DNP-enhanced MAS NMR reveals extended conformations for the DsbA signal peptide within the ribosome exit tunnel.
Keywords: Protein structure, ribosome, dynamic nuclear polarisation, MAS NMR, signal peptide, co-translational folding
Abstract
Proteins are synthesized in cells by ribosomes and, in parallel, prepared for folding or targeting. While ribosomal protein synthesis is progressing, the nascent chain exposes amino-terminal signal sequences or transmembrane domains that mediate interactions with specific interaction partners, such as the signal recognition particle (SRP), the SecA–adenosine triphosphatase, or the trigger factor. These binding events can set the course for folding in the cytoplasm and translocation across or insertion into membranes. A distinction of the respective pathways depends largely on the hydrophobicity of the recognition sequence. Hydrophobic transmembrane domains stabilize SRP binding, whereas less hydrophobic signal sequences, typical for periplasmic and outer membrane proteins, stimulate SecA binding and disfavor SRP interactions. In this context, the formation of helical structures of signal peptides within the ribosome was considered to be an important factor. We applied dynamic nuclear polarization magic-angle spinning nuclear magnetic resonance to investigate the conformational states of the disulfide oxidoreductase A (DsbA) signal peptide stalled within the exit tunnel of the ribosome. Our results suggest that the nascent chain comprising the DsbA signal sequence adopts an extended structure in the ribosome with only minor populations of helical structure.
INTRODUCTION
During biological protein synthesis, peptide bond formation takes place inside the ribosome at the peptidyl transferase center (PTC) within its large subunit. After passing the exit tunnel, newly synthesized proteins emerge at the surface (1) where the interaction partners bind in an orchestrated fashion to the N-terminal signal sequences and/or the protein itself (2–7). N-terminal signal sequences coding for protein secretion through signal recognition particle (SRP)–independent pathways stimulate direct or indirect interactions between the translating ribosome and the motor protein SecA, which is a part of the Sec translocon (2). Recognition by the SRP is postulated to initiate before the nascent chain emerges at the surface of the ribosome (6). In this context, the earliest protein folding events such as helix formation are assumed to take place within the ribosome (5, 8–12) or directly after exiting to the surface (13). This secondary structure may be sensed by the surface-exposed ribosomal protein L23 that forms the major binding site for the trigger factor, SRP, and SecA and extends into the tunnel, potentially interacting there with the nascent protein and communicating its appearance to the surface for triggering subsequent translocation events (6).
The tunnel is 80 to 100 Å long, 10 to 20 Å wide, and may fit about 30 amino acids in an extended conformation or up to 70 amino acids when an α-helical structure is formed (14, 15). It contains a narrow passage of ~30 Å from the PTC, where the ribosomal proteins L4 and L22 are part of the constriction point (16). The protein secretion monitor (SecM), a regulator of the cellular secretion state, interacts with residues at the constriction point via its 17–amino acid–long consensus sequence, leading to a stable arrest of translation (17). The tunnel wall consists predominantly of ribosomal RNA (rRNA) beyond the constriction point, with properties that are thought to promote the nascent chain to traverse undisturbed in a stretched conformation (18, 19). In studies with model peptides, a helical structure in the exit tunnel was detected by single-particle cryo-electron microscopy data at a resolution of 7.1 Å (11).
Here, we probe the structure of the signal sequence MKKIWLALAGLVLAFSASAA- of disulfide oxidoreductase A (DsbA) (20) within the tunnel of the ribosome by dynamic nuclear polarization (DNP) magic-angle spinning (MAS) nuclear magnetic resonance (NMR) (21–27). The sequence was C-terminally fused to the stalling sequence 150-FSTPVWISQAQGIRAGP-166 of SecM (17, 28) (Fig. 1A). The DsbA signal sequence has high propensity for helical structures [PSIPRED; residues 1 to 14; see Fig. 1A (29)], as observed for signal peptides in general (30).
Fig. 1. Expression constructs and protocol.
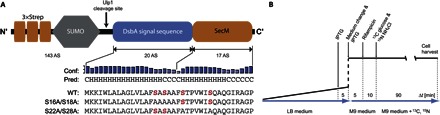
(A) Schematic representation of the nascent chain constructs contained in the investigated stably arrested ribosome-nascent chain complexes (RNCs). After cleavage of the triple Strep-tag by Ubl-specific protease 1 (Ulp1), the remaining nascent chain consists of the signal sequence of DsbA and the SecM stalling sequence; the nascent chain sequences measured in this study are drawn together with their predicted secondary structure (C, coil; H, helical) and the confidence value of the prediction, as provided by PSIPRED (29). Serine residues are depicted in red. WT, wild type. (B) Expression scheme for the production of stably arrested RNCs. IPTG, isopropyl-β-d-thiogalactopyranoside.
