Figure 2B–2C). In addition, higher EPB41L4A-AS2 expression was observed in patients with HER-2 negative status compared with those with positive status (p = 0.036; Figure 2D). In the PAM50 molecular subtype system, higher EPB41L4A-AS2 expression was observed in patients with the Luminal A subtype compared with other subtypes (p < 0.05; Supplementary Figure S2).
Figure 2. EPB41L4A-AS2 expression in different solid tumors in TCGA.
EPB41L4A-AS2 expression: (A) in breast cancer tissues (N = 837) and normal tissues (N = 105), (B) in breast cancer with ER positive (N = 584) or negative (N = 174), (C) in breast cancer with PR positive (N = 508) or negative (N = 247), (D) in breast cancer with HER-2 positive (N = 113) or negative (N = 630), (E) in renal cancer tissues (N = 448) and normal tissues (N = 47), (F) in lung cancer tissues (N = 478) and normal tissues (N = 58). ***p < 0. 001; *p < 0.05.
In addition to breast cancer, we also analyzed other solid tumors such as renal cancer and lung cancer. As expected, low expression of EPB41L4A-AS2 was observed in renal cancer tissues compared with normal tissues (p < 0.001; Figure 2E). Expression of EPB41L4A-AS2 in tumors with high tumor stage or grade was also downregulated compared with those with low tumor stage or grade (p < 0.05; Supplementary Figures S3 and S4). We also saw similar results in lung cancer tissues vs. normal tissues (p < 0.001; Figure 2F). Thus, tumors with more malignant characteristics have lower levels of EPB41L4A-AS2, suggesting that EPB41L4A-AS2 may be a tumor suppressor.
High EPB41L4A-AS2 expression is associated with better overall survival of breast cancer patients
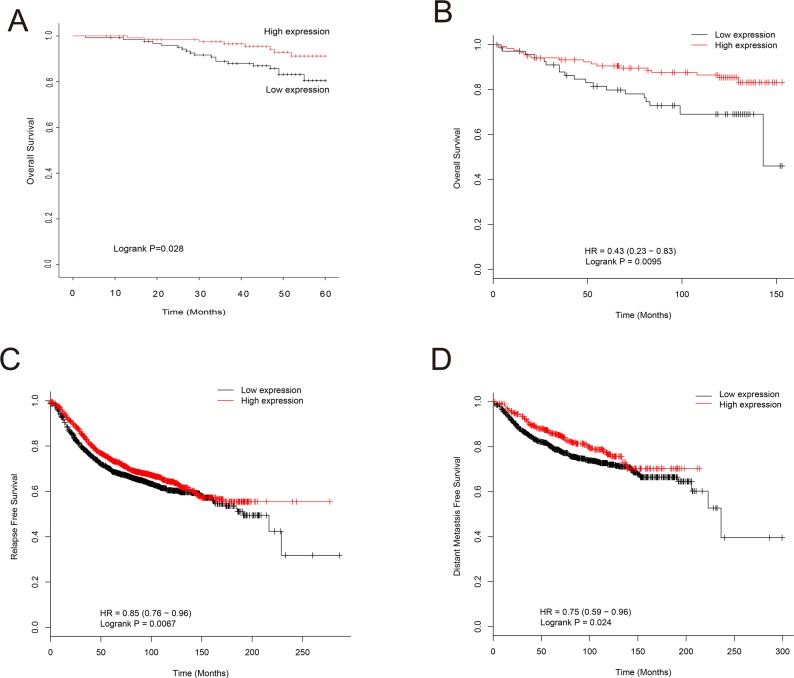
The relationship between EPB41L4A-AS2 expression and prognosis was investigated using both cohorts I and III (from the GEO database). In cohort I, breast cancer patients with low EPB41L4A-AS2 expression had poor overall survival (OS) when compared with patients with high expression (p = 0.028; Figure 3A). This result was validated in cohort III, which consisted of 26 breast cancer datasets containing clinicopathologic information (Supplementary Table S1). High EPB41L4A-AS2 expression was associated with better OS than low expression in patients with ER positive breast cancer, low tumor grade, or uninvolved lymph nodes (all p < 0.05; Supplementary Figures S5–S7).
Figure 3. Association of EPB41L4A-AS2 expression with survival of breast cancer patients.
(A) EPB41L4A-AS2 expression and OS of breast cancer patients in cohort I (N = 250). (B) EPB41L4A-AS2 expression and OS in TP53 wild-type patients in cohort III (N = 187). (C) EPB41L4A-AS2 expression and RFS of breast cancer patients in cohort III (N = 3,554). (D) EPB41L4A-AS2 expression and DMFS of breast cancer patients in cohort III (N = 1,609).
TP53 mutations are common in breast cancers and typically lead to chemoresistance. We next explored the relationship between TP53 mutations and EPB41L4A-AS2 expression in breast cancer patients. A better OS was observed in TP53 wild-type breast cancer patients with high EPB41L4A-AS2 expression, while the converse was found for breast cancer patients with a TP53 mutation (p < 0.01 and p = 0.43, respectively; Figure 3B and Supplementary Figure S8). For relapse-free survival (RFS) and distant metastasis-free survival (DMFS), breast cancer patients with high EPB41L4A-AS2 expression had better prognoses than those with low levels of EPB41L4A-AS2 (p < 0.01, p = 0.024, respectively; Figure 3C–3D). A meta-analysis was also performed to further validate the above results in a larger population (Cohort IV) of 24 datasets in GEO with 3699 breast cancer patients (Supplementary Table 2). We again found that high EPB41L4A-AS2 expression results in a better prognosis for any event of relapse, metastasis, or death in breast cancer patients (p = 0.0175; Supplement Figure S9).
