Abstract
Leukotrienes (LTs) play major roles in lung immune responses, and LTD4 is the most potent agonist for cysteinyl LT1, leading to bronchoconstriction and tissue remodeling. Here, we studied LT crosstalk between myeloid cells and pulmonary epithelial cells. Monocytic cells (Mono Mac 6 cell line, primary dendritic cells) and eosinophils produced primarily LTC4. In coincubations of these myeloid cells and epithelial cells, LTD4 became a prominent product. LTC4 released from the myeloid cells was further transformed by the epithelial cells in a transcellular manner. Formation of LTD4 was rapid when catalyzed by γ-glutamyl transpeptidase (GGT)1 in the A549 epithelial lung cancer cell line, but considerably slower when catalyzed by GGT5 in primary bronchial epithelial cells. When A549 cells were cultured in the presence of IL-1β, GGT1 expression increased about 2-fold. Also exosomes from A549 cells contained GGT1 and augmented LTD4 formation. Serine-borate complex (SBC), an inhibitor of GGT, inhibited conversion of LTC4 to LTD4. Unexpectedly, SBC also upregulated translocation of 5-lipoxygenase (LO) to the nucleus in Mono Mac 6 cells, and 5-LO activity. Our results demonstrate an active role for epithelial cells in biosynthesis of LTD4, which may be of particular relevance in the lung.
Keywords: arachidonic acid, eicosanoid, inflammation, cancer, lung, macrophage/monocytes, extracellular vesicles, γ-glutamyl transpeptidase, 5-lipoxygenase
Leukotrienes (LTs) are mediators of inflammation (1) formed by immune cells in response to pathogens or danger signals. When cells are stimulated, cytosolic phospholipase A2 α (cPLA2α) and 5-lipoxygenase (LO) migrate to the nuclear membrane, arachidonic acid (AA) is released from nuclear membrane phospholipids, and LTA4 is produced (2). Efficient translocation and activation of 5-LO requires two scaffold proteins, 5-LO activating protein (FLAP) and coactosin-like protein (CLP) (3). LTA4 is then further metabolized by cytosolic LTA4 hydrolase to LTB4, or by LTC4 synthase (LTC4S) to LTC4. LTC4S is primarily located at the nuclear membrane together with FLAP, and both proteins are members of the family of membrane bound proteins involved in eicosanoid and glutathione metabolism (MAPEG). LTC4 and its metabolites, LTD4 and LTE4, are jointly referred to as the cysteinyl LTs (CysLTs). CysLTs elicit edema formation, mucus secretion, and smooth muscle contraction, as well as eosinophil trafficking and tissue remodeling, and thus contribute to symptoms in several chronic inflammatory diseases, such as asthma (4). There are at least three different receptors for the CysLTs; in the lung, bronchoconstriction and other effects are mediated via CysLT1. LTD4 has a 10- to 50-fold higher potency for activation of CysLT1 compared with LTC4, while LTE4 is the least active (5, 6). Thus, metabolism of LTC4 to LTD4 may have a considerable impact on CysLT1-mediated effects, also depending on further conversion to LTE4.
γ-Glutamyl transpeptidase (GGT)1 and GGT5 are cell surface enzymes that hydrolyze γ-glutamyl compounds, typically in glutathione metabolism, and also the conversion of LTC4 to LTD4 (7–10). In rat lung, GGT (probably GGT1) was found to be expressed in Clara cells of the bronchioles and in alveolar type II epithelium (11, 12). GGT activity and GGT1 mRNA was also found in the non-small lung cancer cell line, A549 (13, 14). In rat lung, exposure to NO2 increased GGT mRNA, protein, and enzyme activity (15) and GGT deficiency conferred oxidative stress in mouse lung (12). GGT is implicated in redox regulation in both normal and cancer cells (8), and recently, serum levels of GGT were associated particularly with liver cancer and also with lung cancer in males (16). Interestingly, part of the GGT in plasma is bound to exosomes, similar to GGT secreted from Helicobacter pylori-infected cells (17). Exosomes are nano-sized membrane vesicles that can transfer proteins, lipids, and nucleic acids between cells. It has been suggested that a specific function of GGT5 could be transformation of LTC4 to LTD4 (18). However, human GGT5 was less efficient compared with human GGT1 regarding formation of LTD4 in vitro (9). Recently, GGT5 was found in macrophages present in human tissues, most abundant in the lung (19).
Studies on human blood monocytes and peritoneal macrophages have shown that these cells produce mainly LTC4 and only small amounts of LTD4. This was found both in early work (20) and in recent lipidomic analyses (21–24). Likewise, eosinophils (25–27) and mast cells (28–32) produce mainly LTC4, but apparently no or only small amounts of LTD4. Here we show transcellular conversion of leukocyte-derived LTC4 to LTD4 by A549 lung cancer cells expressing both GGT1 and GGT5, and by normal human primary bronchial epithelial cells (PBECs) expressing GGT5. Epithelial cells have an active role in pathogenesis of asthma (33); our findings highlight the role of epithelium in biosynthesis of the bronchoconstrictor, LTD4.
MATERIAL AND METHODS
All materials were purchased from Sigma-Aldrich unless otherwise stated.
Cells
Mono Mac 6 (MM6) cells were cultured in RPMI 1640 medium with glutamine supplemented with 10% FBS, 100 mg/ml streptomycin, 100 U/ml penicillin, 1× nonessential amino acids, and oxalacetic acid, sodium pyruvate, and insulin, as described (34). The cells were maintained within a range of 0.2–1 × 106 cells/ml.
A549 cells were cultured on 10 cm Falcon tissue culture dishes (nr 353003) in Ham’s F12 medium supplemented with 10% FBS, 100 mg/ml streptomycin, and 100 U/ml penicillin. IL-1β was added as indicated. Cells were split regularly before confluence, typically every 3–4 days.
Monocyte-derived dendritic cells (MDDCs) were differentiated from primary monocytes isolated from buffy coats of healthy human donors (Karolinska Hospital Blood Bank). PBMCs, obtained by Ficoll-Paque PREMIUM (GE Healthcare) gradient centrifugation, were seeded in multi-well cell culture plates for 2 h, allowing monocytes to adhere to plastic. The plates were washed twice with PBS to remove lymphocytes. To obtain MDDCs, monocytes were differentiated for 7 days in RPMI 1640 medium with glutamine supplemented with 10% FBS, 100 mg/ml streptomycin, 100 U/ml penicillin, 1× nonessential amino acids, 25 mM HEPES, and rhGM-CSF (10 ng/ml; Sigma) plus IL-4 (10 ng/ml, Invitrogen). On day 3 and day 6, cells were resupplied with the cytokines. To enhance LTC4S activity in MDDCs, TGFβ was added on day 0 at a final concentration of 2 ng/ml (35). Eosinophils were isolated from two female donors. Following Ficoll gradient centrifugation, eosinophils were prepared from the PMNL fraction using an eosinophil isolation kit (Milteny Biotech, Bergisch Gladbach, Germany).
PBECs were harvested in connection with lobectomy, as earlier described (36) (ethical approval 99-357 from Karolinska Institutet research ethics committee North at Karolinska Hospital). The PBECs were cultured at a density of 1–2 × 106 on T75 tissue culture flasks (ThermoFisher Scientific, Waltham, MA) that were precoated with coating buffer [fibronectin (Gibco, Life Technologies, Paisley, UK), BSA fraction V, vitrogen 100 collagen, and PBS without Ca2+/Mg2+]. The cells were maintained in keratinocyte serum-free medium (Gibco) supplied with human recombinant epidermal growth factor (5 ng/ml; Gibco), bovine pituitary extract (50 ug/ml; Gibco), and penicillin and streptomycin antibiotics (BioWhittaker, Lonza, Basel, Switzerland). The cultures were kept in 5% CO2 at 37°C and medium was changed every second day. At confluence, the cells were detached by treatment with trypsin/EDTA solution (0.03/0.01% in PBS without Ca2+/Mg2+; Gibco). The number of cells was counted and the viability (>95%) was assessed using exclusion of trypan blue dye. The PBECs used in this study were at passage 5 and passage 6 from three different donors.
Exosome isolation
Exosomes were isolated as described (35), with minor changes. Briefly, A549 cells were grown to confluence for 48 h in complete medium with exosome-depleted FBS (centrifuged at 100,000 g ON) and IL-1β (5 ng/ml). The cell culture supernatant was collected (typically 400 ml) and centrifuged at 3,000 g for 30 min, 10,000 g for 30 min, and 100,000 g for 2 h. The pellet was washed in PBS and centrifuged again at 100,000 g for 2 h. Finally, the pellet was resuspended in 1 ml of PBS and protein concentration (0.5–1 mg/ml) was measured by Bradford assay (Bio-Rad protein reagent). The average yield was 2.8 μg exosomal protein per 106 A549 cells. Exosomes were further characterized by nanoparticle tracking analysis and FACS, as described (37).
