Abstract
This study examined whether differential DNA methylation is associated with clinical features of more aggressive disease at diagnosis and prostate cancer recurrence in African American men, who are more likely to die from prostate cancer than other populations. Tumor tissues from 76 African Americans diagnosed with prostate cancer who had radical prostatectomy as their primary treatment were profiled for epigenome-wide DNA methylation levels. Long-term follow-up identified 19 patients with prostate cancer recurrence. Twenty-three CpGs were differentially methylated (FDR q≤0.25, mean methylation difference ≥0.10) in patients with vs. without recurrence, including CpGs in GCK, CDKL2, PRDM13, and ZFR2. Methylation differences were also observed between men with metastatic-lethal prostate cancer vs. no recurrence (five CpGs), regional vs. local pathological stage (two CpGs), and higher vs. lower tumor aggressiveness (one CpG). These results indicate that differentially methylated CpG sites identified in tumor tissues of African American men may contribute to prostate cancer aggressiveness.
Keywords: DNA methylation, prostate cancer, tumor tissue, African American, recurrence, aggressiveness
1. Introduction
Men of African ancestry have the highest incidence of and mortality from prostate cancer (PCa) worldwide (1). Compared to European Americans, African American men have a 60% increased incidence of PCa, present with more advanced disease at diagnosis, have a shorter post-treatment period of progression-free survival, and are more than twice as likely to die from PCa (2-4). Factors that may contribute to this health disparity include socioeconomics, environmental exposures and lifestyle, access to healthcare and PCa screening, and the type of treatment received (5-8). In addition, biological differences, including genetics, genomics and epigenomics may play a role, particularly molecular factors that influence tumor aggressiveness (9-11).
DNA methylation is an epigenetic regulator of gene expression that has previously been associated with cancer, including PCa (12-14). Disruption of DNA methylation patterns in PCa is characterized by genome-wide loss of methylation, along with hypermethylation of gene promotor regions (15). Increased methylation in promotor regions, particularly within CpG islands, can be associated with gene silencing. One example is the frequent dysregulation (i.e., promoter region hypermethylation) of the GSTP1 gene in prostate tumor tissue, whereas other examples include hypomethylation of gene regions (e.g., repetitive elements) that may be associated with genomic instability (16,17). Changes in methylation patterns can occur early in carcinogenesis and particular methylation events have been associated with disease outcomes, indicating that methylation biomarkers may be useful for predicting prognosis and guiding treatment decisions (18,19).
There are few investigations that have examined aberrant DNA methylation in African American PCa patients, and these mainly consist of candidate gene studies (20-26). For instance, Tang et al. (25) reported that hypermethylation of RARB was significantly associated with a higher risk of PCa in African American men but not in men of European ancestry. Woodson et al. (20) described hypermethylation of CD44 in tumor tissues from African Americans (43% compared to 25% in European Americans), and found that this was positively correlated with tumor grade. The one publication to date that focused on epigenome-wide DNA methylation in African American PCa patients analyzed only three tumors in this high-risk population. That study found that the promotor regions of several genes, including SNRPN, MST1R, and ABCG5, were hypermethylated in African Americans compared to European Americans (11).
In order to search for differentially methylated CpG sites/gene regions in men of African ancestry, we characterized epigenome-wide DNA methylation profiles using radical prostatectomy (RP) tumor tissue samples from 76 African American men with PCa. We evaluated whether methylation levels of individual CpG sites were associated with PCa recurrence or features of more aggressive PCa, and whether observed methylation differences corresponded with changes in mRNA expression of these genes.
2. Results
The mean age at diagnosis for African American men in the two groups of PCa patients, 58 years for Fred Hutchinson Cancer Research Center (FHCRC) patients and 59 years for Eastern Virginia Medical School (EVMS) patients, was similar (p = 0.21) (Table 1). In order to increase the study power, analyses were performed on the combined group (n=76) and study group was included as a covariate in the models, along with age at diagnosis. In the combined sample, there were 19 recurrences, including 12 individuals who progressed to metastatic disease or who died from PCa. Recurrence was defined as a post-treatment PSA ≥ 0.2 ng/mL, receipt of secondary treatment (e.g., radiation therapy, androgen deprivation therapy, orchiectomy, or chemotherapy), an MRI, CT, bone scan or biopsy that was positive for PCa, if a physician stated that PCa had recurred, or if a patient died from PCa. In addition, 25% of patients had a high Gleason score [7(4+3) or 8-10], 49% had regional pathological stage, and 68% had a high composite tumor aggressiveness status [defined as one or more of the following: PCa recurrence and/or death; Gleason score (7=4+3, 8-10), and/or regional pathological stage]. A total of 477,460 CpG sites were analyzed for differential DNA methylation in patient subgroups defined by outcome events or clinicopathological features at diagnosis of more aggressive tumor biology.
Table 1. Characteristics of the African American prostate cancer patient population with tumor DNA methylation data.
| FHCRC patients (n=44) | EVMS patients (n=32) | Combined study population (n=76) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| No. | % | Mean (SD) | No. | % | Mean (SD) | Pa | No. | % | Mean (SD) | |
| Age at diagnosis (yr) | 58.1 (6.0) | 59.3 (6.1) | 0.21 | 58.6 (6.0) | ||||||
| Gleason score | <0.01 | |||||||||
| 5-6 | 14 | 31.8 | 6 | 18.8 | 20 | 26.3 | ||||
| 7(3+4) | 20 | 45.5 | 17 | 53.1 | 37 | 48.7 | ||||
| 7(4+3) | 6 | 13.6 | 5 | 15.6 | 11 | 14.5 | ||||
| 8-10 | 4 | 9.1 | 4 | 12.5 | 8 | 10.5 | ||||
| Pathological stage | <0.01 | |||||||||
| Local (pT1/T2) | 25 | 56.8 | 14 | 43.8 | 39 | 51.3 | ||||
| Regional (pT3) | 19 | 43.2 | 18 | 56.2 | 37 | 48.7 | ||||
| Recurrence status | <0.01 | |||||||||
| No recurrence | 20 | 45.4 | 18 | 56.3 | 38 | 50.0 | ||||
| Recurrence | 5 | 11.4 | 14 | 43.5 | 19 | 25.0 | ||||
| Mets/PCa Death | 0 | 0.0 | 12 | 37.5 | 12 | 15.8 | ||||
| Unknown | 19 | 43.2 | 0 | 0.0 | 19 | 25.0 | ||||
| Tumor aggressivenessb | <0.01 | |||||||||
| Low | 19 | 43.2 | 10 | 31.3 | 29 | 38.2 | ||||
| High | 25 | 56.8 | 22 | 68.7 | 47 | 61.8 | ||||
Abbreviations: PCa, prostate cancer; SD, standard deviation
P value calculated using a t-test (age) or chi-square test (categorical variables)
Composite variable of tumor aggressiveness (High = one or more of the following: PCa recurrence or death; Gleason sum (7=4+3 or 8-10); and/or regional stage)
In the analysis of patients with vs. without PCa recurrence, there were 11,444 CpGs identified with q ≤0.50, of which 70% were hypermethylated and 30% were hypomethylated in the recurrence group. Forty-one percent of these differentially methylated sites were located in the proximal promotor region of genes [consisting of the region 201 to 1500 base pairs upstream of a transcription start site (TSS1500), within 200 base pairs of a transcription start site (TSS200), 5′untranslated region (UTR), and the 1st exon], 32% were in the gene body, 3% in the 3′ untranslated region, and 24% were intergenic (Fig. 1).
