Abstract
In the present work, the emission characteristics of lipids as a function of the primary ion cluster size and energy were studied using time-of-flight secondary ion mass spectrometry (TOF-SIMS). Characteristic fragmentation patterns for common lipids are described, and changes in secondary ion (SI) yields using various primary ion beams are reported. In particular, emission characteristics were studied for pairs of small polyatomic and nanoparticle primary ion beams (e.g., Bi3+ versus Ar1000+ and Au3+ versus Au400+4) based on the secondary ion yield of characteristic fragment and intact molecular ions as a function of the lipid class. Detailed descriptions of the fragmentation patterns are shown for positive and negative mode TOF-SIMS. Results demonstrate that the lipid structure largely dictates the spectral presence of molecular and/or fragment ions in each ionization mode due to the localization of the charge carrier (head group or fatty acid chain). Our results suggest that the larger the energy per atom for small polyatomic projectiles (Bi3+ and Au3+), the larger the SI yield; in the case of nanoparticle projectiles, the SI increase with primary ion energy (200–500 keV range) for Au400+4 and with the decrease of the energy per atom (10–40 eV/atom range) for Arn=500-2000+ clusters. The secondary ion yield of the molecular ion of lipids from a single standard or from a mixture of lipids does not significantly change with the primary ion identity in the positive ion mode TOF-SIMS and slightly decreases in the negative ion mode TOF-SIMS.
I. INTRODUCTION
Secondary ion mass spectrometry (SIMS) is the gold standard for surface analysis of biological samples with submicron spatial resolution.1–6 Over the years, the primary ion beam of choice has changed as new ion sources have been developed and capabilities by application (e.g., organic versus inorganic surfaces) have been documented.7–9 For example, atomic and small polyatomic projectiles have shown distinct advantages for high spatial resolution, while larger clusters and nanoparticle projectiles have shown enhanced molecular ion emission.7,8,10–12 In addition, for the analysis of biological surfaces, the reduced damaged cross section of some nanoparticle projectiles (e.g., C60, argon, and water clusters) has triggered recent developments for tridimensional biological imaging and profiling.7,13–17 For example, a continuous Ar1000+ beam provides a somewhat “softer” desorption process which reduces the internal energy imparted to desorbed molecules, resulting in significant improvements of molecular ion or pseudomolecular ion yields. These types of molecular ions tend to be more diagnostic for structural characterization and identification of the biological systems from which they are generated.18 It has been reported that for an argon cluster beam, ideal ion yields are achieved when Eatom ≥ 10 eV and ion yields will quickly decline as the Eatom decreases.15 It was also observed that when water molecules are used as primary ion beams, the optimal energy was about 3 eV/atom, which leads authors to believe that further exploration into larger cluster projectiles is possible.15 During the study of peptides using various primary ion energies of an argon cluster, it has been shown that larger peptide fragments were observed with lower energy beams as long as they were above 10 eV/atom; in addition, the fragment intensity tends to decrease with increasing mass at 20 or 40 eV/atom.18
Lipid profiling of biological samples is traditionally based on liquid extraction followed by liquid chromatography coupled to mass spectrometry (MS), with the collision induced dissociation spectra providing the necessary structural identification of the lipid class.19–21 Alternatively, we have recently shown that lipid identification can be performed using direct surface probe analysis [matrix-assisted laser desorption/ionization (MALDI)], coupled to ultrahigh resolution mass spectrometry [Fourier transform ion cyclotron resonance mass spectrometry], followed by statistical analysis of variability and reproducibility across batches using internal standards.22 Lipid assignment from MS data can be performed utilizing the LIPIDMAPS database, where lipids are divided into eight major classes: fatty acyls (FA), glycerolipids, glycerophospholipids, sphingolipids, sterol lipids (ST), prenol lipids, saccharolipids, and polyketides.23–25 Analogous to MALDI probes, SIMS allows for in situ analysis of native biological surfaces, with higher spatial resolution. Due to the nature of molecular ion emission during SIMS analysis (not as soft as MALDI), fragmentation and intact molecular ion emission can be observed, with relative intensities varying with projectile size and energy. That is, the selection of the primary ion and energy determines the energy deposited per surface layer and the desorption volume, which corresponds to the observation of specific secondary ions (SIs).26,27 For example, during the analysis of lipid components from a biological surface, analyte specific fragment ions (lipid head groups and fatty acid fragments) are mainly observed under monoatomic and small polyatomic bombardment (e.g., In, Ga, Cs, Au3+, Bi3+, sources)28 while lipid molecular ions are increased under larger projectile bombardment (e.g., C60 and Au400+4). In a comparison of 40 keV C60+ to 40 keV Ar4000+ by Angerer and coworkers, it was observed that a majority of intact lipids from mouse brain were seen at higher secondary ion yields with the Ar4000+ primary ion, which is the primary ion beam that provided the larger cluster size but lower Eatom.29 This study also analyzed the signal of the pseudomolecular ion of cholesterol [M+H-H2O]+ using the aforementioned primary ion species and observed that similar secondary ion yields were detected for both of the projectiles; however, using Ar4000+, a lower yield in the smaller lipid fragments was observed.29
In the present paper, we study the lipid specific molecular ion emission as a function of the primary ion characteristics utilizing time-of-flight, secondary ion mass spectrometry (TOF-SIMS). In particular, we revisit the molecular ion emission characteristics for two pairs of small polyatomic and nanoparticle primary ion beams (e.g., Bi3+ versus Ar1000+ and Au3+ versus Au400+4) using the secondary ion yield of fragment and intact molecular ions for familiar lipids. Emphasis is made on the relative distribution of lipid-specific fragment ions and molecular ions as a function of the projectile size and energy as well as the matrix effects on the ionization efficiency and secondary ion yields.
II. EXPERIMENT
A. Sample preparation
Lipid standards of sulfatides [131305, Brain, Porcine, (major component 18:1/24:1 ST)], sphingomyelin [860061, Egg, Chicken (major component 18:1/16:0 SM)], 1,2-dipalmitoyl-sn-glycero-3-phosphocholine [850355, (16:0 PC DPPC)], 1-stearoyl-2-oleoyl-sn-glycero-3-phospho-(1′-rac-glycerol) (sodium salt) [840503, (18:0–18:1 PG)], and 1,2-dimyristoyl-sn-glycero-3-phosphoethanolamine [850745, (14:0 PE)] were purchased from Avanti Lipids, Inc. (Alabaster, AL) and used as received. Each standard was dissolved in a dichloromethane: methanol (60:40) solution for a final concentration of 1 mg/mL each. Each standard was deposited onto an ITO slide (Sigma Aldrich, St. Louis, MO) by aerosol spray of 1 ml to guarantee surface homogeneity. The aerosol sprayer was washed with the same solvent solution in between spraying of individual standards. The samples were allowed to dry in a chemical hood prior to SIMS analysis. The same procedure was followed for the preparation of a mixture of lipid standards consisting of sulfatides, SM, DPPC, PE, and PG all equivolume with concentrations of 0.167 mg/ml.