High EPB41L4A-AS2 expression is associated with better overall survival of renal and lung cancer patients
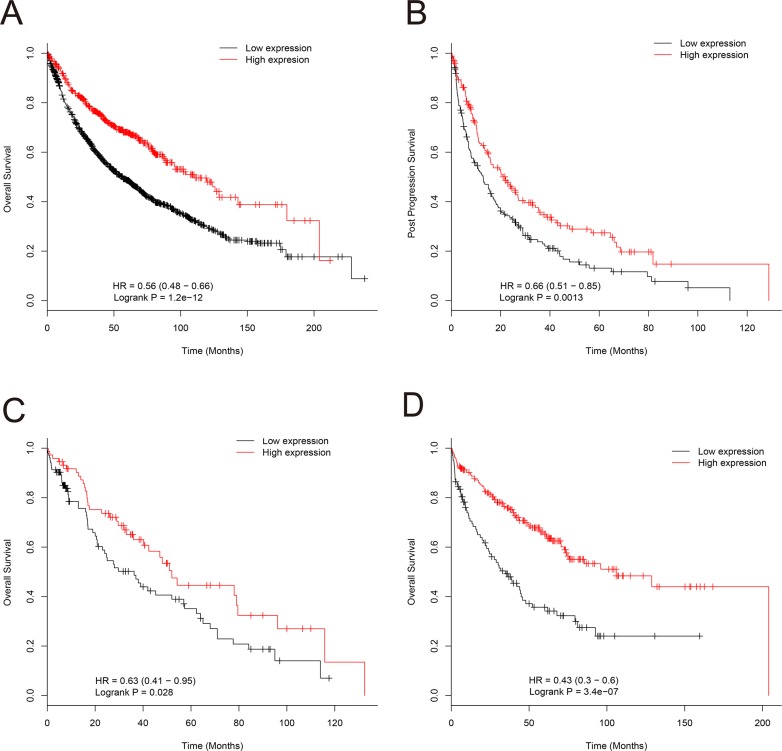
We also examined the relationship between EPB41L4A-AS2 expression and prognosis in cohorts II and III with renal cancer and lung cancer. In cohort II, renal cancer patients with high EPB41L4A-AS2 expression had better OS than those with low EPB41L4A-AS2 expression, which was consistent with our findings in breast cancer patients (p < 0.001, Supplementary Figure S10). In cohort III, lung cancer patients with high EPB41L4A-AS2 expression also had better OS and post-progression survival than those with low EPB41L4A-AS2 expression (p < 0.001and p = 0.0013, respectively; Figure 4A–4B). Subgroup analysis was also performed in lung cancer patients. As expected, lung cancer patients with high EPB41L4A-AS2 expression exhibited better OS in various stratifications, such as chemotherapy status, tumor tissue histology, gender, and smoking history than patients with low EPB41L4A-AS2 expression (all p < 0.05, Figure 4C–4D, Supplementary Figures S11–S16).
Figure 4. Association of EPB41L4A-AS2 expression with lung cancer patient survival in cohort III.
(A) EPB41L4A-AS2 expression and OS of lung cancer patients (N = 1926). (B) EPB41L4A-AS2 expression and post-progression survival of lung cancer patients (N = 344). (C) EPB41L4A-AS2 expression and OS of lung cancer patients with chemotherapy (N = 176), and without chemotherapy (N = 310) (D).
EPB41L4A-AS2 and its co-expressed genes play roles in tumor associated biological processes
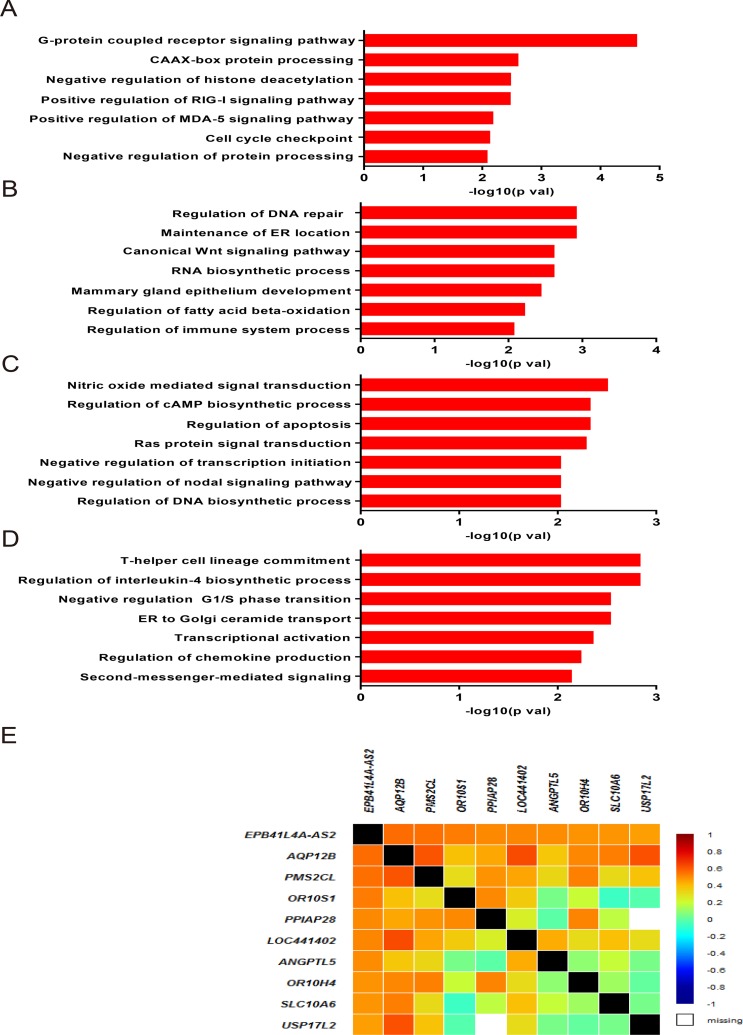
We speculated that EPB41L4A-AS2 and its co-expressed genes could participate in various biological processes in tumor malignancies. Therefore, we investigated co-expressed genes and analyzed them with gene ontology (GO) enrichment analysis for breast cancer patients in cohort IV. EPB41L4A-AS2 expression was correlated with tumor-associated biological processes, including G-protein coupled receptor signaling pathways, CAAX-box protein processing, negative regulation of histone deacetylation, the MDA-5 signaling pathway, the RIG-I signaling pathway, and cell cycle checkpoints (all p < 0.05; Figure 5A). We also performed the same analysis in breast cancer patients with different PAM50 molecular subtypes. Biological processes including DNA repair, maintenance of ER location, RNA biosynthesis, the canonical Wnt signaling pathway, mammary gland epithelial development, fatty acid beta-oxidation, and regulation of immune system processes were enriched in the basal-like subtype (all p < 0.05; Figure 5B). In the luminal subtype, the regulation of apoptosis, Ras signal transduction, endothelial cell migration, cell proliferation, and nodal signaling pathways were detected (all p < 0.05; Figure 5C). In the HER-2 subtype, biological processes involved in the regulation of cell cycle G1/S phase transition, second-messenger-mediated signaling, T-helper cell lineage commitment, interleukin-4 biosynthetic process, ER transport, transcriptional activation, and chemokine production were observed (all p < 0.05; Figure 5D). Representative co-expressed genes are shown in Figure 5E.
