Abstract
The oligonucleotide therapeutics field has seen remarkable progress over the last few years with the approval of the first antisense drug and with promising developments in late stage clinical trials using siRNA or splice switching oligonucleotides. However, effective delivery of oligonucleotides to their intracellular sites of action remains a major issue. This review will describe the biological basis of oligonucleotide delivery including the nature of various tissue barriers and the mechanisms of cellular uptake and intracellular trafficking of oligonucleotides. It will then examine a variety of current approaches for enhancing the delivery of oligonucleotides. This includes molecular scale targeted ligand-oligonucleotide conjugates, lipid- and polymer-based nanoparticles, antibody conjugates and small molecules that improve oligonucleotide delivery. The merits and liabilities of these approaches will be discussed in the context of the underlying basic biology.
AN OVERVIEW OF OLIGONUCLEOTIDE THERAPEUTICS
The initial advent of antisense and siRNA oligonucleotides sparked high hopes for their eventual use in treatment of disease. However, these early expectations remained largely unfulfilled as first generation oligonucleotides failed to meet therapeutic end points in a number of clinical trials. After a period of disappointment, the field of oligonucleotide therapeutics has now been re-invigorated (1). This is due to the convergence of several developments including improved chemistries, better understanding of the basic biology of oligonucleotides, more sophisticated delivery systems and most importantly, increasing success in the clinic. The 2013 approval of the first major antisense drug, Kynamro® (2), an inhibitor of apolipoprotein B expression, was accompanied by promising clinical trials involving siRNA (3) and splice switching oligonucleotides (SSOs) (4). More recently, a number of clinical trials utilizing various types of oligonucleotides have reported impressive results. Some examples might include a use of a receptor-targeted siRNA conjugate (5), strong effects on liver diseases using antisense with novel chemical modifications (6,7), anti-cancer effects with a miRNA (8) and treatment of a neurodegenerative disease via intrathecal administration of a SSO (9). More detailed summaries of selected current clinical studies are provided in several recent reviews (10–13).
Despite these advances at the clinical level, effective delivery of oligonucleotides in vivo remains a major challenge, especially at extra-hepatic sites (13–15). Various strategies are being pursued including chemical modification of the oligonucleotide itself, use of various lipid or polymeric nanocarriers, linking oligonucleotides to receptor targeting agents such as carbohydrates, peptides or aptamers, and use of small molecules to enhance oligonucleotide effectiveness. The intent of the current article is to provide a broad but analytic review of the oligonucleotide delivery area. The emphasis will be on basic biological aspects rather than recent clinical developments. There are an enormous number of publications in this area, far too many to be cited in their entirety. Thus the focus in this review will be on reports that stand out because of their novelty, or that provide important mechanistic information, or that display significant translational potential. This article will also convey the author's personal view on the future evolution of the oligonucleotide delivery area.
BASIC INFORMATION UNDERLYING OLIGONUCLEOTIDE THERAPEUTICS
The scope of the oligonucleotide therapeutics field has expanded substantially over the last few years as additional types of nucleic acids are used and as new targets are addressed. One of the most exciting developments is the realization that thousands of non-coding RNAs play important roles in cellular function (16) and that these entities can be readily manipulated using oligonucleotides (17). A continuing thrust in the field is the pursuit of clinical problems that are not easily addressed with small molecule drugs. Thus there has been emphasis on relatively rare disorders for which no current therapy exists. The various therapeutic approaches currently under investigation involve several types of nucleic acids with different chemistries and mechanisms of action; therefore it seems worthwhile to briefly review some basic aspects of oligonucleotide biology and chemistry.
Basic mechanisms of oligonucleotide actions
Classic single stranded antisense oligonucleotides (ASOs) primarily act in the nucleus by selectively cleaving pre-mRNAs having complementary sites via an RNase H dependent mechanism (18). Although ASOs can also act by translation arrest, they are currently primarily used as ‘gapmers’, having a central region that supports RNase H activity flanked by chemically modified ends that increase affinity and reduce susceptibility to nucleases (19). SSOs are a form of ASO; however they are fully modified so as to ablate RNase H activity and allow interaction with nuclear pre-mRNA during the splicing process. SSOs can be designed to bind to 5′ or 3′ splice junctions or to exonic splicing enhancer or silencer sites. In doing so they can modify splicing in various ways such as promoting alternative use of exons, exon exclusion or exon inclusion (20). SSOs are very flexible tools and are seeing increasing use in therapeutic approaches (21).
RNA interference (RNAi) is a fundamental endogenous mechanism for control of gene expression (22). It can involve selective message degradation, translation arrest or modulation of transcription (23). Both endogenous miRNAs and chemically synthesized externally administered siRNAs utilize Argonaute-containing RISC complexes to regulate gene expression (24,25). With siRNA, selective cleavage of mRNA in the cytosol involves Argonaute 2-containing complexes and requires essentially complete complementarity between the siRNA ‘guide’ strand and the target, usually within the coding region of the message. Because of their selectivity, siRNAs have seen widespread use in the laboratory and there is great interest in their potential therapeutic applications (26). With miRNA, partial complementarity, often in 3′-untranslated regions, leads to translation arrest followed by message degradation; this involves Argonaute proteins and largely takes place in cytoplasmic P-bodies. As mentioned above, miRNA can also regulate transcription within the nucleus by utilizing other forms of RISC complexes (27). Since miRNA recognition involves only partial complementarity, a single miRNA can influence expression of multiple mRNAs. This lack of selectivity can be a problem, but it may also be an advantage in that it can provide coordinate regulation of an entire set of genes. ASOs can act as miRNA antagonists (antagomirs) thus potentially increasing expression of miRNA-regulated genes.
Observations from the ENCODE project indicate that non-coding RNAs (ncRNAs) account for up to 75% of the transcripts from the human genome. There is a bewildering variety of short and long ncRNAs and understanding of their biological functions is still at an early stage (28). However, in many cases non-coding RNAs are involved in negative regulation of gene expression (17). Thus ASOs complementary to a ncRNA sequence can act as antagonists and promote upregulation of expression of genes regulated by the ncRNA. While attempts to therapeutically exploit ncRNAs are just beginning, there is a great deal of interest in the potential of this approach (29).
Another emerging thrust involves possible therapeutic use of chemically modified mRNA. In vitro transcribed mRNAs incorporating modified bases can effectively express proteins in vivo while having reduced effects on the innate immune system (30). In effect this serves as a transient form of gene therapy. This technology may be particularly useful in the context of stem cell therapies (31).
Thus a variety of nucleic acids are now being considered as potential agents for disease therapy. However, there are inevitably problems associated with any therapeutic approach. For antisense and siRNA off-target actions due to partial complementarity remain a concern, although chemical modifications can be helpful in this regard. Further, the extent of this type of problem is easily evaluated using contemporary methods for quantitating mRNA expression such as ‘gene chips’ or RNA-Seq. A more complex issue involves interaction with the innate immune system (32). Exogenous nucleic acids can trigger inflammatory responses via interactions with pattern recognition receptors including membrane-bound Toll-like receptors (TLRs) or cytosolic RIG-I family receptors (33). While undesired effects on innate immunity are a major problem for use of ASOs and siRNAs in therapeutics, the converse aspect is that oligonucleotides can be used to modulate the innate immune system in useful ways by acting as agonists or antagonists of TLRs or RIG-I (34–37). Other problems for oligonucleotides include potentially undesirable interactions with blood components (38), or with intracellular proteins (39), and rapid clearance via the kidney (40).
Chemical modifications of oligonucleotides
Recent advances in oligonucleotide therapeutics have heavily depended on progress in the medicinal chemistry of these molecules. A number of excellent reviews provide a comprehensive account of oligonucleotide chemistry (41–43); thus this section is simply a brief recapitulation designed to set the stage for discussion of the delivery of the various types of oligonucleotides.
Phosphorothioates
The phosphorothioate (PS) backbone modification has been the keystone for contemporary work on ASOs and SSOs (44). Although it creates a modest reduction in binding affinity, in compensation it provides two important advantages. First, it improves stability to nucleases in the blood and tissues. Second, it promotes protein binding and thus supports interactions with albumin and other blood proteins thereby retarding renal clearance. A disadvantage is that there are significant toxicities associated with the protein binding capabilities of PS oligonucleotides (44). The PS modification is fully consistent with RNase H activity.
Neutral backbones
The phosphorodiamidate morpholino oligomer (PMO) and peptide nucleic acid (PNA) modifications provide neutral backbones and high resistance to nucleases; however, they do not support RNase H activity. Thus PMOs, and to a lesser degree PNAs, have primarily seen use as SSOs (45).
2′ modifications
The most widely used alterations at the 2′ sugar position are the 2′-O-Me and 2′-O-(2-methoxyethyl) (MOE) modifications. Both promote an A-form or RNA-like conformation and considerably increase binding affinity to RNA, as well as providing enhanced nuclease resistance. Oligonucleotides fully modified at the 2′ position do not support RNase H activity and thus can be used as SSOs. However, RNase H dependent antisense effects can be achieved by use of ‘gapmers’ that contain a central unmodified section of about seven residues flanked by 2′ modified regions. Kynamro®, the first FDA approved ASO, is a MOE gapmer with a PS backbone. Modification of the 2′ position is also widely used in siRNA with 2′-O-Me and 2′–F being the most common. An important aspect for siRNA is that 2′ modifications can reduce both immunostimulatory effects (33) and off target effects (26).
Bridged rings
The locked nucleic acid (LNA) (46) chemistry as well as constrained ethyl (cEt) and tricyclo-DNA (tc-DNA) modifications involve bridging of the sugar ring. They each promote an RNA-like structure, display nuclease resistance and most importantly, provide dramatic increases in binding affinity. They do not support RNase H activity, but can be used effectively in antisense gapmers or as SSOs.
Novel approaches
Recently several highly novel approaches to oligonucleotide chemistry have been developed. A strategy pursued by Dowdy et al. entailed a complete redesign of the synthesis of siRNA so as to reversibly mask the negative charges of the phosphate backbone thus creating neutral siRNAs (47). Although this did not in itself allow increased delivery to cells, it enhanced binding to serum protein thus reducing renal clearance. Further, the neutral siRNAs could be effectively delivered to tissues by conjugation with a targeting ligand. A development of far reaching significance is the advent of XNAs, polymers formed from building blocks not found in nature that mimic many of the properties of RNA and DNA (48). Although this technology has yet to find therapeutic application, it clearly opens up many exciting possibilities.
CHALLENGES FOR NUCLEIC ACID DELIVERY
The key problem for oligonucleotide-based therapeutics is to deliver the active oligonucleotide to its site of action in the cytosol or nucleus of cells within tissues. There are really two parts to this problem. The first is to convey the oligonucleotide to the tissue of therapeutic interest while minimizing exposure of other tissues. The second is to convey the oligonucleotide to the right intracellular compartment. The delivery problem can be usefully considered in terms of barriers to movement of oligonucleotides within the body. The relative importance of the various barriers will depend on the chemical and physical properties of the oligonucleotide therapeutic being employed. For example, the biodistribution of antisense or siRNA oligonucleotides when used as individual molecules will obviously be quite different from that attained when some type of nanoparticle carrier is used.
Tissue barriers to delivery
The first challenge concerns getting the oligonucleotide to the tissue of therapeutic interest. In this section we will consider several barriers that influence oligonucleotide access to tissue sites, as schematically illustrated in Figure 1.
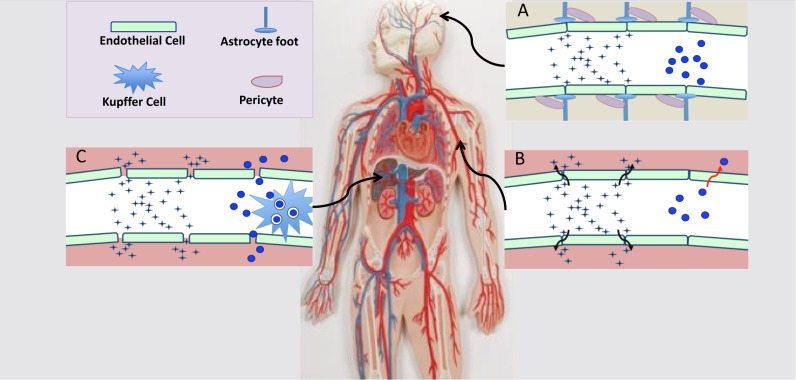
Figure 1.
Tissue barriers to oligonucleotide delivery. Barriers for blood to parenchyma transfer are depicted. The star-shaped forms represent ‘free’ oligonucleotides or molecular scale oligonucleotide conjugates. The blue circles represent oligonucleotides incorporated in nanoparticles. Tissue parenchyma is represented as pink or tan (brain) coloration. (A) Blood brain barrier. The tightly apposed endothelial cells as well as pericytes and astrocyte processes present an essentially impenetrable barrier for both free oligonucleotides and oligonucleotides in nanoparticles. (B) Blood tissue barrier. In many tissues oligonucleotides can readily cross the endothelium by diffusion through paracellular routes. Permeation of nanoparticles is much more limited and may take place via transcytosis. (C) Blood liver barrier. The fenestrated endothelium in liver and spleen is easily permeated by both free oligonucleotides and nanoparticles. However, the liver kupffer cells avidly take up nanoparticles.
The vascular endothelial barrier
In most tissues the capillary lumen is surrounded by a layer of endothelial cells that tightly abut upon each other and are joined together by VE-cadherin containing adherence junctions and by occludin and claudin-containg tight junctions, thus forming a barrier between blood and the parenchymal space (49,50). Molecules in the blood can be transported across the endothelial barrier by two routes. The first is paracellular transport that occurs through the junctions between cells and is limited to molecules of ∼6 nm diameter or less. The second is caveolar-mediated transcytosis that carries albumin and other large proteins across the endothelium within vesicles of about 70 nm. Both forms of transport are tightly regulated by various signaling systems. In most tissues neither transport system is capable of efficiently conveying typical ∼100 nm nanoparticles. However, in some tissues, such as liver and spleen, there are gaps or fenestrations between the endothelial cells, thus allowing egress of larger macromolecules and particles. Endothelial permeability is also increased in sites of inflammation and in some tumors. This last is a basis of the ‘Enhanced Permeation Retention’ (EPR) effect that has evoked much interest among proponents of nanoparticle-based drug delivery for cancer therapy. The concept is that the increased leakiness of tumor vasculature will allow nanoparticles to selectively accumulate at these sites (51). While this is clearly true for a number of rapidly growing xenograft tumors, not all xenografts display a strong EPR effect, and the extent of the effect in human tumors is rather unclear (52,53). Thus there are concerns regarding reliance on the EPR effect as a delivery strategy. In summary, the vascular endothelium allows ready passage of molecules the size of individual oligonucleotides into many tissues, but limits the passage of nanoparticles, except in certain sites such as the liver.
The reticuloendothelial system (RES)
The mononuclear phagocytes of the reticuloendothelial system (RES) provide a key aspect of host defense through their ability to engulf and inactivate pathogens (54). Their highly developed phagocytic capacity also allows them to internalize a wide variety of particulate materials including denatured proteins, apoptotic cell fragments and man-made nanoparticles. The kupffer cells of the liver sinusoids, as well as splenic macrophages, play an especially prominent role in the clearance of nanoparticles from the blood and much effort has been devoted to finding ways to evade uptake by these cells. However, this has been only partially successful despite many attempts to modify nanoparticle surfaces with polyethylene glycol (PEG) or other inert polymers (55,56). Thus the administration of oligonucleotides incorporated into liposomes or other nanoparticles will usually result in a large fraction of the material being taken up by the cells of the RES, particularly the kupffer cells (57). Additionally, the RES also plays an important role in uptake and clearance of individual ‘free’ oligonucleotides. Thus mononuclear phagocytes express a number of cell surface receptors, including integrins and scavenger receptors, which can potentially be involved in uptake (19,58,59). The role of scavenger receptors in uptake of free PS oligonucleotides in vivo is somewhat controversial (60). Nonetheless, such receptors have clearly been implicated in the uptake of morpholino oligonucleotides conjugated with cell penetrating peptides (CPPs) (61). In summary, the phagocytic cells of the RES are important modulators of the biodistribution of both ‘free’ and nanoparticulate oligonucleotides (62).
Renal excretion and effects on pharmacokinetics and biodistribution
The kidney plays an important role in the pharmacokinetics and biodistribution of oligonucleotides. Typically molecules with sizes of 3–6 nm or less can be ultrafiltered by the kidney (63); many types of oligonucleotide fall in this range and thus can be rapidly excreted by the renal route. PS ASOs bind to plasma proteins thus slowing their renal clearance and permitting broad distribution to tissues, with accumulation to the highest levels in liver and kidney (64). However, the kidney is also the primary route of excretion of phosphorothioates although this is a relatively slow process and mainly involves nuclease degradation products. In contrast, siRNA and uncharged oligonucleotides do not bind extensively to plasma proteins (40,65). Thus they are cleared by the kidney much more readily than phosphorothioates and tend to accumulate at lower levels in tissues. For siRNAs, the liver and kidney are the major sites of accumulation (66). Uncharged morpholino oligonucleotides are rapidly cleared by the kidney largely as intact molecules and display lower levels of tissue accumulation than phosphorothioates (65); the kidney and the liver are the primary tissues of distribution for these molecules. Thus renal clearance plays an important role in the pharmacokinetics and biodistribution of essentially all types of ‘free’ oligonucleotides.
The blood brain barrier and therapy of CNS diseases
An exciting and somewhat surprising recent development is the effective use of oligonucleotides for therapy of diseases of the central nervous system (CNS), despite the difficulties in accessing this compartment (67,68). The blood brain barrier (BBB) is comprised of tightly linked endothelial cells supported by a network of pericytes and astrocyte processes and is impervious to molecules as small as sucrose (67). The BBB is also largely impervious to oligonucleotides; as discussed below, there have been many attempts to deliver oligonucleotides across this barrier, but these have met with only modest success. Consequently, the most promising results in addressing CNS diseases have come through direct administration of oligonucleotides, usually by intrathecal injection. When administered by this route oligonucleotides distribute broadly in the CNS and are taken up by both neurons and glial cells (68). A variety of intractable diseases have been approached in this manner at both the pre-clinical and clinical trial levels. For example, chemically modified ASOs have been used to treat models of Huntington's disease (69) as well as familial forms of Alzheimers and ALS (70). Correction of the defect in spinal muscular atrophy involves the use of SSOs (71) and has progressed from animal experiments to phase II clinical trials (72). A particularly impressive recent study in a mouse model of Angelman syndrome used ASOs to reduce levels of a lncRNA resulting in ‘un-silencing’ of the key gene in this disease (73).
Obviously systemic administration is preferable to direct CNS administration and thus there have been many attempts to convey oligonucleotides across the BBB. Perhaps the most promising involve conjugates of PMOs with CPPs (74). There have been several reports of CPP-PMO conjugates reaching the brain (67,75). However, there remain concerns about the possible systemic and CNS toxicities of the polycationic CPPs. Another interesting development is a recent report that systemically administered tricyclic SSOs had an effect in the brain (76). Although the tricyclic modification provides increased affinity it is unclear why these molecules should cross the BBB while other oligonucleotides with similar backbones do not. It has been suggested that the tricyclos aggregate to form nanoparticles, however there are also problems with nanoparticle delivery to the brain. Thus there have been many studies using various nanoparticles seeking to deliver drugs, peptides or oligonucleotides across the BBB (77), but usually with limited success. One potentially interesting approach is to link nanoparticles to transferrin receptor ligands or to anti-receptor antibodies, thus making use of a transferrin receptor-mediated transcytotic route across the vascular endothelium (78–80). However, as yet there are no reports of functional in vivo delivery of oligonucleotides by this approach. Another interesting strategy involves use of a rabies virus peptide to target siRNA nanoparticles to neurons (81,82). However, since the receptors for this peptide are on neurons rather than brain capillary endothelium it is unclear how this would help to traverse the BBB. A further consideration is that even if nanoparticles cross the brain endothelium their relatively large size will restrict their diffusion through the extracellular matrix of brain parenchyma (83) whereas ‘free’ oligonucleotides readily spread throughout the brain. Thus while there are a number of reports in the literature purporting to achieve delivery across the BBB with nanoparticles, it is important to ask whether the BBB was intact in these studies or was it comprised by infection, cancer, inflammation or the toxic properties of the delivery vehicle itself. In summary, systemic delivery of oligonucleotides to the CNS remains a challenge that is largely unresolved.
Receptors and cell-selective targeting
There is increasing interest in ‘targeting’ oligonucleotides to specific cell types within the body. Perhaps the best way to do this is to conjugate the oligonucleotide (or its nanocarrier) to a ligand that interacts selectively with a cell surface receptor. Ideally, one would like to utilize a receptor that is expressed only in a single tissue, that is abundant, that rapidly and extensively internalizes, and for which high affinity ligands are readily available. Obviously, no receptor fully meets this ideal. However, there are many instances where receptor mediated targeting can greatly assist oligonucleotide delivery. Experience with targeting of antibodies and nanoparticles suggests that the key beneficial effect of targeting relates primarily to increased uptake at the cellular level rather than to overall changes in biodistribution (84,85). The paragraphs below briefly describe some of the basic characteristics of several important receptor families, emphasizing the aspects that are relevant to targeted delivery. A later section of the review will discuss studies that use these receptors to target oligonucleotides to specific cells.
Integrins
The integrins comprise a family of heterodimeric cell surface receptors that are differentially expressed on a variety of cell types. The 18 α and 8 β chains give rise to 24 distinct integrins in mammals. Integrins serve both as structural proteins and as components of the signal transduction machinery (86,87). Thus integrins link the cytoskeleton to large extracellular matrix proteins such as fibronectin and laminin. They also directly generate intracellular signals themselves, primarily through focal adhesion kinase (88). As well they can modulate other signaling processes, including the MAP Kinase pathway (89). Integrins are expressed at relatively high levels, typically in the range of hundreds of thousand of copies per cell (90). Integrins are actively internalized by clathrin-dependent and independent endocytotic mechanisms and usually recycle to the cell surface via Rab4- or Rab-11 mediated trafficking pathways (91). While many integrins are rather ubiquitously expressed, there are several examples of tissue or disease state selective expression including alphaIIbbeta3 on platelets, beta2 integrins in leukocytes and alphavbeta3 expression in angiogenic endothelia and in certain tumors (92,93). This last has engendered a great deal of interest in using alphavbeta3 selective ligands for tumor targeting (94). In summary, integrins offer many potential advantages for targeting including relatively high expression levels, rapid recycling and the availability of well-defined peptide and small molecule ligands.
GPCRs
The ∼800 G protein-coupled receptors (GPCRs) comprise the largest receptor family in the human genome (95). The signaling activity of GPCRs is tightly linked with their endocytosis and intracellular trafficking (96). Internalization via clathrin coated pits desensitizes the receptor and reduces signaling via ‘classical’ second messengers such as cyclic adenosine monophosphate (AMP), while activating new signaling pathways involving β-arrestin and c-Src (97). GPCRs often have distinct distributions in tissues or in disease states. For example, overexpression of the gastrin releasing peptide receptor has been observed in a number of cancers and this has been exploited therapeutically (98). There is a huge stockpile of highly selective GPCR ligands since such agents account for about 40% of all clinically utilized drugs (http://www.iuphar-db.org/index.jsp). Thus GPCRs offer some advantages for targeting, particularly in terms of differential tissue expression and ligand availability. However, there are also disadvantages. One is that these receptors are often expressed at relatively low levels compared to other receptor families, typically between 103 and 104 copies per cell (99). Another is that, in most cases, efficient internalization takes place only when the receptor is presented with an agonist ligand but not an antagonist.
RTKs
The human receptor tyrosine kinase (RTK) family is comprised of 58 members grouped into multiple subfamilies. The basic mechanism of activation of RTKs involves binding of the specific growth factor causing receptor oligomerization thus activating the tyrosine kinase and triggering cell signaling (100). The intracellular trafficking of RTKs is a key aspect of their function (101); the EGFR is well studied in this regard and can serve as an example for other receptors. After ligand binding, the EGFR enters early endosomes and is subsequently trafficked to late endosomes (LEs) and then to lysosomes, where both ligand and receptor are degraded thus terminating signaling (100). Differential expression of certain RTKs is observed in various tissues or disease states including over-expression of HER2 in some forms of breast cancer (102), overexpression of Trk family members in neuronal tissues (103), and enhanced expression of VEGFR2 in vascular endothelial cells (104). The expression of RTKs can vary widely, ranging from 103 to 106 copies per cell (105,106). The endogenous ligands for RTKs are all relatively large polypeptides and thus are not ideally suited for delivery approaches. However, there are many high affinity monoclonal antibodies for RTK external domains (107) that can be used as targeting reagents, converted to Fab fragments, or reconfigured as scFv reagents (108). In summary, the RTKs offer a mixed picture for targeting purposes. An advantage is that they are often highly expressed. However, this is offset by the fact that, upon internalization, the receptor is largely degraded rather than recycling to the cell surface. Another disadvantage is the lack of relatively small ligands that can be readily coupled to oligonucleotides or nanocarriers.
TLRs
The 10 members of the TLR family in humans primarily respond to ligands that contain pathogen associated molecular patterns derived from bacterial cell walls or membranes and from bacterial or viral nucleic acids (109,110). The TLRs are comprised of an external ligand binding domain, a single transmembrane segment and a cytosolic TIR (Toll/IL-1R) domain. There are two groups of TLRs; members of the first group reside at the plasma membrane as monomers and respond to lipid and protein ligands by dimerization to initiate signaling. Members of the second group are found as dimers within the endoplasmic reticulum and endosomes and are activated by exogenous nucleic acids in a process that involves conformational alteration of the receptor. Thus TLR 9 is activated by DNA with unmethylated CpGs, TLR 7, 8 are activated by single stranded RNAs, while TLR3 is activated by ds-RNA. The issue of how TLRs discriminate exogenous and endogenous RNA is an area of active investigation. Signal transduction by TLRs involves interaction with cytosolic proteins such as Myd88 and TRIF that also contain TIR domains. Downstream responses include induction of genes for inflammatory cytokines as well induction of interferons. TLRs are most highly expressed in macrophages and dendritic cells but many cell types express at least one member of this receptor family (111). However, there seems to be little quantitative information in the literature regarding TLR expression at the protein level. The plasma membrane TLRs are internalized by clathrin mediated endocytosis; this both downregulates receptor availability and is important to aspects of the signaling process (112). There is little information on the trafficking of the endosomal TLRs subsequent to the ligand binding event. In terms of targeted delivery of oligonucleotides, TLRs offer one important advantage, their ability to bind nucleic acids. Thus the investigator can synthesize ‘chimeric’ oligonucleotides that contain an active segment such as an siRNA and a delivery segment such as a CpG motif.
