Abstract
Background. Methicillin-resistant Staphylococcus aureus (MRSA) USA300 is the leading cause of MRSA infections in the United States and has caused an epidemic of skin and soft-tissue infections. Recurrent infections with USA300 MRSA are common, yet intrahost evolution during persistence on an individual has not been studied. This gap hinders the ability to clinically manage recurrent infections and reconstruct transmission networks.
Methods. To characterize bacterial intrahost evolution, we examined the clinical courses of 4 subjects with 3–6 recurrent USA300 MRSA infections, using patient clinical data, including antibiotic exposure history, and whole-genome sequencing and phylogenetic analysis of all available MRSA isolates (n = 29).
Results. Among sequential isolates, we found variability in diversity, accumulation of mutations, and mobile genetic elements. Selection for antimicrobial-resistant populations was observed through both an increase in the number of plasmids conferring multidrug resistance and strain replacement by a resistant population. Two of 4 subjects had strain replacement with a genetically distinct USA300 MRSA population.
Discussions. During a 5-year period in 4 subjects, we identified development of antimicrobial resistance, intrahost evolution, and strain replacement among isolates from patients with recurrent MRSA infections. This calls into question the efficacy of decolonization to prevent recurrent infections and highlights the adaptive potential of USA300 and the need for effective sampling.
Keywords: Staphylococcus aureus, MRSA, USA300, skin and soft-tissue infections, intrahost, within host, whole-genome sequencing, evolution, phylogenetic, recurrent infections, antibiotic resistance
Methicillin-resistant Staphylococcus aureus (MRSA) pulsed-field gel electrophoresis–type USA300 is the leading cause of MRSA infections in the United States and has caused an epidemic of skin and soft-tissue infections (SSTIs) in multiple settings [1]. Recurrent infections with USA300 are common, but many host and pathogen characteristics underlying the risk of recurrence are not known [2].
Whole-genome sequencing (WGS) has been used to elucidate MRSA infection outbreaks in hospitals [3, 4], epidemiology in households [5], and cloud of variation in an individual [6]. On a larger scale, the geographic origins of common genotypes [7], community transmission [8], and convergent adaptation [9] have been investigated. However, the evolution of MRSA during a single infection has rarely received attention, despite knowledge that substantial diversity can arise [3], and within-host evolution during antimicrobial therapy can result in the development of resistance.
If information about the diversity arising in MRSA strains through within-host evolution were available, it would enhance accurate reconstruction of interhost transmission networks during epidemic investigations [10] and improve management of recurrent infections [11]. Intrahost diversity in colonizing MRSA strains can vary tremendously as a result of the diversity of bacteria acquired through a transmission event (ie, bottleneck size), duration of colonization and/or infection, and selective pressures such as antibiotic exposure [10, 12]. Longitudinal monitoring of intrahost MRSA populations may identify adaptive mutations or acquisition of mobile genetic elements conferring antibiotic resistance or augmented virulence, which would not be recognized in a cross-sectional study.
To characterize bacterial intrahost evolution, we examined the clinical courses of 4 patients with recurrent USA300 MRSA infections, using detailed clinical data, including antibiotic exposure data, from patients and WGS, comparative genomics analysis, and phylogenetic analysis of all available isolates. We demonstrate a unique progression of strain evolution and evidence of heterogeneity in intrahost evolutionary dynamics among patients. We also highlight the importance of comprehensive sampling of intrahost bacterial populations. WGS data would enhance the ability of clinicians to treat MRSA infections by providing information on strain characteristics and distinguishing between new and recurrent infections.
METHODS
Prospective collection of all MRSA isolates from the Clinical Microbiology Laboratory at the University of Chicago has been ongoing. Patients with ≥1 episode of culture-isolated MRSA from an infection treated in outpatient or inpatient settings between November 2003 and April 2009 were identified. An episode was defined as isolation of MRSA from the site of a clinically significant infection, separated by >30 days from any previous isolation of MRSA. We randomly selected 20% of these individuals and characterized the first isolate from each episode by multilocus sequence typing (MLST), staphylococcal cassette chromosome mec (SCCmec) typing, and polymerase chain reaction analysis of the genes encoding Panton-Valentine leukocidin (PVL) [13, 14]. MRSA isolates that were ST8 and carried SCCmec type IV, and the genes encoding PVL were considered to be USA300 [15]. Available isolates from the 4 patients (patients A–D) who had the greatest number of recurrent USA300 episodes underwent WGS (n = 29). This included 2 non-USA300 MRSA isolates. Antibiograms (Vitek, Marcy L'Etoile), sites of infection, and bacteria cultured from polymicrobial infections were recorded for each isolate. Age, race, comorbidities, dates of patient encounters, antibiotic therapy, and other means of treatment were abstracted from medical records. The University of Chicago Institutional Review Board approved the study, and written informed consent or a waiver of consent was obtained from participants.
WGS, genome assembly, and single-nucleotide polymorphism (SNP) calling and annotation were performed as previously described [4]. Briefly, genomic DNA was sequenced on the Illumina MiSeq, using the NexteraXT library preparation kit. Paired-end 250–base pair reads were assembled to reference genome USA300-FPR3757 (accession CP000255). SNPs were called using FreeBayes v0.9.14 and filtered, requiring a depth of coverage of 5 and a minimum alternate allele frequency of 0.75. To confidently call a SNP, we considered only sites with a read depth of ≥5 among all intrahost isolates. Clustered SNPs were investigated through review of BAM files to assess read mapping, and SNP flanking insertions and deletions were removed. The resulting high-quality SNPs were annotated using SNPeff v4.1. Functional categories were assigned using RAST (available at: http://rast.nmpdr.org/) [16]. SNP alignments for each patient and a single alignment of all 27 USA300 isolates were constructed for phylogenetic analysis. Recombination events were investigated using Gubbins [17]. To identify mobile genetic elements, unmapped reads from each reference-based assembly were de novo assembled with SPAdes v3.5.0 and scaffolded with SSPACE v3.0. BLAST was used to identify putative plasmids from contigs, which were ordered and aligned to the highest match, using Mauve v2.4.0. Plasmid genomes were annotated using Prokka v1.11 and visualized using GView.
Maximum likelihood phylogenetic analysis was used to investigate the genomic relatedness of isolates within and among patients. A maximum likelihood phylogeny of the 27 USA300 isolates was inferred by means of RAxML v8.2.8, using an ASC_GTRGAMMA substitution model and 100 bootstrap replicates. For each patient, the temporality of infection, medical interventions, and MRSA intrahost evolution were illustrated with minimum spanning trees.
