Abstract
Animal cells use traction forces to sense the mechanics and geometry of their environment. Measuring these traction forces requires a workflow combining cell experiments, image processing and force reconstruction based on elasticity theory. Such procedures have already been established mainly for planar substrates, in which case one can use the Green's function formalism. Here we introduce a workflow to measure traction forces of cardiac myofibroblasts on non-planar elastic substrates. Soft elastic substrates with a wave-like topology were micromoulded from polydimethylsiloxane and fluorescent marker beads were distributed homogeneously in the substrate. Using feature vector-based tracking of these marker beads, we first constructed a hexahedral mesh for the substrate. We then solved the direct elastic boundary volume problem on this mesh using the finite-element method. Using data simulations, we show that the traction forces can be reconstructed from the substrate deformations by solving the corresponding inverse problem with an L1-norm for the residue and an L2-norm for a zeroth-order Tikhonov regularization. Applying this procedure to the experimental data, we find that cardiac myofibroblast cells tend to align both their shapes and their forces with the long axis of the deformable wavy substrate.
Keywords: cell adhesion, cell mechanics, traction force microscopy, elastic substrates, finite-element method, elasticity theory
1. Introduction
Animal tissue cells sense the mechanical and geometrical features of their environment by applying traction forces to the extracellular matrix. Various studies over recent decades have demonstrated the importance of tissue mechanics for cell behaviour, including cell adhesion, migration, proliferation, differentiation and fate [1–3]. It has already been shown that cells respond sensitively to the rigidity of their environment, with larger spreading area and higher force generation on stiffer substrates [4–6]. Even more dramatically, essential cell processes such as cell differentiation are controlled by substrate stiffness [7–9]. Because environmental stiffness is a passive property of the environment, cells have to actively pull on it to determine its magnitude [10].
Cellular traction forces are used not only to sense extracellular stiffness but also to sense geometrical properties of the environment [11–13]. This is most evident for adhesive patterns, whose geometry also determines cell responses such as cell survival and differentiation [7,14,15]. However, geometry sensing also includes cell sensitivity to topographical features of the environment, which spans several orders of magnitude, ranging from the molecular up to the cellular scale [16–18]. It has been shown that cell differentiation can be controlled also by nanotopography [19,20]. Regarding the cytoskeletal response, it has been found that cells tend to align with grooves on substrates with corresponding nanotopography and microtopography [21–23]. Another physiologically relevant example is topography-driven polarity guidance and directional growth of neurons [24]. Early examples of topography sensing on the micrometre scale are studies that showed the alignment of actin microfilaments in cells adhered to microcylinders [25,26]. Interestingly, it was found that cells tend to align either parallel or orthogonal to the direction of highest curvature, depending on the cell type. Inspired by these observations mechanical cell models have been proposed that suggest the importance of mechanical stress in the cytoskeleton for the detection of and response to curved structures [27,28].
Recently, the combined effect of stiffness and geometry has been studied by engineering topographic features into polyacrylamide (PAA) substrates, which as hydrogels, however, tend to swell in medium and, therefore, change dimensions [29]. Interestingly, it was found that cells align with the topographic features independent of stiffness. However, no attempt has been made to measure cellular traction forces in these experiments. In general, there are many physiological situations in which cells are exposed to mechanical and geometrical cues simultaneously, and, therefore, will use cellular traction forces to sense them both at the same time. An interesting example is podocytes, which are epithelial cells lining the basement membrane of the glomerular capillaries in kidneys, which have a wavy shape [30]. However, a set-up that allows the quantitative measurement of forces in such situations is still missing, despite recent progress in measuring cellular traction forces on planar substrates.
During the last three decades, the measurement of cellular forces (traction force microscopy; TFM) has become a mature research field [31–34]. The standard set-up uses a planar elastic substrate whose deformations are tracked using embedded marker beads. Standard choices for the substrate material are PAA or cross-linked polydimethylsiloxane (PDMS). For sufficiently thick substrates, one then can use the Green's function of an elastic half-space to relate these deformations to traction forces (GF-TFM) [35]. For a thin substrate, the corresponding Green's function is also known [32,36,37]. TFM procedures can be made quite rapid by inverting the elastic equations in Fourier space (Fourier transform traction cytometry; FTTC), which is a special case of GF-TFM [38]. Traditionally, cell forces had been reconstructed only in the x–y-plane of a planar substrate (with the z-direction denoting the normal direction), but, over recent years, the Green's function approach has been extended to also reconstruct the z-forces that cells exert on the substrate [39]. Alternatively, one can use the finite-element method (FEM) to reconstruct these z-forces (FEM-TFM) [40,41]. Within the FEM approach, one does not rely on the analytical form of a Green's function, but uses numerical solutions to the mechanical problem interpolated on a suitable chosen grid [42–44]. Another alternative to GF-TFM is the direct method, in which deformations are directly converted into a stress tensor, from which the traction forces are extracted [45,46].
If one aims at implementing TFM for non-planar substrates, of these three methods (GF-TFM, FEM-TFM and direct TFM) only the second seems feasible. First, it is notoriously hard to analytically calculate Green's functions for non-planar (e.g. wavy) surfaces, thus ruling out GF-TFM. As we will see below, wavy substrates lead to rather noisy displacement data, which are hard to deal with in direct TFM, because it relies on constructing derivatives of the measured data. Therefore, we opted for an approach using FEM-TFM, which needs more computer time than traditional GF-TFM, but offers the same level of robustness. We first developed a novel experimental technique to prepare curved micromoulded PDMS substrates with embedded fluorescent marker beads matching the requirements of TFM application. In contrast with the PAA hydrogels often used for studies of mechanosensing, the micromoulded PDMS substrates used here do not suffer from swelling, and, therefore, present time-independent geometrical cues to the cells. However, because the point spread function in such a substrate varies in space, special procedures have to be used for tracking the marker beads. Because here we focus on measuring traction forces, we did not vary the mechanical stiffness of the substrates. On the computational side, we established a complete workflow to reconstruct cellular traction fields on such non-planar substrates. The core of this technique is a parallelized optimization framework that efficiently implements FEM to reconstruct cellular traction forces in three dimensions. We validated our procedures by reconstructing simulated traction patterns under various experimental conditions. In particular, we show that the use of the L1-norm for the definition of the residue strongly improves our force reconstruction because it deals better with outliers than the L2-norm. We then applied TFM to cardiac myofibroblasts cultured on curved elastic substrates, thus complementing a traditional contact guidance experiment with measurements of cell traction forces. By comparing both polarization of the cytoskeleton and the distribution of cellular traction, we show that cells adjust not only their morphology but also their moments of traction force to geometrical properties of their surroundings.
2. Material and methods
2.1. Experimental procedures
Elastomeric substrates were made of Sylgard 184 Silicone Elastomer Kit (Dow Corning GmbH, Wiesbaden, Germany) as described previously [31]. In brief, both components (base and cross-linker) of the elastomer kit were mixed at various ratios to generate after cross-linking either stamps from vinyl disc masters (10 : 1) or cell culture substrates of stiffness 15 kPa (50 : 1). For stamp manufacturing, round polypropylene rings (diameter 10 mm) were placed on top of the vinyl disc and cross-linked at 40°C for 4 h. Stamps were peeled off and silanized with trichlorosilane. For force analysis, cell culture substrates were equipped with red fluorescent beads (0.1 µm diameter, non-modified beads; Magspheres, CA, USA), resulting in a dense microstructured volume (typical bead distance 4 µm). Base oil was mixed with a 1 : 50 dilution of beads in methanol. The modified base oil was incubated at 60°C for 3 h to evaporate methanol. The desired amount of cross-linker was added to the modified base oil and mixed intensively. The stamp was placed onto an 80 µm coverslip with a drop of silicone oil mixture. Layer thickness was adjusted to 80 µm using glass slices of the same defined thickness (Menzel GmbH, Braunschweig, Germany) as spacers. Cross-linking of silicone oil was performed at 60°C for 16 h. After curing stamps were carefully peeled off and structured elastomeric substrates were glued to the bottom of 3.5 cm Petri dishes to cover predrilled holes [47]. The mechanical properties of all elastomeric mixing ratios were characterized as described, resulting in elasticities as given above and a Poisson's ratio of 0.5.
Cardiac fibroblasts were isolated from 19-day-old Wistar rat embryos as described previously [48,49]. Primary cardiac fibroblasts were cultured for an additional 5 days at 37°C and 5% CO2 in a humidified incubator on standard polystyrene cell culture dishes to induce their differentiation to myofibroblasts, which have more prominent focal adhesions and stress fibres. Cells were trypsinized and subsequently transfected using Nucleofector technology (Lonza-Amaxa Systems, Cologne, Germany). A total of 106 cells were resuspended in 100 µl liposome solution containing 2 µg purified plasmid DNA (GFP-VASP or GFP-Vinculin). After transfection, resuspended myofibroblasts were seeded on fibronectin-coated (2.5 µg cm−2) (BD Bioscience, Franklin Lakes, NJ, USA) silicone rubber wave substrates at a density of 2 × 104 per sample for traction force microscopy. The cells were seeded for at least 24 h before the measurements were taken.
Live cell analyses were performed at 37°C and 5% CO2 using an inverse confocal laser scanning microscope (LSM 710; Zeiss, Germany; software ZEN 2011) equipped with live cell imaging accessories and a 63 × Planapochromat oil immersion objective (Ph3, NA 1.4; Zeiss). Confocal three-dimensional (3D) micrographs were taken using appropriate laser and filter settings for detection of green and red fluorescent light. Z-stacks of approximately 50 µm with optimized overlap were taken. As a reference value, z-stacks without cells (peeled off with a glass micro needle) were performed with the same parameters. Acquisition of a single slice took 8 s and we typically acquired 100 slices with a distance of around 0.4 µm. Thus, the acquisition time for one stack was around 15 min.