Scavenger receptors
This cohort of transmembrane receptors constitutes a functional family rather than one demarcated by common sequence or structure (59,113). Their role is to remove modified or damaged endogenous macromolecules or cells and to clear the body of foreign macromolecules or particles. The various scavenger receptors tend to bind a wide variety of ligands with extensive overlap of binding between different receptors. However, a commonality is the tendency to bind polyanions, probably via patches of cationic residues on the receptor external domain. The scavenger receptors have been grouped into eight sub-families (A-I) of which the class A (SCARA) subfamily is most widely studied. Despite having very short cytoplasmic domains several scavenger receptors are known to participate in signal transduction processes, probably via formation of complexes with other cell surface receptors. Scavenger receptors efficiently internalize via endocytosis or phagocytosis thus conveying their ligands into endomembrane compartments. It is difficult to use these receptors for targeting purposes because of their widespread expression and diverse and overlapping ligand binding abilities. However, whether intended or not, it seems increasingly likely that scavenger receptors play a substantial role in the cellular uptake of both ‘free’ oligonucleotides and nanocarriers (62).
The asialoglycoprotein receptor (ASGR)
This receptor is a C-type lectin that is predominantly displayed on the plasma membranes of hepatocytes (114). Its physiological role is to clear de-sialyated glycoproteins from blood but it also is almost ideal for targeted delivery of materials to the liver. The asialoglycoprotein receptor (ASGR) is expressed at extremely high density (about 5 × 105 copies per hepatocyte) and is rapidly internalized and recycled with a turnover time of about 20 min. The preferred ligand for the ASGR is a triantennary sugar terminating in galactose or N-acetyl galactosamine. Upon ligand binding, the ASGR is accumulated in coated pits where it rapidly internalizes and is then trafficked to early/sorting endosomes. The low pH endomembrane environment causes ligand-receptor dissociation allowing the ASGR to rapidly return to the plasma membrane while the ligand is trafficked to lysosomes. Thus, the ASGR has many desirable aspects for targeted delivery of oligonucleotides including cell-type selectivity, high expression levels, rapid internalization and recycling, and the existence of well-characterized small ligands. Interestingly, despite being the focus of many liver-directed delivery strategies, there has been little consideration of the potential role of the ASGR in signaling. However, it has now become clear that the ASGR is a signaling receptor and indeed is crucially involved in platelet homeostasis through a JAK-STAT signaling pathway that regulates thrombopoietin production (115).
In addition to the receptors mentioned above there are obviously many other receptor families that might be used for oligonucleotide targeting. Interleukin and interferon receptors, Wnt-family receptors, Transforming growth factor (TGF)-β/activin receptors, the immunoglobulin family receptors found in lymphoid cells, and the numerous receptors involved in neuronal cell recognition all come to mind as possibilities. As well, the folate/folate receptor system has been widely used to target ovarian cancer cells (116). Each of these receptors must be considered in terms of tissue selectivity, expression levels, rate of internalization and recycling, and availability of ligands. The properties of several receptor families relevant to oligonucleotide delivery are summarized in Table 1.
Table 1. Receptor properties relevant to oligonucleotide targeting.
| Receptor family | Diversity | Abundance | Internalization and recycling | Tissue selective expression | Ligands |
|---|---|---|---|---|---|
| Integrins | 24 members in humans | Variable-up to several hundred thousand copies per cell | Active internalization and recycling | Most integrins are widely distributed but several show tissue selectivity | Many small peptide and organic molecule ligands |
| GPCRs | ∼800 members | Usually low expression: 103–104 per cell | Internalization and degradation of agonist loaded receptor | Variable, but some show high tissue or disease selectivity | Many highly selective small molecule ligands |
| RTKs | 58 members | Variable 103–106 per cell | Typically ligand induced internalization and degradation | Variable, but some show high tissue or disease selectivity | Small molecule ligands not available |
| TLRs | 10 members | Limited information | Limited information | Widespread but highest expression in macrophages and dendritic cells | Nucleic acids can be used as ligands |
| Scavenger receptors | Many members: eight subfamiles | Variable | Active internalization | Widespread expression | Little selectivity |
| ASGPR | Unique, but member of C-type lectin family | >5 × 105 per cell | Very rapid internalization and recycling | Expressed in hepatocytes | Triantennary carbohydrates |
| Folate receptor | FR-α, member of a small family | Variable | Very rapid internalization and recycling when monovalent conjugates are used | Widely expressed but highly overexpressed in ovarian cancer and additional cancers | Folates |
Some receptor characteristics important for oligonucleotide delivery are listed in the table.
Unintended consequences of targeting
While targeted delivery of oligonucleotides, drugs and imaging agents has been the focus of thousands of publications over the last couple of decades, there has been a surprising lack of emphasis on one of the basic consequences of the targeting process. A ligand that provides effective delivery by virtue of its high affinity binding to a specific receptor will also serve as an agonist or antagonist of that receptor. In doing so it will strongly affect the downstream signal transduction cascade. Thus the net effect will combine both the sequence specific actions of the oligonucleotide and the signaling effects of the targeting agent. It is possible that this may be of modest importance in short term laboratory experiments, but if ligand-conjugated oligonucleotides (or nanoparticles) are to be used for therapy of human disease, the consequences of chronic modulation of key signaling processes must be considered. While the need to be concerned about signaling when dealing with GPCRs or RTKs is rather obvious, recent studies reveal that receptors not usually associated with signaling, including SCARAs and ASGR, can nonetheless participate in important signal transduction cascades.
Cellular uptake, intracellular trafficking and endosomal barriers
Upon reaching the cell surface, ‘free’ oligonucleotides, oligonucleotide conjugates or nanocarriers bearing oligonucleotides all share essentially the same fate; they are internalized by endocytosis and then traffic through multiple membrane-bound intracellular compartments. Thus most of the oligonucleotide accumulated by cells remains separated from the cytosol and nucleus by membrane barriers. The concept of an endosome escape barrier has become prominent in the literature over the last few years and is now generally regarded as perhaps the most important impediment to effective use of oligonucleotides in therapeutics. Several recent reviews have dealt in detail the mechanisms of endocytosis and trafficking and how these impact oligonucleotide pharmacology (117–119). Thus here we will briefly outline some of the basic aspects most relevant to oligonucleotide delivery. Figure 2 schematically depicts some of the processes described below.
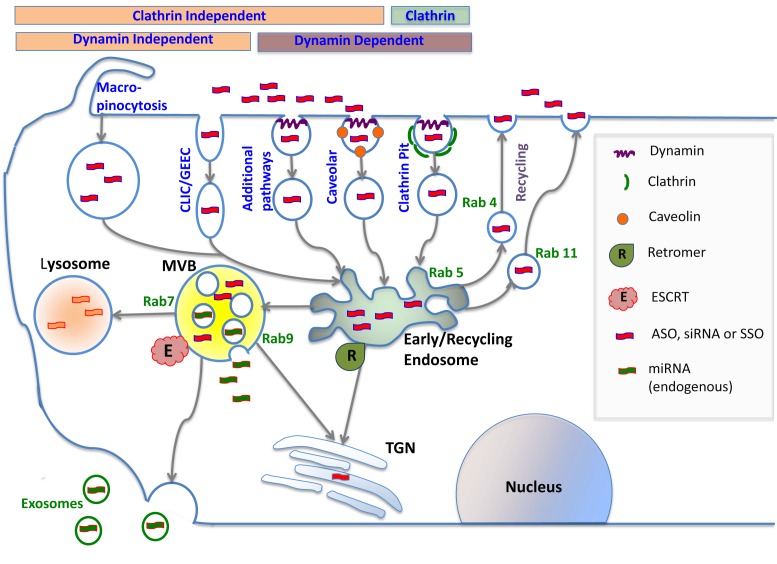
Figure 2.
Cellular uptake and intracellular trafficking of oligonucleotides. Oligonucleotides enter cells via several endocytotic pathways that vary in terms of their dependence on clathrin, caveolin or dynamin. These pathways all initially lead to the early/re-cycling endosome compartment; nonetheless molecules entering via different pathways can traffic to different downstream destinations. Most internalized oligonucleotide accumulates in late endosomes/multivesicular bodies (MVBs) and in lysosomes; however, some trafficking to other membrane bound compartments does occur. Oligonucleotides within endomembrane compartments are pharmacologically inert, but a very small portion of internalized oligonucleotide can spontaneously escape to the cytosol and nucleus. Intracellular trafficking is highly regulated by a large number of proteins and protein complexes. Thus the Rab family of GTPases regulates many aspects of trafficking and individual members serve as markers for distinct endomembrane compartments. The formation of MVBs is regulated by the multi-protein ESCRT complex that has recently been demonstrated to play a key role in the effectiveness of oligonucleotides. This complex also plays a role in exosome formation. The retromer complex may deliver oligonucleotides to the trans-Golgi instead of to lysosomes.
Basic aspects of endocytosis
The simple term endocytosis encompasses a variety of complex events whereby cells take up materials from their surroundings (120). The best-known internalization mechanism is the coated pit pathway that utilizes adaptor proteins, a clathrin network, and the GTPase dynamin to concentrate ligand-bound receptors at the cell surface and then convey them into cells. Several important physiological processes utilize clathrin-mediated endocytosis including uptake of transferrin and low-density lipoproteins, as well as internalization of agonist activated GPCRs (121). Caveolae originate from membrane structures enriched in cholesterol, sphingolipids and the transmembrane protein caveolin (122). While the role of caveolae in endocytosis has been questioned at times, it now seems clear that these compact structures (100 nm) do play a role in the internalization of certain receptors and their ligands. For example, some members of the integrin family as well as certain sodium channels are internalized via the caveolar pathway (123,124). Multiple additional endocytotic pathways occur in cells including ones that do not rely on clathrin, caveolin or dynamin (125). An important example is the CLIC/GEEC pathway that results in the formation of tubular endosomes that make a large contribution to fluid phase endocytosis. Another high volume pathway is macropinocytosis whereby the cells use an actinomyosin-driven process to pinch off and engulf large amounts of extracellular fluid. All of these processes have been implicated to varying degrees in the initial uptake of oligonucleotides. Increasing evidence indicates that the initial route of endocytosis can be an important determinant of oligonucleotide pharmacology and that there are both productive and non-productive paths of cellular uptake (126,127).
Basic aspects of intracellular trafficking
After initial internalization by endocytosis, oligonucleotides, like all internalized materials, must traffic through a complex network of endomembrane compartments each with distinct characteristics and functions. Major membrane-bound compartments include early and recycling endosomes, LEs/multi-vesicular bodies (LEs/MVBs), lysosomes, the Golgi apparatus and the endoplasmic reticulum. Irrespective of the original pathway of endocytosis, most substances entering the cell are initially delivered to early endosomes. Subsequently there are two basic fates for materials that reach the early endosome; they can be shunted to lysosomes for degradation, or they can be recycled to the plasma membrane and cell exterior (128).
Immediately after internalization, the initial endosomal vesicle fuses with early/recycling endosomes (EEs) (129). These are relatively large, tubulated structures usually located near the cell periphery and having a pH of about 6.0–6.5. For many ligand-receptor complexes, the early endosome is the site where the receptor and its ‘cargo’ are dissociated. The free receptor migrates to the tubular region and is eventually returned to the plasma membrane via small shuttle vesicles, while the cargo remains in the central lumen and will eventually be delivered to lysosomes for degradation.
The next stage of the trafficking process involves LEs/MVBs that are the primary stepping-stone on the road to lysosomes (130). These structures are morphologically and biochemically distinct from early endosomes, having a non-tubulated appearance, a perinuclear location and a lumen filed with small intraluminal vesicles (ILVs) that have pinched off from the boundary membrane of this organelle. Although the major task for LEs is to convey internalized material to lysosomes, an interesting detour involves the externalization of the ILVs as exosomes that can convey materials to other cells (131).
Ultimately most material internalized by endocytosis is delivered to lysosomes. These dense organelles are rich in hydrolases that function in the low pH environment (pH 4.5–5.5) that is maintained by an active V-ATPase proton pump, thus allowing the hydrolases to degrade proteins, lipids, carbohydrates and nucleic acids to their constituent building blocks (132). Besides degrading internalized materials, lysosomes are also a key part of the machinery for autophagy whereby cells degrade damaged proteins and organelles and recycle the constituents (133).
It is important to note that the early endosome to lysosome pathway is not linear and that there are several branches and loops. An important one is the retrograde trafficking pathway that links endosomes to the trans-Golgi. A classic example of retrograde transfer is the recapture of mannose-6 phosphate receptors from endosomes to the Golgi, while their hydrolase ligands are delivered to lysosomes (134). Interestingly, several pathogens have ‘hijacked’ this pathway; for example, certain bacterial toxins reach the cytosol by following the retrograde pathway to the trans-Golgi and thence to the endoplasmic reticulum (135).
In summary, the pathways of endocytosis and intracellular trafficking are complex and dynamic. Contemporary imaging technologies (136) are starting to provide detailed insights into these pathways that will be very helpful in understanding the fundamental basis of oligonucleotide pharmacology. However, it remains a challenging problem to link microscopic observations on the intracellular trafficking of a fluorescent oligonucleotide to the pharmacological effects of that molecule. Visualization of the bulk distribution may not reveal minor compartments that are key to biological activity.
The machinery of intracellular trafficking
There are two basic ways by which materials can move through the intracellular trafficking network (128,136,137). One involves relatively small shuttle vesicles that convey both luminal and membrane material between larger endomembrane compartments. The other is endosome maturation whereby one compartment gradually assumes the characteristics of a second compartment.
Although differing in detail, all transfer of material via shuttle vesicles involves several basic steps (138–140). First, coat proteins help to pinch off small vesicles from the donor compartment. Second, the shuttle vesicle moves toward the recipient compartment using actin or tubulin cytoskeletal machinery. Third, a recognition event occurs between the shuttle vesicle and the recipient. Fourth, the shuttle vesicle fuses with the recipient compartment and delivers its contents to the recipient membrane and lumen. This dynamic process is very precisely regulated by a plethora of proteins and multi-protein complexes. Some key examples include the Rab family of small GTPases, SNARE complexes, tethering complexes, the ESCRT complex and the Retromer complex. These various proteins/protein complexes play important functional roles, but they also serve as easily recognizable markers for specific endomembrane compartments.
Members of the numerous (>60) Rab GTPase protein family serve as molecular switches that regulate many aspects of trafficking including vesicle uncoating, movement along cytoskeletal tracks and the ultimate membrane fusion events involving tethers and SNARES (137,141). Rab proteins also serve as excellent markers of individual membrane compartments and trafficking pathways. For example, Rab5 is a marker for early endosomes while Rab7 identifies LEs.
The shuttle vesicle trafficking process involves coat proteins that assist in the initial formation of the vesicle (140,142). The generation of clathrin-coated vesicles at the plasma membrane is a good example, but other types of coats exist including the COPI proteins involved in Golgi to ER transport and the COPII proteins involved in the reverse process. Disjunction of the clathrin-coated vesicle from the donor membrane is accomplished by the dynamin GTPase, but other pinching off mechanisms exist for other types of vesicles.
Tethering proteins impart selectivity to trafficking by promoting preferential interactions between the vesicle and the recipient compartment. The coiled-coil tethers and the multi-subunit tethers comprise the two broad classes of tethering proteins (143,144). By associating with both Rab proteins and SNARES, tethers are thought to physically link the two membranes destined for fusion. However, some tethers clearly have multiple functions including possibly ‘proof-reading’ SNARE complexes to assure fusion of the correct partners.
The final transfer of both the membrane and luminal contents of the shuttle vesicle to the recipient compartment occurs via a fusion process mediated by SNAREs (soluble N-methylmaleimide sensitive factor attachment protein receptors)(145,146). Vesicle SNARES (v-SNARES) interact with SNARES on the target membrane (t-SNARES) forming a four-helix bundle. This undergoes a dramatic conformational change, inducing fusion of the apposed lipid bilayer membranes. Only cognate pairs of v- and t-SNARES will sustain fusion, indicating considerable specificity of the process. Re-segregation of the v-SNARES and t-SNARES is mediated by the ATP-dependent NSF/SNAP protein complex.
An important detour on the pathway between endosomes and lysosomes is retrograde trafficking between early endosomes and the trans-Golgi (134,135,147). This process is driven by the retromer that includes a trimeric complex (Vps26–Vps35–Vps29) that binds to the cytoplasmic tails of potential cargo proteins. It also includes SNX proteins that have PX domains that recognize membrane phosphoinositides and BAR domains that can affect membrane curvature. This results in the tubulation of the EE membrane and eventual formation of shuttle vesicles that traffic to the trans-Golgi.
In addition to the shuttle vesicle mechanism, intracellular trafficking also utilizes processes involving maturation of one major endomembrane compartment into another (128,148). Perhaps the best example is the conversion of EEs to LEs. The Rab5 GTPase plays a key role in the identity and function of early endosomes (137,149). The activating proteins Rabex-5 and Rabaptin-5 stimulate Rab5 on the cytosolic surface of the EE resulting in the recruitment of Rab5 effectors including the tethering factor EEA1 and the PI 3-OH kinase Vps34. Initially this process is self-sustaining allowing the EE to interact with other EEs and to recycle receptors and other membrane constituents to the cell surface. Eventually other proteins are recruited to the EE that drive displacement of Rab5 and association with Rab7. Two sets of effectors, the SAND-1/Mon complex and the HOPS complex, seem to work in tandem in the EE to LE maturation (128). In parallel to the Rab5 to Rab7 conversion, the endosome loses ability to interact with EE partners and instead acquires the ability to associate with LE partners. One of the key aspects of the EE to LE maturation is the formation of ILVs (150). This process helps to concentrate selected proteins and lipids in the LE lumen directing them to lysosomal degradation. The five multi-protein complexes of the ESCRT (endosomal sorting complex required for transport) machinery, ESCRTs 0–III and Vps4–Vta1, recognize ubiquitinated membrane proteins and drive them into invaginations that ultimately form ILVs within the lumen of the LE/MVB (151).
How is all this trafficking complexity linked to oligonucleotide delivery? As an example, in an extremely important recent investigation of basic aspects of oligonucleotide trafficking, Wagenaar et al. (152) used shRNA libraries to identify TSG101, a component of the ESCRT machinery, as having a key role in the uptake and intracellular trafficking of oligonucleotides. Silencing of this gene led to a dramatic increase in the effectiveness of an antisense antagonist of miR-21. This publication establishes two critically important points. First, that the endomembrane trafficking machinery plays a key role in the pharmacology of oligonucleotides, and second that the machinery can be manipulated so as to improve oligonucleotide delivery and actions.
Thus, the complex and dynamic pathways of intracellular trafficking are regulated by an equally complex set of proteins whose interactions vary in time and space throughout the process. While this makes the investigation of subcellular trafficking rather complicated it also opens up many opportunities to manipulate the machinery of trafficking using molecular and chemical probes. As shown by Wagenaar et al., this can have important implications for oligonucleotide pharmacology. Thus increasing understanding of the mechanistic basis of oligonucleotide trafficking will no doubt provide important new avenues to manipulate oligonucleotide delivery.
Breaching the endosomal barrier
While the trafficking machinery is usually quite efficient in moving internalized material to the appropriate intracellular destination, nonetheless opportunities for molecules to escape from endomembrane compartments to the cytosol do exist. Trafficking involves a plethora of membrane fusion and fission events. These events create localized membrane stress that can result in the formation of non-bilayer lipid domains (153). Typically non-bilayer regions can be much more permeable to solutes than bilayer regions (154,155). Thus there is an inherent relationship between the fusion/fission events essential to intracellular trafficking and the potential for leakage of vesicle contents. There are several loci in the intracellular trafficking network that may be particularly susceptible to increases in permeability that would allow release of oligonucleotides to the cytosol. The first is in early/sorting endosomes where there is extensive tubulation and formation of vesicles for return of receptors to the plasma membrane. A second locus is in LE/MVBs where the ESCRT complex distorts the endosome membrane to form ILVs. Third, retrograde traffic from early or LEs to trans-Golgi offers another possibility for membrane instability. Finally, SNARE driven membrane fusions at multiple sites afford opportunities for partial leakage of vesicular contents (156). The role of specific trafficking events in oligonucleotide delivery can be explored using molecular techniques such as siRNA or vectors expressing dominant negative proteins to perturb these events. Another interesting approach is to use cell lines with defects in trafficking processes. There are a growing number of examples of both approaches in the oligonucleotide literature (127,152,157,158). In addition to endogenous escape of oligonucleotides from endosomes, a substantial portion of the recent literature on oligonucleotide delivery is focused on approaches to disrupt or alter the endosomal barrier. Thus cationic lipids and polymers have been used to destabilize the endosome membrane. Titratable peptides or polymers have been used to alter intra-endosomal pH thus affecting endosome stability and trafficking. Recently a variety of small molecules have emerged that seem to assist in endocytosis of oligonucleotides or that selectively permeabilize endosomal compartments leading to oligonucleotide release to the cytosol. It is important to note that once anionic single stranded oligonucleotides reach the cytosol they readily enter the nucleus (159,160).
APPROACHES TO DELIVERY
This section will examine the recent literature on oligonucleotide delivery. Since hundreds of publications on this topic appear yearly there is no attempt to provide a comprehensive account. Rather, after introducing the topic, the focus will be on reports that (i) provide insights into basic mechanisms of oligonucleotide uptake and trafficking, (ii) utilize novel approaches or (iii) seem unusually promising at the pre-clinical or clinical levels.
There are two broad strategies for oligonucleotide delivery. One is to incorporate the oligonucleotide into some form of nanocarrier that then determines the tissue distribution and cellular interactions of the oligonucleotide. The other is to chemically modify the oligonucleotide itself, most commonly with a targeting ligand, while preserving the molecular nature of the conjugate. A fundamental difference between the two approaches lies in the size of the delivery moiety, nanoscale versus molecular scale; this has profound effects on the biodistribution and biological actions of the oligonucleotide. Several of the delivery approaches that will be discussed in detail below are depicted in Figure 3.
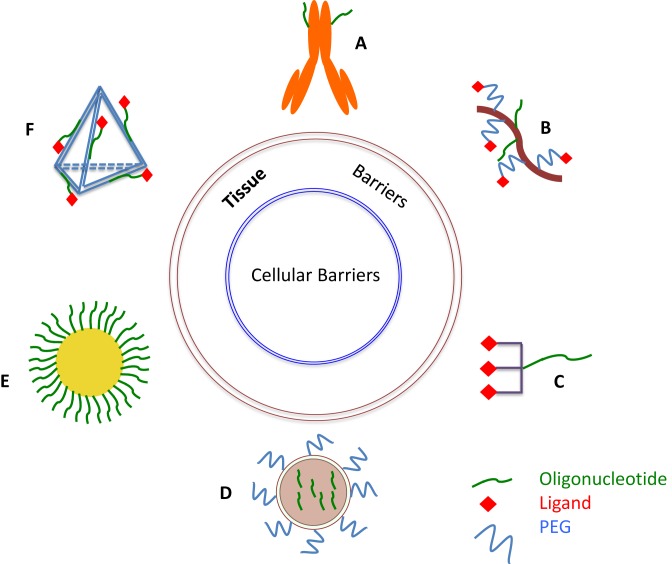
Figure 3.
Oligonucleotide delivery strategies. Several approaches to oligonucleotide delivery are depicted. (A) Antibody-oligonucleotide conjugate. (B) Polymer-oligonucleotide conjugate with PEGylation and targeting ligand. (C) Molecular scale ligand-oligonucleotide conjugate with triantennary carbohydrate ligand. (D) Lipid nanoparticle with PEGylation. (E) Gold nanoparticle with dense oligonucleotide coat. (F) DNA nanostructure with oligonucleotide and targeting ligand incorporated. Images are not to scale.
Delivery at the nanoscale
Lipid nanoparticles
The delivery approach that is both most widely used and most clinically advanced is to complex anionic oligonucleotides with cationic lipids thus forming lipid nanoparticles (LNPs)(161–163). This approach has been especially important for therapeutic use of siRNA (164) and good accounts of current clinical trials using siRNA LNPs are found in recent reviews (10,165). In addition to the cationic lipid, LNPs typically include a neutral lipid such as cholesterol, but a variety of compositions are possible. While simple LNPs are effective as cell culture transfection agents, they cannot be used in vivo because extensive interactions with opsonic proteins in blood lead to rapid clearance by RES phagocytes. LNPs for in vivo use are usually 100–200 nm in size and include a surface coating of a neutral polymer such as PEG to minimize protein binding and uptake by RES cells. This allows greater persistence in the circulation and the opportunity to interact with other cell types. However, because of their size, LNPs can only exit the circulation at sites where the endothelial barrier is fenestrated, particularly liver, spleen and certain tumors having a high EPR effect. Because of this, much of the work with siRNA-LNPs has focused on liver diseases including transthyrethrin-mediated amyloidosis, clotting disorders, liver cancer and disorders of lipid metabolism (11,166).
The action of LNPs involves initial uptake by endocytosis. In some cases this is mediated by the binding of apolipoprotein E and interaction with the LDL receptor (167). Once in endosomes, the cationic lipids of the LNP interact with anionic membrane lipids to disrupt membrane structure through the formation of a non-bilayer lipid phase termed inverted hexagonal (HII). This leads both to increased membrane permeability and to dissolution of the LNP and is the basis for conveying the oligonucleotide to the cytosol (168). However, the interaction of cationic lipids with cellular membranes is also the basis for possible toxicities of LNPs (169,170).
A great deal of effort has been expended on optimizing the delivery properties of LNPs. One problem concerns the PEG coating (which is usually attached to the LNP by a lipid anchor). A dense coat is beneficial for increasing circulation time, but it also reduces uptake by cells such as the hepatocytes that are the intended destination of the oligonucleotide. Several approaches have been tried to attain dynamic control of PEG levels, including use of cleavable linkers or short lipid anchors, with the intent of maintaining a dense coating of PEG in the circulation while allowing release in the cellular environment (161). Studies of how PEG density and characteristics affect the pharmacokinetics, biodistribution and function of LNPs remains an active area of investigation (171).