RESULTS
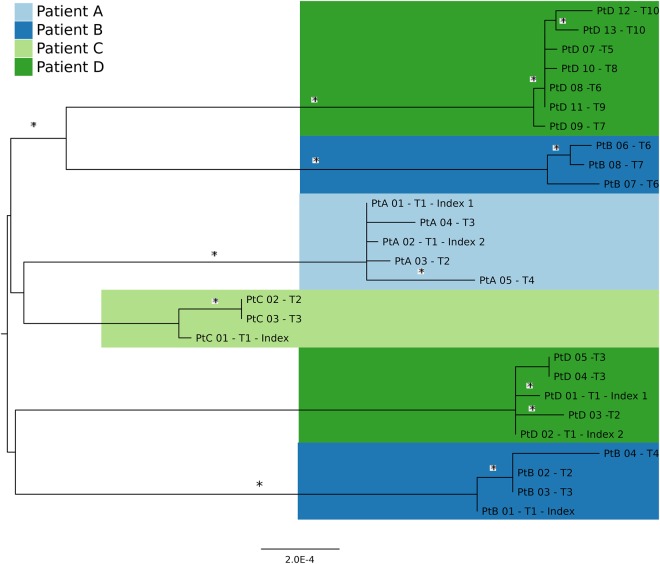
The 4 patients ranged from 1 to 54 years of age. Three had diabetes mellitus type 2. No other comorbidities were identified. Table 1 delineates the clinical course of each patient. A minimum of 3 and maximum of 13 isolates were obtained from the 4 patients (Table 2). MRSA decolonization was not attempted. Among reference-based assemblies, the average genome coverage with a read depth of ≥5 or 10 was 93.7% and 88.3%, respectively (ie, on average, 93.7% of the genome had sufficient depth of coverage to call SNPs confidently). The earliest USA300 populations in patients B and D were putatively replaced by a genetically distinct USA300 population (ie, strain replacement) during their follow-up (Figures 1 and 2). This was inferred from phylogeny data, anatomical sites of recovery and temporality of infection, and MRSA-negative cultures obtained between populations 1 and 2 (Figure 1 and Supplementary Table 1). The 2 groups of isolates from patients B and D formed monophyletic clades and were therefore analyzed as independent populations. In addition, patients B and D both acquired a non-USA300 MRSA strain. Intrapatient mean SNP differences for 6 discrete infections ranged from 2.6 to 7.3 (mean, 4.9), compared with an overall between-patient (interhost) mean difference of 54.5 SNPs (Supplementary Table 2). The progressive accumulation of SNPs was punctuated by periods of little-to-no mutation, followed by episodes of rapid intrahost evolution.
Table 1.
Clinical Histories and Treatment of 4 Patients Treated at the University of Chicago With Recurrent Methicillin-Resistant Staphylococcus aureus USA300 Infections, 2005–2009
| Patient | Time Point | Day | Clinical Description, Presentation, Treatment, Outcome |
|---|---|---|---|
| A | T1 | 0 | Male aged 1 y seen in ED for lower extremity abscess, underwent incision and drainage and received oral clindamycin for 10 d; an additional colonization isolate was obtained from anterior nares culture |
| T2 | 194 | Buttock abscess, underwent incision and drainage in ED and was treated with clindamycin for 10 d | |
| T3 | 235 | Perirectal and abdominal wall abscess, required admission and surgical drainage and debridement, treated with oral clindamycin for 10 d | |
| T4 | 750 | Scalp folliculitis, ED visit, no incision and drainage, treated with clindamycin for 10 d | |
| B | T1 | 0 | Male aged 44 y with a history of diabetes mellitus and an infected diabetic foot ulcer, resulting in amputation below the left knee 2 mo earlier; had SSTI on right forearm, which was treated with incision and drainage in the clinic and received oral amoxicillin/clavulanic acid for 7 d; had received cephalexin within past 6 mo |
| T2 | 58 | SSTI at amputation site below left knee, surgical incision and drainage completed, treated with oral clindamycin and ciprofloxacin | |
| T3 | 133 | SSTI reoccurrence at amputation site below left knee, treated with oral clindamycin and ciprofloxacin | |
| T4 | 424 | SSTI reoccurrence at amputation site below left knee, underwent amputation above the knee, treated with oral moxifloxacin for 14 d | |
| T5 | 1086 | SSTI at right third toe amputation site, which did not improve with intravenous vancomycin, clindamycin, and ciprofloxacin; required transmetatarsal amputation for osteomyelitis and treatment with extended courses of intravenous daptomycin, oral ciprofloxacin, and metronidazole | |
| T6 | 1232 | SSTI at site of accidental burn on right heel, requiring skin grafting; underwent multiple surgeries and treated with intravenous vancomycin and piperacillin/tazobactam | |
| T7 | 1300 | SSTI at right heel, requiring amputation below knee | |
| C | T1 | 0 | Female aged 33 y with hip and buttock abscesses that required surgical incision and drainage, treated with intravenous vancomycin and discharged receiving oral amoxicillin/clavulanic acid, subsequently readmitted for repeat incision and drainage due to failure, treated with linezolid 3 wk later |
| T2 | 131 | SSTI, abscess of upper extremity that required incision and drainage, treated with oral clindamycin and ciprofloxacin; did not respond to this regimen, but intravenous vancomycin resulted in cure | |
| T3 | 225 | SSTI, abscess of breast, seen in outpatient clinic, treated with oral ciprofloxacin and rifampin with resolution | |
| D | T1 | 0 | Male aged 54 y with diabetes mellitus with SSTI of the left fifth toe amputation site, treated with oral amoxicillin/clavulanic acid and trimethoprim/sulfamethoxazole |
| T2 | 104 | SSTI at ulcer on the right foot, progressed to osteomyelitis that required right first metatarsal and sesamoid amputation, treated with intravenous vancomycin and piperacillin/tazobactam for 6 wk | |
| T3 | 112 | Same as for T2 | |
| T4 | 230 | SSTI of right foot ulcer that progressed to osteomyelitis despite treatment with oral antibiotics, required right second toe and left fourth toe amputations | |
| T5 | 326 | SSTI of right toe that did not resolve after wound management and oral antibiotics, underwent right transmetatarsal amputation, treated with oral amoxicillin/clavulanic acid and trimethoprim/sulfamethoxazole for 6 wk | |
| T6 | 327 | ||
| T7 | 390 | SSTI at transmetatarsal amputation stump site, underwent surgical debridement, treated with oral amoxicillin/clavulanic acid and trimethoprim/sulfamethoxazole for 2 wk | |
| T8 | 396 | Same as for T7 | |
| T9 | 447 | SSTI of plantar ulcer on left foot proceeding to osteomyelitis, underwent podiatry care, treated with oral trimethoprim/sulfamethoxazole for 10 d | |
| T10 | 553 | Previous osteomyelitis did not respond to treatment, underwent left transmetatarsal amputation, treated with linezolid, amoxicillin/clavulanic acid, and ciprofloxacin for 14 d |
Time points indicate each infection episode that corresponds to Figure 2.
Abbreviations: ED, emergency department; SSTI, skin and soft-tissue infection.
Table 2.
