Abstract
After allogeneic hematopoietic stem cell transplantation (allo-HSCT), intestinal bacteria modulate risks of infection and graft-versus-host disease (GVHD). Neutropenic fever is common and treated with a choice of clinically equivalent antibiotics that target obligately anaerobic bacteria (anaerobes) to varying degrees. We retrospectively examined 857 allo-HSCT recipients and found that treatment of neutropenic fever with imipenem-cilastatin and piperacillin-tazobactam was associated with increased GVHD-related mortality at 5 years (21.5% in imipenem-cilastatin-treated patients vs. 13.1% in untreated patients, p=0.025, and 19.8% in piperacillin-tazobactam-treated patients vs. 11.9% in untreated patients, p=0.007). However, two other antibiotics also used to treat neutropenic fever, aztreonam and cefepime, were not associated with GVHD-related mortality (p=0.78 and p=0.98, respectively). Analysis of stool microbiota composition showed that piperacillin-tazobactam administration was associated with increased compositional perturbation. Studies in mouse models demonstrated similar effects of these antibiotics, as well as aggravated GVHD mortality with imipenem-cilastatin or piperacillin-tazobactm compared to aztreonam (p<0.01 and p<0.05, respectively). We found pathological evidence for increased GVHD in the colon of imipenem-cilastatin-treated mice (p<0.05), but no differences in short-chain fatty acid concentrations or regulatory T cells numbers. Notably, imipenem-cilastatin treatment of mice with GVHD led to loss of the protective lining of mucus in the colon (p<0.01) and intestinal barrier function was compromised (p<0.05). Sequencing of mouse stool specimens showed expansion of Akkermansia muciniphila (p<0.001), a commensal bacterium with mucus-degrading capabilities, raising the possibility that mucus degradation can contribute to murine GVHD. We demonstrate an underappreciated risk for antibiotics with activity against anaerobes to exacerbate colonic GVHD after transplant.
INTRODUCTION
Systemic bacterial infections are potentially life-threatening and occur commonly in patients undergoing allogeneic hematopoietic stem cell transplantation (allo-HSCT), particularly early after allo-HSCT during the period of neutropenia (1, 2). When these patients develop neutropenic fever, treatment with rapid administration of empiric antibiotics that target gram-negative bacteria has been shown to reduce mortality (3–5). Potential late effects of antibiotic treatment, however, on the frequency and severity of downstream complications following allo-HSCT have not been well studied. In particular, effects of various antibiotics on graft-versus-host disease (GVHD), an inflammatory condition that results from the donor immune system recognizing the transplant recipient's tissues as foreign, is not well understood.
The intestinal microbiota consists of diverse bacterial taxa that contribute to host health through a variety of functions (6, 7). Several studies have indicated that anaerobes in the intestine, including Clostridial species, are important mediators of intestinal homeostasis. An experimental consortium of intestinal bacteria from the order Clostridiales can prevent inflammation by a variety of mechanisms, including induction of intestinal regulatory T cells (8–10). Interestingly, development of GVHD results in major changes in the intestinal microbiota that are characterized by bacterial shifts from Clostridiales to Lactobacillales or Enterobacteriales (11–14). Several studies have demonstrated that commensal bacteria can be potent modulators of the severity of GVHD (11–13, 15). Our group recently reported that the loss of intestinal bacteria from the genus Blautia, a prominent member of Clostridiales from human intestinal flora, is associated with increased mortality from GVHD (16).
When empirically treating neutropenic fever, clinicians can select from several antibiotics. Whereas in some cases choice of antibiotic is dictated by localizing symptoms or known drug allergies, in the majority of patients several options are considered equally effective. Interestingly, despite clinical equivalency in treating neutropenic fever, these antibiotics vary considerably in the strength of their activity against anaerobic commensals. We hypothesized that treatment with an antibiotic that spares anaerobic bacteria may minimize loss of potentially beneficial intestinal commensal bacteria and reduce the risk of severe GVHD. We identified a cohort of 857 recipients of T-cell replete allografts transplanted at our center and evaluated the cohort for associations among individual antibiotic treatments and GVHD-related mortality. Furthermore, utilizing a murine model of allo-HSCT, we examined the impact of four different antibiotics used to treat neutropenic fever on the severity of GVHD.
RESULTS
Treatment with antibiotics with increased activity against anaerobes correlates with increased GVHD-related mortality and altered intestinal microbiota in allo-HSCT patients with neutropenic fever
We began by asking if the use of certain antibiotics in the peri-transplant setting was associated with clinical differences in GVHD-related mortality. At our center, as at many bone marrow transplant (BMT) centers, efforts are made to adhere to practice guidelines, including standard prophylactic and empiric antibiotic treatments. However, there remains considerable heterogeneity in the antibiotics that allo-HSCT patients receive, due to multiple factors including patients allergic or intolerant to certain medications, differences in conditioning regimen intensity, and clinical variability in transplant courses with respect to duration and severity of neutropenia, fevers and infections. We examined 857 adult recipients of allo-HSCT transplanted at our center from May 1992 to July 2015. Recipients of T-cell-depleted allografts were excluded, given their reduced risk for GVHD. The overall incidence of GVHD-related mortality was 14.9% at 5 years. Clinical characteristics are provided in Table 1.
Table 1.
Clinical characteristics of 857 allo-HSCT patients transplanted at MSKCC
| Dates of transplant | May 1992 to July 2015 |
|
| |
| Age (years) | 18 to 77, median 49 |
|
| |
| Gender | Female 341 (40%), male 516 (60%) |
|
| |
| Primary malignancy | NHL 242 (28%), AML 277 (32%), CML 48 (5.6%), MDS/MPD 71 (8.3%), ALL 74 (8.6%), CLL 41 (4.8%), Hodgkin disease 70 (8.2%), multiple myeloma 24 (2.8%), other hematological malignancies 16 (1.9%), other malignancies 8 (0.9%), non-malignant hematological disorders 22 (2.6%) |
|
| |
| Graft source | Peripheral blood 497 (58%), cord blood 197 (23%), bone marrow 163 (19%) |
|
| |
| Conditioning intensity | Standard intensity myeloablative 305 (36%), reduced intensity myeloablative 283 (33%), nonmyeloablative 268 (31%) |
|
| |
| HLA matching | HLA identical 604 (70%), HLA mismatch 253 (30%) |
|
| |
| Antibiotics for neutropenic fever | No antibiotics for neutropenic fever 474 (55%) |
| Imipenem-cilastatin or piperacillin-tazobactam first-line 306 (36%) | |
| Received imipenem-cilastatin only 6 (0.7%) | |
| Received piperacillin-tazobactam only 183 (21%) | |
| Received both 117 (14%) | |
| Also received aztreonam or cefepime 106 (12%) | |
| Aztreonam or cefepime first-line 77 (9%) | |
| Received aztreonam only 18 (2.1%) | |
| Received cefepime only 40 (4.7%) | |
| Received both 19 (2.2%) | |
| Also received imipenem-cilastatin 23 (2.7%) | |
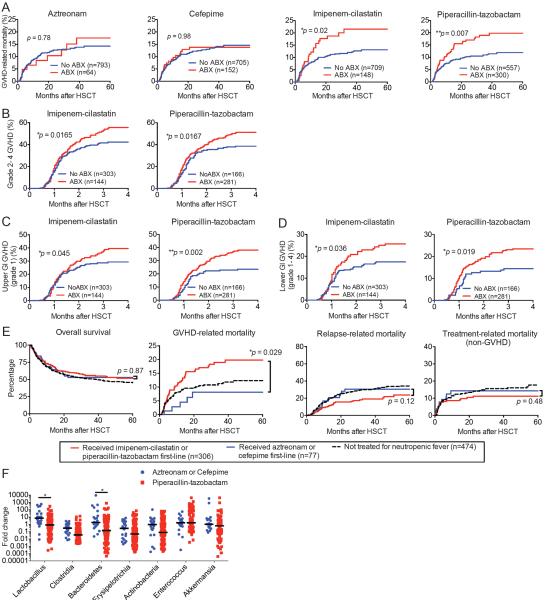
We identified the 12 most frequently administered antibiotics given between days −7 to +28 relative to allo-HSCT (Table 2). The first and second most commonly administered antibiotics in this patient population were intravenous (IV) vancomycin and IV ciprofloxacin, respectively, which was congruent with our institutional standard prophylactic antibiotic regimen being the combination of IV vancomycin and IV ciprofloxacin since 2006 (17). For each antibiotic, we stratified patients into two cohorts: one consisting of patients who had been treated with that antibiotic, and a second cohort who had not been treated. We then compared the cumulative incidence of GVHD-related mortality for the two cohorts and evaluated them for potential differences using the Gray's test. We found that of the twelve antibiotics evaluated in this manner, the two most significantly associated with a difference in GVHD-related mortality were piperacillin-tazobactam (p=0.007) and imipenem-cilastatin (p=0.025) (Table 2). At our institution, piperacillin-tazobactam has been our standard first-line antibiotic for empiric treatment of neutropenic fever since 2002, whereas imipenem-cilastatin is commonly utilized as a second-line treatment for neutropenic fever in patients with microbiological evidence of resistance to first-line treatment, as well as those with persistent fevers or clinical worsening of symptoms. Each treatment was associated with increased incidence of GVHD-related mortality (Fig. 1A). Interestingly, treatment with cefepime and aztreonam, which are both used as alternative first-line treatments for neutropenic fever in patients with penicillin allergies, was not associated with differences in GVHD-related mortality (Fig. 1A, p=0.98 and p=0.78, respectively, and Table 2). We then evaluated the relationship between treatment with each of these four antibiotics and clinical GVHD grades 2-4 (on a standardized scale of 1 to 4, with grade 4 being the most severe (18), and found that treatment with piperacillin-tazobactam and imipenem-cilastatin were both associated with an increased incidence of grade 2-4 clinical GVHD (Fig. 1B, p=0.0167 and p=0.0165, respectively). Additionally, exposure to piperacillin-tazobactam and imipenem-cilastatin was significantly associated with an increased incidence of upper gastrointestinal (GI) GVHD (Fig. 1C and Table 3, p=0.002 and p=0.045, respectively) and lower GI GVHD (Fig. 1D and Table 3, p=0.019 and p=0.036, respectively), which is the most common life-threatening form of GVHD. Notably, piperacillin-tazobactam and imipenem-cilastatin are both highly active against obligately anaerobic bacteria, whereas cefepime and aztreonam have reduced activity against anaerobes (19). Thus, these results suggested that differences in the spectrum of activity of antibiotics used to treat neutropenic fever might modulate the subsequent frequency and severity of GVHD.
Table 2.
