SECTION I INTRODUCTION
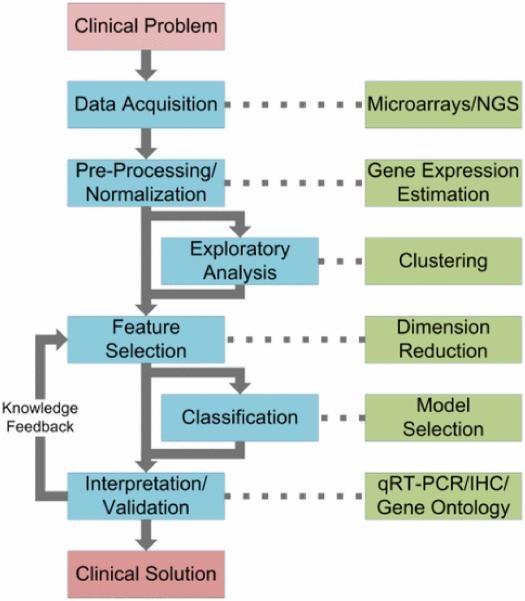
GENOMIC biomarker identification is essential for understanding human disease and for developing diagnostic and prognostic clinical tools. In this review, we focus on cardiovascular genomics to present a bioinformatics pipeline that is driven by clinical problems and produces potential clinical solutions using six steps (see Fig. 1): 1) data acquisition, 2) preprocessing and normalization, 3) exploratory analysis, 4) feature selection, 5) classification, and 6) interpretation and validation. The application of this pipeline to cardiovascular disease (CVD) is important because, despite achievements in cardiovascular genomics, clinically applicable tools for CVD risk prediction, diagnosis, and therapy are still lagging [1]. The goal of this review is to describe an analysis pipeline that researchers may use to more effectively identify CVD biomarkers and propose predictive models. We describe current bioinformatics tools and methods for each step of the pipeline, identify challenges involved in implementing the pipeline, and summarize opportunities remaining for further research. Finally, we illustrate some of the methods described in the pipeline by analyzing publicly available CVD gene expression data.
Fig. 1.
Genomic data analysis pipeline. Beginning with a clinical problem, the aim of the pipeline is to find a clinical solution. This is achieved by identifying biomarkers that can uncover the underlying biological mechanisms of disease or that can predict disease presence or treatment outcome. After data acquisition, analytical steps in the pipeline include 1) preprocessing and normalization, 2) exploratory analysis, 3) feature selection, 4) classification, and 5) interpretation and validation. Exploratory analysis is optional, but can be an informative step prior to feature selection. Classification optionally follows feature selection if the target clinical solution is of a diagnostic or prognostic nature.
In Section II, we review the biology of gene expression and describe how we can use microarray and next-generation sequencing (NGS) technology to measure gene expression. Furthermore, we review data preprocessing and normalization algorithms to address technical and biological variability in the data. In Section III, we review data mining methods for exploratory analysis, feature selection, and classification. Exploratory analysis methods can discover relationships within the data, e.g., clustering subgroups within a set of patient expression profiles. Feature selection methods can reduce data size and identify biomarkers, resulting in improved diagnostic classifier performance. Classifiers can categorize the samples based on features selected. We review methods for building disease classification models and discuss commonly encountered pitfalls. In Section IV, we review computational and experimental techniques for interpretation and validation. While robust algorithms for biomarker identification and classification may produce mathematically valid results, these results must be biologically validated prior to clinical application. The results of validation also provide performance measures and feedback to improve data mining methods, particularly in the feature selection step. Various tools exist for validating analytical results using the literature, annotated biological databases, or clinical experiments. In Section V, we present a case study that analyzes several CVD microarray datasets consisting of samples from diseased patients and healthy control patients. The case study illustrates all steps in the bioinformatics pipeline, with the goal of identifying differentially expressed biomarkers for predicting CVD presence.
SECTION II EXPERIMENTAL METHODS
The functional state of cells may be estimated by quantifying gene expression using genomic assay methods. The first step in gene expression is DNA transcription to produce mRNA. Then, functional proteins are produced from mRNA by translation [2]. Because of posttranscriptional modifications, mRNA levels are not directly correlated with protein levels. Although with better understanding of posttranscriptional agents such as microRNA (i.e., miRNA or noncoding mRNA that affect mRNA levels), the complex role of transcriptional regulation may be better understood, in particular, for diseases such as CVD [3]. As such, numerous reviews have been published, for instance, that focus on the role of miRNA in CVD [4]. Recent studies have also suggested that genomic (as well as proteomic) biomarkers, whether in the form of mRNA or miRNA, may improve the accuracy of cardiovascular risk identification on an individual basis using panels of tests or multimarker (i.e., multigene) assays [5]. Other studies have focused on detecting gene expression changes in tissues involved in various stages of atherosclerotic plaque formation [6], [7], [8].
Emerging technologies such as genome-wide association (i.e., detection of single nucleotide polymorphisms, SNPs), deep sequencing, and miRNA arrays [3], [9], [10] have improved our understanding of the genetic basis of CVD [11]. In this section, we focus on genomic data acquisition and data preprocessing in microarrays and NGS, which may be used for quantification of SNPs, mRNA, or miRNA. Although specific analytical and experimental methods exist for SNPs, mRNA, and miRNA, among others, we limit the scope of this paper to focus on general methods that may be applied to a wide variety of genomic assays. We also describe challenges encountered in microarray technology and potential solutions to these challenges made possible by NGS technology. Furthermore, we discuss current challenges with NGS technology and its potential for replacing or complementing microarray technology.
A. Quantifying Gene Expression With Microarrays
The concept that underlies microarray technology is the specific hybridization, or binding, of labeled nucleotide sequences—i.e., isolated and fluorescently tagged mRNA molecules—to predefined arrays of complementary sequences. The fluorescence intensity of each location on the array indicates the amount of mRNA, or gene expression, from the original biological sample. Several variants of microarray technologies exist, with the most widely used being cDNA and oligonucleotide microarrays [12].
cDNA microarrays, developed by Schena et al., are based on long sequences of cDNA (complementary DNA) that are immobilized onto a substrate (a glass slide or a nylon membrane) in a spotted matrix [13]. Each spot on the array corresponds to a specific gene or transcript selected from a library of gene sequences. Oligonucleotide microarrays consist of short nucleotide sequences instead of the longer sequences of cDNA microarrays. A photolithographic technique developed by Affymetrix enables production of high-density microarrays by building nucleotide sequences directly onto a substrate such as a silicon wafer [14]. This technique limits the length of sequences to about 20–25 nucleotides and reduces the specificity of hybridization. Careful selection of multiple unique probe sequences for each mRNA can improve specificity [12]. After hybridization, optical scanners quantify the fluorescence intensity of each spot on the array to produce raw gene expression signals that require further processing before analysis.
Microarray data preprocessing involves normalization, or summarization, of quantified fluorescent signals to estimate gene expression and to detect or correct data quality issues. Several preprocessing algorithms exist for the popular Affymetrix oligonucleotide arrays such as distribution free weighted [15], factor analysis for robust microarray summarization [16], genechip robust multiarray average [17], Affymetrix MicroArray Suite 5.0 [18], model based expression index [19], probe logarithmic intensity error estimation [20], robust multiarray average (RMA) [21], variance-stabilizing normalization [22], and caCORRECT [23]. Similar normalization methods exist for Illumina arrays [24], [25], [26], [27]. Comparisons of these methods often find that there is some variability in performance, and the choice of a “best” normalization algorithm can vary from project to project [28]. Some studies have found significant differences among normalization methods that can greatly affect downstream analysis [29]. However, a more recent study comparing microarray normalization methods to TaqMan RT-PCR arrays concludes that the differences among multiple methods are not statistically significant [30]. Finally, data quality plays an important role in the adoption of microarrays for clinical applications. Thus, normalization methods often include functions for assessing data quality. For instance, both RMA and caCORRECT can identify and reduce the effect of spatial artifacts in microarray data [21], [23], [31]. caCORRECT also computes a quality score for the purpose of discarding arrays with significant, irreversible technical artifacts. Other metrics such as correlation or quantity of outlier spots may be used for outlier array detection [32]. Similar methods exist for detecting biological microarray outliers using principal component analysis (PCA) [33]. Despite the abundance of robust methods for processing microarray data, concerns with data quality still hinder adoption of the technology for clinical applications.
A recent study by the FDA-led microarray quality control (MAQC) consortium revealed conflicting reports of reproducibility in inter-platform and cross-laboratory microarray studies [34]. For example, Tan et al. reported considerable divergence across different platforms [35], whereas Petersen et al. reported high reproducibility across platforms [36]. In light of these conflicts, the MAQC consortium has conducted a multiplatform and multitest site experimental design to show that microarrays are reproducible [34]. Moreover, the MAQC study consists of a number of more focused studies. For example, Canales et al. have compared five microarray platforms to three quantitative and low-throughput gene expression technologies such as TaqMan, standardized RT-PCR, and QuantiGene assays [37]. They found that the quantitative results of all platforms generally agreed with the exception of a few genes due to weak expression or due to differences in probe sequence. Furthermore, Patterson et al. have compared one-color and two-color microarray platforms in terms of reproducibility, sensitivity, specificity, and accuracy, and found that these platforms are identical in terms of data quality [38].
In addition to issues with reproducibility addressed by the MAQC consortium, other limitations of microarrays including 1) dynamic range (i.e., genes expressed at very low or very high levels are not correctly quantified because of background hybridization or signal saturation), 2) probe design (i.e., genes can only be detected if the correct probes are used, limiting the discovery of new splice variants), and 3) a high level of technical noise that, when combined with small-sample size, can lead to false discoveries [39]. Initial research suggests that emerging NGS technology is capable of addressing these issues and potentially replacing microarrays for some applications.
B. Quantifying Gene Expression With NGS
NGS technology provides potential solutions to current microarray limitations. We briefly describe 1) a general overview of NGS experimental protocols, 2) the benefits of NGS compared to microarray technology in the context of gene expression measurement, and 3) current challenges in NGS bioinformatics.
NGS refers to high-throughput sequencing of short nucleotide chains on a massively parallel scale. Some commercially available NGS platforms include 454 pyrosequencing [40], Applied Biosystems SOLiD sequencing [41], Illumina Genome Analyzer, and Helicos/Heliscope [42], [43]. These platforms differ primarily in experimental protocol, but the basic steps remain the same: 1) after library preparation, DNA fragments are isolated and attached to a matrix and 2) the fragments are amplified and sequenced in parallel. The iterative parallel sequencing steps produce a large dataset of fluorescence images that are converted into sequence reads by base calling algorithms [44], [45]. Applications of NGS include, but are not limited to, complete genome sequencing [46], quantification of gene expression [47], and metagenomic sequencing [48]. In this paper, we focus on current advances in gene expression quantification, or RNA-Seq, using NGS technology.