B. SIMS analysis
Standards were analyzed using Bi3+, Ar1000+, Au3+, and Au400+4 primary ions in positive and negative ionization modes. A commercial IonTOF (Ref. 5) instrument (Chestnut Ridge, NY) containing a hybrid detector with a single microchannel plate, scintillator, and photomultiplier was used for the 25 keV Bi3+ and 20 keV Ar500–2000+ analyses. The 25 keV Bi3+ (0.12 pA) and 20 keV Ar1000+ (0.04 pA) primary ion beams were rastered in sawtooth mode over a 250 × 250 μm2 field of view, and mass spectra were collected for a total dose density of 2 × 1011 ions/cm2. Measurements were obtained in the pulsed mode static SIMS at a frequency of 7.7 kHz. The opening time of the second plate of the dual blanking plate has been reduced to obtain a lower beam current for Bi3+ and subsequently avoid saturation of the detector. Secondary ion yields were normalized to the number of primary ions used to generate the mass spectral peak or total ion dose. A low energy flood gun is also utilized between pulses to ensure the sample surface remains neutral throughout the analysis. An internal calibration was performed using low mass ions and lipid head groups typically present in the sample: C2H3+, C2H5+, C3H7+, C5H12N+, and C5H14NO+ in the positive mode and CH−, CH2−, OH−, CN− Cl−, CNO−, PO2−, PO3−, H2PO4, C4H10PO4− in the negative ion mode. For comparison of primary ion beam (Sec. III B), SI yield as a function of primary ion energy (Sec. III C), and SI yield as a function of chemical environment (Sec. III D), triplicate analyses were performed; error bars are calculated by the standard deviation between the SI yields of each replicate. The mass resolution of each primary ion beam was on average 1500 for Ar1000+ and 4500 for Bi3+ at m/z 400. For Au3+ and Au400+4 analyses, the primary ions were obtained from a 120 kV Pegase Platform,30–32 equipped with a gold liquid metal ion source capable of producing a variety of projectiles (e.g., 150 nA for Au1+, 15 nA for Au3+, and 1 nA for Au400+4 without beam collimation/pulsing at the target).33 The primary ion projectiles were mass-selected using a Wien filter and focused into the TOF-SIMS analysis chamber. The negative mode TOF-SIMS was performed in the analysis chamber 1, where the target voltage is held at −10 kV (total acceleration voltage of up to 130 kV), whereas the positive mode TOF-SIMS was performed in analysis chamber 2, where the target voltage is held at +10 kV (total acceleration voltage of up to 110 kV).33 Further information about the instrumental setup can be found in Refs. 30–33. The average mass resolution for Au3+ and Au400+4 analyses at m/z 400 is 2000 and 450 in chamber 1 (equipped with a reflectron TOF) and in chamber 2 (equipped with a linear TOF), respectively.
III. RESULTS AND DISCUSSION
A. Lipid characterization by class
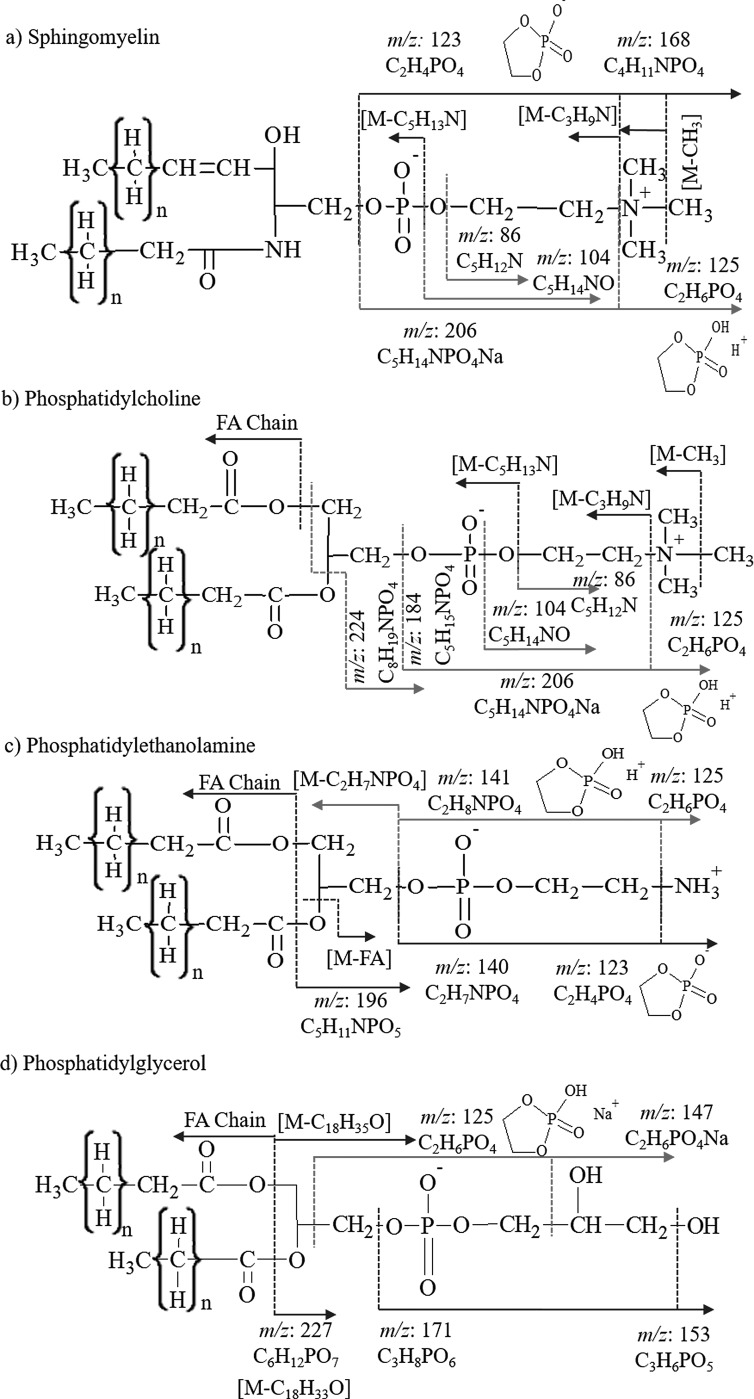
TOF-SIMS analysis can provide sufficient information to identify a lipid of a specific class based on the simultaneous detection of analyte specific fragment and intact molecular and/or pseudomolecular ions (e.g., [M]+, [M+H]+, [M+Na]+, [M]−, and [M-H]−). For example, phospholipids are a class of lipids that are predominantly abundant in biological membranes and consist of two fatty acids, glycerol, phosphate, and an alcohol group. There are several subclasses of phospholipids which differ based on the alcohol moiety present in the molecule (i.e., serine, ethanolamine, choline, glycerol, or inositol). In previous reports, various lipid species, including intact lipids, head group fragments, and fatty acyls, have been identified in cell lines using several mass spectrometry techniques (e.g., ESI-MS/MS, DESI-MS/MS, MALDI-MS/MS, and SIMS). DESI-MS/MS, MALDI-MS/MS, and SIMS have the advantage over traditional ESI-MS/MS in that lipid identification may not require sample extraction protocols and direct analysis can be performed from the biological tissue of interest; while there are major differences between the ionization mechanism of DESI-MS/MS, MALDI-MS/MS, and SIMS, all three techniques can provide spatial information with SIMS providing the highest spatial resolution. A tradeoff is that in the case of TOF-SIMS, the ratio of molecular to fragment ion and the spatial resolution significantly depends on the projectile size and energy as well as on the lipid species of interest and TOF-SIMS analysis mode (positive versus negative). For example, in the positive mode TOF-SIMS, head group fragments corresponding to the sphingomyelin and phosphatidylcholine are identified at m/z: 206 C5H14NPO4Na+, 184 C5H15NPO4+, 104 C5H14NO+, and 86 C5H12N+ via TOF-SIMS, ESI-MS/MS [Figs. 1(a) and 1(b)].1,4,34–41 Both of these lipid classes yield an internal fragment at m/z 125 in the positive ionization mode arising from fragmentation of the head group to yield a cyclic C2H6PO4+ fragment.40,42 The negative mode TOF-SIMS of sphingomyelin and phosphatidylcholine reveals two head group fragments at m/z 123 C2H4PO4− and m/z 168 C4H11NPO4−.43 Three larger fragments of higher mass are detected in sphingomyelin analysis related to the loss of methyl [M-CH3]−, trimethylamine [M-C3H9N]−, and ethyltrimethylammonium [M-C5H13N]− groups from the head group of the lipid [Figs. 1(a) and S1].4,42,62 In addition, fragment ions corresponding to the carboxylic acid chain are typically detected from phosphatidylcholine only [see example in Fig. 1(b)]. Lipids in the sphingomyelin and phosphatidylcholine classes have the same head group and therefore cannot be differentiated from each other exclusively based on the head group fragment detection. There are few differences between the ionization of sphingomyelin and phosphatidylcholine by TOF-SIMS; the major difference is attributed to the head group fragment at m/z 224 which is not observed in sphingomyelin.44–47 A second difference in the TOF-SIMS of PC versus SM is the presence of fatty acid fragments in the phosphatidylcholine lipid profile in the negative mode, which are not observed in sphingomyelin [Figs. 1(a), 1(b), and 2(f)].
