Abstract
Autosomal dominant polycystic kidney disease (ADPKD) often results in ESRD but with a highly variable course. Mutations to PKD1 or PKD2 cause ADPKD; both loci have high levels of allelic heterogeneity. We evaluated genotype-phenotype correlations in 1119 patients (945 families) from the HALT Progression of PKD Study and the Consortium of Radiologic Imaging Study of PKD Study. The population was defined as: 77.7% PKD1, 14.7% PKD2, and 7.6% with no mutation detected (NMD). Phenotypic end points were sex, eGFR, height–adjusted total kidney volume (htTKV), and liver cyst volume. Analysis of the eGFR and htTKV measures showed that the PKD1 group had more severe disease than the PKD2 group, whereas the NMD group had a PKD2-like phenotype. In both the PKD1 and PKD2 populations, men had more severe renal disease, but women had larger liver cyst volumes. Compared with nontruncating PKD1 mutations, truncating PKD1 mutations associated with lower eGFR, but the mutation groups were not differentiated by htTKV. PKD1 nontruncating mutations were evaluated for conservation and chemical change and subdivided into strong (mutation strength group 2 [MSG2]) and weak (MSG3) mutation groups. Analysis of eGFR and htTKV measures showed that patients with MSG3 but not MSG2 mutations had significantly milder disease than patients with truncating cases (MSG1), an association especially evident in extreme decile populations. Overall, we have quantified the contribution of genic and PKD1 allelic effects and sex to the ADPKD phenotype. Intrafamilial correlation analysis showed that other factors shared by families influence htTKV, with these additional genetic/environmental factors significantly affecting the ADPKD phenotype.
Keywords: ADPKD, Genotype/phenotype, Prognostic studies
Autosomal dominant polycystic kidney disease (ADPKD) is a common (frequency 1:400–1:1000) inherited disorder characterized by progressive development of kidney cysts, often resulting in ESRD.1 ADPKD is genetically heterogeneous, with PKD1 and PKD2 accounting for approximately 77% and approximately 13% of patients, respectively, with no mutation detected (NMD) in approximately 8%–10%.2–4 Despite the monogenic nature of the disease, the severity of the renal phenotype and the occurrence of clinically significant extrarenal manifestations, including polycystic liver disease, are highly variable. Studies of ADPKD populations have shown that the causative gene strongly influences the phenotype, with ESRD occurring, on average, approximately 20 years earlier in PKD1 than PKD2 (approximately 55–58 versus approximately 74–80 years old)5,6 and magnetic resonance imaging–determined total kidney volume (TKV) approximately 40% larger in PKD1 than PKD2.7,8
A high level of allelic heterogeneity is found in both PKD1 and PKD2 (1272 and 202 different described pathogenic mutations, respectively).9 Despite this complexity, genotype-phenotype studies have found differences in renal outcomes associated with PKD1 mutation type or position. Previously, the position of the mutation in PKD1 was modestly associated with the severity of renal disease and the occurrence of intracranial aneurysms, with 5′ mutations causing more severe disease.10,11 A more recent, larger study found that truncating PKD1 mutations were associated with more severe renal disease than nontruncating changes (ESRD at 55.6 versus 67.9 years old).6 This indicates that a significant proportion of nontruncating mutations are incompletely penetrant (hypomorphic). Studies of atypical ADPKD patients homozygous (or compound heterozygous) for PKD1 missense mutations or patients with early-onset ADPKD with a truncating and likely hypomorphic allele (or two hypomorphic alleles) in trans also indicate the presence of hypomorphic PKD1 alleles.12–15
Large ADPKD populations with mainly typical renal phenotypes and a wealth of clinical, imaging, and genetic data are now available from observational and clinical trials that can facilitate genotype-phenotype studies. Here, we describe genotype-phenotype and sex studies of patients without ESRD from the HALT PKD Clinical Trial and the Consortium of Radiologic Imaging Study of Polycystic Kidney Disease (CRISP) Observational Study, with a focus on the significance of PKD1 allelic effects.16–20
Results
The ADPKD Populations
The study populations consisted of the HALT PKD Trial and the CRISP Study participants with available DNA samples, which were mutation screened for the coding regions of PKD1 and PKD2 (Concise Methods). Nineteen families with complex genotypes were excluded from the study (Concise Methods and Table 1). Table 1 shows the genetic and clinical details of the total study population used for the eGFR analyses and the height–adjusted total kidney volume (htTKV) population. Details of the eGFR-only group (the total population consists of the htTKV plus eGFR-only populations) are also shown. Comparison of the independent htTKV and eGFR populations showed that the htTKV population was younger with preserved renal function, whereas both groups had high levels of patients who were hypertensive on the basis of the inclusion criteria for the HALT PKD Trial and the CRISP Study16,18 (Table 1). Importantly, the proportions of patients in the key genic and allelic groups (apart from marginally for mutation type) were not different between the two populations. Sex, genic, and allelic groups were analyzed for differences in phenotype measured by eGFR and htTKV. Both measures of renal disease were corrected for age in all analyses, sex for the genic and allelic analyses, and genotype for the sex analyses.
Table 1.
Clinical and genetic characteristics of the studied populations
| Variables | Study Populationsa | |||
|---|---|---|---|---|
| Total | htTKV | eGFR Only | P Valueb | |
| Total patients (%) | 1141 (100) | |||
| Total families (%) | 964 (100) | |||
| Excluded study patients (%) | 22 (2.0) | |||
| Excluded study families (%) | 19 (2.0) | |||
| Included study patients (%) | 1119 (98) | 663 | 456 | |
| Sex (%) | 0.28 | |||
| Men | 540 (48.3) | 311 (46.9) | 229 (50.2) | |
| Women | 579 (51.7) | 352 (53.1) | 227 (49.8) | |
| Included study families (%) | 945 (98) | 581 | 364 | |
| Mean age, yr (SD) | 40.7±10.8 | 35.3±8.7 | 48.5±8.6 | <0.0001 |
| Median eGFRc (quartile 1, quartile 3) | 73.4 (52.1, 95.8) | 91.6 (76.6, 105.5) | 48.4 (38.9, 58.6) | <0.0001 |
| Median htTKV (quartile 1, quartile 3) | 567.8 (387.6, 827.4) | 567.8 (387.6, 827.4) | NA | |
| Hypertension (%) | 1051 (93.9) | 595 (89.7) | 456 (100) | <0.0001 |
| Mean age of onset of hypertension, yr (SD)d | 32.8±10.0 | 30.1±8.9 | 36.3±10.3 | <0.0001 |
| CKD stage (%) | <0.0001 | |||
| Stage 1 | 357 (31.9) | 350 (52.8) | 7 (1.5) | |
| Stage 2 | 388 (34.7) | 295 (44.5) | 93 (20.4) | |
| Stage 3 | 353 (31.5) | 18 (2.7) | 335 (73.5) | |
| Stage 4 | 21 (1.9) | 0 (0.0) | 21 (4.6) | |
| Patients in the genic groups: PKD1, PKD2, and NMD (%) | 0.12 | |||
| Patients with NMD | 85 (7.6) | 58 (8.7) | 27 (5.9) | |
| Families with NMD | 79 (8.4) | 55 (9.5) | 24 (6.6) | |
| Patients with PKD2 (%) | 165 (14.7) | 103 (15.5) | 62 (13.6) | |
| Men (%) | 86 (52.1) | 50 (48.5) | 36 (58.1) | |
| Women (%) | 79 (47.9) | 53 (51.5) | 26 (41.9) | |
| Families with PKD2 (%) | 135 (14.3) | 90 (15.5) | 45 (12.4) | |
| Total patients with PKD1 (%) | 869 (77.7) | 502 (75.7) | 367 (80.5) | |
| Men (%) | 415 (47.8) | 236 (47.0) | 179 (48.8) | |
| Women (%) | 454 (52.2) | 266 (53.0) | 188 (51.2) | |
| Total families with PKD1 (%) | 731 (77.4) | 436 (75.0) | 295 (81.0) | |
| Total patients with PKD1, mutation type (%) | 0.04 | |||
| Frameshift, D/I | 282 (32.5) | 159 (31.7) | 123 (33.5) | |
| Splice | 96 (11.0) | 48 (9.6) | 48 (13.1) | |
| Nonsense | 214 (24.6) | 124 (24.7) | 90 (24.5) | |
| Missense | 223 (25.7) | 130 (25.9) | 93 (25.3) | |
| In-frame, D/I | 54 (6.2) | 41 (8.2) | 13 (3.5) | |
| Patients with PKD1, MSG (%) | 0.65 | |||
| PKD1 truncating, MSG1 | 575 (66.3) | 329 (65.7) | 246 (67.2) | |
| PKD1 nontruncating, MSG2 | 172 (19.8) | 98 (19.6) | 74 (20.2) | |
| PKD1 nontruncating, MSG3 | 120 (13.8) | 74 (14.8) | 46 (12.6) | |
NA, not available; D/I, deletion or insertion.
