ABSTRACT
CsrA family RNA-binding proteins are widely distributed in bacteria and regulate gene expression at the posttranscriptional level. Pseudomonas aeruginosa has a canonical member of the CsrA family (RsmA) and a novel, structurally distinct variant (RsmF). To better understand RsmF binding properties, we performed parallel systematic evolution of ligands by exponential enrichment (SELEX) experiments for RsmA and RsmF. The initial target library consisted of 62-nucleotide (nt) RNA transcripts with central cores randomized at 15 sequential positions. Most targets selected by RsmA and RsmF were the expected size and shared a common consensus sequence (CANGGAYG) that was positioned in a hexaloop region of the stem-loop structure. RsmA and RsmF also selected for longer targets (≥96 nt) that were likely generated by rare PCR errors. Most of the long targets contained two consensus-binding sites. Representative short (single consensus site) and long (two consensus sites) targets were tested for RsmA and RsmF binding. Whereas RsmA bound the short targets with high affinity, RsmF was unable to bind the same targets. RsmA and RsmF both bound the long targets. Mutation of either consensus GGA site in the long targets reduced or eliminated RsmF binding, suggesting a requirement for two tandem binding sites. Conversely, RsmA bound long targets containing only a single GGA site with unaltered affinity. The RsmF requirement for two binding sites was confirmed with tssA1, an in vivo regulatory target of RsmA and RsmF. Our findings suggest that RsmF binding requires two GGA-containing sites, while RsmA binding requirements are less stringent.
IMPORTANCE The CsrA family of RNA-binding proteins is widely conserved in bacteria and plays important roles in the posttranscriptional regulation of protein synthesis. P. aeruginosa has two CsrA proteins, RsmA and RsmF. Although RsmA and RsmF share a few RNA targets, RsmF is unable to bind to other targets recognized by RsmA. The goal of the present study was to better understand the basis for differential binding by RsmF. Our data indicate that RsmF binding requires target RNAs with two consensus-binding sites, while RsmA recognizes targets with just a single binding site. This information should prove useful to future efforts to define the RsmF regulon and its contribution to P. aeruginosa physiology and virulence.
INTRODUCTION
RNA-binding proteins play an integral role in the posttranscriptional regulation of protein synthesis by altering translation initiation, mRNA stability, and/or RNA processing. The CsrA family of RNA-binding proteins regulates carbon metabolism, virulence factor production, and motility in a number of Gram-negative bacteria (1–5). CsrA proteins usually bind sites on target mRNAs that overlap the Shine-Dalgarno sequence to prevent translation initiation (6–8). Although considerable sequence variability exists between natural CsrA-binding sites, a common feature is a core GGA sequence that is usually presented in the loop portion of a stem-loop structure (9, 10). High-affinity interactions between CsrA and RNA targets have been analyzed by two powerful techniques. First, nuclear magnetic resonance (NMR) spectroscopy was used to show that RsmE, a CsrA homolog in Pseudomonas fluorescens, makes optimal contact with the sequence 5′-(A/U)CANGGANG(U/A), where N is any nucleotide (10). RsmE functions as a molecular clamp and gathers the ANGGAN core into a hexaloop, with the flanking nucleotides forming a 3-bp stem (10). The second approach, a systematic evolution of ligands by exponential enrichment (SELEX), was used to identify high-affinity RNA ligands of Escherichia coli CsrA (9). SELEX is a method used to select for RNA ligands that bind proteins of interest (11). Evolution of the ligands is based on repeated cycles of in vitro selection (12). The selection is driven toward optimized RNA targets that bind to the protein of interest with high affinity and specificity (12). The CsrA-binding consensus sequence was determined as RUACARGGAUGU (where R is either A or G); the underlined ACA and GGA motifs are 100% conserved, and the underlined GU is 98% conserved (9). All high-affinity ligands for CsrA had GGA motifs that were presented in the context of hexaloops (80%), tetraloops (15%), or octaloops (5%) with highly conserved stems, indicating the importance of both the primary nucleotide sequence and the secondary structure for high-affinity interactions (9).
The opportunistic pathogen Pseudomonas aeruginosa has two CsrA homologs, RsmA and RsmF (also called RsmN) (13, 14). RsmA and RsmF exist as ∼14-kDa homodimers in solution with two identical RNA-binding sites on either side of the dimer (13–15). Similar to other CsrA family members, the RsmA secondary structure is characterized by five highly conserved β-strands (β1 to β5) and a carboxyl-terminal α-helix. In contrast, the RsmF secondary structure consists of two amino-terminal β-strands (β1 and β2), an internal α-helix, and 3 carboxy-terminal β-strands (β3 to β5) (14, 16–18). While RsmA and RsmF differ in their secondary structure, they contain a conserved arginine residue (R44 in RsmA and R62 in RsmF) that is positioned at the carboxy-terminal end of β5 and is essential for RNA binding activity (13, 17, 19).
The presence of two CsrA proteins appears to be common in the pseudomonads (13, 14, 20). P. fluorescens RsmA and RsmE appear to be functionally redundant, wherein deletion of either results in intermediate phenotypes (i.e., production of biocontrol factors) and deletion of both is required for maximal regulatory effects (20). P. aeruginosa RsmA and RsmF do not appear to be redundant in the same way as seen in P. fluorescens. The RsmA regulon consists of >500 genes and includes genes required for production of pyocyanin, elastase, hydrogen cyanide, and type III and VI secretion systems (3, 21, 22). The regulatory role of RsmF is less clear, and the RsmF regulon has yet to be defined. Whereas many phenotypes are altered in an rsmA mutant, an rsmF mutant shows no alteration of the same phenotypes. For example, biofilm formation is increased in an rsmA mutant but is unchanged in an rsmF mutant relative to the parental strain (13). In an rsmA rsmF double mutant, however, biofilm formation is significantly elevated compared to the rsmA single mutant. RsmF function can also be demonstrated in complementation experiments wherein expression of either RsmA or RsmF in an rsmA rsmF mutant restores biofilm formation to wild-type (wt) levels. RsmA and RsmF share several binding targets in common, including the RsmY and RsmZ regulatory RNAs and the tssA1 leader region. RsmF binding affinities for these RNAs, however, is significantly lower than those seen for RsmA. Binding studies with short RNA oligonucleotides suggest that a putative core GGA sequence in the tssA1 leader region is required for binding by RsmA and RsmF (13). In other cases, RsmF is unable to bind to targets that RsmA interacts with, such as the pslA leader region (13), suggesting that RsmF has differential binding properties relative to RsmA.
In this study, we examine the differences in the RNA-binding properties of RsmA and RsmF using a SELEX approach. SELEX data show that RsmA and RsmF are remarkably similar in their preference for targets with a GGA sequence presented as a hexaloop in a stem-loop structure. Further analyses of the SELEX data suggested that RsmF requires two GGA binding sites, and this was confirmed experimentally with representative targets identified in the SELEX. Further, we utilized selective 2′-hydroxyl acylation analyzed by primer extension and mutational profiling (SHAPE-MaP) (23) to confirm that the untranslated leader sequence of tssA1, a known in vivo target of RsmF, contains two GGA stem-loop structures that are necessary for RsmF binding.
MATERIALS AND METHODS
In vitro selection of RNA ligands.
SELEX was performed as previously described (9, 24, 25). The double-stranded DNA (dsDNA) library was generated as follows. Primer 92978257 (see Table S1 in the supplemental material) (10 pmol), which contains a 15-nucleotide (nt) randomized region flanked on either side by constant regions (Fig. 1A), was used as the template in a PCR with primers 92978255 and 92978256 (240 pmol each) using the FailSafe PCR system (94°C for 1 min, 53°C for 1 min, and 72°C for 0.5 min for a total of 25 cycles). The PCR product was gel purified, and 5 μg was transcribed in vitro using the MEGAshortscript T7 transcription kit (Life Technologies) in the presence of 50 μCi/mmol [α-32P]ATP (PerkinElmer) (to facilitate gel purification) for 4 h at 37°C. The transcription reaction was treated with five units of Turbo DNase I (Life Technologies) for 15 min at 37°C to remove the dsDNA template, and an equal volume of gel loading buffer II (Ambion) was added to the reaction. The reaction mixture was gel purified (5% acrylamide/8 M urea) and quantified. Gel-purified RNA was renatured by heating to 85°C followed by slow cooling to 27°C. The radiolabeled RNA pool (round 0) was split into separate aliquots for enrichment by either RsmAHis6 or RsmFHis6. RsmAHis6 or RsmFHis6, purified by Ni2+ affinity chromatography as previously described (13), was combined with RNA in 500-μl binding reaction mixtures containing 1× electrophoretic mobility shift assay (EMSA) binding buffer (10 mM Tris-HCl [pH 7.5], 10 mM MgCl2, 100 mM KCl, 0.5 mg yeast tRNA, and 7.5% glycerol). Reaction mixtures were incubated at 37°C for 30 min. Ni-nitrilotriacetic acid (Ni-NTA)–agarose (50-μl packed beads) (Qiagen) was equilibrated with 1× binding buffer and was added to the reactions followed by incubation at 37°C for another 10 min. Binding reactions were agitated with a pipette every 2 min to achieve optimal binding of RsmAHis6/RsmFHis6 to the Ni-NTA resin. The reaction mixture was then washed three times with 100 μl of 1× binding buffer to remove unbound and weakly bound RNA ligands. Bound RNA was eluted from the Ni-NTA resin with 100 μl 1× binding buffer containing 2 M imidazole. The RNA was phenol-chloroform extracted and ethanol precipitated. The eluted RNA was converted into cDNA by incubating with 4 μM of the reverse primer (primer 92978256) at 65°C for 5 min and was then placed on ice for 2 min. Transcripts were reverse transcribed in the presence of two units of avian myeloblastosis virus (AMV) reverse transcriptase (Life Technologies) and 2 mM deoxynucleoside triphosphates (dNTPs) (Invitrogen) for 45 min at 42°C. Samples were heated to 70°C for 15 min to inactivate the enzyme. The reaction mixture was then used as the template in a PCR mixture containing 2 μM (each) of the first strand and reverse primers (primers 92978255 and 92978256, respectively), 2 mM dNTP, and 2 units Taq DNA polymerase using the parameters described above. The PCR products were used as the templates for in vitro transcription, and the pooled RNA was subjected to the next round of selection. RsmAHis6 was added at final concentrations of 500 nM in rounds 1 and 2, 250 nM in rounds 3 and 4, 125 nM in rounds 5 and 6, and 62.5 nM in rounds 7 and 8. RsmFHis6 was added at final concentrations of 500 nM in the first four rounds of SELEX, 350 nM in rounds 5 and 6, and 175 nM in rounds 7 and 8. In each round, the RNA was added at a concentration that was eight times greater than the concentration of the respective proteins.