Isolate, Genotype, Anatomical Site, and Antibiogram of 29 Methicillin-Resistant Staphylococcus aureus USA300 Isolates From 4 Patients
| Patient | Isolate, Time Point | Genotype | Site Description | Antibiogram |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cip | Cli | Erm | Gen | Oxc | Rif | Van | D Test | SXT | ||||
| A | 01, T1 | ST8/USA300 | SSTI: thigh abscess | S | S | R | S | R | S | <1 | − | S |
| 02, T2 | ST8/USA300 | Colonization | S | S | R | S | R | S | <1 | − | S | |
| 03, T2 | ST8/USA300 | SSTI: right buttock wound | S | S | R | S | R | S | <1 | − | S | |
| 04, T3 | ST8/USA300 | SSTI: perirectal and abdominal wall abscess | S | S | R | S | R | S | <1 | − | S | |
| 05, T4 | ST8/USA300 | SSTI: scalp folliculitis | S | S | R | S | R | S | <1 | − | S | |
| B | 01, T1 | ST8/USA300 | SSTI: right forearm | S | S | R | S | R | S | <1 | − | S |
| 02, T2 | ST8/USA300 | SSTI: left leg stump (BKA) | S | S | R | S | R | S | <1 | − | S | |
| 03, T3 | ST8/USA300 | SSTI: left leg stump (BKA) | S | S | R | S | R | S | <1 | − | S | |
| 04, T4 | ST8/USA300 | SSTI: left leg stump (AKA) | S | S | R | S | R | S | <1 | − | S | |
| 05, T5 | ST-105 (CC5) | SSTI: right foot wound | R | R | R | S | R | S | <1 | − | S | |
| 06, T6 | ST8/USA300 | SSTI: right leg wound | R | S | R | S | R | S | <1 | − | S | |
| 07, T6 | ST8/USA300 | SSTI: right leg wound | R | S | R | S | R | S | <1 | − | S | |
| 08, T7 | ST8/USA300 | Bone/joint: heel ulcer | R | S | R | S | R | S | <1 | − | S | |
| C | 01, T1 | ST8/USA300 | SSTI: left hip and buttocks | S | R | R | S | R | S | <1 | − | S |
| 02, T2 | ST8/USA300 | SSTI: right upper extremity | S | S | R | S | R | S | <1 | − | S | |
| 03, T3 | ST8/USA300 | SSTI: left breast wound | S | R | R | S | R | S | <1 | − | S | |
| D | 01, T1 | ST8/USA300 | SSTI: left foot wound | S | S | R | S | R | S | <1 | − | S |
| 02, T1 | ST8/USA300 | SSTI: left foot wound | S | S | R | S | R | S | <1 | − | S | |
| 03, T2 | ST8/USA300 | Bone/joint: right foot | S | S | R | S | R | S | <1 | − | S | |
| 04, T3 | ST8/USA300 | SSTI: right foot | S | R | R | R | R | S | <1 | − | S | |
| 05, T4 | ST8/USA300 | SSTI: right foot | S | R | R | R | R | S | <1 | − | S | |
| 06, T4 | ST-59 (CC59) | SSTI: right foot | S | S | R | S | R | S | <1 | − | S | |
| 07, T5 | ST8/USA300 | Bone/joint: right foot | R | S | R | S | R | S | <1 | − | S | |
| 08, T6 | ST8/USA300 | SSTI: right foot TMA site | R | S | R | S | R | S | <1 | − | S | |
| 09, T7 | ST8/USA300 | SSTI: right foot TMA site | R | S | R | S | R | S | <1 | − | S | |
| 10, T8 | ST8/USA300 | SSTI: right foot TMA site | R | S | R | S | R | S | <1 | − | S | |
| 11, T9 | ST8/USA300 | Bone/joint: left foot wound | R | S | R | S | R | S | <1 | − | S | |
| 12, T10 | ST8/USA300 | Bone/joint: left foot wound | R | S | R | S | R | S | <1 | − | S | |
| 13, T10 | ST8/USA300 | SSTI: left flexion tendon | R | S | R | S | R | S | <1 | − | S | |
Vancomycin minimum inhibitory concentrations are included.
Abbreviations: AKA, above knee amputation; BKA, below knee amputation; Cip, ciprofloxacin; Cli, clindamycin; D test, macrolide-inducible clindamycin resistance; Erm, erythromycin; Gen, gentamicin; Oxc, oxacillin; R, resistant; Rif, rifampin; S, susceptible; SSTI, skin and soft tissue infection; SXT, trimethoprim/sulfamethoxazole; TMA, transmetatarsal amputation; Van, vancomycin; −, negative.
Figure 1.
Maximum likelihood phylogenetic tree inferred from single-nucleotide polymorphism (SNP) alignment of 27 methicillin-resistant Staphylococcus aureus USA300 isolates and colored by patient. Phylogeny was inferred using RAxML v8.0.0, using the ASC_GTRGAMMA nucleotide substitution model with ascertainment bias correction and 100 bootstrap replicates. The tree is midpoint rooted, and clades with bootstrap values of >80% are denoted with an asterisk. The phylogeny illustrates that patients B and D had strain replacement events in which a separate USA300 population replaced their incident USA300 population. Separate strain acquisitions are inferred from the temporality of isolate collection and phylogenetic data illustrating distinct, well-supported monophyletic clades for each USA300 population.
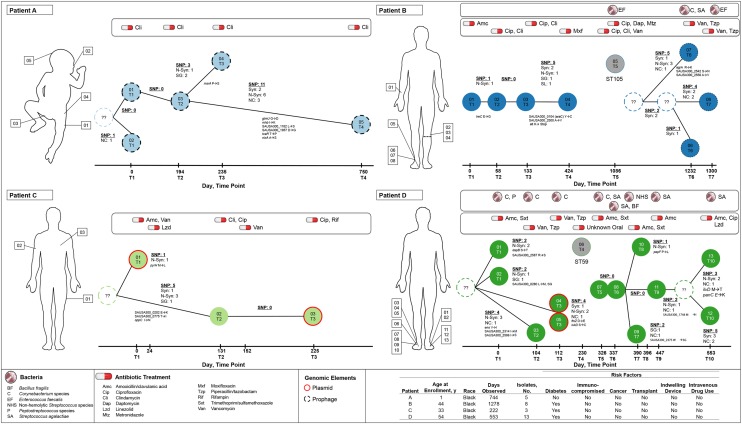
Figure 2.
Time-scaled minimum spanning trees of 4 patients (A–D). The number and annotation of single-nucleotide polymorphisms (SNPs) are labeled on branches linking nodes (unique isolates). The sampling date and time point, which correspond to the clinical histories in Table 1, are specified on the x-axes. Synonymous (syn), nonsynonymous (N-syn), stop gained (SG), start lost (SL), and noncoding (NC) are labeled, as well as the gene name and amino acid change. Node labels correspond to Table 2. Nodes labeled with question marks and dashed outlines represent unsampled intermediate ancestors. Gray nodes are non-USA300 methicillin-resistant Staphylococcus aureus isolates. The red node outline specifies isolates with identified plasmids, and black dashed outlines represent isolates with bacteriophages. This figure is available in black and white in print and in color online.
SNPs were more often in coding regions with significantly more nonsynonymous than synonymous mutations (P = .03; Figure 2). In addition, SNPs that may have impacted protein function occurred more frequently in genes associated with membrane transport (rate ratio, 6.7; 95% confidence interval, 2.97–15.1; Table 3 and Supplementary Table 3). However, there was no pattern of mutations to suggest convergent evolutionary changes of USA300 isolates among patients. Instead, our observations were more consistent with an accumulation of potentially deleterious nonsynonymous mutations. No intrahost recombination events were identified (Supplementary Figure 1). A complete table of annotated SNPs is available in Supplementary File 1.
