Abstract
Objective
To evaluate the impact of mitochondrial DNA (mtDNA) haplogroups on virologic and immunological outcomes of HIV infection.
Design
HAART-naive African American adolescent participants to the Reaching for Excellence in Adolescent Care and Health study.
Methods
The mtDNA haplogroups were inferred from sequenced mtDNA hypervariable regions HV1 and HV2 and their predictive value on HIV outcomes were evaluated in linear mixed models, controlled for human leukocyte antigen (HLA)-B27, HLA-B57 and HLA-B35-Px alleles and other covariates.
Results
We report data showing that the mtDNA L2 lineage, a group composed of L2a, L2b and L2e mtDNA haplogroups in the studied population, is significantly associated (beta=−0.08; Bonferroni-adjusted P=0.004) with decline of CD4+ T cells (median loss of 8 ± 1 cells per month) in HAART-naive HIV-infected individuals of African American descent (n=133). No significant association (P<0.05) with set-point viral load was observed with any of the tested mtDNA haplogroups. The present data concur with previous findings in the AIDS Clinical Trials Group study 384, implicating the L2 lineage with slower CD4+ T-cell recovery after antiretroviral therapy in African Americans.
Conclusions
Whereas the L2 lineage showed an association with unfavorable immunological outcomes of HIV infection, its phylogenetic divergence from J and U5a, two lineages associated with accelerated HIV progression in European Americans, raises the possibility that interactions with common nucleus-encoded variants drive HIV progression. Disentangling the effects of mitochondrial and nuclear gene variants on the outcomes of HIV infection is an important step to be taken toward a better understanding of HIV/AIDS pathogenesis and pharmacogenomics.
Keywords: AIDS, CD4+ cell count, HIV, HLA, mitochondrial haplogroup, viral load
Introduction
Until the turn of the century, the bulk of evidence for the link between HIV progression and genetic variation came from candidate gene approaches that have implicated immunomodulatory molecules encoded by the human leukocyte antigen (HLA) system [1–6], notably HLA-B, along with the CC chemokine receptor and ligand (CCR2, CCR5 and CXCL12 known as SDF-1) genes ([7]; for a review). In recent years, several genome-wide association studies (GWAS) have been conducted to search for additional host genetic factors influencing HIV pathogenesis [8–16]. These genome-wide searches have focused on a restricted and largely overlapping set of phenotypes including set-point viral load (VL), and have resulted in striking converging results on the major histocompatibility complex (MHC) loci HPC5, HLA-B and HLA-C. In an early study [17], we were able to replicate certain of these findings in the predominantly African American Reaching for Excellence in Adolescent Care and Health (REACH) cohort; this has validated the use of set-point VL as a proxy for HIV progression in REACH cohort. Most importantly, in more recent studies that have refined the phenotypes, new susceptibility loci outside the MHC region have been identified ([18]; for a review). These new findings suggest that mechanisms of pathogenesis other than those mediated by the classical innate and adaptive immune pathways might be implicated in HIV progression and that the phenotypic heterogeneity of HIV pathogenesis has a genetic basis yet to be uncovered.
Very little is known about the effect of mitochondrial DNA (mtDNA) variation on HIV disease progression in treatment-naÏve study populations of African descent. Variants of mtDNA carried on population-specific mtDNA haplogroups may affect HIV progression through changes in mitochondrial oxidative phosphorylation, reactive oxygen species production or apoptosis [19]. mtDNA contains two short (about 400 base pairs of length) hypervariable regions (HV1 and HV2) that are part of the noncoding control region of the D-loop. HV1 and HV2 regions are the most rapidly evolving sequences within the D-loop. These highly polymorphic mtDNA regions define population-specific mitochondrial haplogroups, and we have sequenced them in the REACH cohort [20].
Survival studies in the Multicenter AIDS Cohort Study (MACS) and in other cohort studies of the HIV-infected populations of European descent indicated that mtDNA haplogroups J and U5a are associated with accelerated AIDS progression [21]. Because mtDNA haplogroups J and U5a occur on divergent mitochondrial clades, we set out to evaluate the association of mtDNA haplogroups with HIV outcomes in a population of African descent to get more insights into its meaning. In effect, in the absence of recombination, mtDNA variants are expected to be in cytonuclear disequilibrium with the maternally inherited nuclear DNA variants, making association data with mtDNA variants highly prone to confounding by nuclear variants [22]. A straightforward approach to cross-validate the data is to test the association in populations with distinct mtDNA composition. In this case, it would be possible to exclude potential confounding by cytonuclear disequilibrium if a phylogenetically related mtDNA haplogroup, that is a haplogroup sharing the same most recent common ancestor (MRCA), is associated with the phenotype or trait of interest in the validation study population. With the L mtDNA haplogroups representing the bulk of mtDNA diversity in populations of African descent and being quasi-absent in the European populations, the likelihood of confounding by cytonuclear disequilibrium is predicted if the associated mtDNA haplogroup in the validation study population (here African American) is of the L haplogroup category with no shared MRCA with the European lineages implicated in HIV progression.
