Abstract
Human pyruvate kinase isoform M2 (PKM2) is a glycolytic enzyme isoform implicated in cancer. Malignant cancer cells have higher levels of dimeric PKM2, which is regarded as an inactive form of tetrameric pyruvate kinase. This perceived inactivity has fueled controversy over how the dimeric form of pyruvate kinase might contribute to cancer. Here we investigate enzymatic properties of PKM2G415R, a variant derived from a cancer patient, which we show by size-exclusion chromatography and SAXS to be a dimer that cannot form tetramer in solution. Although PKM2G415R binds to FBP, unlike wildtype this PKM2 variant shows no activation by FBP. In contrast, PKM2G415R is activated by the SAICAR, an endogenous metabolite that we previously showed correlates with increased cell proliferation and promotes protein kinase activity of PKM2. Our results demonstrate an important and unexpected enzymatic activity of the PKM2 dimer that likely has a key role in cancer progression.
Keywords: Pyruvate kinase, Quaternary structure, SAICAR
INTRODUCTION
Pyruvate kinase isoform M2 (PKM2) is one of four human pyruvate kinase isoforms (1, 2) that, unlike other pyruvate kinases, is enriched in many types of cancer (2). Highlighting its importance in cell proliferation, replacement of PKM2 with other pyruvate kinase isoforms hinders cancer cell survival and growth (3). Interestingly, PKM2 has long been used as a marker for cancer. In cancerous cells, PKM2 is abundantly found as a dimer, with the level of PKM2 dimer correlated with malignancy and thus has been named tumor PKM2 (4). How PKM2 contributes to malignancy is still controversial, but is believed to arise from a combination of metabolic (5) and cell signaling roles (6).
Although found to exist in different oligomeric forms in cells, like other pyruvate kinases, PKM2 is considered to have maximal pyruvate kinase activity as a tetramer (7). Earlier biochemical studies found the PKM2 dimer to be a nearly inactive (7). The low activity of the dimer was because of a KM for phosphoenolpyruvate (PEP) that is much higher than physiological PEP concentrations, while the tetrameric PKM2 has a low μM KM for PEP (7). This led to the belief that the dimeric PKM2 is a nearly inactive enzyme at physiological PEP concentrations. How a nearly inactive form of pyruvate kinase might contribute to cell proliferation has been unclear. Initially it was thought that the nearly inactive pyruvate kinase allows cancer cells to divert more glycolytic intermediates to anabolic processes (5). However, recent evidence shows that the dimeric PKM2 may have an additional active role in cell growth (8-14).
In addition to its classical role in generating ATP from ADP and the phosphate donor PEP, PKM2 also has been found to phosphorylate protein substrates (9, 10, 12-16). A significant fraction of PKM2 in cancer cells is located in the nucleus (17) and this proportion increases when cancer cell proliferation is stimulated (14). Purification of this nuclear PKM2 showed that it can phosphorylate various nuclear proteins using PEP as a phosphate source (9-11, 16). Mutational studies such as ones using the variant PKM2R399E showed that this dimer-prone mutant promotes PKM2’s unusual protein kinase activity (13-15). This led to the belief that dimeric PKM2 may be responsible for the unusual signaling activity and contribute to cancer. However, a challenging aspect of this model is how dimeric PKM2 could maintain significant activity given its high KM for PEP that, under physiological conditions, would severely limit the ability phosphorylate protein substrates.
Both the oligomeric state and activity of PKM2 are influenced by bound ligands. For decades, PKM2 and several other pyruvate kinase isoforms like PKL and PKR have been known to be induced by fructose-1, 6-bisphosphate (FBP) (18, 19). The binding of FBP shifts PKM2 to a tetrameric oligomeric state and lowers the KM for PEP (7). More recently, we reported that SAICAR, an intermediate of de novo purine nucleotide biosynthesis, activates PKM2 in an isozyme-specific manner (12). SAICAR levels are enriched when cancer cells are in stress conditions, and binding of SAICAR to PKM2 promotes cancer cell survival (12). In addition, the binding of SAICAR induces the protein kinase activity of PKM2 in vitro and in cultured cancer cells (16).