Figure 1.
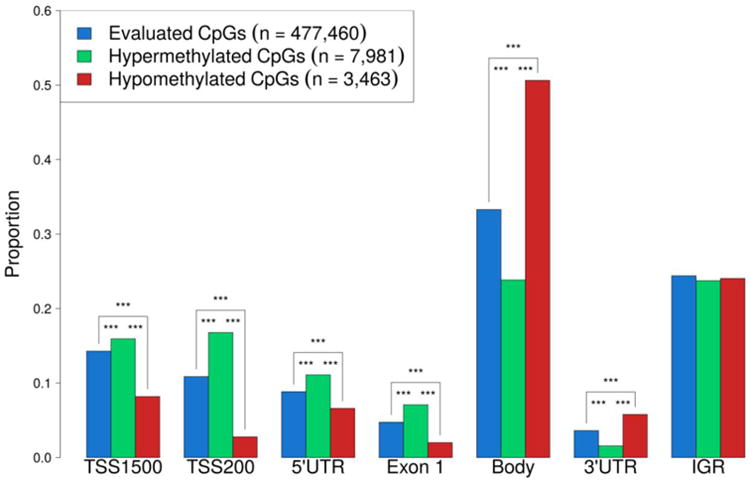
Bar plot showing proportion of CpG sites in each genomic region with FDR q ≤ 0.50 for all evaluated CpGs and significantly hyper- and hypo-methylated CpGs in African American prostate cancer patients with recurrence compared to those with no evidence of recurrence.
We next focused on the CpGs that exhibited the largest differences between patient groups (mean methylation β value difference between groups ≥0.10) using a more stringent false discovery rate (FDR) q-value threshold of 0.25, as these may be the more relevant for assessing tumor biology. Based on these criteria, we identified 23 CpGs that were differentially methylated in men with PCa recurrence compared to men with no evidence of recurrence, with higher methylation levels observed in the men with recurrent PCa for all but three of the CpGs (Fig.2, Table 2). Eleven of these CpGs were located in proximal promotor regions, seven CpGs in gene bodies, and five CpGs in intergenic regions. Genes with differentially methylated CpG sites that were identified in this analysis included GCK, CDKL2, PRDM13, and ZFR2, among others. There were two CpGs in the proximal promotor region of gene RBFOX3 that had higher methylation levels in patients with recurrent PCa. These CpGs were only 46 bp away from each other and their methylation status was significantly correlated (Pearson's r = 0.55, p<0.05). Nearly all of the top-ranked CpGs were surrounded by other CpGs in the same gene and epigenetic region that had an increase or decrease in mean DNA methylation level in the same direction as the top-ranked CpGs, although they did not reach statistical significance. For example, CDKL2 and ZFR2 each had 100% of probed CpGs (6 and 3 CpGs, respectively) in the TSS200/CpG island locations that had higher levels of methylation in the recurrence group. Exceptions included SLC24A20 and RBFOX3, with 33% and 47% of investigated CpGs having higher methylation levels in recurrent patients.
Figure 2.
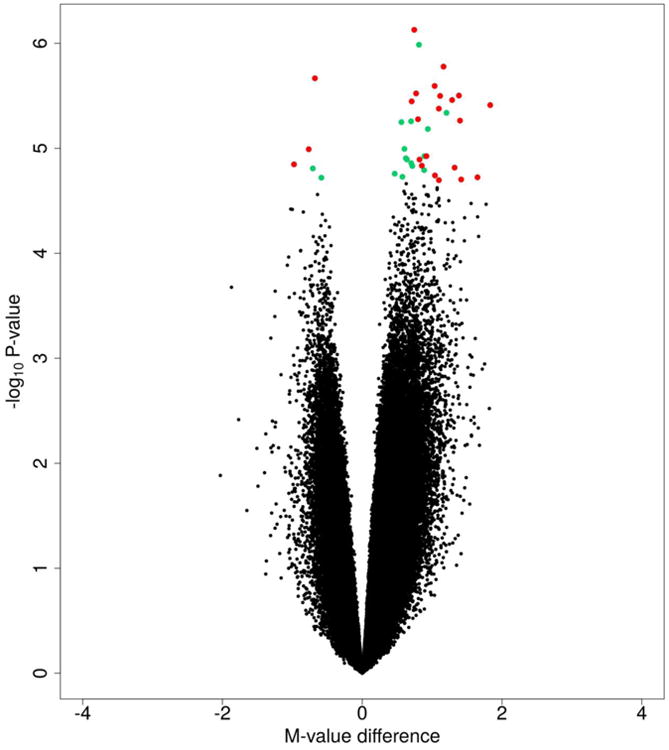
Volcano plot for African American patients with prostate cancer recurrence compared to those with no evidence of recurrence. Green = CpG sites with FDR q ≤ 0.25; Red = CpG sites with FDR q ≤ 0.25 and mean methylation β value difference ≥ 0.10.
