Significance
Antigen presentation genes are exceptionally polymorphic, enhancing immune defense. Polymorphism within additional components of the MHC pathway, particularly the antigen processing genes, may also shape immune responses. Using transcriptome, exome, and whole-genome sequencing to examine immune gene variation in zebrafish, we uncovered several antigen processing genes not found in the reference genome clustered within a deeply divergent haplotype of the core MHC locus. Our data provide evidence that these previously undescribed antigen processing genes retain ancient alternative sequence lineages, likely derived during the formation of the adaptive immune system, and represent the most divergent collection of antigen processing and presentation genes yet identified. These findings offer insights into the evolution of vertebrate adaptive immunity.
Keywords: comparative genomics, proteasome and TAP evolution, major histocompatibility, MHC class I pathway, CG2 clonal zebrafish
Abstract
Antigen processing and presentation genes found within the MHC are among the most highly polymorphic genes of vertebrate genomes, providing populations with diverse immune responses to a wide array of pathogens. Here, we describe transcriptome, exome, and whole-genome sequencing of clonal zebrafish, uncovering the most extensive diversity within the antigen processing and presentation genes of any species yet examined. Our CG2 clonal zebrafish assembly provides genomic context within a remarkably divergent haplotype of the core MHC region on chromosome 19 for six expressed genes not found in the zebrafish reference genome: mhc1uga, proteasome-β 9b (psmb9b), psmb8f, and previously unknown genes psmb13b, tap2d, and tap2e. We identify ancient lineages for Psmb13 within a proteasome branch previously thought to be monomorphic and provide evidence of substantial lineage diversity within each of three major trifurcations of catalytic-type proteasome subunits in vertebrates: Psmb5/Psmb8/Psmb11, Psmb6/Psmb9/Psmb12, and Psmb7/Psmb10/Psmb13. Strikingly, nearby tap2 and MHC class I genes also retain ancient sequence lineages, indicating that alternative lineages may have been preserved throughout the entire MHC pathway since early diversification of the adaptive immune system ∼500 Mya. Furthermore, polymorphisms within the three MHC pathway steps (antigen cleavage, transport, and presentation) are each predicted to alter peptide specificity. Lastly, comparative analysis shows that antigen processing gene diversity is far more extensive than previously realized (with ancient coelacanth psmb8 lineages, shark psmb13, and tap2t and psmb10 outside the teleost MHC), implying distinct immune functions and conserved roles in shaping MHC pathway evolution throughout vertebrates.
Genetic diversity promotes robust immune function. MHC gene polymorphism provides a classic example, because human populations carry hundreds of MHC class I alleles (MHCI), which present antigens to activate an immune response (1). Variation observed between alleles of immune genes may exceed levels explained by simple accumulation of mutations within a species over time. For example, sequence variation within human MHC genes has been traced back 10–50 My (2–4), including allelic variants shared with other primate species. Transspecies polymorphism explains this observation by positing that some alleles survive multiple speciation events, thereby providing descendant species with higher functional sequence diversity (5). Starting with this diversity, balancing selection preserves polymorphism within populations during conditions when no single allele is optimized for all environments, with a disproportionate impact on immune loci (6). Some nonmammalian vertebrates, such as bony fish, frogs, and sharks, maintain MHC polymorphism at even higher levels than mammals (7–10), implying preservation of ancient alleles across different species.
Recent genomic studies have offered considerable insights into the evolution of the vertebrate adaptive immune system by comparing phylogenetically divergent species (11–14). Throughout vertebrates, gene linkage within the MHC region is highly conserved. For example, MHCI and antigen processing genes remain tightly linked in sharks, members of the oldest vertebrate lineage to maintain an MHC-mediated adaptive immune system (15, 16). This tight linkage is also highly conserved in bony fish (17–19) as well as in additional nonmammalian jawed vertebrates, such as frogs (20). Coevolution of MHCI and antigen processing genes is facilitated by their close physical proximity in the genome, leading to coinheritance of alleles throughout the MHC pathway with compatible peptide specificities. Juxtaposition of these genes into compact haplotypes may, thus, provide a foundation for the evolution of MHC pathways functioning with more highly specialized peptide repertoires (21).
In contrast, MHC gene arrangements in mammals generally differ from those of other vertebrates, including much greater distance between the MHCI and antigen processing genes. Mammalian antigen processing genes are instead found in the class II region of the MHC, where they are far removed from the MHCI genes found in the class I region. This physical distance limits the capacity of these genes to coevolve distinct peptide specificities, because increased recombination is likely to deter the specialization of alleles upstream in the MHC pathway because of the potential for downstream incompatibility. Accordingly, compared with nonmammalian vertebrates, mammalian antigen processing genes are much less polymorphic, and mammalian MHC pathway diversity remains instead focused primarily in the MHCI genes. These findings are consistent with an immune strategy favoring the cleavage and transport of a more “generic” peptide repertoire, with greater emphasis on the downstream peptide binding specificities using a collection of diverse MHCI molecules (21–23).
Some notable exceptions to these observations have been reported. For example, rat MHCI genes are found more tightly linked with antigen processing genes than in other rodents, such as mice, consistent with rat haplotypes having more specialized antigen transport (TAP) alleles exhibiting either “restrictive” or “permissive” peptide repertoires (24). Chickens represent another interesting exception, because antigen processing genes from the first stage of the MHC pathway, the inducible proteasome subunits proteasome-β 8–10 (psmb8–10), are altogether missing (25). However, this lack of inducible proteasome subunits in chickens does not seem to have reduced the capacity for polymorphic antigen transport genes, encoding the TAP subunits, to coevolve distinctive peptide specificities with coinherited MHCI alleles (26).
In teleosts (bony fish), MHC arrangements are similar to other nonmammalian vertebrates, but important differences are observed. Teleost antigen processing genes are found adjacent to their MHCI genes, despite the teleost MHCI and MHCII regions being distinctively unlinked (27). Teleost MHCI genes retain ancient lineages, with sequences estimated to be hundreds of millions of years old (9). These lineages are ancient compared with other vertebrates, such as mammals, where MHCI lineages appear to be much younger at millions or tens of millions of years old. Furthermore, the antigen processing gene psmb8 has also been shown to retain ancient sequence lineages in teleosts that are hundreds of millions of years old (28). Finally, an additional set of unique and perhaps teleost-specific proteasome subunit genes was previously linked to teleost MHCI genes (17–19, 29), and one of these genes referred to as psmb10 displayed limited (believed nonfunctional) polymorphism (30). However, few other studies have examined these genes in detail.
We recently characterized classical MHCI genes from six divergent haplotypes found in clonal and selectively bred zebrafish (31) and identified remarkable additional haplotype variability. Unlike in most other species examined, we found that distinct zebrafish MHC haplotypes express altogether different sets of MHCI genes, which are each linked to the divergent antigen processing gene psmb8a or psmb8f. Copy number variation for nearby tap2 and tapbp genes also accompanied these other haplotypic differences. Revealing the full extent of additional haplotype diversity would require genomic sequencing; indeed, to date, such sequences have been lacking for haplotypes containing the ancient psmb8f lineage. Intriguingly, additional putative antigen processing genes from zebrafish have been found only in expression libraries, despite the availability of a high-quality reference genome (32). Such “orphan” sequences may be associated with alternative haplotypes (for example, the core MHC region on zebrafish chromosome 19), where the genes are likely to maintain central roles in immune function. Incorporating these divergent sequences into genomic assemblies has great potential to further improve our understanding of the formation of the adaptive immune system, an event closely associated with the origin of all vertebrates (33, 34).
Here, we report the genomic sequence for the core MHC region of CG2 clonal zebrafish. Our assembly uncovers genes from an alternative haplotype and places them into genomic context. By examining the sequence properties of these divergent zebrafish antigen processing genes, we provide evidence that they represent ancient lineages, including polymorphic residues likely to contribute to immune function. In addition, our comparative analysis across vertebrates yields a more comprehensive understanding of the relationships and extensive diversity found among these antigen processing and presentation genes, revealing additional genes in species such as zebrafish and coelacanths, and evidence of ancient lineages and haplotypes that have shaped the evolution of the MHC pathway.
Results
Whole-Exome Sequencing of Clonal Zebrafish.
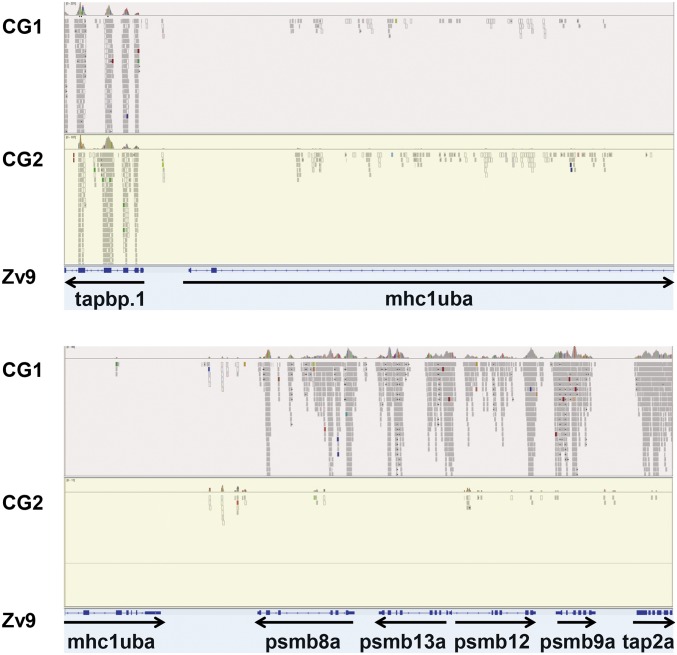
To identify potential variants in immune loci, we first performed whole-exome sequencing of two different lines of homozygous diploid, clonal golden zebrafish: CG1 and CG2 (35, 36). Analysis of our exome data revealed distinctive patterns of haplotypic variation. For example, for both clonal zebrafish exomes, essentially no reads were aligned to the mhc1uba gene on chromosome 19 of the reference genome (Fig. 1). For the CG2 clonal zebrafish line, this absence of aligned reads was also found for additional genes adjacent to mhc1uba. The pattern extended throughout a region of ∼100 kb, encompassing the MHCI genes as well as linked antigen processing genes—psmb8a, psmb9a, psmb12, psmb13a, and tap2a. In contrast, numerous reads from CG1 matched antigen processing exons from the Zv9 reference genome. Therefore, the extended MHC variation seems to be specific for CG2.