In the context of RNA-Seq, NGS technology directly addresses dynamic range and probe design limitations of microarray technology. RNA-Seq measures gene expression by counting the number of short sequences that align to or “hit” a region of a reference genome [47]. In contrast to microarrays, RNA-Seq relies on computational matching of RNA sequences to known gene sequences rather than chemical hybridization of fluorescently labeled RNA fragments to complementary probes. This approach gives NGS a much greater dynamic range compared to microarrays because as little as a single RNA molecule to as many as all RNA molecules can be quantified from a sample (provided that there is sufficient sequencing depth). For example, NGS has been shown to be highly correlated with quantitative real-time polymerase chain reaction (qRT-PCR) over several orders of magnitude, a dynamic range much larger than typical microarrays [49], [50]. Moreover, RNA-Seq is not limited by probe design. That is, all RNA molecules expressed by a cell can be detected and quantified without the requirement of designing a specific probe sequence. Thus, aligning RNA-Seq samples to a whole genome reference can not only quantify known genes, but can also identify new genes and splice variants [51]. Finally, it is widely believed that microarrays suffer from a high level of technical noise that can result in high false discovery rates [39]. Although there have been few studies directly comparing microarray and NGS false discovery rates, initial studies suggest that, for applications such as miRNA detection, microarray and NGS platforms perform similarly [52], [53]. Thus, NGS technology appears to address some, but not all, shortcomings of microarray technology.
Challenges remain in NGS bioinformatics research because of the continuing evolution of the technology and the availability of many different platforms, making it difficult for research in algorithm development. The accuracy of RNA-Seq alignment to reference genomes depends on 1) sequence quality or sequencing error profiles (i.e., substitutions, deletions, and insertions of nucleotide bases during the sequencing phase), 2) the sequence alignment algorithm, and 3) expression normalization after alignment. Each NGS platform has a unique error profile [54]. Thus, many alignment algorithms have been developed to exploit the advantages of each NGS platform (see [55] for a review of sequence alignment algorithms). After sequence alignment, raw “hits” must be converted, summarized, and normalized into interpretable gene expression values using algorithms similar to those of microarray normalization algorithms. However, there is no consensus in the NGS community for selecting these algorithms for applications such as differential gene detection [56]. Thus, several studies have proposed algorithms for normalization, taking into consideration factors such as gene length [57] or assumptions that most genes are not differentially expressed [58].
The development of NGS technology for genomic bioinformatics addresses some of the issues associated with microarray technology. Because of advantages such as improved dynamic range and correlation with qRT-PCR, NGS technology appears poised to replace microarrays. However, for some applications, microarrays may dominate for the foreseeable future—e.g., for assays of small genomes or low-cost screening of large numbers of samples. NGS and microarrays may be considered complimentary, for example, in cases where a deeper analysis of the genome for a subset of samples is desired after screening with microarrays [49]. As with microarray technology in the past, there are analytical and experimental challenges in quantifying NGS data, e.g., handling the large volume of data and understanding the confounding factors of alignment algorithm, normalization algorithm, and NGS platform. Thus, many research opportunities remain in genomic bioinformatics.
SECTION III STATISTICAL ANALYSIS AND DATA MINING
Following acquisition and preprocessing are statistical analysis and data mining for genomic data analysis (see Fig. 1). This includes 1) exploratory analysis, i.e., discovering biologically interesting relationships in the data, 2) feature selection, i.e., mining for biomarkers, and 3) classification, i.e., building statistical models for disease diagnosis and prognosis. In the following sections, we discuss commonly used methods in each step and identify challenging and open research questions, in particular, for cardiovascular genomic bioinformatics.
A. Exploratory Analysis
Exploratory analysis methods can reveal the natural organization of genomic data and identify patterns [59]. Unsupervised clustering methods are one such group of exploratory analysis methods that can identify groups of genes with similar expression patterns, or groups of samples with similar molecular profiles [60]. For example, clustering has revealed related gene groups in several cardiovascular-related studies, including coronary artery disease (CAD) and atherosclerosis [61], [62], [63]. The most common unsupervised clustering method applied to gene expression data is hierarchical clustering [64]. Other methods include Formula-means clustering [65], PCA, [66], biclustering [67], and self-organizing maps (SOMs) [68]. In this section, we briefly describe the underlying concepts for each of these methods.
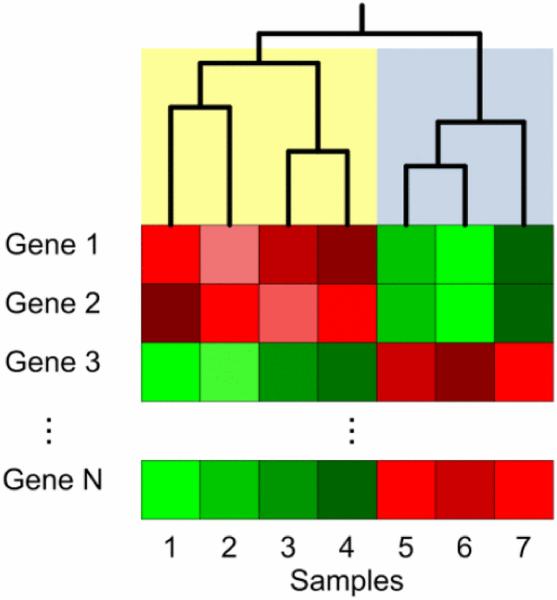
Hierarchical and Formula-means clustering algorithms group samples or features based on similarity metrics such as Euclidean distance, Manhattan distance, Mahalanobis distance, and various correlations [69]. Hierarchical clustering is an agglomerative technique; i.e., initially, there are several single member clusters that are iteratively combined based on distance until only a single cluster remains [70]. Hierarchical clustering is very useful for microarray or NGS analysis because its results can be visualized using dendrograms, or tree-like structures that capture the iterative formation of these data clusters. These visualizations often include heatmaps that represent gene expression, with genes and samples arranged based on hierarchical clustering. Simultaneous clustering of genes and samples is referred to as biclustering, which enables not only identification of biological samples with similar genetic profiles, but also identification of correlated genes [67]. SOMs are artificial neural networks that are well suited for clustering high-dimensional gene expression data. Generally, they are able to represent high-dimensional data in a low-dimensional space, while spatially preserving the similarity of data points [68]. Fig. 2 illustrates both the dendrogram and heatmap resulting from hierarchical clustering. Unlike hierarchical clustering, Formula-means clustering is initialized with a predefined number of clusters. Each sample is then randomly assigned to one of Formula clusters. The algorithm then iteratively reassigns samples to clusters based on distance. After reassigning a sample, the algorithm recomputes the centroids of the altered clusters.
Fig. 2.
Illustration of hierarchical clustering of seven samples, each with Formula genes. Gene expression is usually represented by a heatmap. Two large clusters emerge in this example. One cluster, shaded in yellow, contains samples 1, 2, 3, and 4. The second cluster, shaded in blue, contains samples 5, 6, and 7. Each cluster, in turn, contains smaller clusters of 1 or 2 samples.
Although clustering methods are applicable to large dimensional datasets, reducing data dimensionality may produce more meaningful and less noisy clustering results. For example, a large number of genes in a high-throughput genomic dataset may be irrelevant to the biological problem of interest (e.g., CVD). In such cases, data dimensionality may be reduced by removing such genes from the dataset without losing any important features. In other cases, it may also be desirable (or simpler) to represent a large number of relevant genes in a reduced dimensional space. PCA is a common method for reducing the dimensionality of gene expression data without discarding important information [71]. However, a drawback of PCA is that the results of feature selection or clustering after applying PCA are not easy to interpret because principal components are not a one to one representation of genes. Instead, each principal component is a linear combination of the original data dimensions (i.e., genes). Despite this, PCA remains useful for gene expression analysis [72], [73], [74].
Although clustering and dimensionality reduction methods are useful tools for revealing the underlying structure of unlabeled (or unclassified) data, they are not useful for biomarker identification. Supervised learning methods, i.e., methods that work on labeled data, can more precisely identify specific factors that are capable of differentiating samples in distinct disease groups.
B. Supervised Feature Selection
Many genomic experiments generate microarray or NGS data using samples with known underlying grouping or clustering information. This information can be used to supervise the identification of genes that are differentially expressed. These genes are called biomarkers and are valuable because of their potential in diagnostic screening for diseases such as atherosclerosis [75], [76]. There are numerous approaches in supervised feature selection, and each selects genes based on properties of the data that capture the degree of differential expression. Feature selection methods can be categorized into three groups: filter, wrapper, and embedded methods [77]. Here, we describe basic properties of these feature selection methods and discuss factors that must be taken into consideration when selecting a method. An important factor to consider in feature selection is sample size. We discuss methods for determining minimum sample size to accurately identify differentially expressed genes, and then describe methods that attempt to reduce uncertainty by integrating domain knowledge.
Filter methods identify differentially expressed genes by examining the intrinsic properties of the data [77]. Usually, features are scored using a metric for determining degree of differential expression, and then low-scoring features are removed. In classification applications, filter methods are applied to the data independent of the classifier. Thus, filter methods are fast and scalable to large datasets, especially for univariate methods such as fold-change, significance analysis of microarrays (SAM) [78], and rank products [79]. There is a comprehensive review of univariate filter methods (also known as gene ranking methods) [80]. Univariate filter methods assume independence among features and can possibly lead to lower classification or clustering performance. On the other hand, multivariate filter methods such as the minimum redundancy, maximum relevance (mRMR) method can address this issue by identifying groups of genes that may have low predictive value when acting independently, but are good predictors when acting in concert [81]. However, multivariate methods are hard to optimize in a very large search space (e.g., all combinations of gene sets for tens of thousands of genes). Wrapper methods have been proposed to address this issue. Wrapper methods evaluate feature sets by training and testing a classifier model [77]. That is, the feature search is wrapped around a classifier so that the interaction of features and classifiers is considered during optimization. Wrapper methods can search the feature space in a deterministic (e.g., sequential forward selection [82]), or randomized (e.g., genetic algorithms [83]) manner. One disadvantage of wrapper methods is that they can be very computationally costly. As an improvement, embedded methods are very similar to wrapper methods, but are computationally cost effective because the feature search is built into the classifier construction phase [77]. An example of an embedded method is the support vector machine (SVM) recursive feature elimination method [84].
High-throughput genomic experiments pose unique problems in data analysis. Because of the small-sample size compared to the large feature size, feature selection often suffers from lack of statistical power. Thus, methods have been developed to estimate minimum sample size for achieving a desired sensitivity in detecting differentially expressed genes [85]. Although small-sample size gene expression studies are considered to be a serious problem, it is still not realized by all practitioners [86]. Knowledge-based methods have emerged as an alternative approach for handling small-sample data.
