Abstract
Rice tillering has an important influence on grain yield, and is promoted by nitrogen (N) fertilizer. Several genes controlling rice tillering, which are regulated by poor N supply, have been identified. However, the molecular mechanism associated with the regulation of tillering based on N supply is poorly understood. Here, we report that rice microRNA393 (OsmiR393) is involved in N-mediated tillering by decreasing auxin signal sensitivity in axillary buds. Expression analysis showed that N fertilizer causes up-regulation of OsmiR393, but down-regulation of two target genes (OsAFB2 and OsTB1). In situ expression analysis showed that OsmiR393 is highly expressed in the lateral axillary meristem. OsmiR393 overexpression mimicked N-mediated tillering in wild type Zhonghua 11 (ZH11). Mutation of OsMIR393 in ZH11 repressed N-promoted tillering, which simulated the effects of limited N, and this could not be restored by supplying N fertilizer. Western blot analysis showed that OsIAA6 was accumulated in both OsmiR393-overexpressing lines and N-treated wild type rice, but was reduced in the OsMIR393 mutant. Therefore, we deduced that N-induced OsmiR393 accumulation reduces the expression of OsTIR1 and OsAFB2, which alleviates sensitivity to auxin in the axillary buds and stabilizes OsIAA6, thereby promoting rice tillering.
Rice tillering (branching) is an important agronomic trait, as the number of tillers per plant determines the panicle number, which is a key factor of the rice grain yield1,2. The rice tiller represents a form of shoot branching that only exists in monocotyledonous plants at the jointing stage. Unlike in Arabidopsis, the dominance of the apical meristem in rice is weak, so tillering occurs during the vegetative stage upon inhibition by the apical meristem or in previously activated buds, which is not sufficiently strong. However, when the panicles of the main stems begin to differentiate, the buds formed at the elongated upper internodes become dormant3. The dormant bud is activated and later transformed into an activated, growing tiller. This process is complicated and intricate. Rice tillering is mediated by the interplay of the environment and endogenous signals such as phytohormones. It is already known that auxin, cytokinin (CK), and strigolactones (SLs) are specific regulators involved in bud outgrowth regulation4. Auxin and SLs inhibit bud outgrowth, whereas CK activates and promotes this process5,6. Nitrogen (N) can considerably increase CK levels in rice tiller buds and nodes, and can augment IAAs (early auxin responsive genes) in rice nodes7. The tiller number per plant is strongly affected by N fertilizer availability. High N fertilizer allows rice to produce more tillers7, and the Arabidopsis root system architecture is mediated by N availability8.
More is known of the mechanism by which limited N reduces branching than how abundant N promotes branching. In Arabidopsis, nitrate limitation reduces shoot branching by both inhibiting bud initiation and weakening the basipetal sequence of bud initiation that is caused by flowering9. Auxin was the first phytohormone to be identified as an important regulator of plant branching. Bud outgrowth is strongly inhibited by the apex of a whole plant. If the apex is removed, previously inactivated axillary buds become active and plant branching is initiated10. However, auxin cannot enter axillary meristem cells and therefore, its inhibitory functions are deemed to be indirect11. Auxin suppresses bud development through at least two different processes. First, auxin is synthesized in the plant apex and is transported basipetally towards the root, which is known as polar auxin transport (PAT), and this inhibits bud outgrowth12. Because auxin cannot enter the axillary meristem cells, the second mechanism is regarded as the regulation of other signalling molecules (CK and SLs) by auxin; these proteins can enter axillary meristem cells and regulate axillary bud initiation and outgrowth13,14. The PAT model suggests that polar auxin transport and high auxin concentrations inhibit axillary bud development14,15. The second messenger model suggests that auxin regulates the distribution and activity of CKs and SLs in the axillary meristem to control plant branching. Both of these models are supported by recent studies16.
Several genes have been identified as regulators that control rice tillering and branching. MOC117 controls initiation and outgrowth of axillary meristems at both the vegetative and reproductive stages. LAX1 is a regulator that controls axillary meristem initiation and/or maintenance during rice reproductive development18. OsMADS57 was reported to interact with OsTB1, and targets D14 (Dwarf 14) to control the outgrowth of axillary buds19. OsmiR156 targets OsSPL14 and mediates its degradation to control rice tillering20. In Arabidopsis, auxin and SL signalling are required to coordinate shoot branching with respect to N supply9.
MicroRNAs (miRNAs) are a class of oligonucleotides, 20 to 24 nucleotides long, and are endogenous small RNAs that are involved in post–transcriptional gene regulation in multicellular organisms, through influencing both the stability and translation of mRNAs21. miRNAs have important regulatory roles in the uptake, assimilation, and translocation of nutrients in plants22. miR395 and miR399 can regulate the distribution and homeostasis of sulphur and phosphate, respectively, in Arabidopsis and rice23,24. miR169 has specific roles in the plant’s response to N deficiency25. miR167 and miR393 were reported to regulate NO3− signalling during lateral root development8,26. MIR393 is conserved among different plant species27, and recent studies revealed that it has multiple functions in plant growth and development, such as controlling root architecture8, regulation of leaf development28, antibacterial resistance to pathogen attack27, tolerance to stress29, and maintenance of normal plant growth30. In Arabidopsis, miR393 is encoded by two distinct loci: MIR393a and MIR393b. In aerial tissues such as leaves, miR393 is mainly transcribed from MIR393b, suggesting distinct roles for MIR393a and MIR393b28. Moreover, miR393-guided target cleavage processes generate secondary small interfering RNAs (siRNAs) from miR393 target transcripts. Feedback regulates the targeting of neo-generated siRNA, guiding the cleavage of miR393 targets28. Similar to that in Arabidopsis, the rice OsmiR393 family is encoded by two loci, OsMIR393a and OsMIR393b31,32. Rice OsMIR393a and OsMIR393b also showed different expression patterns, which suggests that conserved mechanisms were adopted in monocots and eudicots during plant development32. To date, the exact mechanism of rice tillering regulation by OsmiR393 has not been reported.
miR393 was shown to target auxin (IAA) receptor genes TIR1 and AFB in different plants, including Arabidopsis and rice31,33,34. TIR1 and AFBs encode F-box proteins, which combine with three other proteins, ASK1, CUL1, and RBX, for assembly into the ubiquitin degradative complex (SCFTIR1) to degrade specific substrates during auxin signaling35,36,37. AUX/IAAs represent a class of proteins that represses auxin signalling. AUX/IAA proteins can bind to and repress ARFs (auxin response factor) to activate downstream auxin-responsive genes. TIR1 and AFBs recognize and bind IAA and degrade AUX/IAA proteins via the SCFTIR1 complex37 to ensure correct auxin signalling.
Here, we present findings showing that N-induced rice tillering is caused by attenuating the sensitivity of tiller buds to auxin through microRNA OsmiR393-mediated auxin signal transduction.
