Summary
Microbial pathogens induce or inhibit death of host cells during infection, with significant consequences for virulence and disease progression. Death of an infected host cell can either facilitate release and dissemination of intracellular pathogens or promote pathogen clearance. Histoplasma capsulatum is an intracellular fungal pathogen that replicates robustly within macrophages and triggers macrophage lysis by unknown means. To identify H. capsulatum effectors of macrophage lysis, we performed a genetic screen and discovered three mutants that grew to wild-type levels within macrophages but failed to elicit host-cell death. Each mutant was defective in production of the previously identified secreted protein Cbp1 (calcium-binding protein 1), whose role in intracellular growth had not been fully investigated. We found that Cbp1 was dispensable for high levels of intracellular growth, but required to elicit a unique transcriptional signature in macrophages, including genes whose induction was previously associated with endoplasmic reticulum stress and host-cell death. Additionally Cbp1 was required for activation of cell-death caspases-3/7, and macrophage death during H. capsulatum infection was dependent on the pro-apoptotic proteins Bax and Bak. Taken together, these findings strongly suggest that the ability of Cbp1 to actively program host-cell death is an essential step in H. capsulatum pathogenesis.
Introduction
Intracellular pathogens use their host cells as a safe place to reside and replicate, often subverting the normal biology of the host in the process. Recently, it has become clear that pathogens can induce or inhibit death of host cells during infection, and that the subsequent consequences for virulence are significant (Labbe & Saleh, 2008). In some cases, death of an infected host cell facilitates release and dissemination of an intracellular pathogen, thereby promoting disease progression. In others, death of an infected host cell eliminates a pathogen niche and promotes pathogen clearance, thereby playing a protective role for the host. Thus understanding the role and mechanism of cell death in the progression of disease is critical to elucidating mechanisms of both virulence and host defense.
We are interested in identifying strategies used by the fungal intracellular pathogen Histoplasma capsulatum to manipulate macrophage viability. H. capsulatum, which infects both healthy and immunocompromised hosts, is thought to be one of the most common causes of fungal respiratory infection in immunocompetent people (Bullock, 1993, Eissenberg & Goldman, 1991, Woods, 2003). In endemic areas, the soil harbors a sporulating mold form of the organism that can be inhaled by a mammalian host. Within the lungs, fungal cells are phagocytosed by alveolar macrophages, one of the primary host cells for H. capsulatum during infection, and switch their growth program to a budding-yeast form during host colonization. In vitro studies examining infection of murine macrophages have demonstrated that the H. capsulatum yeast cells replicate to high levels within phagolysosomes, ultimately lysing their host cells (Porta & Maresca, 2000). Recent work showed that H. capsulatum can trigger apoptosis of host cells (Deepe & Buesing, 2012), but the fungal molecules required to regulate host-cell death are unknown. Furthermore, it is unknown whether lysis of the host cell is actively triggered by H. capsulatum, or whether lysis is secondary to the very high intracellular fungal burden.
A previous screen investigating approximately 2000 mutants was performed to identify H. capsulatum virulence factors (Edwards et al., 2011b). To elucidate mechanisms of Histoplasma-induced host-cell death, we performed a genetic screen targeted to identify H. capsulatum mutants that are unable to kill host cells. We identified a class of mutants that grew to high levels within macrophages but failed to lyse them, indicating that high fungal burden is not sufficient for host-cell death. These mutants were defective in the calcium-binding protein 1 (CBP1) gene, which was previously shown by others to be a highly expressed yeast-specific gene required for macrophage lysis (Batanghari et al., 1998, Kugler et al., 2000, Patel et al., 1998, Sebghati et al., 2000). Our results were surprising because the published defect of the cbp1 mutant in host-cell lysis was assumed to be secondary to a requirement for intracellular growth. Here we use primary murine macrophages to examine the role of Cbp1 in the ability of H. capsulatum to survive, replicate, and lyse host cells during infection. Our observation that the cbp1 mutant grew to high levels within macrophages without eliciting host-cell death provides the first evidence that macrophage death during H. capsulatum infection is not simply a passive consequence of high intracellular fungal burden, but instead reflects an active, Cbp1-dependent process. We also show that Cbp1 is required for robust in vivo growth and for mice to succumb to H. capsulatum infection. Whole-genome transcriptional profiling of infected macrophages revealed that H. capsulatum induces a Cbp1-dependent macrophage transcriptional signature that is associated with cell death, and Cbp1 is required for activation of executioner caspases-3/7 during infection. Finally, we determine that pro-apoptotic Bcl2-family proteins Bax and Bak are required for the normal kinetics and extent of host-cell death during H. capsulatum infection. Taken together, these findings highlight a key role for Cbp1 in the manipulation of macrophage cell death pathways and suggest that induction of macrophage death is an important mechanism of virulence for H. capsulatum.
Results
A genetic screen identifies H. capsulatum mutants defective in macrophage lysis
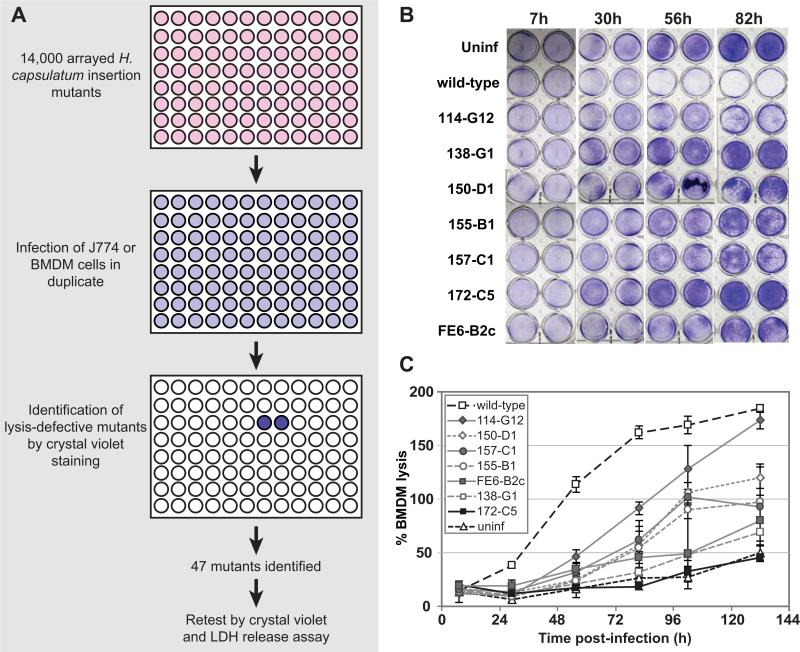
To identify genes that are important for virulence of H. capsulatum during macrophage infection, we performed a forward genetic screen in the highly virulent G217B strain background to isolate insertion mutants that were defective in macrophage lysis. We generated 14,000 individual insertion mutants by Agrobacterium-mediated transformation of the G217B ura5Δ parent strain and arrayed these mutants into 96-well plates. These mutants were tested for their ability to cause host-cell lysis during infection of murine J774.1 cells or bone-marrow derived macrophages (BMDMs) (Figure 1A). In an effort to identify the most robust lysis-defective mutants, we sought those mutants that gave phenotypes in both types of host macrophages. Mutants defective in causing host-cell lysis were identified by fixing and staining infected monolayers with crystal violet. H. capsulatum strains that were capable of wild-type levels of macrophage lysis cleared the macrophage monolayer, resulting in very little crystal violet staining (e.g. Figure 1B). Forty-seven mutants reproducibly failed to clear macrophage monolayers during infections of both J774.1 cells and BMDMs, indicating that they were strong candidates for lysis-defective (LDF) mutants. The lysis defect of the LDF mutants was verified and quantified in BMDMs using a lactate dehydrogenase (LDH) release assay, which measures the release of cytosolic lactate dehydrogenase into the culture supernatant as macrophages lyse. Two of the 47 mutants did not show a quantifiable defect in the LDH release assay, whereas the lysis defect of the remaining 45 mutants ranged in severity (e.g. Figure 1C). Southern blot analysis indicated that three of these mutants had multiple insertion sites in the H. capsulatum genome, whereas the overwhelming majority of mutants were single insertions. Using inverse PCR, we were able to map the site of insertion in 26 mutants; these insertion sites are reported in Table S1. Several mutants had insertions in repeat regions of the genome so the precise site of insertion could not be mapped. The insertion sites of five additional mutants were mapped; however, in these cases, the borders of each insertion did not map to adjacent regions in the genome, consistent with a genomic deletion or rearrangement event that occurred concomitant with insertion of the T-DNA. These types of rearrangements during Agrobacterium-mediated mutagenesis have been observed previously (Kemski et al., 2013). These mutants are not included in Table S1.
Figure 1. A genetic screen identifies Cbp1 as required for lysis of macrophages during H. capsulatum infection.
(A) A schematic of the genetic screen used to identify lysis-defective mutants is shown. 14,000 H. capsulatum insertion mutants were arrayed into 96-well plates and used to infect J774.1 macrophage-like cells or bone-marrow-derived macrophages (BMDMs). Each infection was performed in duplicate. After incubation at 37°C, macrophage monolayers were fixed and stained with crystal violet. Crystal violet staining was observed in duplicate wells containing monolayers that failed to be lysed by a given mutant. (B) BMDMs were mock-infected (uninf), infected with wild-type H. capsulatum (wild-type), or infected with various H. capsulatum insertion mutants. At indicated hours (h) post-infection, the supernatant was collected and the remaining monolayers were fixed and stained with 0.2% crystal violet solution to assess monolayer integrity. Duplicate monolayers are shown for each timepoint. (C) At 7, 30, 56, 82, 102 and 132 hours (h) post-infection for the experiment shown in (B), the collected supernatants were assessed for lactate dehydrogenase activity to monitor BMDM lysis. The average % BMDM lysis of four measurements ± standard deviation is shown.
For further analysis, we focused on mutant FE6-C3 (gene name HCL1, described elsewhere (Isaac et al., 2013)), as well as the three most severe lysis-defective mutants, 172-C5, 138-G1, and UA6-C8. These latter mutants showed lysis profiles that were nearly indistinguishable from uninfected macrophages (Figure 1C and data not shown). Inverse PCR was used to map the location of the Agrobacterium T-DNA insertion in mutant 172-C5. We determined that it contains an insertion 68 nucleotides upstream of the CBP1 ATG (Figure S1A). Northern blot analysis confirmed that CBP1 expression was abrogated in this mutant (Figure S1B). Since 138-G1 and UA6-C8 showed a severe lysis-defective phenotype that was nearly indistinguishable from 172-C5, we hypothesized that they, too, might be defective in CBP1 expression. We found that the 138-G1 mutant has an insertion at a locus unrelated to CBP1 (Table S1), but Northern analysis revealed that this mutant and UA6-C8 fail to express CBP1 (Figure S1C). Additionally, both Southern blot analysis and PCR of genomic DNA revealed a large genomic rearrangement at the CBP1 locus in mutant UA6-C8 (data not shown).
Cbp1 is required for macrophage lysis
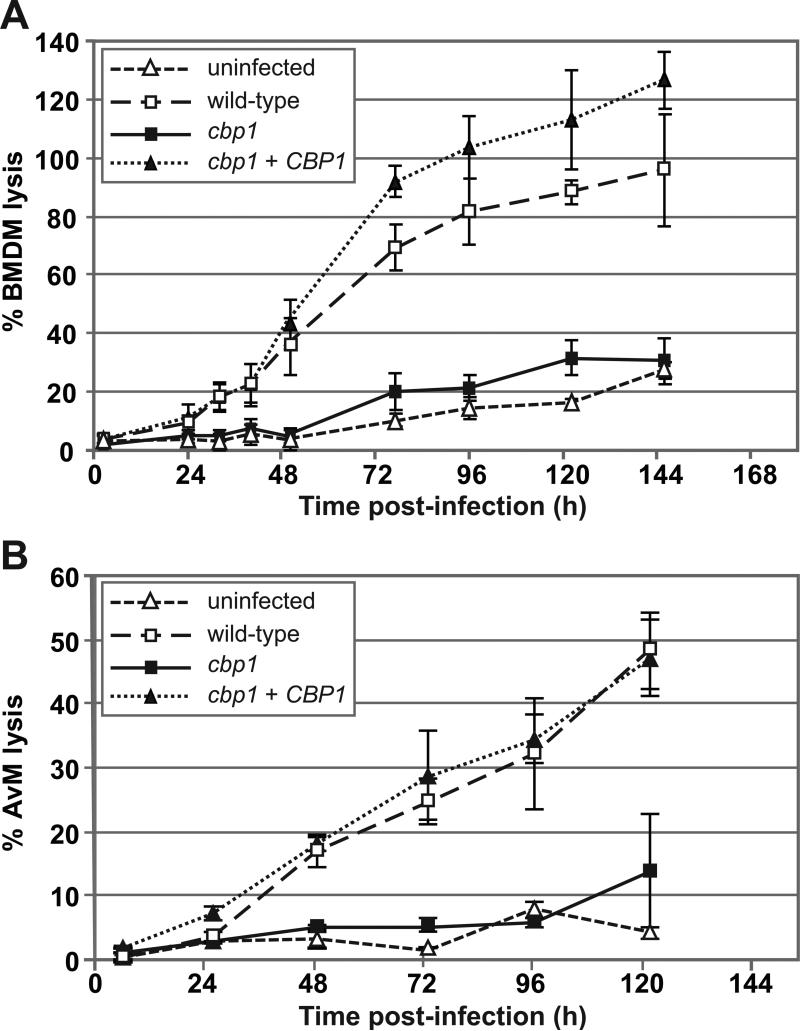
Since 172-C5 contains an unambiguous insertion at the CBP1 locus, we designated this strain as the “cbp1 mutant,” and continued to study it further. To quantify the lysis defect of this mutant, we infected BMDMs with either wild-type H. capsulatum (the G217B ura5Δ strain), the cbp1 mutant, or the cbp1 complemented strain (where the wild-type CBP1 gene was reintroduced into the mutant strain on an episomal plasmid (cbp1 + CBP1)) and performed an LDH release assay. For BMDMs infected with wild-type H. capsulatum, initiation of lysis was observed within 24 hours after infection (Figure 2A). By six days post-infection, these monolayers were completely destroyed and no intact macrophages remained. In contrast, the lysis profile of macrophages infected with the cbp1 mutant was indistinguishable from that of uninfected macrophages. Reintroduction of a wild-type copy of the CBP1 gene into the cbp1 mutant rescued the ability of the mutant to lyse BMDMs, indicating that the lysis defect of the mutant was due to disruption of CBP1.
