Abstract
Chronic HIV infection is associated with accumulation of germinal center T follicular helper cells (GC-Tfh) in the lymphoid tissue. The GC-Tfh cells can be heterogeneous based on the expression of chemokine receptors associated with T helper lineages such as CXCR3 (Th1), CCR4 (Th2), and CCR6 (Th17). However, the heterogeneous nature of GC-Tfh cells in the lymphoid tissue and its association with viral persistence and antibody production during chronic SIV/HIV infection is not known. To address this, here we characterized the expression of CXCR3, CCR4, and CCR6 on GC-Tfh in lymph nodes following SIVmac251 infection in rhesus macaques (RMs). In SIV naïve RM, only a small fraction of GC-Tfh expressed CXCR3, CCR4 and CCR6. However, during chronic SIV infection, the majority of GC-Tfh cells expressed CXCR3, while the proportion of CCR4+ cells did not change and CCR6+ cells decreased. CXCR3+ but not CXCR3− GC-Tfh produced IFN-γ (Th1 cytokine) and IL-21 (Tfh cytokine) while both subsets expressed CD40L following stimulation. Immunohistochemistry analysis demonstrated an accumulation of CD4+ IFN-γ+ T cells within the hyperplastic follicles during chronic SIV infection. CXCR3+ GC-Tfh also expressed higher levels of ICOS, CCR5 and α4β7, contained more copies of SIV DNA compared to CXCR3− GC-Tfh cells. However, both CXCR3+ and CXCR3− GC-Tfh delivered help to B cells in vitro for production of IgG. These data demonstrate that chronic SIV infection promotes expansion of Th1-biased GC-Tfh cells, which are phenotypically and functionally distinct from conventional GC-Tfh cells and contribute to hypergammaglobulinemia and viral reservoirs.
Introduction
Lymphoid organs are the primary compartments for the generation of an effective adaptive immune response. CD4 T cells play a central role in the generation of adaptive immunity by providing help to both CD8 T cells and B cells (1). CD4 T cells comprise multiple subpopulations including Th1, Th2, Th17, Tfh, Th9, Th22, Th-CTL and T-regulatory cells (1–4) based on the function they exert and cytokines they produce. Each of these T helper subsets is tightly regulated by specific transcription factors and cytokines (2). Among the various subsets of CD4+ T cells, follicular CD4 T cells (Tfh) play a major role in providing B cell help for the generation of long-lived memory B cell response. Tfh cells reside within the germinal centers (GCs) and are essential for the formation of GCs where memory B cells proliferate and undergo affinity maturation and Immunoglobulin (Ig) class switching (5–8). Interaction between Tfh and B cells is mediated by many cellular and soluble factors such as IL-21, IL-10, IL-4, CD40L and ICOS (1, 9). Phenotypically, Tfh cells are characterized by the expression of chemokine receptor CXCR5, transcription factor Bcl-6, ICOS and a high level expression of programmed death-1 (PD-1) (9).
Multiple studies including our own have characterized the Tfh cells in the lymph nodes (LNs) during chronic HIV infection in humans (10–13) and SIV infection in rhesus macaques (RMs) (14–18). These studies used different combinations of markers to define Tfh. Despite these differences, general conclusions could be derived. These studies demonstrated a marked increase in Tfh cells despite a decline in total CD4 T cells, and this increase in Tfh frequency has been shown to be associated with increased antibody production (11, 12, 14–16). These Tfh cells express the Tfh transcription factor Bcl-6 and Tfh cell markers CXCR5, PD-1, ICOS and CD40L, and secrete soluble factors such as IL-4, IL-10 and IL-21 (9). IL-21 production in particular, has been reported to be markedly elevated in LN Tfh cells from HIV-infected patients (10, 11, 16, 19) and SIV-infected macaques (16, 20, 21). In addition, a significant fraction of GC-Tfh cells are positive for viral RNA demonstrating that they support active virus replication during chronic HIV and SIV infections (11, 14, 16, 19). The expansion of GC-Tfh cells in HIV infection correlated with an increase in GC B cells and plasma cells and a decrease in memory B cells leading to the hypergammaglobulinemia that is characteristic in these patients (10, 12, 22).
The origin of Tfh cells is still under active investigation. Tfh cells are considered to be of a separate lineage. However, studies have shown that Tfh cells can be generated from Th1 (23), Th2 (24) or other CD4 T cell lineages suggesting a significant flexibility in vivo (25, 26). Recent studies have shown that CXCR5+ CD4 T cells in the blood possess a resting memory phenotype as they do not express ICOS or CD69 (27). These circulating CXCR5+ CD4 T cells express chemokine receptors associated with Th1 (CXCR3) (28, 29), Th2 (CCR4) (30) and Th17 (CCR6) lineage (31). Some of these studies explored the ability of these Th1, Th2 and Th17 like memory Tfh cells to deliver help to B cells in vitro and showed that CXCR3− Tfh but not CXCR3+ Tfh provide B cell help (31, 32). However, studies have also shown the emergence of CXCR3+ ICOS+ CXCR5+ cells (effector CD4 T cells) at day 7 after influenza vaccination (28). Interestingly, ICOS expression was seen on a large proportion of CXCR3+ Tfh cells. ICOS+ but not ICOS− CXCR3+ CXCR5+ provided help to memory B cells in vitro and predicted increase in anti-HA antibody titers at day 28 after immunization (28). In HIV infected individuals, the frequency of CXCR3− PD-1+ CXCR5+ cells in blood was associated with development of neutralizing antibodies (32). These data indicate that the phenotypic characteristics and B cell helper potential of CXCR5+ CD4 T cells in the blood may be specific to the vaccine/infectious agent and the phase (effector vs memory) of analysis. In the context of HIV infection, the data suggest that circulating CXCR3− PD-1+CXCR5+ cells may be promoting the production of highly functional broadly neutralizing antibodies (32).
However, little is known regarding the expression of chemokine receptors associated with non-Tfh cells such as CXCR3, CCR4 and CCR6 on LN resident GC-Tfh cells during chronic SIV/HIV infection. Thus in the present study we investigated the expression of chemokine receptors CXCR3, CCR4 and CCR6 by GC-Tfh cells in the LNs of SIV naïve and chronically SIV-infected RMs and determined the relationship between specific Tfh phenotype and B cell helper function. Our results demonstrated the existence of CXCR3+ GC-Tfh cells and a significant expansion of these cells during chronic SIV infection. These CXCR3+ GC-Tfh cells are functionally distinct from CXCR3− GC-Tfh cells in terms of phenotype and cytokine production. Specifically, CXCR3+ GC-Tfh but not CXCR3− GC-Tfh produced Tfh cytokine IL-21 and Th1 cytokine IFN-γ. In addition, unlike CXCR3− GC-Tfh cells, CXCR3+ GC-Tfh expressed higher levels of CCR5 and α4β7 and contained significantly higher copies of SIV DNA. These data demonstrated an unexpected expansion of CXCR3+ biased GC-Tfh cells during chronic SIV infection, which are phenotypically and functionally distinct from conventional GC-Tfh cells and contribute to hypergammaglobulinemia and viral reservoirs.
Materials & Methods
Ethics statement
All animal experimentations were conducted at the Yerkes National Primate Research Center, which is accredited by American Association of Accreditation of Laboratory Animal Care, International, following guidelines established by the Animal Welfare Act and the NIH for housing and care of laboratory animals. Blood and tissue collections were obtained under anesthesia. RMs were fed standard monkey chow (Jumbo Monkey Diet 5037, Purina Mills, St Louis, MO) supplemented with fresh fruit or vegetable daily. Consumption is monitored and adjustments are made as necessary depending on sex, age, and weight. SIV-infected RMs were singly caged but had visual, auditory, and olfactory contact with at least one social partner, permitting the expression of non-contact social behavior. The YNPRC enrichment plan employs several general categories of enrichment. Appropriate procedures were performed to ensure that potential distress, pain, discomfort and/or injury was limited to that unavoidable in the conduct of the research plan. Ketamine (10 mg/kg) and/or Telazol (4 mg/kg) were used for collection of blood and tissues and analgesics were used when determined appropriate by veterinary medical staff.
Animals
Indian adult rhesus macaques obtained from the Yerkes breeding colony were cared for under the guidelines established by the Animal Welfare Act and the National Institute of Health (NIH) Guide for the Care and Use of Laboratory Animals using protocols approved by the Emory University Institutional Animal Care and Use Committee. SIV infected animals were infected with SIVmac251 intrarectally as described previously (33). Some animals were positive for Mamu A*01 allele and all animals were negative for Mamu B08 and Mamu B17. See supplemental Table 1 for additional details.
Antibodies
Cells were stained with fluorochrome-conjugated Abs specific for CXCR5 (clone MU5UBEE; eBioscience), CD3 (clone SP-34-2; BD Biosciences), CXCR3 (clone IC6; BD Biosciences), CCR6 (clone 11A9), CCR4 (clone 1G1), ICOS (clone C398.4A; ebioscience), CD95 (clone; BD Biosciences), PD-1(Clone EH12.1; Biolegend), CD8 (Clone SK1; BD Bioscience), CD4 (clone; OKT4; biolegend), CCR5 (Clone 3A9, BD Biosciences), α4β7 (Act1, NHP reagent resource), Live Dead-IR stain (Invitrogen), Ki-67 (Clone B56; BD Biosciences), Bcl-6 (clone K112-91), IL-21 (Clone 3A3-N2; BD Biosciences), CD4 (Clone L200; BD Biosciences), Alexa700 conjugated IFNγ (Clone B27; BD Biosciences). FITC conjugated CD40L (Clone TRAP1; BD Biosciences).