Feature selection algorithms can produce widely varying results and it is not clear which algorithms produce the most biologically relevant results. Algorithms that rely on both data and knowledge tend to perform better than those that rely on data alone. Purely data-driven feature selection algorithms require a larger number of patient samples in order to adequately represent the problem space. There are several examples of knowledge integration in the literature that specifically target feature selection [87], [88], [89]. Aerts et al. combined data from several resources to prioritize genes relevant to diseases of interest [90]. Their data sources included gene ontology (GO) databases, published literature, microarray repositories, and sequence information. They extracted “training” genes—genes tagged as differentially expressed in, or related to, the biological problem—from these databases and ranked test genes according to how similar they were in comparison to training genes. Kuffner et al. identified groups of genes that simultaneously correlate to genes mentioned in the relevant literature and to differential components of expression profiles [91]. Kong et al. searched for combinations of genes that are differentially regulated based on multivariate Hotelling's T2 statistic and that also correlate with GO and other pathway databases [92]. Mukherjee and Roberts developed a theoretical framework to compare feature ranking metrics in the presence of control features [93]. Chen et al. modified an independent component analysis (ICA) method for detecting biomarkers using inferred biological knowledge [88]. They showed that their knowledge-guided method improved the efficiency of detecting biomarkers compared to traditional ICA methods. Both of these studies—Mukherjee and Chen—are similar to the study by Phan et al., which used biological knowledge in the form of validated biomarkers to identify the best feature ranking method within a population of methods. The authors showed that a knowledge-guided iterative approach to feature selection improved the efficiency of identifying relevant biomarkers [87]. Alternative approaches to biomarker identification use knowledge in the form of protein or gene interaction networks. These methods give preference to candidate biomarkers that are highly connected to, or are statistically associated with, known interaction networks [94], [95].
Although both exploratory and supervised feature selection methods in bioinformatics have been studied in detail, challenges remain, especially in the context of CVD genomics. Primarily, genetic factors tend to be weakly associated with CVD risk. This supports a CVD “polygenic theory” that multiple genetic variants contribute to overall disease risk [96]. Thus, there is a need for biomarker algorithms that can identify multiple genetic factors acting in concert to predict CVD risk.
C. Classification
Exploratory analysis and feature selection ultimately lead to predictive analysis, an important aspect of diagnostic and prognostic bioinformatics. An increasing number of bioinformatics studies have focused on genetic screening to determine susceptibility to cardiovascular-related diseases [97], [98], [99], [100], [101], [102], [103], [104]. Some of these studies stress the importance of multifactorial tests (i.e., tests that consider multiple genotypes as well as environmental factors) because of the complex nature of the disease [97], [104]. Regardless of the biological feasibility of predicting disease state based on genetics alone, statistical models must be evaluated in a systematic manner [105], [106], [107], [108], [109].
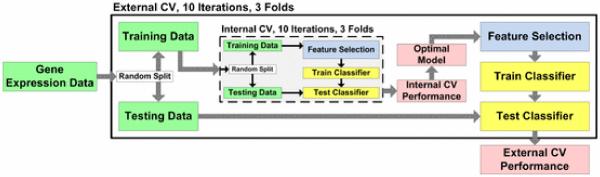
Simon and Quackenbush et al. examined key steps and common pitfalls involved in building clinically predictive models [105], [108]. The most common pitfall is incorrect estimation of the accuracy of prediction models when classifying future samples. The correct method involves proper division of samples into training and testing sets such that information about test samples does not “leak” into the prediction models, resulting in biased performance estimates. Fig. 3 illustrates the correct method for estimating prediction performance using cross validation. Internal cross validation is used for identifying the best protocol for deriving a predictive model from the data. The protocol includes a feature selection method and classifier parameters. External cross validation is used to estimate the performance of the protocol on independent data. A final prediction model can then be derived by applying the protocol to the entire dataset to obtain a specific classifier and features. The final model should be evaluated using an independent testing dataset. Moreover, the predictive performance of the final model should be compared to existing clinical prediction models. For example, the Framingham Risk Score estimates ten-year CVD risk in individuals based primarily on clinical factors and life style. However, up to 20% of CVD patients have none of these traditional risk factors, hence the need for potentially more accurate genetic predictors [110].
Fig. 3.
Example of estimating predictive model performance using Formula-fold cross validation (in this case, threefold). The estimated predictive performance is an average of the validation scores from each of the Formula-folds (and often from multiple iterations of Formula-fold CV). The folds are randomly partitioned training and testing groups. It is important that feature selection and classifier parameter selection occur within the cross validation in order to avoid estimation bias.
Michiels et al. reinforces the recommendations of Simon and Quackenbush after reanalyzing several large cancer prediction studies [106]. Their results indicated that many of these studies predicted no better than random chance because the selection of features greatly depends on the samples. They recommended a method of repeated random sampling to better estimate the mean and variance of prediction error. The best practice protocol, as well as common pitfalls, for building predictive models has been summarized [111], [112], [113]. Most recently, the FDA-led MAQC consortium published a comprehensive study to report issues related to microarray classification for clinical genomic testing and provided guidelines for proper statistical evaluation [114]. The MAQC study examined multiple prediction problems ranging from toxicology to cancer, but did not include cardiovascular-related datasets. Although there exists a large multicenter predictive study, which validated a 23-gene CAD diagnostic test [100], a more comprehensive study for other cardiovascular-related diseases designed in a similar framework as the MAQC study would be informative.
SECTION IV INTERPRETATION AND VALIDATION
Following the biomarker identification process, we must interpret and validate the results in a clinically meaningful way. There are a variety of approaches to interpret and validate selected biomarkers. One way is by using literature, for instance ontologies. Gene ontologies are organized terminologies that classify gene products into three major classes: molecular function, biological process, and cellular component. These vocabularies are by no means exhaustive; instead, they represent significant progress towards a common standard for biological function annotation. The GO Project [115] is a collaborative effort to create a consistent annotation for gene products across various databases. More recently, the Cardiovascular GO Annotation Initiative has identified and annotated over 4000 cardiovascular-related genes (http://www.ucl.ac.uk/cardiovasculargeneontology/). By locating selected biomarkers in the biological context of an ontological tree, we can gather more evidence to support or refute the significance of these biomarkers. GoMiner [116] is one of many software tools available that provides information from GO databases linking biomarkers to biological functions, molecular structures, and pathways. Closely related to ontologies are biomarker databases curated from the literature. These databases may be used to verify the relevance of biomarker identification algorithms or to guide algorithm selection [87]. Databases exist for general human disease genes [117] and for CVD-related genes [118].
Besides ontologies, considerable effort has been dedicated to molecular and cellular pathways, in particular, to model pathways that are potentially involved in pathological expression. Using microarrays and NGS, networks of cellular pathways may be sampled and visualized simultaneously to obtain a systems-level view. As is the case with ontologies, selected biomarkers may be verified by their location in the annotated pathway visualizations. For instance, the Pathway Explorer [119] developed by the Institute of Genomics and Bioinformatics at Graz University of Technology in Austria is a popular web-based tool for visualizing biological pathways derived from databases such as KEGG [120], BioCarta, and GenMapp [121]. The drawback for using annotated pathway visualizations is that, because this field is still in its infancy, the number of available curated pathways is minimal, and thus, these pathways are not always consistently annotated. Some of these pathway tools have been used for CVD studies [122]. Another way to interpret and validate selected biomarkers is by using experimental methods. For instance, we can validate microarray mRNA transcripts using qRT-PCR. By eliminating some variables involved in a microarray experiment, a qRT-PCR experiment can increase the statistical confidence of measured gene expression levels. Clinical studies are a third way to interpret and validate selected biomarkers. A significant amount of due diligence must be dedicated to issues such as cost, practical, and ethical considerations before clinical studies can be conducted.
SECTION V MICROARRAY CASE STUDY
We illustrate the genomic data analysis pipeline with a case study analyzing several CVD microarray datasets from the Gene Expression Omnibus (GEO), a public repository of gene expression data. Table I lists four datasets with unique prediction models (determined at the end of the data analysis pipeline) and accuracy for each dataset.
TABLE I.
CVD-Related Microarray Datasets and Data-Derived Optimal Prediction Models
| Dataset 1 | Dataset 2 | Dataset 3 | Dataset 4 | |
|---|---|---|---|---|
| Description | Blood Gene Exp. CAD | Baseline Macro. Athero. | Monocytes Fam. Hyper. | Monocytes Athero. |
|
| ||||
| GEO Accession | GSE12288 [123] | GSE9874 [2] | GSE6054 [124] | GSE23746 |
|
| ||||
| # of Samples | 222 | 30 | 23 | 95 |
|
| ||||
| Array Platform | Affy. HG U133A | Affy. HG U133A | Affy. HG U133 Plus 2.0 | Illumina Beadchip |
|
| ||||
| Classifier | KNN, K=10 | Linear SVM, Cost=4 | Linear SVM, Cost=1 | Bayesian, Near. Cent. |
|
| ||||
| Feature Selection Method | mRMRQ | T-Test | Fold Change | T-Test |
|
| ||||
| # of Features | 20 | 5 | 1 | 2 |
|
| ||||
| Est. Accuracy | 0.61 ±0.05 | 0.87 ±0.05 | 0.55 ±0.06 | 0.97 ±0.04 |
A. Preprocessing and Normalization
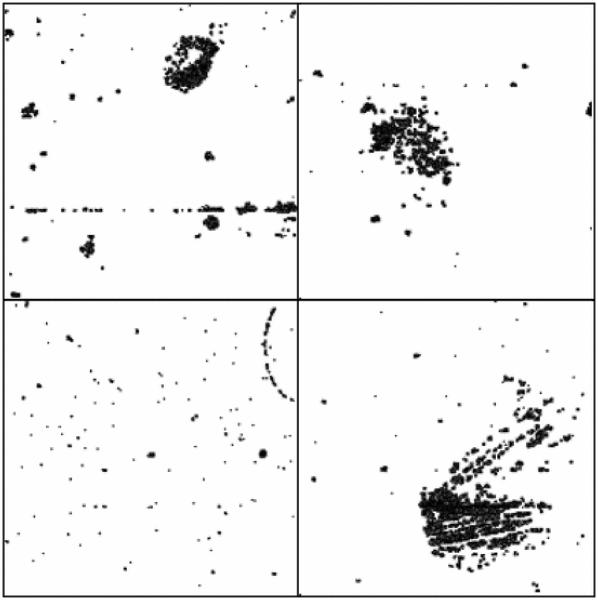
As described in Fig. 1, the first step after data acquisition in the bioinformatics pipeline is preprocessing and normalization. Many of the cardiovascular datasets available in GEO are composed of Affymetrix arrays. In cases where raw data files are available, we use caCORRECT to convert the raw Affymetrix probe expression to gene expression values. caCORRECT detects spatial statistical artifacts on each array and generates images indicating regions of high variance [23]. As illustrated in Fig. 4, caCORRECT identifies several arrays in Dataset 1 (Blood Gene Exp. CAD, Table I) with spatial artifacts. caCORRECT replaces these regions with a mean or median value across all arrays, reducing their overall effect on the final gene expression values. Alternatively, we can identify array outliers using caCORRECT's quality score. However, because of the small-sample sizes in these datasets, we rely on caCORRECT to remove artifacts. For each dataset in Table I, we use caCORRECT when raw Affymetrix data are available. Otherwise, we download preprocessed data from GEO.
