Abstract
The discovery of C9orf72 mutations as the most common genetic cause of amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) has awakened a surge of interest in deciphering how mutations in this mysterious gene cause disease and what can be done to stop it. C9orf72 harbors a hexanucleotide repeat, GGGGCC, in a non-coding region of the gene and a massive expansion of this repeat causes ALS, FTD, or both (FTD/ALS). Many questions lie ahead. What does this gene normally do? What is the consequence of an enormous GGGGCC repeat expansion on that gene’s function? Could that hexanucleotide repeat expansion have additional pathological actions unrelated to C9orf72 function? There has been tremendous progress on all fronts in the quest to define how C9orf72 mutations cause disease. Many new experimental models have been constructed and unleashed in powerful genetic screens. Studies in mouse and human patient samples, including iPS-derived neurons, have provided unprecedented insights into pathogenic mechanisms. Three major hypotheses have emerged and are still being hotly debated in the field. These include 1) loss of function owing to decrease in the abundance of C9orf72 protein and its ability to carryout its still unknown cellular role; 2) RNA toxicity from bidirectionally transcribed sense (GGGGCC) and antisense (GGCCCC) transcripts that accumulate in RNA foci and might sequester critical RNA-binding proteins; 3) proteotoxicity from dipeptide repeat proteins produced by an unconventional form of translation from the expanded nucleotide repeats. Here we review the evidence in favor and against each of these three hypotheses. We also suggest additional experiments and considerations that we propose will help clarify which mechanism(s) are most important for driving disease and therefore most critical for considering during the development of therapeutic interventions.
Keywords: ALS, FTD, C9orf72, RNA, dipeptide repeat protein
Introduction
The recent discovery of a mutation in the C9orf72 gene as the most common genetic cause of FTD and ALS (c9FTD/ALS) has opened up many new and exciting areas of investigation in the quest to understand neurodegenerative disease mechanisms and to develop effective disease-modifying strategies. The C9orf72 gene contains a polymorphic hexanucleotide repeat, GGGGCC, located in an intron. The repeat tract length in unaffected individuals, although variable, is typically between five and ten repeats and almost always fewer than 23 repeats (DeJesus-Hernandez et al., 2011). In c9FTD/ALS cases, the hexanucleotide repeat tract is expanded to hundreds or even thousands of repeats (DeJesus-Hernandez et al., 2011; Renton et al., 2011). This mutation can now explain ~40% of familial ALS and ~5–10% of sporadic cases (Renton et al., 2014). Hence, the major contribution of this mutation to sporadic and inherited ALS and FTD has initiated intense interest in defining the mechanism by which C9orf72 GGGGCC repeat expansions cause neurodegeneration (Ling et al., 2013).
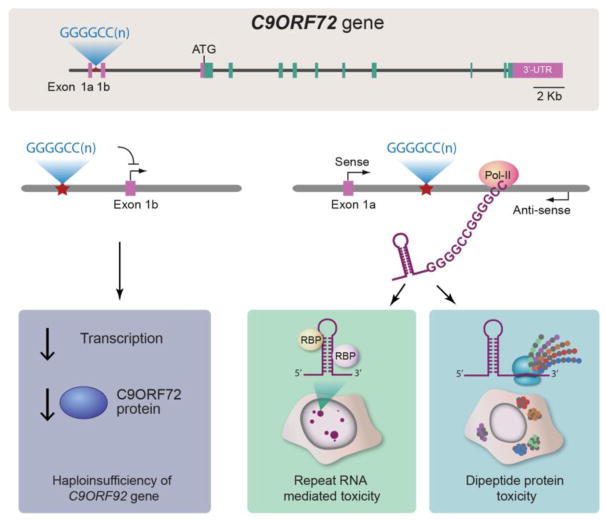
There are currently three major hypotheses to explain how such repeat expansions could be pathogenic (Figure 1). First, the presence of an enormous GGGGCC repeat expansion could cause a downregulation in C9orf72 gene expression, leading to a loss of C9orf72’s still undefined normal cellular function (Figure 1A). Indeed, there is evidence that the presence of this repeat expansion leads to a decrease in C9orf72 expression (DeJesus-Hernandez et al., 2011). Second, an RNA-mediated toxicity mechanism could contribute to disease (Figure 1B). Cells harboring C9orf72 repeat expansions, including patient brain and spinal cord neurons, contain prominent nuclear foci of GGGGCC RNA (DeJesus-Hernandez et al., 2011; Renton et al., 2011) as well as the antisense GGCCCC RNA (Gendron et al., 2013), which could cause the sequestration of essential RNA-binding proteins, including splicing factors, leading to defects in pre-mRNA splicing (Gendron et al., 2014). Third, it has emerged that sense and antisense repeat RNAs are substrates for an unconventional form of translation to generate a series of dipeptide repeat proteins, which accumulate in the brain and spinal cord of C9orf72 mutation carriers and may themselves be what is driving neurodegeneration (Figure 1C).
Figure 1. C9orf72 mutations: three proposed pathomechanisms.
A) The C9orf72 gene harbors a polymorphic hexanucleotide (GGGGCC) repeat in a non-coding region of the gene. Large expansions of this nucleotide repeat cause c9FTD/ALS. There are currently three major hypotheses to explain how such repeat expansions could be pathogenic. B) The large GGGGCC repeat expansion could cause a downregulation in C9orf72 gene expression by interfering with transcription, leading to a decrease in C9orf72 protein and a loss of C9orf72’s function. C) RNA transcripts harboring C9orf72 repeat expansions are produced by both sense and antisense transcription, resulting in the accumulation of nuclear or cytoplasmic foci of GGGGCC RNA as well as the antisense GGCCCC RNA, which could cause the sequestration of essential RNA-binding proteins (RBP), including splicing factors, leading to defects in pre-mRNA splicing by an RNA toxicity mechanism. D) Sense and antisense repeat RNAs are substrates for an unconventional form of translation to generate a series of dipeptide repeat proteins, which accumulate in the brain and spinal cord of C9orf72 mutation carriers and may cause disease by dipeptide repeat protein toxicity mechanism. Figure adapted from (Ling et al., 2013).
These three proposed mechanisms are not completely mutually exclusive, but defining the major disease mechanism will be critical for the development of effective therapeutic interventions. For example, antisense oligonucleotide approaches to target the repeat expansion (i.e., targeting mechanisms 2 and 3 above) are being pursued and anticipated to enter clinical trials in humans in the coming years. However, the success of these trials depends on there not being a major requirement for C9orf72’s normal function because antisense approaches will lower levels of C9orf72 expression (unless they can be engineered to specifically target the mutant allele (or specific RNA isoforms). Even if selectivity can be achieved, could haploinsufficiency contribute to disease? Several recent studies have provided evidence either in support or in opposition to each of the three proposed mechanisms. Here, we provide a review of the evidence in favor and against each of the three hypotheses and we propose additional experiments and analyses to further test each hypothesis and help clarify the role of C9orf72 mutations in ALS and FTD pathogenesis.
Mechanism 1: Loss of function
Genetic discoveries can provide insight into molecular and cellular pathways that open up new areas for mechanistic studies. For example, the identification of mutations in the RNA-binding proteins TDP-43 and FUS/TLS immediately focused attention on RNA metabolism as an important disease mechanism in ALS (Lagier-Tourenne and Cleveland, 2009) and spurred research into RNA processing alterations in ALS, ways to mitigate it, and potential roles for additional RNA-binding proteins. Likewise, mutations in VCP, UBQLN2, SQSTM1, and OPTN quickly focused attention on cellular protein quality control pathways. But C9orf72 mutations were puzzling because they were located in a non-coding region of an uncharacterized human gene (literally, the seventy-second open reading frame on chromosome 9). Nevertheless, efforts were initiated to characterize the function of C9orf72 and to test the hypothesis that loss of this function contributes to disease.
Evidence for C9orf72 loss of function
The C9orf72 gene is transcribed as three distinct transcript variants. In variants 1 and 3 the expanded GGGGCC repeat is located in an intron between two alternatively spliced exons, whereas in variant 2 the repeat is located in the promoter region. Initial reports of the C9orf72 hexanucleotide repeat expansion as a cause of FTD/ALS included evidence of decreased C9orf72 variant 2 transcript levels in cells from mutation carriers (DeJesus-Hernandez et al., 2011; Gijselinck et al., 2012). This decrease in C9orf72 expression could cause disease by haploinsufficiency, if expression of the wild type allele is not sufficient to produce enough functional C9orf72 protein. Other reports have suggested that the expanded GGGGCC repeats might also interfere with the transcription or splicing of the other variants (Mori et al., 2013a); Sareen, 2013; Haeusler, 2014}. Further studies have demonstrated decreased expression levels of C9orf72 in iPS neurons and brain from c9FTD/ALS patients (Almeida et al., 2013; Belzil et al., 2013; Donnelly et al., 2013; Tran et al., 2015; Waite et al., 2014), whereas others have argued that C9orf72 levels are not significantly lowered and in fact the mutant allele is preferentially upregulated or stabilized (Mori et al., 2013a; Sareen et al., 2013).
