Summary
Fascin is an F-actin-bundling protein shown to stabilize filopodia and regulate adhesion dynamics in migrating cells, and its expression is correlated with poor prognosis and increased metastatic potential in a number of cancers. Here, we identified the nuclear envelope protein nesprin-2 as a binding partner for fascin in a range of cell types in vitro and in vivo. Nesprin-2 interacts with fascin through a direct, F-actin-independent interaction, and this binding is distinct and separable from a role for fascin within filopodia at the cell periphery. Moreover, disrupting the interaction between fascin and nesprin-2 C-terminal domain leads to specific defects in F-actin coupling to the nuclear envelope, nuclear movement, and the ability of cells to deform their nucleus to invade through confined spaces. Together, our results uncover a role for fascin that operates independently of filopodia assembly to promote efficient cell migration and invasion.
Graphical Abstract

Highlights
-
•
Fascin binds directly to nesprin-2 at the nuclear envelope
-
•
Fascin-nesprin-2 binding occurs independently of fascin-actin bundling
-
•
The fascin-nesprin-2 complex regulates nuclear movement in migration
-
•
Uncoupling the fascin-nesprin complex reduces nuclear deformation and cell invasion
Fascin is an F-actin-bundling protein that is significantly upregulated in most cancers and correlates with poor prognosis. Jayo et al. show that fascin can associate directly with the nuclear envelope protein nesprin-2. This complex is required for nuclear movement and deformation during cell invasion through 3D matrices.
Introduction
Fascin is an actin-binding protein that is known to regulate the parallel bundling of actin filaments (Vignjevic et al., 2006), stabilize filopodia and invadopodia (Jayo et al., 2012, Li et al., 2010), and regulate adhesion dynamics in migrating cells (Elkhatib et al., 2014). Fascin has received considerable attention in recent years as its expression is very low or absent in normal adult epithelia, but it is dramatically upregulated at both transcript and protein levels in all forms of human carcinomas studied to date (Hashimoto et al., 2005). Thus, fascin is emerging as an excellent prognostic marker and a potential therapeutic target for metastatic disease (Tan et al., 2013, Adams, 2015). Despite this recognized clinical importance, there is still very little molecular detail available defining the mechanisms underpinning fascin-dependent cell invasion, thus significantly limiting strategic approaches for therapeutic design. It is also unclear whether these defined roles for fascin in tumorigenesis rely upon the classical F-actin-bundling function or whether other roles may exist that coordinate cell invasion.
Fascin comprises four tandem β-trefoil domains that form a bilobed structure, with β-hairpin triplets located symmetrically on opposite sides of each lobe that are proposed to act as the actin-binding domains (Sedeh et al., 2010). These actin bundles, whether in the form of filopodia extending beyond the cell edge or microspikes within lamellae of migrating cells or neuronal growth cones, are involved in controlling cell migration in vitro (Adams, 2004) and embryonic development in vivo (Wood and Martin, 2002, Mattila and Lappalainen, 2008, Hashimoto et al., 2011). Invasion of carcinoma cells is a highly coordinated process that depends largely on alterations to cell-cell and cell-extracellular matrix (ECM) adhesion and organization of the actin cytoskeleton (Guo and Giancotti, 2004). Carcinoma cells migrating in 3D ECM and in living tissues assemble membrane protrusions and specialized ECM-degrading adhesions termed invadopodia to enable tunneling through the matrix (Friedl and Wolf, 2003, Condeelis et al., 2005, Li et al., 2010). We and other groups have shown that loss of fascin function in a range of cell types results in reduced assembly of actin protrusions, more stable adhesions, and reduced migration and invasion in vivo (Hashimoto et al., 2007, Kim et al., 2009, Chen et al., 2010, Jayo et al., 2012, Zanet et al., 2012). However, it remains unclear whether these reported functions for fascin depend upon actin bundling within filopodia alone, or whether other roles for fascin exist within normal and metastatic cells that promote motility.
Physicochemical properties of the ECM play an important role in the regulation of cell migration (Charras and Sahai, 2014, Friedl and Alexander, 2011) and cancer cells have been shown to have great plasticity, enabling them to adapt their migratory strategies to external cues (Sanz-Moreno et al., 2008, Wolf et al., 2003, Balzer et al., 2012). Several studies have demonstrated that nuclear size and deformation act as limiting factors of cell migration in physically confined environments (Wolf et al., 2013, Rowat et al., 2013, Davidson et al., 2014). Contractile force generation, cytoskeleton-driven force transmission to the nucleus, and nuclear stiffness (Harada et al., 2014, Lammermann et al., 2008, Lombardi et al., 2011, Alam et al., 2015) can together create a migratory threshold (Isermann and Lammerding, 2013, Swift and Discher, 2014). The linker of the nucleus and cytoskeleton (LINC) complex connects the cytoskeleton to the nuclear inner lamina ((Chang et al., 2015, Meinke and Schirmer, 2015)) and is formed by KASH proteins (Klarsicht, ANC-1, and Syne Homology proteins, nesprins) at the outer nuclear envelope (NE) and SUN proteins (Sad1 and UNC-84) at the inner NE. This complex is known to play an essential role in force transmission (Lombardi et al., 2011, Alam et al., 2015), NE response to physical strain (Guilluy et al., 2014), and nuclear localization in migrating cells (Luxton et al., 2010, Meinke et al., 2014). Mutations in the LINC complex have been associated mainly with muscular dystrophies and cardiomyopathies (Isermann and Lammerding, 2013, Zhang et al., 2007), but despite its role in cell motility, the potential contribution to cancer cell invasion and metastatic disease is currently unknown.
Here, we report that nesprin-2 binds directly to the fascin β-trefoil3 domain through spectrin repeats (SRs) 51–53 located at its C-terminal domain. Nesprin-2 recruits fascin to the NE both in vitro and in vivo, where it regulates F-actin connection to the LINC complex, modulating nuclear localization in fibroblasts and nuclear deformation in cancer cells. These effects are independent of the fascin F-actin-bundling activity, since the disruption of fascin-nesprin-2 interaction by the overexpression of β-trefoil3 domain does not alter F-actin dynamics. Together, these results provide a role for fascin in coupling the NE to the cytoskeleton to assist in coordinated force transmission during migration.
