Abstract
Glucosinolates are amino acids derived secondary metabolites, invariably present in Brassicales, which have huge health and agricultural benefits. Sulphoraphane, the breakdown product of glucosinolate glucoraphanin is known to posses anti-cancer properties. AOP (2-oxoglutarate-dependent dioxygenases) or GSL-ALK enzyme catalyzes the conversion of desirable glucoraphanin to deleterious gluconapin and progoitrin, which are present in very high amounts in most of the cultivable Brassica species including Brassica juncea. In this study we showed that B. juncea encodes four functional homologs of GSL-ALK gene and constitutive silencing of GSL-ALK homologs resulted in accumulation of glucoraphanin up to 43.11 μmoles g−1 DW in the seeds with a concomitant reduction in the anti-nutritional glucosinolates. Glucoraphanin content was found remarkably high in leaves as well as sprouts of the transgenic lines. Transcript quantification of high glucoraphanin lines confirmed significant down-regulation of GSL-ALK homologs. Growth and other seed quality parameters of the transgenic lines did not show drastic difference, compared to the untransformed control. High glucoraphanin lines also showed higher resistance towards stem rot pathogen Sclerotinia sclerotiorum. Our results suggest that metabolic engineering of GSL-ALK has huge potential for enriching glucoraphanin content, and improve the oil quality and vegetable value of Brassica crops.
Glucosinolates (GSL) are nitrogen and sulfur rich secondary metabolites mostly present in the order Brassicales. Upon tissue damage, these compounds are hydrolysed by β-thioglucoside glucohydrolases called myriosinases into various biologically active compounds like isothiocyanates (ITC) and nitriles1. Inherent as defense compounds, glucosinolates protect Brassicaceae plants against a wide range of herbivores, insects and pathogenic invaders2. Glucosinolate breakdown products also impart aroma and flavour to Brassicales which finds its utilization in culinary purposes from time immemorial. Sulphoraphane (4MSOB-ITC), the isothiocyanate derived from 4-methylsulfinylalkyl glucosinolate (glucoraphanin, GRA) is known to induce phase-II detoxification enzymes during tumour progression and thus act as anti-carcinogenic agents3. However degradation products like oxazolidine-2-thione derived from 2-hydroxy-3-butenyl (progoitrin, PRO) glucosinolate have goitrogenic effect on livestock4,5.
Glucoraphanin is the most widely investigated glucosinolate in the area of health benefits. Healing properties of sulphoraphane, in breast, cervical, prostrate, colon and stomach cancer are well established. Studies show that diet rich in sulphoraphane can fight against Helicobacter pylori causing stomach ulcers. Sulphoraphane can also protect against cystic fibrosis, aging, rhinitis, arthritis, asthma and other lung disorders. Hence regular consumption of cruciferous vegetables is highly recommended3,6,7,8,9,10. Among the cruciferous vegetables broccoli contains highest amount of glucoraphanin. Other members of Brassica oleracea like chinese kale, cabbage and brussels sprout also possess significant amounts of glucoraphanin. However, numerous Brassica cultivars grown for vegetable or oil purpose contain less or negligible amount of glucoraphanin. B. juncea is an important oilseed crop cultivated worldwide in central and south Asia, Europe and North America, northern Africa and China. Even though cultivated mainly as an oilseed, leaves of the young plants are consumed as vegetable as well. In general, 3-butenyl (gluconapin, GNA), 2-propenyl (sinigrin, SIN) and 4-pentenyl (glucobrassicin, GBN) glucosinolates are the major aliphatic glucosinolates present in B. juncea11. The presence of high amounts of these glucosinolates in B. juncea is anti-nutritional in nature12. Hence metabolic engineering of B. juncea for enrichment of desirable glucosinolate glucoraphanin and reducing the anti-nutritional glucosinolates seems highly essential to improve the food and feed value of this crop.
Glucosinolate biosynthesis occurs from amino acid precursors through three major processes viz., chain elongation, core structure formation and side chain modifications (Fig. 1). Glucosinolates are classified as aliphatic, aromatic and indolic glucosinolates based on their precursor amino acids1,13. Aliphatic glucosinolates can be further classified into 3C, 4C and 5C based on their side chain length, or methylthioalkyl, methylsulfinyl alkyl, alkenyl and hydroxyalkenyl based on the side chain structure14. The vast glucosinolates diversity is attributed to the side chain elongation and side chain modification reactions. GSL-ELONG gene locus encoding methylthioalkyl malate synthases (MAM) control the initial chain elongation reactions15. A subclade of flavin-monooxygenase (FMOGS-OX1-5) catalyzes the conversion of methylthioalkyl glucosinolates into methylsulfinylalkyl glucosinolates16,17. Two α-ketoglutarate-dependent dioxygenases AOP2 and AOP3 are demonstrated to control the conversion of methylsulfinylalkyl to alkenyl- and hydroxyalkenyl glucosinolates, respectively18,19.
Figure 1. General scheme for aliphatic glucosinolate biosynthesis.
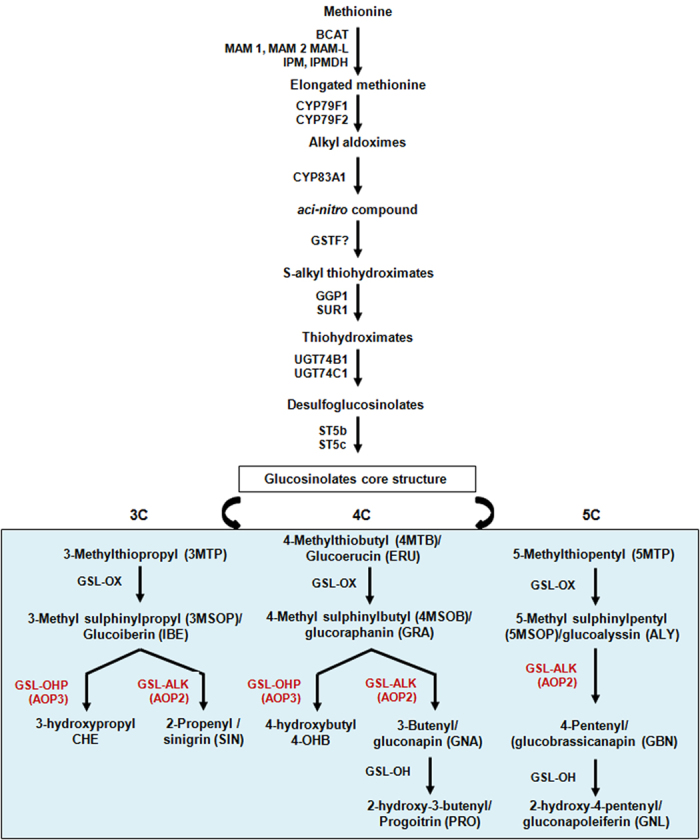
Precursor amino acid (here methionine) undergoes a series of reactions to form the glucosinolate core structure. The core structure further undergoes modifications of its side chain generating diverse glucosinolate structures. The side chain modification step is represented in box. Enzymes catalyzing specific steps are also indicated. Chemical name, trivial name and abbreviations of individual glucosinolates are given in parenthesis. C3, C4 and C5 represent glucosinolates with 3, 4 and 5 carbon chain respectively in the core structure.
Accumulation of glucoraphanin is governed by the GSL-AOP locus which contains the GSL-ALK (AOP2) and GSL-OHP (AOP3) locus. Non functional gene product of this loci results in the accumulation of glucoraphanin20,21. In Arabidopsis thaliana ecotype Columbia, AOP2 gene was shown to be marginally expressed which results in accumulation of glucoraphanin in this accession22. In broccoli, non functional GSL-ALK homolog has been identified which is associated with high glucoraphanin accumulation. In B. rapa three AOP2 genes identified were found to be functional which explains the reason for the absence of glucoraphanin in B. rapa and other related Brassica species14,23,24. The high amount of gluconapin and absence of glucoraphanin in B. juncea necessitates the characterization and manipulation of these gene homologs in B. juncea. Since B. juncea is an allotetraploid, where gene duplication and redundancy make genetic manipulation, the only alternative to achieve high glucoraphanin accumulation is through silencing of the GSL-ALK gene family. Hence the study is undertaken to isolate all homologs of the GSL-ALK gene from B. juncea and its subsequent utilization to develop high glucoraphanin lines which can improve the food and feed value of this oilseed crop as a potential source of anti-cancer compounds.
