Abstract
Saccharomyces cerevisiae preferentially uses glucose as a carbon source, but following its depletion, it can utilize a wide variety of other carbons including nonfermentable compounds such as ethanol. A shift to a nonfermentable carbon source results in massive reprogramming of gene expression including genes involved in gluconeogenesis, the glyoxylate cycle, and the tricarboxylic acid cycle. This review is aimed at describing the recent progress made toward understanding the mechanism of transcriptional regulation of genes responsible for utilization of nonfermentable carbon sources. A central player for the use of nonfermentable carbons is the Snf1 kinase, which becomes activated under low glucose levels. Snf1 phosphorylates various targets including the transcriptional repressor Mig1, resulting in its inactivation allowing derepression of gene expression. For example, the expression of CAT8, encoding a member of the zinc cluster family of transcriptional regulators, is then no longer repressed by Mig1. Cat8 becomes activated through phosphorylation by Snf1, allowing upregulation of the zinc cluster gene SIP4. These regulators control the expression of various genes including those involved in gluconeogenesis. Recent data show that another zinc cluster protein, Rds2, plays a key role in regulating genes involved in gluconeogenesis and the glyoxylate pathway. Finally, the role of additional regulators such as Adr1, Ert1, Oaf1, and Pip2 is also discussed.
Keywords: Saccharomyces cerevisiae, nonfermentable carbon, gluconeogenesis, transcriptional regulator, zinc cluster protein
Introduction
As observed in many unicellular organisms, the budding yeast Saccharomyces cerevisiae preferentially uses glucose over other carbon sources as it can directly enter the glycolytic pathway. However, when glucose is unavailable, alternative carbon sources are used for the production of metabolic energy and cellular biomass. Budding yeast is able to utilize a wide variety of different carbons; for example, other alternative sugars such as galactose, sucrose, maltose, and melbiose as well as nonsugar carbons such as ethanol, lactate, glycerol, acetate, or oleate may be used. The enzymatic pathways required for the specific utilization of these carbon compounds are very well characterized. Quite often, enzymes needed for a specific pathway are produced only when required. This regulation is mainly (but not exclusively) exerted at the transcriptional level. A classical example is the galactose-induced expression of genes required for catabolism of this sugar by the transcriptional activator Gal4 (Lohr et al., 1995). Various groups have reviewed the utilization of alternate carbon sources in S. cerevisiae (Gancedo, 1998; Carlson, 1999; Schüller, 2003; Barnett & Entian, 2005; Gurvitz & Rottensteiner, 2006b; Zaman et al., 2008). This current review is aimed at highlighting the recent progress made toward better understanding the transcriptional regulation of genes involved in the use of nonfermentable carbon sources.
A shift from one carbon source to another is referred to as a diauxic shift, where exhaustion of a preferred carbon source will be followed by considerably reduced growth leading to adaptation for using an alternate supply for carbon. The name diauxic was first described in Escherichia coli for adaptation to the use of lactose upon glucose exhaustion. Another classical example of a diauxic shift is provided by yeast with a shift from a fermentative to a nonfermentative mode of growth. During this transition, a massive reprogramming of expression occurs for genes in various pathways such as carbon metabolism, protein synthesis, and carbohydrate storage (DeRisi et al., 1997). Fitness experiments with pooled deletion strains showed that over 600 genes are required for optimal growth with nonfermen-table carbons such as ethanol (Steinmetz et al., 2002). The upregulation of gluconeogenic gene expression is indispensable for the production of glucose-6-phosphate, which is critical for cell growth. For instance, glucose-6-phosphate is required for nucleotide metabolism, glycosylation, cell wall biosynthesis, and storage of carbohydrates (Barnett & Entian, 2005). The expression of gluconeogenic genes is coregulated with the expression of many respiratory genes, as respiration is necessary in order to obtain energy by oxidative phosphorylation during gluconeogenic processes; as a result, respiratory-deficient mutants are unable to grow on the nonfermentable carbon sources (Hampsey, 1997). Biosynthesis of mitochondrial proteins depends on the presence of oxygen and heme and the availability of a carbon source (Schüller, 2003). For example, the expression of mitochondrial genes is increased in the presence of glycerol as compared with glucose (Roberts & Hudson, 2006).
Metabolism of nonfermentable carbons
Metabolism of glycerol
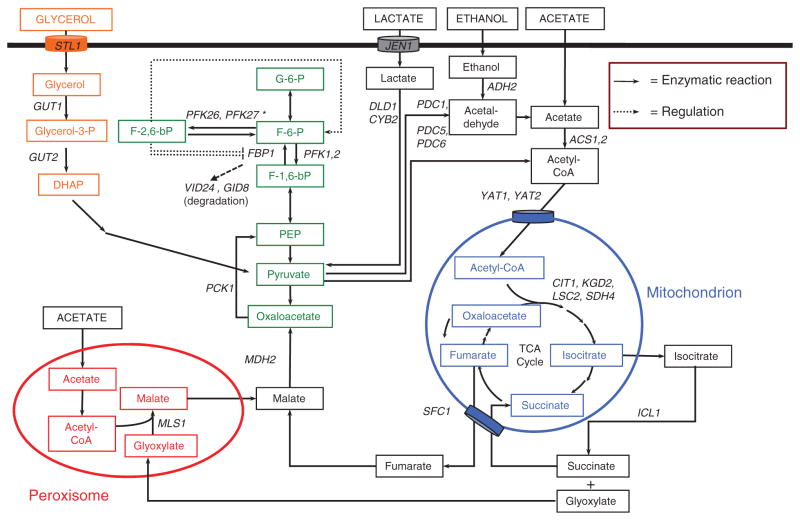
Yeast cells use glycerol as a carbon source as well as for osmoregulation (Hohmann, 2002). Glycerol uptake is mediated by the symporter Stl1 (sugar transporter-like protein) (Ferreira et al., 2005) (Fig. 1). Following its uptake, glycerol is converted to glycerol-3-phosphate by the cytoplasmic kinase Gut1 before entering the mitochondria. The mitochondrial FAD-dependent glycerol-3-phosphate dehydrogenase, encoded by the GUT2 gene, is responsible for the conversion to dihydroacetone phosphate, which can enter the glycolytic or the gluconeogenic pathway. Both GUT1 and GUT2 are expressed with cells grown in the presence of glycerol or ethanol while these genes are repressed in the presence of glucose (Pavlik et al., 1993).
Fig. 1.
Metabolic pathways and genes involved in the utilization of nonfermentable carbons. Metabolic pathways for utilization of nonfermentable carbons are schematically shown as well as key genes involved in this process. The pathway for fatty acid metabolism was omitted (see Hiltunen et al., 2003 for a review). Arrows with full lines correspond to enzymatic reactions while arrows with dashed lines correspond to regulatory steps. STL1 and JEN2 encode membrane transporters for glycerol and lactate, respectively. SFC1 encodes a mitochondrial transporter for fumarate. More information for specific genes can be found at the yeast genome database (http://www.yeastgenome.org).
Metabolism of lactate, ethanol, and acetate
In contrast to glycerol, lactate is taken up in the cells through a specific permease called Jen1 that also transports pyruvate (Casal et al., 1999; reviewed in Casal et al., 2008). JEN1 expression is repressed in the presence of glucose and is induced by lactate. D-Lactate and L-lactate are metabolized to pyruvate by two distinct mitochondrial lactate cytochrome c oxidoreductases, encoded by the DLD1 and CYB2 genes, respectively (Lodi & Ferrero, 1999). Unlike glycerol or lactate, ethanol and acetate are thought to enter the cells by passive diffusion, although an acetate carrier has been identified (Casal et al., 1996) (Fig. 1). Ethanol is also produced routinely in the cell as a consequence of alcoholic fermentation. Following its uptake, ethanol is metabolized to acetaldehyde by alcohol dehydrogenase (encoded by ADH2) and to acetate by aldehyde dehydrogenase (ALD6). Acetate is then transformed to acetyl-CoA by acetyl-CoA synthetase (ACS1).