How could the massive hexanucleotide expansion affect C9orf72 expression levels? One hypothesis is that the G-quadruplex and R-loop structures that the repeat can form (Fratta et al., 2012; Reddy et al., 2013) could lead to abortive transcription of C9orf72 (Haeusler et al., 2014). Alternatively, the GGGGCC repeat could lead to hypermethylation of the C9orf72 locus. Methylation of cytosine (C) in CpG islands is a mechanism to silence gene expression and other nucleotide repeat diseases, such as Friedrich ataxia and fragile X mental retardation syndrome, are associated with repeat-dependent hypermethylation and silencing of gene expression (He and Todd, 2011). The large increase in CpG dinucleotides, by virtue of the GGGGCC expansion, could provide many more CpG islands as substrates for hypermethylation. Indeed, using bisulfite sequencing, a method to directly detect CpG methylation, the C9orf72 locus was hypermethylated in some C9orf72 mutation carriers (Xi et al., 2013). In addition to CpG methylation, histone methylation of lysine residues is another epigenetic modification that can alter gene expression. Trimethylation of histones H3 and H4 at the C9orf72 locus was detected in the blood of C9orf72 mutation carriers (Belzil et al., 2013), providing another mechanism to explain how this mutation could lead to decreases in C9orf72 expression levels. Intuitively, it would seem that hypermethylation of C9orf72 would be deleterious, since it would lead to a decrease in expression of C9orf72 and indeed one report provides evidence that hypermethylation levels correlate with shorter disease duration (Xi et al., 2013). However, other studies have provided conflicting evidence and suggest that hypermethylation of the mutant C9orf72 allele might actually be protective (Liu et al., 2014; McMillan et al., 2015; Russ et al., 2015).
The above results show that expression levels of C9orf72 are reduced in a mutant dependent manner but they do not address the physiological consequences of lowering C9orf72. Initial studies of C9orf72 loss of function in vivo have been performed in model organisms. A null mutation in the C. elegans C9orf72 orthologue resulted in motor neuron degeneration and age-dependent deficits in motility. These mutants were also hypersensitive to environmental stress induced neurodegeneration (Therrien et al., 2013).
Studies of C9orf72 function have also been performed in vertebrates. There is a single C9orf72 orthologue, zC9orf72, present in zebrafish, which is 76% identical to the human protein (Ciura et al., 2013). To lower levels of zC9orf72, zebrafish embryos were injected with three different morpholino antisense oligonucleotides designed to block either the translation or splicing of zC9orf72. These oligonucleotides are like nucleic acids but with important chemical modifications, which increase their stability and make them resistant to cellular nucleases and allow them to evade the innate immune system. As negative controls, embryos were injected with morpholino oligonucleotides designed against zC9orf72 but which harbored five nucleotide mismatches to block effective binding to the zC9orf72 mRNA. The oligonucleotides targeting zC9orf72 resulted in shortened motor axons and defects in axonal arborization in developing larvae. In addition to the axonal phenotypes, targeting zC9orf72 levels elicited motor deficits (reduction in both spontaneous swimming and escape swimming in response to a light touch). These phenotypes could be rescued by co-injecting mRNA encoding human C9orf72. Together, these results provided the first in vivo evidence that loss of C9orf72 function could impair motor neuron function. If these results are validated and extended, the zebrafish model could be a powerful platform for drug screening and to identify genetic modifiers. Importantly, phenotypes obtained using morpholino oligonucleotides in zebrafish should be interpreted with caution, since off-target effects, developmental delays, and other non-specific effects could confound results (Gerety and Wilkinson, 2011). Genome editing using CRISPR/Cas9 works robustly in animal models, including zebrafish, and can be used to engineer stable loss of function mutations in the zC9orf72 gene (Hruscha et al., 2013). On the other hand, phenotypic differences between genetic mutations and gene knockdowns have been observed in zebrafish (Rossi et al., 2015). Thus, a combination of both approaches, together with the appropriate positive and negative controls, will be important in assessing the requirement for C9orf72 function in zebrafish.
Evidence against C9orf72 loss of function
In contrast to the results in C. elegans and zebrafish, studies in mouse have so far not supported a role for C9orf72 loss of function as a cause of FTD/ALS. Administering antisense oligonucleotides (ASOs) targeting mouse C9orf72 by stereotactic intracerebroventricular (ICV) injection reduced C9orf72 mRNA levels to 30–40% of control levels in the spinal cord and brain (Lagier-Tourenne et al., 2013). This effect seemed long-lived and C9orf72 levels remained lowered even several months after the initial ASO injection. C9orf72 depletion in these mice was well tolerated and did not result in any behavioral or motor impairments. Cytoplasmic aggregation of ubiquitinated TDP-43 is the hallmark pathological feature of FTD and ALS, including c9FTD/ALS. TDP-43 remained nuclear in brain and spinal cord sections and ubiquitinated aggregates were not detected in mice with C9orf72 depletion (Lagier-Tourenne et al., 2013). Thus, reducing C9orf72 levels by over 50% in the nervous system for several months does not result in neuropathological or behavioral phenotypes.
Another way to lower levels of C9orf72 in mouse is by gene knockout. A conditional allele of C9orf72 was generated using the Cre/loxP system. These mice were crossed to Nestin-Cre mice, which express Cre recombinase in neurons and glia starting at E10.5 and continuing into adulthood (Tronche et al., 1999). Cre-mediated inactivation of C9orf72 in neurons and glia did not cause deficits in motor neuron numbers or in motor function, including motor performance and grip strength (Koppers et al., 2015). Hallmark ALS pathologies, including ubiquitinated TDP-43 aggregates and gliosis were not detected either. There was no effect on survival even after 24 months. Thus, in two different mouse models, loss of C9orf72 function is not sufficient to cause neurodegeneration and FTD/ALS-related phenotypes.
Two studies of human c9FTD/ALS have provided evidence arguing against a loss of function disease mechanism. First, if hexanucleotide repeat expansion mutations in C9orf72 cause FTD/ALS by a loss of function mechanism then other ways to disable C9orf72 function could also be a cause of disease. However, an analysis of the C9orf72 gene in several hundred ALS patients did not identify deleterious mutations in the coding region of C9orf72 (including nonsense and frameshift mutations) (Harms et al., 2013). Second, since heterozygous C9orf72 mutation is sufficient to cause FTD/ALS, homozygous mutations might be predicted to cause a more severe form of the disease or even a different clinical presentation. However, an analysis of a patient homozygous for the C9orf72 hexanucleotide repeat expansion revealed severe clinical and pathological features that were in the normal disease spectrum seen in heterozygous patients (Fratta et al., 2013). These two studies, while certainly not definitive, are not consistent with a loss-of-function mechanism.
Finally, studies in patient cells have provided somewhat of a formal test for the loss-of-function vs. gain-of-function hypotheses. Several studies have used RNA profiling to characterize gene expression changes associated with C9orf72 mutations. These studies have included fibroblasts (Lagier-Tourenne et al., 2013), iPS-derived neurons (Donnelly et al., 2013), iPS-derived motor neurons (Sareen et al., 2013), and human brain (Prudencio et al., 2015). Each study uncovered a mutant-specific RNA signature (i.e., present in C9orf72 mutant carriers but not healthy controls), albeit different from one cell type to the next. If these alterations in gene expression were caused by a loss of C9orf72 function, then lowering levels of C9orf72 (e.g., by targeting expression with ASOs) would be predicted to either worsen or have no effect on the RNA signature. However, the studies in iPS neurons and the iPS-motor neurons revealed that targeting C9orf72 with ASOs actually improved the signature rather than worsening it (Donnelly et al., 2013; Sareen et al., 2013). Further, lowering C9orf72 in control cells did not recapitulate the RNA signature (Lagier-Tourenne et al.; Sareen et al.). These results are not consistent with C9orf72 mutations causing a loss of function.
