Abstract
Context
We define the scope and needs within the new discipline of computational pathology, a discipline critical to the future of both the practice of pathology and, more broadly, medical practice in general.
Objective
To define the scope and needs of computational pathology.
Data Sources
A meeting was convened in Boston, Massachusetts, in July 2014 prior to the annual Association of Pathology Chairs meeting, and it was attended by a variety of pathologists, including individuals highly invested in pathology informatics as well as chairs of pathology departments.
Conclusions
The meeting made recommendations to promote computational pathology, including clearly defining the field and articulating its value propositions; asserting that the value propositions for health care systems must include means to incorporate robust computational approaches to implement data-driven methods that aid in guiding individual and population health care; leveraging computational pathology as a center for data interpretation in modern health care systems; stating that realizing the value proposition will require working with institutional administrations, other departments, and pathology colleagues; declaring that a robust pipeline should be fostered that trains and develops future computational pathologists, for those with both pathology and non-pathology backgrounds; and deciding that computational pathology should serve as a hub for data-related research in health care systems. The dissemination of these recommendations to pathology and bioinformatics departments should help facilitate the development of computational pathology.
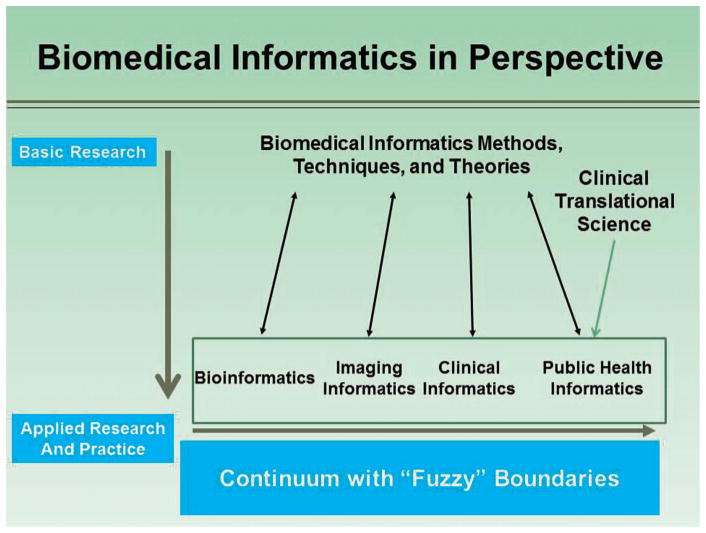
The recent growth of electronic health records ([EHRs]; including the massive amounts of information in clinical laboratory information systems), coupled with powerful new computational approaches for analyzing large data sets, has created an opportunity to establish a new discipline within pathology and laboratory medicine: computational pathology. This proposed discipline leverages information systems architecture, semantic ontology approaches to data management and engineering, and algorithms and mathematic approaches from machine learning and biomedical informatics to focus on unique computational challenges—particularly those that derive from the broad range of unique data streams created in our diagnostic, translational, and basic science laboratories. From the perspective of the field of pathology and laboratory medicine, computational pathology is the natural outcome of the growth and complexity of current clinical laboratories. Its predicted emergence is similar to what happens when any data-generating organization becomes capable of making so many primary observations that users of the service cannot effectively use all of them and/or cannot easily glean the relevant relationships among them. Anatomic pathology and laboratory medicine are at this stage now, and computation is the natural progression of the growth, complexity, and importance of the pathology laboratory. Clearly, pathology departments will take different approaches to frame computational pathology: some will establish it as their own discipline within pathology, whereas others will integrate their efforts into their health system or medical school efforts in the broader field of biomedical informatics. Regardless of the approach, computational pathology could in some ways be considered as a focused discipline within the broad area of biomedical informatics. In fact, as can be seen in the Figure, computational pathology is a direct offshoot of biomedical informatics as defined by the American Medical Informatics Association,1 but with a focused application to the data sets and information generated in pathology departments.
Figure.
Computational pathology fits within the spectrum of biomedical informatics, as depicted by the American Medical Informatics Association (written communication March 19, 2015). Used with permission.
Although attempts have been made to paint broad definitional brushstrokes for this proposed new discipline,2 more precise outlines on how to foster such a discipline remain undrawn. To address this need, a meeting was convened in Boston in July 2014 that was held just prior to the annual Association of Pathology Chairs meeting. The meeting was attended by a variety of pathologists, including individuals highly invested in pathology informatics as well as the chairs of pathology departments (see a full list of attendees in the Appendix).
DEFINITION
As the field of computational pathology evolves, practical working definitions have included:
An approach to diagnosis that incorporates multiple sources of raw data (eg, clinical electronic medical records, laboratory data including “-omics,” and imaging [both radiology and pathology imaging]); extracts biologically and clinically relevant information from these data; uses mathematic models at the molecular, individual, and population levels to generate diagnostic inferences and predictions; and presents this clinically actionable knowledge to customers through dynamic and integrated reports and interfaces, enabling physicians, patients, laboratory personnel, and other health care system stakeholders to make the best possible medical decisions.2
More generally, using computation for the interpretation of multiparameter data to improve health care.
The former, more specific definition is useful in thinking out the parts needed to create computational pathology, whereas the latter, more general definition is more useful in understanding its value propositions, particularly to health care systems.
THE VALUE PROPOSITIONS
The meeting participants firmly believed that there were powerful value propositions for the application of biomedical informatics within pathology departments, supporting the concept of “computational pathology.” An apt analogy was made by one of us (J.R.G.) to meteorology, a discipline that is capable of generating large amounts of primary data from multiple modalities but that has also learned how to use computational approaches to analyze these complex data and present visualizations and interpretations to a broad audience: by turning on the television or visiting a Web site, even briefly, users can easily understand what is happening, what will happen, and what is actionable. Just as meteorologists are the major users of computational meteorology, we envision that pathologists will be the major users of computational pathology, with providers and patients being the users of the actionable knowledge generated by computational pathology—just as the public is already the avid user of information provided by meteorology.