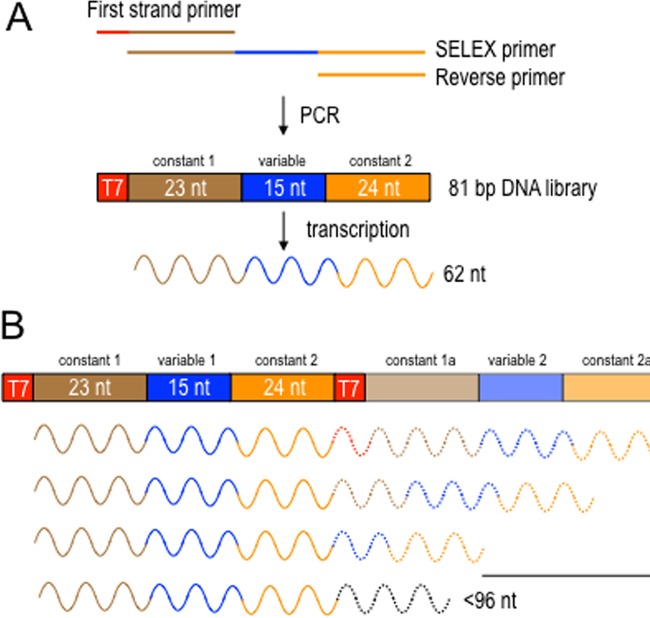
FIG 1.
(A) Diagram of the SELEX strategy showing the three primers used to generate the 81-bp dsDNA library by PCR. The first strand primer incorporated a promoter for T7 RNA polymerase. In vitro transcription yielded a 62-nt RNA library consisting of constant region 1 (23 nt), a 15-nt variable region, and constant region 2 (24 nt). The theoretical diversity of the library is ∼1 × 109 unique sequences. (B) General properties of long RNA targets selected by RsmF in later rounds of selection. All of the long targets contained the original 62 target sequence followed by portions of either the T7 promoter and/or constant region 1. Whereas targets that were <96 nt contained a single variable region, most of the targets that were ≥96 nt had two variable regions (variable region 1 and 2).
Ion torrent sample preparation, sequencing, and data processing.
PCR amplified cDNA pools from rounds 0, 2, 4, 6, and 8 for RsmA and RsmF were RNase A treated to remove residual RNA. Samples were heated to 65°C for 15 min to inactivate the RNase, loaded onto a reaction cleanup column (Qiagen), and eluted in 18 μl Tris-ethylenediaminetetraacetic acid (EDTA), pH 8.0. The eluted product was subjected to fusion SuperMix high-fidelity PCR (Life Technologies) to add unique bar codes for high-throughput sequencing. Fusion PCR was performed with 5 μM reverse (primer 114791840) and forward primers (primer 114205660 for round 0; primers 114345916, 114345918, 114345920, and 114345922 for RsmA rounds 2, 4, 6, and 8, respectively; and primers 114345917, 114345919, 114345921, and 114345923 for RsmF rounds 2, 4, 6, and 8, respectively) (see Table S1 in the supplemental material). The PCR parameters were 94°C for 3 min, 61°C for 0.5 min, and 68°C for 0.5 min for a total of 25 cycles. PCR products were separated on a 2% agarose gel, and bands corresponding to ∼128 bp were excised and gel purified. Samples were analyzed on the Agilent Bioanalyzer for assessment of size distribution and accurate quantification purposes. An equimolar pool of the amplicon libraries was prepared (∼26 pM) and submitted for Ion Torrent sequencing (University of Iowa, Iowa Institute of Human Genetics). Sequences that were obtained from Ion Torrent high-throughput sequencing were sorted into 9 pools based on the barcodes. Ion Torrent sequencing yielded 4,816,625 sequences. Sequences were trimmed with a sliding window of 10 and a Phred quality score of 15 using Trimmomatic 0.33 (26). Sequences of less than 50 nucleotides in length were discarded from subsequent analyses. The remaining sequences were clustered using USEARCH with a 98% identity cutoff and were reverse complemented (27). The constant and variable regions of the clustered sequences were analyzed using a script generated in R (version 3.2.2). Positional clustering of the variable regions and length analysis were performed using R (version 3.2.2), and secondary structures were examined using Mfold (28).
Electrophoretic mobility shift assays.
Gel-purified transcripts from SELEX rounds 0 to 8 were dephosphorylated and 5′-end-labeled using 3,000 μCi/mmol [γ-32P]ATP (PerkinElmer) and T4 polynucleotide kinase (NEB). Unincorporated [γ-32P]ATP was removed using NucAway spin columns (Life Technologies). Radiolabeled RNA was gel purified, eluted overnight, phenol-chloroform extracted, ethanol precipitated, and resuspended in TE (10 mM Tris-HCl [pH 8.0] and 1 mM EDTA) as previously described. Radiolabeled RNA was incubated with the indicated concentration of RsmAHis6 or RsmFHis6 at 37°C for 30 min and analyzed by native gel electrophoresis and phosphorimaging as previously described (13). Binding properties were determined with Prism 6.0e using the binding saturation equation for specific binding with a Hill slope. The apparent Keq represents the molar concentration of RsmA/RsmF required to shift 50% of an RNA probe. The apparent Keq values reported in the text are the average of at least three independent experiments.
The Gibson assembly method (29) was used to generate native and mutant DNA templates with the corresponding gBlocks (Integrated DNA Technologies, Coralville, IA) (see Table S2 in the supplemental material). gBlock adaptor sequences were removed, and the target sequence was amplified using specific primers (132895788 and 132895789). In vitro transcription was performed using the MEGAscript SP6 transcription kit (Life Technologies). RNA templates were end-labeled with [γ -32P]ATP as previously described (16).
SHAPE-MaP analysis.
RNA SHAPE-MaP analysis was performed as described previously (23). An RNA structure cassette containing the 5′ untranslated region (UTR) of tssA1 (TssA1-SHAPE) was synthesized as a gBlock DNA template (see Table S2 in the supplemental material) for in vitro transcription. The cassette carries a 5′ T7 RNA polymerase promoter (TAATACGACTCACTATAGGG) and the tssA1 UTR flanked by a stable UUCG tetraloop (GGCCTTCGGGCCAA) at the 5′ end and two tandem UUCG tetraloops (TCGATCCGGTTCGCCGGATCCAAATCGGGCTTCGGTCCGGTTC) at the 3′ end as previously described (30). RNA was generated using the HiScribe T7 high-yield RNA synthesis kit (NEB), purified using the RNeasy kit (Qiagen), and examined by gel electrophoresis for quality and length of product. Five hundred nanograms of RNA was incubated at 37°C for 5 min and then treated with 100 nM 1-methyl-7-nitroisatoic anhydride (1M7) for 5 min at 37°C. Denatured controls containing 1M7 and formamide along with negative-control reaction mixtures lacking 1M7 were performed in parallel. Following modification, RNA was isolated using G-50 spin columns (GE Healthcare). Total purified RNAs were incubated with 2 pmol of RNA structure cassette-specific primer (GAACCGGACCGAAGCCCG) at 65°C for 5 min, cooled to 4°C, then mixed with 10 mmol dNTP, 0.1 M dithiothreitol (DTT), 500 mM Tris (pH 8.0), 750 mM KCl, and 500 mM MnCl2, and incubated at 42°C for 2 min. Two hundred units of SuperScript II (Thermo Fisher Scientific) was then added, and the reaction mixture was incubated for 180 min, heat inactivated at 70°C for 15 min, and purified over a G-50 column. Use of MnCl2 rather than MgCl2 during reverse transcription allows for read-through at modified bases and incorporation of a noncomplementary nucleotide at modified positions (23). The resulting cDNAs were amplified in two rounds of PCR using Q5 DNA polymerase (NEB) according to the manufacturer recommendations. First-round PCR consisted of six cycles using construct-specific primers (Rnd1Fwd and Rnd1Rev) that incorporate terminal Illumina-specific sequences and random nucleotides to improve sequence cluster identification (see Table S1 in the supplemental material). Second-round PCR consisted of 19 cycles with primers Rnd2Fwd and Rnd2Rev to add additional Illumina-specific adapter sequences and barcodes for sample-specific identification (see Table S1). After each round of PCR, amplicons were purified using the PureLink Micro PCR cleanup kit (Life Technologies). Prior to sequencing, the experimental and control amplicons were pooled at equal concentrations, and the resulting libraries were cleaned using AMPure XP beads (Beckman Coulter) and quantified with a Bioanalyzer (Agilent Technologies) and Qubit (Life Technologies). Sequencing was performed on a MiSeq desktop sequencer (Illumina) using a MiSeq Reagent kit v3 (600-cycle). SHAPE reactivity values for each nucleotide were determined using the ShapeMapper pipeline (23). Structures were determined using RNAstructure to incorporate SHAPE reactivities into the folding free energy model to identify the structure that best agrees with the experimental data (31).