Experiments to further test C9orf72 loss of function
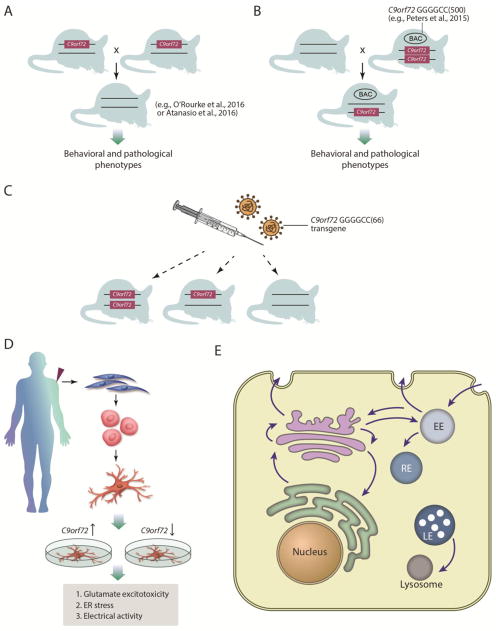
Lowering levels of C9orf72 in C. elegans and zebrafish appears deleterious, whereas conditional inactivation of the gene specifically in motor neurons and glia in mouse does not affect motor neuron function or survival. Several additional studies will be useful to help resolve these discrepancies (Figure 2). The nestin-Cre deletion of murine C9orf72 may not have removed enough of the gene or in all of the right cells and tissues. Indeed, both human C9orf72 and the mouse homolog (3110043O21Rik) are expressed most highly in microglia and macrophages in the brain (Zhang et al., 2014a; Zhang et al., 2016a), thus it will be important to consider potential non-cell-autonomous mechanisms of neurodegeneration (e.g., by using additional Cre driver lines to delete C9orf72 from microglia).
Figure 2. Additional experiments to test C9orf72 loss of function.
A) Mice have been generated in which the β–galactosidase gene replaces exons 2–6 of one of the C9orf72 alleles (Suzuki et al., 2013). These mice could be intercrossed to generate homozygous mutant mice (Atanasio et al., 2016; O’Rourke et al., 2016) and, together with their heterozygous littermates, extensively analyzed for any effects on pathological phenotypes, survival and cognitive or motor behavioral impairments. B) Crossing transgenic mice containing a human BAC with a fragment of the C9orf72 locus harboring ~500 GGGGCC repeats (e.g., Peters et al., 2015) to the C9orf72 knockout mice will test if disease is accelerated by reducing wild type C9orf72 function. C) Injecting the C9orf72 transgene (Chew et al., 2015) into the central nervous system of C9orf72 WT, +/−, or −/− animals will test if disease features are accelerated by the reduction of wild type C9orf72. D) iPS derived from c9FTD/ALS patients have been reported to exhibit phenotypic differences from control neurons, including glutamate excitotoxicity, sensitivity to ER stress, and alterations in electrical activity. If these phenotypes are due to loss of C9orf72 function, then increasing C9orf72 levels should mitigate them and lowering C9orf72 levels should worsen them. E) C9orf72 may function as a guanine nucleotide exchange factor (GEF) to regulate Rab GTPase activity. Rabs orchestrate multiple steps of membrane trafficking within cells and it will be important to define which Rab and thus which trafficking step C9orf72 regulates.
A germline knockout of C9orf72 would allow for the analysis of heterozygous and homozygous mutant animals constitutively lacking C9orf72 expression. Mice have been generated in which the β–galactosidase gene replaces exons 2–6 of one of the C9orf72 alleles (Suzuki et al., 2013). Two very recent studies have used gene targeting to generate homozygous mutant mice (Figure 2A) and extensively analyzed for any effects on survival and cognitive or motor behavioral impairments (Atanasio et al., 2016; O’Rourke et al., 2016). These mice did not develop motor neuron disease but instead developed splenomegaly and several other peripheral pathologies, including marked expansion of myeloid cells and deficits in immune responses and microglial function (Atanasio et al., 2016; O’Rourke et al., 2016). The neuroinflammation seen in these mice is reminiscent of that in human patient tissue. Thus, while these data suggest that loss of C9orf72 function per se is unlikely sufficient to cause motor neuron disease, its requirement for proper microglia function could suggest a possible way that its loss could contribute to disease progression, similar to what is seen in mouse models of familial ALS caused by SOD1 mutations (Boillee et al., 2006; Ilieva et al., 2009).
Another formal test of loss- vs. gain-of-function involves the use of C9orf72 knockout mice and some of the recently described viral-mediated and BAC transgenic c9FTD/ALS models (Chew et al., 2015; O’Rourke et al., 2015; Peters et al., 2015). These models employ expression of human C9orf72 transgenes harboring various GGGGCC repeat lengths either via adeno-associated virus mediated somatic transgenesis (Chew et al., 2015) or in transgenic mice generated from a bacterial artificial chromosomes (BAC) that expresses a fragment of human C9orf72 containing an expanded hexanucleotide repeat (O’Rourke et al., 2015; Peters et al., 2015) or the full length C9orf72 gene harboring an expanded repeat (O’Rourke et al., 2015). These mice exhibit various phenotypes and pathological features reminiscent of c9FTD/ALS (Chew et al., 2015; O’Rourke et al., 2015; Peters et al., 2015). Breeding these mice to C9orf72 knockout mice (heterozygous and homozygous) or injecting the C9orf72 transgene into the central nervous system of the knockout animals will test if disease features are or are not accelerated by the reduction of wild type C9orf72 (Figure 2B, C). If lowering levels of C9orf72 has no effect on the phenotypes of human C9orf72 transgenic mice it would argue directly against the hypothesis that reduced C9orf72 function contributes to c9FTD/ALS. Similar approaches have been used to support a gain-of-function toxicity mechanism caused by SOD1 mutations in familial ALS (Bruijn et al., 1998).
The ASO and RNA signature experiments described above, which we used to argue against a loss-of-function mechanism, could be extended one step further. If C9orf72 mutations cause disease by a loss-of-function then increasing levels of C9orf72 would be predicted to reverse this signature. Experiments to upregulate C9orf72 expression levels (e.g., by transfecting expression constructs) could be used to test this hypothesis in cell lines from C9orf72 mutation carriers (Figure 2D). If lowering C9orf72 levels in these cell lines (Donnelly et al., 2013; Sareen et al., 2013) does not make things worse and increasing C9orf72 levels does not make things better, it would argue against a loss-of-function mechanism.
The normal function of C9orf72 still remains poorly understood and experiments to define this function will facilitate the study of how alterations in that function might contribute to disease. C9orf72 protein has homology to the Differentially Expressed in Normal and Neoplasia (DENN) protein family, which function as guanine nucleotide exchange factors (GEFs) to regulate Rab GTPase activity (Levine et al., 2013; Zhang et al., 2012). Rab GTPases act as molecular switches to orchestrate multiple steps of membrane trafficking within cells (Yoshimura et al., 2010). It will be important to define which Rab(s) C9orf72 regulates since this will provide insight into the particular trafficking step and cellular location (e.g., endosomes, lysosomes, Golgi, etc.) where it likely functions (Figure 2E). Assays to measure these trafficking steps in cells from c9FTD/ALS patients will help to test for C9orf72 loss of function effects.
Mechanism 2: RNA toxicity
A second way that the C9orf72 hexanucleotide repeat expansion could cause disease is by a gain of RNA toxicity mechanism. The initial descriptions of the mutation included evidence that RNA foci containing the GGGGCC repeat accumulated in the brain and spinal cord of c9FTD/ALS patients (DeJesus-Hernandez et al., 2011). With analogy to other nucleotide repeat expansion diseases in which repeat-containing RNA foci accumulate, such as in myotonic dystrophy, it was postulated that these GGGGCC RNA foci could act as kind of landing pads for RNA-binding proteins and splicing factors, sequestering them away from their normal function. Adding to the complexity of the proposed RNA toxicity mechanism, it was subsequently discovered that the antisense GGCCCC repeat RNA was also transcribed from the C9orf72 hexanucleotide repeat and that these antisense RNAs accumulated in distinct foci in c9FTD/ALS patients. Thus, a different suite of RNA-binding proteins, which could bind and be sequestered by the antisense foci was now sought. The race was on to identify these RNA-binding proteins and to determine if and how their loss of function contributes to disease.
