Abstract
We report an atomic-resolution structure for a sensory member of the microbial rhodopsin family, the phototaxis receptor sensory rhodopsin II (NpSRII), which mediates blue-light avoidance by the haloarchaeon Natronobacterium pharaonis. The 2.4 angstrom structure reveals features responsible for the 70- to 80-nanometer blue shift of its absorption maximum relative to those of haloarchaeal transport rhodopsins, as well as structural differences due to its sensory, as opposed to transport, function. Multiple factors appear to account for the spectral tuning difference with respect to bacteriorhodopsin: (i) repositioning of the guanidinium group of arginine 72, a residue that interacts with the counterion to the retinylidene protonated Schiff base; (ii) rearrangement of the protein near the retinal ring; and (iii) changes in tilt and slant of the retinal polyene chain. Inspection of the surface topography reveals an exposed polar residue, tyrosine 199, not present in bacteriorhodopsin, in the middle of the membrane bilayer. We propose that this residue interacts with the adjacent helices of the cognate NpSRII transducer NpHtrII.
Microbial rhodopsins are a family of membrane-embedded photoactive retinylidene proteins found throughout the three domains of life: archaea (1–3), eubacteria (4), and unicellular eukaryotes (5, 6). They share a common design of seven transmembrane helices forming an interior pocket for the chromophore retinal, and their functions are driven by a common photochemical reaction—light-induced retinal isomerization—but they carry out two distinctly different functions: light-driven ion transport and photosensory signaling. Both functional types are found in haloarchaea such as Halobacterium salinarum and Natronobacterium pharaonis. In the haloarchaea, bacteriorhodopsin (BR) and halorhodopsin (HR) are light-driven ion pumps for protons and chloride, respectively (2, 3); and the sensory rhodopsins I and II (SRI and SRII) are phototaxis receptors controlling the cell’s swimming behavior in response to changes in light intensity and color (1). The N. pharaonis phototaxis receptor NpSRII is 27% identical to BR in amino acid sequence (7) and exhibits typically ~40% identity with other sensory rhodopsins; all contain ~80% identity in the residues known to form the retinal binding pocket in BR.
Atomic-resolution crystal structures of BR and HR from H. salinarum have been obtained by x-ray crystallography (8–10). An intermediate resolution (6.9 Å) projection structure of NpSRII derived from electron crystallography of two-dimensional (2D) crystals showed an overall disposition of the seven helices that was similar to that in the two transport rhodopsins (11). Here we report atomic-resolution information for a microbial sensory rhodopsin, the 2.4 Å structure of NpSRII obtained from x-ray diffraction of 3D crystals grown in a cubic lipid phase.
The overall seven-helical structure is similar to those of the transport rhodopsins BR and HR (8–11). The antiparallel β sheet at the BC loop is two residues shorter than in BR (9). The EF loop participates in crystal packing between bilayer sheets, and helix E is fully ordered with a π-bulge kink (10) at position 154. For data collection and refinement statistics, see Table 1.
Table 1.
X-ray data collection, molecular replacement, and refinement statistics. Crystals were grown from a cubic lipid phase (27) using octylglucoside-purified NpSRII (28.0 mg/ml) preincubated with H. salinarum polar lipids (11.2 mg/ml) (11). Ten-microliter aliquots were mixed with 10 μl of monoolein glyceride (Nu-Check Prep., MI) and centrifuged for 1 hour at 11,000g at 22°C. After overnight incubation at 22°C, precipitant was added. The crystals used were thin rods about 5 μm by 20 μm by 200 μm and were examined 5 weeks after the addition of 50 μl of 3.5 M KCl in 50 mM MES (pH 5.3) as precipitant. Addition of exogenous H. salinarum polar lipids was essential for crystal formation. The crystals belong to space group C2221, with a = 87.34 Å, b = 130.81 Å, and c = 50.87 Å, and consist of bilayers stacked in the b direction. The a and c cell dimensions, as well as the packing, are very similar to those reported for 2D crystals of NpSRII (11). Diffraction data were collected on one cryocooled crystal at the microfocus beamline ID13 at ESRF (Grenoble), using a MAR Research charge-coupled device detector. Each image was 1° in φ, with an exposure time of 2 s. Images were reduced, scaled, and merged with the programs DENZO/SCALEPACK (28). Molecular replacement was carried out with the program X-PLOR (29), using the coordinates of the 1.55 Å bacteriorhodopsin structure (PDB code 1C3W, protein without retinal) and yielded an initial R factor of 49.2% (4 to 12 Å). Successive rounds of refinement and model building with the programs CNS (30) and CHAIN (31) using annealed simulated omit and 3|Fo| – 2|Fc| maps resulted in an R factor of 23.3%, and an Rfree of 28.0% for all data between 2.4 and 20 Å without σ cutoff. In many regions near the hydrophobic protein surface, long tubes of density are present for native dihydrophytyl lipids, which as in the case of BR remained tightly bound to the protein through solubilization and cubic lipid phase crystallization. All peptide bonds fall into the allowed regions of the Ramachandran plot.