Fig. 1.
(Color online) Fragmentation schemes of lipid standards of (a) sphingomyelin, (b) phosphatidylcholine, (c) phosphatidylethanolamine, and (d) phosphatidylglycerol for positive (red) and negative (blue) mode TOF-SIMS analyses.
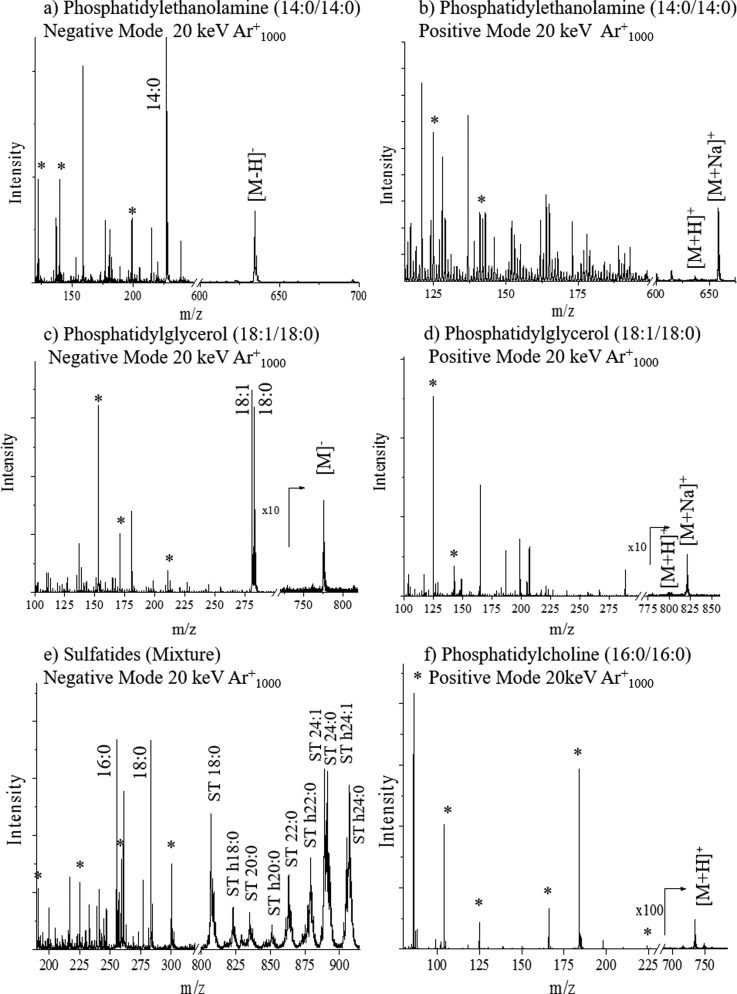
Fig. 2.
Typical TOF-SIMS mass spectra of familiar lipids in positive and negative modes. Characteristic fragment ions (*), fatty acid fragments, and intact molecular ions are denoted in the spectra.
In addition to phosphatidylcholine and sphingomyelin, phosphatidylethanolamine and phosphatidylglycerol fragmentation patterns by TOF-SIMS analysis are described herein. Fragments observed in the phosphatidylethanolamine lipid class include the internal fragments of m/z 125 C2H6PO4+ and m/z 123 C2H4PO4− in positive and negative modes, respectively.40 A tail group fragment after the loss of the head group [M-C2H7NPO4]− and the loss of a fatty acid chain [M-FA Chain]− were detected [Figs. 1(c) and 2(b)]. Two major characteristic fragments of phosphatidylethanolamine were detected at m/z 141 C2H8NPO4+,43 and m/z 140 C2H7NPO4− and m/z 196 C5H11NPO5− (Ref. 43) in positive and negative TOF-SIMS modes, respectively, allowing for identification of the phosphatidylethanolamine class versus all other lipids considered in this study.