Gluconeogenesis
Glycolysis and gluconeogenesis are two opposite pathways for glucose metabolism and multiple levels of regulation insure that only one pathway is active at a time. For example, the gluconeogenic enzymes fructose-1,6-bisphosphatase (FBP1), malate dehydrogenase (MDH2), and phosphoenolpyruvate carboxykinase (PCK1) are subject to degradation in the presence of glucose (Hung et al., 2004; Santt et al., 2008). Interestingly, the enzymatic activity of Pck1 requires acetylation at lysine 514 by the NuA4 acetyltransferase complex. This post-translational modification is essential for the growth of yeast cells on nonfermentable carbon sources (Lin et al., 2009). Allosteric control of enzymatic activity is also observed (Heinisch et al., 1996). Moreover, mRNA stability of some gluconeogenic genes is increased in the presence of a non-fermentable carbon source (Lombardo et al., 1992; Mercado et al., 1994; Andrade et al., 2005). Finally, another important mechanism of regulation is exerted at the transcriptional level. For instance, the expression of the gluconeogenic genes PCK1 and FBP1 as well as genes encoding glyoxylate enzymes ICL1 (isocitrate lyase) and MLS1 (malate synthase) is considerably upregulated during glucose depletion.
A number of enzymes are common to both glycolytic and gluconeogenic pathways while three enzymes are specific to gluconeogenesis, as described hereafter. Oxaloacetate is produced from pyruvate by pyruvate carboxylase encoded by the PYC1 and PYC2 genes. Oxaloacetate is then converted to phosphoenolpyruvate by the PCK1 gene product. A series of reactions allow the production of fructose-1,6-bisphosphate. The gluconeogenic enzyme fructose-1,6-bisphosphatase converts this compound to fructose-6-phosphate, which then yields glucose-6-phosphate by a reaction performed by phosphoglucose isomerase (PGI1).
Metabolism of oleic acid
The presence of oleate as a sole carbon source results in the upregulation of genes encoding enzymes for fatty acid β-oxidation and proteins involved in the enlargement of peroxisomes (reviewed in Hiltunen et al., 2003; Gurvitz & Rottensteiner, 2006a). There is evidence that the transporter Fat1 and the acyl-CoA synthetases Faa1 and Faa4 mediate active intracellular import (and activation) of fatty acids (Black & DiRusso, 2007). A heterodimer of the ATP-binding cassette transporters Pxa1 and Pxa2 is responsible for transport of activated fatty acids into the peroxisome, where β-oxidation takes place (Hiltunen et al., 2003). Enzymes involved in fatty acid oxidation include Fox1/Pox1 (a fatty-acyl coenzyme A oxidase), Fox2 (a protein with dual activity: 3-hydroxyacyl-CoA dehydrogenase and enoyl-CoA hydratase), and Pot1/Fox3 (a 3-ketoacyl-CoA thiolase).
Transcriptional regulators: the zinc cluster proteins
A number of transcriptional regulators implicated in the use of alternate carbon sources have been identified and are listed in Table 1. Many of them belong to the Gal4 family and form a subclass of zinc finger proteins called zinc binuclear cluster or zinc cluster proteins (Vallee et al., 1991). Zinc cluster proteins form one of the largest families of transcriptional regulators in the yeast S. cerevisiae, consisting of over 50 members (MacPherson et al., 2006). They are characterized by the presence of a well-conserved and fungal-specific zinc cluster motif, CysX2CysX6 CysX5–12CysX2CysX6–8Cys, located in the DNA-binding domain (Todd & Andrianopoulos, 1997; MacPherson et al., 2006). The proper folding of this domain is co-ordinated through the binding of the conserved cysteine residues to two zinc atoms. Mutation or deletion of these cysteines, or the absence of zinc, results in the loss of DNA-binding activity (Bai & Kohlhaw, 1991). The zinc cluster motif makes contact with three base pairs, usually CGG triplets, in the major groove of the DNA (Marmorstein et al., 1992; Marmorstein & Harrison, 1994). Altering the spacing between the triplets generates binding sites for different zinc cluster proteins. Variation in the relative orientation of the CGG triplets [inverted (CGG Nx CCG), direct (CGG Nx CGG), or everted (CCG Nx CGG) repeats] further increases the repertoire of binding sites for these regulators (Mac-Pherson et al., 2006). Quite often, zinc cluster proteins bind to DNA as homo- or heterodimers although monomeric binding has also been described (MacPherson et al., 2006). Zinc cluster proteins can act as transcriptional activators or repressors and some of them have been shown to perform both functions (Larochelle et al., 2006; MacPherson et al., 2006; Soontorngun et al., 2007).
Table 1.
Major transcriptional regulators of nonfermentable carbon utilization and their targets
| Transcriptional regulator | Type of DNA-binding domain | Target genes |
|---|---|---|
| Adr1 (alcohol dehydrogenase regulator) | Cys2His2 zinc finger protein | Nonfermentable carbon metabolism (e.g. ADH2, ACS1, GUT1) Peroxisome biogenesis and fatty acids utilization (e.g. POX1, PXA1) |
| Cat8 (CATabolite repression) | Zinc cluster protein | Gluconeogenic genes (e.g. PCK1, FBP1) Glyoxylate cycle genes Transcription factor (SIP4) |
| Ert1 (ethanol regulator of translation) | Zinc cluster protein |
PCK1 Other targets unknown |
| Gsm1 (glucose starvation modulator) | Zinc cluster protein | Gluconeogenesis (PCK1, FBP1) Transcription factor (HAP4) |
| Hap1 (heme activator protein) | Zinc cluster protein | Respiration genes (e.g. CYC1, CYC7) |
| Hap2/3/4/5 (heme activator protein) | CCAAT-binding complex | Respiration genes (e.g. CYC1), TCA cycle |
| Oaf1 (oleate-activated transcription factor) | Zinc cluster protein | Fatty acids utilization (e.g. POX1, FOX3) Peroxisome biogenesis |
| Oaf3 (oleate-activated transcription factor) | Zinc cluster protein | Weak repressor of oleate-responsive genes |
| Pip2 (peroxisome induction pathway) | Zinc cluster protein | Fatty acids utilization (e.g. POX1, FOX3) Peroxisome biogenesis |
| Rds2 (regulator of drug sensitivity) | Zinc cluster protein | Gluconeogenic genes (e.g. PCK1, FBP1) Glyoxylate cycle genes (MLS1, TCA cycle genes) Transcription factors (HAP4, SIP4) |
| Sip4 (Snf1-interacting protein) | Zinc cluster protein | Gluconeogenic genes (e.g. PCK1) |
For references, see text.
TCA, tricarboxylic acid.
A number of zinc cluster regulators play central roles in co-ordinating gene expression during adaptation to different carbon sources. For example, Gal4 and its control of GAL structural genes for galactose catabolism is a classic example of eukaryotic transcriptional regulation (Lohr et al., 1995; Traven et al., 2006). Three other zinc cluster proteins, Mal13, Mal3R, and Mal63, are involved in the control of maltose metabolic genes in some yeast strains (Needleman, 1991). Other zinc cluster proteins described below are involved in the use of nonfermentable carbons.
Role of the zinc cluster proteins Cat8 and Sip4
Schöler & Schüller (1994) previously reported the presence of a carbon source-responsive element (CSRE) in the promoter of ICL1-encoding isocitrate lyase, a key enzyme of the glyoxylate cycle. They showed that the CSRE is an element necessary for ICL1 derepression in the absence of glucose. Additionally, it was shown that the CSRE alone allows for transcription on a heterologous minimal promoter in a carbon source-dependent manner. A number of other genes also contain CSREs in their promoters [consensus sequence: YCCRTTNRNCGG (Roth et al., 2004)]: FBP1, PCK1, MLS1, ACS1, MDH2 (malate dehydrogenase), SFC1 (succinate/fumarate transporter), CAT2 (carnitine acetyltransferase), IDP2 (NADP-dependent isocitrate dehydrogenase), and JEN1 (Schüller, 2003). Activation of genes containing CSREs is mediated, among others, by the zinc cluster proteins Cat8 (CATabolite repression) and Sip4, which was isolated as an Snf1-interacting protein (Hedges et al., 1995; Lesage et al., 1996; Rahner et al., 1996; Vincent & Carlson, 1998). Snf1 is a central serine–threonine kinase in the signaling pathway for glucose-mediated repression.
Other studies showed that both Cat8 and Sip4 bind to CSREs in the promoter of gluconeogenic genes in vitro (Vincent & Carlson, 1998; Rahner et al., 1999). Although these two activators are involved in gluconeogenesis, their relative contribution via the CSRE is different. A substantial reduction in the expression of CSRE-dependent genes was shown in the absence of Cat8, while removal of Sip4 accounted for only a minor reduction in gene activation (Hiesinger et al., 2001). Additionally, cells lacking Cat8, but not Sip4, are unable to grow on nonfermentable carbon sources (Hedges et al., 1995; Rahner et al., 1996).