Total population studied for eGFR analysis and htTKV population studied for htTKV.
htTKV versus eGFR-only populations.
eGFR calculated from serum creatinine measurements using the CKD-EPI equation and expressed as milliliters per minute per 1.73 m2.
Only patients who were hypertensive were included.
Sex Effects on the ADPKD Phenotype
Regression analysis in the total (eGFR) and htTKV populations (Table 1) showed significantly more severe disease in men than women exemplified by lower eGFR and larger htTKV (Figure 1, A and B, Table 2). The same sex difference held true for the PKD1 population with eGFR and htTKV and PKD2 assayed by eGFR (Figure 1, C–E, Table 2). However, no sex difference was seen for htTKV in PKD2 (Supplemental Figure 1, Table 2).
Figure 1.
Men have more severe renal disease in ADPKD. Comparison of men and women in terms of (A, C, and E) renal function measured by eGFR and (B and D) renal structure (htTKV; plotted on a natural log scale [loge]) for the (A and B) total, (C and D) PKD1, and (E) PKD2 populations. Population numbers are indicated in Table 1, and details of the eGFR and htTKV differences and significance are shown in Table 2. Parallel regression lines are plotted for each variable in each comparison, and the data are corrected for age, gene, and mutation type, with the P value indicated.
Table 2.
Significance of sex and genotypes to the eGFR and htTKV phenotypes and ICC analysis: Kidney disease
| Variable and Population | eGFRa Difference (P Value) | htTKV Difference, % (P Value) | Figure | ICC eGFR (P Value) | ICC htTKV (P Value) |
|---|---|---|---|---|---|
| Sex: Womenb | |||||
| Total study | +4.34 (0.0002)c | −22.1 (<0.0001)c | Figure 1, A and B | 0.12 (0.06) | 0.46 (<0.001)c |
| PKD1 | +3.48 (0.009)c | −19.7 (<0.0001)c | Figure 1, C and D | 0.13 (0.07) | 0.40 (<0.001)c |
| PKD2 | +8.47 (0.002)c | −19.7 (0.07) | Figure 1E, Supplemental Figure 1 | 0.09 (0.32) | 0.63 (0.01)c |
| Gene | |||||
| Total study | Figure 2, A and B | 0.12 (0.06) | 0.46 (<0.001)c | ||
| PKD2d | +13.49 (<0.0001)c | −34.3 (<0.0001)c | |||
| NMDd | +9.42 (<0.0001)c | −30.9 (<0.0001)c | |||
| Mutation position: 3′ halfe | |||||
| PKD1 | −2.18 (0.11) | +7.8 (0.06) | Supplemental Figure 2, A and B | 0.13 (0.07) | 0.39 (<0.001)c |
| PKD2 | −2.05 (0.45) | −15.2 (0.12) | Supplemental Figure 2, C and D | 0.07 (0.34) | 0.62 (0.02)c |
| Mutation type: PKD1 | Figure 3, A and B | 0.12 (0.09) | 0.46 (<0.001)c | ||
| Splicef | −1.06 (0.65) | +1.0 (0.94) | |||
| Nonsensef | +0.45 (0.46) | −3.9 (0.46) | |||
| Missensef | +6.16 (0.0004)c | −10.4 (0.06) | |||
| In frame D/If | +5.63 (0.05) | +6.2 (0.46) | |||
| Mutation effect: Nontruncatingg | |||||
| PKD1 | +5.09 (0.0003)c | −5.8 (0.16) | Figure 3, C and D | 0.13 (0.07) | 0.40 (<0.001)c |
| PKD2 | +3.34 (0.34) | −10.4 (0.35) | Supplemental Figure 3 | 0.09 (0.31) | 0.63 (0.01)c |
| Mutation group: PKD1 | Figure 4, A and B | 0.14 (0.06) | 0.39 (<0.001)c | ||
| MSG2h | +2.90 (0.09) | +3.2 (0.71) | |||
| MSG3h | +7.80 (<0.0001)c | −17.3 (0.003)c |
D/I, deletion or insertion.
In milliliters per minute per 1.73 m2 calculated using the CKD-EPI equation.
Versus men.
Significant.
Versus PKD1.
Versus 5′ half.
Versus frameshifting.
Versus truncating.
Versus MSG1.