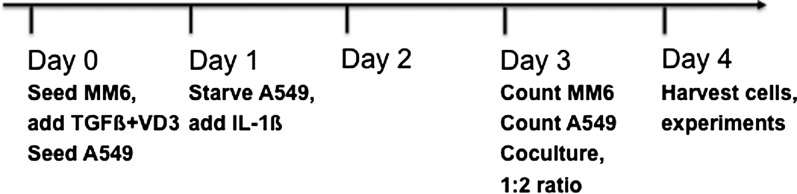
Timeline of cell culture and coculture
MM6 and A549 cells were treated and cocultured as shown in Fig. 1. Briefly, on day 0, MM6 cells were seeded at a density of 0.2 × 106/ml and differentiation was started by addition of TGFβ (2 ng/ml) and 1,25-dihydroxyvitamin D3 (50 nM) (38). A549 cells were seeded at 0.5 × 106 cells per plate. On day 1, A549 cells were washed twice with PBS, and 10 ml of Ham’s F12 medium containing 2% FBS and IL-1β (1 ng/ml) was added to starve and stimulate the cells. On day 3, cocultures were started. First the number of A549 cells on the separate counting plate was determined after detachment with trypsin/EDTA (typically 2–3 × 106 cells). Inspection under the microscope consistently indicated that similar cell numbers were present on the other A549 dishes in that experiment. Also, the MM6 cell count was determined on day 3 (typically 0.3–0.7 × 106 cells/ml). MM6 cell suspension was added to A549 dishes to achieve a 1:2 MM6/A549 ratio. Thus, around 2–3 ml of MM6 cell suspension was added to the medium on A549 dishes (starvation medium kept since day 1). On day 4, all cells were harvested. MM6 cells not subjected to coculture were thus kept in differentiation culture (with TGFβ + VD3) for 4 days, and A549 cells not subjected to coculture were grown in the presence of IL-1β for 3 days. Morphology and trypan blue exclusion indicated that A549 and MM6 cells were in good condition during coculture, there were no signs of reduced cell viability.
Fig. 1.
Timeline of MM6 and A549 cell treatments and coculture.
Cell incubations
On day 4, the MM6 cultures, A549 cultures, and MM6-A549 cocultures were collected. MM6 cells were counted and then centrifuged at 150 g for 5 min. The final pellet was resuspended in 0.5 ml PBS for incubations with LTA4 or PGC buffer for incubations with A23187 (PGC is PBS containing 1 mg/ml glucose and 1 mM CaCl2). A549 cells were washed twice with PBS and then detached using trypsin/EDTA. After counting, the cells were centrifuged at 150 g for 5 min and the final pellet was resuspended in 0.5 ml PBS or PGC. Cocultures were detached by scraping and centrifuged at 150 g for 5 min. The final pellet was resuspended in 0.5 ml PBS or PGC. The coincubations (0.5 ml) contained 2–3 × 106 A549 cells and 1–1.5 × 106 MM6 cells.
Cells were incubated with Ca2+ ionophore A23187 at two different conditions. In condition 1, cells were pretreated with 100 nM PMA for 10 min at 37°C and subsequently incubated with 5 μM A23187 for 10 min at 37°C. In condition 2, cells were incubated with 40 μM AA together with 5 μM A23187 for 10 min at 37°C. The amount of ethanol (solvent for A23187, AA) did not exceed 0.2% (v/v). The reaction was stopped by adding 0.5 ml of methanol containing internal standards (normally 250 pmol PGB2 and 250 pmol 17-OH-C22:4, kind gifts from Mats Hamberg, Karolinska Institutet) and kept on ice or at −20°C for at least 1 h. For the coincubations, formation of eicosanoids is given per million of MM6 cells present.
Cells were incubated with LTA4 (20 μM) for 5 min at 37°C. LTA4 was added in ethanol (1–2 μl). The reactions were stopped as described above. For LTA4 incubations of primary leukocytes together with A549 cells, the incubation time was 15 min.
Analysis of LTs and 5-HETE
After precipitation of proteins, the samples were centrifuged at 10,000 g for 10 min at 4°C and supernatants (approximately 1 ml) collected and added to 2 ml citrate/phosphate buffer (0.1 and 0.2 mM, respectively, pH 5.6). Extraction was performed using a C18 column (Supelco, Belfonte, PA) preconditioned with 3 ml methanol and 1 ml water. After application of the 3 ml sample, the column was washed with 1 ml water and finally oxylipins were eluted with 300 μl methanol followed by 300 μl water. The methanol and water eluates were pooled and aliquots analyzed by reverse-phase HPLC. In test extractions of mock samples (known amounts of PGB2 and LTC4 added to PBS buffer), the recovery of PGB2 was 85% and of LTC4 was 83%. HPLC was performed with a C18 column [Phenomenex Luna, 5 u C18(2) 100A, 2 × 150 mm] eluted at a flow rate of 0.8 ml/min. For LT analysis, the mobile phase was water:methanol:acetonitrile:acetic acid (43:27:30:0.1, pH 5.6 adjusted with NH3) and UV absorbance at 280 nm was monitored. For analysis of the less polar mono-HETEs, the mobile phase was water:acetonitrile:acetic acid (40:60:0.1) and UV absorbance at 235 nm was monitored. LTs and 5-HETE were quantitated from peak areas and extinction coefficients in relation to the internal standards (PGB2 for LTs; 17-OH-C22:4 for 5-HETE).
Conversion of LTC4 to LTD4 by A549 cells
A549 cells were harvested on day 4 (Fig. 1). After centrifugation (150 g for 4 min), around 16 × 106 cells were resuspended in 3 ml PBS and incubated with 0.36–0.56 μM LTC4 (Cayman Chemicals). At the indicated intervals (1, 2, 3, 5, 15, and 30 min), aliquots of 0.5 ml containing 2.7 × 106 cells were removed and added to a tube containing 0.5 ml methanol and 125 pmol of PGB2 in order to stop the reaction. Extraction and analysis were performed as described above. Two incubation series were performed in each experiment.
GGT1 inhibition with serine-borate complex
Serine-borate complex (SBC) is a competitive inhibitor of GGT1 (39). Cells in 0.5 ml of PBS or PGC buffer were treated with 20 μl of a solution of equimolar L-serine (0.25 M) and sodium borate (0.25 M) to give a concentration of 10 mM SBC. Cells were treated with SBC for 5–10 min at 37°C before incubations with LTA4 or PMA plus A23187.
SDS-PAGE and Western blotting
SDS-PAGE and Western blotting were performed essentially as described (34) using Bio-Rad 4–20% ready-made gels and nitrocellulose membranes (Amersham). The following antibodies were used: in-house antisera (rabbit polyclonal) against 5-LO (1:3,000 dilution), FLAP (1:300 dilution), LTA4H (1:2,500 dilution), LTC4S (1:750 dilution), and mPGES-1 (1:3,000 dilution). A mouse monoclonal against GGT1 (used at 1:500 dilution) was from Santa Cruz Biotechnology, Santa Cruz, CA. A rabbit polyclonal against GGT5 (used at 1:200 dilution) was from Abcam, Cambridge, UK. A peroxidase-conjugated primary antibody against β-actin (1:2,000 dilution) and peroxidase-conjugated secondary antibodies (1:5,000 dilution) were from Sigma. Protein bands were detected by enhanced chemiluminescence; after scanning, relative amounts were calculated with ImageJ software.
Subcellular fractionation by detergent lysis
MM6 cells, differentiated with TGFβ and VD3 for 96 h, were subjected to subcellular fractionation after NP-40 lysis, as described (34). Histone H4, a nuclear protein, was used as marker for correct fractionation.
Immunofluorescence microscopy
Differentiated MM6 cells were harvested and resuspended in 1 ml PGC buffer. Cells (1–2 × 106) were resuspended in 1 ml PGC buffer and stimulated, in order, with SBC (10 mM), PMA (100 nM), and A23187 (5 μM) for 10 min, 10 min, and 5 min, respectively. Unstimulated control cells received only vehicle. Samples were immediately chilled on ice for 5 min and centrifuged onto glass slides at 4°C. The cells were then fixed with methanol at −20°C for 4 min and washed four times with PBS. Samples were blocked with PBS containing 1% BSA and 0.1% Tween-20 for 1 h at room temperature and incubated with anti-5-LO antibody overnight at 4°C. Then samples were washed four times with PBS, incubated with Cy3 goat anti-rabbit IgG (Invitrogen) for 1 h at room temperature, and washed five times with PBS. The samples were coated with two drops of aqueous antifade gel mounting medium (Vector Laboratories) with DAPI under coverslips. The fluorescent signal was observed with an Olympus FluoView FV1000 microscope system.
Data analysis
Student’s t-test was performed, P < 0.05 was considered statistically significant.