Fig. 1.
The core MHC region in clonal zebrafish. Aligned reads are shown for representative exomes from clonal golden zebrafish lines (CG1 and CG2) that map to the core MHC region on chromosome 19 of the Zv9 zebrafish reference genome (coordinates 7689327–7748684). Refseq annotated exons are shown below with transcriptional orientations.
Sequencing and de Novo Assembly of a Divergent MHC Locus.
To identify divergent sequences potentially missed by the hybridization-based exome sequencing approach, we performed whole-genome sequencing of CG2 clonal zebrafish. Starting with paired end reads obtained at 25× coverage, we generated genomic scaffolds by de novo assembly. Most of our draft CG2 genome assembly matched the reference genome, with 72,406 scaffolds (of 73,507 with size >1 kb) having an average of 95.4% sequence identity. However, some scaffolds did not align to the reference genome, indicating candidate regions of the CG2 zebrafish genome harboring extensive haplotypic variation.
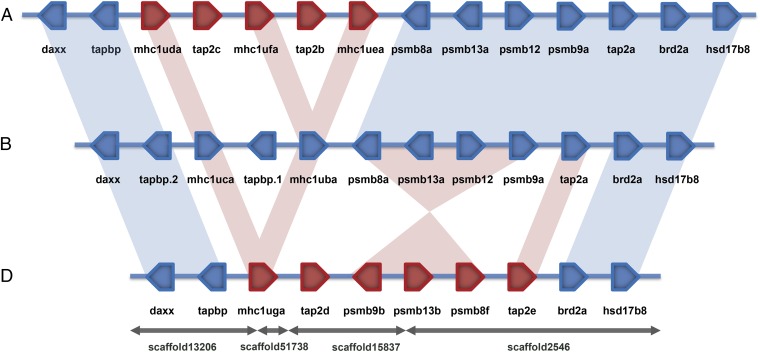
We focused on the chromosome 19 core MHC locus, where considerable haplotypic variation was observed previously (17, 31). BLAST searches of our CG2 genomic database identified scaffolds containing MHC flanking genes, including tapbp and brd2a (Fig. 2). These scaffolds also included mhc1uga and psmb8f that we previously mapped with linkage and expression data (using offspring of MHC haplotype compound heterozygous fish) to the core MHC haplotype D of CG2 clonal zebrafish (31).
Fig. 2.
Comparison of zebrafish core MHC haplotypes. Genes highly conserved with the reference haplotype B are shown in blue, and these highly conserved relationships are illustrated with blue shading. Genes divergent from the reference haplotype B are shown in red, and divergent gene relationships with haplotype B are illustrated with red shading. Haplotype A is part of an alternative haplotypic assembly of sequences derived from AB zebrafish (VEGA59 chromosome AB: 9329627–9629628), haplotype B is from the reference genome derived primarily from Tuebingen zebrafish (Zv9 chromosome 19: 7623109–78026601), and haplotype D was obtained by de novo assembly of genomic sequences from CG2 clonal golden zebrafish (this study).
Scaffold orientation was inferred by using predicted gene models, which were improved by RNA sequencing (RNA-Seq) data, and also, the presence of conserved MHC flanking sequences (SI Appendix, Table S1). Two of three scaffold junctions occurred within introns, with these two junctions together imparting orientation for four of the scaffolds, whereas the two distal scaffolds were anchored using their conserved MHC flanking sequences. These distal regions from the divergent haplotype D assembly, including the tapbp, daxx, brd2a, and hsd17b8 genes, were highly conserved with haplotype B found in the reference genome. The conserved flanking regions together with linkage data for psmb8f and mhc1uga (31) anchor the scaffolds as a divergent MHC haplotype on chromosome 19.
Comparison of Zebrafish Core MHC Haplotypes.
Genomic sequences are available for three zebrafish core MHC haplotypes: A from a prior AB assembly, B from the Zv9 reference genome, and D derived from CG2 clonal zebrafish in this study. All three sequenced zebrafish MHC haplotypes are flanked by conserved chromosome 19 sequences (Fig. 2) as illustrated by highlighting conserved genes daxx and tapbp on the left and brd2a and hsd17b8 on the right. Of eight genes found in between these flanking genes in reference haplotype B, five genes are shared with haplotype A that maintain high levels of sequence identity: psmb8a, psmb13a, psmb12, psmb9a, and tap2a. In contrast, differences between haplotypes A and B are evident for the divergent MHCI gene sequences. Three MHCI genes are found for haplotype A (mhc1uda, mhc1ufa, and mhc1uea) compared with two genes for haplotype B (mhc1uca and mhc1uba). Differences are also observed for the duplicated genes found between the MHCI genes, where two genes are present for haplotype A (tap2c and tap2b) compared with one gene for haplotype B (tapbp.1). Nevertheless, these two haplotypes share highly conserved antigen processing genes psmb8a, psmb13a, psmb12, psmb9a, and tap2a.
In contrast, haplotype D from CG2 zebrafish shares none of eight central genes from haplotype B (Fig. 2). Haplotype D instead carries a single divergent MHCI gene mhc1uga and two divergent tap2 genes tap2d and tap2e as well as an apparent inversion containing the divergent psmb9b, psmb13b, and psmb8f genes. Although each of three zebrafish core MHC haplotypes maintains distinctive genomic sequence arrangements, haplotype D remains most divergent in sequence, including each of the antigen processing genes.
Genes in a Divergent Zebrafish Core MHC Haplotype.
Genomic context for the psmb8f and mhc1uga genes was markedly different from the corresponding region of the reference genome. Analysis of the divergent haplotype D assembly revealed four additional gene sequences that are not present in the reference zebrafish genome: tap2d, psmb9b, psmb13b, and tap2e (Fig. 2). Thus, our assembly incorporates previously unknown or unplaced genes into an alternative haplotype for the core MHC locus on zebrafish chromosome 19.
Each of the genes from haplotype D also had corresponding transcripts identified within the RNA-Seq database derived from CG2 immune tissues (SI Appendix, Table S1). These data provide direct experimental evidence of expression for each gene found within the divergent haplotype D genomic assembly, including transcripts for the tapbp, daxx, brd2a, and hsd17b8 genes as well as the tap2d, psmb9b, psmb13b, tap2e, mhc1uga, and psmb8f genes. Consistent with our alternative haplotype assembly (Fig. 2), no RNA-Seq transcripts were identified from CG2 immune tissues for seven genes associated with the reference core MHC haplotype B: mhc1uba, mhc1uca, psmb8a, psmb9a, psmb12, psmb13a, and tap2a.
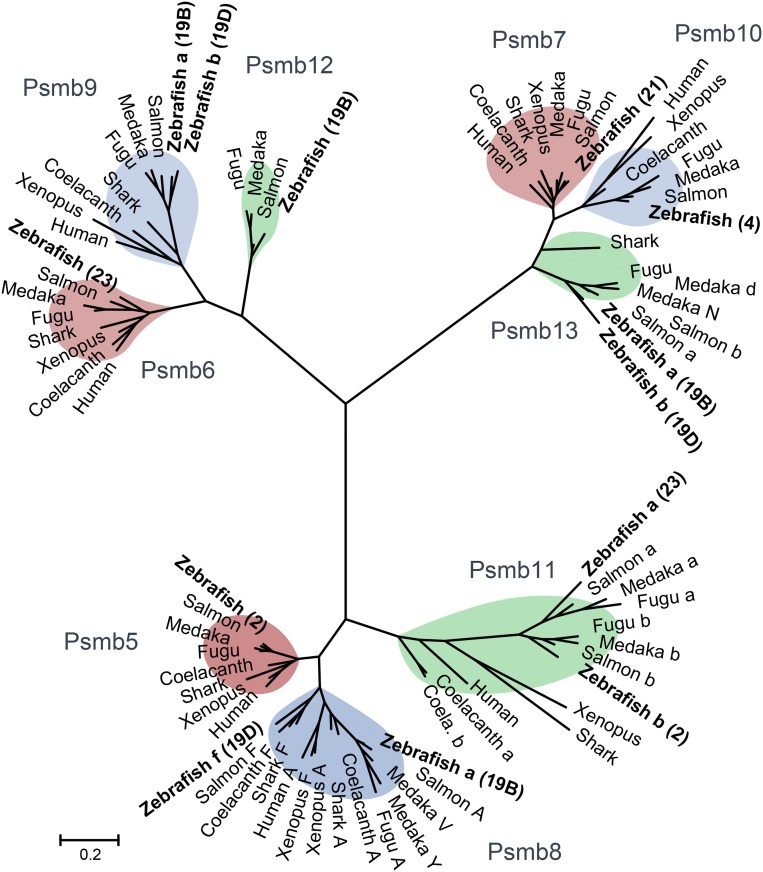
Phylogenetic Analysis of Zebrafish Proteasome Subunits.
All three forms of proteasome subunits (constitutive, immunoproteasome, and thymoproteasome) are conserved in zebrafish (Fig. 3), including single copies found for the three constitutive subunits (Psmb5, Psmb6, and Psmb7). The thymoproteasome subunits Psmb11a and Psmb11b in zebrafish represent teleost-specific gene duplicates associated with an ancient teleost-specific whole-genome duplication (37). Consistent with other largely monomorphic MHC pathway genes that are found outside the core MHC locus, such as tap1 (SI Appendix, Table S2), these three constitutive proteasome and two thymoproteasome subunits, all non-MHC linked, each share 99 to 100% sequence identity between the reference genome and CG2 zebrafish genome assemblies.
Fig. 3.