Results
Isolation and sequence analysis of GSL-ALK gene homologs from B. juncea
The full-length genomic sequences of the GSL-ALK genes were isolated from B. juncea high glucosinolate cultivar Varuna using degenerate primers in our previous study25. Based on the genomic sequences, primers were designed and full-length cDNA sequences were isolated. Four full-length coding sequences were identified from B. juncea. Contributor sub-genomes of the four homologs were ascertained by simultaneous isolation and sequence comparison of the GSL-ALK orthologs from both B. rapa (AA) and B. nigra (BB), the progenitor sub-genomes of B. juncea (AABB). The homologs were named as BjuA.GSL-ALK-1, BjuA.GSL-ALK-2, BjuB.GSL-ALK-1 and BjuB.GSL-ALK-2 (A stands for A-sub-genome and B-stands for B-sub-genome). Comparison of coding sequences with the reported genomic sequences of GSL-ALK gene homologs25 showed the presence of three exons intervened by two introns (Supplementary Table S1). The coding sequences of BjuA.GSL-ALK-1 and BjuB.GSL-ALK-1 genes were found to be 1323 bp whereas the CDS of BjuA.GSL-ALK-2 and BjuB.GSL-ALK-2 was 1320 bp long with a deletion of single codon. The coding sequences of BjuGSL-ALK genes shared around 75% sequence identity with A. thaliana AOP2 (Cvi) gene and >90% identity among themselves (Supplementary Table S2). The deduced amino acid sequence also showed an identity percentage of 87–99% among the four B. juncea GSL-ALK homologs (Supplementary Table S3, Supplementary Fig. S1). The A-sub-genome specific homologs were more similar to each other and similarly the B-sub-genome homologs.
To illustrate the evolutionary relationships among the GSL-ALK gene homologs, phylogenetic tree was constructed using AOP sequences from A. thaliana and Brassica species (Fig. 2). On the maximum likelihood tree, the AOP1, AOP2 and AOP3 genes resolved into two distinct clusters, in which AOP2 and AOP3 were clustered together. The four BjuGSL-ALK genes grouped nicely in the AOP2 clade along with other Brassica homologs. Within the AOP2 clade, the two A-sub-genome specific homologs were grouped together with that of B. rapa (Bra034180) and B. oleracea (AY044424.1, AY044425.1) sequences, whereas the B-sub-genome specific homologs were grouped separately into another subgroup with high bootstrap support. The phylogenetic tree clearly demonstrated that all four GSL-ALK genes isolated from B. juncea were AOP2 type and were evolutionary conserved.
Figure 2. Phylogenetic relationship of GSL-ALK (AOP) genes.
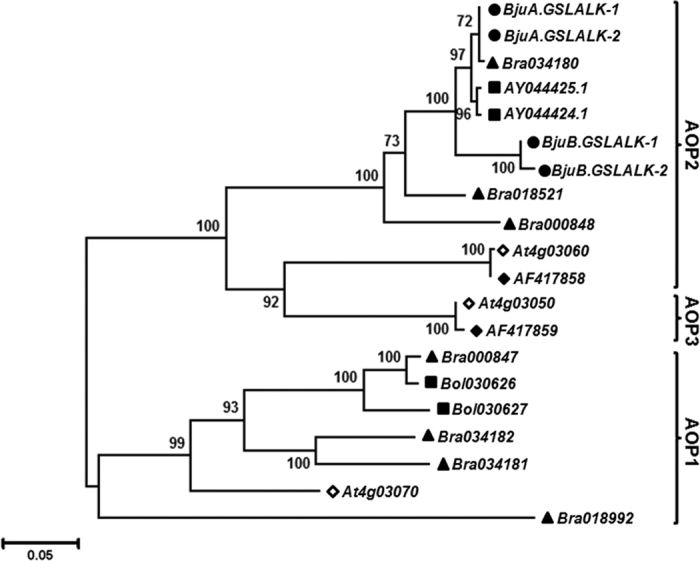
Phylogenetic analysis of GSL-ALK cDNA with the other AOP sequences from Arabidopsis and related Brassica species was performed. The AOP like genes from A. thaliana ecotype Columbia (◇), A. thaliana ecotype Cvi (◆), B. rapa (▴), B. oleraceae (▄) and B. juncea (●) were used to construct the tree. The evolutionary history was inferred by using the Maximum Likelihood method in MEGA545. The percentage of trees in which the associated taxa clustered together is shown next to the branches. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site.
The divergence time analysis of AOP genes based on the synonymous base substitution (Ks) values between Brassica and Arabidopsis (Cvi) genes indicated that both BjuGSL-ALK A- and B- sub-genome specific genes diverged somewhere around 14.8–16.0 MYA (Supplementary Table S4), similar to that observed for B. rapa (Bra034180, Bra018521) and B. olereaceae (AY044424.1, AY044425.1) GSL-ALK gene orthologs. Thus, the multiple BjuGSL-ALK genes seem to have evolved as a result of whole genome triplication events which is hypothesized to have occurred somewhere around 13–17 MYA in ancestral Brassica prototype26 followed by an allopolyploidization of two simpler Brassica genomes.
Knock-down of GSL-ALK gene provides enrichment of glucoraphanin in transgenic B. juncea
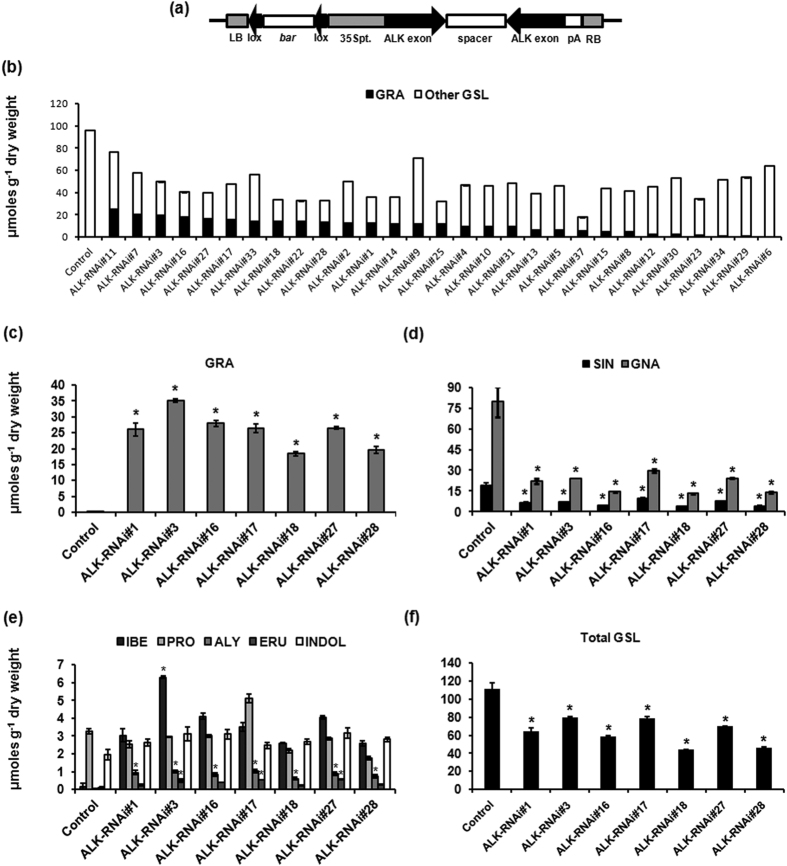
For knock-down of GSL-ALK gene family, intron spliced hairpin RNAi (ihpRNAi) construct (ALK-RNAi) was developed, which consisted of a conserved 325 bp region from third exon of GSL-ALK gene homologs. The conserved knock-down cassette was driven by the CaMV35S promoter to achieve constitutive down regulation of the four GSL-ALK gene homologs in all the tissue types (Fig. 3a). The transformation cassette was introduced into high glucosinolate Indian cultivar of B. juncea (cv. Varuna having a total seed glucosinolate content of ca. 107.52 μmoles g−1 DW) through Agrobacterium mediated genetic transformation. A total of 29 primary transgenic (T0) lines were generated. The transgenic lines were maintained by selfing under the containment field conditions and analyzed for glucosinolate phenotype.
Figure 3. Development of ALK-RNAi transgenic lines of B. juncea.