The value of computational pathology should be thought of relative to the health care system and relative to the field of pathology and laboratory medicine. For health care systems, there is a central role for data in guiding individual and population-based health care. The scope of data and the unique forms and domain-level knowledge required to understand data from pathology information systems, as well as the advent of systems to collect and analyze such data, highlight computational pathology as a discipline driving the interpretation and integration of pathology-based data sets in a modern health care system. Expected benefits would be improved use of data provided by existing diagnostics as well as new testing methods entering the realm of clinical care, such that health care systems can improve their quality of care and also leverage basic and translational medicine initiatives that together push the boundaries of clinical care. Possible examples could include: (1) determining disease likelihood in advance of the patient getting sick, and disease trend in advance of complications setting in (“minimize surprise” using predictive medicine); (2) measuring quality and cost to improve both within the health care system; (3) serving as a hub for data-related research in the health care system; and (4) providing the analytic basis for more precise patient selection in clinical trials.
Realizing this value will require teams of practitioners skilled in modern biomedical informatics who, through their computational and analytic skills, will work alongside health system administrators and other (nonpathology) departments. Pathologists must be “at the table” within our own institutions and with our pathology and nonpathology colleagues across the country.
The value of computational pathology must also be considered relative to the field of pathology and laboratory medicine. The field needs to create a culture that considers the computer and computation as being as central to pathology as the microscope. To do so will require the field to determine ways to bring biomedical informaticists, computer scientists, mathematicians, statisticians, and systems biologists into pathology. This will require not only creating career paths (within our departments as well as nationally in pathology societies) and mentoring systems for nonpathologists, but it will also require clearly articulating the value proposition of computational pathology to such individuals.
Finally, and perhaps most importantly for the field of pathology and laboratory medicine, the group felt it vital to note that the future of computational pathology remains ours to lose; thus, we must view it as essential to the future of the discipline. With this belief in the critical nature of computational pathology in mind, the key conclusions of the meeting are discussed below, specifically for clinical, research, and training aspects.
KEY CONCLUSIONS: THE IMPACT OF COMPUTATIONAL PATHOLOGY ON CLINICAL CARE
The Clinical Value Proposition
Computational pathology has clinical value in all aspects of medicine, via a focus on computational methods that incorporate clinical pathology, anatomic pathology (including digital imaging), and molecular/genomic pathology data sets. Although its application has been more traditionally associated with bioinformatics pipelines for next-generation sequencing as well as image analysis algorithms on whole-slide images, computational pathology will also likely have a widespread impact by deriving clinically meaningful relationships among the observational data collected during routine clinical pathology and anatomic pathology testing. When combined with the ability of computational algorithms to help providers select the most appropriate and efficient laboratory tests, computational pathology will drive faster and more accurate preventive management, diagnosis, and therapeutic intervention for patients. Continuously expanding knowledge about the human genome, genomes of infectious agents, and the combination of an individual’s genome and specific environments, will only be useful in preventive management if computational pathology algorithms are developed to link these data and provide them to physicians and patients in a consumable format. Similarly, rapid and accurate diagnosis of underlying and subclinical conditions based on mild and often ignored changes in laboratory results and other data will avoid delays in diagnosis that lead to worse patient outcomes and increased cost. Once therapy is initiated, computational pathology algorithms can detect the effectiveness and/or complications from therapy to enable more precise management of the individual patient. Development, maintenance, and continuous quality improvement of algorithms used to order the most clinically useful tests, and to derive clinically actionable meaning from the breadth of the patient’s results, will require ongoing specialty expertise from pathologists.
An early successful example of the clinical value proposition for computational pathology is in the clinical adoption of next-generation sequencing for diagnosis of germ line and somatic mutations. Most institutions have seen a significant and tangible value to using computational approaches to interpret sequencing data within the contexts of their pathology laboratories. Indeed, attention to the potential benefits of deep sequencing data has played prominently in both the scientific and lay presses. Nonetheless, there are large opportunities for further gains in clinical utility as the field expands from targeted panels to exomes to genomes. Moreover, computational models from animal data, evolutionary biology, and system biology may provide insights that in turn drive clinical developments.
Other tantalizing examples provide a window into the potential future that computational pathology can bring to all of medicine. For example, computational models have been developed that can predict anemia from algorithmic examination of frequently ignored complete blood count parameters.3 Similarly, via computational algorithms the rate of progression to hepatic fibrosis has been correlated with the hepatitis C virus sequence in a patient.4 Another example is the prediction of pathogenicity of a particular nucleotide variant using complex models (PolyPhen, SIFT, etc).5–9 Inclusion of these tools, as well as retrieval of data from dbSNP, ClinVar, ClinGen, COSMIC, and other databases, will help pathologists and geneticists interpret the clinical significance of variants identified either in a patient’s tumor or in his or her germ line DNA, and will help derive meaning at the population level for new variants discovered during clinical testing.10–16 In hematology, machine learning methods applied to analyzing automated complete blood count parameters have been used to accurately identify patients with myelodysplasia.17 Software is actively being developed that provides the next level of “logistical computational” software, interfaces, and tools that allow user-friendly movement of next-generation sequencing files and data sets through instruments and the bioinformatics pipeline to the laboratory information system; between the laboratory information system and online curated data sources that allow a pathologist to generate an accurate and up-to-date report; and from the laboratory information system to the EHRs as both human-readable and computationally mineable data.11,12,18,19 In addition, computational quality assurance and quality control tools are being incorporated into the analysis of these large and multidimensional data sets to ensure that the data meet quality standards.
However, the delivery of precision medicine in a rapid and affordable manner with the highest degree of accuracy possible will only be realized if institutions and departments support the development of computational pathology as a field at the very foundation of pathology practice.20 President Obama’s recent announcement of the Precision Medicine Initiative further underscores the importance of computational pathology.21 This leads to three key conclusions.
1. The Importance of Being “At The Table” at One’s Institution
Institutional leadership by pathologists in this area is absolutely critical in realizing the future that computational pathology can provide. Implementation of computational pathology will require deeply integrated and foundational changes to existing information architectures. Such changes require the initial and continued support of institutional leadership in making the appropriate financial and infrastructural decisions that forge a pathway for computational and clinically actionable algorithms to succeed. These changes require that institutional leadership partner with biomedical informaticists to leverage existing EHR technology and to implement new uses for EHRs and associated software that can deliver additional value—even if the EHR was not developed with big data analytics as a necessary component. EHRs are gold mines of big data that are not being used effectively for research and discovery. Pathologists who know the importance of computational pathology and its application to current and future patient care through research and discovery can help guide institutional leaders to support investments that lay a foundation for future success.