Analysis of phosphatidylglycerol in the positive mode TOF-SIMS yields the cyclic head group fragment at m/z 125 C2H6PO4+ as well as the sodiated form of that fragment ion at m/z 147 C2H6PO4Na+.48 Fragments at m/z 171 C6H8PO6− and m/z 152 C3H6PO5− are characteristic fragments significant to the phosphatidylglycerol class in the negative mode TOF-SIMS. A m/z 227 C6H12PO7− is also observed corresponding to the loss of both fatty acid chains from the phosphatidylglycerol lipid in the negative mode TOF-SIMS [Figs. 1(d) and 2(c)].49 The sulfatide class is typically analyzed in the negative mode TOF-SIMS. Characteristic fragments are detected for the head group at m/z: 97 HSO4−, 199 C4H7O7S−, 225 C6H9O7S−, 257 C6H9O9S−, 259 C6H11O9S−, and 300 C8H14NO9−. All head group fragments are specific to the sulfatides due to the presence of a sulfur atom. A mixture of sulfatides with varying fatty acid composition results in the observation of a variety of intact molecular ions [Figs. 2(e) and S2].62
Fatty acid chains are typically observed in the negative mode TOF-SIMS in phosphatidylethanolamine, phosphatidylglycerol, phosphatidylcholine, and sulfatides. For example, phosphatidylethanolamine and phosphatidylglycerol analyses show peaks at m/z 227 C14H29O2− and m/z 283 C18H35O− corresponding to the 14:0 and 18:0 fatty acid chain fragments [Figs. 1(c) and 2(a)]. The sulfatide standard contains a mixture of lipids and fatty acid fragments, and peaks are observed in the mixture at m/z 255 (16:0), m/z 283 (18:0) and m/z 311 (20:0) [Figs. 2(e) and S2].62 By combining the information obtained from the head group fragments, fragment fatty acid chains, tail group fragment, loss of a fatty acid chain, and the pseudomolecular ions, it may be possible to directly correlate the TOF-SIMS spectral features to the lipid structure in complex biological matrices. The latter analysis can be simplified further when using coincidence TOF-SIMS techniques during single ion bombardment.50
B. SI yield as a function of the projectile size
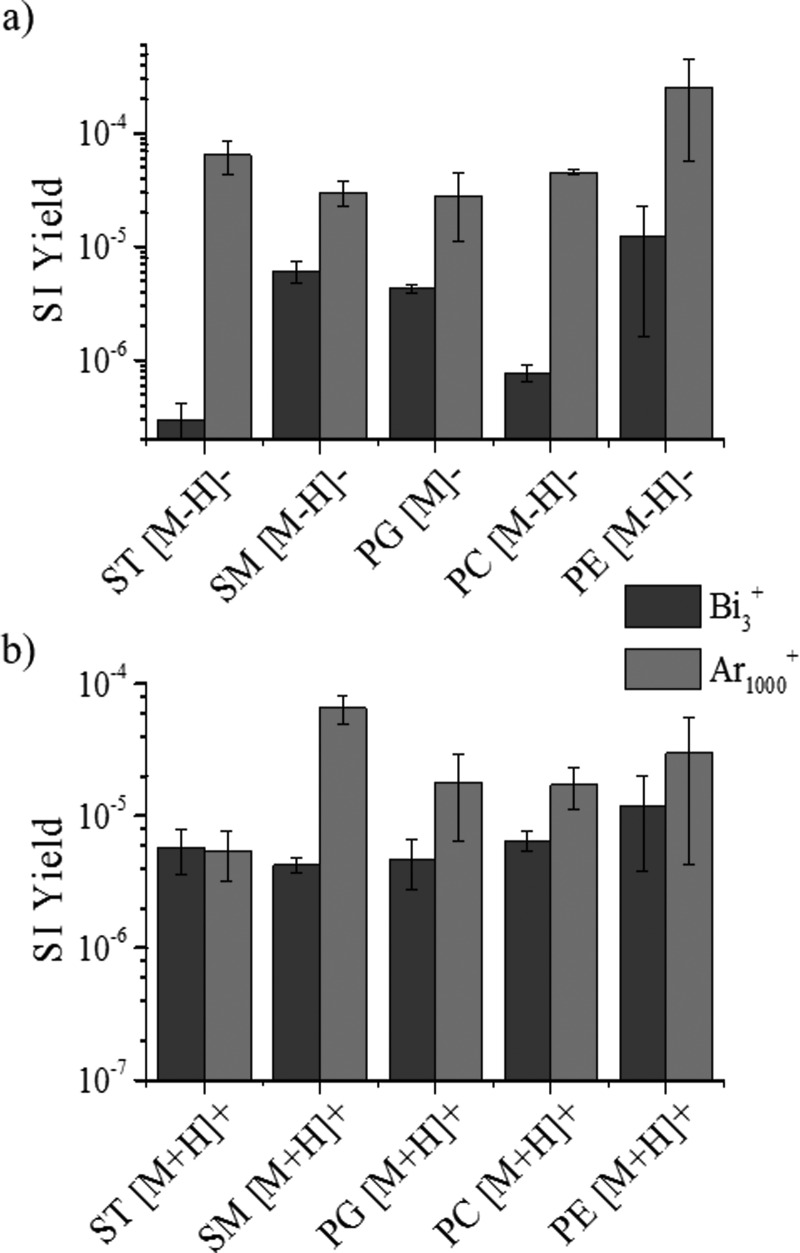
Lipid standards were analyzed as a function of the projectile size (Bi3+ and Au3+, Ar1000+, and Au400+4) and energy. An increase in secondary ion yield (SI yield) was observed between small polyatomic and nanoparticle projectiles throughout all lipid classes in positive and negative ionization modes (see, for example, Bi3+ and Ar1000+ in Fig. 3). In the sulfatide class, over tenfold increase is observed for 20 keV Ar1000+ when compared to 25 keV Bi3+ in the negative mode TOF-SIMS. The [M-H]− SI yields of the other lipids showed an increase when going from Bi3+ to Ar1000+ primary ions, but most of the changes were not as large as those seen in the ST class [Fig. 3(a)]. In the cases of sphingomyelin and phosphatidylethanolamine, the SI yield of the [M-H]− molecular ion had less than an order of magnitude of change in abundance, whereas phosphatidylglycerol and phosphatidylcholine had a more significant abundance increase. Phosphatidylcholine had the second largest increase in SI yield when changing from Bi3+ to Ar1000+ primary ions. SI signal enhancements were previously reported for argon clusters relative to bismuth clusters for the analysis of biological molecules (diadenosine triphosphate and diadenosine tetraphosphate).51 In the positive mode TOF-SIMS, a different trend is observed for SI yield variation with the primary ion projectile size [see Fig. 3(b)]. The only protonated molecular ion showing significant differences in the SI yield corresponds to the sphingomyelin class. The remaining [M+H]+ molecular ions of the lipids do not show significant changes in SI yield using the two different primary ion beams. A variation between the SI yields of the intact lipid molecular ion ([M-H]−/[M+H]+) of each lipid class is observed for both positive and negative mode TOF-SIMS; however, the negative mode TOF-SIMS shows greater variation in the SI yields (Fig. 3). The increase in SI yield with the Ar1000+ beam can be attributed to the softer desorption regime compared to that of Bi3+. The Eatom for each beam is discussed in detail later; however, in the examples shown, the Ar1000+ beam has an energy per atom closest to that deemed the ideal (Eatom = 10 eV/atom).15
Fig. 3.
(Color online) Intact molecular ion SI yield emission using small polyatomic Bi3+ and nanoparticle Ar1000+ primary ion projectiles for familiar lipid standards using TOF-SIMS in (a) negative and (b) positive modes.
The comparison of SI yields of lipids using gold projectiles showed that the nanoparticle projectile Au400+4 may provide over tenfold increase in the SI yield when compared to the small polyatomic projectile Au3+ (Table S1).62 For example, during sulfatide analysis, the intact molecular ion (ST 40:1) showed a 100-fold increase in the secondary ion yield from 50 keV Au3+ to 440 keV Au400+4 and a tenfold increase for the smaller gold beam when compared to 25 keV Bi3+ (Table S1).62 There is a 2–3 order of magnitude increase in the SI yield when using 440 keV Au400+4 versus 20 keV Ar1000+ that can be primarily attributed to the incident energy of the projectile (Table S1).62 Overall, the analysis of lipids using Au400+4 followed the same trend with a 2 order of magnitude increase in SI yield compared to Au3+ (Tables S1 and S2).62 SI yields of 440 keV Au400+4 were 2–3 orders of magnitude higher than those obtained using the Ar1000+ primary ion beam for most lipid classes (e.g., sphingomyelin, phosphatidylglycerol, phosphatidylcholine, and phosphatidylethanolamine, see Table S1).62 It has been previously noted that the estimated Au3+ SI yields would be equal to Bi3+ yields because they are of similar nature and size.52 For a more detailed analysis, the Eatom needs to be considered due to the large differences in energies used for each primary ion beam. For the small polyatomic projectiles, the Eatom are calculated at approximately 8000 and 16 000 eV for Bi3+ and Au3+, respectively. Our results suggest that the larger the Eatom for small polyatomic projectiles, the larger the SI yield. A discussion of the SI yield as a function of the energy and cluster size is provided for the Arn=500–2000+ and Au400+4 nanoparticle projectiles.