Genic Effects on the ADPKD Phenotype
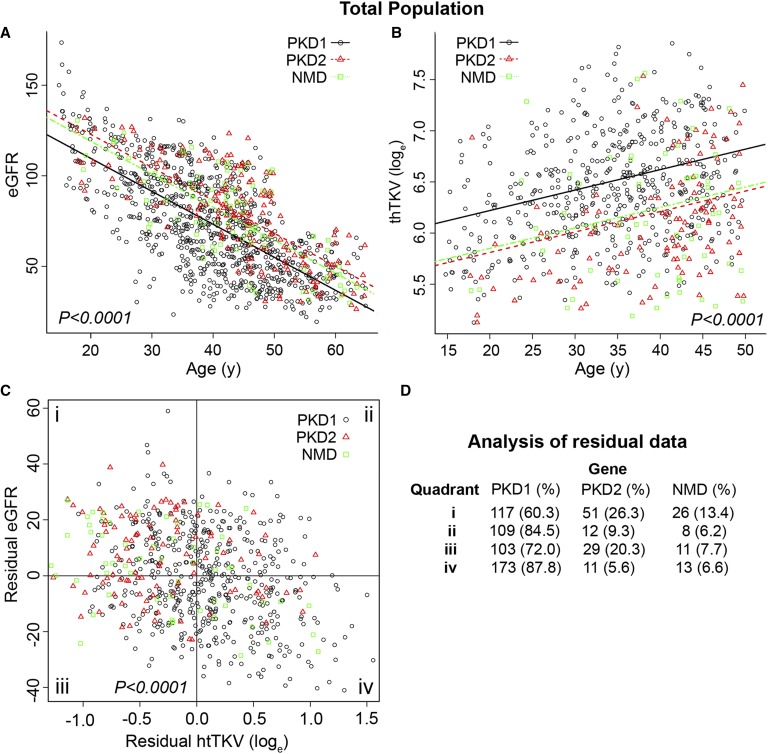
Regression analysis of eGFR for the PKD1, PKD2, and NMD groups indicated lower values for PKD1 compared with PKD2, whereas the NMD group was more similar to PKD2 than PKD1 (Figure 2A, Table 2). Comparison of the PKD1, PKD2, and NMD groups with htTKV showed smaller kidneys in the PKD2 group compared with those in the PKD1 group, again with the NMD group closely matching the PKD2 cohort (Figure 2B, Table 2).
Figure 2.
The mutated gene strongly influences the renal phenotype in ADPKD. The PKD1, PKD2, and NMD genic groups are compared in terms of (A) eGFR and (B) htTKV natural log scale (loge), with overall P values indicated. Population numbers and details of the eGFR and htTKV differences and the individual significances are shown in Tables 1 and 2, respectively. (C) Residual analysis shows the relationship between the eGFR and htTKV measurements in the genic populations, with the corresponding number (percentage) of each population in each of the four quadrants (i–iv) shown in D. The zero point on the x and y axes is where the average age– and sex–corrected residual is equal to zero (no difference between the observed and predicted outcomes).
To illustrate the strong relationship between eGFR and htTKV in the PKD1, PKD2, and NMD groups, age- and sex-corrected residuals for each outcome were calculated and plotted for each patient (Figure 2C). The age- and sex-corrected residual is the difference between the observed and predicted values of the outcome from the regression model. Dividing the data into quadrants showed a significant difference in the distribution of genotypes. A strong enrichment for patients with PKD2 and NMD was seen in the mild quadrant (quadrant i) for each phenotypic measure, whereas there was a strong enrichment for patients with PKD1 in the severe end point quadrant (quadrant iv) (Figure 2D). Additional analysis was performed of quadrant iii, which represented patients with lower than average eGFR (although not severe renal insufficiency) (Table 1) but smaller than average–sized kidneys (Supplemental Table 1). These patients were not different in age, age at onset of hypertension, or proportion of atypical kidneys (as defined by Irazabal et al.21) compared with those in the other quadrants, although predictably, there were fewer patients in the typical, rapidly progressive subclasses.21 Patients with PKD2 were more common, but this was not significant.
To further understand the significance of genic effects on extreme phenotypes, we analyzed the most and least severe decile groups of the age- and sex-corrected eGFR and htTKV data and saw very significant differences in the abundance of the different genic groups (Table 4, genic). Both the eGFR and htTKV data showed a near monopoly of PKD1 in the severe decile, with a relative enrichment for both the PKD2 and NMD populations in the mildest groups.
Table 4.
Analysis of the extreme deciles (10% and 90%) for the genic and PKD1 allelic variables
| Phenotype and Gene | Most Severe 10% | Least Severe 10% | P Value |
|---|---|---|---|
| Genic | |||
| eGFR (%) | <0.0001 | ||
| PKD1 | 106 (95.5) | 61 (55.0) | |
| PKD2 | 2 (1.8) | 37 (33.3) | |
| NMD | 3 (2.7) | 13 (11.7) | |
| htTKV (%) | <0.0001 | ||
| PKD1 | 58 (87.8) | 24 (36.4) | |
| PKD2 | 4 (6.1) | 23 (34.8) | |
| NMD | 4 (6.1) | 19 (28.8) | |
| PKD1 allelic effect | |||
| eGFR (%) | <0.0001 | ||
| Truncating | 67 (77.0) | 41 (47.1) | |
| Nontruncating | 20 (23.0) | 46 (52.9) | |
| htTKV (%) | 0.05 | ||
| Truncating | 36 (72.0) | 27 (52.9) | |
| Nontruncating | 14 (28.0) | 24 (47.1) | |
| PKD1 mutation strength | |||
| eGFR (%) | 0.0002 | ||
| Truncating: MSG1 | 67 (77.0) | 41 (47.1) | |
| Nontruncating: MSG2 | 11 (12.7) | 21 (24.2) | |
| Nontruncating: MSG3 | 9 (10.3) | 25 (28.7) | |
| htTKV (%) | 0.005 | ||
| Truncating: MSG1 | 36 (72.0) | 27 (52.9) | |
| Nontruncating: MSG2 | 11 (22.0) | 8 (15.7) | |
| Nontruncating: MSG3 | 3 (6.0) | 16 (31.4) |
Allelic Effects: Mutation Position in PKD1 and PKD2
Comparison of eGFR and htTKV in the PKD1 groups on the basis of mutation position along the transcript separated at the midway point in the PKD1 coding region (codon 2151) showed no difference with either end point (Supplemental Figure 2, A and B, Table 2). Similar analysis of the PKD2 population separated at the protein midpoint showed no difference in disease severity (Supplemental Figure 2, C and D, Table 2).
Allelic Effects: Mutation Type in PKD1 and PKD2
For the initial analysis of mutation groups in PKD1, five broad categories of mutation type were analyzed: frameshifting deletions or insertions (indels), nonsense, splicing, missense, and in-frame indels. Comparing mutation type with eGFR, the five mutation types clustered as two groups: frameshifting, nonsense, and splicing in one group and missense and in-frame in the other (Figure 3A, Table 2). Given the evidence from the individual PKD1 mutation types, we classified the five subgroups into truncating and nontruncating (Concise Methods has details of these populations) and found that truncating mutations were associated with a lower eGFR (Figure 3C, Table 2). Similar comparisons of the five mutation groups to htTKV (in the smaller and milder population) (Table 1) showed that, although there was a trend for missense mutations to have smaller kidneys, there was no significant difference between the groups (Figure 3B, Table 2). Likewise, when truncating and nontruncating mutations were compared by htTKV, the nontruncating population tended to have smaller kidneys, but this also did not reach significance (Figure 3D, Table 2).
Figure 3.
Truncating PKD1 mutations are associated with worse renal function than nontruncating mutations. Mutations are divided into five different types—frameshifting indels, splicing, nonsense, missense, and in-frame indels (D/I)—and compared with (A) eGFR and (B) htTKV natural log scale (loge), with overall P values indicated. Comparison of mutations predicted to truncate or nontruncate the protein with (C) eGFR and (D) htTKV (loge). Population numbers and details of the eGFR and htTKV differences and the significance are shown in Tables 1 and 2, respectively.
