Abstract
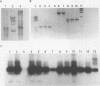
OCT embedded cryostat sections of stored pathological specimens of non-Hodgkin's lymphoma were used to provide RNA. After reverse transcription to produce cDNA, the polymerase chain reaction was performed with primers for standard and variant forms of the CD44 molecule. Using Southern transfer and hybridisation with a probe specific for exon 4 of the CD44 gene, both standard and variant forms were visualised by autoradiography. This method was shown to be applicable to other gene products by using primers specific for the abl and bcr genes. This technique permits retrospective analysis of RNA from small amounts of stored pathological samples.
Full text
PDF
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Cross N. C., Feng L., Bungey J., Goldman J. M. Minimal residual disease after bone marrow transplant for chronic myeloid leukaemia detected by the polymerase chain reaction. Leuk Lymphoma. 1993;11 (Suppl 1):39–43. doi: 10.3109/10428199309047861. [DOI] [PubMed] [Google Scholar]
- Cross N. C., Melo J. V., Feng L., Goldman J. M. An optimized multiplex polymerase chain reaction (PCR) for detection of BCR-ABL fusion mRNAs in haematological disorders. Leukemia. 1994 Jan;8(1):186–189. [PubMed] [Google Scholar]
- Dall P., Heider K. H., Sinn H. P., Skroch-Angel P., Adolf G., Kaufmann M., Herrlich P., Ponta H. Comparison of immunohistochemistry and RT-PCR for detection of CD44v-expression, a new prognostic factor in human breast cancer. Int J Cancer. 1995 Feb 8;60(4):471–477. doi: 10.1002/ijc.2910600408. [DOI] [PubMed] [Google Scholar]
- East J. A., Hart I. R. CD44 and its role in tumour progression and metastasis. Eur J Cancer. 1993;29A(14):1921–1922. doi: 10.1016/0959-8049(93)90442-i. [DOI] [PubMed] [Google Scholar]
- Kaufmann M., Heider K. H., Sinn H. P., von Minckwitz G., Ponta H., Herrlich P. CD44 variant exon epitopes in primary breast cancer and length of survival. Lancet. 1995 Mar 11;345(8950):615–619. doi: 10.1016/s0140-6736(95)90521-9. [DOI] [PubMed] [Google Scholar]
- Mackay C. R., Terpe H. J., Stauder R., Marston W. L., Stark H., Günthert U. Expression and modulation of CD44 variant isoforms in humans. J Cell Biol. 1994 Jan;124(1-2):71–82. doi: 10.1083/jcb.124.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura Y., Tarin D. Significance of CD44 gene products for cancer diagnosis and disease evaluation. Lancet. 1992 Oct 31;340(8827):1053–1058. doi: 10.1016/0140-6736(92)93077-z. [DOI] [PubMed] [Google Scholar]
- Ruiz P., Schwärzler C., Günthert U. CD44 isoforms during differentiation and development. Bioessays. 1995 Jan;17(1):17–24. doi: 10.1002/bies.950170106. [DOI] [PubMed] [Google Scholar]
- Screaton G. R., Bell M. V., Jackson D. G., Cornelis F. B., Gerth U., Bell J. I. Genomic structure of DNA encoding the lymphocyte homing receptor CD44 reveals at least 12 alternatively spliced exons. Proc Natl Acad Sci U S A. 1992 Dec 15;89(24):12160–12164. doi: 10.1073/pnas.89.24.12160. [DOI] [PMC free article] [PubMed] [Google Scholar]