Results
Nitrogen supply promotes OsmiR393 accumulation in rice
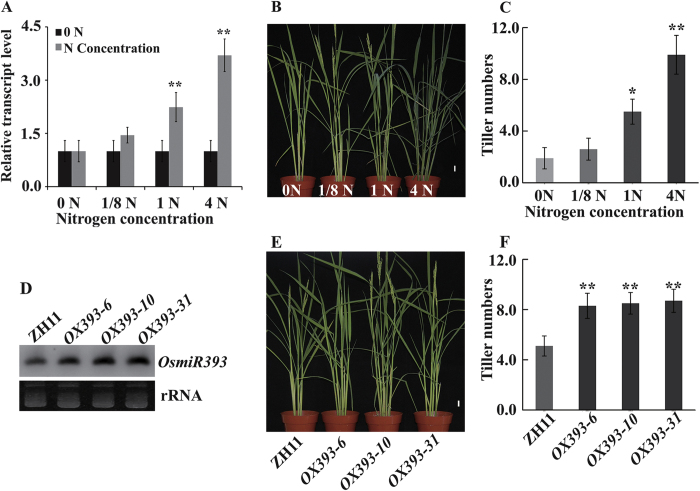
In Arabidopsis, miR393, which targets AFB3, is a unique nitrogen (N) responsive module that mediates root system architecture in response to external and internal N availability8. To investigate whether OsmiR393 expression also responds to exogenous N fertilizer in rice, we analysed the accumulation of OsmiR393 with various levels of NH4NO3 fertilizer as the N source. The level of NH4NO3 (1.43 mM) in the IRRI nutrient solution38 was set to the normal level (1 N) of N fertilizer. OsmiR393 accumulation increased following exposure to elevating levels of N fertilizer, from 0 mM (0 N) to 5.72 mM (4 N) NH4NO3, as assessed by a quantitative reverse transcription polymerase chain reaction (qRT–PCR) (Fig. 1A) and by a small RNA gel blot (Supplemental Fig. S1). Under conditions of 1 N and 4 N NH4NO3, OsmiR393 expression increased by 1.4- and 3.5-fold, respectively, compared to that of the 0 N condition (Fig. 1A). These results showed that OsmiR393 accumulation in rice is induced by high N fertilizer levels, similar to that observed in Arabidopsis8.
Figure 1. Nitrogen fertilizer triggers OsmiR393 accumulation and overexpression resulting in an increased number of tillers.
(A and D), qRT-PCR (A) and small RNA gel blot (D) analysis of OsmiR393 expression level in ZH11 with elevating NH4NO3 levels (A) and in OsmiR393-overexpressing rice lines (OX393) under normal nitrogen (N) levels (1 N) (D). The level of NH4NO3 (1.43 mM) in the IRRI nutrient solution38 was set as the normal (1 N) level of N fertilizer; other N levels represent multiples of the 1 N level. Vertical bars in (A) indicate standard error from three individual repeats. Rice U6 was used as an internal control. The asterisk in (A) indicates a mean fold change of greater than 2 vs the 0 N condition. (B,C) and (E,F), Tiller numbers of ZH11 with elevating NH4NO3 levels (B,C) and in OsmiR393-overexpressing rice plants with a normal N concentration (1 N) (E,F) respectively, when plants were grown in a pot. Scale bar = 2 cm in (B) and (E). Tiller numbers in (C) and (F) were counted and analysed in excel. Vertical bars in (C) and (F) indicate standard error (n ≥ 15 plants for every sample and each experiment was repeated three times). The asterisk indicates significant differences (*P ≤ 0.05 and **P ≤ 0.01) compared to ZH11 at 0 N (C), and at 1 N (F) as determined by a t-test.
Overexpression of OsmiR393 mimics N-promoted rice tillering
High N fertilizer can promote tillering in rice, and we found that OsmiR393-overexpressing rice produced more tillers than wild type rice with normal levels of N fertilizer39. Given that high N fertilizer promotes OsmiR393 accumulation (Fig. 1A, Supplemental Fig. S1), we hypothesized that OsmiR393 is involved in mediating N-mediated rice tillering. To test this hypothesis, we first measured the tiller number in wild type japonica rice cultivar Zhonghua 11 (ZH11) with different levels of N fertilizer (Fig. 1B,C). Tiller numbers in ZH11 were increased with elevating levels of NH4NO3. When grown under low N conditions, specifically, 0 and 0.18 mM NH4NO3 (0 N and 1/8 N, respectively), ZH11 produced 2.6 tillers on average. At normal N levels (1.43 mM NH4NO3; 1 N), it produced 5.5 tillers, which is 2-fold greater than that of the 0 N condition. The tiller number reached approximately 10 when plants were cultivated with high N (5.72 mM NH4NO3; 4 N), which is 4-fold greater than that of the 0 N condition. Furthermore, tillering was analysed in three OsmiR393-overexpressing rice lines39, relative to N content. A small RNA gel blot assay confirmed that OsmiR393 accumulated in the three lines (OX393-6, OX393-10, and OX393-31) compared to expression in wild type ZH11, under normal N conditions (1 N) (Fig. 1D). When grown under normal N (1 N), all three OsmiR393-overexpressing lines produced more tillers than ZH11 (Fig. 1E,F). At 1 N, tiller numbers in the three OsmiR393-overexpressing lines reached approximately 8.3, which was about 1.5-fold higher than that observed in ZH11. These results demonstrate that tillering with OsmiR393 overexpression at normal N levels mimics that of ZH11 at higher N levels. Third, to investigate whether OsmiR393 overexpression at different N levels increases tillers as in ZH11, we treated OX393-6 with a gradient of N levels (Supplemental Fig. S2A,B). Although the tiller number in OX393-6 increased with elevating N levels, it did not reach the maximum observed with ZH11 (Fig. 1B,C). With 4 N conditions, the tiller number reached 7.4 in ZH11, but averaged only 5 in OX393-6. Moreover, at each N level (except 4 N), OX393-6 produced more tillers than ZH11. However, OX393-6 and ZH11 reached approximately the same tiller numbers when they were grown at 4 N. At the 1/8 N level, OX393-6 produced on average 5.2 tillers, whereas ZH11 produced 3.8 tillers. At the 1 N level, OX393-6 produced on an average 6.8 tillers and ZH11 produced 4.4 tillers. At the 4 N level, the tiller number for both OX393-6 and ZH11 reached approximately 9.5. In summary, our data suggests that OsmiR393 overexpression is sufficient to increase tillering at almost all N concentrations, indicating that it is involved in N-promoted tillering. However, OsmiR393 overexpression cannot fully mimic N-induced tillering at 4 N, suggesting that other factors/signalling pathways are involved in this process.
N fertilizer cannot restore the reduced tiller phenotype of an OsMIR393 mutant
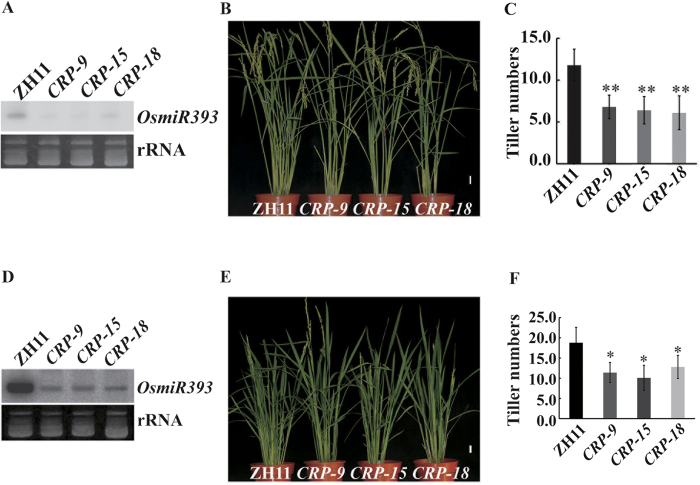
To confirm that OsmiR393 is involved in N-mediated rice tillering, OsMIR393 mutants were generated using a clustered regularly interspaced short palindromic repeats (CRISPR) approach40,41. An sgRNA sequence was designed to target the mature OsmiR393 sequence and ultimately disrupt this gene. Next, the sgRNA was linked to the sgRNA-Cas9 vector42 and transformed into ZH11. Three knock-out OsMIR393 mutant lines were selected and designated as CRP-9, CRP-15, and CRP-18. A small RNA gel blot assay showed that OsmiR393 accumulation was significantly decreased (Fig. 2A). To inspect the tillering phenotype in these OsMIR393 mutants, we grew them in a controlled paddy that was managed conventionally. Contrary to OsmiR393 overexpressing lines, which produced more tillers (Fig. 1E,F), OsMIR393 mutants produced fewer tillers than ZH11 (Fig. 2B,C). ZH11 produced on average 12.2 tillers, whereas all OsMIR393 mutants (CRP-9, CRP-15, and CRP-18) produced approximately 6–7 tillers, approximately half that of ZH11. Combining these results, we concluded that OsmiR393 regulates rice tillering. To investigate whether OsmiR393 is associated with N fertilizer in rice tillering, and if tillering could be restored in OsMIR393 mutants by N fertilizer, we grew OsMIR393 mutants at a 4 N fertilizer level (Fig. 2E,F). OsMIR393 mutants were planted in a controlled paddy with the aforementioned fertilizer level. A high N level (4 N) did not restore OsmiR393 accumulation in OsMIR393 mutants (Fig. 2D), and the tiller number in ZH11 was still much higher than that in OsMIR393 mutants (Fig. 2E,F). This indicated that N fertilizer and OsmiR393 have overlapping functions in rice tillering. However, tiller numbers in OsMIR393 mutants grown at 4 N were still much higher than in those grown at 1 N. OsMIR393 mutants produced on average 11 tillers at the 4 N level (Fig. 2F), and produced approximately 6 tillers at the 1 N fertilizer level (Fig. 2C). This implies that OsmiR393 is required, but not sufficient, for N-mediated rice tillering. Thus, there could be some other mechanisms, distinct from OsmiR393, involved in rice tillering in the presence of N.