We previously differentiated nontruncating mutations into highly likely pathogenic and likely pathogenic on the basis of a scoring algorithm including a substitution score—conservation of the substituted residue in orthologs (Grantham Variation [GV]) and paralogs/defined domains and the chemical difference of the substitution (Grantham Difference)—and a contextual score.3,12–14 Here, we have used a modified substitution score (details are in Concise Methods and Supplemental Table 2) to define PKD1 mutation strength groups (MSGs): strongly predicted nontruncating mutations [MSG2] and weakly predicted nontruncating mutations [MSG3], plus truncating mutations [MSG1]. Comparison of the MSG1 population with the other MSGs using eGFR showed that the MSG3 group was associated with significantly milder disease, whereas MSG2 was not (Figure 4A, Table 2). Comparison of the MSGs by htTKV again showed that, although the MSG2 population was not different from MSG1, MSG3 had significantly smaller kidneys (Figure 4B, Table 2). Residual eGFR and htTKV analysis showed a significant difference of the MSGs in the quadrants (Figure 4, C and D), with enrichment of patients with MSG3 in the mild eGFR and htTKV quadrant i compared with the corresponding severe quadrant iv, whereas patients with MSG2 were more evenly distributed. Analysis of the extreme eGFR and htTKV deciles showed that nontruncating mutations were under-represented in the severe groups (Table 4, PKD1 allelic effect) and that the level of patients with MSG3 was much higher in the mildest deciles (Table 4, PKD1 mutation strength).
Figure 4.
Strongly predicted nontruncating PKD1 mutations are associated with more severe disease than weak nontruncating mutations. PKD1 mutations are divided into three groups—truncating (MSG1), strongly predicted nontruncating (MSG2), and weakly predicted nontruncating (MSG3) (Supplemental Table 2)—and assayed for (A) eGFR and (B) htTKV natural log scale (loge), with overall P values shown. Population numbers and details of the eGFR and htTKV differences and the significance are shown in Tables 1 and 2, respectively. (C) A plot of the residual analysis for the two phenotypic variables and (D) the frequency of each MSG in each quadrant show a significantly different distribution for the MSG3 group.
Analysis of individual mutation types (data not shown) and truncating and nontruncating mutations in PKD2 did not show a difference when compared by eGFR or htTKV (Supplemental Figure 3, Table 2). This is not surprising given the small size of the PKD2 population and the low number of nontruncating mutations.
Genic, Allelic, and Sex Effects on Liver Cyst Volumes
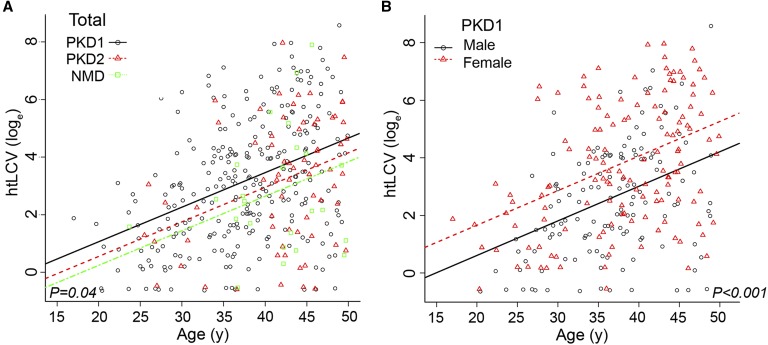
Comparison of height–adjusted liver cyst volume (htLCV) in the three genic groups showed an overall difference with marginal significance, with both patients with PKD2 and patients with NMD having less liver cyst burden than patients with PKD1 (Figure 5A, Table 3). Neither PKD1 mutation type (P=0.97) nor the MSG (P=0.70) was associated with htLCV (data not shown). However, sex significantly influenced htLCV, with women having larger volumes, a difference that was significant for PKD1 but not PKD2 (Figure 5A, Supplemental Figure 4, Table 3).
Figure 5.
Women and marginally PKD1 gene type are associated with larger liver cyst volumes. (A) htLCV plotted natural log scale (loge) against age with the genic groups identified. (B) Analysis of sex shows that women have larger liver cyst volumes in the PKD1 population. Population numbers and details of the eGFR and htTKV differences and the significance are shown in Tables 1 and 3, respectively, with overall P values shown.
Table 3.
Significance of sex and genotypes to the htLCV phenotype and ICC analysis: Liver disease
| Variable and Population | htLCV Difference, % (P Value) | Figure | ICC htLCV (P Value) |
|---|---|---|---|
| Gene | |||
| Total study | Figure 5A | 0.57 (0.002)a | |
| PKD2b | −41.1 (0.05) | ||
| NMDb | −56.4 (0.05) | ||
| Sex: Womenc | |||
| Total study | +53.7 (0.0002)a | ||
| PKD1 | +63.5 (<0.001)a | Figure 5B | 0.51 (0.01)a |
| PKD2 | +20.5 (0.66) | Supplemental Figure 4 |
Significant.
Versus PKD1.
Versus men.
Analyses of Interfamilial Variability
For each of the models mentioned above, we reanalyzed them after accounting for possible interfamilial variability. We used linear mixed models with random effects for family identification and calculated intrafamilial correlation coefficients (ICCs) (Tables 2 and 3). Although most patients in the study were single-study participants within a family, 143 families had two or more individuals in the study, consisting of a total of 317 patients (details are in Concise Methods). Interestingly, no significant ICC values were found for the eGFR end point, but the htTKV and htLCV values were highly significant (Tables 2 and 3). For htTKV and htLCV, this indicates that, after accounting for the effects of age, sex, and genic and allelic predictors, there is substantially less variability within families than between them.
Discussion
ADPKD prognostic information can identify a more severe population that requires closer clinical monitoring, is suitable for clinical trials, and would benefit most from the treatments that are now becoming available. TKV has proved a good early disease predictor, but eGFR has been considered of little value until the last 10–15 years before ESRD because of compensatory mechanisms in the kidney at early disease stages.22 Genetic information (potentially available at an early age) has prognostic potential. The gene mutated has been known to have strong prognostic value for >20 years, but the value of allelic information is only now being realized, and up to now, it was only correlated with age at ESRD.5,6,23,24 Data presented here show the value of genic and PKD1 allelic data by correlating with early measures of disease severity. In addition, the value of predictive estimates of disease penetrance of PKD1 nontruncating mutations is shown to compartmentalize the population.
The total study population (eGFR analysis) was collected from three different populations with different selection criteria,16,18 representing most patients with ADPKD from late teens to >60 years old, whereas the htTKV population consists of younger patients with preserved renal function; genetically, the groups were indistinguishable for the key genetic end points. However, the selection criteria for the HALT PKD Study A and the CRISP Study for younger patients with normal renal function and the HALT PKD Study B for patients with significant renal insufficiency mean that older patients with preserved renal function (and young patients with early decline in renal function) are under-represented in our populations (Figure 1A).16–20 Although a wholly representative population may have more patients with PKD2 and more PKD1 patients with MSG3, the phenotypic diversity of this population has revealed strong genotype-phenotype correlations, although at present, they are not precise enough to inform the outcome of a specific patient.