Figure 2. Cbp1 is required for lysis of primary macrophages.
(A) BMDMs were mock-infected (uninfected) or infected with either wild-type G217B ura5Δ+URA (wild-type), the cbp1+URA mutant (cbp1), or the CBP1 complemented strain (cbp1+CBP1) at an MOI of 2. At 2, 24, 32, 40, 48, 72, 96, 120 and 144 hours post-infection, supernatants were removed from the infected monolayers and lactate dehydrogenase activity was assessed to monitor BMDM lysis. The average % BMDM lysis of four measurements ± standard deviation is shown. (B) Alveolar macrophages (AvMs) were mock-infected (uninfected) or infected with wild-type, cbp1 mutant, or complemented (cbp1+CBP1) yeast cells at an MOI of 5. At 6, 26, 49, 73, 97, and 122 hrs post-infection, supernatants were removed from the infected monolayers and lactate dehydrogenase activity was measured to monitor AvM lysis. The average % AvM lysis of four measurements ± standard deviation is shown.
We next sought to determine if CBP1 played a role in the lysis of alveolar macrophages (AvMs), which are thought to be an initial host cell for H. capsulatum during natural respiratory infection of mammalian hosts. Murine AvMs were isolated by broncheoalveolar lavage and infected with wild-type H. capsulatum, the cbp1 mutant, and the complemented strain. Measurements of the LDH released from these infected macrophages showed that AvM lysis in response to H. capsulatum infection was also dependent on CBP1 (Figure 2B). Taken together, these data demonstrate that CBP1 is required for H. capsulatum lysis of primary macrophages.
CBP1 is dispensable for high intracellular fungal burden
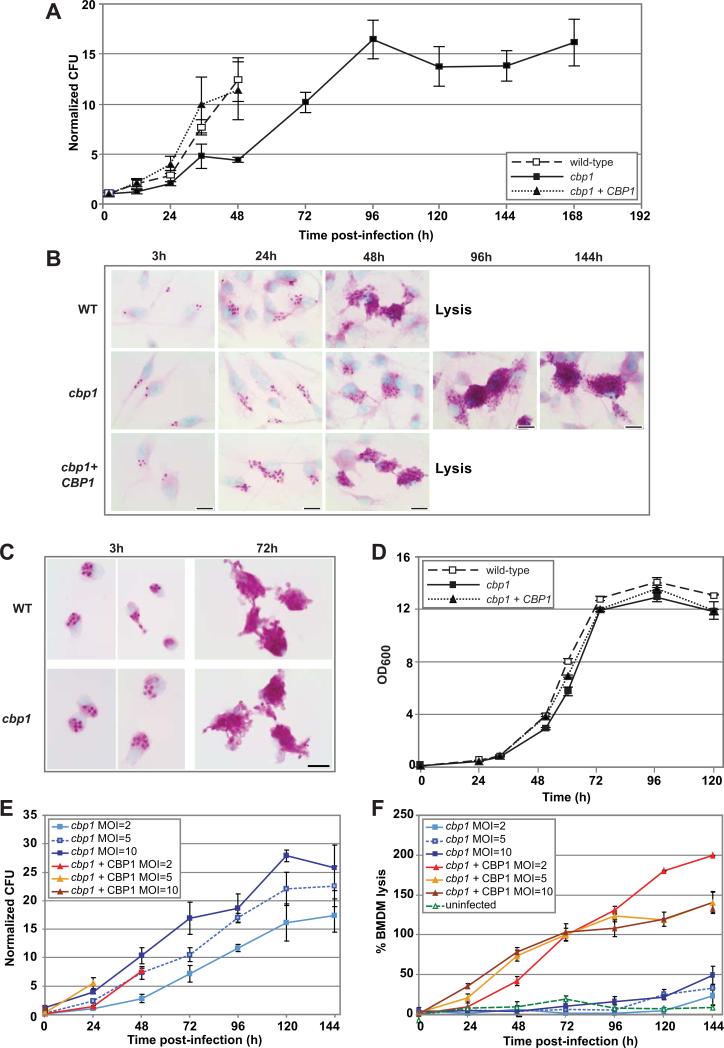
The failure of the cbp1 mutant to lyse macrophages could simply be due to an inability of this mutant to survive or replicate intracellularly. To determine if the cbp1 mutant had an intracellular viability or growth defect, intracellular yeasts were released from BMDMs by osmotic lysis and colony-forming units (CFUs) were enumerated. In the case of infection with the wild-type and complemented strains, measurements were terminated at the onset of Histoplasma-induced macrophage lysis (48 hpi) to avoid measuring the replication of extracellular yeast cells that had been released from infected macrophages. The CFUs of all three strains increased throughout the course of the infection, indicating that CBP1 was not required for the viability of H. capsulatum within host macrophages (Figure 3A). Notably, the cbp1 mutant achieved a high intracellular fungal burden that equaled or exceeded that of wild-type cells at the time of Histoplasma-induced macrophage lysis, indicating that high intracellular fungal burden is not sufficient to trigger host-cell death. When intracellular yeasts were visualized by PAS staining, it was also clear that the cbp1 mutant achieved high fungal burden within macrophages at late time points (Figure 3B). Microscopic analysis of infected AvMs also revealed intracellular survival and replication of the cbp1 mutant (Figure 3C).
Figure 3. Intracellular growth delay of cbp1 mutant is observed in primary macrophages but is not responsible for the cbp1 lysis defect.
(A) BMDMs were infected with either wild-type G217B ura5Δ+URA (WT), the cbp1 mutant + URA (cbp1), or the complemented strain (cbp1+CBP1) at an MOI of 2. At 2, 12, 24, 36, 48, 72, 96, 120, 144, and 168 hpi, BMDMs were osmotically lysed and the lysates were plated for H. capsulatum CFUs. Each measurement is the average of 4 platings (duplicate infections/ duplicate platings) ± standard deviation. (B) BMDMs were infected with either wild-type G217B ura5Δ+URA (WT), the cbp1 mutant + URA (cbp1), or the complemented strain (cbp1+CBP1) at an MOI of 2. At 3, 24, 48, 96, and 144 hpi, the infected monolayers were fixed and H. capsulatum yeast cells were stained with periodic acid/Schiff (PAS) reagent. Representative images at 100x magnification are shown. In the case of infection with the wild-type and complemented strains, the monolayers showed extensive lysis after 48 hpi. Scale bar = 10 μm. (C) AvMs were infected with either wild-type or cbp1 mutant yeast cells at an average MOI of 2. At 3 and 72 hpi, the infected monolayers were fixed and H. capsulatum yeast cells were stained with periodic acid/Schiff (PAS) reagent. Representative images at 100x magnification are shown. Scale bar = 10 μm. (D) In vitro growth curve analysis of wild-type H. capsulatum, the cbp1 mutant, and the complemented strains was performed. Yeast cells were inoculated into HMM at a starting OD600 = 0.1 and subsequent OD600 was monitored over time. The average of three measurements ± standard deviation is shown. (E) and (F) BMDMs were infected with either the cbp1 mutant or the complemented strain (cbp1+CBP1) at an MOI of either 2, 5, or 10. CFU are enumerated in (E) and an LDH release assay to quantify cytotoxicity is shown in (F). Due to continued replication of BMDMs over the course of the experiments, the total LDH at later time points is greater than the total LDH from the 2 hr time point, resulting in an apparent % lysis that is greater than 100%.
Although the cbp1 mutant survived and replicated within macrophages, it exhibited both a growth delay and a reduced growth rate as detected by colony-forming units. The slower rate of replication of the cbp1 mutant was specific to its growth in macrophages, as the mutant grew similarly to wild-type H. capsulatum during in vitro broth culture (Figure 3D). We reasoned that the inability of the cbp1 mutant to lyse macrophages might be due to slower accumulation of intracellular yeasts. To mimic a rapid increase in phagosome burden during infections with the cbp1 mutant, we infected BMDMs with the cbp1 mutant at a high multiplicity of infection (MOI). The resultant CFU and lysis data were compared to data from infections with the complemented strain performed at a lower MOI (Figure 3E and F). In the case of infections with the complemented strain, measurements were terminated at the onset of Histoplasma-induced macrophage lysis 24-48 hpi) to avoid measuring the replication of extracellular yeast cells that had been released from infected macrophages. BMDMs were infected with either the cbp1 mutant or the complemented strain at MOI of 2, 5, and 10. Analysis of colony-forming units showed that infection with the cbp1 mutant at MOI = 5 yielded a similar increase in fungal burden as observed for infection with the complemented strain at MOI = 2, and infection with the cbp1 mutant at MOI = 10 showed a higher rate of increase in fungal burden compared to the complemented strain at MOI = 2 (Figure 3E). Nonetheless, despite an equivalent or higher increase in fungal burden compared to the complemented strain, each of the cbp1 mutant infections failed to trigger BMDM lysis irrespective of MOI, whereas all infections with the complemented strain resulted in lysis (Figure 3F). These data indicate that the requirement for Cbp1 in host-cell lysis is not simply due to a requirement for Cbp1 in intracellular growth rate, and suggest that Cbp1 directly provokes macrophage cell death during infection.
Cbp1 is required for H. capsulatum virulence in the murine model of histoplasmosis
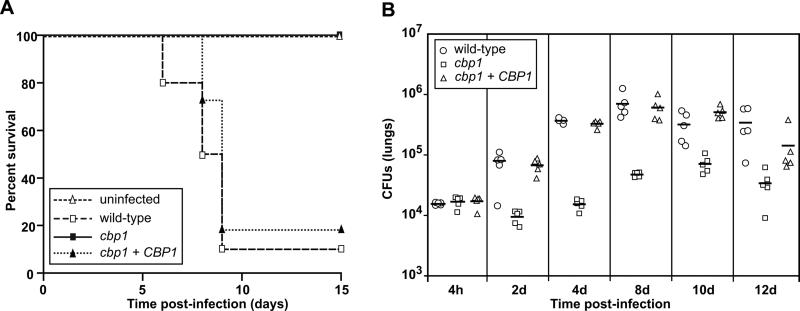
Given the critical role for Cbp1 in macrophage lysis, we hypothesized that Cbp1 might be required for optimal in vivo growth and virulence of H. capsulatum in the mouse model of histoplasmosis. A previous study (Sebghati et al., 2000) generated a cbp1 mutant in a strain background (G186AR) generally thought to be less virulent than the G217B parent strain used in our laboratory, especially at standard infection doses (Mayfield & Rine, 2007, Medoff et al., 1986, Sepulveda et al., 2014, Tewari & Berkhout, 1972). The ability of the G186AR cbp1 mutant to colonize the mouse lung was compromised (Sebghati et al., 2000). To determine the role of Cbp1 in the highly virulent G217B strain, and to conduct the first virulence study examining a role for Cbp1 in lethal infection, we infected female C57Bl/6 mice intranasally with a lethal dose (1.25 × 106 yeast cells/mouse) of either wild-type H. capsulatum, the cbp1 mutant, or the complemented strain, and monitored the mice daily for symptoms of disease, including weight loss, panting, and lack of grooming. Mice infected with wild-type H. capsulatum were symptomatic at five days post-infection and 90% of these mice (n = 10) succumbed to the infection within nine days (Figure 4A). Conversely, all mice infected with the cbp1 mutant survived for the duration of the experiment, even though they transiently displayed symptoms of respiratory distress. Mice infected with the complemented strain showed disease progression similar to that of mice infected with wild-type H. capsulatum, with 81% of the mice (n=11) succumbing to the infection. These results indicate that CBP1 is required for virulence in the mouse model of histoplasmosis.