Figure 2.
OsMIR393 mutants develop fewer tillers (A–C) under 1 N condition and external application (4N) of NH4NO3 does not restore tillers in OsMIR393 mutants (D–F). (A and D), Small RNA gel blot analysis of OsmiR393 expression in OsMIR393 mutants under normal nitrogen (N) levels (1 N) (A) and with 4 N levels of N fertilizer (D). (B and C) Tiller numbers for the OsMIR393 mutant grown in a controlled field and supplied with 1 N fertilizer (NH4NO3). (E and F) Tiller numbers of OsMIR393 mutants grown in a controlled field and supplied with 4 N fertilizer (NH4NO3). Vertical bars in (C) and (F) indicate standard error (n ≥ 15 plants for every sample and each experiment was repeated three times). The asterisk indicates significant differences (*P ≤ 0.05 and **P ≤ 0.01) compared to the ZH11 control, as assessed by a t-test. Scale bar = 3 cm in (B) and (E).
Auxin signal transduction, mediated by OsmiR393, is involved in N-mediated rice tillering
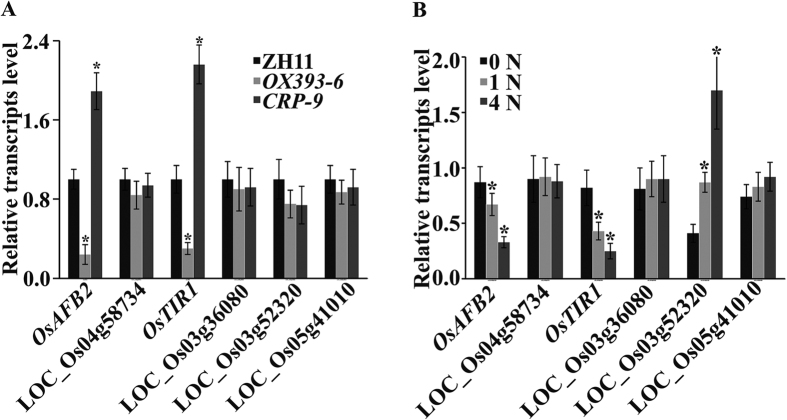
Two auxin receptors OsAFB2 and OsTIR1 have been verified as target genes of OsmiR393, and seven genes were predicted to be candidate OsmiR393 targets in rice32,39. To better understand the actual targets of N fertilizer involved in rice tillering, plant materials were collected comprising regions between the shoot and root containing the SAM (shoot apical meristem), an area of the plant where tillers are formed, for RNA extraction and qRT–PCR detection. qRT-PCR results showed that OsAFB2 and OsTIR1 were down-regulated in the tillering region of OX393-6, and dramatically up-regulated in CRP-9, when compared to levels in ZH11, at N levels of 1 N (Fig. 3A).
Figure 3. Inverse expression of OsmiR393 and its target genes (OsAFB2 and OsTIR1) with elevating nitrogen (N) levels.
(A) qRT-PCR verification of OsmiR393 targets in OsmiR393 overexpressing line (OX393-6), and OsMIR393 mutant (CRP-9) with 1 N fertilizer. (B) qRT–PCR expression analysis of putative OsmiR393 target genes in wild type rice ZH11 with elevating N levels. Results are presented as the means of three experiments. Vertical bars indicate standard error. The asterisk mark indicates a mean fold change greater than 2 or less than 0.5 between ZH11 and the mutants (A), or the elevating N treated ZH11 (B).
To determine whether OsAFB2 and OsTIR1 also decreased with elevating N levels, qRT-PCR assays were conducted in wild type ZH11 (Fig. 3B). In contrast to over-expression of OsmiR393 with elevating N levels (Fig. 1A), OsAFB2 and OsTIR1 transcripts decreased (Fig. 3B). This demonstrated that the two target genes (OsAFB2 and OsTIR1) and OsmiR393 are reciprocally expressed with elevating N levels, indicating that OsAFB2 and OsTIR1 are involved in the response to N supply in rice. However, the expression of another putative target gene (LOC_Os03g52320) was up-regulated in response to high N fertilizer levels in ZH11, suggesting that it was not a target of OsmiR393. LOC_Os03g52320 was predicted to encode a GRF1-interacting factor involved in various aspects of tissue differentiation and organ development43,44. We speculated that its expression was enhanced, either directly or indirectly, by N fertilizer to control rice seedling growth. One putative target, LOC_Os10g39790 was not amplified by PCR, indicating that it was not expressed in the region that we collected.
During auxin signal transduction, TIR1 and AFB2 perceive the auxin signal and degrade repressors of auxin signalling via the SCF (SKP1-CULLIN/cdc53-F-box) complex mediated by the 26S proteasome45. OsmiR393 overexpression and high levels of N, with OsmiR393 accumulation, might desensitize rice plants to exogenous auxin. In contrast, OsMIR393 mutants should become hypersensitive under these conditions. To test this hypothesis, we treated ZH11, OX393-6, and CRP-9 with NAA and observed their responses. The application of 5 mg L–1 NAA typically inhibited the outgrowth of rice tiller buds (Supplemental Fig. S3A, ZH11, and ZH11 mock). However, OX393-6 and 4 N-treated ZH11 plants showed obvious resistance. Tiller buds in CRP-9 were dramatically inhibited, exhibiting hypersensitivity (Supplemental Fig. S3A). This result further confirmed N-mediated rice tillering resulting from attenuated auxin signalling through OsmiR393-mediated cleavage of OsTIR1/OsAFB2. The callus induction rate is an important index reflecting auxin/cytokinin cross-interactions46. To further substantiate our hypothesis, ZH11, OX393-6, and CRP-9 plants were induced to form calluses, and their callus induction rates were compared (Supplemental Fig. S3B). Calluses were induced in ZH11 at a faster rate than in OsmiR393-overexpressing lines, but at a slower rate than in the OsMIR393 mutants. Moreover, adventitious root development is a typical process involved in auxin signaling32,47. The OsmiR393-overexpressing line, OX393-6, showed a significant reduction in adventitious roots, whereas that in CRP-9 increased (Supplemental Fig. S3C,D). The above results implied that auxin signalling interferes with adventitious root formation in OsmiR393-overexpressing lines, but is attenuated in OsMIR393 mutants.