Phenotyping
Approximately 1×106 mononuclear cells isolated from the lymph node were stained with LIVE/DEAD Near-IR Dead Cell stain at room temperature for 15 min in PBS to stain for dead cells. Cells were then washed with FACS wash and stained on the surface using antibodies specific to CD3, CD4, CD95, CXCR5, CXCR3, CD20 and PD-1 for 25 minutes at room temperature. Cells were then fixed with 1x BD FACS Lysing solution for 10min at room temperature, permeabilized with 1x BD Permeablizing solution for 8 min at room temperature, washed with FACS wash. Cells were stained intracellularly using antibody to Ki-67 and Bcl-6 in FACS wash for 30 minutes at room temperature, washed 2x with FACS wash, acquired using LSR-Fortessa with four lasers (405, 488, 532, 633nm) and analyzed using the FlowJo software (Treestar Inc. CA). At least 50,000 events were acquired for each sample as done previously (16). The live memory CD4 T cells (CD3+ CD4+, CD95+) were analyzed for expression of other surface and intracellular markers described above.
Intracellular cytokine staining
Mononuclear cells from fresh or frozen LN samples were suspended in RPMI medium (Gibco, Life Technologies) with 10% FBS (HyClone, Thermo Fisher Scientific), 100 IU/mL of penicillin, and 100 μg/mL of streptomycin (Lonza). Stimulations were conducted in the presence of anti-CD28 antibody and anti-CD49d antibody (1μg/ml; BD Pharmingen). Sorted 50,000 cells were stimulated with 10 ng/ml of PMA and 1ug/mL of Ionomycin in the presence of Brefeldin A (5 μg/mL; Sigma) and GolgiStop (0.5 μL/mL; BD Pharmingen) after 2 hours of stimulation for 4 hours at 37°C in the presence of 5% CO2. At the end of stimulation, cells were washed once with FACS wash (PBS containing 2% FBS and 0.05% of sodium azide) and surface stained with anti-CD3, anti-CXCR5, anti-PD-1, anti-CXCR3, anti-CD95 at room temperature for 20 min. Cells were then fixed with cytofix/cytoperm (BD Pharmingen) for 20min at 4°C and washed with Perm wash (BD Pharmingen). Cells were then incubated for 30 min at 4°C with antibodies specific to IFN-γ, IL-21, CD40L and CD4, washed once with Perm wash, once with FACS wash, and re-suspended in PBS containing 1% formalin. Cells were acquired on LSR-Fortessa with four lasers (405, 488, 532, 633nm) and analyzed using FlowJo software (Treestar Inc. CA). At least 50,000 events were acquired for each sample.
Immunofluorescence and quantitative image analysis (QIA)
Immunofluorescence and QIA were performed as previously described(14) using 4–5 μm paraffin tissue sections. The sections were deparaffinized and rehydrated through sequential ethanol washes. Heat-induced epitope retrieval was performed with DIVA Decloaker in a high-pressure cooker (Biocare Medical). Sections were then blocked with the SNIPER reagent (Biocare Medical) for 15 min and in PBS/0.1% triton-X100/4 % goat serum for 30 min at room temperature. After the blocking step, the sections were incubated with mouse anti-human IFN-γ (clone B27, BD Biosciences), rabbit anti-human IL-21 (Polyclonal Ab; AbD Serotec), and rat anti-human CD3 (clone CD3-12, AbD serotec) antibodies diluted 1:50 to 1:100 in blocking buffer at 4° C overnight. These sections were incubated with secondary antibodies conjugated with fluorescent dyes diluted 1:1000–1:2000 (Jackson ImmunoResearch) in blocking buffer for 30 min at room temperature. Finally, nuclei were stained with Hoechst 33342 (Invitrogen) for 10 min at room temperature, and mounted using warmed glycerol gelatin (Sigma) containing 4 mg/ml n-propyl gallate (Fluka). Every step was followed by three washes with TBS automation buffer (Biocare Medical). For each lymph node tissue section stained with antibodies (IL-21, IFN-γ and CD3), images were collected with an Axio Imager Z1 microscope (Zeiss) using 20x objectives. Lymphoid follicles area in cortex was acquired as much as possible (5 to 52 follicle-like area), based on CD3 and Hoechst staining. GC size and mean fluorescence intensities of cytokines were calculated using AxioVs40 V4.8.1.0 program (Zeiss) and Image J1.43u (NIH).
Cell Sorting
Approximately 100 ×106 mononuclear cells isolated from the LN were stained with Live/dead, anti-CD3, anti-CD4, anti-CD95, anti-CXCR5, anti-CXCR3, anti-CD20 and anti-PD-1 for 25 minutes at 4°C. The memory CD4 T cells (CD95+) were further gated using CXCR5, PD-1 and CXCR3 markers and divided into CXCR3+CXCR5+PD-1++(CXCR3+GC-Tfh) and CXCR3−CXCR5+PD-1++(CXCR3− GC-Tfh) CD4+ T cell populations and were sorted using a FACSAria II (BD). For all sorted subsets, the purity was >95%. Sorted cells were used for in vitro stimulation for cytokine production using PMA/Iono stimulations and for T and B cell co culture experiments.
Antibody measurements
SIV-specific antibody (Ab) responses were assessed in serum by Env-specific enzyme-linked immunosorbent assays (ELISAs) using commercially purchased SIVmac239 gp140 antigen (Cat.No: IT-001-140P, Immune Technology Corp, New York, NY, USA). Briefly, ELISA plates (Costar; Corning Life Sciences, Lowell, MA) were coated with SIV gp140 (0.5 μg/ml) in 10mM Sodium bicarbonate buffer pH 9.3 overnight at 4 °C. Plates were washed and blocked for 1 hour with PBS-Tween, 4% whey and 5% dry milk. SIV infected macaque serum samples were added to duplicate wells in serial 5-fold dilutions and incubated for 2 hours at room temperature (RT). Plates were then washed, and bound Ab was detected using peroxidase-conjugated anti-monkey IgG (Accurate Chemical and Scientific, Westbury, NY) and tetramethylbenzidine substrate (KPL, Gaithersburg, MD). Reactions were stopped with 100 μl 1M H3PO4 and read at 450nm. Each plate included a standard curve generated using goat anti-monkey IgG and known concentrations of rhesus IgG starting from 100ng/ml (both from Accurate Chemical and Scientific Corp.). SIV-specific IgG1 Ab responses were assessed in serum in a similar manner except for detection biotin labelled mouse monoclonal anti-rhesus IgG1 (7H11 Biotin, NHP Reagent Resource) followed with Avidin HRP (Vector Labs) was used. Each plate included a standard curve generated using mouse monoclonal anti-monkey IgG (8F1) for coating and know amounts of purified rhesus IgG1 starting from 10ug/ml (NHP Reagent Resource) for capture.
Total macaque IgG concentrations in culture supernatants were assessed by coating ELISA plates with goat anti-monkey IgG in 50μl/well. Culture supernatants were used either undiluted or 1:2 diluted and bound IgG was detected using peroxidase-conjugated anti-monkey IgG as described above. Total macaque IgG1 Ab responses, the culture supernatants were assessed by coating ELISA plates with 50μl of mouse monoclonal anti rhesus IgG1 (7H11, 2μg/ml, NHP Reagent Resource) in PBS overnight. Diluted or 1:2 diluted culture supernatants were added after blocking. Bound IgG1 was detected using biotin labeled mouse monoclonal anti rhesus IgG (8F1 Biotin, NHP Reagent Resource) followed with Avidin-HRP (Vector Labs). Standard curves were fitted and sample concentrations interpolated as micrograms/nanograms of Ab per milliliter of serum/culture supernatant using SOFTmax 2.3 software (Molecular Devices, Sunnyvale, CA). The concentrations of anti SIV gp140 specific IgG, IgG1, total macaque IgG and IgG1 are relative to our standard curve, not absolute values.
Quantitation of SIV RNA and DNA
The SIV copy number in plasma was determined using a quantitative real-time PCR as described previously (34). For viral load determinations in LN, total DNA was extracted from ~50,000 sorted CXCR3+ and CXCR3− GC-Tfh subsets and data were normalized to albumin as described previously (35). Sorting experiments were performed on cryopreserved LN samples. All PCRs were performed in duplicates with a limit of detection of 60 copies/reaction.
T and B cell co-cultures
Co-culture experiments were performed on frozen LN cells. 5×104 sorted CD4 T cells were cultured with 5×104 sorted autologous total B cells (1:1 ratio) in the presence of SEB (0.5 μg/ml). Supernatants harvested on day 9 were analyzed for total monkey immunoglobulins for IgG and IgG1. For determining the proliferation of total B cells by flow cytometry, cells were harvested on Day 5.
Statistical analyses
Wicoxon test was used for comparisons between two or more subsets from the same animal. Unpaired non-parametric Mann-Whitney test was used for comparisons between SIV uninfected and SIV-infected animals. Spearman rank test was used for all correlations. The boolean analyses were performed using the SPICE software (NIAID, NIH). When box-and-whisker plots are used, the box size represents the limits of the data for the second and third quartiles, with median shown as a bar. The GraphPad Prism statistical analysis program (GraphPad Software) was used to determine P values. P values of less than 0.05 were considered significant.