Evidence for RNA toxicity
The striking appearance of GGGGCC sense and GGCCCC antisense foci in the cells of patients with C9orf72 expansions (DeJesus-Hernandez et al., 2011; Gendron et al., 2013; Lagier-Tourenne et al., 2013; Mori et al., 2013a; Zu et al., 2013) provided an attractive pathogenic mechanism: RNA-binding proteins and splicing factors that recognized GGGGCC and GGCCCC binding sites would be sequestered into these foci, disrupting their normal function. This RNA toxicity mechanism is what underlies myotonic dystrophy type 1 (DM1) and other microsatellite repeat expansion diseases (Echeverria and Cooper, 2012). DM1 is caused by a CTG repeat expansion in the 3′UTR of the DMPK gene (Atanasio et al.; Brook et al., 1992). The transcribed repeat expansion (CUG) accumulates as nuclear RNA foci in DM1 patients (Davis et al., 1997; Taneja et al., 1995) and causes alterations in RNA processing, including alternative splicing (Lin et al., 2006). The RNA-binding protein muscleblind (MBNL) is sequestered in the CUG-repeat containing foci in DM1 models and in DM1 patients (Fardaei et al., 2001; Jiang et al., 2004; Mankodi et al., 2001; Miller et al., 2000). Importantly, upregulation of MBNL was sufficient to rescue phenotypes in a fly DM1 model (de Haro et al., 2006) and Mbnl knockout mice exhibited the same phenotype and RNA processing changes seen in DM1. Taken together, there is compelling evidence that DM1 is caused by an RNA toxicity mechanism, owing to sequestration of the MBNL RNA-binding protein. Indeed, DM1 discoveries have been paradigmatic for how repeat expansion diseases could be caused by RNA toxicity (Echeverria and Cooper, 2012).
When it was discovered that a repeat expansion is the most common cause of ALS and FTD, an RNA-toxicity mechanism was immediately considered and efforts were launched to find the “muscleblind” type of RNA-binding protein that would be sequestered by GGGGCC or GGCCCC repeat foci. Many RNA-binding proteins have been proposed to be sequestered by these repeats but there still remains disagreement about which one, if any, is critical for disease. These include SRSF2, hnRNP H1/F, ALYREF, hnRNPA3, hnRNPA1, hnRNP-H, nucleolin, Pur-α, ASF/SF2, ADARB2, and RanGAP1 (Donnelly et al., 2013; Haeusler et al., 2014; Lee et al., 2013; Mori et al., 2013b; Reddy et al., 2013; Sareen et al., 2013; Xu et al., 2013); Zhang, 2015; Cooper-Knock, 2014}. Future studies along the lines of those described above for DM1, will be needed to determine if loss of function of any of these RNA-binding proteins produces the same molecular alterations caused by C9orf72 mutations and if upregulating their levels reverses these phenotypes.
The RNA toxicity mechanism need not be limited to the nucleus. Indeed, RNA foci have been detected in the cytoplasm of fibroblasts from C9orf72 mutation carriers (Lagier-Tourenne et al., 2013); Sareen, 2013; Donnelly, 2013}. Furthermore, a combination of iPS-derived neurons from patients harboring C9orf72 mutations, studies in primary rodent neurons, and experiments in Drosophila has demonstrated that GGGGCC repeat RNA localizes distally within neurites where it associates with ribonucleoprotein transport granules and interferes with local translation (Schweizer Burguete et al., 2015).
Evidence against RNA toxicity
Because both RNA foci and dipeptide repeat proteins (DPRs) are produced form C9orf72 expansions, it has been difficult to determine the relative contributions of each to pathogenesis. Two recent experiments in model organisms have provided strong evidence against the RNA toxicity mechanism. Several Drosophila models have been generated to study the impact of expression of C9orf72 GGGGCC repeats (Freibaum et al., 2015; Mizielinska et al., 2014; Schweizer Burguete et al., 2015; Tran et al., 2015; Xu et al., 2013; Zhang et al., 2015). Transgenic fly lines can express the expanded repeat in a tissue-specific manner (e.g., just the eye, only in motor neurons, throughout the nervous system, etc.). Expression of GGGGCC repeats in flies produces RNA foci and DPRs (Freibaum et al., 2015; Mizielinska et al., 2014; Tran et al., 2015). The first experiment, by the Isaacs and Partridge groups cleverly used Drosophila to disentangle potential contributions from the C9orf72 repeat RNA and those of the DPRs (Mizielinska et al., 2014). They generated two different fly lines, each containing a long GGGGCC repeat. One of the lines had a pure GGGGCC repeat but for the other one they engineered it to contain regular interruptions with Stop codons to prevent it from being translated. Both pure and interrupted repeats were expressed well and formed RNA foci, however the interrupted one could not be translated to form DPRs whereas the pure one could be a substrate for RAN translation. Expression of pure repeats caused toxicity and early lethality whereas the interrupted ones had no effect (Mizielinska et al., 2014). These experiments provide evidence that the GGGGCC repeats can cause toxicity in vivo through the production of RAN translation products and not from the RNA alone.
A second experiment in Drosophila also argues against an RNA toxicity mechanism and provides important new information (Tran et al., 2015). The authors generated flies with a transgene harboring 160 GGGGCC repeats embedded within an intron. This transgene was expressed, and spliced, and the GGGGCC repeat formed many sense RNA foci in the nucleus. But there was no neurodegeneration, in contrast to flies produced by other labs (e.g., (Mizielinska et al., 2014)). A key difference between the Tran et al. flies and the Mizielinska et al. ones is the presence of the repeat within the intron. The flies in the Mizielinska et al. paper are made to express the GGGGCC expansion in the context of an mRNA with a 3′UTR, which allows it to be efficiently exported to the cytoplasm. This leads to the production of high levels of DPRs and causes neurodegeneration. The flies made by Tran et al. express the repeat from within an intron, have high levels of sense RNA foci in the nucleus, low levels of RAN translation, and no neurodegeneration. This means that accumulation of sense RNA foci in the nucleus is not sufficient to drive neurodegeneration in this fly model. The authors’ C9orf72 intron fly model does not seem to produce antisense RNA foci, which appears to be an important feature of c9FTD/ALS (Cooper-Knock et al., 2015). Before we can conclude that RNA foci in the nucleus do not contribute to neurodegeneration, it will be important in the future to test the effect of a similar level of antisense RNA transcripts in the fly model.
Experiments to further test RNA toxicity
A parsimonious explanation for the findings from the two Drosophila experiments described above is that pathologies seen in the fly C9orf72 models are due in large part (if not mostly) to translation products from the repeat. Whether this is the situation in human cells and mouse remains to be determined. Several attempts have been made to model C9orf72 mutations in mouse. Bacterial artificial chromosomes (BAC) harboring various fragments (Peters et al., 2015) or the full-length (O’Rourke et al., 2015) human C9orf72 locus containing GGGGCC repeat expansions have been generated. All of these mice recapitulate pathological features, especially sense and antisense RNA foci and DPR production (O’Rourke et al., 2015; Peters et al., 2015). However, these mice do not seem to recapitulate the neurodegenerative disease features seen in ALS and FTLD, although future studies to analyze contributions of strain background and other factors are needed.
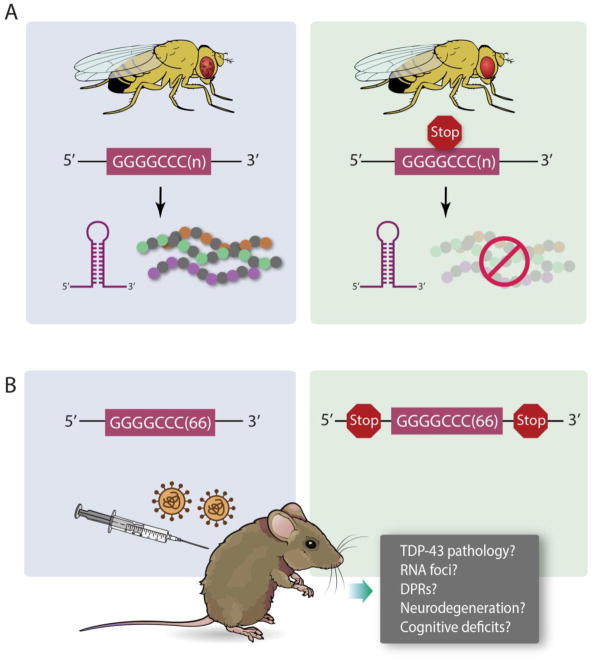
Another approach to model c9FTD/ALS in mouse was attempted by Petrucelli and colleagues (Chew et al., 2015). They used adeno-associated virus (AAV) to deliver a construct containing 66 repeats of GGGGCC (disease-range) or 2 repeats (negative control). They administered these viruses by intracerebroventricular injection into P0 mouse pups and waited for 6 months before performing a battery of behavioral and pathological analyses on these mice. This mouse model recapitulates the cardinal features seen in human disease, including the accumulation of RNA foci transcribed from the sense strand of the GGGGCC repeat, production of RAN translation products (GP, GA, GR) from the sense strand, neuronal loss and astrogliosis, and behavioral and locomotor impairments (Chew et al., 2015). Strikingly, these mice also exhibit robust TDP-43 pathology, a key feature of c9FTD/ALS, not recapitulated in the BAC models (O’Rourke et al., 2015; Peters et al., 2015). Given the ease and reproducibility of this viral vector model, it can now be used, in a way similar to the fly experiments, to test relative roles of RNA and DPRs towards neurodegenerative phenotypes. Constructs could be generated that have Stop codons interrupting the repeats or flanking the repeats, in order to prevent translation but preserve RNA foci formation (Figure 3A). Future iterations of the viral vector approach could also employ cell type specific promoters to express the repeats in specific cell types (e.g., glia vs. neurons).