N fertilizer supply and OsmiR393 overexpression promote axillary bud outgrowth
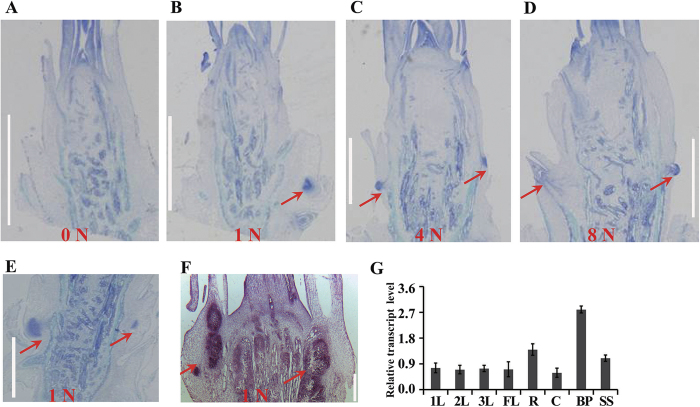
To further investigate the relationship between OsmiR393 overexpression and N-mediated rice tillering, we explored the axillary bud formation process in ZH11 in response to different N levels and in the OsmiR393-overexpressing line, OX393-6, by tissue section (Fig. 4A–E; Supplemental Table S2). After 12 d of cultivation with different levels of N fertilizer, the axillary meristem numbers in ZH11 were inspected. No axillary meristems were observed when ZH11 was grown without N fertilizer (0 N, Fig. 4A), one was observed when ZH11 was grown at 1 N (Fig. 5B), and two were found with 4 N fertilizer (Fig. 4C). With an excessive N level (8 N), only one axillary meristem was observed, while the other had already formed an axillary bud (Fig. 4D). In contrast, the axillary meristem number for OX393-6 grown in 1 N fertilizer was 2 (Fig. 4E), which resembled that of ZH11 with 4 N fertilizer (Fig. 4C). This suggests that OsmiR393 overexpression has the same effect on axillary bud growth as growth with 4 N fertilizer, in ZH11 plants, and that both N fertilizer supply and OsmiR393 expression promote outgrowth of the axillary bud. OsmiR393 expression in rice organs was also analysed by qRT–PCR (Fig. 4G). The results showed that OsmiR393 was expressed in all rice organs, and at a higher level in the roots and booting panicle, indicating that OsmiR393 might play a role in booting panicle development and lateral root growth. To investigate whether OsmiR393 is expressed in the axillary meristem, in situ RNA hybridization experiments were performed (Fig. 4F). The results showed that OsmiR393 was strongly expressed in the lateral axillary meristem (Fig. 4F). These results demonstrate that N fertilizer and OsmiR393 overexpression promote outgrowth of axillary buds, and that OsmiR393 is involved in axillary bud formation.
Figure 4. OsmiR393 affects axillary bud outgrowth and is expressed at a high level in the lateral axillary meristem.
(A–D) Tissue section of wild type ZH11 rice grown with different nitrogen (N) levels using an NH4NO3 fertilizer. (E) Tissue section of OsmiR393-overexpressing line (OX393-6) under normal N levels (1 N) using NH4NO3 fertilizer. Red arrows in (A–E) indicate the formed axillary meristem. Pictures are representative of sections from 20 plants. (F) In situ hybridization of OsmiR393 in the ZH11 stem base with 1 N NH4NO3 fertilizer. The red arrow in (F) indicates intense OsmiR393 expression. Bar = 100 μm in (A–F). (G) qRT–PCR detection of OsmiR393 levels in different organs of ZH11 under normal N levels (1 N) using NH4NO3 fertilizer. Vertical bars indicate standard error from three repeats. 1L, the first leaf; 2L, the second leaf; 3L, the third leaf; FL, flag leaf; R, root; C, culm; BP, booting panicle; SS, shoot sheath.
Figure 5. OsIAA6 protein level in ZH11 plants with different nitrogen (N) levels and in different transgenic rice with 1 N levels.
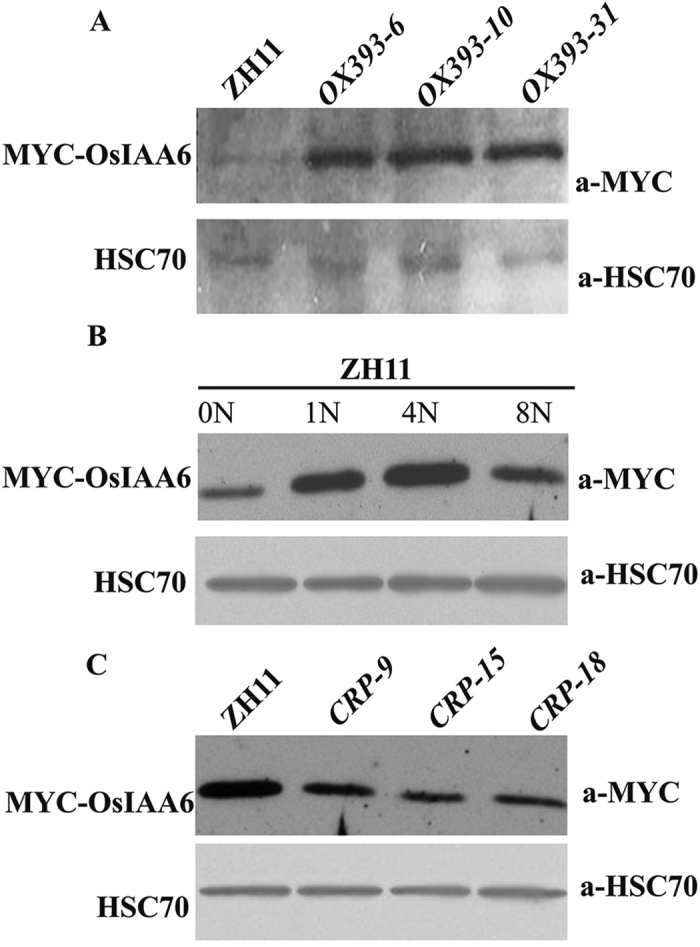
(A–C) Western blot analysis of OsIAA6 in the protoplast of OsmiR393-overexpressing lines (OX393) (A) N-treated ZH11 (B) and OsMIR393 mutants (CRP) (C). Protoplasts were prepared from 2-week old seedlings. HSC70 was used as a loading control.
N fertilizer facilitates rice tillering through preventing the degradation of OsIAA6
TIR1 and AFB2 are two members of the SCF complex, which degrades auxin signalling repressors, to ensure correct auxin signaling45,48. To determine which proteins are degraded by the SCFTIR1 complex, a construct with cMYC fused to OsTIR1 was generated and transformed into ZH11. Subsequently, a co-immunoprecipitation assay was employed and the precipitated proteins were analysed by mass spectrometry. We found that levels of OsIAA6, a protein belonging to the AUX/IAA class of auxin signalling repressors, changed tremendously between 35S:cMYC–TIR1 and OX393-6 plants. OsIAA6 was reported to enhance rice drought tolerance and regulate tiller outgrowth49. Furthermore, AUX/IAA proteins are a substrate of the SCFTIR1 complex50. We suspected that OsmiR393 might block degradation of OsIAA6 by the OsTIR1 and OsAFB2-mediated SCFTIR1 complex to control rice tillering. To investigate whether OsIAA6 expression changed following altered OsmiR393 expression, or in N-treated ZH11, we generated a construct containing an OsIAA6 full-length protein fused to a MYC epitope tag. This construct was transformed into protoplasts of OsmiR393-overexpressing rice plants, OsMIR393 mutants, and N-treated ZH11. Western blot analysis showed that OsIAA6 was remarkably increased in the three OsmiR393 overexpressing lines (Fig. 5A), and in ZH11 with elevating N levels (Fig. 5B), but was reduced in OsMIR393 mutants (Fig. 5C). These results suggest that OsmiR393 might affect auxin signal transduction through regulation of OsIAA6 accumulation.
Discussion
Availability of N fertilizer strongly affects rice tillering7, which is a major determinant of grain yield. In the past decades, several key regulators of rice tillering have been cloned and characterized, including MOC117, TAD151, LAX1 and SPA18, OsTB152, OsmiR15620, and genes in the strigolactone signalling pathway53,54,55,56. However, the mechanisms through which N fertilizer contributes to rice tiller formation remains elusive. In this study, we revealed a novel role for OsmiR393 in affecting rice tillering in response to N fertilizer. Our results show that adding N fertilizer increases the tiller number in rice (Fig. 1B). This is achieved by enhancing the initiation and activation of tiller bud outgrowth (Fig. 4A–D). High N fertilizer causes OsmiR393 accumulation in tiller bud (Figs 1A and 4F; Supplemental Fig. S1). OsmiR393 accumulation then decreases the transduction of auxin signalling (Figs 3 and 5), resulting in decreased sensitivity to auxin in the tiller buds (Supplemental Fig. S3). Consequently, dampening the response to auxin signalling in the tillers with high N fertilizer might promote initiation and outgrowth of the tiller bud.