2. The Importance of Interfacing With Other (Nonpathology) Departments at One’s Institution
The benefits of computational pathology hinge on interpretation of existing data, not all of which are generated by the laboratories. Pharmacy data, radiology data, clinical notes, vital signs, and procedural test results all have an impact on clinical decisions, as does clinical decision support for laboratory orders and algorithmic interpretation of sets of data. For instance, the decision whether or not to transfuse allogeneic red blood cells to a patient may depend not only on the patient’s hemoglobin level but also on the patient’s existing antibody profile, the status of the antibody screen, vital signs, central venous oxygen saturation, clinical diagnoses, the patient’s prior reactions to transfusion, and other data. Automated comparisons of radiologic and pathologic data could be built to detect discrepancies between results, prompting review. No automated or computerized comparisons of radiologic and pathologic data were found in the recent medical literature, likely because both radiology and pathology reports have traditionally been (and still are in most cases) written in free text. However, two tantalizing applications have appeared, one commercial (Montage; http://montagehealthcare.com; accessed April 26, 2015) and one open source (Text Information Extraction System or TIES; http://ties.upmc.com/; accessed April 1, 2015), that have the capacity to encode both radiology and pathology using a common structured language model. This allows easier integration of these two disparate free text corpuses. Free text is notoriously difficult to mine even with the best natural language processing algorithms. Although analysis of strictly pathology laboratory data to drive new meaning from these multiple parameters will certainly benefit patient care, the strength of computational pathology will truly be realized when these models, tools, and data are developed and applied in conjunction with specialists in other areas of medicine.
3. The Importance of Sharing Best Practices Among Pathology Departments Across the Country
The overarching goal of computational pathology, whether it be for clinical or research purposes, is to improve patient care. Pathology and laboratory medicine have a long history of sharing practice standards, laboratory procedures, and clinical practice guidelines to improve the standard of medical care across the country. Discovery and implementation of computational pathology algorithms should be no different. Inclusive collaboration helps accelerate research and discovery, and feedback from institutions that implement novel computational algorithms can be used to refine and strengthen computational models. Examples of standards development for best practices are still relatively new in the area of computational pathology, but several white papers are expected shortly from several major molecular and genomic pathology organizations on the subject of interpretation of next-generation sequencing. It is hoped that the open publication of such guidelines will set a tone of shared best practices in the community, one that will extend eventually beyond next-generation sequencing. In addition, although not worked out in detail at the meeting, suggestions for other mechanisms of sharing best practices included the creation of “playbooks” on how to go about setting up computational approaches, centrally validated “toolboxes” that would be open to participating users, and user groups oriented around particular areas of computational pathology. However, best practices should also be shared with specialties outside of pathology, and lessons learned from nonpathology specialties should also be incorporated into the computational pathology framework. Shared knowledge reduces time between development in the computer lab and implementation in the clinical setting.
KEY CONCLUSIONS: THE IMPACT OF COMPUTATIONAL PATHOLOGY ON RESEARCH
Integrative analyses of large, high-throughput data sets play increasingly important roles in many areas of science and engineering. Computational pathology builds on this base by demonstrating the use of methods, including machine learning as well as statistical and computational modeling, to understand complex disease states. To support the development of novel diagnostics and predictive algorithms for diagnosing disease, selecting appropriate therapies, and assisting in the prediction of patient outcomes, quantitative characterization of combined molecular, structural, and temporal information from multiple biomedical inputs will be necessary. Data integration may take the traditional form of a combination of interpretations or, more intriguingly, a normalization and fusion of quantitative data in metaspaces created by artificial intelligence approaches. Such tools have a ready application in many areas of pathology, including cancer diagnosis, genomic interpretations, and the means to extract new knowledge and information from existing clinical and anatomic pathology data sets. In the case of cancer analyses, the capacity to model interactions among (1) cancer genetics, genomics, proteomics, metabolomics, epigenetics, glycomics, and the expososome; (2) tumor and stromal architectures; and (3) immune interactions, will play crucial roles in the development of improved means to diagnose, manage, and predict long-term outcomes. Furthermore, these characterizations will support studies of tumor initiation, development, heterogeneity, invasion, and metastasis. Such integrated analyses will require data repositories that combine imaging, molecular, and other biomarkers that can be leveraged for a variety of machine learning and image analysis methods; in the near future we anticipate that computational modeling in cancer and other disease states will play an increasingly important role.
A variety of groups have launched projects that embody this research approach. These groups have been able to link morphologic and molecular cancer characterizations and to make use of combined imaging and molecular signatures to substratify patient populations. The National Cancer Institute has recently published a report that articulates approaches taken by the pathology and radiology research community to linking imaging phenotypes with large-scale genomic analyses.22 Characterization of tissue morphology–related phenotypes is a complex effort that requires the development and deployment of pipelines consisting of image analysis, feature extraction, and machine learning algorithms.23 A number of groups have developed tools designed to create, explore, and quantify rich morphologic and molecular characterizations of tissue samples at microscopic resolution and to incorporate other data sets in cancer analyses, including genomic and proteomic analyses.24–26 Such software and methods will enable researchers to assemble and visualize detailed, multiscale descriptions of tissue morphologic changes originating from a wide range of microscopy instruments. These tools will make it possible to efficiently manage, interrogate, and explore microscopic imaging data at multiple scales, and to identify and analyze features across individuals and cohorts.
In other areas of pathology, computational research programs cover a range of activities, from the modeling and extraction of new and clinically relevant information from clinical laboratory data sets3 to developing novel algorithmic approaches for the modeling of complex systems, such as the longitudinal dynamics of the gut microbiota.27,28 Furthermore, biomedical activities developing disease classifiers almost universally include clinical and/or anatomic pathology data points in their models. Such activities commonly employ multidisciplinary teams and offer ready opportunity for pathologists to work closely with statisticians, medical informaticists, and computational biologists, while ensuring that these tools retain a focus on needs within the specialty.
