Antibodies are valued reagents in most modern laboratories and indispensable for many experiments. They can be utilized in a wide range of experimental setups designed to elucidate various scientific questions. In our laboratory, antibodies are used to investigate protein localization, quantity and phosphorylation state, and furthermore to identify various cell types as cell markers or to block protein function.
We have recently published the paper Neuronal Death in the Dorsal Root Ganglion after Sciatic Nerve Injury does not depend on Sortilin (Gurgor et al., 2016), investigating the proposed involvement of p75 neurotrophin receptor (p75NTR) and sortilin in neuronal death. As a part of our paper, the co-localization between the receptors sortilin and p75NTR in the dorsal root ganglion (DRG) neurons was investigated with immunohistochemistry (IHC), finding that approximately 12% of the p75NTR positive neurons were also positive for sortilin. This question has been investigated previously, but with markedly different results, for instance by Arnett et al. (2007) showing that approximately 50% of the p75NTR positive neurons were also positive for sortilin.
As part of the preliminary work for our paper, antibody validation was performed and disclosed serious problems with several available sortilin antibodies. The validation was based on IHC of DRG sections from wildtype (wt) and sortilin knock out (KO) mice. The IHC experiments revealed that the sortilin antibody used by Arnett et al. (2007) provided an apparently identical staining pattern in wt and sortilin KO tissue sections, indicating antibody cross-reactivity with an unknown epitope. Based on further IHC antibody validations we were able to identify an antibody from R&D Biosystems (AF2934) which did not show cross-reactivity when applied to the sortilin KO tissue. The same procedure was performed to identify a selective p75NTR antibody, utilizing DRG section from p75NTR KO mice. The dramatic difference in results obtained by Arnett et al. and our group could therefore result from their use of unvalidated antibodies, resulting in a higher estimate of sortilin-p75NTR colozalization in sensory neurons. Consequently, the hypotheses that p75NTR and sortilin are important for neuronal death in the DRG after a peripheral nerve injury was not supported by our study.
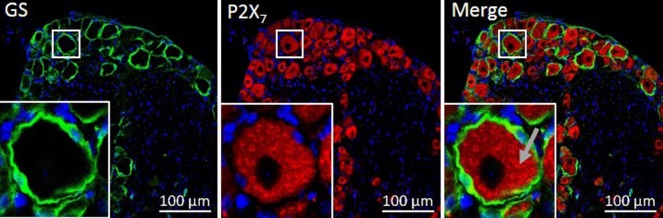
We have previously encountered difficulties identifying antibodies with selectivity, specificity and sensitivity to various proteins of interest. As in the case with the sortilin antibody discussed above, cross-reactivity with unknown epitopes is often the major concern. Another episode from our group demonstrating antibody cross-reactivity was encountered when testing the P2X7 antibody (APR-008) from Alomone labs. The antibody was tested by IHC staining of mouse DRG section where P2X7 is known to be localized solely in the satellite glial cells surrounding the neuronal cell bodies (Kobayashi et al., 2005; Zhang et al., 2005). Surprisingly, a stronger immunoreactivity for P2X7 was seen in the neuronal cell bodies compared with little or no immunoreactivity in the satellite glial cells, clearly indicating highly problematic antibody cross-reactivity (Figure 1). Based on the immunoreactivity profile we can only conclude that this antibody is not P2X7 selective in the mouse DRG, and consequently cannot be used in our experimental setup.
Figure 1.
The P2X7 antibody lacking P2X7 selectivity.
A mouse dorsal root ganglion (DRG) section was stained against the satellite glial cell marker glutamine synthetase (GS) in green and against P2X7 in red. Surprisingly, the merge photo shows a strong P2X7 immunoreactivity in the neurons (gray arrow). P2X7 is solely to be expressed in the satellite glial cells in the DRG. Staining method: For details see Gurgor et al., 2016. Briefly, the mouse DRG sections were incubated overnight with rabbit anti-P2X7 (Alomone, APR-008, 1:200) and goat anti-GS (Santa Cruz Biotechnology, Santa Crzu, CA, USA, sc-6640, 1:200) diluted in tris-base buffer (50 mM tris-base in PBS) with 5% donkey serum. After washing steps the tissue sections were incubated with secondary Alexa Fluor antibodies (1:400) in tris-base buffer with 5% bovine serum albumin for 4 hours. They were washed and stained with Hoechst before mounting and analysis with a Zeiss LSM780 confocal microscope. A secondary antibody control was included by omitting the primary antibodies in the staining protocol. As expected, the secondary antibody control showed no fluorescent signal (see Gurgor et al., 2016).
Reports on commercial antibodies with selectivity problems can also be found in the scientific literature. An illustrative example is the work by Ioannis Prassas et al. (2014), describing the use of a commercial antibody-based assay to detect their protein of interest (CUZD1). Preliminary data indicated that CUZD1 was a candidate protein as a clinical marker for pancreatic cancer. After exhaustive testing of the commercial CUDZ1 assay it was however finally concluded that the assay antibodies did not bind CUZD1 but rather another non-homologue protein already established to be a marker for pancreatic cancer. Without the excessive assay testing by Ioannis Prassa et al. (2014), CUDZ1 could have been announced a new cancer biomarker on a false basis. This again highlights the necessity of testing antibody specificity, avoiding false conclusions which ultimately prevents scientific and medical progress.