Figure 5. Associated biological processes of EPB41L4A-AS2 co-expressed genes in breast cancer.
Gene ontology enrichment analysis for EPB41L4A-AS2 co-expressed genes and correlated biological processes in breast cancer for all patients (A), in basal-like subtype (B), in luminal subtype (C) and in HER-2 subtype (D) in cohort IV. (E) Correlation between EPB41L4A-AS2 and its representative coexpressed genes in cohort IV.
Overexpression of EPB41L4A-AS2 inhibits tumor cell proliferation
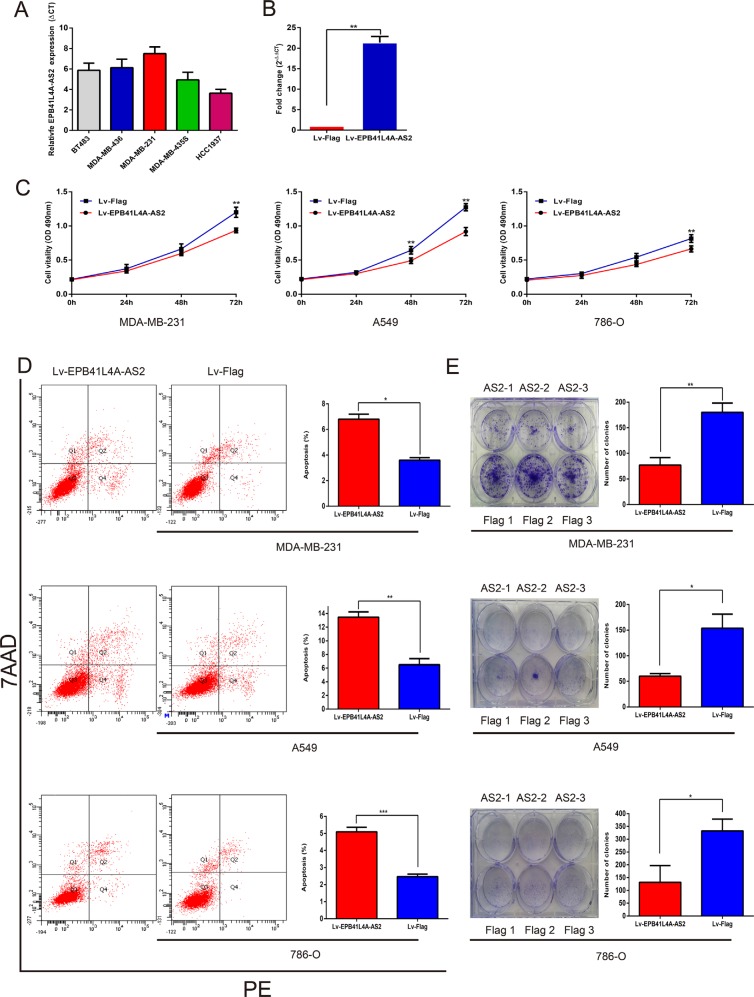
From the GO enrichment analysis, G-protein coupled receptor signaling pathways, histone deacetylation regulation, the RIG-I signaling pathway, and cell cycle checkpoint processes were found to be associated with tumor cell proliferation [39–46]. To elucidate the role of EPB41L4A-AS2 in tumor cell proliferation, we overexpressed the lncRNA using purified lentivirus for EPB41L4A-AS2 or a Flag control. First, we measured the endogenous expression of EPB41L4A-AS2 in breast cancer cell lines (Figure 6A). We selected MDA-MB-231 cells for our EPB41L4A-AS2 overexpression assay, and transfections were successful based on qRT-PCR analysis (Figure 6B). We also overexpressed EPB41L4A-AS2 in the renal cancer cell line 786-O and the lung cancer cell line A549. Cell proliferation assays showed that overexpression of EPB41L4A-AS2 inhibited tumor cell growth compared with the control in all three cancer cell lines (Figure 6C). Furthermore, all three cell lines exhibited increased apoptosis with EPB41L4A-AS2 overexpression as assessed by flow cytometry (Figure 6D). Finally, in a clonogenic assay, overexpression of EPB41L4A-AS2 reduced clone numbers in the three cancer cell lines (Figure 6E). These in vitro experiments suggest that upregulated EPB41L4A-AS2 expression may suppress tumor cell proliferation, which is in agreement with our clinical findings.
Figure 6. Ectopic overexpression of EPB41L4A-AS2 inhibits tumor cell proliferation.
(A) Endogenous expression of EPB41L4A-AS2 was detected in breast cancer cell lines. (B) Validation of ectopic overexpression of EPB41L4A-AS2 by qRT-PCR. Ectopic overexpression of EPB41L4A-AS2 inhibits proliferation (C), promotes apoptosis (D) and decreases colony formation (E) in MDA-MB-231, A549 and 786-O cell lines. qRT-PCR amplification was performed in triplicate and the levels of EPB41L4A-AS2 were normalized against control GAPDH expression. Data are presented as the mean value from three independent experiments ± S.D. ***p < 0. 001; **p < 0. 01; *p < 0.05.
DISCUSSION
Recent analyses of the mammalian transcriptome have revealed that up to 70% of the genome is transcribed into lncRNAs, while only 2% of the genome serves as coding transcripts [47, 48]. Given the uncertainty regarding their roles in cancer and other biological processes, lncRNAs have been regarded as transcriptional noise in the genome [49, 50]. Despite the growing recognition of the importance of lncRNAs in tumor biology, it is still a challenge to scientists to determine the functions of the large number of lncRNAs.