RESULTS
Formation of 5-LO products in coincubations of MM6 and A549 cells
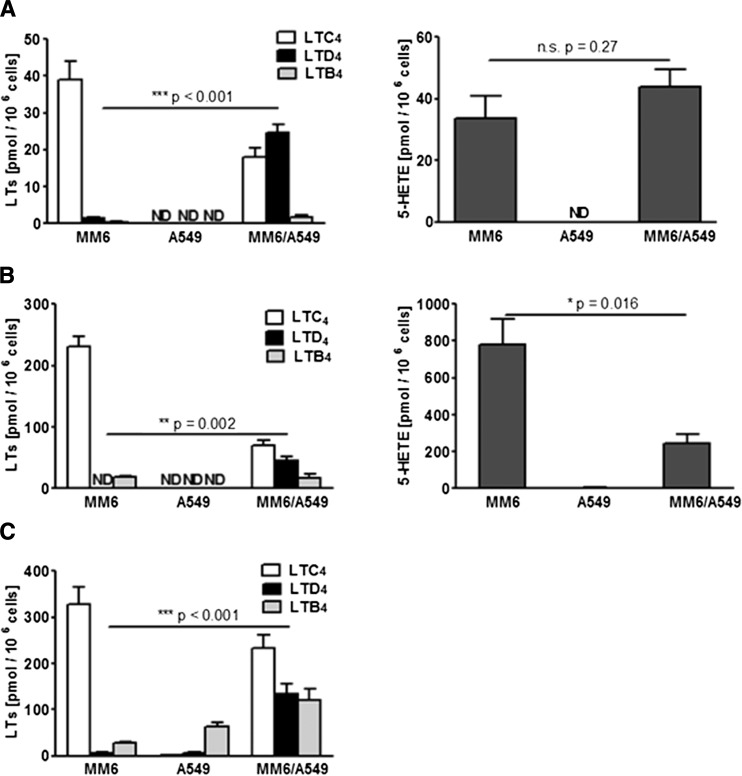
MM6 cells were differentiated with TGFβ and VD3 for 72 h, which leads to upregulation of 5-LO (2), and A549 cells were starved and stimulated with IL-1β for 48 h. The cells were then combined and cocultured for 24 h before different incubations and determinations of LT and 5-HETE biosynthesis, see Fig. 1.
For MM6 cells, it was observed before that priming with PMA strongly upregulated ionophore-induced translocation of 5-LO, as well as product formation from endogenous AA (34). In such incubations of MM6 cells alone, the major 5-LO product was LTC4, closely followed by 5-HETE, see Fig. 2A. Only minute amounts of LTB4 and LTD4 were formed. When MM6 cells and A549 cells were cocultured (cell ratio 1:2) and incubated together (priming with 100 nM PMA for 10 min followed by 5 μM A23187 for 10 min), the major difference was that LTD4 also, but not LTE4, appeared (Fig. 2A). Concomitantly, the recovery of LTC4 was reduced (Fig. 2A). LTB4 was slightly increased; nevertheless, LTB4 was still a minor product. Formation of 5-HETE was similar as incubations of MM6 cells alone, and no other mono-HETEs appeared. Finally, when A549 cells alone were incubated with PMA plus A23187, no LTs or HETEs were detected.
Fig. 2.
Formation of 5-LO products in different incubation conditions. MM6 monocultures, A549 monocultures, and MM6/A549 cocultures (ratio 1:2) were prepared as described in the Materials and Methods (see also Fig 1). On day 4, cells were harvested and different incubations were performed. After solid phase extraction, LTs and 5-HETE were analyzed by HPLC. In coincubations, product formation is given as picomoles per million MM6 cells. A: Cells were primed with PMA (100 nM) for 10 min and then stimulated with ionophore A23187 (5 μM) for 10 min (n = 9). B: Cells were stimulated with ionophore A23187 (5 μM) and AA (40 μM) for 10 min (n = 5). C: Cells were incubated with LTA4 (20 μM) for 5 min (n = 10). Results are given as mean ± SE, Student’s t-test. n.s., not significant; ND, not detected.
In incubations of MM6 cells with ionophore A23187 (5 μM) together with exogenous AA (40 μM), formation of all 5-LO products was strongly increased compared with incubations with PMA plus ionophore (Fig. 2B). When MM6 cells and A549 cells were cocultured (cell ratio 1:2) and incubated together with A23187 plus exogenous AA, again LTD4 appeared, LTE4 was not observed, and LTB4 formation was approximately the same as with MM6 cell monocultures. A clear difference compared with incubations of MM6 cells alone was that the yield of 5-HETE decreased considerably (Fig. 2B). Again, A549 cells alone did not produce LTs or HETEs.
Cells were also incubated with LTA4 (20 μM). With MM6 cells alone, this gave the highest production of LTC4, a very small amount of LTD4, and a minor amount of LTB4 (less than 10% of the LTC4 formation) (Fig. 2C). In coincubations of MM6 cells and A549 cells, LTD4 was produced and formation of LTB4 now also increased. This was most probably due to LTA4 hydrolase activity in A549 cells, as LTB4 formation was also evident in the incubations of A549 cells alone with LTA4 (Fig. 2C).
For increased formation of LTD4, it was required to have MM6 and A549 cells together. In control experiments, MM6 cells were cultured with addition of conditioned medium from IL-1β-treated A549 cells. In subsequent incubations with PMA and ionophore, or with LTA4, there was no increased LTD4 production and no appreciable effect on the other LTs or 5-HETE. Also, when A549 cells were cultured with MM6 cell-conditioned medium (TGFβ and VD3 added), there was no effect on their capacity to transform LTC4 to LTD4 and there was no induced 5-LO activity (data not shown). In the incubations of A549 cells with LTA4, one of the nonenzymatic LTA4 hydrolysis products (12-epi-6-trans-LTB4), particularly, was further metabolized to the 5-oxo derivative with UV maximum at 318 nm (40).
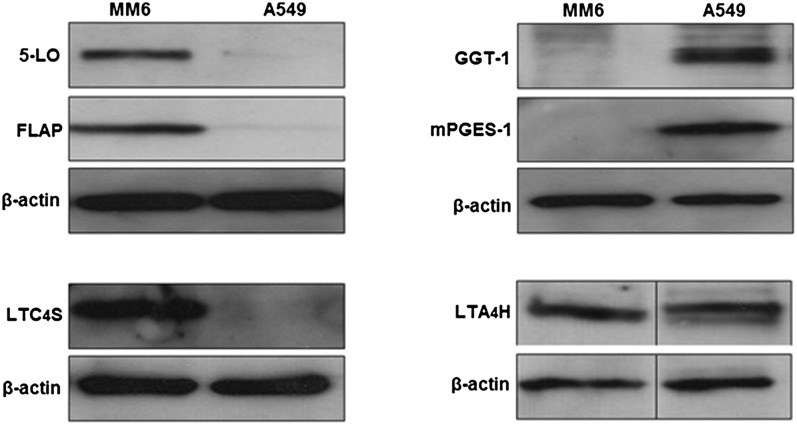
Expression of enzymes for LT biosynthesis in MM6 and A549 cells
MM6 cells were differentiated with TGFβ and VD3, and A549 cells were starved and stimulated with IL-1β. The 10,000 g supernatants from total cell lysates were analyzed by Western blots. The 5-LO, FLAP, and LTC4S were expressed only in differentiated MM6 cells; these proteins were not detected in A549 cells (Fig. 3). On the other hand, GGT1 and mPGES-1 were only found in A549 cells; these proteins were not expressed in MM6 cells. For GGT1, the small subunit was analyzed and at least two bands were observed in the 20–25 kDa range, in accordance with previously observed glycosylation (41). Finally, LTA4 hydrolase was about equally expressed in differentiated MM6 cells and in A549 cells. The exclusive expression of 5-LO and LTC4S in MM6 cells, and of GGT1 in A549 cells, agrees with both cell types being required for biosynthesis of LTD4.
Fig. 3.
MM6 cells were differentiated with TGFβ and VD3, and A549 cells were starved and stimulated with IL-1β. Supernatants (10,000 g) from total cell lysates were analyzed by Western blots, as described in the Materials and Methods. The 50 μg protein samples were applied to the SDS-PAGE gels; β-actin was control for equal loadings. For each protein, similar results were observed in at least three different experiments.
Export of LTC4 from myeloid cells
The relative amount of LTC4 that was further converted to LTD4 was quite different for the three different coincubation conditions, see Fig. 2. In incubations with LTA4, the highest amount of CysLT was formed (360 pmol/106 MM6 cells) and, of this, about 37% was LTD4. In incubations with A23187 and AA, 110 pmol CysLT was formed per 106 MM6 cells and, of this, about 40% was LTD4. Finally, in incubations with PMA plus A23187, 42 pmol CysLT was formed per 106 MM6 cells and, of this, about 60% was LTD4. Cell numbers and incubation times were the same in all these incubations. The result from the incubations with LTA4 indicated a high capacity of the A549 cells to convert LTC4 to LTD4, which should be more than sufficient for all CysLT to be metabolized to LTD4 in the incubations with ionophore plus AA and with PMA plus ionophore. However, this was not the case. This could depend on the release of LTC4 from MM6 cells, which would determine conversion to LTD4 by the A549 cells.
To investigate this, MM6 cells were incubated as described above, but before stopping the reactions, cells and media were separated by quick centrifugation and intra-/extracellular LTC4 was analyzed (Table 1). For MM6 cells incubated with PMA and ionophore or with LTA4, substantial shares of the LTC4 formed remained associated to the cell fractions and would, thus, not be available for metabolism by A549 cells. The degree of extracellular LTC4 (Table 1) agrees with the further conversions to LTD4 in coincubations (compare Fig. 2). For MDDCs and eosinophils, the amount of extracellular LTC4 was about 60%, seemingly compatible with the relative amounts of LTD4 formed in coincubations (Fig. 6). It appears that export of LTC4 from the myeloid cells is one factor determining LTD4 formation in coincubations.