The expression of the transcriptional regulator Cat8 is under the control of the carbon source (Hedges et al., 1995; Randez-Gil et al., 1997). In the presence of glucose, CAT8 expression is repressed by Mig1 (a Cys2His2 zinc finger protein), possibly by direct binding of this regulator to the CAT8 promoter (Hedges et al., 1995; Rahner et al., 1996). A related regulatory mechanism applies to another CSRE-binding protein, Sip4. Derepression of CSRE-containing genes is abolished in a Δcat8Δsip4 deletion mutant, suggesting their role as sole activators specific for the CSRE motif (Roth et al., 2004). However, evidence suggests that they may utilize different CSRE variants and that Sip4 actually recognizes a narrower range of binding sites as compared with Cat8 (Roth et al., 2004). Importantly, Cat8 is an activator of SIP4 transcription and, therefore, indirectly of Sip4 target genes (Haurie et al., 2001; Tachibana et al., 2005). A CSRE-like element is found on the SIP4 promoter, which may explain the carbon source-dependent activation of SIP4 expression (Vincent & Carlson, 1998). In agreement with this hypothesis, a microarray study showed that the transcription of SIP4 is induced approximately ninefold during a diauxic shift (DeRisi et al., 1997). Moreover, deletion of CAT8 results in a reduction of SIP4 mRNA, further arguing for a crosstalk between these two genes (Haurie et al., 2001).
Role of the zinc cluster protein Rds2
Recently, another zinc cluster protein was described as being important for regulating gluconeogenesis (Soontorngun et al., 2007). A number of phenotypes are associated with a deletion of the ORF of YPL133C including sensitivity to calcofluor white and the antifungal drug ketoconazole, and it was named RDS2 (for regulator of drug sensitivity) (Akache et al., 2001; Akache & Turcotte, 2002). Depending on the strain background, impaired growth on glycerol or lactate is also observed with a partial deletion of RDS2 (Akache et al., 2001). ChIP-chip, a technique that relies on chromatin immunoprecipitation (ChIP) and microarray (chip), was used to determine the genome-wide localization of Rds2. Results showed that this factor binds to a limited number of promoters with cells grown in the presence of glucose while it binds to many additional genes when ethanol is used as a carbon source. Strikingly, the genes bound by Rds2 are involved in gluconeogenesis (e.g. PCK1) and related pathways such as the glyoxylate shunt and the tricarboxylic acid cycle. Importantly, it was shown that Rds2 acts as a transcriptional activator of gluconeogenic genes while it is a repressor of the negative regulators of gluconeogenesis. Genes under the positive regulation of Rds2 include PCK1, FBP1, and LSC2. In the absence of RDS2, the expression of GID8 (glucose-induced degradation) is increased with cells grown in the presence of ethanol. Gid8 is a part of a complex involved in the degradation of Fbp1 and Pck1 under glucose conditions (Regelmann et al., 2003; Santt et al., 2008). These results suggest that, following a shift from glucose to ethanol, the expression of GID8 is repressed to prevent degradation of gluconeogenic enzymes by the Gid complex. Similarly, under ethanol conditions, Rds2 is a repressor of the PFK27 gene. Pfk27 catalyzes the production of fructose-2,6-bisphosphate, an allosteric activator of the glycolytic enzyme phosphofructokinase (PFK1,2) and a repressor of Fbp1 (Fig. 1) (Noda et al., 1984; Heinisch et al., 1996). Thus, Rds2 has activator and repressor functions that contribute to the selective activation of gluconeogenesis over glycolysis.
The importance of RDS2 in controlling genes involved in ethanol utilization is further exemplified by the fact that it binds and upregulates the expression of HAP4. The Hap2/3/ 4/5 complex controls the expression of respiration genes via an activating subunit encoded by HAP4, the only subunit whose expression is regulated by a carbon source (Forsburg & Guarente, 1989; DeRisi et al., 1997). This effect may be mediated by a functional CSRE present in the HAP4 promoter (Brons et al., 2002). Moreover, Rds2 binding is also detected at the OPI1 promoter, encoding a negative regulator of the phospholipid biosynthetic pathway. The connection between phospholipids and Rds2 may not be obvious. However, the GUT1 and the GUT2 genes, involved in glycerol utilization, were shown to be negatively regulated by the repressor Opi1 (Grauslund et al., 1999; Grauslund & Ronnow, 2000). Deletion of OPI1 allows derepression of GUT1, as assayed in glucose. Thus, Rds2 may positively regulate the expression of GUT1 and GUT2 indirectly by repressing OPI1 expression in the presence of nonfermen-table carbons (but not in the presence of glucose). Rds2 also binds to the regulatory gene SIP4, raising the possibility that both Cat8 and Rds2 control SIP4 expression. As observed for Cat8 and Sip4, the purified DNA-binding domain of Rds2 binds in vitro to CSREs, and mutations diminishing Cat8 binding also affect the binding of Rds2 (Soontorngun et al., 2007). In summary, Rds2 is a newly characterized transcriptional regulator playing a central role in the regulation of gluconeogenesis in yeast.
Role of the zinc finger protein Adr1
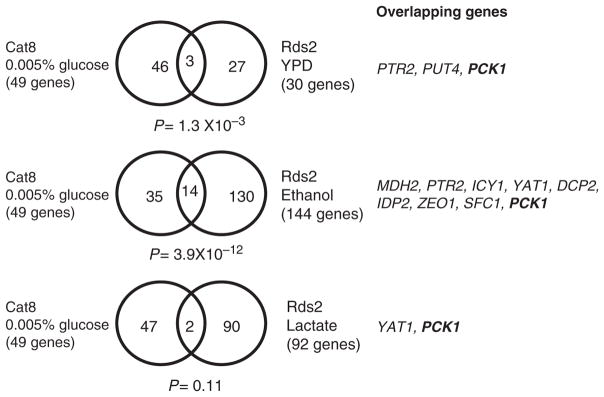
Adr1 is a transcription factor of the Cys2His2 class of zinc finger that binds DNA as a monomer (Thukral et al., 1991; Cheng et al., 1994). Adr1 is involved in regulating genes for utilization of ethanol, glycerol, and lactate (Simon et al., 1991; Young et al., 2003). In fact, the expression of over 100 genes is dependent on Adr1, as shown by microarray analysis (Young et al., 2003). For example, Adr1 regulates the expression of over 30 glucose-repressed genes such as ADH2, encoding an alcohol dehydrogenase acting at the first step of ethanol utilization (Fig. 1). Other genes regulated by Adr1 include ALD4, ACS1, GUT1, and FOX2. Adr1 and Cat8 coregulate some genes such as JEN1, although expression profiling and ChIP-chip data indicate that only a handful (14) of overlapping gene targets is shared between them (Young et al., 2003; Tachibana et al., 2005). Similarly, a comparison of the ChIP-chip data obtained with Cat8 (under low glucose conditions) and Rds2 (ethanol) shows that these factors have only a limited number of common targets that include PCK1, MDH2, and SFC1 (Fig. 2) (Soontorngun et al., 2007).
Fig. 2.
Limited overlap between Rds2 and Cat8 target genes. Cat8 target genes identified by ChIP-chip analysis under low glucose conditions (Tachibana et al., 2005) were compared with those identified for Rds2 under ethanol (Soontorngun et al., 2007) or lactate conditions (N. Soontorngun & B. Turcotte, unpublished data). P-values used for gene selection are indicated below Venn diagrams.
The zinc cluster protein Ert1
A recent study identified the zinc cluster genes AcuM and AcuK as being involved in regulating the transcription of gluconeogenic genes in the filamentous fungi Aspergillus nidulans (Hynes et al., 2007). AcuM appears to be a homologue of Rds2 while AcuK shows a strong similarity to the zinc cluster protein Ybr239c (alias Ert1) in S. cerevisiae. Interestingly, a large-scale two-hybrid study in budding yeast suggested a physical interaction between Rds2 and Ert1 (Ito et al., 2001). To learn more about the role of Ert1, ChIP analysis was performed with this factor and binding was observed at the PCK1 promoter (X.B. Liang & B. Turcotte, unpublished data). Moreover, deletion of ERT1 results in a slight decrease of the expression of PCK1 (X.B. Liang & B. Turcotte, unpublished data). The exact role of this zinc cluster protein remains to be defined. Taken together, the results show that at least four zinc cluster proteins (Cat8, Sip4, Rds2, and Ert1) can bind to the PCK1 promoter. Clearly, a complex regulation is exerted at this gene encoding a key component of gluconeogenesis. The specific role of these factors and their interplay at PCK1 (and other genes) remains to be defined more precisely.