Fig. 4.
caCORRECT identifies spatial artifacts in microarrays from Dataset 1 (Blood Gene Exp. CAD). These spatial artifacts represent regions of high variance. We can qualitatively assess the integrity of each microarray by examining the coverage of the artifact masks. Spatial artifacts resembling scratches or other statistically unlikely shapes can indicate defects in the microarray or substandard experimental procedures.
B. Exploratory Analysis
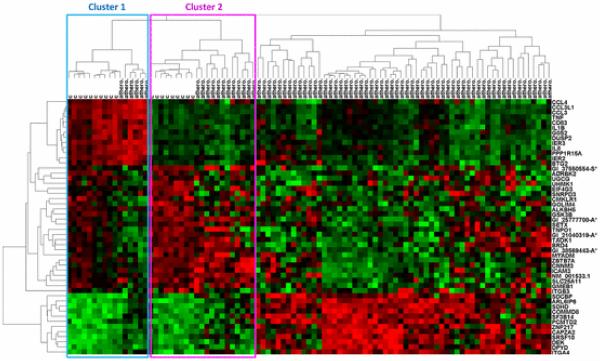
The second step in the data analysis pipeline is exploratory analysis. We use hierarchical clustering software created by Eisen et al. [64] to explore Dataset 4 (Monocytes Athero.). This dataset examines monocyte gene expression in patients with and without atherosclerosis. Because clustering large datasets can be computationally demanding, gene expression datasets are usually filtered to remove uninformative genes prior to clustering. We use SAM to identify the top 50 genes differentially expressed between patients with and without atherosclerosis [78]. Although filtering with SAM, a univariate method, can result in a loss of information (as described in Section III-B), we use it here simply to illustrate an exploratory analysis method. Hierarchical clustering of this reduced dataset (95 samples and 50 genes) using complete linkage and Euclidean distance reveals underlying groups in both the samples and the genes (see Fig. 5). Microarray samples (clustered horizontally) generally fall into three groups: Cluster 1 and cluster 2 contain a mixture of control patients and patients with atherosclerosis. The third cluster contains only patients with atherosclerosis. This clustering suggests that gene expression profiles can reveal underlying groups of atherosclerosis that are not represented in the original sample labels. Relationships revealed in exploratory analysis can guide the feature selection (see Section V-C) and classification (see Section V-D) steps by proposing new labels for the data prior to supervised analysis. Genes (clustered vertically) also tend to group together due to correlated expression. For example, genes in the rightmost cluster of Fig. 5 (including CCL4, CCL3L1, CCL3, TNF, CD83, etc.) tend to have higher expression in the Cluster 1 (mostly control) group of microarray samples.
Fig. 5.
Hierarchical clustering of Dataset 4 (Monocytes Athero.), an atherosclerosis microarray dataset with 95 samples and 50 genes (filtered from 24 350 genes using SAM). Microarray samples, clustered horizontally, fall into three large clusters with varying degrees of atherosclerosis. Clusters 1 and 2 contain a mixture of control and atherosclerosis samples. The remaining samples cluster into a large group containing all atherosclerosis samples. Genes (clustered vertically) also tend to cluster into groups with similar expression. Hierarchical clustering was performed using complete linkage and Euclidean distance.
C. Feature Selection
Supervised feature selection is the third step in the data analysis pipeline. Ranking genes in Dataset 4 (Monocytes Athero.) using supervised feature selection methods identifies several genes common among the ranking methods (see Table II). We used seven filter methods, including fold change, T-test, rank sum, SAM, rank products, and two mRMR methods [78], [79], [81] to rank genes in the dataset. We consider these genes to be candidate biomarkers whose function and clinical utility need to be verified using literature, biological databases, or more specific low-throughput experimental assay methods such as qRT-PCR (see Section V-E).
TABLE II.
Identification of Biomarkers in Dataset 4 Using Several Filter Methods
| Fold Change | T-Test | Rank Sum | SAM | Rank Products | mRMR-D | mRMR-Q |
|---|---|---|---|---|---|---|
| CCL3L1 | GSK3B | ALKBH5 | ALKBH5 | HBB | GSK3B | GSK3B |
| CCL4 | CMKLR1 | GSK3B | GSK3B | CCL3L1 | CMKLR1 | EPC1 |
| IL1B | ALKBH5 | GI_21040319-A* | CCL3L1 | CCL4 | CAMK1D | NAA16 |
| IL8 | ADRBK2 | GI_38569443-A* | IER3 | HBA2 | ARL6IP6 | RHOT1 |
| TNF | ARL6IP6 | GI_25777700-A* | ZBTB7A | IL8 | TMOD3 | GI_42657510-S* |
| GOS2 | SDHD | ARL6IP6 | CCL4 | HLA-DRB3 | ALKBH5 | KCTD18 |
| IER3 | UHMK1 | GMEB1 | CMKLR1 | HLA-DRB1 | KCTD18 | LRRC43 |
| CCL3 | GOLIM4 | CMKLR1 | GOS2 | DEFA1 | UHMK1 | EIF4G3 |
| IER2 | GI_21040319-A* | EIF4G3 | CD83 | IL1B | GI_38569443-A* | KRCC1 |
| CD83 | GMEB1 | SRSF10 | GI_21040319-A* | DEFA3 | GI_21464106-A* | TMSL3 |
| EGR1 | GI_25777700-A* | SLC25A11 | IER2 | HLA-DRB5 | EIF4G3 | GI_42660285-S* |
| DUSP2 | EIF4G3 | DEK | TNF | HLA-DQA1 | GI_42660285-S* | CAMK1D |
| PPP1R15A | SF3B14 | TAF4 | ADRBK2 | IER3 | CD58 | UHMK1 |
| HBB | SNRPD3 | SDHD | IL1B | GOS2 | KRCC1 | CMKLR1 |
| NFKBIA | SRSF10 | CNNM3 | SDHD | HBA1 | GI_5803174-S* | GI_5803174-S* |
| TNFAIP3 | TNPO1 | SETX | GI_25777700-A* | EGR1 | TNPO1 | GI_21464106-A* |
| GI_33386702-S* | PCMTD2 | CAMK1D | ARL6IP6 | GI_33386702-S* | GI_37550554-S* | ALKBH5 |
| GI_31088849-S* | GI_38569443-A* | TGFBRAP1 | CCL3 | TNF | SLC25A11 | SF3B14 |
| BTG2 | DEK | GI_37550554-S* | DUSP2 | GI_31088849-S* | GI_4826967-S* | GI_41120019-S* |
| GI_37550226-S* | SLC25A11 | PLEKHF2 | SDCBP | GZMH | TAF4 | C9orf102 |
Highlighted genes are some examples that occur within the top 20 genes for at least three methods and may be linked to cardiovascular disease.
Microarray probe names are used when gene symbols are not available.
The resulting lists of genes in Table II are evidence that feature selection methods can produce highly variable results. One of the methods to combat this variability involves the use of known, or validated, differentially expressed genes to select an optimal feature selection method. That is, the validated genes should be ranked favorably. Thus, we can validate the expression of some of the genes in Table II and use them to identify a biologically relevant feature selection method. The biologically relevant method should increase the probability of correctly identifying additional biomarkers while reducing the number of false discoveries [87].
D. Classification
Classification, or predictive modeling, is the fourth step in the data analysis pipeline. We analyze all CVD-related microarray datasets in Table I using nested cross validation with several classifiers and feature selection methods with the goal of identifying predictive modeling parameters that produce the highest accuracy. We use four classifiers (SVM, logistic regression, Bayesian, and Formula-nearest neighbors) and seven feature selection methods (fold change, T-test, rank sum, SAM, rank products, and two mRMR methods), varying feature sizes from 1 to 20. We compute the internal and external cross-validation performance as in Fig. 3. Ten iterations and three folds of internal cross validation estimate the predictive performance of each model (i.e., each combination of classifier, feature selection method, and feature size). The best performing model is then used to compute external cross-validation performance. In Fig. 6, we plot internal cross-validation performance and the corresponding external validation performance for each iteration of external validation.
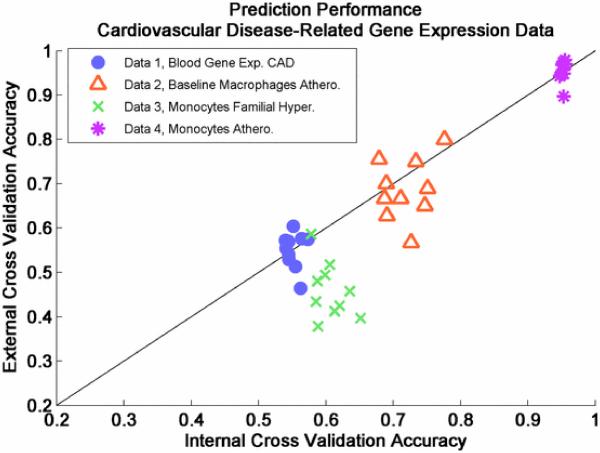
Fig. 6.
Nested cross-validation performance of several cardiovascular-related disease datasets. Predictive performance was optimized using four classifiers (linear SVM, KNN, logistic regression, and Bayesian), seven feature selection methods (fold change, T-test, SAM, rank sum, rank products, mRMRD, and mRMRQ), and feature sizes ranging from 1 to 20. Predictive performance greatly depends on dataset.
As reported in the MAQC Phase II article published in Nature Biotechnology, using an ANOVA test, the predictive performance of models derived from gene expression are most influenced by dataset [114]. That is, regardless of the algorithmic or modeling tools used, the biological endpoint of interest determines the expected prediction performance. Moreover, for some datasets in the MAQC-II study, data analysis teams could only identify good modeling parameters with predictive accuracy similar to random chance, despite a wide variety of methods employed. Predictive analysis of the cardiovascular-related datasets in Table I yields results similar to the MAQC-II study. In Fig. 6, we see that Dataset 1 and Dataset 3 (Blood Gene Exp. CAD and Monocytes Familial Hyper.) are difficult to predict. Biologically, this suggests that gene expression profiles assayed for these datasets cannot discriminate the samples into the groups of interest. On the other hand, gene expression profiles in Dataset 2 (Baseline Macrophages Athero.) are able to predict the presence of atherosclerosis with an accuracy of around 70% to 80% (see Fig. 6, orange). Moreover, gene expression profiles in Dataset 4 (Monocytes Athero.) are able to predict the presence of atherosclerosis with accuracy greater than 90% (see Fig. 6, magenta). As described in Section III-C, a final predictive model for each dataset, including candidate biomarkers, can be derived by applying the optimal protocol (i.e., feature selection method and classifier parameters that result in highest internal cross-validation performance) to the entire dataset. Unfortunately, we cannot evaluate the performance of the final predictive model because of a lack of available independent datasets in this case study.