TABLE 1.
Release of LTC4 from MM6 cells, MDDCs, and eosinophils
| LTC4 (pmol/106 cells) | |||
| Intracellular | Extracellular | Sum | |
| MM6 incubated LTA4 | 163 ± 21 | 90 ± 5 | 253 |
| MM6 incubated PMA + A23187 | 9 ± 1 | 17 ± 1 | 26 |
| MDDCs incubated LTA4 | 23 ± 10 | 33 ± 7 | 56 |
| Eosinophils incubated LTA4 | 28 ± 7 | 44 ± 12 | 72 |
Release of LTC4 from MM6 cells, MDDCs, and eosinophils was analyzed after incubations with LTA4 (20 μM) for 5 min at 37°C (n = 4–5). MM6 cells were incubated with PMA also (100 nM, 10 min) followed by A2387 (5 μM, 10 min) (n = 6). The incubations were stopped by centrifugation at 4°C (3 min at 1,000 g). Supernatants (extracellular) were rapidly removed and added to 0.5 ml methanol containing 250 pmol PGB2, while the cell pellet was resuspended in 1 ml methanol:water (1:1), with 250 pmol PGB2. After extraction, LTC4 was analyzed by HPLC. Results are mean ± SE.
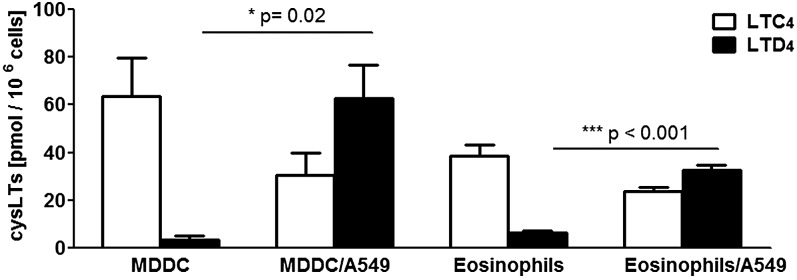
Fig. 6.
CysLT formation in coincubations of A549 cells with MDDCs and eosinophils. The primary cells were mixed in suspension with A549 cells (leukocyte to A549 ratios 1:2 to 1:4) and coincubated for 15 min with LTA4 (20 μM). Before the incubations, the A549 cells had been starved and stimulated with IL-1β for 72 h. After solid phase extraction, LTs were analyzed by HPLC. In coincubations, product formation is given as picomoles per million of MDDCs or eosinophils. The MDDC to A549 cell ratio is 1:4. The eosinophils to A549 cell ratio is 1:2. Leukocytes were prepared from two different donors and incubations were performed in duplicate. Values are mean ± SE, Student’s t-test (n = 4).
Conversion of exogenous LTC4 to LTD4 by A549 cells
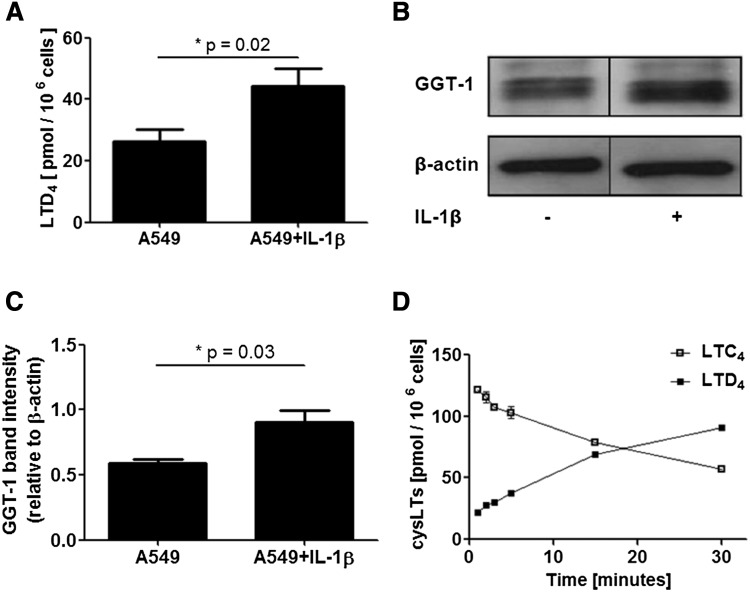
When A549 cells (approximately 3 × 106 cells in 0.5 or 1 ml of PBS) were incubated with exogenous LTC4 (0.36–0.56 μM) for 5 min, LTD4 formation was 25–50 pmol/106 cells. In all these incubations, LTC4 was in excess; more than half of the recovered CysLT was LTC4. Cells treated with IL-1β produced about 2-fold more LTD4 compared with untreated cells (Fig. 4A), and in the Western blot, the GGT1 band intensity was increased (Fig. 4B, C). In a time course experiment, 35 × 106 cells (in 6.5 ml PBS) were incubated with 2.6 nmol of LTC4 (0.4 μM). As shown in Fig. 4D, after 15 min almost half of the substrate had been converted and at 30 min about 60% of the CysLT was LTD4. Practically no LTE4 was formed; at 30 min, less than 1% of the CysLT was LTE4.
Fig. 4.
Conversion of exogenous LTC4 to LTD4 by A549 cells. A: Cells were cultured with or without IL-1β (1 ng/ml) as indicated. On day 4, cells were harvested and incubated with LTC4 (0.36–0.56 μM) for 5 min. Results are given as mean ± SE (n = 6). B: GGT1 Western blot analysis of cells treated (or not) with IL-1β (1 ng/ml) for 72 h. Samples are 40 μg aliquots of 10,000 g supernatants from total cell lysates. C: GGT1 normalized to β-actin band intensity, measured in three independent experiments (n = 3). D: An incubation of A549 cells treated with IL-1β was followed over time. Cells (35 × 106) in 6.5 ml PBS were incubated with 0.4 μM LTC4. At the indicated intervals (1, 2, 3, 5, 15, and 30 min), two aliquots of 0.5 ml were removed and added to separate tubes containing 0.5 ml methanol and 125 pmol of PGB2. Following extractions, CysLTs were analyzed by HPLC. Thus, for each time point two samples were analyzed.
The formation of LTD4 from exogenous LTC4 in these experiments (25–50 pmol/106 A549 cells at 5 min, Fig. 4A, C) is in the same range as the formation of LTD4 in coincubations of MM6 and A549 cells with LTA4 (Fig. 2C). In the coincubations, the sum of CysLT produced per 106 MM6 cells during 5 min was 360 pmol. Of this, 130 pmol was further converted to LTD4 by 2 × 106 A549 cells (MM6 to A549 cell ratio 1:2). Thus, in the coincubations, the formation of LTD4 was 65 pmol/106 A549 cells. The similar yields of LTD4 from exogenously added LTC4 and from LTC4 produced by MM6 cells indicates that there was no special role for MM6-A549 cell to cell interactions in the further metabolism of LTC4 to LTD4.
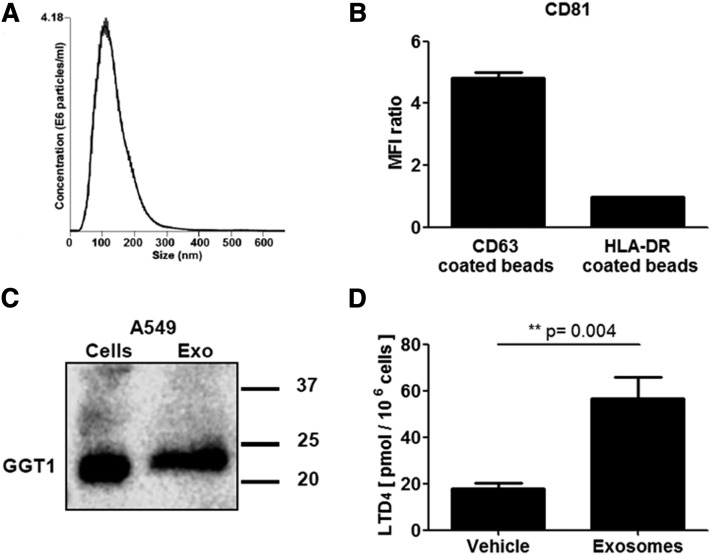
GGT1 in exosomes from A549 cells
Exosomes prepared from A549 cells were of the expected size (Fig. 5A) and carried the tetraspanins, CD63 and CD81, characteristic for exosomes (Fig. 5B). By Western blot, GGT1 was detected in both A549 cells and A549-derived exosomes (Fig. 5C). Conversion of LTA4 was determined in coincubations of MM6 cells with exosomes from A549 cells. When cells and exosomes were mixed and incubated directly with LTA4, the yield of LTD4 increased (Fig. 5D). Exosomes were enriched from A549-conditioned medium, and it should be observed that in the final preparation the exosome concentration was increased considerably (>100-fold). We could not detect GGT1 activity in the control experiments of MM6 together with A549-conditioned medium. Furthermore, when MM6 cells were cultured with exosomes for a long time (24 h) before incubation with LTA4, LTD4 was not increased (data not shown), possibly indicating degradation of exosomal contents.