The zinc cluster protein Gsm1
Other studies suggest that another zinc cluster protein is also implicated in the use of nonfermentable carbon sources. Indeed, an expression profiling study showed that mRNA levels of the zinc cluster gene GSM1 (glucose starvation modulator) are increased 12 times in the presence of glycerol or ethanol, as compared with glucose (Roberts & Hudson, 2006). Moreover, ChIP-chip experiments show that this protein binds, for example, to the HAP4 and IDP2 promoters (van Bakel et al., 2008). Gsm1 regulates the expression of the gluconeogenic genes PCK1 and FBP1 (W.G. Bao & M. Bolotin-Fukuhara, pers. commun.). Interestingly, the expression of GSM1 is decreased in cells lacking HAP2 or HAP4 (Buschlen et al., 2003). These results suggest an interplay between HAP4 and GSM1.
It remains to be seen whether additional factors may be involved in the use of nonfermentable carbons. Its transcriptional regulation involves more regulatory factors than initially anticipated. The roles of specific transcriptional regulators may also differ according to the nonfermentable carbon. For example, ChIP-chip analysis of Rds2 under lactate shows that its targets differ from those identified under ethanol conditions (N. Soontorngun & B. Turcotte, unpublished data). As stated above, Rds2 and Ert1 interact with each other in a two-hybrid assay, suggesting they could form heterodimers at some target promoters, as observed for some zinc cluster proteins involved in conferring drug resistance (Mamnun et al., 2002; Akache et al., 2004) or the Oaf1–Pip2 pair. Putative heterodimers could also be formed by an interaction with the other regulators Sip4, Cat8, and Ert1.
Role of the zinc cluster proteins Oaf1 and Pip2 in oleate utilization
The expression of the genes for fatty acid metabolism and peroxisome biogenesis is regulated by a combination of transcription factors (Smith et al., 2007). For example, the zinc cluster protein Oaf3 is a weak repressor of oleate-responsive genes (Smith et al., 2007). The zinc cluster proteins Oaf1 and Pip2 (Oaf2) have been extensively characterized and shown to mediate the response to oleate by binding as heterodimers to oleate response elements (consensus: CGGN3TNAN9–12CCG) found in the promoters of β-oxidation genes (Rottensteiner et al., 1996, 1997). Although OAF1 and PIP2 coregulate the same genes, their expression is differentially regulated (Rottensteiner et al., 1997). OAF1 expression is constitutive whereas the expression of PIP2 is positively autoregulated (Rottensteiner et al., 1997). ChIP analysis has demonstrated that Oaf1 and Pip2 are found at common promoters (Karpichev et al., 2008). The presence of Pip2 is also required for Oaf1 binding at most promoters tested. Moreover, successive ChIP assays (re-ChIP) with differently tagged Oaf1 and Pip2 have shown that these factors co-occupy the same target promoters. Thus, these data strongly suggest that an Oaf1–Pip2 heterodimer is mainly responsible for the activation of target genes. However, a few target genes (e.g. FOX2, CTA1) appear to be regulated by Oaf1, but not Pip2, suggesting activation by an Oaf1 homodimer (Trzcinska-Danielewicz et al., 2008).
Binding of Oaf1–Pip2 to oleate response elements in vivo is increased by shifting cells from repressing (glucose) to derepressing conditions (glycerol), but is only marginally affected under inducing (oleate) conditions (Karpichev et al., 2008). Thus, under derepressed conditions, Oaf1–Pip2 is constitutively bound to target promoters. Adr1 is also involved in regulating the expression of some genes for fatty acid oxidation and peroxisome biogenesis (Young et al., 2003). ChIP experiments show that this factor is required for optimal binding of Oaf1–Pip2 at some promoters and vice versa (Karpichev et al., 2008). Activation of the Oaf1/Pip2 heterodimer is mediated by direct binding of oleate to Oaf1 (Phelps et al., 2006; Thakur et al., 2009). Moreover, the presence of oleate results in hyperphosphorylation of Oaf1 and correlates with its transcriptional activity. The activation domain of Oaf1 was shown to interact with Med15 (Gal11), a subunit of the mediator complex that links transcriptional activators to general transcription factors and RNA polymerase II (Thakur et al., 2009). From these various observations, a model for the mechanism of activation of Oaf1–Pip2 can be proposed. Under derepressing conditions, Oaf1 becomes phosphorylated by an unknown kinase favoring binding of the heterodimer to target genes, including the promoter of PIP2. Binding of oleate to Oaf1 would trigger a conformational change allowing interaction with Med15 (Gal11) and transcriptional activation. It is unclear as to why the presence of oleate results in hyperphosphorylation of Oaf1. One possibility is that this post-transcriptional modification may favor the interaction with Med15 (Gal11).
Mechanism of activation of transcription factors for utilization of nonfermentable carbons
As stated above, a key factor for the activation of glucose-repressed genes is the kinase Snf1 (also called Cat1) (for reviews, see Hardie et al., 1998; Sanz, 2003, 2007; Hedbacker & Carlson, 2008). Briefly, Snf1 is activated under low glucose conditions and is a part of a complex that includes the activating subunit Snf4 (also called Cat3) and a third partner (Gal83, Sip1, or Sip2) (Erickson & Johnston, 1993; Yang et al., 1994). The exact mechanism of Snf1 activation is still unclear, but it has been shown that the kinases Sak1 (Pak1), Tos3, and Elm1 are upstream effectors of Snf1 (Hong et al., 2003; Sutherland et al., 2003). These kinases phosphorylate Thr210 of the Snf1 activation loop. Pak1 activity is also required for nuclear localization of Snf1 (Hedbacker et al., 2004). Tos3 activity is dispensable for responding to a sharp decrease of glucose levels, but is required for the activation of CSRE-containing genes with cells grown in ethanol/glycerol (Kim et al., 2005). It is still not well understood as to how these kinases become activated under low glucose conditions. Moreover, other levels of regulation of Snf1 activity include autoinhibition (Jiang & Carlson, 1996; Leech et al., 2003) as well as a potential control by dephosphorylation via the protein phosphatase complex I (Glc7/Reg1) (Sanz et al., 2000).
Snf1 has multiple targets such as chromatin (histone H3), transcriptional activators, and repressors (Hedbacker & Carlson, 2008). For example, the transcriptional repressor Mig1 is a target of the Snf1 kinase. Phosphorylated Mig1 dissociates from the corepressor Ssn6-Tup1 protein complex and is exported to the cytoplasm through the exportin Msn5 (DeVit & Johnston, 1999; Smith et al., 1999; Papamichos-Chronakis et al., 2004), resulting in an increased expression of CAT8. Cat8 is also phosphorylated by the Snf1 kinase (Randez-Gil et al., 1997). Convincing studies by Noël-Geoiris’ group have shown that phosphorylation of a single serine residue in Cat8 from S. cerevisiae (or its homologue in Kluyveromyces lactis) is responsible for the activation of this factor (Charbon et al., 2004). Snf1 phosphorylation of Sip4 correlates with its transcriptional activity (Lesage et al., 1996). Similarly, hyperphosphorylation of Rds2 in ethanol is Snf1 dependent (Soontorngun et al., 2007).
As stated above, transcriptional regulators of nonfermentable carbon utilization have distinct and overlapping targets. This observation raises the question of whether a given regulator requires a partner for binding at specific promoters or not. Is activation of target genes controlled at the step of binding of the regulator to specific DNA sequences in target promoters? A number of studies have addressed the interplay among these factors and diverse mechanisms appear to operate according to the factor and the target genes studied. For example, ChIP-chip results show that Rds2 is constitutively bound to the PCK1 promoter, even under glucose conditions where (1) this gene is not expressed, (2) the Snf1 kinase is inactive, and (3) Cat8 is present at very low levels (Soontorngun et al., 2007). Thus, phosphorylation of Rds2 by Snf1 is not required for binding of this factor at some promoters. However, under ethanol conditions, deletion of CAT8 results in a modest decrease in binding of Rds2 at the PCK1 promoter (twofold) while a more pronounced effect (over sixfold) is observed at the FBP1 promoter, as determined by standard ChIP analysis (Soontorngun et al., 2007). These results provide an example of the interplay among these factors.