Pathology also offers unique opportunities to develop broad and scalable infrastructure to support basic, translational, and clinical research programs. Pathology-developed tools, including Crimson29 and caLIMS,30 designed to integrate with the National Institutes of Health roadmap projects i2b231 and the cancer Biomedical Informatics Grid (caBIG),32 allowed both programs to realize their aims and develop high-throughput and cost-effective methods to obtain raw materials and data sets for analyses.
KEY CONCLUSIONS: COMPUTATIONAL PATHOLOGY AND THE NEED FOR EDUCATION AND EXPERTISE
Promoting computational pathology through training and recruitment is a major challenge at all levels of pathology practice and training. Exposure to the field and its underlying concepts needs to occur during medical school, and preferably at the level of undergraduate education, in addition to teaching the needed quantitative skills in pathology residency programs. Integration of computational skills into careers in pathology can lead to a range of personnel, including full-fledged directors of computational pathology in departments, computational “champions” within pathology groups, or simply intelligent consumers of computational diagnostic tools. Flexibility and creative thinking about such career paths will be important. A barrier for many pathology departments to offering such training, either to new trainees or existing staff who want to expand their computational skills, is the lack of expertise within departments. A possible solution to building significant bases of computational knowledge is to look outside of pathology departments, creating strong collaborations with or even directly hiring scientists with quantitative expertise, such as statisticians, computer scientists, and mathematicians. At several universities, pathology departments have teamed up with departments of biomedical engineering, computer science, and/or physics to develop new avenues of integrated research as well as new pipelines for future faculty. These alliances open the door for a role in training nonphysician contributors and enriching the computational environment within pathology departments through close integration of these personnel; the goal is for these individuals to make significant intellectual contributions to the field, rather than simply act as technicians.
Alternatively, it would be ideal for computational pathologists to emerge from our pathology departments with a deep understanding of clinical and/or anatomic pathology, but this is going to be a difficult undertaking in light of the pressures facing the field today (eg, financial constraints, resident time restrictions, educational infrastructure). Nonetheless, the next three sections illustrate some approaches taken by departments to create computational pathologists as part of pathology education, including residency and fellowship training as well as in partnership with other computational disciplines.
Creating a “Computational Culture” in Pathology Departments
Before computational pathology (or even training in computational pathology) can succeed, pathologists and especially residents must decide to engage in, and build careers around, informatics and/or computation. Since 2009, Massachusetts General Hospital (Boston, Massachusetts) and Brigham and Women’s Hospital (Boston, Massachusetts) have sent 10 of their residents in total between the two programs to informatics fellowship programs. This is well above the national average and indicates that departmental culture may play a role in those decisions. Recently, the Partners Pathology Informatics Fellowship Program has identified a number of environmental and structural factors that have a positive impact on a resident’s engagement in pathology informatics and computation,33 which include:
A strong, clear vision, articulated by department leadership, of the importance of computation to the future of pathology;
A strong informatics presence, widely distributed across the department (not limited to a formal division) and continuous over time (not limited to a couple of weeks per year); from a resident’s point of view, such a presence normalizes informatics/computation in pathology because residents see respected pathologists signing out cases, running laboratories, and doing informatics;
Informatics/computational opportunities scaled to the skills/experience of each resident; increasingly, the risk is not that we do not teach the resident who lacks computational interest/skills, but that we fail to inspire the residents who have them;
Long-term, longitudinal resident engagement (with growing responsibility and ownership) in active informatics/computational services, because computational initiatives have a different cadence than most pathology rotations;
Perhaps most importantly, presenting and teaching informatics and computation as an intrinsic part of other pathology domains and subspecialties. Most fellows in the Partners Pathology Informatics Fellowship also train in a traditional anatomic, molecular, or clinical pathology fellowship and plan to use informatics or computation as part of a career in a traditional pathology specialty.34
Integrative Training in Computational Pathology
The Department of Pathology at the University of Alabama at Birmingham (UAB) started a Division of Informatics in 2011 specifically as an embedded academic home for both data- and computation-intensive approaches to the study of disease. Accordingly, a division director was recruited from a bioinformatics department with a research focus on big data, followed by additional faculty coming from computational genomics and from systems biology. The division also included recruits with a hybrid background in computer science and pathology, who had secondary clinical appointments. Additional faculty, from both within and outside of pathology, were extended secondary appointments reflecting their research and training activities, such as the current department chair of electrical and software engineering at UAB.
Within a couple of years of the inception of the Division of Informatics within the Department of Pathology at UAB, it became clear that the role of “education as the interface” was fast becoming the division’s single largest impact on the institution (see a list of courses offered at http://www.uab.edu/cb2/training/courses-training; accessed April 27, 2015). Three critical lessons were learned along the way:
The key role of quantitative approaches to the understanding of disease is for the most part already well understood by a wide range of trainees, from pathology residents to MD-PhD students. Moreover, trainees in computational and applied mathematics programs now understand that there is an interesting new frontier at the biomedical interface. It is therefore not uncommon for informal training programs to be initiated by those trainees, such as through meetups and hackathons. Structuring and enhancing those incipient initiatives, rather than replacing them with an overly prescriptive program, may be the key to computational pathology as a discipline that trains the workforce needed to assemble the systems biomedicine puzzle.2
The health information technology infrastructure of academic medical centers is notoriously resistant to change, and expecting a commercial EHR to be a resource for research may be a deadly trap.35 That resistance to change needs to be understood and framed as a feature of the computational pathology landscape36 and also as providing critical clues for novel health information technology architectures that circumvent its pitfalls.37
Creating an autonomous software development capability offers a particularly effective route to generating the synergies that advance computational pathology as a field. Approximately half of all pathology residents and of MD-PhD students at UAB gain familiarity with the LAMP stack (Linux software stack for data management and development of Web services) and learn how to program through participation in immersive computational pathology training venues. The language chosen varies between trainees (typically between Python, Java, PHP, Matlab, R, and JavaScript), depending on the goals of their projects. There is a specific expectation that the resulting computational artifacts be published whenever possible, as illustrated by the project of one trainee.38 The new skill set typically drives both the mentors and mentees in these training programs to contextualize their “small data” in their individual experiments and about individual patients by using an increasing body of reference public “big data.” The new behavior, compounded by the transdisciplinary networks created by the process of acquiring the new skill sets, is what ultimately improves the way disease is understood at a systems level, and how comprehensive personalized care is delivered.