Sex is shown to be significant by both functional (eGFR) and structural (htTKV) measures of renal disease in both the total population and the PKD1 and PKD2 (eGFR) populations, with men having more severe disease. This has been a controversial subject in ADPKD but is consistent, with more ADPKD men than women reaching ESRD and earlier ESRD in the Genkyst Study PKD1 population.5,6,10,11,25,26 A strong sex difference was found in the PKD2 population, consistent with an older study, and the early end points assayed here may be sensitive to detect this difference.27 TKV and eGFR may not be ideal measures to compare men with women, but because our data included sex correction factors in the Chronic Kidney Disease Epidemiology Collaboration (CKD-EPI) equation and TKV correction by height to account for men having larger kidneys, it suggests that these results likely reflect real sex differences.22,27 The greater liver cyst burden in women is known,28 but we have shown that this is only significant in the larger PKD1 population.
The well known genic difference described as age at ESRD is reflected in our population for the first time by eGFR measurements. Our data show clear (although overlapping) differences between the PKD1 and PKD2 populations. The strong relationship between eGFR and htTKV22 and the discriminatory value of the residual plot when looking at PKD1 and PKD2 populations are shown by the enrichment of patients with PKD2 in the mild quadrant. The analysis of extremes shows an overwhelming enrichment for PKD1 in the most severe decile with both disease measures but more even representation in the mildest decile, although patients with PKD1 are approximately five times more common than patients with PKD2. Nevertheless, the finding of a significant proportion of patients with PKD1 in the mildest deciles hints at the significance of PKD1 allelic effects (see below). In addition, the residual analysis suggests that there is a proportion of patients with smaller than average kidneys but lower than average eGFR. This indicates that larger than average htTKVs alone may not detect all patients with lower than average eGFR, although study of a population with significant renal insufficiency and additional analysis of this population are required before firm conclusions can be drawn.
Interesting new information is described about the approximately 8.5% of patients with NMD, a percentage similar to that of other analyses of large ADPKD populations.3,4 By both eGFR and htTKV, the patients with NMD behaved like patients with PKD2. This population no doubt reflects a heterogeneous etiology, but our data suggest that these patients cannot be explained predominantly by missed fully inactivating mutations at the complex PKD1 locus.29 Although they may represent missed conventional PKD2 mutations, it seems more likely that hypomorphic PKD1 variants not meeting the defined pathogenic threshold, mosaics, and other atypical mutations explain these patients. However, an additional gene associated with mild disease cannot be ruled out.30,31
Previous studies suggested that the position of the PKD1 mutation was associated with the severity of renal disease (5′ was more severe), although that was not confirmed in a recent larger study.6,10 Our eGFR and TKV data also do not show a significant difference related to the position of the mutation in PKD1 (5′ versus 3′ of the midpoint). Given the size of the two studies with negative position data using three different phenotypic end points, it seems unlikely that straightforward PKD1 mutation location effects strongly influence the renal phenotype.
We show here, consistent with the renal survival information from the Genkyst Study6 but using eGFR as the phenotypic measure, that some PKD1 nontruncating mutations are hypomorphic. The mutation groups classed as nontruncating (missense and in-frame indels) have milder disease than the three truncating classes (frameshifting indels, nonsense, and splicing), which is also reflected when these two entire groups are compared. Interestingly, although htTKV is considered a better measure of disease severity early in the disease,22 there was not a significant difference between truncating and nontruncating populations with this outcome measure. This is likely partially because the htTKV population was much smaller (n=663 versus n=1119) and included more patients in early disease stage (Table 1). However, even analysis of the smaller htTKV population by eGFR showed a significant difference between the truncating and nontruncating groups (Supplemental Figure 5), indicating that genetic influences on eGFR and htTKV may differ, perhaps reflecting the different functional and structural features measured. This is not that surprising, because other factors not reflected in renal volume, such as fibrosis, likely play a role in loss of renal function.32
To estimate penetrance information from the PKD1 nontruncating population, we subdivided these patients on the basis of bioinformatics criteria and showed that mutations weakly predicted to be pathogenic (MSG3) behaved differently from the strongly predicted group (MSG2) and truncating changes (MSG1) by both eGFR and htTKV analysis. This shows that bioinformatics mutation assessment can be of value for predicting outcomes at the population level and is emphasized by the distribution of the MSG3 population by the residual and extreme decile analyses. However, this classification was not perfect, with three and nine patients classified as MSG3 in the most severe htTKV and eGFR deciles, respectively, whereas 21 and eight patients classified as MSG2 were in the mildest deciles for these two outcomes, respectively. Analyses of this correlative data, family studies, data from recurrent mutations, and in vitro studies of the PKD1 protein33–35 will enable the classification method to be further refined, increasing its prognostic value.
Analyses of genic, PKD1 allelic, and sex data with the same end points mean that we can estimate the relative contribution of each to the renal phenotype. Using the more complete eGFR data, we can see that gene type is most important followed by the allelic effects (comparing MSG3 with MSG1) and the sex influence.
Despite the importance of PKD1 allelic effects, the truncating PKD1 group is still the largest in the mildest eGFR and htTKV deciles. Some of these patients may be mosaics or have other incompletely penetrant mutations, but it is clear that other factors influence renal disease severity. This is also seen by the spread of values of patients of the same age within allelic groups whether measured by eGFR or htTKV. Calculation of ICC values shows that, beyond the sex and disease gene–related variables analyzed here, htTKV has greater inter- compared with intrafamilial variability, suggesting that it is influenced by other genetic variants and environmental factors shared within the family. For instance, the htTKV ICC of 0.46 found in the total population after correcting for genic effects indicates that the intrafamilial correlation is responsible for 46% of the unexplained variability in htTKV. Because eGFR ICC values did not significantly differ between inter- and intrafamilial populations, in this case, genetic and environmental factors seem to have less influence, perhaps partially reflecting that eGFR is not a precise disease measure early in the disease. Identifying genetic modifiers by genome–wide association studies and sequencing approaches will add additional value as genetic biomarkers in ADPKD.