2.2. Bead tracking
Bead tracking for wavy substrates is challenging because of the long stack acquisition time leading to considerable drift not only between the deformed and the reference states, but even within one stack. We believed that this drift did not result from cell activity, because 24 h after spreading they were quiescent. Moreover, the observed drift was similar at any depth in the substrate. As another special feature of wavy substrates, we observed that, in contrast to planar substrates, the point spread function varied in space.
Our bead tracking routine consists of three steps: determination of image drift, single bead localization and feature vector-based tracking of bead movement. To reduce noise, the stacks were first binomially filtered with a 3 × 3 × 3 filter kernel, voxel sizes 0.1 µm or 0.11 µm lateral and 0.39 µm or 0.56 µm vertical. Then spots (usually four) in the undeformed corners of the image were marked (in the x–y-plane). At these spots, a cuboid (volume of interest; VOI) with 10% of the image size in each dimension was cropped from the first image and cross-correlated with the second image to obtain the drift. Because the drift in each slice of the stack can differ, this VOI is cropped along each slice and so the drift is calculated for every slice separately. The area in which the VOI is cross-correlated with the second stack has to be larger than the maximum shift of the two image stacks. To decrease the calculation time for the cross correlation (CC), the size of the image stack and the VOIs are initially reduced by two levels of a Gaussian pyramid. The positions of the maximum CC values are then used to calculate the drift on the full image to obtain the exact drift. This can be done in a very small search area (5 × 5 × 5 pixels) around the previously calculated positions. To obtain a subpixel accurate positioning of the VOIs, parabolas were fitted through three points (maximum of CC and both neighbours) in the x, y- and z-directions, respectively. The extreme values of the fits are defined as the subpixel accurate VOI positions. The CC and parabola fitting is also done in the first image stack to obtain a subpixel accurate position there. The mean difference between the VOI positions in the first and in the second stack defines the drift, separately for each slice.
To localize beads in each image stack, both stacks were first filtered with a 7 × 7 and a 31 × 31 binomial filter. These filtered images were then subtracted, to obtain a bandpass-filtered stack, with negative grey values set to zero. Then a VOI that represents one bead in three dimensions is selected manually and fitted to a 3D Gaussian. The fit is used as a reference template to find other beads that match the template. The fitted Gaussian is cross-correlated with the first and the second stack. Then the cross-correlated dataset is segmented with a threshold (0.7) and each segment is labelled. At the maximum CC value in each labelled segment, the data are again fitted to a Gaussian and the centre of mass of that fit is defined as the bead position.
Once the beads are localized in both stacks, they have to be correlated to each other. Here, we adapted a feature vector-based tracking method as published previously [43]. Before we apply the tracking procedure, the determined drift is subtracted from the bead positions found in the second stack. Then the distance of a bead (B1) from stack 1 to all other beads in its neighbourhood (50 × 50 pixels in the x–y- and 20 pixels in the z-direction) is calculated. Vectors are created from B1 to the, at most, seven beads with the lowest distance. These vectors are added to beads (B2) in stack 2 (again only in a neighbourhood of 50 × 50 × 20 pixels around the position of B1) and a cubic surrounding (e.g. 7 × 7 × 7 pixels) is created at each end of the vector. If there is a bead located within this surrounding, a hit is counted. The more hits that are counted, the bigger is the probability that bead B1 corresponds to B2. The bead B2 with the most hits (at least three of seven) is assigned to bead B1. If two or more beads B2 have the same number of hits, the one with the lowest deviation at the vector ends is chosen.
2.3. Reconstruction of substrate shape
As a pre-processing step for the traction reconstruction we need to determine the substrate shapes. Although the substrate preparation described above leads to reproducible samples with micrometre accuracy, shape variations and material relaxation after mould removal occasionally change the final shape. The latter effect can in principle be predicted theoretically [50]; however, in practice local shape variations occur and make the use of theoretical shape predictions difficult. For that reason, we determine the substrate shape for each individual TFM dataset by image processing of the relaxed configuration of the marker beads, which are distributed sufficiently homogeneously in the substrate as to carve out the substrate shape.
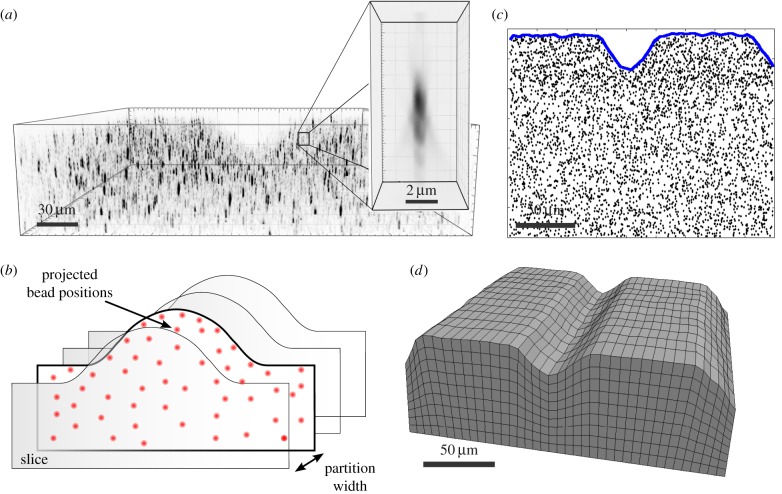
We developed a custom mesh generation program as illustrated in figure 1. In figure 1a, we show an image of the marker beads; the inset shows the strong anisotropy of the point spread function. First, bead locations are partitioned into sections along the long axis of the wavy pattern that are separated by a partition width w. In a second step, bead positions are mapped to each section plane (normal direction pointing towards the partition direction; figure 1b). By doing this, we end up with a set of separated slices associated with two-dimensional (2D) bead distributions. After that, we calculate the 2D hull for each bead distribution using the 2D α-shape algorithm [51] implemented in the open-source computational geometry algorithm library (CGAL) (figure 1c). After determination of the hull for each segmented slice, we again stitch the determined contours together, separated by the partition width w. We then form a 3D hexahedral mesh that approximates the substrate shape. In this particular step, the program uses the open-source mesh generator GMSH.1 A representative result for the described procedure is depicted in figure 1d.
Figure 1.
Reconstruction of substrate shape from bead positions. (a) Image of the marker beads. The inset shows the anisotopic nature of the point spread function. (b) The substrate contour is partitioned into volumeric slices along a lateral axis. Bead displacements within the partitioned volumes are projected to the boundary plane. (c) The 2D α-shape algorithm is applied to projected bead positions in order to determine the envelope. (d) From multiple envelopes a 3D hexahedral substrate mesh is reconstructed. Meshing with gmesh and visualization with paraview. (Online version in colour.)
2.4. Direct elastic problem and finite-element method implementation
Traction reconstruction is established by solving an inverse elastic boundary volume problem (BVP). We first describe the corresponding direct BVP. Because Green's functions are not known for the wavy shapes considered here, we use the FEM. In principle, this allows us to also use nonlinear formulations for large deformations and nonlinear materials. However, here we deal with linear material (PDMS) and small strains, thus the linear formulation is sufficient. We, therefore, have to solve the Navier–Lamé equation for the displacement field u(x) in the computational domain Ω,
| 2.1 |
Here λ and μ are the Lamé coefficients. Traction exerted by cells is applied to the top surface 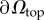 of the substrate volume in the reference state Ω. Accordingly, the traction field t(x) enters as the stress boundary condition,
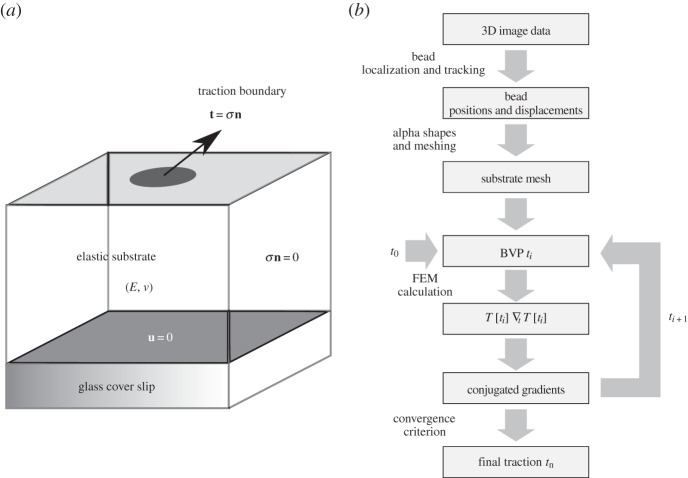
of the substrate volume in the reference state Ω. Accordingly, the traction field t(x) enters as the stress boundary condition, 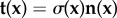 , where n is the normal vector of the unit surface element and σ(x) is the substrate Cauchy stress tensor. As illustrated in figure 2a, we choose appropriate mixed boundary conditions at the remaining parts of the boundary arriving at a well-defined BVP. At the bottom surface we require zero displacement, u(x) = 0, due to rigid coupling between the soft elastomeric substrate and the underlying rigid glass coverslip. As the used mesh represents a cut-out of the substrate, which is largely extended in lateral directions, proper boundary conditions at the side surfaces are applied. In the case of an infinite half-space, an appropriate boundary condition for the side surfaces of the cut-out would be a counter stress of the same magnitude. This, however, would lead to an undesired recursive problem. Here we use vanishing stress boundary conditions, σn = 0, for sufficiently large cut-outs. Based on the knowledge that the displacement field monotonically decays at least as 1/r, the influence of boundary conditions can be neglected for sufficient large cut-outs. Therefore, we occasionally extend the substrate mesh in the lateral directions to ensure the repression of boundary effects.