There has also been progress in optimizing the delivery characteristics of the cationic lipids themselves. A widely used type of nanocarrier for siRNA delivery is the SNALP (stable-nucleic-acid lipid particle), a PEG stabilized LNP. Two important steps for optimizing the cationic lipids were (i) altering the pKa so that the lipids were almost uncharged in the circulation but became charged in the low pH endosome and (ii) using linkages that were readily biodegradable (172,173). This resulted in dramatic improvement in effectiveness, allowing siRNA doses as low as 0.005 mg/kg to achieve significant silencing of hepatic targets in animal models, accompanied by low toxicity (174). Additional work on the chemistry of lipids continues. One powerful approach is the testing of chemical libraries of ‘lipoids’ (lipid like molecules) for their delivery capabilities (175). Another interesting strategy is the design of multifunctional lipids that include a pH responsive head group and well as SH moieties. The multifunctional LNPs provide increased stability in the blood but then promote endosome escape in the low pH and reducing environment of the cell interior (176). Another approach that may avoid some of the toxicities of cationic LNPs involves using siRNA entrapped in neutral liposomes (165,177). There has also been manipulation of the physical structure of lipid delivery systems. Thus ‘cuboplexes’ are novel lipid structures designed to promote interaction with endosome membranes and thus allow escape of siRNA to the cytosol (178). Another interesting approach uses liposomes termed ‘Smarticles ®’ made from dialkyl cationic amino acids; these undergo a pH sensitive conversion to a HII phase in endosomes allowing oligonucleotide escape (179).
The precise mechanisms of uptake, intracellular trafficking and ultimate delivery of oligonucleotides by LNPs has been a matter of some debate (157,180). However, two articles appearing simultaneously in 2013 used advanced imaging techniques to provide unprecedented insights into the intracellular fates of siRNA LNPs. Thus Gilleron et al. (181) found that LNPs were initially taken up by clathrin-mediated endocytosis but further accumulation involved macropinocytosis. The LNPs accumulated in an EE-LE hybrid compartment; however, only 1–2% of the siRNA reached the cytosol. Sahay et al. (158) also demonstrated a role for macropinocytosis. However, they found that much of the siRNA was re-exported from LEs/lysosomes using a process involving the NPC1 lipid transporter protein. A more recent study using highly sensitive fluorescence microscopy techniques documented release of siRNA from individual endosomes (182). Release took place primarily at the EE/LE conversion step rather than from lysosomes. These studies provide important insights into the mechanistic basis of oligonucleotide delivery via LNPs and attest to the overall inefficiency and transience of oligonucleotide delivery even when using effective nanocarrier systems.
An important issue is whether it is possible to target LNPs to particular cells or tissues. An obvious approach is to decorate the LNP surface with antibodies. However, long experience with targeting of drug-loaded liposomes suggests that it is quite difficult to find the right balance between PEG shielding and availability of the antibody (183). Recently however, there have been some interesting reports using antibody-targeted siRNA LNPs. Thus a scFv was used to target LNPs to dendritic cells in vivo. Good ‘knock down’ of several co-stimulatory surface antigens and inhibition of mixed lymphocyte reactions were observed (184). In another study LNPs decorated with antibody to CD20 delivered Bcl-2 ASO to B-cell tumors in vivo with good antitumor effect (185). Additionally an anti CD4 monoclonal was used to target siRNA LNPs to CD4 positive T-cells resulting in knock-down of the intended mRNAs both in cell culture and in vivo (186).
Progress has also been made using small molecule ligands to target LNPs. For example, in a series of publications, Huang et al. have used anisamide as a ligand to assist in the delivery of siRNA LNPs to tumors (187,188). Finally aptamers are also being explored as a targeted delivery approach for LNPs (189). Some of the above mentioned studies on LNP targeting are also of interest because they involve delivery of oligonucleotides to non-hepatic sites.
Finally, there have been some impressive recent pre-clinical studies using LNPs. Thus LNPs prepared using a novel lipid from a chemical library selectively delivered oligonucleotides to the lung. When used with si-KRAS and miR-34a, significant inhibition of lung cancer was attained in a genetically engineered mouse tumor model (190). In another study, siRNA LNPs showed therapeutic efficacy versus Marburg virus (an Ebola relative) in non-human primates (191). These studies are both good examples of non-hepatic delivery using LNPs.
In summary, over the last few years there have been impressive advances in the LNP delivery technology. Current formulations allow highly effective delivery of siRNA to hepatocytes using doses of oligonucleotide and of lipid carrier that display only minimal toxicity. This has allowed several liver-based diseases to be addressed. The greater challenge lies with non-hepatic delivery. Although there have been some interesting reports using LNPs to deliver siRNA to other tissues and to tumors, there remain questions regarding efficacy and toxicity particularly in the context of long term use in human therapy.
Polymeric nanocarriers
Various types of polymeric nanoparticles provide another widely used strategy for oligonucleotide delivery. Although they have not progressed clinically to the same degree as LNPs, it is worth noting that the first use of siRNA to treat human cancer involved a ligand-targeted cyclodextrin-based polymeric nanoparticle (192). Excellent recent reviews provide comprehensive accounts of the numerous publications utilizing this general approach to oligonucleotide delivery (193,194).
There are several types of polymeric nanocarriers. Early studies in this area primarily used well-known, biomedically compatible polymers such as poly (lactic-co-glycolic acid) (PLGA) to form solid nanoparticles through various oil-in-water emulsion techniques. However, since PLGA is anionic, a common approach was to incorporate positive side chains in the polymer or to complex the anionic oligonucleotide with a positively charged moiety such as polyethylene imine (PEI) (195,196). Polymeric micelles, sometimes called ‘core-shell’ nanoparticles, have also been widely used for oligonucleotide delivery. These are formed by self-assembly of amphiphilic polymers in a water environment. Typically a tri-block polymer might be used, including a hydrophobic portion to drive self-assembly, a cationic portion to bind the oligonucleotide and PEG or other neutral polymer to provide a protective coating (197,198). A recent report described a novel self-assembled hybrid nanocarrier comprised of a PLGA core and a lipid-PEG shell. This system provided impressive results in terms of a long circulation lifetime and functionally effective delivery of siRNA to tumor xenografts in mice (199). A third type of polymeric nanocarrier is the nanohydrogel. These nanoparticles have an open, water-filled polymer lattice that can easily incorporate bio-macromolecules such as polypeptides and oligonucleotides, whose release kinetics can be controlled by the degree of cross linking of the lattice. A particularly interesting form is the PRINT nanohydrogel whose size and shape can be precisely controlled by a nano-molding technique (200,201). A virtue of many polymeric nanocarriers is their ready ability to convey both an oligonucleotide and a small molecule drug; this is especially interesting in the context of cancer chemotherapy. Thus there are several promising reports of in vivo co-delivery of siRNA and anticancer drugs (194,197,198).
Another common approach involves the direct complexation of anionic oligonucleotides with cationic molecules that have some degree of endosome escape capability inherent in their chemistry. This would include formation of nanoscale polyplexes involving CPPs (202,203), cationic dendrimers such as PAMAMs (194,204) or linear or cross-linked PEI (205).
CPPs have been extensively studied for oligonucleotide delivery (74,206). Starting with the original Transactivator of Transcription (TAT) and penetratin structures, a wide variety of short polycationic CPPs have been synthesized and used as delivery agents for drugs, peptides, proteins and nucleic acids (203,207). In many cases short amphipathic sequences are also included with the intent of promoting endosome escape. While some work has been done with chemical conjugation of CPPs to anionic oligonucleotides, in most cases delivery is via formation of nanocomplexes. These are usually in the 100–200 nm range with a surface charge that depends on the chemistry of the CPP and the ratio of cationic peptide to oligonucleotide. Perhaps the most advanced CPP for oligonucleotide delivery remains PepFect6 that has been used to deliver siRNA in an animal model resulting in ‘knock down’ of target gene expression in several tissues (208). Additional work on this type of CPP has included the incorporation of endosome disrupting moieties such as quinoline derivatives (209). Another promising study used a cationic peptide derived from bee melittin to form nanocomplexes with siRNA directed against NF-kB; these were used to treat a mouse model of rheumatoid arthritis (210). In addition to their direct use as polyplexes, CPPs have been used to augment the properties of other delivery systems such as LNPs (211). An interesting variant of the CPP approach entailed making a chimera of an RNA binding protein and a CPP. This entity could bind and deliver siRNA in cell culture and in an animal model (212).
Dendrimers are branched polymers with well-defined architectures. By controlling the degree of branching, different ‘generations’ of dendrimer can be made thus varying size and the number of potentially reactive surface moieties. PAMAMs (213), the dendrimer type most commonly used for oligonucleotide delivery, shares with PEI the ability to exert a strong ‘proton sponge’ effect. Thus the titratable amines on the PAMAM bind protons in endosomes and lysosomes leading to increased pumping of protons by the membrane V-ATPase; this is accompanied by Cl− transport, water accumulation and swelling and rupture of the organelle. This makes PAMAM dendrimers very effective agents for nucleic acid delivery (204). Unfortunately the strength of the protein sponge effect, along with lack of biodegradability and multiple non-specific interactions with blood and tissue proteins, all contribute to the substantial toxicities observed with in vivo use of PAMAMs (214). By virtue of their multiple surface amino groups PAMAMs are easily conjugated to a variety of ligands. For example, PEG has been used to reduce toxicity and improve in vivo biodistribution properties (215). It is also possible to simultaneously conjugate both targeting ligands and therapeutically active small molecules to PAMAM dendrimers (216). Thus there is a great deal of flexibility inherent in this delivery system but concerns remain about toxicity.
PEIs are linear or branched polymers that have multiple titratable amino groups. Thus they can readily form nanocomplexes with oligonucleotides (217). Like PAMAM dendrimers, PEI exerts a strong ‘proton sponge’ effect making it quite effective for nucleic acid delivery. It is also readily modified by conjugation with a variety of ligands. For example, a branched PEI modified with PEG and Arg-Gly-Asp (RGD) ligands was used to deliver siRNA to tumors in mice (218). In addition to its direct use as an oligonucleotide carrier, as noted above, PEI is often incorporated into other types of nanoparticles to provide binding for oligonucleotides and as an endosome escape agent. Unfortunately, PEIs are poorly biodegradable.
There has been a great deal of interest in methods for targeting nanoparticles with a plethora of research publications and multiple reviews on this broad topic (219–221). Recent reviews have focused particularly on siRNA or ASO delivery using targeted polymeric nanocarriers (222,223). A widely used approach has been to couple folate to the nanocarrier surface to promote interaction with cancer cells that overexpress the folate receptor (224,225). However, although the first demonstrated siRNA effects in man involved a targeted polymeric nanocarrier (192), there has been little further clinical progress with this approach.
Despite the popularity of polymeric nanoparticles with pharmaceutical scientists, there is relatively little information available about their mechanism of delivery at the cellular level. A recent study showed that the cellular uptake of CPP/oligonucleotide nanocomplexes involves scavenger receptors (226). Another interesting report demonstrated that the NPC1 lipid transport system, previously shown to be involved in LNP processing (158), is also involved in the processing of certain types of polymeric nanoparticles (227). However, a reduction in NPC1 levels had the opposite effect on delivery of siRNA using polymers as in the case of delivery using lipids. There have been only a few comparisons of uptake mechanisms for LNPs and polyplexes (157,228); both of the cited reports suggest that the mechanisms of uptake are different and one report indicated that polyplexes are released from endosomes via a rapid bursting process. A recent study sought to incorporate endosome-disruptive helical peptides into siRNA nanoparticles and demonstrated release into the cytosol; this system was used to knock down TNF-α in an animal model of inflammation (229).
In contrast to the lack of information at the cellular level, there is substantial data on the pharmacokinetics and biodistribution of polymeric nanoparticles (193,230). In mice, as well as in patients, polymeric nanoparticles tend to be cleared quickly (231) with accumulation in the liver predominating (232,233). However, as mentioned above, a recent study with a novel polymeric nanoparticle formulation demonstrated an extended lifetime in circulation and impressively high levels of tumor uptake as compared to liver uptake (199). Thus it may be possible to substantially improve the biodistribution characteristics of polymeric nanoparticles. Nonetheless, polymeric nanoparticles have not yet advanced as far as LNPs in terms of pre-clinical and clinical development.
An interesting offshoot for polymeric nanoparticles involves siRNA delivery via the gastrointestinal tract. Thus siRNA nanoparticles have been used orally to block inflammatory effects mediated by TNF-α (234), or have been administered into the colon to knock down apolipoprotein B mRNA (235), or via the rectum to suppress inflammatory cytokines (236).
Advantages and liabilities of nanoscale delivery
There is an interesting divergence in the oligonucleotide therapeutics field in terms of approaches to delivery of single strand molecules and of siRNA or miRNA. For the most part delivery of ASOs and SSOs, at least at the clinical level, has relied on ‘free’ oligonucleotides. By contrast, with one major exception, siRNA delivery has relied on nanocarriers, particularly SNALPS. The reasons for this divergence are easy to understand. ASOs and SSOs can readily be chemically modified to resist nuclease degradation and, in the case of phosphorothioates, they have a long circulation lifetime and are readily taken up by cells. By contrast, early versions of siRNA had none of these advantages; they were very unstable, rapidly cleared by the kidney, and poorly accumulated by cells. Based on these problems, a huge effort has gone into the development of siRNA nanocarriers. Although a bewildering variety of lipids and polymers have been tried, they almost all share the weakness of being positively charged and therefor highly interactive with anionic biological macromolecules and cells. This inevitably leads to toxicity especially upon chronic use. As discussed above, extensive work on the lipid components of SNALPs has both increased their efficacy for siRNA delivery and reduced their toxicity. Thus SNALP siRNA formulations seem a sound approach for siRNA delivery to the liver. It is not clear that the same can be said about the various types of polymeric nanoparticles. Although attempts have been made to reduce the toxicity of dendrimers, PEI or cationic peptides it is difficult to avoid this entirely because of their inherent chemical nature.
A potential advantage of nanocarrier systems is that a large bolus of oligonucleotide can be delivered during one cellular uptake event. However, the converse is that entry of a nanoparticle into the cells entails the delivery of a large mass of carrier material. Even with the best nanocarriers only a few percent of the total mass is siRNA with the rest being carrier (172). Thus nanoparticle delivery is inevitably associated with the accumulation of substantial amounts of potentially toxic material; this is especially problematic for lipids or polymers that are not readily biodegradable. It seems unlikely that such materials will ever be clinically useful.
A final concern regarding nanocarriers involves their limited biodistribution. As mentioned early in this review, typical nanoparticles with diameters of ∼100 nm are excluded from most tissues by the barrier presented by the capillary endothelium. Accumulation occurs primarily in liver and spleen where the endothelium is fenestrated, and to a lesser degree in certain rapidly growing tumors that have a strong EPR effect. The parenchyma of most other normal tissues are virtually inaccessible, thus limiting the range of therapeutic applications for nanocarriers.
Delivery at the molecular scale: ligand-oligonucleotide conjugates
Partly because of the many concerns about using nanoparticle delivery systems, there has recently been a surge of interest in molecular scale ligand-oligonucleotide conjugates. These offer the possibility of selective delivery to specific cells or tissues via receptor mediated mechanisms coupled with an avoidance of the toxicities often associated with nanocarriers. Another advantage of conjugates is that they are well-defined molecular entities as opposed to the heterogeneity characteristic of nanoparticles. Thus while the conjugate approach is still in its infancy, it seems to offer a promising path forward for oligonucleotide therapeutics.
Conjugation chemistries
There are numerous synthetic paths for the preparation of oligonucleotide conjugates, as discussed at length in recent reviews (237–240). Nonetheless there are two basic strategies for oligonucleotide conjugation. The first is an all solid-phase approach where the oligonucleotide and ligand are linked while attached to a support. The other is to couple the two components while in solution. There are advantages and limitations with both approaches. Solid phase synthesis is highly efficient and facilitates purification. However, the availability of compatible synthons and coupling reagents for both the oligonucleotide and the ligand is often a limitation. Another issue is the need for both ligand and oligonucleotide to be stable under the conditions of synthesis. By contrast, solution phase conjugation initially pursues synthesis of each component under the most efficient conditions followed by the conjugation reaction. However, that reaction may be inefficient and significant post-synthesis purification problems may exist. Various groups have utilized both strategies for conjugation, with the majority preferring solution phase approaches.
There are many possible linkages for oligonucleotide conjugates. Recently there has been much interest in using ‘click’ chemistry since the reactants do not cross-react with common biomolecules and the process is quick and efficient (240–243). However, the bond formed is not bioreversible. More traditional linkages include disulfide bridges and pH sensitive ester linkages that provide reversibility within the cell (240). The issue of reversibility can be an important one depending on the choice of ligand. For smaller ligands such as short peptides or small organic molecules our experience (244) as well as that of others (245) indicates that both reversible and non-reversible linkages permit biological activity of the oligonucleotide. However, for larger ligands such as proteins or polymers reversibility is likely to be more important.
Non-targeted conjugates
Much of the early work on oligonucleotide conjugates involved ligands that were designed to promote cell uptake but lacked specificity. The most notable examples are lipid conjugates and CPP conjugates.
Lipid conjugates
One of the earliest successes in enhancing the delivery of siRNA was through conjugation of the oligonucleotide with cholesterol (246). This significantly promoted uptake by the liver and enabled silencing of hepatic genes. Similar effects were seen with other lipidic conjugates including α-tocopherol and long chain fatty acids (247,248). The cholesterol conjugates associated with serum lipoproteins thus facilitating uptake via lipoprotein receptors. Although this approach was a pioneering one it faced a number of difficulties including the need for relatively large doses to attain efficacy. Thus much of the effort in hepatic delivery has switched to SNALPs or to other types of conjugates.
CPP conjugates
As mentioned above, nanocomplexes of CPPs with anionic oligonucleotides have been widely evaluated as delivery vehicles. However, molecular scale chemical conjugates of CPPs with uncharged oligonucleotides such as PMOs and PNAs have also shown promise. By far the most extensively studied conjugates involve a variety of CPPs linked to PMO SSOs designed to correct the defective production of dystrophin that is the basis of Duchenne muscular dystrophy. Much of this work was recently reviewed in detail (74) and only a brief summary is provided here. The CPPs used in these studies contain tracts of arginine residues interrupted by short hydrophobic sequences. A large number of variants have been tested thus permitting structure activity relationship studies. The CPPs are usually coupled via their COOH terminal to the 3′ end of the PMO, but other configurations are possible. Using the mdx mouse model of muscular dystrophy, strong splice correction and upregulation of dystrophin expression in muscle was seen with several conjugates. Recent versions have also increased dystrophin expression in heart, which is a particularly challenging problem. Other studies include attempts at combining CPPs with targeting peptides derived from phage display so as to increase specificity of delivery. Another interesting variation has been to couple two different oligonucleotides to a single CPP thus addressing two targets at the same time (243). Although the mechanism of delivery of the CPP-PMOs is not entirely clear, recent studies have investigated their subcellular behavior. Interestingly the same CPP-PMO conjugate displayed different uptake and trafficking processes in cultures of skeletal muscle cells versus cardiomyocytes, with uptake and nuclear delivery of the PMO-SSO correlating with biological effect (249). Another interesting study found that CPP-PMO conjugates, as well as tricyclo oligonucleotides, tend to spontaneously form nanoparticles whose uptake was then mediated by scavenger receptors of the SCARA 1 class (61).
In addition to studies in the context of muscular dystrophy, CPP-PMO conjugates have also been explored for anti-viral and anti bacterial properties (250,251). However, this thrust has been supplanted by the use of PMOs having positive piperazine residues incorporated into the backbone (252,253); these seem to be less toxic than the CPP-PMOs in the antiviral context and have shown impressive results in trials against Marburg virus in monkeys and were apparently well tolerated in phase I trials in man (254).
Targeted conjugates
The last couple of years have seen a major shift of emphasis in the oligonucleotide therapeutics area away from lipid or polymer based nanocarriers and toward targeted molecular- or macromolecular-scale conjugates. Thus far the greatest successes have come in delivery to the liver but interesting reports of extra hepatic targeting have also appeared. The sections below recapitulate some of the most important developments in this rapidly growing area; several recent reviews have also dealt with targeted conjugates (240,255,256).
Glycoconjugates: the asialoglycoprotein receptor
The most dramatic advance in oligonucleotide targeting has involved delivery via the ASGR. As mentioned above, this receptor is almost ideal for targeted delivery to the liver, combining tissue specificity, high expression levels and rapid internalization and turnover. Additionally, there exists a deep background on the chemistry of oligonucleotide glycoconjugates that has facilitated progress in this area (257). A major breakthrough came from researchers at Alnylam Pharmaceuticals who developed multivalent N-acetylgalactosamine (GalNac) conjugated siRNAs that bind at nanomolar levels to the ASGR (258). A synthetic scheme was devised that allowed direct solid phase synthesis of the conjugates. The siRNAs used were heavily chemically modified to enhance their in vivo stability. The conjugates were effectively taken up into primary mouse hepatocytes by a receptor-specific mechanism, leading to silencing of targeted genes. Similarly, using a radiolabeled conjugate, excellent uptake into liver was observed in mice; interestingly higher levels were obtained by subcutaneous administration rather than intravenous administration. Conjugates addressing the transthyretin gene effectively reduced expression, with subcutaneous administration being superior to intravenous thus paralleling the uptake studies. Chronic dosing resulted in gene silencing for over 9 months with no adverse effects apparent. This study highlighted several of the virtues of conjugates including use of a molecularly defined entity, high tissue and cell selectivity, the ability to use the subcutaneous route, and lack of substantial toxicity. Although the siRNA doses required were higher than those needed for advanced SNALP systems, nonetheless the advantages of the conjugate approach seem clear.
There have been a number of other studies of ASGR-mediated delivery. The Alnylam group has further optimized the chemistry of the glycoconjugates, providing simpler alternatives for synthesis (259,260). Multivalent glycoconjugates have also been used to target novel uncharged siRNA entities (47) with the attainment of similar functional effects in cells and in vivo. GalNac based conjugates have also been used to deliver ASOs to the liver in mice with good effects on reduction of target gene expression (261). However, since in contrast to siRNAs, the unmodified ASOs also accumulate efficiently in liver, the fold enhancement observed for GalNac conjugated ASOs was not as great as that described for siRNAs. This exciting technology has elicited the interest of many groups and various publications on the chemistry and biology of GalNac oligonucleotide conjugates are starting to appear (262–264).
The successes with glycoconjugates in the laboratory have facilitated their rapid translation to clinical evaluation, with several hepatic genes being addressed. Thus a GalNac-siRNA conjugate for therapy of transthyretin mediated amyloidotic cardiomyopathy is now in Phase 3. Additionally, Phase 1 or 2 studies are currently in progress for siRNA glycoconjugates addressing hemophilia A or B, antitrypsin deficiency and porphyria (Clinical Trials.Gov)(5). While the long-term effectiveness and toxicity of siRNA glycoconjugates remain to be determined, at the present moment this approach seems to offer the most promising avenue for targeted oligonucleotide delivery to the liver.
Peptides
There have been a number of studies with peptide-oligonucleotide conjugates designed to bind to specific cell surface receptors. Much of the activity has focused on cyclic Arg-Gly-Asp (RGD) or other integrin ligands; these have been widely studied in connection with nanoparticle delivery (265) but there are only a limited number of studies of such ligands with conjugates. An early study demonstrated the feasibility of using RGD ligands to increase the cellular uptake and effectiveness of a SSO (244). In another report mono-, bi- tri- and tetra-valent RGD siRNA conjugates were tested in cell culture (266). Interestingly the tri- and tetra-valent conjugates provided stronger silencing than the other conjugates even under conditions where cell uptake was comparable. This suggests that valency may affect the intracellular trafficking of the conjugate in a manner that impinges on effectiveness. The most complete study with RGD ligands involved chemically modified siRNA directed against VEGF-Receptor 2 and conjugated to a PEG-RGD (267). Since VEGF is essential for tumor angiogenesis and since RGD is primarily a ligand for the alphavbeta3 integrin that is overexpressed in angiogenic endothelial cells, this combination seemed well suited to inhibit rapidly growing tumors. The RGD-siRNA conjugates were taken up by cultured endothelial cells via a receptor specific mechanism and reduced VEGF-R2 expression. In tumor bearing mice repeated dosing resulted in a modest reduction in VEGF-R2 expression but a significant reduction in tumor growth. No toxicity was apparent in these studies. Although the in vivo results were only partial it should be noted that the doses used (∼0.7 mg/kg) were rather low as compared to, for example, the doses of the GalNac conjugates described above.
A variety of other peptide ligands and their cognate receptors have been used for targeting. For example, the gastrin-releasing peptide receptor, a member of the GPCR superfamily, has been utilized to enhance delivery of a SSO (268) or a siRNA (269), both in cell culture. A later study used multivalent hybrid peptides that included the targeting sequence as well as several histidine residues designed to promote endosome destabilization via the proton sponge mechanism; however, the inclusion of the histidine produced only a modest improvement (270). Ligands targeting the gastrin-releasing peptide receptor have been extensively used in vivo for delivery of imaging agents and drugs (271) but such in vivo studies have not yet been done with oligonucleotides. Another study used an siRNA conjugate of insulin-like growth factor 1 for delivery in cultured cells (272), but this has not yet been followed up. Thus, at present, in vivo studies with molecular scale peptide-oligonucleotide conjugates lag far behind those with glycoconjugates. It is possible that this may indicate that there have been a number of unreported failures with this approach; however, it is also possible that determined efforts have not yet been made.
Aptamers and other oligonucleotides
Nucleic acid aptamers offer one of the most promising tools for targeted oligonucleotide delivery (273). Aptamers are nucleic acids that form three-dimensional (3D) structures via intra-molecular base pairing. Using the well-known SELEX technique, aptamers can be selected to bind to virtually any receptor with nanomolar to picomolar affinities (274). Although aptamers are usually generated by in vitro transcription and are many tens of bases in length, in some cases they can be redesigned and truncated so as to permit their production by solid phase chemical synthesis. This allows very efficient production of chimeras of the aptamer and the passenger strand of an siRNA, with the guide strand then added by base pairing. The use of aptamer-siRNA conjugates was pioneered by Sullenger et al. who described an aptamer that bound prostate-specific membrane antigen (PSMA) coupled to siRNAs directed toward key pro-survival genes. This chimera could effectively kill tumor cells that expressed PSMA but not those that did not. The chimera also had growth inhibitory effects when injected directly into the tumor (275). In later studies Giangrande et al. optimized the PSMA aptamer and conjugated it to PEG to improve in vivo characteristics (276). The modified aptamers displayed good antitumor activity at moderate doses and without apparent toxicity in a prostate cancer xenograft model. Subsequently, there have been a number of additional studies exploring aptamer-mediated delivery of siRNA in the cancer context. This includes use of bivalent aptamer-siRNA conjugates (277), aptamers that target growth receptors in leukemic cells (278), delivery of cancer inhibitory miRNAs via aptamers (279,280), as well as delivery of siRNAs that sensitize cells to chemotherapeutic agents (281). An interesting approach is to use an aptamer that is itself inhibitory to a growth promoting receptor linked an siRNA that also causes growth inhibition, thus potentially having a dual effect on tumor cells (278,279). While several of these approaches seem promising, thus far they have mostly been restricted to studies in cell culture.