Concise Methods
The Study Population
The study participants consisted of all patients recruited into the HALT PKD Trial or the CRISP Study who signed a consent allowing genetic studies and provided a DNA sample. Institutional review boards at the recruitment sites for these studies (Emory University School of Medicine, Kansas University Medical Center, Mayo Clinic, University of Alabama, Birmingham, University of Colorado Health Sciences Center, Tufts Medical Center, Cleveland Clinic, and Beth Israel Deaconess Medical Center) approved the study. Details of the phenotypic data collected as part of these studies are described elsewhere and summarized in Table 1 with baseline HALT PKD Trial and CRISP Study data used here.16,18 The phenotypic end points used were eGFR calculated from the serum creatinine measurement using the CKD-EPI equation and expressed as milliliters per minute per 1.73 m227 and TKV and liver cyst volume determined by analysis of magnetic resonance images using the stereology method and corrected by height (htTKV/htLCV) to better allow comparison of men and women.22
Mutation Analyses and Categorization of Variants
Details of mutation screening of the CRISP Study cohort have been described,3 and complete mutation details of the HALT PKD Trial population will be described separately (C.M. Heyer and P.C. Harris, unpublished data). The screening protocol for the HALT PKD Trial population was similar to previous descriptions,3 with the entire coding regions of PKD1 and PKD2 plus flanking intronic regions (±50 bp) screened by Sanger sequencing. Patients with no clear pathogenic mutation detected after Sanger sequencing were screened for gross rearrangements using multiplex ligation-dependent probe amplification.36
Mutations were defined as truncating (MSG1; frameshifting indels, nonsense mutations, canonical splicing changes, and in-frame indels ≥5 amino acids) or nontruncating (missense, in-frame indels ≤4 amino acids and noncanonical splicing events). PKD1 nontruncating mutations were further defined as strongly predicted (MSG2) and less strongly predicted (MSG3) using criteria similar to those previously described to obtain a substitution score (SS).3 Details of the PKD1 nontruncating mutations and their classification are shown in Supplemental Table 2, and a brief description of the algorithm is described. For substitutions and small in–frame indels, a multisequence alignment of orthologs consisting of human, dog, rat, mouse, opossum, chicken, frog (Xenopus tropicalis), and the consensus of three fish species (Fugu rubripes, Danio rerio [pkd1a], and Tetraodon nigroviridis) was used to determine the GV, and a matrix comparing the GV and Grantham Difference was used to generate scores from +8 to −8. On this scale, the extremes represent highly nonconservative substitutions at invariant sites in orthologs to conservative substitutions at nonconserved sites in orthologs, respectively.3 In the case of indels, the loss/gain of an amino acid was considered as a highly nonconservative change and assessed per amino acid. Other factors considered were conservation in recognized domains (LRR and flanks, WSC, PKD repeats, C-type lectin, GAIN and GPS, PLAT, and G Protein Peptide Activating Sequence) and homology with PC1 and PC2 paralogs and related sea urchin REJ proteins in other homologous regions (REJ, PC-A, PC-B, and the transmembrane and loop areas) with scores from +6 (invariant) to 0 (no conservation) (Supplemental Table 2). Other factors considered were described substitution of the residue to another amino acid change (+1 per recurrent example) and predicted changes in structure (insertions and deletion of amino acids because of indels, signal peptide, transmembrane, and coiled coil) plus cysteine introductions in extracellular regions and proline substitutions in α-helical regions with scores of +6 (strongly predicted disrupted) to 0 (no predicted disruption) per amino acid. Patients with an MSG score of ≥+8 were defined as MSG2, and those with lower scores but reaching a pathogenic threshold were defined as MSG3 (Supplemental Table 2).
Atypical splicing changes were scored using the BGDP Splice Site Prediction by Neural Network website (http://www.fruitfly.org/seq_tools/splice.html) and assigned scores from +10 to 0 on the basis of the predicted variation from the wild type. Splicing scores of approximately +7 to +10 (MSG2) usually resulted in a BGDP>60% reduction in the predicted wild–type splice site, a >60% strength increase of a cryptic splice site, or a new splice site that scored greater than or equal to the wild-type site, whereas splicing scores from +4 to +7 (MSG3) usually had a BGDP score variation of 60%–30%.
Patients with complex cases, including those who were mosaic, digenic, or diallelic, were excluded from the analysis population, and details will be described elsewhere (C.M. Heyer and P.C. Harris, unpublished data).
Statistical Analyses
Renal function (eGFR) and a structural measure of the kidney (htTKV) were the primary outcomes. We investigated the relationship between these outcomes and sex, gene type (PKD1, PKD2, or NMD), mutation type (truncating or nontruncating), mutation position (5′ or 3′ of the midpoint in the gene), and PKD1 mutation strength (MSG1–MSG3). For the genetic studies, linear regression models were fitted with each of the outcomes as a function of age, sex, and the predictor of interest, whereas for sex analysis, the genotype was substituted for sex. To graphically summarize the results, each of the primary outcomes was plotted against age, with adjusted parallel regression lines corresponding to the predictor of interest (i.e., PKD1, PKD2, and NMD). To assess the relationship between eGFR and htTKV, age- and sex-corrected residuals were calculated for each outcome and plotted against each other. Furthermore, the resulting figures were split into quadrants (defined by the zero [mean] values), and predictors of interest were described within each. Chi-squared tests were used to assess the significance of these relationships. Finally, the age- and sex-corrected residuals were ordered, and cut points for the 10th and 90th percentiles were determined. The predictors of interest were described within each of these extreme groups. For each of the models mentioned above, we reanalyzed them after accounting for possible interfamilial variability. We used linear mixed models with random effects for family identification and calculated ICCs. In total, 143 families were multiplex, including 118 with two participants, 20 with three participants, four with four participants, and one with five participants.
Disclosures
None.
Supplementary Material
Acknowledgments
The patients and coordinators involved in the HALT progression of PKD Study and the Consortium of Radiologic Imaging Study of Polycystic Kidney Disease (CRISP) Study are thanked for their participation and efforts.
Mutation analysis of the HALT PKD Study and the CRISP Study populations was supported by National Institute of Diabetes, Digestive and Kidney Diseases (NIDDK) Grants DK062410S1, DK056957S1, and DK058816 and Mayo Translational Polycystic Kidney Disease Center Grant DK090728. MRC-Holland is thanked for providing multiplex ligation-dependent probe amplification kits at a discount. The CRISP Study and the HALT PKD Study were supported by NIDDK Cooperative Agreements DK056943, DK056956, DK056957, DK056961, DK062410, DK062408, DK062402, DK082230, DK062411, and DK062401; National Center for Research Resources General Clinical Research Centers Cooperative Agreements RR000039, RR000585, RR000054, RR000051, RR023940, and RR001032; and National Center for Advancing Translational Sciences Clinical and Translational Science Awards Cooperative Agreements RR025008, TR000454, RR024150, TR00135, RR025752, TR001064, RR025780, TR001082, RR025758, TR001102, RR033179, and TR000001 to the participating centers.
Other investigators involved in these studies, including Drs. Franz Winklhofer (Kansas University Medical Center), Peter Czarnecki (Beth Israel Deaconess Medical Center), Marie Hogan (Mayo Clinic), Dana Miskulin (Tufts Medical Center), Frederic Rahbari-Oskoui (Emory University School of Medicine), and Lisa Guay-Woodford (Children’s National Hospital), are also thanked.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2015050583/-/DCSupplemental.