Given the apparent switch in substrate specificity upon binding to SAICAR and the prevalence of dimeric PKM2 in cancer, we sought to determine whether there was a connection between SAICAR binding and the oligomeric state of PKM2. Here we report that SAICAR stimulates PKM2 dimer without inducing formation of a PKM2 tetramer. We focused on a PKM2 variant identified in patient-derived tumors, PKM2G415R (20), and show that this variant can bind to FBP but does not form tetramer. FBP cannot activate PKM2G415R, suggesting that FBP-mediated PKM2 activation requires the tetramerization of PKM2. SAICAR, on the other hand, still activates PKM2G415R, and in fact binds better to the dimeric form of PKM2 and improves substrate binding. Our results support an active role for the PKM2 dimer in cancer, and suggest that the allosteric activation of PKM2 by SAICAR may in fact preferentially act via the dimeric form.
EXPERIMENTAL PROCEDURES
Materials
Plasmid encoding GST-PKM2 in pGEX-6P-1 vector (GE Life Sciences) was custom synthesized by GenScript. Oligonucleotides were synthesized by Operon. Site directed mutagenesis was done using QuikChange XL II kits (Stratagene) and confirmed by sequencing from the T7 promoter and from the T7 terminator sites. Lactate dehydrogenase (LDH) was purchased from Roche. SAICAR was purchased from Toronto Research Chemicals. All other chemicals were from Sigma-Aldrich unless specifically noted otherwise.
Protein Expression and Purification
PKM2 wild type and mutants were expressed and purified from Escherichia coli strain Rosetta (DE3) pLysS (Novagen). Cells were grown in Terrific Broth at 37°C, and the protein expression was induced by the addition of isopropyl-β-D-thiogalactopyranoside (IPTG, 0.5 mM) followed by an incubation for 18 hours at 16°C. Following steps were carried out at 4°C or on ice unless noted otherwise. Cells were collected by centrifugation (4,000 g for 20 min), and the pellet was resuspended in buffer A (20 mM Tris-HCl, pH 8.0, 500 mM NaCl, 1 mM EDTA, 1 mM DTT). Cells were lysed by sonication, and debris was removed by centrifugation (45,000 g for 45 min) and then filtration through a 1.1 micron glass fiber filter (Thermo Scientific). The supernatant was loaded onto a 5 ml GSTrap column (GE Life Sciences) pre-equilibrated in buffer A. The resin was washed with buffer A, and the bound protein was eluted with a linear gradient of buffer B (20 mM Tris-HCl, pH 8, 500 mM NaCl, 1 mM EDTA, 1 mM DTT, 10 mM reduced glutathione) over 20 column volumes. Fractions containing target protein were combined and digested with PreScission protease ((21), GE Life Sciences) in buffer C (50 mM Tris-HCl, pH 6.9, 150 mM NaCl, 1 mM EDTA, 1 mM DTT) overnight. Cleaved proteins were purified by gel filtration chromatography on a HiLoad 16/600 Superdex 200 column (GE Healthcare) with buffer D (10 mM HEPES pH7, 150 mM NaCl, 5 mM MgCl2, 0.25 mM tris(2-carboxyethyl)phosphine hydrochloride (TCEP)). The protein concentrations were determined by measuring the absorbance at 280 nm in denaturing conditions, using a calculated extinction coefficient of 29,910 M−1 cm−1 (22, 23). Protein contents were analyzed by SDS-PAGE and Coomassie Blue staining.
Pyruvate Kinase Activity Assay
Enzyme activity was measured in vitro with a lactate dehydrogenase (LDH) coupled assay as previously described (12). PKM2 were diluted to the desired concentration in the assay buffer (50 mM Tris-HCl, pH 7.6, 100 mM KCl, 6.2 mM MgCl2, 1 mM DTT) and incubated with ligands at 37°C for 30 minutes in a UV-transparent 96-well plate. After incubation, ADP (final concentration 2 mM), NADH (final concentration 220 μM), and LDH (final concentration 5 units/ml) were added to the solution. Reactions were initiated by the addition of PEP (final concentration 150 μM for typical experiments). The rate of NADH oxidation was spectroscopically measured at 340 nm using a Tecan Infinite M2 microplate reader.