RESULTS
Samples of unlabeled ribosomes carrying the uniformly 13C,15N–labeled DsbA-SecM peptide were generated by using a construct N-terminally fused with three Strep-tags and a SUMO domain via a SUMO protease cleavage site. To suppress spurious labeling of ribosome residues, a multistep expression protocol, including use of rifampicin after induction, change of medium, affinity purification, and cleavage of the N-terminal affinity tag, was applied (Fig. 1B) (31). Sample purity was tested by SDS–polyacrylamide gel electrophoresis (PAGE) (fig. S1), and the efficiency of labeling of the nascent chain and suppression of spurious labeling was tested by mass spectrometry (fig. S2). To facilitate the distinction of serine signal sets, two double mutants, S16A/S18A and S22A/S28A-DsbA-SecM, were created. The N-terminal DsbA and C-terminal SecM portions of the nascent chain each contain two serine residues. The two serines in the DsbA part of the first case and the serines from the SecM portion in the second case were replaced by alanines (Fig. 1A). These preparations were measured in frozen state at 105 K and embedded into a matrix consisting of 60% deuterated glycerol, 30% D2O, and 10% H2O as solvent and in the presence of 30 mM TOTAPOL (32).
The nascent chain studied here is 37 residues long (~3.8 kD), which poses a selectivity problem, given the much larger protein content of the ribosome (more than 5000 amino acids) if labels are uniformly distributed. Even the background signals from natural abundance 13C spins in the ribosome may override the nascent chain signals in one-dimensional (1D) spectra. However, 2D cross peaks between carbon signals appear at a probability factor that is 10,000 times higher for the nascent chain as compared to the ribosome, assuming 100% 13C labeling of nascent chain sites and 1% natural abundance labeling in the ribosome. Taking into account the numbers of amino acids in the nascent chain and the ribosome, 2D cross peaks involving nascent chain signals will be favored by a factor of 65 compared to the bulk ribosome signals, assuming no 13C labels are spread into ribosomal proteins. To ensure selective labeling, we tuned the expression protocol as described above (Fig. 1B) to avoid labeling of the ribosome while, at the same time, minimizing the expression of unlabeled nascent chain. We analyzed ribosomal fractions not carrying the nascent chain (“empty ribosomes”) found in the flow-through of the affinity column for comparison (figs. S3 and S4). The 1D and 2D carbon spectra of ribosomes carrying the labeled nascent chain showed characteristic protein signals in the carbonyl and aliphatic regions, whereas empty ribosomes from the same purification showed only weak, nearly vanishing signals (fig. S4).
Nascent chain signals were detected by 2D POST-C7 double-quantum to single-quantum (DQ-SQ) spectroscopy (33) that offers efficient suppression of the natural abundance background of the ribosome (Fig. 2). In the spectral range from 40 to 130 parts per million (ppm) in F1 and 10 to 70 ppm in F2, the signals of the nascent chain occur (Fig. 2). The respective backbone and side-chain carbonyl signals appear between the F1 chemical shifts of 200 and 250 ppm (not shown). The spectra also show a large cross peak conglomerate around 130 to 180 ppm in F1 (DQ chemical shifts) and 60 to 110 ppm in F2 (SQ chemical shifts) stemming from the RNA portion of the ribosome. Additional signals near 25 and 70 ppm in the SQ dimension and 95 ppm in the DQ dimension that do not arise from proteins are observed (Fig. 2, left panel, and fig. S5). Similar signals are found in the spectra from mistic (fig. S6), which was measured in the form of a crude membrane preparation. In these preparations, impurities like membrane lipids or components of the bacterial cell wall such as peptidoglycans are difficult to minimize below the detection limits of DNP, in particular because TOTAPOL has an affinity to bind to peptidoglycans (34). Therefore, we conclude that these signals in Fig. 2 arise from components of the cell wall and/or membrane that were especially enhanced. However, residual amounts of peptidoglycans should not interfere with our analysis because bridging tetrapeptides in Escherichia coli are very regular and composed of the amino acids l-alanine, d-alanine, d-glutamine, and meso-diaminopimelic acid. In addition to those signals, peak conglomerates are found around 45 and 70 ppm in the SQ dimension and between 110 and 120 ppm in the DQ dimension of the spectra in Fig. 2 (for annotation, see fig. S5) that arise from copurified DNA. Signals of the C2′-C3′ correlation of adenine and cytosine typically appear in this spectral region.