(a) The T-DNA map of hairpin RNAi cassette of GSL-ALK gene. A 325 bp coding region of the GSL-ALK gene was cloned in sense and antisense orientations on both sides of a spacer intron, to facilitate the formation of a hairpin structure. The plant selection marker gene bar was cloned within the lox P site of bacterial cre-lox P recombination system. (b) Content of glucoraphanin and the other glucosinolates (GSL) of each line in T1 generation seeds, represented as single data point. (c) Glucoraphanin (GRA) content in four day old sprouts of ALK-RNAi lines of B. juncea. (d) The major glucosinolates sinigrin (SIN) and gluconapin (GNA). (e) Other glucosinolates, glucoiberin (IBE), progoitrin (PRO), glucoalyssin (ALY), glucoerucin (ERU) and indolic glucosinolates. (f) Total glucosinolates. Glucosinolate content of ALK-RNAi lines was estimated by HPLC (in μmoles g−1 DW). Three independent experiments were carried out and the average values are represented along with their standard errors. Asterisks on the top of bars indicate significant differences in glucosinolate content compared to the control, wild-type Varuna at P < 0.05, in Fishers LSD test determined by ANOVA.
Transgenic lines developed were analyzed by HPLC to estimate content of glucoraphanin and other glucosinolates in T1 generation. The total seed glucosinolate content in the ALK-RNAi transgenic lines ranged from 17.65–76.30 μmoles g−1 DW and the glucoraphanin content ranged from 0–24.71 μmoles g−1 DW suggesting variable degree of gene silencing efficiency (Fig. 3b; Supplementary Table S5). More than 93% of the transgenic lines showed enhanced accumulation of glucoraphanin (GRA) compared to that of wild-type Varuna plants. Along with the accumulation of glucoraphanin, a concomitant reduction in major glucosinolates was observed, resulting in overall reduction in total glucosinolate content. For instance, gluconapin (GNA), the most predominant and undesirable aliphatic glucosinolate was reduced to levels as low as 7.98 μmoles g−1 DW in the transgenic lines compared to 93.55 μmoles g−1 DW of wild-type level. Similarly sinigrin (SIN), the second major seed glucosinolate was also reduced to 1.94 μmoles g−1 DW in the transgenic lines whereas wild-type plants had ca. 21.08 μmoles g−1 DW (Supplementary Table S5). 4-Methylthiobutyl (Glucoerucin, ERU), the precursor glucosinolate for glucoraphanin, which is otherwise marginally present in wild-type plants was increased up to 2.84 μmoles g−1 DW in the transgenic lines. Content of 3C glucosinolate, 3-Methylsulfinylpropyl (glucoiberin, IBE) and 5C glucosinolate, 5-Methylsulfinyl (glucoalyssin, ALY) were found to be enhanced in most of the transgenic lines (Supplementary Table S5). There was no significant difference in the content of other glucosinolates in the seeds.
Sprouts are shown to accumulate highest levels of glucoraphanin3. Seven representative transgenic lines having seed glucoraphanin content >12.0 μmoles g−1 DW were selected and glucosinolate content in the four day old sprouts were analyzed using HPLC. Glucoraphanin content in the sprouts ranged from 18.39–35.13 μmoles g−1 DW against 0.28 μmoles g−1 DW of the wild-type, thereby showing more than 100 fold increases in the transgenic lines (Fig. 3c). Significant reduction in the major aliphatic glucosinolates like gluconapin and sinigrin were observed in the sprouts (Fig. 3d). Among other glucosinolates, glucoerucin and glucoalyssin were found increased significantly in the sprouts. The minor glucosinolate glucoiberin also showed significant enhancement in few transgenic lines (Fig. 3e). Progoitrin (PRO), an anti-nutritional glucosinolate found in remarkable amounts in sprouts remained unaltered in the transgenic lines (Fig. 3e). Overall, the total glucosinolates content showed a decline to ca. 58% in the sprouts of transgenic lines compared to the wild-type plants (Fig. 3f) suggesting key involvement of GSL-ALK locus towards controlling total glucosinolate content in B. juncea. Further, indolic glucosinolate pool remained almost unaltered in the transgenic lines (Fig. 3e).
Molecular analysis of ALK-RNAi transgenic lines showed reduced levels of GSL-ALK gene transcripts
Molecular characterization of the seven transgenic lines having highest levels of glucoraphanin content was carried out. T-DNA insertion number was ascertained using Southern blot and segregation analysis. Southern blot analysis of the T0 transgenic events showed stable integration of T-DNA cassette mostly in single copy (Fig. 4a). Chi-square analysis of T1 lines segregating for Basta resistance also confirmed single copy insertion of T-DNA cassette in most of the transgenic lines (Supplementary Table S6). However there seems no direct co-relation between the gene copy number and high glucoraphanin phenotype. For example, the high glucoraphanin accumulating line ALK-RNAi#3 was found to have T-DNA integration in more than one copy on southern blot. The variation in glucoraphanin content observed among the selected transgenic lines could be attributed to the position-effect mediated phenomenon.
Figure 4. Molecular characterization of ALK-RNAi transgenic lines.
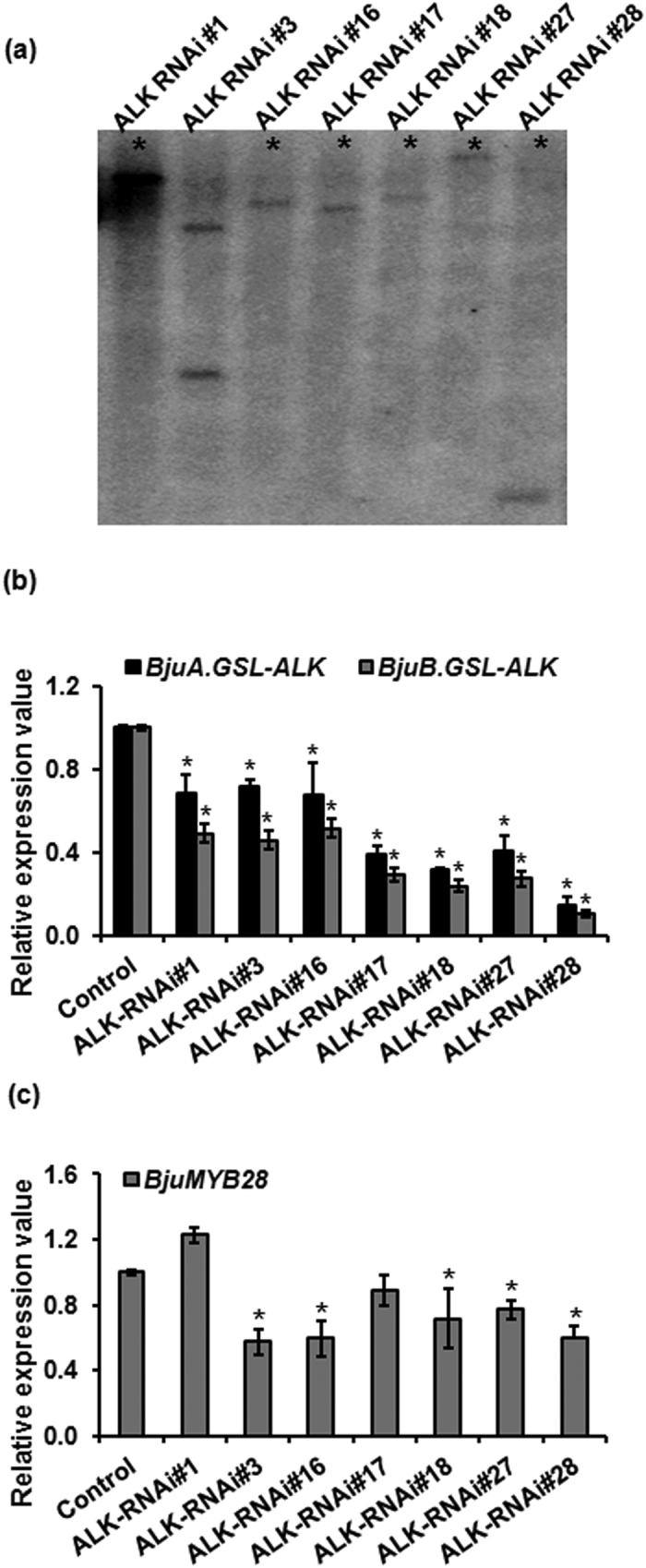
(a) Southern blot analysis of selected ALK-RNAi lines. Asterisks represent single copy insertion of the T-DNA cassette. (b) Steady state mRNA levels of ALK-RNAi lines. qRT-PCR analysis of GSL-ALK genes was performed and the transcript accumulation was measured with reference to control (wild-type Varuna) set at 1. (c) Gene expression analysis of BjuMYB28 genes in ALK-RNAi transgenic lines. Transcript level of BjuMYB28 genes was estimated using qRT-PCR with primers conserved to BjuMYB28 homologs. Asterisks on the top of bars indicate significant differences in gene expression compared to the wild-type control Varuna (set at 1) at P < 0.05, in Fishers LSD test determined by ANOVA.