E. Interpretation and Validation
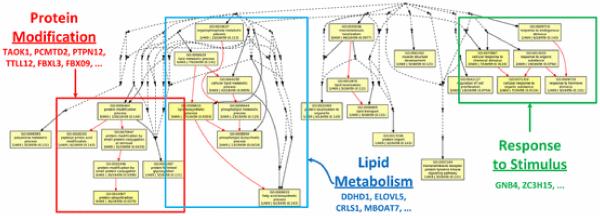
Interpretation and validation of candidate biomarkers is the fifth step of the data analysis pipeline. The final predictive model should generally include biomarkers identified using the optimal feature selection method, as described in the previous section. However, since cross-validation performances are very similar for Dataset 4, we consider the results of several feature selection methods (see Table II) for this example of interpretation and validation. A literature search reveals that some of the candidate biomarkers for prediction of atherosclerosis in Dataset 4 (Monocyte Athero.) (see Table II) are biologically relevant. For example, IL1B has been linked to obesity and CVD [125]. CCL3L1 copy number variations have been linked to immune response, but may also be linked to cardiovascular-related diseases [126]. GSK3B is known as an Alzheimer's disease-related gene, but supports the theory that Alzheimer's disease and CVD may share some biological mechanisms [127]. ALKBH5 and ARL6IP6 have no obvious relation to CVD in the scientific literature, but appear to be highly ranked for several feature selection methods. Such novel or surprising results derived from the genomic data analysis pipeline, after appropriate validation, may provide clues or stimulate further promising research in CVD genomics. GO analysis with GOEAST reveals several biological processes over-represented in the list of genes identified as differentially expressed in Dataset 4 [128]. These biological processes, rendered by the GOEAST application and annotated in Fig. 7, tend to group into categories that include protein modification, lipid metabolism, and response to stimulus. Lipid metabolism is particularly interesting because it has been linked to atherosclerosis and obesity in numerous studies [129].
Fig. 7.
Differentially expressed genes profiled from monocytes of atherosclerosis and control patients are involved in biological processes associated with protein modification, lipid metabolism, and response to stimulus. This hierarchy of biological processes was generated using the GOEAST application that identifies statistically over-represented GO terms.
After interpretation and validation, the resulting prediction models will have been 1) optimized to maximize performance using data-driven and knowledge-driven parameter selection methods, 2) statistically validated using independent external data, and 3) biologically validated to improve the clinical feasibility.
SECTION VI CONCLUSION
Genomic technologies, primarily microarray and NGS, enable us to obtain molecular profiles and to identify biomarkers that can potentially improve clinical diagnostic and prognostic applications. Genomic data generally have a small number of samples (i.e., corresponding to patients) and a relatively large number of features (i.e., corresponding to genes). Because of these data properties, we must make special considerations during data analysis. In this review, we have presented a high-throughput data analysis pipeline that aims to help researchers construct predictive, biologically relevant disease models. This pipeline consists of the following steps: 1) data acquisition, 2) preprocessing and normalization, 3) exploratory analysis, 4) feature selection, 5) classification, and 6) interpretation and validation. For each step, we described common bioinformatics methods and current challenges related to the specific clinical application of predicting CVD risk. Although other reviews also focus on cardiovascular genomics [130], this review is unique because it provides an in-depth view of current bioinformatics methods in each step of the pipeline. Moreover, it provides a broad case study that applies the pipeline to several datasets and presents specific examples for each step.
In the case study, we examined several microarray datasets comparing biological samples of CVD-related conditions (i.e., atherosclerosis) to control samples. The case study highlights some current challenges in genomic bioinformatics for CVD. First, compared to genomic cancer datasets, there are far fewer publicly available CVD-related datasets (see Table I). This limits the ability for bioinformatics researchers to validate algorithmic results using external datasets. Second, even for datasets with a large number of samples, there are very few differentially expressed genes. In general, genetic factors tend to be weakly associated with CVD. This supports a “polygenic theory” that CVD is the result of multiple genetic variants acting in concert. The lack of definitive biomarkers in some of the case study datasets also explains the modest classification performance and reinforces the “polygenic theory” such that multibiomarker panels are necessary to adequately characterize CVD [5]. The case study we presented is far from comprehensive and only serves as an example of the bioinformatics data analysis pipeline. Thus, opportunities remain for bioinformatics researchers to 1) propose more cardiovascular genomic questions and engage biologists and clinicians to generate more data, 2) address the problem of multibiomarker panels that might improve disease prediction accuracy, and 3) conduct a comprehensive study that mirrors the FDA-led MAQC study by examining a large number of datasets representing a broad overview of CVD.
Acknowledgments
This work was supported by grants from the National Institutes of Health (NHLBI 5U01HL080711, Bioengineering Research Partnership R01CA108468, Center of Cancer Nanotechnology Excellence U54CA119338), Georgia Cancer Coalition (Distinguished Cancer Scholar Award to Prof. M. D. Wang), Microsoft Research, and Hewlett-Packard.
Contributor Information
John H. Phan, Email: jhphan@gatech.edu, Department of Biomedical Engineering, Georgia Institute of Technology and Emory University, Atlanta, GA 30332 USA.
Chang F. Quo, Email: cfquo@gatech.edu, Department of Biomedical Engineering, Georgia Institute of Technology and Emory University, Atlanta, GA 30332 USA
May Dongmei Wang, Email: maywang@bme.gatech.edu, M. D. Wang is with the Department of Biomedical Engineering, the Department of Electrical and Computer Engineering, Winship Cancer Institute, and Parker H. Petit Institute of Bioengineering and Biosciences, Georgia Institute of Technology and Emory University, Atlanta, GA 30332 USA.
REFERENCES
- 1.Schnabel RB, Baccarelli A, Lin H, Ellinor PT, Benjamin EJ. Next steps in cardiovascular disease genomic research—Sequencing, epigenetics, and transcriptomics. Clin. Chem. 2012;58(1):113–126. doi: 10.1373/clinchem.2011.170423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Molecular Biology of the Cell. 4th Garland Science; New York: 2002. [Google Scholar]
- 3.Small EM, Olson EN. Pervasive roles of microRNAs in cardiovascular biology. Nature. 2011;469(7330):336–342. doi: 10.1038/nature09783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thum T, Mayr M. Review focus on the role of microRNA in cardiovascular biology and disease. Cardiovasc. Res. 2012;93(4):543–544. doi: 10.1093/cvr/cvs085. [DOI] [PubMed] [Google Scholar]
- 5.Kullo IJ, Cooper LT. Early identification of cardiovascular risk using genomics and proteomics. Nat. Rev. Cardiol. 2010;7(6):309–317. doi: 10.1038/nrcardio.2010.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hägg DA, Jernis M, Wiklund O, Thelle DS, Fagerberg B, Eriksson P, Hamsten A, Olsson B, Carlsson B, Carlsson L, Svensson P. Expression profiling of macrophages from subjects with atherosclerosis to identify novel susceptibility genes. Int. J. Mol. Med. 2008;21(6):697–704. [PubMed] [Google Scholar]
- 7.Seo D, Goldschidt-Clermont P, Velazquez O, Beecham G. Genomics of premature atherosclerotic vascular disease. Curr. Atheroscler. Rep. 2010;12(3):187–193. doi: 10.1007/s11883-010-0104-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yuan Z, Miyoshi T, Bao Y, Sheehan JP, Matsumoto AH, Shi W. Microarray analysis of gene expression in mouse aorta reveals role of the calcium signaling pathway in control of atherosclerosis susceptibility. Am. J. Physiol. Heart Circ. Physiol. 2009;296(5):H1336–H1343. doi: 10.1152/ajpheart.01095.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cipollone F, Felicioni L, Sarzani R, Ucchino S, Spigonardo F, Mandolini C, Malatesta S, Bucci M, Mammarella C, Santovito D. A unique microRNA signature associated with plaque instability in humans. Stroke. 2011;42(9):2556–2563. doi: 10.1161/STROKEAHA.110.597575. [DOI] [PubMed] [Google Scholar]
- 10.Di Stefano V, Zaccagnini G, Capogrossi MC, Martelli F. microRNAs as peripheral blood biomarkers of cardiovascular disease. Vascul. Pharmacol. 2011;55(4):111–118. doi: 10.1016/j.vph.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 11.Chico TJA, Milo M, Crossman DC. The genetics of cardiovascular disease: New insights from emerging approaches. J. Pathol. 2010;220(2):186–197. doi: 10.1002/path.2641. [DOI] [PubMed] [Google Scholar]
- 12.Schulze A, Downward J. Navigating gene expression using microarrays: A technology review. Nat. Cell Biol. 2001;3:E190, E195. doi: 10.1038/35087138. [DOI] [PubMed] [Google Scholar]
- 13.Schena M, Shalon D, Davis R, Brown P. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 14.Lipshutz R, Fodor S, Gingeras T, Lockhart D. High density synthetic oligonucleotide arrays. Nat. Genet. 1999;21:20–24. doi: 10.1038/4447. [DOI] [PubMed] [Google Scholar]
- 15.Chen Z, McGee M, Liu Q, Scheuermann RH. A distribution free summarization method for affymetrix GeneChip§R arrays. Bioinformatics. 2007;23(3):321–327. doi: 10.1093/bioinformatics/btl609. [DOI] [PubMed] [Google Scholar]
- 16.Hochreiter S, Clevert DA, Obermayer K. A new summarization method for affymetrix probe level data. Bioinformatics. 2006;22(8):943–949. doi: 10.1093/bioinformatics/btl033. [DOI] [PubMed] [Google Scholar]
- 17.Wu Z, Irizarry RA. Stochastic models inspired by hybridization theory for short oligonucleotide arrays. J. Comput. Biol. 2005 Jul-Aug;12(6):882–893. doi: 10.1089/cmb.2005.12.882. [DOI] [PubMed] [Google Scholar]
- 18.Hubbell E, Liu WM, Mei R. Robust estimators for expression analysis. Bioinformatics. 2002;18(12):1585–1592. doi: 10.1093/bioinformatics/18.12.1585. [DOI] [PubMed] [Google Scholar]
- 19.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: Expression index computation and outlier detection. Proc. Natl. Acad. Sci. USA. 2001;98(1):31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Affymetrix Technote note: Guide to probe logarithmic intesity error (PLIER) estimation. 2008 [Online]. Available: http://www.affymetrix.com/support/technical/technotes/plier_technote.pdf.