, where n is the normal vector of the unit surface element and σ(x) is the substrate Cauchy stress tensor. As illustrated in figure 2a, we choose appropriate mixed boundary conditions at the remaining parts of the boundary arriving at a well-defined BVP. At the bottom surface we require zero displacement, u(x) = 0, due to rigid coupling between the soft elastomeric substrate and the underlying rigid glass coverslip. As the used mesh represents a cut-out of the substrate, which is largely extended in lateral directions, proper boundary conditions at the side surfaces are applied. In the case of an infinite half-space, an appropriate boundary condition for the side surfaces of the cut-out would be a counter stress of the same magnitude. This, however, would lead to an undesired recursive problem. Here we use vanishing stress boundary conditions, σn = 0, for sufficiently large cut-outs. Based on the knowledge that the displacement field monotonically decays at least as 1/r, the influence of boundary conditions can be neglected for sufficient large cut-outs. Therefore, we occasionally extend the substrate mesh in the lateral directions to ensure the repression of boundary effects.
Figure 2.
Illustration of the direct boundary volume problem (BVP) and the work flow to solve the corresponding inverse problem. (a) Direct elastic BVP with mixed boundary conditions. The substrate elasticity is defined by Young's modulus E and Poisson's ratio v. Boundary conditions: bottom—zero displacement (u = 0), sides—zero stress (σn = 0), top—traction field (t(x) = σn). (b) Work flow of traction reconstruction (inverse BVP). Starting from the 3D image stack of raw data several steps are processed: 3D bead tracking, substrate mesh generation, FEM calculations and numeric traction optimization.
For a given traction pattern t(x), the direct BVP formulated in equation (2.1) is now solved by means of the FEM as typically applied to elastic problems [52–54]. For this purpose, we transform the elastic equations into the weak form and use linear shape functions on a hexahedral lattice to arrive at an algebraic problem (the mathematical details are provided as the electronic supplementary material). The algebraic equations are solved with the conjugated gradient (CG) method. Alternatively, it is also possible to directly invert them by means of, for example, Gauss elimination. The achieved solution can then be used to interpolate the displacement to any position within the domain Ω. For the implementation of our FEM approach, we used the FEM C++ library Deal.II [55]. It provides the essential set of features to achieve an FEM calculation, for instance managing local and global indices, matrix manipulation feature, Gauss quadrature, coordinate transformations and solvers for the linear algebraic system.
2.5. Inverse elastic problem
The inversion of the direct BVP is in general ill-posed, which implies that no unique or sufficiently stable solution exists. Therefore, regularization methods have to be used to render the inverse problem stable. According to Tikhonov & Arsenin [56], the regularized inverse problems can be formulated as a minimization problem and in TFM the corresponding Tikhonov functional 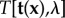 has the general form
has the general form
| 2.2 |
where 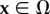 and
and 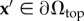 .
. 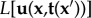 represents an error estimate that assigns a value to differences between calculated and measured displacements (the residue). The larger the deviation, the larger is the scalar value the estimate returns. The second term
represents an error estimate that assigns a value to differences between calculated and measured displacements (the residue). The larger the deviation, the larger is the scalar value the estimate returns. The second term 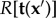 is a penalty functional introduced to recover a well-posed solution (the regularization term) [57].
is a penalty functional introduced to recover a well-posed solution (the regularization term) [57].
We discretized the traction field t(x) according to the FEM mesh in order to set up a finite space of parameters. For this purpose, we use interpolation based on shape functions. Hence, the entire field is characterized by a set of fixed point values ti, with 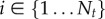 , which represent the degrees of freedom (d.f.) for the considered optimization problem. As we use linear shape functions, fixed point values are associated with nodal positions on the top surface of the FEM mesh
, which represent the degrees of freedom (d.f.) for the considered optimization problem. As we use linear shape functions, fixed point values are associated with nodal positions on the top surface of the FEM mesh 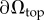 . Thus, the total number of optimization parameters Nt is determined by the number of surface mesh vertices
. Thus, the total number of optimization parameters Nt is determined by the number of surface mesh vertices 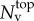 , and the number of space dimensions (Nd = 3),
, and the number of space dimensions (Nd = 3), 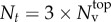 . The discretized version of the Tikhonov functional then reads
. The discretized version of the Tikhonov functional then reads 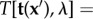
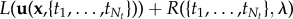 .
.
We next need to define the form of the residue and the regularization term in equation (2.2). Standard TFM uses the least squares estimate 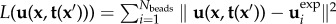 to measure deviations between measured and computed displacements. Additionally, most methods use zeroth-order Tikhonov regularization
to measure deviations between measured and computed displacements. Additionally, most methods use zeroth-order Tikhonov regularization 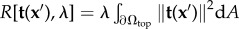 . This enforces a smooth traction solution by penalizing the amount of total force and thus represents the simplest approach to repressing noise-induced fluctuations [35,58,59]. Thus in the standard approach, both terms employ the L2-norm. Here we write a more general form,
. This enforces a smooth traction solution by penalizing the amount of total force and thus represents the simplest approach to repressing noise-induced fluctuations [35,58,59]. Thus in the standard approach, both terms employ the L2-norm. Here we write a more general form,
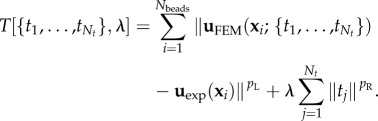 |
2.3 |
The choices p = 1 and p = 2 correspond to the L1- and L2-norms, respectively. Below we will always use the standard choice pR = 2 for the regularization term. For the residue, we will first use the standard choice pL = 2, which corresponds to the least squares estimator. From a statistical point of view, the least squares estimator can be derived as the maximum-likelihood estimate (MLE) for Gaussian distributed errors [60]. Later, we will argue that pL = 1 (L1-norm for the residue) is a better choice in our case, because it deals better with the outliers in our experimental data.
The form written in equation (2.3) implicitly assumes an isotropic error (same error distribution in all spatial directions). In our case, this assumption is not satisfied anymore, due to a reduced resolution in the z-direction (figure 1a). Therefore, we introduce a scaling procedure that weights the estimate contribution due to their relative accuracy. In detail, the z-contribution is weighted with a factor of 1/3 compared with the x- and y-contributions, which are assumed to have the same weight.
Owing to repeated time-consuming FEM calculations of the direct BVP during minimization of the Tikhonov functional, efficient computation is a key issue in solving the inverse problem. The overall computation time depends mainly on the number of traction d.f., Nt. Hence, a major objective was to achieve the best possible local accuracy for a given number of d.f., Nt. In order to achieve this demand we used predefined mesh refinement and adaptive local mesh refinement (h-refinement). Mesh refinement is employed by selectively dividing volume elements into smaller elements, which effectively increases the density of d.f. The idea behind this is to use local variations in the mesh size to concentrate d.f. at regions with higher levels of t. Other regions far away from the traction sources remain coarser at the same time. For the sake of completeness, we want to mention the alternative of local polynomial refinement, which describes local variation of the polynomial degree of used shape functions, called p-refinement. Also combinations of both refinement types in terms of hp-refinement schemes are conceivable. However, as h-refinement and p-refinement have similar effects to the local resolution, we here consider h-refinement only. In practice, the h-refinement in our program is done by an adaptive scheme. Based on reconstruction results on a coarser mesh, it is decided by global thresholding whether an element is refined. Afterwards the reconstruction process is restarted with the interpolated traction field obtained previously. This process is repeated until a desired local resolution is achieved. Alternatively, we can also implement a predefined local refinement based on thresholding of the measured displacement field. For the calculations in this work, we retain adaptive local mesh refinement schemes. This procedure is tested below by reconstruction of simulated data.
2.6. Optimization procedure
The core module of the FEM traction reconstruction program is the implemented multi-dimensional parameter optimization procedure based on the CG method. The implementation follows essentially the Fletcher–Reeves variant of this algorithm as described in [61]. Different from the standard implementation, we parallelized most parts of the procedure, such as the numeric gradient calculation  and the subsequent line minimization. Parallel computing was realized by using the message passing interface (MPI), which is suitable for large-scale distributed computing on computer clusters or sheared memory systems. Although the CG method is a common tool in the field of inverse problems (e.g. [62]), we also tested other optimization methods, namely gradientless downhill-simplex optimization, simple steepest descent and heuristic Monte Carlo optimization with simulated annealing (all described in [61]). We found that the CG method led to the shortest computation times and excellent convergence. Figure 2b shows a schematic representation of our complete workflow.
and the subsequent line minimization. Parallel computing was realized by using the message passing interface (MPI), which is suitable for large-scale distributed computing on computer clusters or sheared memory systems. Although the CG method is a common tool in the field of inverse problems (e.g. [62]), we also tested other optimization methods, namely gradientless downhill-simplex optimization, simple steepest descent and heuristic Monte Carlo optimization with simulated annealing (all described in [61]). We found that the CG method led to the shortest computation times and excellent convergence. Figure 2b shows a schematic representation of our complete workflow.