C. SI yield as a function of the projectile energy
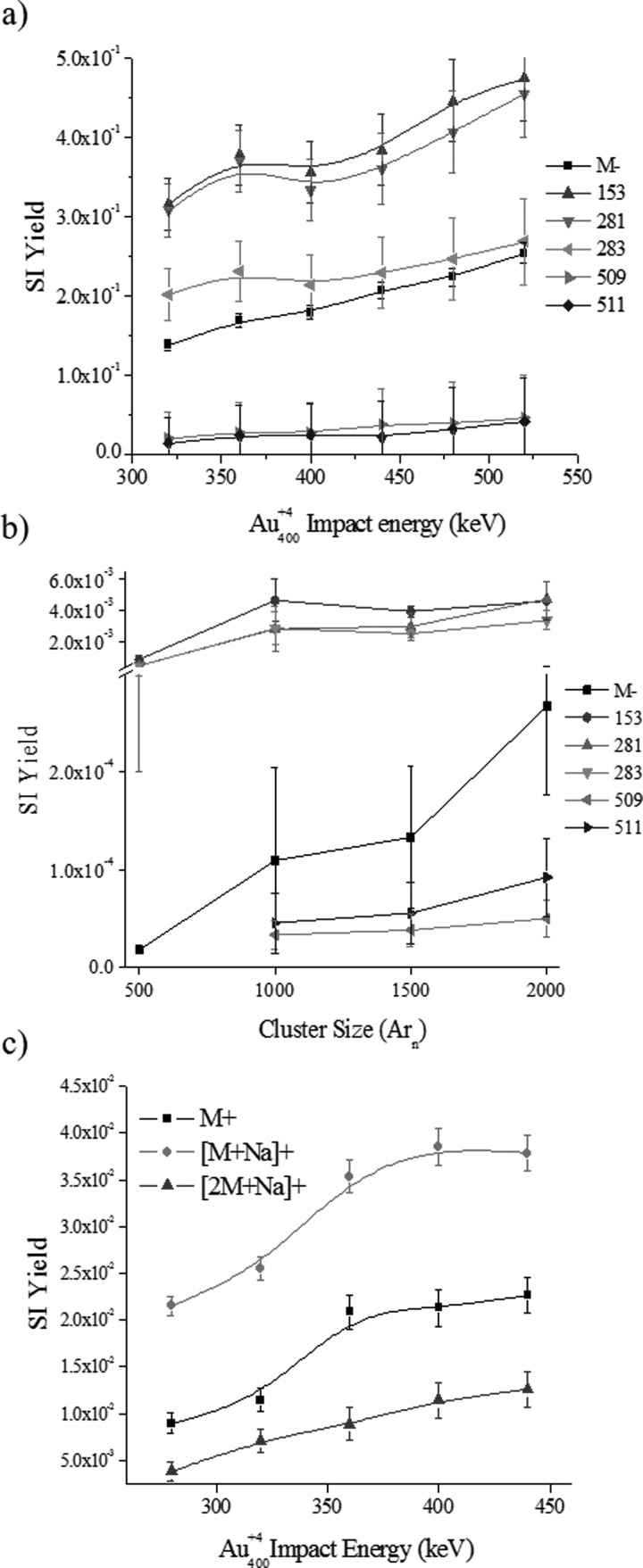
Previous work has shown that high energy, massive gold projectiles are advantageous for biological sample analysis and that the higher the cluster size the higher the SI yield.32 This correlates to data obtained by Vickerman et al. and by Yokoyama et al. where SI yields of both metals and organic substances increases with the primary ion impact energy (up to 120 keV for C60 projectiles), being most notable for higher mass fragments.53,54 Here, we further investigate the effect of the primary ion energy and size on the SI yield for the case of the phosphatidylglycerol (18:0–18:1 PG) lipid standard (see Fig. 4). Two studies are carried out: (1) the influence of the projectile energy on the SI yield for nanoparticle Au400+4 projectiles, and (2) the influence of the projectile size on the SI yield for 20 keV Arn+ (n = 500–2000 atoms) projectiles.
Fig. 4.
(Color online) Secondary ion yields as a function of the nanoparticle Au400+4 projectile energy or Arn+ cluster size for a lipid model target of phosphatidylglycerol (18:0–18:1 PG) (a) Au400+ and (b) Arn+ negative and (c) Au400+ positive mode. Notice the break in vertical axis in (b).
In positive and negative mode TOF-SIMS, as the primary Au4004+ ion energy increases, there is a net SI yield increase of molecular and fragment ions [Figs. 4(a) and 4(c)]. For example, in the negative mode TOF-SIMS, closer inspection shows that small (m/z: 153, 281, and 283) and larger (m/z 509 and 511) mass fragments show a slightly different increasing slope, being a steeper positive slope for the smaller fragments [see Fig. S3(a) and Table S3].62 Notice that analyte-specific fragment ions corresponds to the head group (m/z 153), fatty acid groups (m/z 281 and 283 for 18:1 and 18:0, respectively), and the loss of one fatty acid group (m/z 509 and 511). For the low mass fragments, results show that as the Au4004+ ion energy increases the ratio of molecular to fragment ion increases in a nonlinear fashion. That is, the SI yield of [M]− increases more than the SI yield of the low mass fragments. For the higher mass fragments (m/z 509 and 511), the opposite trend is observed where the ratio of [M]− to larger fragment ions decreases. We interpret the difference in slopes between the low (m/z 153, 281 and 283) and higher (m/z 509 and 511) mass fragments as a decrease of the internal energy distribution of the molecular ions with the projectile energy increase due to a larger emission volume (the larger the volume the lower the energy per emitted species). That is, as the projectile energy increases, the larger desorption volume leads to a lower internal energy distribution which favors the production of higher mass fragments and intact molecular ions. In the positive mode TOF-SIMS, the molecular ion emission also increases with the Au400+4 projectile energies. Closer inspection shows that as the Au4004+ ion energy increases a proportional increase in M+, [M+Na]+, and [2M+Na]+ emission is observed [Fig. 4(c) and Table S5].62 These positive and negative mode TOF-SIMS results suggest that as the Au400+4 Eatom increases the nanoparticle penetration into the sample increases, thus creating a larger emission volume in a way that scales the emission of intact molecular and higher mass fragment ions as a consequence of the lower internal energy of the emitted species.
In a different scenario, an increase of the 20 keV Arn+ cluster size from 500 to 2000 atoms (or decrease in the Eatom) results in an increase in SI yield of M−, head group fragments, and fatty acid fragments [Fig. 4(b)]. The SI yield of the molecular ion is rising at a faster rate than all of the fragment ions as the cluster size ranges from 500 to 2000 argon atoms, being the most significant between Ar1500 and Ar2000 [Fig. 4(b), Tables S4 and S3b]. This SI yield dependence on the Arn+ cluster size is in good agreement with recent observations by Yokoyama et al. using 20 keV Arn=2000–5000 cluster impacts.54 Previous report of Arn cluster impacts at 10–40 eV/atom has also shown that larger fragments are more favorable with the increase in Arn projectile size.18 These results suggest that as the size of the Arn+ projectile increases a larger impact cross section is achieved that enhances the emission volume; the weak forces that hold the Arn cluster together do not lead to penetration/implantation into the surface. Notice that this mechanism is very different from that of the nanoparticle Au400+4 projectiles.