- 21.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 22.Huber W, Von Heydebreck A, Sültmann H, Poustka A, Vingron M. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics. 2002;18(suppl 1):S96–S104. doi: 10.1093/bioinformatics/18.suppl_1.s96. [DOI] [PubMed] [Google Scholar]
- 23.Moffitt R, Yin-Goen Q, Stokes TH, Parry RM, Torrance JH, Phan JH, Young AN, Wang MD. caCORRECT2: Improving the accuracy and reliability of microarray data in the presence of artifacts. BMC Bioinformat. 2011;12(1):383. doi: 10.1186/1471-2105-12-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Du P, Kibbe WA, Lin SM. Lumi: A pipeline for processing Illumina microarray. Bioinformatics. 2008;24(13):1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 25.Lin SM, Du P, Huber W, Kibbe WA. Model-based variance-stabilizing transformation for illumina microarray data. Nucleic Acids Res. 2008;36(2):e11. doi: 10.1093/nar/gkm1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cairns J, Dunning MJ, Ritchie ME, Russell R, Lynch AG. BASH: A tool for managing BeadArray spatial artefacts. Bioinformatics. 2008;24(24):2921–2922. doi: 10.1093/bioinformatics/btn557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xie Y, Wang X, Story M. Statistical methods of background correction for illumina beadarray data. Bioinformatics. 2009;25(6):751–757. doi: 10.1093/bioinformatics/btp040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seo J, Hoffman E. Probe set algorithms: Is there a rational best bet? BMC Bioinformat. 2006;7(1):395. doi: 10.1186/1471-2105-7-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Millenaar F, Okyere J, May S, Van Zanten M, Voesenek L, Peeters A. How to decide? Different methods of calculating gene expression from short oligonucleotide array data will give different results. BMC Bioinformat. 2006;7(1):137. doi: 10.1186/1471-2105-7-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gyorffy B, Molnar B, Lage H, Szallasi Z, Eklund AC. Evaluation of microarray preprocessing algorithms based on concordance with RT-PCR in clinical samples. PLoS One. 2009;4(5):e5645. doi: 10.1371/journal.pone.0005645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song J, Maghsoudi K, Li W, Fox E, Quackenbush J, Liu X. Microarray blob-defect removal improves array analysis. Bioinformatics. 2007;23(8):966–971. doi: 10.1093/bioinformatics/btm043. [DOI] [PubMed] [Google Scholar]
- 32.Yang S, Guo X, Hu H. MOF: An R function to detect outlier microarray. Genomics, Proteomics Bioinformat. 2008;6(3/4):186–189. doi: 10.1016/S1672-0229(09)60006-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shieh AD, Hung YS. Detecting outlier samples in microarray data. Statist. Appl. Genetics Molecular Biol. 2009;8(1):13. doi: 10.2202/1544-6115.1426. [DOI] [PubMed] [Google Scholar]
- 34.Shi L, Reid LH, Jones WD, Shippy R, Warrington JA, Baker SC, Collins PJ, de Longueville F, Kawasaki ES, Lee KY, Luo Y, Sun YA, Willey JC, Setterquist RA, Fischer GM, Tong W, Dragan YP, Dix DJ, Frueh FW, Goodsaid FM, Herman D, Jensen RV, Johnson CD, Lobenhofer EK, Puri RK, Schrf U, Thierry-Mieg J, Wang C, Wilson M, Wolber PK, Zhang L, Amur S, Bao W, Barbacioru CC, Lucas AB, Bertholet V, Boysen C, Bromley B, Brown D, Brunner A, Canales R, Cao XM, Cebula TA, Chen JJ, Cheng J, Chu TM, Chudin E, Corson J, Corton JC, Croner LJ, Davies C, Davison TS, Delenstarr G, Deng X, Dorris D, Eklund AC, Fan XH, Fang H, Fulmer-Smentek S, Fuscoe JC, Gallagher K, Ge W, Guo L, Guo X, Hager J, Haje PK, Han J, Han T, Harbottle HC, Harris SC, Hatchwell E, Hauser CA, Hester S, Hong H, Hurban P, Jackson SA, Ji H, Knight CR, Kuo WP, LeClerc JE, Levy S, Li QZ, Liu C, Liu Y, Lombardi MJ, Ma Y, Magnuson SR, Maqsodi B, McDaniel T, Mei N, Myklebost O, Ning B, Novoradovskaya N, Orr MS, Osborn TW, Papallo A, Patterson TA, Perkins RG, Peters EH, Peterson R, Philips KL, Pine PS, Pusztai L, Qian F, Ren H, Rosen M, Rosenzweig BA, Samaha RR, Schena M, Schroth GP, Shchegrova S, Smith DD, Staedtler F, Su Z, Sun H, Szallasi Z, Tezak Z, Thierry-Mieg D, Thompson KL, Tikhonova I, Turpaz Y, Vallanat B, Van C, Walker SJ, Wang SJ, Wang Y, Wolfinger R, Wong A, Wu J, Xiao C, Xie Q, Xu J, Yang W, Zhang L, Zhong S, Zong Y, Slikker W., Jr. The microarray quality control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat. Biotechnol. 2006 Sep;24(9):1151–1161. doi: 10.1038/nbt1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tan PK, Downey TJ, Spitznagel EL, Jr., Xu P, Fu D, Dimitrov DS, Lempicki RA, Raaka BM, Cam MC. Evaluation of gene expression measurements from commercial microarray platforms. Nucleic Acids Res. 2003;31(19):5676–5684. doi: 10.1093/nar/gkg763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Petersen D, Chandramouli G, Geoghegan J, Hilburn J, Paarlberg J, Kim C, Munroe D, Gangi L, Han J, Puri R. Three microarray platforms: An analysis of their concordance in profiling gene expression. BMC Genomics. 2005;6(1):63. doi: 10.1186/1471-2164-6-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Canales R, Luo Y, Willey J, Austermiller B, Barbacioru C, Boysen C, Hunkapiller K, Jensen R, Knight C, Lee K, Ma Y, Maqsodi B, Papallo A, Peters E, Poulter K, Ruppel P, Samaha R, Shi L, Yang W, Zhang L, Goodsaid F. Evaluation of DNA microarray results with quantitative gene expression platforms. Nat. Biotechnol. 2006;24(9):1115–1122. doi: 10.1038/nbt1236. [DOI] [PubMed] [Google Scholar]
- 38.Patterson T, Lobenhofer E, Fulmer-Smentek S, Collins P, Chu T, Bao W, Fang H, Kawasaki E, Hager J, Tikhonova I, Walker S, Zhang L, Hurban P, De Longueville F, Fuscoe J, Tong W, Shi L, Wolfinger R. Performance comparison of one-color and two-color platforms within the microarray quality control (MAQC) project. Nat. Biotechnol. 2006;24(9):1140–1150. doi: 10.1038/nbt1242. [DOI] [PubMed] [Google Scholar]
- 39.Klebanov L, Yakovlev A. How high is the level of technical noise in microarray data. Biol Direct. 2007;2(1):9. doi: 10.1186/1745-6150-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z. Genome sequencing in open microfabricated high density picoliter reactors. Nature. 2005;437(7057):376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Valouev A, Ichikawa J, Tonthat T, Stuart J, Ranade S, Peckham H, Zeng K, Malek JA, Costa G, McKernan K. A high-resolution, nucleosome position map of C. Elegans reveals a lack of universal sequence-dictated positioning. Genome Res. 2008;18(7):1051–1063. doi: 10.1101/gr.076463.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Metzker ML. Sequencing technologies—The next generation. Nat. Rev. Genet. 2009;11(1):31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 43.Shendure J, Ji H. Next-generation DNA sequencing. Nat. Biotechnol. 2008;26(10):1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- 44.Erlich Y, Mitra PP. Alta-Cyclic: A self-optimizing base caller for next-generation sequencing. Nat. Methods. 2008;5(8):679–682. doi: 10.1038/nmeth.1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rougemont J, Amzallag A, Iseli C, Farinelli L, Xenarios I, Naef F. Probabilistic base calling of Solexa sequencing data. BMC Bioinformat. 2008;9(1):431. doi: 10.1186/1471-2105-9-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wheeler DA, Srinivasan M, Egholm M, Shen Y, Chen L, McGuire A, He W, Chen YJ, Makhijani V, Roth GT. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452(7189):872–876. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- 47.Wang Z, Gerstein M, Snyder M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009;10(1):57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464(7285):59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hurd PJ, Nelson CJ. Advantages of next-generation sequencing versus the microarray in epigenetic research. Brief. Funct. Genomic Proteomic. 2009;8(3):174–183. doi: 10.1093/bfgp/elp013. [DOI] [PubMed] [Google Scholar]
- 50.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456(7221):470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trapnell C, Pachter L, Salzberg SL. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Trapnell S, Pachter L, Salzberg SL. Concordance among digital gene expression, microarrays, and qPCR when measuring differential expression of microRNAs. Biotechniques. 2010;48(3):219–222. doi: 10.2144/000113367. [DOI] [PubMed] [Google Scholar]
- 53.Willenbrock H, Salomon J, Søkilde R, Barken KB, Hansen TN, Nielsen FC, Møller S, Litman T. Quantitative miRNA expression analysis: Comparing microarrays with next-generation sequencing. RNA. 2009;15(11):2028–2034. doi: 10.1261/rna.1699809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McPherson JD. Next-generation gap. Nat. Methods. 2009;6:S2–S5. doi: 10.1038/nmeth.f.268. [DOI] [PubMed] [Google Scholar]
- 55.Li H, Homer N. A survey of sequence alignment algorithms for next-generation sequencing. Brief. Bioinformat. 2010;11(5):473–483. doi: 10.1093/bib/bbq015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Oshlack A, Robinson MD, Young MD. From RNA-seq reads to differential expression results. Genome Biol. 2010;11(12):220. doi: 10.1186/gb-2010-11-12-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee S, Seo CH, Lim B, Yang JO, Oh J, Kim M, Lee B, Kang C. Accurate quantification of transcriptome from RNA-Seq data by effective length normalization. Nucleic Acids Res. 2011;39(2):e9. doi: 10.1093/nar/gkq1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11(3):R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jain AK. Data clustering: 50 years beyond K-means. Pattern Recognit. Lett. 2010;31(8):651–666. [Google Scholar]