Although aberrant expression of lncRNAs has been identified as a marker in various solid tumors, the expression of lncRNA EPB41L4A-AS2 was unknown in malignant diseases. In this study, we demonstrated that EPB41L4A-AS2 expression differed with tumor grade, tumor size, disease stage, receptor status, and molecular subtype in breast cancer. To further validate and confirm our initial result, we downloaded and analyzed data from TCGA. Since the lncRNA, MALAT1, was upregulated in solid tumor tissue compared with the corresponding normal tissue [51–53], we investigated whether EPB41L4A-AS2 expression had a similar expression pattern in both renal cancer and lung cancer. Interestingly, low levels of EPB41L4A-AS2 were observed in renal cancer and lung cancer tissues compared with normal tissues. The expression of EPB41L4A-AS2 in high-staged tumors was also downregulated compared with those with low tumor stage in lung and renal cancers. Aberrant expression of EPB41L4A-AS2 in tumor tissues may indicate that it serves as a tumor suppressor and that its downregulation contributes to tumorigenesis and disease progression.
Because lncRNAs can serve as prognostic markers for patients with malignancies, we next asked whether EPB41L4A-AS2 expression could be a predictor of prognosis in solid tumors. We found that downregulation of EPB41L4A-AS2 was associated with poor survival outcomes, suggesting potential role for this marker in the clinical management of breast cancer. More importantly, our findings were consistent across multiple cohorts and on the basis of more than 20 independent studies, which collectively included thousands of patients. The lncRNA HOTAIR was identified as a prognostic marker not only in breast cancer but also in other solid tumors, such as gastric cancer and lung cancer [54–59]. To determine whether EPB41L4A-AS2 has similar prognostic potential we explored the relationship between its expression and patient prognosis in breast cancer, renal cancer, and lung cancer in another independent cohort. We consistently found that downregulation of EPB41L4A-AS2 was associated with poor survival outcomes in three major cancers.
One classic approach to exploring the putative functions of lncRNAs is through a “guilt-by-association” method [60]. This method associates lncRNAs with biological processes based on a common expression pattern and identifies lncRNAs that are associated with specific cellular processes [61, 62]. Analysis of co-expressed lncRNA genes has revealed co-regulation of genes by lncRNAs, suggesting that lncRNA clusters may regulate biological processes. In this study, we examined genes co-expressed with EPB41L4A-AS2 and performed GO enrichment analysis using cohort IV. Many tumor-associated biological processes, such as G-protein coupled receptor signaling pathways, negative regulation of cell cycle G1/S phase transition, and canonical Wnt signaling were identified. Using the guilt-by-association method, we propose that EPB41L4A-AS2 may regulate tumor cell proliferation by mediating expression of some of these genes that are associated with tumor growth, but this association remains to be investigated in future studies.
Antisense lncRNA is transcribed from the opposite strand to a protein-coding gene [63, 64]. Antisense lncRNAs may act as epigenetic regulators to interfere with the transcription, or increase the mRNA stability, of coding genes [65, 66]. As a novel antisense lncRNA, it would be interesting to investigate whether EPB41L4A-AS2 can mediate the transcription of the sense strand gene, EPB41L4A, or increase its mRNA stability. In this study, our in vitro experiments on breast cancer, renal cancer, and lung cancer cell lines demonstrated that overexpression of EPB41L4A-AS2 inhibited tumor cell proliferation. These results were consistent with the findings of our clinical studies, which underscore the potential involvement of EPB41L4A-AS2 in solid tumors. However, little is known about how and why EPB41L4A-AS2 behaves like a tumor suppressor. Further studies are needed to determine how EPB41L4A-AS2 is involved in mediating tumor biology.
MATERIALS AND METHODS
Patient specimens and clinical assessments
Eligible patients who had a histological diagnosis of breast cancer received neither chemotherapy nor radiotherapy before surgical resection. In total, 250 breast cancer tissues and 50 normal tissues were obtained from the Harbin Medical University Cancer Hospital in Harbin, China. All samples were frozen in liquid nitrogen immediately after surgical resections and only tumors with > 80% tumor cells were selected for RNA extraction. In clinic, breast cancer patients are classified into five main subtypes: luminal A, luminal B (HER2+), luminal B (HER2-), HER2 type, and triple negative based on the immunohistochemical tests for proteins ER, PR, HER2, and Ki-67 [67]. Two independent senior pathologists confirmed the pathological diagnosis and molecular subtype identification of each cancer tissue. This study conformed to the clinical research guidelines and was approved by the research ethics committee of the Harbin Medical University Cancer Hospital. We obtained written informed consent from all patients.
RNA extraction and quantitative real-time PCR
Total RNA was extracted from fresh samples preserved and frozen in liquid nitrogen using TRIzol Reagent (Invitrogen, Carlsbad, CA, USA), according to the manufacturer's instructions. Total RNA (2 μg) was reverse transcribed into cDNA using High-Capacity cDNA Reverse Transcriptase Kits (Applied Biosystems, Foster City, USA). The relative level of EPB41L4A-AS2 to the control gene GAPDH was determined by qRT-PCR using the 7500 Fast Real-Time PCR System (Applied Biosystems). The specific sequences of the primers used were 5′- CGGAGCAGGTGCAATCTGT-3′ (forward) and 5′-TCAAAACTACGTCTGATGCCAAA-3′ (reverse) for EPB41L4A-AS2; and 5′-ACCACAGTCCATGCCATCAC-3′ (forward) and 5′- TCCACCCTGTTGCTGTA-3′ (reverse) for GAPDH. PCR amplification was performed in triplicate, first at 95°C for 10 m, 40 cycles of 95°C for 10 s, and 60°C for 60 s. Quantitative normalization of EPB41L4A-AS2 cDNA was performed in each sample using the expression of GAPDH as an internal control. Relative levels of EPB41L4A-AS2 transcripts vs. GAPDH were determined by the comparative CT (2−ΔCT) method.