Results
GC-Tfh cells show a distinct profile for co-expression of chemokine receptors compared to non-GC-Tfh in SIV naïve RM
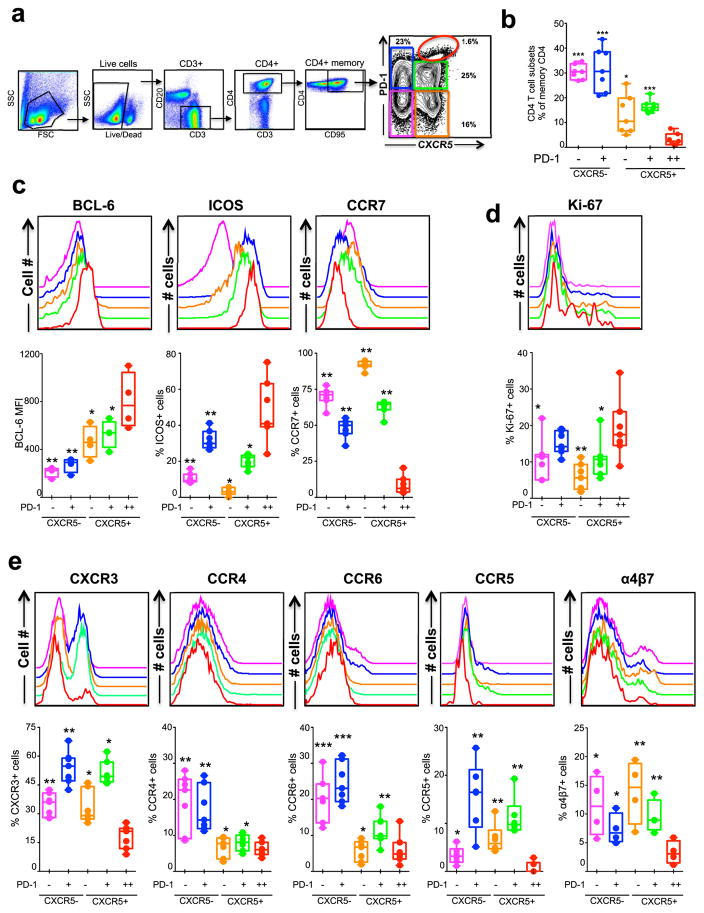
Tfh cells are characterized by the combined expression of CXCR5, ICOS and high levels of PD-1 in mice and humans (9). Similar to humans and mice, few studies defined Tfh in RM based on CXCR5 (16, 18, 36, 37), a key chemokine receptor required for homing of cells to B cell follicle/GC. In addition, it is important to characterize the expression of chemokine receptors that influence the migration and lineage relationship of CD4 T cells. To understand the relative distribution of CXCR5+ and CXCR5− CD4 T cells in the LN of SIV naïve RM, we characterized five populations of memory CD4 T cells (CD3+, CD4+, CD95+) based on CXCR5 and PD-1 expression namely CXCR5−PD-1− (X5− PD-1−), CXCR5−PD-1+ (X5− PD-1+), CXCR5+PD-1− (X5+ PD-1−), CXCR5+PD-1+ (X5+ PD-1+) and CXCR5+PD-1++ (X5+ PD-1++) (Figure 1a). Among these five subsets, the CXCR5− subsets constituted about 50–60% of the total cells and the frequency of X5+PD-1++ was the lowest with about 5% (Figure 1b). The relative distribution of these five subsets within the memory CD4 T cells was comparable to what has been reported for CD4 T cells in LNs of humans (10–12, 29, 38).
Figure 1. Characterization of five memory CD4 T cell subsets defined based on CXCR5 and PD-1 expression in the lymph node (LN) of SIV naïve rhesus macaques (RM).
(a, b) Relative distribution of five memory CD4 T cell subsets (CD95+ CD4+ cells) in the LN defined based on CXCR5 and PD-1 expression. Representative flow plot is shown in (a) and summary for a group (n=7) of SIV-naïve RM is shown in (b). (c) Expression of Tfh markers Bcl-6 and ICOS, and non-Tfh marker CCR7 (n=7). (d) Expression of proliferation marker Ki-67 (n=7). (e) Expression of different chemokine receptors CXCR3, CCR4, CCR6, CCR5 and α4β7 (n=7 except for α4β7 for which n=4). P values shown are for CXCR5+ PD-1++ (GC-Tfh) subset vs other subsets. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. The legend for histogram plots shown in c–e is shown in b.
We then characterized these cells for co-expression of markers associated with Tfh cells such as Bcl-6, ICOS and CCR7. Similar to human LN cells (10–12), the macaque X5+PD-1++ cells expressed highest levels of GC-Tfh markers Bcl-6 and ICOS, and lowest levels of CCR7 suggesting that the X5+ PD-1++ subset represents the macaque GC-Tfh (Figure 1c) hence we will refer to this subset as GC-Tfh hereafter. The Bcl-6 expression was mostly confined to GC-Tfh (Figure 1c) (1, 2). In contrast to Bcl-6 expression, ICOS was expressed on both CXCR5+ and CXCR5− memory CD4 T cells, with particularly high levels on PD-1+/++ cells. However, an opposite trend was observed for CCR7 expression, which was generally low on PD-1+ memory CD4 T cells and was lowest on the GC-Tfh. Of note, the X5−PD-1+ subset expressed relatively higher levels of ICOS and lower levels of CCR7 compared to other non-GC-Tfh subsets suggesting the existence of non-Tfh memory population with potential to provide B cell help. We then investigated the ex vivo proliferation status of these 5 different memory CD4 T cell subsets based on Ki-67 expression and found that about 10–20% of the CXCR5− and CXCR5+ subsets in the LN expressed Ki-67, with the GC-Tfh cells were the most proliferative compared to any other subsets (Figure 1d).
We next studied the expression of additional chemokine receptors CXCR3, CCR4 and CCR6 as the expression of these receptors has been shown to be associated with Th1, Th2 and Th17 CD4 T cell helper lineage, respectively (30, 39) as well as CCR5 and the integrin α4β7 (Figure 1e). A significant fraction of all non-GC-Tfh memory subsets expressed CXCR3 and the expression was higher on PD-1+ subsets than on PD-1− subsets. However, only a small fraction of the GC-Tfh expressed CXCR3. In contrast, CCR4 and CCR6 expression was mostly restricted to CXCR5− memory subsets. Similarly, the expression of CCR5 was mostly restricted to non-GC-Tfh cells and within the non-GC-Tfh, expression of CCR5 was higher on PD-1+ cells compared to PD-1− cells. Only a small fraction of GC-Tfh expressed CCR5, in spite of the fact that these cells are highly infected during chronic SIV infection (40). Similar to the CCR5, the α4β7 expression was restricted to non-GC-Tfh subsets. Interestingly, the expression of α4β7 was relatively higher in PD-1− cells compared to PD-1+ cells. Collectively these results revealed that the GC-Tfh CD4 subset was distinct from non-GC-Tfh with limited/low expression for CXCR3, CCR4, CCR6, CCR7 CCR5 and α4β7. This low expression of other chemokine receptors was not a characteristic of all CXCR5+ subsets as the X5+PD-1+ subset showed strong expression of CXCR3, CCR5 and α4β7 (Figure 1e). These results also revealed that the X5−PD-1+ CD4 subset was distinct from other GC-Tfh and non-Tfh subset as they showed strong expression of all other chemokine receptors studied here.
Rapid enrichment of CXCR3+ GC-Tfh cells during chronic SIV infection
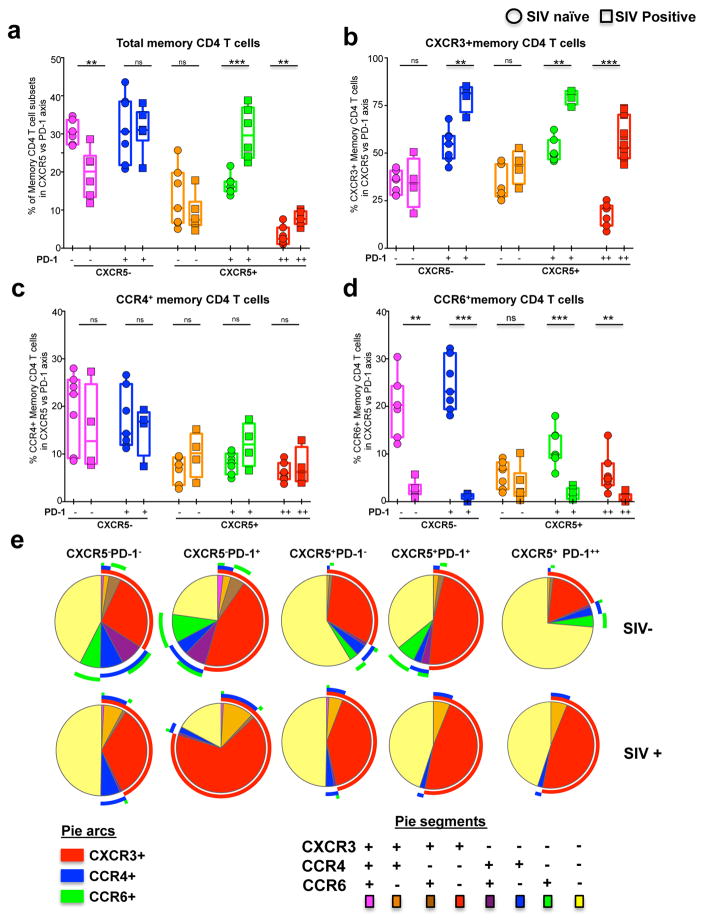
We next investigated the influence of SIV infection on the relative proportions of these five memory CD4 subsets and expression of chemokine receptors CXCR3, CCR4 and CCR6 (Figure 2). For this purpose, we utilized samples from a cohort of SIVmac251 infected RMs. The relative distribution of memory CD4 T cells was significantly altered in LNs of SIV infected RMs compared to SIV uninfected RMs (Figure 2a). The frequency of X5+PD-1+ and GC-Tfh CD4 T cells was higher and the frequency of X5−PD-1− subset was lower in the SIV-infected animals. However, the frequency of X5−PD-1+ and X5+PD-1− CD4+ T cells are comparable. Interestingly, we observed a profound increase for CXCR3 expression on both CXCR5+ and CXCR5− memory CD4+ T cells, and this increase was mostly restricted to the memory CD4 T cells that co-expressed PD-1, including the GC-Tfh (Figure 2b). However, there was no significant difference in the expression of CCR4 on memory CD4 T cells (Figure 2c). In contrast, the frequency of CCR6+ memory CD4 T cells are dramatically lower on all memory CD4 T cell subsets irrespective of their CXCR5 expression demonstrating their preferential depletion during chronic SIV infection (Figure 2d). We performed boolean analysis to understand the co-expression of these chemokine receptors. The results revealed that in SIV naïve animals, the majority of GC-Tfh do not express CXCR3, CCR4 or CCR6, and during chronic SIV infection, the frequency of CXCR3+ single positive cells increase dramatically as the majority of these CXCR3+ cells failed to co-express CCR4 or CCR6 (Figure 2e). These results demonstrated that chronic SIV infection significantly enhances the frequency of GC-Tfh that co-express high levels of Th1 associated chemokine receptor CXCR3.