RESULTS
Target selection by RsmA and RsmF.
Our previous observations that RsmF does not bind some RNA targets bound by RsmA, and that RsmA and RsmF have different binding affinities for RNA targets shared in common (13), led to the hypothesis that RsmA and RsmF have distinct sequence-specific and/or structural requirements for high-affinity binding. A consensus CsrA-binding sequence was previously determined using a SELEX approach (9). We used the same strategy to identify RNA targets that specifically bind RsmA and RsmF and to test the hypothesis that RsmA and RsmF recognize distinct targets. The starting dsDNA library consisted of two constant regions flanking a 15-bp variable region (Fig. 1A). The variable region was randomized at each nucleotide position to generate a library consisting of ∼109 different sequences. A T7 promoter allowed for conversion of the DNA into a 62-nt single-stranded RNA (ssRNA) library. Aliquots (∼1012 transcripts) of the initial RNA library (round 0) were incubated with purified histidine-tagged RsmA or RsmF (∼109 molecules of dimers) and washed, and bound RNA was eluted and converted to cDNA to complete one cycle of selection. A total of eight selection rounds were performed.
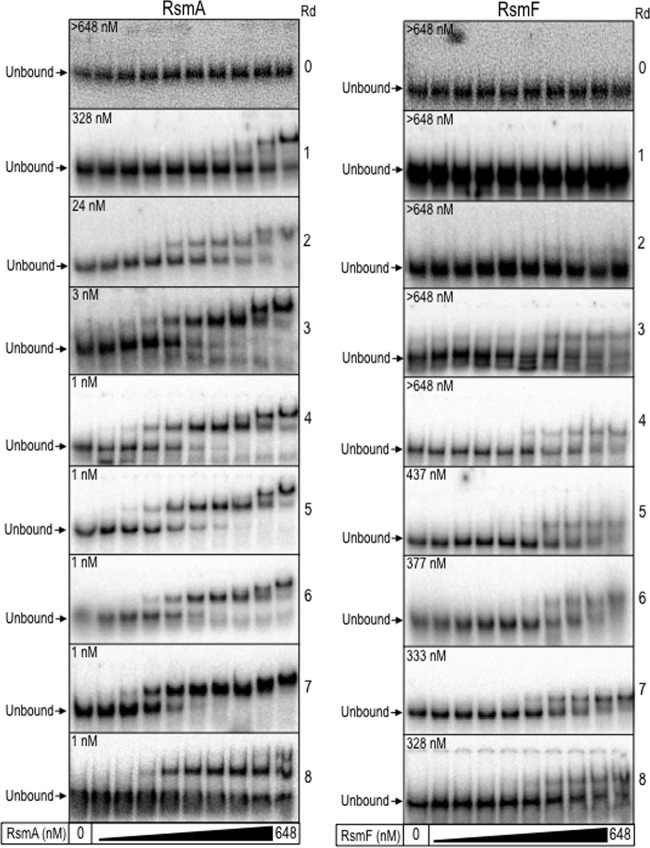
The progress of the SELEX enrichment was tracked using electrophoretic mobility shift assays (EMSAs). Aliquots of the RNA libraries from each round of selection were radiolabeled, incubated with purified RsmA or RsmF, and analyzed by native gel electrophoresis and phosphorimaging (Fig. 2). Neither RsmA nor RsmF demonstrated detectable binding to the initial RNA library (Fig. 2, round 0). After one round of selection, however, the apparent equilibrium binding constant (Keq) for RsmA was 328 nM, and this affinity increased through subsequent rounds of selection. To enrich for high-affinity targets, additional selective pressure was applied by reducing the concentration of RsmA in subsequent rounds (250 nM in rounds 3 and 4, 125 nM in rounds 5 and 6, and 62.5 nM in rounds 7 and 8). Binding affinity plateaued by the fourth round of selection at 1 nM.
FIG 2.
Binding experiments with the RNA libraries from each round (Rd) of selection. Samples of the initial RNA library (Rd 0) and from each subsequent round (Rd 1 to 8) of selection for either RsmA and RsmF were radiolabeled and incubated with purified RsmA or RsmF at the indicated concentrations (0, 0.1, 0.3, 0.9, 2.7, 8, 24, 72, 216, or 648 nM) for 30 min. Samples were immediately analyzed by native polyacrylamide gels and phosphorimaging. The position of the unbound probe is indicated. The apparent equilibrium binding constant (Keq) for RsmA and RsmF following each round of selection is shown on the left side of the images.
In contrast to our findings for RsmA, RsmF required five rounds of selection before a detectable increase in binding affinity (437 nM) was observed (Fig. 2). To enrich for high-affinity ligands, the selective pressure was increased by reducing the concentration of RsmF in rounds 5 and 6 (350 nM) and 7 and 8 (175 nM). EMSAs performed on the RNA library from rounds 7 and 8 showed a maximum binding affinity of 328 nM. This suggested that the enrichment had reached a plateau and that further rounds of selection were not required. Because affinity maturation was completed by round 6 for RsmA and RsmF, several of the analyses described below focus on sequence data from round 6.
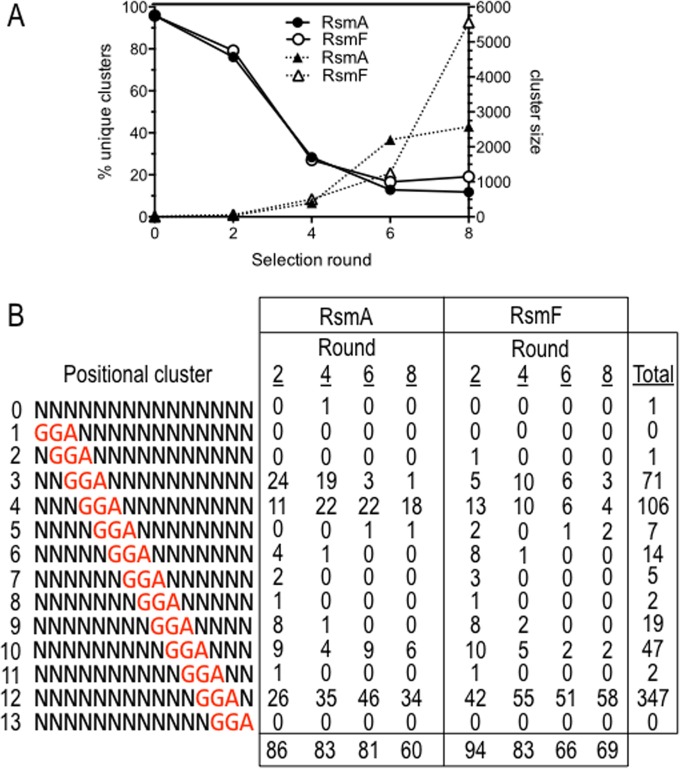
cDNA samples from the initial starting pool (round 0) and rounds 2, 4, 6, and 8 for RsmA and RsmF were barcoded, pooled, and subjected to high-throughput sequencing. A total of ∼4.8 million sequence reads were obtained (Table 1). The sequences were trimmed to remove the T7 promoter on the 5′ end and the adapter sequences on the 3′ end and were then examined for sequence length and quality. Sequences of <50 nt or with a Phred quality score of <15 were eliminated from the analyses, leaving a total of ∼2.3 million sequences (Table 1). The remaining sequences from each round of selection were clustered using a 98% sequence identity cutoff. As expected, the number of unique clusters decreased following each round of selection for RsmA and RsmF and showed an inverse correlation with cluster size (i.e., the number of times each cluster was represented) (Fig. 3A and Table 1). Both of these parameters further indicated enrichment for specific targets.
TABLE 1.
Summary of SELEX sequence data
| Protein and round | No. of total reads | No. of trimmed readsa | No. of unique clustersb | Top 100 cluster sizesc | No. of top 100 clusters with GGAd |
|---|---|---|---|---|---|
| 0 | 473,191 | 230,070 | 220,702 | 3–7 | 11 |
| RsmA 2 | 611,146 | 247,437 | 188,432 | 12–33 | 100 |
| RsmA 4 | 508,727 | 244,808 | 69,653 | 93–399 | 99 |
| RsmA 6 | 372,141 | 188,467 | 24,403 | 264–2,203 | 100 |
| RsmA 8 | 400,410 | 202,539 | 26,823 | 316–2,578 | 100 |
| RsmF 2 | 756,668 | 342,979 | 271,770 | 16–57 | 100 |
| RsmF 4 | 460,964 | 214,983 | 58,138 | 96–504 | 100 |
| RsmF 6 | 368,007 | 183,294 | 30,465 | 208–1,249 | 100 |
| RsmF 8 | 910,371 | 419,300 | 79,960 | 497–5,559 | 100 |
| Total | 4,816,625 | 2,273,877 |
Number of sequences remaining after removing reads that were <50 nt and trimming for sequence quality.