3. Results
3.1. Method validation
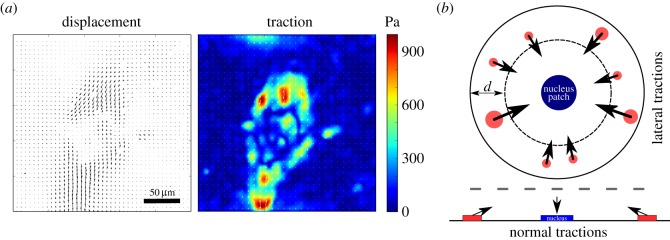
As reported earlier, for 2D TFM our implementation of FEM-TFM gives equivalent results to the standard approach with FTTC [34]. In this case, FTTC is much more efficient and faster. Here, however, we want to address 3D TFM for wavy substrates in which a Green's function is not known, making FEM-TFM the most reasonable approach. In order to validate our method for a 3D situation, we first reconstructed simulated data for planar substrates with both tangential and normal components. We generated these distributions by a simple set of rules that mimic typical cellular force distributions. Figure 3a shows a typical FTTC traction force pattern for a cardiac myofibroblast cultured on a flat 15 kPa PDMS substrate. This cell shows the typical dipole pattern that has been suggested as a minimal model for a contractile cell [10]. Figure 3b illustrates the rules we use to generate artificial traction patterns. A cell is modelled as a circle with traction patches located only in a peripheral annulus of width d. Each of these patches carries a tangential traction stress of f = 3 kPa, which is a typical value found for cells [63,64]. However, previous studies also reported appreciable cellular traction stress in normal directions [39,65,66]. They found a typical pull–push pattern, such that the cell is pulling up at the periphery and pushing down with the cell body. The upward forces might arise from actin fibres being anchored at the dorsal side of the cell or at the upper side of the nucleus, while the downward force might be the reaction force localized by the large and relatively stiff nucleus [67]. Here we include these normal forces in our simulations by adding force in the positive z-direction to our adhesion patches that are counterbalanced by an extra traction patch located at the cell centre. Together, these rules allow us to generate realistic traction distributions that satisfy basic properties of a typical cell-induced 3D traction pattern.
Figure 3.
Simulation of traction patterns. (a) Experimental example: cardiac myofibroblast on a flat 15 kPa PDMS substrate. Substrate displacement field and 2D traction forces obtained from particle imaging velocimetry and regularized Fourier transform traction cytometry (reg-FTTC) [59]. The cell shows a typical dipole pattern with strong inward directed forces at the cell periphery. (b) Illustration of traction pattern simulation. Traction patches are distributed within a ring-shaped area close to the cell edge with normal components that are balanced by a central patch that is consequently pushed into the substrate. Traction magnitude in the lateral direction is set to a constant value of 3 kPa.
The displacement fields resulting from these simulated traction patterns were calculated using the direct BVP introduced above (equation (2.1)) with our FEM implementation. From the resulting displacement field, we sampled Nbead random displacements. To account for uncertainties due to the contribution of experimental noise, we introduced additive random displacement errors uerr that modify each displacement component by a random value. Subsequently, a simulated bead displacement is expressed by ubead = uFEM + uerr. The simulated data allow us to validate and characterize features and limits of our method under well-controlled conditions.
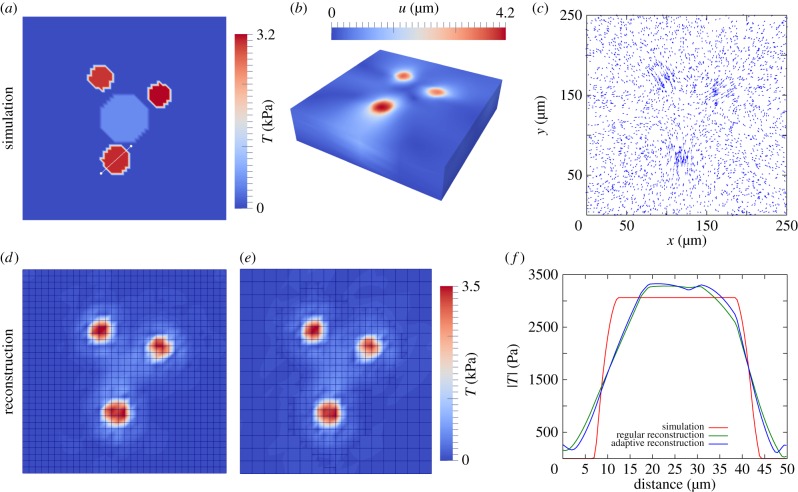
In figure 4a–c, we show the results of a typical simulation without noise (uerr = 0). Figure 4d,e depicts a comparison of reconstruction results on a homogeneous and an adaptively refined mesh, respectively. For the adaptive reconstruction, we started with a two times larger mesh size than the homogeneous mesh. After one refinement step, we subsequently achieved the same mesh size at refined regions, which makes the results comparable. Both results show excellent agreement and reconstruct correctly the original traction pattern. The depicted mesh topology shows that the adaptive refinement automatically adjusts the local mesh size to regions of accumulated traction stress, where we accordingly establish higher local accuracy. By doing this we saved 47% of d.f., which has a positive effect on the computation time. Figure 4f shows the traction profile through a reconstructed traction patch. Both reconstructions led to a slightly smoothed profile compared with the original. This reduced resolution of a sharp edge can be explained by a limiting bead density (in our simulation nbead = 0.048 µm−2) according to the sampling theorem by Nyquist and Shannon. From the simulation results, we conclude that adaptive local mesh refinement has no detrimental effect on the traction reconstruction accuracy. However, it clearly needs much less d.f. and, therefore, is much more computer time efficient.
Figure 4.
Reconstruction of simulated traction forces with the finite-element method (FEM-TFM). (a–c) Simulation of bead displacement. Simulated bead density nbead = 0.048 beads µm−2, which corresponds to a bead number of Nbead = 3000. (d) Traction reconstructed with FEM on regular mesh (3267 d.f.). (e) Reconstruction on locally h-refined mesh (1848 d.f.) works equally well. (f) Comparison of simulation and reconstruction along a traction profile through a force patch (white line in (a)).
When solving an inverse problem the stability of the solution strongly depends on the uncertainties in the provided data [57]. In particular, the problem might be ill posed due to the effect of noise. This has been explicitly shown for the case of traction reconstruction [58]. As mentioned in the introduction, experimental conditions limit the resolution measured displacement fields. The main reasons for this are the limited optical resolution of the microscope and errors in the bead tracking procedures. The first issue can be treated as Gaussian-shaped errors that limit the spatial localization of beads [33]. The latter is less relevant in the planar 2D case because state-of-the-art 2D tracking methods are very accurate with eventually negligible error rates. However, bead localization and tracking in three dimensions is far more challenging, even with modern microscopic set-ups. This is due to anisotropic optical resolution that produces a notably inferior accuracy in the z-direction compared with the corresponding lateral directions. There already exist improved set-ups such as dual objective super-resolution microscopy techniques [68–70]; however, these are not standard and rarely available and because of this no application in TFM has been shown so far. Therefore, we aimed at improving bead tracking in three dimensions and at adapting the traction reconstruction method to data with anisotropic optical resolution. In fact, we realized that due to the substrate topography we have to deal with a locally varying point spread function. That directly affected the tracking of bead movements when using image cross-correlation techniques. Hence, we applied a more robust single bead tracking method than done in standard TFM, using feature vectors introduced earlier for 3D TFM for the same reasons [43]. By using this technique we obtained a significantly improved displacement field; however, the data showed strong and inhomogeneous drift which could not be corrected by a single drift vector (evaluated in one focal plane). This effect can be explained by relatively long image stack acquisition times of approximately 30 min when recording ≈ 100 images per stack. Owing to this long acquisition time we observed non-monotonic drift when comparing two image stacks. In order to solve this problem, we applied drift correction for each individual slice, which successfully cancelled out most of the drift. Nevertheless, the derived displacement data showed anisotropic deviations and occasionally higher densities of outliers.
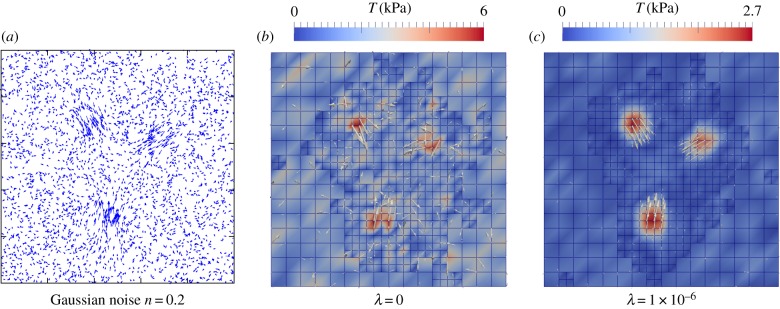
Figure 5a shows the displacement field used in figure 4 with Gaussian noise of strength 20% (measured with respect to maximal displacement). The force reconstruction without regularization is shown in figure 5b and clearly is not very good. Therefore, we next used zeroth-order Tikhonov regularization (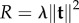 ) to find an approximated reconstruction solution. Here, we determine an optimal regularization parameter by using generalized cross validation (GCV) [71]. The optimal value of λ depends on the noise level and, for the given example, we determined λ = 1 ×10−6. The regularization improves the result significantly, as shown in figure 5c. The penalization of total force induced by zeroth-order Tikhonov regularization, however, led to traction underestimation and edge smoothing.
) to find an approximated reconstruction solution. Here, we determine an optimal regularization parameter by using generalized cross validation (GCV) [71]. The optimal value of λ depends on the noise level and, for the given example, we determined λ = 1 ×10−6. The regularization improves the result significantly, as shown in figure 5c. The penalization of total force induced by zeroth-order Tikhonov regularization, however, led to traction underestimation and edge smoothing.
Figure 5.
Effect of Gaussian noise and the need for regularization. (a) Simulated displacement field from figure 4 with Gaussian noise added (noise level 20%). (b) Reconstruction without regularization does not give a clear traction pattern. (c) Reconstruction with a regularization parameter λ = 1 × 10−6, selected by generalized cross validation (GCV), reconstructs the original traction pattern; however, with a reduced maximal traction magnitude of approximately 11% compared with the simulated pattern.