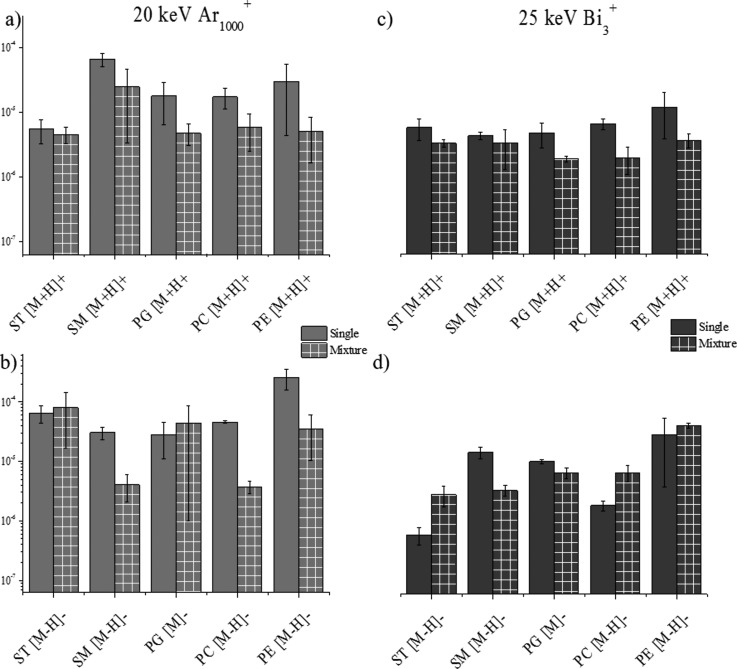
D. SI yield as a function of chemical environment
SIMS is a very useful mass spectrometry tool for the analysis of biological matrices as previously described.2,16,18,32,43,55,56 The chemical environment (matrix) of compounds of interest has a significant impact on the limit of detection, ionization efficiency, and ion suppression for TOF-SIMS and for other ionization sources.57 To evaluate the influence a matrix has on the SI yield, we compared the emission from the single lipid component sample to the emission from a sample containing a mixture of all five-lipid classes at equal concentrations. The protonated and deprotonated molecular ion of each lipid was utilized for comparison in positive and negative mode TOF-SIMS, respectively. The analysis of the sulfatides mixture in negative and positive mode TOF-SIMS using Ar1000+ showed no suppression or enhancement for the deprotonated/protonated molecular ion [Figs. 5(a) and 5(b)]. For Bi3+ analysis, in the negative mode TOF-SIMS, ion enhancement is observed for the sulfatides and in the positive TOF-SIMS no change is observed from the single standard to the mixture [Figs. 5(d) and 5(c)]. Typically, sulfatides are preferentially ionized in negative ionization mode and this could contribute to the fact that opposite trends are observed between the two ionization modes for Bi3 analysis. For a characteristic ion of the sulfatide lipid class (HSO4−), the SI yields of the fragments are on the same order of magnitude in both, alone and as a mixture using Ar1000+ and Bi3+projectiles (data not shown). For the sphingomyelin class of lipids, the positive mode TOF-SIMS shows no suppression in the SI yield of the [M+H]+; however, in the negative mode TOF-SIMS, an order of magnitude decrease is observed between the SI yield of the single lipid and the SI yield of the lipid mixture [Figs. 5(a) and 5(b)]. Using the Bi3+ projectiles, no significant changes are observed between single lipid and lipid mixture analysis of sphingomyelin in the positive ionization mode TOF-SIMS and a slight decrease in SI yield is observed in the negative mode TOF-SIMS [Figs. 5(c) and 5(d)].
Fig. 5.
(Color online) Secondary ion yields of intact molecular ions from a single lipid standard sample and a mixture of lipid standards sample in (a) positive mode Ar1000+, (b) negative mode Ar1000+, (c) positive mode Bi3+, and (d) negative mode Bi3+.
The analysis of phosphatidylglycerol in the positive mode TOF-SIMS demonstrates no significant changes with the Ar1000+ projectiles and a small decrease in SI yield using the Bi3+ projectiles [Figs. 5(a) and 5(c)]. For the analysis of phosphatidylglycerol in the negative mode TOF-SIMS, no ionization suppression or enhancement is observed for either of the primary ion projectiles [Figs. 5(b) and 5(d)]. SI yields of characteristic fragment peaks for phosphatidylglycerol (i.e., m/z 153 and 171) also showed no change between single and mixture conditions (data not shown). As previously mentioned, phosphatidylcholine is a lipid typically analyzed in the positive mode TOF-SIMS; in the positive mode TOF-SIMS, the SI yield of the [M+H]+ ion does not show a major matrix effect and no large changes in ion abundance are observed for Ar1000+ or Bi3+ [Figs. 5(a) and 5(c)]. In the negative mode TOF-SIMS, a different trend is seen for phosphatidylcholine, with suppression in the mixture signal observed using Ar1000+ and an enhancement in SI yield observed for Bi3+ projectiles [Figs. 5(b) and 5(d)]; this difference in trend between the positive and negative mode TOF-SIMS could be attributed to the fact that phosphatidylcholine is preferentially ionized in the positive mode TOF-SIMS. A previous study by Jones et al. of matrix effects in TOF-SIMS used phosphatidylcholine as a complex matrix while looking for targeted drug analytes.57 It was determined that the lipid had a very strong suppression effect on the abundance of the molecular ion of the drug due to its proton affinity.57 In our study, we observed a similar suppression effect on the pseudomolecular ions of the lipids in the mixture, potentially from the presence of phosphatidylcholine [Figs. 5(a) and 5(b)]. For the analysis of phosphatidylethanolamine by Ar1000+, there is less than tenfold suppression of ion signal going from the single lipid to the mixture of lipids [Figs. 5(a) and 5(b)]. No major changes are observed using Bi3+ for the [M-H]− or [M+H]+ from phosphatidylethanolamine [Figs. 5(c) and 5(d)]. No major changes in SI yield are seen for the characteristic peaks (m/z 141 and 196) of the phosphatidylethanolamine lipid class. In general, these matrix studies have shown that matrix effects are minimal on the SI yield of lipid standards when the lipid is analyzed in either ionization polarity. According to previous studies, chemical environment of a sample has a large impact on the SI yield of targeted molecules as shown with the large suppressive effects phosphatidylcholine has on the M and the enhancement effects cholesterol induced when used as a comparative matrix.57,58 More recently, matrix enhanced TOF-SIMS has been developed in order to more easily and efficiently ionize samples within complex biological matrices via TOF-SIMS.57,59–61 Our studies suggest that the secondary ion yield of the molecular ion of lipids from a single standard or from a mixture of lipids does not significantly change with the primary ion identity in the positive ion mode TOF-SIMS and slightly decreases in the negative ion mode TOF-SIMS.