Public data analysis
The genome-wide lncRNA expression profiles of breast cancer, renal cancer, and lung cancer were downloaded from TCGA (https://tcga-data.nci.nih.gov/). The expression of lncRNA EPB41L4A-AS2 was calculated by summarizing the background corrected intensity of all probes mapped to this gene. Gene expression data for meta-analysis and prognosis analysis were selected from the GEO database. Meta-analysis included 24 datasets with 3699 breast cancer patients (Supplementary Table S1). Prognosis analysis included 26 datasets with 3554 breast cancer patients (Supplementary Table S2). The raw CEL files were downloaded from the GEO database (http://www.ncbi.nlm.nih.gov/geo/), and the background was adjusted according to a previously described method [68]. Expression of EPB41L4A-AS2 was dichotomized using study-specific median expression as the cutoff to define “high value” at or above the median versus “low value” below the median. The meta-analysis was conducted with the use of Review Manager (Revman Version 5.3, Copenhagen, Denmark; http://tech.cochrane.org/revman/), whereas gene co-expression and gene ontology term enrichment were analyzed using DAVID as previously described [69, 70].
Cell culture experiments
MDA-MB-231 cells were cultured in L-15 medium (Invitrogen, Carlsbad, CA) containing 10% fetal bovine serum and 100 units/ml of penicillin/streptomycin at 37°C without CO2. A549 and 786-O cells were cultured in a 1640 medium (Invitrogen, Carlsbad, CA) containing 10% fetal bovine serum and 100 units/ml of penicillin/streptomycin at 37°C in an atmosphere containing 5% CO2. All cell lines were obtained from the Chinese Type Culture Collection, Chinese Academy of Sciences. Cells were used in their logarithmic growth phase.
Lentiviral construction and production
The full-length nucleotide sequence of EPB41L4A-AS2 (NR_027706.1) was directly synthesized by Sangon Biotech Company and cloned into pLVX-Puro lentiviral expression vector (Clontech) between EcoRI and XbaI sites using an In-Fusion Cloning kit (Clontech). Lentiviruses were produced in 293T cells. Briefly, 293T cells were transiently transfected with pLVX plasmid and the packaging plasmids pLP1, pLP2, and pLP/VSVG using lipofectamine 2000. Lentiviral particles were harvested by collecting the supernatants 72 h later, which were centrifuged at 1500 g for 5 min and filtered through a 0.45 μm filter to remove cellular debris. Then, the crude lentivirus was concentrated by ultracentrifugation at 22,000 rpm for 2.5 h at 4°C, using a Beckman Ti70 rotor. The pellets were resuspended in DPBS and incubated at 4°C overnight. The purified lentivirus was then stored at −80°C.
Cell proliferation assay
A cell proliferation assay was carried out using the Cell Counting Kit-8 according to the manufacturer's instructions (Beyotime, Shanghai, China). Briefly, 2 × 103 cells were seeded in a 96-well plate. Cell proliferation was assessed for 24, 48, and 72 h. After adding 20 μl WST-1 reagents per well, cultures were incubated for 2 h and the absorbance was measured at 490 nm using a microplate reader (BioTek, VT, United States).
Clonogenic assay
For the clonogenic assay, 500 cells per well were seeded in 6-well plates. Two weeks later, visible colonies were noticed using the naked eye, fixed with 4% formaldehyde, and stained with 0.1% crystal violet. The colonies with a diameter greater than 1 mm were counted.
Flow cytometry
The Annexin-PE Apoptosis detection kit (BD Biosciences, San Jose, CA) was used to examine cell apoptosis according to the manufacturer's instructions. Briefly, cells were harvested by washing twice in cold PBS, and resuspending in 1 × Binding Buffer. Then, 100 μl of the cell solution (1 × 105 cells) was transferred into a 5 ml culture tube, and 5 μl of Annexin V-PE and 5 μl of 7-AAD were added. The cells were gently vortexed and incubated for 15 min at RT (25°C) in the dark. Then, 400 μl of 1 × binding buffer was added to each tube, and apoptosis analysis was performed in a FACScan instrument (Becton Dickinson, Mountain View, CA, USA).
Statistical analysis
Clinical and pathologic associations of EPB41L4A-AS2 were determined using the Chi-square test. A Welch's unpaired t test was used to evaluate the difference of EPB41L4A-AS2 expression among the PAM50 molecular subtypes. A significant p-value indicated that EPB41L4A-AS2 expression was different in at least two subtypes. Kaplan–Meier method and log-rank test were performed to show survival differences according to EPB41L4A-AS2 expression. The time for OS or RFS was calculated as the time from surgery until the occurrence of death and relapse, respectively. DMFS or PPS represented the time for distant metastasis-free survival and post progression survival, respectively. AE represented any event including metastasis, relapse, or death. The differences between groups in our in vitro experiments were analyzed using a Student's t-test. Spearman correlation coefficients were calculated for correlation analysis. All experiments were performed in triplicate, and the SPSS 16.0 software system (SPSS, Chicago, IL) was used for statistical analysis. All statistical tests were two-sided, and p < 0.05 was considered statistically significant.
SUPPLEMENTARY MATERIALS FIGURES AND TABLES
Acknowledgments
The authors would like to thank the contributors to the Cancer Genome Atlas project and Gene Expression Omnibus. We also thank the doctors and the patients who participated in this study. This article is subject to the NIH Public Access Policy.
Footnotes
GRANT SUPPORT
This work was supported by funding from the Specific research fund for public service sector, national health and family planning commission of the people's republic of China (Grant Numbers 201402003), National Natural Science Foundation of China (Grant Numbers 81172498), the National Key Technology Support Program (Grant Number 2013BAI09B00, 2014BAI09B00), the Project of Heilongjiang Province Applied Technology Research and Development (Grant Number GA13C201), the Harbin Medical University Graduate Student Innovation Fund Project (Grant Number YJSCX2014-03HYD), and the Haiyan Fund Project of Harbin Medical University Cancer Hospital (Grant Number JJMS2014-05).
CONFLICTS OF INTEREST
No potential conflicts of interest were disclosed.