Next, we simulated the presence of outliers. We drew the additive random displacement error uerr for each bead either from a Gaussian distribution N(σnoise) or from a box-shaped distribution Hi(wi) limited by the box width w. We decided from which distribution an error contribution is drawn by calculating an equally distributed random variable  over the interval [0,1]. If
over the interval [0,1]. If 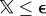 , we draw uerr from the box-shaped distribution, corresponding to an outlier. In the case of
, we draw uerr from the box-shaped distribution, corresponding to an outlier. In the case of 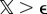 , we calculated a Gaussian distribution uerr. Here,
, we calculated a Gaussian distribution uerr. Here,  corresponds to the fraction of outliers. In general, there are two possible approaches to diminish the effect of outliers for the reconstruction. One is to filter out outliers based on predefined criteria. This demands that appropriate filter limits are set up. Moreover, such a procedure might overlook valuable information in the data. As a second approach one can use robust estimates for the optimization process. We will show in the following that reconstruction can be improved, when the least square estimate in the Tikhonov functional is replaced by a robust measure that is more insensitive to data outliers. We tested different robust MLEs known from optimization theory [60].
corresponds to the fraction of outliers. In general, there are two possible approaches to diminish the effect of outliers for the reconstruction. One is to filter out outliers based on predefined criteria. This demands that appropriate filter limits are set up. Moreover, such a procedure might overlook valuable information in the data. As a second approach one can use robust estimates for the optimization process. We will show in the following that reconstruction can be improved, when the least square estimate in the Tikhonov functional is replaced by a robust measure that is more insensitive to data outliers. We tested different robust MLEs known from optimization theory [60].
The simplest robust estimate uses the L1-norm, 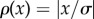 . As the L2 estimate described above, it can be derived analytically from the maximum-likelihood assumption, in this case based on a Laplace distribution
. As the L2 estimate described above, it can be derived analytically from the maximum-likelihood assumption, in this case based on a Laplace distribution 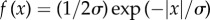 . The essential advantage of this estimate is its weighting of deviations, which compared with the L2 is less sensitive to outlier contributions. As alternative estimates, we implemented also Huber functions or biweighting functions [60]. They require an additional cut-off parameter k to characterize the transition feature of the effective deviation weighting. This indeed requires a good guess about expected outlier strength.
. The essential advantage of this estimate is its weighting of deviations, which compared with the L2 is less sensitive to outlier contributions. As alternative estimates, we implemented also Huber functions or biweighting functions [60]. They require an additional cut-off parameter k to characterize the transition feature of the effective deviation weighting. This indeed requires a good guess about expected outlier strength.
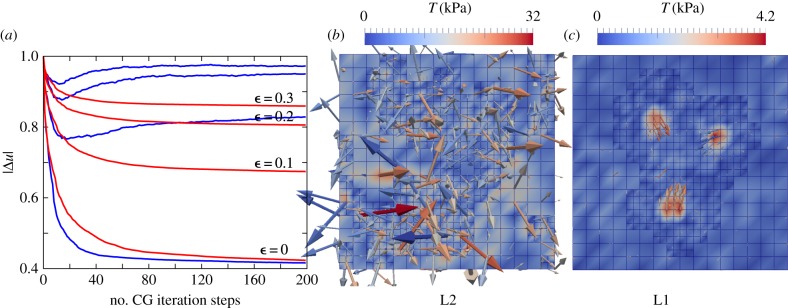
Figure 6 depicts the influence of the estimate used on the quality of the reconstructed traction field. The simulation study considered an isotropic Gaussian-based noise of nnoise = 0.1 and a isotropic box-shaped distribution with w = 10 µm. We investigated the convergence behaviour for the optimization achieved by L1 and L2 estimates with respect to the outlier fractions 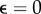 , 0.1, 0.2, 0.3. To compare the results, we introduced the relative displacement deviation
, 0.1, 0.2, 0.3. To compare the results, we introduced the relative displacement deviation 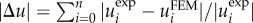 . For
. For 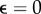 (no outliers), both estimates converge to a similar result; however, the L1 converges slower. If we chose a non-zero fraction of outliers
(no outliers), both estimates converge to a similar result; however, the L1 converges slower. If we chose a non-zero fraction of outliers 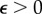 , the convergence dynamics of the optimization procedure starts to differ between L2 and L1. The L1 still shows a monotonic decreasing |Δu|, while in the case of the L2 the curve starts to increase again saturating into a different solution. Corresponding traction field solutions for
, the convergence dynamics of the optimization procedure starts to differ between L2 and L1. The L1 still shows a monotonic decreasing |Δu|, while in the case of the L2 the curve starts to increase again saturating into a different solution. Corresponding traction field solutions for 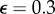 are depicted in figure 6. In the case of using the L2, the reconstruction is strongly influenced by the fraction of outliers. By contrast, the L1 reconstruction leads to satisfying results comparable to the target field.
are depicted in figure 6. In the case of using the L2, the reconstruction is strongly influenced by the fraction of outliers. By contrast, the L1 reconstruction leads to satisfying results comparable to the target field.
Figure 6.
Reconstruction of data with mixed error, including outliers. (a) Mean relative deviation plotted over the number of optimization steps for different outlier fractions 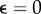 , 0.1, 0.2 ,0.3 and for two different estimates, L2 (blue) and L1 (red). (b) L2-based reconstruction for outlier fraction
, 0.1, 0.2 ,0.3 and for two different estimates, L2 (blue) and L1 (red). (b) L2-based reconstruction for outlier fraction 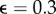 . (c) L1-based reconstruction for outlier fraction
. (c) L1-based reconstruction for outlier fraction 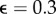 . For all simulations, the Gaussian noise level was 10%.
. For all simulations, the Gaussian noise level was 10%.
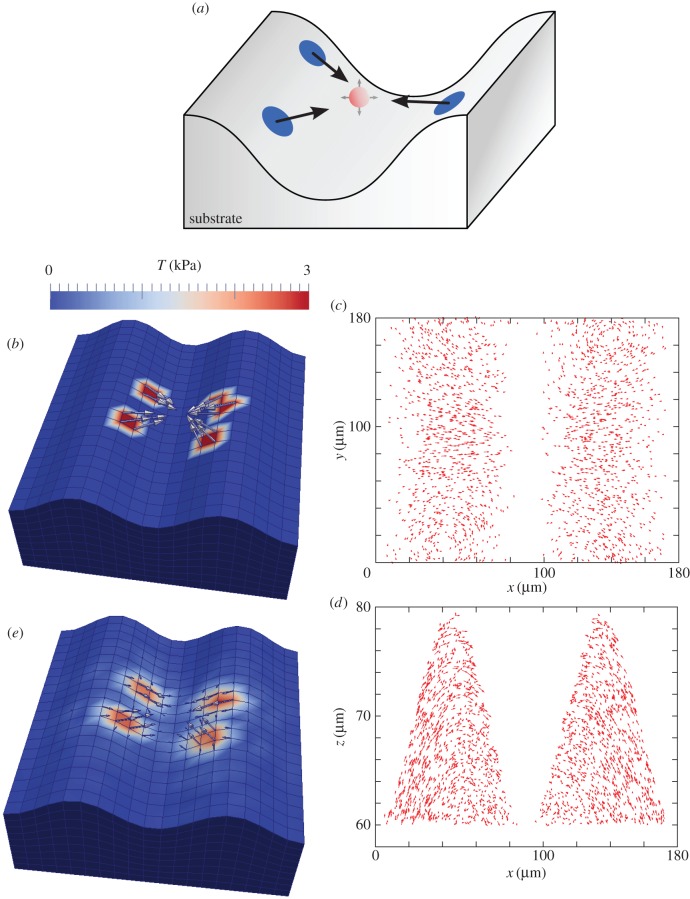
After having established a successful approach for planar substrates with both tangential and normal forces, we finally simulated 3D FEM-TFM with wavy substrates as shown schematically in figure 7a. We first distributed random traction patches at both flanks of the sinusoidal shape, while the traction vectors point at a common virtual point. In a second step, all traction directions were reoriented due to movement of the virtual point in three dimensions, until a balance of overall force was achieved. Figure 7b,c depicts an exemplary simulation result. It shows a traction field with four distinct traction patches with a force density of f = 3 kPa. The corresponding simulated bead displacements are depicted in figure 7c,d as projections to the xy-plane and xz-planes, respectively. Compared with the planar case, the displacements in the z-direction became more prominent (figure 7d). The resulting reconstruction shown in figure 7e shows similar good agreement to the planar case, thereby validating our method also for the experimental set-up to be studied.
Figure 7.
Traction reconstruction simulated on a sinusoidal substrate topography. (a) Illustration of the simulation scheme. All forces point to a virtual point (red) between the two ridges to ensure force balance. (b) Simulated traction field with four traction patches f = 3 kPa. Simulated displacement field with bead volume density 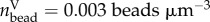 (Nbead = 2000), (c) projected to the xy-plane and (d) projected to the xz-plane. (e) The achieved reconstruction is very close to the original traction pattern.
(Nbead = 2000), (c) projected to the xy-plane and (d) projected to the xz-plane. (e) The achieved reconstruction is very close to the original traction pattern.