Phylogenetic relationships for proteasome subunits from zebrafish and additional vertebrates. Multimeric proteasomes have interchangeable catalytic subunits in most jawed vertebrates. Constitutive proteasome, immunoproteasome, and intermediate proteasome forms each degrade intracellular proteins into the peptide fragments that are presented by MHC molecules at the cell surface. The three ancestral PSMB subunits with distinct catalytic activities (Psmb5, Psmb6, and Psmb7) segregate into three major branches in the phylogenetic tree. These three constitutive proteasome subunits Psmb5 (LMPX), Psmb6 (LMPY), and Psmb7 (LMPZ) are replaced by IFN-inducible immunoproteasome subunits Psmb8 (LMP7), Psmb9 (LMP2), and Psmb10 (MECL-1), respectively, during an immune response. In addition, the thymoproteasome subunit Psmb11 replaces Psmb5 and Psmb8 specifically in the thymus. These more specialized, nonconstitutive, proteasome subunits seem to be specific to jawed vertebrates (87). Deduced amino acid sequences were used to construct maximum likelihood trees. For clarity only, the subunit encoded by the Zv9 reference genome is shown for three constitutive proteasome (Psmb5, Psmb6, and Psmb7) and two thymoproteasome (Psmb11a and Psmb11b) subunits. Chromosome locations for zebrafish subunits are provided in parentheses, including haplotype associations when applicable. Additional phylogenetic trees with bootstrap values and other species are provided in SI Appendix, Figs. S5–S7. Sequences are provided in Dataset S1.
In contrast to the constitutive and thymoproteasome subunits that are more conserved, three MHC-linked immunoproteasome subunits (Psmb8, Psmb9, and Psmb13) have divergent lineages in zebrafish (Table 1). These different genes are maintained in a haplotype-specific manner. Phylogenetic relationships, thus, reveal the presence of ancient lineages for each of three major branches of proteasomal subunits comparing the zebrafish Psmb8f, Psmb9b, and Psmb13b sequences encoded by core MHC haplotype D on chromosome 19 with the Psmb8a, Psmb9a, and Psmb13a sequences encoded by haplotype B.
Table 1.
Comparison of genes from the haplotype 19D assembly with genes from haplotype 19B from the zebrafish reference genome
| Haplotype D gene* | Identity, %† | Haplotype B gene‡ | Chromosome§ |
| daxx | 99 | daxx | 19 |
| tapbp | 98 | tapbp | 19 |
| mhc1uga | 49 | mhc1uba | 19 |
| tap2d | 65 | tap2a | 19 |
| psmb9b | 86 | psmb9a | 19 |
| psmb13b | 71 | psmb13a | 19 |
| psmb8f | 64 | psmb8a | 19 |
| tap2e | 50 | tap2a | 19 |
| brd2a | 100 | brd2a | 19 |
| hsd17b8 | 99 | hsd17b8 | 19 |
Sequences from core MHC locus of CG2 zebrafish assembly (haplotype 19D).
Levels of pairwise percentage identity calculated with BLAST using predicted amino acid sequences.
Most closely matched genes identified from Zv9 zebrafish reference genome (haplotype 19B).
Chromosome location.
Previous studies have shown how the Psmb8a and Psmb8f lineages maintain ancient evolutionary histories approaching 500 My (28). Other proteasomal subunits also maintain distinct lineages, such as the Psmb9a and Psmb9b subunits from different zebrafish core MHC haplotypes (Fig. 3). In addition, Psmb12 is not found in the core MHC haplotype D or the rest of the CG2 genome, providing evidence for presence/absence variation of this subunit in zebrafish (Figs. 1 and 2).
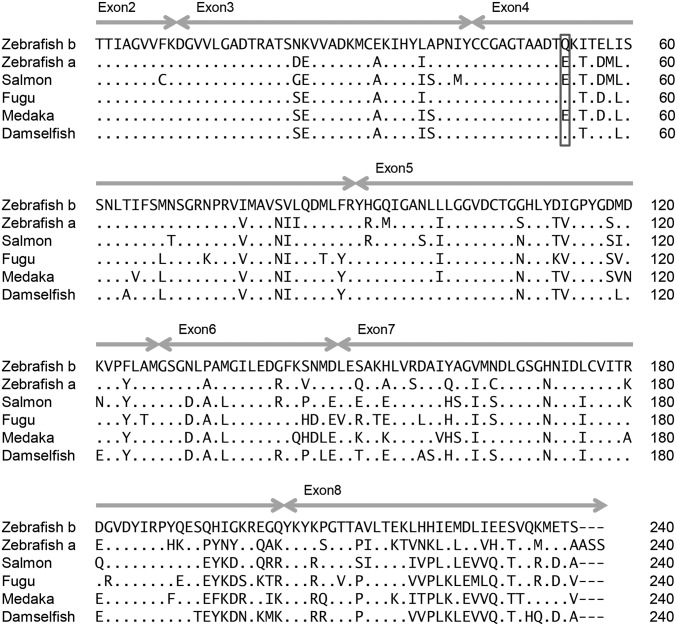
Sequence Properties for the Zebrafish Psmb13b.
Psmb13a and the Psmb13b subunit also maintain ancient lineages. Zebrafish Psmb13b shares levels of divergence with the zebrafish Psmb13a sequence (Fig. 4) that are similar to levels shared with sequences from other teleost species, including salmon (69 to 72% amino acid identity). This divergence pattern, with Psmb13b appearing as the most basal sequence, indicates that Psmb13a and Psmb13b sequences have been independently evolving for ∼300 My, since the time of the last common ancestor of zebrafish and salmonids (38). Comparison of the different zebrafish Psmb13 subunits with sequences from other species, thus, provides clear evidence for ancient lineages.
Fig. 4.
Alignment of Psmb13 amino acid sequences. The deduced amino acid sequence for zebrafish Psmb13b (zebrafish b sequence from CG2 haplotype 19D) is highly divergent from zebrafish Psmb13a (zebrafish a sequence from Zv9 reference genome haplotype 19B). The first residue shown is the start of the mature protein after proteolytic cleavage exposes this essential catalytic residue (T1) of the active site (39). In 3D structures of proteasome subunits, position 53 (highlighted with a box) is located in close proximity to the catalytic site of the enzyme (40). The E53Q substitution is proposed to alter peptide cleavage specificity among divergent zebrafish Psmb13 molecules and also found in additional species. Identity to the sequence is shown with dots, and dashes indicate deletions. Double-headed arrows mark ranges of exons. Accession numbers are provided in SI Appendix, Table S8.
The sequence alignment (Fig. 4) shows that many residues are unique for zebrafish Psmb13b and not found in Psmb13a or sequences identified from other species. However, one potentially important substitution found in Psmb13b is also shared by sequences from additional species. This amino acid substitution at position 53 of the mature proteasomal subunit may influence peptide cleavage specificity (39, 40). At this critical residue, zebrafish Psmb13b has an uncharged glutamine (Q) instead of the charged glutamic acid (E) residue found in most fish species. Notably, sequences from fugu (Takifugu rubripes) and damselfish (Stegastes partitus) also carry the E53Q substitution, suggesting that this is a functionally important polymorphism.
The E53 residue found in zebrafish Psmb13a is also found in other sequences from this family of subunits, including the human PSMB10 immunoproteasome subunit (SI Appendix, Fig. S1). The trypsin-like peptide cleavage activity of PSMB10 remains similar to the constitutive PSMB7 subunit that it replaces on IFN stimulation. The sequences of this family of subunits (including zebrafish Psmb7 and Psmb13a as well as human PSMB7 and PSMB10), for the most part, maintain conserved negatively charged residue E53 or D53, which may provide complementarity within their trypsin-like active sites to the positively charged residues found at the C termini of their cleaved peptides. Thus, the E53Q substitution found in selected subunits, such as zebrafish Psmb13b, may alter the otherwise highly conserved trypsin-like activity of this family of proteasomal subunits.
Phylogenetic Analysis of Zebrafish TAP Subunits.
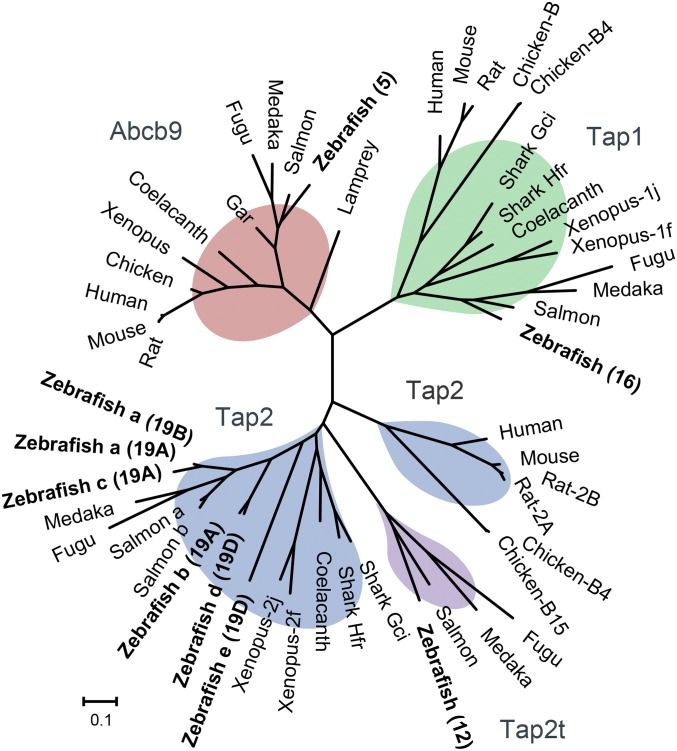
The abcb9 transporter gene (also called TAP-like) is common to all eukaryotes and considered the precursor of the heterodimeric tap1 and tap2 genes that arose during whole-genome duplications in ancestral vertebrates. Also found in jawless fish, such as lamprey (Fig. 5), the abcb9 gene in jawed vertebrates may have more limited function in MHCI antigen processing (41, 42). The ancestral abcb9 gene is largely monomorphic, unlike the polymorphic tap1 and tap2 genes found in many jawed vertebrates. For example, the derived tap1 gene was found to be highly polymorphic in some species, such as Xenopus (43) and chicken (26). Similarly, polymorphic alleles for tap2 have also been found in several species (Fig. 5), with divergent sequences ranging from >95% amino acid identity in chickens (26) to as low as 70% identity in Xenopus. In Xenopus, tap2 lineages evolved transspecifically, shared across species that diverged on the order of 80–100 Mya (43).
Fig. 5.