The gene knock-down efficiency of ALK-RNAi construct was determined at transcriptional level. Steady state transcript accumulation, using real-time qRT-PCR analysis was carried out in the T1, Basta resistant progeny of each transgenic event. Primers, specific to GSL-ALK genes corresponding to the A- and B-sub-genome were used for transcript profiling. A significant reduction in the transcript levels of GSL-ALK gene homologs was observed compared to the control plants in all transgenic lines with both set of primers (Fig. 4b). The level of expression was found to be down regulated to variable extents, with a lowest value of at least five fold lower than that of the wild-type in ALK-RNAi lines. However, no direct relationship between the glucoraphanin content and transcript accumulation was observed. Since the total aliphatic glucosinolate content was also found to be lowered in the transgenic lines, expression of MYB28 transcription factors which is the major positive regulator of aliphatic glucosinolate biosynthesis in B. juncea27 were also assayed in the transgenic lines. Interestingly, expression of MYB28 homologs was also found to be altered across the ALK-RNAi transgenic lines with a significant down regulation observed in most of the transgenic lines (Fig. 4c).
ALK-RNAi transgenic lines were stable for high glucoraphanin content and other seed quality traits in advance generations
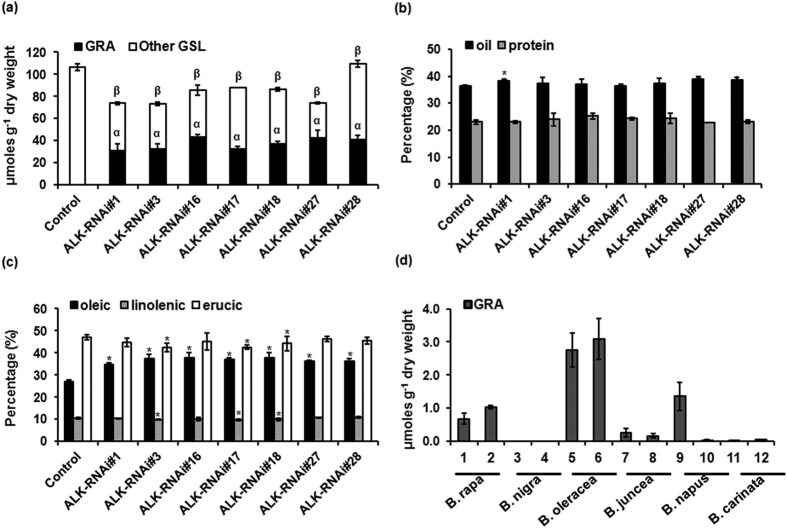
Glucosinolate content were estimated in the T2 seeds of ALK-RNAi lines to check the stability of transgenic lines using HPLC. The glucoraphanin content was found as high as 43.11 μmoles g−1 DW in the seeds. Glucosinolate profile followed a similar trend as observed for the T1 seeds. However total glucosinolate content showed an enhancement due to the enrichment of glucoraphanin in the T2 seeds compared to T1 seeds (Fig. 5a). RNAi induced gene silencing often meet with off- target effects in crop plants. Since B. juncea is cultivated mainly as an oilseed crop, analysis of other seed quality parameters is also imperative. Total oil content, protein content and fatty acid compositions were analyzed in T2 seeds of the ALK-RNAi lines by NIRS. Total oil content of transgenic lines ranged from 36.27–39.02% compared to 36.46% in the wild-type, indicating lesser significant difference (Fig. 5b). Protein content is another important trait as far as B. juncea seed is concerned, as it is highly rich in protein and hence can be a substitute for soybean oil cake in the international market as cattle feed. When the total protein content of the transgenic lines was analyzed, there was no significant difference compared to the wild-type (Fig. 5b). Fatty acid compositions are important determinants of Brassica oil quality. Fatty acid compositions of the transgenic lines were also analyzed. Oleic acid (C18:1) which is known to be beneficial to humans was found to be significantly elevated in the ALK-RNAi transgenic lines. Oleic acid content of transgenic lines ranged from 34.69%–37.92% compared to 27.04% in the wild-type. Contents of other fatty acids namely, linolenic acid (C18:3) and erucic acid (C22:1) also showed significant enhancement in most of the transgenic lines compared to the wild-type which needs further investigation (Fig. 5c). The tested transgenic lines showed neither developmental nor phenotypic aberrations compared to the wild-type plants when grown under containment field conditions.
Figure 5. Characterization of ALK-RNAi transgenic lines for stability and seed quality traits.
(a) Content of glucoraphanin and other glucosinolates in the seeds of ALK-RNAi lines in T2 generation as estimated by HPLC. (b) Total oil and protein content (c) Fatty acid composition (oleic, linolenic and erucic acid as estimated using Near Infra-red Reflectance Spectroscopy (NIRS). Average values of three independent measurements (n = 9) are represented along with standard deviation. Transformation host Varuna was used as control. Asterisk indicate significant difference at P < 0.001 in Fishers LSD as determined by ANOVA. (d) Screening of Brassica germplasm for glucoraphanin content. Glucoraphanin content (μmoles g−1 DW) in four day old sprouts of two cultivars each of six cultivated Brassica species was estimated by HPLC. (1. YID1, 2. Pusa Gold, 3. Sangam, 4. IC-257, 5. Pusa Sharad, 6. Palam Samridhi, 7. Heera, 8. Varuna, 9. HNS-9010, 10. NU-98, 11. BEC-218 and 12. CAR- 52.
Further, we screened few Brassica germplasm to assess and compare the glucoraphanin content between the ALK-RNAi transgenic lines developed and commonly cultivated Brassica germplasm to check the superiority of these transgenic lines. Two representative cultivars from each of Brassica species commonly cultivated in India were screened for glucoraphanin content in 4-day old sprouts. There was only trace amount of glucoraphanin observed among B. nigra, B. carinata and B. juncea cultivars whereas B. oleraceae, B. napus and B. rapa showed somewhat higher amount of glucoraphanin content. B. oleracea cultivars Pusa Sharad (cauliflower) and Palam Samridhi (broccoli) showed maximum glucoraphanin content of around 2.75 and 3.08 μmoles g−1 DW respectively (Fig. 5d). Hence it can be assured that the ALK-RNAi transgenic lines developed in the current study can be a superior source for high glucoraphanin content up to 43.11 μmoles g−1 DW in the seeds. The sprouts are expected to have even higher levels of glucoraphanin.
Susceptibility of ALK-RNAi lines towards Sclerotinia sclerotiorum infection
Glucosinolates are very important for plant fitness. When grown in field, the plant can undergo various biotic stresses in which fungal pathogens are one of the major challenges. The response of ALK-RNAi lines towards the stem rot pathogen Sclerotinia sclerotiorum was conducted since, not only the total glucosinolate content, but its profile also has altered in the transgenic lines. Four transgenic lines showing high glucoraphanin content were taken for analysis. The glucosinolate content and total phenol content of the leaf tissue were analyzed, as composition of both can alter pest status. Glucoraphanin content in leaves ranged from 6.27–22.11 μmoles g−1 DW of the leaf tissue (Table 1). Among other glucosinolates, the 5C glucosinolate, glucoalyssin showed a significant increase. There was no significant difference in the total phenol content of the transgenic lines compared to the wild-type plants (Table 1). Susceptibility of ALK-RNAi lines towards S. sclerotiorum was tested using disc assays using methanolic extracts of leaves. Interestingly, there was significant decrease in the mycelial diameter in the transgenic lines compared to the wild-type plants (Table 1; Supplementary Fig. S2). When compared to the mock control, effect of total glucosinolate in fungal growth retardation was also very evident. Effect of individual glucosinolates on fungal growth (diameter of mycelium) was predicted by linear regression analysis. Striking correlation was observed between the glucosinolate content and S. sclerotiorum growth. Mycelial diameter decreased significantly (p ≤ 0.05) with increase in content of glucoraphanin and glucoalyssin (Table 1). Mycelial diameter decreased by 0.061 cm and 0.378 cm with per unit increase in of glucoraphanin and glucoalyssin respectively. The content of all these glucosinolates were found to be accumulated in the transgenic lines at higher concentrations compared to the wild-type plants.