Table 2. Top-ranked differentially methylated CpGs in African American patients by prostate cancer (PCa) recurrence or mortality statusa.
| CpG ID | Chromosome | Gene | Gene location | Epigenetic location | Mean β recurb | Mean β no recur | Mean β difference ≥ 10% Δβ | Methylation status | FDR q-value ≤ 0.25 | Total number of CpGs analyzed within the same gene and epigenetic location, and % that have a similar methylation profile to the top CpGc |
|---|---|---|---|---|---|---|---|---|---|---|
| Recurrenced vs. no recurrence | ||||||||||
| cg13476133 | 7 | GCK | Body | island | 0.40 | 0.19 | 0.21 | hyper | 0.25 | 5 (80%) |
| cg10344081 | 4 | CDKL2 | TSS200 | island | 0.31 | 0.11 | 0.20 | hyper | 0.16 | 6 (100%) |
| cg16377881 | 6 | PRDM13 | Body | island | 0.44 | 0.24 | 0.20 | hyper | 0.16 | 9 (100%) |
| cg07909759 | 10 | C10orf82 | TSS1500 | island | 0.41 | 0.22 | 0.19 | hyper | 0.16 | 2 (100%) |
| cg07838270 | 16 | intergenic | – | open sea | 0.58 | 0.39 | 0.19 | hyper | 0.16 | – |
| cg04957198 | 10 | PRTFDC1 | TSS1500 | island | 0.50 | 0.32 | 0.18 | hyper | 0.16 | 2 (100%) |
| cg06537894 | 19 | MAST1 | Body | island | 0.34 | 0.16 | 0.18 | hyper | 0.24 | 24 (67%) |
| cg03915940 | 16 | C16orf90 | Body | north shore | 0.60 | 0.76 | -0.16 | hypo | 0.24 | 1 (100%) |
| cg22792646 | 11 | INSC | TSS200 | island | 0.32 | 0.17 | 0.15 | hyper | 0.16 | 5 (100%) |
| cg12077664 | 12 | intergenic | – | island | 0.55 | 0.70 | -0.15 | hypo | 0.16 | – |
| cg04063589 | 5 | NEUROG1 | TSS1500 | island | 0.49 | 0.35 | 0.14 | hyper | 0.16 | 6 (100%) |
| cg25624927 | 3 | SLC25A20 | TSS200 | south shore | 0.20 | 0.07 | 0.14 | hyper | 0.16 | 3 (33%) |
| cg19393677 | 15 | MAP2K5 | Body | open sea | 0.70 | 0.84 | -0.13 | hypo | 0.24 | 16 (100%) |
| cg17531849 | 17 | RBFOX3 | 5′UTR | open sea | 0.36 | 0.22 | 0.13 | hyper | 0.25 | 111 (47%) |
| cg27655158 | 4 | intergenic | – | island | 0.18 | 0.06 | 0.12 | hyper | 0.25 | – |
| cg24265610 | 10 | FLJ41350 | TSS200 | island | 0.31 | 0.20 | 0.12 | hyper | 0.16 | 7 (86%) |
| cg03250870 | 17 | RBFOX3 | 5′UTR | open sea | 0.49 | 0.37 | 0.11 | hyper | 0.24 | 111 (47%) |
| cg11186405 | 13 | intergenic | – | island | 0.29 | 0.18 | 0.11 | hyper | 0.16 | – |
| cg26250461 | 5 | CDH18 | Body | open sea | 0.73 | 0.61 | 0.11 | hyper | 0.25 | 3 (67%) |
| cg07003030 | 20 | FOXA2 | 5′UTR | island | 0.26 | 0.15 | 0.11 | hyper | 0.24 | 5 (100%) |
| cg24770596 | 10 | PSD | Body | island | 0.16 | 0.06 | 0.11 | hyper | 0.16 | 6 (100%) |
| cg15823845 | 19 | ZFR2 | TSS200 | island | 0.22 | 0.12 | 0.10 | hyper | 0.16 | 3 (100%) |
| cg27080194 | 12 | intergenic | – | north shore | 0.22 | 0.12 | 0.10 | hyper | 0.24 | – |
| PCa death vs. no recurrence | ||||||||||
| cg13400512 | 6 | TNXB | Body | island | 0.64 | 0.41 | 0.23 | hyper | 0.23 | 71 (87%) |
| cg25624927 | 3 | SLC25A20 | TSS200 | south shore | 0.23 | 0.07 | 0.17 | hyper | 0.20 | 3 (33%) |
| cg02710173 | X | FLNA | TSS1500 | south shore | 0.68 | 0.83 | -0.16 | hypo | 0.20 | 3 (100%) |
| cg04887544 | 8 | intergenic | – | open sea | 0.71 | 0.86 | -0.15 | hypo | 0.20 | – |
| cg04166618 | 2 | LOC643387 | Body | island | 0.45 | 0.32 | 0.13 | hyper | 0.20 | 6 (67%) |
Abbreviations: PCa, prostate cancer; TSS1500, 201 to 1500 base pairs upstream of transcription start site; TSS200, 200 base pairs upstream of transcription start site; UTR, untranslated region
Results shown are from a model that adjusted for age at diagnosis and study group and are limited to CpGs with an FDR q-value ≤ 0.25 and mean methylation β value difference ≥ 0.10.
Patients with evidence of PCa recurrence or with metastatic-lethal PCa
The % of CpGs in the same region as the top-ranked CpG that did not meet the criteria for differential methylation (FDR q-value ≤ 0.25 and mean methylation β value difference ≥ 0.10), but that had an increase or decrease in mean methylation level in the same direction as the top CpG in patients with PCa recurrence (or metastatic-lethal PCa) vs. no evidence of recurrence.
Patients with recurrence include metastatic-lethal PCa cases
Based on analysis of a subset of the patients who developed PCa metastasis or died from PCa, five CpGs were differentially methylated compared to patients with no evidence of recurrence (Table 2). These were located in four genes (TNXB, SLC25A20, FLNA, and LOC643387) and one intergenic region on chromosome 8. Comparing these results with those from the analysis of PCa recurrence, we found that the CpG in SLC25A20 was also significant for overall PCa recurrence status. CpGs surrounding the top-ranked CpGs had an increase or decrease in methylation level in the same direction as the top CpGs identified in the analyses, including TNXB with 86% of CpGs in the gene body/CpG island having higher methylation levels in patients who died from PCa. SLC25A20 was the exception, with only 33% of CpGs in the same region as the top-ranked CpG having higher mean methylation levels, as previously mentioned for overall recurrence.