Phylogenetic relationships for TAP subunits from zebrafish and additional vertebrates with divergent lineages. Deduced amino acid sequences were used to construct maximum likelihood trees. For the largely monomorphic non–MHC-linked Abcb9, Tap1, and Tap2t subunits, only the copies encoded by the Zv9 reference genome are shown. Bootstrap values are provided in SI Appendix, Fig. S8. Chromosome locations for zebrafish Tap2 subunits are provided in parentheses, including haplotype associations when applicable. Sequences are provided in Dataset S2.
Remarkably, phylogenetic analysis highlights the various zebrafish Tap2 subunits as the most divergent sequences among species with polymorphic Tap2 molecules (Fig. 5). Three major lineages are observed for the MHC-linked zebrafish Tap2 subunits: Tap2a/Tap2c, Tap2b/Tap2d, and Tap2e. The Tap2a subunit encoded by haplotypes A and B is relatively closely related with Tap2c, but Tap2c may actually represent a diverging tandem duplicate. Some relatively unusual substitutions, including T217V and R262D (SI Appendix, Table S3), may imply that Tap2c function is not conserved with that of the other zebrafish Tap2 subunits. Tap2c also has rather uncharacteristic insertions and deletions in its alignment relative to sequences from other tap2 genes. For an additional zebrafish Tap2 lineage, the Tap2b and Tap2d subunits encoded by haplotypes A and D maintain 90% sequence identity, making them as divergent from one another as salmon Tap2a and Tap2b (91% sequence identity). The salmon tap2b gene is found in a duplicated MHCIB region (44) maintained 100 My after a salmonid-specific genome duplication event (45), providing a divergence time estimate consistent with these duplicated salmon tap2 genes now being ∼90% identical.
Perhaps most striking from the tree (Fig. 5) is the deep divergence between the zebrafish Tap2d and Tap2e subunits (sharing only 50% amino acid sequence identity). This level of sequence divergence is comparable with the relationship shared between Xenopus and shark Tap2 subunits (51 to 59% identity) and also, the relationship shared between sequences for other diverse vertebrates (42 to 57% identity), species that have been independently evolving for ∼500 My (46). Sequences derived from polymorphic tap2 alleles from chickens (26) each differ by ∼1–25 residues (>95% amino acid identity), similar to what has been found in rats (47). Xenopus species maintain divergent Tap2 sequences with over 200 amino acid substitutions (70% identity), representing lineages separated by 60–100 My of evolution (48). Therefore, the distinct zebrafish MHC haplotypes encode Tap2 molecules that are much more divergent than those found in other species previously described as maintaining highly polymorphic Tap2 molecules, such as rat, chicken, and Xenopus. These findings implicate independent evolution of tap2 sequences among zebrafish core MHC haplotypes over exceptionally long periods of time, approaching the time to reach common ancestors among major vertebrate lineages.
Several residues have been shown to alter the transport specificity of peptide antigens (47) within Tap2 sequences (SI Appendix, Table S3). Positively charged R262 is associated with restricted peptide transport in rats with a restrictive allele 2B, and R262 is also encoded by the restrictive mouse tap2 gene. In contrast, an uncharged residue Q262 is found in rats carrying a permissive peptide transport allele 2A, similar to the uncharged N262 encoded by the human permissive tap2 gene. Both charged (R262) and uncharged (Q262) amino acids are found among sequences of five zebrafish Tap2 subunits. At a second functional site, a bulky F266 residue is found in mice and rats with restrictive alleles, whereas a less bulky hydrophobic residue L266 is found in humans and rats with permissive tap2 genes. Both bulky (M266) and less bulky (L266) hydrophobic amino acids are also found in different zebrafish Tap2 subunits. At the start of the specificity loop is a third site, which has a T217A polymorphism that contributes to permissive peptide transport in rats. Both T217 and A217 residues are encoded among the divergent Tap2 subunits in zebrafish. These three polymorphisms are shared with functional polymorphisms found within Tap2 molecules from better characterized model organisms, providing evidence of potentially specialized functions for the divergent zebrafish Tap2 molecules. Our findings for proteasome subunit and Tap2 polymorphisms are in addition to other widespread polymorphisms found throughout the predicted peptide binding cleft of the linked MHCI genes (SI Appendix, Tables S9 and S10), which taken together, suggest strong likelihood for coevolution of peptide binding specificity throughout the entire zebrafish MHC pathway.
Proteasome and TAP Diversity Throughout Vertebrates.
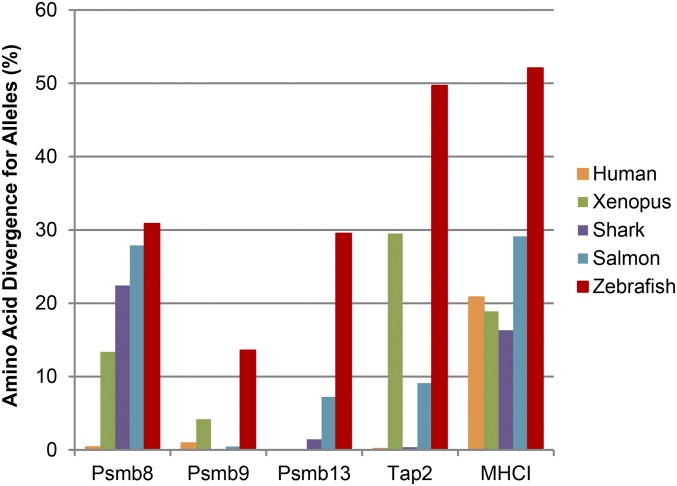
Comparative analysis of antigen processing genes throughout vertebrates yielded a number of surprises. Levels of divergence for alleles of zebrafish antigen processing and presentation genes exceeded levels found in other vertebrate species (Fig. 6). Higher levels of divergence were evident in the zebrafish psmb9, psmb13, tap2, and MHCI genes, particularly for psmb13.
Fig. 6.
Comparison of divergence levels for antigen processing and presentation genes. To compare the levels of divergence encoded by antigen processing and presentation genes from zebrafish and additional species, we searched for divergent alleles across vertebrates using the BLAST algorithm with expressed sequence tag (EST), transcriptome sequence assembly (TSA), and nonredundant (nr) GenBank databases. The most divergent alleles for each gene were identified and cataloged (accession numbers, divergence levels, and species identifiers are provided in SI Appendix, Table S11). Amino acid divergence (percentage) was calculated between these alleles using BLAST to identify those with the lowest amino acid identity, with divergence (%) = 100 (%) − identity (%). In many cases, these results are likely to underestimate diversity because of limitations, such as undersampling of sequences.
We also uncovered divergent psmb8f as well as psmb8a lineages in coelacanths (Fig. 3). These ancient psmb8 lineages cluster separately across sharks, teleosts, and coelacanths, implying that both lineages were present in the ancestors of all vertebrates, including tetrapods, such as humans and Xenopus. This observation supports the hypothesis that the somewhat less divergent psmb8 lineages found in Xenopus were derived as the result of “erosion” of ancestral psmb8f sequences (49), representing fragmented transspecies polymorphism, and are not the result of convergent evolution as originally proposed.
In addition, sharks apparently have maintained a psmb13 ortholog (Fig. 3), which is more closely related to the psmb13 lineage from teleosts than to the psmb10 lineage. However, unlike in teleosts, an additional gene representing the psmb10 lineage may, instead, be absent from sharks. This shark psmb13 gene appears to be largely monomorphic, unlike the salmon and zebrafish psmb13 genes (Fig. 6). Lower sequence diversity would be consistent with a non–MHC-linked (psmb10-like) role for the psmb13 gene in sharks.
Furthermore, paralleling previous findings in tetrapods (50), we determine that teleosts also have retained a non–MHC-linked psmb10 gene (Fig. 3). Teleost psmb10 is found outside of the core MHC, similar to human PSMB10, and these teleost psmb10 genes maintain conserved synteny with human psmb10 outside of their core MHC loci (SI Appendix, Fig. S3). Although previous studies had suggested that the teleost ortholog of human PSMB10 was, instead, MHC-linked (17, 18), our findings clearly establish a non–MHC-linked gene as the true PSMB10 ortholog in teleosts (Fig. 3). By also maintaining a largely monomorphic psmb10 gene, teleosts may have additional capacity to support more specialized functions for their divergent psmb13 genes.
Finally, we find that teleosts also have maintained a distinctive tap2 gene, tap2t, which seems be teleost-specific (Fig. 5). This gene is in addition to their MHC-linked and highly divergent tap2 lineages, indicating that the largely monomorphic non–MHC- linked gene, tap2t (SI Appendix, Fig. S4), may have additional conserved functions. In summary, teleosts maintain much higher diversity in their antigen processing genes than other vertebrates examined, including ancient sequence lineages across each of the MHC-linked antigen processing genes as well as conserved ancient paralogs tap2t (rather than only tap2), psmb12 (rather than only psmb9), and psmb13 (rather than only psmb10).
Discussion
In this study, we performed comparative genomic analysis of the core MHC region of zebrafish. Based on our de novo assembly of an alternative haplotype, we identified three antigen processing genes (tap2d, psmb13b, and tap2e) as well as additional MHC haplotype diversity. This diversity includes copy number differences for the tap2, psmb12, and tapbp genes and an inversion containing the three immunoproteasome genes. In addition to these genomic structural differences, ancient lineages are maintained for psmb8, psmb9, psmb13, and tap2. Taken together, these findings represent the most extensive diversity yet identified within the antigen processing genes of any species. Evidence of allelic variation for some antigen processing genes had been lacking (23), despite examination of numerous species across major vertebrate lineages. Therefore, our work addresses previously unrecognized gaps in our understanding of the evolution of vertebrate MHC regions, including identification of deeply divergent lineages for additional classes of proteasome and TAP subunits.
We have shown previously that zebrafish antigen presentation genes (MHCI) maintain copy number differences and divergent lineages among MHC haplotypes (31, 51). These findings for zebrafish antigen processing genes (including tap2, psmb12, and psmb13), thus, parallel and also greatly expand on the diversity that we previously described for the tightly linked MHCI genes. Unlike copy number differences in closely related genes found at many other loci, these antigen processing genes (tap2, tapbp, psmb8, psmb9, psmb12, and psmb13) and MHCI genes, although linked, are at most only weakly related to one another by sequence. Nevertheless, these genes remain functionally united by their various roles within a common antigen processing and presentation pathway.