Results
Fascin Interacts with Nesprin-2
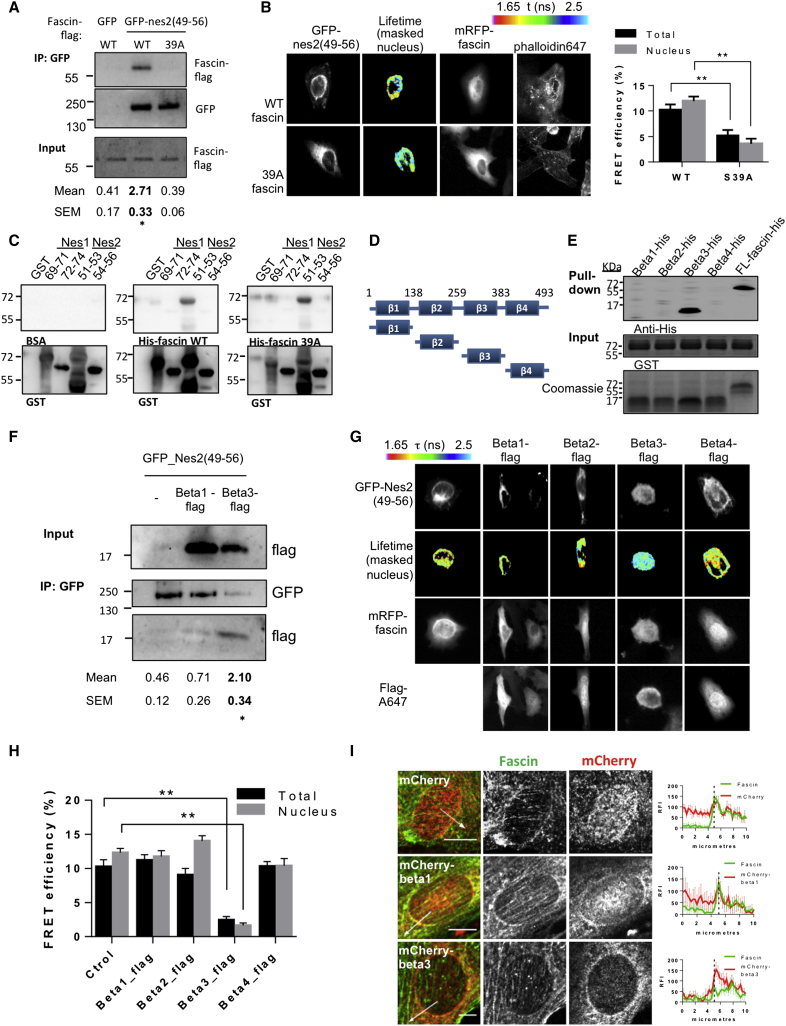
To define potential unexplored regulators of fascin function, we performed mass spectrometry analysis of specific isolated proteins in complex with GFP-fascin immunoprecipitated from MDA MB 231 human breast carcinoma cells. Within the reproducible hits that were significantly represented in the resulting peptide analysis, we identified nesprin-1, a member of the LINC complex (Figure S1A). We have previously shown that fascin localizes to the nucleus and the nuclear periphery both during Drosophila development and in mammalian cells (Groen et al., 2015). The function associated with this specific localization remains unknown; therefore, we chose to further characterize the potential interaction with the nesprin family of proteins. Initial experiments did not reveal a detectable interaction with endogenous full-length nesprin-1, but as this analysis was performed at low detergent concentration, we cannot exclude that this was due to retained co-association between other cytoskeletal-associated NE proteins in complex with fascin. Based on our previously published data on fascin subcellular localization, we then focused on the C-terminal domains of the proteins nesprin-1 and -2, as these regions are present in all nesprin-1 and -2 isoforms anchored to the NE and encompass the most conserved regions across the whole sequence (Autore et al., 2013) (Figure S1B). Co-immunoprecipitation (coIP) experiments with either GFP-nes1(66–74) or GFP-nes2(49–56) demonstrated that fascin was able to specifically form a complex with the nesprin-2 C-terminal region (Figure 1A). We confirmed this interaction in intact cells by quantifying fluorescence resonance energy transfer (FRET) using fluorescence lifetime imaging microscopy (FLIM) between GFP-tagged nesprins and monomeric RFP (mRFP)-tagged fascin. This analysis demonstrated a specific and direct interaction in whole cells between GFP-nes2(49–56), but not GFP-nes1(66–74) or the inner NE protein SUN2 (Figure 1B). To define the region of nesprin-2 that associates with fascin, we used recombinant glutathione S-transferase (GST)-tagged regions of tandem SRs from nesprin-1 and nesprin-2 as bait to pull down endogenous fascin from lysates of NIH 3T3 fibroblasts. Western blotting revealed that fascin was only found in a complex with SR51–53 from nesprin-2 (Figure 1C). We verified that fascin directly interacted with SR51–53 of nesprin-2 through in vitro binding assays (Figure 1D) and far-western blot analysis of GST-nesprin SR proteins probed with His-tagged fascin (Figure S1C). Moreover, GFP-mini-N2G-ΔSR3-50 containing the SR51–53 region (schematic representation in Figure S1B) showed specific coIP with endogenous fascin in NIH 3T3cells (Figure 1E). Collectively these data demonstrate that fascin forms a complex specifically with nesprin-2 in vitro and in cells, and this binding occurs directly and through SR51–53 in nesprin-2.
Figure 1.
Fascin Interacts with the C-Terminal Region of Nesprin-2
(A) Co-immunoprecipitation (coIP) of GFP, GFP-nes1(66–74) and GFP-nes2(49–56) with FLAG-tagged fascin expressed in HEK293T cells. Densitometry values normalized to GFP IP protein content from three independent experiments ± SEM are depicted below. ∗p < 0.05 compared with GFP only.
(B) HeLa cells co-expressing GFP-nes1(66–74), GFP-nes2(49–56), or GFP-SUN2 with mRFP-fascin. Center panels show pseudocolored FLIM images. Graph shows mean FRET efficiency of GFP-tagged proteins and mRFP-tagged fascin. n = 18 cells from three experiments ± SEM. ∗∗p < 0.01 compared with GFP-SUN2.
(C) Western blot of endogenous fascin pulled down from NIH 3T3 fibroblast lysates with GST alone and GST-tagged nesprin-1 and nesprin-2 SR triplets. Densitometry values normalized to GST ± SEM from three experiments are shown below. ∗∗p < 0.01 compared with GST.
(D) Western blot of recombinant His-tagged fascin pulled down with GST-tagged nesprin-1 and nesprin-2 SR triplets. Mean densitometry values normalized to GST ± SEM from three experiments are shown below. ∗∗p < 0.01 compared with GST.
(E) CoIP of endogenous fascin with GFP, GFP-mini-N2G-ΔSR3-54, and GFP-mini-N2G-ΔSR3-50 expressed in NIH 3T3. Densitometry values normalized to GFP ± SEM from three experiments are shown below. ∗p < 0.05 compared with GFP.
(F) Representative single confocal slice showing co-localization of endogenous fascin and nesprin-2 in NIH 3T3 acquired by structured illumination microscopy. Arrowheads show enrichment of fascin at the NE; arrows show fascin localized at fibers over the nucleus. Scale bars, 2.5 μm.
(G) Endogenous fascin and nesprin-2 in NIH 3T3 expressing a non-targeting scrambled SCR shRNA (SCR), nesprin-2-targeting shRNA (Nes2 KD), or fascin-targeting shRNA (Fascin KD). Graphs show fluorescence line scan analysis averaged from at least 15 cells in two experiments. Yellow arrows represent sites of line scan analysis. Scale bars, 10 μm.
See also Figure S1.
We next wanted to determine whether fascin localization at the NE was common across different cell types and conditions. Confocal images of NIH 3T3 cells demonstrated recruitment of fascin at the NE where it colocalized with nesprin-2 (Figure 1F). Interestingly, fascin localization also showed significant enrichment within the perinuclear area together with nesprin-2 in human cancer cells grown as xenograft tumors in nude mice (Figure S1D) and with the NE marker protein SUN-2 in spontaneous colon carcinomas (Figure S1E). We have previously shown that fascin localizes to the nucleus of nurse cells of the developing Drosophila ovarian follicle (Groen et al., 2015) (Figure S1G), suggesting that the role of fascin at the NE is evolutionarily conserved. To determine whether nesprin-2 was responsible for fascin localization at the NE or vice versa, we knocked down each protein in fibroblasts and analyzed their respective localization by confocal microscopy. Nesprin-2 knockdown (KD), including giant and smaller isoforms (Figure S1F), resulted in loss of fascin localization at the NE, but nesprin-2 localization was unaffected by fascin KD (Figure 1H). Similarly in Drosophila embryos, complete disruption of the LINC complex through homozygous Klaroid mutation (Technau and Roth, 2008) or loss of only the nesprin homolog MSP-300 (Yu et al., 2006) disrupted fascin localization to the NE (Figure S1G), while loss of fascin did not affect LINC complex localization (Figure S1H). Together, these data demonstrate that fascin localizes to the NE in a nesprin-2-dependent manner.
Fascin-Actin Bundling Is Not Required for Nesprin Binding
Fascin-dependent F-actin bundling is known to be partly controlled by phosphorylation of Ser39 by protein kinase C (PKC) such that mutation of Ser39 to a phospho-dead alanine mimic results in constitutive F-actin bundling by fascin (Ono et al., 1997, Adams et al., 1999, Anilkumar et al., 2003). To determine whether fascin-nesprin-2 binding was dependent upon fascin-F-actin bundling, we performed coIP and FRET/FLIM analysis of wild-type (WT) or S39A fascin and nes2(49–56). Both assays demonstrated that significantly lower levels of S39A fascin bound to nes2(49–56) compared with WT fascin (Figures 2A and 2B). Analysis of total FRET efficiency and that associated to the NE showed that most of the interaction was restricted to the perinuclear area (Figure 2B). S39A fascin localization was associated with a more prominent cytoplasmic recruitment and less enrichment at the NE in HeLa cells (Figure 2B), and a similar mutation in Drosophila fascin (S52A) also reduced fascin localization to the NE (Figure S2A). We thus hypothesized that non-S39 phosphorylated fascin may conformationally restrict the interaction with nesprin-2. To test this in a cell-free in vitro system, we used far-western blot assays with recombinant fascin mutants and nesprin-2 and nesprin-1 most C-terminal GST-tagged SR triplets. S39A mutant fascin did not show significantly impaired binding to SR51–53 (Figure 2C), suggesting that any conformational change in this region does not significantly alter binding. This further implies that the stable F-actin bundling and recruitment to filopodia by this mutant may be incompatible with fascin localization to the NE and interactions with nesprin-2.
Figure 2.
Fascin-Actin Bundling Is Not Required for Nesprin Binding
(A) CoIP of GFP and GFP-nes2(49–56) with FLAG-tagged WT fascin and S39A mutant co-expressed in HEK293T cells. Densitometry values normalized to GFP IP ± SEM from three experiments are shown below. ∗p < 0.05 compared with WT FLAG-fascin.