Table 3.
Annotation and Function of Notable Single-Nucleotide Polymorphisms (SNPs)
| Patient | SNP Position | Gene | Codon | Effect | Amino Acid Change | Gene Name/Function | Category | Association |
|---|---|---|---|---|---|---|---|---|
| A | 534 272 | glmU | gGc/gAc | NS | G208D | Bifunctional N-acetylglucosamine-1-phosphate uridyltransferase | Cell wall and capsule | Key component of cell wall biosynthesis [18] |
| 941 455 | glpQ | Cag/Tag | SG | Q125STOP | Glycerophosphoryl diester phosphodiesterase | Carbohydrates | … | |
| 1 246 149 | rnhB | aTa/aAa | NS | L64K | Ribonuclease HII | RNA metabolism | Degrades RNA of DNA-RNA hybrids | |
| 1 302 306 | SAUSA300_1182 | tTg/tCg | NS | L511S | Pyruvate ferredoxin oxidoreductase, α subunit | Respiration | … | |
| 1 449 721 | msrA | Cca/Tca | NS | P23S | Methionine sulfoxide reductase A | Protein metabolism | Bacterial oxidative stress response [19] | |
| 2 130 440 | SAUSA300_1975 | gAt/gGt | NS | D140G | Aerolysin/leukocidin family protein | Virulence, disease, and defense | … | |
| 2 476 503 | tcaR | Act/Cct | NS | T99P | Transcriptional regulator, TcaR | Virulence, disease, and defense | Important role in biofilm formation [20] and expression of virulence factors, including sarS and spa [21] | |
| 2 857 261 | nixA | Gca/Tca | NS | A285S | High-affinity nickel transporter | Membrane transport | Necessary for urease activity [22] | |
| B | Population 1 | |||||||
| 16 135 | SAUSA300_0012 | atG/atA | SL | M1I | Homoserine O-acetyltransferase | Amino acids and derivatives | Metabolism | |
| 115 262 | SAUSA300_0104 | tAt/tGt | NS | Y569C | Transcriptional regulator, AraC | Regulation and cell signaling | … | |
| 507 017 | treC | gAa/gGa | NS | E437G | α-phosphotrehalase | Virulence, disease, and defense | … | |
| 1 046 867 | atl | Aaa/Taa | SG | K374STOP | Bifunctional autolysin | Virulence, disease, and defense | Cell wall metabolism and biofilm development | |
| 2 705 337 | SAUSA300_2500 | gCa/gTa | NS | A259V | 4,4′-diaponeurosporenoate glycosyl transferase | Membrane transport | Metabolism of terpenoids and polyketides | |
| Population 2 | ||||||||
| 2 149 492 | agrA | aGa/aAa | NS | R170K | Accessory gene regulator protein A | Regulation and cell signaling | Regulates expression of several virulence genes and may have a role in biofilm production [23] | |
| 2 746 919 | SAUSA300_2542 | aGt/aAt | NS | S323N | Putative AMP-binding enzyme | Regulation and cell signaling | May be linked to daptomycin nonsusceptibility [24] | |
| 2 766 651 | SAUSA300_2556 | gCg/gTg | NS | A519V | ABC transporter protein | Membrane transport | Effluxing cytotoxic drugs across the cell membrane [25] | |
| C | 238 924 | SAUSA300_0202 | Gaa/Aaa | NS | E273K | Peptide ABC transporter permease | Membrane transport | Peptide ABC transporter permease [26] |
| 975 709 | oppC | aTt/aAt | NS | I211N | Oligopeptide ABC transporter, permease protein | Membrane transport | Peptide ABC transporter permease [26] | |
| 975 798 | oppC | Aaa/Taa | SG | K241STOP | Oligopeptide ABC transporter, permease protein | Membrane transport | Peptide ABC transporter permease [26] | |
| 1 263 354 | pyrH | Atg/Ttg | NS | M207L | Uridylate kinase | Cofactors, vitamins, prosthetic groups, pigments | Involved in cell wall and RNA biosynthesis [27] | |
| D | Population 1 | |||||||
| 848 576 | eno | Gta/Ata | NS | V119I | Phosphopyruvate hydratase | Amino acids and derivatives | Enhances the activation of plasminogen, assisting in tissue dissemination [28] | |
| 1 181 644 | ftsZ | gaC/gaA | NS | D31E | Cell division protein, FtsZ | Cell division and cell cycle | Essential for cell division [29] | |
| 1 420 581 | dapB | Tca/Aca | NS | S211T | Dihydrodipicolinate reductase | Amino acids and derivatives | Essential component for lysine biosynthesis [30] | |
| 1 955 851 | lukD | tCt/tGt | NS | S150C | Leukotoxin, LukD | Virulence, disease, and defense | Well-known cytotoxic virulence gene responsible for cell lyses [31] | |
| 2 808 635 | SAUSA300_2587 | Cgt/Agt | NS | R57S | Accessory secretory protein, Asp1 | Membrane transport | Cell-wall-associated protein involved in the pathogenesis [32] | |
| Population 2 | ||||||||
| 1 405 837 | pepF | cCt/cTt | NS | P11L | Oligoendopeptidase F | Protein metabolism | Protein turnover [33] | |
| 2 165 813 | ilvD | aTg/aCg | NS | M313T | Dihydroxy-acid dehydratase | Amino acids and derivatives | Dehydratase required for isoleucine metabolism and isoleucyl tRNA limitation [34, 35] | |
| 2 554 758 | SAUSA300_2375 | tGg/tAg | SG | W547STOP | Multidrug ABC transporter ATP-binding/permease | Membrane transport | ABC transporter ATP-binding permease [25] | |
| 2 734 666 | panC | Gaa/Aaa | NS | E203K | Pantoate–β-alanine ligase | Cofactors, vitamins, prosthetic groups, pigments | Essential component of CoA synthesis and has been reported as a putative antimicrobial target [36] | |
SNPs resulting in a putative change in protein function are listed by patient. The annotation includes the location in respect to the USA300_FPR3757 reference genome (accession CP000255), gene name, and location and effect of the amino acid change. Significantly more mutations were observed in genes associated with membrane transport. NS mutations occurring in genes coding for hypothetical proteins were excluded from this table but can be found in Supplementary File 1.
Abbreviations: NS, nonsynonymous; SG, stop gained; SL, stop lost; tRNA, transfer RNA.
Assessment of de novo assemblies of unmapped reads demonstrated that all isolates possessed complete or partial fragments of plasmids pUSA01-ISMMS (CP007177) and pUSA02-ISMMS (CP007178). Contigs from unmapped reads of isolate PtD-04 were aligned to these reference plasmids and manually curated to fully circularize the molecules, which were then annotated. This produced 31-kb and 3-kb ISMMS-like plasmids with 48 and 4 coding sequences, respectively (Figure 3). The larger plasmid contained several genes coding for antibiotic resistance to aminoglycosides, macrolides, lincosamides, bacitracin, and β-lactams. Additionally, 4 isolates from 2 patients harbored plasmids carrying constitutively expressed ermC, and 1 patient had 2 isolates with a transposon carrying aac(6′)-aph(2″) conferring clindamycin and aminoglycoside resistance, respectively.