Figure 4. Cbp1 is a virulence factor that is required for growth of H. capsulatum in vivo.
(A) Kaplan-Meir plots/survival curves of female C57Bl/6 mice intranasally infected with 1.25 × 106 CFUs of either the wild-type G217B ura5Δ+URA (wild-type) (n=10), the cbp1 mutant + URA (cbp1) (n=10), or the cbp1+CBP1 (n=11) strain. p < 0.0001 (log rank test, wild-type vs cbp1) (B) Female C57Bl/6 mice were infected intranasally with a sublethal dose of either wild-type, cbp1 mutant, or cbp1+CBP1 (5 × 104 CFUs). At various time points post-infection, lungs were harvested and homogenized. The homogenate was then plated to determine H. capsulatum CFUs. Horizonal bars represent the average CFUs/group. p < 0.0001 (ANOVA with Bonferroni post-tests correction, wild-type vs cbp1.
To determine if the cbp1 mutant is avirulent due to a failure to survive in the mouse, we monitored pulmonary fungal burden over time in mice infected with the wild-type, cbp1 mutant, or complemented strains. Female C57Bl/6 mice were infected intranasally with a sub-lethal dose (5 × 104 yeast cells/mouse) of each strain and CFUs were enumerated from lungs at various times post-infection (Figure 4B). The pulmonary fungal burden of mice infected with either the wild-type or complemented strains increased steadily between 4 hpi and 8 dpi, resulting in a 40-50-fold increase in CFUs. In contrast, levels of the cbp1 mutant were static over the first 4 days of infection, after which there was a modest 5-fold increase in CFUs by 10 dpi. These data indicate that Cbp1 is required for robust growth within the mouse lung. Interestingly, as observed previously (Edwards et al., 2011a, Sebghati et al., 2000), the cbp1 mutant was not cleared from the mouse. Consistent with the CFU analysis, histological examination of lung sections at 8 dpi revealed a markedly decreased fungal burden and inflammatory infiltrate in the lungs of mice infected with the cbp1 mutant when compared to mice infected with the wild-type or complemented strains (Figure S2). We reproducibly observed that infection with the cbp1 mutant yielded limited numbers of focal lesions, as shown in Figure S2, consistent with limited growth within and/or escape of the mutant from host cells. Taken together, a compelling hypothesis that arises from this work is that the ability of Cbp1 to trigger macrophage lysis correlates with increased fungal burden and lethal infection in the mouse model of histoplasmosis.
Cbp1 is required for a specific transcriptional signature of macrophages infected with H. capsulatum
To take an unbiased approach to further explore the effect of Cbp1 on host macrophages during infection, we performed expression profiling on infected macrophages with the hope of uncovering host pathways that might lead to Cbp1-dependent host-cell death. Since whole-genome expression profiling of the host response to H. capsulatum had not been performed before, we began by examining the transcriptional profile of macrophages infected with wild-type G217B cells. To distinguish between host transcriptional responses that were a consequence of passive pattern recognition versus those that were induced by intracellular growth and replication of H. capsulatum, we compared the transcriptional response of BMDMs infected with (1) live G217B cells, which undergo robust intracellular replication, or (2) UV-treated (UVT) G217B cells, which are rapidly degraded following phagocytosis (Figure S3). The transcriptional profile of BMDMs infected at an MOI of 10 was assessed at seven time points post-infection ranging from 2 hpi, an early time point following phagocytosis of H. capsulatum, to 24 hpi, which was 6 hours after visible lysis had initiated. A mock-infected time course was performed in parallel with these experiments to identify those transcripts whose induction or repression was specific to H. capsulatum infection rather than due to the culture conditions during infection. Microarray data from several replicate time courses was subjected to Significance Analysis of Microarrays (SAM) (Tusher et al., 2001) to identify 152 genes that were reproducibly induced (129 genes) or repressed (23 genes) at least 2-fold during infection with wild-type H. capsulatum yeasts when compared with mock-infected macrophages (Table S2).
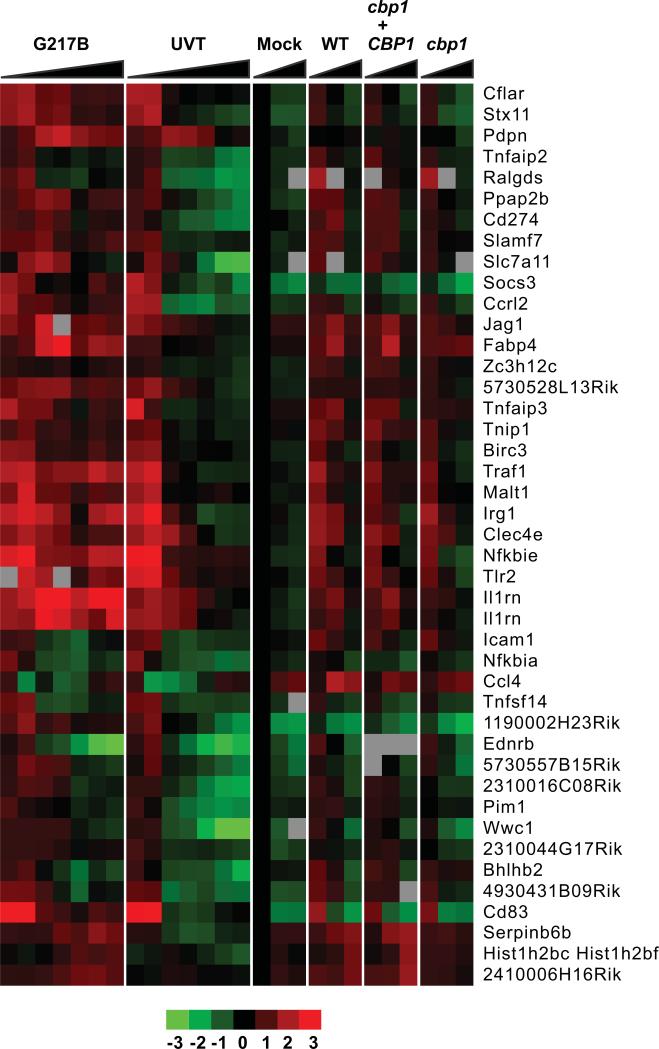
A subset of the induced genes were canonical inflammatory response genes, which were upregulated at early time points following infection (1-3 hpi) with both live and UVT cells (Figure 5). Notably, this inflammatory response was sustained at multiple time points after infection with live G217B, but was only transiently induced by UVT cells and disappeared concomitant with their degradation. This gene set included well-characterized inflammatory mediators (CCRL2, CCL4/MIP1β, and CXCL2/MIP2α) and genes involved in cell migration and/or adhesion (ICAM1, PDPN).
Figure 5. H. capsulatum induces a Cbp1-independent general inflammatory response in infected macrophages.
BMDMs were infected with H. capsulatum at an MOI of 5 and gene expression was examined by microarray analysis at several time points post-infection. Select clusters of macrophage genes whose expression is induced or repressed at least 2-fold following infection with H. capsulatum yeasts are shown. Each column corresponds to the following time points: G217B: 2, 3, 6, 8, 12, 20, 24 hpi; UVT (UV-treated G217B): 2, 3, 6, 8, 12, 20, 24 hpi; Mock: 3, 6, 12 hpi; WT (G217B ura5Δ+URA): 3, 6, 12 hpi; cbp1+ CBP1: 3, 6, 12 hpi; cbp1 (cbp1+URA): 3, 6, 12 hpi. Red indicates an increase in gene expression relative to mock-infected samples, green indicates a decrease, black indicates no change, and gray indicates missing data. SAM analysis was performed on data from multiple time courses as described in Materials and Methods. For clarity, only one representative time course for each infection is shown. Individual clusters are separated by horizontal white lines. The general (Cbp1-independent) inflammatory response is shown.
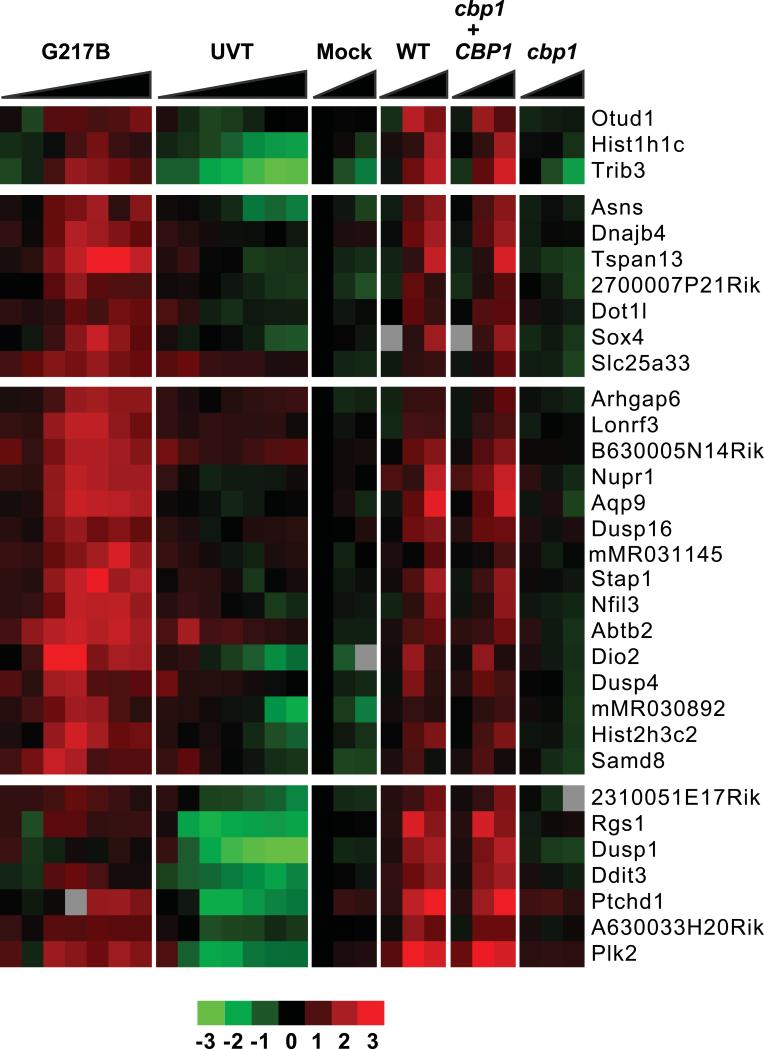
We next compared the transcriptional response of BMDMs infected with the cbp1 mutant, as well as its isogenic wild-type (G217B ura5Δ) and complemented strains. BMDMS were infected at an MOI of 10, and harvested for transcriptional profiling at 3, 6, and 12 hpi, before appreciable lysis by wild-type and complemented strains could occur. Of the 23 genes that are repressed during infection with wild-type H. capsulatum, 3 were dependent on Cbp1 (Table S2). The cbp1 mutant induced a similar canonical inflammatory response as the wild-type and complemented strains, indicating that Cbp1 is not required for the ability of the host to recognize H. capsulatum (Figure 5). Strikingly, of the 129 genes whose transcription is induced in response to H. capsulatum infection, SAM analysis indicated that 44 genes were dependent on Cbp1 for their induction in infected macrophages (Table S2). Of these, 32 showed at least a two-fold difference in induction in macrophages infected with wild-type vs. cbp1 mutant yeasts (these genes are depicted in Figure 6). Several of the Cbp1-dependent genes (Table S2) play important roles in mammalian cell biology and/or host-fungal interactions. For example, Prostaglandin-endoperoxide synthase 2 (Ptgs2; also known as Cox2) facilitates synthesis of prostaglandins from arachidonic acid. Interestingly, treating H. capsulatum-infected mice with prostaglandin inhibitors results in decreased fungal burden and increased host survival (Pereira et al., 2013), suggesting that H. capsulatum-mediated stimulation of COX2 could promote pathogenesis.
Figure 6. H. capsulatum induces a Cbp1-dependent macrophage transcriptional response program.
Data from the same experiments described in the Figure 5 legend are shown. The Cbp1-dependent transcriptional response is depicted.