In the present study, we evaluated the association of mtDNA haplogroups with virologic (set-point VL) and immunologic (CD4+ T-cell trajectory) outcomes in the REACH participants of African American descent who were HAART-naive at baseline, and we report positive associations of CD4+ T-cell trajectories with macro-haplogroup L2.
Methods
Study population
The study population consisted of eligible participants of self-reported African American descent in REACH, an observational cohort study of HIV-1-seropositive and high-risk seronegative adolescents (12–18 years old) recruited from 15 clinical sites in 13 US cities between 1996 and 2000. All infected patients had a positive ELISA, with a confirmatory western blot performed before REACH enrollment. Participants were followed on a quarterly basis for epidemiologic, clinical, and laboratory evaluations, including documentation of demographic and risk behaviors, collection of medical history and various biological samples, along with tests for HIV-1 infection, other sexually transmitted infections, CD4+ T-cell count, and immunological outcomes.
The parent study and this substudy conformed to the procedures for informed consent (parental permission was obtained whenever required) approved by institutional review boards at all sponsoring organizations and to human experimentation guidelines set forth by the US Department of Health and Human Services.
Study design
Nearly all of the selected youth were HIV-1-seroprevalent at study entry, but the duration of infection was relatively short as judged by sexual debut and other risk behaviors [23]. The inclusion criteria were availability of good-quality DNA, non-Hispanic African American ethnicity and longitudinal measures of VL and CD4+ T-cell count. The exclusion criteria were HAART treatment at baseline and data points after treatment initiation. Two independent measures of HIV-related outcomes were used to gauge the likelihood of disease progression.
Set-point viral load
Estimation of set-point VL, a known predictor of HIV progression, has been described in a previous study in REACH [17] based on the same sample. Briefly, we used the covariance structure model of the first 2–4 sequential VL measures, with intervisit VL variation not greater than 0.5 log10 copies per ml of plasma, as steady set-point values. Of note, in most cases, duration of infection, as inferred from young age (median at 17), self-reported sexual debut (not earlier than age 13), and other risk behaviors (no vertical transmission), should be no more than 5 years when VL measures were taken. As a result, stable VL at two or more HAART-naive clinical visits was treated as a proxy for set-point VL. Of the 192 eligible participants, 121 (102 women and 19 men) had measurements of set-point VL and among them, a group of 99 individuals of African American descent was sampled for the study of set-point VL outcome.
CD4+ T-cell count trajectory
CD4+ T-cell count trajectories during HAART-free periods were estimated from baseline to the clinical visits at which HAART was initiated [median 5; interquartile range (IQR) (3, 10)]. Longitudinal measurements of CD4+ T-cell count were available for additional participants of African or non-African descent allowing different sensitivity analyses. These study samples consisted of the above set-point VL sample (n=99), an expanded non-Hispanic African American group (n=133) sampled for the study of the CD4+ trajectory and a slightly larger sample that included all ethnic groups (n=166) (Supplementary Fig. S1, http://links.lww.com/QAD/A542).
Mitochondrial DNA sequencing
The experimental design and bioinformatics treatment of nucleotide sequence data have been described [20]. Briefly, DNA extracted from buffy coat samples (Qiagen DNA purification kit) was used as templates in PCR to amplify the hypervariable regions HV1 and HV2 as described [20].
DNA typing
Genotyping of several nuclear loci including HLA-B and HLA-C loci have been reported in previous REACH studies [24,25].
Mitochondrial DNA haplogroup assessment and quality control
Following bidirectional DNA sequencing of HV1 and HV2 regions, the quality of forward and reverse sequences was calculated in CodonCode Aligner 3.7.1.1, and sequences with scores 300 or less were discarded. Consensus sequences were assembled using default conditions and poor-quality regions were re-sequenced at least once. Regions that were not resolved by the sequencing assay (runs of ‘n’ at both ends of the sequences or around the stretch of repetitive ‘C’ nucleotides in the HVI sequence) were coded as missing. Only unique haplotypes were included in the reconstruction of the phylogenetic tree, as duplicate haplotypes do not affect tree structure or support, thus reducing computation burden. All analyses were run assuming an unequal transition/transversion ratio and an equal mutation rate across sites. The likelihood analysis consisted of 100 separate heuristic searches and 1000 bootstrap replicates were performed using the GTRCAT model of evolution. All sequence designations are based on comparisons to the revised Cambridge reference sequence (rCRS) [26].
The standard MitoTool [27] and mthap [28] programs were used to infer mtDNA haplogroup membership on the basis of HV1 and HV2 nucleotide sequences that passed the quality control filters above. Combined classification data from MitoTool and mthap, and from our reconstructed phylogenetic tree (not shown), were used to cross-validate the inferred haplogroups and resolve discrepancies. Specifically, phylogenetically defined branches and clades were used to assign a likely haplogroup to individuals for whom classification by MitoTool and mthap have failed or produced ambiguous haplogroups (more than one possible haplogroup per individual).