Table 1. Glucosinolate (GSL in μmoles g−1 DW), total phenol content (TPC in GAE 100 mg−1 tissue) and mycelial growth (cm) of S. sclerotiorum in methanolic extracts of ALK-RNAi lines.
| Line | IBE | GRA | SIN | ALY | GNA | GBN | Indolic GSL | Total GSL | TPC | Mycelial Diameter |
|---|---|---|---|---|---|---|---|---|---|---|
| Mock | — | — | — | — | — | — | — | — | — | 9.00 ± 0.24 |
| Control | 0.06 ± 0.02 | 0.07 ± 0.05 | 8.67 ± 0.18 | 0.10 ± 0.01 | 57.12 ± 1.82 | 3.47 ± 0.02 | 1.02 ± 0.31 | 70.76 ± 2.11 | 0.93 ± 0.09 | 6.32 ± 0.25a |
| ALK-RNAi#1 | 0.44 ± 0.02 | 6.27 ± 0.43* | 3.06 ± 0.09 | 1.62 ± 0.09 | 16.02 ± 1.64 | 0.17 ± 0.10 | 0.47 ± 0.13* | 28.26 ± 1.81* | 0.97 ± 0.08 | 5.61 ± 0.13ab |
| ALK-RNAi#3 | 1.49 ± 0.20* | 16.08 ± 1.96* | 1.59 ± 0.15 | 2.66 ± 0.19* | 6.74 ± 0.55 | 0.63 ± 0.39 | 1.08 ± 0.34 | 30.53 ± 3.66* | 0.90 ± 0.16 | 5.72 ± 0.42ab |
| ALK-RNAi#16 | 2.01 ± 0.16* | 18.22 ± 1.78* | 2.04 ± 0.45 | 3.09 ± 0.19* | 6.23 ± 0.35 | 0.13 ± 0.38 | 1.19 ± 0.06 | 32.97 ± 2.46* | 0.70 ± 0.01 | 5.33 ± 0.34ab |
| ALK-RNAi#27 | 2.31 ± 0.07* | 22.11 ± 0.31* | 1.68 ± 0.12 | 4.45 ± 0.15* | 6.85 ± 1.52 | 0.21 ± 0.26 | 0.78 ± 0.40 | 38.54 ± 1.26* | 0.73 ± 0.05 | 5.43 ± 0.31ab |
Concentration of individual glucosinolates and total phenol content in the leaves of ALK-RNAi lines (T1) used for fungal infectivity assay is given. Asterisk indicates significant increase in glucosinolate compared to the wild-type control. Lettera on top indicate significant difference in mycelial diameter compared to mock (methanol) and lettersab represent significant difference from both mock and wild-type control. Statistical analysis was conducted using one-way ANOVA following Fishers LSD test of significance at P < 0.05.
Discussion
Brassicaceae is a large family of agriculturally important plants which consists of mustards, cabbages, broccoli, turnips, radish, kale, cresses, and their many relatives. Plants of this family are well known for the presence of the secondary metabolites called glucosinolates. Genetic analysis in Arabidopsis and Brassica crops has shown that QTLs like GS-ELONG, GS-OX, GS-AOP and GS-OH are important in determining glucosinolate content and its component variability18,19,20,28. GSL-ELONG locus controls chain length and is composed of MAM1/MAM2 and MAM3 genes. FMOGS-OX1-5is localized in GS-OX QTL and converts methylthioalkyl glucosinolates to the corresponding methylsulfinylalkyl glucosinolates17. QTL GS-AOP is composed of GS-ALK and GS-OH loci. AOP2 is localized within GS-ALK whereas AOP3 is localized within GS-OH21. These two loci are tightly linked and are responsible for converting methylsulfinylalkyl glucosinolates into alkenyl or hydroxyalkyl glucosinolates16,20. The presence or absence of either of the loci determines the type of glucosinolate produced in a particular species.
Glucoraphanin is the widely studied glucosinolate which has many health protective properties including cancer prevention. The occurrence of glucoraphanin is associated with GSL-ALK gene expression as evident by previous studies in Arabidopsis, B. oleracea, Chinese kale etc.14,22,23,24,29. However there is no Indian type Brassica accession known for high glucoraphanin content. In B. juncea the high level of accumulation of 4C glucosinolate, gluconapin which is the reaction product of glucoraphanin, indicate that the GSL-ALK/AOP2 gene is fully functional in B. juncea. Enhancement of glucoraphanin through conventional breeding seems quite challenging as the conventional breeding strategies are not simpler in polyploid crops like B. juncea and B. napus where gene multiplicity and redundancy create complex genetic interactions. The presence of very high content of gluconapin also make the Indian B. juncea a promising candidate for achieving high glucoraphanin content through the silencing of GSL-ALK gene family. Hence, in the current study we isolated the GSL-ALK gene homologs from B. juncea and utilized it for the development of high glucoraphanin lines.
B. juncea is formed by the hybridization between the diploid species B. rapa and B. nigra which diverged 13–17 mya from the ancestral Arabidopsis lineage26. Whole genome triplication events followed by genome shrinkage shaped up the genome architecture of today’s Brassica species. Hence multiple copies of a given gene are expected which we could observe in the current study. Four GSL-ALK homologs, two of which representing A-sub-genome and other two representing B-sub-genome based on their sequence identity were isolated. Sequence analysis and phylogeny clearly showed that the B. juncea, GSL-ALK genes are the true orthologs of AtAOP2 genes, which are required for the alkeylation of parent methylsulfinyl glucosinolates. In B. oleracea and B. rapa three copies of AOP2 gene were identified29 whereas B. napus have up to nine members of AOP- like gene family30. We could isolate only two expressed members corresponding to A- sub-genome from B. juncea. The coding sequences of the four GSL-ALK homologs were found highly similar which shows that these genes are evolutionarily conserved. Similar level of conservation we could also observe while working with other glucosinolate genes like MYB28, CYP83A1 and GSL-ELONG27,31. The coding sequences of B. juncea GSL-ALK genes encodes full-length protein, upon translation which further supports the assumption that these genes might be fully functional in B. juncea. This can further explain the presence of high amount of gluconapin and trace amount of glucoraphanin in B. juncea as well as the two progenitor genomes namely, B. rapa and B. nigra.
Knock-down of GSL-ALK gene have been shown to enhance glucoraphanin content in B. napus and Chinese kale30,32. RNAi based silencing of the GSL-ALK gene family in B. napus did not show significant alteration in the total glucosinolates content in the transgenic lines. However, the detrimental glucosinolate progoitrin was drastically reduced and glucoraphanin was increased to a high concentration in the seeds of F1 progeny of a cross between B. napus ALK-RNAi line and a double haploid line of high glucosinolate containing rapeseed30. The achievement of high glucoraphanin in F1 generation of B. napus makes the approach labour-intensive, time-consuming, and more importantly event-dependent, following a complex genetic control of glucosinolate trait in advance segregating generations. Moreover, gluconapin was found decreased only when the gene silencing was very strong. Similarly, in antisense mediated down regulation of GSL-ALK gene in Chinese kale, even though glucoraphanin content was increased, gluconapin content did not declin and surprisingly showed an enhancement32. But in our current study along with enhancement of glucoraphanin, we could observe a drastic decline in the concentrations of gluconapin and sinigrin as well as the total glucosinolate content in the ALK-RNAi lines in the initial T1 generation itself, which was also stable in advance T2 generation. Furthermore, the level of glucoraphanin accumulated in the selected B. juncea ALK-RNAi transgenic lines was even higher than the content of glucoraphanin reported in florets of Benefort’e® broccoli33. Other 4C glucosinolate, glucoerucin which comes earlier in the pathway was found to be enhanced in the transgenic lines. The accumulation of 3C and 5C alkyl glucosinolate substrates namely, glucoiberin and glucoalyssin respectively in the ALK-RNAi lines further confirm that the RNAi construct developed in the study could effectively silence the target gene in B. juncea. Our study hence clearly demonstrates that RNAi based silencing of GSL-ALK gene homologs not only produces high amount of glucoraphanin in B. juncea, but also reduces un-desirable glucosinolates including gluconapin and sinigrin to a significantly lower level. The high glucoraphanin trait was found stable in the subsequent T2 generation with even higher levels of glucoraphanin accumulation.