| Data reduction resolution range | 2.4–20.0 Å | 2.40–2.44 Å |
|---|---|---|
| Total observations | 80,761 | |
| Unique structure factors | 10,704 | 520 |
| Rmerge (%) | 9.5 | 44.0 |
| Average I/σ(I) | 5.7 | 1.3 |
| Completeness (%) | 92.2 | 92.0 |
| Mosaicity (°) | 0.86 | |
| Refinement resolution range | 2.4–20.0 Å | |
| R factor (%) for working set, no σ cutoff | 23.3 | |
| Rfree (%) for 8.7% of the unique structure factors | 28.0 | |
| Deviation from ideal bond lengths (Å) | 0.0086 | |
| Deviation from ideal bond angles (°) | 1.29 |
Among the known photosensory pigments, a unique property of retinylidene proteins, in microbial rhodopsins as well as higher animal visual pigments, is the tuning of the absorption spectrum of the retinal chromophore over a wide range of the visible region. BR, HR, and SRI in H. salinarum absorb green-orange light, with absorption maxima at 570 to 590 nm, whereas SRII exhibits blue-shifted absorption maxima, at 487 and 497 nm for the H. salinarum and N. pharaonis homologs [HsSRII (12) and NpSRII (13, 14)], respectively. A protonated retinylidene Schiff base compound in methanol/Cl− exhibits maximal absorption at 440 nm (12). Interactions between the retinal chromophore and its protein environment typically shift the absorption to longer wavelengths (the “opsin shift”) (15), resulting in absorption maxima at 497 nm in NpSRII and 568 nm in BR. In the better understood case, BR, three contributing factors have been identified (16): (i) the protein forces the conformation of the C6–C7 single bond in the retinal to be 6s-trans, allowing ring/chain coplanarity; (ii) the positive charge on the protonated Schiff base is only weakly stabilized by a complex counterion provided by the protein environment; and (iii) a less well-defined third factor, which is thought to involve the interaction of polar or polarizable protein groups with the chromophore, destabilizes the ground state or stabilizes the excited state.
The retinal-binding pocket of NpSRII provides insights into structural changes responsible for its blue shift with respect to BR (Figs. 1 and 2). The ring and polyene chain of NpSRII are coplanar as in BR, and therefore we expect a similar opsin shift contribution from this factor. In the case of HsSRII, it has been suggested that ring/chain coplanarity alone is sufficient to explain its relatively small opsin shift to 487 nm (12).
Fig. 1.
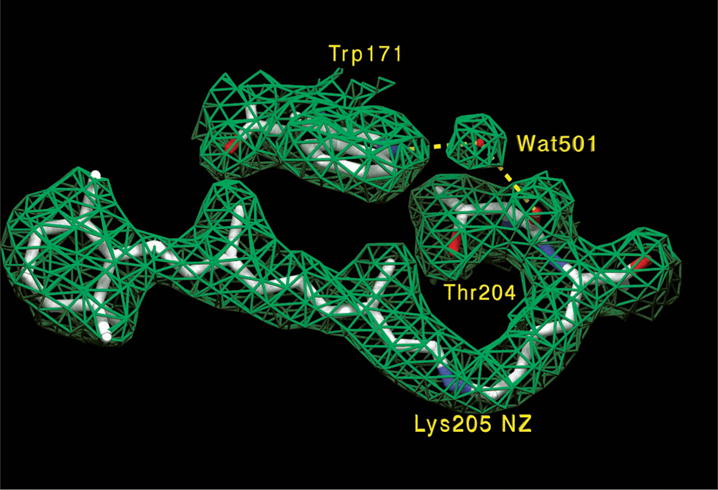
Electron density map (2|Fo| – |Fc|, contoured at 1σ) and corresponding molecular model of the retinal binding pocket. As in BR and HR, the all-trans retinal polyene chain is coplanar with the beta-ionone ring, which is in the 6s-trans conformation. Wat501 is bridging helices F (Trp171) and G (Thr204 carbonyl). In addition, Wat501 is also hydrogen-bonding with Thr167 OH (not shown).