Fig. 5.
GGT1 in exosomes from A549 cells treated with IL-1β. A: Size range of A549 exosomes determined by nanoparticle tracking analysis. B: A549-derived exosomes carry CD81 and CD63. Beads coated with CD63 or HLA-DR antibody were added to exosomes (corresponding to 5 μg of exosomal proteins) in 500 μl PBS and incubated overnight at room temperature. After washing, the beads were incubated with FITC-labeled CD81 antibody and subjected to FACS analysis. C: A549 cells were starved and stimulated with IL-1β. An aliquot (20 μg) of a 10,000 g supernatant (from total cell lysate) and exosomes corresponding to 16 μg of protein were analyzed for GGT1 by Western blot. D: Formation of LTD4 in coincubations of MM6 cells (1 × 106) and A549 exosomes (100 μg of exosomal protein) in 1 ml PBS. Incubated with LTA4 (10 μM) for 15 min (n = 6).
SBC inhibits formation of LTD4 in MM6/A549 coincubations
GGT is inhibited by SBC (39) and SBC was 8-fold more potent to inhibit purified GGT1 (Ki 0.5 mM) compared with purified GGT5 (Ki 4.2 mM) (9). When MM6 and A549 cells were coincubated with LTA4, both LTC4 and LTD4 were formed. In cells pretreated with SBC (10 mM, 10 min) before addition of LTA4, formation of LTD4 was undetectable (supplemental Fig. S1A). A similar result was obtained when cells were stimulated with PMA plus ionophore A23187 (supplemental Fig. S1B). These observations support that, in the MM6/A549 coincubations, LTC4 is converted to LTD4 by GGT1. Interestingly, in the incubations with PMA plus A23187, SBC also had another effect: the total formation of CysLTs (LTC4) increased in comparison to CysLTs in cells not receiving SBC (LTC4 plus LTD4). This was not observed for cells incubated with LTA4.
SBC upregulates 5-LO activity and translocation in MM6 cells
To investigate the SBC-induced upregulation of CysLTs in cells incubated with PMA plus A23187, MM6 cells alone were pretreated with SBC for 5 min. When these cells were stimulated with PMA plus ionophore A23187, pretreatment with SBC increased formation of both LTC4 and 5-HETE 2- to 3-fold (supplemental Fig. S2A, B). Human monocytes isolated from peripheral blood showed a similar increase in 5-HETE formation when pretreated with SBC (supplemental Fig. S2D). This indicated that SBC could upregulate 5-LO activity. One factor determining the activity of 5-LO in cells is translocation to the nuclear membrane. This was investigated first by subcellular fractionation. Thus, MM6 cells were lysed with NP-40 and 5-LO was analyzed in nuclear and nonnuclear fractions by Western blot. As shown in supplemental Fig. S2C, in unstimulated control cells, 5-LO was recovered in the soluble nonnuclear fraction, while stimulation with PMA plus A23187 resulted in 5-LO in both nuclear and nonnuclear fractions. When cells were pretreated with SBC (5 min) before stimulation with PMA plus A23187, the majority of 5-LO was recovered in the nuclear fraction. When MM6 cells were treated with SBC alone, there was no change in the subcellular localization of 5-LO (data not shown). MM6 cells were analyzed also by immunocytochemistry. As shown in supplemental Fig. S3, pretreatment with SBC seemed to give a more pronounced accumulation of 5-LO to the perinuclear membrane, as compared with stimulation with only PMA plus A23187. These findings indicate that SBC can prime the cells for more efficient translocation of 5-LO, possibly due to effects of GGT on cellular redox status.
Formation of CysLT in coincubations of A549 cells with MDDCs and eosinophils
To validate the results in primary cells, MDDCs and eosinophils were prepared from two donors each. These primary cells were mixed in suspension with scraped A549 cells (leukocyte to A549 ratios 1:2 to 1:4) and coincubated for 15 min with LTA4 (20 μM). Prior to incubations, the A549 cells had been starved and stimulated with IL-1β for 72 h. As shown in Fig. 6, the dominating CysLT formed in the leukocytes alone was LTC4, while in the coincubations, the yield of LTD4 increased substantially. In the incubations of MDDCs and eosinophils (both known to express 15-LO) with LTA4, considerable amounts of lipoxins (four isomers) were also formed.
Formation of CysLT in coincubations of PBECs with MM6 cells
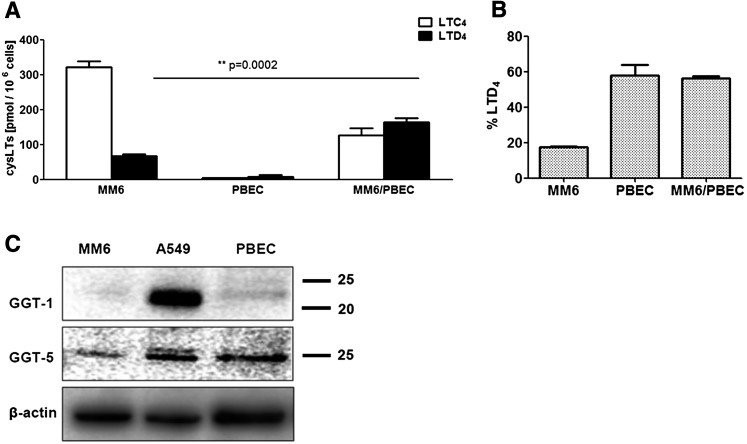
To further validate the biological relevance, we determined whether normal human PBECs could contribute to formation of LTD4. In preliminary experiments, adherent PBECs (3 × 106 cells, under DMEM) were incubated with LTC4 (0.35 μM). After 1 h, about 6% of the CysLT recovered in the cell culture medium was LTD4; after 6 h, this had increased to 33%. Similar results were obtained for PBECs treated with IL-1β (10 ng/ml, 24 h). In these incubations 11-trans isomers of LTC4 and LTD4 appeared, but further conversion to LTE4 was not observed. Also, in coincubations of PBECs and MM6 cells (incubated with LTA4), a long incubation time was needed for LTD4 to appear (Fig. 7A). In these 6 h incubations, conversion of LTC4 to LTD4 was also observed in MM6 cells alone; however, the presence of PBECs also increased the yield of LTD4 considerably (Fig. 7B). The slow metabolism of LTC4 is compatible with expression of GGT5 in both MM6 cells and in PBECs, as found by Western blot (Fig. 7C). Human bronchial epithelium can express 5-LO pathway enzymes and formation of LTC4 has been demonstrated in 6 h incubations (42). We also found formation of CysLt from LTA4 in PBECs (about 10 pmol/106 cells, 6 h incubation time) (Fig. 7A) and most of this was LTD4.
Fig. 7.
CysLT formation in coincubations of PBECs with MM6 cells. PBECs were cultured as described in the Materials and Methods, yielding approximately 3 × 106 PBECs in each T75 tissue culture flask. The cell culture medium was removed and replaced with 4 ml of DMEM (20 mM glutamine) containing differentiated MM6 cells (0.5 × 106 cells). The MM6 to PBEC ratio was 1:6. LTA4 (10 μM) was added and the incubation was kept in the cell culture cabinet for 6 h. After solid phase extraction, LTs were analyzed by HPLC. A: Yields of LTC4 and LTD4 in the incubations of MM6 cells alone, PBECs alone, and in coincubations (n = 4). In coincubations, product formation is given as picomoles per million MM6 cells. B: Percentage of LTD4 in the respective incubations. Values are mean ± SE, Student’s t-test (n = 4). C: Western blot analysis of GGT1 and GGT5. MM6 cells were differentiated with TGFβ and VD3. A549 cells were starved and stimulated with IL-1β. PBECs were cultured as described in the Materials and Methods with epidermal growth factor and bovine pituitary extract. Supernatants (10,000 g) from total cell lysates were analyzed by Western blots as described in the Materials and Methods. Protein samples (25 ug) were applied to the SDS-PAGE gel; β-actin served as loading control.
DISCUSSION
We determined LT biosynthesis in coincubations of monocytic cells (MM6, MDDCs) and eosinophils with epithelial A549 cells or primary epithelial cells. Our study confirms and extends a role for epithelial cells in LTD4 biosynthesis in the lung environment. Conversions of exogenous LTC4 by epithelial cells has been studied previously (43, 44), but to our knowledge, metabolism of leukocyte-derived LTC4 was not demonstrated before in coincubations of monocytic and epithelial cells. The non-small cell lung cancer cell line, A549, transformed leukocyte-derived LTC4 to LTD4 within minutes; while for normal PBECs, long incubation times (6 h) were required. The slow LTC4 to LTD4 conversion by PBECs is in accordance with these cells expressing GGT5, while A549 cells also express GGT1, which is considerably faster (9). Treatment with proinflammatory IL-1β upregulated the capacity of A549 cells for conversion of LTC4 to LTD4, and GGT1 was also present in exosomes from these cells.