- 60.Freyhult E, Landfors M, Önskog J, Hvidsten T, Rydén P. Challenges in microarray class discovery: A comprehensive examination of normalization, gene selection and clustering. BMC Bioinformat. 2010;11(1):503. doi: 10.1186/1471-2105-11-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Michael E, James W, Philip B, Whittemore T, Szilard V, William K, Geoffrey G, Stephen E, Naheem T, Ron W. Development of a blood-based gene expression algorithm for assessment of obstructive coronary artery disease in non-diabetic patients. BMC Med. Genomics. 2011;4(26) doi: 10.1186/1755-8794-4-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tabibiazar R, Wagner RA, Ashley EA, King JY, Ferrara R, Spin JM, Sanan DA, Narasimhan B, Tibshirani R, Tsao PS. Signature patterns of gene expression in mouse atherosclerosis and their correlation to human coronary disease. Physiol. Genomics. 2005;22(2):213–226. doi: 10.1152/physiolgenomics.00001.2005. [DOI] [PubMed] [Google Scholar]
- 63.Hägg S, Skogsberg J, Lundström J, Noori P, Nilsson R, Zhong H, Maleki S, Shang MM, Brinne B, Bradshaw M. Multi-organ expression profiling uncovers a gene module in coronary artery disease involving transendothelial migration of leukocytes and LIM domain binding 2: The Stockholm atherosclerosis gene expression (STAGE) study. PLoS Genet. 2009;5(12):e1000754. doi: 10.1371/journal.pgen.1000754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA. 1998;95(25):14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu FX. Genetic weighted k-means algorithm for clustering large-scale gene expression data. BMC Bioinformat. 2008;9(Suppl 6):S12. doi: 10.1186/1471-2105-9-S6-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ringnér M. What is principal component analysis? Nat. Biotechnol. 2008;26(3):303–304. doi: 10.1038/nbt0308-303. [DOI] [PubMed] [Google Scholar]
- 67.Preli A, Bleuler S, Zimmermann P, Wille A, Bühlmann P, Gruissem W, Hennig L, Thiele L, Zitzler E. A systematic comparison and evaluation of biclustering methods for gene expression data. Bioinformatics. 2006;22(9):1122–1129. doi: 10.1093/bioinformatics/btl060. [DOI] [PubMed] [Google Scholar]
- 68.Tamayo P, Slonim D, Mesirov J, Zhu Q, Kitareewan S, Dmitrovsky E, Lander ES, Golub TR. Interpreting patterns of gene expression with self-organizing maps: Methods and application to hematopoietic differentiation. Proc. Nat. Acad. Sci. USA. 1999;96(6):2907. doi: 10.1073/pnas.96.6.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.D’haeseleer P. How does gene expression clustering work? Nat. Biotechnol. 2005;23:1499–1501. doi: 10.1038/nbt1205-1499. [DOI] [PubMed] [Google Scholar]
- 70.Quackenbush J. Computational analysis of microarray data. Nat. Rev. Genet. 2001;2:418–427. doi: 10.1038/35076576. [DOI] [PubMed] [Google Scholar]
- 71.Wang A, Gehan E. Gene selection for microarray data analysis using principle component analysis. Stat. Med. 2005;24:2069–2087. doi: 10.1002/sim.2082. [DOI] [PubMed] [Google Scholar]
- 72.Hubert M, Engelen S. Robust PCA and classification in biosciences. Bioinformatics. 2004;20(11):1728–1736. doi: 10.1093/bioinformatics/bth158. [DOI] [PubMed] [Google Scholar]
- 73.Reich D, Price AL, Patterson N. Principal component analysis of genetic data. Nat. Genet. 2008;40(5):491–491. doi: 10.1038/ng0508-491. [DOI] [PubMed] [Google Scholar]
- 74.Lu J, Kerns RT, Peddada SD, Bushel PR. Principal component analysis-based filtering improves detection for Affymetrix gene expression arrays. Nucleic Acids Res. 2011;13:e86. doi: 10.1093/nar/gkr241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chiou KR, Charng MJ, Chang HM. Array-based resequencing for mutations causing familial hypercholesterolemia. Atherosclerosis. 2011;216(2):383–389. doi: 10.1016/j.atherosclerosis.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 76.Civeira F, Ros E, Jarauta E, Plana N, Zambon D, Puzo J, Martinez de Esteban JP, Ferrando J, Zabala S, Almagro F. Comparison of genetic versus clinical diagnosis in familial hypercholesterolemia. Am. J. Cardiol. 2008;102(9):1187–1193. doi: 10.1016/j.amjcard.2008.06.056. [DOI] [PubMed] [Google Scholar]
- 77.Saeys Y, Inza I, Larran~aga P. A review of feature selection techniques in bioinformatics. Bioinformatics. 2007;23(19):2507–2517. doi: 10.1093/bioinformatics/btm344. [DOI] [PubMed] [Google Scholar]
- 78.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA. 2001;98(9):5116–5112. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Breitling R, Armengaud P, Amtmann A, Herzyk P. Rank products: A simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett. 2004;573(1/3):83–92. doi: 10.1016/j.febslet.2004.07.055. [DOI] [PubMed] [Google Scholar]
- 80.Kadota K, Shimizu K. Evaluating methods for ranking differentially expressed genes applied to microarray quality control data. BMC Bioinformat. 2011;12(1):227. doi: 10.1186/1471-2105-12-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Peng H, Long F, Ding C. Feature selection based on mutual information: Criteria of max-dependency, max-relevance, and min-redundancy. IEEE Trans. Pattern Anal. Mach. Intell. 2005 Aug.27(8):1226–1238. doi: 10.1109/TPAMI.2005.159. [DOI] [PubMed] [Google Scholar]
- 82.Gheyas IA, Smith LS. Feature subset selection in large dimensionality domains. Pattern Recognit. 2010;43(1):5–13. [Google Scholar]
- 83.Hong JH, Cho SB. Efficient huge-scale feature selection with speciated genetic algorithm. Pattern Recognit. Lett. 2006;27(2):143–150. [Google Scholar]
- 84.Zhang X, Lu X, Shi Q, Xu X, Leung H, Harris L, Iglehart J, Miron A, Liu J, Wong W. Recursive SVM feature selection and sample classification for mass-spectrometry and microarray data. BMC Bioinformat. 2006;7(1):197. doi: 10.1186/1471-2105-7-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lin WJ, Hsueh HM, Chen JJ. Power and sample size estimation in microarray studies. BMC Bioinformat. 2010;11(1):48. doi: 10.1186/1471-2105-11-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Klebanov L, Yakovlev A. Is there an alternative to increasing the sample size in microarray studies? Bioinformation. 2007;1(10):429–431. doi: 10.6026/97320630001429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Phan J, Yin-Goen Q, Young AN, Wang MD. Improving the efficiency of biomarker identification using biological knowledge. Pac. Symp. Biocomput. 2009;14:427–438. [PMC free article] [PubMed] [Google Scholar]
- 88.Chen L, Xuan J, Wang C, Shih I, Wang Y, Zhang Z, Hoffman E, Clarke R. Knowledge-guided multi-scale independent component analysis for biomarker identification. BMC Bioinformat. 2008;9(1):416. doi: 10.1186/1471-2105-9-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Phan J, Moffitt RA, Stokes TH, Liu J, Young AN, Nie S, Wang MD. Convergence of biomarkers, bioinformatics and nanotechnology for individualized cancer treatment. Trends Biotechnol. 2009;27(6):350–358. doi: 10.1016/j.tibtech.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Aerts S, Lambrechts D, Maity S, Van Loo P, Coessens B, De Smet F, Tranchevent L-C, De Moor B, Marynen P, Hassan B, Carmeliet P, Moreau Y. Gene prioritization through genomic data fusion. Nat. Biotechnol. 2006;24(5):537–544. doi: 10.1038/nbt1203. [DOI] [PubMed] [Google Scholar]
- 91.Kuffner R, Fundel K, Zimmer R. Expert knowledge without the expert: Integrated analysis of gene expression and literature to derive active functional contexts. Bioinformatics. 2005;21(suppl 2) doi: 10.1093/bioinformatics/bti1143. no. ii259–ii267. [DOI] [PubMed] [Google Scholar]
- 92.Kong S, Pu W, Park P. A multivariate approach for integrating genome-wide expression data and biological knowledge. Bioinformatics. 2006;22(19):2373–2380. doi: 10.1093/bioinformatics/btl401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mukherjee S, Roberts S. A theoretical analysis of the selection of differentially expressed genes. J Bioinform. Comput. Biol. 2005;3(3):627–643. doi: 10.1142/s0219720005001211. [DOI] [PubMed] [Google Scholar]
- 94.Yousef M, Ketany M, Manevitz L, Showe L, Showe M. Classification and biomarker identification using gene network modules and support vector machines. BMC Bioinformat. 2009;10(1):337. doi: 10.1186/1471-2105-10-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jin G, Zhou X, Wang H, Zhao H, Cui K, Zhang XS, Chen L, Hazen SL, Li K, Wong STC. The knowledge-integrated network biomarkers discovery for major adverse cardiac events. J. Proteome Res. 2008;7(9):4013–4021. doi: 10.1021/pr8002886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Simonson M, Wills A, Keller M, McQueen M. Recent methods for polygenic analysis of genome-wide data implicate an important effect of common variants on cardiovascular disease risk. BMC Med. Genomics. 2011;12(1):146. doi: 10.1186/1471-2350-12-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Humphries SE, Ridker PM, Talmud PJ. Genetic testing for cardiovascular disease susceptibility: A useful clinical management tool or possible misinformation? Arterioscler. Thromb. Vasc. Biol. 2004;24(4):628–636. doi: 10.1161/01.ATV.0000116216.56511.39. [DOI] [PubMed] [Google Scholar]
- 98.Robin NH, Tabereaux PB, Benza R, Korf BR. Genetic testing in cardiovascular disease. J. Am. Coll. Cardiol. 2007;50(8):727–737. doi: 10.1016/j.jacc.2007.05.015. [DOI] [PubMed] [Google Scholar]
- 99.Vasan RS. Biomarkers of cardiovascular disease: Molecular basis and practical considerations. Circulation. 2006;113(19):2335–2362. doi: 10.1161/CIRCULATIONAHA.104.482570. [DOI] [PubMed] [Google Scholar]