We were intrigued by the realization that a number of the Cbp1-dependent genes are also known to be induced during endoplasmic reticulum (ER) stress, suggesting that Cbp1 may directly or indirectly induce this stress response pathway in host cells during infection. For example, Cbp1 was required for expression of the stress-regulated transcription factor NUPR1/p8, which is induced in human glioma cells during ER stress (Salazar et al., 2009). In those experiments, NUPR1/p8 stimulates expression of the transcription factors ATF4 (activating transcription factor 4) and DDIT3/CHOP (DNA-damage inducible factor 3/C-EBP homologous protein), which in turn stimulate expression of the pseudo-kinase tribbles homolog 3 (TRB3), (Carracedo et al., 2006, Salazar et al., 2009). Similarly, in human cells treated with tunicamycin and other ER stress-inducing agents, ATF4 and CHOP coordinately regulate expression of TRB3 (Ohoka et al., 2005). We observed that ATF4, DDIT3/CHOP, and TRB3 were also induced during infection of macrophages with H. capsulatum, and that their induction was Cbp1-dependent (although ATF4 did not meet our criteria of two-fold induction during infection with wild-type H. capsulatum and thus was not included in the final data set). Notably, activation of TRB3 expression by ATF4 and DDIT3/CHOP has been correlated with apoptosis in pancreatic islet cells (Bromati et al., 2011).
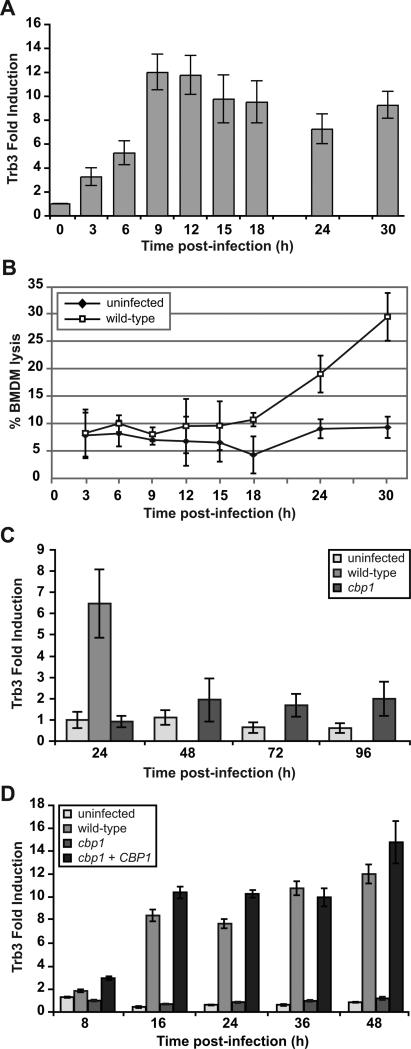
Given its role in stimulating apoptosis and autophagy under some conditions (Shang et al., 2009, Ohoka et al., 2005, Carracedo et al., 2006, Ord & Ord, 2005, Qing et al., 2012), and cell survival in other contexts (Shimizu et al., 2012, Ord et al., 2012), we were particularly interested in Cbp1-dependent induction of TRB3. According to our microarray data, infection of BMDMs with wild-type H. capsulatum stimulated 4.5-fold induction of TRB3 at 12 hpi compared to an early mock-infected time point, whereas its expression was repressed by 3.6-fold in cbp1 mutant-infected macrophages compared to mock-infected cells. To determine the kinetics of TRB3 induction relative to host-cell lysis, we simultaneously monitored induction of TRB3 transcript by qRT-PCR and release of LDH from infected BMDMs. We observed approximately 12-fold induction of TRB3 expression by 9-12 hpi at MOI of 5 (Figure 7A), which was at least fifteen hours prior to the onset of detectable macrophage lysis by wild-type H. capsulatum (Figure 7B). In macrophages infected with the cbp1 mutant, appreciable TRB3 induction was never observed even at late time points after infection (Figure 7C). Additionally, Cbp1-dependent induction of TRB3 was also observed at a lower MOI of 2 (Figure 7D). Taken together, our results suggest that Cbp1 can mediate host-cell death by inducing TRB3 expression.
Figure 7. Induction of TRB3 mRNA is dependent on CBP1 and precedes macrophage lysis.
(A) BMDMs were infected with G217B yeast cells at an MOI of 5. At various time points post-infection, total RNA was isolated from BMDMs and TRB3 expression was monitored by quantitative RT-PCR. Fold induction was calculated relative to the uninfected 0-hour sample. (B) To monitor macrophage lysis, supernatants from the same infected macrophages assayed in (A) were subjected to LDH release analysis. (C) BMDMs were either mock-infected (uninfected) or infected with wild-type G217B ura5Δ+URA (wild-type) or cbp1+URA mutant cells (cbp1) at an MOI of 10. Lysis of the macrophage monolayer infected with wild-type cells was complete by 48 hpi. At the indicated times post-infection, total RNA was isolated from BMDMs and TRB3 expression was monitored by quantitative RT-PCR. Fold induction was calculated relative to the mock-infected 24-hour sample. (D) BMDMs were either mock-infected (uninfected) or infected with wild-type G217B ura5Δ+URA (wild-type), cbp1+URA mutant cells (cbp1), or the complemented strain (cbp1+CBP1) at an MOI of 2. At the indicated times post-infection, total RNA was isolated from BMDMs and TRB3 expression was monitored by quantitative RT-PCR. Fold induction was calculated relative to the mock-infected 0-hour sample.
Activation of caspase-3/7 by Histoplasma capsulatum is dependent on Cbp1
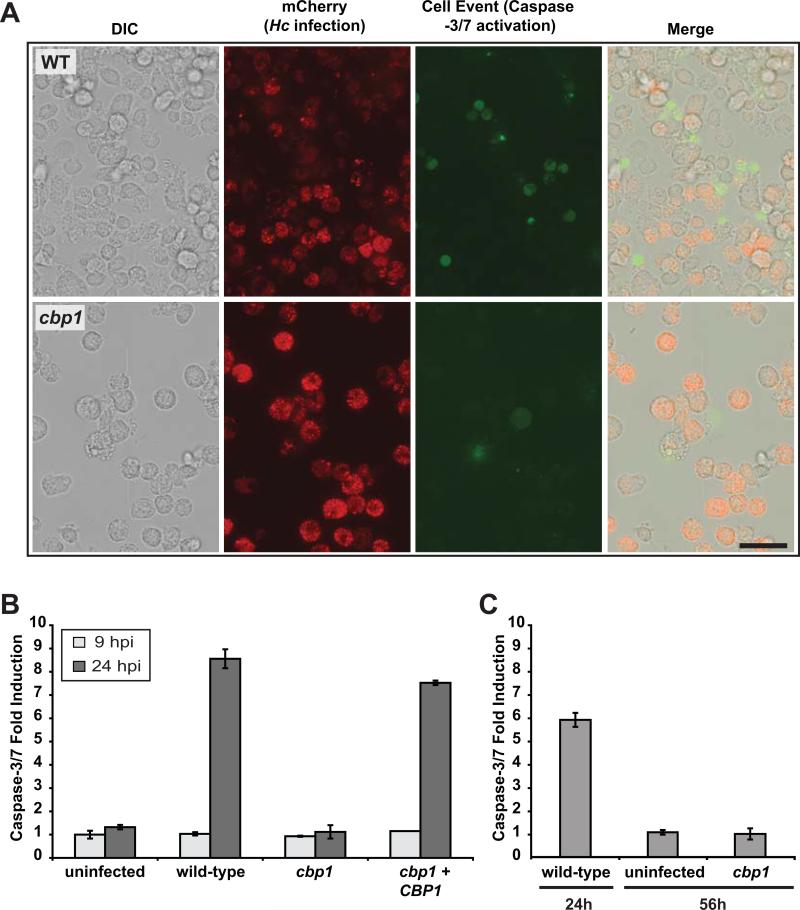
Previous work indicated that GM-CSF-derived bone marrow macrophages produce tumor necrosis factor α (TNFα), which triggers extrinsic apoptotic pathways to activate caspase-3/7 during H. capsulatum infection (Deepe & Buesing, 2012). In contrast, in our CSF-derived macrophages, we have never been able to detect TNFα production during H. capsulatum infection (data not shown). Nonetheless, we hypothesized that Cbp1 could play a role in triggering apoptosis, and we favor the model that Cbp1 causes an intrinsic cellular stress, triggers Trb3 induction, and activates proapoptotic pathways, resulting in caspase-3/7 activation. Caspase-3 and -7 play a critical role in the execution phase of apoptosis by targeting a number of cellular substrates for cleavage (Salvesen & Riedl, 2008). We first tested whether we could observe Cbp1-dependent caspase-3/7 activation in BMDMs using a microscope-based assay. To more easily detect H. capsulatum within macrophages, we transformed the wild-type and cbp1 mutant strains with a construct encoding a secreted fluorescent protein (mCherry fused to an H. capsulatum signal sequence). We observed mCherry fluorescence marking H. capsulatum-containing phagosomes in infected macrophages (Figure 8A). To detect active caspase-3/7, we used CellEvent caspase-3/7 detection reagent, which is a four amino-acid peptide (DEVD) conjugated to a nucleic acid binding dye. In cells containing active caspase-3/7, the peptide is cleaved from the dye, which then binds DNA and fluoresces. BMDMs were infected with either the wild-type or cbp1 mutant strain, and infected macrophages were treated with CellEvent caspase-3/7 detection reagent at 48 hpi. We observed significant intracellular fungal burden for infections with both wild-type and cbp1 mutant strains as indicated by the mCherry signal (Figure 8A). However, appreciable CellEvent signal was only detected in BMDMs infected with wild-type H. capsulatum. We also monitored caspase-3/7 activation in host-cell lysates (Figure 8B). We observed a robust increase (7-8 fold over mock-infected) in caspase-3/7 activity at 24 hours post-infection in BMDMs harboring wild-type strains of H. capsulatum, but not in BMDMs infected with cbp1 mutant yeasts. We failed to detect induction of caspase-3/7 activity in BMDMs infected with the cbp1 mutant even late in infection, thus ruling out the possibility that caspase-3/7 activation was simply delayed in the absence of CBP1 (Figure 8C). These data indicate that H. capsulatum stimulates caspase-3/7 activation in macrophages in a Cbp1-dependent manner.
Figure 8. Cbp1 is required to promote caspase-3/7 activation in BMDMs infected with H. capsulatum yeast cells.
(A) Wild-type G217B ura5Δ or cbp1 mutant cells, each transformed with a construct expressing secreted mCherry, were used to infect BMDMs. Two days post-infection, infected BMDMs were incubated with CellEvent caspase-3/7 detection reagent. Scale bar corresponds to 50 μm. (B) BMDMs were either mock-infected (uninfected) or infected with either wild-type G217B ura5Δ+URA (wild-type), cbp1 mutant +URA (cbp1), or CBP1 complemented (cbp1+CBP1) yeast cells. At 9 and 24 hpi, BMDM lysates (containing 100 μg of total protein) were combined with DEVD-pNA colorimetric substrate, and cleavage of the substrate by active caspase-3/7 was monitored. Fold induction relative to mock-infected BMDMs at 9 hpi is shown and is representative of two independent experiments. (C) BMDMs were either mock-infected (uninfected) or infected with wild-type or cbp1 mutant cells. At the indicated time post-infection, BMDM lysates (containing 100 μg of total protein) were combined with DEVD-pNA colorimetric substrate, and cleavage of the substrate by active caspase-3/7 was monitored. Fold infection relative to mock-infected at 24 hpi is graphed. Although infection with wild-type cells resulted in a strong signal by 24 hpi followed by host-cell lysis, cbp1-infected cells did not show appreciable caspase-3/7 activation even at 56hpi.
Histoplasma-mediated cell death is dependent on apoptosis but not necrosis or pyroptosis
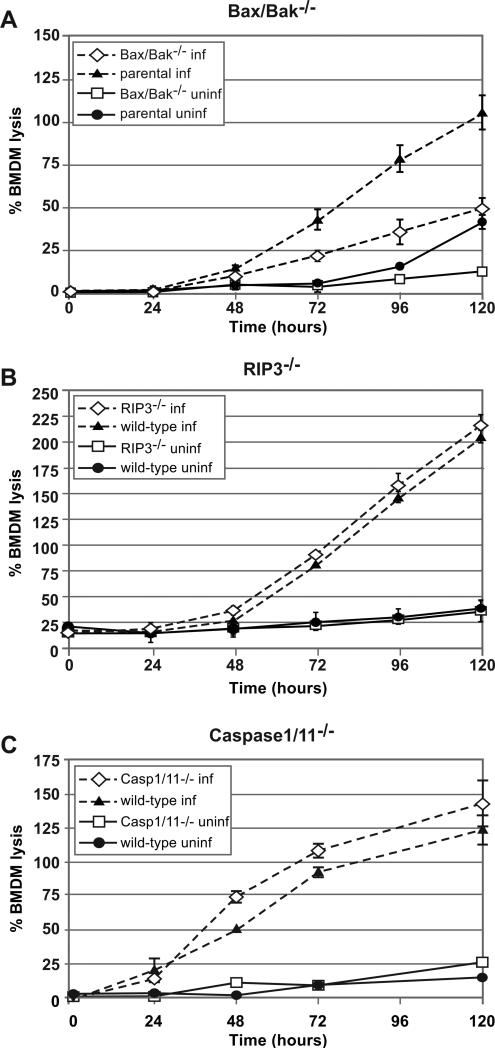
We next performed a definitive genetic test to determine whether apoptosis plays a role in H. capsulatum-mediated host-cell lysis. Although membrane integrity is usually preserved during apoptosis, LDH release has been observed for macrophages treated with agents that trigger apoptosis, such as gliotoxin (Brennan & Cookson, 2000), leaving open the possibility that BMDMs could die from apoptosis during H. capsulatum infection. To test the hypothesis that H. capsulatum triggers apoptosis in our macrophage model, we generated BMDMs that were deficient in pro-apoptotic Bcl-2 family proteins Bak and Bax, which form pores in the mitochondrial membrane during intrinsic apoptosis (Labbe & Saleh, 2008). We observed that Bak−/− Bax−/− mutant macrophages showed a markedly decreased rate and extent of lysis during H. capsulatum infection compared to parental macrophages (Figure 9A). In contrast, macrophages deficient in RIP3, which is required for many types of necrosis (Sridharan & Upton, 2014), or caspase-1 and -11, which are required for pyroptosis (Shin & Brodsky, 2015), showed no resistance to H. capsulatum-mediated host-cell death (Figure 9B, C). These data indicate that intrinsic apoptosis contributes to host-cell death during infection of macrophages with H. capsulatum.