(B) FRET/FLIM analysis of HeLa cells co-expressing GFP-nes2(49–53) and mRFP-tagged WT and S39A fascin. Center panels show pseudocolored images of nuclear-masked FRET values. Graph shows mean FRET efficiency values both in the whole cell or masked nuclear area (n = 18 cells per condition). ∗∗p < 0.01 compared with WT mRFP-fascin.
(C) Far-western blot of GST and GST-tagged nesprin-1 and nesprin-2 SR triplets incubated with BSA as control or purified recombinant WT and S39A His-tagged fascin detected using an anti-His antibody.
(D) Schematic of fascin structure and β-trefoil domain constructs.
(E) Pull-down of GST-nes2(51–53) with His-tagged full-length (FL) fascin or His-tagged β-trefoil subdomains. Representative of three experiments.
(F) CoIP of GFP-nes2(49–56) with FLAG-tagged fascin β-trefoil3 domain expressed in HEK293T cells. Representative of three experiments, densitometry values normalized to GFP-nes2(49–56) IP levels ± SEM are shown below. ∗p < 0.05 compared with GFP.
(G) Example micrographs of cells co-expressing GFP-nes2(49–56) and mRFP-fascin together with different FLAG-tagged β-trefoil domains. Second row shows pseudocolored images of GFP-nes2(49–56) FLIM images. Final row shows FLAG-tagged β-trefoil domain localization.
(H) Graph of mean FRET efficiency of GFP-nes2(49–56) and mRFP-fascin co-expressed with β-trefoil domains from three experiments. ∗∗p < 0.01 compared with cells co-expressing only GFP-nes2(49–56) and mRFP-fascin.
(I) Endogenous fascin and mCherry-tagged β-trefoil constructs in NIH 3T3. Fascin and mCherry were visualized using specific antibodies. Graphs show averaged fluorescence line scan analysis performed from >15 cells from three experiments. White arrows indicate position of line scan analysis. Scale bars, 10 μm.
See also Figure S2.
To further define the region(s) on fascin required for the interaction with nesprin-2, we generated four constructs that mapped to the four distinct β-trefoil domains in fascin (Figure 2D). In vitro pull-down assays with purified GST-nes2(51–53) and recombinant full-length fascin and β-trefoil subdomains revealed that this interaction occurred specifically through the β-trefoil3 subdomain (Figure 2E). GFP-nes2(49–56) immunoprecipitations also showed a complex formation with overexpressed β-trefoil3 subdomain but not β-trefoil1 (Figure 2F), and fascin-nes2(51–53) interaction could be competed in vitro with increasing amounts of recombinant β-trefoil3 subdomain (Figure S2B). We then co-expressed FLAG-tagged β-trefoil subdomains with GFP-nes2(49–56) and mRFP-fascin and measured fascin-nesprin-2 binding by FRET/FLIM in HeLa cells. Quantification of FRET values demonstrated that overexpression of the β-trefoil3 significantly inhibited nesprin-2-fascin binding, whereas the other three domains had no effect on formation of the complex (Figures 2G and 2H). Moreover, expression of mCherry-tagged versions of β-trefoil3, but not 1, 2, or 4, led to displacement of endogenous fascin from the NE in NIH 3T3cells (Figure 2I). Conversely, expressing mCherry-tagged SR(51–53) domain in NIH 3T3cells displaced fascin localization from the NE (Figure S2D).
The formin FHOD1 has been reported to interact with nesprin-2 at its N-terminal domain (amino acids 1,130–1,724), and plays a role creating an additional binding site for actin cables to the NE during nuclear movement. To determine potential interdependence of these two actin-binding proteins in connecting the actin cytoskeleton and LINC complex, we first explored the role of FHOD1 in the interaction of fascin with nesprin-2. Immunoprecipitation assays showed that silencing FHOD1 did not affect fascin binding to the GFP-mini-N2G-ΔSR3-50 construct (Figure S2E). Moreover, FRET between GFP-Nes2(49–56) with mRFP-fascin after silencing FHOD1 showed no change in the level of interaction (Figures S2F–S2H). Taken together, these results demonstrate that fascin binds directly to the region of nesprin-2 encompassing SRs 51–53 through its third β-trefoil domain. This interaction is required for fascin localization at the NE and is independent from the nesprin-2 interaction with FHOD1.
The Fascin-Nesprin-2 Complex Couples F-Actin to the Nuclear Envelope
We next sought to determine the functional consequences of fascin localization to the NE and its association with nesprin-2. Nesprin-2 has been shown to be important for active positioning of the nucleus during cell migration. Starved fibroblasts require stimulation with serum factors such as LPA to establish their nucleo-centrosomal axis during cell polarization after scratch wounding (Gomes et al., 2005). This process relies in the rearward displacement of actin cables that associate with nesprin-2 at the transmembrane actin-associated (TAN) lines. These structures are responsible for coupling the nuclei to the moving actin cables, inducing the nuclear positioning at the cell rear while the centrosome remains stationary at the cell centroid (Luxton et al., 2010, Kutscheidt et al., 2014, Lombardi et al., 2011). We therefore hypothesized that the fascin-nesprin complex may play a role in nuclear positioning during migration. To test this, we first determined whether fascin localized to TAN lines in NIH 3T3cells. Confocal microscopy and fluorescence intensity line scan analysis demonstrated that overexpressed myc-tagged fascin showed significant overlap with both F-actin and GFP-mini-N2G-ΔSR3-54 in cables positioned over the nucleus (Figure 3A), endogenous fascin colocalized with GFP-mini-N2GΔSR3-50 (Figure S3A), and GFP-fascin and F-actin cable displacement both correlated with nuclear movement during cell polarization (Figure S3B). Moreover, stable KD of fascin in NIH 3T3cells (Figure S3C) resulted in a significant reduction in nuclear rearward movement during polarization, and this defect was restored by re-expression of human WT GFP-fascin but not S39A mutant (Figures 3B and 3C) correlating with fascin binding to nesprin-2. The reduction in rearward nuclear displacement was also coupled with a reduction in the speed of actin fiber moving over the nucleus (Figures 3D and 3E; Movie S1). This demonstrates that fascin localization at the NE is required for not only efficient F-actin-nuclear coupling but also normal actin rearward movement during cell polarization.
Figure 3.
Fascin Couples F-Actin and the Nuclear Envelope
(A) GFP-mini-N2G-Δ3-54, myc-tagged fascin, and F-actin in TAN lines. Starved NIH 3T3 monolayer was wounded, constructs microinjected, and cells stimulated with 10 μM LPA for 1 hr before fixation. Scale bar, 10 μm. Graph shows fluorescence line scan analysis of white arrows shown in the left panel. Yellow arrow indicates points of colocalization. Black arrows on graph show points of GFP-mini-N2G-Δ3-50, myc-fascin, and F-actin co-localization at TAN lines.
(B) NIH 3T3 fibroblasts during nucleo-centrosomal axis formation 2 hr after scratch wound and LPA stimulation. Upper panels show merged channels for DNA, tubulin, and F-actin. Lower panels show GFP and GFP-fascin (WT and S39A mutant) channels in the same cells. Representative of four experiments. Scale bar, 20 μm.
(C) Quantification of relative nuclear and centrosomal position respective to the cell centroid in SCR and fascin KD fibroblasts expressing GFP, and GFP-fascin (WT and S39A) after LPA stimulation. Mean of four independent experiments, at least 90 cells per group. ∗∗p < 0.01, ∗∗∗p < 0.001 compared with SCR GFP.
(D) Quantification of actin fibers retrograde displacement speed during LPA-induced fibroblast polarization. Each value represents the speed of a single fiber, two fibers from >10 cells per condition from three experiments. ∗∗p < 0.01, ∗∗∗∗p < 0.0001 compared with SCR.
(E) Kymographs of actin fiber retrograde displacement during LPA-induced fibroblast polarization in fascin KD cells expressing GFP, GFP-fascin WT, or S39A mutant. Wound located at top of kymographs. Arrows indicate example actin cables and dashed lines represent nuclear outline. Representative of three experiments. Scale bars, 10 μm.
(F) NIH 3T3 expressing mCherry, mCherry-β-trefoil1, and mCherry-β-trefoil3 after scratch wound and LPA stimulation. Upper panels show DNA (DAPI), tubulin, and F-actin (phalloidin). Lower panels show mCherry and mCherry-β-trefoil expression in the same cells. Representative of four experiments. Scale bar, 20 μm.