OsmiR393 responses to N fertilizer
Arabidopsis miR393 is induced by nitrate, and the nitrate-responsive miR393/AFB3 regulatory module mediates root system architecture8. In this study, the addition of N fertilizer also caused the accumulation of OsmiR393 in rice seedlings (Fig. 1A; Supplemental Fig. S1). This result suggests that in plants, a similar mechanism has been adopted in monocots and dicots, to respond to environmental changes in N levels. Over time, expression of its target genes (OsTIR1 and OsAFB2) was repressed (Fig. 3B). This suggests that OsmiR393 is also involved in the response to exogenous N in rice. However, unlike Arabidopsis miR393, wherein expression induced by NO3− was specifically localized to the root8, the expression of OsmiR393 in rice was induced by NH4NO3 in the leaf and root tissue (Figs 1A and 4F). This discrepancy between rice and Arabidopsis might be due to different N treatments, or it might imply that there are functional differences for miR393 between dicots and monocots.
OsmiR393 affects rice tillering
miR393 is a plant-conserved miRNA that participates in many processes during plant development. Recent studies have demonstrated that miR393 plays a role in the establishment of root system architecture in response to nitrate8, the regulation of leaf development28, and auxin signaling57, and is involved in antibacterial resistance in response to pathogens27. In this study, overexpression of OsmiR393 resulted in an increased number of tillers, compared to that in wild type plants (Fig. 1), which was similar to previous findings39. In contrast, an OsMIR393 mutant produced fewer tillers than wild type rice (Fig. 2). Furthermore, supplementation with N did not rescue this defect in OsMIR393 mutants. These results suggest that OsMIR393 is a regulator that participates in modulating rice tillering.
In addition, OsmiR393 altered tillering in response to N Fertilizer. OsmiR393 was induced by NH4NO3 treatment in rice (Fig. 1A), and was highly expressed in the bud meristem (Fig. 4F). When OsmiR393 was overexpressed, the increase in tiller numbers induced by N fertilizer (Supplemental Fig. 2) was not as high as that observed in wild type rice (Fig. 1B,C). However, with the same N level, OsmiR393-overexpressing rice produced more tillers than wild type rice (Fig. 1E,B compared to Supplemental Fig. S2). In contrast, the mutation of OsMIR393 resulted in fewer tillers than that in wild type with high and normal N fertilizer levels (Fig. 2). These results suggest that OsMIR393 is involved in the regulation of rice tillering in response to N fertilizer. Rice varieties exhibit high diversity with respect to N response. According to the degree of responsiveness, rice varieties were classified into two groups, less-responsive varieties (such as, ZH11, QZL2, and Balila) and highly-responsive varieties (such as Minghui 63 and Nanjing 6)58. Highly-responsive rice varieties tend to produce more tillers than less-responsive varieties under the same level of N fertilizer58. OsmiR393 expression was compared (Supplemental Fig. S4) in these two groups. However, the expression of OsmiR393 was not obviously different. Different DEP1 alleles confer variable N responses among less responsive and highly responsive rice cultivars, and DEP1 has been the subject of artificial selection during Oryza sativa spp. japonica rice domestication58. This result might imply that OsmiR393 is a general regulator of rice tillering that responds to N fertilizer, but is not involved in the N-response variability observed among different rice varieties.
OsmiR393 affects auxin signal transduction in response to N fertilizer treatment
Plants synthesize auxin in the growing apex, wherein most of their energy is expended59. Auxin flows from the stem towards the root, basipetally, a phenomenon known as polar auxin transport (PAT). This PAT inhibits axillary bud growth in terms of apical dominance60. It is well known that plants establish their architecture and apical dominance through the effects of PAT.
In this study, we revealed that N-fertilizer promotes OsmiR393 accumulation (Fig. 1A) and decrease expression of OsTIR1 and OsAFB2 (Fig. 3B), and increases tiller number (Fig. 1B) in wild type ZH11 rice. This N-promoted tillering is mimicked by OsmiR393 overexpression (Fig. 1E). Moreover, high N-treatment in wild type ZH11 and overexpression of OsmiR393 resulted in a decrease in OsmiR393-target gene (OsTIR1 and OsAFB2) expression (Fig. 3). The reduced expression of OsTIR1 and OsAFB2 might attenuate auxin perception and signalling, and alleviate the effects of apical dominance. TIR1 and AFB2 are two important substrate recognition subunits of the well-known SKP1-CULLIN-F-box (SCF) ubiquitin ligase complex, which functions in auxin perception to promote the 26S proteasome-mediated degradation of Aux/IAA transcriptional repressors47. In addition, the response of MIR393 to environmental signals is conserved across plant species, through the auxin signalling pathway61,62. It has been shown that auxin cannot enter axillary meristem cells11; however, auxin from the top buds could inhibit export of auxin that is synthesized in the axillary buds11,12. This would increase the auxin concentration in the axillary buds, and inhibit bud outgrowth. High OsmiR393 accumulation after N treatment (Fig. 1A) might decrease sensitivity in the axillary buds to high levels of auxin.
We proved that OsmiR393 overexpression and N treatment prevents degradation of OsIAA6 (Fig. 5), which is an important regulator of axillary bud formation49,63. This strongly supports the hypothesis that OsmiR393 influences axillary bud formation following N fertilizer treatment through auxin signal transduction. Auxin signalling components have been conserved throughout land plant evolution, and have evolved to control specific developmental processes32. Plant genomes encode large numbers of F-box proteins (FBPs), and there are approximately 700 FBPs in Arabidopsis64. The role of miR393/TIR1/AFBs in the plant’s response to auxin represents a ubiquitous model for adaption and acclimation. Therefore, we concluded that N fertilizer promotes OsmiR393 accumulation, which interferes with auxin signalling, finally triggering tiller production in rice.
N fertilizer can promote cytokinin biosynthesis/signalling and stimulates rice to produce more tillers65,66. There is evidence to suggest that exogenous application of N fertilizer can promote cytokinin biosynthesis67 and stimulate lateral bud outgrowth68. In Arabidopsis, the expression of AtIPT3 and AtIPT5 is in response to N69,70; these genes are key determinants of CK biosynthesis that respond to rapid changes in NO3− availability. Our present results showed that OsmiR393 also responds to N fertilizer, interferes with auxin signalling, and attenuates apical dominance, ultimately leading to tiller production in rice. We did not analyse whether these two N-mediated tillering pathways overlap. Thus, the effect of N fertilizer on rice tillering might be through these two pathways, or in parallel, or through other undiscovered pathways. This also explains why exogenous N-treated OsMIR393 mutants still produced more tillers than wild type plants in normal N conditions (Fig. 2B,C,E,F).
Materials and Methods
Plant materials and growth conditions
Oryza sativa japonica Zhonghua11 (ZH11) was used as the wild type and the source for transgenic plants. Plant materials used in this study were ZH11, three OsmiR393 overexpressing lines (OX393-6, OX393-10, and OX393-31)39, three OsMIR393 knock-out lines (CRP-9, CRP-15, and CRP-18), and the distinct N responsive varieties (QZL2, Balila, Minghui 63, and Nanjing 6)58. Seeds were dried at 37 °C for 24 h before germination at 25 °C, and were supplemented with water in the dark for 48 h. For phenotypic observations, germinated seeds were grown under controlled field conditions or in boxes filled with sand supplemented with hydroponic medium. For nutrient treatments, plants were grown in hydroponic cultures using the International Rice Research Institute (IRRI) liquid culture medium recipe (IRRI nutrient solution)38. Except for modified NH4NO3, all nutrients were kept at the same concentration in all cultures. The NH4NO3 content was based on that of the IRRI nutrient solution, which was designated as the normal level; other N fertilizer levels were based on this concentration. For example, NH4NO3 content in 4-fold N fertilizer was quadruple that of the normal level. For paddy culture, seeds were germinated and sown on soil and then conventionally managed.