Figure 9. Histoplasma-mediated cell death is dependent on apoptosis but not necrosis or pyroptosis.
Macrophages lacking the pro-apoptotic factors Bax and Bak (A), necrosis factor RIP3 (B), or pyroptosis proteins caspase-1 and caspase-11 (C) were either mock-infected (uninf) or infected with wild-type G217B ura5Δ+URA cells (inf). At the indicated hours post-infection, supernatants were removed from the infected monolayers and lactate dehydrogenase activity was assessed to monitor BMDM lysis. The average % BMDM lysis of four measurements ± standard deviation is shown for all samples at each timepoint. In some instances, however, the standard deviation is below the limit of resolution of the graph and is not apparent (i.e. uninfected BMDMs).
Discussion
H. capsulatum is an intracellular fungal pathogen that grows to high levels within the macrophage phagosome. In resting macrophages, the ultimate result of colonization with H. capsulatum is destruction of the host cell (Porta & Maresca, 2000, Deepe & Buesing, 2012). It was unknown whether death of the infected macrophage is simply a passive consequence of high fungal burden, or if it is actively induced by a fungal factor. Thus, when we began our genetic screen, it was unclear if it would be possible to identify mutants that grow to high levels within macrophages but fail to lyse them. Of the 14,000 insertion mutants that we screened, 3 were able to grow to high levels within macrophages and yet displayed an extremely strong lysis defect. All 3 mutants were deficient in expression of the CBP1 gene. We show that mutants lacking Cbp1 are able to survive and replicate within primary macrophages, reaching a level comparable to or exceeding that achieved by wild-type cells. Despite the high fungal burden achieved by the cbp1 mutant, infected macrophages fail to lyse, demonstrating for the first time that macrophage lysis is not an inevitable result of intracellular fungal load, but rather is the result of an active process mediated directly or indirectly by Cbp1. Additionally, we show that Cbp1 is required for robust in vivo growth and virulence, hence correlating the ability of Cbp1 to trigger host-cell death in vitro with a productive and lethal infection in vivo. Whole-genome expression profiling of infected macrophages revealed that Cbp1 is required for the induction of a unique transcriptional signature in host cells, including the expression of ER stress-responsive genes, some of which are known to modulate cell death. Additionally, we showed that Cbp1 is required for the activation of executioner caspases-3/7 during macrophage infection. Taken together, these data lead to the provocative model that H. capsulatum utilizes Cbp1 to actively trigger macrophage death, thereby facilitating dissemination of the fungus and optimal growth during animal infection.
Cbp1 is required for normal growth kinetics in vivo
Though Cbp1 is dispensable for survival of H. capsulatum in macrophages and mice, our data indicate that it modulates intracellular growth in addition to its role in host-cell lysis. Although the cbp1 mutant is able to replicate to high levels in macrophages, the mutant yeasts exhibit a reduced accumulation rate, with a doubling time approximately twice that of wild-type cells. Our data suggest that the lower rate of cbp1 accumulation is due to decreased growth rate rather than increased clearance because the mutant yeasts appeared to be viable: quantification of cbp1 yeast cells released from infected macrophages using microscopy gave numbers equivalent to enumeration of viable cells by CFU assay, indicating that the majority of intracellular cbp1 yeasts are alive. Additionally, there was no microscopic evidence of yeast-cell degradation inside macrophages.
The reason for the reduced intracellular growth rate of the cbp1 mutant is unclear. Cbp1 was originally identified as a highly abundant secreted calcium-binding protein present in the culture supernatant of H. capsulatum yeast cells (Batanghari & Goldman, 1997). Initial studies showed that Cbp1 was required for the ability of H. capsulatum to grow under calcium-limiting conditions in vitro (Sebghati et al., 2000), but the relevance of these data to intracellular growth is not known. Additionally, it has been hypothesized that Cbp1 could alter the character of the phagosome to facilitate growth of H. capsulatum (Beck et al., 2009).
In mice, we observed that Cbp1 was required for robust in vivo growth, although the host was unable to clear cbp1 mutant cells. In contrast to wild-type yeast cells, which increased approximately 50-fold over the first eight days post-infection, levels of the cbp1 mutant cells were essentially constant for the first four days post-infection, after which only a modest increase in cell number was observed. We favor the hypothesis that the inability of the cbp1 mutant to lyse infected macrophages and spread through the mouse lung contributes to its net replication defect. Additionally, we performed the first experiments to evaluate host survival during infection with the cbp1 mutant and observed an extremely strong virulence defect for the mutant compared to infection with either the parental or complemented strains.
H. capsulatum actively promotes host-cell death
Of particular interest from this work is the discovery that H. capsulatum actively promotes host-cell death by a Cbp1-dependent pathway. Past studies hypothesized that cbp1 mutants fail to lyse macrophages because of a presumed requirement for Cbp1 in intracellular growth (Sebghati et al., 2000). By showing that Cbp1 is dispensable for achieving high fungal burden, our work strongly suggests that H. capsulatum utilizes Cbp1 to trigger macrophage lysis. There are an increasing number of studies of host-cell death triggered by fungi (Johnston & May, 2013, Skeldon & Saleh, 2011), though little is known about fungal molecules that manipulate the viability of mammalian host cells. Since Histoplasma species and the closely related human fungal pathogen Paracoccidioides brasiliensis are the only organisms that contain homologs of Cbp1 (Beck et al., 2009), it could be that Cbp1 triggers host-cell death by a mechanism that is specific to H. capsulatum and related fungi. The timing of host-cell lysis may also be regulated such that it only occurs after high intracellular fungal burden has been achieved. Cbp1 is produced at high levels by yeast cells in culture as well as by intracellular yeasts, but perhaps a threshold level of Cbp1 has to accumulate before lysis ensues (Batanghari & Goldman, 1997, Batanghari et al., 1998, Patel et al., 1998). This model is consistent with the observation that time to lysis decreases as MOI increases.
Transcriptional profiling of infected macrophages reveals a Cbp1-independent inflammatory response
The global transcriptional response of macrophages to infection with wild-type or cbp1 mutant H. capsulatum was compared to identify host pathways that are manipulated by Cbp1. Additionally, these experiments provided a comprehensive molecular analysis of the interaction between H. capsulatum and host macrophages. The majority (two-thirds) of the transcriptional response of macrophages to infection with H. capsulatum is independent of Cbp1. Wild-type yeasts, UV-treated (UVT) wild-type yeasts, and cbp1 mutant yeasts all trigger a classic inflammatory response. Presumably this response reflects recognition of H. capsulatum pathogen-associated molecular patterns by host pattern recognition receptors. We found that the genes encoding the pattern recognition receptors TLR2 and CLEC7A/Dectin1 are upregulated in macrophages infected with H. capsulatum (Table S2). This observation is intriguing because several studies show that Dectin1 and TLR2 synergize to mediate recognition and downstream signaling in response to fungi (Brown et al., 2003, Gantner et al., 2003, Gersuk et al., 2006, Luther et al., 2007, Goodridge et al., 2007) as well as Mycobacteria (Yadav & Schorey, 2006, Shinet al., 2008).
Cbp1 triggers a specific gene expression response in host macrophages that could promote host-cell lysis
Transcriptional profiling of the infected macrophage revealed that Cbp1 is required for the induction of 44 genes. Given the requirement for Cbp1 in host-cell lysis, we were intrigued to discover that at least two of these genes, NUPR1/p8 and TRB3, are known to play roles in stress response and induction of cell death. NUPR1/p8 encodes a stress-regulated transcription factor with homology to the HMG/IY family of chromosomal binding proteins (Mallo et al., 1997, Encinar et al., 2001). Its activity is induced in glioma cells that are stimulated to undergo apoptosis by treatment with cannabinoids, resulting in expression of the transcription factors ATF4 and DDIT3/CHOP. In turn, ATF4 and DDIT3/CHOP induce TRB3 expression, which is required for activation of autophagy and apoptosis (Carracedo et al., 2006, Salazar et al., 2009). Notably, although the molecular function of TRB3 is not well understood, its expression has been shown to induce death in a number of cell types (Ohoka et al., 2005, Carracedo et al., 2006, Bromati et al., 2011).
Interestingly, a number of the Cbp1-dependent genes (NUPR1/p8, DDIT3/CHOP, ATF4, TRB3, and CHAC1) are known to be induced during ER stress in a variety of cell types (Goruppi & Iovanna, 2010, Su & Kilberg, 2008, Mungrue et al., 2009, Ohoka et al., 2005). ER signaling is becoming increasingly linked to innate immune responses (Martinon et al., 2010). Higher eukaryotic cells can initiate a three-pronged unfolded protein response pathway to counter ER stress, including PERK-dependent induction of DDIT3/CHOP and ATF4, IRE1-dependent splicing of XBP1, and ATF6 processing and activation (reviewed in (Todd et al., 2008)). Notably, it is well established that unmitigated ER stress can trigger caspase activation and cell death (reviewed in (Merksamer & Papa, 2010)). These data, along with our exploration of caspase-3/7 activation and the role of Bax and Bak in H. capsulatum-mediated host-cell death, suggest the model that Cbp1 initiates an ER stress response, resulting in TRB3 induction, Bax/Bak oligomerization, caspase activation, and apoptotic cell death. The pathways implicated by this study provide our first molecular understanding of how H. capsulatum is able to manipulate macrophage viability so effectively during infection.
Experimental Procedures
Strains and Culture Conditions
H. caspsulatum strain G217B ura5Δ (WU15) was a kind gift of William Goldman (University of North Carolina, Chapel Hill). For all studies involving the cbp1 mutant, “wild-type” refers to G217B ura5Δ transformed with a URA5-containing episomal vector, “cbp1” refers to G217B ura5Δ cbp1::T-DNA (mutant 172-C5) transformed with the same URA5-containing episomal vector, and “complemented” strain refers to G217B ura5Δ cbp1::T-DNA transformed with the URA5-containing episomal plasmid bearing the wild-type CBP1 gene (see below). Yeast cells were grown in liquid Histoplasma macrophage media (HMM) or on HMM agarose plates (Worsham & Goldman, 1988). Liquid cultures were grown in an orbital shaker at 37°C with 5% CO2. Stock cultures were maintained by passaging every 2-3 days at a 1:25 dilution. Plates were incubated in a humidified chamber at 37°C with 5% CO2.
For macrophage infections with individual strains, an overnight, mid-log culture of yeast cells (OD600 = 5-7) was prepared. Approximately 18 hours prior to the infection, a two-day late log/stationary phase culture (OD600 = 10-12) was diluted 1:5 into HMM media. The diluted cells were then incubated at 37°C with 5% CO2 overnight to obtain mid-log cultures at the time of infection. Culture ODs were measured using an Eppendorf BioPhotometer.
Insertional mutagenesis of H. capsulatum
H. capsulatum insertion mutants were generated using Agrobacterium-mediated transformation of WU15 as previously described (Nguyen & Sil, 2008). Transformants (hygromycin resistant (HygR)) were selected on HMM/uracil plates containing 400 μg/ml uracil, 200 μg/ml hygromycin B and 200 μM cefotaxime.