Figure 2. Enrichment of CXCR3+ CD4 T cells in the LN during chronic SIV infection.
(a) Relative distribution of five memory CD4 T cell subsets in the LN defined based on CXCR5 and PD-1 expression before and after (24 weeks) SIV infection. Expression of CXCR3 (b), CCR4 (c) and CCR6 (d) on five memory CD4 T cell subsets defined based on CXCR5 and PD-1 expression before and 24 weeks after SIV infection. (e) Pie charts summarizing the co-expression of chemokine receptors CXCR3, CCR4 and CCR6 on different memory CD4 T cell subsets. Red arcs identify CXCR3+ populations, blue arcs identify CCR4+ population, green arcs identify CCR6+ population. Data for seven SIV− and SIV+ animals are shown.
CXCR3+ GC-Tfh cells are phenotypically and functionally distinct from CXCR3− GC-Tfh cells
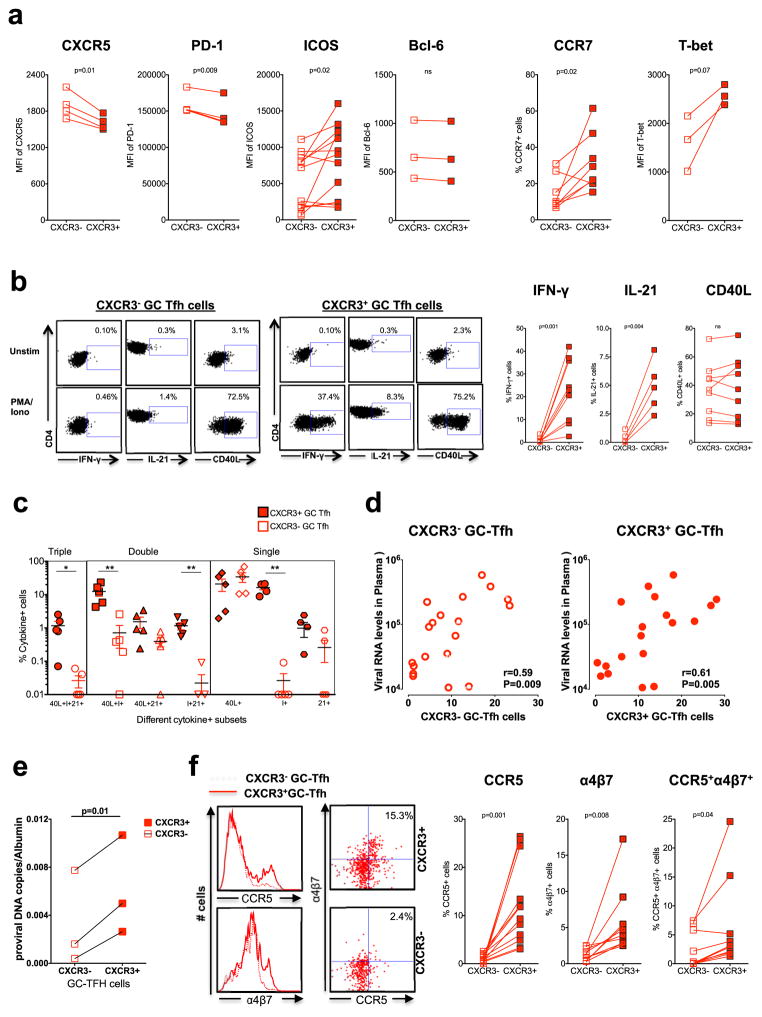
Previous studies showed that Th1 cells express CXCR3 and CXCR3 ligand mediated interaction facilitates the migration of cells towards inflammatory sites such as interfollicular and medullary zones of LNs (41). We next investigated whether CXCR3+ GC-Tfh cells are phenotypically or functionally distinct from the CXCR3− GC-Tfh cells. We compared the relative expression of Tfh markers CXCR5, PD-1, and ICOS, and Th1 markers CCR7 and T-box transcription factor T-bet within the GC-Tfh compartment (Figure 3a). CXCR3+ GC-Tfh cells expressed significantly lower levels of CXCR5 and PD-1 compared to CXCR3− GC-Tfh cells, although the difference is small. However, the CXCR3+ GC-Tfh cells expressed higher levels of CCR7, ICOS and T-bet (trend). The expression of Bcl-6 was comparable. These results suggested that CXCR3 expression identifies Th1-like GC-Tfh cells in GCs.
Figure 3. CXCR3+ GC-Tfh cells are phenotypically and functionally distinct from CXCR3− GC-Tfh cells.
(a) Expression of Tfh and Th1 markers on CXCR3+ and CXCR3− GC-Tfh cells (b) Expression of IFN-γ, IL-21 and CD40L by sorted CXCR3+ and CXCR3− GC-Tfh cells following stimulation with PMA/Ionomycin. Representative flow plots are shown on the left. (c) Boolean analysis determining co-expression of IFN-γ, IL-21 and CD40L. (d) Correlation between CXCR3+ GC-Tfh or CXCR3− GC-Tfh and plasma viral RNA levels at 24 weeks post SIV infection. (e) Copies of SIV proviral DNA in sorted CXCR3+ and CXCR3− GC-Tfh cells. (f) Expression of CCR5 and α4β7 on CXCR3+ and CXCR3− GC-Tfh cells.
To assess the functional cytokine profile, we sorted CXCR3+ and CXCR3− GC-Tfh cells and measured the production of IFN-γ, IL-21 and CD40L following stimulation with PMA/Ionomycin. CXCR3+ but not CXCR3− GC-Tfh cells predominantly produced IFN-γ and IL-21 although the expression of CD40L was comparable between the two subsets (Figure 3b). To assess the polyfunctional capacity of CXCR3+ and CXCR3− GC-Tfh cells in the LN during chronic SIV infection we performed boolean cytokine analysis. Interestingly, the CXCR3+ GC-Tfh cell populations comprised significantly higher proportion of triple producers (IFN-γ/IL-21/CD40L), double producers (IFN-γ/CD40L; IFN-γ/IL-21) and single producers (IFN-γ)(Figure 3c). These data clearly suggest that CXCR3+ GC-Tfh cells are functionally different compared with CXCR3− GC-Tfh cell populations.
To understand the relationship between the subsets and viral replication further, we studied the association between the frequency of CXCR3+ and CXCR3− GC-Tfh and plasma viremia and determined the levels of SIV DNA in these subsets. The frequency of CXCR3+ as well as CXCR3− GC-Tfh cells showed a direct correlation with plasma viremia (Figure 3d) but a higher proportion of CXCR3+ GC-Tfh compared to CXCR3− GC-Tfh contained SIV DNA (Figure 3e). Since the CXCR3+ GC-Tfh cells contained higher levels of SIV DNA, we compared the relative expression of HIV co-receptor CCR5 and HIV binding protein α4β7 on CXCR3+ and CXCR3− GC-Tfh cells. Interestingly the CXCR3+ GC-Tfh cells expressed higher levels of CCR5 (p=0.001), α4β7 (p=0.008) and CCR5+α4β7+ double positive cells (p=0.04) compared to CXCR3− GC-Tfh cells (Figure 3f) suggesting the increased susceptibility of CXCR3+ GC-Tfh cells to HIV/SIV infection. Thus, our results clearly demonstrate that CXCR3+ and CXCR3− GC-Tfh cells are phenotypically and functionally different within GCs suggesting the existence of two different populations in the SIV infected GC-Tfh compartment and support the notion that CXCR3+ GC-Tfh cells could represent a Tfh1 (42, 43) subset in the GCs during chronic SIV infection.