Five forms of proteasome assemblies have been described in mammals: constitutive proteasome, immunoproteasome, two forms of intermediate (“mixed”) proteasome, and thymoproteasome (52). The number of different proteasome compositions seems limited by constraints of cooperative subunit assembly (53) (e.g., psmb8 before psmb9), because otherwise, at least twice this number of different assemblies might be expected. In zebrafish, a significantly larger number of distinct subunits (at least 12 variants vs. only 7 variants in mammals) offers potential for many additional proteasome assemblies. Even after accounting for cooperative assembly constraints, as many as 30 distinct combinations (25 unique to fish) could form in zebrafish that inherit two alternative MHC haplotypes (SI Appendix, Table S4). Moreover, based on predicted cleavage properties for the different subunits, different peptide repertoires are likely associated with these distinct zebrafish proteasome assemblies. Thus, even if most of these subunit combinations remain purely hypothetical, the additional proteasome subunits may nevertheless support a much greater diversity of peptide repertoires in zebrafish.
Widespread variation found among these different MHC pathway genes may be related to the specialization of antigen repertoires between haplotypes. This hypothesis is supported by several findings, including polymorphism that may lead to reduced trypsin-like activity for the Psmb13b subunit (Fig. 4). Furthermore, we identified additional polymorphism within the specificity loop of zebrafish Tap2 subunits (SI Appendix, Table S3), with substitutions identical to those shown in other species to control antigen transport specificity. These substitutions may be either permissive or restrictive to the transport of tryptic-like cleavage products having positively charged C termini. Polymorphisms associated with each of these linked zebrafish genes may help reinforce one another’s functions by promoting compatible peptide antigen repertoires, as was previously observed for linked tap2 and MHCI genes that coevolve distinctive peptide specificities in the rat and chicken (24, 26). Divergent sequences for the various zebrafish antigen processing genes may, therefore, be related to specialized functions, such as has been proposed for ancient transspecies polymorphism in psmb8 also found in other species.
The psmb8a and psmb8f sequences from different zebrafish MHC haplotypes have been diverging for approximately 0.5 billion y (28). Here, we provide genomic context for psmb8f, which to date, has been studied primarily through amplicons and/or expressed transcripts. Our comparative genomic analysis shows that additional divergent sequences extend far beyond the boundaries of the zebrafish psmb8f gene, covering ∼100 kb of the MHC region. Surprisingly, distinct zebrafish MHC haplotypes maintain large regions of nearly unalignable sequence (SI Appendix, Fig. S2), comprising divergent gene lineages, copy number differences, and other structural changes. Despite this extensive sequence divergence, the psmb8f haplotype still retains representatives from all of the MHCI pathway genes (except psmb12), apparently leaving the integrity of this pathway intact.
We identified a chromosomal inversion (containing three divergent proteasome subunit genes for haplotype D) that may help further suppress recombination throughout this region. A similar mechanism by which chromosomal inversion suppresses recombination has been proposed for mouse MHC haplotypes (54). Because of stable haplotypes, coinherited genes may have accumulated their genetic diversity primarily because of this shared genomic location, maintaining deep lineages that parallel the divergent psmb8f and mhc1uga genes. Conversely, these genes may have developed their tight linkage primarily because of their cooperative and exclusive roles in enhancing shared MHC pathway function (23, 55). Maintaining a stable haplotypic structure would then help avoid sequence exchange events, such as recombination, that would interfere with coinherited gene function (56, 57) and thus, maintain efficiency of the MHC pathway.
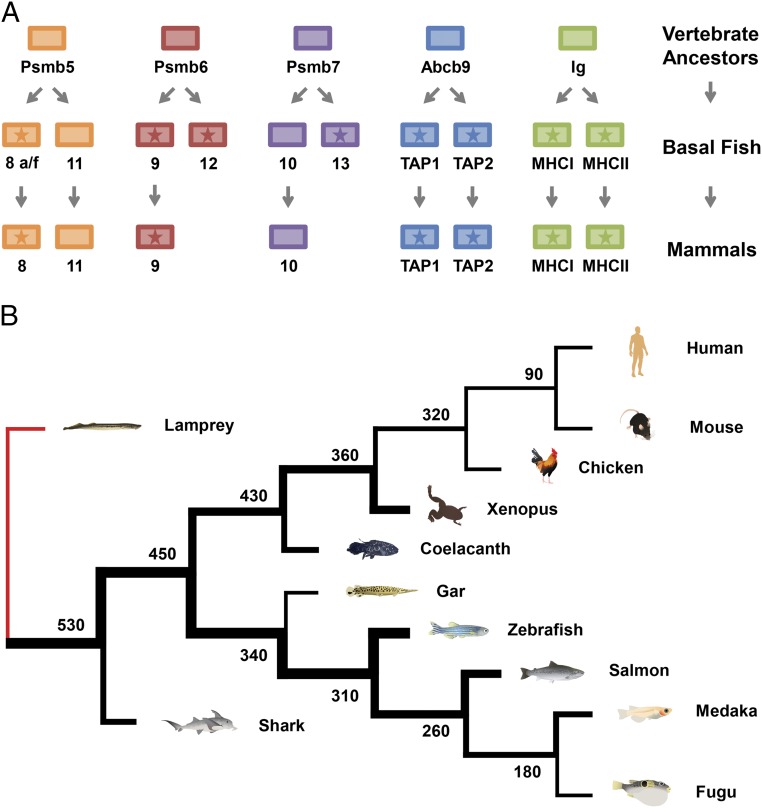
Our results support a model where ancient whole-genome duplications produced a collection of precursor antigen processing and presentation genes in the ancestors of jawed vertebrates (Fig. 7A). After two rounds of whole-genome duplication, psmb5 provided the precursors for psmb8a, psmb8f, and psmb11. Although psmb5 was maintained as a constitutive proteasomal subunit, the three derived genes experienced reduced functional constraints as the paralogous psmb8 genes gained IFN response and psmb11 became thymus-specific. Similarly, constitutive psmb6 duplicated to produce precursors for IFN-inducible psmb9 as well as psmb12. In addition, psmb7 served as the precursor for IFN-inducible psmb10 and also, psmb13. These scenarios were mirrored by the abcb9 gene, which yielded the heterodimeric tap1 and tap2. Additionally, the Ig domain served as a foundation for formation of both MHCI and MHCII genes. A large proportion of these genes has maintained core MHC linkage throughout vertebrates, reflecting not only presumed primordial linkage important for evolution of the MHC pathway but also, continued linkage and coevolution optimizing MHC pathway function. An alternative model might consider psmb12 and psmb13 to be teleost-specific, similar to tap2t or psmb11a. However, evidence of shark sequences related to teleost psmb13 suggests that these psmb13 sequences are much older than teleosts. In addition, relative to the other proteasome genes (Fig. 3), the divergent nature of the psmb12 and psmb13 genes (surpassing even the divergent psmb8 lineages that are shared across all major branches of vertebrates), instead, argues for a much more basal position for the psmb12 and psmb13 genes in ancestral vertebrates, most similar to the position of psmb11.
Fig. 7.
Evolution of antigen processing and presentation genes throughout vertebrates. (A) The ancestors of jawed vertebrates underwent two rounds of whole-genome duplication. Many of the duplicated genes subsequently acquired roles in immune function, such as peptide binding/specificity, whereas early linkage within the proto-MHC likely facilitated coevolution of these genes to optimize MHC pathway function. Referred to here as basal fish, the last common ancestors of sharks, ray-finned fish, and lobe-finned fish maintained a higher number of MHC pathway genes than many extant vertebrates, particularly compared with mammals. This loss of mammalian MHC pathway diversity apparently included psmb12 and psmb13 as well as psmb8f. Genes with stars map to the core MHC region. Linkage differences are now found in some lineages, likely as a derived condition; for example, in teleost fish, the MHCII and tap1 genes are no longer linked to the core MHC. (B) Lampreys as jawless fish completely lack an MHC pathway; however, sharks in the oldest surviving jawed vertebrate lineage had already evolved a complete MHC pathway similar to that of other jawed vertebrates. Ray-finned fish (e.g., zebrafish) as well as lobe-finned fish (e.g., coelacanths) lineages are shown to have maintained varying amounts of MHC pathway diversity, appearing more concentrated initially. Thicker lines represent greater sequence diversity for genes within the MHC pathway estimated by comparing available sequence lineages relative to other vertebrates examined. Sequence information is provided in SI Appendix, Tables S11–S13. Numbers at branch points indicate divergence time estimates in millions of years (38).
Accordingly, some of these genes, including psmb13, and allelic lineages, such as psmb8f, were subsequently lost in the mammalian branch of the tetrapod lineage. In certain vertebrate lineages, additional subsets of MHC pathway genes have been selectively lost, including MHCII genes in Atlantic cod (58) and nonconstitutive proteasome subunits in chickens (25). Nevertheless, immune systems within these species have apparently compensated for these genetic losses through other strategies, such as amplifying their MHCI gene number (58) or maintaining a larger number of MHCI and TAP alleles (26). Mammalian MHCI gene function may also have been shaped by the early loss of genes, such as psmb13 and psmb8f. These losses combined with physical separation limiting the coevolution of their MHCI genes and antigen processing genes likely helped select for their hallmark collections of highly polymorphic classical MHC genes.
The exceptionally high levels of allelic divergence for genes, such as psmb8 (Fig. 6), found across a full range of nonmammalian vertebrate species, including zebrafish, sharks, and coelacanths, exemplify the sharing of ancestral duplicated genes and ancient alleles for genes throughout the MHC pathway. Ancient sequence lineages for these genes have since been “eroded” to various extents across different vertebrate branches as well as occasionally lost in branches, such as mammals (Fig. 7B). The amount of remaining MHC pathway diversity may, thus, have had an important role in shaping vertebrate immunity.