Effects of antibiotic exposure on increased 5-year GVHD-related mortality
| Antibiotic | Untreated 5-year GVHD-related mortality (n) | Treated 5-year GVHD related mortality (n) | P-value |
|---|---|---|---|
| Atovaquone (PO) | 14.1% (772) | 19.1% (85) | 0.496 |
| Aztreonam (IV) | 14.2% (793) | 17.5% (64) | 0.777 |
| Cefepime (IV) | 14.6% (705) | 13.8% (152) | 0.980 |
| Ciprofloxacin (IV) | 14.4% (535) | 14.7% (322) | 0.862 |
| Imipenem-cilastatin (IV) | 13.1% (709) | 21.5% (148) | 0.025 |
| Metronidazole (IV) | 14.0% (779) | 18.6% (78) | 0.197 |
| Metronidazole (PO) | 14.0% (801) | 20.8% (56) | 0.206 |
| Piperacillin-tazobactam (IV) | 11.9% (557) | 19.8% (300) | 0.007 |
| Sulfamethoxazole-trimethoprim (IV) | 14.8% (769) | 12.9% (88) | 0.625 |
| Sulfamethoxazole-trimethoprim (PO) | 14.6% (727) | 12.8% (130) | 0.522 |
| Vancomycin (IV) | 13.4% (408) | 15.6% (449) | 0.579 |
| Vancomycin (PO) | 14.4% (796) | 17.3% (61) | 0.942 |
Fig. 1.
Clinical use of imipenem-cilastatin and piperacillin-tazobactam is associated with increased GVHD-related mortality. (A to D) A retrospective cohort of 857 adult patients was identified who received non-T cell depleted allo-HSCT at our center from 1992 to 2015. (A) GVHD-related mortality in patients exposed to aztreonam, cefepime, imipenem-cilastatin or piperacillin-tazobactam. (B) Analyses of overall grade 2-4 GVHD are shown. (C) Analyses of upper GI grade 1 GVHD are shown. (D) Analyses of lower GI grade 1–4 GVHD are shown. (E) A subset of patients who had been treated for neutropenic fever was stratified according to whether they received first-line treatment with imipenem-cilastatin or piperacillin-tazobactam, versus aztreonam or cefepime. Outcomes indicated were depicted by Kaplan-Meier plots and curves compared by the logrank test. *, P < 0.05; **, P < 0.01 in A–E. (F) Intestinal microbiota composition analysis using 16S rRNA sequencing prior to and after beginning treatment with the indicated antibiotics in allo-HSCT recipients. *, P < 0.05 after multiple comparison with Holm-Sidak correction.
Table 3.
Association between antibiotic treatment and increased GVHD in target organs
| Antibiotic | GVHD Target Organ (p value) | |||
|---|---|---|---|---|
| Skin (Stage 1–4) | Liver (Stage 1–4) | Upper GI (Stage 1) | Lower GI (Stage 1–4) | |
| Aztreonam | 0.153 | 0.107 | 0.601 | 0.702 |
| Cefepime | 0.133 | 0.183 | 0.332 | 0.514 |
| Imipenem-cilastatin | 0.188 | 0.200 | 0.045 | 0.036 |
| Piperacillin-tazobactam | 0.180 | 0.165 | 0.002 | 0.019 |
To further evaluate an association between antibiotic spectrum of activity and GVHD-related mortality, we examined a subset of patients who all received treatment for neutropenic fever, and compared patients who were treated with first-line antibiotics imipenem-cilastatin or piperacillin-tazobactam (n=306) with patients who were treated with the first-line antibiotics aztreonam or cefepime (n=77). We found that the first group exhibited increased GVHD-related mortality (p=0.029), although we did not detect a change in overall survival (Fig. 1E, p=0.87). Univariate analyses including previously identified GVHD risk factors revealed that antibiotic treatment with either aztreonam or cefepime was significantly associated with a decreased risk of GVHD-related mortality in this data set (Table 4), suggesting that the type of antibiotic exposure may be a predictor of GVHD-related mortality. We also performed a multivariate analysis evaluating the association of type of antibiotic exposure with GVHD-related mortality while adjusting for GVHD risk factors associated with GVHD-related mortality in our patient group using a significance criteria of p<0.05. We found that antibiotic exposure to either aztreonam or cefepime remained significantly correlated with reduced GVHD-related mortality (p=0.048) after adjusting for conditioning regimen intensity (Table 4). These results support the possibility that selecting antibiotics that preserve anaerobic commensal bacteria may reduce the risk of GVHD. An alternative hypothesis would be that patients with a history of allergies to penicillins (who are thus more likely to receive cephalosporins or aztreonam instead of penicillins and carbapenems) may be protected against GVHD, although this seems to have less biological plausibility.
Table 4.
Univariate and multivariate analyses of factors for GVHD-related mortality
| Univariate | Multivariate | |||
|---|---|---|---|---|
| HR (95% CI) | P-value | HR (95% CI) | P-value | |
| Antibiotic treatment group | ||||
| Imipenem/Piperacillin-tazobactam | reference | reference | ||
| Aztreonam/Cefepime | 0.38 (0.15–0.94) | 0.037 | 0.4 (0.16–0.99) | 0.048 |
| Age > 40 | 1.35 (0.69–2.64) | 0.38 | ||
| Race | ||||
| White/Black | reference | |||
| Asian/Hispanic | 1.19 (0.56–2.53) | 0.65 | ||
| Other | 1.89 (0.64–5.57) | 0.25 | ||
| Source | ||||
| PBSC | reference | |||
| BM | 1.27 (0.44–3.72) | 0.66 | ||
| Cord | 1.21 (0.69–2.11) | 0.51 | ||
| Donor | ||||
| Sibling Identical | reference | |||
| Unrelated Identical | 1.47 (0.64–3.35) | 0.36 | ||
| Non-identical | 2.08 (0.96–4.47) | 0.062 | ||
| Intensity | ||||
| Ablative | reference | reference | ||
| Reduced | 1.03 (0.49–2.19) | 0.93 | 1.04 (0.49–2.2) | 0.920 |
| Nonablative | 2.37 (1.15–4.91) | 0.020 | 2.3 (1.11–4.76) | 0.026 |
| TBI (any) | 1.72 (0.92–3.22) | 0.087 | ||
HR; hazard ratio, PBSC; peripheral blood stem cells, BM; bone marrow, TBI; total body irradiation
We then asked how the degree of antibiotic activities against anaerobes would affect the intestinal bacterial composition of allo-HSCT patients. We utilized 16S rRNA gene deep sequencing analysis of our stool specimen bank, which consisted of specimens prospectively collected weekly from patients undergoing allo-HSCT at our center beginning in 2009. From this specimen bank, we identified pairs of stool specimens from allo-HSCT patients collected prior and subsequent to initiation of a specific antibiotic treatment. Representative cases of patients treated for neutropenic fever, as well as patients who did not require therapeutic antibiotics (but did receive prophylactic antibiotics), are shown in fig.S1A to F. We identified 22 instances where microbiota analyses were available from patients who had begun aztreonam or cefepime therapy, and 106 instances where patients had begun piperacillin-tazobactam therapy. We quantified and compared the fold change in abundance of major subsets of intestinal microbiota with these two treatments, and found that piperacillin-tazobactam therapy was associated with a greater loss of Bacteroidetes and Lactobacillus, whereas the fold change in abundance of Enterococcus, Akkermansia and Erysipelotrichia was similar; Clostridia and Actinobacteria trended towards showing a decrease after piperacillin-tazobactam treatment, although this did not meet strict statistical significance after correction for multiple comparisons (Fig. 1F). Overall, these results demonstrated that piperacillin-tazobactam treatment resulted in larger losses of several major subsets of intestinal bacterial populations. We were unfortunately unable to assess the effects of imipenem-cilastatin in our patients, because at our center imipenem-cilastatin is almost uniformly administered to allo-HSCT patients as second-line therapy for neutropenic fever following prior treatment with piperacillin-tazobactam, cefepime, or aztreonam.
Treatment with piperacillin-tazobactam or imipenem-cilastatin disrupts the intestinal microbiota and exacerbates GVHD in mice
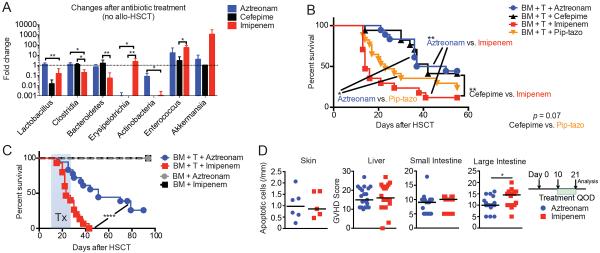
To further explore potential causality and mechanisms of the effects of antibiotics with anaerobic activity on GVHD, we utilized murine models of allo-HSCT. We first treated healthy C57BL/6 mice with aztreonam, cefepime, imipenem-cilastatin and piperacillin-tazobactam. Mice were treated by subcutaneous (SC) injections of each antibiotic twice a day for two days (500 mg/kg for piperacillin-tazobactam and 100 mg/kg for others) and stool specimens were collected, followed by 16S rRNA gene amplification and sequence analysis. We found that systemic treatment with imipenem-cilastatin reduced the abundance of Clostridiales and increased that of Erysipelotrichia and Enterococcus, while treatment with aztreonam or cefepime spared Clostridiales (Fig. 2A). Interestingly, treatment with piperacillin-tazobactam resulted in no amplifiable bacterial DNA after two days of treatment, indicating near-complete decontamination in mice.
Fig. 2.
Imipenem-cilastatin treatment, compared to aztreonam treatment, suppresses anaerobic commensals and elevates GVHD severity in mice. (A) Intestinal microbiota composition analysis using 16S rRNA sequencing prior to and after treatment with the indicated antibiotics in healthy C57BL/6 mice. Mice were treated with subcutaneous (SC) injections of each antibiotic twice a day for two days (100 mg/kg) and stool specimens were collected the following day. Values represent mean ± SEM (n = 6–7). *, P < 0.05; **, P < 0.01 by multiple comparisons corrected by Holm-Sidak test. Data are combined from two independent experiments. (B to D) Lethally irradiated 129S1 recipients were transplanted with MHC-matched C57BL/6 T-cell depleted bone marrow (“BM” in the figure) cells and 1 × 106 C57BL/6 T cells (“T” in the figure). Recipients were treated with aztreonam, cefepime, imipenem-cilastatin or piperacillin-tazobactam (pip-tazo) (100 mg/kg, SC, 3 times a week from day 10 to day 24 following allo-HSCT). (B) Comparison of overall survival, with combined data from two independent experiments (n = 17–18). *, P < 0.05; **, P < 0.01 by Mantel-Cox logrank test. (C) Comparison with control mice with T-cell depleted BM only (no GVHD). Overall survival, with combined data from three independent experiments (n = 15–43). ****, P < 0.0001 by Mantel-Cox logrank test. Tx indicates the period of antibiotics treatment in B and C. (D) Mice with GVHD treated with antibiotics were sacrificed on day 21 and GVHD histology scores in target organs were quantified by a blinded pathologist. Data are combined from three independent experiments (n = 5–20). *, P < 0.05 by Mann-Whitney U test.