Fig. 2.
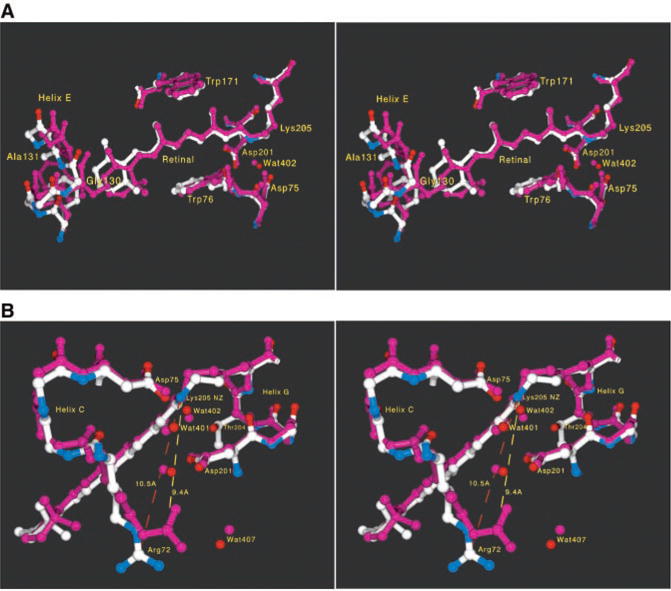
Comparison of NpSRII and BR structures. The coordinates were aligned with the Iterative Magic Fit procedure of Swiss-PDBViewer version 3.7b2 (33), yielding a root mean square deviation of 0.96 Å for 816 main-chain atoms (BR is shown in purple; NpSRII is shown in white, blue, and red). (A) Stereo view of the retinal binding pocket. There is a change in the tilt of the whole polyene chain, presumably largely due to extra room near the end of the ring (C3 and C4 atoms) because of a combination of smaller side chains on residues 130/131 (Gly130/Ala131 in NpSRII, Ser141/Thr142 in BR) and a local outward bowing of the backbone of helices E and F by up to 1.3 Å [see (C)]. The Ser141/Thr142 pair in BR, not labeled in the figure, contributes two hydroxyl groups, which are evident as purple protruding from helix E toward the ring. And despite equivalent retinal stereochemical restraints in refinement, the distance from the Schiff base nitrogen (NZ) to retinal C4 increases from 14.0 to 14.6 Å, resulting in a more linear (or less bent) polyene chain in NpSRII. The relative positions of the protonated Schiff base nitrogen and the two negatively charged aspartate carboxylates of Asp75 and Asp201 are nearly unchanged. However, the electron density for the water molecule found between the Schiff base and these two carboxylates (Wat402) (9, 32) is very weak, suggesting that this water is either more mobile or not fully occupied in NpSRII. In contrast, waters 401, 406, and 501 are well ordered (9). The π bulge of helix G at residue 204 is also present as it is in BR (9) and HR (10), despite the bulkier side chain (Thr204) at this position. (B) Stereo view of the Schiff base and the complex counterion. The guanidinium of Arg72 moves 1.1 Å away (10.5 versus 9.4 Å) from the Schiff base nitrogen and also reorients its Ne hydrogen. This movement becomes possible because several side chains in the region between Arg72 and the extracellular side (below Arg72 in the figure) have reduced volumes (Phe208 → Ile197, Glu194 → Pro183, and Glu204 → Asp192). (C) Plot of distance versus residue number. The average main-chain distance between NpSRII and BR is plotted as a function of residue number (BR numbering). The largest deviations are at the shortened antiparallel β sheet in the BC loop (near residue 70) and at the COOH-terminus, which is being displaced by a detergent molecule.
Although one of the important means of modulating the absorption of a protonated retinal Schiff base is to vary its distance to its counterion (15), the two aspartate carboxyls Asp75 and Asp201 in NpSRII are in nearly identical positions as in BR. Instead, several changes in the structure combine to cause the blue shift from BR by minimizing factors (ii) and (iii), which shift BR absorption to longer wavelengths. The first is a displacement of the guanidinium group of Arg72 by 1.1 Å, coupled with a rotation away from the Schiff base in NpSRII. This increase in distance reduces the influence of Arg72 on the counterion, thus strengthening the Schiff base/counterion interaction.