Number of unique sequence clusters for the trimmed reads using a ≥98% sequence identify cutoff.
Cluster size range (number of times each sequence cluster was represented) for the top 100 most frequent clusters.
Number of the top 100 most frequent clusters that contain a GGA sequence within the variable region. The frequency of GGA sequences within the variable region expected by chance in round 0 is 18.5%.
FIG 3.
Summary of the primary SELEX sequencing data. (A) Graph showing that the percentage of unique sequence clusters decreased with each round of selection for RsmA and RsmF and was inversely correlated with cluster size (the number of times each cluster was represented). (B) Diagram of the positional clustering strategy used for the analysis of targets that were ≤96 nt. Each target was clustered based on the location of the GGA sequence within the 15-nt variable region. One target lacking a GGA sequence was placed in cluster 0. The reported values are for targets of ≤96 nt found in the top 100 most frequent clusters for rounds 2, 4, 6, and 8 for RsmA and RsmF. Only targets with 15-nt variable regions were included in the analyses.
We next examined the top 100 clusters from each round of the selection. Most of the RNA targets in the initial pool (round 0) were 62 nt in length, and this trend remained the same through selection rounds 2 to 8 for RsmA and RsmF (see Fig. S1 in the supplemental material). Also apparent were longer RNA targets that increased in frequency with each successive round of selection (Table 2). Most of the targets that were <96 nt in length consisted of the original 62-nt sequence followed by a 3′ extension, with portions of constant regions 1 and/or 2 (Fig. 1B). Targets that were <96 nt generally lacked a second variable region, although a few exceptions were observed. By selection round 8, targets of ≥96 nt represented 36% and 30% of those selected by RsmA and RsmF, respectively (Table 2). Most targets of ≥96 nt consisted of a complete 62-nt target sequence at the 5′ end followed by either an incomplete or complete second target sequence (Fig. 1B). Targets of ≥96 nt were quite variable, but most fell into one of three general classes: (i) two complete RNA targets linked together by the T7 promoter sequence, (ii) two linked targets lacking the T7 sequence and partially truncated in constant region 1a, and (iii) two targets linked together lacking the T7 sequence and constant region 1a and partially truncated in variable region 2 (Fig. 1B).
TABLE 2.
Summary of target sizes for each round of selection
| Protein and round | % of targetsa of: |
No. of non-15-mer clustersb | |
|---|---|---|---|
| <96 nt | ≥96 nt | ||
| 0 | 87 | 0 | 13 |
| RsmA 2 | 86 | 5 | 9 |
| RsmA 4 | 83 | 1 | 16 |
| RsmA 6 | 81 | 3 | 16 |
| RsmA 8 | 60 | 36 | 4 |
| RsmF 2 | 94 | 0 | 6 |
| RsmF 4 | 83 | 7 | 10 |
| RsmF 6 | 66 | 27 | 7 |
| RsmF 8 | 69 | 30 | 1 |
Percentages of the targets with the indicated sequence lengths in the top 100 most frequent clusters, limited to targets with a 15-nt variable region.
Number of clusters in the top 100 with variable regions of 14 or 16 nt in length.
Analyses of the short (<96 nt) target sequences.
Previous studies found that a core GGA sequence is critical for RNA binding by CsrA, RsmA, and RsmF (9, 13). For this reason, each of the top 100 variable regions in targets of <96 nt were scanned for a GGA sequence. Each variable region selected by RsmA and RsmF in rounds 2, 4, 6, and 8 contained a GGA sequence, with the exception of one target in round 4 for RsmA (Table 1). In contrast, GGA sequences were present in only 11 of the top 100 targets from the initial library (round 0) prior to selection. Targets of <96 nt from each round of selection were further clustered based on the position in which the GGA sequence appeared within the 15-nt variable region (i.e., positional clusters) (Fig. 3B). By round 6, positional clusters 4 and 12 accounted for >80% of the targets selected by RsmA. RsmF also preferentially selected targets with the GGA at positions 3, 4, 10, and 12, with the strongest preference for positional cluster 12 (77% by round 6). Note that positional clustering was only applied to targets that were <96 nt in length and with 15-nt variable regions. As indicated in Table 2, 14- and 16-nt variable regions were also present in each round of the selection (Table 2). Aside from a single nucleotide insertion or deletion within the variable region, targets with 14- or 16-nt variable regions were indistinguishable from those with a 15-nt variable region and contained a GGA sequence that was usually presented in the loop portion of a stem-loop structure (discussed below).
In addition to the requirement for a GGA sequence, presentation of the GGA sequence is also important and is optimal for CsrA and RsmA when presented in the loop portion of a stem-loop structure (9, 10). To examine the structural context of the GGA sequences, predicted structures for targets of <96 nt from rounds 0 and 6 were determined using Mfold (28). Of the 11 targets from round 0 that contained a GGA sequence, only three (27%) of the predicted structures had the GGA sequence located in the loop of a stem-loop structure, and those GGA sequences were in positional clusters 7 or 9. GGA sequences at those positions were infrequent in the targets selected by RsmA and RsmF (Fig. 3B). In contrast, most of the structures with the lowest free energy from round 6 for RsmA (80/81 of the short targets) and RsmF (71/73) had the GGA sequence presented in the loop portion of stem-loop structures. In a few cases, it was the structure with the second lowest free energy rather than the lowest that had the GGA presented in a loop. In those cases, we used the second structure because the difference in free energy between the two was negligible. The predicted secondary structures for targets in positional clusters 3, 4, 10, and 12 for RsmA and RsmF were variable. Most of the structures presented the GGA sequence in the context of a hexaloop (68% and 65% for RsmA and RsmF, respectively). The next most common presentation was a tetraloop (29% and 28% for RsmA and RsmF, respectively). All structures with tetraloops had either an A-U or G-U base pair at the top of the stem, possibly allowing for flexibility in the presentation of the GGA sequence in the tetraloop and hexaloop conformations.
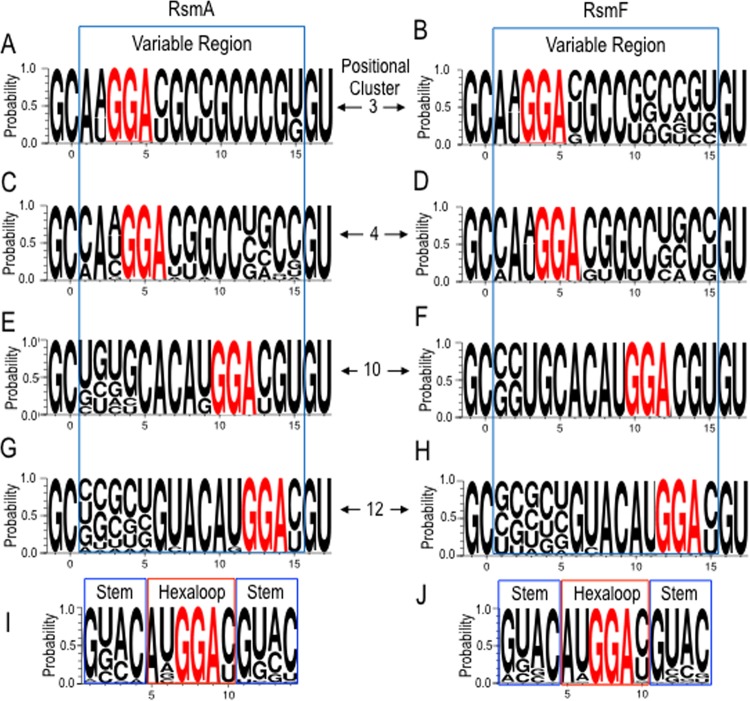
To survey the diversity surrounding the GGA sequences, the RNA targets in positional clusters 3, 4, 10, and 12 from round 6 for RsmA and RsmF were examined using WebLogo 3 (32). The most striking finding was the similarity in the sequences surrounding the GGA for both RsmA and RsmF, irrespective of positional clustering (Fig. 4A to H). The consensus sequences for RsmA and RsmF (5′-CANGGAYG, where N is any nucleotide, the GGA is 100% conserved, and Y is a either cytosine or uracil) were identical and consistent with the previously reported consensus sequences for E. coli CsrA (RUACARGGAUGU) and P. fluorescens RsmE (WCANGGANGN) (9, 10). Clusters 3 and 10 showed the most disparity between RsmA and RsmF, with more variability in cluster 3 at positions 10 to 15 for RsmF and in cluster 10 at positions 1 to 4 for RsmF (Fig. 4A versus B). Comparison of the positional clusters to one another and between RsmA and RsmF also revealed striking similarities. The common features are a 4-bp stem of variable sequence and a highly conserved hexaloop. This was confirmed by generating a WebLogo using all of the RNA sequences from positional clusters 3, 4, 10, and 12 for RsmA or RsmF (Fig. 4I and J). The consensus sequences showed remarkable complementarity in the stem positions (positions 1 to 14, 2 to 13, 3 to 14, and 4 to 11) and the consensus hexaloop sequence of ANGGAY.
FIG 4.
WebLogo3 illustrations of the RsmA and RsmF consensus binding sites. (A to H) Consensus sites for positional clusters 3, 4, 10, and 12 for RsmA and RsmF, limited to targets that were ≤96 nt. The 15-nt variable regions are shown within the blue boxes, and the conserved GGA sequences are highlighted in red font. The sequence outside the blue box is derived from the constant regions that flank the variable region. (I and J) WebLogo3 illustrations combining all of the sequences from positional clusters 3, 4, 10, and 12 for RsmA and RsmF. The GGA sequences are highlighted in red font, the conserved hexaloops are shown in red boxes, and nucleotides that base pair to form a 4-bp stem are shown in blue boxes.
