Significance
The DNA checkpoint kinases ATAXIA TELANGIECTASIA MUTATED (ATM) and ATM AND RAD3-RELATED play crucial roles in the maintenance of genome stability, safeguarding cellular survival and the faithful transmission of genetic information. Here we show that ATM is an important factor that influences seed quality by linking progression through germination with genome integrity. Our findings provide insight into the roles of DNA damage responses, identifying their importance in regulating germination, a process critical for plant survival in the natural environment and crop production.
Keywords: DNA repair, seed vigor, DNA damage response, dormancy cycling, soil seed bank
Abstract
Genome integrity is crucial for cellular survival and the faithful transmission of genetic information. The eukaryotic cellular response to DNA damage is orchestrated by the DNA damage checkpoint kinases ATAXIA TELANGIECTASIA MUTATED (ATM) and ATM AND RAD3-RELATED (ATR). Here we identify important physiological roles for these sensor kinases in control of seed germination. We demonstrate that double-strand breaks (DSBs) are rate-limiting for germination. We identify that desiccation tolerant seeds exhibit a striking transcriptional DSB damage response during germination, indicative of high levels of genotoxic stress, which is induced following maturation drying and quiescence. Mutant atr and atm seeds are highly resistant to aging, establishing ATM and ATR as determinants of seed viability. In response to aging, ATM delays germination, whereas atm mutant seeds germinate with extensive chromosomal abnormalities. This identifies ATM as a major factor that controls germination in aged seeds, integrating progression through germination with surveillance of genome integrity. Mechanistically, ATM functions through control of DNA replication in imbibing seeds. ATM signaling is mediated by transcriptional control of the cell cycle inhibitor SIAMESE-RELATED 5, an essential factor required for the aging-induced delay to germination. In the soil seed bank, seeds exhibit increased transcript levels of ATM and ATR, with changes in dormancy and germination potential modulated by environmental signals, including temperature and soil moisture. Collectively, our findings reveal physiological functions for these sensor kinases in linking genome integrity to germination, thereby influencing seed quality, crucial for plant survival in the natural environment and sustainable crop production.
Maintenance of genome integrity is indispensable for cellular survival and transmission of genetic information to the next generation; however, constant exposure of DNA to environmental and cellular oxidative stresses results in damage that can arrest growth and result in mutagenesis or cell death. Consequently, organisms have evolved powerful DNA repair and DNA damage signaling mechanisms. In plants, as in other eukaryotes, the cellular response to DNA damage is orchestrated by the phosphoinositide-3-kinase–related protein kinases (PIKKs) ATAXIA TELANGIECTASIA MUTATED (ATM) and ATM AND RAD3-RELATED (ATR) (1). In response to genotoxic stresses, these checkpoint kinases activate DNA repair factors, delay or halt cell cycle progression, and promote endocycles or programmed cell death (1–4). ATM is activated by double-strand DNA breaks (DSBs), a highly toxic form of DNA damage that results in chromosome fragmentation (1, 5), and plants mutated in ATM display hypersensitivity to DSBs induced by gamma radiation or radiomimetics (6). ATM also mediates a strong transcriptional up-regulation of hundreds of genes, including the SIAMESE/SIAMESE-RELATED cell cycle inhibitors SMR5 and SMR7 (1, 7). ATR is activated by single-strand regions of DNA, arising during DNA replication or processing of DSBs (1, 8). Although much of our knowledge of plant DNA damage responses has come from the use of genotoxins that would not be encountered naturally, the physiological roles of these DNA damage response pathways are less well characterized.
Seeds represent a stage of the life cycle in which plants experience particularly high levels of genotoxic stress (9). Maturing orthodox (desiccation-tolerant) seeds enter a period of desiccation during which their moisture content decreases to approximately 10–15%, which reduces cellular activity in the embryo to minimal levels, enabling maintenance of a viable but quiescent embryo for prolonged periods (10). Reduced cellular maintenance in the quiescent state, in combination with cycle(s) of desiccation and rehydration, are associated with high levels of damage by reactive oxygen species, resulting in deterioration of proteins, DNA, and cellular structures (11). Genome damage is exacerbated by adverse environmental conditions (typically high temperature and relative humidity) that cause increased oxidative damage and seed aging (12–15). This results in a loss of seed vigor, manifested as decreasing rapidity and uniformity of germination, leading to significant losses in yield in crop species (16), and ultimately culminates in loss of viability (17).
Delayed germination is accompanied by an extended period of pregerminative DNA repair initiated in the earliest stages of imbibition (17). This is observed as high levels of de novo DNA synthesis several hours before entry of cells into S phase and completion of germination, marked by the emergence of the young root (radicle) from the seed coat (17, 18). Seeds also can remain in a dormant state in the soil seed bank, often experiencing cycles of hydration and dehydration. Germination occurs only when dormancy has declined and seeds become sensitive to environmental conditions favorable for growth (10, 19, 20). DNA repair activities are operative in imbibed dormant seeds, enhancing seed longevity by preserving genome integrity during prolonged stasis in the soil seed bank and ensuring optimal germination when environmental conditions are favorable (17, 21).
In agriculture, rapid germination (high seed vigor) and high seed viability are important traits ensuring efficient seedling establishment, which is key to plant survival and crop production. Loss of seed viability is preceded by a progressively increasing lag period in imbibed nondormant seeds before germination. Cytological studies have established a correlation between loss of vigor and viability on seed aging and accumulation of DNA damage, reporting increased frequencies of both chromosome breakage and rearrangements and abnormal, mutated seedlings on germination of aged seed (11). Significantly, even high-quality seeds display a background level of chromosomal breaks (i.e., DSBs) (12, 22).
Consistent with those studies, our recent work has identified a strong DSB-specific transcriptional response, indicative of high levels of genotoxic stress, initiated early in seed imbibition even in unaged seeds (23). In addition, the increased sensitivity to seed aging of Arabidopsis mutants deficient in DSB repair suggests that the repair of chromosomal breaks is fundamental to seed germination (23). The observation that DNA repair activity can influence the rate of seed germination indicates that seed vigor and viability are linked to levels of genomic damage.
Here we sought to identify the molecular link between progression of germination and genome integrity in seeds. We show that the persistence of DSBs is sufficient to delay the completion of germination. We identify ATR as a determinant of seed longevity, and reveal ATM as a key factor influencing seed vigor and viability, integrating genome integrity with progression through germination in response to damage accumulated during the dry quiescent state. In the natural hydrated dormant state of seeds in the soil seed bank, ATM and ATR transcript levels display dynamic regulation with changes in environmental conditions. Taken together, these results reveal critical functions for DNA damage responses in regulating seed quality that are vital for plant survival and crop productivity.