(G) Quantification of relative nuclear and centrosomal position with respect to the cell centroid in fibroblasts expressing mCherry and mCherry-tagged β-trefoil domains 2 hr after LPA stimulation. Average of three experiments, >90 cells per condition. ∗∗p < 0.01 compared with SCR mCherry.
(H) Quantification of actin fiber retrograde displacement speed during LPA-induced polarization in fibroblasts expressing mCherry and mCherry-tagged β-trefoil domains. Each value represents the speed of a single fiber, >30 cells per condition in three experiments. ns, not significant compared with SCR mCherry.
(I) Kymographs of dorsal actin fiber displacement during LPA-induced fibroblast polarization in fibroblasts expressing mCherry and mCherry-tagged β-trefoil domains. Arrows indicate example actin fibers. Dashed lines indicate nuclear outline. Representative of three experiments. Scale bars, 10 μm.
To further confirm that this fascin-dependent defect in nuclear movement was dependent upon fascin-nesprin binding, we expressed mCherry-tagged β-trefoil domains 1 or 3 of fascin in NIH 3T3 and analyzed nuclear movement. Our data demonstrated that β-trefoil3, which specifically competes for fascin-nesprin-2 binding, but not β-trefoil1, resulted in a significant reduction in nuclear rearward movement during fibroblast polarization (Figures 3F and 3G). This was specific to fascin-nesprin disruption, as no further defect in nuclear movement was seen in fascin KD cells expressing β-trefoil3 compared with mCherry alone (Figure S3D). The lack of rescue of phenotype in fascin-depleted cells expressing β-trefoil3 also demonstrates that this domain is not sufficient to regulate nesprin-2-F-actin linkage. Expression of β-trefoil3 did not alter the speed of retrograde actin fiber displacement, suggesting that these defects seen in fascin KD or S39A-fascin-expressing cells are due to global fascin-F-actin coupling (Figures 3H and 3I; Movie S2). Importantly, β-trefoil domains of fascin expressed in fibroblasts did not localize to F-actin or change stress fibers or filopodia formation (Figures S3E and S3F). This demonstrated that the expression of single β-trefoils does not affect F-actin cytoskeleton and that the role of fascin at the NE is uncoupled and mechanistically distinct from that at the cell periphery. These data combined demonstrate that fascin-nesprin binding at the NE is specifically required for nuclear polarization and movement during polarization.
The Fascin-Nesprin-2 Complex Contributes to Nuclear Morphology and Deformability
Work from our laboratory and others has previously shown that fascin contributes to invasion of human cancer cells (Li et al., 2010, Hashimoto et al., 2007, Schoumacher et al., 2010). We confirmed that KD of fascin leads to significantly reduced invasion of MDA MB 231 cells through 3D collagen gels, an effect that was restored upon re-expression of small hairpin RNA (shRNA)-resistant GFP-fascin (Figures 4A and S4A). Previous reports have suggested that this pro-invasive effect is due to fascin-dependent stabilization of filopodia and possibly invadopodia. However, a number of recent studies have shown that nuclear deformation and movement in tumor cells are required to enable efficient cell navigation through changing ECM environments (Davidson et al., 2014, Zwerger et al., 2011, Wolf et al., 2013). We therefore reasoned that our discovery of fascin-dependent nuclear movement could represent a new way for fascin to control cell invasion independently of F-actin bundling. To explore this further, we examined the morphology and volume of nuclei of control or fascin KD cells in stiff 2D surfaces and embedded in 3D collagen. Quantification revealed that in 3D matrices, but not on 2D surfaces, nuclear volume was significantly reduced in fascin-depleted cells, and that this was restored in cells rescued with shRNA-resistant GFP-fascin (Figures 4B–4D) suggesting that fascin is required to maintain nuclear architecture in 3D environments. Acto-myosin force generation and transmission are required to induce nuclear shape change in a range of migrating cells (Lammermann et al., 2008, Wolf et al., 2013), but intrinsic nuclear mechanical properties can modify cell responses to force, acting as a limiting factor of nuclear deformability and, thus, migration (Harada et al., 2014, Davidson et al., 2014). To determine whether fascin loss correlated with altered nuclear stiffness, we used atomic force microscopy (AFM). To avoid overall contribution from cytoplasmic dorsal actin fibers to nuclear stiffness, we averaged several local measurements performed with a quadratic pyramidal-tipped cantilever per nuclei instead of a broader spherical tip (Figures S4B–S4D, see Experimental Procedures). In MDA MB 231 cells plated on a rigid surface, KD of fascin did not change nuclear stiffness (Figure 4E). Spatial confinement on 2D matrices is known to modulate nuclear size and volume, but the contribution of the LINC complex and microfilaments to this process remain unclear at present (Khatau et al., 2009, Li et al., 2015). Our results show that fascin KD only affects nuclear volume in cells embedded in 3D matrices and indicates that fascin contributes to nuclear deformation under conditions of 3D spatial confinement. This suggests that fascin plays a role in the transduction of mechanical forces to nesprin-2 and NE without affecting bulk mechanical properties, and that this plays a more profound role in 3D environments.
Figure 4.
Fascin Contributes to Nuclear Morphology and Deformability
(A) SCR, fascin KD, and fascin KD re-expressing GFP-fascin MDA MB 231 cells invading collagen gels. mCherry-H2BK histone is shown as nuclear marker. Dashed line represents the 45-μm invasion depth used as a threshold for invasion quantification. Scale bars, 50 μm. Graph shows invasion levels averaged from five fields per condition in four independent experiments. ∗p < 0.05 compared with SCR GFP.
(B and C) Nuclear volume quantification of SCR, fascin KD, and fascin KD re-expressing GFP-fascin MDA MB 231 cells on 2D surface (B) or in a collagen gel (C) from three experiments. ∗p < 0.05, ∗∗∗p < 0.001 compared with SCR.
(D) Images of SCR, fascin KD, and fascin KD re-expressing RFP-fascin MDA MB 231 cells embedded in collagen gels. Upper row shows maximum-intensity projection of F-actin (phalloidin), nucleus (SYTO16), and collagen fibers (reflection). Lower row shows 3D reconstruction of nuclei.
(E) Quantification of nuclear stiffness by AFM on cells growing on 2D surfaces. Values represent Young's modulus from single cells averaged from four measurements in the same nucleus performed in duplicate. Results from at least 15 cells per condition in two experiments.
(F) Schematic representation of microfluidic device used to challenge nuclear deformation. Channel height was constant (5 μm) while the width ranged from 20 to 4 μm.
(G) Kymographs of SCR, fascin KD, and fascin KD re-expressing GFP-fascin MDA MB 231 cells translocating into the 7-μm-width microchannel. Dashed line represents microchannel entry. Scale bars, 50 μm.
(H) Percentage of SCR, fascin KD, and fascin KD re-expressing GFP-fascin MDA MB 231 cells translocating nuclei inside 7-μm-width microchannels of the microfluidic device depicted in (F). Averaged from >22 cells per channel in three experiments. ∗p < 0.05.
(I) Quantification MDA MB 231 cells expressing mCherry or mCherry-β-trefoil3 nuclear translocation inside microfluidic device. Average of >20 cells in three experiments. ∗p < 0.05.
(J) Quantification of the invasion of SCR and fascin KD MDA MB 231 cells expressing mCherry or mCherry-β-trefoil3 through collagen gels. Average from five fields per conditions in three experiments. ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001 compared with SCR mCherry.
(K) SCR and fascin KD MDA MB 231 cells expressing either mCherry or mCherry-β-trefoil3 invading through collagen gels. Representative stacks from three experiments with SYTO16 signal as nuclear marker. Dashed line shows 45-μm invasion depth used as threshold for invasion. Scale bars, 50 μm.
See also Figure S4.
To study this further in an environment with controlled spatial constrictions, we used a microfluidic device with arrays of channels of different sizes (Figure 4F) (Malboubi et al., 2015). Similar devices have been previously used to study the ability of MDA MB 231 cells to invade in 3D, showing that nesprin-2 plays an important role in nuclear translocation through confined spaces (Thomas et al., 2015). In our system, control MDA MB 231 cells (SCR) showed equally efficient nuclear translocation through channel widths of 15 μm, 10 μm, and 7 μm, but decreased significantly in 5-μm wide gaps, where only ∼30% of nuclei entered the channels (Figures 4G and 4H). Stable fascin KD cells, however, showed a significant reduction in nuclear translocation into channels with widths smaller than 10 μm, suggesting that loss of fascin limits nuclear deformation, a phenotype that was rescued by the stable re-expression of shRNA-resistant GFP-fascin (Figures 4G and 4H). Nuclear translocation through microchannels of different widths fits to a sigmoidal distribution in which the inflection point of the curve, where 50% of the cells translocate, can be used as a relative cut-off size for cell translocation. In accordance with our previously published data (Malboubi et al., 2015), control MDA MB 231 showed a cut-off size of 6.99 μm, while in fascin KD cells this increased to 9.77 μm. Conversely, stable expression of shRNA-resistant GFP-fascin induced a decrease in the cut-off size to 6.45 μm, close to the control levels (Figures S4E and S4F). Our experimental setup did not allow us to discriminate rescued cells overexpressing fascin levels above the WT, and thus we cannot discard that this partially enhanced effect in nuclear translocation might be due to the levels of fascin achieved in the fascin KD cell line re-expressing GFP-fascin (Figure S4A). These data suggest that fascin regulates nuclear translocation into spatially confined environments (Figures S4E and S4F).