Second, a blue shift is expected (15, 17) from the removal of two hydroxyls near the β-ionone ring from Ser141 and Thr142 in BR, replaced with nonpolar residues Gly130 and Ala131 in NpSRII. A possible third contributor to the color difference is the change in tilt and slant of the retinal polyene chain, altering the interaction of the conjugated π system with the binding pocket, thereby modulating ground- and excited-state energy levels (Fig. 2A).
Mutagenic substitution of 10 residues, in or near the retinal-binding pocket with their corresponding BR residues, including Gly130 to Ser141 and Ala131 to Thr142, produced only a ~25-nm red shift of the NpSRII absorption maximum (18, 19). Our interpretation is that these substitutions do not produce the full 71-nm red shift to the BR value, because they do not result in the movements of Arg72 and the other retinal pocket residues from their altered positions, which are determined by residue interactions outside the pocket as well as by helix backbone differences between NpSRII and the transport rhodopsins (Fig. 2C). Arg72 is repositioned as a consequence of several factors, including movement of its helix backbone by 0.9 Å and the cavity created by changes from BR: Phe208 → Ile197, Glu194 → Pro183, and Glu204 → Asp192 (Fig. 2B). Hence the spectral tuning results from precise positioning of retinal binding pocket residues and the guanidinium of Arg72, which cannot be deduced from primary structure alone but requires atomic-resolution tertiary structure information.
Wegener et al. (20) have used electron paramagnetic resonance spectroscopy to measure the accessibility and static and transient mobility of nitroxide labels placed at the positions of eight residues on the cytoplasmic side of helices F and G in NpSRII. They concluded that Lys157 and Ser158 are oriented toward the aqueous phase; Leu159, Tyr160, Ile211, and Leu213 face the protein interior; and Phe210 and Ala212 are located at the protein/lipid bilayer interface. Each of these assignments is confirmed by the crystal structure except Ile211, which is on the surface of helix G facing the hydrophobic portion of the bilayer. A possible explanation of the contrasting assignment of Ile211 is that the spin-label nitroxide distorts the structure. Further, light-induced changes in the spin labels indicated movement of helix F with negligible movement of helix G (20). The disconnection of the two helices was suggested as possibly being due to the predicted absence of the π bulge in helix G (20), but we observe the π bulge to be present in the NpSRII structure, indicating that the apparent dissociation of helixes F and G in the spin-labeling study must be attributable to other factors.
The sensory rhodopsins do not pump ions when complexed with their cognate transducers, but in some cases light-driven vectorial translocation of protons does occur in the absence of transducer (21). Substantial proton transport by NpSRII required factors that are expected to increase cytoplasmic-side proton conductivity (the presence of sodium azide and the Phe86 → Asp86 mutation), and both wild-type NpSRII and HsSRII circulate protons primarily to and from the extracellular side. In the structure, the low proton conductivity on the cytoplasmic side is evident in that the Asp96-Thr46 pair in the cytoplasmic channel of BR is replaced by the Phe86-Leu40 pair and presents a hydrophobic barrier (22). The greater hydrophobicity of the cytoplasmic side is attested by the presence of a detergent molecule (β-octylglucoside) that is inserted with its C8 tail perpendicular to the bilayer into the middle of the seven-helix bundle, almost reaching the side chain of Phe86.