An analysis of clinical features of more aggressive disease at diagnosis identified two CpGs that were 96 bp away from each other in the gene body of EBF1 and which were hypermethylated in patients with regional stage disease compared to men with localized disease (Table 3). The methylation status of these two CpGs was significantly correlated (Pearson's r = 0.79, p < 0.05). One CpG in an intergenic region on chromosome 7 was hypermethylated in men with more aggressive disease features (i.e., high aggressiveness status). No significant methylation differences were found in an analysis of patients stratified by high (7=4+3, 8-10) vs. low (5-6, 7=3+4) Gleason score.
Table 3. Top-ranked differentially methylated CpGs in African American patients according to features of more aggressive prostate cancera.
| CpG ID | Chromosome | Gene | Gene location | Epigenetic location | Mean β group 1b | Mean β group 2c | Mean β difference ≥ 10% Δβ | Methylation status | FDR q-value ≤ 0.25 | Total number of CpGs analyzed within the same gene and epigenetic location, and % that have a similar methylation profile to the top CpGd |
|---|---|---|---|---|---|---|---|---|---|---|
| Regional vs. local stage | ||||||||||
| cg11891579 | 5 | EBF1 | Body | island | 0.58 | 0.41 | 0.18 | hyper | 0.03 | 7 (100%) |
| cg13295238 | 5 | EBF1 | Body | island | 0.48 | 0.30 | 0.17 | hyper | 0.03 | 7 (100%) |
| High vs. low tumor aggressivenesse | ||||||||||
| cg18845377 | 7 | intergenic | – | island | 0.25 | 0.15 | 0.10 | hyper | 0.06 | – |
Abbreviations: PCa, prostate cancer; TSS1500, 201 to 1500 base pairs upstream of transcription start site; TSS200, 200 base pairs upstream of transcription start site; UTR, untranslated region
Results are from models that adjusted for age at diagnosis and study group and limited to CpGs with an FDR q-value ≤ 0.25 and mean methylation β value difference ≥ 0.10.
Patients with regional pathological stage or high tumor aggressiveness
Patients with local pathological stage or low tumor aggressiveness
The % of CpGs in the same region as the top-ranked CpG that did not meet the criteria for differential methylation (FDR q-value ≤ 0.25 and mean methylation β value difference ≥ 0.10), but that had an increase or decrease in mean methylation level in the same direction as the top CpG for patients with regional vs. local stage.
Composite variable of tumor aggressiveness (High = one or more of the following: PCa recurrence or death; Gleason score (7=4+3 or 8-10); and/or regional stage)
We next examined whether any of these differentially methylated CpGs were associated with altered mRNA expression levels in the respective genes. Gene expression data were not available for C16orf90, FLJ41350, TNXB, or LOC643387. The FOXA2 gene had data for 3 transcripts available for analysis; GCK, INSC, and MAP2K5 each had data for 2 transcripts, and the remaining genes had data for one transcript each. There were significant correlations for four of the 17 genes with differentially methylated CpGs, including: three FOXA2 transcripts (Spearman's rank correlation rho = -0.27, P=0.05; rho = -0.32, P = 0.02; rho = -0.50, P = 1.4×10-4); two GCK transcripts (rho = -0.30, P = 0.03; rho = -0.48, P = 2.9×10-4;); CDKL2 (rho = -0.27, P = 0.04); and NEUROG1 (rho = -0.37, P = 5.5×10-3). For genes FOXA2, CDKL2, and NEUROG1, increased promotor region methylation was correlated with decreased mRNA levels; for GCK, CpG methylation of an island within the gene body was also negatively correlated with gene expression. We did not observe significant differences in mRNA expression levels for the other top-ranked genes as identified by differentially methylated CpG sites.
As a secondary analysis, the CpGs identified in the African American men as being associated with PCa recurrence or features of more aggressive tumor behavior were evaluated in our cohort of European American prostate cancer patients (n=479) in order to evaluate whether the results were population-specific (Suppl. Table 1). Three of the differentially methylated CpGs identified in the recurrence group (in ZFR2, C10orf82, and INSC) and one CpG related to the high composite aggressiveness variable (on chromosome 7) also had significantly different methylation levels at p ≤ 0.05 in the European American PCa patients (Suppl. Table 2). However, none of these had mean β differences ≥ 0.10 between the recurrence vs. non-recurrence groups, suggesting that differential methylation of these CpG sites may be unique to African American patients.
3. Discussion
African American men are at increased risk of developing more aggressive PCa and they are more likely to die from the disease compared to European American men. Few studies, however, have examined the relationship with tumor DNA methylation, and how this may relate to PCa racial disparity (1,5). This epigenome-wide study identified several differentially methylated CpG sites in tumor tissue from African American patients that were related to more aggressive features of PCa. Most of these CpG sites were not associated with recurrence or advanced disease in a comparison group of European Americans, suggesting that these methylation changes may be specific to the African American patients. These differentially methylated CpGs observed in African Americans but not in European American patients may contribute to differences in prostate cancer outcomes between these two populations.
Of interest for PCa biology in African Americans is a CpG in the gene body of GCK (Glucokinase, involved in glucose metabolism) that had a higher level of methylation in the recurrence group, and was associated with decreased mRNA levels. A genetic variant in this gene that increases serum glucose level was previously described as being associated with PCa (27). It is possible that metabolic dysregulation may contribute to more aggressive PCa in African American patients. CpGs in other metabolic-related genes were found to have higher methylation levels in the recurrence group: one CpG in the promotor region of FOXA2 and CDKL2, respectively, that was associated with decreased mRNA levels; one CpG in the promotor region of SLC25A20, and one in the body of MAST1; while one CpG in the body of MAP2K5 was hypomethylated in patients with PCa recurrence. These genes play roles in ATP-binding and protein kinase activity and are thus important regulators of cellular processes that contribute to oncogenesis, including cell proliferation, cellular metabolism, apoptosis, and DNA damage repair.
Also of interest are five metal ion binding genes (classified according to the DAVID gene ontology database) that were identified in this study as having differentially methylated CpG sites in patients with vs. without PCa recurrence. This includes ZFR2 (zinc finger RNA binding protein 2), a gene with higher mean methylation levels of CpGs in the transcription start site (TSS200) of the gene promotor region in patients with PCa recurrence. The other genes include MAST1, CDH18, MAP2K5, and PRDM13, all with differential methylation of CpGs in the gene body. Metal ions are essential for life, as they serve as oxygen carriers, regulate glucose metabolism, and serve as structural frameworks for proteins such as zinc finger motifs, which regulate gene functions. Altered metal ion levels have been described in tumor tissues, including lower levels of zinc in PCa (28,29). Lower zinc level increases metabolic efficiency and cell proliferation, and has been associated with both incident PCa and biochemical recurrence (29-31).