Size-exclusion Chromatography
Size exclusion chromatography was carried out using Superdex S200 10/300 GL column (GE Healthcare). Purified PKM2WT or PKM2G415R (0.20 mL in buffer D) was loaded onto the column and eluted with buffer D (0.3 mL/min at 4°C). Samples with FBP or SAICAR were incubated for one hour at 4°C or at 18°C before loading.
Small Angle X-Ray Scattering
Size exclusion chromatography small angle X-ray scattering (SEC-SAXS) experiments were performed at BioCAT (beamline 18-ID, Advanced Photon Source at Argonne National Laboratory (24)). The instrument used for the experiment used a 12 KeV (1.03 Å) X-ray beam and a Pilatus 3 1M detector (Dectris). Samples were passed through a Superdex 200 Increase 10/300 GL column (GE Life Sciences), and the eluted solution was directed to a 1.5 mm quartz capillary sample cell at a rate of 0.75 mL/min. The sample-to-detector distance was ~ 3.5 m, which resulted in a q-range of 0.004-0.38 Å−1 (0.5 sec exposure every 2 sec). Data were corrected for background scattering by subtracting the buffer signal. Radius of gyration (Rg) values were calculated from Guinier approximations and P(R) curves using PRIMUS (25).
Isothermal Titration Calorimetry (ITC)
ITC measurements were performed by using a Microcal ITC200 calorimeter at 25.0°C. Protein solutions were dialyzed against buffer D. Ligands were dissolved in buffer D. Both protein and ligand solutions were degassed in vacuum before use. PKM2WT or PKM2G415R solution (300 μM, 0.22 mL) was placed in the sample cell. FBP or SAICAR solution (3 mM, 20 injections of 2 μL /injection) was injected with 180 s intervals. The baseline-corrected data were analyzed with Microcal Origin software.
RESULTS
PKM2G415R mutant does not form tetramer in solution
To isolate the potential activities for a dimeric form of PKM2, we sought to find a PKM2 variant unable to form a tetramer. Given that PKM2 has long been known to preferentially form a dimer in cancers, known as a “tumor dimer”, we analyzed the locations of mutations in PKM2 isolated from patient-derived tumors (20). One amino acid change, a glycine-to-arginine mutation at position 415 (G415R), is located at the dimer-dimer interface of the PKM2 tetramer (Figure 1A). Based on its location at the interface and the introduction of two opposing positive charges, we speculated that this change might disrupt tetramer formation.
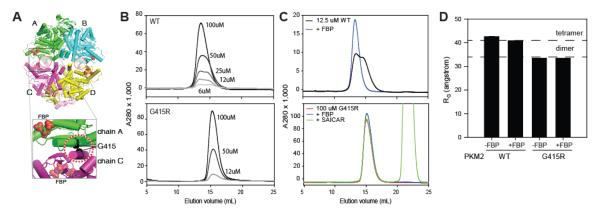
Figure 1.
PKM2G415R is constitutively a dimer. (A) Structure of the PKM2 tetramer (PDB code 3ME3; (29)). The Cα of residue G415 is shown as a black sphere in each of the four chains. (B) PKM2WT (top) but not PKM2G415R (bottom) displays a concentration dependent gel-filtration elution changes. (C) FBP does not alter the oligomeric state of PKM2G415R. Gel filtration experiments show PKM2WT to have an FBP-dependent shift, whereas PKM2G415R was unaffected. (D) Small-angle X-ray scattering experiments suggest that PKM2G415R is constitutively a dimer. Values for the radius of gyration (Rg) of PKM2WT and PKM2G415R were calculated from Guiner plots, using scattering data taken with PKM2 in the presence and absence of FBP. For reference, the Rg values calculated from a PKM2 tetramer crystallographic structure (PDB code 3ME3, chains A-D) or a hypothetical dimer (chains A and B only) are shown as dashed lines.