Figure 3. Additional experiments to test C9orf72 RNA toxicity.
A) Drosophila has been used to disentangle the contributions of C9orf72 RNA toxicity and dipeptide repeat proteins (Mizielinska et al., 2014). Flies expressing a GGGGCC expanded repeat produce RNA foci and dipeptide repeat proteins (DPR), and exhibit neurodegenerative phenotypes (e.g., rough eye). Engineering stop codons into the GGGGCC transgene maintains RNA foci but abolishes DPR production, and mitigates the degenerative phenotypes. B) The new viral vector transgenic mouse model (Chew et al., 2015) could be used in a way similar to the fly experiments, to test relative roles of RNA and DPRs towards neurodegenerative phenotypes. Constructs could be generated that have Stop codons interrupting the repeats or flanking the repeats, in order to prevent translation but preserve RNA foci formation. These mice could be assessed for pathological features (TDP-43, RNA foci, DPRs) as well as neurodegeneration and cognitive deficits.
Mechanism 3: Dipeptide repeat protein toxicity
A third potential mechanism has emerged based on the finding that the bidirectionally transcribed pathogenic repeat expansion can be translated, even in the absence of an ATG start codon and even though it is located in a non-coding region of C9orf72 (Ash et al., 2013; Mori et al., 2013c; Zu et al., 2013). RAN (repeat-associated non-ATG) translation, originally discovered by Ranum and colleagues to occur in spinocerebellar ataxia type 8 (SCA8) and DM1, which are caused by nucleotide repeat expansions (Zu et al., 2011), seems to be generalizable to other nucleotide repeat expansion diseases, including Fragile X tremor ataxia syndrome (Todd et al., 2013), Huntington disease (Banez-Coronel et al., 2015) and now c9FTD/ALS (Ash et al., 2013; Mori et al., 2013c; Zu et al., 2013). This unconventional translation occurs in all reading frames and results in the production of six dipeptide repeat proteins in c9FTD/ALS: glycine-alanine (GA) and glycine-arginine (GR) from sense GGGGCC transcripts, proline-arginine (PR) and proline-alanine (PA) from antisense GGCCCC transcripts, and glycine-proline (GP) from both sense and antisense transcripts. These dipeptide repeat proteins (DPRs) are themselves aggregation-prone and accumulate throughout the central nervous system (Ash et al., 2013; Gendron et al., 2013; Mori et al., 2013a; Mori et al., 2013c; Zu et al., 2013). Are these DPRs benign bystanders or do they contribute to neurodegeneration? And if they are toxic, are certain DPRs more toxic than others? What are the cellular pathways that DPRs affect and how do these impairments contribute to disease?
Evidence for dipeptide repeat protein toxicity
There is evidence that RAN translation products are components of pathology in c9FTD/ALS (Ash et al., 2013; Gendron et al., 2013; Mori et al., 2013a; Mori et al., 2013c; Zu et al., 2013). Moving from pathology to potentially pathogenesis, several groups reported experiments demonstrating that C9orf72 DPRs are toxic and can cause neurodegeneration (Kwon et al., 2014; May et al., 2014; Mizielinska et al., 2014; Wen et al., 2014; Yamakawa et al., 2014; Zhang et al., 2014b; Zu et al., 2013; Yang, 2015). For instance, the Petrucelli group has reported that expression of GA proteins in the absence of RNA foci in primary neurons leads to impaired proteasome activity, induction of endoplasmic reticulum stress, and neurotoxicity in the absence of foci formation (Zhang et al., 2014b). GA-induced neurotoxicity has also been associated with loss of Unc119 function (May et al., 2014). GA has the ability to form toxic amyloids and may even be able to spread from cell to cell in a prion-like manner (Chang et al., 2016). Transgenic mice generated to produce abundant GA pathology exhibit neurodegeneration and behavioral deficits, possibly because of sequestration of HR23 proteins, which are involved in proteasomal degradation (Zhang et al., 2016b). Other experiments have focused attention on the arginine-rich DPRs (GR and PR). The addition of recombinant PR or GR polymers to HeLa cells or human astrocytes caused numerous RNA processing alterations and toxicity (Kwon et al., 2014). The DPRs were able to rapidly enter the nucleus and localize to nucleoli (sites of rRNA processing). Expression of PR repeats within human motor neurons was also toxic (Wen et al., 2014). GR and PR were also toxic in vivo because expressing 50 repeats of GR or PR caused toxicity and early lethality in Drosophila (Mizielinska et al., 2014; Wen et al., 2014). Thus, in model systems and cell culture, DPRs are sufficient to cause toxicity. Whether this is directly related to pathologies seen in human disease is still unresolved.
A series of recent papers has implicated nucleocytoplasmic transport impairments caused by C9orf72 mutations ((Boeynaems et al., 2016; Freibaum et al., 2015; Jovicic et al., 2015; Zhang et al., 2015)) and reviewed in (Fox and Tibbetts, 2015; van Blitterswijk and Rademakers, 2015)). Transport of RNA and protein cargos to and from the nucleus is a highly regulated fundamental cellular process (Burns and Wente, 2012). Defects in nucleocytoplasmic transport could explain how TDP-43 and potentially other RNA-binding proteins might accumulate in the cytoplasm in c9FTD/ALS. Whereas all four groups agree on the cellular defect and remarkably converged on the same pathway using vastly different approaches and models, there is disagreement over the cause of the defect. Zhang et al. say it’s the sense RNA that is toxic, Freibaum et al. say that their phenotypes can be caused by toxic RNAs, DPRs, or some combination of both, and Jovičić et al. and Boeynaems et al. say it’s the DPRs that are causing the defects. The Zhang et al. and Freibaum et al. studies use systems that produce both RNA and DPRs, whereas the experiments by Jovičić et al. and Boeynaems et al. use models, yeast and Drosophila, respectively, which only express DPRs. Given this, the fact that all groups identified the same types of genes involved in nucleocytoplasmic transport as modifiers of C9orf72 phenotypes, suggests that the defects in these models were likely caused by the DPRs. Moreover, the studies by Tran et al., also using Drosophila, suggests that the DPRs are responsible for the neurodegenerative phenotypes and the RNA foci are, if anything, actually protective (Tran et al., 2015).
Evidence against dipeptide repeat protein toxicity
The experimental data in model systems demonstrate that DPRs can be toxic but they do not prove that these are what drive disease in humans. If DPRs are major drivers of neurodegeneration in human c9FTD/ALS then a prediction is that one or more of the DPRs should accumulate at high levels in the most affected regions of the central nervous system. And perhaps the abundance of DPR pathology should correlate with disease severity. However, several studies of postmortem samples from C9orf72 mutation carriers have so far mostly failed to correlate the abundance and localization of DPR pathology with neurodegeneration and clinical phenotypes (Davidson et al., 2014; Mackenzie et al., 2014; Mackenzie et al., 2015; Schludi et al., 2015), although one study did identify a correlation between GP levels and cognitive performance (Gendron et al., 2015). In terms of relative abundance, it seems that GA- and GP-positive inclusions are the most abundant with GR being less abundant, and the PA and PR DPRs produced by RAN translation of the antisense transcript being exceptionally rare (Mackenzie et al., 2015).
Thus, there appears to be a disconnect between the striking toxicities elicited by some of the DPRs in model systems and cell culture and the apparent lack of clinicopathological evidence by analysis of human postmortem samples. One interpretation is that DPRs are not the major pathomechanisms associated with C9orf72 mutations (Mackenzie et al., 2015). But could some of the DPRs that are difficult to detect in postmortem analysis be so highly toxic (e.g., PR) that they do not accumulate to high enough levels before causing neuron death? Likewise, could DPRs exist in multiple conformations or strains, some toxic and others benign, as do other neurodegenerative disease proteins (e.g., tau (Clavaguera et al., 2013), α–syn (Guo et al., 2013), and Aβ (Aguzzi and Gitler, 2013; Lu et al., 2013)) and, if so, are the existing antibodies used to detect DPR pathology only detecting certain conformations but not other potentially more toxic ones?