Increased promotor region methylation level has been associated with decreased gene expression (12,13). In this study we identified three genes, CDKL2, FOXA2, and NEUROG1 (associated with colorectal cancer (32)) that had higher promotor region methylation and a corresponding decrease in mRNA level in the patients with more aggressive PCa. It is not clear what effect methylation of CpGs in the gene body may have on gene expression, as it has been variously associated with both increased and decreased gene expression (33). Here we found increased methylation of a CpG in the gene body of GCK that was correlated with reduced mRNA levels. We did not see a correlation between methylation and gene expression for the other genes identified in this study as having differentially methylated CpGs, however, there are many other factors besides methylation that can influence gene expression, including transcriptional activators and repressors, RNA polymerases, microRNAs, histone acetylases and deacetylases, etc. In our study, we also identified several intergenic CpGs that were related to PCa recurrence, and it is possible that they may play a regulatory role, for example as gene enhancers.
For the comparison of African American patients with lethal PCa vs. no recurrence, a hypermethylated CpG was identified in the body of TNXB, a gene in the HLA region on chromosome 6 that encodes an extracellular matrix glycoprotein that appears to enhance tumor growth (34,35). A hypomethylated CpG was identified in the promotor region of FLNA, a gene that is important in cell proliferation, migration, and adhesion, as well as in tumor progression (36,37). In addition, hypermethylation of CpGs in the promotor of SLC25A20 (involved in metabolism) and in the gene body of LOC643387 (associated with neurodegenerative disorders) was also identified in the tumor tissues of men who died from PCa. EBF1, a transcriptional activator, was the only gene with aberrantly methylated CpG sites that was identified when comparing regional vs. localized disease stage in African Americans. These results point to candidate markers that can be further evaluated for their contribution to aggressive disease in African American PCa patients.
To our knowledge, this is the largest study of epigenome-wide DNA methylation profiles of African American PCa patients. The only other published study of epigenome-wide methylation results for African Americans included only three PCa patients (11). They reported differential promotor region CpG methylation levels in African American vs. European American patients (n=3) for a selected set of genes, and identified three genes (SNRPN, MST1R, and ABCG5) with higher promotor region CpG methylation levels in the African American patients that also corresponded with lower mRNA expression levels. We tested if there were significant differences in methylation level of CpGs in 23 of their 25 highlighted genes (CpG data were not available for IPO9 or TERF2IP in our samples). We found significant (p≤0.05) differences in methylation level according to ancestry for two of these genes; STOX7, and SNRPN (SNRPN methylation results, however, did not correspond with differences in mRNA expression). Other studies examined selected candidate genes for differential methylation levels in African American patients, with the advantage of not having to adjust for multiple testing on an epigenome-wide scale, and they reported significant results for several genes including APC, AR, CD44, NKX2-5, PMEPA1, RARB, RARB2, SPARC, and TIMP3 (20,21,24-26). Our study replicated results for PMEPA1 and TIMP3, with higher promotor region CpG methylation levels in the African American patients. We did not replicate results for the other CpGs in candidate genes. While our study supported some results reported by earlier studies, there were also several differences, which may in part be due to the examination of different CpG sites across studies. Other reasons for these discrepancies may relate to differences in sample size (e.g., Devaney et al. included only 3 African American PCa patients), heterogeneity of African American samples between studies (e.g., population substructure among African Americans from different geographic regions within the US), and differences in clinical or pathological features among patient groups.
In conclusion, this study identified several genes with differentially methylated CpG sites in tumor tissue from African American patients with more vs. less aggressive PCa features, and the majority of these changes appear to be specific to the African American patient population. Some of the genes with differentially methylated CpGs identified here have previously been associated with PCa and/or other cancers, while other genes influence biological processes related to tumor aggressiveness, including cellular metabolism, proliferation/cell cycle, DNA damage repair, and apoptosis. However, future work with a larger sample of African American patients is needed to replicate these results and elucidate the specific molecular mechanisms by which these differentially methylated CpGs contribute to prostate cancer progression.
4. Methods
4.1 Patient population
The 76 African American patients in this study were previously enrolled in either population-based PCa studies at the Fred Hutchinson Cancer Research Center (FHCRC) (n=44) or were treated at Eastern Virginia Medical School (EVMS) (n=32). All patients had radical prostatectomy as primary therapy. EVMS patients were selected based on recurrence status, with the aim of studying similar numbers of patient with recurrent vs. non-recurrent PCa. IRB approval for this study was granted by FHCRC, and participants signed statements of informed consent. For the FHCRC cohort, men were aged 40-64 years and diagnosed between 1993 and 1996 (38), or were 35-74 years of age and diagnosed between 2002 and 2005 (39). Patient baseline information and clinicopathological data were collected during in-person interviews and from the Seattle-Puget Sound SEER cancer registry, and follow-up information was obtained from two surveys completed by patients in 2004-2005 and in 2010-2011. Patients were classified as having PCa recurrence if they had a post-treatment PSA ≥ 0.2 ng/mL, received secondary treatment (e.g., radiation therapy, androgen deprivation therapy, orchiectomy, or chemotherapy at least 12 or more months after prostatectomy), had an MRI, CT, bone scan or biopsy that was positive for PCa, were told by a physician that their PCa had recurred, or died from PCa. Over an average follow-up period of 8.3 years, 5 African American patients in the FHCRC cohort had PCa recurrence and 20 had no evidence of recurrence. Recurrence status could not be determined for 19 men who did not complete either follow-up survey. The other population included a selected group of PCa patients who were diagnosed between 1992 and 2009 and were treated at EVMS. There were 18 patients with no evidence of PCa recurrence and 14 men with recurrence (including 12 patients who developed metastasis or died from PCa) in the EVMS cohort. Clinical data were available for these patients from a database of PCa patients treated at EVMS, and patients were followed for prostate cancer outcomes over an average period of 9.0 years.