Additional recent studies have provided insights into the mechanism of activation of these nonfermentable gene regulators. For example, binding of Adr1 to the ADH2 promoter as well as to other target genes (CTA1, ACS1, GUT1, and POT1) is Snf1 dependent (Young et al., 2002). However, the cyclin-dependent kinase Pho85, but not Snf1, appears to be indirectly involved in Adr1 phosphorylation and inactivation (Kacherovsky et al., 2008). Phosphorylation of Ser98, located in the DNA-binding domain of Adr1, is important for controlling the activity of this factor. For example, mimicking phosphorylation by mutating Ser98 to Asp decreases the binding affinity of Adr1, as assayed in vitro by an electrophoretic mobility shift assay and in vivo by ChIP (Kacherovsky et al., 2008). As expected from the binding studies, the Asp98 mutant is transcriptionally inactive (Kacherovsky et al., 2008).
A double deletion of the histone deacetylase genes HDA1 and RPD3 allows, even under repressive conditions, constitutive binding of Adr1 and Cat8 at target promoters such as ADH2 (Tachibana et al., 2007). Both Adr1 and Cat8 require the mediator complex as well as the chromatin remodeling complexes SWI/SNF and SAGA (Spt-Ada-Gcn5-acetyl transferase) for transcriptional activation (Biddick et al., 2008). In a Δhda1Δrpd3 strain, binding of these cofactors is observed while only marginal transcriptional activity is observed in the absence of Snf1 activation. In fact, binding of Adr1 and Cat8 is reduced in a triple deletion strain Δhda1Δrpd3Δsnf1 (Tachibana et al., 2007). Other results show that Snf1 mediates its effect after the binding of RNA polymerase II. Finally, a fusion of the DNA-binding domain of Adr1 to the Med15 (Gal11) component of the mediator bypasses the requirement for Snf1, SWI/SNF, and SAGA for the activation of ADH2 (Young et al., 2008). Taken together, Young’s results suggest that the promoter of ADH2 is accessible to Adr1, but that, under normal (repressing) conditions, Adr1 lacks the ability to interact with coactivators such as a mediator.
A model for the regulatory network of regulators of nonfermentable carbons
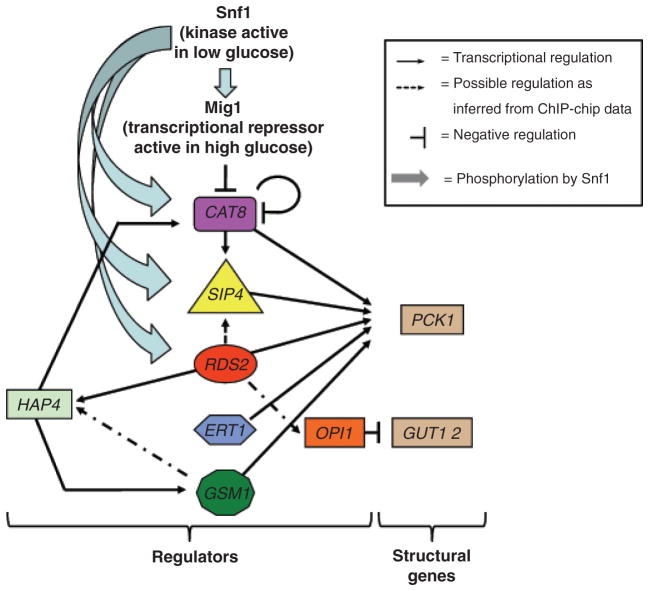
As stated above, the various transcriptional regulators of nonfermentable carbon metabolism have distinct and overlapping functions. Recent studies using genome-wide expression profiling and location analysis have provided additional useful information on the interplay among these transcription factors. Even though some of the experiments were not performed under the same conditions and may not be directly comparable, a model for the network of regulators of nonfermentable carbons is proposed in Fig. 3 that integrates various data. The expression of Rds2 does not vary significantly according to the carbon source and its activation correlates with its phosphorylation by the Snf1 kinase (Soontorngun et al., 2007). Rds2 (and potentially Gsm1, as suggested by the ChIP analysis) increases the expression of HAP4 encoding the limiting and activating subunit of the Hap2/3/4/5 complex (Soontorngun et al., 2007; van Bakel et al., 2008). The expression of GSM1 is increased in nonfermentable carbons (Roberts & Hudson, 2006) by the Hap2/3/4/5 complex (Buschlen et al., 2003), providing a putative autoregulatory loop between HAP4 and GSM1.
Fig. 3.
A model for the regulatory network of regulators of nonfermentable carbons. Low glucose levels activate the Snf1 kinase, resulting in phosphorylation and inactivation of the Mig1 repressor. Cat8, Sip4, and Rds2 are also substrates of Snf1. Rds2 and probably Gsm1 are activators of HAP4, whose gene product is a part of a complex involved in the positive control of CAT8 and GSM1. Cat8 and most likely Rds2 are positive regulators of SIP4. CAT8 expression is probably autoregulated. Cat8, Sip4, Rds2, Ert1, and Gsm1 are all transcriptional regulators of PCK1 encoding a key gluconeogenic enzyme. ChIP analysis showed that Rds2 binds to the OPI1 gene encoding a repressor of GUT1 and GUT2 expression involved in glycerol metabolism. See text for more details.
Inactivation of Mig1 by Snf1 relieves the repression of CAT8 expression, allowing the Hap2/3/4/5 complex to positively regulate the expression of CAT8. In agreement with this model, the expression of a CAT8-lacZ reporter was reduced five times when assayed in low glucose with a Δhap2 strain (Rahner et al., 1996). Increased Cat8 levels and its activation by phosphorylation allow positive regulation of SIP4, which is probably also mediated by Rds2 because binding of this activator was detected at the SIP4 promoter by ChIP (Soontorngun et al., 2007). Remarkably, Cat8, Sip4, Rds2 Ert1, and Gsm1 all regulate the expression of PCK1. Regulation of Snf1 activity provides a means to control the whole network. In addition, Cat8 may provide a negative feedback loop in this system because expression of a CAT8-lacZ reporter is increased when assayed in a Δcat8 strain (Rahner et al., 1996).
In recent years, significant progress has been made toward understanding the mechanism of transcriptional regulation of nonfermentable carbon utilization in S. cerevisiae. However, many questions remain to be answered. What is the exact mechanism of regulation of Snf1 activity? What is the exact role of Gsm1 and Ert1? Are there additional transcriptional regulators involved in this process?
Acknowledgments
We are grateful to Monique Bolotin-Fukuhara (Université Paris-Sud) for communicating results before publication. We also thank Dr Geoffrey Hendy and Karen Hellauer for comments on the manuscript. This work was supported by a grant from the Natural Sciences and Engineering Research Council of Canada to B.T. F.R. holds a new investigator award from the Canadian Institutes for Health Research.