REFERENCES
- 1.DeSantis C, Ma J, Bryan L, Jemal A. Breast cancer statistics, 2013. CA Cancer J Clin. 2014;64:52–62. doi: 10.3322/caac.21203. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 3.Pagani O, Senkus E, Wood W, Colleoni M, Cufer T, Kyriakides S, Costa A, Winer EP, Cardoso F, Force E-MT. International guidelines for management of metastatic breast cancer: can metastatic breast cancer be cured? J Natl Cancer Inst. 2010;102:456–463. doi: 10.1093/jnci/djq029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stevens RG, Brainard GC, Blask DE, Lockley SW, Motta ME. Breast cancer and circadian disruption from electric lighting in the modern world. CA Cancer J Clin. 2014;64:207–218. doi: 10.3322/caac.21218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang CC, Tu SH, Lien HH, Huang CS, Huang CJ, Lai LC, Tsai MH, Chuang EY. Refinement of breast cancer risk prediction with concordant leading edge subsets from prognostic gene signatures. Breast Cancer Res Treat. 2014;147:353–370. doi: 10.1007/s10549-014-3104-6. [DOI] [PubMed] [Google Scholar]
- 6.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 7.Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown JA, Bulkley D, Wang J, Valenstein ML, Yario TA, Steitz TA, Steitz JA. Structural insights into the stabilization of MALAT1 noncoding RNA by a bipartite triple helix. Nat Struct Mol Biol. 2014;21:633–640. doi: 10.1038/nsmb.2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 10.Fukuoka H, Takahashi Y. The role of genetic and epigenetic changes in pituitary tumorigenesis. Neurol Med Chir (Tokyo) 2014;54:943–957. doi: 10.2176/nmc.ra.2014-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McHugh CA, Chen CK, Chow A, Surka CF, Tran C, McDonel P, Pandya-Jones A, Blanco M, Burghard C, Moradian A, Sweredoski MJ, Shishkin AA, Su J, et al. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature. 2015;521:232–236. doi: 10.1038/nature14443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yarmishyn AA, Kurochkin IV. Long noncoding RNAs: a potential novel class of cancer biomarkers. Front Genet. 2015;6:145. doi: 10.3389/fgene.2015.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Okugawa Y, Grady WM, Goel A. Epigenetic Alterations in Colorectal Cancer: Emerging Biomarkers. Gastroenterology. 2015;149:1204–1225. e1212. doi: 10.1053/j.gastro.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meseure D, Drak Alsibai K, Nicolas A, Bieche I, Morillon A. Long Noncoding RNAs as New Architects in Cancer Epigenetics, Prognostic Biomarkers, and Potential Therapeutic Targets. Biomed Res Int. 2015;2015:320214. doi: 10.1155/2015/320214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jin K, Luo G, Xiao Z, Liu Z, Liu C, Ji S, Xu J, Liu L, Long J, Ni Q, Yu X. Noncoding RNAs as potential biomarkers to predict the outcome in pancreatic cancer. Drug Des Devel Ther. 2015;9:1247–1255. doi: 10.2147/DDDT.S77597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cui W, Qian Y, Zhou X, Lin Y, Jiang J, Chen J, Zhao Z, Shen B. Discovery and characterization of long intergenic non-coding RNAs (lincRNA) module biomarkers in prostate cancer: an integrative analysis of RNA-Seq data. BMC genomics. 2015;16:S3. doi: 10.1186/1471-2164-16-S7-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang J, Zhang KY, Liu SM, Sen S. Tumor-associated circulating microRNAs as biomarkers of cancer. Molecules. 2014;19:1912–1938. doi: 10.3390/molecules19021912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Veltri WR. Non-coding RNAs as biomarkers for metastatic prostate cancer. Lancet Oncol. 2014;15:1412–1413. doi: 10.1016/S1470-2045(14)71124-6. [DOI] [PubMed] [Google Scholar]
- 19.Ronnau CG, Verhaegh GW, Luna-Velez MV, Schalken JA. Noncoding RNAs as novel biomarkers in prostate cancer. Biomed Res Int. 2014;2014:591703. doi: 10.1155/2014/591703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prensner JR, Zhao S, Erho N, Schipper M, Iyer MK, Dhanasekaran SM, Magi-Galluzzi C, Mehra R, Sahu A, Siddiqui J, Davicioni E, Den RB, Dicker AP, et al. RNA biomarkers associated with metastatic progression in prostate cancer: a multi-institutional high-throughput analysis of SChLAP1. Lancet Oncol. 2014;15:1469–1480. doi: 10.1016/S1470-2045(14)71113-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee B, Mazar J, Aftab MN, Qi F, Shelley J, Li JL, Govindarajan S, Valerio F, Rivera I, Thurn T, Tran TA, Kameh D, Patel V, et al. Long noncoding RNAs as putative biomarkers for prostate cancer detection. J Mol Diagn. 2014;16:615–626. doi: 10.1016/j.jmoldx.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bussemakers MJ, van Bokhoven A, Verhaegh GW, Smit FP, Karthaus HF, Schalken JA, Debruyne FM, Ru N, Isaacs WB. DD3: a new prostate-specific gene, highly overexpressed in prostate cancer. Cancer Res. 1999;59:5975–5979. [PubMed] [Google Scholar]
- 23.Crawford ED, Rove KO, Trabulsi EJ, Qian J, Drewnowska KP, Kaminetsky JC, Huisman TK, Bilowus ML, Freedman SJ, Glover WL, Jr, Bostwick DG. Diagnostic performance of PCA3 to detect prostate cancer in men with increased prostate specific antigen: a prospective study of 1,962 cases. J Urol. 2012;188:1726–1731. doi: 10.1016/j.juro.2012.07.023. [DOI] [PubMed] [Google Scholar]
- 24.Wang XS, Zhang Z, Wang HC, Cai JL, Xu QW, Li MQ, Chen YC, Qian XP, Lu TJ, Yu LZ, Zhang Y, Xin DQ, Na YQ, et al. Rapid identification of UCA1 as a very sensitive and specific unique marker for human bladder carcinoma. Clin Cancer Res. 2006;12:4851–4858. doi: 10.1158/1078-0432.CCR-06-0134. [DOI] [PubMed] [Google Scholar]
- 25.Zhang XW, Bu P, Liu L, Zhang XZ, Li J. Overexpression of long non-coding RNA PVT1 in gastric cancer cells promotes the development of multidrug resistance. Biochem Biophys Res Commun. 2015;462:227–232. doi: 10.1016/j.bbrc.2015.04.121. [DOI] [PubMed] [Google Scholar]