To test this hypothesis, human PKM2G415R was recombinantly expressed and purified from E. coli and compared to human PKM2WT by gel filtration chromatography (Figure 1B and 1C). At low μM protein concentrations, PKM2WT eluted at two major peak positions, which we interpret as tetrameric and dimeric species. We observed that either increasing concentrations of PKM2WT or addition of FBP predominantly shifted the elution profile to the larger species, consistent with the larger species corresponding to a tetramer and smaller species a dimer. In contrast to wildtype, PKM2G415R eluted in a single peak corresponding to the smaller dimeric species, and failed to shift to the larger tetrameric profile at high (100 μM) protein concentration or with the addition of FBP.
To independently verify that these gel filtration elution profiles reflect differences in oligomeric states, radius of gyration (Rg) values of wild-type and variant PKM2 were determined using SAXS (26) (Figure 1D). To minimize aggregation, SAXS measurements were recorded following an inline size-exclusion column. To maximize signal-to-noise, PKM2 variants were applied to the column at 100 μM concentrations. PKM2WT had an Rg value of 43 Å (Figure 1D and Supplementary Figure S1), consistent with the Rg value calculated from crystallographic structures of tetrameric PKM2 (41 Å). The binding of FBP did not significantly alter the Rg of PKM2WT (41 Å), which is expected at the high concentrations used in these experiments. On the other hand, PKM2G415R showed a much smaller Rg value (33 Å; Figure 1D and Supplementary Figure S1). Guinier plots of these SAXS data showed that the molecular weight of PKM2G415R in solution is approximately one half of the PKM2WT tetramer in solution (Figure S1). The Rg was not affected by FBP and was nearly identical to the Rg value expected from a half of tetrameric PKM2 (34 Å). These SAXS results showed that PKM2G415R indeed remains as a dimer even at high protein concentration or in the presence of FBP.
The PKM2G415R dimer is in its tense form and cannot be activated by FBP
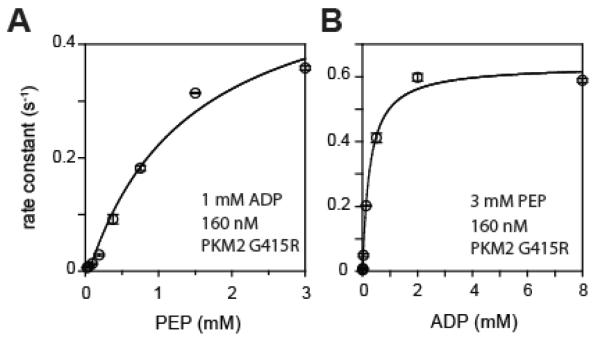
To assess the activation and properties of the PKM2 dimer, we measured the pyruvate kinase activity of PKM2G415R with varying ADP and PEP concentrations (Figure 2). Previous work found the dimeric form of PKM2WT to have relatively low activity, and be considered to be in a tense form (7). Here, we found PKM2G415R to have a weak affinity to PEP (KM 1.2 mM PEP; kcat 0.57 s−1) and a low millimolar KM for ADP (0.25 mM ADP). These values are comparable to the tense form reported for wildtype PKM2 dimer (7, 12).
Figure 2.
Pyruvate kinase activity of PKM2G415R. (A) With titration of PEP, the PKM2G415R variant shows a KM of 1.2 mM PEP and kcat 0.57 sec−1. (B) With titration of ADP, PKM2G415R shows a KM of 0.25 mM ADP.
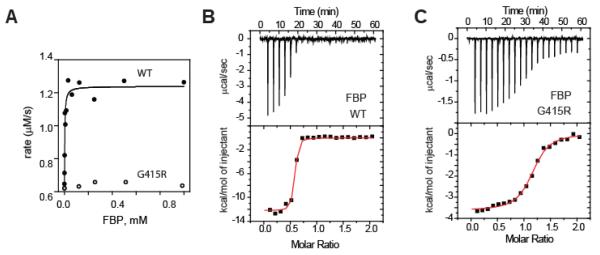
FBP is well-known for promoting the tetramerization and coincident activation of PKM2 (6, 27). Our SAXS and size-exclusion experiments showed that FBP is unable to convert PKM2G415R to a tetrameric form (Fig 1). To see the extent that activity might be impacted, we compared pyruvate kinase activities of PKM2WT and PKM2G415R at varying FBP concentrations (Figure 3A). In contrast to the clear activation of PKM2WT, PKM2G415R appeared to be unaffected by the presence of FPB and remain in the tense form.