A fundamental question is how the common design of microbial rhodopsin proteins has been adapted to carry out their two distinct functions. Transport rhodopsins function independently of interaction with other proteins and translocate ions through an intramolecular channel. Sensory rhodopsins, on the other hand, transmit signals by protein-protein interaction with transducer proteins that control cytoplasmic enzymatic activity (7, 23, 24). In HsSRI and HsSRII, the receptors form a molecular complex with their cognate integral membrane transducer proteins, HtrI and HtrII, respectively, which in turn modulate a cytoplasmic phosphorylation pathway that controls the cell’s motility apparatus (1). Chimera experiments with the H. salinarum transducers showed that the interaction specificities of SRI with HtrI and SRII with HtrII are determined by the transmembrane helices of the Htr subunits (25), indicating that interaction occurs within or near the membrane. A fragment of NpHtrII lacking most of the cytoplasmic domain interacts with NpSRII in vitro, supporting the idea that transmembrane helix-helix interactions occur also in the N. pharaonis pair (20). Bearing on the possible location of the transducer-binding surface of the receptor, the structure of NpSRII reveals that a tyrosine residue protrudes from the lipid-facing surface of helix G (Fig. 3). Unlike threonine or serine hydroxyls, which can hydrogen-bond to main-chain carbonyls of their own helix, the tyrosine phenol group is too long and rigid to allow the formation of such hydrogen bonds. To avoid an unpaired polar hydroxyl in the middle of the bilayer, in the 3D crystals, and presumably in the 2D crystals as well, the Tyr199 hydroxyl forms an intermolecular hydrogen bond to the main-chain carbonyl of a transmembrane helix from a neighboring molecule in the same bilayer (Ala125 in helix E). Tyr199 is conserved in the three known SRII sequences, whereas this residue is a phenylalanine in the two known SRI sequences (1). The need to hydrogen-bond to an adjacent protein makes Tyr199 an excellent candidate for transducer binding in the SRII-HtrII complex in N. pharaonis membranes. The hydrophobic surface of NpSRII displays three distinct faces: face I is defined by helices F, G, and A; face II by helices A (the CP half), B, C, D, and E (the EC half); and face III by helices E and F. Tyr199 is located prominently in the middle of face I, the face that also contains both helices known to undergo large motions during the BR photocycle (26). It is thus likely that face I provides the majority of the interaction with the photosignal transducer. Other residues on the outside of helix G also show clear grouping between the SRI and SRII subfamilies, possibly contributing to transducer binding specificity.
Fig. 3.
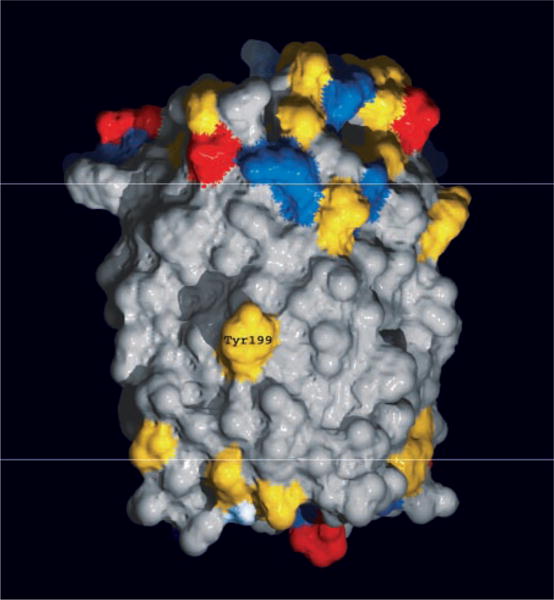
Exposed conserved tyrosine in middle of the bilayer. The NpSRII surface (CP side, top; EC side, bottom) is colored according to amino acid type (red, negatively charged; blue, positively charged; yellow, polar; gray, hydrophobic). Residue Tyr199 is situated in the middle of the bilayer (whose hydrophobic portion lies between the white horizontal lines) near a π bulge on the outside of helix G one-and-a-half helix turns from the lysine (Lys205) to which the retinal is attached.
Acknowledgments
We thank R. R. Birge for discussions regarding spectral tuning in NpSRII and BR, C. Riekel for access to the microfocus beamline at the European Synchrotron Radiation Facility (ESRF), and J.-P. Cartailler for assistance with Fig. 2C. Supported by NIH grants R01-GM59970 (H.L.), R01-GM29498 (J.K.L.), and R01-GM27750 (J.L.S.); Department of Energy grant DEFG03-86ER13525 (J.K.L.); and a Robert A. Welch Foundation award (J.L.S.). The atomic coordinates of NpSRII have been deposited in the Protein Data Bank (PDB) with the entry name of 1JGJ.