To further determine whether this defect in fascin-dependent cell movement and adaptation was due to fascin-nesprin binding, we analyzed WT cells expressing mCherry alone versus mCherry-tagged β-trefoil3 domains of fascin in the microfluidic chambers. Our analysis demonstrated that specifically competing the fascin-nesprin-2 complex with β-trefoil3 resulted in a significant impairment of nuclear translocation through gaps smaller than 10 μm wide compared with control or mCherry-expressing cells (Figures 4I, S4E, and S4F). β-Trefoil3 expression in MDA MB 231 cells or fibroblasts did not induce any change in actin cytoskeleton organization, morphology, or focal adhesion formation cells, all features closely related to acto-myosin contractility (Figure S4J). Fascin KD is known to increase non-muscle myosin II activity and cell contractility (Elkhatib et al., 2014), which, together with nesprin-2 tethering to actin cytoskeleton, favors cancer cell invasion and nuclear translocation into spatially confined 3D matrices (Thomas et al., 2015). This phenomenon could partially counteract the negative effects of fascin depletion in cell invasion and mask some of these observed effects. In our experimental setup, we specifically disengaged fascin binding to nesprin-2 from other fascin-dependent effects. The larger channel cut-off size for β-trefoil3-expressing cells further suggests that acute disruption of the nesprin-fascin complex severely impairs the transmission of cytoplasmic forces and, thus, nuclear deformability.
In line with this notion, our data demonstrated that silencing nesprin-2 in MDA MB 231 cells resulted in impaired invasion through 3D gels (Figures S4H and S4I). We therefore sought to determine whether disruption of the fascin-nesprin complex, and subsequent defects in nuclear deformation might play a role in cancer cell invasion through 3D ECM. Analysis of MDA MB 231 cells showed that β-trefoil3 expression resulted in a significant reduction in invasion into collagen gels (Figures 4J and 4K). Moreover, expression of β-trefoil3 in fascin KD cells did not alter invasion levels, demonstrating that this effect was specific, but also that this domain of fascin is not sufficient to enable coupling of nesprin-2 and F-actin to permit efficient nuclear displacement and cell invasion (Figures 4J and 4K). Importantly, focal adhesion formation and migration speed of cells expressing of β-trefoil3 on 2D surfaces was not significantly altered (Figures S4J and S4K), further suggesting that the inhibition of invasion in 3D is due to nuclear deformation and force transmission defects and not an overall loss of actin-based motility. Together, these results show that fascin plays a key role in coordinating the link between nesprin-2 and actin that in turn facilitates nuclear deformation and the ability of tumor cells to navigate through complex 3D matrices.
Discussion
Here we have uncovered a filopodia-independent role for fascin in controlling cell invasion through coupling nesprin-2 to F-actin at the NE. Our data also identify a non-F-actin-bundling functional role for fascin in cells. These findings support a model in which fascin can occupy at least two distinct roles in cells to regulate migration and invasion: (1) F-actin bundling, through two distinct F-actin-binding sites within filopodia at the cell periphery; and (2) F-actin and nesprin-2 binding at the NE to couple F-actin retrograde flow and nuclear translocation in 3D environments. The notion of these distinct and separable functions for fascin is supported by our ability to uncouple filopodia formation from nuclear movement through overexpression of the β-trefoil3 domain of fascin.
Fascin has a number of known binding partners that play a role in regulating F-actin bundling, stability, or localization (Anilkumar et al., 2003, Zhang et al., 2009, Jayo et al., 2012). Our data demonstrate that nesprin-2 is a fascin-interacting protein that recruits it to a yet uncharacterized subcellular localization. Interestingly this interaction is direct, and occurs through the β-trefoil3 domain, but constitutive actin bundling and filopodia formation competes with this interaction in whole cells. The β-trefoil3 domain contains a number of residues shown to be important for actin bundling (Zanet et al., 2012, Yang et al., 2013, Jansen et al., 2011), but it is unclear whether it sits on an actin-binding site or plays a structural regulatory role (Jansen et al., 2011, Yang et al., 2013). Our β-trefoil3 overexpression approach did not substantially alter actin microfilament cytoskeleton while disrupting the interaction with nesprin-2, showing that the nature of this interaction is structurally distinct from the binding to the actin filament and that a larger structural domain is needed for F-actin filament binding. In this context, constitutive F-actin bundling by S39A-fascin impeded an efficient interaction with nesprin-2, showing that PKC-mediated phosphorylation at S39 is required for both normal actin dynamics and F-actin cytoskeleton connections with the LINC complex. Future studies of this molecular switch will shed further light on how fascin acts as a linker between F-actin bundles and nesprin-2.
Actin-dependent nuclear movement in fibroblasts relies on the interaction of actin filaments with the N-terminal site of the nesprin-2 giant isoform (Luxton et al., 2010). More recently, an interaction with FHOD1 formin, another actin-binding protein, was reported to play a role in TAN-line formation and nuclear movement (Kutscheidt et al., 2014). These findings and data presented here support the emerging concept that the actin-NE connection is supported by more than one F-actin-nesprin-2 interaction site through its actin-binding domain. FHOD1 interacts with nesprin-2 through a domain close to its N-terminal region, and we have shown that FHOD1 and fascin-nesprin-2 interactions are not interdependent. These additional binding sites may play a role in strengthening the link and bearing a higher strain exerted by the cytoskeleton under certain conditions (Antoku et al., 2015). Our data would suggest that this is even more evident in the context of migration into 3D environments where the nuclear deformability acts as a limiting factor (Wolf et al., 2013, Davidson et al., 2014, Rowat et al., 2013). Several factors modulate the nuclear deformability in a wide range of cells, such us contractility (Lammermann et al., 2008), nuclear stiffness (Harada et al., 2014), and nucleo-cytoskeletal force transduction (Lombardi et al., 2011, Alam et al., 2015). In our model, while loss of fascin did not induce any change in the mechanical properties of the nuclei, disruption of fascin-nesprin interactions led to a severe defect in nuclear translocation into physically confined environments. This function of fascin appears to be essential in this process, since despite the increased myosin activity induced by fascin depletion (Elkhatib et al., 2014), this is not sufficient to sustain normal cell translocation rates.
Together, these findings suggest a role for fascin as a mechano-transducer of cytoplasmic forces and provide evidence of its role in nuclear deformation and movement during cell migration. As fascin is a prognostic marker in many different cancer types and a potential therapeutic target, our data may provide new avenues to target fascin for anti-metastatic therapy.
Experimental Procedures
Antibodies, DNA Constructs, Cell Lines, Fly Stocks, and Mouse Xenograft Models
See Supplemental Experimental Procedures for details.
Mass Spectrometry Identification of Fascin-Interacting Proteins
For the proteomic analysis of fascin-interacting proteins. GFP and GFP-fascin were transfected into fascin KD MDA MB 231 cells, cells were lysed in a buffer (0.5% Triton X-100, 150 mM NaCl, 15 mM Tris [pH 7.4], and propidium iodide) and GFP-fascin-containing complexes were immunoprecipitated overnight with a polyclonal anti-GFP antibody-coated Protein AG agarose beads. Complexes were washed, denatured, and resolved by SDS-PAGE. Single bands of proteins interacting specifically with GFP-fascin were excised and sent for mass spectrometry analysis (University of Aberdeen).
Protein Production
His-fascin was prepared as previously described (Zanet et al., 2012). For GST-tagged nesprin constructs, protein production was induced in BL21 Escherichia coli strains overnight at 16°C with 200 mM isopropyl β-D-1-thiogalactopyranoside. Pellets were resuspended in 10 mM phosphate buffer, 300 mM NaCl, 1 mM DTT, and 25 mM imidazole in the presence of protein inhibitors, sonicated, and cleared by centrifugation. GST-tagged proteins were bound to glutathione Sepharose beads for 2 hr, washed, and proteins eluted in 10 mM phosphate buffer, 150 mM NaCl, 1 mM DTT, 25 mM imidazole, and 3 mg/mL reduced glutathione (pH 7.5) for 30 min at 4°C. Proteins were dialyzed overnight and quantified by SDS-PAGE.