References
- Akache B, Turcotte B. New regulators of drug sensitivity in the family of yeast zinc cluster proteins. J Biol Chem. 2002;277:21254–21260. doi: 10.1074/jbc.M202566200. [DOI] [PubMed] [Google Scholar]
- Akache B, Wu KQ, Turcotte B. Phenotypic analysis of genes encoding yeast zinc cluster proteins. Nucleic Acids Res. 2001;29:2181–2190. doi: 10.1093/nar/29.10.2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akache B, MacPherson S, Sylvain MA, Turcotte B. Complex interplay among regulators of drug resistance genes in Saccharomyces cerevisiae. J Biol Chem. 2004;279:27855–27860. doi: 10.1074/jbc.M403487200. [DOI] [PubMed] [Google Scholar]
- Andrade RP, Kotter P, Entian KD, Casal M. Multiple transcripts regulate glucose-triggered mRNA decay of the lactate transporter JEN1 from Saccharomyces cerevisiae. Biochem Bioph Res Co. 2005;332:254–262. doi: 10.1016/j.bbrc.2005.04.119. [DOI] [PubMed] [Google Scholar]
- Bai YL, Kohlhaw GB. Manipulation of the ‘zinc cluster’ region of transcriptional activator LEU3 by site-directed mutagenesis. Nucleic Acids Res. 1991;19:5991–5997. doi: 10.1093/nar/19.21.5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett JA, Entian KD. A history of research on yeasts 9: regulation of sugar metabolism. Yeast. 2005;22:835–894. doi: 10.1002/yea.1249. [DOI] [PubMed] [Google Scholar]
- Biddick RK, Law GL, Chin KKB, Young ET. The transcriptional coactivators SAGA, SWI/SNF, and mediator make distinct contributions to activation of glucose-repressed genes. J Biol Chem. 2008;283:33101–33109. doi: 10.1074/jbc.M805258200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black PN, DiRusso CC. Yeast acyl-CoA synthetases at the crossroads of fatty acid metabolism and regulation. Biochim Biophys Acta. 2007;1771:286–298. doi: 10.1016/j.bbalip.2006.05.003. [DOI] [PubMed] [Google Scholar]
- Brons JF, De Jong M, Valens M, Grivell LA, Bolotin-Fukuhara M, Blom J. Dissection of the promoter of the HAP4 gene in S. cerevisiae unveils a complex regulatory framework of transcriptional regulation. Yeast. 2002;19:923–932. doi: 10.1002/yea.886. [DOI] [PubMed] [Google Scholar]
- Buschlen S, Amillet JM, Guiard B, Fournier A, Marcireau C, Bolotin-Fukuhara M. The S. cerevisiae HAP complex, a key regulator of mitochondrial function, coordinates nuclear and mitochondrial gene expression. Comp Funct Genom. 2003;4:37–46. doi: 10.1002/cfg.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson M. Glucose repression in yeast. Curr Opin Microbiol. 1999;2:202–207. doi: 10.1016/S1369-5274(99)80035-6. [DOI] [PubMed] [Google Scholar]
- Casal M, Cardoso H, Leao C. Mechanisms regulating the transport of acetic acid in Saccharomyces cerevisiae. Microbiology. 1996;142:1385–1390. doi: 10.1099/13500872-142-6-1385. [DOI] [PubMed] [Google Scholar]
- Casal M, Paiva S, Andrade RP, Gancedo C, Leao C. The lactate-proton symport of Saccharomyces cerevisiae is encoded by JEN1. J Bacteriol. 1999;181:2620–2623. doi: 10.1128/jb.181.8.2620-2623.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal M, Paiva S, Queiros O, Soares-Silva I. Transport of carboxylic acids in yeasts. FEMS Microbiol Rev. 2008;32:974–994. doi: 10.1111/j.1574-6976.2008.00128.x. [DOI] [PubMed] [Google Scholar]
- Charbon G, Breunig KD, Wattiez R, Vandenhaute J, Noël-Georis I. Key role of Ser562/661 in Snf1-dependent regulation, of Cat8p in Saccharomyces cerevisiae and Kluyveromyces lactis. Mol Cell Biol. 2004;24:4083–4091. doi: 10.1128/MCB.24.10.4083-4091.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C, Kacherovsky N, Dombek KM, Camier S, Thukral SK, Rhim E, Young ET. Identification of potential target genes for Adr1p through characterization of essential nucleotides in Uas1. Mol Cell Biol. 1994;14:3842–3852. doi: 10.1128/mcb.14.6.3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRisi JL, Iyer VR, Brown PO. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- DeVit MJ, Johnston M. The nuclear exportin Msn5 is required for nuclear export of the Mig1 glucose repressor of Saccharomyces cerevisiae. Curr Biol. 1999;9:1231–1241. doi: 10.1016/s0960-9822(99)80503-x. [DOI] [PubMed] [Google Scholar]
- Erickson JR, Johnston M. Genetic and molecular characterization of GAL83: its interaction and similarities with other genes involved in glucose repression in Saccharomyces cerevisiae. Genetics. 1993;135:655–664. doi: 10.1093/genetics/135.3.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira C, van Voorst F, Martins A, Neves L, Oliveira R, Kielland-Brandt MC, Lucas C, Brandt A. A member of the sugar transporter family, Stl1p is the glycerol/H+ symporter in Saccharomyces cerevisiae. Mol Biol Cell. 2005;16:2068–2076. doi: 10.1091/mbc.E04-10-0884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsburg SL, Guarente L. Identification and characterization of HAP4: a third component of the CCAAT-bound HAP2/HAP3 heteromer. Gene Dev. 1989;3:1166–1178. doi: 10.1101/gad.3.8.1166. [DOI] [PubMed] [Google Scholar]
- Gancedo JM. Yeast carbon catabolite repression. Microbiol Mol Biol R. 1998;62:334–361. doi: 10.1128/mmbr.62.2.334-361.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grauslund M, Ronnow B. Carbon source-dependent transcriptional regulation of the mitochondrial glycerol-3-phosphate dehydrogenase gene, GUT2, from Saccharomyces cerevisiae. Can J Microbiol. 2000;46:1096–1100. doi: 10.1139/w00-105. [DOI] [PubMed] [Google Scholar]
- Grauslund M, Lopes JM, Ronnow B. Expression of GUT1, which encodes glycerol kinase in Saccharomyces cerevisiae, is controlled by the positive regulators Adr1p, Ino2p and Ino4p and the negative regulator Opi1p in a carbon source-dependent fashion. Nucleic Acids Res. 1999;27:4391–4398. doi: 10.1093/nar/27.22.4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiard B. Structure, expression and regulation of a nuclear gene encoding a mitochondrial protein: the yeast L(+)-lactate cytochrome c oxidoreductase (cytochrome b2) EMBO J. 1985;4:3265–3272. doi: 10.1002/j.1460-2075.1985.tb04076.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurvitz A, Rottensteiner H. The biochemistry of oleate induction: transcriptional upregulation and peroxisome proliferation. BBA-Mol Cell Res. 2006a;1763:1392–1402. doi: 10.1016/j.bbamcr.2006.07.011. [DOI] [PubMed] [Google Scholar]
- Gurvitz A, Rottensteiner H. The biochemistry of oleate induction: transcriptional upregulation and peroxisome proliferation. BBA-Mol Cell Res. 2006b;1763:1392–1402. doi: 10.1016/j.bbamcr.2006.07.011. [DOI] [PubMed] [Google Scholar]
- Hampsey M. A review of phenotypes in Saccharomyces cerevisiae. Yeast. 1997;13:1099–1133. doi: 10.1002/(SICI)1097-0061(19970930)13:12<1099::AID-YEA177>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Hardie DG, Carling D, Carlson M. The AMP-activated/ Snf1 protein kinase subfamily – metabolic sensors of the eukaryotic cell. Annu Rev Biochem. 1998;67:821–855. doi: 10.1146/annurev.biochem.67.1.821. [DOI] [PubMed] [Google Scholar]
- Haurie V, Perrot M, Mini T, Jeno P, Sagliocco F, Boucherie H. The transcriptional activator Cat8p provides a major contribution to the reprogramming of carbon metabolism during the diauxic shift in Saccharomyces cerevisiae. J Biol Chem. 2001;276:76–85. doi: 10.1074/jbc.M008752200. [DOI] [PubMed] [Google Scholar]
- Hedbacker K, Carlson M. SNF1/AMPK pathways in yeast. Front Biosci. 2008;13:2408–2420. doi: 10.2741/2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedbacker K, Hong SP, Carlson M. Pak1 protein kinase regulates activation and nuclear localization of Snf1-Gal83 protein kinase. Mol Cell Biol. 2004;24:8255–8263. doi: 10.1128/MCB.24.18.8255-8263.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges D, Proft M, Entian KD. CAT8 a new zinc cluster-encoding gene necessary for derepression of gluconeogenic enzymes in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:1915–1922. doi: 10.1128/mcb.15.4.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinisch JJ, Boles E, Timpel C. A yeast phosphofructokinase insensitive to the allosteric activator fructose 2,6-bisphosphate – glycolysis metabolic regulation allosteric control. J Biol Chem. 1996;271:15928–15933. doi: 10.1074/jbc.271.27.15928. [DOI] [PubMed] [Google Scholar]