Experiments to further test dipeptide repeat protein toxicity
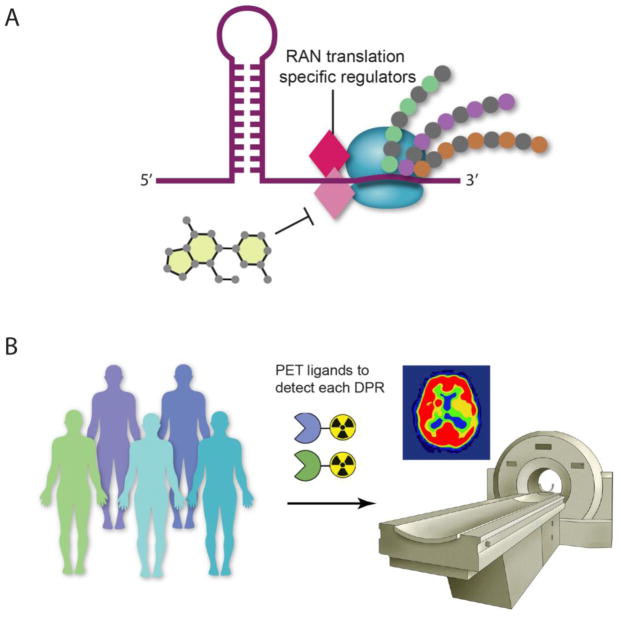
The ultimate test of DPR toxicity as a mechanism in c9FTD/ALS will require a way to specifically block RAN translation (with genetic or chemical approaches), without affecting the sequence or structure of the repeat. This will require a detailed understanding of RAN translation mechanisms. What are the regulators and other machinery that recognize extended repeat sequences and can these be targeted to specifically inhibit RAN translation? The development of specific RAN translation inhibitors will allow all of these hypotheses to be tested in cell lines that express the C9orf72 repeat expansion (sense and antisense) in the context of the endogenous location (Figure 4A).
Figure 4. Additional experiments to test C9orf72 dipeptide repeat protein toxicity.
A) To specifically block RAN translation will require elucidating RAN translation mechanisms and identifying RAN translation-specific regulators. These putative regulators will be new targets for the development of small molecule inhibitors to specifically inhibit RAN translation. B) The development of positron emission tomography (PET) ligands to detect DPR pathology in vivo would allow longitudinal studies of C9orf72 mutation carriers to help resolve the role of DPRs in disease pathogenesis and to eventually be used in clinical trial settings to assess efficacy of candidate therapeutics.
To resolve the apparent disconnect between data from cell culture and model systems with that from histopathological examination of postmortem samples, imaging modalities to detect DPRs in the brain of living C9orf72 mutation carriers would empower such studies. In Alzheimer disease, compounds that preferentially bind amyloid fibrils have been used as positron emission tomography (PET) ligands, enabling in vivo imaging of amyloid pathology (Mitsis et al., 2014). The development of similar molecular beacons to detect DPR pathology in vivo would allow longitudinal studies of C9orf72 mutation carriers to help resolve the role of DPRs in disease pathogenesis and to eventually be used in clinical trial settings to assess efficacy of candidate therapeutics. Meanwhile, powerful and highly sensitive immunoassays are currently being developed to detect DPRs in blood or cerebrospinal fluid (CSF) as a way to measure DPR levels in human patients at early and late stages of disease progression (Su et al., 2014).
Concluding remarks
The discovery five years ago of C9orf72 mutations as the most common cause of ALS and FTD (DeJesus-Hernandez et al., 2011; Renton et al., 2011) has revolutionized the ALS and FTD research field leading to many new and exciting model systems, hypotheses, and even proposed therapeutic strategies. Ultimately, when it comes to therapies, it may actually not be important to distinguish between RNA toxicity and DPRs, since therapies targeting the C9orf72 mutation (e.g., ASOs) will affect both RNA and protein (Donnelly et al., 2013; Lagier-Tourenne et al., 2013; Sareen et al., 2013)and can be designed to not interfere with expression of the wild type allele or to specifically target certain RNA isoforms (Donnelly et al., 2013; Lagier-Tourenne et al., 2013; Sareen et al., 2013). If haploinsufficiency plays an important role in the disease, then such C9orf72 lowering strategies may not be effective. Thus, it is of high priority to fully define the cellular function of C9orf72 and rigorously test the impact of C9orf72 loss of function phenotypes in mouse models and human patient-derived cells.
There are intense discussions about which of the three mechanisms causes disease and, as detailed above, there has been compelling evidence in support of and against each of the proposed mechanisms. It is important to consider that it is possible (perhaps probable) that a combination of multiple mechanisms may actually be what causes disease. For example, perhaps reduced levels or function of C9orf72 could sensitize neurons and increase neuronal vulnerability to other facets of C9orf72 pathology (e.g., RNA foci or DPRs). Looking forward, the field now has a powerful collection of model systems, experimental reagents, and analysis methods in hand to further clarify pathogenic mechanisms and to eventually develop effective therapeutic strategies.
Highlights.
Three C9orf72 disease mechanisms are reviewed
Evidence in favor of each mechanism presented
Evidence against each mechanism presented
Suggestions for experiments to test each mechanism further presented
Acknowledgments
We thank Nicholas Kramer, Ana Jovičić and Leonard Petrucelli for critically reading the manuscript and providing useful suggestions. We thank Lili Guo for expert graphical assistance with figures.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Aaron D. Gitler, Email: agitler@stanford.edu.
Hitomi Tsuiji, Email: hitomitsuiji@phar.nagoya-cu.ac.jp.
References
- Aguzzi A, Gitler AD. A template for new drugs against Alzheimer’s disease. Cell. 2013;154:1182–4. doi: 10.1016/j.cell.2013.08.049. [DOI] [PubMed] [Google Scholar]
- Almeida S, et al. Modeling key pathological features of frontotemporal dementia with C9ORF72 repeat expansion in iPSC-derived human neurons. Acta Neuropathol. 2013;126:385–99. doi: 10.1007/s00401-013-1149-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ash PE, et al. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron. 2013;77:639–46. doi: 10.1016/j.neuron.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atanasio A, et al. C9orf72 ablation causes immune dysregulation characterized by leukocyte expansion, autoantibody production, and glomerulonephropathy in mice. Sci Rep. 2016;6:23204. doi: 10.1038/srep23204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banez-Coronel M, et al. RAN Translation in Huntington Disease. Neuron. 2015;88:667–77. doi: 10.1016/j.neuron.2015.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belzil VV, et al. Reduced C9orf72 gene expression in c9FTD/ALS is caused by histone trimethylation, an epigenetic event detectable in blood. Acta Neuropathol. 2013;126:895–905. doi: 10.1007/s00401-013-1199-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeynaems S, et al. Drosophila screen connects nuclear transport genes to DPR pathology in c9ALS/FTD. Sci Rep. 2016;6:20877. doi: 10.1038/srep20877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boillee S, et al. Onset and progression in inherited ALS determined by motor neurons and microglia. Science. 2006;312:1389–92. doi: 10.1126/science.1123511. [DOI] [PubMed] [Google Scholar]
- Brook JD, et al. Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3′ end of a transcript encoding a protein kinase family member. Cell. 1992;68:799–808. doi: 10.1016/0092-8674(92)90154-5. [DOI] [PubMed] [Google Scholar]
- Bruijn LI, et al. Aggregation and motor neuron toxicity of an ALS-linked SOD1 mutant independent from wild-type SOD1. Science. 1998;281:1851–4. doi: 10.1126/science.281.5384.1851. [DOI] [PubMed] [Google Scholar]
- Burns LT, Wente SR. Trafficking to uncharted territory of the nuclear envelope. Curr Opin Cell Biol. 2012;24:341–9. doi: 10.1016/j.ceb.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YJ, et al. Glycine-Alanine Dipeptide Repeat from C9orf72 Hexanucleotide Expansions Forms Toxic Amyloids Possessing Cell-to-cell Transmission Property. J Biol Chem. 2016 doi: 10.1074/jbc.M115.694273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew J, et al. Neurodegeneration. C9ORF72 repeat expansions in mice cause TDP-43 pathology, neuronal loss, and behavioral deficits. Science. 2015;348:1151–4. doi: 10.1126/science.aaa9344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciura S, et al. Loss of function of C9orf72 causes motor deficits in a zebrafish model of amyotrophic lateral sclerosis. Ann Neurol. 2013;74:180–7. doi: 10.1002/ana.23946. [DOI] [PubMed] [Google Scholar]