Self-report of population of origin and ethnicity was substantiated by discriminant analysis of a validated set of 150 ancestry informative markers (AIMs) typed in a previous REACH study [25].
Statistical analysis
Association with set-point VL was tested in univariate models comparing estimated mean set-point VL by each mtDNA macrohaplogroup (or mtDNA haplogroup in sensitivity analyses) to that of all other haplogroups (carrier status), and adjusting for the effects of sex, baseline age and HLA alleles known to delay (HLA-B*57 or HLA-B*27) or accelerate (HLA-B*35 Px) HIV progression and for multiple comparisons (Tukey–Kramer adjustment). Set-point VL means were estimated in linear mixed models using an autoregressive covariance structure to accommodate the steady VL across the set-point phase and to account for the correlated data within patients. As noted above, the follow-up time in this HIV cohort of youth is relatively short (4 years). Therefore significant differences (P<0.05) in the least squares mean of log10 set-point VL between carriers and noncarriers of the test haplogroup that were larger than 0.75 log10 copies per ml of plasma (1.5-fold higher than the defined steady set-point value) were deemed meaningful.
The change of CD4+ T-cell count over time (CD4+ T-cell count trajectory) was assessed by the interaction term (haplogroup × time) in linear mixed models with adjustment for the above covariates. A random intercept effect was included in the models to account for the group differences in the CD4+ T-cell counts at baseline.
All analyses were performed in SAS (SAS Institute, Cary, North Carolina, USA) using normalized data (square root of CD4+ T-cell count and log10 of set-point viral load). Bonferroni-corrected thresholds (8.3×10−3 – 6.2×10−3 range for the 6–8 tests performed) were used to assess the significance of the associations and two-sided P values from t tests are reported.
Results
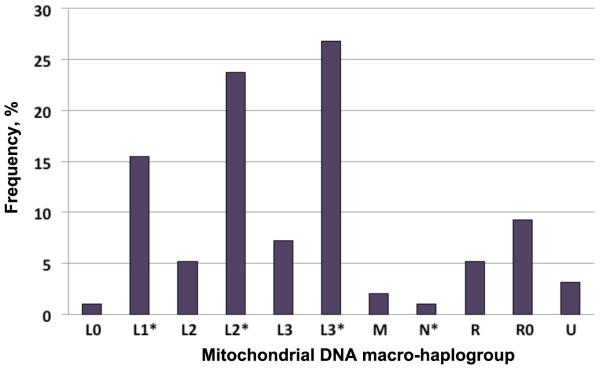
The demographic and clinical characteristics of the African American REACH population are depicted in Table 1. Principal component analysis (PCA) based on a validated set of 150 nuclear AIMs typed in the predominantly African American REACH cohort previously enabled us to identify and exclude study participants of admixed population descent, namely, other than non-Hispanic African or European descent [20]. In that study (n=343), from which the present study sample was derived, we observed 17 out of the 18 mtDNA haplogroups reported so far for populations of African American descent, indicating a good representation of the African American population diversity in our studied REACH sample. Because the present study sample was a random selection based on DNA availability, no major distortion in the distribution of the common African American mtDNA haplogroups was observed (Fig. 1). To increase the study statistical power, mtDNA haplogroups sharing the same MRCA defined according to the global human mtDNA phylogenetic tree (Supplementary Fig. S2, http://links.lww.com/QAD/A542) were grouped together as macrohaplogroups. Haplogroups defined with different resolutions by MitoTool and mthap (e.g. L2 for L2a) represented the majority of the observed ambiguities (about 7%); with the adopted grouping procedure, these haplogroups felt under the same macrohaplogroups. As reported in the early study [20], about 2.9% of self-reported Caucasian descents were re-classified by PCA as African American descents.
Table 1.
Demographic and clinical characteristics of a subset of 133 non-Hispanic African Americans of the REACH cohort sampled for the mitochondrial study*
| Characteristics | Female | Male | all |
|---|---|---|---|
|
|
|
|
|
| Number of participants count (%) |
114 (85.7) | 19 (14.3) | 133 (100) |
| Age at baseline, years median (IQR) |
17 (16, 18) | 17 (17, 18) | 17 (17, 18) |
| Follow-up medical visits**
median (IQR) |
5 (3, 11) | 5 (4, 7) | 5 (3, 10) |
| CD4+ T cells/mm3
median (IQR) |
546 (412, 745) | 502 (349, 576) | 543 (404, 713) |
| HIV-1 Log10 RNA copies/ml median (IQR) |
3.4 (2.5, 4.1) | 4.0 (3.3, 4,7) | 3.5 (2.6, 4.2) |
Footnote to Table 1
Only participants who were HAART-naïve at baseline and subsequent follow-medical visits were sampled.
Median number of quarterly follow-up HAART-naive medical visits. (IQR) Interquartile range.
Fig. 1. Mitochondrial DNA diversity in the African American group of the Reaching for Excellence in Adolescent Care and Health cohort.