- Hiesinger M, Roth S, Meissner E, Schuller HJ. Contribution of Cat8 and Sip4 to the transcriptional activation of yeast gluconeogenic genes by carbon source-responsive elements. Curr Genet. 2001;39:68–76. doi: 10.1007/s002940000182. [DOI] [PubMed] [Google Scholar]
- Hiltunen JK, Mursula AM, Rottensteiner H, Wierenga RK, Kastaniotis AJ, Gurvitz A. The biochemistry of peroxisomal beta-oxidation in the yeast Saccharomyces cerevisiae. FEMS Microbiol Rev. 2003;27:35–64. doi: 10.1016/S0168-6445(03)00017-2. [DOI] [PubMed] [Google Scholar]
- Hohmann S. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol Mol Biol R. 2002;66:300–372. doi: 10.1128/MMBR.66.2.300-372.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SP, Leiper FC, Woods A, Carling D, Carlson M. Activation of yeast Snf1 and mammalian AMP-activated protein kinase by upstream kinases. P Natl Acad Sci USA. 2003;100:8839–8843. doi: 10.1073/pnas.1533136100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung GC, Brown CR, Wolfe AB, Liu JJ, Chiang HL. Degradation of the gluconeogenic enzymes fructose-1,6-bisphosphatase and malate dehydrogenase is mediated by distinct proteolytic pathways and signaling events. J Biol Chem. 2004;279:49138–49150. doi: 10.1074/jbc.M404544200. [DOI] [PubMed] [Google Scholar]
- Hynes MJ, Szewczyk E, Murray SL, Suzuki Y, Davis MA, Lewis HMS. Transcriptional control of gluconeogenesis in Aspergillus nidulans. Genetics. 2007;176:139–150. doi: 10.1534/genetics.107.070904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. P Natl Acad Sci USA. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang R, Carlson M. Glucose regulates protein interactions within the yeast Snf1 protein kinase complex. Gene Dev. 1996;10:3105–3115. doi: 10.1101/gad.10.24.3105. [DOI] [PubMed] [Google Scholar]
- Kacherovsky N, Tachibana C, Amos E, Fox D, III, Young ET. Promoter binding by the Adr1 transcriptional activator may be regulated by phosphorylation in the DNA-binding region. PLoS One. 2008;3:e3213. doi: 10.1371/journal.pone.0003213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpichev IV, Durand-Heredia JM, Luo Y, Small GM. Binding characteristics and regulatory mechanisms of the transcription factors controlling oleate-responsive genes in Saccharomyces cerevisiae. J Biol Chem. 2008;283:10264–10275. doi: 10.1074/jbc.M708215200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MD, Hong SP, Carlson M. Role of Tos3, a Snf1 protein kinase kinase, during growth of Saccharomyces cerevisiae on nonfermentable carbon sources. Eukaryot Cell. 2005;4:861–866. doi: 10.1128/EC.4.5.861-866.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larochelle M, Drouin S, Robert F, Turcotte B. Oxidative stress-activated zinc cluster protein Stb5 has dual activator/ repressor functions required for pentose phosphate pathway regulation and NADPH production. Mol Cell Biol. 2006;26:6690–6701. doi: 10.1128/MCB.02450-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leech A, Nath N, McCartney RR, Schmidt MC. Isolation of mutations in the catalytic domain of the Snf1 kinase that render its activity independent of the Snf4 subunit. Eukaryot Cell. 2003;2:265–273. doi: 10.1128/EC.2.2.265-273.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesage P, Yang X, Carlson M. Yeast SNF1 protein kinase interacts with SIP4, a C6 zinc cluster transcriptional activator: a new role for SNF1 in the glucose response. Mol Cell Biol. 1996;16:1921–1928. doi: 10.1128/mcb.16.5.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YY, Lu JY, Zhang J, et al. Protein acetylation microarray reveals that NuA4 controls key metabolic target regulating gluconeogenesis. Cell. 2009;136:1073–1084. doi: 10.1016/j.cell.2009.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodi T, Ferrero I. Isolation of the DLD gene of Saccharomyces cerevisiae encoding the mitochondrial enzyme D-lactate ferricytochrome c oxidoreductase. Mol Gen Genet. 1999;238:315–324. doi: 10.1007/BF00291989. [DOI] [PubMed] [Google Scholar]
- Lohr D, Venkov P, Zlatanova J. Transcriptional regulation in the yeast GAL gene family – a complex genetic network. FASEB J. 1995;9:777–787. doi: 10.1096/fasebj.9.9.7601342. [DOI] [PubMed] [Google Scholar]
- Lombardo A, Cereghino GP, Scheffler IE. Control of mRNA turnover as a mechanism of glucose repression in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:2941–2948. doi: 10.1128/mcb.12.7.2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacPherson S, Larochelle M, Turcotte B. A fungal family of transcriptional regulators: the zinc cluster proteins. Microbiol Mol Biol R. 2006;70:583–604. doi: 10.1128/MMBR.00015-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamnun YM, Pandjaitan R, Mahe Y, Delahodde A, Kuchler K. The yeast zinc finger regulators Pdr1p and Pdr3p control pleiotropic drug resistance (PDR) as homo- and heterodimers in vivo. Mol Microbiol. 2002;46:1429–1440. doi: 10.1046/j.1365-2958.2002.03262.x. [DOI] [PubMed] [Google Scholar]
- Marmorstein R, Harrison SC. Crystal structure of a PPR1–DNA complex: DNA recognition by proteins containing a Zn2Cys6 binuclear cluster. Gene Dev. 1994;8:2504–2512. doi: 10.1101/gad.8.20.2504. [DOI] [PubMed] [Google Scholar]
- Marmorstein R, Carey M, Ptashne M, Harrison SC. DNA recognition by GAL4: structure of a protein–DNA complex. Nature. 1992;356:408–414. doi: 10.1038/356408a0. [DOI] [PubMed] [Google Scholar]
- Mercado JJ, Smith R, Sagliocco FA, Brown AJP, Gancedo JM. The levels of yeast gluconeogenic mRNAs respond to environmental factors. Eur J Biochem. 1994;224:473–481. doi: 10.1111/j.1432-1033.1994.00473.x. [DOI] [PubMed] [Google Scholar]
- Needleman R. Control of maltase synthesis in yeast. Mol Microbiol. 1991;5:2079–2084. doi: 10.1111/j.1365-2958.1991.tb02136.x. [DOI] [PubMed] [Google Scholar]
- Noda T, Hoffschulte H, Holzer H. Characterization of fructose 1,6-bisphosphatase from bakers’ yeast. J Biol Chem. 1984;259:7191–7197. [PubMed] [Google Scholar]
- Papamichos-Chronakis M, Gligoris T, Tzamarias D. The Snf1 kinase controls glucose repression in yeast by modulating interactions between the Mig1 repressor and the Cyc8-Tupl co-repressor. EMBO Rep. 2004;5:368–372. doi: 10.1038/sj.embor.7400120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlik P, Simon M, Schuster T, Ruis H. The glycerol kinase (GUT1) Gene of Saccharomyces cerevisiae – cloning and characterization. Curr Genet. 1993;24:21–25. doi: 10.1007/BF00324660. [DOI] [PubMed] [Google Scholar]
- Phelps C, Gburcik V, Suslova E, Dudek P, Forafonov F, Bot N, MacLean M, Fagan RJ, Picard D. Fungi and animals may share a common ancestor to nuclear receptors. P Natl Acad Sci USA. 2006;103:7077–7081. doi: 10.1073/pnas.0510080103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahner A, Scholer A, Martens E, Gollwitzer B, Schuller HJ. Dual influence of the yeast Cat1p (Snf1p) protein kinase on carbon source-dependent transcriptional activation of gluconeogenic genes by the regulatory gene CAT8. Nucleic Acids Res. 1996;24:2331–2337. doi: 10.1093/nar/24.12.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahner A, Hiesinger M, Schuller HJ. Deregulation of gluconeogenic structural genes by variants of the transcriptional activator Cat8p of the yeast Saccharomyces cerevisiae. Mol Microbiol. 1999;34:146–156. doi: 10.1046/j.1365-2958.1999.01588.x. [DOI] [PubMed] [Google Scholar]
- Randez-Gil F, Bojunga N, Proft M, Entian KD. Glucose derepression of gluconeogenic enzymes in Saccharomyces cerevisiae correlates with phosphorylation of the gene activator Cat8p. Mol Cell Biol. 1997;17:2502–2510. doi: 10.1128/mcb.17.5.2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regelmann J, Schule T, Josupeit FS, Horak J, Rose M, Entian KD, Thumm M, Wolf DH. Catabolite degradation of fructose-1,6-bisphosphatase in the yeast Saccharomyces cerevisiae: a genome-wide screen identifies eight novel GID genes and indicates the existence of two degradation pathways. Mol Biol Cell. 2003;14:1652–1663. doi: 10.1091/mbc.E02-08-0456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts GG, Hudson AP. Transcriptome profiling of Saccharomyces cerevisiae during a transition from fermentative to glycerol-based respiratory growth reveals extensive metabolic and structural remodeling. Mol Genet Genomics. 2006;276:170–186. doi: 10.1007/s00438-006-0133-9. [DOI] [PubMed] [Google Scholar]