3.2. Morphology and traction forces of cardiac myofibroblasts on wavy substrates
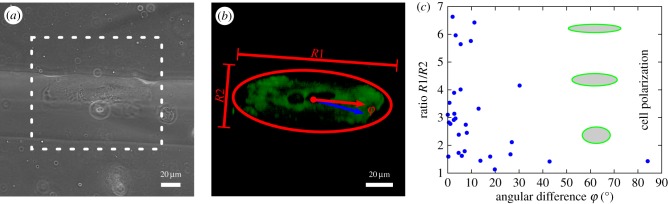
We finally investigated myofibroblasts adhered to elastic wavy PDMS substrates with a focus on their morphology and traction forces. For our context, the choice of PDMS has several advantages over the PAA system, which is often employed for planar 2D TFM. In particular, it does not swell due to water uptake, it has superior optical properties, and it can be reproducibly moulded into wave structures on a microscopic scale of several tens of micrometres. We first quantified the effect of substrate topography on cell morphology and cytoskeletal polarization. For this purpose, we evaluated a dataset of stack images of 30 cells adhered to waves of approximately 70 µm width and approximately 30 µm height. A representative phase contrast image is shown in figure 8a. Such images were used to determine the orientation of the topographic features. The cells were fluorescently stained for cell adhesion-related molecules to measure their internal organization. We determined the direction and degree of cell polarization by fitting a 3D ellipse to the image and subsequently evaluated the orientation and eccentricity of the ellipse (figure 8b). We finally correlated the two measures. Figure 8c shows the eccentricity as a function of the angle between the substrate and the cell orientations. The plot confirms that cells tend to align with the long axis of the substrate. Two-thirds of the cells show a ratio R1/R2 (semi-major over semi-minor) larger than 2 and approximately 80% are aligned with the wave within the angular range of 0–15°. This indicates that even the cells that are close to round have sensed the topographic features of the wavy substrate. From our ellipse analysis we further found that they tend to adhere most often to the wave flanks and less to the valleys or hill tops of the height-modulated structures.
Figure 8.
Cell polarization and orientation. (a) Phase contrast image of a cardiac myofibroblast aligned with substrate topography. (b) Illustration of ellipse fitting to analyse cell orientation and shape in terms of the ratio between lengths of semi-major R1 and semi-minor R2 from images of vinculin-transfected cells. The angular difference is defined as angle φ between the cell orientation (red arrow) and the substrate orientation (blue arrow). (c) Cell shape plotted versus the angular difference φ.
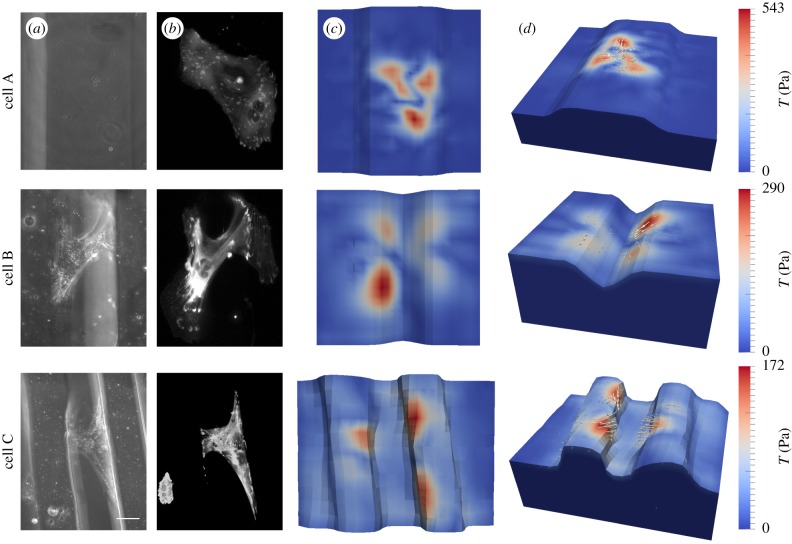
In addition to the morphological study we applied the FEM traction reconstruction to an exemplary set of cells. Here, we wanted to investigate the influence of environmental topography on cellular contractility. The long acquisition time for displacement data, however, made it very difficult to conduct a full statistical analysis, and, therefore, in figure 9 we only show a few typical examples of cardiac myofibroblasts cultured on wavy substrates with a Young's modulus of E = 15 kPa. In figure 9, two different topographies are used, micro-grooves and wave shapes. For the substrates in cases A/B/C we used 70/142/91 slices with a z-distance between two slices of 0.55/0.39/0.30 µm. We tracked 6205/9614/13 613 beads, with an average distance between beads of 5.40/5.91/2.90 µm. For the reconstruction, we used the adaptive mesh refinement approach and started to calculate a traction solution on a coarser mesh (approx. 50 optimization steps). Afterwards the algorithm refined approximately 40% of the computational grid cells at the surface based on obtained traction magnitudes. We subsequently achieved local mesh sizes of dm = 7/10/6 µm for the corresponding cells A/B/C (figure 9). We further used an L1 estimate for the residue and a zeroth-order Tikhonov regularization with L2-norm (same regularization parameter for all data). As explained above, the z-direction is weighted only with a factor of 1/3 due to its poor resolution compared with the two lateral directions.
Figure 9.
Traction reconstruction for cardiac fibroblasts cultivated on three different topographies. (a) Phase contrast image, (b) VASP-GFP fluorescence image, (c) top view and (d) 3D view of the reconstructed traction field, respectively. Scale bar, 50 µm.
Cell A was adhered on top of a slightly curved surface between two grooves (dimensions: h = 25 µm, w = 120 µm). Compared with the others it shows the largest spread area and a roughly homogeneous distribution of adhesion sites (observed in a vasodilator-stimulated phosphoprotein (VASP) fluorescence image). For this cell, we obtained the largest maximal traction as Tmax = 543 Pa. Cell B occupied the edge region of two opposite groove flanks (groove dimensions: h = 30 µm, w = 70 µm). FAs were concentrated to a limited region at the edge and the cell spanned the groove with actin SFs bridging free space. The maximal traction was Tmax = 290 Pa. Strikingly the traction vectors revealed that most of the forces were balanced along the edges and not at the opposite groove flanks. Cell C spanned the gap between two wave-shaped hills (wave shape: h = 30 µm, w = 25 µm with separation distance d = 60 µm). FAs were more concentrated at vertical parts of the edge region with a similar total area to cell B. We derived the lowest maximal traction Tmax = 172 Pa for this cell. As for cell B, however, most of the forces seemed to be balanced along the right wave instead of across the gap. Therefore, we conclude that, even if cells span two neighbouring hills, they still tend to balance their forces along the ridges, in agreement with the earlier observation that their cytoskeleton is on average organized in this direction.
4. Discussion
Here we have described experimental and theoretical methods to reconstruct cellular traction patterns of cells adhering to non-planar (wavy) substrates. This involves novel micromoulding techniques to prepare curved elastic substrates with embedded fluorescent beads, improved image-processing procedures for bead tracking and a completely novel 3D TFM workflow to achieve traction reconstruction using FEM and optimization procedures. We successfully checked the validity of our method by first reconstructing simulated data considering different experimental conditions.
As shown §3.1, the presence of outliers plays a crucial role in proper traction force reconstruction for our 3D data on topographic substrates. By replacing the typically used L2- by an L1-error estimate, we demonstrated a way to prevent problems in traction reconstruction originated from outliers. The L1 estimate is a much more robust measure in the presence of outliers in noisy data. This can be essentially traced back to a reduced weighting of outliers (linear weighting) during the reconstruction process. Compared with other known robust estimates, the L1-norm needs no additional cut-off parameters and converges to the L2 solution for outlier-free data with Gaussian error distributions [60]. We note that this approach aims to improve traction field solutions for non-smooth and noisy 3D data. It has no essential influence on the regularization scheme. Here we stay with the standard L2-norm, which seems appropriate for our case of dense marker beads. Recently, it has been suggested that the L1-norm is favourable for the regularization term in the case of high-resolution 2D TFM [72]. Although we expect that this approach is not required in our case of rather dense bead positioning (typical bead difference of 4 µm and cell size around 100 µm), for future TFM applications it seems very interesting to explore which combination of norms for the residue and the regularization term are most favourable in which situation.
We also emphasize the need for adaptations in the image-processing procedures on wavy substrates. Because the point spread function for the marker beads can vary locally in this case, we cannot use the standard cross-correlation approach for bead tracking, but resort to a template-matching procedure based on feature vectors [43]. We also take into account the anisotropy in the point spread function by using a reduced weight for the z-component in the residue. Together with the use of the L1-norm for the residue, this ensures a very good quality of our traction force reconstruction as validated by computer simulations. Our analysis is facilitated by the use of topographic substrates with cylinder symmetry, which allows us to use the slicing and meshing procedures shown in figure 1.
We next investigated the degree and orientation of cellular polarity for cardiac myofibroblasts, which have been exposed to a wavy substrate geometry. We found that these cells strongly align and polarize perpendicular to the direction of maximal curvature, which has been reported before for other types of fibroblasts on rigid microcylinders. We note that even round cells tend to align with the substrates' features, presumably because the feature dimensions are such that every cell is affected by the topographical cues. In addition to this observation we determined traction force maps for an exemplary set of cells and demonstrated the successful reconstruction of the 3D forces. We found that cells also balance their forces along the orientation of the pattern, even if at the same time they form a bridge between neighbouring hills. Because our reconstruction is strongly regularized due to the noise issues with wavy substrates, the absolute values of the measured traction stresses are expected to be lower than the actual values, as shown earlier with model-based traction force microscopy [73]. In fact, we expect the real traction stress to be closer to the kPa range as used for the simulated data.
In the future, further advances in experimental techniques as described here (including faster acquisition of the image data, for example with a light sheet microscope) should make a full statistical analysis feasible. It would also be interesting to conduct these experiments with other cell types whose physiological function depends on a curved or corrugated environment, such as podocytes filtering the blood in kidney glomeruli.
Supplementary Material
Endnote
GMSH is a three-dimensional finite-element mesh generator with built-in pre- and postprocessing facilities.
Competing interests
We declare we have no competing interests.