These three lessons share an inescapable tandem feature: low capital costs with long-term commitment. Investing in quantitative sciences embedded within pathology has relatively small capital costs, an attractive trait in an era of diminishing funding. Nonetheless, the benefits of those investments (eg, Roth and Almeida39) can only be achieved after a persistent commitment to develop all components, from training to research.
Pathology Departments Form “Computational Pathology” Collaborations
One approach for advancing computational pathology is to enlist the collaboration of faculty based in other departments who have skills complementary to those of pathology faculty. Stanford University is fortunate to have outstanding departments of computer science (in the School of Engineering) and statistics (in the School of Humanities and Sciences). One example of how faculty with appointments in those departments have created and helped apply methods enabling work in the broad field of computational pathology is the development, by statistics faculty and faculty in health science and policy in the School of Medicine, of methods to analyze large complex data sets, such as those produced by the application of microarray40 or RNA sequencing41 approaches, which can then be applied to problems in pathology.42–44 Another is the application of machine learning approaches by pathologists, in collaboration with faculty in the Department of Computer Science, to develop the C-Path (computational pathologist) system to measure a large set of quantitative features of breast cancer epithelium and stroma, including standard morphometric descriptors of image objects as well as higher-level contextual, relational, and global image features, and to use these measurements to construct a prognostic model that implicates stromal morphologic structure as a previously unrecognized prognostic determinant for breast cancer.45 The State University of New York at Buffalo has recently formed a “structural science learning center” to apply computational approaches to anatomy and gross pathology, thus integrating the work of anatomists, pathologists, biomechanical engineers, and surgeons in the quantitative evaluation of the human body.
The Role of Pathology Organizations in Computational Pathology Education
Pathology societies also have a critical role to play in the development of a culture of computational pathology. Our societies are unique in that taken as a whole, they serve learners in pathology at all levels of sophistication and in all circumstances of professional practice; they often integrate education across undergraduate, graduate, and continuing medical education domains. Computational pathology as an emerging change agent for medical practice should have a central role in the educational programs of all pathology societies. The universal digital capture of all modes of data used in pathology labs and the algorithmic analysis of those data are fundamental principles that pathology societies must endorse and convey. Pathology societies through their national and international programs can help cross-generational learners in pathology understand how to work with computers as their fundamental laboratory tool. Computational features of anatomic pathology, clinical pathology, and experimental pathology must be synthesized, packaged, and disseminated by our medical societies to medical and graduate students, to residents and fellows, and to practitioners. Our pathology societies, by embracing the paradigm of computational pathology in all of their programs, can truly lead the way in changing the practice of pathology from a culture of functioning as a back-office resource to a culture of serving as the central repository of meaningful data and decision support analysis in medicine.
Creating a Pipeline of Future Talent for Computational Pathology
It is critical for the future of pathology that the field focuses on bringing the best and the brightest trainees to our discipline, including such new initiatives as computational pathology. Doing this must begin as early as possible in the career path of innovative students. For example, the University of Pittsburgh has begun to recruit high school students to its pathology and biomedical informatics programs as part of the University of Pittsburgh Cancer Institute Summer Academy (see http://www.upci.upmc.edu/summeracademy/; accessed April 27, 2015). The Computer Science, Biology and Biomedical Informatics (CoSBBI) track began in 2012 and has resulted in an influx of highly talented medical school–focused students who learn how to program and spend 8 weeks doing a mentored research project with the faculty in biomedical and pathology informatics.46 Three of the CoSBBI scholars have been Intel Science and Engineering Fair finalists, and 4 students have now chosen bioinformatics undergraduate programs as their collegiate majors. Educating students (and their parents) about the job opportunities that exist at the interface of informatics, computer science, and health care could be a tremendous catalyst in fostering computational pathology. The future is bright for students who learn critical thinking in a research laboratory and consider computational pathology as a career path, and exposing such students to the field at the earliest points possible (high school, college, graduate and/or medical school) may be an optimal method for ensuring the future of this highly interdisciplinary specialty.
POSSIBLE FUNDING MODELS
Considerable funding will be required to enable the success of computational pathology. Investments in infrastructure, such as hardware and commercial software, the hiring and training of appropriate personnel, and the development of custom software, are needed. The group felt strongly that an integrated and durable national movement to share such resources as much as possible would maximize opportunity and minimize the duplication of infrastructures. Multi-institutional sharing of analytics, infrastructure, and even training programs were all considered possibilities. It was recognized that local investments would certainly be needed, even in the setting of shared national resources. A variety of sources were discussed as possibilities to seed fund local and emerging national components of a computational pathology program.
Departmental sources (practice plan excess margins, sundry accounts) may be available to some groups, although this source is becoming less available in an era of decreasing clinical revenues.
Many larger health care systems, in the compelling interests of population management and cost control, should be willing to contribute to establishing such programs, particularly in the setting of Accountable Care Organizations and similar health care financing structures. Clear evidence of utility will have to be demonstrated for this to be a successful funding source. Nonetheless, a compelling existing example is Crimson,29 a tool developed by pathologists and supported by the Partners HealthCare system that has provided a highly cost-effective means of identifying and acquiring samples for clinical and translational studies (see “Research,” above).
Given the growing recognition of the importance of large data sets and of training people to use such data sets, grants are becoming increasingly available from the government for both developing analytics and training. A more diverse grant base of funding can include entities such as the US Department of Defense, the National Science Foundation, the Defense Advanced Research Projects Agency, and foundations, because the standard R01-funded single investigator is not typical for computational faculty, whether in pathology or in other disciplines. National Institutes of Health funding of primary computational proposals often falls within larger program initiatives and thus requires the development of collaborative teams in which pathology-based faculty and personnel can be in positions to develop and drive projects, either as the principal investigator or through collaborative efforts.
Industrial partners may be powerful allies in developing components of a computational pathology system, notably in the areas in which the companies are invested; although piecemeal from the vantage point of computational pathology as a whole, this approach can nonetheless bring important resources to bear.
Because computational pathology forms the underpinnings of personalized (or precision) medicine, nucleating philanthropic opportunities around this topic may be a way to get pathology departments more directly involved in philanthropy.
Last but not least, developing some of the components of computational pathology may yield intellectual property and patents that may lead to additional revenue sources.