GGT enzymes are present on epithelial cells in many tissues (7). GGT activity can also be expressed in leukocytes, e.g., in U937 cells (45), and accordingly, LTD4 was formed in short-term (A23187, 10 min) incubations of U937 cells differentiated with DMSO (46). LTD4 was also produced in RBL-1 cells (47). GGT1 was not detectable in MM6 cells, in line with the very small amounts of LTD4 in 15 min incubations of MM6 cells alone. This is in agreement with several previous studies on human blood monocytes and human peritoneal macrophages using incubation times up to 60 min [see (20) and references therein]. Also, in more recent lipidomic analyses of mouse peritoneal macrophages (21) and RAW264.7 murine macrophages (22, 23), LTC4 and 5-HETE (but not LTD4) were major 5-LO products. In a recent study of macrophages, FACS-sorted from a mouse lung tumors, LTC4 was more than 10-fold more abundant than LTD4 (24). In all these studies, incubation times were at most 60 min. Likewise, eosinophils (25–27) and mast cells (28–32) produce mainly LTC4, but apparently no or only small amounts of LTD4. Interestingly, in mast cells cocultured with 3T3 fibroblasts [lacking GGT (48)], LTC4 was also the major CysLT produced (29). Our results with MM6, MDDCs, and eosinophils support the concept that LTC4 is the predominating CysLT formed in short-term (5–15 min) incubations of these cells. At the same time, LTD4 was formed in long-term incubations of MM6 cells, in agreement with these cells (Fig. 7), as human macrophages (19), expressing GGT5. However, in previous experiments with lung tissue, induced formation of CysLT including LTD4 was quick (10–15 min) [see (49)], suggesting involvement of GGT1. Thus, it appears that GGT1 can be expressed in normal human lung; candidates are Clara cells and alveolar type II epithelium, which were previously found to express GGT in rat lung (11, 12). The A549 cell line has alveolar type II-like characteristics (50). However, human PBECs, which differ from alveolar epithelium in several respects (36, 50), did not express GGT1 (Fig. 7C).
In normal human lung, the CysLT1 receptor was first observed in smooth muscle fibers and in interstitial macrophages (51). In bronchial biopsies from chronic obstructive pulmonary disease (COPD) patients, CysLT1 was expressed in mast cells, monocytes/macrophages, and neutrophils, and CysLT1 expression was associated with exacerbations of COPD (52). CysLT1 mediates bronchoconstriction (4) and, via CysLT1, LTD4 also upregulated expression of TGFβ (in A549 cells and in normal human bronchial epithelial cells) and furin (in monocytic THP-1 cells), both implicated in airway remodeling (53, 54). Of the three CysLTs, LTD4 is the most efficient CysLT1 agonist, while LTE4 is the weakest (5, 6). Thus, further conversion of LTC4 to LTD4, but not to LTE4 (as shown here by A549 and PBECs), could have a considerable effect on all the effects mediated by CysLT1. However, in a study of human tracheal epithelial cells, conversion of LTD4 to LTE4 was found (43). This shows that CysLT metabolism can be different in epithelial (and other) cells from different anatomical sites. It can most probably also be influenced by other factors, such as cell differentiation states, inflammation, and disease.
LTB4 formation was not increased in coincubations of MM6 and A549 cells when stimulated with A23187 plus AA. In these coincubations, a similar minor amount of LTB4 was produced, as in MM6 cells alone. At the same time, A549 cells incubated with exogenous LTA4 produced substantial amounts of LTB4 (Fig. 2). Thus, monocytic MM6 cells do not appear to export LTA4, in the manner shown before for neutrophils (55), but MM6 cells did release LTC4 (Table 1). In a previous study where A549 cells were coincubated with PMNL, CysLTs were formed, presumably by transcellular metabolism of LTA4 (56). However, in our incubations of A549 cells with LTA4, CysLT formation was negligible and LTC4S was not detectable in A549 cell extracts by Western blot. This may be attributed to A549 cell line heterogeneity (57).
We found previously that when MM6 cells were differentiated in the presence of zymosan, formation of PGE2 was induced, which in turn downregulated the activity of LTC4S (38). Thus, we hypothesized that, in cocultures (24 h), PGE2 produced by A549 cells might downregulate MM6 cell CysLT formation. The A549 cells had been starved and stimulated with IL-1β for 48 h, which upregulates PGE2 formation (58). However, as evident from the LTA4 incubations, the activity of LTC4S was not decreased. The concentration of PGE2 in medium from these cocultures was 3–5 nM, determined by LC-MS. This may not be sufficient to downregulate LTC4S in MM6 cells, because exogenous PGE2 (100 nM) reduced conversion of LTA4 to LTC4 only by half (38). Previously, treatment of macrophages with apoptotic cells led to downregulation of 5-LO pathways via mechanisms involving TGFβ and PGE2 (59).
MM6 and A549 cells have been combined before in coculture models of pulmonary inflammation; e.g., when cocultures of A549 and MM6 cells were exposed to ultrafine particles, cytokine release (IL-6 and IL-8) was increased in comparison to monocultures of the respective cells [see (50) and references therein]. Also, growth of mycobacteria in MM6 cells was reduced when MM6 were in coculture with A549 cells, which was attributed to A549-derived cytokines (particularly TNFα) (60). Our results on transcellular conversion of LTC4 to LTD4 are in line with an increased inflammatory response due to cell interactions and to interactions between myeloid cells and epithelium-derived exosomes. Exosomes have the potential to travel to distant sites and could possibly contribute to spreading of an inflammatory reaction to other sites in the body.
The coculture of MM6 cells with A549 cells and exosomes may also be considered as a model of cell interactions and transcellular LTD4 biosynthesis in a tumor milieu. LTs and the monohydroxy acid, 5-HETE, can have growth factor-like effects, promoting cancer cell growth and survival. Interestingly, quite different effects of LTC4 and LTD4 were published for intestinal epithelial cells and CaCo-2 cells; LTD4 promoted proliferation, while this was not found for LTC4 (61). This effect of LTD4 involved increased expression of COX-2 and PGE2 (62, 63). Accordingly, in a mouse model of lung cancer, the production of CysLTs and PGE2 increased considerably during tumor growth (24). In addition to GGT influencing cancer cell redox status (8), its role in LTD4 formation can be another tumor promoting effect of GGT. Thus, not only in pulmonary inflammation, but also in cancer, conversion of LTC4 to LTD4 by transcellular metabolism, possibly involving tumor-associated monocytic cells and epithelial cancer cells, may be important for the final LT effects and outcome.
The results suggest that pulmonary epithelium may be a source of LTD4 in the lung. Monocytic cells (macrophages, dendritic cells), mast cells, and eosinophils are fundamental players in pulmonary inflammation, with established roles in diseases such as asthma and COPD. These cells are major sources of CysLTs, but mainly LTC4, as also observed in this study. This leads us to speculate that transcellular metabolism of LTC4 involving healthy or cancerous pulmonary epithelium may contribute substantially to biosynthesis of LTD4.
Supplementary Material
Acknowledgments
The authors thank Göran Månsson at the Karolinska Institutet Click facility for help with immunofluorescence microscopy, and Casper Wahlund for help with NanoSight analyses.