References
- 1.Torres VE, Harris PC, Pirson Y: Autosomal dominant polycystic kidney disease. Lancet 369: 1287–1301, 2007 [DOI] [PubMed] [Google Scholar]
- 2.Harris PC, Torres VE: Polycystic kidney disease. Annu Rev Med 60: 321–337, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rossetti S, Consugar MB, Chapman AB, Torres VE, Guay-Woodford LM, Grantham JJ, Bennett WM, Meyers CM, Walker DL, Bae K, Zhang QJ, Thompson PA, Miller JP, Harris PC CRISP Consortium : Comprehensive molecular diagnostics in autosomal dominant polycystic kidney disease. J Am Soc Nephrol 18: 2143–2160, 2007 [DOI] [PubMed] [Google Scholar]
- 4.Audrézet MP, Cornec-Le Gall E, Chen JM, Redon S, Quéré I, Creff J, Bénech C, Maestri S, Le Meur Y, Férec C: Autosomal dominant polycystic kidney disease: Comprehensive mutation analysis of PKD1 and PKD2 in 700 unrelated patients. Hum Mutat 33: 1239–1250, 2012 [DOI] [PubMed] [Google Scholar]
- 5.Hateboer N, v Dijk MA, Bogdanova N, Coto E, Saggar-Malik AK, San Millan JL, Torra R, Breuning M, Ravine D: Comparison of phenotypes of polycystic kidney disease types 1 and 2. European PKD1-PKD2 Study Group. Lancet 353: 103–107, 1999 [DOI] [PubMed] [Google Scholar]
- 6.Cornec-Le Gall E, Audrézet MP, Chen JM, Hourmant M, Morin MP, Perrichot R, Charasse C, Whebe B, Renaudineau E, Jousset P, Guillodo MP, Grall-Jezequel A, Saliou P, Férec C, Le Meur Y: Type of PKD1 mutation influences renal outcome in ADPKD. J Am Soc Nephrol 24: 1006–1013, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grantham JJ, Chapman AB, Torres VE: Volume progression in autosomal dominant polycystic kidney disease: The major factor determining clinical outcomes. Clin J Am Soc Nephrol 1: 148–157, 2006 [DOI] [PubMed] [Google Scholar]
- 8.Harris PC, Bae K, Rossetti S, Torres VE, Grantham JJ, Chapman A, Guay-Woodford L, King BF, Wetzel LH, Baumgarten D, Kenney PJ, Consugar M, Klahr S, Bennett WM, Meyers CM, Zhang Q, Thompson PA, Zhu F, Miller JP, Consortium C: Cyst number but not the rate of cystic growth is associated with the mutated gene in autosomal dominant polycystic kidney disease. J Am Soc Nephrol 17: 3013–3019, 2006 [DOI] [PubMed] [Google Scholar]
- 9.Autosomal Dominant Polycystic Kidney Disease Mutation Database. Available at: http://pkdb.mayo.edu. Accessed October 15, 2015
- 10.Rossetti S, Burton S, Strmecki L, Pond GR, San Millán JL, Zerres K, Barratt TM, Ozen S, Torres VE, Bergstralh EJ, Winearls CG, Harris PC: The position of the polycystic kidney disease 1 (PKD1) gene mutation correlates with the severity of renal disease. J Am Soc Nephrol 13: 1230–1237, 2002 [DOI] [PubMed] [Google Scholar]
- 11.Rossetti S, Chauveau D, Kubly V, Slezak JM, Saggar-Malik AK, Pei Y, Ong AC, Stewart F, Watson ML, Bergstralh EJ, Winearls CG, Torres VE, Harris PC: Association of mutation position in polycystic kidney disease 1 (PKD1) gene and development of a vascular phenotype. Lancet 361: 2196–2201, 2003 [DOI] [PubMed] [Google Scholar]
- 12.Rossetti S, Kubly VJ, Consugar MB, Hopp K, Roy S, Horsley SW, Chauveau D, Rees L, Barratt TM, van't Hoff WG, Niaudet P, Torres VE, Harris PC: Incompletely penetrant PKD1 alleles suggest a role for gene dosage in cyst initiation in polycystic kidney disease. Kidney Int 75: 848–855, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vujic M, Heyer CM, Ars E, Hopp K, Markoff A, Orndal C, Rudenhed B, Nasr SH, Torres VE, Torra R, Bogdanova N, Harris PC: Incompletely penetrant PKD1 alleles mimic the renal manifestations of ARPKD. J Am Soc Nephrol 21: 1097–1102, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bergmann C, von Bothmer J, Ortiz Brüchle N, Venghaus A, Frank V, Fehrenbach H, Hampel T, Pape L, Buske A, Jonsson J, Sarioglu N, Santos A, Ferreira JC, Becker JU, Cremer R, Hoefele J, Benz MR, Weber LT, Buettner R, Zerres K: Mutations in multiple PKD genes may explain early and severe polycystic kidney disease. J Am Soc Nephrol 22: 2047–2056, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Audrézet MP, Corbiere C, Lebbah S, Morinière V, Broux F, Louillet F, Fischbach M, Zaloszyc A, Cloarec S, Merieau E, Baudouin V, Deschênes G, Roussey G, Maestri S, Visconti C, Boyer O, Abel C, Lahoche A, Randrianaivo H, Bessenay L, Mekahli D, Ouertani I, Decramer S, Ryckenwaert A, Cornec-Le Gall E, Salomon R, Ferec C, Heidet L: Comprehensive PKD1 and PKD2 mutation analysis in prenatal autosomal dominant polycystic kidney disease [published online ahead of print July 2, 2015]. J Am Soc Nephrol doi:ASN.2014101051 [DOI] [PMC free article] [PubMed]
- 16.Chapman AB, Guay-Woodford LM, Grantham JJ, Torres VE, Bae KT, Baumgarten DA, Kenney PJ, King BF JrGlockner JF, Wetzel LH, Brummer ME, O'Neill WC, Robbin ML, Bennett WM, Klahr S, Hirschman GH, Kimmel PL, Thompson PA, Miller JP Consortium for Radiologic Imaging Studies of Polycystic Kidney Disease cohort: Renal structure in early autosomal-dominant polycystic kidney disease (ADPKD) : The Consortium for Radiologic Imaging Studies of Polycystic Kidney Disease (CRISP) cohort. Kidney Int 64: 1035–1045, 2003 [DOI] [PubMed]
- 17.Grantham JJ, Torres VE, Chapman AB, Guay-Woodford LM, Bae KT, King BF Jr., Wetzel LH, Baumgarten DA, Kenney PJ, Harris PC, Klahr S, Bennett WM, Hirschman GN, Meyers CM, Zhang X, Zhu F, Miller JP CRISP Investigators : Volume progression in polycystic kidney disease. N Engl J Med 354: 2122–2130, 2006 [DOI] [PubMed] [Google Scholar]