Funding
This work was supported by MechanoSys (BMBF grant no. 0315501) and the EcTop programme of the Cluster of Excellence Cellular Networks. BH, RM and USS thank the Isaac Newton Institute for Mathematical Sciences for its hospitality during the programme ‘Coupling geometric partial differential equations with physics for cell morphology, motility and pattern formation’ (CGPW02) supported by EPSRC grant no. EP/K032208/1. USS is a member of the Interdisciplinary Center for Scientific Computing (IWR) at Heidelberg.
References
- 1.Vogel V, Sheetz M. 2006. Local force and geometry sensing regulate cell functions. Nat. Rev. Mol. Cell Biol. 7, 265–275. ( 10.1038/nrm1890) [DOI] [PubMed] [Google Scholar]
- 2.Chen CS. 2008. Mechanotransduction—a field pulling together? J. Cell Sci. 121, 3285–3292. ( 10.1242/jcs.023507) [DOI] [PubMed] [Google Scholar]
- 3.Humphrey JD, Dufresne ER, Schwartz MA. 2014. Mechanotransduction and extracellular matrix homeostasis. Nat. Rev. Mol. Cell Biol. 15, 802–812. ( 10.1038/nrm3896) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pelham RJ, Wang Y-L. 1997. Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc. Natl Acad. Sci. USA 94, 13 661–13 665. ( 10.1073/pnas.94.25.13661) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Engler A, Bacakova L, Newman C, Hategan A, Griffin M, Discher D. 2004. Substrate compliance versus ligand density in cell on gel responses. Biophys. J. 86, 617–628. ( 10.1016/S0006-3495(04)74140-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nisenholz N, Rajendran K, Dang Q, Chen H, Kemkemer R, Krishnan R, Zemel A. 2014. Active mechanics and dynamics of cell spreading on elastic substrates. Soft Matter 10, 7234–7246. ( 10.1039/C4SM00780H) [DOI] [PubMed] [Google Scholar]
- 7.McBeath R, Pirone DM, Nelson CM, Bhadriraju K, Chen CS. 2004. Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev. Cell 6, 483–495. ( 10.1016/S1534-5807(04)00075-9) [DOI] [PubMed] [Google Scholar]
- 8.Engler AJ, Sen S, Sweeney HL, Discher DE. 2006. Matrix elasticity directs stem cell lineage specification. Cell 126, 677–689. ( 10.1016/j.cell.2006.06.044) [DOI] [PubMed] [Google Scholar]
- 9.Wen JH, Vincent LG, Fuhrmann A, Choi YS, Hribar KC, Taylor-Weiner H, Chen S, Engler AJ. 2014. Interplay of matrix stiffness and protein tethering in stem cell differentiation. Nat. Mater. 13, 979–987. ( 10.1038/nmat4051) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwarz US, Safran SA. 2013. Physics of adherent cells. Rev. Mod. Phys. 85, 1327–1381. ( 10.1103/RevModPhys.85.1327) [DOI] [Google Scholar]
- 11.Schwarz US, Bischofs IB. 2005. Physical determinants of cell organization in soft media. Med. Eng. Phys. 27, 763–772. ( 10.1016/j.medengphy.2005.04.007) [DOI] [PubMed] [Google Scholar]
- 12.Kollmannsberger P, Bidan CM, Dunlop JWC, Fratzl P. 2011. The physics of tissue patterning and extracellular matrix organisation: how cells join forces. Soft Matter 7, 9549–9560. ( 10.1039/c1sm05588g) [DOI] [Google Scholar]
- 13.Kraning-Rush CM, Reinhart-King CA. 2012. Controlling matrix stiffness and topography for the study of tumor cell migration. Cell Adhes. Migration 6, 274–279. ( 10.4161/cam.21076) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen CS. 1997. Geometric control of cell life and death. Science 276, 1425–1428. ( 10.1126/science.276.5317.1425) [DOI] [PubMed] [Google Scholar]
- 15.Kilian KA, Bugarija B, Lahn BT, Mrksich M. 2010. Geometric cues for directing the differentiation of mesenchymal stem cells. Proc. Natl Acad. Sci. USA 107, 4872–4877. ( 10.1073/pnas.0903269107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Curtis A, Wilkinson C. 1997. Topographical control of cells. Biomaterials 18, 1573–1583. ( 10.1016/S0142-9612(97)00144-0) [DOI] [PubMed] [Google Scholar]
- 17.Bettinger CJ, Langer R, Borenstein JT. 2009. Engineering substrate topography at the micro- and nanoscale to control cell function. Angew. Chem. Int. Ed. Engl. 48, 5406–5415. ( 10.1002/anie.200805179) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim D-H, Provenzano PP, Smith CL, Levchenko A. 2012. Matrix nanotopography as a regulator of cell function. J. Cell Biol. 197, 351–360. ( 10.1083/jcb.201108062) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dalby MJ, Gadegaard N, Tare R, Andar A, Riehle MO, Herzyk P, Wilkinson CDW, Oreffo ROC. 2007. The control of human mesenchymal cell differentiation using nanoscale symmetry and disorder. Nat. Mater. 6, 997–1003. ( 10.1038/nmat2013) [DOI] [PubMed] [Google Scholar]
- 20.Yim EKF, Pang SW, Leong KW. 2007. Synthetic nanostructures inducing differentiation of human mesenchymal stem cells into neuronal lineage. Exp. Cell Res. 313, 1820–1829. ( 10.1016/j.yexcr.2007.02.031) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dalby MJ, Riehle MO, Yarwood SJ, Wilkinson CDW, Curtis ASG. 2003. Nucleus alignment and cell signaling in fibroblasts: response to a micro-grooved topography. Exp. Cell Res. 284, 272–280. ( 10.1016/S0014-4827(02)00053-8) [DOI] [PubMed] [Google Scholar]
- 22.Biela SA, Su Y, Spatz JP, Kemkemer R. 2009. Different sensitivity of human endothelial cells, smooth muscle cells and fibroblasts to topography in the nano-micro range. Acta Biomater. 5, 2460–2466. ( 10.1016/j.actbio.2009.04.003) [DOI] [PubMed] [Google Scholar]
- 23.Ulbrich S, Friedrichs J, Valtink M, Murovski S, Franz CM, Müller DJ, Funk RHW, Engelmann K. 2011. Retinal pigment epithelium cell alignment on nanostructured collagen matrices. Cells Tissues Organs 194, 443–456. ( 10.1159/000323653) [DOI] [PubMed] [Google Scholar]
- 24.Corey JM, Feldman EL. 2003. Substrate patterning: an emerging technology for the study of neuronal behavior. Exp. Neurol. 184, 89–96. ( 10.1016/S0014-4886(03)00392-3) [DOI] [PubMed] [Google Scholar]
- 25.Svitkina TM, Rovensky YA, Bershadsky AD, Vasiliev JM. 1995. Transverse pattern of microfilament bundles induced in epitheliocytes by cylindrical substrata. J. Cell Sci. 108, 735–745. [DOI] [PubMed] [Google Scholar]
- 26.Levina EM, Domnina LV, Rovensky YA, Vasiliev JM. 1996. Cylindrical substratum induces different patterns of actin microfilament bundles in nontransformed and in ras-transformed epitheliocytes. Exp. Cell Res. 229, 159–165. ( 10.1006/excr.1996.0354) [DOI] [PubMed] [Google Scholar]
- 27.Biton YY, Safran SA. 2009. The cellular response to curvature-induced stress. Phys. Biol. 6, 046010 ( 10.1088/1478-3975/6/4/046010) [DOI] [PubMed] [Google Scholar]
- 28.Sanz-Herrera JA, Moreo P, Garca-Aznar JM, Doblaré M. 2009. On the effect of substrate curvature on cell mechanics. Biomaterials 30, 6674–6686. ( 10.1016/j.biomaterials.2009.08.053) [DOI] [PubMed] [Google Scholar]
- 29.Charest JM, Califano JP, Carey SP, Reinhart-King CA. 2012. Fabrication of substrates with defined mechanical properties and topographical features for the study of cell migration. Macromol. Biosci. 12, 12–20. ( 10.1002/mabi.201100264) [DOI] [PubMed] [Google Scholar]
- 30.Endlich N, Endlich K. 2006. Stretch, tension and adhesion—adaptive mechanisms of the actin cytoskeleton in podocytes. Eur. J. Cell Biol. 85, 229–234. ( 10.1016/j.ejcb.2005.09.006) [DOI] [PubMed] [Google Scholar]
- 31.Cesa CM, Kirchgessner N, Mayer D, Schwarz U, Hoffmann B, Merkel R. 2007. Micropatterned silicone elastomer substrates for high resolution analysis of cellular force patterns. Rev. Sci. Instrum. 78, 034301 ( 10.1063/1.2712870) [DOI] [PubMed] [Google Scholar]
- 32.Style RW, Boltyanskiy R, German GK, Hyland C, MacMinn CW, Mertz AF, Wilen LA, Xu Y, Dufresne ER. 2014. Traction force microscopy in physics and biology. Soft Matter 10, 4047–4055. ( 10.1039/c4sm00264d) [DOI] [PubMed] [Google Scholar]
- 33.Plotnikov SV, Sabass B, Schwarz US, Waterman CM. 2014. High-resolution traction force microscopy, vol. 123. Amsterdam, The Netherlands: Elsevier Inc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwarz US, Soiné JRD. 2015. Traction force microscopy on soft elastic substrates: a guide to recent computational advances. Biochim. Biophys. Acta 1853, 3095–3104. ( 10.1016/j.bbamcr.2015.05.028) [DOI] [PubMed] [Google Scholar]