- Roth S, Kumme J, Schuller HJ. Transcriptional activators Cat8 and Sip4 discriminate between sequence variants of the carbon source-responsive promoter element in the yeast Saccharomyces cerevisiae. Curr Genet. 2004;45:121–128. doi: 10.1007/s00294-003-0476-2. [DOI] [PubMed] [Google Scholar]
- Rottensteiner H, Kal AJ, Filipits M, Binder M, Hamilton B, Tabak HF, Ruis H. Pip2p: a transcriptional regulator of peroxisome proliferation in the yeast Saccharomyces cerevisiae. EMBO J. 1996;15:2924–2934. [PMC free article] [PubMed] [Google Scholar]
- Rottensteiner H, Kal AJ, Hamilton B, Ruis H, Tabak HF. A heterodimer of the Zn2Cys6 transcription factors Pip2p and Oaf1p controls induction of genes encoding peroxisomal proteins in Saccharomyces cerevisiae. Eur J Biochem. 1997;247:776–783. doi: 10.1111/j.1432-1033.1997.00776.x. [DOI] [PubMed] [Google Scholar]
- Santt O, Pfirrmann T, Braun B, Juretschke J, Kimmig P, Scheel H, Hofmann K, Thumm M, Wolf DH. The yeast GID complex, a novel ubiquitin ligase (E3) involved in the regulation of carbohydrate metabolism. Mol Biol Cell. 2008;19:3323–3333. doi: 10.1091/mbc.E08-03-0328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz P. Snf1 protein kinase: a key player in the response to cellular stress in yeast. Biochem Soc T. 2003;31:178–181. doi: 10.1042/bst0310178. [DOI] [PubMed] [Google Scholar]
- Sanz P. Yeast as a model system to study glucose-mediated signalling and response. Front Biosci. 2007;12:2358–2371. doi: 10.2741/2238. [DOI] [PubMed] [Google Scholar]
- Sanz P, Alms GR, Haystead TAJ, Carlson M. Regulatory interactions between the Reg1-Glc7 protein phosphatase and the Snf1 protein kinase. Mol Cell Biol. 2000;20:1321–1328. doi: 10.1128/mcb.20.4.1321-1328.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöler A, Schüller HJ. A carbon source-responsive promoter element necessary for activation of the isocitrate lyase gene ICL1 is common to genes of the gluconeogenic pathway in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:3613–3622. doi: 10.1128/mcb.14.6.3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüller HJ. Transcriptional control of nonfermentative metabolism in the yeast Saccharomyces cerevisiae. Curr Genet. 2003;43:139–160. doi: 10.1007/s00294-003-0381-8. [DOI] [PubMed] [Google Scholar]
- Simon M, Adam G, Rapatz W, Spevak W, Ruis H. The Saccharomyces cerevisiae ADR1 gene is a positive regulator of transcription of genes encoding peroxisomal proteins. Mol Cell Biol. 1991;11:699–704. doi: 10.1128/mcb.11.2.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith FC, Davies SP, Wilson WA, Carling D, Hardie DG. The SNF1 kinase complex from Saccharomyces cerevisiae phosphorylates the transcriptional repressor protein Mig1p in vitro at four sites within or near regulatory domain 1. FEBS Lett. 1999;453:219–223. doi: 10.1016/s0014-5793(99)00725-5. [DOI] [PubMed] [Google Scholar]
- Smith JJ, Ramsey SA, Marelli M, Marzolf B, Hwang D, Saleem RA, Rachubinski RA, Aitchison JD. Transcriptional responses to fatty acid are coordinated by combinatorial control. Mol Syst Biol. 2007;3:115. doi: 10.1038/msb4100157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soontorngun N, Larochelle M, Drouin S, Robert F, Turcotte B. Regulation of gluconeogenesis in Saccharomyces cerevisiae is mediated by activator and repressor functions of Rds2. Mol Cell Biol. 2007;27:7895–7905. doi: 10.1128/MCB.01055-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz LM, Scharfe C, Deutschbauer AM, Mokranjac D, Herman ZS, Jones T, Chu AM, Giaever G, Prokisch H, Oefner PJ, Davis RW. Systematic screen for human disease genes in yeast. Nat Genet. 2002;31:400–404. doi: 10.1038/ng929. [DOI] [PubMed] [Google Scholar]
- Sutherland CM, Hawley SA, McCartney RR, Leech A, Stark MJR, Schmidt MC, Hardie DG. Elm1p is one of three upstream kinases for the Saccharomyces cerevisiae SNF1 complex. Curr Biol. 2003;13:1299–1305. doi: 10.1016/s0960-9822(03)00459-7. [DOI] [PubMed] [Google Scholar]
- Tachibana C, Yoo JY, Tagne JB, Kacherovsky N, Lee TI, Young ET. Combined global localization analysis and transcriptome data identify genes that are directly coregulated by Adr1 and Cat8. Mol Cell Biol. 2005;25:2138–2146. doi: 10.1128/MCB.25.6.2138-2146.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana C, Biddick R, Law GL, Young ET. A poised initiation complex is activated by SNF1. J Biol Chem. 2007;282:37308–37315. doi: 10.1074/jbc.M707363200. [DOI] [PubMed] [Google Scholar]
- Thakur JK, Arthanari H, Yang F, Chau KH, Wagner G, Naar AM. Mediator subunit Gal11p/MED15 is required for fatty acid-dependent gene activation by yeast transcription factor Oaf1p. J Biol Chem. 2009;284:4422–4428. doi: 10.1074/jbc.M808263200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thukral SK, Morrison ML, Young ET. Alanine scanning site-directed mutagenesis of the zinc fingers of transcription factor ADR1: residues that contact DNA and that transactivate. P Natl Acad Sci USA. 1991;88:9188–9192. doi: 10.1073/pnas.88.20.9188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd RB, Andrianopoulos A. Evolution of a fungal regulatory gene family: the Zn(II)2Cys6 binuclear cluster DNA binding motif. Fungal Genet Biol. 1997;21:388–405. doi: 10.1006/fgbi.1997.0993. [DOI] [PubMed] [Google Scholar]
- Traven A, Jelicic B, Sopta M. Yeast Gal4: a transcriptional paradigm revisited. EMBO Rep. 2006;7:496–499. doi: 10.1038/sj.embor.7400679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trzcinska-Danielewicz J, Ishikawa T, Micialkiewicz A, Fronk J. Yeast transcription factor Oaf1 forms homodimer and induces some oleate-responsive genes in absence of Pip2. Biochem Bioph Res Co. 2008;374:763–766. doi: 10.1016/j.bbrc.2008.07.105. [DOI] [PubMed] [Google Scholar]
- Vallee BL, Coleman JE, Auld DS. Zinc fingers, zinc clusters, and zinc twists in DNA-binding protein domains. P Natl Acad Sci USA. 1991;88:999–1003. doi: 10.1073/pnas.88.3.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bakel H, van Werven FJ, Radonjic M, Brok MO, van Leenen D, Holstege FCP, Marc Timmers HT. Improved genome-wide localization by ChIP-chip using double-round T7 RNA polymerase-based amplification. Nucleic Acids Res. 2008;36:e21. doi: 10.1093/nar/gkm1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent O, Carlson M. Sip4, a Snf1 kinase-dependent transcriptional activator, binds to the carbon source-responsive element of gluconeogenic genes. EMBO J. 1998;17:7002–7008. doi: 10.1093/emboj/17.23.7002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XL, Jiang R, Carlson M. A family of proteins containing a conserved domain that mediates interaction with the yeast Snf1 protein kinase complex. EMBO J. 1994;13:5878–5886. doi: 10.1002/j.1460-2075.1994.tb06933.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young ET, Kacherovsky N, Van Riper K. Snf1 protein kinase regulates Adr1 binding to chromatin but not transcription activation. J Biol Chem. 2002;277:38095–38103. doi: 10.1074/jbc.M206158200. [DOI] [PubMed] [Google Scholar]
- Young ET, Dombek KM, Tachibana C, Ideker T. Multiple pathways are co-regulated by the protein kinase Snf1 and the transcription factors Adr1 and Cat8. J Biol Chem. 2003;278:26146–26158. doi: 10.1074/jbc.M301981200. [DOI] [PubMed] [Google Scholar]
- Young ET, Tachibana C, Chang HWE, Dombek KM, Arms EM, Biddick R. Artificial recruitment of mediator by the DNA-binding domain of Adr1 overcomes glucose repression of ADH2 expression. Mol Cell Biol. 2008;28:2509–2516. doi: 10.1128/MCB.00658-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaman S, Lippman SI, Zhao X, Broach JR. How Saccharomyces responds to nutrients. Annu Rev Genet. 2008;42:27–81. doi: 10.1146/annurev.genet.41.110306.130206. [DOI] [PubMed] [Google Scholar]