Stable integration of T-DNA cassette is necessary to achieve consistent gene silencing phenomena in transgenic plants. Measurement of steady state mRNA levels of GSL-ALK gene in the transgenic lines showed significant down regulation of these transcripts reflecting the effectiveness of the RNAi construct against members of GSL-ALK gene family. Interestingly, in our study we observed a significant decline in total glucosinolates content also, suggesting that GSL-ALK gene might exert a regulatory role towards glucosinolate biosynthesis in B. juncea. It has been proposed that AOP2 gene could be involved in a positive feedback regulation of glucosinolate biosynthesis34,35. Hence we measured the transcript levels of MYB28 genes, the major positive transcriptional regulators of aliphatic glucosinolates and found that there was a significant reduction the transcript levels in most of the ALK-RNAi transgenic lines. Very recently it has been showed that, this feedback mechanism exists and is dependent on the glucosinolate transcriptional regulators MYB28 and MYB29 in A. thaliana36, which is somewhat similar to that observed in our study. However, the level of down regulation of BjuMYB28 homologs was not very huge which could be due to the multiplicity of these genes. The highly complex glucosinolate biosynthesis in the polyploid B. juncea could be controlled by both MYB-dependent and independent regulatory networks, which needs detailed investigation.
Glucosinolates are primarily defense metabolites of the Brassicaceae plants. Altering the glucosinolate content and profile can alter the plant defense mechanism. In general, indolic glucosinolates are regarded as the critical determinants for pest and disease resistance whereas aliphatic glucosinolates play major role for the overall fitness of the plant37. However, contribution of individual glucosinolates towards particular pest and pathogen is not discovered in detail. In the current study, even though the indolic glucosinolates remained almost unaltered, the profile and content of aliphatic glucosinolates was altered. In Arabidopsis it has been shown that sulphoraphane, is an important defense arsenal against many pathogens in vitro38. Susceptibility of glucosinolate biosynthesis mutant, gsm1-1 which was found to be largely deficient in two of the major antimicrobial compounds, including sulphoraphane was found responsive only to Fusarium oxysporum while showing no change in susceptibility towards other fungi like Alternaria brassicicola, Plectosphaerella cucumerina, Botrytis cinerea, Peronospora parasitica, and the bacteria Erwinia carotovora and Pseudomonas syringae39.
Sclerotinia sclerotiorum (Lib.) de Bary is a necrotrophic plant pathogen causing stem rot in Brassica species mainly in temperate areas of the world40,41. Recently the pathogen has emerged as a major pest in B. juncea in India also. Stem rot caused by S. sclerotiorum leads to severe yield loss in mustards42,43. The growth of S. sclerotiorum was found to be negatively correlated with glucoraphanin and glucoalyssin content. The resistance of these lines towards S. sclerotiorum infection can be attributed to high glucoraphanin and glucoalyssin in the transgenic lines. Studies by Stotz et al.44 suggested that accumulation of long chain glucosinolates is important for defense against S. sclerotiorum even though the role of sulphoraphane glucosinolates was not ruled out. Since indolic glucosinolate pool is not much affected, we do not expect much compromise on the defense response of the B. juncea ALK-RNAi transgenic lines. The susceptibility of the transgenic ALK-RNAi lines developed in the current research, against different pests/pathogens of Brassica species needs to be further studied to evaluate the potential for commercialization of the high glucoraphanin lines of B. juncea.
Overall, the study is an excellent example of translational research in Brassica crops. High glucoraphanin accumulation was achieved in all parts of the plant through constitutive down regulation of GSL-ALK/AOP2 gene. The sprouts can be consumed in salads or leaves can be used for vegetable purpose, which can provide significant levels of the precursor anti-cancer glucosinolate, glucoraphanin to humans. Besides, industrial production of high amounts of glucoraphanin can also be achieved from seeds and sprouts of these high glucoraphanin lines of B. juncea. Hence in the global market, the commercial value of B. juncea can be raised not only as an oilseed but also as a health promoting food crop.
Methods
Plant materials and growth conditions
B. juncea (cv. Varuna) was grown in a growth chamber maintained at 10 h light/14 h dark cycle with a light intensity of 300 μmol m−2 s−1, temperature of 22 °C day/15 °C night, and relative humidity of 70%. B. juncea transgenic plants were selected by spraying herbicide Basta having active ingredient phosphinothricin (Bayer crop sciences, Mumbai, India) and maintained in the containment net-house facility available at NIPGR as per the guidelines of Department of Biotechnology, Government of India. For screening for glucoraphanin content in Indian grown Brassicas, B. rapa (YID1, Pusa Gold), B. nigra (Sangam, IC-257), B. oleracea (Pusa Sharad- cauliflower, Palam Samridhi- broccoli), B. juncea (Heera, Varuna), B. napus (HNS-9010, NU-98) and B. carinata (BEC-218, CAR-52) were used.
Isolation of genomic and cDNA sequences of GSL-ALK homologs from B. juncea
The isolation of full-length genomic DNA sequences of the GSL-ALK genes was earlier reported by Bisht et al.25. The coding sequences of the GSL-ALK genes were amplified based on the primers designed using genomic sequences and PCR amplifications were carried out in B. juncea (cv. Varuna), B. nigra (cv. IC-257) and B. rapa (cv. YID1). RT- PCR products were cloned into pGEM-T Easy cloning vector (Promega, Madison, Wisconsin, USA) and sequenced to check correctness of the sequence. Progenitor genomes of BjuGSL-ALK genes were assigned based on the sequence comparison with the homologs obtained from B. nigra and B. rapa. All sequence analysis was performed using Lasergene core suite (DNASTAR, Madison, Wisconsin, USA). A list of primers used in the current study is summarized in Supplementary Table S7.
Phylogenetic analysis and estimation of divergence time
Coding sequences AOP genes of A. thaliana ecotype Columbia (At4g03070, At4g03050), B. rapa (Bra018992, Bra034180, Bra034181, Bra034182, Bra000847, Bra000848, Bra018521,) B. oleracea (Bol030626, Bol030626) were retrieved from BRAD database (http://brassicadb.org/brad) whereas sequence of B. oleracea (AY044424.1, AY044425.1), A. thaliana ecotype Cape Verdi Islands (Cvi) (AF417859, AF417859) and Columbia (At4g03060) were retrieved from NCBI and TAIR databases. Multiple sequence alignment of the full-length coding sequences was generated using ClustalW and the evolutionary tree was constructed using Maximum Likelihood method in MEGA545.
To determine the divergence time of AOP homologs, coding DNA sequences from Brassica species were aligned using ClustalW. Synonymous (Ks) and non-synonymous (Ka) base substitution were calculated by DnaSP v5 software46. The divergence time of AOP genes was calculated using the equation, divergence time in MYA, T = Ks/(2×[1.5 × 10−8] where 1.5 × 10−8 substitution per site per year is the synonymous mutation rate reported for Brassica genus47.
Generation of ALK-RNAi transformation construct and knock-down lines in B. juncea
Binary vector pPZP200lox:bar having 35Sde-bar-ocspA fragment conferring resistance to herbicide Basta was used for generating transgenic plants. Approximately 325 bp fragment (949 bp downstream of ATG start codon) of BjuGSL-ALK genes was amplified and cloned in both sense and antisense orientation on either side of BjuMYB28 intron to generate an intron spliced hairpin RNAi cassette12. The Exon(sense)-int2-Exon(antisense)-35SpA cassette driven by CaMV35S promoter was cloned into the binary vector pPZP200lox:35Sde-bar-ocspA to create the final transformation cassette. The cassette was mobilized into A. tumefaciens strain GV3101 and later into B. juncea cv. Varuna using the protocol described earlier13. Primers used for the construct design are provided in Supplementary Table S7.
Glucosinolate analysis using HPLC
The seeds, sprouts and leaves of transgenic lines were analyzed for their individual glucosinolate contents using HPLC as per the protocols described earlier12. p-hydroxylbenzyl glucosinolate (Sinalbin), purified from Sinapis alba (kindly provided by Dr. Michael Reichelt, Max Plank Institute of Chemical Ecology, Jena Germany) was used as the internal standard. Concentrations of individual glucosinolate were calculated relative to the area of the internal standard peak applying their relative response factors and expressed in μmoles g−1 dry weight (DW) of the tissue. Data from three independent experiments were averaged and standard deviation was calculated. Test of significance were carried out using one way ANOVA following Fishers LSD.