Results
DSBs Are Rate-Limiting for Germination in Arabidopsis.
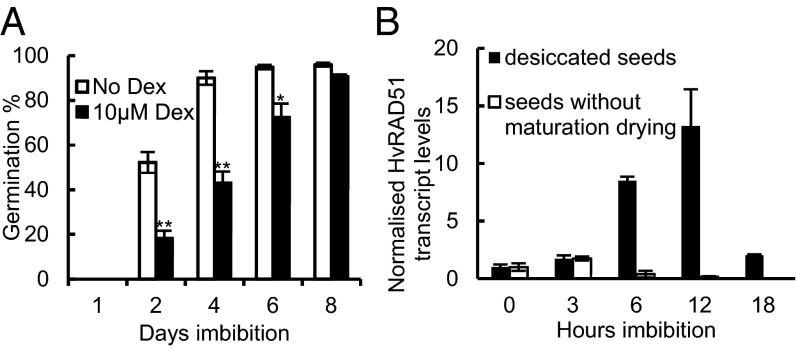
Our previous studies identified hypersensitivity to seed aging in mutants deficient in the DSB repair enzymes DNA LIGASE 6 (LIG6) and LIG4, suggesting aging-induced DSBs as a major factor that delays the progression to germination (23). To establish the rate-limiting effects of DSBs on germination, we analyzed transgenic Arabidopsis lines in which endonuclease-induced DSBs can be created by treating plants with dexamethasone. Induction of the endonuclease I-PpoI, which cleaves the Arabidopsis genome within the conserved 5S rRNA genes in vitro, resulted in an increase in cell death in seedling root tips, indicative of DNA damage (3) (SI Appendix, Fig. S1 A–D). On I-PpoI induction in seeds, germination was delayed, with the mean germination time (MGT, a measure of germination vigor) increasing from 3.1 d to 4.5 d (P < 0.05) (Fig. 1A). This finding demonstrates that the accumulation of DSBs in the seed genome is rate-limiting for germination, and that their persistence adversely affects germination vigor.
Fig. 1.
Seed desiccation is required for induction of the plant DNA damage response, and DSBs are rate-limiting for germination. (A) Germination of two independent lines holding the I-PpoI construct was analyzed with and without the induction of DSBs by dexamethasone. Seeds were plated onto germination paper prewetted with 10 µM dexamethasone or 0.5% DMSO (control) and stratified at 4 °C for 48 h before transfer to 20 °C. Error bars indicate the SEM of three replicates of 50 seeds each. Significant differences in mean values are indicated. *P < 0.05, **P < 0.01, t test. (B) Embryos were isolated from desiccated barley grain cv Maris Otter or from undesiccated barley grain (30 days postanthesis) that were capable of germination but had not undergone maturation drying, and placed on 1% agar plates. Quantitative PCR (qPCR) analysis of HvRAD51 expression was performed using cDNA synthesized from RNA isolated from imbibing embryos. Control qPCR was performed using primers specific to HvACTIN2.
ATM Is Activated in Early Imbibition of Orthodox Seeds.
In previous work, we identified marked activation of the DNA damage transcriptional response within 3 h of Arabidopsis seed imbibition, indicating that high levels of genome damage are incurred even under ideal conditions of seed development, storage, and imbibition (23). Activation of the plant transcriptional response to DSBs is dependent predominantly on ATM and includes genes involved in DNA metabolism, chromatin structure, and repair (1). Consistent with the role of ATM in controlling DNA damage- responsive gene expression, we found no significant induction of RAD51 recombinase transcripts in atm mutant seeds on imbibition, in contrast to wild type seeds (SI Appendix, Fig. S2).
Analysis of HvRAD51 transcript levels during seed imbibition of wild type barley (Hordeum vulgare) also showed high levels of gene induction at 12 h of imbibition (Fig. 1B), demonstrating that early induction of the DNA damage response is a conserved component of germination in desiccation-tolerant seeds. However, the transcriptional DNA damage response was absent in the germination of physiologically mature barley embryos that had not undergone maturation drying on the mother plant (Fig. 1B). This signifies that the cellular response to genome damage in germination is directly activated in response to high levels of DNA damage accumulated by the seeds during the cycle of desiccation, embryo quiescence, and rehydration.
Regulation of DNA Damage-Responsive Genes in Seeds.
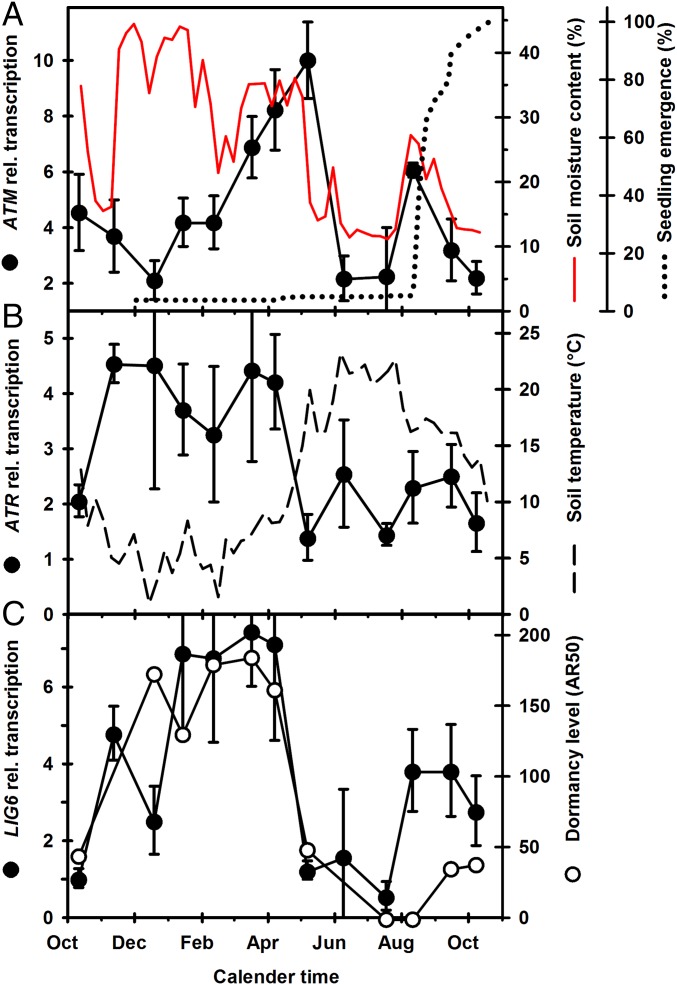
To further investigate the role of DNA repair in seeds, we compared the transcriptional profiles of the major DNA damage response genes in published seed array data using the Arabidopsis eFP browser (24). This analysis revealed similar patterns in the transcript levels of LIG6, ATR, and ATM in hydrated seeds undergoing laboratory-based dormancy cycling (SI Appendix, Table S1) (25, 26). Particularly high expression levels of these genes were observed in seeds in a prolonged hydrated state of dormancy, from 1 to 2 mo, as would be experienced by seeds overwintering in the soil. Significantly, transcriptional regulation of ATM and ATR has not been previously reported in plants, and this led us to look at the response of these genes in seeds experiencing the seasonally changing soil seed bank environment. Fluctuations in soil temperature and moisture content elicit transcriptional responses in seeds and influence dormancy and germination potential (19). We identified large seasonal fluctuations in transcript levels of ATM, ATR, and LIG6 (Fig. 2). In spring, ATM transcript levels in overwintered seeds increased fivefold during prolonged hydration in the soil, then declined sharply in May as soil moisture was lost and before seedlings emerged (Fig. 2A). Seasonal transcript profiles of ATR and LIG6 were positively correlated with each other (P < 0.05) and with levels of dormancy (AR50; P < 0.01 and < 0.05, respectively), but negatively correlated with soil temperature (P < 0.01 and < 0.05, respectively) (Fig. 2 B and C). These results are consistent with the idea of ATM and ATR regulating genome maintenance during dormancy cycling in the soil seed bank.