4.2 Sample preparation and DNA methylation profiling
Formalin-fixed paraffin-embedded (FFPE) tumor tissue blocks from RP samples were obtained for patients ascertained from each institution, and were used to prepare hematoxylin and eosin (H&E) stained slides. Slides were reviewed by pathologists to confirm the presence and location of prostate adenocarcinoma, and two 1-mm cores were taken from areas containing ≥ 75% tumor cells from the dominant lesion for each patient. DNA was extracted using the RecoverAll Total Nucleic Acid Isolation Kit (Ambion/Applied Biosciences, Austin, TX), quantified (PicoGreen), and stored at -80°C. DNA samples were aliquoted onto 96-well plates and shipped to Illumina, Inc. (San Diego, CA) for DNA methylation profiling.
Samples were bisulfite converted using the EZ DNA Methylation Kit (Zymo Research, Irvine, CA) according to the manufacturer's instructions, and the Infinium® 450K Human Methylation450 BeadChip array (Illumina, Inc.) was used to measure DNA methylation levels at individual CpG sites (40). Blind duplicate (FHCRC=16, EVMS=7) and replicate (FHCRC=2, EVMS=3) samples from each patient population were included on the plates, along with Illumina controls and negative controls.
4.3 Data processing and statistical analysis
DNA methylation data were analyzed using R and the Bioconductor package minfi (41). Samples were considered failed if they had fewer than 95% of CpG sites with detection P-values <0.05. Individual CpG sites were filtered out if they had an average detection P-value >0.01. Based on these criteria, there were 477,460 CpGs sites available for further analysis. The data were normalized using SWAN (42), and ComBat (43) was used to remove potential batch effects. There were high correlations between duplicates (Pearson's r=0.96 to 0.99 for FHCRC; r=0.98 to 0.99 for EVMS) and technical replicates (r≥0.99 for FHCRC; r=0.98 for EVMS). Methylation β values that measure methylation intensity and range from 0 (unmethylated) to 1 (completely methylated) and methylation M values (the logit transformation of β values, and which approximate a normal distribution) were calculated for each CpG site (44).
To identify differentially methylated CpG sites, linear regression models were implemented in the limma Bioconductor package (45), using M values with age and study population included as covariates. The primary analysis focused on identifying differentially methylated CpGs for African American patients with high vs. low Gleason score, regional vs. local pathological tumor stage, PCa recurrence vs. no recurrence, and high vs. low tumor aggressiveness based on a composite variable [high = one or more of the following: PCa recurrence and/or death; Gleason score (7=4+3, 8-10), and/or regional pathological stage].
Genes that were identified as having a significantly differentially methylated CpG site(s) were evaluated for whether there was evidence for corresponding changes in mRNA levels for the FHCRC and EVMS patients. Gene expression profiling was done using the Whole-Genome DASL® HT Assay, Illumina, Inc., as previously described (46). Batch effects were removed using ComBat (43) and gene expression data were quantile normalized and log2 transformed using R statistical computing software (47). Spearman correlations between DNA methylation and mRNA expression levels were calculated using R (47).
A secondary analysis took the top-ranked CpGs identified in African Americans and evaluated whether these CpG sites were significantly differentially methylated in our European American PCa patients (n=479). These men were part of the FHCRC PCa studies (38,39) and their sample collection and DNA methylation profiling has been previously described (14).
Supplementary Material
Highlights.
Tumor DNA methylation is profiled in African American prostate cancer patients
Differentially methylated CpGs are identified for more aggressive disease features
CpG methylation level in GCK, CDKL2, PRDM13, and ZFR2 is associated with recurrence
DNA methylation status is associated with altered mRNA level for several genes
Results may advance knowledge of tumor aggressiveness African American patients
Acknowledgments
We thank Drs. Beatrice Knudson and Antonio Hurado-Coll for their assistance with the pathology, and Illumina, Inc. for supplying and performing the DNA methylation and gene expression arrays. Financial support was provided by grants from the National Cancer Institute: R01 CA056678, R01 CA092579, K05 CA175147, and P50 CA097186; with additional support from the Fred Hutchinson Cancer Research Center, the Intramural Program of the National Human Genome Research Institute, and the Prostate Cancer Foundation. Milan Geybels is the recipient of a Dutch Cancer Society Fellowship (BUIT 2014– 6645).
Abbreviations
- EVMS
Eastern Virginia Medical School
- FDR
False discovery rate
- FFPE
Formalin-fixed paraffin-embedded
- FHCRC
Fred Hutchinson Cancer Research Center
- H&E
Hematoxylin and eosin
- PCa
Prostate cancer
- RP
Radical prostatectomy
- TSS1500
201 to 1500 base pairs upstream of transcription start site
- TSS200
200 base pairs upstream of transcription start site
- UTR
untranslated region
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rebbeck TR, Devesa SS, Chang B, Bunker CH, Cheng I, Cooney KA, Eeles R, Fernandez P, Giri VN, Gueye SM, Haiman CA, Henderson B, Heyns CF, Hu JJ, Ingles SA, Isaacs W, Jalloh M, John EM, Kibel AS, Kidd LC, Layne P, Leach RJ, Neslund-Dudas C, Okobia MN, Ostrander EA, Park JY, Patrick AL, Phelan C, Ragin C, Roberts R, Rybicki BA, Stanford J, Strom SS, Thompson IM, Witte JS, Xu J, Yeboah E, Hsing AW, Zeigler-Johnson C. Global patterns of prostate cancer incidence, aggressiveness, and mortality in men of african descent. Prostate Cancer. 2013 [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65(1):5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 3.O'Keefe EB, Meltzer JP, Bethea TN. Health disparities and cancer: racial disparities in cancer mortality in the United States, 2000-2010. Frontiers in public health. 2015;3:51. doi: 10.3389/fpubh.2015.00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chornokur G, Dalton K, Borysova ME, Kumar NB. Disparities at presentation, diagnosis, treatment, and survival in African American men, affected by prostate cancer. Prostate. 2011;71(9):985–997. doi: 10.1002/pros.21314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taksler GB, Keating NL, Cutler DM. Explaining racial differences in prostate cancer mortality. Cancer. 2012;118(17):4280–4289. doi: 10.1002/cncr.27379. [DOI] [PubMed] [Google Scholar]