We next investigated the effects of antibiotic treatment in a clinically relevant MHC-matched minor antigen-mismatched allo-HSCT mouse model (C57BL/6 into 129S1). Lethally irradiated 129S1 recipients were transplanted with C57BL/6 T-cell depleted bone marrow (TCD-BM) cells and 1 × 106 C57BL/6 splenic T cells. Recipients were treated with either aztreonam, cefepime, imipenem-cilastatin, or piperacillin-tazobactam 100 mg/kg SC three times per week starting on day 10 after allo-HSCT. Remarkably, we observed significantly increased mortality in piperacillin-tazobactam (p<0.05, compared to aztreonam) and imipenem-cilastatin-treated (p<0.01, compared to aztreonam) recipients within 2 weeks of starting treatment (Fig. 2B). These results suggested that in the setting of allo-HSCT, treatment with piperacillin-tazobactam or imipenem-cilastatin aggravated GVHD severity.
We performed additional murine experiments comparing the differences between aztreonam and imipenem-cilastatin, given that these two antibiotic treatments produced the most distinct phenotypes in terms of GVHD severity. We found that imipenem-cilastatin treatment of mice with GVHD reproducibly resulted in shortened survival compared to aztreonam treatment (Fig. 2C). Importantly, control recipients who underwent allo-HSCT in the absence of GVHD showed 100% survival with both antibiotic treatments, indicating that imipenem-cilastatin itself did not have adverse effects on bone marrow engraftment or survival after myeloablative irradiation. Histological examination of GVHD target organs on day 21 after allo-HSCT (11 days after starting antibiotic therapy) revealed that increased GVHD pathology was present in mice treated with imipenem-cilastatin. Interestingly, this was localized to the colon (Fig. 2D and fig. S2), whereas other common GVHD target organs including the skin, liver and small intestine showed no significant differences in the degree of inflammation and damage.
Imipenem-cilastatin treatment boosts donor CD4+ T cells, IL-23 production, and granulocyte number in mouse colon
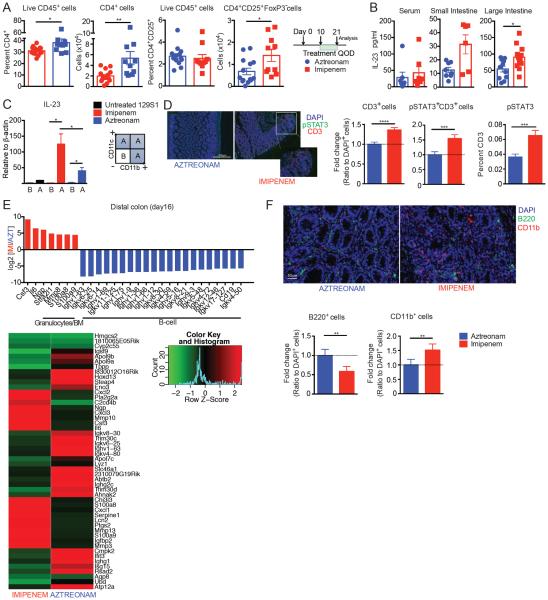
To explore additional mechanistic aspects by which imipenem-cilastatin treatment can aggravate GVHD after allo-HSCT, we characterized immune cell populations in the colons of mice with GVHD on day 21. Flow cytometric analysis of the colon lamina propria leukocytes showed increased donor effector CD4+ T cells (CD25+ FoxP3−) in imipenem-treated recipients (Fig. 3A). We did not observe any significant differences in CD8+ T cells, CD4+FoxP3+ regulatory T cells, or other immune populations that can play a role in the setting of GVHD in the colon (fig. S3 and Table S1). We further performed multiplex cytokine analysis on serum, and homogenates of small intestine and colon and found that interleukin-23 (IL-23) was significantly increased in the colon of recipients treated with imipenem-cilastatin (p<0.05, Fig. 3B and fig. S4). IL-23 signaling has been identified as an important mediator of GVHD specifically in the colon (20, 21), corroborating our finding that imipenem-cilastatin treatment is associated with increased colonic GVHD and elevated IL-23 within the colon. Prior studies have identified two colonic myeloid populations, macrophages (CD11b+CD11c−) and dendritic cells (CD11b+CD11c+) to be important sources of IL-23 in the setting of GVHD (20, 21). To determine the source of increased IL-23 in the setting of imipenem-cilastatin treatment, we utilized a mixture of CD11b and CD11c magnetic beads to enrich colonic lamina propria leukocytes into two fractions, one CD11b- and CD11c-depleted (CD11b−CD11c−) and a second enriched for both CD11b- and CD11c-expressing cells (CD11b+ and/or CD11c+). Quantitative real time PCR showed that IL-23 was highly expressed in the population enriched for CD11b and CD11c in imipenem-cilastatin-treated mice (Fig. 3C), indicating that imipenem-cilastatin treatment augmented IL-23 expression in macrophages and/or dendritic cells in recipients with GVHD. We also looked for evidence of increased IL-23 signaling and found that donor T cells infiltrating the colon of imipenem-cilastatin-treated recipients indeed showed significantly higher levels of phosphorylated signaling adapter molecule signal transducer and activator of transcription 3 (STAT3) (p<0.001, Fig. 3D). Although the IL-23 pathway is only one of many pathways that signal through STAT3, together these results support the possibility that IL-23 signaling in donor T cells is increased in the colon of GVHD mice treated with imipenem-cilastatin.
Fig. 3.
Treatment with imipenem-cilastatin in GVHD mice results in elevated numbers of donor CD4+ T cells, and increased production of IL-23 in the colon. (A to F) Lethally irradiated 129S1 recipients were transplanted with C57BL/6 T-cell depleted bone marrow cells with 1× 106 C57BL/6 T cells. Recipients were treated with imipenem-cilastatin or aztreonam as described in Fig. 2. (A) Colonic lamina propria-infiltrating leukocytes from recipients were analyzed on day 21 by flow cytometry. Data are combined from two independent experiments. Values represent mean ± SEM (n = 10–14). *, P < 0.05; **, P < 0.01 by Mann-Whitney U test. (B) Concentration of IL-23 in serum, whole small intestine homogenate, and whole colon homogenate. Data are combined from two independent experiments. Values represent mean ± SEM (n = 10–12). *, P < 0.05 by Mann-Whitney U test. The individual plots in the graphs indicate individual animals sacrificed at the time of the analysis in A and B. (C) Lamina propria-infiltrating leukocytes from the colon on day 21 were enriched for CD11b and CD11c simultaneously using a mixture of magnetic beads; IL-23 transcripts were quantified by real time PCR. Data are representative of two independent experiments. Values represent mean ± SEM (n = 3). *, P < 0.05 by Mann-Whitney U test. (D) Immunofluorescence staining was used to quantify pSTAT3, CD3, and DAPI-positive cells in colonic tissue collected on day 21 (the white bar indicates 500 μm). (E) RNA sequencing analysis of the distal colon on day 16 after allo-HSCT. The top 50 regulated genes are shown in the heatmap panel. (F) Immunofluorescence staining was used to quantify CD11b, B220, and DAPI-positive cells in colonic tissue collected on day 21 (the white bar indicates 50 μm). Data are representative of two independent experiments. Values represent mean ± SEM (n = 7–8). **, P < 0.01; ***, P < 0.001; ****; P < 0.0001 in D and F by Mann-Whitney U test.
Next, we performed RNA-sequencing of transcripts purified from colonic tissue of recipients treated with imipenem-cilastatin or aztreonam collected on day 16 after allo-HSCT. We observed increased expression of Ngp, Stfa2l1, Mmp8, S100a8, and S100a9 in imipenem-cilastatin-treated recipients (Fig. 3E). These genes are highly expressed by granulocytes, suggesting that there may be greater infiltration of the colon by granulocytes in allo-HSCT recipients with GVHD treated with imipenem-cilastatin vs. aztreonam, which we were able to directly confirm by histology (Fig. 3F). Importantly, neutrophil infiltration in the intestine has recently been found to contribute to GVHD pathophysiology (22). We additionally observed that the colon of recipients with GVHD treated with aztreonam vs. imipenem-cilastatin showed enrichment for transcripts normally expressed by B cells (Fig. 3E), suggesting increased numbers of B cells within the colon, and this was also confirmed by histology (Fig. 3F).
Imipenem-cilastatin treatment of mice with GVHD resulted in an increase in the Akkermansia strain in the colon
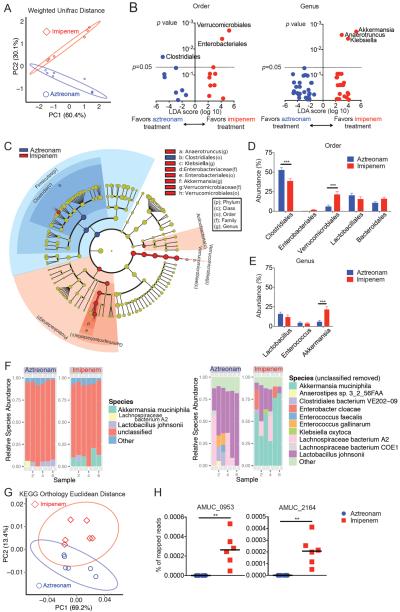
16S rRNA gene sequencing of stool specimens from mice with GVHD followed by principal component analysis indicated that aztreonam and imipenem-cilastatin therapy led to distinct patterns of gut microbiota composition (Fig. 4A). The taxa that best explained the differences between these groups, as assessed by linear discriminate analysis of effect size (LEfSe) (23) are depicted in Fig. 4B and C. Allo-HSCT mouse recipients with GVHD treated with imipenem-cilastatin compared to aztreonam showed a loss of Clostridiales. Interestingly, we also observed a consistent expansion of Akkermansia muciniphila in these animals (Fig. 4, D and E and fig. S5). Clostridiales have notably been identified as major producers of short-chain fatty acids (SCFA) (10, 24), which are bacterial fermentation products that play an important role in maintaining colonic homeostasis and health (25, 26). Surprisingly, despite large differences in the abundance of Clostridiales, we observed no significant changes in the amounts of SCFAs in the colon comparing specimens from recipients treated with aztreonam or imipenem-cilastatin (fig. S6)
Fig. 4.