Fig. 2.
Transcript profiles of ATM, ATR, and LIG6 in relation to seasonal changes in the soil seed bank environment. (A) ATM transcript levels in seeds recovered from the soil over 12 mo from October 2007. Soil moisture (%) profile at seed depth (5 cm). Also shown is seedling emergence (% of total emerged) in the field following monthly soil disturbance (n = 4). (B) ATR transcript levels and soil temperature (°C) at seed depth. (C) LIG6 transcript levels and dormancy level, measured as the dry after-ripening time (days) required to achieve 50% germination at 20 °C in light (AR50). Error bars indicate the SEM. n = 3. Soil temperature, soil moisture content, AR50, and emergence data are from Footitt et al. (19).
Germination of atm and atr Mutant Seeds Is Resistant to Accelerated Aging.
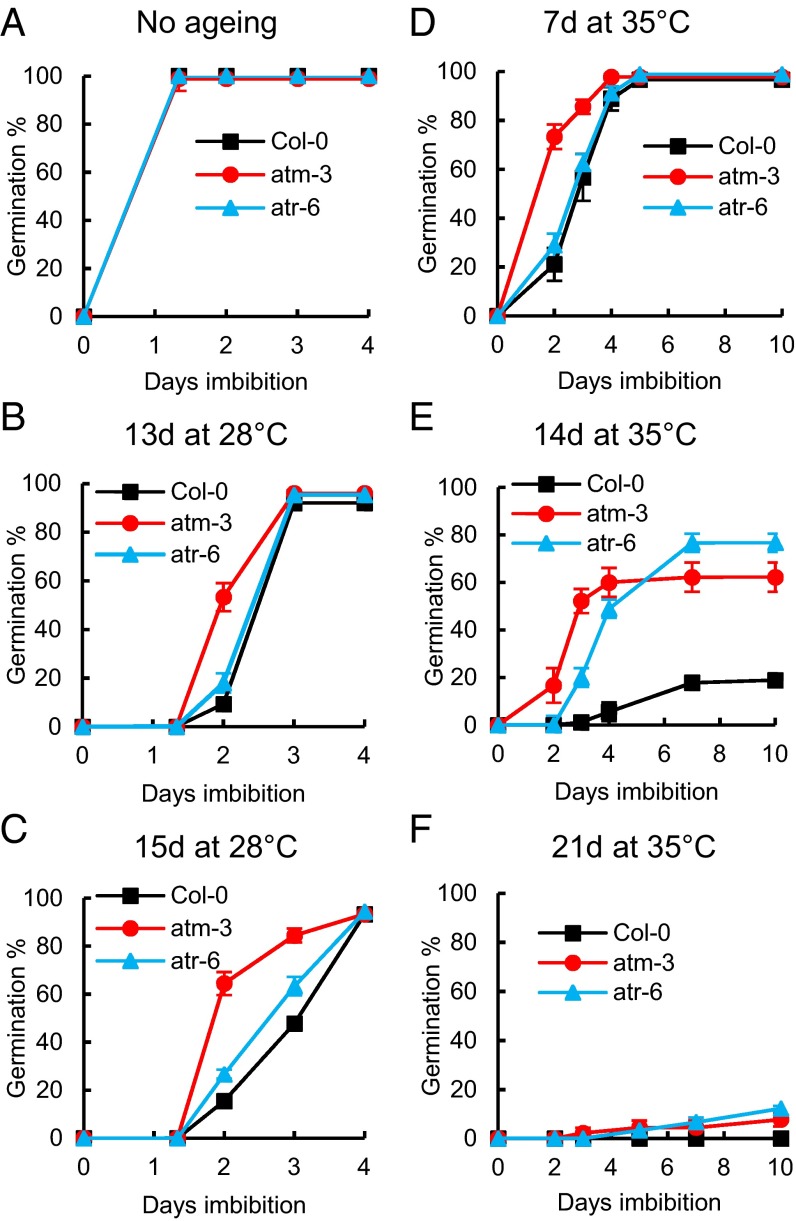
We tested the hypothesis that ATM and ATR integrate the sensing of DNA integrity with the onset of germination by analyzing the germination phenotypes of multiple independent alleles of atm and atr mutant lines (27). Accelerated aging is widely used to simulate natural aging by incubating seeds at elevated temperature and relative humidity (RH) (28). Here we used three different aging regimes—28 °C and 84.6% RH, 35 °C and 83% RH, and 49 °C and 80.6% RH (29)—with the lower temperature and humidity treatments representing conditions typically experienced by seeds in the natural environment (30). In the absence of seed aging, germination of wild type (Col-0) Arabidopsis seeds and the mutant lines atm-3 and atr-6 displayed no significant differences (Fig. 3A); however, atm mutant seeds germinated markedly more rapidly than wild type seeds after aging at all temperature and RH regimes investigated (Fig. 3 B–F and SI Appendix, Figs. S3–S5).
Fig. 3.