Estimation of total protein, oil content and fatty acid composition
Total protein, oil content and fatty acid (oleic acid, linoleic acid, erucic acid) composition of transgenic seeds were determined using Near Infrared Reflectance Spectroscopy48 (NIRS, DS-2500, FOSS, Denmark) as per manufacturer’s instructions. Briefly, intact B. juncea seed samples were placed in the NIRS sample holder (3 cm diameter round cell) until it was three-fourths full, and their spectra were registered as an individual file, in the range from 400 to 2500 nm, at 0.5 nm interval. Using the program Global Calibration Development software (WINISI III, Infrasoft International, LLC, Port Matilda, PA), different calibration equations were developed on the calibration set (n ~ 550) having variable ranges of these seed quality parameters. Calibration equations were then validated on known seed samples, and subsequently used for the experimental test samples. Seeds of wild-type Varuna were used as control. Statistical analyses were conducted using one-way ANOVA applying Fishers LSD test of significance.
Southern blot analysis for identification of single copy insertions
Single copy insertions of the T-DNA cassette were identified by Southern blot analysis according to the protocol described earlier12. Briefly, genomic DNA was isolated using CTAB method and ca. 20 μg of purified genomic DNA digested with EcoRV was used for blotting. Selection marker gene, bar (390 bp) was used as the probe which was labelled with dCTP [32Pα] using Amersham Megaprime DNA labelling systems (GE Healthcare, Littlechalfont, Buckinghamshire, UK) according to the manufacturer’s instructions.
Real-time quantitative RT-PCR (qRT-PCR) analysis
The transcript levels of GSL-ALK genes were analyzed using quantitative real-time RT-PCR in ABI 7900HT Fast Real-time PCR machine (Applied Biosystems, Foster city, CA, USA) following SYBR green protocol. Total RNA was isolated using Spectrum Total RNA Isolation Kit (Sigma-Aldrich, St. Louis, Missouri, USA). Two micrograms each of total RNA were reverse-transcribed using high capacity first strand cDNA synthesis kit (Applied Biosystems, Foster city, CA, USA) according to manufacturer’s instructions. The BjuACTIN2 gene was used as endogenous control49. Three independent sets of experiments were performed with three technical replicates each to finalize the data. Primers used for qRT-PCR analysis are tabulated in Supplementary Table S7.
Estimation of total phenolic content
Total phenolic content were estimated using Folin-ciocalteu (FC) reagent50,51. Exactly 100 mg lyophilized plant tissues were extracted twice with 2 ml each of hot 70% methanol. The extract was diluted 10 fold and used for the assay. For the assay 250 μl of the diluted sample was aliquoted into assay vials in duplicates and 1.25 ml of FC reagent (10%) was added to each vial. To this one ml of Na2CO3 (7.5%) was added and the contents were mixed thoroughly by vortexing. The reaction was allowed to stand for one hour at room temperature and readings were taken in a spectrophotometer (SmartSpec Plus, Bio-Rad laboratories) at 765 nm. Gallic acid was used as the standard and the total phenolics content was expressed as gallic acid equivalent (GAE) 100 mg−1 dry weight of the tissue.
Fungal infectivity assays
Stem rot pathogen of B. juncea, S. sclerotiorum (Delhi isolate-I, kindly provided by Dr. Jagreet Kaur, University of Delhi, India) was used for the susceptibility assays. Disc assay earlier described by Sotelo et al.40 was used with minor modifications. PDA plates (90 mm) were prepared with 100 μl each of leaf methanolic extracts of 30 day old transgenic (T1) lines and the control plants. Fungal pathogen was grown in separate PDA plates for three days and 6 mm disc of the PDA medium containing the actively growing mycelium was placed on the centre of each plates. PDA plate with 70% methanol was used as negative control (mock). Plates were incubated at 22 °C and growth diameter of mycelium (in cm) was measured after three days. The experiment was repeated four times with three technical replicates each time. Glucosinolates profiles of the tested plant samples were analyzed by HPLC. To examine the relationship between glucosinolates content and mycelial diameter, the data were fitted with simple linear model (y = a + bx). Linear regression analysis was performed in SPSS software package v.17.0 for Windows (SPSS Inc. released in 2008), by considering glucosinolates content as independent variable and mycelial diameter as dependent variable.
Additional Information
How to cite this article: Augustine, R. and Bisht, N. C. Biofortification of oilseed Brassica juncea with the anti-cancer compound glucoraphanin by suppressing GSL-ALK gene family. Sci. Rep. 5, 18005; doi: 10.1038/srep18005 (2015).
Supplementary Material
Acknowledgments
The work was supported by IYBA-2012 grant (BT/06/IYBA/2012) of Department of Biotechnology India to NCB. RA was funded with short term research fellowship from NIPGR, India. We are thankful to Dr. Prabodh K. Bajpai for his assistance in statistical analysis using SPSS software and Dr. Pritam Kalia (Division of Vegetable Science, IARI, New Delhi, India) for providing B. oleracea seeds. We are grateful to central instrumentation facility and plant growth facility at NIPGR. Technical assistance from Vinod Kumar is also acknowledged.
Footnotes
Author Contributions R.A. and N.C.B. designed the research, R.A. conducted, analyzed and interpreted data, R.A. and N.C.B. wrote and approved the paper.
References
- Halkier B. A. & Gershenzon J. Biology and biochemistry of glucosinolates. Annu. Rev. Plant Biol. 57, 303–333 (2006). [DOI] [PubMed] [Google Scholar]
- Hopkins R. J., van Dam N. M. & van Loon J. J. A. Role of glucosinolates in insect plant relationships and multitrophic interactions. Annu. Rev. Entomol. 54, 57–83 (2009). [DOI] [PubMed] [Google Scholar]
- Fahey J. W., Zhang Y. & Talalay P. Broccoli sprouts: an exceptionally rich source of inducers of enzymes that protect against chemical carcinogens. Proc. Natl. Acad. Sci. USA 94, 10367–10372 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartea M. E. & Velasco P. Glucosinolates in Brassica foods: bioavailability in food and significance for human health. Phytochem. Rev. 7, 213–229 (2008). [Google Scholar]
- Mawson R., Heaney R. K., Zdunczyk Z. & Kozlowska H. Rapeseed meal-glucosinolates and their antinutritional effects. Part II. Flavour and palatability. Mol. Nutr. Food Res. 37, 336–344 (1993). [DOI] [PubMed] [Google Scholar]
- Clarke J. D., Hsu A., Yu Z., Dashwood R. H. & Ho E. Differential effects of sulforaphane on histone deacetylases, cell cycle arrest and apoptosis in normal prostate cells versus hyperplastic and cancerous prostate cells. Mol. Nutr. Food Res. 55, 999–1009 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahey J. W. et al. Protection of humans by plant glucosinolates: efficiency of conversion of glucosinolates to isothiocyanates by the gastrointestinal microflora. Cancer Prev. Res. 5, 603–611 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahey J. W. & Talalay P. Antioxidant functions of sulforaphane: a potent inducer of phase II detoxication enzymes. Food Chem. Toxicol. 37, 973–979 (1999). [DOI] [PubMed] [Google Scholar]
- Shapiro T. A., Fahey J. W., Wade K. L., Stephenson K. K. & Talalay P. Chemoprotective glucosinolates and isothiocyanates of broccoli sprouts: metabolism and excretion in humans. Cancer Epidemiol. Biomarkers Prev. 10, 501–508 (2001). [PubMed] [Google Scholar]
- Yanaka A. et al. Dietary sulforaphane-rich broccoli sprouts reduce colonization and attenuate gastritis in Helicobacter pylori infected mice and humans. Cancer Prev. Res. 2, 353–360 (2009). [DOI] [PubMed] [Google Scholar]
- Sodhi Y. S. et al. Genetic analysis of total glucosinolate in crosses involving a high glucosinolate Indian variety and a low glucosinolate line of Brassica juncea. Plant Breed. 121, 508–511 (2002). [Google Scholar]
- Augustine R., Mukhopadhyay A. & Bisht N. C. Targeted silencing of BjMYB28 transcription factor gene directs development of low glucosinolate lines in oilseed Brassica juncea. Plant Biotech. J. 11, 855–866; doi: 10.1111/pbi.12078 (2013a). [DOI] [PubMed] [Google Scholar]