- 6.Steck SE, Arab L, Zhang H, Bensen JT, Fontham ET, Johnson CS, Mohler JL, Smith GJ, Su JL, Trump DL, Woloszynska-Read A. Association between Plasma 25-Hydroxyvitamin D, Ancestry and Aggressive Prostate Cancer among African Americans and European Americans in PCaP. PLoS One. 2015;10(4):e0125151. doi: 10.1371/journal.pone.0125151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ziehr DR, Mahal BA, Aizer AA, Hyatt AS, Beard CJ, D'Amico AV, Choueiri TK, Elfiky A, Lathan CS, Martin NE, Sweeney CJ, Trinh QD, Nguyen PL. Income inequality and treatment of African American men with high-risk prostate cancer. Urol Oncol. 2015;33(1):18 e17–13. doi: 10.1016/j.urolonc.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 8.Mahal BA, Ziehr DR, Aizer AA, Hyatt AS, Lago-Hernandez C, Choueiri TK, Elfiky AA, Hu JC, Sweeney CJ, Beard CJ, D'Amico AV, Martin NE, Kim SP, Lathan CS, Trinh QD, Nguyen PL. Racial disparities in an aging population: the relationship between age and race in the management of African American men with high-risk prostate cancer. Journal of geriatric oncology. 2014;5(4):352–358. doi: 10.1016/j.jgo.2014.05.001. [DOI] [PubMed] [Google Scholar]
- 9.Powell IJ, Bollig-Fischer A. Minireview: the molecular and genomic basis for prostate cancer health disparities. Molecular endocrinology (Baltimore, Md) 2013;27(6):879–891. doi: 10.1210/me.2013-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Castro P, Creighton CJ, Ozen M, Berel D, Mims MP, Ittmann M. Genomic profiling of prostate cancers from African American men. Neoplasia. 2009;11(3):305–312. doi: 10.1593/neo.81530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Devaney JM, Wang S, Furbert-Harris P, Apprey V, Ittmann M, Wang BD, Olender J, Lee NH, Kwabi-Addo B. Genome-wide differentially methylated genes in prostate cancer tissues from African-American and Caucasian men. Epigenetics. 2015;10(4):319–328. doi: 10.1080/15592294.2015.1022019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349(21):2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 13.Baylin SB, Herman JG. DNA hypermethylation in tumorigenesis: epigenetics joins genetics. Trends Genet. 2000;16(4):168–174. doi: 10.1016/s0168-9525(99)01971-x. [DOI] [PubMed] [Google Scholar]
- 14.Stott-Miller M, Zhao S, Wright JL, Kolb S, Bibikova M, Klotzle B, Ostrander EA, Fan JB, Feng Z, Stanford JL. Validation study of genes with hypermethylated promoter regions associated with prostate cancer recurrence. Cancer Epidemiol Biomarkers Prev. 2014;23(7):1331–1339. doi: 10.1158/1055-9965.EPI-13-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perry AS, Watson RW, Lawler M, Hollywood D. The epigenome as a therapeutic target in prostate cancer. Nature reviews Urology. 2010;7(12):668–680. doi: 10.1038/nrurol.2010.185. [DOI] [PubMed] [Google Scholar]
- 16.Lee WH, Morton RA, Epstein JI, Brooks JD, Campbell PA, Bova SG, Hsieh WS, Issacs WB, Nelson WG. Cytidine methylation of regulatory sequences near the p-class glutathione S-transferase gene accompanies human prostatic carcinogenesis. Proc Natl Acad Sci USA. 1994;91:11733–11737. doi: 10.1073/pnas.91.24.11733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cho NY, Kim BH, Choi M, Yoo EJ, Moon KC, Cho YM, Kim D, Kang GH. Hypermethylation of CpG island loci and hypomethylation of LINE-1 and Alu repeats in prostate adenocarcinoma and their relationship to clinicopathological features. The Journal of pathology. 2007;211(3):269–277. doi: 10.1002/path.2106. [DOI] [PubMed] [Google Scholar]
- 18.Richiardi L, Fiano V, Vizzini L, De Marco L, Delsedime L, Akre O, Tos AG, Merletti F. Promoter methylation in APC, RUNX3, and GSTP1 and mortality in prostate cancer patients. J Clin Oncol. 2009;27(19):3161–3168. doi: 10.1200/JCO.2008.18.2485. [DOI] [PubMed] [Google Scholar]
- 19.Richiardi L, Fiano V, Grasso C, Zugna D, Delsedime L, Gillio-Tos A, Merletti F. Methylation of APC and GSTP1 in non-neoplastic tissue adjacent to prostate tumour and mortality from prostate cancer. PLoS One. 2013;8(7):e68162. doi: 10.1371/journal.pone.0068162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Woodson K, Hayes R, Wideroff L, Villaruz L, Tangrea J. Hypermethylation of GSTP1, CD44, and E-cadherin genes in prostate cancer among US Blacks and Whites. Prostate. 2003;55(3):199–205. doi: 10.1002/pros.10236. [DOI] [PubMed] [Google Scholar]
- 21.Woodson K, Hanson J, Tangrea J. A survey of gene-specific methylation in human prostate cancer among black and white men. Cancer Lett. 2004;205(2):181–188. doi: 10.1016/j.canlet.2003.11.027. [DOI] [PubMed] [Google Scholar]
- 22.Enokida H, Shiina H, Urakami S, Igawa M, Ogishima T, Pookot D, Li LC, Tabatabai ZL, Kawahara M, Nakagawa M, Kane CJ, Carroll PR, Dahiya R. Ethnic group-related differences in CpG hypermethylation of the GSTP1 gene promoter among African-American, Caucasian and Asian patients with prostate cancer. Int J Cancer. 2005;116(2):174–181. doi: 10.1002/ijc.21017. [DOI] [PubMed] [Google Scholar]
- 23.Das PM, Ramachandran K, Vanwert J, Ferdinand L, Gopisetty G, Reis IM, Singal R. Methylation mediated silencing of TMS1/ASC gene in prostate cancer. Molecular cancer. 2006;5:28. doi: 10.1186/1476-4598-5-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kwabi-Addo B, Wang S, Chung W, Jelinek J, Patierno SR, Wang BD, Andrawis R, Lee NH, Apprey V, Issa JP, Ittmann M. Identification of differentially methylated genes in normal prostate tissues from African American and Caucasian men. Clin Cancer Res. 2010;16(14):3539–3547. doi: 10.1158/1078-0432.CCR-09-3342. [DOI] [PubMed] [Google Scholar]