Germination of atm and atr mutant seeds is resistant to accelerated aging. Germination of atm-3, atr-6, and Col-0 seeds was analyzed after accelerated aging at 28 °C or 35 °C over a saturated solution of KCl. Shown is germination of unaged seeds (A), seeds after accelerated aging at 28 °C (B and C), and seeds after accelerated aging at 35 °C (D–F). Seeds were stratified at 4 °C for 48 h before being transferred to 22 °C/light and then scored for radicle emergence on each day postimbibition. Error bars indicate the SEM of three replicates of 50 seeds each.
Progressively increased seed aging resulted in severely delayed germination in Col-0 seeds compared with atm mutant seeds; after 14 d of aging at 35 °C, the MGT was 2.9 d for atm, compared with 6.4 d for Col-0 (P < 0.01) (SI Appendix, Fig. S3). Thus, our data indicate that ATM controls the time to germination in aged seeds, observed as an extended delay to radicle emergence with increasing seed deterioration. Progressive loss of vigor on seed aging eventually culminates in loss in viability. Both the atm and atr mutants exhibited increased levels of germination compared with wild type controls after aging (Fig. 3 E and F; SI Appendix, Fig. S5). Although Col-0 seeds displayed low viability after 14 d of aging at 35 °C (19%), atm and atr lines retained significantly higher levels of germination (62% and 77%, respectively; P < 0.01) (Fig. 3E). Seeds that failed to germinate were resistant to dormancy-breaking treatments, including gibberellin, fluridone, and cold imbibition at 4 °C. Lack of viability was confirmed by staining with 2,3,5-triphenyltetrazolium chloride (31) (SI Appendix, Fig. S5 I and J). The increased germination of atm and atr mutant seeds after aging was confirmed in the independent mutant alleles atm-2 and atr-2 (1) (SI Appendix, Fig. S5 G and H). Taken together, these results establish both ATM and ATR as molecular determinants of viability and reveal ATM as a key controller of germination in response to the deteriorative effects incurred during seed aging.
ATM Controls Cell Cycle Progression in Response to Seed Aging.
In response to DNA damage, ATM and ATR activate checkpoints that delay or arrest cell cycle progression to promote repair processes, which has the effect of delaying growth (2, 32). We hypothesized that the rapid germination of atm mutant seeds is due to loss of the checkpoint activity that slows germination in the presence of DNA damage. In Arabidopsis seeds, cells in the embryo are arrested in G1 phase, and resumption of cell cycle activity and S phase (DNA replication) is required for normal germination (33, 34).
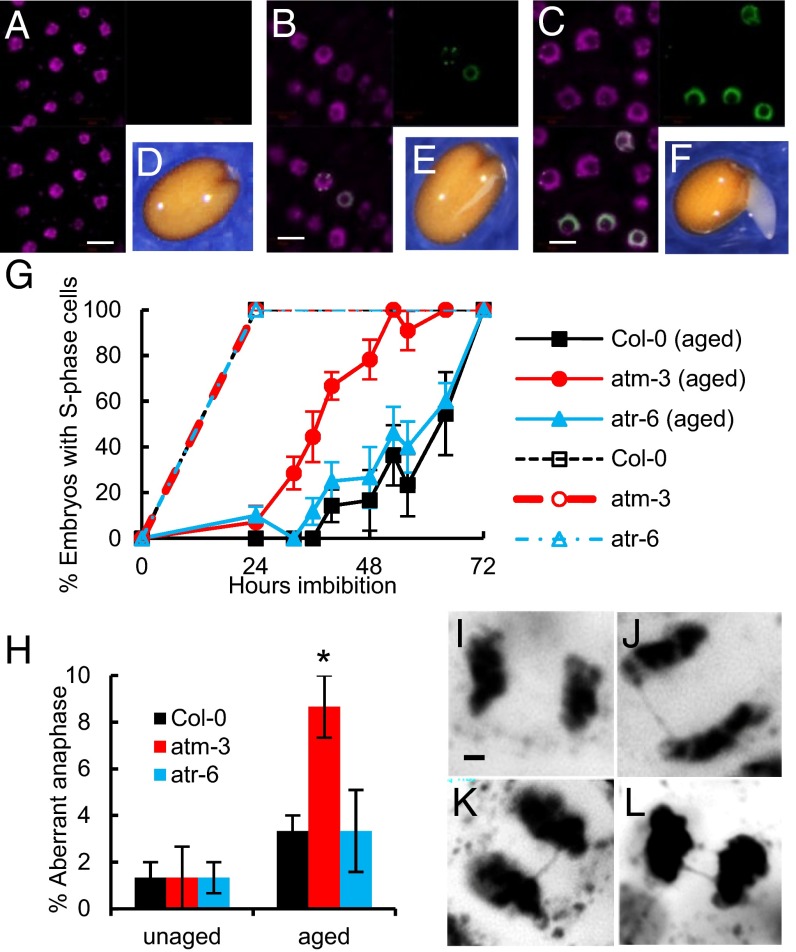
We investigated whether an aging-induced delay to cell cycle activation was evident in wild type seeds, but not atm or atr mutant lines. Cell cycle initiation was determined by incorporation of the thymidine nucleoside analog 5-ethynyl-2′-deoxyuridine (EdU) to monitor the onset of DNA replication in wild type, atm, and atr lines. In wild type seeds, few embryos contained cells that were replicating DNA until around the time of seed coat rupture, when labeled nuclei are first visible in root meristem cells (Fig. 4 A–F and SI Appendix, Fig. S6). We observed no significant difference in the timing of S phase in unaged wild type, atm, and atr seeds (Fig. 4G); however, when using seeds aged for 7 d at 35 °C and for 14 d at 35 °C, we found that DNA replication was initiated substantially earlier in atm mutant seeds than in the Col-0 or atr lines (Fig. 4G), consistent with the faster germination of the aged atm lines. This indicates that ATM, but not ATR, activity delays onset of S phase in the wild type lines, coinciding with a loss in germination vigor. At 14 d, we found increased numbers of S-phase cells in both atm- and atr-deficient seeds, correlated with the increased germination observed in these mutants (SI Appendix, Fig. S7). We conclude that ATM delays initiation of the cell cycle in aged seeds, and that both ATM and ATR contribute to the inhibition of DNA replication in aged seeds.
Fig. 4.