- 35.Dembo M, Wang Y-L. 1999. Stresses at the cell-to-substrate interface during locomotion of fibroblasts. Biophys. J. 76, 2307–2316. ( 10.1016/S0006-3495(99)77386-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Merkel R, Kirchgessner N, Cesa CM, Hoffmann B. 2007. Cell force microscopy on elastic layers of finite thickness. Biophys. J. 93, 3314–3323. ( 10.1529/biophysj.107.111328) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Del Alamo JC, Meili R, Alonso-Latorre B, Rodrguez-Rodrguez J, Aliseda A, Firtel RA, Lasheras JC. 2007. Spatio-temporal analysis of eukaryotic cell motility by improved force cytometry. Proc. Natl Acad. Sci. USA 104, 13 343–13 348. ( 10.1073/pnas.0705815104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Butler JP, Tolić-Nørrelykke IM, Fabry B, Fredberg JJ. 2002. Traction fields, moments, and strain energy that cells exert on their surroundings. Am. J. Physiol. Cell Physiol. 282, C595–C605. ( 10.1152/ajpcell.00270.2001) [DOI] [PubMed] [Google Scholar]
- 39.Delanoë-Ayari H, Rieu JP, Sano M. 2010. 4D Traction force microscopy reveals asymmetric cortical forces in migrating dictyostelium cells. Phys. Rev. Lett. 105, 2–5. ( 10.1103/PhysRevLett.105.248103) [DOI] [PubMed] [Google Scholar]
- 40.Hur SS et al. 2012. Roles of cell confluency and fluid shear in 3-dimensional intracellular forces in endothelial cells. Proc. Natl Acad. Sci. USA 109, 11 110–11 115. ( 10.1073/pnas.1207326109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Legant WR, Choi CK, Miller JS, Shao L, Gao L, Betzig E, Chen CS. 2013. Multidimensional traction force microscopy reveals out-of-plane rotational moments about focal adhesions. Proc. Natl Acad. Sci. USA 110, 881–886. ( 10.1073/pnas.1207997110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang Z, Lin J-S, Chen J, Wang JH-C. 2006. Determining substrate displacement and cell traction fields—a new approach. J. Theor. Biol. 242, 607–616. ( 10.1016/j.jtbi.2006.05.005) [DOI] [PubMed] [Google Scholar]
- 43.Legant WR, Miller JS, Blakely BL, Cohen DM, Genin GM, Chen CS. 2010. Measurement of mechanical tractions exerted by cells in three-dimensional matrices. Nat. Methods 7, 969–971. ( 10.1038/nmeth.1531) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Steinwachs J, Metzner C, Skodzek K, Lang N, Thievessen I, Mark C, Münster S, Aifantis KE, Fabry B. 2015. Three-dimensional force microscopy of cells in biopolymer networks. Nat. Methods 13, 171–176. ( 10.1038/nmeth.3685) [DOI] [PubMed] [Google Scholar]
- 45.Maskarinec SA, Franck C, Tirrell DA, Ravichandran G. 2009. Quantifying cellular traction forces in three dimensions. Proc. Natl Acad. Sci. USA, 106, 22 108–22 113. ( 10.1073/pnas.0904565106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Toyjanova J, Bar-Kochba E, López-Fagundo C, Reichner J, Hoffman-Kim D, Franck C. 2014. High resolution, large deformation 3D traction force microscopy. PLoS ONE 9, e90976 ( 10.1371/journal.pone.0090976) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ulbricht A, et al. 2013. Cellular mechanotransduction relies on tension-induced and chaperone-assisted autophagy. Curr. Biol. 23, 430–435. ( 10.1016/j.cub.2013.01.064) [DOI] [PubMed] [Google Scholar]
- 48.Hersch N, Wolters B, Dreissen G, Springer R, Kirchgeßner N, Merkel R, Hoffmann B. 2013. The constant beat: cardiomyocytes adapt their forces by equal contraction upon environmental stiffening. Biol. Open 2, 351–361. ( 10.1242/bio.20133830) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hampe N, Jonas T, Wolters B, Hersch N, Hoffmann B, Merkel R. 2014. Defined 2-D microtissues on soft elastomeric silicone rubber using lift-off epoxy-membranes for biomechanical analyses. Soft Matter 10, 2431–2443. ( 10.1039/c3sm53123f) [DOI] [PubMed] [Google Scholar]
- 50.Gordan OD, Persson BNJ, Cesa CM, Mayer D, Hoffmann B, Dieluweit S, Merkel R. 2008. On pattern transfer in replica molding. Langmuir 24, 6636–6639. ( 10.1021/la800728x) [DOI] [PubMed] [Google Scholar]
- 51.Edelsbrunner H, Kirkpatrick D, Seidel R. 1983. On the shape of a set of points in the plane. IEEE Trans. Inform. Theory 29, 551–559. ( 10.1109/TIT.1983.1056714) [DOI] [Google Scholar]
- 52.Braess D. 2007. Finite elemente theorie: schnelle Löser und Anwendungen in der Elastizitätstheorie, 4th edn Berlin, Germany: Springer. [Google Scholar]
- 53.Ameen M. 2005. Computational elasticity. Harrow, UK: Alpha Science Int. Ltd. [Google Scholar]
- 54.Bonet J, Wood RD. 1997. Nonlinear continuum mechanics for finite element analysis. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 55.Bangerth W, Hartmann R, Kanschat G. 2007. deal.II—A general-purpose object-oriented finite element library. ACM Trans. Math. Softw. 33, 24. ( 10.1145/1268776.1268779) [DOI] [Google Scholar]
- 56.Tikhonov AN, Arsenin VY. 1977. Solution of ill-posed problems. New York, NY: John Wiley & Sons. [Google Scholar]
- 57.Vogel CR. 2002. Computational methods for inverse problems. Philadelphia, PA: Society for Industrial and Applied Mathematics. [Google Scholar]
- 58.Schwarz US, Balaban NQ, Riveline D, Bershadsky A, Geiger B, Safran SA. 2002. Calculation of forces at focal adhesions from elastic substrate data: the effect of localized force and the need for regularization. Biophys. J. 83, 1380–1394. ( 10.1016/S0006-3495(02)73909-X) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sabass B, Gardel ML, Waterman CM, Schwarz US. 2008. High resolution traction force microscopy based on experimental and computational advances. Biophys. J. 94, 207–220. ( 10.1529/biophysj.107.113670) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Maronna RA, Martin RD, Yohai VJ. 2006. Robust statistics—theory and methods. New York, NY: John Wiley & Sons, Ltd. [Google Scholar]
- 61.Press WH, Teukolsky SA, Vetterling WT, Flannery DP (eds). 2007. Numerical recipes, 3rd edn Cambridge, UK: Cambridge University Press. [Google Scholar]
- 62.Lukyanenko DV, Yagola AG. 2012. Using parallel computing for solving multidimensional ill-posed problems. In Computational methods for applied inverse problems (eds Wang Y, Yagola AG, Changchun Y), pp. 49–81. Boston, MA: De Gruyter. [Google Scholar]
- 63.Balaban NQ et al. 2001. Force and focal adhesion assembly: a close relationship studied using elastic micropatterned substrates. Nat. Cell Biol. 3, 466–472. ( 10.1038/35074532) [DOI] [PubMed] [Google Scholar]
- 64.Fu J, Wang Y-K, Yang MT, Desai RA, Yu X, Liu Z, Chen CS. 2010. Mechanical regulation of cell function with geometrically modulated elastomeric substrates. Nat. Methods 7, 733–736. ( 10.1038/nmeth.1487) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hur SS, Zhao Y, Li Y-S, Botvinick E, Chien S. 2009. Live cells exert 3-dimensional traction forces on their substrata. Cell. Mol. Bioeng. 2, 425–436. ( 10.1007/s12195-009-0082-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kristal-Muscal R, Dvir L, Weihs D. 2013. Metastatic cancer cells tenaciously indent impenetrable, soft substrates. New J. Phys. 15, 035022 ( 10.1088/1367-2630/15/3/035022) [DOI] [Google Scholar]
- 67.Dvir L, Nissim R, Alvarez-Elizondo MB, Weihs D. 2015. Quantitative measures to reveal coordinated cytoskeleton-nucleus reorganization during in vitro invasion of cancer cells. New J. Phys. 17, 043010 ( 10.1088/1367-2630/17/4/043010) [DOI] [Google Scholar]
- 68.Shtengel G, et al. 2009. Interferometric fluorescent super-resolution microscopy resolves 3D cellular ultrastructure. Proc. Natl Acad. Sci. USA 106, 3125–3130. ( 10.1073/pnas.0813131106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xu K, Babcock HP, Zhuang X. 2012. Dual-objective STORM reveals three-dimensional filament organization in the actin cytoskeleton. Nat. Methods 9, 185–188. ( 10.1038/nmeth.1841) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Aquino D, Schönle A, Geisler C, Middendorff CV, Wurm CA, Okamura Y, Lang T, Hell SW, Egner A. 2011. Two-color nanoscopy of three-dimensional volumes by 4Pi detection of stochastically switched fluorophores. Nat. Methods 8, 353–359. ( 10.1038/nmeth.1583) [DOI] [PubMed] [Google Scholar]
- 71.Hansen PC. 1992. Analysis of discrete ill-posed problems by means of the L-curve. SIAM Rev. 34, 561–580. ( 10.1137/1034115) [DOI] [Google Scholar]
- 72.Han SJ, Oak Y, Groisman A, Danuser G. 2015. Traction microscopy to identify force modulation in subresolution adhesions. Nat. Methods 12, 653–656. ( 10.1038/nmeth.3430) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Soiné JRD, Brand CA, Stricker J, Oakes PW, Gardel ML, Schwarz US. 2015. Model-based traction force microscopy reveals differential tension in cellular actin bundles. PLoS Comput. Biol. 11, e1004076 ( 10.1371/journal.pcbi.1004076) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.