- Clavaguera F, et al. Brain homogenates from human tauopathies induce tau inclusions in mouse brain. Proc Natl Acad Sci U S A. 2013;110:9535–40. doi: 10.1073/pnas.1301175110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper-Knock J, et al. Antisense RNA foci in the motor neurons of C9ORF72-ALS patients are associated with TDP-43 proteinopathy. Acta Neuropathol. 2015;130:63–75. doi: 10.1007/s00401-015-1429-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson YS, et al. Brain distribution of dipeptide repeat proteins in frontotemporal lobar degeneration and motor neurone disease associated with expansions in C9ORF72. Acta Neuropathol Commun. 2014;2:70. doi: 10.1186/2051-5960-2-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis BM, et al. Expansion of a CUG trinucleotide repeat in the 3′ untranslated region of myotonic dystrophy protein kinase transcripts results in nuclear retention of transcripts. Proc Natl Acad Sci U S A. 1997;94:7388–93. doi: 10.1073/pnas.94.14.7388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haro M, et al. MBNL1 and CUGBP1 modify expanded CUG-induced toxicity in a Drosophila model of myotonic dystrophy type 1. Hum Mol Genet. 2006;15:2138–45. doi: 10.1093/hmg/ddl137. [DOI] [PubMed] [Google Scholar]
- DeJesus-Hernandez M, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–56. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly CJ, et al. RNA toxicity from the ALS/FTD C9ORF72 expansion is mitigated by antisense intervention. Neuron. 2013;80:415–28. doi: 10.1016/j.neuron.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echeverria GV, Cooper TA. RNA-binding proteins in microsatellite expansion disorders: mediators of RNA toxicity. Brain Res. 2012;1462:100–11. doi: 10.1016/j.brainres.2012.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fardaei M, et al. In vivo co-localisation of MBNL protein with DMPK expanded-repeat transcripts. Nucleic Acids Res. 2001;29:2766–71. doi: 10.1093/nar/29.13.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox BW, Tibbetts RS. Neurodegeneration: Problems at the nuclear pore. Nature. 2015;525:36–7. doi: 10.1038/nature15208. [DOI] [PubMed] [Google Scholar]
- Fratta P, et al. C9orf72 hexanucleotide repeat associated with amyotrophic lateral sclerosis and frontotemporal dementia forms RNA G-quadruplexes. Sci Rep. 2012;2:1016. doi: 10.1038/srep01016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fratta P, et al. Homozygosity for the C9orf72 GGGGCC repeat expansion in frontotemporal dementia. Acta Neuropathol. 2013;126:401–9. doi: 10.1007/s00401-013-1147-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freibaum BD, et al. GGGGCC repeat expansion in C9orf72 compromises nucleocytoplasmic transport. Nature. 2015;525:129–33. doi: 10.1038/nature14974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron TF, et al. Antisense transcripts of the expanded C9ORF72 hexanucleotide repeat form nuclear RNA foci and undergo repeat-associated non-ATG translation in c9FTD/ALS. Acta Neuropathol. 2013;126:829–44. doi: 10.1007/s00401-013-1192-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron TF, et al. Mechanisms of toxicity in C9FTLD/ALS. Acta Neuropathol. 2014;127:359–76. doi: 10.1007/s00401-013-1237-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron TF, et al. Cerebellar c9RAN proteins associate with clinical and neuropathological characteristics of C9ORF72 repeat expansion carriers. Acta Neuropathol. 2015;130:559–73. doi: 10.1007/s00401-015-1474-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerety SS, Wilkinson DG. Morpholino artifacts provide pitfalls and reveal a novel role for pro-apoptotic genes in hindbrain boundary development. Dev Biol. 2011;350:279–89. doi: 10.1016/j.ydbio.2010.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gijselinck I, et al. A C9orf72 promoter repeat expansion in a Flanders-Belgian cohort with disorders of the frontotemporal lobar degeneration-amyotrophic lateral sclerosis spectrum: a gene identification study. Lancet Neurol. 2012;11:54–65. doi: 10.1016/S1474-4422(11)70261-7. [DOI] [PubMed] [Google Scholar]
- Guo JL, et al. Distinct alpha-synuclein strains differentially promote tau inclusions in neurons. Cell. 2013;154:103–17. doi: 10.1016/j.cell.2013.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeusler AR, et al. C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature. 2014;507:195–200. doi: 10.1038/nature13124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms MB, et al. Lack of C9ORF72 coding mutations supports a gain of function for repeat expansions in amyotrophic lateral sclerosis. Neurobiol Aging. 2013;34:2234 e13–9. doi: 10.1016/j.neurobiolaging.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, Todd PK. Epigenetics in nucleotide repeat expansion disorders. Semin Neurol. 2011;31:470–83. doi: 10.1055/s-0031-1299786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruscha A, et al. Efficient CRISPR/Cas9 genome editing with low off-target effects in zebrafish. Development. 2013;140:4982–7. doi: 10.1242/dev.099085. [DOI] [PubMed] [Google Scholar]
- Ilieva H, Polymenidou M, Cleveland DW. Non-cell autonomous toxicity in neurodegenerative disorders: ALS and beyond. J Cell Biol. 2009;187:761–72. doi: 10.1083/jcb.200908164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, et al. Myotonic dystrophy type 1 is associated with nuclear foci of mutant RNA, sequestration of muscleblind proteins and deregulated alternative splicing in neurons. Hum Mol Genet. 2004;13:3079–88. doi: 10.1093/hmg/ddh327. [DOI] [PubMed] [Google Scholar]
- Jovicic A, et al. Modifiers of C9orf72 dipeptide repeat toxicity connect nucleocytoplasmic transport defects to FTD/ALS. Nat Neurosci. 2015;18:1226–9. doi: 10.1038/nn.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppers M, et al. C9orf72 ablation in mice does not cause motor neuron degeneration or motor deficits. Ann Neurol. 2015;78:426–38. doi: 10.1002/ana.24453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon I, et al. Poly-dipeptides encoded by the C9orf72 repeats bind nucleoli, impede RNA biogenesis, and kill cells. Science. 2014;345:1139–45. doi: 10.1126/science.1254917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagier-Tourenne C, Cleveland DW. Rethinking ALS: the FUS about TDP-43. Cell. 2009;136:1001–4. doi: 10.1016/j.cell.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagier-Tourenne C, et al. Targeted degradation of sense and antisense C9orf72 RNA foci as therapy for ALS and frontotemporal degeneration. Proc Natl Acad Sci U S A. 2013;110:E4530–9. doi: 10.1073/pnas.1318835110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YB, et al. Hexanucleotide repeats in ALS/FTD form length-dependent RNA foci, sequester RNA binding proteins, and are neurotoxic. Cell Rep. 2013;5:1178–86. doi: 10.1016/j.celrep.2013.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine TP, et al. The product of C9orf72, a gene strongly implicated in neurodegeneration, is structurally related to DENN Rab-GEFs. Bioinformatics. 2013;29:499–503. doi: 10.1093/bioinformatics/bts725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, et al. Failure of MBNL1-dependent post-natal splicing transitions in myotonic dystrophy. Hum Mol Genet. 2006;15:2087–97. doi: 10.1093/hmg/ddl132. [DOI] [PubMed] [Google Scholar]
- Ling SC, Polymenidou M, Cleveland DW. Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron. 2013;79:416–38. doi: 10.1016/j.neuron.2013.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu EY, et al. C9orf72 hypermethylation protects against repeat expansion-associated pathology in ALS/FTD. Acta Neuropathol. 2014;128:525–41. doi: 10.1007/s00401-014-1286-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu JX, et al. Molecular structure of beta-amyloid fibrils in Alzheimer’s disease brain tissue. Cell. 2013;154:1257–68. doi: 10.1016/j.cell.2013.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie IR, Frick P, Neumann M. The neuropathology associated with repeat expansions in the C9ORF72 gene. Acta Neuropathol. 2014;127:347–57. doi: 10.1007/s00401-013-1232-4. [DOI] [PubMed] [Google Scholar]
- Mackenzie IR, et al. Quantitative analysis and clinico-pathological correlations of different dipeptide repeat protein pathologies in C9ORF72 mutation carriers. Acta Neuropathol. 2015;130:845–61. doi: 10.1007/s00401-015-1476-2. [DOI] [PubMed] [Google Scholar]
- Mankodi A, et al. Muscleblind localizes to nuclear foci of aberrant RNA in myotonic dystrophy types 1 and 2. Hum Mol Genet. 2001;10:2165–70. doi: 10.1093/hmg/10.19.2165. [DOI] [PubMed] [Google Scholar]