The combination of these approaches, if pursued aggressively, should lead to reasonable levels of funding.
SUMMARY
The July 2014 meeting of pathology department–based informaticists and chairs was a productive interchange of ideas and an interesting set of discussions that involved two major stakeholder groups. The participants made a set of recommendations for next steps in the development of computational pathology, as follows:
Value propositions for the field of pathology should be based on the view that bringing the skills of computational and systems biology to pathology will create vast new opportunities, and that if this is not done by pathologists, it will be done by others.
The field of computational pathology should be defined clearly and succinctly and its “value propositions” clearly articulated.
Value propositions for health care systems should be based on the central role of data-driven analyses in guiding care for the individual and across populations. In this manner, the scope of pathology-based data sets and analytical tools establishes computational pathology as a central field for data interpretation in a modern health care system. Examples include applications in predictive medicine: determining disease likelihood in advance of a patient getting sick and disease trend in advance of complications setting in (“minimize surprise”), and measuring quality and cost to improve both within the health care system.
Pathologists must take active roles in communicating value propositions with health system administrations, other departments, and our pathology colleagues across the country.
A culture should be created that views the computer/computation as a central tool, much like the microscope/microscopy in anatomic pathology.
A robust pipeline should be fostered that develops future computational pathologists, regardless of whether entry occurs from high school to fellowship or from outside the field of pathology. For nonpathologists, career paths and mentoring systems (local or virtual) need to be defined and the career value proposition explained.
Computational pathology should serve as a hub for data-related research in the health care system, and it should be based around the central role of data in guiding individual and population health care.
With the dissemination of these recommendations to many pathology and bioinformatics departments, we anticipate taking the next steps forward in the development of the field of computational pathology.
POSTSCRIPT: THE POTENTIAL IMPACT OF REGULATION ON COMPUTATIONAL PATHOLOGY
Between the meeting held in July 2014 and the publication of this manuscript, the US Food and Drug Administration (FDA) published two draft guidances on its intent to remove the enforcement discretion (ie, no ongoing enforcement by the FDA) that is currently in place for most laboratory-developed tests (LDTs) performed by laboratories certified by the Clinical Laboratory Improvements Amendments.47–49 The public comment period was open from October 3, 2014, until February 2, 2015, and a public workshop was held in Bethesda, Maryland, on January 8–9, 2015.50 In the guidance regarding a new framework of oversight for LDTs, the FDA cited the development of “high-tech instrumentation and software to generate results and interpretations” as one of the major reasons for potentially “increased risk for patients in the absence of appropriate oversight” compared with the so-called traditional LDTs that were in use prior to 1976. Under this proposed guidance, the FDA also stated that in “considering whether to exercise enforcement discretion for Traditional LDTs,” it would consider several factors, one of which was whether the LDT was interpreted without the use of automated instrumentation or software for interpretation.
The FDA has cited some tests using computational algorithms that have resulted in patient harm.51 The presence of such harm caused by tests using these complex algorithms further strengthens the need for the involvement of experts in computational pathology in clinical patient care, research, and discovery. It also highlights the need to expand the number of experts in the field through training programs, because a lack of access to such experts may have allowed errors in algorithm development to go unnoticed. However, if these guidances become finalized, any test using computational pathology algorithms will be required to undergo premarket review process (most likely either premarket approval or 510(k) clearance) by the FDA, with the per-test cost of this process likely being prohibitive to most departments. Moreover, during the recent FDA workshop on this topic, much concern was raised about the increased cost of testing, which widens health care disparities, limits patient access to tests, and lengthens the already significant delay between discovery and clinical implementation of new tests and algorithms. Similarly, research and discovery could be stifled if investors are discouraged from funding the development of such new technologies. In combination, the potential new barriers that would arise from the proposed FDA changes would undoubtedly slow the eventual reality of computational pathology in clinical care, and are therefore a cause for substantial concern.
Acknowledgments
We thank Priscilla Markwood of the Association of Pathology Chairs for her tremendous help in facilitating the meeting.
APPENDIX
Meeting Attendees
Abul Abbas, MD (University of California at San Francisco); Jonas Almeida, PhD (University of Alabama, Birmingham; now at Stony Brook University, Stony Brook, New York); Ulysses Balis, MD (University of Michigan, Ann Arbor); Michael J. Becich, MD, PhD (University of Pittsburgh, Pittsburgh, Pennsylvania); Andrew Beck, MD, PhD (Beth Israel Deaconess Medical Center, Boston, Massachusetts, and Harvard Medical School, Boston, Massachusetts); Bruce Beckwith, MD (North Shore Medical Center, Salem, Massachusetts, and Harvard Medical School); Marianne Boswell (Massachusetts General Hospital, Boston, and Harvard Medical School); Jonathan Braun, MD, PhD (University of California at Los Angeles); Lynn Bry, MD, PhD (Brigham and Women’s Hospital, Boston, Massachusetts, and Harvard Medical School); Alexis Carter, MD (Emory University, Atlanta, Georgia); Robert Daber, PhD (University of Pennsylvania, Philadelphia); Anand Dighe, MD, PhD (Massachusetts General Hospital and Harvard Medical School); Michael Feldman, MD, PhD (University of Pennsylvania); Stephen J. Galli, MD (Stanford University, Stanford, California); Georg Gerber, MD, PhD, MPH (Brigham and Women’s Hospital and Harvard Medical School); John R. Gilbertson, MD (Massachusetts General Hospital and Harvard Medical School); Jeffrey A. Golden, MD (Brigham and Women’s Hospital and Harvard Medical School); Metin Gurcan, PhD (Ohio State University, Columbus); Peter Jensen, MD (University of Utah, Salt Lake City); David N. Louis, MD (Massachusetts General Hospital and Harvard Medical School); Bob McGonnagle (College of American Pathologists, Northfield, Illinois); Jon Morrow, MD, PhD (Yale University, New Haven, Connecticut); Rakesh Nagarajan, MD, PhD (Washington University, St Louis, Missouri); Tristram G. Parslow, MD, PhD (Emory University); John D. Pfeifer, MD, PhD (Washington University); Michael B. Prystowsky, MD, PhD (Albert Einstein College of Medicine, Bronx, New York); Kevin Roth, MD, PhD (University of Alabama, Birmingham); Jeffrey Saffitz, MD, PhD (Beth Israel Deaconess Medical Center and Harvard Medical School); Joel Saltz, MD, PhD (Stony Brook University); John Sinard, MD, PhD (Yale University); John E. Tomaczewski, MD (State University of New York at Buffalo); James Versalovic, MD, PhD (Texas Children’s Hospital, Houston); Geoff Smith, MD (Emory University); James M. Musser, MD, PhD (The Methodist Hospital, Houston, Texas); and Donald S. Karcher, MD (George Washington University, Washington, DC).