- Sønderby I. E., Geu-Flores F. & Halkier B. A. Biosynthesis of glucosinolates-gene discovery and beyond. Trends Plant Sci. 15, 283–90 (2010). [DOI] [PubMed] [Google Scholar]
- Li G. & Quiros C. F. In planta side-chain glucosinolate modification in Arabidopsis by introduction of dioxygenase Brassica homolog BoGSLALK. Theor. Appl. Genet. 106, 1116–1121 (2003). [DOI] [PubMed] [Google Scholar]
- Textor S., Kraker de J. W., Hause B., Gershenzon J. & Tokuhisa J. G. MAM3 catalyzes the formation of all aliphatic glucosinolate chain lengths in Arabidopsis. Plant Physiol. 144, 60–71(2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen B. G., Kliebenstein D. J. & Halkier B. A. Identification of a flavin-monooxygenase as the S-oxygenating enzyme in aliphatic glucosinolate biosynthesis in Arabidopsis. Plant J. 50, 902–910 (2007). [DOI] [PubMed] [Google Scholar]
- Li J., Hansen B. G., Ober J. A., Kliebenstein D. J. & Halkier B. A. Subclade of flavin-monooxygenases involved in aliphatic glucosinolate biosynthesis. Plant Physiol. 148, 1721–1733 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giamoustaris A. & Mithen R. Genetics of aliphatic glucosinolates IV Side-chain modification in Brassica oleracea. Theor. Appl. Genet. 93, 1006–1010 (1996). [DOI] [PubMed] [Google Scholar]
- Kliebenstein D. J. et al. Genetic control of natural variation in Arabidopsis glucosinolate accumulation. Plant. Physiol. 126, 811–825 (2001b). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mithen R., Clarke J., Lister C. & Dean C. Genetics of aliphatic glucosinolates. III. Side chain structure of aliphatic glucosinolates in Arabidopsis thaliana. Heredity 74, 210–215 (1995). [Google Scholar]
- Kliebenstein D. J., Lambrix V. M., Reichelt M., Gershenzon J. & Mitchell-Olds T. Gene duplication in the diversification of secondary metabolism: tandem 2-oxoglutarate-dependent dioxygenases control glucosinolate biosynthesis in Arabidopsis. Plant Cell 13, 681–693 (2001a). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neal C. S., Fredericks D. P., Griffiths C. A. & Neale A. D. The characterization of AOP2: a gene associated with biosynthesis of aliphatic alkenyl glucosinolates in Arabidopsis thaliana. BMC Plant Biol. 10, 170–186 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M., Li G., Yang B., Mc Combie R. W. & Quiros C. F. Comparative analysis of a Brassica BAC clone containing several major aliphatic glucosinolate genes with its corresponding Arabidopsis sequence. Genome 47, 666–679 (2004). [DOI] [PubMed] [Google Scholar]
- Liu S. et al. The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat. Commun. 5, 3930 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisht N. C. et al. Fine mapping of loci involved with glucosinolate biosynthesis in oilseed mustard (Brassica juncea) using genomic information from allied species. Theor. Appl. Genet. 118, 413–421 (2009). [DOI] [PubMed] [Google Scholar]
- Mun J.-H. et al. Genome-wide comparative analysis of the Brassica rapa gene space reveals genome shrinkage and differential loss of duplicated genes after whole genome triplication. Genome. Biol. 10, 111 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augustine R., Majee M., Gershenzon J. & Bisht N. C. Four genes encoding MYB28, a major transcriptional regulator of aliphatic glucosinolate pathway, are differentially expressed in the allopolyploid Brassica juncea. J. Exp. Bot. 64, 4907–4921 (2013b). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkin I. et al. Genetics of aliphatic glucosinolates. II. Hydroxylation of alkenyl glucosinolates in Brassica napus. Heredity 72, 594–598 (1994). [Google Scholar]
- Wang H. et al. Glucosinolate biosynthetic genes in Brassica rapa. Gene 487, 135–142 (2011). [DOI] [PubMed] [Google Scholar]
- Liu Z. et al. Reducing progoitrin and enriching glucoraphanin in Brassica napus seeds through silencing of the glucosinolate-ALK gene family. Plant Mol. Biol. 79, 179–189 (2012). [DOI] [PubMed] [Google Scholar]
- Meenu, Augustine R., Majee M., Pradhan A. K. & Bisht N. C. Genomic origin, expression differentiation and regulation of multiple genes encoding CYP83A1, a key enzyme for core glucosinolate biosynthesis, from the allotetraploid Brassica juncea. Planta 241, 651- 665 (2015). [DOI] [PubMed] [Google Scholar]
- Qian, H. et al. Variation of glucosinolates and quinone reductase activity among different varieties of Chinese kale and improvement of glucoraphanin by metabolic engineering. Food Chem. 168, 321–326 (2015). [DOI] [PubMed] [Google Scholar]
- Traka M. H. et al. Genetic regulation of glucoraphanin accumulation in Benefort’e broccoli. New Phytologist 198, 1085–1095 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burow M., Halkier B. A. & Kliebenstein D. J. Regulatory networks of glucosinolates shape Arabidopsis thaliana fitness. Curr. Opin. Plant Biol. 13, 348–353 (2010). [DOI] [PubMed] [Google Scholar]
- Wentzell A. M. et al. Linking metabolic QTLs with network and cis-eQTLs controlling biosynthetic pathways. PLoS Genetics 3, e162; doi: 10.1371/journal.pgen.0030162 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burow, M. et al. The glucosinolate biosynthetic gene AOP2 mediates feed-back regulation of jasmonic acid signaling in Arabidopsis. Mol. Plant 8, 1201–1212 (2015). [DOI] [PubMed] [Google Scholar]
- Kim J. H. & Jander G. Myzus persicae (green peach aphid) feeding on Arabidopsis induces the formation of a deterrent indole glucosinolate. Plant J. 49, 1008–1019 (2007). [DOI] [PubMed] [Google Scholar]
- Tierens K. F. et al. Study of the role of antimicrobial glucosinolate-derived isothiocyanates in resistance of Arabidopsis to microbial pathogens. Plant Physiol. 125, 1688–1699 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednarek P. et al. A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science 323, 101–106 (2009). [DOI] [PubMed] [Google Scholar]
- Sotelo T., Lema M., Soengas P., Cartea M. E. & Velasco P. In vitro activity of glucosinolates and their degradation products against brassica-pathogenic bacteria and fungi. Appl. Environ. Microbiol. 81, 432–440 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J. et al. Patterns of differential gene expression in Brassica napus cultivars infected with Sclerotinia sclerotiorum. Mol. Plant Pathol. 10, 635–649 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbetti M. J. et al. Comparative genotype reactions to Sclerotinia sclerotiorum within breeding populations of Brassica napus and B. juncea from India and China. Euphytica 197, 47–59 (2014). [Google Scholar]
- Chattopadhyay C., Meena P. D., Sastry R. K. & Meena R. L. Relationship among pathological and agronomic attributes for soilborne diseases of three oilseed crops. Indian J. Plant Prot. 31, 127–128 (2003). [Google Scholar]
- Stotz H. U. et al. Role of camalexin, indole glucosinolates, and side chain modification of glucosinolate-derived isothiocyanates in defense of Arabidopsis against Sclerotinia sclerotiorum. Plant J. 67, 81–93 (2011). [DOI] [PubMed] [Google Scholar]
- Tamura K. et al. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P. & Rozas J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452 (2009). [DOI] [PubMed] [Google Scholar]
- Koch M. A., Haubold B. & Mitchell-Olds T. Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis, and related genera (Brassicaceae). Mol. Biol. Evol. 17, 1483–1498 (2000). [DOI] [PubMed] [Google Scholar]
- Kumar S., Chauhan J. S. & Kumar A. Screening for erucic acid and glucosinolate content in rapeseed-mustard seeds using near infrared reflectance spectroscopy. J. Food Sci. Technol. 47, 690–692 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandna R., Augustine R. & Bisht N. C. Evaluation of candidate reference genes for gene expression normalization in Brassica juncea using real time quantitative RT-PCR. PLoS One. 7, e36918; doi: 10.1371/journal.pone.0036918 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anesini C., Ferraro G. E. & Filip R. Total polyphenol content and antioxidant capacity of commercially available tea (Camellia sinensis) in Argentina. J. Agric. Food Chem. 56, 9225–9229 (2006). [DOI] [PubMed] [Google Scholar]
- Singleton V. L., Rudolf O. & Lamuela-Raventós R. M. Analysis of total phenols and other oxidation substrates and antioxidants by means of folin–ciocalteu reagent. Method. Enzymol. 299, 152–178 (1999). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.