- May S, et al. C9orf72 FTLD/ALS-associated Gly-Ala dipeptide repeat proteins cause neuronal toxicity and Unc119 sequestration. Acta Neuropathol. 2014;128:485–503. doi: 10.1007/s00401-014-1329-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan CT, et al. C9orf72 promoter hypermethylation is neuroprotective: Neuroimaging and neuropathologic evidence. Neurology. 2015;84:1622–30. doi: 10.1212/WNL.0000000000001495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JW, et al. Recruitment of human muscleblind proteins to (CUG)(n) expansions associated with myotonic dystrophy. EMBO J. 2000;19:4439–48. doi: 10.1093/emboj/19.17.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsis EM, et al. Tauopathy PET and amyloid PET in the diagnosis of chronic traumatic encephalopathies: studies of a retired NFL player and of a man with FTD and a severe head injury. Transl Psychiatry. 2014;4:e441. doi: 10.1038/tp.2014.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizielinska S, et al. C9orf72 repeat expansions cause neurodegeneration in Drosophila through arginine-rich proteins. Science. 2014;345:1192–4. doi: 10.1126/science.1256800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K, et al. Bidirectional transcripts of the expanded C9orf72 hexanucleotide repeat are translated into aggregating dipeptide repeat proteins. Acta Neuropathol. 2013a;126:881–93. doi: 10.1007/s00401-013-1189-3. [DOI] [PubMed] [Google Scholar]
- Mori K, et al. hnRNP A3 binds to GGGGCC repeats and is a constituent of p62-positive/TDP43-negative inclusions in the hippocampus of patients with C9orf72 mutations. Acta Neuropathol. 2013b;125:413–23. doi: 10.1007/s00401-013-1088-7. [DOI] [PubMed] [Google Scholar]
- Mori K, et al. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science. 2013c;339:1335–8. doi: 10.1126/science.1232927. [DOI] [PubMed] [Google Scholar]
- O’Rourke JG, et al. C9orf72 BAC Transgenic Mice Display Typical Pathologic Features of ALS/FTD. Neuron. 2015;88:892–901. doi: 10.1016/j.neuron.2015.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rourke JG, et al. C9orf72 is required for proper macrophage and microglial function in mice. Science. 2016;351:1324–9. doi: 10.1126/science.aaf1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters OM, et al. Human C9ORF72 Hexanucleotide Expansion Reproduces RNA Foci and Dipeptide Repeat Proteins but Not Neurodegeneration in BAC Transgenic Mice. Neuron. 2015;88:902–9. doi: 10.1016/j.neuron.2015.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prudencio M, et al. Distinct brain transcriptome profiles in C9orf72-associated and sporadic ALS. Nat Neurosci. 2015;18:1175–82. doi: 10.1038/nn.4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy K, et al. The disease-associated r(GGGGCC)n repeat from the C9orf72 gene forms tract length-dependent uni- and multimolecular RNA G-quadruplex structures. J Biol Chem. 2013;288:9860–6. doi: 10.1074/jbc.C113.452532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renton AE, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–68. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renton AE, Chio A, Traynor BJ. State of play in amyotrophic lateral sclerosis genetics. Nat Neurosci. 2014;17:17–23. doi: 10.1038/nn.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi A, et al. Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature. 2015;524:230–3. doi: 10.1038/nature14580. [DOI] [PubMed] [Google Scholar]
- Russ J, et al. Hypermethylation of repeat expanded C9orf72 is a clinical and molecular disease modifier. Acta Neuropathol. 2015;129:39–52. doi: 10.1007/s00401-014-1365-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sareen D, et al. Targeting RNA foci in iPSC-derived motor neurons from ALS patients with a C9ORF72 repeat expansion. Sci Transl Med. 2013;5:208ra149. doi: 10.1126/scitranslmed.3007529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schludi MH, et al. Distribution of dipeptide repeat proteins in cellular models and C9orf72 mutation cases suggests link to transcriptional silencing. Acta Neuropathol. 2015;130:537–55. doi: 10.1007/s00401-015-1450-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer Burguete A, et al. GGGGCC microsatellite RNA is neuritically localized, induces branching defects, and perturbs transport granule function. Elife. 2015;4 doi: 10.7554/eLife.08881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Z, et al. Discovery of a biomarker and lead small molecules to target r(GGGGCC)-associated defects in c9FTD/ALS. Neuron. 2014;83:1043–50. doi: 10.1016/j.neuron.2014.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N, et al. The mouse C9ORF72 ortholog is enriched in neurons known to degenerate in ALS and FTD. Nat Neurosci. 2013;16:1725–7. doi: 10.1038/nn.3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taneja KL, et al. Foci of trinucleotide repeat transcripts in nuclei of myotonic dystrophy cells and tissues. J Cell Biol. 1995;128:995–1002. doi: 10.1083/jcb.128.6.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therrien M, et al. Deletion of C9ORF72 results in motor neuron degeneration and stress sensitivity in C. elegans. PLoS One. 2013;8:e83450. doi: 10.1371/journal.pone.0083450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd PK, et al. CGG repeat-associated translation mediates neurodegeneration in fragile X tremor ataxia syndrome. Neuron. 2013;78:440–55. doi: 10.1016/j.neuron.2013.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran H, et al. Differential Toxicity of Nuclear RNA Foci versus Dipeptide Repeat Proteins in a Drosophila Model of C9ORF72 FTD/ALS. Neuron. 2015;87:1207–14. doi: 10.1016/j.neuron.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tronche F, et al. Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat Genet. 1999;23:99–103. doi: 10.1038/12703. [DOI] [PubMed] [Google Scholar]
- van Blitterswijk M, Rademakers R. Neurodegenerative disease: C9orf72 repeats compromise nucleocytoplasmic transport. Nat Rev Neurol. 2015;11:670–2. doi: 10.1038/nrneurol.2015.219. [DOI] [PubMed] [Google Scholar]
- Waite AJ, et al. Reduced C9orf72 protein levels in frontal cortex of amyotrophic lateral sclerosis and frontotemporal degeneration brain with the C9ORF72 hexanucleotide repeat expansion. Neurobiol Aging. 2014;35:1779 e5–1779 e13. doi: 10.1016/j.neurobiolaging.2014.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen X, et al. Antisense Proline-Arginine RAN Dipeptides Linked to C9ORF72-ALS/FTD Form Toxic Nuclear Aggregates that Initiate In Vitro and In Vivo Neuronal Death. Neuron. 2014;84:1213–25. doi: 10.1016/j.neuron.2014.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Z, et al. Hypermethylation of the CpG island near the G4C2 repeat in ALS with a C9orf72 expansion. Am J Hum Genet. 2013;92:981–9. doi: 10.1016/j.ajhg.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, et al. Expanded GGGGCC repeat RNA associated with amyotrophic lateral sclerosis and frontotemporal dementia causes neurodegeneration. Proc Natl Acad Sci U S A. 2013;110:7778–83. doi: 10.1073/pnas.1219643110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakawa M, et al. Characterization of the dipeptide repeat protein in the molecular pathogenesis of c9FTD/ALS. Hum Mol Genet. 2014 doi: 10.1093/hmg/ddu576. [DOI] [PubMed] [Google Scholar]
- Yoshimura S, et al. Family-wide characterization of the DENN domain Rab GDP-GTP exchange factors. J Cell Biol. 2010;191:367–81. doi: 10.1083/jcb.201008051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, et al. Discovery of Novel DENN Proteins: Implications for the Evolution of Eukaryotic Intracellular Membrane Structures and Human Disease. Front Genet. 2012;3:283. doi: 10.3389/fgene.2012.00283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, et al. The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature. 2015;525:56–61. doi: 10.1038/nature14973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci. 2014a;34:11929–47. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, et al. Purification and Characterization of Progenitor and Mature Human Astrocytes Reveals Transcriptional and Functional Differences with Mouse. Neuron. 2016a;89:37–53. doi: 10.1016/j.neuron.2015.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YJ, et al. Aggregation-prone c9FTD/ALS poly(GA) RAN-translated proteins cause neurotoxicity by inducing ER stress. Acta Neuropathol. 2014b;128:505–24. doi: 10.1007/s00401-014-1336-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YJ, et al. C9ORF72 poly(GA) aggregates sequester and impair HR23 and nucleocytoplasmic transport proteins. Nat Neurosci. 2016b doi: 10.1038/nn.4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu T, et al. Non-ATG-initiated translation directed by microsatellite expansions. Proc Natl Acad Sci U S A. 2011;108:260–5. doi: 10.1073/pnas.1013343108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu T, et al. RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc Natl Acad Sci U S A. 2013;110:E4968–77. doi: 10.1073/pnas.1315438110. [DOI] [PMC free article] [PubMed] [Google Scholar]