ATM induces a delay in DNA replication on seed aging. Shown is the timing of DNA replication (S phase) in the germination of aged and unaged seeds. (A–F) The timing of S phase in unaged wild type seeds was determined using EdU labeling and fluorescence microscopy. (A and D) Before seed coat rupture, little EdU labeling is observed. (B and E) Rupture of the seed coat occurs at around 24 h, coincident with the onset of S phase, with several EdU-positive nuclei evident in embryos. (C and F) As seeds germinate, progressively increasing numbers of EdU-positive nuclei are observed in cells of the expanding radicle. (Top Left) DAPI-stained nuclei. (Top Right) EdU-labeled nuclei. (Bottom Left) Image merge. (Scale bar: 10 μm.) (Bottom Right) Representative stage of germination. (G) Analysis of S-phase initiation in aged and unaged wild type, atm, and atr mutant seeds. Quantification of numbers of embryos containing one or more EdU-positive (S phase) cells in unaged seeds (broken lines) and seeds aged for 7 d at 35 °C and 83% RH (continuous lines). Aged seeds displayed loss of vigor but retained high viability, as shown in Fig. 3D. (H) Frequency of anaphase bridges in germinating seeds. Seeds were either unaged or aged for 7 d at 35 °C and 83% RH. Error bars show the SE of three replicates of 50 anaphases each. *P < 0.05, t test. (I–L) Airy scanning confocal images showing examples of anaphases: (I) normal anaphase from unaged Col-0; (J–L) anaphase bridges from aged Col-0 (J), aged atm-3 (K), and aged atr-6 (L). (Scale bar: 1 μm.)
Severe Chromosomal Abnormalities Are Elevated in atm Mutant Seeds.
To investigate the hypothesis that ATM integrates germination progression with genome integrity, we determined the frequencies of chromosomal abnormalities in aged and unaged Col-0, atm mutant, and atr mutant seeds (Fig. 4 H–L). Anaphase bridges were scored after commencement of cell division in radicle cells of germinating seeds. These bridges represent inaccurate chromosomal break repairs by the host cell recombination machinery. Consistent with the induction of the ATM-dependent DNA damage response, even in high-quality seeds, we observed a low frequency (1–2%) of abnormalities in unaged Col-0, atm, and atr seeds (Fig. 4H), but still significantly higher than that reported for other stages of the Arabidopsis lifecycle (35). After aging (7 d at 35 °C and 83% RH), this frequency increased to 4% in Col-0 and atr seeds, indicating that high levels of genome damage accumulate in the dry quiescent state and can be tolerated by the germinating seeds (Fig. 4H). However, substantially higher frequencies (9%) were observed in aged atm seeds, lending further support to the function of ATM in controlling the advancement of germination to promote repair of the high levels of genome damage sustained by extended periods in the dry quiescent state.
The Cell Cycle Inhibitor SMR5 Regulates Germination in Response to Aging.
We further investigated the mechanism by which control of germination is integrated with genome surveillance by analyzing the downstream effectors of ATR and ATM signaling. ATR activates a DNA replication stress checkpoint through transcriptional induction of the cell cycle regulator WEE1 (8, 36). However, in our analysis of seed aging, WEE1 transcript levels did not differ significantly among Col-0, atm, or atr mutant seeds, and, furthermore, wee1 mutants did not exhibit the aging resistant phenotype of atr seeds (SI Appendix, Fig. S8). These findings indicate that the effects of seed aging on germination are not attributable to replication stress (1, 8, 36).
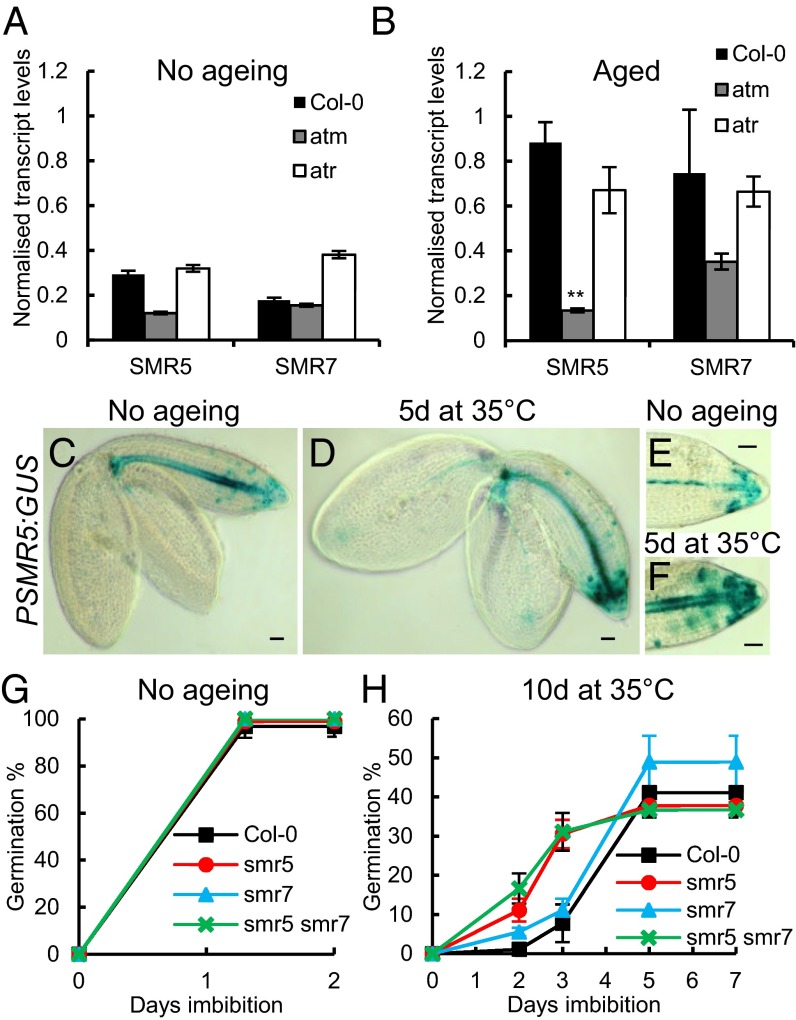
In response to genotoxic stress, ATM induces SMR5 and SMR7, members of a family of SIAMESE/SIAMESE-RELATED cell cycle inhibitors. Both genes display ATM-dependent transcriptional induction in response to genotoxic stresses and regulate cell cycle activity and growth under stress conditions (7, 37). In seeds, SMR5 transcripts declined rapidly during imbibition, reaching low levels by the time of germination (SI Appendix, Fig. S9 A and C); however, transcript analysis of aged seeds (7 d at 35 °C) at 48 h of imbibition revealed significantly higher SMR5 levels in wild type lines, but not in atm lines (Fig. 5 A and B). This finding is consistent with a role for SMR5 in regulating the germination of aged seeds, given the known functions of this cell cycle inhibitor in genotoxic stress responses (7). In the soil seed bank, SMR5 and ATM transcript levels were highly correlated (P < 0.05; SI Appendix, Fig. S10). SMR5 GUS-reporter analysis localized promoter activity to regions of the radicle where S phase is initiated (Fig. 5 C–F).