Figure 3.
PKM2G415R binds to FBP but is not activated. (A) Pyruvate kinase activity of PKM2WT (filled circles) and PKM2G415R (empty circles) in the presence of FBP. (B and C) Isothermal titration calorimetric analysis of FBP binding to (B) PKM2WT (0.54 sites, Kd 0.21 μM, ΔH: −12.2 kcal/mole) and (C) PKM2G415R (1.0 sites, Kd 1.0 μM, ΔH −4.7 kcal/mole).
A simple possibility to explain these results could be that the G415R substitution interferes with FBP binding. To determine whether FBP still binds to PKM2G415R, the interaction of FBP with PKM2 was analyzed by isothermal titration calorimetry (ITC; Figure 3B-C). For PKM2WT, we observed an apparent binding isotherm consistent with a sub-μM KD (Figure 3B). The enthalpy contribution to the binding suggests that FBP induced formation of the PKM2 tetramer (calculated buried surface area 2400 Å2). Interestingly, for PKM2G415R, titration of FBP promoted a clear change in measured heat released, with a KD approximately 5-fold higher than that of PKM2WT (Figure 3C). Compared to the FBP-PKM2WT interaction, the enthalpy contribution was much smaller and consistent of FBP binding (buried surface area 326 Å2) without drastic changes in PKM2 quarternary structure. Taken together, these results demonstrate that PKM2G415R can bind to FBP, but fails to exhibit activation in concert with conversion to a tetrameric form.
The PKM2G415R dimer can be activated by SAICAR without forming tetramer
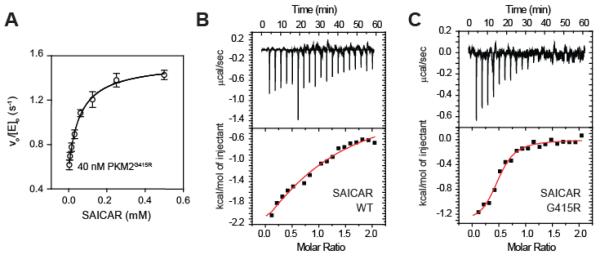
Our previous work showed that PKM2WT can be activated by SAICAR (12). To see if activation might be intrinsically coupled to formation of the PKM2 tetramer, we monitored the distribution of size exclusion peaks after incubation with SAICAR. The addition of SAICAR had no measurable effects on oligomeric state for either PKM2WT or PKM2G415R (Fig 1). This raised the possibility that SAICAR primarily exerts its influence on PKM2 without altering its oligomerization state. Given the inability of PKM2G415R to tetramerize, we therefore used this cancer variant to investigate whether SAICAR could stimulate the dimeric form of PKM2. Remarkably, pyruvate kinase assays performed showed a distinct 2-fold activation by SAICAR (EC50 60 ± 20 μM; 1.0 ± 0.4 SAICAR per PKM2 monomer; Figure 4A). Importantly, the EC50 is approximately 5-fold lower than the EC50 (0.3 mM) observed with PKM2WT activation by SAICAR (12), suggesting that SAICAR binds to PKM2G415R better than to PKM2WT.
Figure 4.
SAICAR still binds to PKM2G415R and activates it. (A) Normalized pyruvate kinase activity of PKM2G415R in the presence of SAICAR. (B and C) Isothermal titration calorimetric analysis of SAICAR binding to (B) PKM2WT (1.0 site per PKM2WT monomer; Kd 300 μM; ΔH −5 kcal/mole) and (C) PKM2G415R (0.48 site per PKM2G415R monomer; Kd 12 μM, ΔH −1.4 kcal/mole).
Tighter binding of SAICAR to PKM2G415R compared with PKM2WT was further supported by ITC experiments (Figure 4B-C). Compared with PKM2WT, which bound to SAICAR with an affinity of 300 ± 90 μM, PKM2G415R bound to SAICAR significantly better, with a KD of 12 ± 3 μM. Taken together, these results demonstrate that SAICAR binds to PKM2 dimer and activates it without inducing tetramer.