Mice treated with imipenem-cilastatin in the setting of GVHD show increased abundance of Akkermansia. (A) Stool specimens obtained from mice treated with imipenem-cilastatin or aztreonam were collected on day 21 and analyzed by 16S rRNA gene sequencing (as in Fig. 2), followed by principal coordinate analysis of weighted and normalized UniFrac distances. Proportion of variance accounted for by each principal component is indicated. (B and C) Differential taxonomic abundance between aztreonam treated and imipenem-cilastatin treated recipients was analyzed by (B) linear discriminate analysis coupled with effect size measurements (LEfSe) and (C) by LEfSe projected as a cladogram. Data are representative of more than five independent experiments in A to C. (D and E) Shown are comparisons of bacterial abundance at the phylogenetic levels of (D) order, and (E) genus. Data are combined from 6 independent experiments (n = 32–36). ***, P < 0.001 by multiple comparisons, corrected by Holm-Sidak test. (F to H) Stool specimens collected from mice with GVHD treated with antibiotics were collected on day 21, and evaluated by metagenomic shotgun sequence analysis. (F) Comparison of bacterial species abundance determined by taxonomy. Numbers 1 through 6 along the x-axis represent the individual subjects. (G) Principal component analysis of quantification of sequence reads from KEGG gene orthologs comparing specimens from mice treated with aztreonam and imipenem-cilastatin. (H) Quantification of gene sequences by homology was performed on stool specimens collected on day 21. Amuc_0953, a sulfatase, and Amuc_2164 a glycosyl hydrolase, are two predicted secreted mucolytic genes found in the genome of Akkermansia muciniphila ATCC BAA-835, isolated from human feces. **, P < 0.01 by Mann-Whitney U test.
In order to acquire greater resolution of the bacterial composition between aztreonam-treated and imipenem-cilastatin-treated murine specimens, we performed metagenomic shotgun sequencing with stool collected on day 21 after allo-HSCT. We found that, concordant with the 16S sequencing results, imipenem-cilastatin but not aztreonam treatment resulted in an increased abundance of Akkermansia muciniphilia (Fig. 4F). However, as the largest percentage of reads from the analysis were determined to be unclassified, it is possible that additional significant differences in bacterial species composition exist between the two antibiotic-treatment types. Metagenomic shotgun sequencing analysis also revealed differences in gene content between microbiota specimens from mice treated with aztreonam and imipenem-cilastatin, depicted by principal component analysis of gene orthologs (Fig. 4G). LEfSe analysis of gene pathways indicated that the microbiota genes in mice treated with imipenem-cilastatin were enriched for processes including lipopolysaccharide synthesis, and relatively depleted in several processes including D-alanine metabolism (fig. S7). Interestingly, lipopolysaccharide is well known for inducing a pro-inflammatory cascade in many disease processes including GVHD (27), while reductions in D-alanine content of lipotechoic acid can enhance the anti-inflammatory properties of Lactobacilli (28, 29).
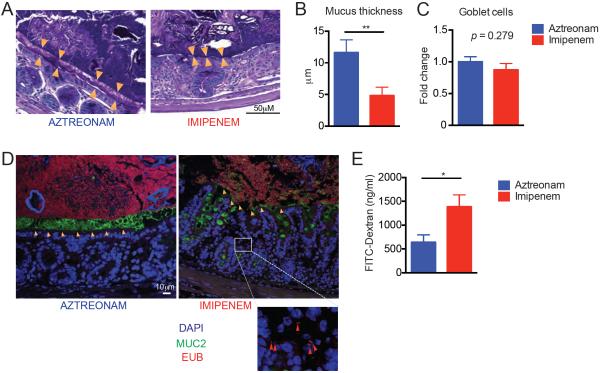
As mentioned above, we detected an increase in Akkermansia muciniphila in the flora of imipenem-cilastatin-treated mice using 16S rRNA deep sequencing (Fig. 4E). This bacterium has the ability to degrade mucus as a carbohydrate source (30, 31). Utilizing our metagenomic shotgun sequencing results, we asked if genes predicted to encode secretory mucolytic enzymes were differentially present in mice treated with each antibiotic. The identification and characterization of bacterial mucolytic enzymes is still a young field, but a recent study examined the whole genomic sequence of Akkermansia muciniphila ATCC BAA-835, isolated from human feces (31). The authors identified two strong candidates for mucus degradation: Amuc_0953, a sulfatase, and Amuc_2164, a glycosyl hydrolase, which both contained predicted secretory signal peptide cleavage sites as well as predicted mucin-binding domains. We quantified the presence of sequences with homology to these two genes and found that both were markedly enriched in specimens from imipenem-cilastatin-treated mice (Fig. 4H). We then sought to characterize the mucus layer of the colon in antibiotic-treated transplanted mice. Using Periodic acid–Schiff staining, we observed a marked reduction in the thickness of the mucus layer in recipients treated with imipenem-cilastatin on day 21 compared to aztreonam-treated recipients (Fig. 5, A and B). No differences in the numbers of mucus-producing goblet cells between aztreonam- and imipenem-cilastatin-treated recipients were seen suggesting that mucus production was not impaired (Fig. 5C). Moreover, by utilizing a general bacterial 16S rRNA probe (EUB338) (32) coupled with Muc2 staining, we directly visualized the inner mucus layer in the colon of antibiotic-treated recipients and confirmed a dramatic thinning of the mucus layer of mice treated with imipenem-cilastatin. Strikingly, we also observed dissemination of bacteria past the colonic epithelial barrier in histopathology specimens from imipenem-cilastatin-treated mice (Fig. 5D), whereas this was not seen in aztreonam-treated mice. We further assessed epithelial barrier function in recipients by performing oral administration of fluorescein isothiocyanate (FITC)-dextran, a poorly absorbed carbohydrate that can enter the bloodstream if the epithelial barrier has been compromised. Consistent with the pathological findings mentioned above, we detected greater FITC in the serum of imipenem-cilastatin treated mice as early as 5 days following initiation of antibiotics (Fig. 5E), indicating greater loss of gut integrity in these recipients. Taken together, these results demonstrate that imipenem-cilastatin treatment can exacerbate GVHD through a combination of factors including compromised barrier function with thinning of the protective mucus layer and reduced numbers of colonic B cells, increased infiltration of granulocytes, elevated IL-23 production, and increased numbers and activation of donor effector CD4+ T cells.
Fig. 5.
Mice treated with imipenem-cilastatin in the setting of GVHD result in the loss of the colonic mucus layer, and impaired intestinal barrier function. (A to C) Colon tissues from mouse recipients were fixed by water-free Methanol-Carnoy's fixative on day 21, stained with Periodic acid-Schiff (PAS) stain and visualized by light microscopy. Orange triangles in A indicate the location of the inner mucus layer. Quantification of mucus layer thickness is shown in B. Number of goblet cells are shown in C. Data are representative of two independent experiments. Values represent mean ± SEM (n = 10). **, P < 0.01 by Mann-Whitney U test. (D) Immunostaining for Muc2 (green) of the colon sections with general bacterial 16S rRNA gene FISH probe EUB338 (red) counterstained with Hoechst (blue). Data are representative of two independent experiments (n = 10). Orange arrowheads indicate inner mucus layer; red arrowheads indicate bacteria penetrating beyond the mucus layer and colonic epithelium. The white scale bar indicates 10 μm. (E) Allo-HSCT recipients of bone marrow and T cells treated with aztreonam or imipenen-cilastatin were challenged with oral gavage of FITC-dextran on day 15 post-transplant. The graph shows plasma FITC-dextran concentrations. Data are representative of two independent experiments. Values represent mean ± SEM (n = 6–8). *, P < 0.05 by Mann-Whitney U test.
DISCUSSION
In the past, the use of broad-spectrum antibiotics in allo-HSCT recipients had been thought to be protective against GVHD. Broad-spectrum combinations were administered with the goal of complete gut decontamination, and this was associated with reduced GVHD in mouse models (33) and in some (34, 35), though not all clinical studies (36–38). Similarly, the addition of metronidazole to ciprofloxacin resulted in reduced GVHD in a small randomized study (39), lending support to the hypothesis that intestinal bacteria contribute to GVHD pathophysiology.
A series of recent studies, however, have described a different association, in which allo-HSCT recipients who sustain more pronounced microbiota injury are more likely to develop severe GVHD (12, 14, 16, 40). Microbiota injury has been observed in several ways, including expansion of commensal Enterococcus species (12), loss of overall diversity (14), reduction in commensals from the genus Blautia, a member of the order Clostridiales (16), and most recently low concentrations of indole, a byproduct of tryptophan metabolism produced by intestinal bacteria that can be quantified in the urine in the form of 3-indoxyl sulfate (40). Consistent with these reports, in the current study we demonstrate that use of antibiotics with a broader spectrum of activity, such as piperacillin-tazobactam or imipenem-cilastatin, leads to increased microbiota injury and increased GVHD severity.
A full explanation has yet to be revealed for the seeming inconsistencies between early studies and more recent studies, but one possible contribution could be the rise of antibiotic-resistant bacteria including resistant enterococci, which can make successful gut decontamination difficult to achieve. Increases in the frequency of colonization with resistant organisms have been observed in allo-HSCT recipients over time (41, 42). A recent study found that gut decontamination was unsuccessful in nearly half of patients where it was attempted. Interestingly, successfully decontaminated patients had a much lower incidence of acute GVHD compared to unsuccessfully decontaminated patients (35).
An important limitation of our clinical analysis is the retrospective, single-center nature of the approach. While our results demonstrate associations between antibiotic treatment and clinical GVHD outcomes, we have not addressed or demonstrated causality. It would thus be interesting to see if the findings can be confirmed retrospectively by other transplant centers, and, more importantly, confirmed in a prospective clinical trial.
In this study, we demonstrate that different antibiotics used to treat neutropenic fever in allo-HSCT recipients have different effects on intestinal microbiota composition, both in patients and in mouse models. We also identified several important changes produced by imipenem-cilastatin treatment in mice with GVHD, including severity of GVHD in the colon, inflammatory changes, and breakdown of the protective colonic mucus barrier. Interestingly, we found that the colons with GVHD pathology from imipenem-cilastatin-treated mice showed increased infiltration of effector CD4+ T cells, whereas there was no increase in CD8+ T lymphocytes, which are classically associated with acute GVHD and thought to be the primary mediators of alloreactive cytotoxicity (43). This could be related to the increase we observed in IL-23 expressed by colonic myeloid cells, since IL-23R is highly expressed by colon-infiltrating CD4+ T cells in settings of intestinal inflammation, including both colitis (44) and GVHD (20). Consistent with this, others have found that CD4+ T cells are also capable of mediating alloreactivity, a phenomenon that to a degree is model-dependent (45, 46).