Vector construction for rice transformation
To generate the cMYC-OsTIR1 construct, the coding region of OsTIR1 was amplified by PCR, cloned into the pEASY–Blunt vector (Transgene, China), and subcloned into the pE3n vector71. The cMYC-OsTIR1 fragment was then inserted into the pCAMBIA2300 vector downstream of the maize (Zea mays) Ubiquitin promoter after KpnI and BamHI digestion. To generate the cMYC-OsIAA6 construct, the coding region of OsIAA6 was amplified by PCR, cloned into the pEASY-Blunt vector (Transgene, China), and subcloned into the pE3n vector. The cMYC-OsIAA6 fragment was then inserted into the pCAMBIA2300 vector downstream of the maize (Zea mays) Ubiquitin promoter after BamHI and NotI digestion. All of the primers used to generate the aforementioned constructs are listed in Supplemental Table S1, and all of the constructs were confirmed by sequencing. The constructed vectors were transformed by Agrobacterium tumefaciens strain EHA105. Wild type ZH11 calli were used as the recipients for Agrobacterium-mediated transformation as described72.
RNA extraction and quantitative RT–PCR
Total RNA was extracted using an RNA extraction kit (Invitrogen, China) and digested with DNase I (Takara, China) according to the manufacturers’ instructions. The RNA quality and integrity were analysed by agarose gel electrophoresis and the RNA concentration was determined using a biophotometer (METASH, B–500, China). cDNA was synthesized from total RNA using AMV Reverse Transcriptase (Promega, China). Small RNA was extracted using an RNAiso kit for small RNA (Takara, China) and digested with DNase I (Takara, China) according to the product manuals. Reverse transcription was performed with a cDNA Synthesis Kit (Promega, China) in combination with a stem-loop RT-PCR technique73. Quantitative RT-PCR was performed on a 7500 RT-qPCR system (Applied Biosystems, USA) with SYBR Green Real-time PCR Master Mix (Toyobo, China) according to the manufacturer’s instructions. Gene expression was normalized to that of rice ACTIN1. Primers used for qRT-PCR are presented in Supplemental Table S1.
Small RNA gel blot assay
Small RNA gel blot analysis was performed as described in Liu et al.74. Briefly, total RNA was extracted from rice seedlings, and total RNA samples (approximately 20 μg) were separated on denaturing 15% polyacrylamide gels and transferred electrophoretically to Hybond-N+ membranes (http://www.gelifesciences.com/). The gel was stained with ethidium bromide before transfer to confirm equal loading. Hybridizations were performed at 42 °C in PerfectHyb Plus buffer with DNA oligonucleotide probes labelled by T4 polynucleotide kinase (New England Biolabs, https://www.neb.com/). Hybridization signals were detected with a phosphorimager (GE Healthcare Life Sciences, http://www.gelifesciences.com). The sequences of the probes are provided in Supplemental Table S1.
Tissue sectioning and in situ hybridization
Seedlings of ZH11 plants were grown with different N fertilizer levels, and OX393-6 was grown with normal levels of N fertilizer for 15 d, after which SAM tissues and axillary buds were fixed and sectioned at a thickness of 7 mm. The sections were then stained with toluidine blue for light microscopic analysis (Zeiss, Germany). In situ hybridization was performed as described75. The OsmiR393 probe was synthesized and labelled with digoxigenin. Shoots containing SAM and axillary meristems were used for hybridization assays. Slides were photographed under a microscope (Zeiss, Germany).
CRISPR-mediated mutation of OsMIR393
An sgRNA: AAGGATCAATGCGATCCCTTTGG was designed to target OsmiR393. The sgRNA was inserted into the AarI site of the p2300-rCas9-U3-gRNA vector, which contains a rice-codon optimized Cas9 driven by a 2× 35S promoter42, and the sgRNA was activated by an OsU3 promoter. The primer used for cloning is shown in Supplemental Table S1 (CRISPR-F and CRISPR-R). After introducing the CRISPR/Cas plasmid into rice varietal ZH11 through Agrobacterium-mediated transformation72, the T0 generation mutants were screened with G418 (Invitrogen). All regenerated T0 transgenic plants were genotyped using the primer (CRISPR-g-F and CRISPR-g-R) to select positive transgenic lines, and a small RNA gel blot analysis was performed to subsequently detect OsmiR393 expression and confirm the knock-out of OsmiR393 in mutated lines.
Co–IP and mass spectrum analysis
To validate the protein interaction profile of OsTIR1 in vivo, a Co-IP assay was employed using an immunoprecipitation kit (Sigma-Aldrich, China). The cDNA for OsTIR1 was amplified using the primers cMYC-TIR1-F and cMYC-TIR1-R. Amplified cDNA was inserted into pRT107–6XMyc between the BamHI and KpnI sites to generate the expression vector 2×35S:6XMyc-OsTIR1. The co–immunoprecipitation procedure was performed in accordance with the manufacturer’s instructions using an anti-c-Myc antibody (Sigma-Aldrich, China). Mass spectrum analysis was performed by the Boxin biotechnology company in Guangzhou. Primers used are listed in Supplemental Table S1.
Protoplast isolation and western blot analysis
To generate the cMYC-OsIAA6 sequence, the OsIAA6 coding sequence was amplified with the gene specific primers, cMYC-OsIAA6-F and cMYC-OsIAA6-R containing the restriction enzyme sites BamHI and NotI, and ligated into a rice transformation vector for constitutive expression. Constructs were introduced into rice protoplasts of 2-week seedlings as previously reported76. The generated constructs were then transfected into the isolated protoplasts using polyethyleneglycol-mediated transformation76. To examine OsIAA6 protein expression, a western blot was performed as described in Niu et al.77.
Tiller bud and callus induction with NAA treatments
Five-leaf seedlings were sprayed with 5 mg/L of NAA, and the length of tiller buds was measured as indicated. Callus induction with 2 mg/L of NAA was performed as previously described39.
Tiller count and data analysis
Rice plants were planted in previously described conditions and grown for approximately 30 days before the headings and tillers were counted; all data were analysed in Excel using a t–test.
Sequence data from this study can be found in the GenBank/EMBL data libraries under accession numbers OsAFB2 (LOC_Os04g32460), OsTIR1 (LOC_Os05g05800), OsIAA6 (LOC_Os01g53880), and ACTIN1 (LOC_Os03g50885).
Additional Information
How to cite this article: Li, X. et al. MicroRNA393 is involved in nitrogen-promoted rice tillering through regulation of auxin signal transduction in axillary buds. Sci. Rep. 6, 32158; doi: 10.1038/srep32158 (2016).
Supplementary Material
Acknowledgments
This work was supported by Guangdong Science and Technology Department of China (grant nos. 2015B020231009 and 2014A020208081), Guangdong Agriculture Department of China (grant no. 2014492), and the National Natural Science Foundation of China (grant no. 31371604/31272240). We thank Xiexiu Zhang of the South China Botanical Garden (SCBG) for technical support. We also thank Dr. Hao Chen, Ze Yun and Dr. Xun Zeng of SCBG for comments on the manuscripts.
Footnotes
Author Contributions X.L. and K.X. performed most of the experiments; Z.L., K.C. and C.G. provided CRISPR technical assistance to X.L. X.L. and M.Z. designed the experiments and analyzed the data; X.L. and M.Z. conceived the project and wrote the article with contributions of all the authors.