Chronic SIV infection results in the accumulation of IL-21+ and IFN-γ+ cells within the follicles
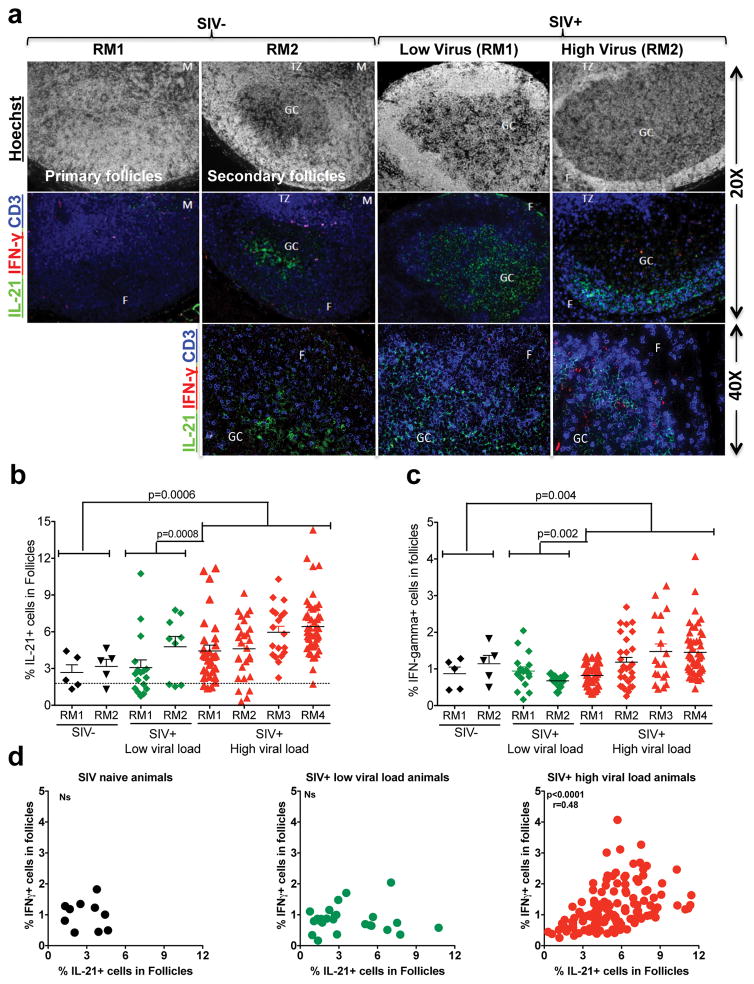
SIV infection induces follicular hyperplasia, which is associated with aberrant accumulation of Tfh cells producing IL-21 within GCs (44). Since we observed an enrichment of phenotypically and functionally different CXCR3+ GC-Tfh cells that produce IFN-γ during chronic SIV infection, we hypothesized that the GCs of SIV infected animals may also have higher levels of IFN-γ during chronic infection. Hence, we investigated the presence of cytokines IFN-γ and IL-21 within the GCs in SIV naïve (n=2) and SIV infected RMs with low (n=2) and high (n=4) plasma viral RNA levels using immunohistochemistry (Figure 4). As shown before (35, 45), the number of hyperplastic follicles with GCs was significantly higher in chronically SIV infected animals compared to SIV-naïve animals (Figure 4a, data not shown). Consistent with our previous findings (16, 44), the frequency of IL-21+ cells in the follicles was significantly higher in SIV-infected animals with high viral load relative to SIV-naïve animals (Figure 4b). However, consistent with higher CXCR3 expression by flow cytometry, we also observed significantly higher density of CD3+ IFNγ+ cells in the follicles of SIV-infected animals with high viral load relative to SIV naïve animals (Figure 4c). In a limited number of RMs, we confirmed that the IFN+ cells are mostly CD4+ (Supplemental Figure 1a) and CD8− (Supplemental Figure 1b). This increase for IL-21+ or IFNγ+ cells was not observed in SIV infected animals with lower viral load. Interestingly the density of IL-21+ and IFN-γ+ cells in GCs correlated positively in high viral load animals but not in low viral load or SIV naïve animals demonstrating that both IL-21+ and IFN-γ+ cells were expanded during chronic SIV infection (Figure 4d). Collectively, these data demonstrate that chronic uncontrolled SIV infection is associated with high levels of GC-Tfh with Th1 phenotype leading to enrichment of IFN-γ+ cells within the follicles. These results provide important clues regarding GC-related hyperimmune responses in the context of disease progression during chronic SIV infection that may help to develop therapeutic strategies to limit lymphoid dysfunction during chronic SIV/HIV infection.
Figure 4. Chronic SIV infection results in accumulation of IL-21+ and IFN-γ+ cells within the follicles.
(a) Representative IHC images showing IFN-γ+ and IL-21+ cells in the follicles, T cell zone, and medulla of LN of SIV-naïve and SIV+ RMs with low (<100 copies) and high (>10,000 copies) viral load. (b) Expression of IL-21 in follicles. (c) Expression of IFNγ in follicles. Each dot indicates the percentage of cytokine intensity in one follicle. (d) Correlation between the expression of IFN-γ+ and IL-21+ cells in follicles.
CXCR3+ GC-Tfh cells help B cells for antibody production during chronic SIV infection
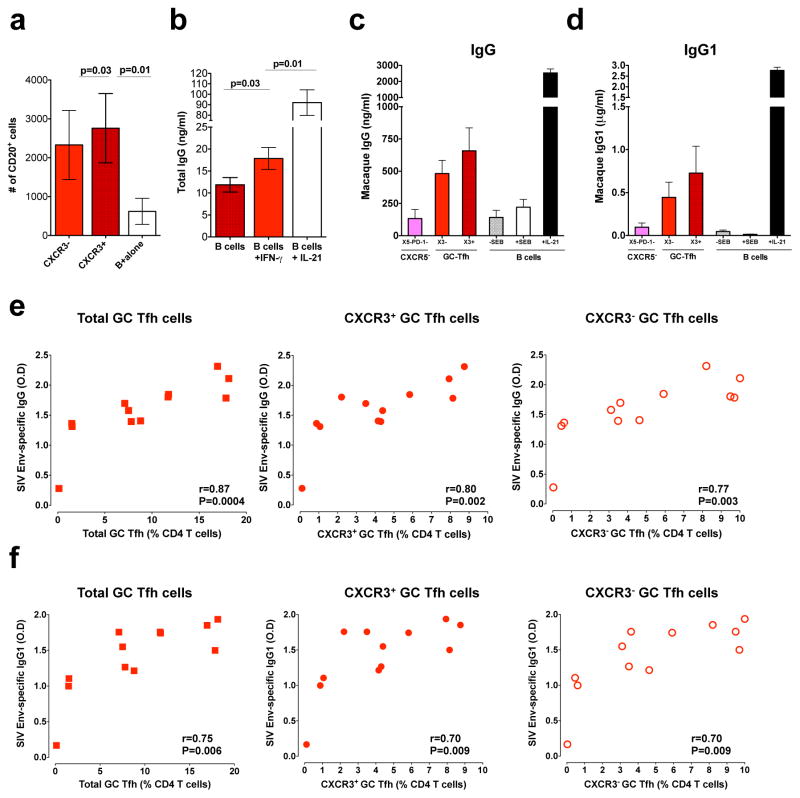
Studies have demonstrated that the differentiation of GC B cells into plasma cells is a Tfh-dependent process and that in fact, a large proportion of Tfh functions are dedicated to B cell differentiation, survival and proliferation. In addition, GC-Tfh cells have been shown to regulate antibody responses during HIV/SIV infection (12, 15, 16, 38, 46). To understand the nature of B cell help provided by CXCR3+ and CXCR3− GC-Tfh cells, we sorted CXCR3+ and CXCR3− GC-Tfh and CXCR5− cells and co-cultured the various sorted subsets with autologous B cells in the presence and absence of staphylococcal enterotoxin B (SEB) for 5–9 days and measured B cell numbers and antibody production (15). We observed a significant increase in B cell numbers in the presence of CD4 T cells, and interestingly, similar increases were observed in the presence of both CXCR3+ and CXCR3− GC-Tfh cells (Figure 5a) despite differences in their production of IFNγ and IL-21 (Figure 3). To test the effects of IFN-γ on IgG production by B cells, we sorted total B cells from three SIV infected macaques and cultured them in the presence of IFN-γ (5ng/ml) for 9 days and measured IgG levels in supernatants. Interestingly, addition of IFN-γ marginally increased the production of total IgG (Figure 5b). However in B cell co-culture experiments, both CXCR3+ and CXCR3− GC-Tfh cells induced production of IgG and IgG1 after 9 days of stimulation (Figure 5c, 5d). Consistent with their potential to help B cells in vitro, we observed a strong direct association between the frequency of CXCR3+ or CXCR3− GC-Tfh and the concentration of SIV envelope-specific IgG (Figure 5e) or IgG1 in the serum (Figure 5f). These results suggest that both CXCR3+ and CXCR3− GC-Tfh cells contribute to the increased accumulation of SIV-specific IgG and IgG1 during chronic SIV infection normally referred to as hypergammaglobulinemia (10).
Figure 5. Both CXCR3+ and CXCR3− GC-Tfh cells help B cells equally irrespective of their functional difference.