Immunoprecipitation, Pull-down, Western Blots, and Far-Western Blots
See Supplemental Experimental Procedures for details.
Immunofluorescence
Details of immunostaining procedures for cells, tissues, and fly samples are provided in Supplemental Experimental Procedures.
FLIM Analysis
FRET efficiency was quantified from HeLa cells transiently expressing donor and acceptor fluorophores by measuring time-domain fluorescence lifetime with a multi-photon microscope system (TE2000, Nikon). In brief, cells were fixed in 3.6% formaldehyde for 15 min, permeabilized with 0.1% Triton X-100, and quenched with 1 mg/mL sodium borohydride for 10 min at room temperature. Cells were mounted or immunostained for FLAG detection. Fluorescence lifetime was measured as described previously (Zanet et al., 2012), and histogram data show mean FRET efficiency from at least 12 cells per condition in three independent experiments. When indicated, FRET efficiency was calculated from the area masked around the cell nucleus, using TRI2 analysis software (Paul Barber, University of Oxford).
Averaged Fluorescence Line Scan Analysis at the Nuclear Envelope
Quantification of protein localization at the NE was performed from single confocal slides of several cells by using Fiji software (Schindelin et al., 2012). In brief, a 10-μm line was drawn from the nucleus to the cytoplasm, while the middle point of the line was made to coincide with the nucleus-cytoplasm boundary using the DAPI channel as nuclear marker. Fluorescence values over the line were plotted using the “plot profile” function, and the fluorescence values over different cells were later normalized. At least 15 cells from three independent experiments were analyzed for the graphs. Fluorescence intensity plots of Drosophila nurse cell nuclei were generated from a single slice of 63× confocal images using ImageJ software (Abramoff et al., 2004). In brief, the image background was subtracted using ImageJ (50-pixel rolling-ball radius), then a line segment was drawn across one of the nurse cell nuclei and the plot profile function was used to generate a fluorescence intensity plot for each desired channel. The raw data files generated by these plot profiles were analyzed in Microsoft Excel, where each plot line was normalized to the peak value within that plot, creating intensity plots where the maximum observed fluorescence of a given line is represented by a value of 1.0; the relative fluorescence intensity. The NE was marked as the set of data points in which the wheat-germ agglutinin value was at least 0.5.
Fibroblast Polarization, Nuclear Localization, and Actin Retrograde Flow
Fibroblast polarization was quantified as previously described (Chang et al., 2013, Kutscheidt et al., 2014). In brief, a monolayer of NIH 3T3 fibroblasts was starved for 48 hr; later a scratch wound was performed with a sterilized pipette tip, and cells were treated with 10 μM lysophosphatidic acid (LPA). Two hours later cells were fixed and stained for tubulin, F-actin, and DNA, confocal snapshots were taken on a Nikon Eclipse Ti-E inverted microscope and images were uploaded and analyzed by custom-made software to define cell boundaries and relative position of the nucleus and centrosome with respect to the cell centroid (Chang et al., 2013). At least ten fields per condition per experiment were taken and analyzed.
For the study of retrograde movement of actin fibers, fibroblasts were plated on 35-mm imaging dishes (IBIDI) and cells were imaged for 2 hr directly after LPA addition under a 60× oil objective in a Nikon Eclipse Ti-E inverted microscope equipped with an SU-×1 Spinning-Disk Confocal and an Andor Ixon3 EM-CCD camera. Fiji software was used to quantify the speed of displacement of the fibers (Schindelin et al., 2012). In brief, time-lapse acquisition files were rotated so that the actin flow was reoriented in an up-down direction. Kymographs of a 10-μm-wide region of interest were then generated, and the coordinates of the fibers over time were annotated to calculate the displacement rate over time. At least two fibers per cell and ten cells per condition in three independent experiments were quantified.
Quantification of Filopodia and Area Covered by Stress Fibers
Filopodia number and length quantification was performed as described previously (Zanet et al., 2012, Jayo et al., 2012). Fascin-containing filopodia were quantified in live fibroblasts growing in complete medium expressing both GFP-fascin and mCherry-β-trefoil domains. Snapshots from a single confocal plane by a cell were taken from several cells and quantification was performed using Fiji software (Schindelin et al., 2012). Finger-like, thin, and straight projections protruding from the cell edge with increased GFP-fascin content were considered as filopodia and measured by drawing a straight line with Fiji from the cell edge to the tip. The “Measure” command was later used for the quantification of the length of each filopodia.
The area covered by F-actin fibers in fibroblasts was quantified from snapshots of time-lapse movies acquired at the same time range from fibroblasts expressing GFP-lifeact after 1 hr of 10 μM LPA stimulation. Using Fiji software, the area covered by F-actin fibers was drawn and measured, and the results were plotted as the percentage of total area covered by fibers.
Inverted Transwell Invasion Assay
For the invasion assays in Matrigel, a previously described method was used (Scott et al., 2011). For collagen invasion assays, the described method was partially adapted: 1.6 mg/mL collagen gel supplemented with 10 μg/mL fibronectin and 2% fetal bovine serum (FBS). More details are provided in Supplemental Experimental Procedures.
Nuclear Volume and Stiffness Measurements
MDA MB 231 cells were plated on 2D coverslips or embedded in 1.6 mg/mL collagen gels supplemented with 10 μg/mL fibronectin and 2% FBS. When the gels polymerized, Ham’s F-12 medium supplemented with 10% FBS was added on top and the cells were allowed to migrate for 24 hr. Gels were then fixed in 3.6% formaldehyde, permeabilized with 0.1% Triton X-100, and incubated with phalloidin-647 and SYTO-16 overnight at 4°C. Imaging was performed with a Nikon A1R Confocal system using a 40× water immersion objective. Reconstruction of 3D stacks was performed using NIS Elements software (Nikon Instruments), and nuclear volume was quantified using the “3D objects counter” function in Fiji (Schindelin et al., 2012). Quantification of nuclear stiffness was performed as previously described (Harris and Charras, 2011). More details are provided in Supplemental Experimental Procedures.
Microfluidic Device and Nuclear Deformation Experiments
The microfluidic device used in this study was manufactured as described previously (Malboubi et al., 2015). More details are provided in Supplemental Experimental Procedures.
Statistical Analysis
All experiments were repeated at least three times unless indicated otherwise. All statistical tests were performed using GraphPad Prism software (GraphPad). First, the outliers were removed following the ROUT method for outlier removal from non-linear regressions. When the populations followed a parametric distribution, ANOVA analysis followed by Dunnett's multiple comparison test or Student's t test for two-groups mean comparison was used. Otherwise, data not following normal distributions were analyzed with a non-parametric Kruskal-Wallis test followed by Dunn's multiple comparison test. Data are expressed as mean ± SEM. Significance was measured as ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001.
Author Contributions
All experiments were conceived by M.P. and A.J., with input from G.G.G., G.C., T.T., and S.A. Experiments were performed and analyzed primarily by A.J., with input on FRET/FLIM from M.P., on adhesion from K.P., on microchannel device experiments from M.M., and on AFM experiments from G.C. Nuclear movement assays in fibroblasts were performed by A.J. and S.A., with input from W.C. Nesprin-2 western blots, TAN lines localization, and nesprin-2 KD invasion assays were performed by S.A. C.G. and T.T. performed the Drosophila experiments. E.O.-Z. analyzed the tumor samples. M.P. and A.J. prepared the figures and wrote the manuscript.
Acknowledgments
We thank Brian Stramer and Andrei Luchici for helpful discussions about actin retrograde flow. We are very grateful to Danijela Vignjevic and John Marshall for providing the mouse colon carcinoma and MDA MB 231 xenograft samples, respectively. We also thank the Nikon Imaging Center@King's (John Harris, Dan Matthews, and Isma Ali) for help with microscopy. A.J. was funded by the Medical Research Council (MRC; MR/J000647/1). M.P. was funded by the Royal Society. M.M. and G.C. were supported by Wellcome Trust grant WT092825.
Published: August 22, 2016
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, four figures, one table, and two movies and can be found with this article online at http://dx.doi.org/10.1016/j.devcel.2016.07.021.