- 25.Tang D, Kryvenko ON, Mitrache N, Do KC, Jankowski M, Chitale DA, Trudeau S, Rundle A, Belinsky SA, Rybicki BA. Methylation of the RARB gene increases prostate cancer risk in black Americans. J Urol. 2013;190(1):317–324. doi: 10.1016/j.juro.2013.01.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sharad S, Ravindranath L, Haffner MC, Li H, Yan W, Sesterhenn IA, Chen Y, Ali A, Srinivasan A, McLeod DG, Yegnasubramanian S, Srivastava S, Dobi A, Petrovics G. Methylation of the PMEPA1 gene, a negative regulator of the androgen receptor in prostate cancer. Epigenetics. 2014;9(6):918–927. doi: 10.4161/epi.28710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murad AS, Smith GD, Lewis SJ, Cox A, Donovan JL, Neal DE, Hamdy FC, Martin RM. A polymorphism in the glucokinase gene that raises plasma fasting glucose, rs1799884, is associated with diabetes mellitus and prostate cancer: findings from a population-based, case-control study (the ProtecT study) International journal of molecular epidemiology and genetics. 2010;1(3):175–183. [PMC free article] [PubMed] [Google Scholar]
- 28.Kolenko V, Teper E, Kutikov A, Uzzo R. Zinc and zinc transporters in prostate carcinogenesis. Nature reviews Urology. 2013;10(4):219–226. doi: 10.1038/nrurol.2013.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sarafanov AG, Todorov TI, Centeno JA, Macias V, Gao W, Liang WM, Beam C, Gray MA, Kajdacsy-Balla AA. Prostate cancer outcome and tissue levels of metal ions. Prostate. 2011;71(11):1231–1238. doi: 10.1002/pros.21339. [DOI] [PubMed] [Google Scholar]
- 30.Lucarelli G, Rutigliano M, Galleggiante V, Giglio A, Palazzo S, Ferro M, Simone C, Bettocchi C, Battaglia M, Ditonno P. Metabolomic profiling for the identification of novel diagnostic markers in prostate cancer. Expert Rev Mol Diagn. 2015:1–14. doi: 10.1586/14737159.2015.1069711. [DOI] [PubMed] [Google Scholar]
- 31.Rishi I, Baidouri H, Abbasi JA, Bullard-Dillard R, Kajdacsy-Balla A, Pestaner JP, Skacel M, Tubbs R, Bagasra O. Prostate cancer in African American men is associated with downregulation of zinc transporters. Appl Immunohistochem Mol Morphol. 2003;11(3):253–260. doi: 10.1097/00129039-200309000-00009. [DOI] [PubMed] [Google Scholar]
- 32.Herbst A, Rahmig K, Stieber P, Philipp A, Jung A, Ofner A, Crispin A, Neumann J, Lamerz R, Kolligs FT. Methylation of NEUROG1 in serum is a sensitive marker for the detection of early colorectal cancer. The American journal of gastroenterology. 2011;106(6):1110–1118. doi: 10.1038/ajg.2011.6. [DOI] [PubMed] [Google Scholar]
- 33.Keil KP, Vezina CM. DNA methylation as a dynamic regulator of development and disease processes: spotlight on the prostate. Epigenomics. 2015;7(3):413–425. doi: 10.2217/epi.15.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yuan Y, Nymoen DA, Stavnes HT, Rosnes AK, Bjorang O, Wu C, Nesland JM, Davidson B. Tenascin-X is a novel diagnostic marker of malignant mesothelioma. Am J Surg Pathol. 2009;33(11):1673–1682. doi: 10.1097/PAS.0b013e3181b6bde3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng SL, Hsing AW, Sun J, Chu LW, Yu K, Li G, Gao Z, Kim ST, Isaacs WB, Shen MC, Gao YT, Hoover RN, Xu J. Association of 17 prostate cancer susceptibility loci with prostate cancer risk in Chinese men. Prostate. 2010;70(4):425–432. doi: 10.1002/pros.21076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robertson SP, Twigg SR, Sutherland-Smith AJ, Biancalana V, Gorlin RJ, Horn D, Kenwrick SJ, Kim CA, Morava E, Newbury-Ecob R, Orstavik KH, Quarrell OW, Schwartz CE, Shears DJ, Suri M, Kendrick-Jones J, Wilkie AO Group OP-sDCC. Localized mutations in the gene encoding the cytoskeletal protein filamin A cause diverse malformations in humans. Nat Genet. 2003;33(4):487–491. doi: 10.1038/ng1119. [DOI] [PubMed] [Google Scholar]
- 37.Kim H, Sengupta A, Glogauer M, McCulloch CA. Filamin A regulates cell spreading and survival via beta1 integrins. Experimental cell research. 2008;314(4):834–846. doi: 10.1016/j.yexcr.2007.11.022. [DOI] [PubMed] [Google Scholar]
- 38.Stanford JL, Wicklund KG, McKnight B, Daling JR, Brawer MK. Vasectomy and risk of prostate cancer. Cancer Epidemiol Biomarkers Prev. 1999;8(10):881–886. [PubMed] [Google Scholar]
- 39.Agalliu I, Salinas CA, Hansten PD, Ostrander EA, Stanford JL. Statin use and risk of prostate cancer: results from a population-based epidemiologic study. Am J Epidemiol. 2008;168(3):250–260. doi: 10.1093/aje/kwn141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bibikova M, Barnes B, Tsan C, Ho V, Klotzle B, Le J, Delano D, Zhang L, Schroth GP, Gunderson KL, Fan JB, Shen R. High density DNA methylation array with single CpG site resolution. Genomics. 2011;98(4):288–295. doi: 10.1016/j.ygeno.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 41.Aryee MJ, J AE, Corrada-Bravo H, Ladd-Acosta C, Feinberg AP, Hansen KD, Irizarry RA. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30(May 15):1363–1369. doi: 10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maksimovic J, Gordon L, Oshlack A. SWAN: Subset-quantile within array normalization for Illumina infinium HumanMethylation450 BeadChips. Genome biology. 2012;13(6):R44. doi: 10.1186/gb-2012-13-6-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8(1):118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 44.Du P, Zhang X, Huang CC, Jafari N, Kibbe WA, Hou L, Lin SM. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC bioinformatics. 2010;11:587. doi: 10.1186/1471-2105-11-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43 doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rubicz R, Zhao S, April C, Wright JL, Kolb S, Coleman I, Lin DW, Nelson PS, Ostrander EA, Feng Z, Fan JB, Stanford JL. Expression of cell cycle-regulated genes and prostate cancer prognosis in a population-based cohort. Prostate. 2015;75(13):1354–1362. doi: 10.1002/pros.23016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Team RDC. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2012. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