Sorted CXCR3+ GC-Tfh and CXCR3− GC-Tfh cells from chronic SIV infected macaques were co-cultured with autologous sorted B cells. (a) Total numbers of viable B cells at day 5 from the T and B co-culture experiment. (b) IgG production by sorted B cells (B cell alone) from SIV infected RM in the presence of IFN-γ (5ng/ml) or IL-21 (10ng/ml). (c) IgG (n=4) and (d) IgG1 (n=3) concentrations at day 9 in T and B co-culture experiments. (e) Correlation between SIV-envelop specific IgG (at 1: 9000 dilution of sera) antibody levels and total, CXCR3+ or CXCR3− GC-Tfh cells at 24 weeks post SIV infection. (f) Correlation between SIV-envelop specific IgG1 (at 1: 450 dilution of sera) antibody levels and total, CXCR3+ or CXCR3− GC-Tfh cells at 24 weeks post SIV infection.
Discussion
There is limited knowledge on the exact identity of various subsets of memory CD4+ T cells that are present in the LN of SIV naïve and SIV infected RMs. Recent studies identified the chemokine receptors CXCR3, CCR4, and CCR6 (47, 48) as surface markers for select lineages of CD4+ T cells with distinct cytokine profiles and lineage-specific transcription factor expression (1) and established a link between CD4 T cell trafficking potential and immunologic function. Furthermore, recent progress in understanding the biology of human and mouse Tfh cells has delineated functionally distinct subsets of memory Tfh cells in the circulation (31, 49, 50), suggesting that the human blood memory Tfh cells co-express chemokine receptors associated with other T helper cell lineages such as CXCR3 (Th1), CCR4 (Th2), and CCR6 (Th17) (28, 31). However, whether the analyses of blood memory Tfh subsets faithfully reflect GC-Tfh responses in lymphoid organs of mice, macaques and humans remains to be established. Here we took advantage of the availability of LN samples from a well-characterized cohort of SIV naive and SIV infected RMs to delineate the different lineages of lymphoid memory CD4 T cell subsets and their function in RMs. Our results revealed that in SIV-naïve RM, the majority of GC-Tfh cells do not express chemokine receptors such as CXCR3, CCR4, CCR6, CCR7 and including the HIV co-receptor CCR5. This was in contrast to any other memory CD4 T cell subset in the LN including other CXCR5+ cells that expressed a diverse combination of these chemokine receptors. These results highlight an important finding that the majority of GC-Tfh in the SIV-naïve RM is not polarized to Th1, Th2 or Th17 lineage.
Our results showed that chronic SIV infection modulates the phenotype of GC-Tfh cells such that they express high levels of Th1 lineage associated chemokine receptor CXCR3, suggesting the emergence of Th1 biased Tfh (Tfh1) cells. Consistent with this phenotype, the CXCR3+ Tfh but not CXCR3− Tfh produced the Th1 cytokine IFN-γ, and the GC of chronically SIV-infected non-controller RM had a significantly higher levels of IFN-γ producing CD4 T cells compared to SIV-naïve and SIV-infected controller RM. The immune mechanisms that contributed to induction of CXCR3+ GC-Tfh are not completely clear. A similar phenotype was observed for GC-Tfh in mice infected with LCMV clone-13 (23). In addition, this phenotype may not be specific to chronic infections as we recently observed this phenotype on GC-Tfh after DNA/MVA vaccination of RM in the presence of SIV envelope protein boost in alum (35). This raises the possibility that the local inflammation alone can induce this phenotype on GC-Tfh cells. Our results are also consistent with a recent study in macaques demonstrating that Th1 transcriptional factor T-bet is up regulated in Tfh cells isolated from the LN and spleen during chronic SIV infection (16, 18, 36, 37). In addition to GC-Tfh, we observed an increase for CXCR3 expression on all other memory CD4 T cells in the LN indicating that this effect is not restricted to GC-Tfh.
Studies in humans have delineated the susceptibility to HIV infection in different lineages of CD4 T cells during chronic HIV infection using CCR5 expression and compared with their level of integrated HIV DNA (45). In SIV naïve animals, Tfh cells particularly the GC-Tfh cells express very low levels of CCR5 (16, 40), yet they were found to harbor high levels of SIV DNA during chronic infection, raising some interesting questions about their infection kinetics/mechanism. However, GC-Tfh cells, when analyzed based on the expression of Th1 marker CXCR3, we found that CXCR3+ GC-Tfh cells express higher levels of CCR5 and α4β7, and harbor higher copies of SIV DNA indicating an increased permissiveness of these cells to SIV infection. Moreover, the data presented here demonstrate a marked expansion of these CXCR3+ GC-Tfh cells during SIV infection. Similar findings were reported in the blood of HIV infected individuals as well as mouse model of HIV infection where CXCR3+ CD4 T cells preferentially expressed the HIV-co-receptor CCR5 (45, 51). Hence, we speculate that CXCR3+ CCR5+ α4β7+ GC-Tfh subsets may have the capacity to maintain a dynamic viral reservoir in GCs under anti-retroviral therapy. Thus, we established a link between Th1 and Tfh subsets during SIV infection demonstrating their distribution, phenotype and function, and the potential dynamics of SIV infection in this compartment. These findings may open novel therapeutic approaches for targeting the viral reservoirs in both the CXCR3+ and CXCR3− GC-Tfh subsets.
In an attempt to understand the contribution of CXCR3+ GC-Tfh to antiviral humoral immunity, we determined the B cell helper potential of these cells and compared with CXCR3− GC-Tfh cells, and found that both CXCR3+ and CXCR3− GC-Tfh cells possess B helper potential. Consistent with this in vitro finding, both CXCR3+ and CXCR3− GC-Tfh cells had the capacity to help B cell proliferation equally when co-cultured with autologous B cells and showed a direct association with SIV-specific serum IgG and IgG1 during chronic SIV infection. This is consistent with the ability of these cells to express molecules associated with B cell help such as ICOS and CD40L. This was in contrast to what has been reported for circulating CXCR5+ CD4 T cells in human blood where only CXCR3− CXCR5+ cells were shown to help B cells in vitro not the CXCR3+CXCR5+ cells (49). However, another human study demonstrated that circulating CXCR3+CXCR5+ICOS+ cells induced following seasonal influenza vaccination help memory B cells and correlate with the development of flu-specific antibody response (28, 52). It is important to note that in all of these studies, the B cell helper potential seems to correlate with the ability of CXCR5+ cells to express ICOS and CD40L, and there may be differences in the expression of these molecules on CXCR3+ and CXCR3− cells between humans and macaques, and between blood and LN.
We think that a potential influence of CXCR3+ Tfh on HIV/SIV pathogenesis during chronic infection could be their ability to induce autoantibodies, a profound feature of chronic HIV/SIV infections. A recent study showed that excessive IFN-γ signaling in GC due to excessive production of IFN-γ by T cells leads to accumulation of GC-Tfh cells and sustained GC reaction and formation of autoantibodies (43). Further, studies have also shown that IFN-γ contributes to the induction of B lymphocyte stimulating factor (BAFF) and Th1 polarized CD4 T cells contribute to production of autoantibodies even in the absence CD40-CD40L signaling (43, 53), which has been shown to be deficient during chronic HIV (54) and SIV infections (55, 56).
In the present study both CXCR3+ and CXCR3− GC-Tfh cells are associated with the increased amount of SIV-envelope specific antibodies during chronic SIV infection. However a recent study in HIV infected individuals described a subset of circulating memory Tfh cells that are negative for CXCR3 (CXCR3−CXCR5+CD4+) which help B cells and are present in high numbers in the HIV infected individuals who generate broadly neutralizing antibodies (29). Taking this into account, we further speculate that both CXCR3+ and CXCR3− GC-Tfh cells help B cells, however the CXCR3+ GC-Tfh in addition may contribute to the induction of autoantibodies through production of IFN-γ as discussed above. In addition, the higher CCR5 and α4β7 expression by the CXCR3+ GC-Tfh may contribute to sustained replication of virus during chronic infection and higher viral reservoirs during anti-retroviral therapy. A better understanding of early differentiation process and the developmental relationship between these cells in the LN during HIV/SIV infection is critical for the establishment of reliable CD4 T cell correlates for monitoring infection or vaccine-associated antibody responses during HIV/SIV infection.
Supplementary Material
Acknowledgments
We thank Dr. Joshy Jacob for his suggestion in performing B cell co-culture assays using sorted cells. We thank the animal care and veterinary staff of the YNPRC, Benton Lawson and Melon Tekola Nega for the virology assays, Dr. Kiran Gill, Dr. Barbara Cervasi, for their help with flow cytometry and flow sorting. We also thank the NIH AIDS Research and Reference Reagent Program for the provision of peptides.
This work was supported by National Institutes of Health Grants R01AI074471, R01AI071852, P01AI088575, U19Al109633 and RC2CA149086 to RRA; Yerkes National Primate Research Center Base Grant P51 RR00165 and Emory Center for AIDS Research Grant P30 AI050409.