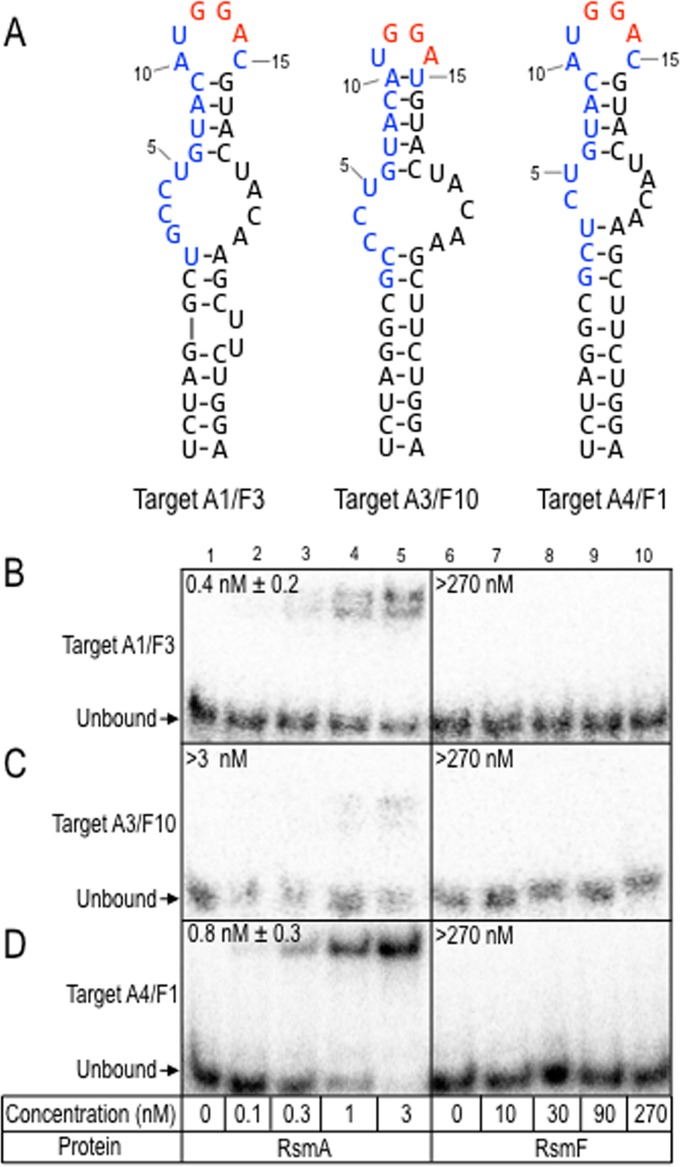
To examine RsmA and RsmF binding to short targets, three representative targets found in the top 10 list for RsmA and RsmF from round 6 were synthesized and used in EMSA experiments. The predicted secondary structures of the targets are shown in Fig. 5A. The radiolabeled RNA probes consisted of the 15-nt variable region flanked by 19 nt from constant region 1 and 24 nt from constant region 2. RsmA bound two of the three targets (A1/F3 and A4/F1) with high affinity (Keq < 1 nM), and weak binding to the third target (A3/F10) was also detected (Fig. 5B to D). Binding by RsmF was not detected for any of the probes, even when using high protein concentrations (720 nM) (Fig. 5B to D and data not shown). In summary, our analyses of the short RNA targets suggest that neither the primary sequence nor the secondary structure account for the differential binding properties of RsmA and RsmF (13).
FIG 5.
RsmA and RsmF binding to targets that were ≤96 nt. (A) Predicted secondary structures for targets A1/F3, A3/F10, and A4/F1, selected by RsmA and RsmF in round 6. Each target was in the top 10 list for RsmA and RsmF as indicated by the numeric designations. (B to D) Target RNAs were radiolabeled and incubated with the indicated concentrations of RsmA and RsmF for 30 min. Samples were analyzed by native gel electrophoresis and phosphorimaging. The positions of the unbound probes are indicated, and the apparent equilibrium binding constant is indicated in each panel.
Analyses of the long target sequences (≥96 nt).
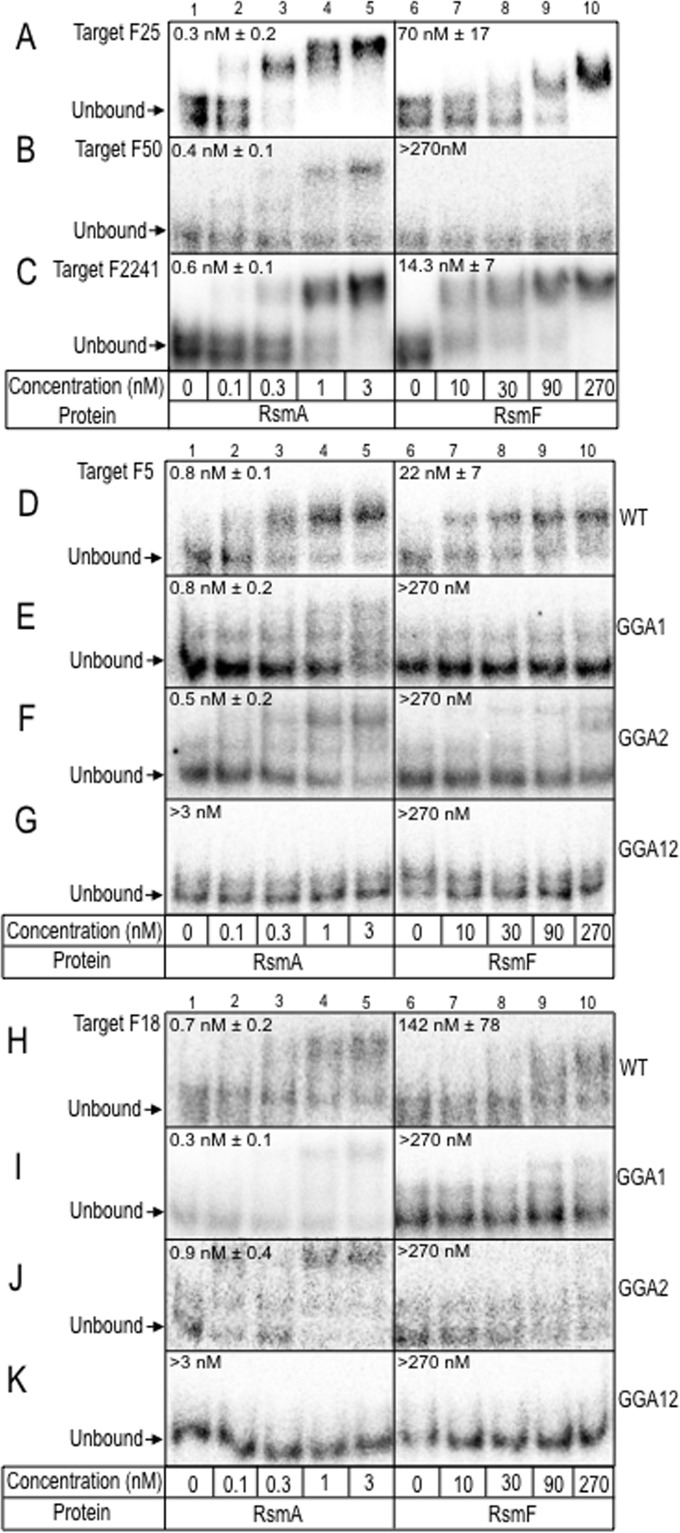
Although most of the top 100 targets selected by RsmF in round 6 were <96 nt, 27 of the targets were ≥96 nt (Table 2). Mfold predictions revealed that 26 of the long targets had two variable regions with stem-loop structures that presented GGA sequences in the context of either a tetraloop or a hexaloop. The GGA sequences were in positional clusters 3, 4, or 12, with the strongest preference for positional cluster 12 in both variable regions (see Fig. S2 and S3 in the supplemental material). The consensus sequences for positional clusters 3, 4, and 12 were similar to those seen for the short targets selected by RsmF (Fig. 4; see also Fig. S3).