Supplemental Information
References
- Abramoff M.D., Magalhaes P.J., Ram S.J. Image processing with ImageJ. Biophotonics Int. 2004;11:36–42. [Google Scholar]
- Adams J.C. Roles of fascin in cell adhesion and motility. Curr. Opin. Cell Biol. 2004;16:590–596. doi: 10.1016/j.ceb.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Adams J.C. Fascin-1 as a biomarker and prospective therapeutic target in colorectal cancer. Expert Rev. Mol. Diagn. 2015;15:41–48. doi: 10.1586/14737159.2015.976557. [DOI] [PubMed] [Google Scholar]
- Adams J.C., Clelland J.D., Collett G.D., Matsumura F., Yamashiro S., Zhang L. Cell-matrix adhesions differentially regulate fascin phosphorylation. Mol. Biol. Cell. 1999;10:4177–4190. doi: 10.1091/mbc.10.12.4177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam S.G., Lovett D., Kim D.I., Roux K., Dickinson R.B., Lele T.P. The nucleus is an intracellular propagator of tensile forces in NIH 3T3 fibroblasts. J. Cell Sci. 2015;128:1901–1911. doi: 10.1242/jcs.161703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anilkumar N., Parsons M., Monk R., Ng T., Adams J.C. Interaction of fascin and protein kinase Calpha: a novel intersection in cell adhesion and motility. EMBO J. 2003;22:5390–5402. doi: 10.1093/emboj/cdg521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoku S., Zhu R., Kutscheidt S., Fackler O.T., Gundersen G.G. Reinforcing the LINC complex connection to actin filaments: the role of Fhod1 in tan line formation and nuclear movement. Cell Cycle. 2015;14:2200–2205. doi: 10.1080/15384101.2015.1053665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Autore F., Pfuhl M., Quan X., Williams A., Roberts R.G., Shanahan C.M., Fraternali F. Large-scale modelling of the divergent spectrin repeats in nesprins: giant modular proteins. PLoS One. 2013;8:E63633. doi: 10.1371/journal.pone.0063633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balzer E.M., Tong Z., Paul C.D., Hung W.C., Stroka K.M., Boggs A.E., Martin S.S., Konstantopoulos K. Physical confinement alters tumor cell adhesion and migration phenotypes. FASEB J. 2012;26:4045–4056. doi: 10.1096/fj.12-211441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang W., Folker E.S., Worman H.J., Gundersen G.G. Emerin organizes actin flow for nuclear movement and centrosome orientation in migrating fibroblasts. Mol. Biol. Cell. 2013;24:3869–3880. doi: 10.1091/mbc.E13-06-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang W., Worman H.J., Gundersen G.G. Accessorizing and anchoring the linc complex for multifunctionality. J. Cell Biol. 2015;208:11–22. doi: 10.1083/jcb.201409047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charras G., Sahai E. Physical influences of the extracellular environment on cell migration. Nat. Rev. Mol. Cell Biol. 2014;15:813–824. doi: 10.1038/nrm3897. [DOI] [PubMed] [Google Scholar]
- Chen L., Yang S., Jakoncic J., Zhang J.J., Huang X.Y. Migrastatin analogues target fascin to block tumour metastasis. Nature. 2010;464:1062–1066. doi: 10.1038/nature08978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condeelis J., Singer R.H., Segall J.E. The great escape: when cancer cells hijack the genes for chemotaxis and motility. Annu. Rev. Cell Dev. Biol. 2005;21:695–718. doi: 10.1146/annurev.cellbio.21.122303.120306. [DOI] [PubMed] [Google Scholar]
- Davidson P.M., Denais C., Bakshi M.C., Lammerding J. Nuclear deformability constitutes a rate-limiting step during cell migration in 3-D environments. Cell Mol. Bioeng. 2014;7:293–306. doi: 10.1007/s12195-014-0342-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkhatib N., Neu M.B., Zensen C., Schmoller K.M., Louvard D., Bausch A.R., Betz T., Vignjevic D.M. Fascin plays a role in stress fiber organization and focal adhesion disassembly. Curr. Biol. 2014;24:1492–1499. doi: 10.1016/j.cub.2014.05.023. [DOI] [PubMed] [Google Scholar]
- Friedl P., Alexander S. Cancer invasion and the microenvironment: plasticity and reciprocity. Cell. 2011;147:992–1009. doi: 10.1016/j.cell.2011.11.016. [DOI] [PubMed] [Google Scholar]
- Friedl P., Wolf K. Tumour-cell invasion and migration: diversity and escape mechanisms. Nat. Rev. Cancer. 2003;3:362–374. doi: 10.1038/nrc1075. [DOI] [PubMed] [Google Scholar]
- Gomes E.R., Jani S., Gundersen G.G. Nuclear movement regulated by Cdc42, mrck, myosin, and actin flow establishes Mtoc polarization in migrating cells. Cell. 2005;121:451–463. doi: 10.1016/j.cell.2005.02.022. [DOI] [PubMed] [Google Scholar]
- Groen C.M., Jayo A., Parsons M., Tootle T.L. Prostaglandins regulate nuclear localization of fascin and its function in nucleolar architecture. Mol. Biol. Cell. 2015;26:1901–1917. doi: 10.1091/mbc.E14-09-1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilluy C., Osborne L.D., Van Landeghem L., Sharek L., Superfine R., Garcia-Mata R., Burridge K. Isolated nuclei adapt to force and reveal a mechanotransduction pathway in the nucleus. Nat. Cell Biol. 2014;16:376–381. doi: 10.1038/ncb2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W., Giancotti F.G. Integrin signalling during tumour progression. Nat. Rev. Mol. Cell Biol. 2004;5:816–826. doi: 10.1038/nrm1490. [DOI] [PubMed] [Google Scholar]
- Harada T., Swift J., Irianto J., Shin J.W., Spinler K.R., Athirasala A., Diegmiller R., Dingal P.C., Ivanovska I.L., Discher D.E. Nuclear lamin stiffness is a barrier to 3D migration, but softness can limit survival. J. Cell Biol. 2014;204:669–682. doi: 10.1083/jcb.201308029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris A.R., Charras G.T. Experimental validation of atomic force microscopy-based cell elasticity measurements. Nanotechnology. 2011;22:345102. doi: 10.1088/0957-4484/22/34/345102. [DOI] [PubMed] [Google Scholar]
- Hashimoto Y., Skacel M., Adams J.C. Roles of fascin in human carcinoma motility and signaling: prospects for a novel biomarker? Int. J. Biochem. Cell Biol. 2005;37:1787–1804. doi: 10.1016/j.biocel.2005.05.004. [DOI] [PubMed] [Google Scholar]
- Hashimoto Y., Parsons M., Adams J.C. Dual actin-bundling and protein kinase C-binding activities of fascin regulate carcinoma cell migration downstream of rac and contribute to metastasis. Mol. Biol. Cell. 2007;18:4591–4602. doi: 10.1091/mbc.E07-02-0157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto Y., Kim D.J., Adams J.C. The roles of fascins in health and disease. J. Pathol. 2011;224:289–300. doi: 10.1002/path.2894. [DOI] [PubMed] [Google Scholar]
- Isermann P., Lammerding J. Nuclear mechanics and mechanotransduction in health and disease. Curr. Biol. 2013;23:R1113–R1121. doi: 10.1016/j.cub.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen S., Collins A., Yang C., Rebowski G., Svitkina T., Dominguez R. Mechanism of actin filament bundling by fascin. J. Biol. Chem. 2011;286:30087–30096. doi: 10.1074/jbc.M111.251439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayo A., Parsons M., Adams J.C. A novel rho-dependent pathway that drives interaction of Fascin-1 with P-Lin-11/Isl-1/Mec-3 kinase (Limk) 1/2 to promote fascin-1/actin binding and filopodia stability. BMC Biol. 2012;10:72. doi: 10.1186/1741-7007-10-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatau S.B., Hale C.M., Stewart-Hutchinson P.J., Patel M.S., Stewart C.L., Searson P.C., Hodzic D., Wirtz D. A perinuclear actin cap regulates nuclear shape. Proc. Natl. Acad. Sci. USA. 2009;106:19017–19022. doi: 10.1073/pnas.0908686106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M.Y., Oskarsson T., Acharyya S., Nguyen D.X., Zhang X.H., Norton L., Massague J. Tumor self-seeding by circulating cancer cells. Cell. 2009;139:1315–1326. doi: 10.1016/j.cell.2009.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutscheidt S., Zhu R., Antoku S., Luxton G.W., Stagljar I., Fackler O.T., Gundersen G.G. Fhod1 interaction with Nesprin-2g mediates tan line formation and nuclear movement. Nat. Cell Biol. 2014;16:708–715. doi: 10.1038/ncb2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammermann T., Bader B.L., Monkley S.J., Worbs T., Wedlich-Soldner R., Hirsch K., Keller M., Forster R., Critchley D.R., Fassler R., Sixt M. Rapid leukocyte migration by integrin-independent flowing and squeezing. Nature. 2008;453:51–55. doi: 10.1038/nature06887. [DOI] [PubMed] [Google Scholar]