Figure 3.
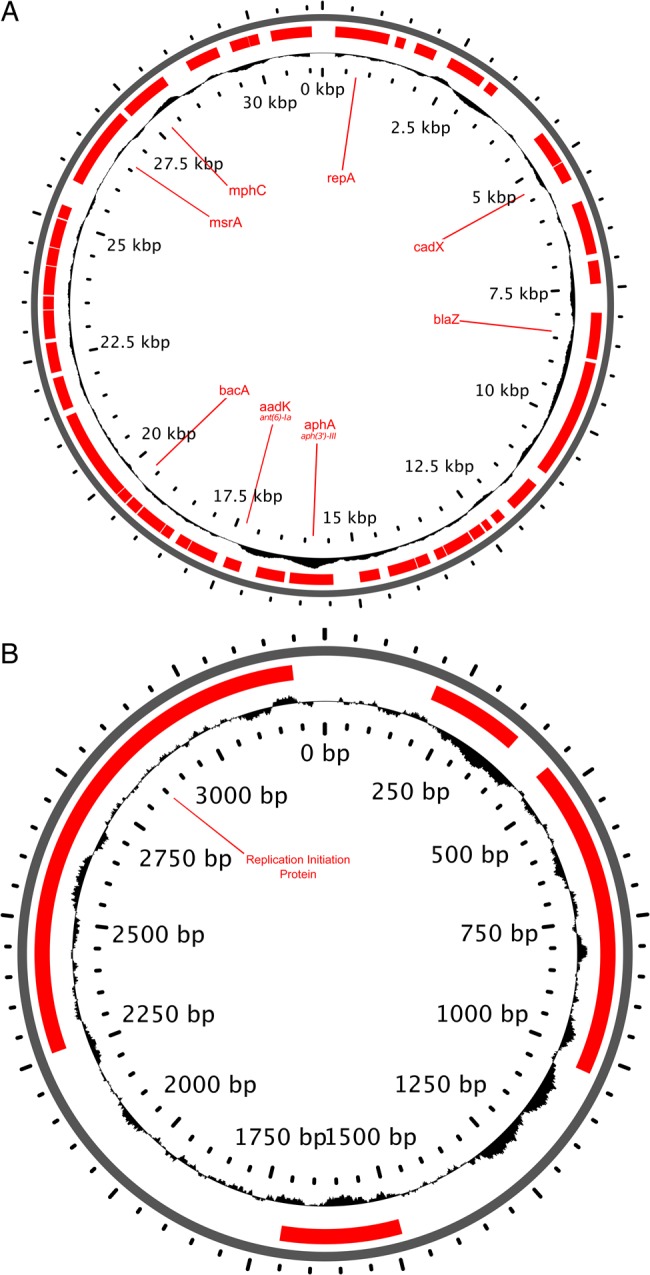
Genome features for both large (31 kb) and small (3 kb) ISMMS-like plasmids identified in isolates 04 and 05 from patient D. Notable resistance and virulence genes are labeled on coding sequences (red), and the GC content is represented on the plot of the inside ring. This figure is available in black and white in print and in color online.
Patient A
Patient A, a previously healthy 19-month-old male at the first time point, had 5 USA300 MRSA strains isolated in 744 days. A colonization isolate and an isolate from a lower extremity abscess collected at the first time point differed by a single SNP (Figure 2). Multiple anatomic sites were involved in subsequent infections (Table 2). Between the time of recovery of the first isolate and 194 days later, no SNPs were identified. However, isolates PtA 04 and PtA 05 possessed 3 and 11 SNPs, respectively, without shared variation (Figure 2).
Half of all mutations that accrued during the microevolution of MRSA in patient A were nonsynonymous, including those in msrA, glmU, tcaR, and nikA (Table 3). Nickel transporter (encoded by nikA) is a virulence factor playing a role in colonization, and mutations in nikA can affect metabolism [22]. In all isolates, we identified a 44-kb class II bacteriophage with homology to bacteriophage 85 (GenBank accession: AY954953) [37].
Patient B
Patient B was 44 years old at the time of recovery of the first isolate, with 8 MRSA isolates collected during the subsequent 1278 days. He had diabetes mellitus and an infected foot ulcer, resulting in an amputation below the left knee. A second infection on his right forearm and new infections at the stump site required multiple courses of antimicrobials and surgical debridement. These first 4 isolates formed a distinct population that evolved linearly, accruing 6 SNPs in 424 days. Patient B underwent a right third toe amputation >1 year later and subsequently developed an infection at the surgical site. Despite intravenous vancomycin treatment, he required a transmetatarsal amputation. A non-USA300 MRSA strain, MLST ST105, was isolated from that site on day 1086. The ST105 strain was resistant to fluoroquinolones and clindamycin, which the patient had previously received to treat the earlier susceptible USA300 population (Table 2). The ST105 strain infection was cured after an extended course of daptomycin, ciprofloxacin, and metronidazole. A second USA300 population was identified approximately 5 months later from a complicated polymicrobial right heel ulcer. Prior to the identification of this population, MRSA-negative culture results cultured from the right foot wound site on day 1114, 3 months prior to the identification of the second USA300 population. The second USA300 group of isolates (PtB 06–08) had greater SNP divergence from the first group (PtB 01–04) than did the between-patient (ie, interhost) populations (Supplementary Table 1). The second USA300 population (PtB 06–08) was fluoroquinolone resistant (gyrA Ser84 → Leu; Supplementary Figure 2) but clindamycin susceptible, and all 3 isolates carried prophages with similarity to bacteriophage 85. While the first population was not observed after day 424, more-comprehensive sampling would be required to confirm elimination.
Within the first USA300 population, SNPs included a nonsense mutation in the gene coding for autolysin (atl) and a nonsynonymous mutation in AraC (Table 3). Autolysin plays a role in cell wall metabolism, biofilm formation, and antibiotic-induced lysis [38]. The second USA300 population possessed greater intrahost nucleotide diversity. Isolate PtB 07 possessed 3 nonsynonymous mutations in loci linked to virulence and antibiotic nonsusceptibility (Table 2), including accessory gene regulator protein A (agrA), which regulates expression of several virulence factor genes [23]. The R170K mutation of agrA maintained the same amino acid charge but varied in size, and visualization of the protein structure suggested a potential conformational change near a binding site that may influence function (Supplementary Figure 3). The subsequent isolate collected from the same anatomic site 2 months later did not possess any of these 3 nonsynonymous mutations, suggesting that isolates PtB 06 and 08 shared an intermediate, unsampled, common ancestor with an unknown time of coalescence (Figure 2).
Patient C
Patient C was 33 years old at the time of collection of the first isolate, with 3 MRSA isolates collected during the subsequent 222 days. She had uncontrolled diabetes mellitus. After treatment failure with amoxicillin/clavulanic acid for the index hip/buttock SSTIs, she was treated with linezolid. On day 131, isolate PtC 02 was obtained from an upper extremity abscess. After failure of oral clindamycin and ciprofloxacin, she received 3 weeks of intravenous vancomycin. On day 225, she presented with a breast abscess, which was treated successfully with oral ciprofloxacin and rifampin. Only 5 SNP sites separated the 3 isolates. PtC 01 and PtC 03 were clindamycin resistant and possessed a 2.4-kb pPV141-like plasmid that constitutively expressed ermC (Supplementary Figure 4). As inferred from SNP diversity and mutation rate estimates, isolates collected on day 0 and 131 shared an ancestor likely predating the acquisition of the USA300 population. SNPs were identified in 2 genes involved in amino acid/nickel transport and cell metabolism (Table 3).