References
- 1.O’Shea JJ, Paul WE. Mechanisms underlying lineage commitment and plasticity of helper CD4+ T cells. Science. 2010;327:1098–1102. doi: 10.1126/science.1178334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pepper M, Jenkins MK. Origins of CD4(+) effector and central memory T cells. Nature immunology. 2011;12:467–471. doi: 10.1038/ni.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bluestone JA, Mackay CR, O’Shea JJ, Stockinger B. The functional plasticity of T cell subsets. Nature reviews Immunology. 2009;9:811–816. doi: 10.1038/nri2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sallusto F, Geginat J, Lanzavecchia A. Central memory and effector memory T cell subsets: function, generation, and maintenance. Annual review of immunology. 2004;22:745–763. doi: 10.1146/annurev.immunol.22.012703.104702. [DOI] [PubMed] [Google Scholar]
- 5.Kim CH, Rott LS, Clark-Lewis I, Campbell DJ, Wu L, Butcher EC. Subspecialization of CXCR5+ T cells: B helper activity is focused in a germinal center-localized subset of CXCR5+ T cells. The Journal of experimental medicine. 2001;193:1373–1381. doi: 10.1084/jem.193.12.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Breitfeld D, Ohl L, Kremmer E, Ellwart J, Sallusto F, Lipp M, Forster R. Follicular B helper T cells express CXC chemokine receptor 5, localize to B cell follicles, and support immunoglobulin production. The Journal of experimental medicine. 2000;192:1545–1552. doi: 10.1084/jem.192.11.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moser B, Schaerli P, Loetscher P. CXCR5(+) T cells: follicular homing takes center stage in T-helper-cell responses. Trends in immunology. 2002;23:250–254. doi: 10.1016/s1471-4906(02)02218-4. [DOI] [PubMed] [Google Scholar]
- 8.Fazilleau N, McHeyzer-Williams LJ, Rosen H, McHeyzer-Williams MG. The function of follicular helper T cells is regulated by the strength of T cell antigen receptor binding. Nature immunology. 2009;10:375–384. doi: 10.1038/ni.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crotty S. Follicular helper CD4 T cells (TFH) Annual review of immunology. 2011;29:621–663. doi: 10.1146/annurev-immunol-031210-101400. [DOI] [PubMed] [Google Scholar]
- 10.Lindqvist M, van Lunzen J, Soghoian DZ, Kuhl BD, Ranasinghe S, Kranias G, Flanders MD, Cutler S, Yudanin N, Muller MI, Davis I, Farber D, Hartjen P, Haag F, Alter G, Schulze zur Wiesch J, Streeck H. Expansion of HIV-specific T follicular helper cells in chronic HIV infection. J Clin Invest. 2012;122:3271–3280. doi: 10.1172/JCI64314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perreau M, Savoye AL, De Crignis E, Corpataux JM, Cubas R, Haddad EK, De Leval L, Graziosi C, Pantaleo G. Follicular helper T cells serve as the major CD4 T cell compartment for HIV-1 infection, replication, and production. J Exp Med. 2013;210:143–156. doi: 10.1084/jem.20121932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cubas RA, Mudd JC, Savoye AL, Perreau M, van Grevenynghe J, Metcalf T, Connick E, Meditz A, Freeman GJ, Abesada-Terk G, Jr, Jacobson JM, Brooks AD, Crotty S, Estes JD, Pantaleo G, Lederman MM, Haddad EK. Inadequate T follicular cell help impairs B cell immunity during HIV infection. Nature medicine. 2013;19:494–499. doi: 10.1038/nm.3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pallikkuth S, Sharkey M, Babic DZ, Gupta S, Stone GW, Fischl MA, Stevenson M, Pahwa S. Peripheral T Follicular Helper Cells Are the Major HIV Reservoir Within Central Memory CD4 T Cells in Peripheral Blood from chronic HIV infected individuals on cART. J Virol. 2015 doi: 10.1128/JVI.02883-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hong JJ, Amancha PK, Rogers K, Ansari AA, Villinger F. Spatial alterations between CD4(+) T follicular helper, B, and CD8(+) T cells during simian immunodeficiency virus infection: T/B cell homeostasis, activation, and potential mechanism for viral escape. J Immunol. 2012;188:3247–3256. doi: 10.4049/jimmunol.1103138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Petrovas C, Yamamoto T, Gerner MY, Boswell KL, Wloka K, Smith EC, Ambrozak DR, Sandler NG, Timmer KJ, Sun X, Pan L, Poholek A, Rao SS, Brenchley JM, Alam SM, Tomaras GD, Roederer M, Douek DC, Seder RA, Germain RN, Haddad EK, Koup RA. CD4 T follicular helper cell dynamics during SIV infection. J Clin Invest. 2012;122:3281–3294. doi: 10.1172/JCI63039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mylvaganam GH, Velu V, Hong JJ, Sadagopal S, Kwa S, Basu R, Lawson B, Villinger F, Amara RR. Diminished viral control during simian immunodeficiency virus infection is associated with aberrant PD-1hi CD4 T cell enrichment in the lymphoid follicles of the rectal mucosa. Journal of immunology. 2014;193:4527–4536. doi: 10.4049/jimmunol.1401222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chowdhury A, Del Rio PM, Tharp GK, Trible RP, Amara RR, Chahroudi A, Reyes-Teran G, Bosinger SE, Silvestri G. Decreased T Follicular Regulatory Cell/T Follicular Helper Cell (TFH) in Simian Immunodeficiency Virus-Infected Rhesus Macaques May Contribute to Accumulation of TFH in Chronic Infection. J Immunol. 2015 doi: 10.4049/jimmunol.1402701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu H, Wang X, Malam N, Aye PP, Alvarez X, Lackner AA, Veazey RS. Persistent SIV infection drives differentiation, aberrant accumulation, and latent infection of germinal center follicular T helper cells. J Virol. 2015 doi: 10.1128/JVI.02471-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Petrovas C, Koup RA. T follicular helper cells and HIV/SIV-specific antibody responses. Curr Opin HIV AIDS. 2014;9:235–241. doi: 10.1097/COH.0000000000000053. [DOI] [PubMed] [Google Scholar]
- 20.Onabajo OO, George J, Lewis MG, Mattapallil JJ. Rhesus macaque lymph node PD-1(hi)CD4+ T cells express high levels of CXCR5 and IL-21 and display a CCR7(lo)ICOS+Bcl6+ T-follicular helper (Tfh) cell phenotype. PloS one. 2013;8:e59758. doi: 10.1371/journal.pone.0059758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu Y, Weatherall C, Bailey M, Alcantara S, De Rose R, Estaquier J, Wilson K, Suzuki K, Corbeil J, Cooper DA, Kent SJ, Kelleher AD, Zaunders J. Simian immunodeficiency virus infects follicular helper CD4 T cells in lymphoid tissues during pathogenic infection of pigtail macaques. J Virol. 2013;87:3760–3773. doi: 10.1128/JVI.02497-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pissani F, Streeck H. Emerging concepts on T follicular helper cell dynamics in HIV infection. Trends in immunology. 2014;35:278–286. doi: 10.1016/j.it.2014.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fahey LM, Wilson EB, Elsaesser H, Fistonich CD, McGavern DB, Brooks DG. Viral persistence redirects CD4 T cell differentiation toward T follicular helper cells. The Journal of experimental medicine. 2011;208:987–999. doi: 10.1084/jem.20101773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Glatman Zaretsky A, Taylor JJ, King IL, Marshall FA, Mohrs M, Pearce EJ. T follicular helper cells differentiate from Th2 cells in response to helminth antigens. The Journal of experimental medicine. 2009;206:991–999. doi: 10.1084/jem.20090303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crotty S. T Follicular Helper Cell Differentiation, Function, and Roles in Disease. Immunity. 2014;41:529–542. doi: 10.1016/j.immuni.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Milpied PJ, McHeyzer-Williams MG. High-affinity IgA needs TH17 cell functional plasticity. Nature immunology. 2013;14:313–315. doi: 10.1038/ni.2567. [DOI] [PubMed] [Google Scholar]