- Li A., Dawson J.C., Forero-Vargas M., Spence H.J., Yu X., Konig I., Anderson K., Machesky L.M. The actin-bundling protein fascin stabilizes actin in invadopodia and potentiates protrusive invasion. Curr. Biol. 2010;20:339–345. doi: 10.1016/j.cub.2009.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Lovett D., Zhang Q., Neelam S., Kuchibhotla R.A., Zhu R., Gundersen G.G., Lele T.P., Dickinson R.B. Moving cell boundaries drive nuclear shaping during cell spreading. Biophys. J. 2015;109:670–686. doi: 10.1016/j.bpj.2015.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardi M.L., Jaalouk D.E., Shanahan C.M., Burke B., Roux K.J., Lammerding J. The interaction between nesprins and sun proteins at the nuclear envelope is critical for force transmission between the nucleus and cytoskeleton. J. Biol. Chem. 2011;286:26743–26753. doi: 10.1074/jbc.M111.233700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luxton G.W., Gomes E.R., Folker E.S., Vintinner E., Gundersen G.G. Linear arrays of nuclear envelope proteins harness retrograde actin flow for nuclear movement. Science. 2010;329:956–959. doi: 10.1126/science.1189072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malboubi M., Jayo A., Parsons M., Charras G. An open access microfluidic device for the study of the physical limits of cancer cell deformation during migration in confined environments. Microelectron. Eng. 2015;144:42–45. doi: 10.1016/j.mee.2015.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattila P.K., Lappalainen P. Filopodia: molecular architecture and cellular functions. Nat. Rev. Mol. Cell Biol. 2008;9:446–454. doi: 10.1038/nrm2406. [DOI] [PubMed] [Google Scholar]
- Meinke P., Schirmer E.C. Linc'ing form and function at the nuclear envelope. FEBS Lett. 2015;589:2514–2521. doi: 10.1016/j.febslet.2015.06.011. [DOI] [PubMed] [Google Scholar]
- Meinke P., Mattioli E., Haque F., Antoku S., Columbaro M., Straatman K.R., Worman H.J., Gundersen G.G., Lattanzi G., Wehnert M., Shackleton S. Muscular dystrophy-associated Sun1 and Sun2 variants disrupt nuclear-cytoskeletal connections and myonuclear organization. PLoS Genet. 2014;10:E1004605. doi: 10.1371/journal.pgen.1004605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono S., Yamakita Y., Yamashiro S., Matsudaira P.T., Gnarra J.R., Obinata T., Matsumura F. Identification of an actin binding region and a protein kinase C phosphorylation site on human fascin. J. Biol. Chem. 1997;272:2527–2533. doi: 10.1074/jbc.272.4.2527. [DOI] [PubMed] [Google Scholar]
- Rowat A.C., Jaalouk D.E., Zwerger M., Ung W.L., Eydelnant I.A., Olins D.E., Olins A.L., Herrmann H., Weitz D.A., Lammerding J. Nuclear envelope composition determines the ability of neutrophil-type cells to passage through micron-scale constrictions. J. Biol. Chem. 2013;288:8610–8618. doi: 10.1074/jbc.M112.441535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz-Moreno V., Gadea G., Ahn J., Paterson H., Marra P., Pinner S., Sahai E., Marshall C.J. Rac activation and inactivation control plasticity of tumor cell movement. Cell. 2008;135:510–523. doi: 10.1016/j.cell.2008.09.043. [DOI] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoumacher M., Goldman R.D., Louvard D., Vignjevic D.M. Actin, microtubules, and vimentin intermediate filaments cooperate for elongation of invadopodia. J. Cell Biol. 2010;189:541–556. doi: 10.1083/jcb.200909113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott R.W., Crighton D., Olson M.F. Modeling and imaging 3-dimensional collective cell invasion. J. Vis. Exp. 2011 doi: 10.3791/3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedeh R.S., Fedorov A.A., Fedorov E.V., Ono S., Matsumura F., Almo S.C., Bathe M. Structure, evolutionary conservation, and conformational dynamics of Homo sapiens Fascin-1, an F-Actin crosslinking protein. J. Mol. Biol. 2010;400:589–604. doi: 10.1016/j.jmb.2010.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift J., Discher D.E. The nuclear lamina is mechano-responsive to ECM elasticity in mature tissue. J. Cell Sci. 2014;127:3005–3015. doi: 10.1242/jcs.149203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan V.Y., Lewis S.J., Adams J.C., Martin R.M. Association of Fascin-1 with mortality, disease progression and metastasis in carcinomas: a systematic review and meta-analysis. BMC Med. 2013;11:52. doi: 10.1186/1741-7015-11-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Technau M., Roth S. The Drosophila Kash domain proteins Msp-300 and Klarsicht and the sun domain protein Klaroid have No essential function during oogenesis. Fly (Austin) 2008;2:82–91. doi: 10.4161/fly.6288. [DOI] [PubMed] [Google Scholar]
- Thomas D.G., Yenepalli A., Denais C.M., Rape A., Beach J.R., Wang Y.L., Schiemann W.P., Baskaran H., Lammerding J., Egelhoff T.T. Non-muscle myosin IIB is critical for nuclear translocation during 3D invasion. J. Cell Biol. 2015;210:583–594. doi: 10.1083/jcb.201502039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignjevic D., Kojima S., Aratyn Y., Danciu O., Svitkina T., Borisy G.G. Role of fascin in filopodial protrusion. J. Cell Biol. 2006;174:863–875. doi: 10.1083/jcb.200603013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf K., Mazo I., Leung H., Engelke K., Von Andrian U.H., Deryugina E.I., Strongin A.Y., Brocker E.B., Friedl P. Compensation mechanism in tumor cell migration: mesenchymal-amoeboid transition after blocking of pericellular proteolysis. J. Cell Biol. 2003;160:267–277. doi: 10.1083/jcb.200209006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf K., Te Lindert M., Krause M., Alexander S., Te Riet J., Willis A.L., Hoffman R.M., Figdor C.G., Weiss S.J., Friedl P. Physical limits of cell migration: control by ECM space and nuclear deformation and tuning by proteolysis and traction force. J. Cell Biol. 2013;201:1069–1084. doi: 10.1083/jcb.201210152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood W., Martin P. Structures in focus—filopodia. Int. J. Biochem. Cell Biol. 2002;34:726–730. doi: 10.1016/s1357-2725(01)00172-8. [DOI] [PubMed] [Google Scholar]
- Yang S., Huang F.K., Huang J., Chen S., Jakoncic J., Leo-Macias A., Diaz-Avalos R., Chen L., Zhang J.J., Huang X.Y. Molecular mechanism of fascin function in filopodial formation. J. Biol. Chem. 2013;288:274–284. doi: 10.1074/jbc.M112.427971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Starr D.A., Wu X., Parkhurst S.M., Zhuang Y., Xu T., Xu R., Han M. The Kash domain protein Msp-300 plays an essential role in nuclear anchoring during Drosophila oogenesis. Dev. Biol. 2006;289:336–345. doi: 10.1016/j.ydbio.2005.10.027. [DOI] [PubMed] [Google Scholar]
- Zanet J., Jayo A., Plaza S., Millard T., Parsons M., Stramer B. Fascin promotes filopodia formation independent of its role in actin bundling. J. Cell Biol. 2012;197:477–486. doi: 10.1083/jcb.201110135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Bethmann C., Worth N.F., Davies J.D., Wasner C., Feuer A., Ragnauth C.D., Yi Q., Mellad J.A., Warren D.T. Nesprin-1 and -2 are involved in the pathogenesis of Emery Dreifuss muscular dystrophy and are critical for nuclear envelope integrity. Hum. Mol. Genet. 2007;16:2816–2833. doi: 10.1093/hmg/ddm238. [DOI] [PubMed] [Google Scholar]
- Zhang J., Fonovic M., Suyama K., Bogyo M., Scott M.P. Rab35 controls actin bundling by recruiting Fascin as an effector protein. Science. 2009;325:1250–1254. doi: 10.1126/science.1174921. [DOI] [PubMed] [Google Scholar]
- Zwerger M., Ho C.Y., Lammerding J. Nuclear mechanics in disease. Annu. Rev. Biomed. Eng. 2011;13:397–428. doi: 10.1146/annurev-bioeng-071910-124736. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.