Screen to identify lysis-defective H. capsulatum mutants
14,000 insertion mutants were screened for their ability to lyse BMDMs and J774.1 macrophages. Individual insertion mutants and the WU15 parental strain were inoculated into the wells of a deep-well 96-well plate (Corning 3598) containing 600 μL HMM/hyg/uracil or HMM/uracil, respectively. These plates were then sealed with Neptune Bioseal Breathable Tape (Continental Life Products 2424.S) and incubated at 37°C with 5% CO2 for four days in a Multitron shaker (Appropriate Technical Resources/Infors). Each culture was then diluted 1:25 into fresh medium in a fresh deep-well 96-well plate and incubated as above for an additional two days. Approximately 18-20 hours before infection, 20 μL of each culture was added to 115 μL of HMM/uracil in a standard 96-well plate (BD Falcon 353027) and incubated as above. At the time of infection, the optical density of each well was measured using the Spectramax Plus 384-well plate reader (Molecular Devices). The median optical density was calculated for each plate, converted to cell number, and macrophages were infected at a multiplicity of infection (MOI) of 10 based on the median H. capsulatum concentration. Either J774.1 cells (at 5.5 × 104 macrophages per well) or BMDMs (8 × 104 macrophages/well) were seeded in a 96-well plate approximately 16 hrs before infection. Each infection was set up in duplicate, yielding two infection plates per one plate of H. capsulatum insertion mutants. Infected macrophages were washed one hr after addition of H. capsulatum. Wells were supplemented with fresh medium at 2 days post-infection (dpi) and 4 dpi if the infection was still in progress. J774.1 monolayers were fixed with by incubating with 150 μL of 10% formaldehyde in PBS for 5 min at 5 dpi whereas BMDM monolayers were fixed at 3-4 dpi due to altered kinetics of infection compared to J774.1 cells. Fixed monolayers were stored in PBS at room temperature for future crystal violet staining. At the time of staining, PBS was removed from each well and replaced with 75 μL of 0.2% w/v crystal violet, 2% v/v EtOH. After 5 min, the crystal violet solution was removed and cells were incubated in PBS for an additional 5 min. The PBS was then removed and wells were air-dried.
Identification of the cbp1 mutant
Forty-seven mutants were defective for macrophage lysis, including 172-C5, 138-G1, and UA6-C8, all of which had a severe lysis phenotype. Inverse PCR was used to map the location of the Agrobacterium T-DNA insertion in mutant 172-C5 as described above, which revealed the presence of an insertion 68 nucleotides upstream of the CBP1 open reading frame. This mutant was designated as the cbp1 mutant and complemented by introduction of the wild-type CBP1 gene as described below.
Complementation of the cbp1 mutant
Transformation
An episomal complementation plasmid, pDTI22, containing the CBP1 ORF, 2764 bp of 5’ flanking sequence and 554 bp of 3’ sequence was generated (Figure S1A). 100 ng of PacI digested pDTI22 was transformed into the cbp1 mutant as previously described (Woods et al., 1998), creating the cbp1+CBP1 strain. The cbp1 mutant was also transformed with pLH211 as a vector control, creating the cbp1+URA5 strain. The ura5Δ parental strain, WU15, was also transformed with pLH211, generating the ura5Δ+URA5 strain (wild-type control). Transformants were selected on HMM-agarose plates.
In vitro H. capsulatum growth
Two-day, late log cultures of the ura5Δ+URA5, the cbp1+URA5, and the cbp1+CBP1 strains were used to inoculate 30 ml HMM media to a starting OD600=0.1. At 20, 26, 47, 69, 97, 124, and 169 hours post inoculation, 3 × 1 ml samples were removed from each culture, vortexed for 30 seconds to break up clumps, and analyzed to determine their OD600.
Macrophage culture
Bone marrow derived macrophages (BMDMs) from 6-8 week old female C57Bl/6 (Charles River Laboratories, Jackson Laboratories) mice (Jackson Laboratories) were isolated using differentiation of bone marrow with murine macrophage colony-stimulating factor (CSF-1), as described previously (Hwang et al., 2008, Isaac et al., 2013). Alveolar macrophages (AvMs) were obtained by broncho-alveolar lavage (BAL) from the lungs of 12- to 15- week-old female C57Bl/6 mice (Charles River Laboratories). 20 mL of BAL fluid (5 mM EDTA in PBS without Ca2+ and Mg2+) were collected from each mouse. The cells were pelleted and resuspended in AKT lysis solution (150 mM NH4Cl, 10 mM KHCO3, 0.1 mM EDTA in ddH2O, pH 7.2-7.4). The remaining cells were pelleted and resuspended in AvM media, which consists of Dulbecco's Modified Eagle Medium (DMEM) High Glucose (UCSF Cell Culture Facility), 10% Fetal Bovine Serum (Hyclone, Thermo Fisher, www.hyclone.com), penicillin and streptomycin (UCSF Cell Culture Facility).
Cytotoxicity/LDH release assay
Bone marrow derived macrophages (BMDMs)
In 24-well tissue-culture treated dishes, 2 × 105 BMDMs were infected, in duplicate, with H. capsulatum strains at an MOI of 2 or 5. In preparation for the infections, logarithmically growing H. capsulatum cultures were pelleted, resuspended in DMEM without phenol red, sonicated for 3 seconds on setting 2 using a Fisher Scientific Sonic Dismembrator Model 100, and counted by hemacytometer. After a 2-hour incubation period, the media was removed from the infected BMDMs, the monolayers were washed twice with DMEM without phenol red and 750 μl BMM without phenol red (DMEM High Glucose without phenol red (UCSF Cell Culture Facility), 20% Fetal Bovine Serum, 10% v/v CMG supernatant (conditioned medium obtained from CMG cells (3T3 fibroblasts transfected with the murine CSF-1 cDNA, kindly provided by Mary Nakamura, UCSF), 2 mM glutamine, 110 mg/mL sodium pyruvate, penicillin and streptomycin) was added to each well. The infected macrophages were then incubated at 37°C with 5% CO2. Approximately 48 hours post-infection, 250 μl fresh BMM was added to each well.
At various time points post-infection, LDH levels in the infected-macrophage supernatants were measured to monitor BMDM lysis as previously described (Isaac et al., 2013). The % BMDM lysis at each time point was calculated as the percentage of the total LDH from lysed uninfected cells at 2 hours post-infection. Due to continued replication of BMDMs over the course of the experiment, the total LDH at later time points is greater than the total LDH from the 2 hr time point, resulting in an apparent % lysis that is greater than 100%.
Alveolar macrophage (AvMs)
In 96-well tissue-culture treated dishes, 1.5 × 105 AvMs were infected, in duplicate, with H. capsulatum strains at an MOI of 5. After a 6-hour incubation period, the media was removed from the infected AvMs, the monolayers were washed twice with DMEM without phenol red and 200 μl AvM media without phenol red (DMEM without phenol red, 10% Fetal Bovine Serum, penicillin and streptomycin) was added to each well. The infected macrophages were then incubated at 37°C with 5% CO2. At various time points post-infection, LDH levels in the infected macrophage supernatants were measured to monitor AvM lysis. To measure the total LDH, at 6 hours post-infection, one mock-infected macrophage monolayer was lysed with 200 μl of lysis solution (1% Triton X-100 in DMEM-phenol red). 50 μl of clarified culture supernatant or mock-infected lysate was transferred to a 96-well plate and LDH release was assessed as described above for BMDMs.
Intracellular Replication Assay
In 24-well tissue culture treated dishes, 2 × 105 BMDMs were infected, in duplicate, with H. capsulatum strains at an MOI of 2. After a 2-hour incubation period, the media was removed, the monolayers were washed twice with DMEM and 1 ml BMM was added to each well. The infected macrophages were then incubated at 37°C with 5% CO2. The media was changed at 48 hours post-infection and every day thereafter. At various time points post-infection, the media was removed from each well and 500 μl ddH2O was added. After 5 minutes of incubation, the macrophages were mechanically lysed by vigorous pipetting. The lysate was collected, sonicated, diluted in HMM and plated for H. capsulatum colony forming units (CFUs) on HMM-agarose plates at 37°C. Colonies were counted 14 days later. The relative CFU at time × is calculated as (CFUtx)/(CFUt0).
Microscopic analysis of intracellular growth and dissemination
AvMs
2 × 105 AvMs per well were seeded in 24-well tissue-culture treated dishes containing 15 mm glass coverslips. Approximately 18 hours later, these cells were infected with 4 × 105 yeast cells per well and incubated for 3 hours. The media was then removed from the infected AvMs, the monolayers were washed twice with AvM media and 750 μl AvM media was added to each well. The infected macrophages were then incubated at 37°C with 5% CO2. An additional 250 μl of AvM media was added to infected cells at 48 hours post-infection.
At various time points post-infection, the media was removed from each well and the monolayers were fixed with 500 μl PAS fixative (3.7% formaldehyde in 100% ethanol) for 1 minute. The fixative was removed and cells were washed twice with PBS before being stored in 500 μl PBS at 4°C. Slides were then stained with Periodic acid-Schiff reagent (to highlight fungal cells) and methyl green (to counterstain macrophage nuclei) as previously described (Webster & Sil, 2008). Microscopic images were taken using the Leica DM 1000 microscope.
BMDMs
3.25 × 105 BMDMs per well were seeded in 12-well tissue-culture treated dishes containing 15 mm glass coverslips. Approximately 18 hours later, these BMDMs were infected with 8.1 × 104 yeast cells per well, centrifuged for 5 minutes at 800 × g, and incubated for 2 hours. The media was then removed from the infected BMDMs, the monolayers were washed twice with DMEM and 2 ml BMM was added to each well. The infected macrophages were then incubated at 37°C with 5% CO2. The media was changed at 48 hours post-infection and every day thereafter. At various time points post-infection the media was removed from each well and the monolayers were fixed with 1 ml PAS fixative (3.7% formaldehyde in 100% ethanol) for 1 minute. The fixative was then removed and cells were washed twice with PBS before being stored in 2 ml PBS at 4°C. Coverslips were then stained with Periodic acid-Schiff reagent (to highlight fungal cells) and methyl green (to counterstain macrophage nuclei) as previously described (Webster & Sil, 2008). Microscopic images were taken with a Leica DM 1000 microscope.
Mouse infections
9 week-old female C57Bl/6 mice (Charles River Laboratories) were anesthetized with isoflurane and infected intranasally with wild-type H. capsulatum (G217B ura5Δ+URA), the cbp1 mutant (cbp1+URA) or the complemented stain (cbp1+CBP1). In preparation for infection, mid-logarithmic cultures of these H. capsulatum strains were washed once with PBS, sonicated for 3 seconds on setting 2 using a Fisher Scientific Sonic Dismembrator Model 100, and counted by hemacytometer to determine cell concentration. To monitor mouse survival, fifteen mice were infected intranasally with 1.25 × 106 yeast cells in approximately 25 μl PBS. At 4 hours post-infection, the lungs and spleens were harvested and homogenized from 5 mice infected with each strain. These homogenates were plated for H. capsulatum CFUs on Brain-Heart Infusion agar (BHI) plates and incubated at 30°C. The remaining mice were monitored daily for symptoms of disease (i.e. weight loss, lack of activity/response to stimulus, panting, lack of grooming). Mice were sacrificed after they exhibited 3 days of sustained weight loss greater than 15% of their maximum weight in conjunction with one other symptom of disease. For the in vivo colonization assay, mice were infected with 5 × 104 yeast cells. At 4 hrs, 2, 4, 8, 10, and 12 days post-infection, the lungs and spleens were harvested from 5 mice infected with each H. capsulatum strain. These organs were homogenized and plated for CFUs on BHI plates at 30°C. Statistical analysis for survival and colonization experiments was performed using GraphPad Prism 5.
All mouse experiments were performed in compliance with the NIH Guide for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committee at the University of California San Francisco.
Microarray hybridizations
In 6-well tissue culture treated dishes, 7 × 105 BMDMs were infected, in duplicate, with various H. capsulatum strains at an MOI of 5 or 10. UV Treated (UVT) yeasts were prepared by subjecting G217B to UV light (UV Stratalinker 1800, setting 1200 (120 J/m2) for 1 hour; these yeasts failed to generate CFUs when plated on HMM agarose. After one hour of co-culture, BMDMs were washed twice with BMM and 2 ml BMM was added to each well. BMDM total RNA was isolated at various time points post-infection using the RNeasy Mini Extraction Kit (Qiagen) and amplified using the Amino Allyl MessageAmp II aRNA Amplification Kit (Applied Biosystems). Microarray probes were generated by coupling aRNA to Cy5 or Cy3 monofunctional dyes (Amersham). Probes were hybridized to MEEBO (mouse exonic evidence-based oligonucleotide) microarrays (http://alizadehlab.stanford.edu/) using a pooled reference strategy in which Cy5-labeled aRNA samples corresponding to each time point were competitively hybridized against a Cy-3 labeled reference pool containing equal amounts of all infected and uninfected samples. Arrays were hybridized for 36 hours at 63°C and scanned on a GenePix 4000B scanner (Axon Instruments).
Microarray data analysis
Arrays were gridded using Spotreader (Niles Scientific) and results files were generated using GenePix Pro, version 6.0 (Molecular Devices). Poor quality features as identified by visual inspection were flagged and excluded from further analysis. Normalization was performed using Nomad 2.0 (http://derisilab.ucsf.edu/microarray/software.html). Ratio of medians (635/532) were extracted for each array feature, excluding features for which the sum of medians for the 635 nm and 532 nm channels was >= 500 intensity units. To extract relative gene expression ratios, these data were transformed relative to the mock-infected 2- or 3-hour time points in each experiment.