RNA sequencing, followed by fluorescence microscopy, demonstrated increased colonic B cells in mice treated with aztreonam vs. imipenem-cilastatin in the setting of GVHD. It is likely that this preservation of colonic B cells in aztreonam-treated mice is a reflection of reduced GVHD severity. Loss of B cells is an indicator of GVHD involvement of the bone marrow and has been observed both in mice and patients (47–49). Persistent B cell aplasia can signal ongoing GVHD (49); thus increased colonic B cells in aztreonam-treated mice may reflect improved reconstitution of B cells in the setting of reduced GVHD, or alternatively could reflect improved preservation of normal intestinal immune homeostasis in the colon of mice treated with aztreonam. Notably, B cells, via production of secretory IgA antibodies, are important contributors to maintaining intestinal barrier function (50).
Bacterial composition analysis revealed that treatment of patients and mice with imipenem-cilastatin resulted in reduced colonization by Clostridiales. Recent studies in gnotobiotic mice have shown that members of Clostridiales can mediate anti-inflammatory properties in the intestine (8, 10, 51). Mechanisms include induction of regulatory T cells via biological activity of short-chain fatty acid metabolites such as butyrate that are frequently produced in large amounts by members of Clostridiales (24–26). Surprisingly, in our system, we found no effects of antibiotics on numbers of regulatory T cells or concentrations of short-chain fatty acids. It is possible that in mice afflicted with GVHD, poor oral intake compromised the ability of Clostridiales to produce butyrate. Alternatively, heavy infiltration of donor effector T cells into the colon, or an increase in IL-23 could have disrupted normal physiological induction and recruitment of regulatory T cells.
In mice with GVHD, imipenem-cilastatin treatment led to a relative expansion of Akkermansia muciniphila, a common commensal bacterium found in the intestinal tract of humans, mice and other animals. Notably, this bacterium is unusual in its ability to utilize mucin as a source of carbon and nitrogen both in vitro (30) and in vivo (52). Associated with expansion of Akkermansia muciniphila, we found that mice with GVHD exhibited loss of the colonic mucus layer and compromised intestinal barrier function. In contrast to our findings, beneficial effects of this bacterium in promoting intestinal epithelial integrity have been reported, including in the setting of murine obesity (53) and murine DSS-induced colitis (54). On the other hand, Akkermansia muciniphila has been found to aggravate intestinal inflammation in the setting of murine Salmonella typhimurium infection (55) and a recent study found that eliminating Akkermansia muciniphila with antibiotics could help to reinforce the mucus layer (56). Taken together, it appears the definitive role of Akkermansia muciniphila in intestinal homeostasis has yet to be completely determined and could be setting-dependent. Why Akkermansia expands in the colon of transplanted mice treated with imipenem-cilastatin is unclear; to our knowledge, Akkermansia isolates have not been noted to be resistant to imipenem-cilastatin or other related antibiotics. Competitive interactions between Akkermansia and Clostridiales have also not been described, although studies have seen similar expansions of Akkermansia following treatment of mice with other antibiotics that inhibit Clostridiales, such as clindamycin (57). Whether expansion of Akkermansia could play a role in exacerbating GVHD in humans is also an open question.
In conclusion, our results suggest that selecting antibiotics with a more limited spectrum of activity (especially against anaerobes) could prevent microbiota injury and thus reduce GVHD in the colon and GVHD-associated mortality.
MATERIALS AND METHODS
Study Design
The subjects analyzed retrospectively for impact of antibiotics on clinical outcomes consisted of 857 adult patients undergoing allo-HSCT at Memorial Sloan Kettering Cancer Center (MSKCC) from 1992 to 2015. Patients who received conventional grafts (non-T-cell depleted) were included in this analysis; patients who received ex vivo T-cell depleted grafts or peri-transplant alemtuzumab were excluded. Stool specimens were collected and stored weekly over the course of the transplant hospitalization since 2009. The study was approved by the Institutional Review Board at MSKCC. All study subjects provided written informed consent for biospecimen collection and analysis. GVHD was diagnosed clinically, confirmed pathologically by biopsy whenever feasible, and classified according to historical consensus criteria (18). Cause of death was determined using a standard algorithm (58).
Transplantation Practices
As per our institutional practice, patients received ciprofloxacin prophylaxis, and those undergoing more intense conditioning than nonmyeloablative regimens also received intravenous vancomycin prophylaxis starting day −2 through day 7 (59). Antibiotic prophylaxis against Pneumocystis jiroveci (trimethoprim-sulfamethoxazole, aerosolized pentamadine, or atovaquone) was given at the discretion of the transplant physician.
Analysis of Specimens
For each stool specimen, DNA was purified using a phenol-chloroform extraction technique with mechanical disruption (bead-beating) based on a previously described protocol (60). Specimens were analyzed using the Illumina MiSeq platform to sequence the V4–V5 region of the 16S rRNA gene. Sequence data were compiled and processed using mothur version 1.34 (61), screened and filtered for quality (62). Operational taxonomic units (OTUs) were classified to the species level (63) using a modified form of the Greengenes reference database (64). Principal component analysis was performed upon a weighted and normalized Unifrac (65) distance matrix of OTU abundance in R software. The ellipses indicate a 95% confidence level with an alpha of 0.2, and were drawn using the stat_ellipse() function in the ggplot2 package (H. Wickham. ggplot2: elegant graphics for data analysis. Springer New York, 2009. URL https://cran.r-project.org/web/packages/ggplot2/ggplot2.pdf). Data from this study has been stored in the NCBI Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra).
Characterizing antibiotic effects on intestinal microbiota
We considered 2,209 specimens collected weekly between days −21 and +21 before or after allo HSCT from 633 patients undergoing initial allo HSCT at our center between September 2009 and May 2015. For this analysis, recipients of CD34-selected allografts were not excluded. We first confirmed that sulfamethoxazole/trimethoprim IV or oral, atovaquone oral, vancomycin IV, and ciprofloxacin IV produced only minor perturbations to the intestinal microbiota (data not shown). We then applied a uniform criteria to systematically identify the first antibiotic administered to each patient (besides the ones that had been determined to minimally perturb the flora), and asked if there was: 1) a stool specimen collected with 10 days prior to starting the antibiotic, and 2) a second stool specimen collected 4 to 10 days following starting the antibiotic, while the antibiotic was still being administered, and prior to initiation of another antibiotic. This approach identified 22 instances where specimens were collected flanking initiation of aztreonam (n=6) or cefepime (n=16), as well as 106 instances of specimens collected flanking initiation of piperacillin-tazobactam. Notably, this is a small number of instances identified relative to the total number of patients evaluated, which is a testament to how difficult it can be to identify instances where only a single antibiotic change was initiated between 2 stool specimens when collecting weekly specimens.
Statistical Analysis
The incidence of GVHD-related mortality was estimated using cumulative incidence functions, treating relapse and death unrelated to GVHD as competing events, and compared across types of antibiotics received using Gray's test. A Fine-Gray model was used to further adjust for other clinical factors. Overall survival was estimated using Kaplan-Meier methodology and compared across factors using the log rank test. Comparisons of bacterial abundance were performed using the Mann-Whitney U for unpaired tests. For mouse experiments, data were presented as mean ± SEM or the median of the fold-change. Mann-Whitney U test was used to compare all continuous endpoints in the mouse experiments. In all analyses, statistical significance was defined as P < 0.05 based on a 2-sided test. In the experiments with multiple comparisons, we corrected the results by Holm-Sidak or Dunn's test (α = 0.05; Fig. 1F, Fig. 2A, Fig. 4D and E). Statistical analyses were performed using R version 3.1.0 (The R Foundation for Statistical Computing, Vienna, Austria) and GraphPad Prism version 6.00 for Machintosh, (GraphPad Software, San Diego, California, USA).
Mice and bone marrow transplantation and assessment of GVHD
Female C57BL/6, C57BL/6/Ly5.1, and 129S1/SvImJ mice were obtained from the Jackson Laboratory (Bar Harbor, Maine, USA). Mice used for experiments were 6–9 weeks old. Mouse allo-HSCT experiments were performed as previously described (66), with 1100 cGy split-dosed lethal irradiation of 129S1 recipients transplanted with bone marrow (5 × 106), T-cell depleted (TCD) with anti-Thy-1.2 and low-TOX-M rabbit complement (Cedarlane Laboratories). Donor T-cells were prepared by harvesting donor splenocytes and enriching T-cells by Miltenyi MACS purification of CD5 (routinely >90% purity). Prior to the beginning of each experiment, we separated each cage of mice from the vendor to be divided as equally as possible amongst the various treatment groups. This helped to minimize the chances of incorrectly attributing differences observed in the microbiota to treatment effects, when in reality they are due to cage effects. In our models, we utilized subcutaneous injections for antibiotics treatment because of the following reasons; 1. when repeated injections of medications are administered to mice with the goal of attaining systemic levels, the most common modalities are intraperitoneal and subcutaneous injections, 2. intravenous injections in general are possible for a very limited number of injections, but not for repeated injections. Mice treated with the same antibiotic were co-housed and monitored daily for survival and weekly for GVHD clinical scores (67). Small intestine, large intestine, liver, and skin samples were evaluated histologically for evidence of GVHD and scored as previously described (68).
Cytokines analyses
Blood was collected into microcentrifuge tubes, allowed to clot and centrifuged, and the serum was collected. Small intestine and colon tissues were homogenized and the supernatant was used to determine cytokine levels. ProcartaPlex Multiplex Immunoassay was conducted per manufacturer's instructions (Affymetrix). Results were acquired with a Luminex 200 instrument and analyzed with xPONENT software (Luminex Corporation).
Antibodies and Flow Cytometry
All antibodies were obtained from BD Biosciences–Pharmingen. For cell analysis of surface markers, cells were stained for 20 minutes at 4°C in phosphate-buffered saline (PBS) with 0.5% BSA (PBS/BSA) after Fc block, washed, and resuspended in DAPI in PBS/BSA. Cell surface staining was followed by intracellular staining with the eBioscience kit per the manufacturer's instructions. Dead cells were excluded with LIVE/DEAD Fixable Dead Cell Stain kit (Invitrogen). All flow cytometry was performed on an LSR II (BD Biosciences) and analyzed with FlowJo (TreeStar Software).
Quantitative PCR for IL-23
Reverse transcription-PCR was performed with a QuantiTect reverse transcription kit (QIAGEN). For real-time PCR, primer and probe sets were obtained from Applied Biosystems (IL-23a: Mm00518984_m1). PCR was performed using a StepOnePlus (Applied Biosystems) with TaqMan Universal PCR Master Mix (Applied Biosystems). Relative amount of IL-23a mRNA was calculate by the comparative ΔC(t) method.
FITC-Dextran Assay
Mice were kept without food and water for 4 hours and then FITC-dextran (#46944-500MG, Sigma) was administered by oral gavage at a concentration of 40 mg/ml in PBS in 400 μl (16mg) per mouse (~800mg/kg). Four hours later, plasma was collected from peripheral blood, then mixed 1:1 with PBS and analyzed on a plate reader at an excitation wavelength of 485 nm and an emission wavelength of 535 nm.