Footnotes
Abbreviations:
- AA
- arachidonic acid
- COPD
- chronic obstructive pulmonary disease
- CysLT
- cysteinyl leukotriene
- FLAP
- 5-lipoxygenase activating protein
- GGT
- γ-glutamyl transpeptidase
- LO
- lipoxygenase
- LT
- leukotriene
- LTC4S
- LTC4 synthase
- MDDC
- monocyte-derived dendritic cell
- MM6
- Mono Mac 6
- PBEC
- primary bronchial epithelial cell
- SBC
- serine-borate complex
This work was supported by the Swedish Research Council (03X-217), Else Kröner-Fresenius-Stiftung (Else Kröner-Fresenius-Graduiertenkolleg), the Swedish Heart-Lung Foundation, Hesselman’s Foundation, the Stockholm County Council, the Cancer and Allergy Research Foundation, and Karolinska Institutet.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Samuelsson B., Dahlén S-E., Lindgren J-Å., Rouzer C. A., and Serhan C. N.. 1987. Leukotrienes and lipoxins: Structures, biosynthesis, and biological effects. Science. 237: 1171–1176. [DOI] [PubMed] [Google Scholar]
- 2.Rådmark O., Werz O., Steinhilber D., and Samuelsson B.. 2015. 5-Lipoxygenase, a key enzyme for leukotriene biosynthesis in health and disease. Biochim. Biophys. Acta. 1851: 331–339. [DOI] [PubMed] [Google Scholar]
- 3.Basavarajappa D., Wan M., Lukic A., Steinhilber D., Samuelsson B., and Radmark O.. 2014. Roles of coactosin-like protein (CLP) and 5-lipoxygenase-activating protein (FLAP) in cellular leukotriene biosynthesis. Proc. Natl. Acad. Sci. USA. 111: 11371–11376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peters-Golden M., and Henderson W. R. Jr. 2007. Leukotrienes. N. Engl. J. Med. 357: 1841–1854. [DOI] [PubMed] [Google Scholar]
- 5.Lynch K. R., O’Neill G. P., Liu Q., Im D. S., Sawyer N., Metters K. M., Coulombe N., Abramovitz M., Figueroa D. J., Zeng Z., et al. 1999. Characterization of the human cysteinyl leukotriene CysLT1 receptor. Nature. 399: 789–793. [DOI] [PubMed] [Google Scholar]
- 6.Sarau H. M., Ames R. S., Chambers J., Ellis C., Elshourbagy N., Foley J. J., Schmidt D. B., Muccitelli R. M., Jenkins O., Murdock P. R., et al. 1999. Identification, molecular cloning, expression, and characterization of a cysteinyl leukotriene receptor. Mol. Pharmacol. 56: 657–663. [DOI] [PubMed] [Google Scholar]
- 7.Heisterkamp N., Groffen J., Warburton D., and Sneddon T. P.. 2008. The human gamma-glutamyltransferase gene family. Hum. Genet. 123: 321–332. [DOI] [PubMed] [Google Scholar]
- 8.Corti A., Franzini M., Paolicchi A., and Pompella A.. 2010. Gamma-glutamyltransferase of cancer cells at the crossroads of tumor progression, drug resistance and drug targeting. Anticancer Res. 30: 1169–1181. [PubMed] [Google Scholar]
- 9.Wickham S., West M. B., Cook P. F., and Hanigan M. H.. 2011. Gamma-glutamyl compounds: substrate specificity of gamma-glutamyl transpeptidase enzymes. Anal. Biochem. 414: 208–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murphy R. C., and Gijon M. A.. 2007. Biosynthesis and metabolism of leukotrienes. Biochem. J. 405: 379–395. [DOI] [PubMed] [Google Scholar]
- 11.Joyce-Brady M., Takahashi Y., Oakes S. M., Rishi A. K., Levine R. A., Kinlough C. L., and Hughey R. P.. 1994. Synthesis and release of amphipathic gamma-glutamyl transferase by the pulmonary alveolar type 2 cell. Its redistribution throughout the gas exchange portion of the lung indicates a new role for surfactant. J. Biol. Chem. 269: 14219–14226. [PubMed] [Google Scholar]
- 12.Jean J. C., Liu Y., Brown L. A., Marc R. E., Klings E., and Joyce-Brady M.. 2002. Gamma-glutamyl transferase deficiency results in lung oxidant stress in normoxia. Am. J. Physiol. Lung Cell. Mol. Physiol. 283: L766–L776. [DOI] [PubMed] [Google Scholar]
- 13.Kang Y. J., Feng Y., and Hatcher E. L.. 1994. Glutathione stimulates A549 cell proliferation in glutamine-deficient culture: the effect of glutamate supplementation. J. Cell. Physiol. 161: 589–596. [DOI] [PubMed] [Google Scholar]
- 14.Zaman K., Hanigan M. H., Smith A., Vaughan J., Macdonald T., Jones D. R., Hunt J. F., and Gaston B.. 2006. Endogenous S-nitrosoglutathione modifies 5-lipoxygenase expression in airway epithelial cells. Am. J. Respir. Cell Mol. Biol. 34: 387–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takahashi Y., Oakes S. M., Williams M. C., Takahashi S., Miura T., and Joyce-Brady M.. 1997. Nitrogen dioxide exposure activates gamma-glutamyl transferase gene expression in rat lung. Toxicol. Appl. Pharmacol. 143: 388–396. [DOI] [PubMed] [Google Scholar]
- 16.Mok Y., Son D. K., Yun Y. D., Jee S. H., and Samet J. M.. 2016. γ-Glutamyltransferase and cancer risk: the Korean cancer prevention study. Int. J. Cancer. 138: 311–319. [DOI] [PubMed] [Google Scholar]
- 17.Franzini M., Corti A., Fierabracci V., and Pompella A.. 2014. Helicobacter, gamma-glutamyltransferase and cancer: further intriguing connections. World J. Gastroenterol. 20: 18057–18058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han B., Luo G., Shi Z. Z., Barrios R., Atwood D., Liu W., Habib G. M., Sifers R. N., Corry D. B., and Lieberman M. W.. 2002. Gamma-glutamyl leukotrienase, a novel endothelial membrane protein, is specifically responsible for leukotriene D(4) formation in vivo. Am. J. Pathol. 161: 481–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanigan M. H., Gillies E. M., Wickham S., Wakeham N., and Wirsig-Wiechmann C. R.. 2015. Immunolabeling of gamma-glutamyl transferase 5 in normal human tissues reveals that expression and localization differ from gamma-glutamyl transferase 1. Histochem. Cell Biol. 143: 505–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laviolette M., Carreau M., Coulombe R., Cloutier D., Dupont P., Rioux J., Braquet P., and Borgeat P.. 1988. Metabolism of arachidonic acid through the 5-lipoxygenase pathway in normal human peritoneal macrophages. J. Immunol. 141: 2104–2109. [PubMed] [Google Scholar]
- 21.Kita Y., Takahashi T., Uozumi N., Nallan L., Gelb M. H., and Shimizu T.. 2005. Pathway-oriented profiling of lipid mediators in macrophages. Biochem. Biophys. Res. Commun. 330: 898–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buczynski M. W., Stephens D. L., Bowers-Gentry R. C., Grkovich A., Deems R. A., and Dennis E. A.. 2007. TLR-4 and sustained calcium agonists synergistically produce eicosanoids independent of protein synthesis in RAW264.7 cells. J. Biol. Chem. 282: 22834–22847. [DOI] [PubMed] [Google Scholar]
- 23.Norris P. C., and Dennis E. A.. 2012. Omega-3 fatty acids cause dramatic changes in TLR4 and purinergic eicosanoid signaling. Proc. Natl. Acad. Sci. USA. 109: 8517–8522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poczobutt J. M., Gijon M., Amin J., Hanson D., Li H., Walker D., Weiser-Evans M., Lu X., Murphy R. C., and Nemenoff R. A.. 2013. Eicosanoid profiling in an orthotopic model of lung cancer progression by mass spectrometry demonstrates selective production of leukotrienes by inflammatory cells of the microenvironment. PLoS One. 8: e79633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weller P. F., Lee C. W., Foster D. W., Corey E. J., Austen K. F., and Lewis R. A.. 1983. Generation and metabolism of 5-lipoxygenase pathway leukotrienes by human eosinophils: predominant production of leukotriene C4. Proc. Natl. Acad. Sci. USA. 80: 7626–7630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Owen W. F. Jr., Soberman R. J., Yoshimoto T., Sheffer A. L., Lewis R. A., and Austen K. F.. 1987. Synthesis and release of leukotriene C4 by human eosinophils. J. Immunol. 138: 532–538. [PubMed] [Google Scholar]
- 27.Bandeira-Melo C., and Weller P. F.. 2003. Eosinophils and cysteinyl leukotrienes. Prostaglandins Leukot. Essent. Fatty Acids. 69: 135–143. [DOI] [PubMed] [Google Scholar]
- 28.Razin E., Mencia-Huerta J. M., Stevens R. L., Lewis R. A., Liu F. T., Corey E., and Austen K. F.. 1983. IgE-mediated release of leukotriene C4, chondroitin sulfate E proteoglycan, beta-hexosaminidase, and histamine from cultured bone marrow-derived mouse mast cells. J. Exp. Med. 157: 189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levi-Schaffer F., Austen K. F., Caulfield J. P., Hein A., Gravallese P. M., and Stevens R. L.. 1987. Co-culture of human lung-derived mast cells with mouse 3T3 fibroblasts: morphology and IgE-mediated release of histamine, prostaglandin D2, and leukotrienes. J. Immunol. 139: 494–500. [PubMed] [Google Scholar]
- 30.Heavey D. J., Ernst P. B., Stevens R. L., Befus A. D., Bienenstock J., and Austen K. F.. 1988. Generation of leukotriene C4, leukotriene B4, and prostaglandin D2 by immunologically activated rat intestinal mucosa mast cells. J. Immunol. 140: 1953–1957. [PubMed] [Google Scholar]
- 31.Macchia L., Hamberg M., Kumlin M., Butterfield J. H., and Haeggstrom J. Z.. 1995. Arachidonic acid metabolism in the human mast cell line HMC-1: 5-lipoxygenase gene expression and biosynthesis of thromboxane. Biochim. Biophys. Acta. 1257: 58–74. [DOI] [PubMed] [Google Scholar]