Histogram shows the composition of mtDNA macrohaplogroups in the study sample. Macrohaplogroups define major human lineages consisting of closely related haplogroups (Supplementary Fig. S2, http://links.lww.com/QAD/A542). In the present study sample, L1* comprised two common mtDNA haplogroups (L1b and L1c), L2* (L2a, L2b and L2e) and L3* (L3b, L3d, L3e, L3f, L3h and L3x). The non-L macrohaplogroups comprised other mtDNA haplogroups as indicated in Fig. S2. mtDNA, mitochondrial DNA; REACH, Reaching for Excellence in Adolescent Care and Health.
Fitting linear mixed models to the set-point viral load data with adjustments for the indicated covariates and for multiple comparisons (Tukey–Kramer adjustment) showed that none of the tested macrohaplogroups predicted significantly (least square mean difference > threshold 0.75 log10 copies/ml and P<0.05) low or high set-point viral load (Table 2). A borderline association with higher set-point viral load [least square mean difference=0.44 log10 copies/ml, standard error (SE)=0.27, P=0.105] was, however, observed with the R0 macrohaplogroup, which in our study is composed of the West-Eurasian H, V, HV haplogroups and their MRCA R0 haplogroup (Fig. S2). Expansion of the study sample to all ethnic groups (n=99) resulted in a slight increase of the association strength between R0 and set-point VL (beta= =0.48, SE=0.25, P=0.069) (not shown).
Table 2.
Mitochondrial haplogroups and virologic and immunological HIV outcomes in 91 HAART-naïve African American adolescents of the REACH study
| linear mixed model¶ | ||||||||
|---|---|---|---|---|---|---|---|---|
|
|
||||||||
| setpoint viral load | CD4+ T cell count trajectory | |||||||
|
|
|
|||||||
| Mitochondrial macrohaplogroup |
frequency | coefficient | (SE) | p § | frequency | coefficient | (SE) | p §† |
|
|
|
|||||||
| L1* | 17 | −0.29 | (0.19) | 0.143 | 17 | 0.007 | (0.024) | 0.815 |
| L2 | 6 | 0.15 | (0.30) | 0.618 | 6 | −0.150 | (0.095) | 0.121 |
| L2* | 22 | −0.02 | (0.18) | 0.925 | 22 | −0.070 | (0.024) | 0.002 |
| L3 | 6 | −0.34 | (0.30) | 0.249 | 6 | 0.051 | (0.075) | 0.499 |
| L3* | 30 | 0.11 | (0.16) | 0.479 | 30 | 0.016 | (0.021) | 0.450 |
| R0 | 8 | 0.44 | (0.27) | 0.105 | 6 | 0.015 | (0.042) | 0.731 |
| U | − | − | − | − | 4 | 0.040 | (0.034) | 0.429 |
| L2 + L2* | 28 | 0.15 | (0.30) | 0.618 | 28 | −0.081 | (0.023) | 0.0005 |
Footnote to Table 2
Macro-haplogroups as defined in the histogram of Figure 1. Note that non-L haplogroups H, HV, V and root R0 were grouped as R0, and the U and K haplogroups as U.
Models adjusted for the effects of gender, baseline age and HLA-B.
p-values adjusted for multiple comparisons (Tukey-Kramer adjustment). Note that only clades or macrohaplogroups with a frequency greater than 3% are shown. SE=Standard Error of the mean.
Parameter estimates for the covariates in the model with L2 & L2* as predictors were for HLA-B*35 (beta= −1.62; SE=1.81; p= 0.370), HLA-B*57 or HLA-B*27 (beta=3.01; SE=1.44; p=0.037), gender (beta= −1.45; SE=1.30; p =0.265) and for age (beta= 0.066; SE=0.25; p=0.795).
We further tested whether mtDNA haplogroups influence the CD4+ T-cell count trajectory over HAART-naÏve follow-up medical visits [median 6; IQR (4, 10); range (2, 17)] and observed that the L2* macrohaplogroup (L2a, L2b and L2e) predicted a significant decline (beta=−0.07, SE=0.024, P=0.002) of CD4+ T-cell counts in African Americans (Table 2). Consistently, the root L2 haplogroup was the only other haplogroup associated, albeit marginally (P=0.12), with a negative and steeper slope of CD4+ T-cell count (beta=−0.15, SE=0.095). With L2 being the MRCA to the haplogroups grouped under L2*, we evaluated the combined effect of L2 and L2* and observed a stronger (beta=−0.081, SE=0.023) and more significant (adjusted P=0.0005) association with the decline of CD4+ T-cell count over the follow-up interval in carriers vs. noncarriers. Specifically, the higher rate of CD4+ T-cell loss in the combined group L2 and L2* was 8 ± 1 cells per month.