Fig. 2. POST-C7 DQ-SQ spectra of the ribosome embedded nascent chain S16A/S18A-DsbA-SecM (left panel) and S22A/S28A-DsbA-SecM (right panel).

Averaged chemical shifts for helical (red squares), strand-like (blue squares), or coil-like (green triangles) geometries are shown (35). Well-separated cross peak patterns of amino acids that do not strongly overlap with other residue types are labeled accordingly. Acquisition in the direct and indirect dimension was 20 ms (1582 points) and 2 ms (128 points), respectively. The data were zero-filled to 4096 points with −25 Hz of Lorentz broadening and a Gaussian offset of 0.08 in the direct dimension and were zero-filled to 1024 points with a sine-squared function with a sine-bell shift of 2 applied in the indirect dimension.
The purity of the interpreted signal pattern was cross-checked by analyzing the spectra of the samples with empty ribosomes in detail. As an example, they did not show cross peaks due to serine even at very low plot levels [see the area between 55 and 60 ppm in F2 and that between 115 and 125 ppm in F1 (fig. S7, A and B), and for comparison, see the respective areas for the S16A/S18A sample and the virus capsid (fig. S7, C and D)]. The absence of cross peaks in the spectra of the empty ribosomes is further exemplified by the cross sections taken at the S16/S18 Cα/Cβ DQ chemical shift (fig. S7E). This highlights that the serine signals discussed are not correlated with the signals of impurities.
It is noteworthy that the sample reproducibility was tested at the beginning of this investigation, with a number of ribosome preparations expressing the wild-type peptide and the mutants. Some of the spectra showed spreading of labels into the ribosome, which is apparent from diagnostic aromatic RNA and additional serine and threonine signals, leading to broader and more intense cross peak patterns for those amino acids. We further recorded spectra of ribosomes carrying the chains of other proteins, with expected differences in signal patterns.
The chemical shifts of the nascent chain cross peak patterns observed in the spectra of the two variants were compared to averaged chemical shifts for helical, strand-like, or coil-like geometries according to statistics from the Protein Chemical shift Database (RefDB; http://refdb.wishartlab.com) (35). Furthermore, we specifically focused on cross peak patterns of amino acids that do not strongly overlap with others residue types, such as serines and alanines, as well as isoleucines, valines, and prolines (Fig. 2). The respective cross peak patterns of serines are particularly well separated from those of other amino acids. The signals of most other amino acids appear as superpositions of Cα/Cβ cross peaks in the regions ranging from 80 to 105 ppm in F1 and around 50 to 65 ppm (for the Cα) as well as 20 to 45 ppm (for the Cβ) in F2 (Fig. 2). Comparing the location of the signals to the secondary structure–specific average chemical shifts of each of the 20 amino acid types (see red squares, blue squares, and green triangles in Fig. 2), it appears that most of the residues are involved in a predominant strand- or coil-like secondary structure. Most of the helix-indicating average shift positions (red symbols) superimpose only with the edges of the cross peak patterns (36), in agreement with the observation of a predominantly stretched or random coil conformation. Spectral regions indicative for Ile, Pro, and Val in helical conformation are not populated at all (cf. Fig. 2, A and B; see the respective red squares and boxed areas indicating the distribution); hence, we conclude that these types of amino acids are located in sections that do not form the α-helical secondary structure in the DsbA-SecM constructs. A closer analysis of the segment involving the C-terminal part of the DsbA signal peptide -SASAA- is of interest because of its high alanine content. We therefore analyzed the chemical shifts of the two serine pairs individually by including spectra of the mutants S16A/S18A-DsbA-SecM (Fig. 2, left panel) and S22A/S28A-DsbA-SecM (Fig. 2, right panel). The mutation of serine residues in the SecM part was found to be neutral for stalling efficiency in previous studies (28). Both mutant nascent chain spectra provide evidence for multiple signal sets for serines, monitored via the remaining serine pair. This confirms the presence of at least two conformations for each of the two serine residues left in the mutants, presumably because of the much slower motional processes at the measurement temperature of 105 K. Substituting both Ser22 and Ser28 with alanine, the remaining signals of Ser16 and Ser18 indicate the presence of a mixture of conformers. A minor component, contributing to less than 20% of the total number of serine signals, indicates serine residues adapting helical secondary structure (Fig. 2B, red squares and boxes). However, when substituting Ser16 and Ser18 by alanine, a serine signal pattern without any indication of an α-helical structure is observed. This supports the fact that the remaining serine residues Ser22 and Ser28 in the SecM part of the expressed peptide (Fig. 2, left panel) are in a nonhelical conformation. Inspection of alanine cross peaks around 73 ppm in F1 and 52 ppm (Cα) and 20 ppm (Cβ) in F2 leads to similar conclusions. There, largest cross peak intensities are in spectral areas indicative for alanines in an extended conformation. However, in the spectrum of the mutant S16A/S18A, a little increase of intensity is observed toward chemical shifts typical for alanines in α-helical environments (Fig. 2, left panel). This is in accordance with the observed 20% helical structure monitored by the chemical shifts of Ser16 and Ser18 in this segment. In summary, all observed isoleucine, proline, serine, and valine signals show chemical shifts typical for extended conformations, with small amounts of helicity indicated by additional signal sets of serines and alanines.
For cross-checking these results, the observed signal pattern of the nascent chain was compared to DNP spectra of two other proteins, the protein mistic (37) and the HIV capsid protein (38) (figs. S6 and S8). The spectra of mistic (fig. S6) show intensity in the area where signals of Ile, Pro, and Val in α-helical environments are expected, reflecting its α-helical structure. Likewise, the capsid protein from HIV (fig. S8) shows populations of the three residue types in α-helical conformation; however, they are much weaker because the HIV capsid protein has a smaller proportion of helical secondary structure.
On the basis of the detected secondary structure in which the analyzed residues Ile, Pro, Ser, and Val are involved, two different models were constructed (Fig. 3). One represents the predominant, extended form (Fig. 3, left panel), and the other counts for the helical populations involving the sequence motif -SASAA- (Fig. 3, right panel). In both forms, the SecM part of the sequence is fully extended, with the consequence that the conserved stalling motif Val-Trp-Ile is then situated at the constriction point. Similarly, the chemical shifts of the respective signals deriving from the signal sequence indicate that neither isoleucine nor valine residues take part in helical conformations, and minor populations of helical structure are observed in the region of Ser16 and Ser18. The respective alanine-rich subsequence -SASAA- is located just below the constriction point, where the tunnel is slightly wider; hence, the formation of a short α helix by this fragment would be sterically feasible (Fig. 3, right). Both models differ by the amount of residues that are emerging at the surface. The small helical segment detected in our case leads to a difference of five amino acids that are available for contacts with interaction partners in case of the extended form.
Fig. 3. Representation of the stalled nascent chain interacting with ribosomal proteins along the exit tunnel.

The isotopically labeled portions are the SecM stalling sequence and the DsbA signal sequence, both depicted as stick models. DsbA is shown in a stretched conformation (left panel) and in a partial α-helical conformation (right panel). Residues for which chemical shifts are indicative for a stretched conformation are color-coded light blue, whereas S16 and S18 (adapting an α-helical conformation) are shown in red (right panel). Two of the three proteins that form the constriction point (L4 and L22) and the tRNA (transfer RNA) are shown in blue and are labeled accordingly.
DISCUSSION
These results contrast to earlier investigations where it was assumed that the DsbA signal peptide would already form an α helix in the nascent state (39). The possibility of helix formation inside the tunnel was demonstrated earlier by means of investigations by applying electron microscopy (11) to ribosomes harboring a dipeptidyl aminopeptidase B–derived peptide containing repeats of the five residues Glu-Ala-Ala-Ala-Lys (EAAAK). This peptide has strong propensity to form an α helix in solution (11) because each Glu can stabilize one repeat of a standard helix by forming a salt bridge to the Lys, leading to >80% helicity in aqueous solvent. In our case, a peptide that is composed of amino acids with mostly short side chains, but fewer helix-stabilizing interactions, was investigated. The DsbA signal peptide is used as a tool for triggering the secretion of expressed proteins; hence, the presented findings concerning the mechanistic background of protein secretion are intriguing. A conformational change of L23 triggered by helix formation of the DsbA signal sequence inside the tunnel seems unlikely, with potential consequences on SRP binding.