IV. SUMMARY AND CONCLUSIONS
In this paper, we provide detailed information on the TOF-SIMS fragmentation pattern for sphingomyelins, phosphatidylcholine, phosphatidylglycerol, sulfatides, and phosphatidylethanolamine lipid classes. Typical mass spectra for common lipids are shown and discussed based on their fragmentation patterns. Changes in secondary ion yields were analyzed as a function of the primary ion (Bi3+ versus Ar1000+ and Au3+ versus Au400+4) using TOF-SIMS. For the case of lipid analysis, the results suggest that for polyatomic projectiles (Bi3+ and Au3+), the increase in the primary ion energy leads to an increase in the SI yield. However, larger SI yields are obtained for molecular ions using nanoparticle projectiles. Two different trends were observed in the case of the nanoparticle projectiles (Arn+ and Au400+4) that may be related to their intramolecular forces. For example, in the case of Au400+4 projectiles, as the projectile energy increases, a larger SI yield is observed for fragment and molecular ions, with small variation on their relative ratio. That is, the larger the Eatom for Au400+4 projectiles the larger the SI yield. In the case of the Arn+ projectiles, the lower the Eatom the larger the SI yield. We interpret these effects as consequence of two ways to increase the desorption volume: (1) larger incident energy for Au400+4 projectiles leads to a larger penetration depth and emission volume, and (2) a larger Arn+ cluster size (larger number of atoms) yields a larger impact cross section and emission volume. Overall, the matrix studies showed that the sample composition has a minimal effect on the desorption yields of intact molecular ions of familiar lipids and primary ion identity shows no significant effects of the secondary ion yield in the positive ion mode TOF-SIMS and a slight decrease in the negative ion mode TOF-SIMS. This work further provides more information on the main factors that affect the SI yield as well as characteristic patterns that allow lipid analysis in biological environments using TOF-SIMS.
ACKNOWLEDGMENTS
This work was supported by NIGMS grant GM106414 to FF-L. The authors acknowledge the support of Emile Schweikert and Serge Della-Negra during the acquisition of the Au3+/Au400+4 data in the 120 kV Pegasus platform at Texas A&M University and for helpful discussions.
References
- 1. Passarelli M. K. and Winograd N., Biochim. Biophys. Acta , 976 (2011). 10.1016/j.bbalip.2011.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. DeBord J. D., Smith D. F., Anderton C. R., Heeren R. M., Pasa-Tolic L., Gomer R. H., and Fernandez-Lima F. A., PLoS One , e99319 (2014). 10.1371/journal.pone.0099319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nie H.-Y., Francis J., Taylor A., Walzak M., Chang W., MacFabe D., and Lau W., Appl. Surf. Sci. , 1079 (2008). 10.1016/j.apsusc.2008.05.149 [DOI] [Google Scholar]
- 4. Saleem M. and Galla H.-J., Biochim. Biophys. Acta , 730 (2010). 10.1016/j.bbamem.2009.10.011 [DOI] [PubMed] [Google Scholar]
- 5. Touboul D., Brunelle A., Halgand F., De La Porte S., and Laprévote O., J. Lipid Res. , 1388 (2005). 10.1194/jlr.M500058-JLR200 [DOI] [PubMed] [Google Scholar]
- 6. Touboul D., Halgand F., Brunelle A., Kersting R., Tallarek E., Hagenhoff B., and Laprévote O., Anal. Chem. , 1550 (2004). 10.1021/ac035243z [DOI] [PubMed] [Google Scholar]
- 7. Rabbani S., Barber A. M., Fletcher J. S., Lockyer N. P., and Vickerman J. C., Anal. Chem. , 3793 (2011). 10.1021/ac200288v [DOI] [PubMed] [Google Scholar]
- 8. Ninomiya S., Ichiki K., Yamada H., Nakata Y., Seki T., Aoki T., and Matsuo J., Rapid Commun. Mass Spectrom. , 1601 (2009). 10.1002/rcm.4046 [DOI] [PubMed] [Google Scholar]
- 9. Yamada I., Matsuo J., Toyoda N., and Kirkpatrick A., Mater. Sci. Eng., R , 231 (2001). 10.1016/S0927-796X(01)00034-1 [DOI] [Google Scholar]
- 10. Lechene C. et al. , J. Biol. , 20.1 (2006). 10.1186/jbiol42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Braun R. M., Beyder A., Xu J., Wood M. C., Ewing A. G., and Winograd N., Anal. Chem. , 3318 (1999). 10.1021/ac9902042 [DOI] [PubMed] [Google Scholar]
- 12. Boxer S. G., Kraft M. L., and Weber P. K., Annu. Rev. Biophys. , 53 (2009). 10.1146/annurev.biophys.050708.133634 [DOI] [PubMed] [Google Scholar]
- 13. Schweikert E., van Stipdonk M. J., and Harris R. D., Rapid Commun. Mass Spectrom. , 1987 (1996). [DOI] [Google Scholar]
- 14. Sheraz née Rabbani S., Barber A., Fletcher J. S., Lockyer N. P., and Vickerman J. C., Anal. Chem. , 5654 (2013). 10.1021/ac4013732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sheraz née Rabbani S., Razo I. B., Kohn T., Lockyer N. P., and Vickerman J. C., Anal. Chem. , 2367 (2015). 10.1021/ac504191m [DOI] [PubMed] [Google Scholar]
- 16. Fletcher J. S., Lockyer N. P., and Vickerman J. C., Mass Spectrom. Rev. , 142 (2011). 10.1002/mas.20275 [DOI] [PubMed] [Google Scholar]
- 17. Gode D. and Volmer D. A., Analyst , 1289 (2013). 10.1039/c2an36337b [DOI] [PubMed] [Google Scholar]
- 18. Aoyagi S., Fletcher J. S., Sheraz S., Kawashima T., Razo I. B., Henderson A., Lockyer N. P., and Vickerman J. C., Anal. Bioanal. Chem. , 6621 (2013). 10.1007/s00216-013-7139-z [DOI] [PubMed] [Google Scholar]
- 19. Wolf C. and Quinn P. J., Prog. Lipid Res. , 15 (2008). 10.1016/j.plipres.2007.09.001 [DOI] [PubMed] [Google Scholar]
- 20. Wenk M. R., Cell , 888 (2010). 10.1016/j.cell.2010.11.033 [DOI] [PubMed] [Google Scholar]
- 21. Layre E. and Moody D. B., Biochimie , 109 (2013). 10.1016/j.biochi.2012.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schenk E. R., Nau F., Thompson C. J., Tse-Dinh Y.-C., and Fernandez-Lima F., J. Mass Spectrom. , 88 (2015). 10.1002/jms.3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fahy E. et al. , J. Lipid Res. , 839 (2005). 10.1194/jlr.E400004-JLR200 [DOI] [PubMed] [Google Scholar]
- 24. Fahy E. et al. , J. Lipid Res. , S9 (2009). 10.1194/jlr.R800095-JLR200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schmelzer K., Fahy E., Subramaniam S., and Dennis E. A., Methods Enzymol. , 171 (2007). 10.1016/S0076-6879(07)32007-7 [DOI] [PubMed] [Google Scholar]
- 26. Wucher A., Appl. Surf. Sci. , 6482 (2006). 10.1016/j.apsusc.2006.02.070 [DOI] [Google Scholar]