RNA Sequencing and Analysis
Mice were sacrificed on day 16 after receiving a total of three times of antibiotics treatment. The distal colon was removed and pooled (n = 4; both for aztreonam and imipenem-cilastatin treatement groups), followed by RNA isolation using TrizolLS. RNA was prepared using RiboMinus (LifeTechnologies). The library was sequenced using the Ion Proton System (LifeTechnologies). Aligned RNA was analyzed for fold change. Differential gene expression was assessed in imipenem-cilastatin vs. aztreonam treated mice.
Metagenomic Shotgun Sequencing and Analysis
Paired-end raw reads from shotgun sequencing were trimmed using Trimmomatic 0.32 (69) using a maximum mismatch of 2, minimum terminal base score of 30, and the Illumina TruSeq adapter sequences. The remaining clipped reads were taxonomically assigned using Kraken (70). Briefly, trimmed and filtered reads were taxonomically classified by k-mer resemblance to bacterial, viral and fungal k-mer profiles generated from the NCBI Genome and Chromosome collections (accessed November 12, 2014). Unclassified reads were further interrogated with BLAST (nt database, March 24, 2015) and non-microbial reads were discarded. Functional analysis was conducted on quality filtered reads using HUMAnN v0.99 (71), which determines the abundance of genes and pathways in a given metagenomic community. To identify those functional categories that were differentially represented between the aztreonam- and imipenem-cilastatin-treated subject specimens, we employed LEfSe (23); a validated tool that identifies differentially abundant biomarkers such as genes, pathways or organisms between microbial communities.
Analysis of the thickness of colon mucus
Recipients were sacrificed on day 21 and 10 mm long segments of colon together with fecal content were carefully collected and soaked into a water-free Methanol-Carnoy's fixative (60% dry methanol, 30% chloroform and 10% acetic acid) (72) overnight. The tissues were then washed in methanol, embedded in paraffin, and then 5 μm sections were placed on glass slides. Slides were deparaffinized, and stained with standard Periodic acid-Schiff method, and assessed by light microscope (73).
Immunostaining and fluorescence in situ hybridization (FISH) of colon tissues
Formalin-fixed colons from recipients were stained with anti-mouse CD3 antibody A0452 (DAKO), pSTAT3 antibody 9135 (Cell Signaling), CD11b antibody ab133357 (Abcam), B220 antibody 550286 (BD Pharmingen), versus isotype control. Immunofluorescence secondary staining was performed with AF488 for pSTAT3 and B220, and AF594 for CD3 and CD11b. Pieces of colon with fecal material were fixed in Carnoy and bacterial FISH (EUB338) (32) and immunostainings were done with MUC2C3 antiserum and DNA by Hoechst 34580 (Life technologies) as previously described (74, 75). The dilutions of our primary antibodies used were as follows. B220 (0.3 μg/ml), CD11b (0.5 μg/ml), pSTAT3 (0.1 μg/ml), CD3 (1.2 μg/ml), EUB338 (10 μg/ml), and MUC2 (2 μg/ml).
Analysis of short-chain fatty acids (SCFAs)
Recipients were sacrificed on day 21 after allo-HSCT and stool specimens from cecum were collected. SCFA levels were determined by 1H-NMR spectroscopy (76, 77) utilizing Unity Inova 600 NMR spectrometer (Varian, Palo Alto, CA, USA).
Supplementary Material
Accessible Summary.
“Antibiotics for allogeneic transplant – a double-edged sword”
Patients undergoing allogeneic hematopoietic stem cell transplantation often receive antibiotics for infections, which can unfortunately also kill intestinal bacteria. These bacteria generally do not cause disease and are thought to suppress inflammation. We examined the records of 857 patients and found that certain antibiotics were linked with developing graft-versus-host disease (GVHD), which can cause intestinal inflammation. Using a mouse model, we found that these antibiotics may select for bacteria that consume intestinal mucus and lead to loss of this important layer of protection for intestinal tissues.
ACKNOWLEDGMENTS
Funding: This research was supported by National Institutes of Health award numbers R01-HL069929 (M.v.d.B.), R01-AI100288 (M.v.d.B.), R01-AI080455 (M.v.d.B.), R01-AI101406 (M.v.d.B.), R00-CA176376 (J.A.D.), and R01-HL124112 (R.J.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Support was also received from the Radiation Effects Research Foundation (RERF-NIAID) (M.v.d.B.), American Society for Blood and Marrow Transplantation (Y.S.), C.J. Martin fellowship from the Australian National Health and Medical Research Council (J.A.D), a Scholar Award from the American Society of Hematology (J.A.D), and the Mechtild Harf Research Grant from the DKMS Foundation for Giving Life (J.A.D). The Experimental Therapeutics Center of Memorial Sloan Kettering Cancer Center funded by Mr. William H. Goodwin and Mrs. Alice Goodwin, The Lymphoma Foundation, Alex's Lemonade Stand, The Geoffrey Beene Cancer Research Center at Memorial Sloan Kettering Cancer Center, and the Susan and Peter Solomon Divisional Genomics Program. We appreciate the invaluable help of the Laboratory of Comparative Pathology and the Molecular Cytology Core Facility (supported by Cancer Support Grant NCI P30-CA008748) of Memorial Sloan Kettering Cancer Center. Technical services provided by the MSKCC Small-Animal Imaging Core Facility, supported in part by NIH Cancer Center Support Grant No 2 P30 CA008748-48, are also gratefully acknowledged.
Footnotes
Author contributions: Y.S. designed and performed experiments, analyzed data and wrote the paper, M.D.D, J.U.P., S.M.P., E.V., J.J.T., A.E.S., O.M.S., L.F.Y., J.G., S.R.L., H.V.J., K.F.A., K.A.P.R., K.X., M.C., H.P., S.C., J.A.D., A.M.H., and B.G. performed experiments, F.R. analyzed RNA sequencing data and performed statistical analyses, G.F.M., C.G., and C.L. performed pathological GVHD analysis, S.M.D. and Y.T. analyzed human data, S.B.F., E.L.M. and A.S.B. analyzed metagenomic shotgun sequencing data, E.P. supervised the study, M.v.d.B. provided experimental design and supervised the study, edited the manuscript and supervised the study, R.R.J. designed experiments, analyzed data, wrote the paper and supervised the study.
Competing interests: MSKCC has filed patent applications related to this work (PCT/US2015/062734 entitled INTESTINAL MICROBIOTA AND GVHD; inventors include M.v.d.B., R.R.J., E.P., and Y.T.).
Data and materials availability: The 16S, RNA sequencing, and metagenomic shotgun sequencing data for this study has been deposited in the European Nucleotide Archive (PRJEB12959).
REFERENCES
- 1.Meyers JD. Infection in bone marrow transplant recipients. The American journal of medicine. 1986;81:27–38. doi: 10.1016/0002-9343(86)90511-5. [DOI] [PubMed] [Google Scholar]
- 2.Wingard JR. Opportunistic infections after blood and marrow transplantation. Transplant infectious disease : an official journal of the Transplantation Society. 1999;1:3–20. doi: 10.1034/j.1399-3062.1999.10102.x. [DOI] [PubMed] [Google Scholar]
- 3.LaRocco MT, Burgert SJ. Infection in the bone marrow transplant recipient and role of the microbiology laboratory in clinical transplantation. Clinical microbiology reviews. 1997;10:277–297. doi: 10.1128/cmr.10.2.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hiemenz JW. Management of infections complicating allogeneic hematopoietic stem cell transplantation. Seminars in hematology. 2009;46:289–312. doi: 10.1053/j.seminhematol.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 5.Freifeld AG, Bow EJ, Sepkowitz KA, Boeckh MJ, Ito JI, Mullen CA, Raad II, Rolston KV, Young JA, Wingard JR. A. Infectious Diseases Society of, Clinical practice guideline for the use of antimicrobial agents in neutropenic patients with cancer: 2010 update by the infectious diseases society of america. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2011;52:e56–93. doi: 10.1093/cid/cir073. [DOI] [PubMed] [Google Scholar]
- 6.Maynard CL, Elson CO, Hatton RD, Weaver CT. Reciprocal interactions of the intestinal microbiota and immune system. Nature. 2012;489:231–241. doi: 10.1038/nature11551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buffie CG, Pamer EG. Microbiota-mediated colonization resistance against intestinal pathogens. Nature reviews. Immunology. 2013;13:790–801. doi: 10.1038/nri3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y, Taniguchi T, Takeda K, Hori S, Ivanov II, Umesaki Y, Itoh K, Honda K. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Narushima S, Sugiura Y, Oshima K, Atarashi K, Hattori M, Suematsu M, Honda K. Characterization of the 17 strains of regulatory T cell-inducing human-derived Clostridia. Gut microbes. 2014;5:333–339. doi: 10.4161/gmic.28572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, Fukuda S, Saito T, Narushima S, Hase K, Kim S, Fritz JV, Wilmes P, Ueha S, Matsushima K, Ohno H, Olle B, Sakaguchi S, Taniguchi T, Morita H, Hattori M, Honda K. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500:232–236. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- 11.Eriguchi Y, Takashima S, Oka H, Shimoji S, Nakamura K, Uryu H, Shimoda S, Iwasaki H, Shimono N, Ayabe T, Akashi K, Teshima T. Graft-versus-host disease disrupts intestinal microbial ecology by inhibiting Paneth cell production of alpha-defensins. Blood. 2012;120:223–231. doi: 10.1182/blood-2011-12-401166. [DOI] [PubMed] [Google Scholar]