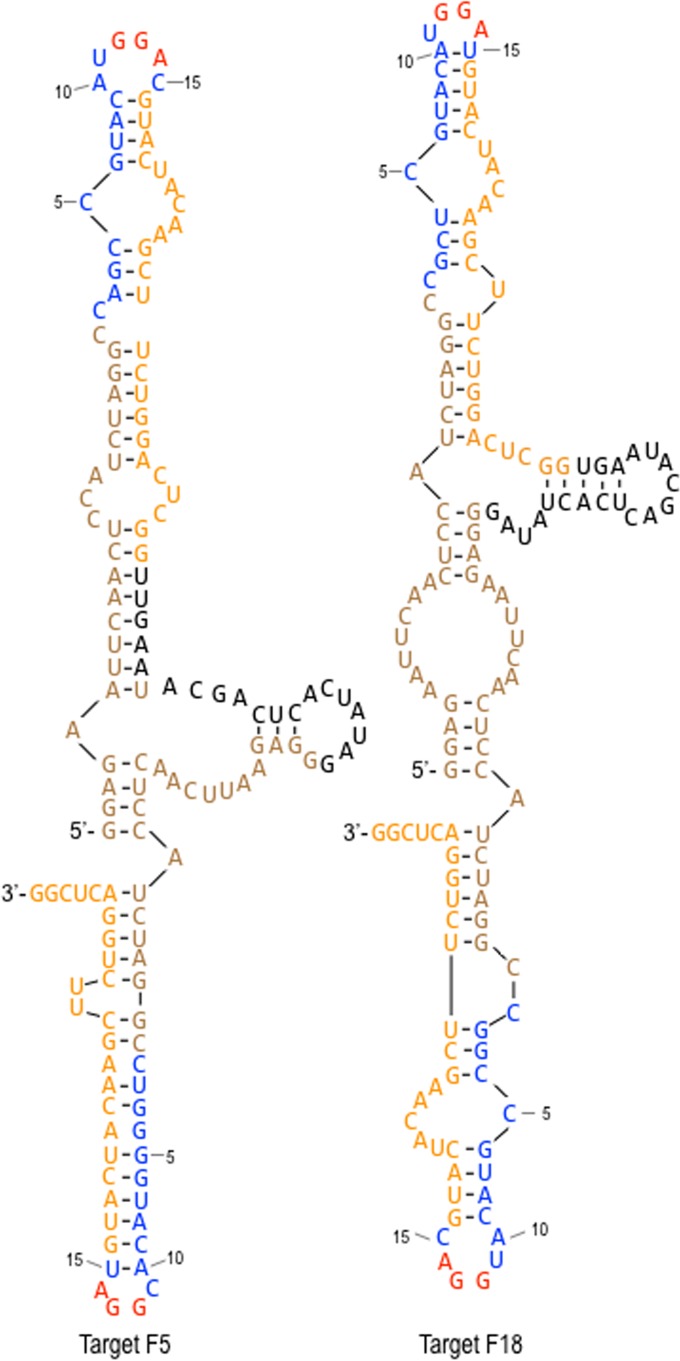
To test for RsmF binding, five long targets were synthesized and tested in EMSA experiments. Four of the targets were chosen from the top 100 list for RsmF (targets F5, F18, F25, and F50) (see Fig. S2 in the supplemental material). The fifth target, chosen from a less abundant cluster (F2241), was also included in the study. The secondary structure predictions for each target are shown in Fig. 6 and in Fig. S4 in the supplemental material. RsmA bound to each of the targets with high affinity (<1 nM), and RsmF bound to four of the five with affinities ranging from 14 to 142 nM (Fig. 7A to D and H). Binding of RsmF to target F50 was not detected. Relative to the other targets, which present the GGA sequences as either tetraloops or hexaloops, the first GGA in target F50 is in a 10-nt loop region (see Fig. S4). Although RsmF binding affinity was reduced relative to RsmA, the finding that RsmF bound to 4 of the 5 long targets while failing to bind short targets (Fig. 5) led to the hypothesis that optimal RsmF binding requires two sites and that RsmA is more tolerant of targets with only a single site. To test this idea, we mutagenized the GGA sequences in each of the long targets bound by RsmF and tested for binding. Mutation of either or both core GGA sequences in targets F5, F18, and F2241 eliminated or significantly reduced RsmF binding (>270 nM) (Fig. 7E, F, I, and J; see also Fig. S5F and G in the supplemental material). The findings for target F25 differed slightly in that a mutation of GGA1 had no effect on RsmF binding, while mutation of GGA2 or both GGA sites prevented RsmF binding (Fig. S5B to D). Unlike RsmF, RsmA bound all of the single GGA mutant targets with high affinity. Neither RsmA nor RsmF demonstrated detectable binding to the targets in which both GGA sequences were mutated (Fig. 7G and K; Fig. S5D and H). These findings are consistent with the hypothesis that detectable binding by RsmF requires two GGA-containing sites.
FIG 6.
Predicted secondary structures for targets F5 and F18, selected by RsmF in round 6. Constant regions 1 and 1a are shown in brown typeface, variable regions 1and 2 are blue, constant regions 2 and 2a are orange, the T7 promoter sequence is black, and the consensus GGA sequences are shown in red.
FIG 7.
Binding by RsmF requires two GGA sequences. Shown are EMSA experiments using probes for corresponding targets F25 (A), F50 (B), F2241 (C), F5 (D), and F18 (H). Mutant probes in which the first GGA sequence (GGA1), second sequence (GGA2), or both (GGA12) were changed to CCU in targets F5 and F18 are shown in panels E to G and I to K, respectively. Each probe was incubated with the indicated concentration of RsmA or RsmF for 30 min and then analyzed by native gel electrophoresis and phosphorimaging. The positions of the unbound probes are indicated by arrows, and the apparent equilibrium binding constant is indicated in each panel.
High-affinity binding by RsmF requires both binding sites in tssA1.
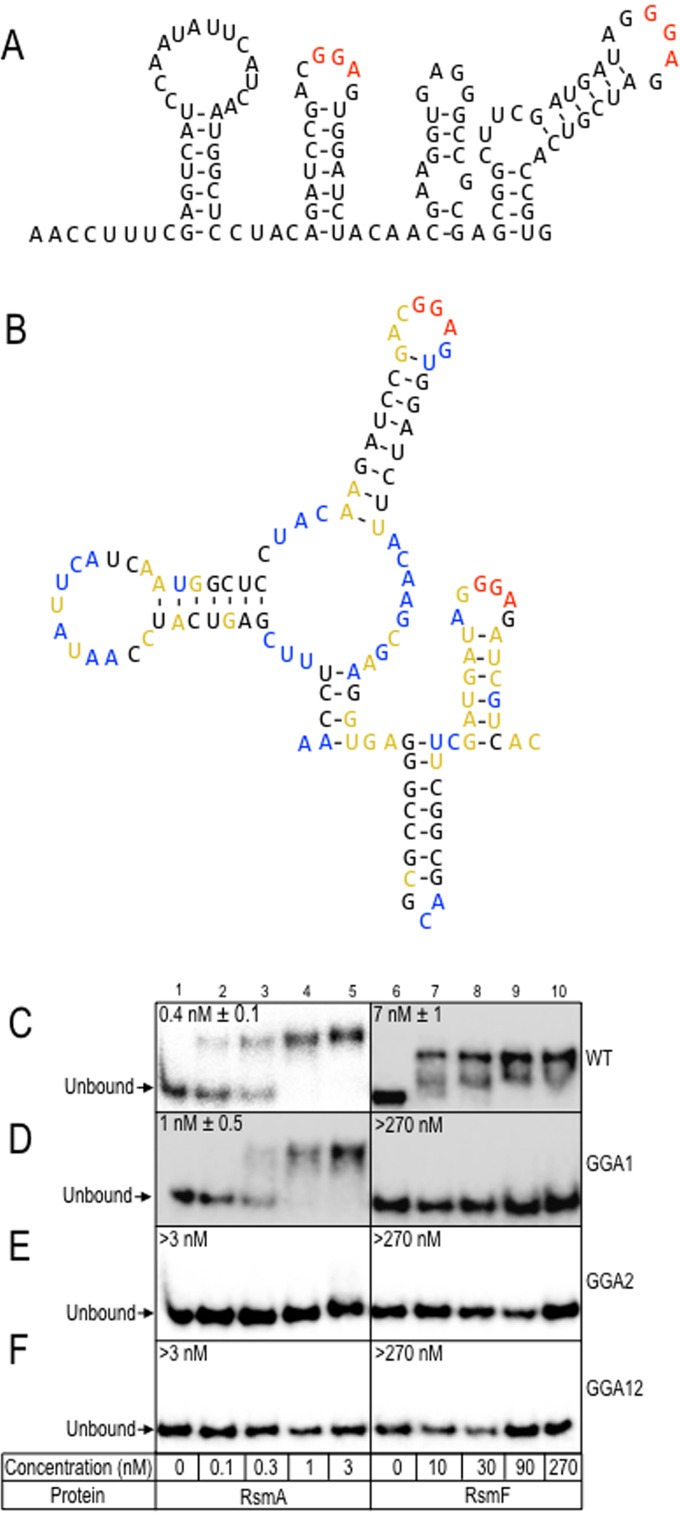
Only a few direct targets of RsmF have been identified to date. The highest affinity target known for RsmF is the leader sequence of tssA1, which encodes a component of a type VI secretion system (33). We previously reported that RsmA and RsmF bind a tssA1 RNA probe with affinities of 0.6 nM and 4 nM, respectively (13). Another study proposed that the tssA1 leader region contains two GGA sequences, both presented in stem-loop structures based on previous Mfold predictions (Fig. 8A) (21). The second GGA sequence overlaps the predicted ribosome-binding site and likely accounts for the inhibition of TssA1 translation upon RsmA or RsmF binding (13, 21). To experimentally verify the structure of the tssA1 leader region, we performed a SHAPE-MaP (selective 2′-hydroxyl acylation analyzed by primer extension and mutational profiling) analysis (23, 30, 34). SHAPE-MaP determines the reactivity of the 1M7 chemical probe for each nucleotide in the RNA (28). Higher reactivity indicates more flexibility, which is generally indicative of unpaired nucleotides. When we incorporate SHAPE-MaP data into the free energy of RNA folding, we predict a slightly different minimum free energy structure from the Mfold prediction (Fig. 8B). Both GGA-containing loop regions are highly reactive, indicating that they are unpaired and accessible for RsmA and RsmF binding. The SHAPE-MaP data also show that the nucleotides in the stem of the hairpin containing the second GGA sequence have medium (yellow) reactivity, indicating that this stem is flexible.
FIG 8.