To identify genes that exhibit significant differential regulation between mock-infected and infected time courses, Significance Analysis of Microarrays (SAM) (Tusher et al., 2001) was performed on a comprehensive input data set that contained three mock-infected time courses, five time courses of infection with wild-type G217B yeasts, and two time courses of infection with cbp1 mutant yeasts. Visual inspection of the original data showed that expression of some genes is consistently restricted to early time points, and expression of others is restricted to later time points during infection. To ensure inclusion of these classes of genes, we performed three unpaired SAM analyses in which early (2 and 3 hours), intermediate (6 and 8 hours), and late (12-24 hours) infected time points were compared to mock infected samples; this analysis yielded 152 significant genes, corresponding to 173 different array oligos. To identify genes that exhibit differential regulation in macrophages infected with wild-type or cbp1 mutant yeasts, three unpaired SAM analyses were performed: 3 hr wild-type versus 3 hr cbp1, 6 hr wild-type versus 6 hr cbp1, and 12 hr wild-type versus 12 hr cbp1. Genes identified by SAM analysis that were induced or repressed by at least 2-fold relative to mock-infected samples were combined, filtered to remove duplicate data, and were hierarchically clustered and visualized in Java Treeview, version 1.1.0 (Saldanha, 2004).
The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession number GSE23378 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE23378).
Quantitative RT-PCR
BMDM infections were performed as described under “Cytotoxicity/LDH release assay”. Total RNA from infected BMDMs was generated as described above and treated with DNase using the RNase-free DNase set for on-column digestion (Qiagen). To generate cDNA, 1 μg input total RNA was reversed transcribed using oligo-dT primer and Affinityscript Multi-temperature Reverse Transcriptase (Stratagene) according to the manufacturer's protocol. Quantitative PCR was performed, in triplicate, on 1:4 dilutions of cDNA template as previously described (Hwang et al., 2008). Reactions were performed on the Mx3000P machine and analyzed using MxPro software (Stratagene). Abundance of Trb3 mRNA was normalized to hypoxanthine phophoribosyl transferase (HPRT) levels. Cycling parameters were as follows: 95°C for 10 min, followed by 40 cycles of 95°C (30s), 55°C (60s), and 72°C (30s), followed by dissociation curve analysis. Gene-specific primer sequences are as follows: TRB3 (mTrb3-f: 5’-TGCAGGAAGAAACCGTTGGAG; mTrb3-r: 5’-CTCAGGCTCATCTCTCACTCG) and HPRT (mHPRT1-f: 5’-AGGTTGCAAGCTTGCTGGT; mHPRT1-r: 5’-TGAAGTACTCATTATAGTCAAGGGCA).
To monitor Trb3 levels in the low MOI infection, the following protocol was used for RNA extraction: Samples were lysed with QIAzol (Qiagen). After addition of chloroform, total RNA was extracted from the aqueous phase using EconoSpin solumns (Epoch Life Science) and then subjected to on-column RNase-free DNase treatment (Qiagen). 1.5 μg of total RNA was used in cDNA synthesis using Superscript RT II (Life Technologies-Invitrogen) following manufacturer's instructions.
Caspase-3/7 activation assays
To monitor caspase-3 activation in individual macrophages, BMDMs were seeded in a 96 well glass bottom plate (In Vitro Scientific, P96-1.5H-N) at a density of 4×104 cells per well on the day prior to infection. WT and cbp1 H. capsulatum strains expressing mCherry fused to a signal sequence (SPChe) were diluted in HMM to reach exponential phase of growth at the time of infection. On the day of infection, H. capsulatum cells were resuspended in BMM, sonicated for 3 seconds on setting 2 using a Fisher Scientific Sonic Dismembrator Model 100, and counted by hemacytometer. BMDMs were infected at an MOI of 20. At 2 days post-infection, BMDMs were incubated with CellEvent Caspase-3/7 Green ReadyProbes reagent (Life Technologies, R37111) in BMM at 37°C and 5% CO2 for 2 hours. Cells were then imaged with a CSU-X1 spinning disk confocal on a Nikon Eclipse Ti inverted microscope with an Andora Clara digital camera using CFI APO 60x Oil TIRF objective. Images were collected using differential interference contrast (DIC), 488nm excitation, and 561nm excitation, and analyzed with NIS-Elements software 4.10 (Nikon).
To monitor caspase-3/7 activation in macrophage lysates, 1.5 × 107 BMDMs were infected with ura5Δ+URA5, the cbp1+URA5, and cbp1+CBP1 yeast strains at an MOI of 10 in 15 cm tissue culture-treated dishes (Corning). After three hours of co-culture, BMDMs were washed twice and 25 ml BMM was added. At 9 and 24 hours post-infection, media was removed, cells were scraped into 25 ml cold PBS and centrifuged for 10 minutes at 250×g. Pellets were resuspended in lysis buffer and total protein concentration of lysates were determined by BCA Assay (Pierce). Caspase-3 assays (R and D Systems) were performed, in duplicate, according to the manufacturers protocol on 100 μg total protein.
Mutant macrophage infections
Bax/Bak−/− infections: Baxfl/fl Bak−/− mice (Takeuchi et al., 2005) were a kind gift from Scott Oakes (UCSF). These mice were crossed to the LyzM Cre mouse (B6.129P2-Lyz2tm1(cre)Ifo/J) purchased from Jackson labs (Jackson Stock ID 004781) to generate LyzM Cre+/− Baxfl/+ Bak−/− mice. These mice were then crossed back to Baxfl/fl Bak−/− mice to generate LyzM Cre+/− Baxfl/fl Bak−/− mice. BMDMs from 6-8 week old female Baxfl/fl Bak−/− mice (referred to as parental control in Figure 9) or LyzM Cre+/− Baxfl/fl Bak−/− mice (referred to as Bak/Bax−/− in Figure 9) were differentiated as described above and the absence of Bax and Bak in cells derived from LyzM Cre+/− Baxfl/fl Bak−/− mice was confirmed by qPCR analysis of genomic DNA. Infections with H. capsulatum were performed at an MOI of 1 and LDH release assays were performed as described above. RIP3−/− infections: RIP3−/− mice (Newton et al., 2004) were a kind gift of Vishva Dixit (Genentech). BMDMs from 6-8 week old male WT C57Bl/6 (Charles River Laboratories) and RIP3−/− mice were differentiated as described above. Infections with H. capsulatum were performed at an MOI of 1 and LDH release assays were performed as described above.
caspase-1−/− caspase-11−/− infections: Isogenic wild-type and caspase-1−/− caspase-11−/− bone-marrow-derived macrophages (Monack et al., 2000) were a kind gift of Denise Monack (Stanford). Infections with H. capsulatum were performed at an MOI of 2 and LDH release assays were performed as described above.
Supplementary Material
Acknowledgements
We are grateful to Daniel Portnoy, Denise Monack, Jeffery Cox, Joseph DeRisi, Russell Vance, Charlie Kim, Sajeev Batra, Greg Barton, Allison Miller, Suzanne Noble, and members of the Noble and Sil laboratories for useful discussion as this work progressed. We greatly appreciate the gift of mice from Vishva Dixit and Scott Oakes and the gift of macrophages from Denise Monack. We thank Johnny Tse for technical assistance. We thank Sinem Beyhan, Sarah Gilmore, and Mark Voorhies for helpful comments on the manuscript. We appreciate the assistance of Jennifer Bolen of the UCSF Mouse Pathology Core facility for preparation of the tissue sections. We thank M. Paige Nittler, Katie Hermens, Sajeev Batra, and the Bay Area Innate Immunity PO1 group for the production of MEEBO arrays. This work was supported by UCSF Immunology Training Grant (T32 AI07334) support to CAB and BCE; Microbial Pathogenesis and Host Defense Training Grant (NIH T32 A1060537) support to CAB, DTI, and BCE; an NSF graduate research fellowship to AC; a UCSF NIGMS fellowship to DTI (1R25GM56847); an American Lung Association Senior Research Training Fellowship (RT-195877-N) to YNL; NIH (R01AI066224 and PO1AI063302) and an HHMI Early Career Scientist Award (http://www.hhmi.org/research/ecs/) to AS; the Sandler Program in Basic Sciences and the Howard Hughes Medical Institute Biomedical Research Support Program Grant (5300246) to the UCSF School of Medicine.
References
- Batanghari JW, Deepe GS, Jr., Di Cera E, Goldman WE. Histoplasma acquisition of calcium and expression of CBP1 during intracellular parasitism. Molecular microbiology. 1998;27:531–539. doi: 10.1046/j.1365-2958.1998.00697.x. [DOI] [PubMed] [Google Scholar]
- Batanghari JW, Goldman WE. Calcium dependence and binding in cultures of Histoplasma capsulatum. Infect Immun. 1997;65:5257–5261. doi: 10.1128/iai.65.12.5257-5261.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck MR, Dekoster GT, Cistola DP, Goldman WE. NMR structure of a fungal virulence factor reveals structural homology with mammalian saposin B. Molecular microbiology. 2009;72:344–353. doi: 10.1111/j.1365-2958.2009.06647.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan MA, Cookson BT. Salmonella induces macrophage death by caspase-1-dependent necrosis. Molecular microbiology. 2000;38:31–40. doi: 10.1046/j.1365-2958.2000.02103.x. [DOI] [PubMed] [Google Scholar]
- Bromati CR, Lellis-Santos C, Yamanaka TS, Nogueira TC, Leonelli M, Caperuto LC, Gorjao R, Leite AR, Anhe GF, Bordin S. UPR induces transient burst of apoptosis in islets of early lactating rats through reduced AKT phosphorylation via ATF4/CHOP stimulation of TRB3 expression. Am J Physiol Regul Integr Comp Physiol. 2011;300:R92–100. doi: 10.1152/ajpregu.00169.2010. [DOI] [PubMed] [Google Scholar]
- Brown GD, Herre J, Williams DL, Willment JA, Marshall AS, Gordon S. Dectin-1 mediates the biological effects of beta-glucans. The Journal of experimental medicine. 2003;197:1119–1124. doi: 10.1084/jem.20021890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullock WE. Interactions between human phagocytic cells and Histoplasma capsulatum. Arch Med Res. 1993;24:219–223. [PubMed] [Google Scholar]
- Carracedo A, Lorente M, Egia A, Blazquez C, Garcia S, Giroux V, Malicet C, Villuendas R, Gironella M, Gonzalez-Feria L, Piris MA, Iovanna JL, Guzman M, Velasco G. The stress-regulated protein p8 mediates cannabinoid-induced apoptosis of tumor cells. Cancer Cell. 2006;9:301–312. doi: 10.1016/j.ccr.2006.03.005. [DOI] [PubMed] [Google Scholar]
- Deepe GS, Jr., Buesing WR. Deciphering the pathways of death of Histoplasma capsulatum-infected macrophages: implications for the immunopathogenesis of early infection. J Immunol. 2012;188:334–344. doi: 10.4049/jimmunol.1102175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic acids research. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards JA, Alore EA, Rappleye CA. The yeast-phase virulence requirement for alpha-glucan synthase differs among Histoplasma capsulatum chemotypes. Eukaryot Cell. 2011a;10:87–97. doi: 10.1128/EC.00214-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards JA, Zemska O, Rappleye CA. Discovery of a role for Hsp82 in Histoplasma virulence through a quantitative screen for macrophage lethality. Infect Immun. 2011b;79:3348–3357. doi: 10.1128/IAI.05124-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissenberg LG, Goldman WE. Histoplasma variation and adaptive strategies for parasitism: new perspectives on histoplasmosis. Clin Microbiol Rev. 1991;4:411–421. doi: 10.1128/cmr.4.4.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Encinar JA, Mallo GV, Mizyrycki C, Giono L, Gonzalez-Ros JM, Rico M, Canepa E, Moreno S, Neira JL, Iovanna JL. Human p8 is a HMG-I/Y-like protein with DNA binding activity enhanced by phosphorylation. The Journal of biological chemistry. 2001;276:2742–2751. doi: 10.1074/jbc.M008594200. [DOI] [PubMed] [Google Scholar]
- Gantner BN, Simmons RM, Canavera SJ, Akira S, Underhill DM. Collaborative induction of inflammatory responses by dectin-1 and Toll-like receptor 2. The Journal of experimental medicine. 2003;197:1107–1117. doi: 10.1084/jem.20021787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gersuk GM, Underhill DM, Zhu L, Marr KA. Dectin-1 and TLRs permit macrophages to distinguish between different Aspergillus fumigatus cellular states. J Immunol. 2006;176:3717–3724. doi: 10.4049/jimmunol.176.6.3717. [DOI] [PubMed] [Google Scholar]