- 12.Holler E, Butzhammer P, Schmid K, Hundsrucker C, Koestler J, Peter K, Zhu W, Sporrer D, Hehlgans T, Kreutz M, Holler B, Wolff D, Edinger M, Andreesen R, Levine JE, Ferrara JL, Gessner A, Spang R, Oefner PJ. Metagenomic analysis of the stool microbiome in patients receiving allogeneic stem cell transplantation: loss of diversity is associated with use of systemic antibiotics and more pronounced in gastrointestinal graft-versus-host disease. Biology of blood and marrow transplantation : journal of the American Society for Blood and Marrow Transplantation. 2014;20:640–645. doi: 10.1016/j.bbmt.2014.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jenq RR, Ubeda C, Taur Y, Menezes CC, Khanin R, Dudakov JA, Liu C, West ML, Singer NV, Equinda MJ, Gobourne A, Lipuma L, Young LF, Smith OM, Ghosh A, Hanash AM, Goldberg JD, Aoyama K, Blazar BR, Pamer EG, van den Brink MR. Regulation of intestinal inflammation by microbiota following allogeneic bone marrow transplantation. The Journal of experimental medicine. 2012;209:903–911. doi: 10.1084/jem.20112408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taur Y, Jenq RR, Perales MA, Littmann ER, Morjaria S, Ling L, No D, Gobourne A, Viale A, Dahi PB, Ponce DM, Barker JN, Giralt S, van den Brink M, Pamer EG. The effects of intestinal tract bacterial diversity on mortality following allogeneic hematopoietic stem cell transplantation. Blood. 2014;124:1174–1182. doi: 10.1182/blood-2014-02-554725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shono Y, Docampo MD, Peled JU, Perobelli SM, Jenq RR. Intestinal microbiota-related effects on graft-versus-host disease. International journal of hematology. 2015;101:428–437. doi: 10.1007/s12185-015-1781-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jenq R, Taur Y, Devlin S, Ponce D, Goldberg J, Ahr KF, Littmann ER, Ling L, Gobourne AC, Miller LC, Docampo MD, Peled JU, Arpaia N, Cross J, Peets TK, Lumish MA, Shono Y, Dudakov JA, Poeck H, Hanash AM, Barker JN, Perales MA, Giralt SA, Pamer EG, van den Brink MR. Intestinal Blautia Is Associated with Reduced Death from Graft-versus-Host Disease. Biology of blood and marrow transplantation : journal of the American Society for Blood and Marrow Transplantation. 2015 doi: 10.1016/j.bbmt.2015.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seo SK, Xiao K, Huang YT, Jongwutiwes U, Chung D, Maloy M, Giralt S, Barker JN, Jakubowski AA, Papanicolaou GA. Impact of peri-transplant vancomycin and fluoroquinolone administration on rates of bacteremia in allogeneic hematopoietic stem cell transplant (HSCT) recipients: a 12-year single institution study. The Journal of infection. 2014;69:341–351. doi: 10.1016/j.jinf.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rowlings PA, Przepiorka D, Klein JP, Gale RP, Passweg JR, Henslee-Downey PJ, Cahn JY, Calderwood S, Gratwohl A, Socie G, Abecasis MM, Sobocinski KA, Zhang MJ, Horowitz MM. IBMTR Severity Index for grading acute graft-versus-host disease: retrospective comparison with Glucksberg grade. British journal of haematology. 1997;97:855–864. doi: 10.1046/j.1365-2141.1997.1112925.x. [DOI] [PubMed] [Google Scholar]
- 19.Solomkin JS, Mazuski JE, Bradley JS, Rodvold KA, Goldstein EJ, Baron EJ, O'Neill PJ, Chow AW, Dellinger EP, Eachempati SR, Gorbach S, Hilfiker M, May AK, Nathens AB, Sawyer RG, Bartlett JG. Diagnosis and management of complicated intra-abdominal infection in adults and children: guidelines by the Surgical Infection Society and the Infectious Diseases Society of America. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2010;50:133–164. doi: 10.1086/649554. [DOI] [PubMed] [Google Scholar]
- 20.Das R, Chen X, Komorowski R, Hessner MJ, Drobyski WR. Interleukin-23 secretion by donor antigen-presenting cells is critical for organ-specific pathology in graft-versus-host disease. Blood. 2009;113:2352–2362. doi: 10.1182/blood-2008-08-175448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Das R, Komorowski R, Hessner MJ, Subramanian H, Huettner CS, Cua D, Drobyski WR. Blockade of interleukin-23 signaling results in targeted protection of the colon and allows for separation of graft-versus-host and graft-versus-leukemia responses. Blood. 2010;115:5249–5258. doi: 10.1182/blood-2009-11-255422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwab L, Goroncy L, Palaniyandi S, Gautam S, Triantafyllopoulou A, Mocsai A, Reichardt W, Karlsson FJ, Radhakrishnan SV, Hanke K, Schmitt-Graeff A, Freudenberg M, von Loewenich FD, Wolf P, Leonhardt F, Baxan N, Pfeifer D, Schmah O, Schonle A, Martin SF, Mertelsmann R, Duyster J, Finke J, Prinz M, Henneke P, Hacker H, Hildebrandt GC, Hacker G, Zeiser R. Neutrophil granulocytes recruited upon translocation of intestinal bacteria enhance graft-versus-host disease via tissue damage. Nature medicine. 2014;20:648–654. doi: 10.1038/nm.3517. [DOI] [PubMed] [Google Scholar]
- 23.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. Metagenomic biomarker discovery and explanation. Genome biology. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, Nakanishi Y, Uetake C, Kato K, Kato T, Takahashi M, Fukuda NN, Murakami S, Miyauchi E, Hino S, Atarashi K, Onawa S, Fujimura Y, Lockett T, Clarke JM, Topping DL, Tomita M, Hori S, Ohara O, Morita T, Koseki H, Kikuchi J, Honda K, Hase K, Ohno H. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 25.Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, Glickman JN, Garrett WS. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, Liu H, Cross JR, Pfeffer K, Coffer PJ, Rudensky AY. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cooke KR, Gerbitz A, Crawford JM, Teshima T, Hill GR, Tesolin A, Rossignol DP, Ferrara JL. LPS antagonism reduces graft-versus-host disease and preserves graft-versus-leukemia activity after experimental bone marrow transplantation. The Journal of clinical investigation. 2001;107:1581–1589. doi: 10.1172/JCI12156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duncker SC, Wang L, Hols P, Bienenstock J. The D-alanine content of lipoteichoic acid is crucial for Lactobacillus plantarum-mediated protection from visceral pain perception in a rat colorectal distension model. Neurogastroenterology and motility : the official journal of the European Gastrointestinal Motility Society. 2008;20:843–850. doi: 10.1111/j.1365-2982.2008.01085.x. [DOI] [PubMed] [Google Scholar]
- 29.Grangette C, Nutten S, Palumbo E, Morath S, Hermann C, Dewulf J, Pot B, Hartung T, Hols P, Mercenier A. Enhanced antiinflammatory capacity of a Lactobacillus plantarum mutant synthesizing modified teichoic acids. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:10321–10326. doi: 10.1073/pnas.0504084102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Derrien M, van Passel MW, van de Bovenkamp JH, Schipper RG, de Vos WM, Dekker J. Mucin-bacterial interactions in the human oral cavity and digestive tract. Gut microbes. 2010;1:254–268. doi: 10.4161/gmic.1.4.12778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Passel MW, Kant R, Zoetendal EG, Plugge CM, Derrien M, Malfatti SA, Chain PS, Woyke T, Palva A, de Vos WM, Smidt H. The genome of Akkermansia muciniphila, a dedicated intestinal mucin degrader, and its use in exploring intestinal metagenomes. PloS one. 2011;6:e16876. doi: 10.1371/journal.pone.0016876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, Koren O, Ley R, Wakeland EK, Hooper LV. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science. 2011;334:255–258. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Bekkum DW, Roodenburg J, Heidt PJ, van der Waaij D. Mitigation of secondary disease of allogeneic mouse radiation chimeras by modification of the intestinal microflora. Journal of the National Cancer Institute. 1974;52:401–404. doi: 10.1093/jnci/52.2.401. [DOI] [PubMed] [Google Scholar]
- 34.Storb R, Prentice RL, Buckner CD, Clift RA, Appelbaum F, Deeg J, Doney K, Hansen JA, Mason M, Sanders JE, Singer J, Sullivan KM, Witherspoon RP, Thomas ED. Graft-versus-host disease and survival in patients with aplastic anemia treated by marrow grafts from HLA-identical siblings. Beneficial effect of a protective environment. The New England journal of medicine. 1983;308:302–307. doi: 10.1056/NEJM198302103080602. [DOI] [PubMed] [Google Scholar]
- 35.Vossen JM, Guiot HF, Lankester AC, Vossen AC, Bredius RG, Wolterbeek R, Bakker HD, Heidt PJ. Complete suppression of the gut microbiome prevents acute graft-versus-host disease following allogeneic bone marrow transplantation. PloS one. 2014;9:e105706. doi: 10.1371/journal.pone.0105706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Petersen FB, Buckner CD, Clift RA, Nelson N, Counts GW, Meyers JD, Thomas ED. Infectious complications in patients undergoing marrow transplantation: a prospective randomized study of the additional effect of decontamination and laminar air flow isolation among patients receiving prophylactic systemic antibiotics. Scandinavian journal of infectious diseases. 1987;19:559–567. doi: 10.3109/00365548709032423. [DOI] [PubMed] [Google Scholar]
- 37.Passweg JR, Rowlings PA, Atkinson KA, Barrett AJ, Gale RP, Gratwohl A, Jacobsen N, Klein JP, Ljungman P, Russell JA, Schaefer UW, Sobocinski KA, Vossen JM, Zhang MJ, Horowitz MM. Influence of protective isolation on outcome of allogeneic bone marrow transplantation for leukemia. Bone marrow transplantation. 1998;21:1231–1238. doi: 10.1038/sj.bmt.1701238. [DOI] [PubMed] [Google Scholar]
- 38.Russell JA, Chaudhry A, Booth K, Brown C, Woodman RC, Valentine K, Stewart D, Ruether JD, Ruether BA, Jones AR, Coppes MJ, Bowen T, Anderson R, Bouchard M, Rallison L, Stotts M, Poon MC. Early outcomes after allogeneic stem cell transplantation for leukemia and myelodysplasia without protective isolation: a 10-year experience. Biology of blood and marrow transplantation : journal of the American Society for Blood and Marrow Transplantation. 2000;6:109–114. doi: 10.1016/s1083-8791(00)70073-5. [DOI] [PubMed] [Google Scholar]
- 39.Beelen DW, Elmaagacli A, Muller KD, Hirche H, Schaefer UW. Influence of intestinal bacterial decontamination using metronidazole and ciprofloxacin or ciprofloxacin alone on the development of acute graft-versus-host disease after marrow transplantation in patients with hematologic malignancies: final results and long-term follow-up of an open-label prospective randomized trial. Blood. 1999;93:3267–3275. [PubMed] [Google Scholar]
- 40.Weber D, Oefner PJ, Hiergeist A, Koestler J, Gessner A, Weber M, Hahn J, Wolff D, Stammler F, Spang R, Herr W, Dettmer K, Holler E. Low urinary indoxyl sulfate levels early after ASCT reflect a disrupted microbiome and are associated with poor outcome. Blood. 2015 doi: 10.1182/blood-2015-04-638858. [DOI] [PubMed] [Google Scholar]