A novel approach to use of aptamers in cancer immunotherapy was developed by Gilboa et al. Cancer cells use the nonsense-mediated decay (NMD) pathway to blunt expression of tumor antigens that might be recognized by the host immune system. A PMSA aptamer was used to deliver siRNAs that inhibit the NMD pathway to tumors thus enhancing the immune response to the tumor and causing significant inhibition of primary and metastatic tumor growth (282). More recently this group used a somewhat similar strategy to stimulate anti-tumor immunity by selective reduction of mTOR in CD8+ T-cells using a CD8 aptamer linked to an siRNA that silences TORC1, a component of the mTOR complex (283). Other investigators have also successfully used aptamer-siRNA chimeras to regulate tumor immunity (284)
There has also been a great deal of work on the use of aptamer-siRNA conjugates in HIV therapeutics with much of this pioneered by Rossi et al. (285). Thus aptamers that recognize the viral envelop protein gp120 were linked to siRNAs targeting viral genes such as tat/rev. These chimeric molecules afforded significant suppression of HIV in infected cells in culture as well as in HIV infected humanized mice (286,287). This group also developed a design approach that they call the ‘sticky bridge’ that allows facile assembly of siRNAs and aptamers. An interesting offshoot of work in this area involves using aptamer-siRNA chimeras to block genital transmission of HIV. This entails use of a vaginal gel containing CD4-binding aptamers linked to siRNAs that target HIV (288).
Although not derived by SELEX, another aptamer-like chimera involves CpG oligonucleotides linked to siRNAs. As mentioned above, TLR9 strongly binds to certain types of unmethylated CpG-rich DNA oligonucleotides. This has been developed as a strategy for siRNA delivery to normal and malignant hematopoietic cells (289) and dendritic cells (290). In the original study CpG oligonucleotides were chemically coupled to an siRNA targeting Stat3. The conjugates were effectively taken up by TLR9 positive cells resulting in silencing of Stat3. Since suppression of Stat 3 is thought to promote antitumor immunity, the CpG-siStat3 conjugates were tested in vivo and shown to inhibit growth in several tumor models (291).
Aptamers represent a powerful strategy for the targeted delivery of oligonucleotides. Nonetheless a decade after the initiation of this approach aptamer-oligonucleotide conjugates or chimeras have not progressed to clinical trials. There may be several reasons for this. Thus the relatively large size and complexity of typical RNA aptamers render them particular susceptible to nuclease degradation. The substitution of DNA-based aptamers may help with this issue (219). Further, although aptamers can be designed to bind to virtually any cell surface receptor, not all receptors effectively deliver cargo to the cell interior. Thus an interesting new development is the evolution of SELEX type approaches for identifying aptamers that are efficiently internalized into cells (292,293). In summary, aptamer mediated delivery of oligonucleotides remains a work in progress with exciting potential that has yet to be fully realized.
Antibodies
Monoclonal antibodies are potentially a powerful tool for targeting of oligonucleotides. However, it has become apparent that there are many complexities that will need to be addressed before this technology is ready for clinical development. Antibody-mediated siRNA delivery was first described by Lieberman et al. (294). In this study an immunoglobulin Fab-protamine chimera was created that targeted the HIV gp160 envelop protein. This was complexed with siRNAs addressed to key viral genes such as gag and could inhibit HIV in infected T-cells. In tumor cells ectopically expressing gp160, complexes of the chimera with siRNAs addressing key growth related genes could inhibit tumor cell growth both in culture and in vivo. Similar studies were also conducted with a scFv-protamine chimera that targeted the ErbB2 cell surface receptor. Another study used a scFv-protamine chimera that targeted Her2 to deliver growth inhibitory siRNAs to Her2 positive breast cancer cells causing retardation of tumor growth in an orthotopic breast cancer model (295). Utilizing a similar approach, another group developed a chemical conjugate of an anti-CD7 scFV with a positively charged arginine peptide; this was then complexed with anti HIV siRNAs. The complexes were used to treat HIV infected humanized mice resulting in alleviation of the disease (296). These initially promising studies shared a major problem; the antibody–siRNA complexes formed were probably ill-defined multimeric aggregates rather than defined molecular species. Thus further pharmaceutical development of these entities would be difficult.
More recently an extremely thorough study has delineated both the potential and the difficulties associated with antibody mediated delivery of siRNA (297). The authors used so-called THIOMABs previously developed for drug-antibody conjugates. This allowed chemical linkage of siRNAs at precisely defined positions on the antibody and resulted in the creation of molecularly defined conjugates. THIOMABs targeting a number of different cell surface receptors were tested where the receptors varied in terms of abundance and routes and rapidity of uptake and intracellular trafficking. While two receptors (TENB2 and NaPi2b) provided moderate silencing, the others did not. Nonetheless, the various THIOMABs and their linked siRNAs all seemed to accumulate in lysosomes indicating that there is not a simple correlation between overall subcellular localization and effect. The TENB2 antibody conjugate was tested in a mouse tumor model; however much of the material only penetrated a short distance from the vasculature. This study highlights several important issues. First, it is very difficult to identify cell surface receptors that can efficiently deliver oligonucleotides (although it should be noted that both of the effective receptors were very abundant). Second, the bulk intracellular trafficking pathways as visualized by fluorescence microscopy may not be relevant and that minor trafficking pathways may be functionally important. Third, conjugates of intact immunoglobulins with siRNA (∼180 000 MW) may have difficulties penetrating tissues. This study also emphasizes that endosome escape is a critical aspect of delivery. Thus the antibody-oligonucleotide delivery area is still evolving. It would be of great interest to bring the THIOMAB approach to much smaller protein targeting reagents such as scFvs or various molecular scaffolds based on DARPINs or fibronectin type III repeats (298).
Other macromolecular-scale conjugates
A very promising approach to oligonucleotide delivery involves so-called ‘dynamic polyconjugates’ (299). In the original conception these were comprised of a polymer with inherent endosome destabilizing properties linked simultaneously to a PEG shield, a carbohydrate ligand targeting the ASGPR and siRNA (300). The PEG linkage was designed to be degraded at low pH thus unmasking the polymer within the endosome; the siRNA was attached by a reducible disulfide linkage. These polyconjugates attained effective silence of genes in hepatocytes in culture and in mouse liver. In a later version (301) a membrane destabilizing peptide (mellitin like peptide, MLP) was conjugated to a ligand for ASGPR and to PEG (via a pH sensitive linker). The siRNA was conjugated to cholesterol thus promoting uptake by lipoprotein receptors. The concept was that the siRNA and the MLP, although taken up by different receptors, would both accumulate in an acidic endosome/lysosome compartment thus unleashing the MLP and causing endosomal destabilization and siRNA release. This version demonstrated impressive silencing effects in a mouse model of hepatitis B. The polyconjugate approach has progressed rapidly and is now in clinical trials; as well, new versions are being developed that may permit non-hepatic targeting (302).
Another form of macromolecular oligonucleotide conjugate involves linking oligonucleotides and targeting ligands to a serum protein. For example, RGD-PEG ligands have been conjugated to albumin, which was then further conjugated with a SSO via a bioreversible linkage (303). In another version the SSO was first conjugated to a ligand (RGD) and then to albumin (304). In both cases there was efficient receptor-selective delivery of the oligonucleotide to cells. It is possible to conjugate 10–15 oligonucleotides to each albumin thus providing a very high load of active agent. The conjugates are about 12 nm in diameter and thus should be large enough to avoid renal clearance but still small enough to penetrate into most tissues. For example, a RGD-SSO-albumin conjugate readily penetrated into a 3D tumor spheroid whereas a conventional nanoparticle could not (305). While this system does not yet include an endosome-destabilizing component it should be relatively easy to incorporate this function. Although still at an early stage of development, protein based oligonucleotide carriers seem to offer a simple and flexible delivery platform.
Small organic molecule conjugates
The pharmaceutical industry possesses a vast number of small organic molecule ligands—drugs or drug candidates—that demonstrate high affinity and selectivity for various receptors. Nonetheless there has been only limited progress to date with small molecule-oligonucleotide conjugates. Anisamide is a ligand that binds to the sigma receptor. Mono- and multivalent anisamide-SSO conjugates were synthesized and tested for biological activity in cells (306). Trivalent but not monovalent conjugates provided effective delivery of the SSO. Anandamide, a ligand for cannabinoid receptors, was conjugated to siRNA via click chemistry (242). These conjugates provided effective functional delivery of siRNA to several cell types. The folate receptor is believed to be upregulated in many types of cancer cells and folate has been extensively used as a targeting ligand for drugs and nanoparticles (307). A folate-PEG-siRNA conjugate was prepared by click chemistry (308). However, although efficiently taken up by cells expressing the folate receptor, the conjugate failed to achieve knock down of the targeted mRNA unless an additional transfection agent was used. Potentially, small organic ligands offer enormous versatility for selective delivery of oligonucleotides. However, their use has been constrained by several problems. First, it is often difficult to prepare an oligonucleotide conjugate without disrupting the ability of the small molecule to interact with its receptor. Second, these materials are relatively small in size and would be rapidly cleared in the kidney. Third, it is difficult to build endosome-disrupting capabilities into these conjugates.
Advantages and liabilities of molecular scale delivery
Molecular scale ligand-oligonucleotide conjugates offer several advantages in terms of delivery considerations. First, they are well-defined molecular entities that can be precisely characterized by standard techniques such as nuclear magnetic resonance and mass spectrometry. This contrasts with most nanoparticles that are heterogeneous in size and comprised of complex mixtures of lipids or polymers. The defined nature of molecular conjugates may be a key determinant in their future development as pharmaceutically and clinically acceptable entities. The second advantage involves their broad biodistribution. Thus, as with ‘free’ oligonucleotides, conjugates readily pass across the capillary endothelium and efficiently penetrate tissue parenchyma. A good example of this is the observation that intrathecally administered ASOs can diffuse widely in the brain (68). Once again this contrasts with nanocarriers that have a much more restricted biodistribution and that may have difficulty in diffusing through the dense extracellular matrix of many tissues. A third potential advantage may be greater selectivity. A single ligand–oligonucleotide conjugate can interact with its cognate receptor via a precise ‘key in lock’ mechanism. By contrast, even liganded nanoparticles may display non-specific binding to cell surfaces unless the nanoparticle surface is extremely well masked by PEGylation or similar means. To be fair, oligonucleotide conjugates can also display non-specific interactions to some degree. For example, the propensity of phosphorothioates to promiscuously stick to proteins may blunt the selectivity of targeted conjugates that use this backbone. Nonetheless, this issue seems more daunting for nanoparticle carriers. Finally, because of their relatively simple composition and lack of high positive charge density, there seems less opportunity for conjugates to generate non-specific toxicities than is the case for typical nanocarriers.
There are also some disadvantages to molecular scale oligonucleotide conjugates. A major one is their small size that allows rapid renal clearance. This can be countered to some degree by linking a PEG chain to the conjugate thus increasing its hydrodynamic radius, but nonetheless remains an issue. A second potential problem involves the ‘payload’. When an oligonucleotide-laden nanocarrier is taken up by a cell, hundreds or thousands of copies of the oligonucleotide are potentially available. By contrast, each uptake event of a molecular conjugate conveys only a single oligonucleotide; thus, many events are required to accumulate sufficient oligonucleotide for a pharmacological effect. In this context, it is interesting to compare effects of the siRNA glycoconjugates described above with those of recent types of siRNA LNPs. While both approaches can effectively silence expression of hepatic genes, the LNP system can do so a doses of 0.1 mg/kg as compared to >5 mg/kg for the glycoconjugate (175,259). A final issue concerns stability. Clearly one of the advantages of nanocarriers is their ability to protect the oligonucleotide load from nucleases; this is less true of conjugates. For conjugates based on highly stable backbones such as PMOs or LNAs this is not a great concern; however, even stabilized forms of siRNA remain somewhat susceptible to nucleases.
Macromolecular scale conjugates occupy a mid ground between nanocarriers and simple conjugates. Size considerations are very important here. For example, the ‘dynamic polyconjugates’ described above are designed so as to reduce renal clearance but still obtain a broad tissue distribution. Nonetheless size constraints may affect tissue penetration, as was the case for the THIOMAB antibody conjugates. Macromolecular conjugates share with nanocarriers the ability to convey multiple oligonucleotides in a single uptake event, but may have less toxic potential, particularly if no polycationic components are used.
In summary then, there are advantages of molecular or macromolecular scale oligonucleotide conjugates over nanocarriers that may be critical in certain therapeutic situations. However, nanocarriers may provide a useful approach in certain instances, with hepatic delivery being one important possibility.
Unusual approaches
Here, we discuss three oligonucleotide delivery approaches that are quite different from the ones mentioned above. Spherical nucleic acids (SNAs) and DNA nanostructures represent two novel nanotechnology-based approaches, while exosome-based delivery utilizes an important endogenous biological mechanism.
Spherical nucleic acids (SNAs)
Mirkin et al. have developed novel nanocarriers based on the dense absorbtion of oligonucleotides to gold nanoparticles via metal-thiol bonds. In two very thorough studies, SNAs were shown to be taken up into a variety of cell types by a process that involves scavenger receptors, caveolae and trafficking to EEs and LEs, but not lysosomes (309,310). The nucleic acid is gradually degraded and released from the cell while the core is not. SNAs have now been evaluated in several therapeutic contexts (311–313). For example, gold-siRNA nanoparticles were able to reduce target mRNA expression in glioblastoma cells in culture. Further, in a glioblastoma xenograft model, good distribution of the nanoparticles within the tumor was observed both after local and systemic administration, suggesting that SNAs can penetrate the tumor-compromised BBB. Finally an SNA directed against a key survival factor for glioblastoma was effective in reducing tumor growth (311). Interestingly, although anti-tumor effects were seen, the overall biodistribution of SNA was similar to that observed with other nanoparticles, with liver predominating. SNAs seem to provide an interesting and flexible technology. It is possible to adapt these nanoparticles for targeting (314) and to use materials other than gold as the core (315). A possible major impediment for this approach is that the SNAs seem to suffer from the same clearance and biodistribution issues as conventional lipid or polymer nanoparticles. However, further manipulation of particle size or of the PEGylation of the particles may be helpful.
Exosomes
Virtually all cells shed a variety of small membranous vesicles. Exosomes, which are ∼30–100 nm in diameter, represent a particular type of vesicle that is generated via the ESCRT machinery. Thus the membrane lipid and protein composition of exosomes closely parallels those of the LE/MVB compartment. Additionally exosomes contain a sampling of cytosolic small molecules, proteins and nucleic acids including both mRNAs and miRNAs (316). Exosomes shed into the extracellular environment can interact with cells in a variety of ways including binding to and stimulating receptors, transferring membrane lipids and proteins, and most interestingly in terms of this review, delivering RNAs (317,318). Because of these multiple interactions there has been a great deal of recent interest in translational aspects of exosomes for both diagnostic and therapeutic purposes and a number of clinical trials involving these vesicles are now underway (ClinTrials.gov). Recent studies have used sophisticated imaging techniques to document the dynamic exosome-mediated transfer of both membrane proteins and mRNA between cells thus validating this concept (319).
Pioneering work on exosome-mediated delivery of siRNA was initiated by Wood et al. (320). They expressed a fusion protein of LAMP2b, a LE/lysosome marker, with a rabies virus peptide sequence that binds the acetylcholine receptor. Exosomes containing the fusion protein were isolated and then loaded with siRNAs. The exosomes were efficiently taken up by neuronal cells in culture resulting in ‘knockdown’ of the siRNA target. Surprisingly, the neuronal-targeted exosomes also delivered the siRNA to the brain causing reduction of the cognate mRNA and protein. There have been a number of additional studies from various groups regarding the delivery of both miRNAs and siRNAs using exosomes, with promising results obtained both in cell culture and in vivo (321–323).
Thus exosomes provide an exciting new avenue for delivery of oligonucleotides. An appealing aspect is the ability to express chimeric proteins on the exosome membrane thus directly engineering targeting capabilities into these vesicles. However, a number of challenges remain. Despite a great deal of effort (324), the production and characterization of exosomes remains poorly defined. This is clearly an impediment to the development of pharmaceutically acceptable delivery strategies using exosomes.
DNA nanostructures
DNA nanostructures represent an exciting new avenue for biotechnology research and development (325,326). DNA can be organized into precisely designed two- and 3D structures by hybridization of complementary sequences. Several synthetic strategies are possible including the ‘DNA origami’ approach wherein long strands of DNA are structured using short DNA ‘staples’, as well as other strategies; powerful computational tools are available to aid in precise design of the intended structures. Delivery of oligonucleotides is one of the many potential biomedical applications of DNA nanostructures, but one that has elicited considerable recent interest. One of the unique features of DNA nanostructures is that targeting capabilities based on nucleic acid aptamers can be directly incorporated into the nanostructure. In an elegant example of this approach aptamers were built into a DNA ‘nanobox’. When the aptamers engaged their cognate cellular ligands the box opened to release entrapped contents (327). Other studies have also used aptamers for the delivery of DNA nanostructures (328,329). In a similar fashion, antisense or siRNA molecules can be incorporated into the design of the nanostructure. One very thorough study created ∼30 nm tetrahedral nanostructures incorporating siRNA designed to act on luciferase message; the nanostructures were further conjugated with folate. These entities were efficiently taken up in cell culture by tumor cells expressing folate receptor and could cause reduction of luciferase mRNA. In vivo the nanostructures were rapidly cleared, but surprisingly and unlike most nanoparticles, they avoided capture by the liver. Partial siRNA effects were attained in a tumor xenograft model (330). In an interesting variant of the nanostructure approach, rolling circle transcription was used to generate RNAs containing multiple siRNA hairpins; these were complexed with short, ligand-modified DNA oligonucleotides to form nucleic acid nanoparticles that were active in cells and in vivo (331). An interesting feature is that stable nanocomplexes were formed in the absence of any polycationic materials, thus avoiding potential toxicities. In another interesting approach CpG oligonucleotides were incorporated into DNA nanostructures resulting in enhanced immuno-stimulation of macrophages in cell culture (332) and in vivo (333).
Certainly DNA nanostructures have exciting potential for the future. They can be precisely engineered at the nanoscale, can readily incorporate both functional nucleic acid sequences as well as aptameric or small molecule targeting ligands, and can potentially be made sufficiently small (<20 nm) so as to access non-hepatic tissues. However, to date there is little experience with the in vivo biodistribution and stability properties of these entities. As anionic polymers they would presumably interact with scavenger receptors and possibly TLRs thus creating signaling cascades that may be detrimental.
Small molecules that enhance oligonucleotide effects
An important new thrust for oligonucleotide therapeutics involves the concept of using small organic molecules to enhance the pharmacological effects of antisense and siRNA. Conceptually small molecules might influence any aspect of oligonucleotide behavior including cellular uptake, intracellular trafficking, access to the cytosol and nucleus and the function of the final effectors such as the RISC complex, RNaseH or the splicesosome. However, most of the molecules discovered to date affect some aspect of oligonucleotide uptake or trafficking.
An early example of this thrust involved a compound called Retro-1. This compound originated from a high throughput screen seeking molecules that could block the actions of certain bacterial and plant toxins that utilize the retrograde trafficking pathway (334). Although there was no obvious connection between oligonucleotides and bacterial toxins, Retro-1 was tested for its ability to enhance the effects of oligonucleotides. Somewhat surprisingly this molecule provided a modest (∼10-fold) augmentation of SSO and ASO effects in the absence of any conventional transfection agent (335). Further experiments showed that the functional effects of Retro-1 were correlated with selective release of the SSO from the LE compartment. Retro-1 treatment also provided a small enhancement of the in vivo effects of an SSO in a mouse tumor model. However, there were some liabilities associated with this compound in that concentrations near 100 uM were needed and further that the lipophilicity of the compound made in vivo experiments difficult. Thus efforts commenced to find compounds superior to Retro-1.
An unbiased high-throughput screen was developed to seek oligonucleotide-enhancing compounds. The screen utilized a cell line stably transfected with a luciferase reporter whose expression could be increased by successful delivery of an SSO. Approximately 150 000 compounds were tested (in the absence of conventional transfection agents) leading to the discovery of three structurally distinct compound families. The screen and characterization of one family of compounds have recently been reported (336). One of the hits from the screen (UNC7938) provided ∼200-fold enhancement of SSO effects in cell culture when used at a non-toxic concentration (10 uM). This molecule could also substantially enhance the effects of ASOs and siRNA. By expressing GFP chimeras of proteins that are known markers of particular endomembrane compartments it was established that the functional effects of UNC7938 were correlated with selective release of oligonucleotide from LEs, similar to the case of Retro-1. In an in vivo model using transgenic mice with an EGFP reporter inducible by SSOs, significant enhancing effects of UNC7938 were found in several tissues including heart, kidney and liver. Thus this report illustrated the possibility of discovering small molecules that can profoundly enhance the effectiveness of oligonucleotides both in cell culture and in vivo. Work on the initial ‘hits’ from this screen is continuing with the goal of finding analogs with greater potency and less toxicity.
Other groups have also utilized screening of compound libraries to seek oligonucleotide enhancing small molecules. In one study a small library of drug-like molecules was screened for their effects on a highly modified siRNA in a dual luciferase reporter system (337). One hit compound, a known drug called Guanabenz, provided a substantial increase in potency and efficacy of the modified siRNA in cell culture when used in the 50–80 uM range. In mechanistic studies it was determined that Guanabenz increases internalization; this molecule is positively charged and apparently forms a molecular complex with the siRNA that promotes cellular uptake. Thus the actions of Guanabenz may be similar to previously reported small cationic molecules that bind to siRNA and enhance its uptake by cells (338). Several compounds related to Guanabenz were also examined to evaluate structure activity relationships. Interestingly, Guanabenz had little effect on forms of oligonucleotide other than the highly modified siRNA, thus potentially limiting its utility. No in vivo data was provided in this study.
Another interesting report described effects of small molecules on two siRNA delivery approaches, namely siRNA LNPs and cholesterol conjugated siRNA (339). Approximately 45 000 compounds were screened in a GFP-expressing cell line using a sophisticated automated fluorescence microscopy approach. The study first determined that the LNPs and the cholesterol-siRNA entered cells by two distinct endocytotic pathways. Upon screening it was determined that 25 compounds improved effects of LNPs while 28 improved effects of cholesterol-siRNA, while only two compounds influenced both. Further mechanistic studies placed the compounds into two groups; those that affect uptake and those that affect endosome escape. By altering the timing of the addition of the test compounds, it was determined that some compounds acted directly on cells while others bound to LNPs to exert their effects. The set of compounds that acted via endosome escape seemed to do so by several different mechanisms, suggesting that there are multiple ways to enhance delivery to the cytosol. Most of the compounds reported in this study were active in the 10 uM range. This study did not examine in vivo effects of the hit compounds.
A very interesting report described the actions of a known drug, dantroline, on enhancing the effects of a SSO in the Duchene muscular dystrophy model (340). However, as dantroline acts directly on the muscle Ryanodine receptor (RyR1), its effects may be limited to muscle cells. Another study used a plant-derived sterol-glycoside to enhance the delivery of siRNA by lipid- or polypeptide-based nanocarriers (341). In the case of the LNPs the small molecule could be directly incorporated into the nanocarrier; this might be very advantageous in terms of making sure that the enhancing molecule and the oligonucleotide are in the same compartment.
In summary, several reports have shown that it is possible to identify small molecules that substantially enhance the pharmacological effects of oligonucleotides. Most of these molecules act either by promoting uptake of the oligonucleotide or by allowing its escape from endosomes. Although it has been known for many years that high concentrations of classic lysosomotropic small molecules like chloroquine can cause endosome escape, many of the recently discovered small molecules achieve the same end but by mechanisms quite distinct from chloroquine (336,339). The small molecule enhancers described to date are largely initial hits from high-throughput screens. Typically such hits have less than ideal characteristics and substantial medicinal chemistry work is required to develop related molecules with greater potency, reduced toxicity and improved pharmacokinetic and biodistribution properties. Although this entails a great deal of effort it may be worthwhile especially if small molecule enhancers can improve oligonucleotide effects in tissues other than the liver. The possible role of small molecule enhancers in the clinical development of oligonucleotides might be viewed as being akin to other drug adjuncts such as the use of beta-lactamase inhibitors with penicillins.
Several particularly interesting recent publications on oligonucleotide delivery are listed in Table 2.
Table 2. Selected recent publications on oligonucleotide delivery.
| Delivery approach | Summary | Significance | Reference |
|---|---|---|---|
| LNPs | Tested a large library of ‘lipoids’ for siRNA delivery in cells and in vivo | Discovered structural and pKa features for design of effective LNPs | (175) |
| Polymer NPs | Developed a hybrid lipid-polymer NP for siRNA delivery | Polymer NPs had excellent pharmacokinetic characteristics and were effective in silencing in a tumor model | (199) |
| Antibody-siRNA conjugates | THIOMAB monoclonals with siRNA conjugation at specific sites were used to test siRNA delivery | Highlights the variability and difficulty in using antibody-siRNA conjugates | (297) |
| Molecular scale ligand–oligonucleotide conjugates | Developed trivalent glycoconjugates targeting the ASGPR; these provided effective siRNA delivery to the liver | Exemplifies the advantages of conjugates including defined molecular properties, selectivity and efficacy | (258) |
| Macromolecular scale conjugates | Developed a targeted polymer-PEG-siRNA conjugate that was effective in delivery to the liver | This is the initial report on ‘dynamic polyconjugates’ which seem to be a promising approach for delivery | (300) |
| Delivery to the CNS | ASOs were used to antagonize a lncRNA involved in a neurodevelopmental disorder | Exemplifies effective use of ASOs in the CNS | (73) |
| DNA nanostructures | Developed tetrahedral DNA nanostructures incorporating siRNA with folate for targeting; these were effective in silencing both in cells and in a xenograft model | Provides a thorough study of the potential of using DNA nanostructures for oligonucleotide delivery | (330) |
| Basic studies: description of oligonucleotide uptake and subcellular trafficking | These three studies used advanced microscopic techniques to quantitatively analyze the uptake, trafficking and delivery of siRNAs | Provide important insights into the intracellular fate of oligonucleotides | (158,181,182) |
| Basic studies: role of the trafficking machinery in oligonucleotide pharmacology | Used shRNA libraries to identify genes important in determining the effectiveness of oligonucleotides; found that TSG101, an ESCRT component, is vital | Demonstrates that the intracellular trafficking machinery is a key determinant of oligonucleotide pharmacology | (152) |
| Small molecules for oligonucleotide delivery | Used high-throughput screening to identify compounds that dramatically enhance the pharmacological effectiveness of oligonucleotides both in cells and in vivo; these compounds act by releasing oligonucleotides from unproductive entrapment in late endosomes | Demonstrates that small molecules can be used to manipulate the intracellular trafficking of oligonucleotides in a beneficial manner | (336) |
A limited number of publications have been selected as excellent examples of oligonucleotide delivery strategies or of basic studies on delivery.