References and Notes
- 1.Hoff WD, Jung KH, Spudich JL. Annu Rev Biophys Biomolec Struct. 1997;26:223. doi: 10.1146/annurev.biophys.26.1.223. [DOI] [PubMed] [Google Scholar]
- 2.Oesterhelt D. Curr Opin Struct Biol. 1998;8:489. doi: 10.1016/s0959-440x(98)80128-0. [DOI] [PubMed] [Google Scholar]
- 3.Lanyi JK. J Phys Chem. 2000;104:11441. [Google Scholar]
- 4.Béjà O, et al. Science. 2000;289:1902. doi: 10.1126/science.289.5486.1902. [DOI] [PubMed] [Google Scholar]
- 5.Bieszke JA, Spudich EN, Scott KL, Borkovich KA, Spudich JL. Biochemistry. 1999;38:14138. doi: 10.1021/bi9916170. [DOI] [PubMed] [Google Scholar]
- 6.Spudich JL, Yang CH, Jung KH, Spudich EN. Annu Rev Cell Dev Biol. 2000;16:365. doi: 10.1146/annurev.cellbio.16.1.365. [DOI] [PubMed] [Google Scholar]
- 7.Seidel R, et al. Proc Natl Acad Sci USA. 1995;92:3036. doi: 10.1073/pnas.92.7.3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Belrhali H, et al. Structure. 1999;7:909. doi: 10.1016/s0969-2126(99)80118-x. [DOI] [PubMed] [Google Scholar]
- 9.Luecke H, Schobert B, Richter HT, Cartailler JP, Lanyi JK. J Mol Biol. 1999;291:899. doi: 10.1006/jmbi.1999.3027. [DOI] [PubMed] [Google Scholar]
- 10.Kolbe M, Besir H, Essen LO, Oesterhelt D. Science. 2000;288:1390. doi: 10.1126/science.288.5470.1390. [DOI] [PubMed] [Google Scholar]
- 11.Kunji ERS, Spudich EN, Grisshammer R, Henderson R, Spudich JL. J Mol Biol. 2001;308:279. doi: 10.1006/jmbi.2001.4565. [DOI] [PubMed] [Google Scholar]
- 12.Takahashi T, et al. Biochemistry. 1990;29:8467. doi: 10.1021/bi00488a038. [DOI] [PubMed] [Google Scholar]
- 13.Hirayama J, et al. Biochemistry. 1992;31:2093. doi: 10.1021/bi00122a029. [DOI] [PubMed] [Google Scholar]
- 14.Chizov I, et al. Biophys J. 1998;75:999. doi: 10.1016/S0006-3495(98)77588-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakanishi K, et al. Photochem Photobiol. 1979;29:657. doi: 10.1111/j.1751-1097.1979.tb07745.x. [DOI] [PubMed] [Google Scholar]
- 16.Yan B, et al. J Biol Chem. 1995;270:29668. doi: 10.1074/jbc.270.50.29668. [DOI] [PubMed] [Google Scholar]
- 17.Spudich JL, et al. Biophys J. 1986;49:479. doi: 10.1016/S0006-3495(86)83657-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shimono K, Iwamoto M, Sumi M, Kamo N. Photochem Photobiol. 2000;72:141. doi: 10.1562/0031-8655(2000)072<0141:eotcaa>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 19.Kamo N, Shimono K, Iwamoto M, Sudo Y. Biochemistry (Moscow) doi: 10.1023/a:1013187403599. in press. [DOI] [PubMed] [Google Scholar]
- 20.Wegener AA, Chizhov I, Engelhard M, Steinhoff HJ. J Mol Biol. 2000;301:881. doi: 10.1006/jmbi.2000.4008. [DOI] [PubMed] [Google Scholar]
- 21.HtrI-free HsSRI was shown to exhibit single photon–induced proton pumping out of cell envelope vesicles at pH > 7 [; Bogomolni RA, et al. Proc Natl Acad Sci USA. 1994;91:10188. doi: 10.1073/pnas.91.21.10188. [DOI] [PMC free article] [PubMed] [Google Scholar]; ]. In cell envelope vesicles, HsSRII exhibits only electroneutral light-induced circulation of protons to and from the extracellular medium [; Sasaki J, Spudich JL. Biophys J. 1999;77:2145. doi: 10.1016/S0006-3495(99)77055-4. [DOI] [PMC free article] [PubMed] [Google Scholar]; NpSRII was found to have some proton transport activity in such vesicles [; Sudo Y, Iwamoto M, Shimono K, Sumi M, Kamo N. Biophys J. 2001;80:916. doi: 10.1016/S0006-3495(01)76070-5. [DOI] [PMC free article] [PubMed] [Google Scholar]; as well as in black lipid films [; Schmies G, et al. Biophys J. 2000;78:959. doi: 10.1016/S0006-3495(00)76654-9. [DOI] [PMC free article] [PubMed] [Google Scholar]; ]. When expressed in Xenopus oocytes, HsSRII exhibited weak proton transport and NpSRII did not show any stationary