- 27.Schmitt N, Bentebibel SE, Ueno H. Phenotype and functions of memory Tfh cells in human blood. Trends Immunol. 2014;35:436–442. doi: 10.1016/j.it.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bentebibel SE, Lopez S, Obermoser G, Schmitt N, Mueller C, Harrod C, Flano E, Mejias A, Albrecht RA, Blankenship D, Xu H, Pascual V, Banchereau J, Garcia-Sastre A, Palucka AK, Ramilo O, Ueno H. Induction of ICOS+CXCR3+CXCR5+ TH cells correlates with antibody responses to influenza vaccination. Science translational medicine. 2013;5:176ra132. doi: 10.1126/scitranslmed.3005191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Locci M, Havenar-Daughton C, Landais E, Wu J, Kroenke MA, Arlehamn CL, Su LF, Cubas R, Davis MM, Sette A, Haddad EK, Poignard P, Crotty S A V. I. P. C. P. I. International. Human circulating PD-(+)1CXCR3(−)CXCR5(+) memory Tfh cells are highly functional and correlate with broadly neutralizing HIV antibody responses. Immunity. 2013;39:758–769. doi: 10.1016/j.immuni.2013.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rivino L, Messi M, Jarrossay D, Lanzavecchia A, Sallusto F, Geginat J. Chemokine receptor expression identifies Pre-T helper (Th)1, Pre-Th2, and nonpolarized cells among human CD4+ central memory T cells. The Journal of experimental medicine. 2004;200:725–735. doi: 10.1084/jem.20040774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boswell KL, Paris R, Boritz E, Ambrozak D, Yamamoto T, Darko S, Wloka K, Wheatley A, Narpala S, McDermott A, Roederer M, Haubrich R, Connors M, Ake J, Douek DC, Kim J, Petrovas C, Koup RA. Loss of circulating CD4 T cells with B cell helper function during chronic HIV infection. PLoS Pathog. 2014;10:e1003853. doi: 10.1371/journal.ppat.1003853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Locci M, Havenar-Daughton C, Landais E, Wu J, Kroenke MA, Arlehamn CL, Su LF, Cubas R, Davis MM, Sette A, Haddad EK, Poignard P, Crotty S A. V. I. P. C. P. I. International. Human circulating PD-1+CXCR3−CXCR5+ memory Tfh cells are highly functional and correlate with broadly neutralizing HIV antibody responses. Immunity. 2013;39:758–769. doi: 10.1016/j.immuni.2013.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kwa S, Sadagopal S, Shen X, Hong JJ, Gangadhara S, Basu R, Victor B, Iyer SS, LaBranche CC, Montefiori DC, Tomaras GD, Villinger F, Moss B, Kozlowski PA, Amara RR. CD40L-Adjuvanted DNA/MVA SIV Vaccine Enhances Protection Against Neutralization Resistant Mucosal SIV Infection. Journal of virology. 2015;89:4690–4695. doi: 10.1128/JVI.03527-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Velu V, Titanji K, Zhu B, Husain S, Pladevega A, Lai L, Vanderford TH, Chennareddi L, Silvestri G, Freeman GJ, Ahmed R, Amara RR. Enhancing SIV-specific immunity in vivo by PD-1 blockade. Nature. 2009;458:206–210. doi: 10.1038/nature07662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iyer SS, Gangadhara S, Victor B, Gomez R, Basu R, Hong JJ, Labranche C, Montefiori DC, Villinger F, Moss B, Amara RR. Codelivery of Envelope Protein in Alum with MVA Vaccine Induces CXCR3-Biased CXCR5+ and CXCR5− CD4 T Cell Responses in Rhesus Macaques. Journal of immunology. 2015;195:994–1005. doi: 10.4049/jimmunol.1500083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moukambi F, Rabezanahary H, Rodrigues V, Racine G, Robitaille L, Krust B, Andreani G, Soundaramourty C, Silvestre R, Laforge M, Estaquier J. Early Loss of Splenic Tfh Cells in SIV-Infected Rhesus Macaques. PLoS Pathog. 2015;11:e1005287. doi: 10.1371/journal.ppat.1005287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miles B, Miller SM, Folkvord JM, Kimball A, Chamanian M, Meditz AL, Arends T, McCarter MD, Levy DN, Rakasz EG, Skinner PJ, Connick E. Follicular regulatory T cells impair follicular T helper cells in HIV and SIV infection. Nat Commun. 2015;6:8608. doi: 10.1038/ncomms9608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu H, Wang X, Lackner AA, Veazey RS. PD-1(HIGH) Follicular CD4 T Helper Cell Subsets Residing in Lymph Node Germinal Centers Correlate with B Cell Maturation and IgG Production in Rhesus Macaques. Frontiers in immunology. 2014;5:85. doi: 10.3389/fimmu.2014.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Acosta-Rodriguez EV, Rivino L, Geginat J, Jarrossay D, Gattorno M, Lanzavecchia A, Sallusto F, Napolitani G. Surface phenotype and antigenic specificity of human interleukin 17-producing T helper memory cells. Nature immunology. 2007;8:639–646. doi: 10.1038/ni1467. [DOI] [PubMed] [Google Scholar]
- 40.Fukazawa Y, Park H, Cameron MJ, Lefebvre F, Lum R, Coombes N, Mahyari E, Hagen SI, Bae JY, Reyes MD, 3rd, Swanson T, Legasse AW, Sylwester A, Hansen SG, Smith AT, Stafova P, Shoemaker R, Li Y, Oswald K, Axthelm MK, McDermott A, Ferrari G, Montefiori DC, Edlefsen PT, Piatak M, Jr, Lifson JD, Sekaly RP, Picker LJ. Lymph node T cell responses predict the efficacy of live attenuated SIV vaccines. Nat Med. 2012;18:1673–1681. doi: 10.1038/nm.2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Groom JR, Richmond J, Murooka TT, Sorensen EW, Sung JH, Bankert K, von Andrian UH, Moon JJ, Mempel TR, Luster AD. CXCR3 chemokine receptor-ligand interactions in the lymph node optimize CD4+ T helper 1 cell differentiation. Immunity. 2012;37:1091–1103. doi: 10.1016/j.immuni.2012.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pepper M, Pagan AJ, Igyarto BZ, Taylor JJ, Jenkins MK. Opposing signals from the Bcl6 transcription factor and the interleukin-2 receptor generate T helper 1 central and effector memory cells. Immunity. 2011;35:583–595. doi: 10.1016/j.immuni.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee SK, Silva DG, Martin JL, Pratama A, Hu X, Chang PP, Walters G, Vinuesa CG. Interferon-gamma excess leads to pathogenic accumulation of follicular helper T cells and germinal centers. Immunity. 2012;37:880–892. doi: 10.1016/j.immuni.2012.10.010. [DOI] [PubMed] [Google Scholar]
- 44.Hong JJ, Amancha PK, Rogers KA, Courtney CL, Havenar-Daughton C, Crotty S, Ansari AA, Villinger F. Early lymphoid responses and germinal center formation correlate with lower viral load set points and better prognosis of simian immunodeficiency virus infection. Journal of immunology. 2014;193:797–806. doi: 10.4049/jimmunol.1400749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gosselin A, Monteiro P, Chomont N, Diaz-Griffero F, Said EA, Fonseca S, Wacleche V, El-Far M, Boulassel MR, Routy JP, Sekaly RP, Ancuta P. Peripheral blood CCR4+CCR6+ and CXCR3+CCR6+CD4+ T cells are highly permissive to HIV-1 infection. J Immunol. 2010;184:1604–1616. doi: 10.4049/jimmunol.0903058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Blackburn MJ, Zhong-Min M, Caccuri F, McKinnon K, Schifanella L, Guan Y, Gorini G, Venzon D, Fenizia C, Binello N, Gordon SN, Miller CJ, Franchini G, Vaccari M. Regulatory and Helper Follicular T Cells and Antibody Avidity to Simian Immunodeficiency Virus Glycoprotein 120. J Immunol. 2015;195:3227–3236. doi: 10.4049/jimmunol.1402699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Annunziato F, Cosmi L, Liotta F, Maggi E, Romagnani S. Human Th1 dichotomy: origin, phenotype and biologic activities. Immunology. 2014 doi: 10.1111/imm.12399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sallusto F, Lenig D, Mackay CR, Lanzavecchia A. Flexible programs of chemokine receptor expression on human polarized T helper 1 and 2 lymphocytes. The Journal of experimental medicine. 1998;187:875–883. doi: 10.1084/jem.187.6.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morita R, Schmitt N, Bentebibel SE, Ranganathan R, Bourdery L, Zurawski G, Foucat E, Dullaers M, Oh S, Sabzghabaei N, Lavecchio EM, Punaro M, Pascual V, Banchereau J, Ueno H. Human blood CXCR5(+)CD4(+) T cells are counterparts of T follicular cells and contain specific subsets that differentially support antibody secretion. Immunity. 2011;34:108–121. doi: 10.1016/j.immuni.2010.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hale JS, Youngblood B, Latner DR, Mohammed AU, Ye L, Akondy RS, Wu T, Iyer SS, Ahmed R. Distinct memory CD4+ T cells with commitment to T follicular helper- and T helper 1-cell lineages are generated after acute viral infection. Immunity. 2013;38:805–817. doi: 10.1016/j.immuni.2013.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Allam A, Majji S, Peachman K, Jagodzinski L, Kim J, Ratto-Kim S, Wijayalath W, Merbah M, Kim JH, Michael NL, Alving CR, Casares S, Rao M. TFH cells accumulate in mucosal tissues of humanized-DRAG mice and are highly permissive to HIV-1. Sci Rep. 2015;5:10443. doi: 10.1038/srep10443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pallikkuth S, Parmigiani A, Silva SY, George VK, Fischl M, Pahwa R, Pahwa S. Impaired peripheral blood T-follicular helper cell function in HIV-infected nonresponders to the 2009 H1N1/09 vaccine. Blood. 2012;120:985–993. doi: 10.1182/blood-2011-12-396648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gorbacheva V, Fan R, Wang X, Baldwin WM, 3rd, Fairchild RL, Valujskikh A. IFN-gamma production by memory helper T cells is required for CD40-independent alloantibody responses. J Immunol. 2015;194:1347–1356. doi: 10.4049/jimmunol.1401573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang R, Fichtenbaum CJ, Hildeman DA, Lifson JD, Chougnet C. CD40 ligand dysregulation in HIV infection: HIV glycoprotein 120 inhibits signaling cascades upstream of CD40 ligand transcription. Journal of immunology. 2004;172:2678–2686. doi: 10.4049/jimmunol.172.4.2678. [DOI] [PubMed] [Google Scholar]
- 55.Klatt NR, Vinton CL, Lynch RM, Canary LA, Ho J, Darrah PA, Estes JD, Seder RA, Moir SL, Brenchley JM. SIV infection of rhesus macaques results in dysfunctional T- and B-cell responses to neo and recall Leishmania major vaccination. Blood. 2011;118:5803–5812. doi: 10.1182/blood-2011-07-365874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brice GT, Mayne AE, Leighton K, Villinger F, Allan JS, Ansari AA. Studies of CD40L expression by lymphoid cells from experimentally and naturally SIV-infected nonhuman primate species. J Med Primatol. 1999;28:49–56. doi: 10.1111/j.1600-0684.1999.tb00250.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.