- 32.Lundström S. L., Saluja R., Adner M., Haeggström J. Z., Nilsson G., and Wheelock C. E.. 2013. Lipid mediator metabolic profiling demonstrates differences in eicosanoid patterns in two phenotypically distinct mast cell populations. J. Lipid Res. 54: 116–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martinez F. D., and Vercelli D.. 2013. Asthma. Lancet. 382: 1360–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Werz O., Klemm J., Samuelsson B., and Radmark O.. 2001. Phorbol ester up-regulates capacities for nuclear translocation and phosphorylation of 5-lipoxygenase in Mono Mac 6 cells and human polymorphonuclear leukocytes. Blood. 97: 2487–2495. [DOI] [PubMed] [Google Scholar]
- 35.Esser J., Gehrmann U., D’Alexandri F. L., Hidalgo-Estevez A. M., Wheelock C. E., Scheynius A., Gabrielsson S., and Radmark O.. 2010. Exosomes from human macrophages and dendritic cells contain enzymes for leukotriene biosynthesis and promote granulocyte migration. J. Allergy Clin. Immunol. 126: 1032–1040. [DOI] [PubMed] [Google Scholar]
- 36.von Scheele I., Larsson K., and Palmberg L.. 2010. Budesonide enhances Toll-like receptor 2 expression in activated bronchial epithelial cells. Inhal. Toxicol. 22: 493–499. [DOI] [PubMed] [Google Scholar]
- 37.Nordin J. Z., Lee Y., Vader P., Mager I., Johansson H. J., Heusermann W., Wiklander O. P., Hallbrink M., Seow Y., Bultema J. J., et al. 2015. Ultrafiltration with size-exclusion liquid chromatography for high yield isolation of extracellular vesicles preserving intact biophysical and functional properties. Nanomedicine. 11: 879–883. [DOI] [PubMed] [Google Scholar]
- 38.Esser J., Gehrmann U., Salvado M. D., Wetterholm A., Haeggstrom J. Z., Samuelsson B., Gabrielsson S., Scheynius A., and Radmark O.. 2011. Zymosan suppresses leukotriene C(4) synthase activity in differentiating monocytes: antagonism by aspirin and protein kinase inhibitors. FASEB J. 25: 1417–1427. [DOI] [PubMed] [Google Scholar]
- 39.Tate S. S., and Meister A.. 1978. Serine-borate complex as a transition-state inhibitor of gamma-glutamyl transpeptidase. Proc. Natl. Acad. Sci. USA. 75: 4806–4809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Powell W. S., Gravelle F., and Gravel S.. 1992. Metabolism of 5(S)-hydroxy-6,8,11,14-eicosatetraenoic acid and other 5(S)-hydroxyeicosanoids by a specific dehydrogenase in human polymorphonuclear leukocytes. J. Biol. Chem. 267: 19233–19241. [PubMed] [Google Scholar]
- 41.West M. B., Chen Y., Wickham S., Heroux A., Cahill K., Hanigan M. H., and Mooers B. H.. 2013. Novel insights into eukaryotic gamma-glutamyltranspeptidase 1 from the crystal structure of the glutamate-bound human enzyme. J. Biol. Chem. 288: 31902–31913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jame A. J., Lackie P. M., Cazaly A. M., Sayers I., Penrose J. F., Holgate S. T., and Sampson A. P.. 2007. Human bronchial epithelial cells express an active and inducible biosynthetic pathway for leukotrienes B4 and C4. Clin. Exp. Allergy. 37: 880–892. [DOI] [PubMed] [Google Scholar]
- 43.Yamaya M., Sekizawa K., Yamauchi K., Hoshi H., Sawai T., and Sasaki H.. 1995. Epithelial modulation of leukotriene-C4-induced human tracheal smooth muscle contraction. Am. J. Respir. Crit. Care Med. 151: 892–894. [DOI] [PubMed] [Google Scholar]
- 44.Zaitsu M., Hamasaki Y., Tsuji K., Matsuo M., Fujita I., Aoki Y., Ishii E., and Kohashi O.. 2003. Dexamethasone accelerates catabolism of leukotriene C4 in bronchial epithelial cells. Eur. Respir. J. 22: 35–42. [DOI] [PubMed] [Google Scholar]
- 45.del Bello B., Paolicchi A., Comporti M., Pompella A., and Maellaro E.. 1999. Hydrogen peroxide produced during gamma-glutamyl transpeptidase activity is involved in prevention of apoptosis and maintainance of proliferation in U937 cells. FASEB J. 13: 69–79. [DOI] [PubMed] [Google Scholar]
- 46.Kargman S., Rousseau P., Reid G. K., Rouzer C. A., Mancini J. A., Rands E., Dixon R. A., Diehl R. E., Léveillé C., Nathaniel D., et al. 1993. Leukotriene synthesis in U937 cells expressing recombinant 5-lipoxygenase. J. Lipid Mediat. 7: 31–45. [PubMed] [Google Scholar]
- 47.Orning L., Hammarstrom S., and Samuelsson B.. 1980. Leukotriene D: a slow reacting substance from rat basophilic leukemia cells. Proc. Natl. Acad. Sci. USA. 77: 2014–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wickham S., Regan N., West M. B., Thai J., Cook P. F., Terzyan S. S., Li P. K., and Hanigan M. H.. 2013. Inhibition of human gamma-glutamyl transpeptidase: development of more potent, physiologically relevant, uncompetitive inhibitors. Biochem. J. 450: 547–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dahlén S. E., Hansson G., Hedqvist P., Bjorck T., Granström E., and Dahlén B.. 1983. Allergen challenge of lung tissue from asthmatics elicits bronchial contraction that correlates with the release of leukotrienes C4, D4, and E4. Proc. Natl. Acad. Sci. USA. 80: 1712–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Klein S. G., Hennen J., Serchi T., Blomeke B., and Gutleb A. C.. 2011. Potential of coculture in vitro models to study inflammatory and sensitizing effects of particles on the lung. Toxicol. In Vitro. 25: 1516–1534. [DOI] [PubMed] [Google Scholar]
- 51.Figueroa D. J., Breyer R. M., Defoe S. K., Kargman S., Daugherty B. L., Waldburger K., Liu Q., Clements M., Zeng Z., O’Neill G. P., et al. 2001. Expression of the cysteinyl leukotriene 1 receptor in normal human lung and peripheral blood leukocytes. Am. J. Respir. Crit. Care Med. 163: 226–233. [DOI] [PubMed] [Google Scholar]
- 52.Zhu J., Bandi V., Qiu S., Figueroa D. J., Evans J. F., Barnes N., Guntupalli K. K., and Jeffery P. K.. 2012. Cysteinyl leukotriene 1 receptor expression associated with bronchial inflammation in severe exacerbations of COPD. Chest. 142: 347–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bossé Y., Thompson C., McMahon S., Dubois C. M., Stankova J., and Rola-Pleszczynski M.. 2008. Leukotriene D4-induced, epithelial cell-derived transforming growth factor β1 in human bronchial smooth muscle cell proliferation. Clin. Exp. Allergy. 38: 113–121. [DOI] [PubMed] [Google Scholar]
- 54.Thompson C., McMahon S., Bosse Y., Dubois C. M., Stankova J., and Rola-Pleszczynski M.. 2008. Leukotriene D4 up-regulates furin expression through CysLT1 receptor signaling. Am. J. Respir. Cell Mol. Biol. 39: 227–234. [DOI] [PubMed] [Google Scholar]
- 55.Folco G., and Murphy R. C.. 2006. Eicosanoid transcellular biosynthesis: from cell-cell interactions to in vivo tissue responses. Pharmacol. Rev. 58: 375–388. [DOI] [PubMed] [Google Scholar]
- 56.Clària J., Lee M. H., and Serhan C. N.. 1996. Aspirin-triggered lipoxins (15-epi-LX) are generated by the human lung adenocarcinoma cell line (A549)-neutrophil interactions and are potent inhibitors of cell proliferation. Mol. Med. 2: 583–596. [PMC free article] [PubMed] [Google Scholar]
- 57.Watanabe N., Dickinson D. A., Krzywanski D. M., Iles K. E., Zhang H., Venglarik C. J., and Forman H. J.. 2002. A549 subclones demonstrate heterogeneity in toxicological sensitivity and antioxidant profile. Am. J. Physiol. Lung Cell. Mol. Physiol. 283: L726–L736. [DOI] [PubMed] [Google Scholar]
- 58.Jakobsson P. J., Thoren S., Morgenstern R., and Samuelsson B.. 1999. Identification of human prostaglandin E synthase: a microsomal, glutathione-dependent, inducible enzyme, constituting a potential novel drug target. Proc. Natl. Acad. Sci. USA. 96: 7220–7225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Freire-de-Lima C. G., Xiao Y. Q., Gardai S. J., Bratton D. L., Schiemann W. P., and Henson P. M.. 2006. Apoptotic cells, through transforming growth factor-beta, coordinately induce anti-inflammatory and suppress pro-inflammatory eicosanoid and NO synthesis in murine macrophages. J. Biol. Chem. 281: 38376–38384. [DOI] [PubMed] [Google Scholar]
- 60.Sato K., Tomioka H., Shimizu T., Gonda T., Ota F., and Sano C.. 2002. Type II alveolar cells play roles in macrophage-mediated host innate resistance to pulmonary mycobacterial infections by producing proinflammatory cytokines. J. Infect. Dis. 185: 1139–1147. [DOI] [PubMed] [Google Scholar]
- 61.Ohd J. F., Wikstrom K., and Sjolander A.. 2000. Leukotrienes induce cell-survival signaling in intestinal epithelial cells. Gastroenterology. 119: 1007–1018. [DOI] [PubMed] [Google Scholar]
- 62.Yudina Y., Parhamifar L., Bengtsson A. M., Juhas M., and Sjolander A.. 2008. Regulation of the eicosanoid pathway by tumour necrosis factor alpha and leukotriene D4 in intestinal epithelial cells. Prostaglandins Leukot. Essent. Fatty Acids. 79: 223–231. [DOI] [PubMed] [Google Scholar]
- 63.Cabral M., Martin-Venegas R., and Moreno J. J.. 2015. Leukotriene D4-induced Caco-2 cell proliferation is mediated by prostaglandin E2 synthesis. Physiol. Rep. 3: e12417. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.