- 27. Wehbe N., Fallavier M., Della-Negra S., Depauw J., Brunelle A., and Andersen H. H., Nucl. Instrum. Methods Phys. Res., Sect. B , 2596 (2010) 10.1016/j.nimb.2010.05.105. [DOI] [Google Scholar]
- 28. Pacholski M. L., Cannon D. M., Ewing A. G., and Winograd N., Rapid Commun. Mass Spectrom. , 1232 (1998). [DOI] [PubMed] [Google Scholar]
- 29. Angerer T. B., Blenkinsopp P., and Fletcher J. S., Int. J. Mass Spectrom. , 591 (2015). 10.1016/j.ijms.2014.05.015 [DOI] [Google Scholar]
- 30. Della-Negra S., Arianer J., Depauw J., Verkhoturov S., and Schweikert E., Surf. Interface Anal. , 66 (2011). 10.1002/sia.3478 [DOI] [Google Scholar]
- 31. Fernandez-Lima F., Eller M., DeBord J., Verkhoturov S., Della-Negra S., and Schweikert E., Nucl. Instrum. Methods Phys. Res., Sect. B , 270 (2012). 10.1016/j.nimb.2011.07.092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fernandez-Lima F. A., Post J., DeBord J. D., Eller M. J., Verkhoturov S. V., Della-Negra S., Woods A. S., and Schweikert E. A., Anal. Chem. , 8448 (2011). 10.1021/ac201481r [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bouneau S., Della-Negra S., Depauw J., Jacquet D., Le Beyec Y., Mouffron J. P., Novikov A., and Pautrat M., Nucl. Instrum. Methods Phys. Res., Sect. B , 579 (2004). 10.1016/j.nimb.2004.06.005 [DOI] [Google Scholar]
- 34. Roddy T. P., Cannon D. M., Meserole C. A., Winograd N., and Ewing A. G., Anal. Chem. , 4011 (2002). 10.1021/ac0255734 [DOI] [PubMed] [Google Scholar]
- 35. Yang H. J., Sugiura Y., Ishizaki I., Sanada N., Ikegami K., Zaima N., Shrivas K., and Setou M., Surf. Interface Anal. , 1606 (2010). 10.1002/sia.3581 [DOI] [Google Scholar]
- 36. Yang H.-J., Ishizaki I., Sanada N., Zaima N., Sugiura Y., Yao I., Ikegami K., and Setou M., Med. Mol. Morphol. , 158 (2010). 10.1007/s00795-009-0487-2 [DOI] [PubMed] [Google Scholar]
- 37. Fletcher J. S., Lockyer N. P., Vaidyanathan S., and Vickerman J. C., Anal. Chem. , 2199 (2007). 10.1021/ac061370u [DOI] [PubMed] [Google Scholar]
- 38. Sjövall P., Lausmaa J., Nygren H., Carlsson L., and Malmberg P., Anal. Chem. , 3429 (2003). 10.1021/ac0207675 [DOI] [PubMed] [Google Scholar]
- 39. Kurczy M. E., Kozole J., Parry S., Piehowski P., Winograd N., and Ewing A., Appl. Surf. Sci. , 1158 (2008). 10.1016/j.apsusc.2008.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ostrowski S. G., Szakal C., Kozole J., Roddy T. P., Xu J., Ewing A. G., and Winograd N., Anal. Chem. , 6190 (2005). 10.1021/ac0508189 [DOI] [PubMed] [Google Scholar]
- 41. Ismaiel O. A., Halquist M. S., Elmamly M. Y., Shalaby A., and Karnes H. T., J. Chromatogr. B , 333 (2008). 10.1016/j.jchromb.2008.08.032 [DOI] [PubMed] [Google Scholar]
- 42. Domingues P., Amado F. M., Santana-Marques M. G. O., and Ferrer-Correia A., J. Am. Soc. Mass Spectrom. , 1189 (1998). 10.1016/S1044-0305(98)00087-7 [DOI] [Google Scholar]
- 43. Brügger B., Erben G., Sandhoff R., Wieland F. T., and Lehmann W. D., Proc. Natl. Acad. Sci. U. S. A. , 2339 (1997). 10.1073/pnas.94.6.2339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zheng L., McQuaw C. M., Ewing A. G., and Winograd N., J. Am. Chem. Soc. , 15730 (2007). 10.1021/ja0741675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Roddy T. P., Cannon D. M., Ostrowski S. G., Ewing A. G., and Winograd N., Anal. Chem. , 4087 (2003). 10.1021/ac026353t [DOI] [PubMed] [Google Scholar]
- 46. Touboul D., Brunelle A., and Laprévote O., Rapid Commun. Mass Spectrom. , 703 (2006). 10.1002/rcm.2362 [DOI] [PubMed] [Google Scholar]
- 47. Tahallah N., Brunelle A., De La Porte S., and Laprévote O., J. Lipid Res. , 438 (2008). 10.1194/jlr.M700421-JLR200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hsu F.-F. and Turk J., J. Am. Soc. Mass Spectrom. , 437 (2000). 10.1016/S1044-0305(99)00150-6 [DOI] [PubMed] [Google Scholar]
- 49. Hsu F.-F. and Turk J., J. Am. Soc. Mass Spectrom. , 1036 (2001). 10.1016/S1044-0305(01)00285-9 [DOI] [PubMed] [Google Scholar]
- 50. Park M. A., Gibson K. A., Quinones K., and Schweikert M. A., Science , 988 (1990). 10.1126/science.248.4958.988 [DOI] [PubMed] [Google Scholar]
- 51. Shon H. K., Cho Y. L., Lim C. S., Choi J. S., Chung S. J., and Lee T. G., Surf. Interface Anal. , 189 (2014). 10.1002/sia.5514 [DOI] [Google Scholar]
- 52. Touboul D., Kollmer F., Niehuis E., Brunelle A., and Laprévote O., J. Am. Soc. Mass Spectrom. , 1608 (2005). 10.1016/j.jasms.2005.06.005 [DOI] [PubMed] [Google Scholar]
- 53. Fletcher J. S., Conlan X. A., Jones E. A., Biddulph G., Lockyer N. P., and Vickerman J. C., Anal. Chem. , 1827 (2006). 10.1021/ac051624w [DOI] [PubMed] [Google Scholar]
- 54. Yokoyama Y. et al. , Anal. Chem. , 3592 (2016). 10.1021/acs.analchem.5b04133 [DOI] [PubMed] [Google Scholar]
- 55. Bich C., Havelund R., Moellers R., Touboul D., Kollmer F., Niehuis E., Gilmore I. S., and Brunelle A., Anal. Chem. , 7745 (2013). 10.1021/ac4009513 [DOI] [PubMed] [Google Scholar]
- 56. Annesley T. M., Clin. Chem. , 1041 (2003). 10.1373/49.7.1041 [DOI] [PubMed] [Google Scholar]
- 57. Jones E., Lockyer N., and Vickerman J., Appl. Surf. Sci. , 6727 (2006). 10.1016/j.apsusc.2006.02.158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Jones E. A., Fletcher J. S., Thompson C. E., Jackson D. A., Lockyer N. P., and Vickerman J. C., Appl. Surf. Sci. , 6844 (2006). 10.1016/j.apsusc.2006.02.145 [DOI] [Google Scholar]
- 59. Altelaar A. F. M., van Minnen J., Jiménez C. R., Heeren R. M. A., and Piersma S. R., Anal. Chem. , 735 (2005). 10.1021/ac048329g [DOI] [PubMed] [Google Scholar]
- 60. Cheng J. and Winograd N., Anal. Chem. , 3651 (2005). 10.1021/ac048131w [DOI] [PubMed] [Google Scholar]
- 61. Fletcher J. S., Henderson A., Biddulph G. X., Vaidyanathan S., Lockyer N. P., and Vickerman J. C., Appl. Surf. Sci. , 1264 (2008). 10.1016/j.apsusc.2008.05.253 [DOI] [Google Scholar]
- 62.See supplementary material at http://dx.doi.org/10.1116/1.4961461E-JVTBD9-34-317605 for additional tables and figures referred to in the text.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- See supplementary material at http://dx.doi.org/10.1116/1.4961461E-JVTBD9-34-317605 for additional tables and figures referred to in the text.