Fig. 5.
ATM controls germination in an SMR5-dependent pathway. (A and B) SMR5 and SMR7 transcript levels in seeds at 48 h of imbibition in unaged Col-0, atm-3, and atr-6 seeds (A) and Col-0, atm-3, and atr-6 seeds aged at 35 °C and 83% RH for 7 d (B) to provide loss of vigor but 100% viability. (C–F) GUS-reporter analysis of SMR5 promoter activity in wild type unaged seeds and seeds aged at 35 °C and 83% RH for 5 d. (G and H) Analysis of smr5, smr7, smr5 smr7, and wild type seed germination performance after accelerated aging. Shown are the viability and vigor of unaged seeds (G) and of seeds after 10 d accelerated aging at 35 °C and 83% RH (H). Seeds were plated and stratified at 4 °C for 48 h before being transferred to an environmental growth chamber at 22 °C, and then scored for radicle emergence at each day postimbibition. Error bars represent the SEM of three replicates of 50 seeds each. **P < 0.01, t test.
Because both SMR5 and SMR7 are involved in plant cell cycle control, with SMR5 in particular required for the reduction in cell division under conditions of elevated oxidative stress (7), we tested single and double smr5 and smr7 mutant lines for sensitivity to seed aging. After accelerated aging, smr7 mutants were not significantly different from wild type lines, whereas both smr5 and the smr5 smr7 lines displayed an aging-induced delay to germination relative to wild type lines (Fig. 5 G and H and SI Appendix, Fig. S11). We conclude that SMR5 plays a key role in controlling the delay to germination observed on seed aging.
Discussion
The evolution of desiccation-tolerant seeds represents a highly successful survival strategy, effectively prolonging embryo viability and providing resistance to adverse environmental stresses until favorable conditions for germination are encountered. Nonetheless, extended periods in the dry quiescent state are associated with the deterioration of biological macromolecules and a progressive delay to germination, which culminates in loss of viability (17). There is considerable intraspecific and interspecific variation in seed longevity, and understanding the genetic basis of seed quality is important for maintaining crop production in changing climates (31). Here we identify important functions for DNA checkpoint kinases in linking genome integrity to germination, thereby influencing seed quality, which is crucial for plant survival in the natural environment and for sustainable crop production. We establish that both ATM and ATR are determinants of seed longevity, and demonstrate that ATM controls the progression of seed germination, integrating genome surveillance with cell cycle activation.
Seed aging is associated with an increasing incidence of chromosomal aberrations (Fig. 4 H–L), and even seed lots with high germination rates display levels of genome damage not seen at other stages of plant development (12, 22). Consistent with this, during the early imbibition phase, seeds display a large and rapid induction of the ATM-mediated transcriptional DSB response (23). We show that activation of the DNA damage response requires previous desiccation and quiescence of the embryo, rather than being a developmentally programmed part of germination per se. The quiescent desiccated state is associated with accumulation of high levels of DSBs, as observed in the desert-dwelling bacterium Deinococcus radiodurans (38). In seeds, the accumulation of genome damage on aging is further compounded by telomere loss (39), a source of chromosomal breaks that also may contribute to loss of vigor. Here, using enzymatic induction of DSBs by the meganuclease I-PpoI during seed imbibition, we provide direct evidence that low levels of chromosomal breaks limit the progression of germination in Arabidopsis.
Plants have evolved robust DNA damage response and repair pathways to counteract the detrimental effects of genotoxic stress, coordinated by the eukaryotic DNA checkpoint kinases ATM and ATR. Here we identify that ATM and ATR function as major factors influencing germination in response to seed aging. We demonstrate that ATM delays the progression of germination by regulating the initiation of DNA replication through up-regulation of the cell cycle inhibitor SMR5 (7). Chemical inhibition of the cell cycle was previously shown to be sufficient to slow germination in Arabidopsis (34), and here we establish a physiological role for DNA damage in cell cycle checkpoints in seeds. Progressive aging culminates in loss of seed viability, and a component of this viability loss is attributable to both ATM and ATR. Whereas ATR induces transcription of the cell cycle regulator WEE1 in response to DNA replication stress (8, 36), WEE1 is not required for the ATR response to genotoxic stress in plants (1, 2, 40). Here we establish that ATR function in seeds is not dependent on WEE1, consistent with the idea that DNA damage rather than replicative stress limits germination.
The increased germination of aged atm and atr mutant seeds relative to wild type seeds is associated with higher levels of S-phase activity, which is consistent with increased DNA replication in the presence of DNA damage and increased genome instability. Our cytological analysis has revealed high frequencies of chromosomal aberrations in atm mutant seeds, identifying a critical role for ATM in safeguarding the genome of the germinating embryo. Furthermore, seedlings germinated from aged atm mutant seeds exhibit lower survival and slower development of true leaves compared with their aged wild type counterparts (SI Appendix, Fig. S12). This demonstrates the requirement for ATM activation in imbibed seeds to ensure that germination does not proceed until damage to the genome is repaired, thereby promoting successful establishment and growth of the young seedlings.
In the natural environment, seeds in the soil seed bank typically undergo several wet–dry cycles at or near the soil surface, or remain continually hydrated deeper in the soil. Imbibed dormant seeds display DNA repair processes (21), which are often active over several imbibition and redrying cycles (41). These seasonal fluctuations in hydration and temperature in the soil seed bank lead to dynamic regulation of ATM, SMR5, and ATR transcript levels. Active genome surveillance and DNA damage signaling in hydrated dormant seeds provides highly effective mechanisms for protecting genomic integrity, which is important for the long-term stability of plant communities.
In conclusion, we show that ATM and ATR are important factors in controlling germination in plants. We identify major roles for ATM in integrating genome surveillance with germination through regulation of cell cycle activities, providing insight into the physiological functions of DNA damage response mechanisms in plant development. Our findings further establish critical functions for ATM in safeguarding genome stability in germinating seeds and in promoting seedling growth and establishment, key determinants of crop yield (42). Understanding the mechanisms that regulate germination provides important insight into the molecular basis of seed vigor and viability, traits that are of critical agronomic and ecological importance.
Materials and Methods
Seed Propagation and Germination.