Zebrafish are a relatively well-studied model organism, endowed with one of the highest quality “finished” vertebrate reference genomes (32). Zebrafish excel as a model to uncover gene function (59, 60) and are now advancing research in many fields, including stem cell biology (61, 62), cancer (63, 64), infectious disease (65, 66), and autoimmune disease studies (67, 68). In this context, we consider the identification of expressed genes in our zebrafish MHC assembly to be noteworthy. Based on earlier studies, genomic sequences were recognized as missing for psmb8f and mhc1uga (31) as well as for additional orphan genes such as psmb9b (32), a gene previously linked to an alternative MHC haplotype assembly (17). However, our examination of the genomic context for psmb8f also revealed three additional expressed genes: psmb13b, tap2d, and tap2e. These results indicate that sampling of immune genes may be far from saturated, even for well-studied model organisms, such as zebrafish.
Genes from divergent MHC pathways might be expected to strongly influence immune responses, particularly susceptibility to infectious disease, such as has been found in studies examining the impact of much more limited polymorphism (56, 69, 70). Zebrafish are now frequently used to model human disease; therefore, future studies could address the impact of these ancient immune sequences in settings such as viral infections. These studies may also inform our understanding of other populations such as humans, in whom these ancient genes have been lost, because this loss may have had important consequences for subsequent evolution of immune function.
Methods
Array-Based Capture and Whole-Exome Sequencing of Clonal Zebrafish.
CG1 and CG2 represent two homozygous diploid (clonal) zebrafish lines (35, 36) derived by in-crossing isogenic offspring after two consecutive rounds of parthenogenesis (71). Each line was cloned from outbred AB zebrafish (72) carrying the golden allele (73). CG1 and CG2 are homozygous for distinct core MHC haplotypes C and D, respectively, among six divergent MHC haplotypes identified within laboratory AB stocks (31).
Genomic DNA samples from CG1 and CG2 (using four individuals from each line) were prepared for whole-exome analysis using previously described methods (59). Briefly, standard Illumina primers were used to amplify libraries and barcode different DNA samples, which were pooled before target enrichment. Agilent SureSelect Target Enrichment relied on 120-bp baits designed against all annotated protein coding genes from the Ensembl Zv9 release of the zebrafish genome. Captured DNA was then sequenced on an Illumina HiSeq2000 instrument using a single lane. Reads were aligned with the Burrows–Wheeler Aligner against the Zv9 reference genome.
SNPs were called by using a combination of GATK Unified Genotyper, SAMtools mpileup, and QCALL. SNPs were identified as either heterozygous or homozygous after passing filters from all three SNP callers with additional quality controls as described (59). Approximately 400,000–500,000 SNPs were identified for each clonal line (SI Appendix, Table S5). However, within each of two clonal lines, the vast majority (>98.2%) of SNPs were called as homozygous, consistent with double-haploid derivation of isogenic zebrafish (71).
Whole-Genome Sequencing and de Novo Assembly.
Genomic DNA from a CG2 clonal zebrafish was sequenced using an Illumina HiSeq2500 instrument, producing over 38 Gb 2 × 100 paired end read data with ∼25× sequence coverage. Illumina adapters were removed using SeqPrep (74) version 1.1, and reads were filtered for quality with Trimmomatic (75) version 0.3. After filtering and clipping, read quality was assessed using FastQC (76). De novo assembly was generated using the SOAPdenovo2 (77) algorithm with optimized parameters (kmer value of 59). The resulting de novo assembly had an N50 (median scaffold size of genomic assembly) value of 34 kb, with 5.7% Ns (unknown bases) and scaffolds covering ∼82% of the genome. Scaffolds larger than 1 kb were aligned against the zebrafish Zv9 reference genome assembly using the nucmer tool from MUMmer (78) version 3.23. We used Augustus (79) to generate gene models from our genomic scaffolds as well as Webscipio (80) to help improve gene annotation. Within the core MHC locus, conserved flanking genes, including daxx and tapbp, anchor CG2 genomic scaffold 13,206, which also includes the 5′ portion of mhc1uga. The 3′ portion of mhc1uga is included within scaffold 51,738. Similarly, brd2a and hsd17b8 within the core MHC locus anchor scaffold 2,546, which also includes tap2e and psmb8f as well as the 3′ portion of psmb13b. The 5′ portion of psmb13b is found within scaffold 15,837, which also includes the psmb9 and tap2d genes.
RNA-Seq Transcriptome Assembly and Sequence Analysis.
Generation of the CG2 RNA-Seq library was described previously (51). Briefly, kidney, spleen, intestine, and gill were dissected and pooled to purify RNA from immune tissues of CG2 clonal zebrafish. Paired end 2 × 100-bp reads were generated with an Illumina HiSeq2000 instrument and assembled using Trinity (81). Amino acid sequences were aligned using MUSCLE (82). Phylogenetic trees were constructed using the maximum likelihood method within the MEGA6 program (83) and bootstrapped with 500 replicates. Pairwise amino acid identity was calculated using BLAST (84). Exome data were visualized using the IGV Viewer (85). Transcripts associated with zebrafish core MHC haplotype D are provided in Dataset S3, predicted amino acid sequences for the CG2 haplotype D antigen processing gene transcripts are provided in Dataset S4, and genomic scaffold sequences identified from haplotype D are provided in Dataset S5.
Nomenclature for Proteasome Subunits.
Nomenclature for proteasome and TAP genes has remained inconsistent across species and studies, particularly for genes not found in the mammalian lineage. Here, we provide systematic nomenclature that encompasses identified as well as additional proteasome genes (SI Appendix, Table S6). This nomenclature takes into account phylogenomic analysis, including conserved syntenies, and is based on original gene nomenclature proposals. All zebrafish gene names have been approved by the zebrafish nomenclature committee.
An MHC-linked zebrafish gene in the psmb6/9 family, first described as psmb11 (29), has also been called psmb9l (18). However, the name psmb11 is currently problematic, because it conflicts with nomenclature for distinct vertebrate genes also called psmb11 (37) (e.g., zebrafish psmb11a and psmb11b that are found outside the core MHC). These two latter zebrafish genes belong to the conserved psmb5/8/11 family (Fig. 3), where subunits follow the (psmb X, X + 3, and X + 6) numbering schema across vertebrates. Psmb9l is another name used for the former MHC-linked gene, but this “psmb9-like” gene is actually highly divergent from the psmb9 lineage, contributing a distinctive third lineage branch across teleosts (Fig. 3). Furthermore, the appended letter for psmb9l may become confusing when in use with genes with other appended letters, such as psmb9a. These considerations led us to propose the name psmb12 (SI Appendix, Table S6), recognizing the psmb6/9/12 gene family. Our proposed name reflects the status of psmb12 as the most divergent branch within this family and provides parallel (psmb Y, Y + 3, and Y + 6) nomenclature while using the next gene name available.
Another proteasome subunit gene discovered in the teleost core MHC was originally described as psmb12 (29), recognizing that, although clearly related, it is also quite distinct from tetrapod psmb10. Subsequently, this gene was annotated as psmb10 in fugu (18) and more recently, other fish species. However, the psmb10 assignment for this gene is justified only if another conserved, non–MHC-linked psmb10 ortholog is truly absent from bony fish genomes. Surprisingly, we found evidence of a conserved non–MHC-linked psmb10 gene throughout bony fish species (Fig. 3). This finding implies that psmb10 was already found outside of the MHC in the common ancestors of tetrapods and bony fish, which is inconsistent with a more derivative tetrapod psmb10 translocation event suggested in previous models. Thus, our nomenclature assigns psmb10 as an immunoproteasome subunit within most vertebrate species, including bony fish, where it is unlinked to the MHC.
We propose that the MHC-linked zebrafish gene formerly known as psmb12 or psmb10 be renamed psmb13 (SI Appendix, Table S6). The psmb13 gene is conserved across teleosts and to a lesser degree, sharks (Fig. 3), suggesting ancient conserved function for this gene that may curiously now be missing from other vertebrates, such as humans. Our proposal, thus, assigns psmb13 as the third divergent lineage belonging to the psmb7/10/13 gene family following parallel nomenclature structure (psmb Z, Z + 3, and Z + 6). In summary, the psmb7/10/13 gene family forms the third and final lineage trifurcation among the psmb5–13 genes (Fig. 3).
Nomenclature for TAP Subunits.
We also propose names for seven zebrafish TAP genes (SI Appendix, Table S7) based on the original nomenclature (17), including tap2a and tap2b (rather than abcb3l1 and abcb3, respectively). Tap2, tap1, and ancestral abcb9 form an ancient lineage trifurcation across jawed vertebrates (Fig. 5). We propose that zebrafish abcb2 be renamed tap1 (SI Appendix, Table S7) to maintain consistency with nomenclature used for orthologous genes, such as in humans (86).
Furthermore, within the larger tap2 branch, several genes cluster together as distinct from the tap2 genes found in the MHC region of their respective teleost species (Fig. 5). Our proposed name for the identified zebrafish gene is tap2t, reflecting recognition of this gene as a teleost-specific member of the larger tap2 family (Fig. 5). Tap2t is not linked to the core MHC region but has conserved synteny among teleosts (SI Appendix, Fig. S4). Similar to the other Tap2 subunits in zebrafish, Tap2t conserves key residues within a specificity loop predicted to interact with peptides (SI Appendix, Table S3). In summary, this zebrafish proteasome and TAP gene nomenclature remains consistent with that in humans as well as additional vertebrates (44). We believe that this proposal will help further promote nomenclature consistency across species, facilitating future comparative studies.
Supplementary Material
Acknowledgments
We thank Ian Sealy for help with exome data, Betsy Scholl for bioinformatics support, Sergei Revskoy for clonal zebrafish, and Daniel Ocampo Daza for vertebrate illustrations. We thank Wilfredo Marin and Haley Engel at the University of Chicago for excellent care of our zebrafish colony. We also thank Pieter Faber from the University of Chicago Genomics core and Cancer Center Support Grant (P30 CA014599) for sequencing support. This work was funded, in part, by a Chicago Biomedical Consortium Postdoctoral Research Grant with support from the Searle Funds at the Chicago Community Trust and, also, the University of Chicago Cancer Research Foundation Auxiliary Board. D.J.W. was supported by the National Evolutionary Synthesis Center (National Science Foundation Grant EF-0905606).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. P.P. is a Guest Editor invited by the Editorial Board.