- 41.Taur Y, Xavier JB, Lipuma L, Ubeda C, Goldberg J, Gobourne A, Lee YJ, Dubin KA, Socci ND, Viale A, Perales MA, Jenq RR, van den Brink MR, Pamer EG. Intestinal domination and the risk of bacteremia in patients undergoing allogeneic hematopoietic stem cell transplantation. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2012;55:905–914. doi: 10.1093/cid/cis580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ubeda C, Taur Y, Jenq RR, Equinda MJ, Son T, Samstein M, Viale A, Socci ND, van den Brink MR, Kamboj M, Pamer EG. Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. The Journal of clinical investigation. 2010;120:4332–4341. doi: 10.1172/JCI43918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chakraverty R, Eom HS, Sachs J, Buchli J, Cotter P, Hsu R, Zhao G, Sykes M. Host MHC class II+ antigen-presenting cells and CD4 cells are required for CD8-mediated graft-versus-leukemia responses following delayed donor leukocyte infusions. Blood. 2006;108:2106–2113. doi: 10.1182/blood-2006-03-007427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yen D, Cheung J, Scheerens H, Poulet F, McClanahan T, McKenzie B, Kleinschek MA, Owyang A, Mattson J, Blumenschein W, Murphy E, Sathe M, Cua DJ, Kastelein RA, Rennick D. IL-23 is essential for T cell-mediated colitis and promotes inflammation via IL-17 and IL-6. The Journal of clinical investigation. 2006;116:1310–1316. doi: 10.1172/JCI21404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shlomchik WD. Graft-versus-host disease. Nature reviews. Immunology. 2007;7:340–352. doi: 10.1038/nri2000. [DOI] [PubMed] [Google Scholar]
- 46.Blazar BR, Murphy WJ, Abedi M. Advances in graft-versus-host disease biology and therapy. Nature reviews. Immunology. 2012;12:443–458. doi: 10.1038/nri3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mensen A, Johrens K, Anagnostopoulos I, Demski S, Oey M, Stroux A, Hemmati P, Westermann J, Blau O, Wittenbecher F, Movassaghi K, Szyska M, Thomas S, Dorken B, Scheibenbogen C, Arnold R, Na IK. Bone marrow T-cell infiltration during acute GVHD is associated with delayed B-cell recovery and function after HSCT. Blood. 2014;124:963–972. doi: 10.1182/blood-2013-11-539031. [DOI] [PubMed] [Google Scholar]
- 48.Shono Y, Shiratori S, Kosugi-Kanaya M, Ueha S, Sugita J, Shigematsu A, Kondo T, Hashimoto D, Fujimoto K, Endo T, Nishio M, Hashino S, Matsuno Y, Matsushima K, Tanaka J, Imamura M, Teshima T. Bone marrow graft-versus-host disease: evaluation of its clinical impact on disrupted hematopoiesis after allogeneic hematopoietic stem cell transplantation. Biology of blood and marrow transplantation : journal of the American Society for Blood and Marrow Transplantation. 2014;20:495–500. doi: 10.1016/j.bbmt.2013.12.568. [DOI] [PubMed] [Google Scholar]
- 49.Shono Y, Ueha S, Wang Y, Abe J, Kurachi M, Matsuno Y, Sugiyama T, Nagasawa T, Imamura M, Matsushima K. Bone marrow graft-versus-host disease: early destruction of hematopoietic niche after MHC-mismatched hematopoietic stem cell transplantation. Blood. 2010;115:5401–5411. doi: 10.1182/blood-2009-11-253559. [DOI] [PubMed] [Google Scholar]
- 50.Lindner C, Thomsen I, Wahl B, Ugur M, Sethi MK, Friedrichsen M, Smoczek A, Ott S, Baumann U, Suerbaum S, Schreiber S, Bleich A, Gaboriau-Routhiau V, Cerf-Bensussan N, Hazanov H, Mehr R, Boysen P, Rosenstiel P, Pabst O. Diversification of memory B cells drives the continuous adaptation of secretory antibodies to gut microbiota. Nature immunology. 2015 doi: 10.1038/ni.3213. [DOI] [PubMed] [Google Scholar]
- 51.Stefka AT, Feehley T, Tripathi P, Qiu J, McCoy K, Mazmanian SK, Tjota MY, Seo GY, Cao S, Theriault BR, Antonopoulos DA, Zhou L, Chang EB, Fu YX, Nagler CR. Commensal bacteria protect against food allergen sensitization. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:13145–13150. doi: 10.1073/pnas.1412008111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Derrien M, Van Baarlen P, Hooiveld G, Norin E, Muller M, de Vos WM. Modulation of Mucosal Immune Response, Tolerance, and Proliferation in Mice Colonized by the Mucin-Degrader Akkermansia muciniphila. Frontiers in microbiology. 2011;2:166. doi: 10.3389/fmicb.2011.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, Guiot Y, Derrien M, Muccioli GG, Delzenne NM, de Vos WM, Cani PD. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:9066–9071. doi: 10.1073/pnas.1219451110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kang CS, Ban M, Choi EJ, Moon HG, Jeon JS, Kim DK, Park SK, Jeon SG, Roh TY, Myung SJ, Gho YS, Kim JG, Kim YK. Extracellular vesicles derived from gut microbiota, especially Akkermansia muciniphila, protect the progression of dextran sulfate sodium-induced colitis. PloS one. 2013;8:e76520. doi: 10.1371/journal.pone.0076520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ganesh BP, Klopfleisch R, Loh G, Blaut M. Commensal Akkermansia muciniphila exacerbates gut inflammation in Salmonella Typhimurium-infected gnotobiotic mice. PloS one. 2013;8:e74963. doi: 10.1371/journal.pone.0074963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ijssennagger N, Belzer C, Hooiveld GJ, Dekker J, van Mil SW, Muller M, Kleerebezem M, van der Meer R. Gut microbiota facilitates dietary heme-induced epithelial hyperproliferation by opening the mucus barrier in colon. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:10038–10043. doi: 10.1073/pnas.1507645112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Buffie CG, Jarchum I, Equinda M, Lipuma L, Gobourne A, Viale A, Ubeda C, Xavier J, Pamer EG. Profound alterations of intestinal microbiota following a single dose of clindamycin results in sustained susceptibility to Clostridium difficile-induced colitis. Infection and immunity. 2012;80:62–73. doi: 10.1128/IAI.05496-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Copelan E, Casper JT, Carter SL, van Burik JA, Hurd D, Mendizabal AM, Wagner JE, Yanovich S, Kernan NA. A scheme for defining cause of death and its application in the T cell depletion trial. Biology of blood and marrow transplantation : journal of the American Society for Blood and Marrow Transplantation. 2007;13:1469–1476. doi: 10.1016/j.bbmt.2007.08.047. [DOI] [PubMed] [Google Scholar]
- 59.Jaffe D, Jakubowski A, Sepkowitz K, Sebti R, Kiehn TE, Pamer E, Papanicolaou GA. Prevention of peritransplantation viridans streptococcal bacteremia with early vancomycin administration: a single-center observational cohort study. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2004;39:1625–1632. doi: 10.1086/425612. [DOI] [PubMed] [Google Scholar]
- 60.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M, Henrissat B, Heath AC, Knight R, Gordon JI. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Applied and environmental microbiology. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schloss PD, Gevers D, Westcott SL. Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PloS one. 2011;6:e27310. doi: 10.1371/journal.pone.0027310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Applied and environmental microbiology. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Applied and environmental microbiology. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lozupone C, Hamady M, Knight R. UniFrac--an online tool for comparing microbial community diversity in a phylogenetic context. BMC bioinformatics. 2006;7:371. doi: 10.1186/1471-2105-7-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shono Y, Tuckett AZ, Ouk S, Liou HC, Altan-Bonnet G, Tsai JJ, Oyler JE, Smith OM, West ML, Singer NV, Doubrovina E, Pankov D, Undhad CV, Murphy GF, Lezcano C, Liu C, O'Reilly RJ, van den Brink MR, Zakrzewski JL. A small-molecule c-Rel inhibitor reduces alloactivation of T cells without compromising antitumor activity. Cancer discovery. 2014;4:578–591. doi: 10.1158/2159-8290.CD-13-0585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cooke KR, Kobzik L, Martin TR, Brewer J, Delmonte J, Jr., Crawford JM, Ferrara JL. An experimental model of idiopathic pneumonia syndrome after bone marrow transplantation: I. The roles of minor H antigens and endotoxin. Blood. 1996;88:3230–3239. [PubMed] [Google Scholar]
- 68.Zakrzewski JL, Kochman AA, Lu SX, Terwey TH, Kim TD, Hubbard VM, Muriglan SJ, Suh D, Smith OM, Grubin J, Patel N, Chow A, Cabrera-Perez J, Radhakrishnan R, Diab A, Perales MA, Rizzuto G, Menet E, Pamer EG, Heller G, Zuniga-Pflucker JC, Alpdogan O, van den Brink MR. Adoptive transfer of T-cell precursors enhances T-cell reconstitution after allogeneic hematopoietic stem cell transplantation. Nature medicine. 2006;12:1039–1047. doi: 10.1038/nm1463. [DOI] [PubMed] [Google Scholar]
- 69.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wood DE, Salzberg SL. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome biology. 2014;15:R46. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Abubucker S, Segata N, Goll J, Schubert AM, Izard J, Cantarel BL, Rodriguez-Mueller B, Zucker J, Thiagarajan M, Henrissat B, White O, Kelley ST, Methe B, Schloss PD, Gevers D, Mitreva M, Huttenhower C. Metabolic reconstruction for metagenomic data and its application to the human microbiome. PLoS computational biology. 2012;8:e1002358. doi: 10.1371/journal.pcbi.1002358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Johansson ME, Larsson JM, Hansson GC. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(Suppl 1):4659–4665. doi: 10.1073/pnas.1006451107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chromek M, Arvidsson I, Karpman D. The antimicrobial peptide cathelicidin protects mice from Escherichia coli O157:H7-mediated disease. PloS one. 2012;7:e46476. doi: 10.1371/journal.pone.0046476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Salzman NH, de Jong H, Paterson Y, Harmsen HJ, Welling GW, Bos NA. Analysis of 16S libraries of mouse gastrointestinal microflora reveals a large new group of mouse intestinal bacteria. Microbiology. 2002;148:3651–3660. doi: 10.1099/00221287-148-11-3651. [DOI] [PubMed] [Google Scholar]
- 75.Swidsinski A, Weber J, Loening-Baucke V, Hale LP, Lochs H. Spatial organization and composition of the mucosal flora in patients with inflammatory bowel disease. Journal of clinical microbiology. 2005;43:3380–3389. doi: 10.1128/JCM.43.7.3380-3389.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jacobs DM, Deltimple N, van Velzen E, van Dorsten FA, Bingham M, Vaughan EE, van Duynhoven J. (1)H NMR metabolite profiling of feces as a tool to assess the impact of nutrition on the human microbiome. NMR in biomedicine. 2008;21:615–626. doi: 10.1002/nbm.1233. [DOI] [PubMed] [Google Scholar]
- 77.Hong YS, Ahn YT, Park JC, Lee JH, Lee H, Huh CS, Kim DH, Ryu do H, Hwang GS. 1H NMR-based metabonomic assessment of probiotic effects in a colitis mouse model. Archives of pharmacal research. 2010;33:1091–1101. doi: 10.1007/s12272-010-0716-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.