Re-evaluation of the associations in an expanded sample (Fig. S1) of non-Hispanic African Americans (n=133) showed similar trends for L2 and L2*, and their combined effects (Table 3). Comparable results were obtained when the study sample was expanded (n=166) to include individuals of European (n=9), Hispanic African (n=11) and other (n=13) population descents (Supplementary Table S1, http://links.lww.com/QAD/A542). In this case, however, the significance of the association with L2* (beta=−0.046, SE=0.020, P=0.024) and with the combination of L2 and L2* (beta=−0.052, SE=0.020, P=0.009) was lost after Bonferroni correction. The sole haplogroup that remained significantly (P<0.05) associated with the CD4+ T-cell count outcome was the root L3 haplogroup, which predicted no decline of the CD4+ T-cell count (beta=0.177, SE=0.063, P=0.005).
Table 3.
Mitochondrial haplogroups and immunological HIV outcomes in HAART-naïve non-Hispanic African American adolescents of the REACH study
| CD4+ trajectory (n=133) | linear mixed model¶ | ||
|---|---|---|---|
|
|
|
||
| coefficient | (SE) | p §† | |
| L0 (n=5) | 0.063 | (0.037) | 0.086 |
| L1* (n=27) | 0.035 | (0.026) | 0.183 |
| L2 (n=7) | −0.162 | (0.101) | 0.109 |
| L2* (n=31) | −0.061 | (0.022) | 0.007 |
| L3 (n=9) | 0.029 | (0.074) | 0.696 |
| L3* (n=36) | −0.002 | (0.020) | 0.915 |
| R0 (n=9) | −0.007 | (0.031) | 0.831 |
| U (n=4) | 0.029 | (0.036) | 0.427 |
| L2 + L2* (n=38) | −0.068 | (0.022) | 0.002 |
Footnote to Table 3
macro-haplogroups as defined in the histogram of Figure 1. Note that non-L haplogroups H, HV, V and root R0 were grouped as R0, and the U and K haplogroups as U.
models adjusted for the effects of gender, baseline age and HLA-B.
p-values adjusted for multiple comparisons (Tukey-Kramer adjustment). Note that only clades or macro-haplogroups with a frequency greater than 3% are shown. SE=Standard error of the mean.
Parameter estimates for the covariates in the model with L2 & L2* as predictors were for HLA-B*35 (beta= −1.21; SE=1.46; p= 0.408), HLA-B*57 or HLA-B*27 (beta=3.87; SE=1.52; p=0.011), gender (beta= − 1.80; SE=1.24; p=0.147) and for age at baseline (beta= −0.28; SE=0.240; p=0.238).
Discussion
We report the first data on the association of mtDNA variation with the virologic and immunological outcomes in HAART-naive HIV-infected African Americans. The data indicated that carriage of the African lineage L2 is associated with unfavorable immunological outcomes of HIV infection. Specifically, we report that compared to the other mtDNA lineages, L2 is associated with a higher loss of CD4+ T-cells (8±1 cells per month) in HAART-naive non-Hispanic African American adolescents. We have also shown that except for the marginal and sole association of R0 with increased set-point VL, none of the other mtDNA lineages affected this outcome. Most importantly, the present data are consistent with those of the AIDS Clinical Trials Group (ACTG) study, which reported a decreased likelihood of CD4+ T-cell count recovery (≥100 cells per cubic millimiter CD4+ T-cell count increase) at week 48 of antiretroviral therapy (ART) among carriers of L2 [29].
As can be expected from a study population of adolescents, the narrow age range of the REACH participants (median 17 years, IQR 17, 18) and the slower HIV progression in young vs. older participants [30] likely precluded detection of an effect of this covariate, if any, on the CD4+ trajectory. Consistent with the published data in REACH and other HIV cohort studies of natural history of HIV HLA -B*57, -B*27 and -B*35 had the expected effects on the CD4+ trajectory in the model testing the L2 lineage in the largest sample (Table S1). Because these HLA-B effects were controlled for in all our models, the reported associations with the L2 lineage are independent of this important AIDS restriction gene.
Nonetheless, the observation of strict effects of mtDNA variation on set-point VL or CD4+ T-cell count trajectory and not on both outcomes in the present study (the virologic outcome was not studied in the ACTG study) raises questions. First of all, whereas our main hypothesis was that specific mitochondrial lineages predict favorable or unfavorable virologic or immunological outcomes in the African American population in the absence of ART, the corollary hypothesis is that observed associations might reflect confounding by the nuclear background if the predictor mitochondrial lineages are different or share no MRCA among ethnic groups.