Data deposition: Exome data for CG1 clonal zebrafish have been deposited in the NCBI Sequence Read Archive (SRA; accession nos. ERS216437, ERS216444, ERS216451, and ERS216458), and exome data for CG2 clonal zebrafish have been deposited in the NCBI SRA (accession nos. ERS216465, ERS216472, ERS216479, and ERS216486). The CG2 immune tissue RNA-Seq data (nonnormalized and normalized) have been deposited in the NCBI SRA (accession no. SRP057116). The nonnormalized and normalized CG2 immune tissue transcript assembly data have been deposited in the Transcriptome Shotgun Assembly (TSA; accession nos. GDQH00000000 and GDQQ00000000). Genomic sequencing data from CG2 clonal zebrafish have been deposited in the NCBI SRA (accession no. SRP062426), and the CG2 genomic assembly has been deposited in the NCBI Whole Genome Shotgun database (accession no. LKPD00000000). Transcripts associated with zebrafish core MHC haplotype D are provided in in Dataset S3, predicted amino acid sequences for the CG2 haplotype D antigen processing gene transcripts are provided in Dataset S4, and genomic scaffold sequences identified from haplotype D are provided in Dataset S5.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1607602113/-/DCSupplemental.
References
- 1.Shiina T, Hosomichi K, Inoko H, Kulski JK. The HLA genomic loci map: Expression, interaction, diversity and disease. J Hum Genet. 2009;54(1):15–39. doi: 10.1038/jhg.2008.5. [DOI] [PubMed] [Google Scholar]
- 2.Klein J, Sato A, Nagl S, O’hUigín C. Molecular trans-species polymorphism. Annu Rev Ecol Syst. 1998;29:1–21. [Google Scholar]
- 3.Piontkivska H, Nei M. Birth-and-death evolution in primate MHC class I genes: Divergence time estimates. Mol Biol Evol. 2003;20(4):601–609. doi: 10.1093/molbev/msg064. [DOI] [PubMed] [Google Scholar]
- 4.Raymond CK, et al. Ancient haplotypes of the HLA Class II region. Genome Res. 2005;15(9):1250–1257. doi: 10.1101/gr.3554305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klein J, Sato A, Nikolaidis N. MHC, TSP, and the origin of species: From immunogenetics to evolutionary genetics. Annu Rev Genet. 2007;41:281–304. doi: 10.1146/annurev.genet.41.110306.130137. [DOI] [PubMed] [Google Scholar]
- 6.Barreiro LB, Quintana-Murci L. From evolutionary genetics to human immunology: How selection shapes host defence genes. Nat Rev Genet. 2010;11(1):17–30. doi: 10.1038/nrg2698. [DOI] [PubMed] [Google Scholar]
- 7.Okamura K, Ototake M, Nakanishi T, Kurosawa Y, Hashimoto K. The most primitive vertebrates with jaws possess highly polymorphic MHC class I genes comparable to those of humans. Immunity. 1997;7(6):777–790. doi: 10.1016/s1074-7613(00)80396-9. [DOI] [PubMed] [Google Scholar]
- 8.Flajnik MF, et al. Two ancient allelic lineages at the single classical class I locus in the Xenopus MHC. J Immunol. 1999;163(7):3826–3833. [PubMed] [Google Scholar]
- 9.Shum BP, et al. Modes of salmonid MHC class I and II evolution differ from the primate paradigm. J Immunol. 2001;166(5):3297–3308. doi: 10.4049/jimmunol.166.5.3297. [DOI] [PubMed] [Google Scholar]
- 10.Aoyagi K, et al. Classical MHC class I genes composed of highly divergent sequence lineages share a single locus in rainbow trout (Oncorhynchus mykiss) J Immunol. 2002;168(1):260–273. doi: 10.4049/jimmunol.168.1.260. [DOI] [PubMed] [Google Scholar]
- 11.Jaratlerdsiri W, et al. Comparative genome analyses reveal distinct structure in the saltwater crocodile MHC. PLoS One. 2014;9(12):e114631. doi: 10.1371/journal.pone.0114631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Venkatesh B, et al. Elephant shark genome provides unique insights into gnathostome evolution. Nature. 2014;505(7482):174–179. doi: 10.1038/nature12826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grimholt U, et al. A comprehensive analysis of teleost MHC class I sequences. BMC Evol Biol. 2015;15(1):32. doi: 10.1186/s12862-015-0309-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ng JHJ, et al. Evolution and comparative analysis of the bat MHC-I region. Sci Rep. 2016;6:21256. doi: 10.1038/srep21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ohta Y, McKinney EC, Criscitiello MF, Flajnik MF. Proteasome, transporter associated with antigen processing, and class I genes in the nurse shark Ginglymostoma cirratum: Evidence for a stable class I region and MHC haplotype lineages. J Immunol. 2002;168(2):771–781. doi: 10.4049/jimmunol.168.2.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flajnik MF, Ohta Y, Namikawa-Yamada C, Nonaka M. Insight into the primordial MHC from studies in ectothermic vertebrates. Immunol Rev. 1999;167(1):59–67. doi: 10.1111/j.1600-065x.1999.tb01382.x. [DOI] [PubMed] [Google Scholar]
- 17.Michalová V, Murray BW, Sültmann H, Klein J. A contig map of the Mhc class I genomic region in the zebrafish reveals ancient synteny. J Immunol. 2000;164(10):5296–5305. doi: 10.4049/jimmunol.164.10.5296. [DOI] [PubMed] [Google Scholar]
- 18.Clark MS, Shaw L, Kelly A, Snell P, Elgar G. Characterization of the MHC class I region of the Japanese pufferfish (Fugu rubripes) Immunogenetics. 2001;52(3-4):174–185. doi: 10.1007/s002510000285. [DOI] [PubMed] [Google Scholar]
- 19.Matsuo MY, Asakawa S, Shimizu N, Kimura H, Nonaka M. Nucleotide sequence of the MHC class I genomic region of a teleost, the medaka (Oryzias latipes) Immunogenetics. 2002;53(10-11):930–940. doi: 10.1007/s00251-001-0427-3. [DOI] [PubMed] [Google Scholar]
- 20.Ohta Y, Goetz W, Hossain MZ, Nonaka M, Flajnik MF. Ancestral organization of the MHC revealed in the amphibian Xenopus. J Immunol. 2006;176(6):3674–3685. doi: 10.4049/jimmunol.176.6.3674. [DOI] [PubMed] [Google Scholar]
- 21.Kaufman J. Antigen processing and presentation: Evolution from a bird’s eye view. Mol Immunol. 2013;55(2):159–161. doi: 10.1016/j.molimm.2012.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Siddle HV, et al. MHC-linked and un-linked class I genes in the wallaby. BMC Genomics. 2009;10(1):310. doi: 10.1186/1471-2164-10-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ohta Y, Flajnik MF. Coevolution of MHC genes (LMP/TAP/class Ia, NKT-class Ib, NKp30-B7H6): lessons from cold-blooded vertebrates. Immunol Rev. 2015;267(1):6–15. doi: 10.1111/imr.12324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Joly E, et al. Co-evolution of rat TAP transporters and MHC class I RT1-A molecules. Curr Biol. 1998;8(3):169–172. doi: 10.1016/s0960-9822(98)70065-x. [DOI] [PubMed] [Google Scholar]
- 25.Erath S, Groettrup M. No evidence for immunoproteasomes in chicken lymphoid organs and activated lymphocytes. Immunogenetics. 2015;67(1):51–60. doi: 10.1007/s00251-014-0814-1. [DOI] [PubMed] [Google Scholar]
- 26.Walker BA, et al. The dominantly expressed class I molecule of the chicken MHC is explained by coevolution with the polymorphic peptide transporter (TAP) genes. Proc Natl Acad Sci USA. 2011;108(20):8396–8401. doi: 10.1073/pnas.1019496108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bingulac-Popovic J, et al. Mapping of mhc class I and class II regions to different linkage groups in the zebrafish, Danio rerio. Immunogenetics. 1997;46(2):129–134. doi: 10.1007/s002510050251. [DOI] [PubMed] [Google Scholar]
- 28.Tsukamoto K, Miura F, Fujito NT, Yoshizaki G, Nonaka M. Long-lived dichotomous lineages of the proteasome subunit beta type 8 (PSMB8) gene surviving more than 500 million years as alleles or paralogs. Mol Biol Evol. 2012;29(10):3071–3079. doi: 10.1093/molbev/mss113. [DOI] [PubMed] [Google Scholar]
- 29.Murray BW, Sültmann H, Klein J. Analysis of a 26-kb region linked to the Mhc in zebrafish: Genomic organization of the proteasome component β/transporter associated with antigen processing-2 gene cluster and identification of five new proteasome β subunit genes. J Immunol. 1999;163(5):2657–2666. [PubMed] [Google Scholar]
- 30.Tsukamoto K, et al. Dichotomous haplotypic lineages of the immunoproteasome subunit genes, PSMB8 and PSMB10, in the MHC class I region of a Teleost Medaka, Oryzias latipes. Mol Biol Evol. 2009;26(4):769–781. doi: 10.1093/molbev/msn305. [DOI] [PubMed] [Google Scholar]
- 31.McConnell SC, Restaino AC, de Jong JLO. Multiple divergent haplotypes express completely distinct sets of class I MHC genes in zebrafish. Immunogenetics. 2014;66(3):199–213. doi: 10.1007/s00251-013-0749-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Howe K, et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature. 2013;496(7446):498–503. doi: 10.1038/nature12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Flajnik MF, Kasahara M. Comparative genomics of the MHC: Glimpses into the evolution of the adaptive immune system. Immunity. 2001;15(3):351–362. doi: 10.1016/s1074-7613(01)00198-4. [DOI] [PubMed] [Google Scholar]
- 34.Flajnik MF, Kasahara M. Origin and evolution of the adaptive immune system: Genetic events and selective pressures. Nat Rev Genet. 2010;11(1):47–59. doi: 10.1038/nrg2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith AC, et al. High-throughput cell transplantation establishes that tumor-initiating cells are abundant in zebrafish T-cell acute lymphoblastic leukemia. Blood. 2010;115(16):3296–3303. doi: 10.1182/blood-2009-10-246488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mizgirev IV, Revskoy S. A new zebrafish model for experimental leukemia therapy. Cancer Biol Ther. 2010;9(11):895–902. doi: 10.4161/cbt.9.11.11667. [DOI] [PubMed] [Google Scholar]
- 37.Sutoh Y, et al. Comparative genomic analysis of the proteasome β5t subunit gene: Implications for the origin and evolution of thymoproteasomes. Immunogenetics. 2012;64(1):49–58. doi: 10.1007/s00251-011-0558-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hedges S, Kumar S. The Timetree of Life. Oxford Univ Press; New York: 2009. [Google Scholar]