- 26.Wang Y, Zhang D, Wu K, Zhao Q, Nie Y, Fan D. Long noncoding RNA MRUL promotes ABCB1 expression in multidrug-resistant gastric cancer cell sublines. Mol Cell Biol. 2014;34:3182–3193. doi: 10.1128/MCB.01580-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takahashi K, Yan IK, Wood J, Haga H, Patel T. Involvement of extracellular vesicle long noncoding RNA (linc-VLDLR) in tumor cell responses to chemotherapy. Mol Cancer Res. 2014;12:1377–1387. doi: 10.1158/1541-7786.MCR-13-0636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Godinho MF, Wulfkuhle JD, Look MP, Sieuwerts AM, Sleijfer S, Foekens JA, Petricoin EF, 3rd, Dorssers LC, van Agthoven T. BCAR4 induces antioestrogen resistance but sensitises breast cancer to lapatinib. Br J Cancer. 2012;107:947–955. doi: 10.1038/bjc.2012.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li T, Xie J, Shen C, Cheng D, Shi Y, Wu Z, Deng X, Chen H, Shen B, Peng C, Li H, Zhan Q, Zhu Z. Upregulation of long noncoding RNA ZEB1-AS1 promotes tumor metastasis and predicts poor prognosis in hepatocellular carcinoma. Oncogene. 2016;35:1575–84. doi: 10.1038/onc.2015.223. [DOI] [PubMed] [Google Scholar]
- 30.Cheng Z, Guo J, Chen L, Luo N, Yang W, Qu X. A long noncoding RNA AB073614 promotes tumorigenesis and predicts poor prognosis in ovarian cancer. Oncotarget. 2015;6:25381–25389. doi: 10.18632/oncotarget.4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bo H, Gong Z, Zhang W, Li X, Zeng Y, Liao Q, Chen P, Shi L, Lian Y, Jing Y, Tang K, Li Z, Zhou Y, et al. Upregulated long non-coding RNA AFAP1-AS1 expression is associated with progression and poor prognosis of nasopharyngeal carcinoma. Oncotarget. 2015;6:20404–20418. doi: 10.18632/oncotarget.4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang EB, Kong R, Yin DD, You LH, Sun M, Han L, Xu TP, Xia R, Yang JS, De W, Chen J. Long noncoding RNA ANRIL indicates a poor prognosis of gastric cancer and promotes tumor growth by epigenetically silencing of miR-99a/miR-449a. Oncotarget. 2014;5:2276–2292. doi: 10.18632/oncotarget.1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quagliata L, Matter MS, Piscuoglio S, Arabi L, Ruiz C, Procino A, Kovac M, Moretti F, Makowska Z, Boldanova T, Andersen JB, Hammerle M, Tornillo L, et al. Long noncoding RNA HOTTIP/HOXA13 expression is associated with disease progression and predicts outcome in hepatocellular carcinoma patients. Hepatology. 2014;59:911–923. doi: 10.1002/hep.26740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu Y, Wang J, Qian J, Kong X, Tang J, Wang Y, Chen H, Hong J, Zou W, Chen Y, Xu J, Fang JY. Long noncoding RNA GAPLINC regulates CD44-dependent cell invasiveness and associates with poor prognosis of gastric cancer. Cancer Res. 2014;74:6890–6902. doi: 10.1158/0008-5472.CAN-14-0686. [DOI] [PubMed] [Google Scholar]
- 35.Hu Y, Chen HY, Yu CY, Xu J, Wang JL, Qian J, Zhang X, Fang JY. A long non-coding RNA signature to improve prognosis prediction of colorectal cancer. Oncotarget. 2014;5:2230–2242. doi: 10.18632/oncotarget.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kogo R, Shimamura T, Mimori K, Kawahara K, Imoto S, Sudo T, Tanaka F, Shibata K, Suzuki A, Komune S, Miyano S, Mori M. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011;71:6320–6326. doi: 10.1158/0008-5472.CAN-11-1021. [DOI] [PubMed] [Google Scholar]
- 37.Meng L, Ward AJ, Chun S, Bennett CF, Beaudet AL, Rigo F. Towards a therapy for Angelman syndrome by targeting a long non-coding RNA. Nature. 2015;518:409–412. doi: 10.1038/nature13975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ota T, Suzuki Y, Nishikawa T, Otsuki T, Sugiyama T, Irie R, Wakamatsu A, Hayashi K, Sato H, Nagai K, Kimura K, Makita H, Sekine M, et al. Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat Genet. 2004;36:40–45. doi: 10.1038/ng1285. [DOI] [PubMed] [Google Scholar]
- 39.Feigin ME, Xue B, Hammell MC, Muthuswamy SK. G-protein-coupled receptor GPR161 is overexpressed in breast cancer and is a promoter of cell proliferation and invasion. Proc Natl Acad Sci U S A. 2014;111:4191–4196. doi: 10.1073/pnas.1320239111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang Y, Vasilatos SN, Boric L, Shaw PG, Davidson NE. Inhibitors of histone demethylation and histone deacetylation cooperate in regulating gene expression and inhibiting growth in human breast cancer cells. Breast Cancer Res Treat. 2012;131:777–789. doi: 10.1007/s10549-011-1480-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jin ZT, Li K, Li M, Ren ZG, Wang FS, Zhu JY, Leng XS, Yu WD. G-protein coupled receptor 34 knockdown impairs the proliferation and migration of HGC-27 gastric cancer cells in vitro. Chin Med J (Engl) 2015;128:545–549. doi: 10.4103/0366-6999.151114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klein DK, Hoffmann S, Ahlskog JK, O'Hanlon K, Quaas M, Larsen BD, Rolland B, Rosner HI, Walter D, Kousholt AN, Menzel T, Lees M, Johansen JV, et al. Cyclin F suppresses B-Myb activity to promote cell cycle checkpoint control. Nat Commun. 2015;6:5800. doi: 10.1038/ncomms6800. [DOI] [PubMed] [Google Scholar]
- 43.Li XY, Jiang LJ, Chen L, Ding ML, Guo HZ, Zhang W, Zhang HX, Ma XD, Liu XZ, Xi XD, Chen SJ, Chen Z, Zhu J. RIG-I modulates Src-mediated AKT activation to restrain leukemic stemness. Mol Cell. 2014;53:407–419. doi: 10.1016/j.molcel.2013.12.008. [DOI] [PubMed] [Google Scholar]