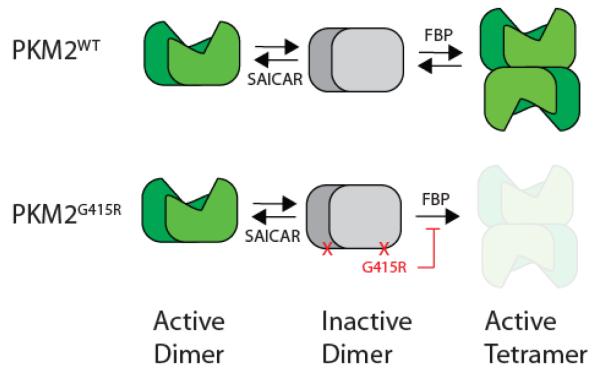
Taken together, these results show that there are two different modes of activating inactive dimeric PKM2 (Figure 5). First, dimeric PKM2 can be activated by becoming tetrameric PKM2. This oligomerization state change is induced by FBP. Alternatively, inactive PKM2 dimer can be activated by forming an active PKM2 dimer. SAICAR binding is an example where inactive PKM2 dimer becomes an active one without having its oligomerization state altered. The PKM2G415R variant from patient-derived tumor shows that the tetramer formation is dispensable and that it can be activated by an alternative mechanism.
Figure 5.
Different modes of PKM2 activation by FBP and SAICAR.
DISCUSSION AND CONCLUSIONS
The relevance of PKM2 in cancer has been demonstrated in multiple systems (6, 27, 28). Interestingly, highly proliferating cells have higher levels of dimeric PKM2, which was widely considered to be an inactive form of pyruvate kinase. Many different models have been proposed to explain how nearly inactive pyruvate kinase can contribute to malignant cell growth. The results presented here show that dimeric PKM2 should not be considered a constitutively inactive form, but one that can be specifically activated by ligands such as SAICAR.
Our findings open new possibilities for regulation and activation of PKM2. Whereas tetramerization is one route to PKM2 activation, the PKM2 dimer is not merely an inactive pool but represents a physiologically important target for specific activation. Our findings that SAICAR bound more tightly to the dimeric PKM2G415R variant suggests that the dimeric form can be targeted by unique regulatory ligands that allow for richer responses to the metabolic state of the cell. We speculate that the pressure for high proliferative growth in cancer cells selects for variants of PKM2 such as G415R that stabilize the dimeric state, which in turn favor ligand binding and biochemical properties of the PKM2 dimer. With dual substrate specificity, the ability of PKM2 to phosphorylate protein targets has been linked to cellular proliferation (6, 10, 13, 16, 28). We previously reported that SAICAR-bound PKM2 can phosphorylate proteins much more efficiently than the FBP-bound form (16). The higher activity we report here for dimeric PKM2 in the presence of SAICAR is consistent with the previous suggestion that protein kinase activity is accomplished by the dimeric form (6). We anticipate that other means of controlling the dimertetramer equilibrium, such as through post-translational modifications, will also be uncovered, and that proper allosteric regulation of the PKM2 dimer will prove to be an important contributor to cellular adaptation.
Supplementary Material
ACKNOWLEDGMENT
We thank Shelby Winans for the preparation of the PKM2G415R plasmid, and Cynthia Wolberger and Beverly Wendland for allowing us to use their equipment.
Funding Sources
This work was supported by National Institute of Health grants (R21CA181751 to G.D.B. and Y.-S.L.; R01CA168658 to Y.-S.L.). This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DEAC02-06CH11357. SAXS experiment was supported by grant 9 P41 GM103622 from the National Institute of General Medical Sciences of the National Institutes of Health. Use of the Pilatus 3 1M detector was provided by grant 1S10OD018090-01 from NIGMS.
ABBREVIATIONS
- PKM2
pyruvate kinase isoform M2
- FBP
fructose-1,6-bisphosphate
- SAICAR
succinyl-5-aminoimidazole-4-carboxamide-1-ribose-5′-phosphate
- ITC
isothermal titration calorimetry
- SAXS
small angle X-ray scattering
Footnotes
Supporting Information. Figure S1 showing additional SAXS results. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
M. Y. prepared all reagents and performed biochemical experiments; S. C., J.M.T., and L. P. carried out SAXS experiments and analysis; M.Y., Y.-S.L., and G.D.B. designed experiments and analyzed data; Y.-S.L. and G.D.B. conceived the project; M.Y., Y.-S.L., and G.D.B. wrote the manuscript.