photocurrent [; Schmies G, Engelhard M, Wood PG, Nagel G, Bamberg E. Proc Natl Acad Sci USA. 2001;98:1555. doi: 10.1073/pnas.031562298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.The cytoplasmic region of NpSRII lacks a nucleation site for the development of a hydrogen-bonded network of water molecules [; Luecke H, et al. J Mol Biol. 2000;300:1237. doi: 10.1006/jmbi.2000.3884. [DOI] [PubMed] [Google Scholar]; Also, the extracellular region lacks proton release machinery; that is, a pair of glutamic acid residues connected to the Schiff base region by a 3D hydrogen-bonded network of side chains and water (9) [; Rammelsberg R, Huhn G, Lübben M, Gerwert K. Biochemistry. 1998;37:5001. doi: 10.1021/bi971701k. [DOI] [PubMed] [Google Scholar]; that prevents the return of the proton from the extracellular side in bacteriorhodopsin [; Balashov SP, Imasheva ES, Govindjee R, Ebrey TG. Biophys J. 1996;70:473. doi: 10.1016/S0006-3495(96)79591-7. [DOI] [PMC free article] [PubMed] [Google Scholar]; Richter H-T, Brown LS, Needleman R, Lanyi JK. Biochemistry. 1996;35:4054. doi: 10.1021/bi952883q. [DOI] [PubMed] [Google Scholar]; A similar situation arises in the Asp96 → Asn96 mutant of BR at a pH below the pK for proton release; but unlike NpSRII, this protein exhibits transport. In BR, although at pH < 6 protonation equilibrium between the Schiff base and its counterion ensures that the proton can pass to the extracellular side [; Brown LS, Dioumaev AK, Needleman R, Lanyi JK. Biophys J. 1998;75:1455. doi: 10.1016/S0006-3495(98)74064-0. [DOI] [PMC free article] [PubMed] [Google Scholar]; at the low pH the reprotonation from the cytoplasmic side is strongly accelerated [; Miller A, Oesterhelt D. Biochem Biophys Acta. 1990;1020:57. [Google Scholar]; In contrast, the cytoplasmic region of NpSRII is likely to be a permanent barrier to protons. If directionality is conferred on the movement of the transported ion by the changing ion conductivities of the extracellular and cytoplasmic regions [; Brown LS, Dioumaev AK, Needleman R, Lanyi JK. Biochemistry. 1998;37:3982. doi: 10.1021/bi9728396. reprotonation of the Schiff base in NpSRII will be, of necessity, from the extracellular rather than the cytoplasmic side. [DOI] [PubMed] [Google Scholar]
- 23.Yao VJ, Spudich JL. Proc Natl Acad Sci USA. 1992;89:11915. doi: 10.1073/pnas.89.24.11915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang W, Brooun A, Mueller MM, Alam M. Proc Natl Acad Sci USA. 1996;93:8230. doi: 10.1073/pnas.93.16.8230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang XN, Zhu J, Spudich JL. Proc Natl Acad Sci USA. 1999;96:19722. doi: 10.1073/pnas.96.3.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Subramaniam S, Henderson R. Nature. 2000;406:653. doi: 10.1038/35020614. [DOI] [PubMed] [Google Scholar]
- 27.Landau EM, Rosenbusch JP. Proc Natl Acad Sci USA. 1996;93:14532. doi: 10.1073/pnas.93.25.14532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Otwinowski Z. In: Data Collection and Processing. Sawyer L, Isaacs N, Bailey S, editors. SERC Daresbury Laboratory; Warrington, UK: 1993. pp. 56–62. [Google Scholar]
- 29.Brunger AT. X-PLOR, Version 3.1: A System for X-Ray Crystallography and NMR. Yale Univ. Press; New Haven, CT: 1992. [Google Scholar]
- 30.Brunger AT, et al. Acta Crystallogr D. 1998;54:905. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 31.Sack JS. J Mol Graphics. 1988;6:224. [Google Scholar]
- 32.Luecke H, Richter HT, Lanyi JK. Science. 1998;280:1934. doi: 10.1126/science.280.5371.1934. [DOI] [PubMed] [Google Scholar]
- 33.Guex N, Peitsch MC. Electrophoresis. 1997;18:2714. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]