- Goodridge HS, Simmons RM, Underhill DM. Dectin-1 stimulation by Candida albicans yeast or zymosan triggers NFAT activation in macrophages and dendritic cells. J Immunol. 2007;178:3107–3115. doi: 10.4049/jimmunol.178.5.3107. [DOI] [PubMed] [Google Scholar]
- Goruppi S, Iovanna JL. Stress-inducible protein p8 is involved in several physiological and pathological processes. The Journal of biological chemistry. 2010;285:1577–1581. doi: 10.1074/jbc.R109.080887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang LH, Mayfield JA, Rine J, Sil A. Histoplasma Requires SID1, a Member of an Iron-Regulated Siderophore Gene Cluster, for Host Colonization. PLoS Pathogens. 2008;4:e1000044. doi: 10.1371/journal.ppat.1000044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaac DT, Coady A, Van Prooyen N, Sil A. The 3-hydroxy-methylglutaryl coenzyme A lyase HCL1 is required for macrophage colonization by human fungal pathogen Histoplasma capsulatum. Infect Immun. 2013;81:411–420. doi: 10.1128/IAI.00833-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston SA, May RC. Cryptococcus interactions with macrophages: evasion and manipulation of the phagosome by a fungal pathogen. Cell Microbiol. 2013;15:403–411. doi: 10.1111/cmi.12067. [DOI] [PubMed] [Google Scholar]
- Kemski MM, Stevens B, Rappleye CA. Spectrum of T-DNA integrations for insertional mutagenesis of Histoplasma capsulatum. Fungal biology. 2013;117:41–51. doi: 10.1016/j.funbio.2012.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugler S, Young B, Miller VL, Goldman WE. Monitoring phase-specific gene expression in Histoplasma capsulatum with telomeric GFP fusion plasmids. Cell Microbiol. 2000;2:537–547. doi: 10.1046/j.1462-5822.2000.00078.x. [DOI] [PubMed] [Google Scholar]
- Labbe K, Saleh M. Cell death in the host response to infection. Cell Death Differ. 2008;15:1339–1349. doi: 10.1038/cdd.2008.91. [DOI] [PubMed] [Google Scholar]
- Luther K, Torosantucci A, Brakhage AA, Heesemann J, Ebel F. Phagocytosis of Aspergillus fumigatus conidia by murine macrophages involves recognition by the dectin-1 beta-glucan receptor and Toll-like receptor 2. Cell Microbiol. 2007;9:368–381. doi: 10.1111/j.1462-5822.2006.00796.x. [DOI] [PubMed] [Google Scholar]
- Mallo GV, Fiedler F, Calvo EL, Ortiz EM, Vasseur S, Keim V, Morisset J, Iovanna JL. Cloning and expression of the rat p8 cDNA, a new gene activated in pancreas during the acute phase of pancreatitis, pancreatic development, and regeneration, and which promotes cellular growth. The Journal of biological chemistry. 1997;272:32360–32369. doi: 10.1074/jbc.272.51.32360. [DOI] [PubMed] [Google Scholar]
- Martinon F, Chen X, Lee AH, Glimcher LH. TLR activation of the transcription factor XBP1 regulates innate immune responses in macrophages. Nat Immunol. 2010;11:411–418. doi: 10.1038/ni.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayfield JA, Rine J. The genetic basis of variation in susceptibility to infection with Histoplasma capsulatum in the mouse. Genes and immunity. 2007;8:468–474. doi: 10.1038/sj.gene.6364411. [DOI] [PubMed] [Google Scholar]
- Medoff G, Maresca B, Lambowitz AM, Kobayashi G, Painter A, Sacco M, Carratu L. Correlation between pathogenicity and temperature sensitivity in different strains of Histoplasma capsulatum. J Clin Invest. 1986;78:1638–1647. doi: 10.1172/JCI112757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merksamer PI, Papa FR. The UPR and cell fate at a glance. J Cell Sci. 2010;123:1003–1006. doi: 10.1242/jcs.035832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monack DM, Hersh D, Ghori N, Bouley D, Zychlinsky A, Falkow S. Salmonella exploits caspase-1 to colonize Peyer's patches in a murine typhoid model. The Journal of experimental medicine. 2000;192:249–258. doi: 10.1084/jem.192.2.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mungrue IN, Pagnon J, Kohannim O, Gargalovic PS, Lusis AJ. CHAC1/MGC4504 is a novel proapoptotic component of the unfolded protein response, downstream of the ATF4-ATF3-CHOP cascade. J Immunol. 2009;182:466–476. doi: 10.4049/jimmunol.182.1.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton K, Sun X, Dixit VM. Kinase RIP3 is dispensable for normal NF-kappa Bs, signaling by the B-cell and T-cell receptors, tumor necrosis factor receptor 1, and Toll-like receptors 2 and 4. Molecular and cellular biology. 2004;24:1464–1469. doi: 10.1128/MCB.24.4.1464-1469.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen VQ, Sil A. Temperature-induced switch to the pathogenic yeast form of Histoplasma capsulatum requires Ryp1, a conserved transcriptional regulator. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:4880–4885. doi: 10.1073/pnas.0710448105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohoka N, Yoshii S, Hattori T, Onozaki K, Hayashi H. TRB3, a novel ER stress-inducible gene, is induced via ATF4-CHOP pathway and is involved in cell death. Embo J. 2005;24:1243–1255. doi: 10.1038/sj.emboj.7600596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ord D, Ord T. Characterization of human NIPK (TRB3, SKIP3) gene activation in stressful conditions. Biochem Biophys Res Commun. 2005;330:210–218. doi: 10.1016/j.bbrc.2005.02.149. [DOI] [PubMed] [Google Scholar]
- Ord T, Ord D, Kuuse S, Plaas M. Trib3 is regulated by IL-3 and affects bone marrow-derived mast cell survival and function. Cell Immunol. 2012;280:68–75. doi: 10.1016/j.cellimm.2012.11.011. [DOI] [PubMed] [Google Scholar]
- Patel JB, Batanghari JW, Goldman WE. Probing the yeast phase-specific expression of the CBP1 gene in Histoplasma capsulatum. J Bacteriol. 1998;180:1786–1792. doi: 10.1128/jb.180.7.1786-1792.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira PA, Trindade BC, Secatto A, Nicolete R, Peres-Buzalaf C, Ramos SG, Sadikot R, Bitencourt Cda S, Faccioli LH. Celecoxib improves host defense through prostaglandin inhibition during Histoplasma capsulatum infection. Mediators Inflamm. 2013;2013:950981. doi: 10.1155/2013/950981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porta A, Maresca B. Host response and Histoplasma capsulatum/macrophage molecular interactions. Med Mycol. 2000;38:399–406. doi: 10.1080/mmy.38.6.399.406. [DOI] [PubMed] [Google Scholar]
- Qing G, Li B, Vu A, Skuli N, Walton ZE, Liu X, Mayes PA, Wise DR, Thompson CB, Maris JM, Hogarty MD, Simon MC. ATF4 regulates MYC-mediated neuroblastoma cell death upon glutamine deprivation. Cancer Cell. 2012;22:631–644. doi: 10.1016/j.ccr.2012.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappleye CA, Eissenberg LG, Goldman WE. Histoplasma capsulatum alpha-(1,3)-glucan blocks innate immune recognition by the beta-glucan receptor. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:1366–1370. doi: 10.1073/pnas.0609848104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar M, Carracedo A, Salanueva IJ, Hernandez-Tiedra S, Lorente M, Egia A, Vazquez P, Blazquez C, Torres S, Garcia S, Nowak J, Fimia GM, Piacentini M, Cecconi F, Pandolfi PP, Gonzalez-Feria L, Iovanna JL, Guzman M, Boya P, Velasco G. Cannabinoid action induces autophagy-mediated cell death through stimulation of ER stress in human glioma cells. J Clin Invest. 2009;119:1359–1372. doi: 10.1172/JCI37948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldanha AJ. Java Treeview--extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- Salvesen GS, Riedl SJ. Caspase mechanisms. Adv Exp Med Biol. 2008;615:13–23. doi: 10.1007/978-1-4020-6554-5_2. [DOI] [PubMed] [Google Scholar]
- Sebghati TS, Engle JT, Goldman WE. Intracellular parasitism by Histoplasma capsulatum: fungal virulence and calcium dependence. Science. 2000;290:1368–1372. doi: 10.1126/science.290.5495.1368. [DOI] [PubMed] [Google Scholar]
- Sepulveda VE, Williams CL, Goldman WE. Comparison of phylogenetically distinct Histoplasma strains reveals evolutionarily divergent virulence strategies. mBio. 2014;5:e01376–01314. doi: 10.1128/mBio.01376-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang YY, Wang ZH, Zhang LP, Zhong M, Zhang Y, Deng JT, Zhang W. TRB3, upregulated by ox-LDL, mediates human monocyte-derived macrophage apoptosis. FEBS J. 2009;276:2752–2761. doi: 10.1111/j.1742-4658.2009.06998.x. [DOI] [PubMed] [Google Scholar]
- Shimizu K, Takahama S, Endo Y, Sawasaki T. Stress-inducible caspase substrate TRB3 promotes nuclear translocation of procaspase-3. PLoS One. 2012;7:e42721. doi: 10.1371/journal.pone.0042721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin DM, Yang CS, Yuk JM, Lee JY, Kim KH, Shin SJ, Takahara K, Lee SJ, Jo EK. Mycobacterium abscessus activates the macrophage innate immune response via a physical and functional interaction between TLR2 and dectin-1. Cell Microbiol. 2008;10:1608–1621. doi: 10.1111/j.1462-5822.2008.01151.x. [DOI] [PubMed] [Google Scholar]
- Shin S, Brodsky IE. The inflammasome: Learning from bacterial evasion strategies. Seminars in immunology. 2015;27:102–110. doi: 10.1016/j.smim.2015.03.006. [DOI] [PubMed] [Google Scholar]
- Skeldon A, Saleh M. The inflammasomes: molecular effectors of host resistance against bacterial, viral, parasitic, and fungal infections. Front Microbiol. 2011;2:15. doi: 10.3389/fmicb.2011.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridharan H, Upton JW. Programmed necrosis in microbial pathogenesis. Trends in microbiology. 2014;22:199–207. doi: 10.1016/j.tim.2014.01.005. [DOI] [PubMed] [Google Scholar]
- Su N, Kilberg MS. C/EBP homology protein (CHOP) interacts with activating transcription factor 4 (ATF4) and negatively regulates the stress-dependent induction of the asparagine synthetase gene. The Journal of biological chemistry. 2008;283:35106–35117. doi: 10.1074/jbc.M806874200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi O, Fisher J, Suh H, Harada H, Malynn BA, Korsmeyer SJ. Essential role of BAX,BAK in B cell homeostasis and prevention of autoimmune disease. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:11272–11277. doi: 10.1073/pnas.0504783102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tewari RP, Berkhout FJ. Comparative pathogenicity of albino and brown types of Histoplasma capsulatum for mice. J Infect Dis. 1972;125:504–508. doi: 10.1093/infdis/125.5.504. [DOI] [PubMed] [Google Scholar]
- Todd DJ, Lee AH, Glimcher LH. The endoplasmic reticulum stress response in immunity and autoimmunity. Nat Rev Immunol. 2008;8:663–674. doi: 10.1038/nri2359. [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, LeBert V, Hung CY, Galles K, Saijo S, Lin X, Cole GT, Klein BS, Wuthrich M. C-type lectin receptors differentially induce th17 cells and vaccine immunity to the endemic mycosis of North America. J Immunol. 2014;192:1107–1119. doi: 10.4049/jimmunol.1302314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster RH, Sil A. Conserved factors Ryp2 and Ryp3 control cell morphology and infectious spore formation in the fungal pathogen Histoplasma capsulatum. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:14573–14578. doi: 10.1073/pnas.0806221105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods JP. Knocking on the right door and making a comfortable home: Histoplasma capsulatum intracellular pathogenesis. Curr Opin Microbiol. 2003;6:327–331. doi: 10.1016/s1369-5274(03)00080-8. [DOI] [PubMed] [Google Scholar]
- Woods JP, Heinecke EL, Goldman WE. Electrotransformation and expression of bacterial genes encoding hygromycin phosphotransferase and beta-galactosidase in the pathogenic fungus Histoplasma capsulatum. Infect Immun. 1998;66:1697–1707. doi: 10.1128/iai.66.4.1697-1707.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worsham PL, Goldman WE. Quantitative plating of Histoplasma capsulatum without addition of conditioned medium or siderophores. J Med Vet Mycol. 1988;26:137–143. [PubMed] [Google Scholar]
- Yadav M, Schorey JS. The beta-glucan receptor dectin-1 functions together with TLR2 to mediate macrophage activation by mycobacteria. Blood. 2006;108:3168–3175. doi: 10.1182/blood-2006-05-024406. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.