The rationale for this latter hypothesis is that by virtue of cytonuclear disequilibrium [31] (another form of linkage disequilibrium), any variants in the mitochondrial genomes can be proxy for nuclear variants and vice versa [22]. Reported associations of mitochondrial lineages with HIV outcomes may point to true mtDNA variant effects, uncontrolled nuclear variant effects or joint effects of both types of variants. One approach that potentially helps distinguish these alternatives is to conduct replication studies in populations with divergent mtDNA composition. In consideration of the above hypothesis, it is interesting to note that R0, the West-Eurasian mtDNA haplogroup that shares MRCAR with J and U5a, was the only haplogroup that showed an association, albeit marginal, with high set-point VL. The marginal association of this non-L haplogroup in our study population may simply be due to the moderate statistical power. Our queries of reference mtDNA databases [28] showed that the only diagnostic position shared solely by J, U5 and L2* (a mixture of L2a, L2b and L2e in our sample) is G13708A, a nonsynonymous polymorphism in the ND5 (NADH dehydrogenase, subunit 5) gene; however, this ND5 variant is present in only two carriers of the rare L2e haplogroup. Thus, whereas low levels of shared variants are predictable for divergent mitochondrial lineages, we cannot fully assess functional polymorphisms in the mitochondrial genome.
Because both the MACS and REACH studies controlled for HLA (and non-HLA loci as well in MACS) known to influence HIV progression, other AIDS-restriction genes become potential targets. Among such targets those encoding the plethoric (n > 966) class of nuclear-encoded mitochondrial proteins (NEMPs) contains strong candidates and some of them have been investigated in MACS [32]. As previously reported in some severe encephalomyopathies [33] and cardiomyopathies [34], functional NEMP variants may become clinically relevant if combined with incompatible mitochondrial lineages. Similarly, de-novo mutations in mtDNA genes (heteroplasmy) may become pathogenic in individuals with predisposing mutations in NEMP genes. Alternatively, with the emerging roles for the mitochondrial genome in nuclear gene expression [35,36], polymorphisms affecting the interplay between nuclear and mitochondrial genes may increase the risk for HIV progression through attenuation or exhaustion of protective cellular mechanisms co-regulated by the two genomes such as apoptosis. Whereas neither refinement of the phenotypes nor consideration of alternative clinical end points is an option in the REACH study, the observed association of set-point VL with mtDNA haplogroups sharing common root with those implicated in AIDS progression in Caucasians may reflect sharing of specific nuclear genetic factors underlying unmeasured subphenotypes. In other words, mtDNA haplogroups may breakdown nuclear genetic heterogeneity influencing the spectrum of HIV disease progression phenotypes and thus increase the power to detect new susceptibility loci.
We are aware of the present study limitations. First, ART treatments received prior to study entry or later during follow-ups precluded the inclusion of the entire African American group for whom mtDNA typing is available (n=347). Second, the follow-up time in REACH is short (4 years) to allow time to event studies or pharmacogenomic studies. Third, study samples of admixed population descent can make difficult the interpretation of association data due to the maternal inheritance of mtDNA. However, as shown here, such studies can also provide clues to refine hypotheses.
In conclusion, we observed a significant association of mitochondrial L2 lineages with the decline of CD4+ T-cell numbers and a marginal association of R0 lineages with slightly elevated set-point VL in HAART-naÏve study population of African American descent. Future studies in larger HIV cohorts are required to validate the present findings and most importantly to investigate further mitochondrial and nuclear genomes for functional variants shared by carriers of L2, J and U5a mtDNA haplogroups.
Because of the genetic hypervariability of the D-loop region, most common mtDNA haplogroups can be tagged by unique diagnostic positions. If validated, this approach would offer an opportunity for developing affordable genetic tests to identify early in their asymptomatic phase those individuals at risk for unfavorable immunological outcomes of HIV infection. Detecting genetic markers validated as predictive of unfavorable HIV outcomes may improve the management of this condition, particularly in resource limited settings.
Supplementary Material
Acknowledgements
We thank the REACH participants as well as the investigators and staff involved in the study (listed in J Adolesc Health 2001; 29:S5–S6) for their valuable contributions. This research was funded in part by the University of Alabama at Birmingham School of Public Health (B.A.). The REACH study (1996–2001) was supported by the National Institute of Child Health and Human Development (U01-HD32842), with supplemental funding from the NIAID, the National Institute on Drug Abuse, and the National Institute of Mental Health.
B.A. supervised the study design, data analyses and writing of the manuscript. S.S. provided analytic support for the study design and data analyses. H.W.W. performed quality control of the data and statistical analyses. J.T. and R.A.K. provided immunogenetic expertise support with laboratory methods. C.W. assisted with clinical data of the REACH participants. All the authors participated in the writing and the reading of the article and have seen and approved the final version of the manuscript.
Footnotes
Conflicts of interest
The authors of the present study have no conflicts of interest to declare.