- 100.Rosenberg S, Elashoff MR, Beineke P, Daniels SE, Wingrove JA, Tingley WG, Sager PT, Sehnert AJ, Yau M, Kraus WE. Multicenter validation of the diagnostic accuracy of a blood-based gene expression test for assessing obstructive coronary artery disease in non-diabetic patients. Ann. Intern. Med. 2010;153(7):425–434. doi: 10.7326/0003-4819-153-7-201010050-00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Paynter NP, Chasman DI, Buring JE, Shiffman D, Cook NR, Ridker PM. Cardiovascular disease risk prediction with and without knowledge of genetic variation at chromosome 9p21. 3. Ann. Intern. Med. 2009;150(2):65–72. doi: 10.7326/0003-4819-150-2-200901200-00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kathiresan S, Melander O, Anevski D, Guiducci C, Burtt NP, Roos C, Hirschhorn JN, Berglund G, Hedblad B, Groop L. Polymorphisms associated with cholesterol and risk of cardiovascular events. N. Engl. J. Med. 2008;358(12):1240–1249. doi: 10.1056/NEJMoa0706728. [DOI] [PubMed] [Google Scholar]
- 103.Arnett DK, Baird AE, Barkley RA. Relevance of genetics and genomics for prevention and treatment of cardiovascular disease. Circulation. 2007;115(22):2878–2901. doi: 10.1161/CIRCULATIONAHA.107.183679. [DOI] [PubMed] [Google Scholar]
- 104.Humphries SE, Yiannakouris N, Talmud PJ. Cardiovascular disease risk prediction using genetic information (gene scores): Is it really informative? Curr. Opin. Lipidol. 2008;19(2):128–132. doi: 10.1097/MOL.0b013e3282f5283e. [DOI] [PubMed] [Google Scholar]
- 105.Quackenbush J. Microarray analysis and tumor classification. N. Engl. J. Med. 2006;354(23):2463–2472. doi: 10.1056/NEJMra042342. [DOI] [PubMed] [Google Scholar]
- 106.Michiels S, Koscielny S, Hill C. Prediction of cancer outcome with microarrays: A multiple random validation strategy. Lancet. 2005;365(9458):488–492. doi: 10.1016/S0140-6736(05)17866-0. [DOI] [PubMed] [Google Scholar]
- 107.Ntzani E, Ioannidis J. Predictive ability of DNA microarrays for cancer outcomes and correlates: An empirical assessment. Lancet. 2003;362(9394):1439–1444. doi: 10.1016/S0140-6736(03)14686-7. [DOI] [PubMed] [Google Scholar]
- 108.Simon R. Diagnostic and prognostic prediction using gene expression profiles in high-dimensional microarray data. Brit. J. Cancer. 2003;89:1599–1604. doi: 10.1038/sj.bjc.6601326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lee J, Lee JB, Park M, Song SH. An extensive comparison of recent classification tools applied to microarray data. Comput. Stat. Data Anal. 2004;48(4):869–885. [Google Scholar]
- 110.Thanassoulis G, Vasan RS. Genetic cardiovascular risk prediction will we get there? Circulation. 2010;122(22):2323–2334. doi: 10.1161/CIRCULATIONAHA.109.909309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Simon R, Radmacher MD, Dobbin K, McShane LM. Pitfalls in the use of DNA microarray data for diagnostic and prognostic classification. J. Natl. Cancer Inst. 2003;95(1):14–18. doi: 10.1093/jnci/95.1.14. [DOI] [PubMed] [Google Scholar]
- 112.Ambroise C, McLachlan G. Selection bias in gene extraction on the basis of microarray gene-expression data. Proc. Natl. Acad. Sci. USA. 2002;99(10):6562–6566. doi: 10.1073/pnas.102102699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Varma S, Simon R. Bias in error estimation when using cross-validation for model selection. BMC Bioinformat. 2006;7:91. doi: 10.1186/1471-2105-7-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Shi L, Campbell G, Jones WD, Campagne F, Wen Z, Walker SJ, Su Z, Chu T-M, Goodsaid FM, Pusztai L, Shaughnessy JD, Oberthuer A, Thomas RS, Paules RS, Fielden M, Barlogie B, Chen W, Du P, Fischer M, Furlanello C, Gallas BD, Ge X, Megherbi DB, Symmans WF, Wang MD, Zhang J, Bitter H, Brors B, Bushel PR, Bylesjo M, Chen M, Cheng J, Cheng J, Chou J, Davison TS, Delorenzi M, Deng Y, Devanarayan V, Dix DJ, Dopazo J, Dorff KC, Elloumi F, Fan J, Fan S, Fan X, Fang H, Gonzaludo N, Hess KR, Hong H, Huan J, Irizarry RA, Judson R, Juraeva D, Lababidi S, Lambert CG, Li L, Li Y, Li Z, Lin SM, Liu G, Lobenhofer EK, Luo J, Luo W, McCall MN, Nikolsky Y, Pennello GA, Perkins RG, Philip R, Popovici V, Price ND, Qian F, Scherer A, Shi T, Shi W, Sung J, Thierry-Mieg D, Thierry-Mieg J, Thodima V, Trygg J, Vishnuvajjala L, Wang SJ, Wu J, Wu Y, Xie Q, Yousef WA, Zhang L, Zhang X, Zhong S, Zhou Y, Zhu S, Arasappan D, Bao W, Lucas AB, Berthold F, Brennan RJ, Buness A, Catalano JG, Chang C, Chen R, Cheng Y, Cui J, Czika W, Demichelis F, Deng X, Dosymbekov D, Eils R, Feng Y, Fostel J, Fulmer-Smentek S, Fuscoe JC, Gatto L, Ge W, Goldstein DR, Guo L, Halbert DN, Han J, Harris SC, Hatzis C, Herman D, Huang J, Jensen RV, Jiang R, Johnson CD, Jurman G, Kahlert Y, Khuder SA, Kohl M, Li J, Li L, Li M, Li Q-Z, Li S, Li Z, Liu J, Liu Y, Liu Z, Meng L, Madera M, Martinez-Murillo F, Medina I, Meehan J, Miclaus K, Moffitt RA, Montaner D, Mukherjee P, Mulligan GJ, Neville P, Nikolskaya T, Ning B, Page GP, Parker J, Parry RM, Peng X, Peterson RL, Phan JH, Quanz B, Ren Y, Riccadonna S, Roter AH, Samuelson FW, Schumacher MM, Shambaugh JD, Shi Q, Shippy R, Si S, Smalter A, Sotiriou C, Soukup M, Staedtler F, Steiner G, Stokes TH, Sun Q, Tan P-Y, Tang R, Tezak Z, Thorn B, Tsyganova M, Turpaz Y, Vega SC, Visintainer R, Frese JV, Wang C, Wang E, Wang J, Wang W, Westermann F, Willey JC, Woods M, Wu S, Xiao N, Xu J, Xu L, Yang L, Zeng X, Zhang J, Zhang L, Zhang M, Zhao C, Puri RK, Scherf U, Tong W, Wolfinger RD. The microarray quality control (MAQC)-II study of common practices for the development and validation of microarray-based predictive models. Nat. Biotechnol. 2010;28(8):827–838. doi: 10.1038/nbt.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R, Eilbeck K, Lewis S, Marshall B, Mungall C. The gene ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32(Database issue):D258–D261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zeeberg B, Feng W, Wang G, Wang M, Fojo A, Sunshine M, Narasimhan S, Kane D, Reinhold W, Lababidi S, Bussey K, Riss J, Barrett J, Weinstein J. GoMiner: A resource for biological interpretation of genomic and proteomic data. Genome Biol. 2003;4:R28. doi: 10.1186/gb-2003-4-4-r28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Amberger J, Bocchini CA, Scott AF, Hamosh A. McKusick’s online Mendelian inheritance in man (OMIM§R ) Nucleic Acids Res. 2009;37(suppl 1):D793–D796. doi: 10.1093/nar/gkn665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Liu H, Liu W, Liao Y, Cheng L, Liu Q, Ren X, Shi L, Tu X, Wang QK, Guo AY. CADgene: A comprehensive database for coronary artery disease genes. Nucleic Acids Res. 2011;39(suppl 1):D991–D996. doi: 10.1093/nar/gkq1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Mlecnik B, Scheideler M, Hackl H, Hartler J, Sanchez-Cabo F, Trajanoski Z. PathwayExplorer: Web service for visualizing high-throughput expression data on biological pathways. Nucleic Acids Res. 2005;33(suppl 2):W633–W637. doi: 10.1093/nar/gki391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010;38(suppl 1):D355–D360. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Dahlquist KD, Salomonis N, Vranizan K, Lawlor SC, Conklin BR. GenMAPP, a new tool for viewing and analyzing microarray data on biological pathways. Nat. Genet. 2002;31(1):19–20. doi: 10.1038/ng0502-19. [DOI] [PubMed] [Google Scholar]
- 122.Wheelock CE, Wheelock ÅM, Kawashima S, Diez D, Kanehisa M, van Erk M, Kleemann R, Haeggström JZ, Goto S. Systems biology approaches and pathway tools for investigating cardiovascular disease. Mol. Biosyst. 2009;5(6):588–602. doi: 10.1039/b902356a. [DOI] [PubMed] [Google Scholar]
- 123.Sinnaeve PR, Donahue MP, Grass P, Seo D, Vonderscher J, Chibout SD, Kraus WE, Sketch M, Nelson C, Ginsburg GS. Gene expression patterns in peripheral blood correlate with the extent of coronary artery disease. PLoS Genet. 2009;4(9):e7037. doi: 10.1371/journal.pone.0007037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Mosig S, Rennert K, Büttner P, Krause S, Lütjohann D, Soufi M, Heller R, Funke H. Monocytes of patients with familial hypercholesterolemia show alterations in cholesterol metabolism. BMC Med. Genomics. 2008;1(1):60. doi: 10.1186/1755-8794-1-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Enquobahrie DA, Rice K, Williams OD, Williams MA, Gross MD, Lewis CE, Schwartz SM, Siscovick DS. IL1B genetic variation and plasma C-reactive protein level among young adults: The CARDIA study. Atherosclerosis. 2009;202(2):513–520. doi: 10.1016/j.atherosclerosis.2008.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Mamtani M, Matsubara T, Shimizu C, Furukawa S, Akagi T, Onouchi Y, Hata A, Fujino A, He W, Ahuja SK. Association of CCR2-CCR5 haplotypes and CCL3L1 copy number with Kawasaki disease, coronary artery lesions, and IVIG responses in Japanese children. PLoS One. 2010;5(7):e11458. doi: 10.1371/journal.pone.0011458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Carter C. Convergence of genes implicated in Alzheimer’s disease on the cerebral cholesterol shuttle: APP, cholesterol, lipoproteins, and atherosclerosis. Neurochem. Int. 2007;50(1):12–38. doi: 10.1016/j.neuint.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 128.Zheng Q, Wang XJ. GOEAST: A web-based software toolkit for Gene Ontology enrichment analysis. Nucleic Acids Res. 2008;36(suppl 2):W358–W363. doi: 10.1093/nar/gkn276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Caesar R, Fåk F, Bäckhed F. Effects of gut microbiota on obesity and atherosclerosis via modulation of inflammation and lipid metabolism. J. Intern. Med. 2010;268(4):320–328. doi: 10.1111/j.1365-2796.2010.02270.x. [DOI] [PubMed] [Google Scholar]
- 130.Cappola TP, Margulies KB. Functional genomics applied to cardiovascular medicine. Circulation. 2011;124(1):87–94. doi: 10.1161/CIRCULATIONAHA.111.027300. [DOI] [PMC free article] [PubMed] [Google Scholar]