Footnotes
Drs Tomaszewski and Bry are paid members of the Scientific Advisory Board for Inspirata Inc (Tampa, Florida) and own stock options. Dr Galli is a paid member of the Board of Directors of Atossa Genetics (Seattle, Washington) and is the owner of stock and stock options in the company. The other authors have no relevant financial interest in the products or companies described in this article.
References
- 1.Kulikowski CA, Shortliffe EH, Currie LM, et al. AMIA Board white paper: definition of biomedical informatics and specification of core competencies for graduate education in the discipline. J Am Med Inform Assoc. 2012;19(6):931–938. doi: 10.1136/amiajnl-2012-001053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Louis DN, Gerber GK, Baron JM, et al. Computational pathology: an emerging definition. Arch Pathol Lab Med. 2014;138(9):1133–1138. doi: 10.5858/arpa.2014-0034-ED. [DOI] [PubMed] [Google Scholar]
- 3.Higgins JM, Mahadevan L. Physiological and pathological population dynamics of circulating human red blood cells. Proc Natl Acad Sci U S A. 2010;107(47):20587–20592. doi: 10.1073/pnas.1012747107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lara J, Lopez-Labrador F, Gonzalez-Candelas F, Berenguer M, Khudyakov YE. Computational models of liver fibrosis progression for hepatitis C virus chronic infection. BMC Bioinformatics. 2014;15(suppl 8):S5. doi: 10.1186/1471-2105-15-S8-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet. 2013;Chapter 7(Unit7):20. doi: 10.1002/0471142905.hg0720s76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flanagan SE, Patch AM, Ellard S. Using SIFT and PolyPhen to predict loss-of-function and gain-of-function mutations. Genet Test Mol Biomarkers. 2010;14(4):533–537. doi: 10.1089/gtmb.2010.0036. [DOI] [PubMed] [Google Scholar]
- 7.Hu J, Ng PC. SIFT Indel: predictions for the functional effects of amino acid insertions/deletions in proteins. PLoS One. 2013;8(10):e77940. doi: 10.1371/journal.pone.0077940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sim NL, Kumar P, Hu J, Henikoff S, Schneider G, Ng PC. SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012;40(Web Server issue):W452–W457. doi: 10.1093/nar/gks539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zou M, Baitei EY, Alzahrani AS, et al. Mutation prediction by PolyPhen or functional assay, a detailed comparison of CYP27B1 missense mutations. Endocrine. 2011;40(1):14–20. doi: 10.1007/s12020-011-9489-7. [DOI] [PubMed] [Google Scholar]
- 10.dbSNP: Short Genetic Variations [database online] Bethesda, MD: National Center for Biotechnology Information; [Accessed September 10, 2014]. http://www.ncbi.nlm.nih.gov/projects/SNP/snp_tableList.cgi. [Google Scholar]
- 11.NCBI Resource Coordinators. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2014;42(Database issue):D7–D17. doi: 10.1093/nar/gkt1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Catalogue of Somatic Mutations in Cancer (COSMIC) [database online] Cambridge, UK: Wellcome Trust Sanger Institute; 2014. [Accessed August 5, 2014]. http://cancer.sanger.ac.uk/cancergenome/projects/cosmic/ [Google Scholar]
- 13.Bamford S, Dawson E, Forbes S, et al. The COSMIC (Catalogue of Somatic Mutations in Cancer) database and website. Br J Cancer. 2004;91(2):355–358. doi: 10.1038/sj.bjc.6601894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Landrum MJ, Lee JM, Riley GR, et al. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014;42(Database issue):D980–D985. doi: 10.1093/nar/gkt1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Riggs ER, Wain KE, Riethmaier D, et al. Towards a universal clinical genomics database: the 2012 International Standards for Cytogenomic Arrays Consortium Meeting. Hum Mutat. 2013;34(6):915–919. doi: 10.1002/humu.22306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.The ClinGen Resource [database online] Bethesda, MD: National Institutes of Health; [Accessed April 1, 2015]. http://clinicalgenome.org/ [Google Scholar]
- 17.Raess PW, van de Geijn GJ, Njo TL, et al. Automated screening for myelodysplastic syndromes through analysis of complete blood count and cell population data parameters. Am J Hematol. 2014;89(4):369–374. doi: 10.1002/ajh.23643. [DOI] [PubMed] [Google Scholar]
- 18.Aronson SJ, Clark EH, Babb LJ, et al. The GeneInsight Suite: a platform to support laboratory and provider use of DNA-based genetic testing. Hum Mutat. 2011;32:532–536. doi: 10.1002/humu.21470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilcox AR, Neri PM, Volk LA, et al. A novel clinician interface to improve clinician access to up-to-date genetic results. J Am Med Inform Assoc. 2013;21:e117–e121. doi: 10.1136/amiajnl-2013-001965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Desmond-Hellman S, Sawyers CL, Cox DR, et al. Toward Precision Medicine: Building a Knowledge Network for Biomedical Research and a New Taxonomy of Disease. 1. Washington, DC: The National Academies Press; 2015. [PubMed] [Google Scholar]
- 21.National Institutes of Health. [Accessed February 23, 2015];Precision Medicine Initiative Web site. http://www.nih.gov/precisionmedicine/
- 22.Colen R, Foster I, Gatenby R, et al. NCI Workshop Report: clinical and computational requirements for correlating imaging phenotypes with genomics signatures. Transl Oncol. 2014;7(5):556–569. doi: 10.1016/j.tranon.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cooper LA, Kong J, Gutman DA, et al. Integrated morphologic analysis for the identification and characterization of disease subtypes. J Am Med Inform Assoc. 2012;19(2):317–323. doi: 10.1136/amiajnl-2011-000700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boca SM, Bravo HC, Caffo B, Leek JT, Parmigiani G. A decision-theory approach to interpretable set analysis for high-dimensional data. Biometrics. 2013;69(3):614–623. doi: 10.1111/biom.12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leiserson MD, Vandin F, Wu HT, et al. Pan-cancer network analysis identifies combinations of rare somatic mutations across pathways and protein complexes. Nat Genet. 2015;47(2):106–114. doi: 10.1038/ng.3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Madabhushi A, Doyle S, Lee G, et al. Integrated diagnostics: a conceptual framework with examples. Clin Chem Lab Med. 2010;48(7):989–998. doi: 10.1515/CCLM.2010.193. [DOI] [PubMed] [Google Scholar]
- 27.Gerber GK. The dynamic microbiome. FEBS Lett. 2014;588(22):4131–4139. doi: 10.1016/j.febslet.2014.02.037. [DOI] [PubMed] [Google Scholar]
- 28.Gerber GK, Onderdonk AB, Bry L. Inferring dynamic signatures of microbes in complex host ecosystems. PLoS Comput Biol. 2012;8(8):e1002624. doi: 10.1371/journal.pcbi.1002624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murphy S, Churchill S, Bry L, et al. Instrumenting the health care enterprise for discovery research in the genomic era. Genome Res. 2009;19(9):1675–1681. doi: 10.1101/gr.094615.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.National Cancer Institute. [Accessed May 25, 2015];caLIMS v2 Software [wiki] https://wiki.nci.nih.gov/display/caLIMS2/caLIMS+v2+Software.