References
- Xing Y. & Zhang Q. Genetic and molecular bases of rice yield. Annu. Rev. Plant Biol. 61, 421–442, (2010). [DOI] [PubMed] [Google Scholar]
- Liang W. H., Shang F., Lin Q. T., Lou C. & Zhang J. Tillering and panicle branching genes in rice. Gene 537, 1–5, 10.1016/j.gene.2013.11.058 (2014). [DOI] [PubMed] [Google Scholar]
- Wang Y. & Li J. The plant architecture of rice (Oryza sativa). Plant Mol. Biol. 59, 75–84, (2005). [DOI] [PubMed] [Google Scholar]
- Dun E. A., de Saint Germain A., Rameau C. & Beveridge C. A. Antagonistic action of strigolactone and cytokinin in bud outgrowth control. Plant Physiol. 158, 487–498, 10.1104/pp.111.186783 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dun E. A., Brewer P. B. & Beveridge C. A. Strigolactones: discovery of the elusive shoot branching hormone. Trends Plant Sci. 14, 364–372, 10.1016/j.tplants.2009.04.003 (2009). [DOI] [PubMed] [Google Scholar]
- Leyser O. The control of shoot branching: an example of plant information processing. Plant Cell Environ. 32, 694–703, (2009). [DOI] [PubMed] [Google Scholar]
- Xu J. et al. The interaction between nitrogen availability and auxin, cytokinin, and strigolactone in the control of shoot branching in rice (Oryza sativa L.). Plant Cell Environ. 34, 1647–1662, (2015). [DOI] [PubMed] [Google Scholar]
- Vidal E. A. et al. Nitrate–responsive miR393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 107, 4477–4482, 10.1073/pnas.0909571107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong M. et al. Auxin and strigolactone signaling are required for modulation of Arabidopsis shoot branching by nitrogen supply. Plant Physiol. 166, 384–395, 10.1104/pp.114.242388 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ongaro V. & Leyser O. Hormonal control of shoot branching. J. Exp. Biol. 59, 67–74, 10.1093/jxb/erm134 (2008). [DOI] [PubMed] [Google Scholar]
- Booker J., Chatfield S. & Leyser O. Auxin acts in xylem–associated or medullary cells to mediate apical dominance. Plant Cell 15, 495–507 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C. & Bangerth F. Auto–inihibition of indole–acetic acid transport in the shoot of two–branched pea (Pisum sativum) plants and its relationship to correlative dominance. Physiol. Plant. 106, 415–420 (1999). [Google Scholar]
- Bangerth F. Response of cytokinin concentration in the xylem exudates of bean (Phaseolus vulgaris L.) plants to decapitation and auxin treatment, and relationship to apical dominance. Planta 194, 439–442 (1994). [Google Scholar]
- Brewer P. B., Dun E. A., Ferguson B. J., Rameau C. & Beveridge C. A. Strigolactone acts downstream of auxin to regulate bud outgrowth in pea and Arabidopsis. Plant Physiol. 150, 482–493, 10.1104/pp.108.134783 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balla J., Kalousek P., Reinohl V., Friml J. & Prochazka S. Competitive canalization of PIN–dependent auxin flow from axillary buds controls pea bud outgrowth. Plant J. 65, 571–577, (2011). [DOI] [PubMed] [Google Scholar]
- Domagalska M. A. & Leyser O. Signal integration in the control of shoot branching. Nat. Rev. Mol. Cell Biol. 12, 211–221, 10.1038/nrm3088 (2011). [DOI] [PubMed] [Google Scholar]
- Li X. et al. Control of tillering in rice. Nature 422, 618–621, 10.1038/nature01518 (2003). [DOI] [PubMed] [Google Scholar]
- Komatsu K. et al. LAX and SPA: major regulators of shoot branching in rice. Proc. Natl. Acad. Sci. USA 100, 11765–11770, 10.1073/pnas.1932414100 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S. et al. The interaction between OsMADS57 and OsTB1 modulates rice tillering via DWARF14. Nat Commun. 4, 1566, 10.1038/ncomms2542 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y. et al. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 42, 541–544, 10.1038/ng.591 (2010). [DOI] [PubMed] [Google Scholar]
- Rogers K. & Chen X. Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell 25, 2383–2399, 10.1105/tpc.113.113159 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo H. F. & Chiou T. J. The role of microRNAs in phosphorus deficiency signaling. Plant Physiol. 156, 1016–1024, 10.1104/pp.111.175265 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G., Yang F. & Yu D. MicroRNA395 mediates regulation of sulfate accumulation and allocation in Arabidopsis thaliana. Plant J. 62, 1046–1057, (2010). [DOI] [PubMed] [Google Scholar]
- Hu B. et al. MicroRNA399 is involved in multiple nutrient starvation responses in rice. Front. Plant Sci. 6, 188 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M., Ding H., Zhu J. K., Zhang F. & Li W. X. Involvement of miR169 in the nitrogen–starvation responses in Arabidopsis. New Phytol. 190, 906–915, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita N. et al. IAA–Ala Resistant3, an evolutionarily conserved target of miR167, mediates Arabidopsis root architecture changes during high osmotic stress. Plant Cell 24, 3590–3602, 10.1105/tpc.112.097006 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro L. et al. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312, 436–439, 10.1126/science.1126088 (2006). [DOI] [PubMed] [Google Scholar]
- Si–Ammour A. et al. miR393 and secondary siRNAs regulate expression of the TIR1/AFB2 auxin receptor clade and auxin–related development of Arabidopsis leaves. Plant Physiol. 157, 683–691, 10.1104/pp.111.180083 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R. & Zhu J. K. Novel and stress–regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 16, 2001–2019, 10.1105/tpc.104.022830 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z. H. et al. Regulation of auxin response by miR393–targeted transport inhibitor response protein 1 is involved in normal development in Arabidopsis. Plant Mol. Biol. 77, 619–629, (2011). [DOI] [PubMed] [Google Scholar]
- Jones–Rhoades M. W. & Bartel D. P. Computational identification of plant microRNAs and their targets, including a stress–induced miRNA. Mol. Cell 14, 787–799, 10.1016/j.molcel.2004.05.027 (2004). [DOI] [PubMed] [Google Scholar]
- Bian H. et al. Distinctive expression patterns and roles of the miRNA393/TIR1 homolog module in regulating flag leaf inclination and primary and crown root growth in rice (Oryza sativa). New Phytol. 196, 149–161, (2012). [DOI] [PubMed] [Google Scholar]
- Dharmasiri N., Dharmasiri S. & Estelle M. The F–box protein TIR1 is an auxin receptor. Nature 435, 441–445, 10.1038/nature03543 (2005). [DOI] [PubMed] [Google Scholar]
- Kepinski S. & Leyser O. The Arabidopsis F–box protein TIR1 is an auxin receptor. Nature 435, 446–451, 10.1038/nature03542 (2005). [DOI] [PubMed] [Google Scholar]
- Gray W. M., Kepinski S., Rouse D., Leyser O. & Estelle M. Auxin regulates SCF(TIR1)–dependent degradation of AUX/IAA proteins. Nature 414, 271–276, 10.1038/35104500 (2001). [DOI] [PubMed] [Google Scholar]
- Lechner E., Achard P., Vansiri A., Potuschak T. & Genschik P. F–box proteins everywhere. Curr. Opin. Plant Biol. 9, 631–638, 10.1016/j.pbi.2006.09.003 (2006). [DOI] [PubMed] [Google Scholar]
- Calderon Villalobos L. I. et al. A combinatorial TIR1/AFB–Aux/IAA co–receptor system for differential sensing of auxin. Nat. Chem. Biol. 8, 477–485, 10.1038/nchembio.926 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S., Forno D. A., Cook J. H. & Gomez K. A. Routine procedures for growing rice plants in culture solution. In: Yoshida S., Forno D. A., Cook J. H., Gomez K. A. eds. Laboratory Manual for Physiological Studies of Rice: International Rice Research Institute, Los Banos, Philippines, 61–66 (1976).