CONCLUSIONS
This is a very exciting time for the field of oligonucleotide therapeutics as a range of new approaches and potential new clinical applications have emerged. In addition to its traditional emphasis on ‘knocking down’ gene expression using ASOs or siRNA, the field has embraced new strategies including altering splicing patterns with SSOs and upregulating gene expression using antagomirs to miRNAs and ncRNAs. A key issue for the future is whether the CRISPR-Cas9 gene editing approach will move from its current reliance on viral vectors to greater use of non-viral delivery systems and thus benefit from the technologies developed for oligonucleotide delivery (162,342). One of the main arguments for oligonucleotide-based therapeutics has always been the ability to address targets that are not ‘druggable’ with conventional agents. This has now become even more important with the emergence of a plethora of ncRNAs that are involved in regulating many aspects of cell function. Clearly short oligonucleotide antagonists are the best tools for regulating the ncRNA regulators.
In this promising landscape, delivery remains a key obstacle. The issue is not simply to get oligonucleotides into tissues, but to get them to the intracellular sites where they actually function. It has been known for some time that the biodistribution of an oligonucleotide at the chemical level may have little to do with its functional biodistribution, which is controlled by endogenous cellular activities (343). Fortunately, challenging delivery issues are being addressed using the large variety of innovative approaches that have been described in this review. While no single delivery strategy will be optimal in all situations, there are some clear distinctions between the various approaches.
Currently ligand-oligonucleotide conjugates seem to offer the greatest potential for future development. The fact that they are distinct molecular entities makes them attractive for large-scale pharmaceutical development. Their relatively small size assures a broad biodistribution, although this may be tempered somewhat by rapid renal clearance. Finally, the use of high affinity, highly selective ligands allow the possibility of very precise targeting to particular cells or tissues. Despite these positive aspects there remain many issues and unknowns. The exemplar of molecular scale conjugates has been the triantennary GalNac conjugates that interact with the ASGPR. However, their success may be predicated on a number of factors that are unique to this situation. This would include: (i) the liver being a highly perfused tissue thus allowing rapid conjugate uptake to overwhelm renal clearance; (ii) the ASGPR being very abundant and with a rapid turnover; (iii) a poorly defined propensity of the liver endomembrane system to be somewhat ‘leaky’ thus allowing oligonucleotide release to the cytosol. The question remains whether comparable effects can be achieved in non-hepatic tissues. Other receptors, for example members of the integrin family, are expressed at levels similar to ASGPR but may not recycle as rapidly. To attain success in selective delivery to non-hepatic tissues it will be essential to have a deep understanding of the biology of the receptors being used for targeting; this would include their abundance, turnover and pathways of intracellular trafficking. Another key issue will be the valency of the ligand and its topology. Receptor cross-linking can often accelerate internalization but it may also disturb the delicate balance between receptor recycling and degradation. Finally, more information about how ligand-oligonucleotide conjugates traffic within the cellular endomembrane machinery will be essential in optimizing their design.
Lipid and polymer-based nanocarriers remain important tools for oligonucleotide delivery. As discussed above, there are two major liabilities associated with nanoparticles. The first is their limited biodistribution and the second is toxicity due to use of polycationic components. However, both of these may be amenable to further refinement. An emerging aspect is the development of unconventional nanocarriers such as SNAs and DNA nanostructures. One intriguing aspect of both of these technologies is that they can explore a size range substantially smaller than the traditional 100 nm nanoparticle that may lead to improved biodistribution. In the case of DNA nanostructures clever computer-assisted design will allow the incorporation of both aptameric targeting ligands and pharmacologically active oligonucleotides into the structure itself. DNA nanostructures also have the advantage of being precisely defined entities, unlike conventional nanoparticles.
Targeted macromolecular scale carriers based on proteins or small non-toxic polymers may incorporate many of the best features of both traditional nanocarriers and molecular scale conjugates. Thus they can include several ligands to provide a high avidity for the receptor, they can carry multiple copies of the oligonucleotide, and they can be designed to be small enough to penetrate the parenchyma of many tissues. Additionally it has been possible to incorporate endsome-lytic capabilities into such conjugates (344). However, the more features built into the conjugate the greater the possibility of toxicity and the greater the difficulty in scale-up and production.
Finally, the emergence of several types of small molecules that enhance oligonucleotide effectiveness potentially provides on exciting new thrust in the oligonucleotide therapeutics field. The field has tended to eschew medicinal chemistry or chemical biology approaches, viewing itself as an alternative to those more conventional paths of drug development. However, it is becoming clear that small molecules can play a helpful role both in understanding and in improving the pharmacological effects of oligonucleotides. The events of endocytosis and intracellular trafficking present a complex drama supported by a cast of hundreds if not thousands of proteins. Each of those proteins provides a potential target for a small molecule that may influence the trafficking process. Thus conceivably it may be possible to find small molecules that enhance the actions of oligonucleotides by affecting their endocytosis and trafficking in many different ways. This might include altering receptor internalization and recycling, affecting the rapidity or extent of endosome maturation, causing release of contents from particular endomembrane compartments or steering the trafficking flow to alternative compartments. There has been relatively little work done on the chemical biology of intracellular trafficking and it seems quite possible to design high-throughput screens to identify small molecules that affect distinct aspect of trafficking.
In summary, a variety of creative approaches have addressed the challenge of oligonucleotide delivery. It seems likely that many of the issues in the oligonucleotide therapeutics arena can be dealt with by judicious application of delivery strategies that currently exist in mature form or that are rapidly emerging. However, the creation of broadly applicable delivery approaches that are highly effective and without significant toxicity in humans remains to be attained. Additionally these approaches must be cost-effective in the context of an increasingly pressured health care system.
FUNDING
NIH [CA 151964 to RLJ]. Funding for open access charge: UNC trust funds (to R.L.J.).
Conflict of interest statement. The author is a founder of Initos Pharmaceuticals LLC, a small company that works in the area of oligonucleotide delivery. The content of this manuscript entirely reflects the author's personal views on the field and does not reflect the views of the company.
REFRENCES
- 1.Haussecker D., Kay M.A. RNA interference. Drugging RNAi. Science. 2015;347:1069–1070. doi: 10.1126/science.1252967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hair P., Cameron F., McKeage K. Mipomersen sodium: first global approval. Drugs. 2013;73:487–493. doi: 10.1007/s40265-013-0042-2. [DOI] [PubMed] [Google Scholar]
- 3.Suhr O.B., Coelho T., Buades J., Pouget J., Conceicao I., Berk J., Schmidt H., Waddington-Cruz M., Campistol J.M., Bettencourt B.R., et al. Efficacy and safety of patisiran for familial amyloidotic polyneuropathy: a phase II multi-dose study. Orphanet J. Rare Dis. 2015;10:109. doi: 10.1186/s13023-015-0326-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mendell J.R., Rodino-Klapac L.R., Sahenk Z., Roush K., Bird L., Lowes L.P., Alfano L., Gomez A.M., Lewis S., Kota J., et al. Eteplirsen for the treatment of Duchenne muscular dystrophy. Ann. Neurol. 2013;74:637–647. doi: 10.1002/ana.23982. [DOI] [PubMed] [Google Scholar]
- 5.Vaishnaw A. Oligonucleotide Therapeutics Society. Netherlands: Leiden; 2015. [Google Scholar]
- 6.Janssen H.L., Reesink H.W., Lawitz E.J., Zeuzem S., Rodriguez-Torres M., Patel K., van der Meer A.J., Patick A.K., Chen A., Zhou Y., et al. Treatment of HCV infection by targeting microRNA. N. Engl. J. Med. 2013;368:1685–1694. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- 7.Buller H.R., Bethune C., Bhanot S., Gailani D., Monia B.P., Raskob G.E., Segers A., Verhamme P., Weitz J.I., Investigators F.-A.T. Factor XI antisense oligonucleotide for prevention of venous thrombosis. N. Engl. J. Med. 2015;372:232–240. doi: 10.1056/NEJMoa1405760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lammers P. Oligonucleotide Therapeutics Society. Netherlands: Leiden; 2015. [Google Scholar]
- 9.Zanetta C., Nizzardo M., Simone C., Monguzzi E., Bresolin N., Comi G.P., Corti S. Molecular therapeutic strategies for spinal muscular atrophies: current and future clinical trials. Clin. Ther. 2014;36:128–140. doi: 10.1016/j.clinthera.2013.11.006. [DOI] [PubMed] [Google Scholar]
- 10.Lorenzer C., Dirin M., Winkler A.M., Baumann V., Winkler J. Going beyond the liver: progress and challenges of targeted delivery of siRNA therapeutics. J. Control. Release. 2015;203:1–15. doi: 10.1016/j.jconrel.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 11.Wittrup A., Lieberman J. Knocking down disease: a progress report on siRNA therapeutics. Nat. Rev. Genet. 2015;16:543–552. doi: 10.1038/nrg3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu S.Y., Lopez-Berestein G., Calin G.A., Sood A.K. RNAi therapies: drugging the undruggable. Sci. Transl. Med. 2014;6:240ps247. doi: 10.1126/scitranslmed.3008362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bobbin M.L., Rossi J.J. RNA interference (RNAi)-based therapeutics: delivering on the promise? Annu. Rev. Pharmacol. Toxicol. 2016;56:103–122. doi: 10.1146/annurev-pharmtox-010715-103633. [DOI] [PubMed] [Google Scholar]
- 14.Roberts T.C., Ezzat K., El Andaloussi S., Weinberg M.S. Synthetic SiRNA delivery: progress and prospects. Methods Mol. Biol. 2016;1364:291–310. doi: 10.1007/978-1-4939-3112-5_23. [DOI] [PubMed] [Google Scholar]
- 15.Nguyen J., Szoka F.C. Nucleic acid delivery: the missing pieces of the puzzle? Acc. Chem. Res. 2012;45:1153–1162. doi: 10.1021/ar3000162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pennisi E. Genomics. ENCODE project writes eulogy for junk DNA. Science. 2012;337:1159–1161. doi: 10.1126/science.337.6099.1159. [DOI] [PubMed] [Google Scholar]
- 17.Wahlestedt C. Targeting long non-coding RNA to therapeutically upregulate gene expression. Nat. Rev. Drug Discov. 2013;12:433–446. doi: 10.1038/nrd4018. [DOI] [PubMed] [Google Scholar]
- 18.Crooke S.T. Antisense strategies. Curr. Mol. Med. 2004;4:465–487. doi: 10.2174/1566524043360375. [DOI] [PubMed] [Google Scholar]
- 19.Bennett C.F., Swayze E.E. RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu. Rev. Pharmacol. Toxicol. 2010;50:259–293. doi: 10.1146/annurev.pharmtox.010909.105654. [DOI] [PubMed] [Google Scholar]
- 20.Kole R., Krainer A.R., Altman S. RNA therapeutics: beyond RNA interference and antisense oligonucleotides. Nat. Rev. Drug Discov. 2012;11:125–140. doi: 10.1038/nrd3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Disterer P., Kryczka A., Liu Y., Badi Y.E., Wong J.J., Owen J.S., Khoo B. Development of therapeutic splice-switching oligonucleotides. Hum. Gene Ther. 2014;25:587–598. doi: 10.1089/hum.2013.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mello C.C., Conte D., Jr Revealing the world of RNA interference. Nature. 2004;431:338–342. doi: 10.1038/nature02872. [DOI] [PubMed] [Google Scholar]
- 23.Hammond S.M. An overview of microRNAs. Adv. Drug Deliv. Rev. 2015;87:3–14. doi: 10.1016/j.addr.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meister G., Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- 25.Meister G. Argonaute proteins: functional insights and emerging roles. Nat. Rev. Genet. 2013;14:447–459. doi: 10.1038/nrg3462. [DOI] [PubMed] [Google Scholar]
- 26.Burnett J.C., Rossi J.J. RNA-based therapeutics: current progress and future prospects. Chem. Biol. 2012;19:60–71. doi: 10.1016/j.chembiol.2011.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holoch D., Moazed D. RNA-mediated epigenetic regulation of gene expression. Nat. Rev. Genet. 2015;16:71–84. doi: 10.1038/nrg3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.St Laurent G., Wahlestedt C., Kapranov P. The Landscape of long noncoding RNA classification. Trends Genet. 2015;31:239–251. doi: 10.1016/j.tig.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ling H., Fabbri M., Calin G.A. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat. Rev. Drug Discov. 2013;12:847–865. doi: 10.1038/nrd4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kormann M.S., Hasenpusch G., Aneja M.K., Nica G., Flemmer A.W., Herber-Jonat S., Huppmann M., Mays L.E., Illenyi M., Schams A., et al. Expression of therapeutic proteins after delivery of chemically modified mRNA in mice. Nat. Biotechnol. 2011;29:154–157. doi: 10.1038/nbt.1733. [DOI] [PubMed] [Google Scholar]
- 31.Mandal P.K., Rossi D.J. Reprogramming human fibroblasts to pluripotency using modified mRNA. Nat. Protoc. 2013;8:568–582. doi: 10.1038/nprot.2013.019. [DOI] [PubMed] [Google Scholar]
- 32.Yin Q., Fu T.M., Li J., Wu H. Structural biology of innate immunity. Annu. Rev. Immunol. 2015;33:393–416. doi: 10.1146/annurev-immunol-032414-112258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sioud M. Deciphering the code of innate immunity recognition of siRNAs. Methods Mol. Biol. 2009;487:41–59. doi: 10.1007/978-1-60327-547-7_2. [DOI] [PubMed] [Google Scholar]
- 34.Goubau D., Schlee M., Deddouche S., Pruijssers A.J., Zillinger T., Goldeck M., Schuberth C., Van der Veen A.G., Fujimura T., Rehwinkel J., et al. Antiviral immunity via RIG-I-mediated recognition of RNA bearing 5′-diphosphates. Nature. 2014;514:372–375. doi: 10.1038/nature13590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krieg A.M. CpG still rocks! Update on an accidental drug. Nucleic Acid Ther. 2012;22:77–89. doi: 10.1089/nat.2012.0340. [DOI] [PubMed] [Google Scholar]
- 36.Pyle A. Oligonucleotide Therapeutics Society. Netherlands: Leiden; 2015. [Google Scholar]
- 37.Kandimalla E.R., Bhagat L., Wang D., Yu D., Sullivan T., La Monica N., Agrawal S. Design, synthesis and biological evaluation of novel antagonist compounds of Toll-like receptors 7, 8 and 9. Nucleic Acids Res. 2013;41:3947–3961. doi: 10.1093/nar/gkt078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jason T.L., Koropatnick J., Berg R.W. Toxicology of antisense therapeutics. Toxicol. Appl. Pharmacol. 2004;201:66–83. doi: 10.1016/j.taap.2004.04.017. [DOI] [PubMed] [Google Scholar]
- 39.Liang X.H., Sun H., Shen W., Crooke S.T. Identification and characterization of intracellular proteins that bind oligonucleotides with phosphorothioate linkages. Nucleic Acids Res. 2015;43:2927–2945. doi: 10.1093/nar/gkv143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Geary R.S., Norris D., Yu R., Bennett C.F. Pharmacokinetics, biodistribution and cell uptake of antisense oligonucleotides. Adv. Drug Deliv. Rev. 2015;87:46–51. doi: 10.1016/j.addr.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 41.Deleavey G.F., Damha M.J. Designing chemically modified oligonucleotides for targeted gene silencing. Chem. Biol. 2012;19:937–954. doi: 10.1016/j.chembiol.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 42.Watts J.K., Corey D.R. Silencing disease genes in the laboratory and the clinic. J. Pathol. 2012;226:365–379. doi: 10.1002/path.2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sharma V.K., Watts J.K. Oligonucleotide therapeutics: chemistry, delivery and clinical progress. Future Med. Chem. 2015;16:221–242. doi: 10.4155/fmc.15.144. [DOI] [PubMed] [Google Scholar]
- 44.Eckstein F. Phosphorothioates, essential components of therapeutic oligonucleotides. Nucleic Acid Ther. 2014;24:374–387. doi: 10.1089/nat.2014.0506. [DOI] [PubMed] [Google Scholar]
- 45.Jarver P., O'Donovan L., Gait M.J. A chemical view of oligonucleotides for exon skipping and related drug applications. Nucleic Acid Ther. 2014;24:37–47. doi: 10.1089/nat.2013.0454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Campbell M.A., Wengel J. Locked vs. unlocked nucleic acids (LNA vs. UNA): contrasting structures work towards common therapeutic goals. Chem. Soc. Rev. 2011;40:5680–5689. doi: 10.1039/c1cs15048k. [DOI] [PubMed] [Google Scholar]
- 47.Meade B.R., Gogoi K., Hamil A.S., Palm-Apergi C., van den Berg A., Hagopian J.C., Springer A.D., Eguchi A., Kacsinta A.D., Dowdy C.F., et al. Efficient delivery of RNAi prodrugs containing reversible charge-neutralizing phosphotriester backbone modifications. Nat. Biotechnol. 2014;32:1256–1261. doi: 10.1038/nbt.3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pinheiro V.B., Taylor A.I., Cozens C., Abramov M., Renders M., Zhang S., Chaput J.C., Wengel J., Peak-Chew S.Y., McLaughlin S.H., et al. Synthetic genetic polymers capable of heredity and evolution. Science. 2012;336:341–344. doi: 10.1126/science.1217622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Komarova Y., Malik A.B. Regulation of endothelial permeability via paracellular and transcellular transport pathways. Annu. Rev. Physiol. 2010;72:463–493. doi: 10.1146/annurev-physiol-021909-135833. [DOI] [PubMed] [Google Scholar]
- 50.Tarbell J.M. Shear stress and the endothelial transport barrier. Cardiovasc. Res. 2010;87:320–330. doi: 10.1093/cvr/cvq146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maeda H., Nakamura H., Fang J. The EPR effect for macromolecular drug delivery to solid tumors: Improvement of tumor uptake, lowering of systemic toxicity, and distinct tumor imaging in vivo. Adv. Drug Deliv. Rev. 2013;65:71–79. doi: 10.1016/j.addr.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 52.Smith B.R., Cheng Z., De A., Rosenberg J., Gambhir S.S. Dynamic visualization of RGD-quantum dot binding to tumor neovasculature and extravasation in multiple living mouse models using intravital microscopy. Small. 2010;6:2222–2229. doi: 10.1002/smll.201001022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Prabhakar U., Maeda H., Jain R.K., Sevick-Muraca E.M., Zamboni W., Farokhzad O.C., Barry S.T., Gabizon A., Grodzinski P., Blakey D.C. Challenges and key considerations of the enhanced permeability and retention effect for nanomedicine drug delivery in oncology. Cancer Res. 2013;73:2412–2417. doi: 10.1158/0008-5472.CAN-12-4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gordon S., Taylor P.R. Monocyte and macrophage heterogeneity. Nat. Rev. Immunol. 2005;5:953–964. doi: 10.1038/nri1733. [DOI] [PubMed] [Google Scholar]
- 55.Yang Q., Jones S.W., Parker C.L., Zamboni W.C., Bear J.E., Lai S.K. Evading immune cell uptake and clearance requires PEG grafting at densities substantially exceeding the minimum for brush conformation. Mol. Pharm. 2014;11:1250–1258. doi: 10.1021/mp400703d. [DOI] [PubMed] [Google Scholar]
- 56.Suk J.S., Xu Q., Kim N., Hanes J., Ensign L.M. PEGylation as a strategy for improving nanoparticle-based drug and gene delivery. Adv. Drug Deliv. Rev. 2015;99:28–51. doi: 10.1016/j.addr.2015.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tseng Y.C., Mozumdar S., Huang L. Lipid-based systemic delivery of siRNA. Adv. Drug Deliv. Rev. 2009;61:721–731. doi: 10.1016/j.addr.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Benimetskaya L., Loike J.D., Khaled Z., Loike G., Silverstein S.C., Cao L., Khoury J., Cai T.Q., Stein C.A. Mac-1 (CD11b/CD18) is an oligodeoxynucleotide-binding protein. Nat. Med. 1997;3:414–420. doi: 10.1038/nm0497-414. [DOI] [PubMed] [Google Scholar]
- 59.Yu X., Guo C., Fisher P.B., Subjeck J.R., Wang X.Y. Scavenger receptors: emerging roles in cancer biology and immunology. Adv. Cancer Res. 2015;128:309–364. doi: 10.1016/bs.acr.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Butler M., Crooke R.M., Graham M.J., Lemonidis K.M., Lougheed M., Murray S.F., Witchell D., Steinbrecher U., Bennett C.F. Phosphorothioate oligodeoxynucleotides distribute similarly in class A scavenger receptor knockout and wild-type mice. J. Pharmacol. Exp. Ther. 2000;292:489–496. [PubMed] [Google Scholar]
- 61.Ezzat K., Aoki Y., Koo T., McClorey G., Benner L., Coenen-Stass A., O'Donovan L., Lehto T., Garcia-Guerra A., Nordin J., et al. Self-assembly into nanoparticles is essential for receptor mediated uptake of therapeutic antisense oligonucleotides. NANO Lett. 2015;15:4364–4373. doi: 10.1021/acs.nanolett.5b00490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shannahan J.H., Bai W., Brown J.M. Implications of scavenger receptors in the safe development of nanotherapeutics. Receptors Clin. Investig. 2015;2:e811. doi: 10.14800/rci.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Haraldsson B., Nystrom J., Deen W.M. Properties of the glomerular barrier and mechanisms of proteinuria. Physiol. Rev. 2008;88:451–487. doi: 10.1152/physrev.00055.2006. [DOI] [PubMed] [Google Scholar]
- 64.Geary R.S. Antisense oligonucleotide pharmacokinetics and metabolism. Expert Opin. Drug Metab. Toxicol. 2009;5:381–391. doi: 10.1517/17425250902877680. [DOI] [PubMed] [Google Scholar]
- 65.Dirin M., Winkler J. Influence of diverse chemical modifications on the ADME characteristics and toxicology of antisense oligonucleotides. Expert Opin. Biol. Ther. 2013;13:875–888. doi: 10.1517/14712598.2013.774366. [DOI] [PubMed] [Google Scholar]
- 66.Kang L., Wang R.F., Yan P., Liu M., Zhang C.L., Yu M.M., Cui Y.G., Xu X.J. Noninvasive visualization of RNA delivery with 99mTc-radiolabeled small-interference RNA in tumor xenografts. J. Nuclear Med. 2010;51:978–986. doi: 10.2967/jnumed.109.069906. [DOI] [PubMed] [Google Scholar]
- 67.Evers M.M., Toonen L.J., van Roon-Mom W.M. Antisense oligonucleotides in therapy for neurodegenerative disorders. Adv. Drug Deliv. Rev. 2015;87:90–103. doi: 10.1016/j.addr.2015.03.008. [DOI] [PubMed] [Google Scholar]
- 68.Southwell A.L., Skotte N.H., Bennett C.F., Hayden M.R. Antisense oligonucleotide therapeutics for inherited neurodegenerative diseases. Trends Mol. Med. 2012;18:634–643. doi: 10.1016/j.molmed.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 69.Southwell A.L., Skotte N.H., Kordasiewicz H.B., Ostergaard M.E., Watt A.T., Carroll J.B., Doty C.N., Villanueva E.B., Petoukhov E., Vaid K., et al. In vivo evaluation of candidate allele-specific mutant huntingtin gene silencing antisense oligonucleotides. Mol. Ther. 2014;22:2093–2106. doi: 10.1038/mt.2014.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lagier-Tourenne C., Baughn M., Rigo F., Sun S., Liu P., Li H.R., Jiang J., Watt A.T., Chun S., Katz M., et al. Targeted degradation of sense and antisense C9orf72 RNA foci as therapy for ALS and frontotemporal degeneration. Proc. Natl. Acad. Sci. U.S.A. 2013;110:E4530–E4539. doi: 10.1073/pnas.1318835110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rigo F., Hua Y., Krainer A.R., Bennett C.F. Antisense-based therapy for the treatment of spinal muscular atrophy. J. Cell Biol. 2012;199:21–25. doi: 10.1083/jcb.201207087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kaczmarek A., Schneider S., Wirth B., Riessland M. Investigational therapies for the treatment of spinal muscular atrophy. Expert Opin. Investig. Drugs. 2015;24:867–881. doi: 10.1517/13543784.2015.1038341. [DOI] [PubMed] [Google Scholar]