Arabidopsis plants were raised in growth chambers under constant humidity (30%), with 16h light and 8h dark cycles at 23 °C. Col-0, atr-6 (SALK_054383) and atm-3 mutants (SALK_089805) were obtained from the NASC. For each experimental replicate, seeds from all lines were harvested simultaneously and stored at 15 °C and 15% humidity for 2 mo to allow after-ripening. Germination tests and accelerated aging were performed as described previously (23, 43) and mean time to germination was calculated as described in ref (44). Arabidopsis lines were as previously described atm-3 (27), lig6 lig4 (23), atr-2 and atm-2 (1), wee1 (36) and pSMR5:GUS, smr5, smr7 and smr5 smr7 (7). Viability staining performed as reported previously (31). The atr-6 mutant allele is presented in SI Appendix, Fig. S13.
Nucleic Acid Purification and Cloning.
DNA procedures and bacterial manipulations were by established protocols (36). Real-time RT-PCR analysis was performed on a CFX96 thermocycler (Bio-Rad), as described previously (45), using SYBR Green Supermix (Bio-Rad). A plant codon-optimized I-PpoI gene was synthesized (Genscript; SI Appendix, Fig. S14) and cloned into pBI-ΔGR and the expression cassette subcloned into pCB1300 carrying a I-PpoI recognition site to create pPPOΔGR (SI Appendix, Fig. S15). Propidium iodide (PI) staining and EdU labeling were performed as described previously (46). Full methods are described in SI Appendix.
Dormancy Analysis.
Seed production, harvest, and storage, along with details of seed burial in and recovery from field soils and postrecovery seed handling, were described previously (19, 20, 25, 26, 47). Further details are provided in SI Appendix.
Supplementary Material
Acknowledgments
We thank Lieven de Veylder (Ghent University) for SMR GUS lines and mutants and Stan Matthews and Alison Powell (University of Aberdeen) for comments on the manuscript. Financial support was provided from the UK Biotechnology and Biological Sciences Research Council (Grants BB/H012346/1 and 24/JF/20608, to W.M.W. and C.E.W., and BB/I022201/1, to S.F. and W.E.F-.S.), the UK Department for the Environment, Food, & Rural Affairs (W.E.F.-S.), and FP7-KBBE EcoSeed (W.M.W., W.E.F.-S., and C.E.W.). The LSM 880 laser scanning microscope was funded by the Wellcome Trust (WT104918MA).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1608829113/-/DCSupplemental.
References
- 1.Culligan KM, Robertson CE, Foreman J, Doerner P, Britt AB. ATR and ATM play both distinct and additive roles in response to ionizing radiation. Plant J. 2006;48(6):947–961. doi: 10.1111/j.1365-313X.2006.02931.x. [DOI] [PubMed] [Google Scholar]
- 2.Adachi S, et al. Programmed induction of endoreduplication by DNA double-strand breaks in Arabidopsis. Proc Natl Acad Sci USA. 2011;108(24):10004–10009. doi: 10.1073/pnas.1103584108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fulcher N, Sablowski R. Hypersensitivity to DNA damage in plant stem cell niches. Proc Natl Acad Sci USA. 2009;106(49):20984–20988. doi: 10.1073/pnas.0909218106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Furukawa T, et al. A shared DNA damage-response pathway for induction of stem-cell death by UVB and by gamma irradiation. DNA Repair (Amst) 2010;9(9):940–948. doi: 10.1016/j.dnarep.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 5.Falck J, Coates J, Jackson SP. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature. 2005;434(7033):605–611. doi: 10.1038/nature03442. [DOI] [PubMed] [Google Scholar]
- 6.Garcia V, et al. AtATM is essential for meiosis and the somatic response to DNA damage in plants. Plant Cell. 2003;15(1):119–132. doi: 10.1105/tpc.006577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yi D, et al. The Arabidopsis SIAMESE-RELATED cyclin-dependent kinase inhibitors SMR5 and SMR7 regulate the DNA damage checkpoint in response to reactive oxygen species. Plant Cell. 2014;26(1):296–309. doi: 10.1105/tpc.113.118943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cools T, et al. The Arabidopsis thaliana checkpoint kinase WEE1 protects against premature vascular differentiation during replication stress. Plant Cell. 2011;23(4):1435–1448. doi: 10.1105/tpc.110.082768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Waterworth WM, Drury GE, Bray CM, West CE. Repairing breaks in the plant genome: The importance of keeping it together. New Phytol. 2011;192(4):805–822. doi: 10.1111/j.1469-8137.2011.03926.x. [DOI] [PubMed] [Google Scholar]
- 10.Bewley JD. Seed germination and dormancy. Plant Cell. 1997;9(7):1055–1066. doi: 10.1105/tpc.9.7.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Waterworth WM, Bray CM, West CE. The importance of safeguarding genome integrity in germination and seed longevity. J Exp Bot. 2015;66(12):3549–3558. doi: 10.1093/jxb/erv080. [DOI] [PubMed] [Google Scholar]
- 12.Dourado AM, Roberts EH. Chromosome aberrations induced during storage in barley and pea seeds. Ann Bot (Lond) 1984;54:767–779. [Google Scholar]
- 13.El-Maarouf-Bouteau H, Mazuy C, Corbineau F, Bailly C. DNA alteration and programmed cell death during ageing of sunflower seed. J Exp Bot. 2011;62(14):5003–5011. doi: 10.1093/jxb/err198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bailly C. Active oxygen species and antioxidants in seed biology. Seed Sci Res. 2004;14(02):93–107. [Google Scholar]
- 15.Job C, Rajjou L, Lovigny Y, Belghazi M, Job D. Patterns of protein oxidation in Arabidopsis seeds and during germination. Plant Physiol. 2005;138(2):790–802. doi: 10.1104/pp.105.062778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Powell AA, Matthews S. Seed aging/repair hypothesis leads to new testing methods. Seed Tech. 2012;34(1):15–25. [Google Scholar]
- 17.Elder R, Osborne D. Function of DNA synthesis and DNA repair in the survival of embryos during early germination and in dormancy. Seed Sci Res. 1993;3:43–53. [Google Scholar]
- 18.Ashraf M, Bray CM. DNA synthesis in osmoprimed leek (Allium porrum L.) seeds and evidence for repair and replication. Seed Sci Res. 1993;3(01):15–23. [Google Scholar]