MATERIALS AND METHODS
Stably arrested RNCs were expressed in vivo in E. coli strain BL21(DE3). To ensure a high percentage of isotopic 13C,15N labeling of the nascent chain with very little background labeling of the ribosome, the large-scale production and purification were performed in vivo following the protocol published by Rutkowska et al. (31). This method has been shown to selectively label the nascent chain with an efficiency of between 75 and 86%, with neither ribosomal proteins nor rRNA labeled. We used the 17–amino acid–long C-terminal stalling sequence (150-FSTPVWISQAQGIRAGP-166) of SecM (17), which was fused to the signal sequence of DsbA, giving the nascent chain (DsbA-SecM) a total length of 37 amino acids. For purification, the DsbA-SecM peptide was coexpressed with three N-terminal StrepII-tags and a SUMO domain (Fig. 1A) (40). A StrepTactin column was used to isolate RNCs from empty ribosomes, as described by Rutkowska et al. (41). The Strep-tag and SUMO domain were removed by incubation with Ubl-specific protease 1 (overnight at 4°C) followed by high-salt sucrose cushion centrifugation [25% sucrose, 1 M KOAc, 12 mM Mg(OAc)2, 50 mM Hepes (pH 7.5), and 1 mM dithiothreitol]. Sample purity before and after each purification step was tested by SDS-PAGE (fig. S1). Mass spectrometry was used to confirm labeling efficiency.
Approximately 10 mg (4 nmol) of RNCs was dissolved in 600 μl of buffer, containing 60% deuterated glycerol, 30% D2O, 10% H2O (v/v/v), 50 mM Hepes (pH 7.5), 100 mM potassium acetate, 12 mM magnesium acetate, and 30 mM TOTAPOL (polarizing agent) (42). The RNCs were then ultracentrifuged into a 3.2-mm zirconium oxide MAS rotor and stored at −80°C until DNP measurements were performed.
Spectra were recorded on a Bruker AVANCE III 400-MHz wide-bore spectrometer equipped with a Bruker DNP probe and a Bruker 264-GHz gyrotron as the microwave source. The 1D spectra were recorded with 1024 scans, and the 2D spectra were recorded with 2048 scans per slice and 128 increments in the indirect dimension, yielding a total measurement time of approximately 4 days per spectrum (with a recycle delay of 1.4 s). Cross polarization contact times were 0.75 ms in all cases, and the total mixing time in the 2D experiments (C7) was 0.9 ms. The MAS frequency was set to 8889 Hz, and the temperature was stabilized to 105 K using a Bruker cryo-NMR heat exchanger. The resulting signal enhancement was between 15 and 20. All spectra were plotted with the lowest contour level at two times the noise level.
Supplementary Material
Acknowledgments
We acknowledge C. Spahn and U. Akbey for helpful discussions. Funding: This work was supported by the Deutsche Forschungsgemeinschaft, SFB 740, and the European project iNEXT. Author contributions: N.R., K.D., G.K., and B.B. prepared and analyzed samples. S.L., W.T.F., M.A.G., A.L., B.-J.v.R., and H.O. recorded and analyzed NMR spectra. G.K., B.B., and H.O. planned the experiments. S.L., G.K., B.-J.v.R., and H.O. wrote the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/2/8/e1600379/DC1
fig. S1. SDS-PAGE of intermediate purification steps.
fig. S2. Mass spectrometry of ribosomes.
fig. S3. 1D 13C spectra of the ribosome embedded nascent chain.
fig. S4. Superposition of the DQ-SQ spectra of ribosomes carrying the investigated nascent chains with those of empty ribosomes.
fig. S5. DQ-SQ spectra of the mutant nascent chains with annotated impurities.
fig. S6. Superposition of the DQ-SQ spectra of ribosomes carrying the investigated nascent chains with those of mistic.
fig. S7. Serine cross peak region from spectra of the flow-through containing empty ribosomes, of the mutant S16A/S18A, and of the HIV capsid.
fig. S8. Superposition of the DQ-SQ spectra of ribosomes carrying the investigated nascent chains with those of the HIV capsid protein.