- 73.Meng L., Ward A.J., Chun S., Bennett C.F., Beaudet A.L., Rigo F. Towards a therapy for Angelman syndrome by targeting a long non-coding RNA. Nature. 2015;518:409–412. doi: 10.1038/nature13975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Boisguerin P., Deshayes S., Gait M.J., O'Donovan L., Godfrey C., Betts C.A., Wood M.J., Lebleu B. Delivery of therapeutic oligonucleotides with cell penetrating peptides. Adv. Drug Deliv. Rev. 2015;87:52–67. doi: 10.1016/j.addr.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Du L., Kayali R., Bertoni C., Fike F., Hu H., Iversen P.L., Gatti R.A. Arginine-rich cell-penetrating peptide dramatically enhances AMO-mediated ATM aberrant splicing correction and enables delivery to brain and cerebellum. Hum. Mol. Genet. 2011;20:3151–3160. doi: 10.1093/hmg/ddr217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Goyenvalle A., Griffith G., Babbs A., El Andaloussi S., Ezzat K., Avril A., Dugovic B., Chaussenot R., Ferry A., Voit T., et al. Functional correction in mouse models of muscular dystrophy using exon-skipping tricyclo-DNA oligomers. Nat. Med. 2015;21:270–275. doi: 10.1038/nm.3765. [DOI] [PubMed] [Google Scholar]
- 77.Alyautdin R., Khalin I., Nafeeza M.I., Haron M.H., Kuznetsov D. Nanoscale drug delivery systems and the blood-brain barrier. Int. J. Nanomed. 2014;9:795–811. doi: 10.2147/IJN.S52236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Clark A.J., Davis M.E. Increased brain uptake of targeted nanoparticles by adding an acid-cleavable linkage between transferrin and the nanoparticle core. Proc. Natl. Acad. Sci. U.S.A. 2015;112:12486–12491. doi: 10.1073/pnas.1517048112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bien-Ly N., Yu Y.J., Bumbaca D., Elstrott J., Boswell C.A., Zhang Y., Luk W., Lu Y., Dennis M.S., Weimer R.M., et al. Transferrin receptor (TfR) trafficking determines brain uptake of TfR antibody affinity variants. J. Exp. Med. 2014;211:233–244. doi: 10.1084/jem.20131660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Youn P., Chen Y., Furgeson D.Y. A myristoylated cell-penetrating peptide bearing a transferrin receptor-targeting sequence for neuro-targeted siRNA delivery. Mol. Pharm. 2014;11:486–495. doi: 10.1021/mp400446v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kumar P., Wu H., McBride J.L., Jung K.E., Kim M.H., Davidson B.L., Lee S.K., Shankar P., Manjunath N. Transvascular delivery of small interfering RNA to the central nervous system. Nature. 2007;448:39–43. doi: 10.1038/nature05901. [DOI] [PubMed] [Google Scholar]
- 82.Hwang do W., Son S., Jang J., Youn H., Lee S., Lee D., Lee Y.S., Jeong J.M., Kim W.J., Lee D.S. A brain-targeted rabies virus glycoprotein-disulfide linked PEI nanocarrier for delivery of neurogenic microRNA. Biomaterials. 2011;32:4968–4975. doi: 10.1016/j.biomaterials.2011.03.047. [DOI] [PubMed] [Google Scholar]
- 83.Chauhan V.P., Stylianopoulos T., Boucher Y., Jain R.K. Delivery of molecular and nanoscale medicine to tumors: transport barriers and strategies. Annu. Rev. Chem. Biomol. Eng. 2011;2:281–298. doi: 10.1146/annurev-chembioeng-061010-114300. [DOI] [PubMed] [Google Scholar]
- 84.Cheng Z., Al Zaki A., Hui J.Z., Muzykantov V.R., Tsourkas A. Multifunctional nanoparticles: cost versus benefit of adding targeting and imaging capabilities. Science. 2012;338:903–910. doi: 10.1126/science.1226338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bander N.H. Antibody-drug conjugate target selection: critical factors. Methods Mol. Biol. 2013;1045:29–40. doi: 10.1007/978-1-62703-541-5_2. [DOI] [PubMed] [Google Scholar]
- 86.Hynes R.O. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110:673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 87.Margadant C., Monsuur H.N., Norman J.C., Sonnenberg A. Mechanisms of integrin activation and trafficking. Curr. Opin. Cell Biol. 2011;23:607–614. doi: 10.1016/j.ceb.2011.08.005. [DOI] [PubMed] [Google Scholar]
- 88.Sulzmaier F.J., Jean C., Schlaepfer D.D. FAK in cancer: mechanistic findings and clinical applications. Nat. Rev. Cancer. 2014;14:598–610. doi: 10.1038/nrc3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Juliano R.L. Signal transduction by cell adhesion receptors and the cytoskeleton: functions of integrins, cadherins, selectins, and immunoglobulin-superfamily members. Annu. Rev. Pharmacol. Toxicol. 2002;42:283–323. doi: 10.1146/annurev.pharmtox.42.090401.151133. [DOI] [PubMed] [Google Scholar]
- 90.Mulgrew K., Kinneer K., Yao X.T., Ward B.K., Damschroder M.M., Walsh B., Mao S.Y., Gao C., Kiener P.A., Coats S., et al. Direct targeting of alphavbeta3 integrin on tumor cells with a monoclonal antibody, Abegrin. Mol. Cancer Ther. 2006;5:3122–3129. doi: 10.1158/1535-7163.MCT-06-0356. [DOI] [PubMed] [Google Scholar]
- 91.Caswell P.T., Vadrevu S., Norman J.C. Integrins: masters and slaves of endocytic transport. Nat. Rev. Mol Cell Biol. 2009;10:843–853. doi: 10.1038/nrm2799. [DOI] [PubMed] [Google Scholar]
- 92.Desgrosellier J.S., Cheresh D.A. Integrins in cancer: biological implications and therapeutic opportunities. Nat. Rev. Cancer. 2010;10:9–22. doi: 10.1038/nrc2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Seguin L., Desgrosellier J.S., Weis S.M., Cheresh D.A. Integrins and cancer: regulators of cancer stemness, metastasis, and drug resistance. Trends Cell Biol. 2015;25:234–240. doi: 10.1016/j.tcb.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Millard M., Odde S., Neamati N. Integrin targeted therapeutics. Theranostics. 2011;1:154–188. doi: 10.7150/thno/v01p0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Giguere P.M., Kroeze W.K., Roth B.L. Tuning up the right signal: chemical and genetic approaches to study GPCR functions. Curr. Opin. Cell Biol. 2014;27:51–55. doi: 10.1016/j.ceb.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hanyaloglu A.C., von Zastrow M. Regulation of GPCRs by endocytic membrane trafficking and its potential implications. Annu. Rev. Pharmacol. Toxicol. 2008;48:537–568. doi: 10.1146/annurev.pharmtox.48.113006.094830. [DOI] [PubMed] [Google Scholar]
- 97.Whalen E.J., Rajagopal S., Lefkowitz R.J. Therapeutic potential of beta-arrestin- and G protein-biased agonists. Trends Mol. Med. 2011;17:126–139. doi: 10.1016/j.molmed.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cornelio D.B., Roesler R., Schwartsmann G. Gastrin-releasing peptide receptor as a molecular target in experimental anticancer therapy. Ann. Oncol. 2007;18:1457–1466. doi: 10.1093/annonc/mdm058. [DOI] [PubMed] [Google Scholar]
- 99.Juliano R.L., Carver K., Cao C., Ming X. Receptors, endocytosis, and trafficking: the biological basis of targeted delivery of antisense and siRNA oligonucleotides. J. Drug Target. 2013;21:27–43. doi: 10.3109/1061186X.2012.740674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lemmon M.A., Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141:1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Parachoniak C.A., Park M. Dynamics of receptor trafficking in tumorigenicity. Trends Cell Biol. 2012;22:231–240. doi: 10.1016/j.tcb.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 102.Shepard H.M., Jin P., Slamon D.J., Pirot Z., Maneval D.C. Herceptin. Handb. Exp. Pharmacol. 2008:183–219. doi: 10.1007/978-3-540-73259-4_9. [DOI] [PubMed] [Google Scholar]
- 103.Brodeur G.M., Minturn J.E., Ho R., Simpson A.M., Iyer R., Varela C.R., Light J.E., Kolla V., Evans A.E. Trk receptor expression and inhibition in neuroblastomas. Clin. Cancer Res. 2009;15:3244–3250. doi: 10.1158/1078-0432.CCR-08-1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Koch S., Claesson-Welsh L. Signal transduction by vascular endothelial growth factor receptors. Cold Spring Harb Perspect. Med. 2012;2:a006502. doi: 10.1101/cshperspect.a006502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Imai Y., Leung C.K., Friesen H.G., Shiu R.P. Epidermal growth factor receptors and effect of epidermal growth factor on growth of human breast cancer cells in long-term tissue culture. Cancer Res. 1982;42:4394–4398. [PubMed] [Google Scholar]
- 106.Napione L., Pavan S., Veglio A., Picco A., Boffetta G., Celani A., Seano G., Primo L., Gamba A., Bussolino F. Unraveling the influence of endothelial cell density on VEGF-A signaling. Blood. 2012;119:5599–5607. doi: 10.1182/blood-2011-11-390666. [DOI] [PubMed] [Google Scholar]
- 107.Krause D.S., Van Etten R.A. Tyrosine kinases as targets for cancer therapy. N. Engl. J. Med. 2005;353:172–187. doi: 10.1056/NEJMra044389. [DOI] [PubMed] [Google Scholar]
- 108.Tohidkia M.R., Barar J., Asadi F., Omidi Y. Molecular considerations for development of phage antibody libraries. J. Drug Target. 2012;20:195–208. doi: 10.3109/1061186X.2011.611517. [DOI] [PubMed] [Google Scholar]
- 109.Gay N.J., Symmons M.F., Gangloff M., Bryant C.E. Assembly and localization of Toll-like receptor signalling complexes. Nat. Rev. Immunol. 2014;14:546–558. doi: 10.1038/nri3713. [DOI] [PubMed] [Google Scholar]
- 110.O'Neill L.A., Golenbock D., Bowie A.G. The history of Toll-like receptors - redefining innate immunity. Nat. Rev. Immunol. 2013;13:453–460. doi: 10.1038/nri3446. [DOI] [PubMed] [Google Scholar]
- 111.Takeda K., Kaisho T., Akira S. Toll-like receptors. Annu. Rev. Immunol. 2003;21:335–376. doi: 10.1146/annurev.immunol.21.120601.141126. [DOI] [PubMed] [Google Scholar]
- 112.McGettrick A.F., O'Neill L.A. Localisation and trafficking of Toll-like receptors: an important mode of regulation. Curr. Opin. Immunol. 2010;22:20–27. doi: 10.1016/j.coi.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 113.Canton J., Neculai D., Grinstein S. Scavenger receptors in homeostasis and immunity. Nat. Rev. Immunol. 2013;13:621–634. doi: 10.1038/nri3515. [DOI] [PubMed] [Google Scholar]
- 114.D'Souza A.A., Devarajan P.V. Asialoglycoprotein receptor mediated hepatocyte targeting—strategies and applications. J. Control. Release. 2015;203:126–139. doi: 10.1016/j.jconrel.2015.02.022. [DOI] [PubMed] [Google Scholar]
- 115.Grozovsky R., Begonja A.J., Liu K., Visner G., Hartwig J.H., Falet H., Hoffmeister K.M. The Ashwell-Morell receptor regulates hepatic thrombopoietin production via JAK2-STAT3 signaling. Nat. Med. 2015;21:47–54. doi: 10.1038/nm.3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Low P.S., Kularatne S.A. Folate-targeted therapeutic and imaging agents for cancer. Curr. Opin. Chem. Biol. 2009;13:256–262. doi: 10.1016/j.cbpa.2009.03.022. [DOI] [PubMed] [Google Scholar]
- 117.Varkouhi A.K., Scholte M., Storm G., Haisma H.J. Endosomal escape pathways for delivery of biologicals. J. Control. Release. 2011;151:220–228. doi: 10.1016/j.jconrel.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 118.Juliano R.L., Ming X., Nakagawa O. Cellular uptake and intracellular trafficking of antisense and siRNA oligonucleotides. Bioconjug. Chem. 2012;23:147–157. doi: 10.1021/bc200377d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Juliano R.L., Carver K. Cellular uptake and intracellular trafficking of oligonucleotides. Adv. Drug Deliv. Rev. 2015;87:35–45. doi: 10.1016/j.addr.2015.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Doherty G.J., McMahon H.T. Mechanisms of endocytosis. Annu. Rev. Biochem. 2009;78:857–902. doi: 10.1146/annurev.biochem.78.081307.110540. [DOI] [PubMed] [Google Scholar]
- 121.Marchese A., Paing M.M., Temple B.R., Trejo J. G protein-coupled receptor sorting to endosomes and lysosomes. Annu. Rev. Pharmacol. Toxicol. 2008;48:601–629. doi: 10.1146/annurev.pharmtox.48.113006.094646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lajoie P., Nabi I.R. Lipid rafts, caveolae, and their endocytosis. Int. Rev. Cell Mol. Biol. 2010;282:135–163. doi: 10.1016/S1937-6448(10)82003-9. [DOI] [PubMed] [Google Scholar]
- 123.Karjalainen M., Kakkonen E., Upla P., Paloranta H., Kankaanpaa P., Liberali P., Renkema G.H., Hyypia T., Heino J., Marjomaki V. A Raft-derived, Pak1-regulated entry participates in alpha2beta1 integrin-dependent sorting to caveosomes. Mol. Biol. Cell. 2008;19:2857–2869. doi: 10.1091/mbc.E07-10-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Parton R.G., del Pozo M.A. Caveolae as plasma membrane sensors, protectors and organizers. Nat. Rev. Mol. Cell Biol. 2013;14:98–112. doi: 10.1038/nrm3512. [DOI] [PubMed] [Google Scholar]
- 125.Mayor S., Parton R.G., Donaldson J.G. Clathrin-independent pathways of endocytosis. Cold Spring Harb. Perspect. Biol. 2014;6 doi: 10.1101/cshperspect.a016758. doi:10.1101/cshperspect.a016758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Alam M.R., Ming X., Dixit V., Fisher M., Chen X., Juliano R.L. The biological effect of an antisense oligonucleotide depends on its route of endocytosis and trafficking. Oligonucleotides. 2010;20:103–109. doi: 10.1089/oli.2009.0211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Koller E., Vincent T.M., Chappell A., De S., Manoharan M., Bennett C.F. Mechanisms of single-stranded phosphorothioate modified antisense oligonucleotide accumulation in hepatocytes. Nucleic Acids Res. 2011;39:4795–4807. doi: 10.1093/nar/gkr089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Huotari J., Helenius A. Endosome maturation. EMBO J. 2011;30:3481–3500. doi: 10.1038/emboj.2011.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Goldenring J.R. Recycling endosomes. Curr. Opin. Cell Biol. 2015;35:117–122. doi: 10.1016/j.ceb.2015.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hanson P.I., Cashikar A. Multivesicular body morphogenesis. Annu. Rev. Cell Dev. Biol. 2012;28:337–362. doi: 10.1146/annurev-cellbio-092910-154152. [DOI] [PubMed] [Google Scholar]
- 131.Alenquer M., Amorim M.J. Exosome biogenesis, regulation, and function in viral infection. Viruses. 2015;7:5066–5083. doi: 10.3390/v7092862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Luzio J.P., Pryor P.R., Bright N.A. Lysosomes: fusion and function. Nat. Rev. Mol. Cell Biol. 2007;8:622–632. doi: 10.1038/nrm2217. [DOI] [PubMed] [Google Scholar]
- 133.Marino G., Madeo F., Kroemer G. Autophagy for tissue homeostasis and neuroprotection. Curr. Opin. Cell Biol. 2011;23:198–206. doi: 10.1016/j.ceb.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 134.Johannes L., Wunder C. Retrograde transport: two (or more) roads diverged in an endosomal tree? Traffic. 2011;12:956–962. doi: 10.1111/j.1600-0854.2011.01200.x. [DOI] [PubMed] [Google Scholar]
- 135.Sandvig K., Skotland T., van Deurs B., Klokk T.I. Retrograde transport of protein toxins through the Golgi apparatus. Histochem. Cell Biol. 2013;140:317–326. doi: 10.1007/s00418-013-1111-z. [DOI] [PubMed] [Google Scholar]
- 136.Kerr M., Teasdale R.D. Live imaging of endosome dynamics. Semin. Cell Dev. Biol. 2014;31:11–19. doi: 10.1016/j.semcdb.2014.03.027. [DOI] [PubMed] [Google Scholar]
- 137.Pfeffer S.R. Rab GTPase regulation of membrane identity. Curr. Opin. Cell Biol. 2013;25:414–419. doi: 10.1016/j.ceb.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Angers C.G., Merz A.J. New links between vesicle coats and Rab-mediated vesicle targeting. Semin. Cell Dev. Biol. 2011;22:18–26. doi: 10.1016/j.semcdb.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Barlan K., Rossow M.J., Gelfand V.I. The journey of the organelle: teamwork and regulation in intracellular transport. Curr. Opin. Cell Biol. 2013;25:483–488. doi: 10.1016/j.ceb.2013.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Cai H., Reinisch K., Ferro-Novick S. Coats, tethers, Rabs, and SNAREs work together to mediate the intracellular destination of a transport vesicle. Dev. Cell. 2007;12:671–682. doi: 10.1016/j.devcel.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 141.Hutagalung A.H., Novick P.J. Role of Rab GTPases in membrane traffic and cell physiology. Physiol. Rev. 2011;91:119–149. doi: 10.1152/physrev.00059.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Brandizzi F., Barlowe C. Organization of the ER-Golgi interface for membrane traffic control. Nat. Rev. Mol. Cell Biol. 2013;14:382–392. doi: 10.1038/nrm3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Solinger J.A., Spang A. Tethering complexes in the endocytic pathway: CORVET and HOPS. FEBS J. 2013;280:2743–2757. doi: 10.1111/febs.12151. [DOI] [PubMed] [Google Scholar]
- 144.Hong W., Lev S. Tethering the assembly of SNARE complexes. Trends Cell Biol. 2014;24:35–43. doi: 10.1016/j.tcb.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 145.Malsam J., Kreye S., Sollner T.H. Membrane fusion: SNAREs and regulation. Cell Mol. Life Sci. 2008;65:2814–2832. doi: 10.1007/s00018-008-8352-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kummel D., Ungermann C. Principles of membrane tethering and fusion in endosome and lysosome biogenesis. Curr. Opin. Cell Biol. 2014;29:61–66. doi: 10.1016/j.ceb.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 147.Attar N., Cullen P.J. The retromer complex. Adv. Enzyme Regul. 2010;50:216–236. doi: 10.1016/j.advenzreg.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 148.Scott C.C., Vacca F., Gruenberg J. Endosome maturation, transport and functions. Semin. Cell Dev. Biol. 2014;31:2–10. doi: 10.1016/j.semcdb.2014.03.034. [DOI] [PubMed] [Google Scholar]
- 149.Zerial M., McBride H. Rab proteins as membrane organizers. Nat. Rev. Mol. Cell Biol. 2001;2:107–117. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
- 150.Bissig C., Gruenberg J. Lipid sorting and multivesicular endosome biogenesis. Cold Spring Harb. Perspect. Biol. 2013;5:a016816. doi: 10.1101/cshperspect.a016816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Henne W.M., Buchkovich N.J., Emr S.D. The ESCRT pathway. Dev. Cell. 2011;21:77–91. doi: 10.1016/j.devcel.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 152.Wagenaar T.R., Tolstykh T., Shi C., Jiang L., Zhang J., Li Z., Yu Q., Qu H., Sun F., Cao H., et al. Identification of the endosomal sorting complex required for transport-I (ESCRT-I) as an important modulator of anti-miR uptake by cancer cells. Nucleic Acids Res. 2015;43:1204–1215. doi: 10.1093/nar/gku1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Wickner W., Schekman R. Membrane fusion. Nat. Struct. Mol. Biol. 2008;15:658–664. doi: 10.1038/nsmb.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Allen T.M., Hong K., Papahadjopoulos D. Membrane contact, fusion, and hexagonal (HII) transitions in phosphatidylethanolamine liposomes. Biochemistry. 1990;29:2976–2985. doi: 10.1021/bi00464a013. [DOI] [PubMed] [Google Scholar]
- 155.Ortiz A., Killian J.A., Verkleij A.J., Wilschut J. Membrane fusion and the lamellar-to-inverted-hexagonal phase transition in cardiolipin vesicle systems induced by divalent cations. Biophys. J. 1999;77:2003–2014. doi: 10.1016/S0006-3495(99)77041-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Wang T., Smith E.A., Chapman E.R., Weisshaar J.C. Lipid mixing and content release in single-vesicle, SNARE-driven fusion assay with 1-5 ms resolution. Biophys. J. 2009;96:4122–4131. doi: 10.1016/j.bpj.2009.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Ming X., Sato K., Juliano R.L. Unconventional internalization mechanisms underlying functional delivery of antisense oligonucleotides via cationic lipoplexes and polyplexes. J. Control. Release. 2011;153:83–92. doi: 10.1016/j.jconrel.2011.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Sahay G., Querbes W., Alabi C., Eltoukhy A., Sarkar S., Zurenko C., Karagiannis E., Love K., Chen D., Zoncu R., et al. Efficiency of siRNA delivery by lipid nanoparticles is limited by endocytic recycling. Nat. Biotechnol. 2013;31:653–658. doi: 10.1038/nbt.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Sixou S., Szoka F.C., Jr, Green G.A., Giusti B., Zon G., Chin D.J. Intracellular oligonucleotide hybridization detected by fluorescence resonance energy transfer (FRET) Nucleic Acids Res. 1994;22:662–668. doi: 10.1093/nar/22.4.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Lorenz P., Misteli T., Baker B.F., Bennett C.F., Spector D.L. Nucleocytoplasmic shuttling: a novel in vivo property of antisense phosphorothioate oligodeoxynucleotides. Nucleic Acids Res. 2000;28:582–592. doi: 10.1093/nar/28.2.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Huang L., Liu Y. In vivo delivery of RNAi with lipid-based nanoparticles. Annu. Rev. Biomed. Eng. 2011;13:507–530. doi: 10.1146/annurev-bioeng-071910-124709. [DOI] [PubMed] [Google Scholar]
- 162.Yin H., Kanasty R.L., Eltoukhy A.A., Vegas A.J., Dorkin J.R., Anderson D.G. Non-viral vectors for gene-based therapy. Nat. Rev. Genet. 2014;15:541–555. doi: 10.1038/nrg3763. [DOI] [PubMed] [Google Scholar]
- 163.Li J., Wang Y., Zhu Y., Oupicky D. Recent advances in delivery of drug-nucleic acid combinations for cancer treatment. J. Control. Release. 2013;172:589–600. doi: 10.1016/j.jconrel.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Leung A.K., Tam Y.Y., Cullis P.R. Lipid nanoparticles for short interfering RNA delivery. Adv. Genet. 2014;88:71–110. doi: 10.1016/B978-0-12-800148-6.00004-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Ozcan G., Ozpolat B., Coleman R.L., Sood A.K., Lopez-Berestein G. Preclinical and clinical development of siRNA-based therapeutics. Adv. Drug Deliv. Rev. 2015;87:108–119. doi: 10.1016/j.addr.2015.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Lammers P. Oligonucleotide Therapeutics Society. Netherlands: Leiden; 2015. [Google Scholar]
- 167.Akinc A., Querbes W., De S., Qin J., Frank-Kamenetsky M., Jayaprakash K.N., Jayaraman M., Rajeev K.G., Cantley W.L., Dorkin J.R., et al. Targeted delivery of RNAi therapeutics with endogenous and exogenous ligand-based mechanisms. Mol. Ther. 2010;18:1357–1364. doi: 10.1038/mt.2010.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Zelphati O., Szoka F.C., Jr Mechanism of oligonucleotide release from cationic liposomes. Proc. Natl. Acad. Sci. U.S.A. 1996;93:11493–11498. doi: 10.1073/pnas.93.21.11493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Xue H.Y., Liu S., Wong H.L. Nanotoxicity: a key obstacle to clinical translation of siRNA-based nanomedicine. Nanomedicine. 2014;9:295–312. doi: 10.2217/nnm.13.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Akhtar S. Cationic nanosystems for the delivery of small interfering ribonucleic acid therapeutics: a focus on toxicogenomics. Expert Opin. Drug Metab. Toxicol. 2010;6:1347–1362. doi: 10.1517/17425255.2010.518611. [DOI] [PubMed] [Google Scholar]
- 171.Mui B.L., Tam Y.K., Jayaraman M., Ansell S.M., Du X., Tam Y.Y., Lin P.J., Chen S., Narayanannair J.K., Rajeev K.G., et al. Influence of polyethylene glycol lipid desorption rates on pharmacokinetics and pharmacodynamics of siRNA lipid nanoparticles. Mol. Ther. 2013;2:e139. doi: 10.1038/mtna.2013.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Semple S.C., Akinc A., Chen J., Sandhu A.P., Mui B.L., Cho C.K., Sah D.W., Stebbing D., Crosley E.J., Yaworski E., et al. Rational design of cationic lipids for siRNA delivery. Nat. Biotechnol. 2010;28:172–176. doi: 10.1038/nbt.1602. [DOI] [PubMed] [Google Scholar]