- 18.Chapman AB, Torres VE, Perrone RD, Steinman TI, Bae KT, Miller JP, Miskulin DC, Rahbari Oskoui F, Masoumi A, Hogan MC, Winklhofer FT, Braun W, Thompson PA, Meyers CM, Kelleher C, Schrier RW: The HALT polycystic kidney disease trials: Design and implementation. Clin J Am Soc Nephrol 5: 102–109, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schrier RW, Abebe KZ, Perrone RD, Torres VE, Braun WE, Steinman TI, Winklhofer FT, Brosnahan G, Czarnecki PG, Hogan MC, Miskulin DC, Rahbari-Oskoui FF, Grantham JJ, Harris PC, Flessner MF, Bae KT, Moore CG, Chapman AB HALT-PKD Trial Investigators : Blood pressure in early autosomal dominant polycystic kidney disease. N Engl J Med 371: 2255–2266, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Torres VE, Abebe KZ, Chapman AB, Schrier RW, Braun WE, Steinman TI, Winklhofer FT, Brosnahan G, Czarnecki PG, Hogan MC, Miskulin DC, Rahbari-Oskoui FF, Grantham JJ, Harris PC, Flessner MF, Moore CG, Perrone RD HALT-PKD Trial Investigators : Angiotensin blockade in late autosomal dominant polycystic kidney disease. N Engl J Med 371: 2267–2276, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Irazabal MV, Rangel LJ, Bergstralh EJ, Osborn SL, Harmon AJ, Sundsbak JL, Bae KT, Chapman AB, Grantham JJ, Mrug M, Hogan MC, El-Zoghby ZM, Harris PC, Erickson BJ, King BF, Torres VE CRISP Investigators : Imaging classification of autosomal dominant polycystic kidney disease: A simple model for selecting patients for clinical trials. J Am Soc Nephrol 26: 160–172, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chapman AB, Bost JE, Torres VE, Guay-Woodford L, Bae KT, Landsittel D, Li J, King BF, Martin D, Wetzel LH, Lockhart ME, Harris PC, Moxey-Mims M, Flessner M, Bennett WM, Grantham JJ: Kidney volume and functional outcomes in autosomal dominant polycystic kidney disease. Clin J Am Soc Nephrol 7: 479–486, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ravine D, Walker RG, Gibson RN, Forrest SM, Richards RI, Friend K, Sheffield LJ, Kincaid-Smith P, Danks DM: Phenotype and genotype heterogeneity in autosomal dominant polycystic kidney disease. Lancet 340: 1330–1333, 1992 [DOI] [PubMed] [Google Scholar]
- 24.Cornec-Le Gall E, Audrézet MP, Rousseau A, Hourmant M, Renaudineau E, Charasse C, Morin MP, Moal MC, Dantal J, Wehbe B, Perrichot R, Frouget T, Vigneau C, Potier J, Jousset P, Guillodo MP, Siohan P, Terki N, Sawadogo T, Legrand D, Menoyo-Calonge V, Benarbia S, Besnier D, Longuet H, Férec C, Le Meur Y: The PROPKD Score: A new algorithm to predict renal survival in autosomal dominant polycystic kidney disease [published online ahead of print July 6, 2015]. J Am Soc Nephrol doi:ASN.2015010016 [DOI] [PMC free article] [PubMed]
- 25.Gabow PA, Johnson AM, Kaehny WD, Kimberling WJ, Lezotte DC, Duley IT, Jones RH: Factors affecting the progression of renal disease in autosomal-dominant polycystic kidney disease. Kidney Int 41: 1311–1319, 1992 [DOI] [PubMed] [Google Scholar]
- 26.Magistroni R, He N, Wang K, Andrew R, Johnson A, Gabow P, Dicks E, Parfrey P, Torra R, San-Millan JL, Coto E, Van Dijk M, Breuning M, Peters D, Bogdanova N, Ligabue G, Albertazzi A, Hateboer N, Demetriou K, Pierides A, Deltas C, St George-Hyslop P, Ravine D, Pei Y: Genotype-renal function correlation in type 2 autosomal dominant polycystic kidney disease. J Am Soc Nephrol 14: 1164–1174, 2003 [DOI] [PubMed] [Google Scholar]
- 27.Levey AS, Stevens LA, Schmid CH, Zhang YL, Castro AF 3rd, Feldman HI, Kusek JW, Eggers P, Van Lente F, Greene T, Coresh J CKD-EPI (Chronic Kidney Disease Epidemiology Collaboration) : A new equation to estimate glomerular filtration rate. Ann Intern Med 150: 604–612, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hogan MC, Abebe K, Torres VE, Chapman AB, Bae KT, Tao C, Sun H, Perrone RD, Steinman TI, Braun W, Winklhofer FT, Miskulin DC, Rahbari-Oskoui F, Brosnahan G, Masoumi A, Karpov IO, Spillane S, Flessner M, Moore CG, Schrier RW: Liver involvement in early autosomal-dominant polycystic kidney disease. Clin Gastroenterol Hepatol 13: 155–64.e6, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hughes J, Ward CJ, Peral B, Aspinwall R, Clark K, San Millán JL, Gamble V, Harris PC: The polycystic kidney disease 1 (PKD1) gene encodes a novel protein with multiple cell recognition domains. Nat Genet 10: 151–160, 1995 [DOI] [PubMed] [Google Scholar]
- 30.Paul BM, Consugar MB, Ryan Lee M, Sundsbak JL, Heyer CM, Rossetti S, Kubly VJ, Hopp K, Torres VE, Coto E, Clementi M, Bogdanova N, de Almeida E, Bichet DG, Harris PC: Evidence of a third ADPKD locus is not supported by re-analysis of designated PKD3 families. Kidney Int 85: 383–392, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harris PC, Rossetti S: Molecular diagnostics for autosomal dominant polycystic kidney disease. Nat Rev Nephrol 6: 197–206, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grantham JJ, Mulamalla S, Swenson-Fields KI: Why kidneys fail in autosomal dominant polycystic kidney disease. Nat Rev Nephrol 7: 556–566, 2011 [DOI] [PubMed] [Google Scholar]
- 33.Hopp K, Ward CJ, Hommerding CJ, Nasr SH, Tuan HF, Gainullin VG, Rossetti S, Torres VE, Harris PC: Functional polycystin-1 dosage governs autosomal dominant polycystic kidney disease severity. J Clin Invest 122: 4257–4273, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gainullin VG, Hopp K, Ward CJ, Hommerding CJ, Harris PC: Polycystin-1 maturation requires polycystin-2 in a dose-dependent manner. J Clin Invest 125: 607–620, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cai Y, Fedeles SV, Dong K, Anyatonwu G, Onoe T, Mitobe M, Gao JD, Okuhara D, Tian X, Gallagher AR, Tang Z, Xie X, Lalioti MD, Lee AH, Ehrlich BE, Somlo S: Altered trafficking and stability of polycystins underlie polycystic kidney disease. J Clin Invest 124: 5129–5144, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Consugar MB, Wong WC, Lundquist PA, Rossetti S, Kubly VJ, Walker DL, Rangel LJ, Aspinwall R, Niaudet WP, Ozen S, David A, Velinov M, Bergstralh EJ, Bae KT, Chapman AB, Guay-Woodford LM, Grantham JJ, Torres VE, Sampson JR, Dawson BD, Harris PC CRISP Consortium : Characterization of large rearrangements in autosomal dominant polycystic kidney disease and the PKD1/TSC2 contiguous gene syndrome. Kidney Int 74: 1468–1479, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.