RsmF binding to the tssA1 leader region requires both GGA sequences. Mfold (A) and SHAPE-MaP-directed (B) structures for the tssA1 leader region. SHAPE-MaP reactivity data are projected onto the structures with the coloring scheme: <0.45 is black, 0.45 < x < 0.8 is orange, and >0.8 is blue. Higher reactivity indicates lower base-pairing potential as well as higher flexibility. The consensus GGA sequences are shown in red. (C to F) EMSA experiments using a probe corresponding to the wt tssA1 leader region (C, wt) and probes with CCU substitutions in the first (D, GGA1), second (E, GGA2), or both (F, GGA12) GGA sequences. The binding affinities for RsmA and RsmF for each probe are provided on the left and right side of the images, respectively.
To determine whether RsmF binding requires both tssA1 binding sites, we generated a series of probes in which the first, second, or both GGA sequences were changed to CCT. Consistent with the previous study (13), RsmA and RsmF bound the native tssA1 probe with high affinity (Fig. 8C). Whereas RsmA also bound the probe with the GGA1 substitution, RsmF was unable to bind the same probe (Fig. 8D). Neither RsmA nor RsmF were able to bind probes with substitutions in the GGA2 site or the double mutants (GGA12) (Fig. 8E and F). Although we expected that RsmA might bind the GGA2 mutant, the overall data support our hypothesis that high-affinity binding by RsmF requires two binding sites.
In silico predictions of RsmA and RsmF binding sites.
Based on our observations that RsmF binding requires two binding sites, we used an in silico approach to search for candidate RsmF targets in the P. aeruginosa genome. To generate as many candidates as possible, we searched for GGA sequences rather than the full CANGGAYG consensus sequence. We first generated a library of 5′ untranslated regions from genes containing multiple GGA sequences based on transcriptome sequencing (RNA-seq) data from a previous experiment with P. aeruginosa strain PA14 grown at 37°C (35). Each of those sequences was folded (Mfold) to identify candidates with two or more GGA sequences presented in predicted stem-loop structures (28). Seven of the most promising candidates were selected based on predicted secondary structure, loop size, and matches to the CANGGAYG consensus (see Fig. S6 in the supplemental material) and were tested for binding by RsmA and RsmF. Despite our in silico predictions, RsmA bound to only four of the probes and RsmF bound to only one of the probes (see Fig. S7 in the supplemental material), which suggests that current in silico modeling is a poor predictor of RNA structural properties or that additional targeting properties remain to be identified.
DISCUSSION
The previous observations that (i) RsmA and RsmF bind with similar affinity to the tssA1 leader sequence, (ii) RsmF binds to the RsmY and RsmZ regulatory RNAs with reduced affinity relative to RsmA, and (iii) several RsmA targets are not recognized by RsmF led us to investigate the basis for differential binding (13). Our initial hypothesis was that discriminatory nucleotides at one or more positions surrounding the core GGA sequence resulted in more restrictive binding by RsmF. The SELEX data, however, indicate that RsmA and RsmF share a consensus binding site (5′-CANGGAYG). Our second hypothesis was that differences in the secondary structure result in differential binding, but examination of the predicted Mfold structures revealed no significant differences between targets selected by RsmA and RsmF. The finding that RsmF bound to targets with two predicted binding sites led to a third hypothesis that binding by RsmF requires two sites while a single site is sufficient for RsmA binding. This was experimentally verified for several long targets selected by RsmF and with the tssA1 leader sequence, wherein disruption of a single binding site disrupted RsmF binding while having little effect on RsmA binding. It is important to remember that RsmA and RsmF are both dimers in solution and that each dimer has two functional binding sites. Our finding that a single site supports strong binding by RsmA is consistent with previous work with mixed CsrA heterodimers composed of one RNA-binding competent monomer and one RNA binding-defective monomer. Both wt CsrA and the mixed heterodimer bound with similar affinity to several RNA targets containing two binding sites (36). It has been proposed that targets with multiple binding sites have the potential for cooperative binding interactions (9). Additionally, multiple sites, which are common on many CsrA targets, are thought to allow for the binding of a dimer to a high-affinity site, thereby increasing the local concentration to enhance binding to a second lower affinity site(s). The Hill coefficients for RsmA and RsmF binding to the tssA1 probe are 1.5 and 1.6, respectively (see Fig. S8 in the supplemental material), and are suggestive of cooperative binding. Our data suggest that this feature is likely essential for RsmF activity and that targets with multiple binding sites may be preferential targets of RsmF. Consistent with this, the only known in vivo targets of RsmF are RsmY, RsmZ, and tssA1, each of which contains ≥2 predicted binding sites.
The strong selection imposed in a SELEX experiment raises the possibility of identifying targets with binding affinities greater than optimal for the biological system. The binding affinities for two of the top RsmA targets were ∼1 nM (Fig. 5). Experimental data from this study and others have demonstrated binding of P. aeruginosa RsmA to 12 different sites on a total of 9 target RNAs (see Fig. S8 in the supplemental material). The available binding affinities for those targets are in the low to mid nanomolar range (0.3 to 55 nM). The affinity of RsmF for the long targets (33 to 108 nM) is also within the range of experimentally determined affinities for the tssA1 and PA14_16030 leader regions (4 and 80 nM, respectively). We conclude that the binding affinities for the targets identified by SELEX are generally within the normal physiological range for RsmA and RsmF targets in vitro.
SELEX experiments may also identify sequences that are not functional sites in the organism of interest. The consensus binding site derived from our studies (5′-CANGGAYG) is similar to the E. coli CsrA sequence (5′-CARGGAUG) determined by SELEX (9). Most characterized CsrA target sites resemble the SELEX-derived consensus (37). Likewise, each of the experimentally verified sites for RsmA and RsmF resembles the RsmA/RsmF consensus sequence, with matches ranging from 4 to 8 nt to consensus (see Fig. S9 in the supplemental material). These data indicate that RsmA and RsmF target sites will conform to the simple rule of matching the SELEX-derived consensus and that this will have some value in predicting target sites. Sequence alone, however, would seem to be a poor predictor of RsmA and RsmF targets. Our attempt to identify RsmF targets using a bioinformatic approach was met with limited success. By examining the 5′ untranslated leader region of genes expressed at 37°C (∼35% of the genome) plus 10 to 20 nt of the coding sequence, we identified 815 candidates with ≥2 GGA sequences. Secondary structure predictions narrowed the candidate field to 35, and 7 of the most promising candidates were selected for further analyses. Even from the pool of 35, however, we were forced to accept suboptimal candidates by allowing the GGA to be presented in loop regions ranging from 4 to 10 nt in length and in single-stranded regions (see Fig. S6 in the supplemental material). RsmF bound to only one of the seven candidates. Perhaps even more telling is that RsmA, with less restrictive binding properties, only bound to three of the candidates. Accurate predictions of RsmA and RsmF binding sites from primary sequence information remain imprecise. This is perhaps not surprising given that in vitro targets can deviate from consensus and that the GGA sequence is not always presented in an optimal hexaloop stem-loop structure. Although the SELEX experiments for RsmA/RsmF and CsrA indicate preferential binding to hexaloops, Lapouge et al. found that pentaloops are the preferred structure for RsmA/CsrA in vivo (38). The limitations of secondary structure prediction and unknown tertiary structural requirements are also factors that likely contributed to our low success rate. The broad application of methodologies such as SHAPE-MaP to more accurately determine genome-wide RNA structures may facilitate improvements in the prediction of in vivo RNA targets of RsmA and RsmF. Finally, it is possible that the in vitro binding assay lacks a factor(s) that facilitates high-affinity binding of RsmA and/or RsmF to some target RNAs.
Supplementary Material
ACKNOWLEDGMENTS
We thank Tony Romeo, Christopher Vakulskas, Teresa Przytycka, and Jan Hoinka for invaluable discussions throughout the course of this study. The 1M7 reagent was a gift from Kevin M. Weeks, Department of Chemistry, University of North Carolina at Chapel Hill.
This work was supported by the National Institutes of Health under grant number AI097264 to M.C.W. and T.L.Y. and grant numbers GM101237, HL111527, and HG008133 to A.L., by the Intramural Research Program of the National Institutes of Health, National Library of Medicine (TMP), and by a postdoctoral fellowship from the Cystic Fibrosis Foundation to C.J.G. K.H.S. was supported by NIH training grants T32GM082729 and 5T32AI007511-19.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00343-16.