Patient D
Patient D, over a 553-day period, had 13 isolates collected and had the greatest diversity among sampled strains. Subsequently, patient D, 54 years old with a history of diabetes mellitus, developed multiple foot ulcers resulting in toe amputations and then bilateral transmetatarsal amputations. We observed 2 distinct USA300 populations, obtained during 2 discrete periods, comprising isolates PtD 01–05 and PtD 07–13, which were separated by a mean between-population SNP difference of 71, compared with an overall between-patient mean difference of 54.5 (Figure 2). The first population was isolated from both infection sites during initial infection episodes and was not subsequently recovered after day 112, despite multiple cultures of the same anatomical sites. In addition, MRSA-negative results were yielded by culture of a specimen collected from the left foot wound site on day 252, approximately 2 months prior to the identification of the second USA300 population. These findings were consistent with strain replacement of the incident USA300 population. A non-USA300, MLST ST59 MRSA strain was cultured between periods of isolation of the 2 USA300 populations.
Isolates PtD 01 and PtD 02 were from an infection of the left fifth toe amputation site and treatment with oral antimicrobials. Subsequently, an ulcer developed on the right foot, progressing to osteomyelitis requiring right first metatarsal and sesamoid amputation and treatment with intravenous antimicrobials for 6 weeks. Isolates PtD 03–05, in addition to other species, were collected from this infection site. The first USA300 population had little genetic variation, reflected by polytomies in the maximum likelihood phylogeny (Figure 1), suggesting that the same population existed concurrently on multiple anatomical sites during recurrent infections. With the exception of identical isolates PtD 04 and 05, the population was segregated by several unshared SNPs, 67% (8 of 12) of which were nonsynonymous, with mutations in genes coding for enolase, dihydrodipicolinate reductase, and accessory secretory protein (Asp1; Table 3). Isolates PtD 04 and 05 were clindamycin and gentamicin resistant by automated testing; the earlier isolates from the same population were susceptible to both. This resistance was associated with a 4.2-kb pSES22-like plasmid carrying constitutively expressed ermC and an aac(6′)-aph(2″)-possessing transposon, conferring clindamycin and aminoglycoside resistance, respectively (Supplementary Figures 5 and 6). The 2 isolates also shared 2 nonsynonymous mutations in ftsZ and LukD.
On day 230, patient D developed a right foot ulcer that progressed to osteomyelitis despite oral antimicrobial therapy. A MRSA ST59 strain, unrelated to ST8 USA300 MRSA, was isolated from this site. A second USA300 population resistant to fluoroquinolones was later identified at the same right foot wound site, requiring transmetatarsal amputation and 6 weeks of an oral antimicrobial. Fluoroquinolone resistance was linked to the Ser84 → Leu mutation in DNA gyrase (gyrA; Supplementary Figure 2). Afterward, a postoperative infection developed at the stump site. Two more isolates were obtained, PtD 09 and PtD 10. One month later, a new plantar ulcer on the left foot progressed to osteomyelitis. After antimicrobial treatment failure, a transmetatarsal amputation was performed. Linezolid, amoxicillin/clavulanic acid, and ciprofloxacin were administered. This series of infections was polymicrobial. The second population of USA300 had a progressive accumulation of mutations surrounded by a cloud of diversity; however, sparse variation was observed during the first 4 months of recurrent infections caused by this population.
Isolate PtD 13 possessed nonsynonymous mutations in ilvD and panC. The gene ilvD codes for dihydroxy-acid dehydratase, which is required for isoleucine metabolism and isoleucyl transfer RNA limitation. Mutations in this pathway have been associated with reduced linezolid susceptibility [34], suggesting that linezolid therapy may have selected for this mutant.
DISCUSSION
We found significant variation among patients in the diversity of the MRSA population, accumulation of mutations, and presence of plasmids and bacteriophages. MRSA USA300 antimicrobial resistance developed in 2 patients through strain replacement, mobile genetic element acquisition, and a mutation putatively generated by antimicrobial pressure. Two of the 4 patients had 2 temporally and genetically distinct USA300 populations with evolving antibiotic resistance, demonstrating that MRSA decolonization after an initial SSTI would likely not have prevented recurrence of disease in these cases.
Intrahost bacterial population dynamics are governed by bottleneck size and the evolutionary rate, which is a function of host (eg, immune response), pathogen (eg, virulence), and environmental (eg, antibiotic exposure or presence of other bacteria) factors [39]. The bottleneck size may range from a single bacterium to a diverse population separated by tens of SNPs, and evolutionary rate estimates for S. aureus have ranged from 5.6 to 9.5 mutations per year [40, 41]. Once a population is acquired, the intrahost population size may fluctuate in association with pathogenesis or the seeding of other anatomical sites [12]. The observed intrahost diversity will therefore reflect these processes, as well as the duration of colonization prior to symptomatic infection and subsequent sampling.
For the 5 instances in which multiple contemporaneous isolates were available, pair-wise differences ranged from 0 to 8 SNPs, and all but 1 of these isolate pairs were from the same anatomic site. This is lower than previous reports of intrahost diversity of 30–40 SNPs [42, 43]. However, previous estimates were largely based on isolates from asymptomatically colonized individuals with unknown durations of colonization. In our study, we observed a cohesive microbial population among isolates collected from recurrent infections across multiple anatomical sites, consistent with previous studies of colonized individuals [12, 44]. However, the true range of bacterial diversity transmitted to a susceptible individual remains unknown.
The rate of diversification varied among patients and infection episodes, perhaps related to antimicrobial therapy or, possibly, in some cases, the presence of other bacteria. All patients in our study received antimicrobial therapy for each infection episode. In MRSA populations A, B(1), and C, we observed a progressive accumulation of mutations emerging from a homogenous population with low diversity. In contrast, in populations B(2), D(1), and D(2), we observed higher rates of diversification and a greater frequency of segregating nonsynonymous polymorphisms. In patient D, higher rates of diversification among concurrently collected isolates from 2 anatomic sites suggested the existence of site-specific subpopulations or a large transmission bottleneck. Interestingly, among all patients, we observed prolonged periods during which no SNPs were observed among sampled isolates, similar to the findings of others [8]. These periods were sometimes followed by a rapid increase in mutations, a phenomenon previously associated with a transition from colonization to invasive infection [12]. It is worth noting that the increase observed at the last time point of patient D(2) preceded the development of osteomyelitis; however, multiple isolates from each infection episode would be required to comprehensively inform population level dynamics. Patients B and D experienced multiple polymicrobial infections, and the presence of other bacteria can impact the evolutionary dynamics of S. aureus. It is known, for example, that cooperative (eg, Enterococcus faecalis) or competitive (eg, Corynebacterium species) interactions exist [45]. We observed substantial intrahost evolutionary rate heterogeneity, which further cautions against the use of SNP cutoffs in determining recent transmission events.