- 173.Jayaraman M., Ansell S.M., Mui B.L., Tam Y.K., Chen J., Du X., Butler D., Eltepu L., Matsuda S., Narayanannair J.K., et al. Maximizing the potency of siRNA lipid nanoparticles for hepatic gene silencing in vivo. Angew. Chem. 2012;51:8529–8533. doi: 10.1002/anie.201203263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Tam Y.Y., Chen S., Cullis P.R. Advances in Lipid Nanoparticles for siRNA Delivery. Pharmaceutics. 2013;5:498–507. doi: 10.3390/pharmaceutics5030498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Whitehead K.A., Dorkin J.R., Vegas A.J., Chang P.H., Veiseh O., Matthews J., Fenton O.S., Zhang Y., Olejnik K.T., Yesilyurt V., et al. Degradable lipid nanoparticles with predictable in vivo siRNA delivery activity. Nat. Commun. 2014;5:4277. doi: 10.1038/ncomms5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Gujrati M., Vaidya A., Lu Z.R. Multifunctional pH-sensitive amino lipids for siRNA delivery. Bioconjug. Chem. 2015;27:19–35. doi: 10.1021/acs.bioconjchem.5b00538. [DOI] [PubMed] [Google Scholar]
- 177.Villares G.J., Zigler M., Wang H., Melnikova V.O., Wu H., Friedman R., Leslie M.C., Vivas-Mejia P.E., Lopez-Berestein G., Sood A.K., et al. Targeting melanoma growth and metastasis with systemic delivery of liposome-incorporated protease-activated receptor-1 small interfering RNA. Cancer Res. 2008;68:9078–9086. doi: 10.1158/0008-5472.CAN-08-2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Kim H., Leal C. Cuboplexes: topologically active siRNA delivery. ACS Nano. 2015;9:10214–10226. doi: 10.1021/acsnano.5b03902. [DOI] [PubMed] [Google Scholar]
- 179.Adami R.C., Seth S., Harvie P., Johns R., Fam R., Fosnaugh K., Zhu T., Farber K., McCutcheon M., Goodman T.T., et al. An amino acid-based amphoteric liposomal delivery system for systemic administration of siRNA. Mol. Ther. 2011;19:1141–1151. doi: 10.1038/mt.2011.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Lu J.J., Langer R., Chen J. A novel mechanism is involved in cationic lipid-mediated functional siRNA delivery. Mol. Pharm. 2009;6:763–771. doi: 10.1021/mp900023v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Gilleron J., Querbes W., Zeigerer A., Borodovsky A., Marsico G., Schubert U., Manygoats K., Seifert S., Andree C., Stoter M., et al. Image-based analysis of lipid nanoparticle-mediated siRNA delivery, intracellular trafficking and endosomal escape. Nat. Biotechnol. 2013;31:638–646. doi: 10.1038/nbt.2612. [DOI] [PubMed] [Google Scholar]
- 182.Wittrup A., Ai A., Liu X., Hamar P., Trifonova R., Charisse K., Manoharan M., Kirchhausen T., Lieberman J. Visualizing lipid-formulated siRNA release from endosomes and target gene knockdown. Nat. Biotechnol. 2015;33:870–876. doi: 10.1038/nbt.3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Torchilin V.P. Recent advances with liposomes as pharmaceutical carriers. Nat. Rev. Drug Discov. 2005;4:145–160. doi: 10.1038/nrd1632. [DOI] [PubMed] [Google Scholar]
- 184.Katakowski J.A., Mukherjee G., Wilner S.E., Maier K.E., Harrison M.T., DiLorenzo T.P., Levy M., Palliser D. Delivery of siRNAs to dendritic cells using DEC205-targeted lipid nanoparticles to inhibit immune responses. Mol. Ther. 2015;24:146–155. doi: 10.1038/mt.2015.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Meissner J.M., Toporkiewicz M., Czogalla A., Matusewicz L., Kuliczkowski K., Sikorski A.F. Novel antisense therapeutics delivery systems: In vitro and in vivo studies of liposomes targeted with anti-CD20 antibody. J. Control. Release. 2015;220:515–528. doi: 10.1016/j.jconrel.2015.11.015. [DOI] [PubMed] [Google Scholar]
- 186.Ramishetti S., Kedmi R., Goldsmith M., Leonard F., Sprague A.G., Godin B., Gozin M., Cullis P.R., Dykxhoorn D.M., Peer D. Systemic gene silencing in primary T lymphocytes using targeted lipid nanoparticles. ACS Nano. 2015;9:6706–6716. doi: 10.1021/acsnano.5b02796. [DOI] [PubMed] [Google Scholar]
- 187.Lecaros R.L., Huang L., Lee T.C., Hsu Y.C. Nanoparticle delivered VEGF-A siRNA enhances photodynamic therapy for head and neck cancer treatment. Mol. Ther. 2016;24:106–116. doi: 10.1038/mt.2015.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Zhang Y., Schwerbrock N.M., Rogers A.B., Kim W.Y., Huang L. Codelivery of VEGF siRNA and gemcitabine monophosphate in a single nanoparticle formulation for effective treatment of NSCLC. Mol. Ther. 2013;21:1559–1569. doi: 10.1038/mt.2013.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Wilner S.E., Levy M. Synthesis and characterization of aptamer-targeted SNALPs for the delivery of siRNA. Methods Mol. Biol. 2016;1380:211–224. doi: 10.1007/978-1-4939-3197-2_18. [DOI] [PubMed] [Google Scholar]
- 190.Xue W., Dahlman J.E., Tammela T., Khan O.F., Sood S., Dave A., Cai W., Chirino L.M., Yang G.R., Bronson R., et al. Small RNA combination therapy for lung cancer. Proc. Natl. Acad. Sci. U.S.A. 2014;111:E3553–E3561. doi: 10.1073/pnas.1412686111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Thi E.P., Mire C.E., Ursic-Bedoya R., Geisbert J.B., Lee A.C., Agans K.N., Robbins M., Deer D.J., Fenton K.A., MacLachlan I., et al. Marburg virus infection in nonhuman primates: therapeutic treatment by lipid-encapsulated siRNA. Sci. Transl. Med. 2014;6:250ra116. doi: 10.1126/scitranslmed.3009706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Davis M.E., Zuckerman J.E., Choi C.H., Seligson D., Tolcher A., Alabi C.A., Yen Y., Heidel J.D., Ribas A. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature. 2010;464:1067–1070. doi: 10.1038/nature08956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Merkel O.M., Kissel T. Quo vadis polyplex? J. Control. Release. 2014;190:415–423. doi: 10.1016/j.jconrel.2014.06.009. [DOI] [PubMed] [Google Scholar]
- 194.Zhao J., Feng S.S. Nanocarriers for delivery of siRNA and co-delivery of siRNA and other therapeutic agents. Nanomedicine. 2015;10:2199–2228. doi: 10.2217/nnm.15.61. [DOI] [PubMed] [Google Scholar]
- 195.Babu A., Wang Q., Muralidharan R., Shanker M., Munshi A., Ramesh R. Chitosan coated polylactic acid nanoparticle-mediated combinatorial delivery of cisplatin and siRNA/Plasmid DNA chemosensitizes cisplatin-resistant human ovarian cancer cells. Mol. Pharm. 2014;11:2720–2733. doi: 10.1021/mp500259e. [DOI] [PubMed] [Google Scholar]
- 196.Patil Y.B., Swaminathan S.K., Sadhukha T., Ma L., Panyam J. The use of nanoparticle-mediated targeted gene silencing and drug delivery to overcome tumor drug resistance. Biomaterials. 2010;31:358–365. doi: 10.1016/j.biomaterials.2009.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Biswas S., Deshpande P.P., Navarro G., Dodwadkar N.S., Torchilin V.P. Lipid modified triblock PAMAM-based nanocarriers for siRNA drug co-delivery. Biomaterials. 2013;34:1289–1301. doi: 10.1016/j.biomaterials.2012.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Zheng C., Zheng M., Gong P., Deng J., Yi H., Zhang P., Zhang Y., Liu P., Ma Y., Cai L. Polypeptide cationic micelles mediated co-delivery of docetaxel and siRNA for synergistic tumor therapy. Biomaterials. 2013;34:3431–3438. doi: 10.1016/j.biomaterials.2013.01.053. [DOI] [PubMed] [Google Scholar]
- 199.Zhu X., Xu Y., Solis L.M., Tao W., Wang L., Behrens C., Xu X., Zhao L., Liu D., Wu J., et al. Long-circulating siRNA nanoparticles for validating Prohibitin1-targeted non-small cell lung cancer treatment. Proc. Nat. Acad. Sci. U.S.A. 2015;112:7779–7784. doi: 10.1073/pnas.1505629112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Ma D., Tian S., Baryza J., Luft J.C., DeSimone J.M. Reductively Responsive hydrogel nanoparticles with uniform size, shape, and tunable composition for systemic siRNA delivery in vivo. Mol. Pharm. 2015;12:3518–3526. doi: 10.1021/acs.molpharmaceut.5b00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Xu J., Wong D.H., Byrne J.D., Chen K., Bowerman C., DeSimone J.M. Future of the particle replication in nonwetting templates (PRINT) technology. Angew. Chem. 2013;52:6580–6589. doi: 10.1002/anie.201209145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Lonn P., Dowdy S.F. Cationic PTD/CPP-mediated macromolecular delivery: charging into the cell. Expert Opin. Drug Deliv. 2015;12:1627–1636. doi: 10.1517/17425247.2015.1046431. [DOI] [PubMed] [Google Scholar]
- 203.Kauffman W.B., Fuselier T., He J., Wimley W.C. Mechanism matters: a taxonomy of cell penetrating peptides. Trends Biochem. Sci. 2015;40:749–764. doi: 10.1016/j.tibs.2015.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Biswas S., Torchilin V.P. Dendrimers for siRNA Delivery. Pharmaceuticals. 2013;6:161–183. doi: 10.3390/ph6020161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Neuberg P., Kichler A. Recent developments in nucleic acid delivery with polyethylenimines. Adv. Genet. 2014;88:263–288. doi: 10.1016/B978-0-12-800148-6.00009-2. [DOI] [PubMed] [Google Scholar]
- 206.Lehto T., Kurrikoff K., Langel U. Cell-penetrating peptides for the delivery of nucleic acids. Expert Opin. Drug Deliv. 2012;9:823–836. doi: 10.1517/17425247.2012.689285. [DOI] [PubMed] [Google Scholar]
- 207.Copolovici D.M., Langel K., Eriste E., Langel U. Cell-penetrating peptides: design, synthesis, and applications. ACS Nano. 2014;8:1972–1994. doi: 10.1021/nn4057269. [DOI] [PubMed] [Google Scholar]
- 208.Andaloussi S.E., Lehto T., Mager I., Rosenthal-Aizman K., Oprea II, Simonson O.E., Sork H., Ezzat K., Copolovici D.M., Kurrikoff K., et al. Design of a peptide-based vector, PepFect6, for efficient delivery of siRNA in cell culture and systemically in vivo. Nucleic Acids Res. 2011;39:3972–3987. doi: 10.1093/nar/gkq1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Kurrikoff K., Gestin M., Langel U. Recent in vivo advances in cell-penetrating peptide-assisted drug delivery. Expert Opin. Drug Deliv. 2016;13:373–387. doi: 10.1517/17425247.2016.1125879. [DOI] [PubMed] [Google Scholar]
- 210.Zhou H.F., Yan H., Pan H., Hou K.K., Akk A., Springer L.E., Hu Y., Allen J.S., Wickline S.A., Pham C.T. Peptide-siRNA nanocomplexes targeting NF-kappaB subunit p65 suppress nascent experimental arthritis. J. Clin. Investig. 2014;124:4363–4374. doi: 10.1172/JCI75673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Hayashi Y., Hatakeyama H., Kajimoto K., Hyodo M., Akita H., Harashima H. Multifunctional Envelope-Type Nano Device: Evolution from nonselective to active targeting system. Bioconjug. Chem. 2015;26:1266–1276. doi: 10.1021/acs.bioconjchem.5b00184. [DOI] [PubMed] [Google Scholar]
- 212.Eguchi A., Meade B.R., Chang Y.C., Fredrickson C.T., Willert K., Puri N., Dowdy S.F. Efficient siRNA delivery into primary cells by a peptide transduction domain-dsRNA binding domain fusion protein. Nat. Biotechnol. 2009;27:567–571. doi: 10.1038/nbt.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Shcharbin D., Shakhbazau A., Bryszewska M. Poly(amidoamine) dendrimer complexes as a platform for gene delivery. Expert Opin Drug Deliv. 2013;10:1687–1698. doi: 10.1517/17425247.2013.853661. [DOI] [PubMed] [Google Scholar]
- 214.Labieniec-Watala M., Watala C. PAMAM dendrimers: destined for success or doomed to fail? Plain and modified PAMAM dendrimers in the context of biomedical applications. J. Pharm. Sci. 2015;104:2–14. doi: 10.1002/jps.24222. [DOI] [PubMed] [Google Scholar]
- 215.Duncan R., Izzo L. Dendrimer biocompatibility and toxicity. Adv. Drug Deliv. Rev. 2005;57:2215–2237. doi: 10.1016/j.addr.2005.09.019. [DOI] [PubMed] [Google Scholar]
- 216.Yuan A., Hu Y., Ming X. Dendrimer conjugates for light-activated delivery of antisense oligonucleotides. RSC Adv. 2015;5:35195–35200. doi: 10.1039/C5RA04091D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Gunther M., Lipka J., Malek A., Gutsch D., Kreyling W., Aigner A. Polyethylenimines for RNAi-mediated gene targeting in vivo and siRNA delivery to the lung. Eur. J. Pharm. Biopharm. 2011;77:438–449. doi: 10.1016/j.ejpb.2010.11.007. [DOI] [PubMed] [Google Scholar]
- 218.Schiffelers R.M., Ansari A., Xu J., Zhou Q., Tang Q., Storm G., Molema G., Lu P.Y., Scaria P.V., Woodle M.C. Cancer siRNA therapy by tumor selective delivery with ligand-targeted sterically stabilized nanoparticle. Nucleic Acids Res. 2004;32:e149. doi: 10.1093/nar/gnh140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Sun H., Zu Y. Aptamers and their applications in nanomedicine. Small. 2015;11:2352–2364. doi: 10.1002/smll.201403073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Dahlman J.E., Kauffman K.J., Langer R., Anderson D.G. Nanotechnology for in vivo targeted siRNA delivery. Adv. Genet. 2014;88:37–69. doi: 10.1016/B978-0-12-800148-6.00003-1. [DOI] [PubMed] [Google Scholar]
- 221.Fang Z., Wan L.Y., Chu L.Y., Zhang Y.Q., Wu J.F. ‘Smart’ nanoparticles as drug delivery systems for applications in tumor therapy. Expert Opin. Drug Deliv. 2015;12:1943–1953. doi: 10.1517/17425247.2015.1071352. [DOI] [PubMed] [Google Scholar]
- 222.Kauffman K.J., Webber M.J., Anderson D.G. Materials for non-viral intracellular delivery of messenger RNA therapeutics. J. Control. Release. 2015 doi: 10.1016/j.jconrel.2015.12.032. doi:10.1016/j.jconrel.2015.12.032. [DOI] [PubMed] [Google Scholar]
- 223.Wagner E. Polymers for nucleic acid transfer-an overview. Adv. Genet. 2014;88:231–261. doi: 10.1016/B978-0-12-800148-6.00008-0. [DOI] [PubMed] [Google Scholar]
- 224.Jones S.K., Lizzio V., Merkel O. Folate receptor targeted delivery of siRNA and paclitaxel to ovarian cancer cells via folate conjugated triblock co-polymer to overcome TLR4 driven chemotherapy resistance. Biomacromolecules. 2015;17:76–87. doi: 10.1021/acs.biomac.5b01189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Ohyama A., Higashi T., Motoyama K., Arima H. In vitro and in vivo tumor-targeting siRNA delivery using folate-PEG-appended dendrimer (G4)/alpha-cyclodextrin conjugates. Bioconjug. Chem. 2016;27:521–532. doi: 10.1021/acs.bioconjchem.5b00545. [DOI] [PubMed] [Google Scholar]
- 226.Lindberg S., Regberg J., Eriksson J., Helmfors H., Munoz-Alarcon A., Srimanee A., Figueroa R.A., Hallberg E., Ezzat K., Langel U. A convergent uptake route for peptide- and polymer-based nucleotide delivery systems. J. Control. Release. 2015;206:58–66. doi: 10.1016/j.jconrel.2015.03.009. [DOI] [PubMed] [Google Scholar]
- 227.Eltoukhy A.A., Sahay G., Cunningham J.M., Anderson D.G. Niemann-Pick C1 affects the gene delivery efficacy of degradable polymeric nanoparticles. ACS Nano. 2014;8:7905–7913. doi: 10.1021/nn501630h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Rehman Z., Hoekstra D., Zuhorn I.S. Mechanism of polyplex- and lipoplex-mediated delivery of nucleic acids: real-time visualization of transient membrane destabilization without endosomal lysis. ACS Nano. 2013;7:3767–3777. doi: 10.1021/nn3049494. [DOI] [PubMed] [Google Scholar]
- 229.He H., Zheng N., Song Z., Kim K.H., Yao C., Zhang R., Zhang C., Huang Y., Uckun F.M., Cheng J., et al. Suppression of hepatic inflammation via systemic siRNA delivery by membrane-disruptive and endosomolytic helical polypeptide hybrid nanoparticles. ACS Nano. 2016;10:1859–1870. doi: 10.1021/acsnano.5b05470. [DOI] [PubMed] [Google Scholar]
- 230.Bartlett D.W., Su H., Hildebrandt I.J., Weber W.A., Davis M.E. Impact of tumor-specific targeting on the biodistribution and efficacy of siRNA nanoparticles measured by multimodality in vivo imaging. Proc. Natl. Acad. Sci. U.S.A. 2007;104:15549–15554. doi: 10.1073/pnas.0707461104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Zuckerman J.E., Gritli I., Tolcher A., Heidel J.D., Lim D., Morgan R., Chmielowski B., Ribas A., Davis M.E., Yen Y. Correlating animal and human phase Ia/Ib clinical data with CALAA-01, a targeted, polymer-based nanoparticle containing siRNA. Proc. Natl. Acad. Sci. U.S.A. 2014;111:11449–11454. doi: 10.1073/pnas.1411393111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Nelson C.E., Kintzing J.R., Hanna A., Shannon J.M., Gupta M.K., Duvall C.L. Balancing cationic and hydrophobic content of PEGylated siRNA polyplexes enhances endosome escape, stability, blood circulation time, and bioactivity in vivo. ACS Nano. 2013;7:8870–8880. doi: 10.1021/nn403325f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Zheng M., Librizzi D., Kilic A., Liu Y., Renz H., Merkel O.M., Kissel T. Enhancing in vivo circulation and siRNA delivery with biodegradable polyethylenimine-graft-polycaprolactone-block-poly(ethylene glycol) copolymers. Biomaterials. 2012;33:6551–6558. doi: 10.1016/j.biomaterials.2012.05.055. [DOI] [PubMed] [Google Scholar]
- 234.Chu S., Tang C., Yin C. Effects of mannose density on in vitro and in vivo cellular uptake and RNAi efficiency of polymeric nanoparticles. Biomaterials. 2015;52:229–239. doi: 10.1016/j.biomaterials.2015.02.044. [DOI] [PubMed] [Google Scholar]
- 235.Murakami M., Nishina K., Watanabe C., Yoshida-Tanaka K., Piao W., Kuwahara H., Horikiri Y., Miyata K., Nishiyama N., Kataoka K., et al. Enteral siRNA delivery technique for therapeutic gene silencing in the liver via the lymphatic route. Sci. Rep. 2015;5:17035. doi: 10.1038/srep17035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Frede A., Neuhaus B., Klopfleisch R., Walker C., Buer J., Muller W., Epple M., Westendorf A.M. Colonic gene silencing using siRNA-loaded calcium phosphate/PLGA nanoparticles ameliorates intestinal inflammation in vivo. J. Control. Release. 2016;222:86–96. doi: 10.1016/j.jconrel.2015.12.021. [DOI] [PubMed] [Google Scholar]
- 237.Phelps K., Morris A., Beal P.A. Novel modifications in RNA. ACS Chem. Biol. 2012;7:100–109. doi: 10.1021/cb200422t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Juliano R.L., Ming X., Nakagawa O. The chemistry and biology of oligonucleotide conjugates. Acc. Chem. Res. 2012;45:1067–1076. doi: 10.1021/ar2002123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Lonnberg H. Solid-phase synthesis of oligonucleotide conjugates useful for delivery and targeting of potential nucleic acid therapeutics. Bioconjug. Chem. 2009;20:1065–1094. doi: 10.1021/bc800406a. [DOI] [PubMed] [Google Scholar]
- 240.Winkler J. Oligonucleotide conjugates for therapeutic applications. Ther. Deliv. 2013;4:791–809. doi: 10.4155/tde.13.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Yamada T., Peng C.G., Matsuda S., Addepalli H., Jayaprakash K.N., Alam M.R., Mills K., Maier M.A., Charisse K., Sekine M., et al. Versatile site-specific conjugation of small molecules to siRNA using click chemistry. J. Org. Chem. 2011;76:1198–1211. doi: 10.1021/jo101761g. [DOI] [PubMed] [Google Scholar]
- 242.Willibald J., Harder J., Sparrer K., Conzelmann K.K., Carell T. Click-modified anandamide siRNA enables delivery and gene silencing in neuronal and immune cells. J. Am. Chem. Soc. 2012;134:12330–12333. doi: 10.1021/ja303251f. [DOI] [PubMed] [Google Scholar]
- 243.Shabanpoor F., McClorey G., Saleh A.F., Jarver P., Wood M.J., Gait M.J. Bi-specific splice-switching PMO oligonucleotides conjugated via a single peptide active in a mouse model of Duchenne muscular dystrophy. Nucleic Acids Res. 2015;43:29–39. doi: 10.1093/nar/gku1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Alam M.R., Dixit V., Kang H., Li Z.B., Chen X., Trejo J., Fisher M., Juliano R.L. Intracellular delivery of an anionic antisense oligonucleotide via receptor-mediated endocytosis. Nucleic Acids Res. 2008;36:2764–2776. doi: 10.1093/nar/gkn115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Turner J.J., Arzumanov A.A., Gait M.J. Synthesis, cellular uptake and HIV-1 Tat-dependent trans-activation inhibition activity of oligonucleotide analogues disulphide-conjugated to cell-penetrating peptides. Nucleic Acids Res. 2005;33:27–42. doi: 10.1093/nar/gki142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Soutschek J., Akinc A., Bramlage B., Charisse K., Constien R., Donoghue M., Elbashir S., Geick A., Hadwiger P., Harborth J., et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432:173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- 247.Nishina K., Unno T., Uno Y., Kubodera T., Kanouchi T., Mizusawa H., Yokota T. Efficient in vivo delivery of siRNA to the liver by conjugation of alpha-tocopherol. Mol. Ther. 2008;16:734–740. doi: 10.1038/mt.2008.14. [DOI] [PubMed] [Google Scholar]
- 248.Wolfrum C., Shi S., Jayaprakash K.N., Jayaraman M., Wang G., Pandey R.K., Rajeev K.G., Nakayama T., Charrise K., Ndungo E.M., et al. Mechanisms and optimization of in vivo delivery of lipophilic siRNAs. Nat. Biotechnol. 2007;25:1149–1157. doi: 10.1038/nbt1339. [DOI] [PubMed] [Google Scholar]
- 249.Lehto T., Castillo Alvarez A., Gauck S., Gait M.J., Coursindel T., Wood M.J., Lebleu B., Boisguerin P. Cellular trafficking determines the exon skipping activity of Pip6a-PMO in mdx skeletal and cardiac muscle cells. Nucleic Acids Res. 2014;42:3207–3217. doi: 10.1093/nar/gkt1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Bai H., You Y., Yan H., Meng J., Xue X., Hou Z., Zhou Y., Ma X., Sang G., Luo X. Antisense inhibition of gene expression and growth in gram-negative bacteria by cell-penetrating peptide conjugates of peptide nucleic acids targeted to rpoD gene. Biomaterials. 2012;33:659–667. doi: 10.1016/j.biomaterials.2011.09.075. [DOI] [PubMed] [Google Scholar]
- 251.Opriessnig T., Patel D., Wang R., Halbur P.G., Meng X.J., Stein D.A., Zhang Y.J. Inhibition of porcine reproductive and respiratory syndrome virus infection in piglets by a peptide-conjugated morpholino oligomer. Antiviral Res. 2011;91:36–42. doi: 10.1016/j.antiviral.2011.04.012. [DOI] [PubMed] [Google Scholar]
- 252.Warren T.K., Shurtleff A.C., Bavari S. Advanced morpholino oligomers: a novel approach to antiviral therapy. Antiviral Res. 2012;94:80–88. doi: 10.1016/j.antiviral.2012.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Warren T.K., Whitehouse C.A., Wells J., Welch L., Charleston J.S., Heald A., Nichols D.K., Mattix M.E., Palacios G., Kugleman J.R., et al. Delayed time-to-treatment of an antisense morpholino oligomer is effective against lethal marburg virus infection in cynomolgus macaques. PLoS Negl. Trop. Dis. 2016;10:e0004456. doi: 10.1371/journal.pntd.0004456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Heald A.E., Charleston J.S., Iversen P.L., Warren T.K., Saoud J.B., Al-Ibrahim M., Wells J., Warfield K.L., Swenson D.L., Welch L.S., et al. AVI-7288 for marburg virus in nonhuman primates and humans. N. Engl. J. Med. 2015;373:339–348. doi: 10.1056/NEJMoa1410345. [DOI] [PubMed] [Google Scholar]
- 255.Lee S.H., Kang Y.Y., Jang H.E., Mok H. Current preclinical small interfering RNA (siRNA)-based conjugate systems for RNA therapeutics. Adv. Drug Deliv. Rev. 2015 doi: 10.1016/j.addr.2015.10.009. doi:10.1016/j.addr.2015.10.009. [DOI] [PubMed] [Google Scholar]
- 256.Ming X., Laing B. Bioconjugates for targeted delivery of therapeutic oligonucleotides. Adv. Drug Deliv. Rev. 2015;87:81–89. doi: 10.1016/j.addr.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Spinelli N., Defrancq E., Morvan F. Glycoclusters on oligonucleotide and PNA scaffolds: synthesis and applications. Chem. Soc. Rev. 2013;42:4557–4573. doi: 10.1039/c2cs35406c. [DOI] [PubMed] [Google Scholar]
- 258.Nair J.K., Willoughby J.L., Chan A., Charisse K., Alam M.R., Wang Q., Hoekstra M., Kandasamy P., Kel'in A.V., Milstein S., et al. Multivalent N-acetylgalactosamine-conjugated siRNA localizes in hepatocytes and elicits robust RNAi-mediated gene silencing. J. Am. Chem. Soc. 2014;136:16958–16961. doi: 10.1021/ja505986a. [DOI] [PubMed] [Google Scholar]
- 259.Matsuda S., Keiser K., Nair J.K., Charisse K., Manoharan R.M., Kretschmer P., Peng C.G., A V.K.i., Kandasamy P., Willoughby J.L., et al. siRNA conjugates carrying sequentially assembled trivalent N-acetylgalactosamine linked through nucleosides elicit robust gene silencing in vivo in hepatocytes. ACS Chem. Biol. 2015;10:1181–1187. doi: 10.1021/cb501028c. [DOI] [PubMed] [Google Scholar]
- 260.Rajeev K.G., Nair J.K., Jayaraman M., Charisse K., Taneja N., O'Shea J., Willoughby J.L., Yucius K., Nguyen T., Shulga-Morskaya S., et al. Hepatocyte-specific delivery of siRNAs conjugated to novel non-nucleosidic trivalent N-acetylgalactosamine elicits robust gene silencing in vivo. Chembiochem. 2015;16:903–908. doi: 10.1002/cbic.201500023. [DOI] [PubMed] [Google Scholar]