- Xia K. et al. OsTIR1 and OsAFB2 downregulation via OsmiR393 overexpression leads to more tillers, early flowering and less tolerance to salt and drought in rice. PloS One 7, e30039, 10.1371/journal.pone.0030039 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M. et al. A programmable dual–RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821, 10.1126/science.1225829 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. & Kim J. S. A guide to genome engineering with programmable nucleases. Nat. Rev. Genet. 15, 321–334, 10.1038/nrg3686 (2014). [DOI] [PubMed] [Google Scholar]
- Shan Q. et al. Targeted genome modification of crop plants using a CRISPR–Cas system. Nat. Biotechnol. 31, 686–688, 10.1038/nbt.2650 (2013). [DOI] [PubMed] [Google Scholar]
- van der Knaap E., Kim J. H. & Kende H. A novel gibberellin–induced gene from rice and its potential regulatory role in stem growth. Plant Physiol. 122, 695–704 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Rice J. H., Chen N., Baum T. J. & Hewezi T. Synchronization of developmental processes and defense signaling by growth regulating transcription factors. PloS One 9, e98477, 10.1371/journal.pone.0098477 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R. & Estelle M. Diversity and specificity: auxin perception and signaling through the TIR1/AFB pathway. Curr. Opin. Plant Biol. 21, 51–58, 10.1016/j.pbi.2014.06.006 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyaboga E. N., Njiru J. M. & Tripathi L. Factors influencing somatic embryogenesis, regeneration, and Agrobacterium–mediated transformation of cassava (Manihot esculenta Crantz) cultivar TME14. Front. Plant Sci. 6, 411, 10.3389/fpls.2015.00411(2015) . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez L. et al. Phenotypic plasticity of adventitious rooting in Arabidopsis is controlled by complex regulation of AUXIN RESPONSE FACTOR transcripts and microRNA abundance. Plant Cell 21, 3119–3132, 10.1105/tpc.108.064758 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X., Chen J. & Yang Z. Auxin regulation of cell polarity in plants. Curr. Opin. Plant Biol. 28, 144–153, 10.1016/j.pbi.2015.10.009 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung H., Lee D. K., Choi Y. D. & Kim J. K. OsIAA6, a member of the rice Aux/IAA gene family, is involved in drought tolerance and tiller outgrowth. Plant Sci. 236, 304–312, 10.1016/j.plantsci.2015.04.018 (2015). [DOI] [PubMed] [Google Scholar]
- Leyser O. Auxin signalling: the beginning, the middle and the end. Curr. Opin. Plant Biol. 4, 382–386 (2001). [DOI] [PubMed] [Google Scholar]
- Xu C. et al. Degradation of MONOCULM 1 by APC/C (TAD1) regulates rice tillering. Nat Commun. 3, 750, 10.1038/ncomms1743 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda T. et al. The OsTB1 gene negatively regulates lateral branching in rice. Plant J. 33, 513–520 (2003). [DOI] [PubMed] [Google Scholar]
- Lin H. et al. DWARF27, an iron–containing protein required for the biosynthesis of strigolactones, regulates rice tiller bud outgrowth. Plant Cell 21, 1512–1525, 10.1105/tpc.109.065987 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z. et al. Genome–wide binding analysis of the transcription activator ideal plant architecture1 reveals a complex network regulating rice plant architecture. Plant Cell 25, 3743–3759, 10.1105/tpc.113.113639 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso C. et al. Natural variation of rice strigolactone biosynthesis is associated with the deletion of two MAX1 orthologs. Proc. Natl. Acad. Sci. USA 111, 2379–2384, 10.1073/pnas.1317360111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang D. et al. Strigolactones regulate rice tiller angle by attenuating shoot gravitropism through inhibiting auxin biosynthesis. Proc. Natl. Acad. Sci. USA 111, 11199–11204, 10.1073/pnas.1411859111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry G. et al. Complex regulation of the TIR1/AFB family of auxin receptors. Proc. Natl. Acad. Sci. USA 106, 22540–22545, 10.1073/pnas.0911967106 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H. et al. Heterotrimeric G proteins regulate nitrogen–use efficiency in rice. Nat. Genet. 46, 652–656, 10.1038/ng.2958 (2014). [DOI] [PubMed] [Google Scholar]
- Smit M. E. & Weijers D. The role of auxin signaling in early embryo pattern formation. Curr. Opin. Plant Biol. 28, 99–105, 10.1016/j.pbi.2015.10.001 (2015). [DOI] [PubMed] [Google Scholar]
- Dun E. A., Ferguson B. J. & Beveridge C. A. Apical dominance and shoot branching. divergent opinions or divergent mechanisms? Plant Physiol. 142, 812–819, 10.1104/pp.106.086868 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windels D. & Vazquez F. miR393: integrator of environmental cues in auxin signaling? Plant Signal Behav. 6, 1672–1675, 10.4161/psb.6.11.17900 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windels D. et al. miR393 is required for production of proper auxin signalling outputs. PloS One 9, e95972, 10.1371/journal.pone.0095972 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu G. et al. OsPIN5b modulates rice (Oryza sativa) plant architecture and yield by changing auxin homeostasis, transport and distribution. Plant J. 83, 913–925, 10.1111/tpj.12939 (2015). [DOI] [PubMed] [Google Scholar]
- Yu H. et al. Untethering the TIR1 auxin receptor from the SCF complex increases its stability and inhibits auxin response. Nature Plants 1, 10.1038/nplants.2014.30 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu–Sato S., Tanaka M. & Mori H. Auxin–cytokinin interactions in the control of shoot branching. Plant Mol. Biol. 69, 429–435, (2009). [DOI] [PubMed] [Google Scholar]
- Ding C., You J., Chen L., Wang S. & Ding Y. Nitrogen fertilizer increases spikelet number per panicle by enhancing cytokinin synthesis in rice. Plant Cell Rep. 33, 363–371, (2014). [DOI] [PubMed] [Google Scholar]
- Hirose N. et al. Regulation of cytokinin biosynthesis, compartmentalization and translocation. J. Exp. Bot. 59, 75–83, 10.1093/jxb/erm157 (2008). [DOI] [PubMed] [Google Scholar]
- Barazesh S. & McSteen P. Hormonal control of grass inflorescence development. Trends Plant Sci. 13, 656–662, 10.1016/j.tplants.2008.09.007 (2008). [DOI] [PubMed] [Google Scholar]
- Miyawaki K., Matsumoto–Kitano M. & Kakimoto T. Expression of cytokinin biosynthetic isopentenyltransferase genes in Arabidopsis: tissue specificity and regulation by auxin, cytokinin, and nitrate. Plant J. 37, 128–138 (2004). [DOI] [PubMed] [Google Scholar]
- Takei K. et al. AtIPT3 is a key determinant of nitrate–dependent cytokinin biosynthesis in Arabidopsis. Plant Cell Physiol. 45, 1053–1062, 10.1093/pcp/pch119 (2004). [DOI] [PubMed] [Google Scholar]
- Exner V., Hirsch–Hoffmann M., Gruissem W. & Hennig L. PlantDB – a versatile database for managing plant research. Plant Methods 4, 1, (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y. J. et al. Establishment of highefficiency Agrobacterium–mediated genetic transformation system of Mudanjiang. Acta Agron. Sin. 28, 294–300 (2002). [Google Scholar]
- Chen C. et al. Real–time quantification of microRNAs by stem–loop RT–PCR. Nucleic Acids Res. 33, e179, 10.1093/nar/gni178 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B. et al. Loss of function of OsDCL1 affects microRNA accumulation and causes developmental defects in rice. Plant Physiol. 139, 296–305, 10.1104/pp.105.063420 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y. Y. et al. Activation of the WUS gene induces ectopic initiation of floral meristems on mature stem surface in Arabidopsis thaliana. Plant Mol. Biol. 57, 773–784, (2005). [DOI] [PubMed] [Google Scholar]
- Zhang Y. et al. A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast–related processes. Plant Methods 7, 30, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu L., Zhang Y., Pei Y., Liu C. & Cao X. Redundant requirement for a pair of PROTEIN ARGININE METHYLTRANSFERASE4 homologs for the proper regulation of Arabidopsis flowering time. Plant Physiol. 148, 490–503, 10.1104/pp.108.124727 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.