During recurrent infections, selective pressures may precipitate phenotypic changes, including antibiotic resistance, small colony variants, or mitigated virulence [46]. To investigate adaptive changes to the USA300 genome, we assessed nonsynonymous SNPs. The average intrahost rate of nonsynonymous mutations was nearly 3 times that of synonymous mutations, suggesting incomplete purifying selection leading to the persistence of putative deleterious mutations [43]. Over a longer time scale, many of these mutations would likely be removed through purifying selection. Among nonsynonymous SNPs, mutations more often occurred in regions of the genome responsible for membrane transport, such as ABC transporter genes involved in allocrite transport and environmental stress responses [26]. Nonsynonymous mutations were also frequent in genes responsible for metabolism, virulence, and defense, consistent with an adaptive response to the oxidative stress of antibiotic exposure or host immune response [47]. These adaptations may have a large evolutionary impact on the emergence of antibiotic resistance or even enhanced biofilm production.
We found variations in the presence of plasmids, transposons, and prophage, which are known to facilitate acquisition of novel genetic material [48]. All USA300 isolates possessed ISMMS-like plasmids. Four harbored additional plasmids conferring clindamycin resistance [49]. A similar ISMMS-like plasmid was prevalent among community-derived USA300 strains from New York, suggesting an evolutionary advantage of enhanced antibiotic and heavy metal resistance [8]. Prophages, when present, were found among all isolates from a patient's USA300 population.
S. aureus may undergo marked evolutionary change during a relatively short period that may have direct implications for pathogenesis, risk of subsequent infection, and changes in antimicrobial resistance. Selection for antimicrobial-resistant populations was observed through gain of multiple, diverse mobile genetic elements conferring multidrug resistance and strain replacement by a resistant population. Patients B and D had 3 separate acquisitions of MRSA, involving multiple body sites and changing antibiotic resistance profiles. In contrast, in another study among patients with cystic fibrosis, S. aureus isolates from the same lineage were recovered repeatedly during chronic infections [50]. Individuals with chronic nonhealing wounds, however, may be at risk for acquisition of multiple MRSA strains of the same MLST in high-prevalence settings. The antibiotic pressure during ongoing treatment likely precipitated fluoroquinolone and aminoglycoside resistance in patients B and D. Additionally, we identified a plasmid conferring clindamycin resistance in the first and last isolates but not the second isolate from patient C. However, treatment failure of the PtC T2 infection episode with oral clindamycin and ciprofloxacin and the presence of the plasmid in the subsequent isolate suggest that some population of USA300 may still have harbored the plasmid at the intermediate time point. This necessitates effective sampling of intrahost bacterial populations, which, combined with timely genomic information, may better inform clinical management.
In conclusion, in this pilot study, we did not identify evidence for convergent evolution to suggest specific patterns associated with persistent carriage of USA300 MRSA. Therefore, it is not clear whether persistence of colonization and risk of recurrent infection is primarily related to MRSA strain or host characteristics. However, we do highlight the adaptive potential of MRSA on relatively short time scales, which may have implications for clinical decision-making.
Supplementary Data
Supplementary materials are available at http://jid.oxfordjournals.org. Consisting of data provided by the author to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the author, so questions or comments should be addressed to the author.
Notes
Financial support. This work was supported by the National Institute of Allergy and Infectious Diseases, National Institutes of Health (grant K23 AI095361 to M. Z. D.); and the Centers for Disease Control and Prevention (grant R01 CCR523379 to R. S. D. and S. B.-V.).
Potential conflicts of interest. All authors: No reported conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.King MD, Humphrey BJ, Wang YF, Kourbatova EV, Ray SM, Blumberg HM. Emergence of community-acquired methicillin-resistant Staphylococcus aureus USA 300 clone as the predominant cause of skin and soft-tissue infections. Ann Intern Med 2006; 144:309–17. [DOI] [PubMed] [Google Scholar]
- 2.Montgomery CP, David MZ, Daum RS. Host factors that contribute to recurrent staphylococcal skin infection. Curr Opin Infect Dis 2015; 28:253–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harris SR, Cartwright EJ, Török ME et al. . Whole-genome sequencing for analysis of an outbreak of meticillin-resistant Staphylococcus aureus: a descriptive study. Lancet Infect Dis 2012; 3099:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Azarian T, Cook RL, Johnson JA et al. . Whole-genome sequencing for outbreak investigations of methicillin-resistant Staphylococcus aureus in the neonatal intensive care unit: time for routine practice? Infect Control Hosp Epidemiol 2015; 36:777–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alam MT, Read TD, Petit RA et al. . Transmission and microevolution of USA300 MRSA in U.S. households: evidence from whole-genome sequencing. MBio 2015; 6:e00054–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paterson GK, Harrison EM, Murray GGR et al. . Capturing the cloud of diversity reveals complexity and heterogeneity of MRSA carriage, infection and transmission. Nat Commun 2015; 6:6560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stegger M, Wirth T, Andersen PS et al. . Origin and evolution of European community-acquired methicillin-resistant Staphylococcus aureus. MBio 2014; 5:e01044–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Uhlemann A-C, Dordel J, Knox JR et al. . Molecular tracing of the emergence, diversification, and transmission of S. aureus sequence type 8 in a New York community. Proc Natl Acad Sci 2014; 111:6738–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baines SL, Holt KE, Schultz MB et al. . Convergent adaptation in the dominant global hospital clone ST239 of methicillin-resistant Staphylococcus aureus. MBio 2015; 6:e00080–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Worby CJ, Lipsitch M, Hanage WP. Within-host bacterial diversity hinders accurate reconstruction of transmission networks from genomic distance data. PLoS Comput Biol 2014; 10:e1003549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Price J, Gordon NC, Crook D, Llewelyn M, Paul J. The usefulness of whole genome sequencing in the management of Staphylococcus aureus infections. Clin Microbiol Infect 2013; 19:784–9. [DOI] [PubMed] [Google Scholar]
- 12.Young BC, Golubchik T, Batty EM et al. . Evolutionary dynamics of Staphylococcus aureus during progression from carriage to disease. Proc Natl Acad Sci U S A 2012; 109:4550–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Enright M, Day N, Davies C. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol 2000; 38:1008–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.International Working Group on the Classification of Staphylococcal Cassette Chromosome Elements (IWG-SCC). Classification of staphylococcal cassette chromosome mec (SCCmec): guidelines for reporting novel SCCmec elements. Antimicrob Agents Chemother 2009; 53:4961–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.David MZ, Taylor A, Lynfield R et al. . Comparing pulsed-field gel electrophoresis with multilocus sequence typing, spa typing, staphylococcal cassette chromosome mec (SCCmec) typing, and PCR for panton-valentine leukocidin, arcA, and opp3 in methicillin-resistant Staphylococcus aureus isolates. J Clin Microbiol 2013; 51:814–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Overbeek R, Olson R, Pusch GD et al. . The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res 2014; 42(D1):D206–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Croucher NJ, Page AJ, Connor TR et al. . Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res 2015; 43:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Komatsuzawa H, Fujiwara T, Nishi H et al. . The gate controlling cell wall synthesis in Staphylococcus aureus. Mol Microbiol 2004; 53:1221–31. [DOI] [PubMed] [Google Scholar]