- 261.Prakash T.P., Graham M.J., Yu J., Carty R., Low A., Chappell A., Schmidt K., Zhao C., Aghajan M., Murray H.F., et al. Targeted delivery of antisense oligonucleotides to hepatocytes using triantennary N-acetyl galactosamine improves potency 10-fold in mice. Nucleic Acids Res. 2014;42:8796–8807. doi: 10.1093/nar/gku531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Prakash T.P., Yu J., Migawa M.T., Kinberger G.A., Wan W.B., Ostergaard M.E., Carty R.L., Vasquez G., Low A., Chappell A., et al. Comprehensive structure-activity relationship of triantennary N-acetylgalactosamine conjugated antisense oligonucleotides for targeted delivery to hepatocytes. J. Med. Chem. 2016;59:2718–2733. doi: 10.1021/acs.jmedchem.5b01948. [DOI] [PubMed] [Google Scholar]
- 263.Watanabe A., Nakajima M., Kasuya T., Onishi R., Kitade N., Mayumi K., Ikehara T., Kugimiya A. Comparative characterization of hepatic distribution and mRNA reduction of antisense oligonucleotides conjugated with triantennary N-acetyl galactosamine and lipophilic ligands targeting apolipoprotein B. J. Pharmacol. Exp. Ther. 2016;357:320–330. doi: 10.1124/jpet.115.230300. [DOI] [PubMed] [Google Scholar]
- 264.Yamamoto T., Sawamura M., Wada F., Harada-Shiba M., Obika S. Serial incorporation of a monovalent GalNAc phosphoramidite unit into hepatocyte-targeting antisense oligonucleotides. Bioorg. Med. Chem. 2016;24:26–32. doi: 10.1016/j.bmc.2015.11.036. [DOI] [PubMed] [Google Scholar]
- 265.Juliano R.L., Ming X., Nakagawa O., Xu R., Yoo H. Integrin targeted delivery of gene therapeutics. Theranostics. 2011;1:211–219. doi: 10.7150/thno/v01p0211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Alam M.R., Ming X., Fisher M., Lackey J.G., Rajeev K.G., Manoharan M., Juliano R.L. Multivalent cyclic RGD conjugates for targeted delivery of small interfering RNA. Bioconjug. Chem. 2011;22:1673–1681. doi: 10.1021/bc200235q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Liu X., Wang W., Samarsky D., Liu L., Xu Q., Zhang W., Zhu G., Wu P., Zuo X., Deng H., et al. Tumor-targeted in vivo gene silencing via systemic delivery of cRGD-conjugated siRNA. Nucleic Acids Res. 2014;42:11805–11817. doi: 10.1093/nar/gku831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Ming X., Alam M.R., Fisher M., Yan Y., Chen X., Juliano R.L. Intracellular delivery of an antisense oligonucleotide via endocytosis of a G protein-coupled receptor. Nucleic Acids Res. 2010;38:6567–6576. doi: 10.1093/nar/gkq534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Sioud M., Mobergslien A. Efficient siRNA targeted delivery into cancer cells by gastrin-releasing peptides. Bioconjug. Chem. 2012;23:1040–1049. doi: 10.1021/bc300050j. [DOI] [PubMed] [Google Scholar]
- 270.Nakagawa O., Ming X., Carver K., Juliano R. Conjugation with receptor-targeted histidine-rich peptides enhances the pharmacological effectiveness of antisense oligonucleotides. Bioconjug. Chem. 2014;25:165–170. doi: 10.1021/bc400500h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Yu Z., Ananias H.J., Carlucci G., Hoving H.D., Helfrich W., Dierckx R.A., Wang F., de Jong I.J., Elsinga P.H. An update of radiolabeled bombesin analogs for gastrin-releasing peptide receptor targeting. Curr. Pharm. Des. 2013;19:3329–3341. doi: 10.2174/1381612811319180015. [DOI] [PubMed] [Google Scholar]
- 272.Cesarone G., Edupuganti O.P., Chen C.P., Wickstrom E. Insulin receptor substrate 1 knockdown in human MCF7 ER+ breast cancer cells by nuclease-resistant IRS1 siRNA conjugated to a disulfide-bridged D-peptide analogue of insulin-like growth factor 1. Bioconjug. Chem. 2007;18:1831–1840. doi: 10.1021/bc070135v. [DOI] [PubMed] [Google Scholar]
- 273.Dassie J.P., Giangrande P.H. Current progress on aptamer-targeted oligonucleotide therapeutics. Ther. Deliv. 2013;4:1527–1546. doi: 10.4155/tde.13.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Nimjee S.M., Rusconi C.P., Sullenger B.A. Aptamers: an emerging class of therapeutics. Annu. Rev. Med. 2005;56:555–583. doi: 10.1146/annurev.med.56.062904.144915. [DOI] [PubMed] [Google Scholar]
- 275.McNamara J.O. 2nd, Andrechek E.R., Wang Y., Viles K.D., Rempel R.E., Gilboa E., Sullenger B.A., Giangrande P.H. Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat. Biotechnol. 2006;24:1005–1015. doi: 10.1038/nbt1223. [DOI] [PubMed] [Google Scholar]
- 276.Dassie J.P., Liu X.Y., Thomas G.S., Whitaker R.M., Thiel K.W., Stockdale K.R., Meyerholz D.K., McCaffrey A.P., McNamara J.O. 2nd, Giangrande P.H. Systemic administration of optimized aptamer-siRNA chimeras promotes regression of PSMA-expressing tumors. Nat. Biotechnol. 2009;27:839–849. doi: 10.1038/nbt.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Wullner U., Neef I., Eller A., Kleines M., Tur M.K., Barth S. Cell-specific induction of apoptosis by rationally designed bivalent aptamer-siRNA transcripts silencing eukaryotic elongation factor 2. Curr. Cancer Drug Targets. 2008;8:554–565. doi: 10.2174/156800908786241078. [DOI] [PubMed] [Google Scholar]
- 278.Zhou J., Tiemann K., Chomchan P., Alluin J., Swiderski P., Burnett J., Zhang X., Forman S., Chen R., Rossi J. Dual functional BAFF receptor aptamers inhibit ligand-induced proliferation and deliver siRNAs to NHL cells. Nucleic Acids Res. 2013;41:4266–4283. doi: 10.1093/nar/gkt125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Catuogno S., Rienzo A., Di Vito A., Esposito C.L., de Franciscis V. Selective delivery of therapeutic single strand antimiRs by aptamer-based conjugates. J. Control. Release. 2015;210:147–159. doi: 10.1016/j.jconrel.2015.05.276. [DOI] [PubMed] [Google Scholar]
- 280.Esposito C.L., Cerchia L., Catuogno S., De Vita G., Dassie J.P., Santamaria G., Swiderski P., Condorelli G., Giangrande P.H., de Franciscis V. Multifunctional aptamer-miRNA conjugates for targeted cancer therapy. Mol. Ther. 2014;22:1151–1163. doi: 10.1038/mt.2014.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Thiel K.W., Hernandez L.I., Dassie J.P., Thiel W.H., Liu X., Stockdale K.R., Rothman A.M., Hernandez F.J., McNamara J.O. 2nd, Giangrande P.H. Delivery of chemo-sensitizing siRNAs to HER2+-breast cancer cells using RNA aptamers. Nucleic Acids Res. 2012;40:6319–6337. doi: 10.1093/nar/gks294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Pastor F., Kolonias D., Giangrande P.H., Gilboa E. Induction of tumour immunity by targeted inhibition of nonsense-mediated mRNA decay. Nature. 2010;465:227–230. doi: 10.1038/nature08999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Berezhnoy A., Castro I., Levay A., Malek T.R., Gilboa E. Aptamer-targeted inhibition of mTOR in T cells enhances antitumor immunity. J. Clin. Investig. 2014;124:188–197. doi: 10.1172/JCI69856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Herrmann A., Priceman S.J., Swiderski P., Kujawski M., Xin H., Cherryholmes G.A., Zhang W., Zhang C., Lahtz C., Kowolik C., et al. CTLA4 aptamer delivers STAT3 siRNA to tumor-associated and malignant T cells. J. Clin. Investig. 2014;124:2977–2987. doi: 10.1172/JCI73174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Zhou J., Rossi J. Cell-type-specific aptamer and aptamer-small interfering RNA conjugates for targeted human immunodeficiency virus type 1 therapy. J. Investig. Med. 2014;62:914–919. doi: 10.1097/JIM.0000000000000103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Neff C.P., Zhou J., Remling L., Kuruvilla J., Zhang J., Li H., Smith D.D., Swiderski P., Rossi J.J., Akkina R. An aptamer-siRNA chimera suppresses HIV-1 viral loads and protects from helper CD4(+) T cell decline in humanized mice. Sci. Transl. Med. 2011;3:66ra66. doi: 10.1126/scitranslmed.3001581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Zhou J., Li H., Li S., Zaia J., Rossi J.J. Novel dual inhibitory function aptamer-siRNA delivery system for HIV-1 therapy. Mol. Ther. 2008;16:1481–1489. doi: 10.1038/mt.2008.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Wheeler L.A., Vrbanac V., Trifonova R., Brehm M.A., Gilboa-Geffen A., Tanno S., Greiner D.L., Luster A.D., Tager A.M., Lieberman J. Durable knockdown and protection from HIV transmission in humanized mice treated with gel-formulated CD4 aptamer-siRNA chimeras. Mol. Ther. 2013;21:1378–1389. doi: 10.1038/mt.2013.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Zhang Q., Hossain D.M., Nechaev S., Kozlowska A., Zhang W., Liu Y., Kowolik C.M., Swiderski P., Rossi J.J., Forman S., et al. TLR9-mediated siRNA delivery for targeting of normal and malignant human hematopoietic cells in vivo. Blood. 2013;121:1304–1315. doi: 10.1182/blood-2012-07-442590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Braun F.C., van den Brandt J., Thomas S., Lange S., Schrank J., Gand C., Przybylski G.K., Schmoeckel K., Broker B.M., Schmidt C.A., et al. In vivo silencing of A20 via TLR9-mediated targeted SiRNA delivery potentiates antitumor immune response. PloS One. 2015;10:e0135444. doi: 10.1371/journal.pone.0135444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Kortylewski M., Swiderski P., Herrmann A., Wang L., Kowolik C., Kujawski M., Lee H., Scuto A., Liu Y., Yang C., et al. In vivo delivery of siRNA to immune cells by conjugation to a TLR9 agonist enhances antitumor immune responses. Nat. Biotechnol. 2009;27:925–932. doi: 10.1038/nbt.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Thiel W.H., Thiel K.W., Flenker K.S., Bair T., Dupuy A.J., McNamara J.O. 2nd, Miller F.J., Giangrande P.H. Cell-internalization SELEX: method for identifying cell-internalizing RNA aptamers for delivering siRNAs to target cells. Methods Mol. Biol. 2015;1218:187–199. doi: 10.1007/978-1-4939-1538-5_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Yan A., Levy M. Cell internalization SELEX: in vitro selection for molecules that internalize into cells. Methods Mol. Biol. 2014;1103:241–265. doi: 10.1007/978-1-62703-730-3_18. [DOI] [PubMed] [Google Scholar]
- 294.Song E., Zhu P., Lee S.K., Chowdhury D., Kussman S., Dykxhoorn D.M., Feng Y., Palliser D., Weiner D.B., Shankar P., et al. Antibody mediated in vivo delivery of small interfering RNAs via cell-surface receptors. Nat. Biotechnol. 2005;23:709–717. doi: 10.1038/nbt1101. [DOI] [PubMed] [Google Scholar]
- 295.Yao Y.D., Sun T.M., Huang S.Y., Dou S., Lin L., Chen J.N., Ruan J.B., Mao C.Q., Yu F.Y., Zeng M.S., et al. Targeted delivery of PLK1-siRNA by ScFv suppresses Her2+ breast cancer growth and metastasis. Sci. Transl. Med. 2012;4:130ra148. doi: 10.1126/scitranslmed.3003601. [DOI] [PubMed] [Google Scholar]
- 296.Kumar P., Ban H.S., Kim S.S., Wu H., Pearson T., Greiner D.L., Laouar A., Yao J., Haridas V., Habiro K., et al. T cell-specific siRNA delivery suppresses HIV-1 infection in humanized mice. Cell. 2008;134:577–586. doi: 10.1016/j.cell.2008.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Cuellar T.L., Barnes D., Nelson C., Tanguay J., Yu S.F., Wen X., Scales S.J., Gesch J., Davis D., van Brabant Smith A., et al. Systematic evaluation of antibody-mediated siRNA delivery using an industrial platform of THIOMAB-siRNA conjugates. Nucleic Acids Res. 2015;43:1189–1203. doi: 10.1093/nar/gku1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Pluckthun A. Designed ankyrin repeat proteins (DARPins): binding proteins for research, diagnostics, and therapy. Annu. Rev. Pharmacol. Toxicol. 2015;55:489–511. doi: 10.1146/annurev-pharmtox-010611-134654. [DOI] [PubMed] [Google Scholar]
- 299.Rozema D.B., Blokhin A.V., Wakefield D.H., Benson J.D., Carlson J.C., Klein J.J., Almeida L.J., Nicholas A.L., Hamilton H.L., Chu Q., et al. Protease-triggered siRNA delivery vehicles. J. Control. Release. 2015;209:57–66. doi: 10.1016/j.jconrel.2015.04.012. [DOI] [PubMed] [Google Scholar]
- 300.Rozema D.B., Lewis D.L., Wakefield D.H., Wong S.C., Klein J.J., Roesch P.L., Bertin S.L., Reppen T.W., Chu Q., Blokhin A.V., et al. Dynamic PolyConjugates for targeted in vivo delivery of siRNA to hepatocytes. Proc. Natl. Acad. Sci. U.S.A. 2007;104:12982–12987. doi: 10.1073/pnas.0703778104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Wooddell C.I., Rozema D.B., Hossbach M., John M., Hamilton H.L., Chu Q., Hegge J.O., Klein J.J., Wakefield D.H., Oropeza C.E., et al. Hepatocyte-targeted RNAi therapeutics for the treatment of chronic hepatitis B virus infection. Mol. Ther. 2013;21:973–985. doi: 10.1038/mt.2013.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Given B. Oligonucleotide Therapeutics Society. Netherlands: Leiden; 2015. [Google Scholar]
- 303.Kang H., Alam M.R., Dixit V., Fisher M., Juliano R.L. Cellular delivery and biological activity of antisense oligonucleotides conjugated to a targeted protein carrier. Bioconjug. Chem. 2008;19:2182–2188. doi: 10.1021/bc800270w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Ming X., Carver K., Wu L. Albumin-based nanoconjugates for targeted delivery of therapeutic oligonucleotides. Biomaterials. 2013;34:7939–7949. doi: 10.1016/j.biomaterials.2013.06.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Carver K., Ming X., Juliano R.L. Multicellular tumor spheroids as a model for assessing delivery of oligonucleotides in three dimensions. Mol. Ther. 2014;3:e153. doi: 10.1038/mtna.2014.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Nakagawa O., Ming X., Huang L., Juliano R.L. Targeted intracellular delivery of antisense oligonucleotides via conjugation with small-molecule ligands. J. Am. Chem. Soc. 2010;132:8848–8849. doi: 10.1021/ja102635c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Low P.S., Henne W.A., Doorneweerd D.D. Discovery and development of folic-acid-based receptor targeting for imaging and therapy of cancer and inflammatory diseases. Acc. Chem. Res. 2008;41:120–129. doi: 10.1021/ar7000815. [DOI] [PubMed] [Google Scholar]
- 308.Dohmen C., Frohlich T., Lachelt U., Rohl I., Vornlocher H.P., Hadwiger P., Wagner E. Defined folate-PEG-siRNA conjugates for receptor-specific gene silencing. Mol. Ther. 2012;1:e7. doi: 10.1038/mtna.2011.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Choi C.H., Hao L., Narayan S.P., Auyeung E., Mirkin C.A. Mechanism for the endocytosis of spherical nucleic acid nanoparticle conjugates. Proc. Natl. Acad. Sci. U.S.A. 2013;110:7625–7630. doi: 10.1073/pnas.1305804110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Wu X.A., Choi C.H., Zhang C., Hao L., Mirkin C.A. Intracellular fate of spherical nucleic acid nanoparticle conjugates. J. Am. Chem. Soc. 2014;136:7726–7733. doi: 10.1021/ja503010a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Jensen S.A., Day E.S., Ko C.H., Hurley L.A., Luciano J.P., Kouri F.M., Merkel T.J., Luthi A.J., Patel P.C., Cutler J.I., et al. Spherical nucleic acid nanoparticle conjugates as an RNAi-based therapy for glioblastoma. Sci. Transl. Med. 2013;5:209ra152. doi: 10.1126/scitranslmed.3006839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Randeria P.S., Seeger M.A., Wang X.Q., Wilson H., Shipp D., Mirkin C.A., Paller A.S. siRNA-based spherical nucleic acids reverse impaired wound healing in diabetic mice by ganglioside GM3 synthase knockdown. Proc. Natl. Acad. Sci. U.S.A. 2015;112:5573–5578. doi: 10.1073/pnas.1505951112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Radovic-Moreno A.F., Chernyak N., Mader C.C., Nallagatla S., Kang R.S., Hao L., Walker D.A., Halo T.L., Merkel T.J., Rische C.H., et al. Immunomodulatory spherical nucleic acids. Proc. Natl. Acad. Sci. U.S.A. 2015;112:3892–3897. doi: 10.1073/pnas.1502850112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Zhang K., Hao L., Hurst S.J., Mirkin C.A. Antibody-linked spherical nucleic acids for cellular targeting. J. Ame. Chem. Soc. 2012;134:16488–16491. doi: 10.1021/ja306854d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Young K.L., Scott A.W., Hao L., Mirkin S.E., Liu G., Mirkin C.A. Hollow spherical nucleic acids for intracellular gene regulation based upon biocompatible silica shells. Nano Lett. 2012;12:3867–3871. doi: 10.1021/nl3020846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Ibrahim A., Marban E. Exosomes: fundamental biology and roles in cardiovascular physiology. Annu. Rev. Physiol. 2016;78:67–83. doi: 10.1146/annurev-physiol-021115-104929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.El Andaloussi S., Lakhal S., Mager I., Wood M.J. Exosomes for targeted siRNA delivery across biological barriers. Adv. Drug Deliv. Rev. 2013;65:391–397. doi: 10.1016/j.addr.2012.08.008. [DOI] [PubMed] [Google Scholar]
- 318.Baumann V., Winkler J. miRNA-based therapies: strategies and delivery platforms for oligonucleotide and non-oligonucleotide agents. Future Med. Chem. 2014;6:1967–1984. doi: 10.4155/fmc.14.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Lai C.P., Kim E.Y., Badr C.E., Weissleder R., Mempel T.R., Tannous B.A., Breakefield X.O. Visualization and tracking of tumour extracellular vesicle delivery and RNA translation using multiplexed reporters. Nat. Commun. 2015;6:7029. doi: 10.1038/ncomms8029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Alvarez-Erviti L., Seow Y., Yin H., Betts C., Lakhal S., Wood M.J. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat. Biotechnol. 2011;29:341–345. doi: 10.1038/nbt.1807. [DOI] [PubMed] [Google Scholar]
- 321.Akao Y., Iio A., Itoh T., Noguchi S., Itoh Y., Ohtsuki Y., Naoe T. Microvesicle-mediated RNA molecule delivery system using monocytes/macrophages. Mol. Ther. 2011;19:395–399. doi: 10.1038/mt.2010.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Liu Y., Li D., Liu Z., Zhou Y., Chu D., Li X., Jiang X., Hou D., Chen X., Chen Y., et al. Targeted exosome-mediated delivery of opioid receptor Mu siRNA for the treatment of morphine relapse. Sci. Rep. 2015;5:17543. doi: 10.1038/srep17543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Zhang Y., Liu D., Chen X., Li J., Li L., Bian Z., Sun F., Lu J., Yin Y., Cai X., et al. Secreted monocytic miR-150 enhances targeted endothelial cell migration. Mol. Cell. 2010;39:133–144. doi: 10.1016/j.molcel.2010.06.010. [DOI] [PubMed] [Google Scholar]
- 324.Batrakova E.V., Kim M.S. Using exosomes, naturally-equipped nanocarriers, for drug delivery. J. Control. Release. 2015;219:396–405. doi: 10.1016/j.jconrel.2015.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Linko V., Ora A., Kostiainen M.A. DNA nanostructures as smart drug-delivery vehicles and molecular devices. Trends Biotechnol. 2015;33:586–594. doi: 10.1016/j.tibtech.2015.08.001. [DOI] [PubMed] [Google Scholar]
- 326.Pinheiro A.V., Han D., Shih W.M., Yan H. Challenges and opportunities for structural DNA nanotechnology. Nat. Nanotechnol. 2011;6:763–772. doi: 10.1038/nnano.2011.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Douglas S.M., Bachelet I., Church G.M. A logic-gated nanorobot for targeted transport of molecular payloads. Science. 2012;335:831–834. doi: 10.1126/science.1214081. [DOI] [PubMed] [Google Scholar]
- 328.Zhang H., Ma Y., Xie Y., An Y., Huang Y., Zhu Z., Yang C.J. A controllable aptamer-based self-assembled DNA dendrimer for high affinity targeting, bioimaging and drug delivery. Sci. Rep. 2015;5:10099. doi: 10.1038/srep10099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Charoenphol P., Bermudez H. Aptamer-targeted DNA nanostructures for therapeutic delivery. Mol. Pharm. 2014;11:1721–1725. doi: 10.1021/mp500047b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Lee H., Lytton-Jean A.K., Chen Y., Love K.T., Park A.I., Karagiannis E.D., Sehgal A., Querbes W., Zurenko C.S., Jayaraman M., et al. Molecularly self-assembled nucleic acid nanoparticles for targeted in vivo siRNA delivery. Nat. Nanotechnol. 2012;7:389–393. doi: 10.1038/nnano.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Jang M., Kim J.H., Nam H.Y., Kwon I.C., Ahn H.J. Design of a platform technology for systemic delivery of siRNA to tumours using rolling circle transcription. Nat. Commun. 2015;6:7930. doi: 10.1038/ncomms8930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Mohri K., Kusuki E., Ohtsuki S., Takahashi N., Endo M., Hidaka K., Sugiyama H., Takahashi Y., Takakura Y., Nishikawa M. Self-assembling DNA dendrimer for effective delivery of immunostimulatory CpG DNA to immune cells. Biomacromolecules. 2015;16:1095–1101. doi: 10.1021/bm501731f. [DOI] [PubMed] [Google Scholar]
- 333.Sellner S., Kocabey S., Nekolla K., Krombach F., Liedl T., Rehberg M. DNA nanotubes as intracellular delivery vehicles in vivo. Biomaterials. 2015;53:453–463. doi: 10.1016/j.biomaterials.2015.02.099. [DOI] [PubMed] [Google Scholar]
- 334.Stechmann B., Bai S.K., Gobbo E., Lopez R., Merer G., Pinchard S., Panigai L., Tenza D., Raposo G., Beaumelle B., et al. Inhibition of retrograde transport protects mice from lethal ricin challenge. Cell. 2010;141:231–242. doi: 10.1016/j.cell.2010.01.043. [DOI] [PubMed] [Google Scholar]
- 335.Ming X., Carver K., Fisher M., Noel R., Cintrat J.C., Gillet D., Barbier J., Cao C., Bauman J., Juliano R.L. The small molecule Retro-1 enhances the pharmacological actions of antisense and splice switching oligonucleotides. Nucleic Acids Res. 2013;41:3673–3687. doi: 10.1093/nar/gkt066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Yang B., Ming X., Cao C., Laing B., Yuan A., Porter M.A., Hull-Ryde E.A., Maddry J., Suto M., Janzen W.P., et al. High-throughput screening identifies small molecules that enhance the pharmacological effects of oligonucleotides. Nucleic Acids Res. 2015;43:1987–1996. doi: 10.1093/nar/gkv060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Osborn M.F., Alterman J.F., Nikan M., Cao H., Didiot M.C., Hassler M.R., Coles A.H., Khvorova A. Guanabenz (Wytensin) selectively enhances uptake and efficacy of hydrophobically modified siRNAs. Nucleic Acids Res. 2015;43:8664–8672. doi: 10.1093/nar/gkv942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Gooding M., Adigbli D., Edith Chan A.W., Melander R.J., MacRobert A.J., Selwood D.L. A bifurcated proteoglycan binding small molecule carrier for siRNA delivery. Chem. Biol. Drug Des. 2014;84:24–35. doi: 10.1111/cbdd.12295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Gilleron J., Paramasivam P., Zeigerer A., Querbes W., Marsico G., Andree C., Seifert S., Amaya P., Stoter M., Koteliansky V., et al. Identification of siRNA delivery enhancers by a chemical library screen. Nucleic Acids Res. 2015;43:7984–8001. doi: 10.1093/nar/gkv762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Kendall G.C., Mokhonova E.I., Moran M., Sejbuk N.E., Wang D.W., Silva O., Wang R.T., Martinez L., Lu Q.L., Damoiseaux R., et al. Dantrolene enhances antisense-mediated exon skipping in human and mouse models of Duchenne muscular dystrophy. Sci. Transl. Med. 2012;4:164ra160. doi: 10.1126/scitranslmed.3005054. [DOI] [PubMed] [Google Scholar]
- 341.Weng A., Manunta M.D., Thakur M., Gilabert-Oriol R., Tagalakis A.D., Eddaoudi A., Munye M.M., Vink C.A., Wiesner B., Eichhorst J., et al. Improved intracellular delivery of peptide- and lipid-nanoplexes by natural glycosides. J. Control. Release. 2015;206:75–90. doi: 10.1016/j.jconrel.2015.03.007. [DOI] [PubMed] [Google Scholar]
- 342.Li L., He Z.Y., Wei X.W., Gao G.P., Wei Y.Q. Challenges in CRISPR/CAS9 delivery: potential roles of nonviral vectors. Hum. Gene Ther. 2015;26:452–462. doi: 10.1089/hum.2015.069. [DOI] [PubMed] [Google Scholar]
- 343.Roberts J., Palma E., Sazani P., Orum H., Cho M., Kole R. Efficient and persistent splice switching by systemically delivered LNA oligonucleotides in mice. Mol. Ther. 2006;14:471–475. doi: 10.1016/j.ymthe.2006.05.017. [DOI] [PubMed] [Google Scholar]
- 344.Sebestyen M.G., Wong S.C., Trubetskoy V., Lewis D.L., Wooddell C.I. Targeted in vivo delivery of siRNA and an endosome-releasing agent to hepatocytes. Methods Mol. Biol. 2015;1218:163–186. doi: 10.1007/978-1-4939-1538-5_10. [DOI] [PubMed] [Google Scholar]