References
- 1.Saah AJ, Hoover DR, Weng S, Carrington M, Mellors J, Rinaldo CR, Jr, et al. Association of HLA profiles with early plasma viral load, CD4+ cell count and rate of progression to AIDS following acute HIV-1 infection. Multicenter AIDS Cohort Study. AIDS. 1998;12:2107–2113. doi: 10.1097/00002030-199816000-00005. [DOI] [PubMed] [Google Scholar]
- 2.Keet IP, Tang J, Klein MR, LeBlanc S, Enger C, Rivers C, et al. Consistent associations of HLA class I and II and transporter gene products with progression of human immunodeficiency virus type 1 infection in homosexual men. J Infect Dis. 1999;180:299–309. doi: 10.1086/314862. [DOI] [PubMed] [Google Scholar]
- 3.Hendel H, Caillat-Zucman S, Lebuanec H, Carrington M, O’Brien S, Andrieu JM, et al. New class I and II HLA alleles strongly associated with opposite patterns of progression to AIDS. J Immunol. 1999;162:6942–6946. [PubMed] [Google Scholar]
- 4.Magierowska M, Theodorou I, Debre P, Sanson F, Autran B, Riviere Y, et al. Combined genotypes of CCR5, CCR2, SDF1, and HLA genes can predict the long-term nonprogressor status in human immunodeficiency virus-1-infected individuals. Blood. 1999;93:936–941. [PubMed] [Google Scholar]
- 5.Tang J, Costello C, Keet IP, Rivers C, Leblanc S, Karita E, et al. HLA class I homozygosity accelerates disease progression in human immunodeficiency virus type 1 infection. AIDS Res Hum Retroviruses. 1999;15:317–324. doi: 10.1089/088922299311277. [DOI] [PubMed] [Google Scholar]
- 6.Carrington M, Nelson GW, Martin MP, Kissner T, Vlahov D, Goedert JJ, et al. HLA and HIV-1: heterozygote advantage and B*35-Cw*04 disadvantage. Science. 1999;283:1748–1752. doi: 10.1126/science.283.5408.1748. [DOI] [PubMed] [Google Scholar]
- 7.Ioannidis JP, Rosenberg PS, Goedert JJ, Ashton LJ, Benfield TL, Buchbinder SP, et al. Effects of CCR5-Delta32, CCR2-64I, and SDF-1 3’A alleles on HIV-1 disease progression: an international meta-analysis of individual-patient data. Ann Intern Med. 2001;135:782–795. doi: 10.7326/0003-4819-135-9-200111060-00008. [DOI] [PubMed] [Google Scholar]
- 8.Fellay J, Shianna KV, Ge D, Colombo S, Ledergerber B, Weale M, et al. A whole-genome association study of major determinants for host control of HIV-1. Science. 2007;317:944–947. doi: 10.1126/science.1143767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dalmasso C, Carpentier W, Meyer L, Rouzioux C, Goujard C, Chaix ML, et al. Distinct genetic loci control plasma HIV-RNA and cellular HIV-DNA levels in HIV-1 infection: the ANRS Genome Wide Association 01 study. PLoS One. 2008;3:e3907. doi: 10.1371/journal.pone.0003907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Le Clerc S, Limou S, Coulonges C, Carpentier W, Dina C, Taing L, et al. Genomewide association study of a rapid progression cohort identifies new susceptibility alleles for AIDS (ANRS Genomewide Association Study 03) J Infect Dis. 2009;200:1194–1201. doi: 10.1086/605892. [DOI] [PubMed] [Google Scholar]
- 11.Loeuillet C, Deutsch S, Ciuffi A, Robyr D, Taffe P, Munoz M, et al. In vitro whole-genome analysis identifies a susceptibility locus for HIV-1. PLoS Biol. 2008;6:e32. doi: 10.1371/journal.pbio.0060032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fellay J, Ge D, Shianna KV, Colombo S, Ledergerber B, Cirulli ET, et al. Common genetic variation and the control of HIV-1 in humans. PLoS Genet. 2009;5:e1000791. doi: 10.1371/journal.pgen.1000791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Siddiqui RA, Sauermann U, Altmuller J, Fritzer E, Nothnagel M, Dalibor N, et al. X chromosomal variation is associated with slow progression to AIDS in HIV-1-infected women. Am J Hum Genet. 2009;85:228–239. doi: 10.1016/j.ajhg.2009.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pelak K, Goldstein DB, Walley NM, Fellay J, Ge D, Shianna KV, et al. Host determinants of HIV-1 control in African Americans. J Infect Dis. 2010;201:1141–1149. doi: 10.1086/651382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Limou S, Coulonges C, Herbeck JT, van Manen D, An P, Le Clerc S, et al. Multiple-cohort genetic association study reveals CXCR6 as a new chemokine receptor involved in long-term nonprogression to AIDS. J Infect Dis. 2010;202:908–915. doi: 10.1086/655782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joubert BR, Lange EM, Franceschini N, Mwapasa V, North KE, Meshnick SR. A whole genome association study of mother-to-child transmission of HIV in Malawi. Genome Med. 2010;2:17. doi: 10.1186/gm138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shrestha S, Aissani B, Song W, Wilson CM, Kaslow RA, Tang J. Host genetics and HIV-1 viral load set-point in African-Americans. AIDS. 2009;23:673–677. doi: 10.1097/QAD.0b013e328325d414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aouizerat BE, Pearce CL, Miaskowski C. The search for host genetic factors of HIV/AIDS pathogenesis in the postgenome era: progress to date and new avenues for discovery. Curr HIV/AIDS Rep. 2011;8:38–44. doi: 10.1007/s11904-010-0065-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wallace DC, Fan W. Energetics, epigenetics, mitochondrial genetics. Mitochondrion. 