- 19.Footitt S, Douterelo-Soler I, Clay H, Finch-Savage WE. Dormancy cycling in Arabidopsis seeds is controlled by seasonally distinct hormone-signaling pathways. Proc Natl Acad Sci USA. 2011;108(50):20236–20241. doi: 10.1073/pnas.1116325108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Footitt S, Huang Z, Clay HA, Mead A, Finch-Savage WE. Temperature, light and nitrate sensing coordinate Arabidopsis seed dormancy cycling, resulting in winter and summer annual phenotypes. Plant J. 2013;74(6):1003–1015. doi: 10.1111/tpj.12186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Villiers TA. Seed aging: chromosome stability and extended viability of seeds stored fully imbided. Plant Physiol. 1974;53(6):875–878. doi: 10.1104/pp.53.6.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roberts EH. Cytological, genetical and metabolic changes associated with loss of seed viability. In: Roberts EH, editor. Viability of Seeds. Chapman and Hall; London: 1972. pp. 253–306. [Google Scholar]
- 23.Waterworth WM, et al. A plant DNA ligase is an important determinant of seed longevity. Plant J. 2010;63(5):848–860. doi: 10.1111/j.1365-313X.2010.04285.x. [DOI] [PubMed] [Google Scholar]
- 24.Winter D, et al. An “electronic fluorescent pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One. 2007;2(8):e718. doi: 10.1371/journal.pone.0000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cadman CSC, Toorop PE, Hilhorst HWM, Finch-Savage WE. Gene expression profiles of Arabidopsis Cvi seeds during dormancy cycling indicate a common underlying dormancy control mechanism. Plant J. 2006;46(5):805–822. doi: 10.1111/j.1365-313X.2006.02738.x. [DOI] [PubMed] [Google Scholar]
- 26.Finch-Savage WE, Cadman CS, Toorop PE, Lynn JR, Hilhorst HW. Seed dormancy release in Arabidopsis Cvi by dry after-ripening, low temperature, nitrate and light shows common quantitative patterns of gene expression directed by environmentally specific sensing. Plant J. 2007;51(1):60–78. doi: 10.1111/j.1365-313X.2007.03118.x. [DOI] [PubMed] [Google Scholar]
- 27.Waterworth WM, et al. NBS1 is involved in DNA repair and plays a synergistic role with ATM in mediating meiotic homologous recombination in plants. Plant J. 2007;52(1):41–52. doi: 10.1111/j.1365-313X.2007.03220.x. [DOI] [PubMed] [Google Scholar]
- 28.Rajjou L, et al. Proteome-wide characterization of seed aging in Arabidopsis: A comparison between artificial and natural aging protocols. Plant Physiol. 2008;148(1):620–641. doi: 10.1104/pp.108.123141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Winston PW, Bates DH. Saturated solutions for the control of humidity in biological research. Ecology. 1960;41(1):232–237. [Google Scholar]
- 30.Finch-Savage WE, Rowse HR, Dent KC. Development of combined imbibition and hydrothermal threshold models to simulate maize (Zea mays) and chickpea (Cicer arietinum) seed germination in variable environments. New Phytol. 2005;165(3):825–838. doi: 10.1111/j.1469-8137.2004.01272.x. [DOI] [PubMed] [Google Scholar]
- 31.Nguyen TP, Keizer P, van Eeuwijk F, Smeekens S, Bentsink L. Natural variation for seed longevity and seed dormancy are negatively correlated in Arabidopsis. Plant Physiol. 2012;160(4):2083–2092. doi: 10.1104/pp.112.206649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Preuss SB, Britt AB. A DNA-damage-induced cell cycle checkpoint in Arabidopsis. Genetics. 2003;164(1):323–334. doi: 10.1093/genetics/164.1.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barrôco RM, et al. The role of the cell cycle machinery in resumption of postembryonic development. Plant Physiol. 2005;137(1):127–140. doi: 10.1104/pp.104.049361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Masubelele NH, et al. D-type cyclins activate division in the root apex to promote seed germination in Arabidopsis. Proc Natl Acad Sci USA. 2005;102(43):15694–15699. doi: 10.1073/pnas.0507581102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Amiard S, Depeiges A, Allain E, White CI, Gallego ME. Arabidopsis ATM and ATR kinases prevent propagation of genome damage caused by telomere dysfunction. Plant Cell. 2011;23(12):4254–4265. doi: 10.1105/tpc.111.092387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De Schutter K, et al. Arabidopsis WEE1 kinase controls cell cycle arrest in response to activation of the DNA integrity checkpoint. Plant Cell. 2007;19(1):211–225. doi: 10.1105/tpc.106.045047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van Leene J, et al. Targeted interactomics reveals a complex core cell cycle machinery in Arabidopsis thaliana. Mol Syst Biol. 2010;6:397. doi: 10.1038/msb.2010.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zahradka K, et al. Reassembly of shattered chromosomes in Deinococcus radiodurans. Nature. 2006;443(7111):569–573. doi: 10.1038/nature05160. [DOI] [PubMed] [Google Scholar]
- 39.Bucholc M, Buchowicz J. Synthesis of extrachromosomal DNA and telomere-related sequences in germinating wheat embryos. Seed Sci Res. 1992;2(03):141–146. [Google Scholar]
- 40.Culligan K, Tissier A, Britt A. ATR regulates a G2-phase cell-cycle checkpoint in Arabidopsis thaliana. Plant Cell. 2004;16(5):1091–1104. doi: 10.1105/tpc.018903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boubriak I, Kargiolaki H, Lyne L, Osborne DJ. The requirement for DNA repair in desiccation tolerance of germinating embryos. Seed Sci Res. 1997;7(02):97–106. [Google Scholar]
- 42.Finch-Savage WE, Bassel GW. Seed vigour and crop establishment: Extending performance beyond adaptation. J Exp Bot. 2016;67(3):567–591. doi: 10.1093/jxb/erv490. [DOI] [PubMed] [Google Scholar]
- 43.Hay FR, Mead A, Manger K, Wilson FJ. One-step analysis of seed storage data and the longevity of Arabidopsis thaliana seeds. J Exp Bot. 2003;54(384):993–1011. doi: 10.1093/jxb/erg103. [DOI] [PubMed] [Google Scholar]
- 44.Ranal MA, Santana DGD, Ferreira WR, Mendes-Rodrigues C. Calculating germination measurements and organizing spreadsheets. Revista Brasil Bot. 2009;32(4):849–855. [Google Scholar]
- 45.Waterworth WM, Drury GE, Blundell-Hunter G, West CE. Arabidopsis TAF1 is an MRE11-interacting protein required for resistance to genotoxic stress and viability of the male gametophyte. Plant J. 2015;84(3):545–557. doi: 10.1111/tpj.13020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Amiard S, et al. Distinct roles of the ATR kinase and the Mre11-Rad50-Nbs1 complex in the maintenance of chromosomal stability in Arabidopsis. Plant Cell. 2010;22(9):3020–3033. doi: 10.1105/tpc.110.078527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dekkers BJ, et al. Identification of reference genes for RT-qPCR expression analysis in Arabidopsis and tomato seeds. Plant Cell Physiol. 2012;53(1):28–37. doi: 10.1093/pcp/pcr113. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.