REFERENCES
- (1).Filipp FV. Cancer metabolism meets systems biology: Pyruvate kinase isoform PKM2 is a metabolic master regulator. J. Carcinog. 2013;12:14. doi: 10.4103/1477-3163.115423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Macintyre AN, Rathmell JC. PKM2 and the Tricky Balance of Growth and Energy in Cancer. Mol. Cell. 2011;42:713–714. doi: 10.1016/j.molcel.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, Fleming MD, Schreiber SL, Cantley LC. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- (4).Mazurek S, Boschek CB, Hugo F, Eigenbrodt E. Pyruvate kinase type M2 and its role in tumor growth and spreading. Semin. Cancer Biol. 2005;15:300–308. doi: 10.1016/j.semcancer.2005.04.009. [DOI] [PubMed] [Google Scholar]
- (5).Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Yang W, Lu Z. Pyruvate kinase M2 at a glance. J. Cell Sci. 2015;128:1655–1660. doi: 10.1242/jcs.166629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Dombrauckas JD, Santarsiero BD, Mesecar AD. Structural basis for tumor pyruvate kinase M2 allosteric regulation and catalysis. Biochemistry. 2005;44:9417–9429. doi: 10.1021/bi0474923. [DOI] [PubMed] [Google Scholar]
- (8).Gao X, Wang H, Yang JJ, Chen J, Jie J, Li L, Zhang Y, Liu ZR. Reciprocal regulation of protein kinase and pyruvate kinase activities of pyruvate kinase M2 by growth signals. J. Biol. Chem. 2013;288:15971–15979. doi: 10.1074/jbc.M112.448753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Yang W, Zheng Y, Xia Y, Ji H, Chen X, Guo F, Lyssiotis CA, Aldape K, Cantley LC, Lu Z. ERK1/2-dependent phosphorylation and nuclear translocation of PKM2 promotes the Warburg effect. Nat. Cell Biol. 2012;14:1295–1304. doi: 10.1038/ncb2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Yang W, Xia Y, Hawke D, Li X, Liang J, Xing D, Aldape K, Hunter T, Alfred Yung WK, Lu Z. PKM2 phosphorylates histone H3 and promotes gene transcription and tumorigenesis. Cell. 2012;150:685–696. doi: 10.1016/j.cell.2012.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Yang W, Xia Y, Cao Y, Zheng Y, Bu W, Zhang L, You MJ, Koh MY, Cote G, Aldape K, Li Y, Verma IM, Chiao PJ, Lu Z. EGFR-induced and PKCepsilon monoubiquitylation-dependent NF-kappaB activation upregulates PKM2 expression and promotes tumorigenesis. Mol. Cell. 2012;48:771–784. doi: 10.1016/j.molcel.2012.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Keller KE, Tan IS, Lee YS. SAICAR stimulates pyruvate kinase isoform M2 and promotes cancer cell survival in glucose-limited conditions. Science. 2012;338:1069–1072. doi: 10.1126/science.1224409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Gao X, Wang H, Yang JJ, Liu X, Liu ZR. Pyruvate kinase M2 regulates gene transcription by acting as a protein kinase. Mol. Cell. 2012;45:598–609. doi: 10.1016/j.molcel.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Yang W, Xia Y, Ji H, Zheng Y, Liang J, Huang W, Gao X, Aldape K, Lu Z. Nuclear PKM2 regulates beta-catenin transactivation upon EGFR activation. Nature. 2011;480:118–122. doi: 10.1038/nature10598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Yang W, Lu Z. Nuclear PKM2 regulates the Warburg effect. Cell Cycle. 2013;12:3154–3158. doi: 10.4161/cc.26182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Keller KE, Doctor ZM, Dwyer ZW, Lee YS. SAICAR induces protein kinase activity of PKM2 that is necessary for sustained proliferative signaling of cancer cells. Mol. Cell. 2014;53:700–709. doi: 10.1016/j.molcel.2014.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Stetak A, Veress R, Ovadi J, Csermely P, Keri G, Ullrich A. Nuclear translocation of the tumor marker pyruvate kinase M2 induces programmed cell death. Cancer Res. 2007;67:1602–1608. doi: 10.1158/0008-5472.CAN-06-2870. [DOI] [PubMed] [Google Scholar]