- 39.Ferrington DA, Gregerson DS. Immunoproteasomes: Structure, function, and antigen presentation. Prog Mol Biol Transl Sci. 2012;109:75–112. doi: 10.1016/B978-0-12-397863-9.00003-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huber EM, et al. Immuno- and constitutive proteasome crystal structures reveal differences in substrate and inhibitor specificity. Cell. 2012;148(4):727–738. doi: 10.1016/j.cell.2011.12.030. [DOI] [PubMed] [Google Scholar]
- 41.Uinuk-ool TS, et al. Identification and characterization of a TAP-family gene in the lamprey. Immunogenetics. 2003;55(1):38–48. doi: 10.1007/s00251-003-0548-y. [DOI] [PubMed] [Google Scholar]
- 42.Herget M, Tampé R. Intracellular peptide transporters in human—compartmentalization of the “peptidome.”. Pflugers Arch. 2007;453(5):591–600. doi: 10.1007/s00424-006-0083-4. [DOI] [PubMed] [Google Scholar]
- 43.Ohta Y, et al. Two highly divergent ancient allelic lineages of the transporter associated with antigen processing (TAP) gene in Xenopus: Further evidence for co-evolution among MHC class I region genes. Eur J Immunol. 2003;33(11):3017–3027. doi: 10.1002/eji.200324207. [DOI] [PubMed] [Google Scholar]
- 44.Lukacs MF, et al. Genomic organization of duplicated major histocompatibility complex class I regions in Atlantic salmon (Salmo salar) BMC Genomics. 2007;8(1):251–266. doi: 10.1186/1471-2164-8-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Berthelot C, et al. The rainbow trout genome provides novel insights into evolution after whole-genome duplication in vertebrates. Nat Commun. 2014;5(3657):3657. doi: 10.1038/ncomms4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Blair JE, Hedges SB. Molecular phylogeny and divergence times of deuterostome animals. Mol Biol Evol. 2005;22(11):2275–2284. doi: 10.1093/molbev/msi225. [DOI] [PubMed] [Google Scholar]
- 47.Deverson EV, et al. Functional analysis by site-directed mutagenesis of the complex polymorphism in rat transporter associated with antigen processing. J Immunol. 1998;160(6):2767–2779. [PubMed] [Google Scholar]
- 48.Ohta Y, et al. Identification and genetic mapping of Xenopus TAP2 genes. Immunogenetics. 1999;49(3):171–182. doi: 10.1007/s002510050478. [DOI] [PubMed] [Google Scholar]
- 49.Huang C-H, Tanaka Y, Fujito NT, Nonaka M. Dimorphisms of the proteasome subunit beta type 8 gene (PSMB8) of ectothermic tetrapods originated in multiple independent evolutionary events. Immunogenetics. 2013;65(11):811–821. doi: 10.1007/s00251-013-0729-2. [DOI] [PubMed] [Google Scholar]
- 50.Kasahara M, et al. Chromosomal localization of the proteasome Z subunit gene reveals an ancient chromosomal duplication involving the major histocompatibility complex. Proc Natl Acad Sci USA. 1996;93(17):9096–9101. doi: 10.1073/pnas.93.17.9096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dirscherl H, Yoder JA. A nonclassical MHC class I U lineage locus in zebrafish with a null haplotypic variant. Immunogenetics. 2015;67(9):501–513. doi: 10.1007/s00251-015-0862-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McCarthy MK, Weinberg JB. The immunoproteasome and viral infection: A complex regulator of inflammation. Front Microbiol. 2015;6:21. doi: 10.3389/fmicb.2015.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guillaume B, et al. Two abundant proteasome subtypes that uniquely process some antigens presented by HLA class I molecules. Proc Natl Acad Sci USA. 2010;107(43):18599–18604. doi: 10.1073/pnas.1009778107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hammer MF, Schimenti J, Silver LM. Evolution of mouse chromosome 17 and the origin of inversions associated with t haplotypes. Proc Natl Acad Sci USA. 1989;86(9):3261–3265. doi: 10.1073/pnas.86.9.3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kaufman J. Co-evolution with chicken class I genes. Immunol Rev. 2015;267(1):56–71. doi: 10.1111/imr.12321. [DOI] [PubMed] [Google Scholar]
- 56.Kaufman J. What chickens would tell you about the evolution of antigen processing and presentation. Curr Opin Immunol. 2015;34:35–42. doi: 10.1016/j.coi.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 57.Tuncel J, et al. EURATRANS Consortium Natural polymorphisms in Tap2 influence negative selection and CD4:CD8 lineage commitment in the rat. PLoS Genet. 2014;10(2):e1004151. doi: 10.1371/journal.pgen.1004151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Star B, et al. The genome sequence of Atlantic cod reveals a unique immune system. Nature. 2011;477(7363):207–210. doi: 10.1038/nature10342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kettleborough RNW, et al. A systematic genome-wide analysis of zebrafish protein-coding gene function. Nature. 2013;496(7446):494–497. doi: 10.1038/nature11992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Phillips JB, Westerfield M. Zebrafish models in translational research: Tipping the scales toward advancements in human health. Dis Model Mech. 2014;7(7):739–743. doi: 10.1242/dmm.015545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Barbosa JS, et al. Neurodevelopment. Live imaging of adult neural stem cell behavior in the intact and injured zebrafish brain. Science. 2015;348(6236):789–793. doi: 10.1126/science.aaa2729. [DOI] [PubMed] [Google Scholar]
- 62.Tamplin OJ, et al. Hematopoietic stem cell arrival triggers dynamic remodeling of the perivascular niche. Cell. 2015;160(1-2):241–252. doi: 10.1016/j.cell.2014.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.White R, Rose K, Zon L. Zebrafish cancer: The state of the art and the path forward. Nat Rev Cancer. 2013;13(9):624–636. doi: 10.1038/nrc3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yen J, White RM, Stemple DL. Zebrafish models of cancer: Progress and future challenges. Curr Opin Genet Dev. 2014;24:38–45. doi: 10.1016/j.gde.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Goody MF, Sullivan C, Kim CH. Studying the immune response to human viral infections using zebrafish. Dev Comp Immunol. 2014;46(1):84–95. doi: 10.1016/j.dci.2014.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cronan MR, Tobin DM. Fit for consumption: Zebrafish as a model for tuberculosis. Dis Model Mech. 2014;7(7):777–784. doi: 10.1242/dmm.016089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Quintana FJ, et al. Adaptive autoimmunity and Foxp3-based immunoregulation in zebrafish. PLoS One. 2010;5(3):e9478. doi: 10.1371/journal.pone.0009478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cusick MF, Libbey JE, Trede NS, Eckels DD, Fujinami RS. Human T cell expansion and experimental autoimmune encephalomyelitis inhibited by Lenaldekar, a small molecule discovered in a zebrafish screen. J Neuroimmunol. 2012;244(1-2):35–44. doi: 10.1016/j.jneuroim.2011.12.024. [DOI] [PubMed] [Google Scholar]
- 69.Grimholt U, et al. MHC polymorphism and disease resistance in Atlantic salmon (Salmo salar); facing pathogens with single expressed major histocompatibility class I and class II loci. Immunogenetics. 2003;55(4):210–219. doi: 10.1007/s00251-003-0567-8. [DOI] [PubMed] [Google Scholar]
- 70. International HIV Controllers Study, et al. (2010) The major genetic determinants of HIV-1 control affect HLA Class I peptide presentation. Science 330(6010):1551–1557. [DOI] [PMC free article] [PubMed]
- 71.Mizgirev I, Revskoy S. Generation of clonal zebrafish lines and transplantable hepatic tumors. Nat Protoc. 2010;5(3):383–394. doi: 10.1038/nprot.2010.8. [DOI] [PubMed] [Google Scholar]
- 72.ZFIN The Zebrafish Model Organism Database 2016 Wild-Type Line AB. Available at https://zfin.org/ZDB-GENO-960809-7. Accessed April 5, 2016.
- 73.Lamason RL, et al. SLC24A5, a putative cation exchanger, affects pigmentation in zebrafish and humans. Science. 2005;310(5755):1782–1786. doi: 10.1126/science.1116238. [DOI] [PubMed] [Google Scholar]
- 74.St. John J. 2013 SeqPrep. Available at https://github.com/jstjohn/SeqPrep. Accessed September 11, 2014.
- 75.Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Andrews S. 2012 FastQC. Available at: www.bioinformatics.babraham.ac.uk/projects/fastqc/. Accessed September 11, 2014.
- 77.Luo R, et al. SOAPdenovo2: An empirically improved memory-efficient short-read de novo assembler. Gigascience. 2012;1(1):18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kurtz S, et al. Versatile and open software for comparing large genomes. Genome Biol. 2004;5(2):R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Stanke M, Diekhans M, Baertsch R, Haussler D. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics. 2008;24(5):637–644. doi: 10.1093/bioinformatics/btn013. [DOI] [PubMed] [Google Scholar]
- 80.Hatje K, et al. Cross-species protein sequence and gene structure prediction with fine-tuned Webscipio 2.0 and Scipio. BMC Res Notes. 2011;4(1):265. doi: 10.1186/1756-0500-4-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Grabherr MG, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29(7):644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Robinson JT, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29(1):24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gray KA, Yates B, Seal RL, Wright MW, Bruford EA. Genenames.org: The HGNC resources in 2015. Nucleic Acids Res. 2015;43(Database issue):D1079–D1085. doi: 10.1093/nar/gku1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kandil E, et al. Isolation of low molecular mass polypeptide complementary DNA clones from primitive vertebrates. Implications for the origin of MHC class I-restricted antigen presentation. J Immunol. 1996;156(11):4245–4253. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.