The general lack of antibody validation in the scientific community has led to various initiatives on antibody validation from both companies and researchers. Professor of Pathology David Rimm from Yale School of Medicine is an antibody validation advocate due to personal experience with scientific reproducibility problems (Baker, 2015). His laboratory utilized an antibody-based test in the attempt to choose a treatment strategy for melanoma patients. The first experiments looked promising but after ordering new antibody vials the results became impossible to reproduce. Contacting the suppliers was not informative as the newly purchased antibody vials were allegedly identical to the previously purchased functional batch. Consequently, the project was abandoned after many years of research. The experience did however inspire Professor David Rimm and his colleagues to write the paper Antibody Validation (Bordeaux et al., 2010) to guide others with antibody validation issues. He subsequently elaborated on the paper at the 1st International Antibody Validation Forum 2014 (Rimm, 2010), where he also suggests the following validation tests:
Tests for sensitivity: IHC to test if endogenous immunoreactivity can be detected in the expected cellular compartment. Test of a cell line series with different degrees of expression for correlation with western blot analysis (dose-response analysis). Use of a quantitative method to determine signal-to-noise ratio and thereby finding the best working concentration of the antibody.
Test for specificity: Test on non-expressing cell lines with and without transfection of the target protein, or testing a highly expressing cell line with and without siRNA knockdown treatment.
Test for reproducibility: Test for lot-to-lot and run-to-run reproducibility. Possibly test a second antibody against the same protein.
These suggestions are suitable to ensure a thorough validation process. Yet, other tests can be used to validate an antibody. In our article Neuronal Death in the Dorsal Root Ganglion after Sciatic Nerve Injury does not Depend on Sortilin (Gurgor et al., 2016) we used tissue from KO mice to validate the antibodies‘ specificity. KO controls are the ultimately control for specificity as they allow testing of the same tissue as the experimental samples. This ensures that specificity controls contain an identical profile of proteins, glycosylations and splice variants, providing a validation experiment with high reliability. However, KO animals are not always available, forcing researchers to settle for less. In these circumstances following the approach by Professor David Rimm will be optimal. Unfortunately, unreliable or insufficient methods are often applied instead, e.g., the use of control peptides which are particular bad controls for selectivity and specificity. The control peptides are often supplied when purchasing antibodies from certain vendors and are the immunogens used to produce the selected antibody. The control peptides are recommended as control by pre-incubation with the antibody before using it in the wanted application. If such a control still provides a signal it is obviously a problematic antibody. However, the lack of signal cannot be used to conclude antibody specificity for the target protein. It only demonstrates that the antibody is able to bind its immunogen but not that it cannot bind other unrelated proteins, giving rise to cross-reactivity and hence lack of selectivity.
Antibody validation is extremely important to support continuous progress in scientific and medical research, however it can be time consuming and expensive. To ease the burden, we can help each other by sharing our antibody validation experiments and antibody experiences in an easily approachable manner. This idea is supported by initiatives such as the website http://www.pabmabs.com where antibody experiences are shared, rated and easily accessed by a search function. Actively contributing to user based resources such as http://www.pabmabs.com will surely facilitate scientific progress by preventing expensive and time consuming experimental detours.
References
- 1.Arnett MG, Ryals JM, Wright DE. Pro-NGF, sortilin, and p75NTR: potential mediators of injury-induced apoptosis in the mouse dorsal root ganglion. Brain Res. 2007;1183:32–42. doi: 10.1016/j.brainres.2007.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker M. Reproducibility crisis: Blame it on the antibodies. Nature. 2015;521:274–276. doi: 10.1038/521274a. [DOI] [PubMed] [Google Scholar]
- 3.Bordeaux J, Welsh A, Agarwal S, Killiam E, Baquero M, Hanna J, Anagnostou V, Rimm D. Antibody validation. Biotechniques. 2010;48:197–209. doi: 10.2144/000113382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gurgor P, Pallesen LT, Johnsen L, Ulrichsen M, de Jong IE, Vaegter CB. Neuronal death in the dorsal root ganglion after sciatic nerve injury does not depend on sortilin. Neuroscience. 2016;319:1–8. doi: 10.1016/j.neuroscience.2016.01.036. [DOI] [PubMed] [Google Scholar]
- 5.Kobayashi K, Fukuoka T, Yamanaka H, Dai Y, Obata K, Tokunaga A, Noguchi K. Differential expression patterns of mRNAs for P2X receptor subunits in neurochemically characterized dorsal root ganglion neurons in the rat. J Comp Neurol. 2005;481:377–390. doi: 10.1002/cne.20393. [DOI] [PubMed] [Google Scholar]
- 6.Prassas I, Brinc D, Farkona S, Leung F, Dimitromanolakis A, Chrystoja CC, Brand R, Kulasingam V, Blasutig IM, Diamandis EP. False biomarker discovery due to reactivity of a commercial ELISA for CUZD1 with cancer antigen CA125. Clin Chem. 2014;60:381–388. doi: 10.1373/clinchem.2013.215236. [DOI] [PubMed] [Google Scholar]
- 7.Rimm D. Use and Abuse of Antibodies in the Lab and in the Clinic. 2010 [Google Scholar]
- 8.Zhang XF, Han P, Faltynek CR, Jarvis MF, Shieh CC. Functional expression of P2X7 receptors in non-neuronal cells of rat dorsal root ganglia. Brain Res. 2005;1052:63–70. doi: 10.1016/j.brainres.2005.06.022. [DOI] [PubMed] [Google Scholar]