REFERENCES AND NOTES
- 1.Frank J., Zhu J., Penczek P., Li Y., Srivastava S., Verschoor A., Radermacher M., Grassucci R., Lata R. K., Agrawal R. K., A model of protein synthesis based on cryo-electron microscopy of the E. coli ribosome. Nature 376, 441–444 (1995). [DOI] [PubMed] [Google Scholar]
- 2.Huber D., Rajagopalan N., Preissler S., Rocco M. A., Merz F., Kramer G., Bukau B., SecA interacts with ribosomes in order to facilitate posttranslational translocation in bacteria. Mol. Cell 41, 343–353 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Kramer G., Boehringer D., Ban N., Bukau B., The ribosome as a platform for co-translational processing, folding and targeting of newly synthesized proteins. Nat. Struct. Mol. Biol. 16, 589–597 (2009). [DOI] [PubMed] [Google Scholar]
- 4.Kramer G., Rauch T., Rist W., Vorderwülbecke S., Patzelt H., Schulze-Specking A., Ban N., Deuerling E., Bukau B., L23 protein functions as a chaperone docking site on the ribosome. Nature 419, 171–174 (2002). [DOI] [PubMed] [Google Scholar]
- 5.Halic M., Blau M., Becker T., Mielke T., Pool M. R., Wild K., Sinning I., Beckmann R., Following the signal sequence from ribosomal tunnel exit to signal recognition particle. Nature 444, 507–511 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Bornemann T., Jöckel J., Rodnina M. V., Wintermeyer W., Signal sequence–independent membrane targeting of ribosomes containing short nascent peptides within the exit tunnel. Nat. Struct. Mol. Biol. 15, 494–499 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Holtkamp W., Lee S., Bornemann T., Senyushkina T., Rodnina M. V., Wintermeyer W., Dynamic switch of the signal recognition particle from scanning to targeting. Nat. Struct. Mol. Biol. 19, 1332–1337 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Lu J., Deutsch C., Folding zones inside the ribosomal exit tunnel. Nat. Struct. Mol. Biol. 12, 1123–1129 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Lee H. C., Bernstein H. D., The targeting pathway of Escherichia coli presecretory and integral membrane proteins is specified by the hydrophobicity of the targeting signal. Proc. Natl. Acad. Sci. U.S.A. 98, 3471–3476 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhushan S., Hoffmann T., Seidelt B., Frauenfeld J., Mielke T., Berninghausen O., Wilson D. N., Beckmann R., SecM-stalled ribosomes adopt an altered geometry at the peptidyl transferase center. PLOS Biol. 9, e1000581 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bhushan S., Gartmann M., Halic M., Armache J.-P., Jarasch A., Mielke T., Berninghausen O., Wilson D. N., Beckmann R., α-Helical nascent polypeptide chains visualized within distinct regions of the ribosomal exit tunnel. Nat. Struct. Mol. Biol. 17, 313–317 (2010). [DOI] [PubMed] [Google Scholar]
- 12.Frauenfeld J., Gumbart J., van der Sluis E. O., Funes S., Gartmann M., Beatrix B., Mielke T., Berninghausen O., Becker T., Schulten K., Beckmann R., Cryo-EM structure of the ribosome–SecYE complex in the membrane environment. Nat. Struct. Mol. Biol. 18, 614–621 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsu S.-T. D., Fucini P., Cabrita L. D., Launay H., Dobson C. M., Christodoulou J., Structure and dynamics of a ribosome-bound nascent chain by NMR spectroscopy. Proc. Natl. Acad. Sci. U.S.A. 104, 16516–16521 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Picking W. D., Picking W. L., Odom O. W., Hardesty B., Fluorescence characterization of the environment encountered by nascent polyalanine and polyserine as they exit Escherichia coli ribosomes during translation. Biochemistry 31, 2368–2375 (1992). [DOI] [PubMed] [Google Scholar]
- 15.Voss N. R., Gerstein M., Steitz T. A., Moore P. B., The geometry of the ribosomal polypeptide exit tunnel. J. Mol. Biol. 360, 893–906 (2006). [DOI] [PubMed] [Google Scholar]