- 19.Singh VK, Moskovitz J. Multiple methionine sulfoxide reductase genes in Staphylococcus aureus: expression of activity and roles in tolerance of oxidative stress. Microbiology 2003; 149(Pt 10):2739–47. [DOI] [PubMed] [Google Scholar]
- 20.Jefferson KK, Pier DB, Goldmann DA, Pier GB. The Teicoplanin-Associated Locus Regulator (TcaR) and the Intercellular Adhesin Locus Regulator (IcaR) Are Transcriptional Inhibitors of the ica Locus in Staphylococcus aureus. J Bacteriol 2004; 186:2449–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCallum N, Bischoff M, Maki H, Wada A, Berger-Bachi B. TcaR, a putative MarR-like regulator of sarS expression. J Bacteriol 2004; 186:2966–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hiron A, Posteraro B, Carrière M et al. . A nickel ABC-transporter of Staphylococcus aureus is involved in urinary tract infection. Mol Microbiol 2010; 77:1246–60. [DOI] [PubMed] [Google Scholar]
- 23.Abdelnour A, Arvidson S, Bremell T, Ryden C, Tarkowski A. The accessory gene regulator (agr) controls Staphylococcus aureus virulence in a murine arthritis model. Infect Immun 1993; 61:3879–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berti AD, Baines SL, Howden BP et al. . Heterogeneity of genetic pathways toward daptomycin nonsusceptibility in Staphylococcus aureus determined by adjunctive antibiotics. Antimicrob Agents Chemother 2015; 59:2799–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dawson RJP, Locher KP. Structure of a bacterial multidrug ABC transporter. Nature 2006; 443:180–5. [DOI] [PubMed] [Google Scholar]
- 26.Garmory HS, Titball RW. ATP-binding cassette transporters are targets for the development of antibacterial vaccines and therapies. Infect Immun 2004; 72:6757–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hari Prasad O, Nanda Kumar Y, Reddy OVS, Chaudhary A, Sarma PVGK. Cloning, expression, purification and characterization of UMP kinase from Staphylococcus aureus. Protein J 2012; 31:345–52. [DOI] [PubMed] [Google Scholar]
- 28.Mölkänen T, Tyynelä J, Helin J, Kalkkinen N, Kuusela P. Enhanced activation of bound plasminogen on Staphylococcus aureus by staphylokinase. FEBS Lett 2002; 517:72–8. [DOI] [PubMed] [Google Scholar]
- 29.Vollmer W. The prokaryotic cytoskeleton: a putative target for inhibitors and antibiotics? Appl Microbiol Biotechnol 2006; 73:37–47. [DOI] [PubMed] [Google Scholar]
- 30.Kuroda M, Kuroda H, Oshima T, Takeuchi F, Mori H, Hiramatsu K. Two-component system VraSR positively modulates the regulation of cell-wall biosynthesis pathway in Staphylococcus aureus. Mol Microbiol 2004; 49:807–21. [DOI] [PubMed] [Google Scholar]
- 31.Laabei M, Uhlemann A-C, Lowy FD et al. . Evolutionary trade-offs underlie the multi-faceted virulence of Staphylococcus aureus. PLoS Biol 2015; 13:e1002229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Siboo IR, Chaffin DO, Rubens CE, Sullam PM. Characterization of the accessory Sec system of Staphylococcus aureus. J Bacteriol 2008; 190:6188–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kanamaru K, Stephenson S, Perego M. Overexpression of the PepF oligopeptidase inhibits sporulation initiation in Bacillus subtilis. J Bacteriol 2002; 184:43–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gao W, Chua K, Davies JK et al. . Two novel point mutations in clinical Staphylococcus aureus reduce linezolid susceptibility and switch on the stringent response to promote persistent infection. PLoS Pathog 2010; 6:e1000944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anderson KL, Roberts C, Disz T et al. . Characterization of the Staphylococcus aureus heat shock, cold shock, stringent, and SOS responses and their effects on log-phase mRNA turnover. J Bacteriol 2006; 188:6739–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choudhry AE, Mandichak TL, Broskey JP et al. . Inhibitors of pantothenate kinase: novel antibiotics for staphylococcal iInfections. Antimicrob Agents Chemother 2003; 47:2051–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alibayov B, Baba-Moussa L, Sina H, Zdeňková K, Demnerová K. Staphylococcus aureus mobile genetic elements. Mol Biol Rep 2014; 41:5005–18. [DOI] [PubMed] [Google Scholar]
- 38.Grilo IR, Ludovice AM, Tomasz A, de Lencastre H, Sobral RG. The glucosaminidase domain of Atl - the major Staphylococcus aureus autolysin - has DNA-binding activity. Microbiologyopen 2014; 3:247–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Didelot X, Walker AS, Peto TE, Crook DW, Wilson DJ. Within-host evolution of bacterial pathogens. Nat Rev Microbiol 2016; 14:150–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harris SR, Feil EJ, Holden MTG et al. . Evolution of MRSA during hospital transmission and intercontinental spread. Science 2010; 327:469–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nübel U, Dordel J, Kurt K et al. . A timescale for evolution, population expansion, and spatial spread of an emerging clone of methicillin-resistant Staphylococcus aureus. PLoS Pathog 2010; 6:e1000855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Price JR, Golubchik T, Cole K et al. . Whole-genome sequencing shows that patient-to-patient transmission rarely accounts for acquisition of Staphylococcus aureus in an intensive care unit. Clin Infect Dis 2014; 58:609–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Golubchik T, Batty EM, Miller RR et al. . Within-host evolution of Staphylococcus aureus during asymptomatic carriage. PLoS One 2013; 8:e61319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Popovich KJ, Snitkin E, Green SJ et al. . Genomic epidemiology of USA300 methicillin-resistant Staphylococcus aureus in an Urban community. Clin Infect Dis 2015; 62:37–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nair N, Biswas R, Götz F, Biswas L. Impact of Staphylococcus aureus on pathogenesis in polymicrobial infections. Infect Immun 2014; 82:2162–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tuchscherr L, Medina E, Hussain M et al. . Staphylococcus aureus phenotype switching: an effective bacterial strategy to escape host immune response and establish a chronic infection. EMBO Mol Med 2011; 3:129–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 2007; 130:797–810. [DOI] [PubMed] [Google Scholar]
- 48.Kwan T, Liu J, DuBow M, Gros P, Pelletier J. The complete genomes and proteomes of 27 Staphylococcus aureus bacteriophages. Proc Natl Acad Sci U S A 2005; 102:5174–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McDougal LK, Fosheim GE, Nicholson A et al. . Emergence of resistance among USA300 methicillin-resistant Staphylococcus aureus isolates causing invasive disease in the United States. Antimicrob Agents Chemother 2010; 54:3804–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McAdam PR, Holmes A, Templeton KE, Fitzgerald JR. Adaptive evolution of Staphylococcus aureus during chronic endobronchial infection of a cystic fibrosis patient. PLoS One 2011; 6:e24301. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.