2010;10:12–31. doi: 10.1016/j.mito.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson DC, Shrestha S, Wiener HW, Makowsky R, Kurundkar A, Wilson CM, et al. Mitochondrial DNA diversity in the African American population. Mitochondrial DNA. 2013 doi: 10.3109/19401736.2013.840591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hendrickson SL, Hutcheson HB, Ruiz-Pesini E, Poole JC, Lautenberger J, Sezgin E, et al. Mitochondrial DNA haplogroups influence AIDS progression. AIDS. 2008;22:2429–2439. doi: 10.1097/QAD.0b013e32831940bb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aissani B. Confounding by linkage disequilibrium. J Hum Genet. 2014;59:110–115. doi: 10.1038/jhg.2013.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilson CM, Ellenberg JH, Douglas SD, Moscicki AB, Holland CA, Reach Project Of The Adolescent Medicine HIVARN CD8+CD38+ T cells but not HIV type 1 RNA viral load predict CD4+ T cell loss in a predominantly minority female HIV+ adolescent population. AIDS Res Hum Retroviruses. 2004;20:263–269. doi: 10.1089/088922204322996482. [DOI] [PubMed] [Google Scholar]
- 24.Tang J, Wilson CM, Meleth S, Myracle A, Lobashevsky E, Mulligan MJ, et al. Host genetic profiles predict virological and immunological control of HIV-1 infection in adolescents. AIDS. 2002;16:2275–2284. doi: 10.1097/00002030-200211220-00007. [DOI] [PubMed] [Google Scholar]
- 25.Makowsky R, Yan Q, Wiener HW, Sandel M, Aissani B, Tiwari HK, et al. The utility of mitochondrial and y chromosome phylogenetic data to improve correction for population stratification. Front Genet. 2012;3:301. doi: 10.3389/fgene.2012.00301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet. 1999;23:147. doi: 10.1038/13779. [DOI] [PubMed] [Google Scholar]
- 27.Ruiz-Pesini E, Lott MT, Procaccio V, Poole JC, Brandon MC, Mishmar D, et al. An enhanced MITOMAP with a global mtDNA mutational phylogeny. Nucleic Acids Res. 2007;35:D823–828. doi: 10.1093/nar/gkl927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Oven M, Kayser M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum Mutat. 2009;30:E386–394. doi: 10.1002/humu.20921. [DOI] [PubMed] [Google Scholar]
- 29.Grady BJ, Samuels DC, Robbins GK, Selph D, Canter JA, Pollard RB, et al. Mitochondrial genomics and CD4 T-cell count recovery after antiretroviral therapy initiation in AIDS clinical trials group study 384. J Acquir Immune Defic Syndr. 2011;58:363–370. doi: 10.1097/QAI.0b013e31822c688b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Touloumi G, Hatzakis A, Rosenberg PS, O’Brien TR, Goedert JJ. Effects of age at seroconversion and baseline HIV RNA level on the loss of CD4+ cells among persons with hemophilia. Multicenter Hemophilia Cohort Study. AIDS. 1998;12:1691–1697. doi: 10.1097/00002030-199813000-00018. [DOI] [PubMed] [Google Scholar]
- 31.Asmussen MA, Arnold J, Avise JC. Definition and properties of disequilibrium statistics for associations between nuclear and cytoplasmic genotypes. Genetics. 1987;115:755–768. doi: 10.1093/genetics/115.4.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hendrickson SL, Lautenberger JA, Chinn LW, Malasky M, Sezgin E, Kingsley LA, et al. Genetic variants in nuclear-encoded mitochondrial genes influence AIDS progression. PLoS One. 2010;5:e12862. doi: 10.1371/journal.pone.0012862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Potluri P, Davila A, Ruiz-Pesini E, Mishmar D, O’Hearn S, Hancock S, et al. A novel NDUFA1 mutation leads to a progressive mitochondrial complex I-specific neurodegenerative disease. Mol Genet Metab. 2009;96:189–195. doi: 10.1016/j.ymgme.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Strauss KA, DuBiner L, Simon M, Zaragoza M, Sengupta PP, Li P, et al. Severity of cardiomyopathy associated with adenine nucleotide translocator-1 deficiency correlates with mtDNA haplogroup. Proc Natl Acad Sci U S A. 2013;110:3453–3458. doi: 10.1073/pnas.1300690110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boland ML, Chourasia AH, Macleod KF. Mitochondrial dysfunction in cancer. Front Oncol. 2013;3:292. doi: 10.3389/fonc.2013.00292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Horan MP, Gemmell NJ, Wolff JN. From evolutionary bystander to master manipulator: the emerging roles for the mitochondrial genome as a modulator of nuclear gene expression. Eur J Hum Genet. 2013;21:1335–1337. doi: 10.1038/ejhg.2013.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.