- 16.Ban N., Nissen P., Hansen J., Moore P. B., Steitz T. A., The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289, 905–920 (2000). [DOI] [PubMed] [Google Scholar]
- 17.Nakatogawa H., Ito K., Secretion monitor, SecM, undergoes self-translation arrest in the cytosol. Mol. Cell 7, 185–192 (2001). [DOI] [PubMed] [Google Scholar]
- 18.Lu J., Kobertz W. R., Deutsch C., Mapping the electrostatic potential within the ribosomal exit tunnel. J. Mol. Biol. 371, 1378–1391 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Lu J., Deutsch C., Electrostatics in the ribosomal tunnel modulate chain elongation rates. J. Mol. Biol. 384, 73–86 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schierle C. F., Berkmen M., Huber D., Kumamoto C., Boyd D., Beckwith J., The DsbA signal sequence directs efficient, cotranslational export of passenger proteins to the Escherichia coli periplasm via the signal recognition particle pathway. J. Bacteriol. 185, 5706–5713 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hall D. A., Maus D. C., Gerfen G. J., Inati S. J., Becerra L. R., Dahlquist F. W., Griffin R. G., Polarization-enhanced NMR spectroscopy of biomolecules in frozen solution. Science 276, 930–932 (1997). [DOI] [PubMed] [Google Scholar]
- 22.Hu K.-N., Yu H.-h., Swager T. M., Griffin R. G., Dynamic nuclear polarization with biradicals. J. Am. Chem. Soc. 126, 10844–10845 (2004). [DOI] [PubMed] [Google Scholar]
- 23.Bajaj V. S., Mak-Jurkauskas M. L., Belenky M., Herzfeld J., Griffin R. G., Functional and shunt states of bacteriorhodopsin resolved by 250 GHz dynamic nuclear polarization–enhanced solid-state NMR. Proc. Natl. Acad. Sci. U.S.A. 106, 9244–9249 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Debelouchina G. T., Bayro M. J., van der Wel P. C. A., Caporini M. A., Barnes A. B., Rosay M., Maas W. E., Griffin R. G., Dynamic nuclear polarization-enhanced solid-state NMR spectroscopy of GNNQQNY nanocrystals and amyloid fibrils. Phys. Chem. Chem. Phys. 12, 5911–5919 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maly T., Debelouchina G. T., Bajaj V. S., Hu K.-N., Joo C.-G., Mak-Jurkauskas M. L., Sirigiri J. R., van der Wel P. C. A., Herzfeld J., Temkin R. J., Griffin R. G., Dynamic nuclear polarization at high magnetic fields. J. Chem. Phys. 128, 052211 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gelis I., Vitzthum V., Dhimole N., Caporini M. A., Schedlbauer A., Carnevale D., Connell S. R., Fucini P., Bodenhausen G., Solid-state NMR enhanced by dynamic nuclear polarization as a novel tool for ribosome structural biology. J. Biomol. NMR 56, 85–93 (2013). [DOI] [PubMed] [Google Scholar]
- 27.Reggie L., Lopez J. J., Collinson I., Glaubitz C., Lorch M., Dynamic nuclear polarization-enhanced solid-state NMR of a 13C-labeled signal peptide bound to lipid-reconstituted Sec translocon. J. Am. Chem. Soc. 133, 19084–19086 (2011). [DOI] [PubMed] [Google Scholar]
- 28.Nakatogawa H., Ito K., The ribosomal exit tunnel functions as a discriminating gate. Cell 108, 629–636 (2002). [DOI] [PubMed] [Google Scholar]
- 29.Buchan D. W. A., Minneci F., Nugent T. C. O., Bryson K., Jones D. T., Scalable web services for the PSIPRED Protein Analysis Workbench. Nucleic Acids Res. 41, W349–W357 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.von Heijne G., Gavel Y., Topogenic signals in integral membrane proteins. Eur. J. Biochem. 174, 671–678 (1988). [DOI] [PubMed] [Google Scholar]
- 31.Rutkowska A., Beerbaum M., Rajagopalan N., Fiaux J., Schmieder P., Kramer G., Oschkinat H., Bukau B., Large-scale purification of ribosome-nascent chain complexes for biochemical and structural studies. FEBS Lett. 583, 2407–2413 (2009). [DOI] [PubMed] [Google Scholar]
- 32.van der Wel P. C. A., Hu K.-N., Lewandowski J., Griffin R. G., Dynamic nuclear polarization of amyloidogenic peptide nanocrystals: GNNQQNY, a core segment of the yeast prion protein Sup35p. J. Am. Chem. Soc. 128, 10840–10846 (2006). [DOI] [PubMed] [Google Scholar]