- 44.Piunti A, Rossi A, Cerutti A, Albert M, Jammula S, Scelfo A, Cedrone L, Fragola G, Olsson L, Koseki H, Testa G, Casola S, Helin K, et al. Polycomb proteins control proliferation and transformation independently of cell cycle checkpoints by regulating DNA replication. Nat Commun. 2014;5:3649. doi: 10.1038/ncomms4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shio S, Kodama Y, Ida H, Shiokawa M, Kitamura K, Hatano E, Uemoto S, Chiba T. Loss of RUNX3 expression by histone deacetylation is associated with biliary tract carcinogenesis. Cancer Sci. 2011;102:776–783. doi: 10.1111/j.1349-7006.2011.01848.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Szabo A, Fekete T, Koncz G, Kumar BV, Pazmandi K, Foldvari Z, Hegedus B, Garay T, Bacsi A, Rajnavolgyi E, Lanyi A. RIG-I inhibits the MAPK-dependent proliferation of BRAF mutant melanoma cells via MKP-1. Cell Signal. 2016;28:335–47. doi: 10.1016/j.cellsig.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 47.Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, Bell I, Cheung E, Drenkow J, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 48.Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, Kodzius R, Shimokawa K, Bajic VB, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 49.Lipovich L, Johnson R, Lin CY. MacroRNA underdogs in a microRNA world: evolutionary, regulatory, and biomedical significance of mammalian long non-protein-coding RNA. Biochim Biophys Acta. 2010;1799:597–615. doi: 10.1016/j.bbagrm.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 50.Chen LL, Carmichael GG. Long noncoding RNAs in mammalian cells: what, where, and why? Wiley Interdiscip Rev RNA. 2010;1:2–21. doi: 10.1002/wrna.5. [DOI] [PubMed] [Google Scholar]
- 51.Wei Y, Niu B. Role of MALAT1 as a Prognostic Factor for Survival in Various Cancers: A Systematic Review of the Literature with Meta-Analysis. Dis Markers. 2015;2015:164635. doi: 10.1155/2015/164635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang L, Bai HS, Deng Y, Fan L. High MALAT1 expression predicts a poor prognosis of cervical cancer and promotes cancer cell growth and invasion. Eur Rev Med Pharmacol Sci. 2015;19:3187–3193. [PubMed] [Google Scholar]
- 53.Zhang J, Zhang B, Wang T, Wang H. LncRNA MALAT1 overexpression is an unfavorable prognostic factor in human cancer: evidence from a meta-analysis. Int J Clin Exp Med. 2015;8:5499–5505. [PMC free article] [PubMed] [Google Scholar]
- 54.Loewen G, Jayawickramarajah J, Zhuo Y, Shan B. Functions of lncRNA HOTAIR in lung cancer. J Hematol Oncol. 2014;7:90. doi: 10.1186/s13045-014-0090-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gokmen-Polar Y, Vladislav IT, Neelamraju Y, Janga SC, Badve S. Prognostic impact of HOTAIR expression is restricted to ER-negative breast cancers. Sci Rep. 2015;5:8765. doi: 10.1038/srep08765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gu Y, Chen T, Li G, Yu X, Lu Y, Wang H, Teng L. LncRNAs: emerging biomarkers in gastric cancer. Future Oncol. 2015;11:2427–2441. doi: 10.2217/fon.15.175. [DOI] [PubMed] [Google Scholar]
- 57.Wang S, Wang Z. Prognostic value of long noncoding RNA HOTAIR in digestive system malignancies. J Gastroenterol Hepatol. 2015;30:1123–1133. doi: 10.1111/jgh.12940. [DOI] [PubMed] [Google Scholar]
- 58.Yu X, Li Z. Long non-coding RNA HOTAIR: A novel oncogene (Review) Mol Med Rep. 2015;12:5611–5618. doi: 10.3892/mmr.2015.4161. [DOI] [PubMed] [Google Scholar]
- 59.Zhao W, Dong S, Duan B, Chen P, Shi L, Gao H, Qi H. HOTAIR is a predictive and prognostic biomarker for patients with advanced gastric adenocarcinoma receiving fluorouracil and platinum combination chemotherapy. Am J Transl Res. 2015;7:1295–1302. [PMC free article] [PubMed] [Google Scholar]
- 60.Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guo X, Gao L, Liao Q, Xiao H, Ma X, Yang X, Luo H, Zhao G, Bu D, Jiao F, Shao Q, Chen R, Zhao Y. Long non-coding RNAs function annotation: a global prediction method based on bi-colored networks. Nucleic Acids Res. 2013;41:e35. doi: 10.1093/nar/gks967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liao Q, Liu C, Yuan X, Kang S, Miao R, Xiao H, Zhao G, Luo H, Bu D, Zhao H, Skogerbo G, Wu Z, Zhao Y. Large-scale prediction of long non-coding RNA functions in a coding-non-coding gene co-expression network. Nucleic Acids Res. 2011;39:3864–3878. doi: 10.1093/nar/gkq1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, Suzuki H, Carninci P, Hayashizaki Y, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 64.Pang KC, Frith MC, Mattick JS. Rapid evolution of noncoding RNAs: lack of conservation does not mean lack of function. Trends Genet. 2006;22:1–5. doi: 10.1016/j.tig.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 65.Morris KV, Vogt PK. Long antisense non-coding RNAs and their role in transcription and oncogenesis. Cell cycle. 2010;9:2544–2547. doi: 10.4161/cc.9.13.12145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yu W, Gius D, Onyango P, Muldoon-Jacobs K, Karp J, Feinberg AP, Cui H. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–206. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Goldhirsch A, Winer EP, Coates AS, Gelber RD, Piccart-Gebhart M, Thurlimann B, Senn HJ, Panel m. Personalizing the treatment of women with early breast cancer: highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2013. Ann Oncol. 2013;24:2206–2223. doi: 10.1093/annonc/mdt303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Risueno A, Fontanillo C, Dinger ME, De Las Rivas J. GATExplorer: genomic and transcriptomic explorer; mapping expression probes to gene loci, transcripts, exons and ncRNAs. BMC bioinformatics. 2010;11:221. doi: 10.1186/1471-2105-11-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 70.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4 doi: 10.2202/1544-6115.1128. Article 17. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.