- 31.National Center for Biomedical Computing. [Accessed December 14, 2014];i2b2: Informatics for Integrating Biology & the Bedside Web site. https://www.i2b2.org/
- 32.National Cancer Institute. [Accessed December 14, 2014];caBIG Web site. http://cabig.cancer.gov/
- 33.Garcia CA, Baron JM, Beckwith BA, et al. Environmental components and methods for engaging pathology residents in informatics training. J Pathol Inform. 2015;6:42. [PMC free article] [PubMed] [Google Scholar]
- 34.Levy BP, McClintock DS, Lee RE, et al. Different tracks for pathology informatics fellowship training: experiences of and input from trainees in a large multisite fellowship program. J Pathol Inform. 2012;3:30. doi: 10.4103/2153-3539.100362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mandl KD, Kohane IS. Escaping the EHR trap–the future of health IT. N Engl J Med. 2012;366(24):2240–2242. doi: 10.1056/NEJMp1203102. [DOI] [PubMed] [Google Scholar]
- 36.Park S, Parwani AV, Aller R, et al. The history of pathology informatics: a global perspective. J Pathol Inform. 2013;4(1):7–42. doi: 10.4103/2153-3539.112689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilkinson SR, Almeida JS. QMachine: commodity supercomputing in web browsers. BMC Bioinformatics. 2014;15:176. doi: 10.1186/1471-2105-15-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cain MD, Siebert JR, Iriabho E, Gruneberg A, Almeida JS, Faye-Petersen OM. Development of novel software to generate anthropometric norms at perinatal autopsy. Pediatr Dev Pathol. 2015;18(3):203–209. doi: 10.2350/14-11-1578-OA.1. [DOI] [PubMed] [Google Scholar]
- 39.Roth KA, Almeida JS. Coming into focus: computational pathology as the new big data microscope. Am J Pathol. 2015;185(3):600–601. doi: 10.1016/j.ajpath.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 40.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98(9):5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li J, Witten DM, Johnstone IM, Tibshirani R. Normalization, testing, and false discovery rate estimation for RNA-sequencing data. Biostatistics. 2012;13(3):523–538. doi: 10.1093/biostatistics/kxr031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alizadeh AA, Eisen MB, Davis RE, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403(6769):503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 43.Pollack JR, Sorlie T, Perou CM, et al. Microarray analysis reveals a major direct role of DNA copy number alteration in the transcriptional program of human breast tumors. Proc Natl Acad Sci U S A. 2002;99(20):12963–12968. doi: 10.1073/pnas.162471999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sorlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98(19):10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Beck AH, Sangoi AR, Leung S, et al. Systematic analysis of breast cancer morphology uncovers stromal features associated with survival. Sci Transl Med. 2011;3(108):108ra113. doi: 10.1126/scitranslmed.3002564. [DOI] [PubMed] [Google Scholar]
- 46.Dutta-Moscato J, Gopalakrishnan V, Lotze MT, Becich MJ. Creating a pipeline of talent for informatics: STEM initiative for high school students in computer science, biology, and biomedical informatics. J Pathol Inform. 2014;5:12. doi: 10.4103/2153-3539.129448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.US Food and Drug Administration. [Accessed May 25, 2015];Draft guidance for industry, Food and Drug Administration staff, and clinical laboratories: framework for regulatory oversight of laboratory developed tests (LDTs) 2015 http://www.fda.gov/downloads/medicaldevices/deviceregulationandguidance/guidancedocuments/ucm416685.pdf.
- 48.US Food and Drug Administration. [Accessed February 23, 2015];Draft guidance for industry, Food and Drug Administration staff, and clinical laboratories: FDA notification and medical device reporting for laboratory developed tests (LDTs) 2014 Oct 3; http://www.fda.gov/downloads/MedicalDevices/DeviceRegulationandGuidance/GuidanceDocuments/UCM416684.pdf.
- 49.Clinical Laboratory Improvement Amendments of 1988. [Accessed February 23, 2015];Laboratory Requirements, 42 CFR §493. http://www.gpo.gov/fdsys/pkg/CFR-2012-title42-vol5/pdf/CFR-2012-title42-vol5-part493.pdf.
- 50.US Food and Drug Administration. Public Workshop - Framework for Regulatory Oversight of Laboratory Developed Tests (LDTs); January 8–9, 2015; [Accessed February 23, 2015]. http://www.fda.gov/MedicalDevices/NewsEvents/WorkshopsConferences/ucm423537.htm. [Google Scholar]
- 51.US Food and Drug Administration. [Accessed February 23, 2015];Warning letters: FDA inspections, compliance, enforcement, and criminal investigations. http://www.fda.gov/ICECI/EnforcementActions/WarningLetters/default.htm.