- 33.Hohwy M., Jakobsen H. J., Edén M., Levitt M. H., Nielsen N. C., Broadband dipolar recoupling in the nuclear magnetic resonance of rotating solids: A compensated C7 pulse sequence. J. Chem. Phys. 108, 2686 (1998). [Google Scholar]
- 34.Takahashi H., Ayala I., Bardet M., De Paëpe G., Simorre J.-P., Hediger S., Solid-state NMR on bacterial cells: Selective cell wall signal enhancement and resolution improvement using dynamic nuclear polarization. J. Am. Chem. Soc. 135, 5105–5110 (2013). [DOI] [PubMed] [Google Scholar]
- 35.Zhang H., Neal S., Wishart D. S., RefDB: A database of uniformly referenced protein chemical shifts. J. Biomol. NMR 25, 173–195 (2003). [DOI] [PubMed] [Google Scholar]
- 36.Wishart D. S., Sykes B. D., Richards F. M., Relationship between nuclear magnetic resonance chemical shift and protein secondary structure. J. Mol. Biol. 222, 311–333 (1991). [DOI] [PubMed] [Google Scholar]
- 37.Jacso T., Franks W. T., Rose H., Fink U., Broecker J., Keller S., Oschkinat H., Reif B., Characterization of membrane proteins in isolated native cellular membranes by dynamic nuclear polarization solid-state NMR spectroscopy without purification and reconstitution. Angew. Chem. Int. Ed. Engl. 51, 432–435 (2012). [DOI] [PubMed] [Google Scholar]
- 38.Gupta R., Lu M., Hou G., Caporini M. A., Rosay M., Maas W., Struppe J., Suiter C., Ahn J., Byeon I.-J. L., Franks W. T., Orwick-Rydmark M., Bertarello A., Oschkinat H., Lesage A., Pintacuda G., Gronenborn A. M., Polenova T., Dynamic nuclear polarization enhanced MAS NMR spectroscopy for structural analysis of HIV-1 protein assemblies. J. Phys. Chem. B 2016, 329–339 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.von Heijne G., Protein targeting signals. Curr. Opin. Cell Biol. 2, 604–608 (1990). [DOI] [PubMed] [Google Scholar]
- 40.Andréasson C., Fiaux J., Rampelt H., Mayer M. P., Bukau B., Hsp110 is a nucleotide-activated exchange factor for Hsp70. J. Biol. Chem. 283, 8877–8884 (2008). [DOI] [PubMed] [Google Scholar]
- 41.Rutkowska A., Mayer M. P., Hoffmann A., Merz F., Zachmann-Brand B., Schaffitzel C., Ban N., Deuerling E., Bukau B., Dynamics of trigger factor interaction with translating ribosomes. J. Biol. Chem. 283, 4124–4132 (2008). [DOI] [PubMed] [Google Scholar]
- 42.Song C., Hu K.-N., Joo C.-G., Swager T. M., Griffin R. G., TOTAPOL: A biradical polarizing agent for dynamic nuclear polarization experiments in aqueous media. J. Am. Chem. Soc. 128, 11385–11390 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Perkins D. N., Pappin D. J. C., Creasy D. M., Cottrell J. S., Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 (1999). [DOI] [PubMed] [Google Scholar]
- 44.Tegha-Dunghu J., Neumann B., Reber S., Krause R., Erfle H., Walter T., Held M., Rogers P., Hupfeld K., Ruppert T., Ellenberg J., Gruss O. J., EML3 is a nuclear microtubule-binding protein required for the correct alignment of chromosomes in metaphase. J. Cell Sci. 121, 1718–1726 (2008). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/2/8/e1600379/DC1
fig. S1. SDS-PAGE of intermediate purification steps.
fig. S2. Mass spectrometry of ribosomes.
fig. S3. 1D 13C spectra of the ribosome embedded nascent chain.
fig. S4. Superposition of the DQ-SQ spectra of ribosomes carrying the investigated nascent chains with those of empty ribosomes.
fig. S5. DQ-SQ spectra of the mutant nascent chains with annotated impurities.
fig. S6. Superposition of the DQ-SQ spectra of ribosomes carrying the investigated nascent chains with those of mistic.
fig. S7. Serine cross peak region from spectra of the flow-through containing empty ribosomes, of the mutant S16A/S18A, and of the HIV capsid.
fig. S8. Superposition of the DQ-SQ spectra of ribosomes carrying the investigated nascent chains with those of the HIV capsid protein.