REFERENCES
- 1.Liu MY, Yang H, Romeo T. 1995. The product of the pleiotropic Escherichia coli gene csrA modulates glycogen biosynthesis via effects on mRNA stability. J Bacteriol 177:2663–2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wei BL, Brun-Zinkernagel AM, Simecka JW, Pruss BM, Babitzke P, Romeo T. 2001. Positive regulation of motility and flhDC expression by the RNA-binding protein CsrA of Escherichia coli. Mol Microbiol 40:245–256. doi: 10.1046/j.1365-2958.2001.02380.x. [DOI] [PubMed] [Google Scholar]
- 3.Pessi G, Williams F, Hindle Z, Heurlier K, Holden MT, Camara M, Haas D, Williams P. 2001. The global posttranscriptional regulator RsmA modulates production of virulence determinants and N-acylhomoserine lactones in Pseudomonas aeruginosa. J Bacteriol 183:6676–6683. doi: 10.1128/JB.183.22.6676-6683.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sterzenbach T, Nguyen KT, Nuccio SP, Winter MG, Vakulskas CA, Clegg S, Romeo T, Baumler AJ. 2013. A novel CsrA titration mechanism regulates fimbrial gene expression in Salmonella Typhimurium. EMBO J 32:2872–2883. doi: 10.1038/emboj.2013.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Williams JW, Ritter AL, Stevens AM. 2012. CsrA modulates luxR transcript levels in Vibrio fischeri. FEMS Microbiol Lett 329:28–35. doi: 10.1111/j.1574-6968.2012.02499.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baker CS, Morozov I, Suzuki K, Romeo T, Babitzke P. 2002. CsrA regulates glycogen biosynthesis by preventing translation of glgC in Escherichia coli. Mol Microbiol 44:1599–1610. doi: 10.1046/j.1365-2958.2002.02982.x. [DOI] [PubMed] [Google Scholar]
- 7.Baker CS, Eory LA, Yakhnin H, Mercante J, Romeo T, Babitzke P. 2007. CsrA inhibits translation initiation of Escherichia coli hfq by binding to a single site overlapping the Shine-Dalgarno sequence. J Bacteriol 189:5472–5481. doi: 10.1128/JB.00529-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yakhnin H, Pandit P, Petty TJ, Baker CS, Romeo T, Babitzke P. 2007. CsrA of Bacillus subtilis regulates translation initiation of the gene encoding the flagellin protein (Hag) by blocking ribosome binding. Mol Microbiol 64:1605–1620. doi: 10.1111/j.1365-2958.2007.05765.x. [DOI] [PubMed] [Google Scholar]
- 9.Dubey AK, Baker CS, Romeo T, Babitzke P. 2005. RNA sequence and secondary structure participate in high-affinity CsrA-RNA interaction. RNA 11:1579–1587. doi: 10.1261/rna.2990205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schubert M, Lapouge K, Duss O, Oberstrass FC, Jelesarov I, Haas D, Allain FH. 2007. Molecular basis of messenger RNA recognition by the specific bacterial repressing clamp RsmA/CsrA. Nat Struct Mol Biol 14:807–813. doi: 10.1038/nsmb1285. [DOI] [PubMed] [Google Scholar]
- 11.Ellington AD, Szostak JW. 1990. In vitro selection of RNA molecules that bind specific ligands. Nature 346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 12.Goringer HU, Homann M, Lorger M. 2003. In vitro selection of high-affinity nucleic acid ligands to parasite target molecules. Int J Parasitol 33:1309–1317. doi: 10.1016/S0020-7519(03)00197-8. [DOI] [PubMed] [Google Scholar]
- 13.Marden JN, Diaz MR, Walton WG, Gode CJ, Betts L, Urbanowski ML, Redinbo MR, Yahr TL, Wolfgang MC. 2013. An unusual CsrA family member operates in series with RsmA to amplify posttranscriptional responses in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 110:15055–15060. doi: 10.1073/pnas.1307217110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morris ER, Hall G, Li C, Heeb S, Kulkarni RV, Lovelock L, Silistre H, Messina M, Camara M, Emsley J, Williams P, Searle MS. 2013. Structural rearrangement in an RsmA/CsrA ortholog of Pseudomonas aeruginosa creates a dimeric RNA-binding protein, RsmN. Structure 21:1659–1671. doi: 10.1016/j.str.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rife C, Schwarzenbacher R, McMullan D, Abdubek P, Ambing E, Axelrod H, Biorac T, Canaves JM, Chiu HJ, Deacon AM, DiDonato M, Elsliger MA, Godzik A, Grittini C, Grzechnik SK, Hale J, Hampton E, Han GW, Haugen J, Hornsby M, Jaroszewski L, Klock HE, Koesema E, Kreusch A, Kuhn P, Lesley SA, Miller MD, Moy K, Nigoghossian E, Paulsen J, Quijano K, Reyes R, Sims E, Spraggon G, Stevens RC, van den Bedem H, Velasquez J, Vincent J, White A, Wolf G, Xu Q, Hodgson KO, Wooley J, Wilson IA. 2005. Crystal structure of the global regulatory protein CsrA from Pseudomonas putida at 2.05 A resolution reveals a new fold. Proteins 61:449–453. doi: 10.1002/prot.20502. [DOI] [PubMed] [Google Scholar]
- 16.Mercante J, Suzuki K, Cheng X, Babitzke P, Romeo T. 2006. Comprehensive alanine-scanning mutagenesis of Escherichia coli CsrA defines two subdomains of critical functional importance. J Biol Chem 281:31832–31842. doi: 10.1074/jbc.M606057200. [DOI] [PubMed] [Google Scholar]
- 17.Heeb S, Kuehne SA, Bycroft M, Crivii S, Allen MD, Haas D, Camara M, Williams P. 2006. Functional analysis of the post-transcriptional regulator RsmA reveals a novel RNA-binding site. J Mol Biol 355:1026–1036. doi: 10.1016/j.jmb.2005.11.045. [DOI] [PubMed] [Google Scholar]
- 18.Gutierrez P, Li Y, Osborne MJ, Pomerantseva E, Liu Q, Gehring K. 2005. Solution structure of the carbon storage regulator protein CsrA from Escherichia coli. J Bacteriol 187:3496–3501. doi: 10.1128/JB.187.10.3496-3501.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duss O, Michel E, Diarra dit Konte N, Schubert M, Allain FH. 2014. Molecular basis for the wide range of affinity found in Csr/Rsm protein-RNA recognition. Nucleic Acids Res 42:5332–5346. doi: 10.1093/nar/gku141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reimmann C, Valverde C, Kay E, Haas D. 2005. Posttranscriptional repression of GacS/GacA-controlled genes by the RNA-binding protein RsmE acting together with RsmA in the biocontrol strain Pseudomonas fluorescens CHA0. J Bacteriol 187:276–285. doi: 10.1128/JB.187.1.276-285.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brencic A, Lory S. 2009. Determination of the regulon and identification of novel mRNA targets of Pseudomonas aeruginosa RsmA. Mol Microbiol 72:612–632. doi: 10.1111/j.1365-2958.2009.06670.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burrowes E, Baysse C, Adams C, O'Gara F. 2006. Influence of the regulatory protein RsmA on cellular functions in Pseudomonas aeruginosa PAO1, as revealed by transcriptome analysis. Microbiology 152:405–418. doi: 10.1099/mic.0.28324-0. [DOI] [PubMed] [Google Scholar]
- 23.Siegfried NA, Busan S, Rice GM, Nelson JA, Weeks KM. 2014. RNA motif discovery by SHAPE and mutational profiling (SHAPE-MaP). Nat Methods 11:959–965. doi: 10.1038/nmeth.3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ulrich H, Magdesian MH, Alves MJ, Colli W. 2002. In vitro selection of RNA aptamers that bind to cell adhesion receptors of Trypanosoma cruzi and inhibit cell invasion. J Biol Chem 277:20756–20762. doi: 10.1074/jbc.M111859200. [DOI] [PubMed] [Google Scholar]
- 25.Vo NV, Oh JW, Lai MM. 2003. Identification of RNA ligands that bind hepatitis C virus polymerase selectively and inhibit its RNA synthesis from the natural viral RNA templates. Virology 307:301–316. doi: 10.1016/S0042-6822(02)00095-8. [DOI] [PubMed] [Google Scholar]
- 26.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 28.Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA III, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 30.Wilkinson KA, Merino EJ, Weeks KM. 2006. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat Protoc 1:1610–1616. doi: 10.1038/nprot.2006.249. [DOI] [PubMed] [Google Scholar]
- 31.Deigan KE, Li TW, Mathews DH, Weeks KM. 2009. Accurate SHAPE-directed RNA structure determination. Proc Natl Acad Sci U S A 106:97–102. doi: 10.1073/pnas.0806929106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crooks GE, Hon G, Chandonia JM, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res 14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hood RD, Singh P, Hsu F, Guvener T, Carl MA, Trinidad RR, Silverman JM, Ohlson BB, Hicks KG, Plemel RL, Li M, Schwarz S, Wang WY, Merz AJ, Goodlett DR, Mougous JD. 2010. A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe 7:25–37. doi: 10.1016/j.chom.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hajdin CE, Bellaousov S, Huggins W, Leonard CW, Mathews DH, Weeks KM. 2013. Accurate SHAPE-directed RNA secondary structure modeling, including pseudoknots. Proc Natl Acad Sci U S A 110:5498–5503. doi: 10.1073/pnas.1219988110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wurtzel O, Yoder-Himes DR, Han K, Dandekar AA, Edelheit S, Greenberg EP, Sorek R, Lory S. 2012. The single-nucleotide resolution transcriptome of Pseudomonas aeruginosa grown in body temperature. PLoS Pathog 8:e1002945. doi: 10.1371/journal.ppat.1002945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mercante J, Edwards AN, Dubey AK, Babitzke P, Romeo T. 2009. Molecular geometry of CsrA (RsmA) binding to RNA and its implications for regulated expression. J Mol Biol 392:511–528. doi: 10.1016/j.jmb.2009.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vakulskas CA, Potts AH, Babitzke P, Ahmer BM, Romeo T. 2015. Regulation of bacterial virulence by Csr (Rsm) systems. Microbiol Mol Biol Rev 79:193–224. doi: 10.1128/MMBR.00052-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lapouge K, Sineva E, Lindell M, Starke K, Baker CS, Babitzke P, Haas D. 2007. Mechanism of hcnA mRNA recognition in the Gac/Rsm signal transduction pathway of Pseudomonas fluorescens. Mol Microbiol 66:341–356. doi: 10.1111/j.1365-2958.2007.05909.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.