- (18).Taylor CB, Bailey E. Activation of liver pyruvate kinase by fructose 1,6-diphosphate. Biochem. J. 1967;102:32C–33C. doi: 10.1042/bj1020032c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Kahn A, Marie J, Boivin P. Pyruvate kinase isozymes in man. II. L type and erythrocyte-type isozymes. Electrofocusing and immunologic studies. Hum. Genet. 1976;33:35–46. doi: 10.1007/BF00447284. [DOI] [PubMed] [Google Scholar]
- (20).Israelsen WJ, Dayton TL, Davidson SM, Fiske BP, Hosios AM, Bellinger G, Li J, Yu Y, Sasaki M, Horner JW, Burga LN, Xie J, Jurczak MJ, DePinho RA, Clish CB, Jacks T, Kibbey RG, Wulf GM, Di Vizio D, Mills GB, Cantley LC, Vander Heiden MG. PKM2 isoform-specific deletion reveals a differential requirement for pyruvate kinase in tumor cells. Cell. 2013;155:397–409. doi: 10.1016/j.cell.2013.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Walker PA, Leong LEC, Ng PWP, Tan SH, Waller S, Murphy D, Porter AG. Efficient and Rapid Affinity Purification of Proteins Using Recombinant Fusion Proteases. Biotechnology. 1994;12:601–605. doi: 10.1038/nbt0694-601. [DOI] [PubMed] [Google Scholar]
- (22).Edelhoch H. Spectroscopic determination of tryptophan and tyrosine in proteins. Biochemistry. 1967;6:1948–1954. doi: 10.1021/bi00859a010. [DOI] [PubMed] [Google Scholar]
- (23).Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. Proteomics Protocols Handbook. Humana Press; Totowa: 2005. pp. 571–607. [Google Scholar]
- (24).Mathew E, Mirza A, Menhart N. Liquid-chromatography-coupled SAXS for accurate sizing of aggregating proteins. J. Synchrotron. Radiat. 2004;11:314–318. doi: 10.1107/S0909049504014086. [DOI] [PubMed] [Google Scholar]
- (25).Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J. Appl. Crystallogr. 2003;36:1277–1282. [Google Scholar]
- (26).Kikhney AG, Svergun DI. A practical guide to small angle X-ray scattering (SAXS) of flexible and intrinsically disordered proteins. FEBS Lett. 2015;589:2570–2577. doi: 10.1016/j.febslet.2015.08.027. [DOI] [PubMed] [Google Scholar]
- (27).Israelsen WJ, Vander Heiden MG. Pyruvate kinase: Function, regulation and role in cancer. Semin. Cell Dev. Biol. 2015;43:43–51. doi: 10.1016/j.semcdb.2015.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Dong G, Mao Q, Xia W, Xu Y, Wang J, Xu L, Jiang F. PKM2 and cancer: The function of PKM2 beyond glycolysis. Oncol. Lett. 2016;11:1980–1986. doi: 10.3892/ol.2016.4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Anastasiou D, Yu Y, Israelsen WJ, Jiang JK, Boxer MB, Hong BS, Tempel W, Dimov S, Shen M, Jha A, Yang H, Mattaini KR, Metallo CM, Fiske BP, Courtney KD, Malstrom S, Khan TM, Kung C, Skoumbourdis AP, Veith H, Southall N, Walsh MJ, Brimacombe KR, Leister W, Lunt SY, Johnson ZR, Yen KE, Kunii K, Davidson SM, Christofk HR, Austin CP, Inglese J, Harris MH, Asara JM, Stephanopoulos G, Salituro FG, Jin S, Dang L, Auld DS, Park HW, Cantley LC, Thomas CJ, Vander Heiden MG. Pyruvate kinase M2 activators promote tetramer formation and suppress tumorigenesis. Nat. Chem. Biol. 2012;8:839–847. doi: 10.1038/nchembio.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.